Abstract
Background and Purpose
Cardioembolic stroke due to paroxysmal atrial fibrillation (AF) may account for 1 out of 4 cryptogenic strokes (CS) and transient ischemic attacks (TIAs). The purpose of this pilot study was to search for biomarkers potentially predicting incident AF in patients with ischemic stroke or TIA.
Methods
Plasma samples were collected from patients aged 18 years and older with ischemic stroke or TIA due to AF (n = 9) and large artery atherosclerosis (LAA) with ipsilateral carotid stenosis (n = 8) and age- and sex-matched controls (n = 10). Analyses were performed with the Olink technology simultaneously measuring 184 biomarkers of cardiovascular disease. For bioinformatics, acquired data were analyzed using gene set enrichment analysis (GSEA). Selected proteins were validated using ELISA. Individual receiver operating characteristic (ROC) curves and odds ratios from logistic regression were calculated. A randomForest (RF) model with out-of-bag estimate was applied for predictive modeling.
Results
GSEA indicated enrichment of proteins related to inflammatory response in the AF group. Interleukin (IL)-6, growth differentiation factor (GDF)-15, and pentraxin-related protein PTX3 were the top biomarkers on the ranked list for the AF group compared to the LAA group and the control group. ELISA validated increased expression of all tested proteins (GDF-15, PTX3, and urokinase plasminogen activator surface receptor [U-PAR]), except for IL-6. 19 proteins had the area under the ROC curve (AUC) over 0.85 including all of the proteins with significant evolution in the logistic regression. AUCs were very discriminant in distinguishing patients with and without AF (LAA and control group together). GDF-15 alone reached AUC of 0.95. Based on RF model, all selected participants in the tested group were classified correctly, and the most important protein in the model was GDF-15.
Conclusions
Our results demonstrate an association between inflammation and AF and that multiple proteins alone and in combination may potentially be used as indicators of AF in CS and TIA patients. However, further studies including larger samples sizes are needed to support these findings. In the ongoing NOR-FIB study, we plan further biomarker assessments in patients with CS and TIA undergoing long-term cardiac rhythm monitoring with insertable cardiac monitors.
Keywords: Atrial fibrillation, Ischemic stroke, Biomarkers, Inflammation, Cryptogenic stroke
Introduction
Despite a modern diagnostic workup, up to 30% of ischemic strokes and transient ischemic attacks (TIAs) remain without a definitive etiology and are defined as cryptogenic [1]. Approximately 25% of cryptogenic strokes (CSs) are caused by silent paroxysmal atrial fibrillation (AF) [2]. Cardioembolic strokes due to AF are especially severe and show the highest rates of mortality and permanent disability [3]. AF detection is, therefore, very important in order to optimize secondary prevention and reduce the risk of recurrent stroke. Continuous ECG monitoring shows higher AF detection rates than intermittent monitoring [4]. There is, however, no consensus regarding the optimal duration and timing of cardiac rhythm monitoring. Based on the detection rates of AF with insertable cardiac monitors from earlier studies [5], the identification of reliable biomarkers associated with AF would allow more targeted selection of patients for prolonged cardiac rhythm monitoring. To our knowledge, studies combining monitoring of CS and TIA patients with insertable cardiac monitors and the identification of AF-specific biomarkers from a broad spectrum of molecular biomarkers have not been performed.
Numerous cardiovascular biomarkers have been studied in relation to endothelial damage, coagulation, fibrosis, and inflammatory processes in AF [6, 7]. So far, the results have been conflicting [8, 9]. Further studies are required to address the clinical implications of cardiovascular biomarkers in the prediction of AF-related thromboembolism.
The purpose of this pilot study was to assess a panel of potential biomarkers to identify candidate markers associated with AF, and whether they can be used to discriminate patients with ischemic stroke due to AF.
Patients and Methods
The study population consisted of 3 participant groups; 8 patients with ischemic stroke and 1 with TIA due to AF, 8 patients with ischemic stroke due to large artery atherosclerosis (LAA) with ipsilateral carotid stenosis, and 10 age- and sex-adjusted healthy blood donors as control group (Table 1). Participants were enrolled at the Oslo University Hospital and Østfold Hospital Trust, Norway. Blood samples were collected from patients aged ≥18 years admitted for acute ischemic stroke or TIA due to AF within 7 days from the index stroke in the period from October 2015 until February 2017. From patients with ischemic stroke due to LAA, blood was collected and processed within 2 days after symptom onset as a part of an earlier carotid plaque study [10]. Control samples were collected in the same period [10]. Plasma derived from collected blood samples was stored at −80oC until further analyses.
Table 1.
Clinical characteristics of the test groups
| AF group (n = 9) | LAA group (n = 8) | Control group (n = 10) | |
|---|---|---|---|
| Age, years | 74.4±7.5 | 63.4±12.8 | 63.1±8.4 |
| Males, n (%) | 3 (33.3) | 5 (62.5) | 5 (50) |
| Current smoking, n (%) | 2 (22.2) | 2 (25) | 0 |
| Hypertension, n (%) | 6 (66.6) | 4 (50) | − |
| Diabetes mellitus, n (%) | 2 (22.2) | 3 (37.5) | − |
| Coronary artery disease, n (%) | 4 (44.4) | 1 (12.5) | − |
| Congestive heart failure, n (%) | 2 (22.2) | 0 | − |
| Statin treatment, n (%) | 3 (33.3) | 5 (62.5) | − |
| CRP, mg/L | 6.2±5.4 | 5.1±4.2 | 1.9±3.0 |
| Leukocyte count, ×109/L | 9.1±2.4 | 7.4±2.3 | 5.7±1.9 |
| Platelets, ×109/L | 240±52 | 233±69 | − |
| Total cholesterol, mmol/L | 4.7±0.8 | 3.9±1.2 | 5.8±0.7 |
| LDL cholesterol, mmol/L | 2.9±0.7 | 2.2±1.1 | 3.8±0.9 |
| HDL cholesterol, mmol/L | 1.2±0.3 | 1.4±0.5 | 1.8±0.4 |
| Triglycerides, mmol/L | 1.2±0.4 | 1.6±0.9 | 1.3±0.7 |
| HbA1c, % | 6.2±0.7 | 6.0±1.3 | 5.3±0.4 |
Values are presented as numbers (%) or means ± SD. AF, atrial fibrillation; LAA, large artery atherosclerosis.
Biomarker Analysis with the Olink Proteomics Technology
Biomarkers were assessed by proximity extension assay using the Olink Proseek Multiplex CVD II and CVD III panels testing simultaneously 184 biomarkers (Olink Bioscience, Uppsala, Sweden) [11].
Gene Set Enrichment Analysis of Biomarkers
Gene set enrichment analysis (GSEA) [12] was applied to the Olink dataset against the Gene Ontology Consortium database (c5.all.v6.1) downloaded from MiSigDB (http://www.broadinstitute.org/gsea/msigdb/). Phenotype permutation for 1,000 times was used. Plasma concentrations of selected proteins from the ranked list were validated using commercially available ELISA kits (R&D Systems, Minneapolis, MN, USA).
Statistical Analyses
The t test statistical approach was chosen to rank proteins and complete the GSEA analysis. Values of p< 0.05 were considered significant.
For predictive modeling, R software (version 3.3.3) was used with several packages: CMA (version 1.24.0), randomForest (RF; version 4.6-14), ROCR (version 1.0.5), and Caret (version 6.0-80). Values below limits of detection (LOD) were substituted with LOD values in all data evaluations. An individual receiver operating characteristic (ROC) curve was performed before the construction of predictive modeling. An area under the ROC curve (AUC) >0.85 was considered to be discriminative. An odds ratio based on logistic regression was performed for each variable. For building the class prediction models, the Classification for MicroArrays (CMA) package [13] with leave-one-out cross-validation, due to a low number of patients, was utilized. An RF model was used for analysis. The results based on the RF package were given as out-of-bag estimate (OOB), which is a method to measure the RF prediction error. All estimations were done on 80% of the data (training data), and 20% of the data were used for the model validation as test data; a stratified sampling method was used to select test and training data to represent all groups in the sets. The number of variables tried at each split was 14. Confusion matrices have been generated to assess the predictive performance of the model.
Results
The characteristics of the 3 different groups of participants are presented in Table 1. In the AF group, mean age was higher, as well as the number of women and patients with hypertension and coronary artery disease compared to the LAA group, while the LAA group had a male preponderance and more statin users than the AF group. Across all 27 samples, 177 of 184 biomarkers were detected in all samples.
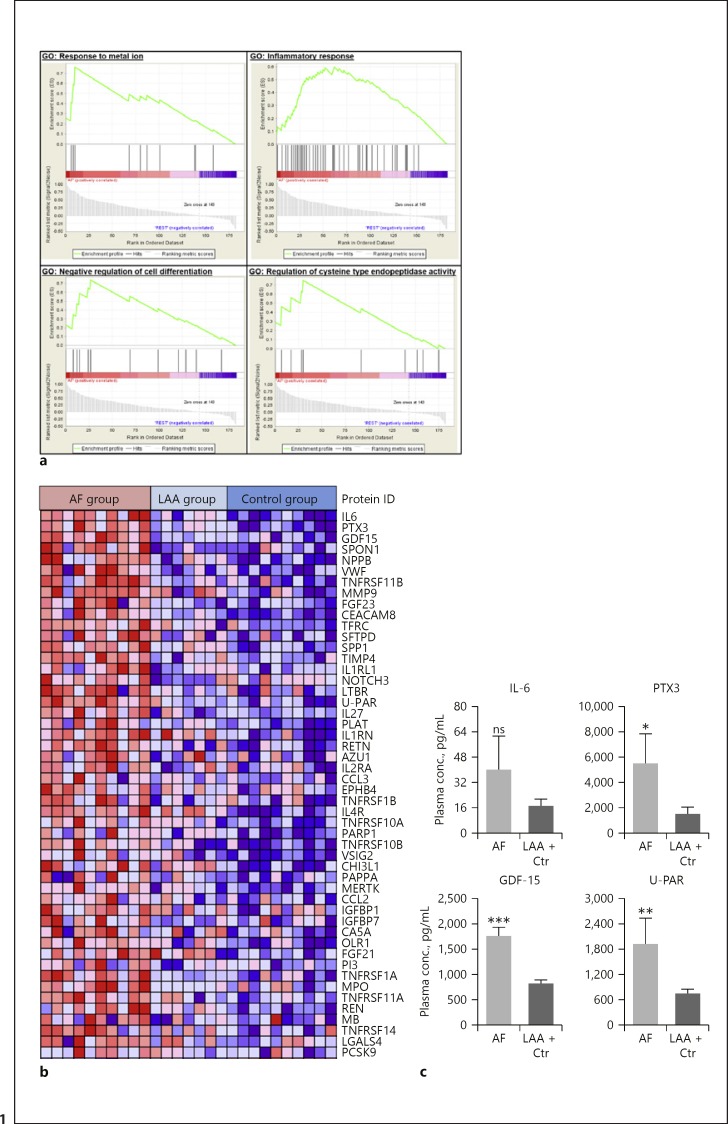
GSEA was utilized to identify biological processes positively correlated with AF. Four gene sets were significantly enriched in the AF group based on the ranked protein list (Fig. 1a) with the gene ontology term “inflammatory response” being particularly strong. Interleukin (IL)-6, pentraxin-related protein PTX3, and growth differentiation factor (GDF)-15 were the top-ranked proteins. Spondin-1 (SPON1) and matrix metalloproteinase-9 (MMP-9) were also found at the top of the ranked list (Fig. 1b). To validate increased expression of the biomarkers, selected markers were subjected to ELISA (IL-6, PTX3, GDF-15, and urokinase plasminogen activator surface receptor [U-PAR]). All proteins were significantly elevated in the AF group compared to the other groups, except for IL-6 (Fig. 1c).
Fig. 1.
Gene set enrichment analysis (GSEA) of differentially expressed proteins in patients with atrial fibrillation (AF) compared to patients with large artery atherosclerosis (LAA) and healthy controls. The protein expression data were analyzed using GSEA. a Significantly enriched gene sets are shown. In every thumbnail, the enrichment score (ES) is plotted as a function of the position within the ranked list and is shown as a green line (top). Black bars indicate the position of the proteins within the ranked list. The ranked list metrics (gray) illustrate the correlation between the signal-to-noise values of all individually ranked proteins. b A heat map representation of the top 50 positively correlated proteins to the AF group. c Plasma concentrations of selected proteins (IL-6, PTX3, GDF-15, and U-PAR) were validated using ELISA (n = 7–18; ns, not significant; * p < 0.05, ** p< 0.01, *** p < 0.005,AF vs. LAA + controls). Means ± SEM.
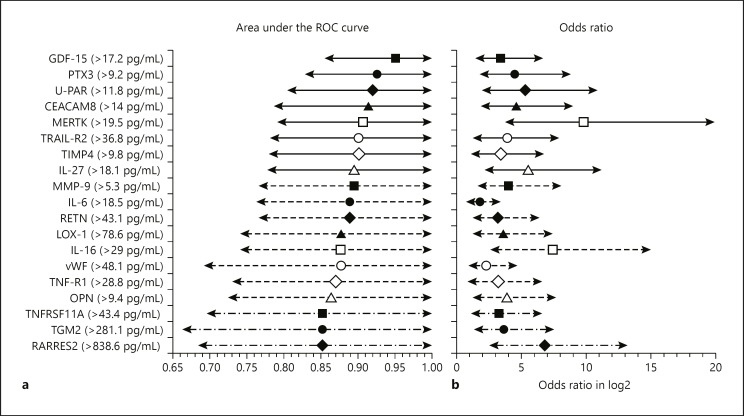
To distinguish patients with and without AF (LAA and control group together), individual AUCs were calculated to estimate the model accuracy for each biomarker; 19 proteins had AUC >0.85, with GDF-15, PTX3, and U-PAR being the top 3 biomarkers (Fig. 2a). These individual AUC values were very discriminative, and GDF-15 alone reached an AUC of 0.95 (Fig. 2a). Additionally, after performing logistic regression and acquiring odds ratios to determine the evolution of the biomarkers between the 2 groups, 43 biomarkers had a significant evolution (p < 0.05), including all of the biomarkers with individual AUC >0.85 (Fig. 2b). Presented odds ratios represent the odds for AF if the biomarker level exceeds the cutoff value specific for each variable. The log2 values of odds ratios and confidence intervals together with the cutoff values for the biomarkers with individual AUC >0.85 are presented in Figure 2b.
Fig. 2.
Forest plots for individual area under the receiver operating characteristic (ROC) curve (AUC) values and odds ratios (ORs) for 19 biomarkers with AUC >0.85. a Forest plot for AUC values with 95% confidence intervals (CIs). b Forest plot for the ORs with 95% CIs representing the odds of having AF if the level of the individual biomarker is over the cutoff value. Presented values are log2 transformed. Individual cutoff values for each biomarker are also shown.
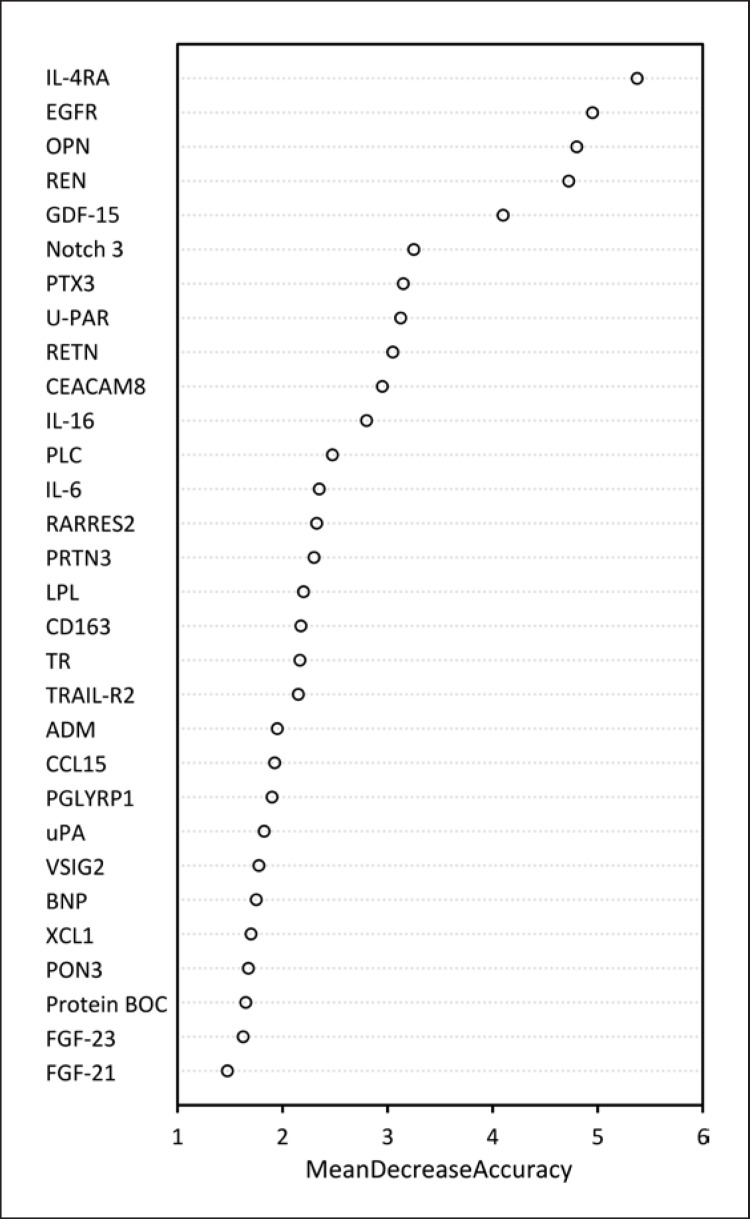
Giving the RF model results as OOB estimate, a model stratifying patients into groups was established. Estimated accuracy of the training data was 1.00 (95% CI 0.845–1.00) and 1.00 of the test data (95% CI 0.478–1.00). Sensitivities and specificities were 1.00 for each category, as all selected participants in the tested group were classified correctly. Biomarkers included in this model are listed in Figure 3 which shows how the model accuracy changes after excluding different biomarkers from the model. The more important the biomarker is in the model, the higher is the mean decrease in accuracy after exclusion of the biomarker. The highest decrease is observed for GDF-15.
Fig. 3.
Variable importance plot shows the weight of each biomarker in the model and how the accuracy of the model is affected after the exclusion of the biomarker from the model. The most important biomarkers in the model are listed on the top.
Discussion
Several biomarkers were strongly associated with AF in this pilot study. A large number of proteins positively correlated to AF was discovered by GSEA with strong inflammatory responses in the AF group. The discriminating power of these biomarkers was further emphasized by their individual AUC, with GDF-15 displaying the highest value. The importance of this biomarker was implied in the RF model as well as the utility of combining multiple biomarkers for a good patient stratification. Our findings indicate that there are numerous biomarkers associated with AF in stroke patients that may potentially be useful for the detection of AF in CS and TIA patients. They also demonstrate that there is a possibility of using individual biomarkers, but their combined use is particularly important for the identification of AF patients. These findings are in line with those of other studies focusing on discovering biomarkers for both AF itself and cardioembolic stroke due to AF [6, 14].
The association between inflammation and AF has been shown in both animal models and human studies [7], demonstrating increased levels of inflammatory biomarkers such as high-sensitivity C-reactive protein, PTX3, IL-6 [15], as well as IL-27 [16]. It has been shown that high plasma levels of IL-6 are not just correlated with the presence and duration of AF but also with an increased left atrial diameter [17]. Furthermore, an association between inflammation and early AF recurrence has been observed in patients with short-lasting persistent AF [18]. GDF-15, a marker of fibrosis which is another important pathomechanism in AF, has been shown to be independently associated with paroxysmal AF [19]. MMP-9 also plays an important role in atrial structural remodeling and dilatation [20, 21]. Hemostatic markers such as the von Willebrand factor were also suggested as a prognostic biomarker for AF [6].
In line with previous studies [6, 15], the results of the present study support the hypothesis that a variety of molecular biomarkers can potentially be used as indicators of underlying AF in CS patients. In earlier studies, the relationship between AF and biomarker levels were mostly investigated by testing individual biomarkers [8, 22] or a specific biomarker selection based on their functional similarities [15]. To our knowledge, this is the first study using a multiple-biomarker approach in patients with stroke of different etiologies. Lind et al. [14] found multiple biomarkers, including GDF-15 and IL-6, to be associated with AF in a large cohort study but not in stroke patients. However, the biomarkers that were positively correlated with AF in our pilot study represent a heterogenic group of proteins with wide variety of functions. This includes not just participation in inflammation and fibrosis but also in immune responses, organ injury, aging, apoptosis, endothelial dysfunction, and thrombogenesis, demonstrating that AF is a multifactorial disease [23].
However, many of the biomarkers identified in our study have also been connected with noncardiac diseases [24]. As Piek et al. [24] demonstrated the challenge to find specific biomarkers of heart failure, we also suppose that the biomarkers investigated in our study are probably not AF or cardiac specific; they likely represent the different underlying pathological processes in AF [8]. As shown in our model, we suppose that the combination of different molecular biomarkers would probably increase their specificity for cardiac diseases, especially AF, even though the individual AUCs were also discriminative. We suppose that these results could be translated to cryptogenic stroke patients and be helpful to differentiate patients with occult AF. The ability of the best candidate biomarkers to correctly identify patients with underlying paroxysmal AF will be tested in the main study. This includes analyzing blood samples taken within 14 days after cryptogenic stroke or TIA and after 1 year of continuous cardiac rhythm monitoring with ICMs.
The weaknesses of this study are the small sample size and the different time to sampling after symptom onset. The different time to sampling may have caused some differences found between groups due to different kinetics of different biomarkers. However, the levels of most of the biomarkers associated with ischemic injury peak in the first days after stroke [25]. Underlying medical conditions, drugs, and other factors may as well have influenced the biomarker levels. However, this was an explorative study and adjusting for variety of possible confounders in such robust biomarker panels would not be able to perform. The major strengths of our study are the patient selection and the high precision and specificity of the method used to analyze the blood samples [11]. Even though proximity extension assay is well established in clinical research, it has not been widely used in stroke patients. We chose, therefore, ELISA to validate the increased expression of some chosen proteins.
Conclusions
The current pilot study indicates that multiple proteins - either alone or in combination - may potentially be used as biomarkers for the detection of AF in CS and TIA patients and that simultaneous testing of multiple biomarkers is indicated for this purpose. In the ongoing NOR-FIB study (ClinicalTrials.gov identifier: NCT02937077), we plan further biomarker assessments using a larger group of patients with CS and TIA undergoing long-term cardiac rhythm monitoring with insertable cardiac monitors.
Statement of Ethics
The study was approved by the Regional Committee for Medical and Health Research Ethics (REK 2013/2371), and a written consent was obtained from all participants.
Disclosure Statement
The authors have no conflicts of interest to declare.
Funding Sources
The analyses and operating costs were funded by the Oslo University Hospital and Østfold Hospital Trust, Norway.
Author Contributions
A.T.L. was responsible for collecting blood samples from patients with AF, literature search, statistical evaluation of the data, and writing the manuscript. X.Y.K. was responsible for blood sample analysis and writing the manuscript. B.R.-T. collected blood samples from AF patients for the analysis and helped with correction of the manuscript. D.A. and D.R. were involved in writing the manuscript and corrections. M.S. and K.S. collected blood samples from patients with atherosclerosis and healthy donors, and helped with correction of the manuscript. V.B. developed the protocol for blood sampling and analyzed blood samples. M.C. performed the statistical analyses. E.S. and H.F. helped with writing the manuscript. B.H. was responsible for blood sample analysis and composing the manuscript. A.H.A. is the leader of the NOR-FIB project; she collected blood samples and was involved in writing and correcting the manuscript.
Acknowledgment
We thank Ellen Lund Sagen, BSc, for her help with the blood sample analysis, and Prof. Seval Kul, PhD, for statistical support.
References
- 1.Li L, Yiin GS, Geraghty OC, Schulz UG, Kuker W, Mehta Z, et al. Oxford Vascular Study Incidence, outcome, risk factors, and long-term prognosis of cryptogenic transient ischaemic attack and ischaemic stroke: a population-based study. Lancet Neurol. 2015 Sep;14((9)):903–13. doi: 10.1016/S1474-4422(15)00132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sposato LA, Cipriano LE, Saposnik G, Ruíz Vargas E, Riccio PM, Hachinski V. Diagnosis of atrial fibrillation after stroke and transient ischaemic attack: a systematic review and meta-analysis. Lancet Neurol. 2015 Apr;14((4)):377–87. doi: 10.1016/S1474-4422(15)70027-X. [DOI] [PubMed] [Google Scholar]
- 3.Pistoia F, Sacco S, Tiseo C, Degan D, Ornello R, Carolei A. The Epidemiology of Atrial Fibrillation and Stroke. Cardiol Clin. 2016 May;34((2)):255–68. doi: 10.1016/j.ccl.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Gladstone DJ, Spring M, Dorian P, Panzov V, Thorpe KE, Hall J, et al. EMBRACE Investigators and Coordinators Atrial fibrillation in patients with cryptogenic stroke. N Engl J Med. 2014 Jun;370((26)):2467–77. doi: 10.1056/NEJMoa1311376. [DOI] [PubMed] [Google Scholar]
- 5.Haeusler KG, Tütüncü S, Schnabel RB. Detection of Atrial Fibrillation in Cryptogenic Stroke. Curr Neurol Neurosci Rep. 2018 Aug;18((10)):66. doi: 10.1007/s11910-018-0871-1. [DOI] [PubMed] [Google Scholar]
- 6.Llombart V, Garcia-Berrocoso T, Bustamante A, Fernandez-Cadenas I, Montaner J. Cardioembolic stroke diagnosis using blood biomarkers. Curr Cardiol Rev. 2013 Nov;9((4)):340–52. doi: 10.2174/1573403X10666140214122633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu YF, Chen YJ, Lin YJ, Chen SA. Inflammation and the pathogenesis of atrial fibrillation. Nat Rev Cardiol. 2015 Apr;12((4)):230–43. doi: 10.1038/nrcardio.2015.2. [DOI] [PubMed] [Google Scholar]
- 8.Howlett PJ, Hatch FS, Alexeenko V, Jabr RI, Leatham EW, Fry CH. Diagnosing Paroxysmal Atrial Fibrillation: Are Biomarkers the Solution to This Elusive Arrhythmia? BioMed Res Int. 2015;2015:910267. doi: 10.1155/2015/910267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang KW, Hsu JC, Toomu A, Fox S, Maisel AS. Clinical Applications of Biomarkers in Atrial Fibrillation. Am J Med. 2017 Dec;130((12)):1351–7. doi: 10.1016/j.amjmed.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 10.Lunde NN, Holm S, Dahl TB, Elyouncha I, Sporsheim B, Gregersen I, et al. Increased levels of legumain in plasma and plaques from patients with carotid atherosclerosis. Atherosclerosis. 2017 Feb;257:216–23. doi: 10.1016/j.atherosclerosis.2016.11.026. [DOI] [PubMed] [Google Scholar]
- 11.Assarsson E, Lundberg M, Holmquist G, Björkesten J, Thorsen SB, Ekman D, et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One. 2014 Apr;9((4)):e95192. doi: 10.1371/journal.pone.0095192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005 Oct;102((43)):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slawski M, Daumer M, Boulesteix AL. CMA: a comprehensive Bioconductor package for supervised classification with high dimensional data. BMC Bioinformatics. 2008 Oct;9((1)):439. doi: 10.1186/1471-2105-9-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lind L, Sundström J, Stenemo M, Hagström E, Ärnlöv J. Discovery of new biomarkers for atrial fibrillation using a custom-made proteomics chip. Heart. 2017 Mar;103((5)):377–82. doi: 10.1136/heartjnl-2016-309764. [DOI] [PubMed] [Google Scholar]
- 15.Masson S, Aleksova A, Favero C, Staszewsky L, Bernardinangeli M, Belvito C, et al. GISSI-AF investigators Predicting atrial fibrillation recurrence with circulating inflammatory markers in patients in sinus rhythm at high risk for atrial fibrillation: data from the GISSI atrial fibrillation trial. Heart. 2010 Dec;96((23)):1909–14. doi: 10.1136/hrt.2009.191460. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y, Zeng J, Zhang R, Zeng L, Li Y, Wei H, et al. Effect of Interleukin-27 Genetic Variants on Atrial Fibrillation Susceptibility. Genet Test Mol Biomarkers. 2017 Feb;21((2)):97–101. doi: 10.1089/gtmb.2016.0219. [DOI] [PubMed] [Google Scholar]
- 17.Guo Y, Lip GY, Apostolakis S. Inflammation in atrial fibrillation. J Am Coll Cardiol. 2012 Dec;60((22)):2263–70. doi: 10.1016/j.jacc.2012.04.063. [DOI] [PubMed] [Google Scholar]
- 18.Smit MD, Maass AH, De Jong AM, Muller Kobold AC, Van Veldhuisen DJ, Van Gelder IC. Role of inflammation in early atrial fibrillation recurrence. Europace. 2012 Jun;14((6)):810–7. doi: 10.1093/europace/eur402. [DOI] [PubMed] [Google Scholar]
- 19.Shao Q, Liu H, Ng CY, Xu G, Liu E, Li G, et al. Circulating serum levels of growth differentiation factor-15 and neuregulin-1 in patients with paroxysmal non-valvular atrial fibrillation. Int J Cardiol. 2014 Mar;172((2)):e311–3. doi: 10.1016/j.ijcard.2013.12.173. [DOI] [PubMed] [Google Scholar]
- 20.Nakano Y, Niida S, Dote K, Takenaka S, Hirao H, Miura F, et al. Matrix metalloproteinase-9 contributes to human atrial remodeling during atrial fibrillation. J Am Coll Cardiol. 2004 Mar;43((5)):818–25. doi: 10.1016/j.jacc.2003.08.060. [DOI] [PubMed] [Google Scholar]
- 21.Sonmez O, Ertem FU, Vatankulu MA, Erdogan E, Tasal A, Kucukbuzcu S, et al. Novel fibro-inflammation markers in assessing left atrial remodeling in non-valvular atrial fibrillation. Med Sci Monit. 2014 Mar;20:463–70. doi: 10.12659/MSM.890635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerasuolo JO, Cipriano LE, Sposato LA. The complexity of atrial fibrillation newly diagnosed after ischemic stroke and transient ischemic attack: advances and uncertainties. Curr Opin Neurol. 2017 Feb;30((1)):28–37. doi: 10.1097/WCO.0000000000000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirchhof P, Benussi S, Kotecha D, Ahlsson A, Atar D, Casadei B, et al. ESC Scientific Document Group 2016 ESC Guidelines for the management of atrial fibrillation developed in collaboration with EACTS. Eur Heart J. 2016 Oct;37((38)):2893–962. doi: 10.1093/eurheartj/ehw210. [DOI] [PubMed] [Google Scholar]
- 24.Piek A, Du W, de Boer RA, Silljé HH. Novel heart failure biomarkers: why do we fail to exploit their potential? Crit Rev Clin Lab Sci. 2018 Jun;55((4)):246–63. doi: 10.1080/10408363.2018.1460576. [DOI] [PubMed] [Google Scholar]
- 25.Worthmann H, Kempf T, Widera C, Tryc AB, Goldbecker A, Ma YT, et al. Growth differentiation factor 15 plasma levels and outcome after ischemic stroke. Cerebrovasc Dis. 2011;32((1)):72–78. doi: 10.1159/000328233. [DOI] [PubMed] [Google Scholar]