Abstract
Background
Cancer remains the number one cause of disease-related mortality in children, and despite advances in the molecular understanding of leukemia and targeted therapies, refractory leukemia remains a leading cause of death. It therefore is essential to further define features, e.g., FLT3 alterations and KMT2A rearrangements, associated with inferior survival early to augment or alter therapeutic strategies to improve outcomes.
Methods
To gain insights into the genetic drivers predictive of aggressive clinical behavior among pediatric leukemia patients, we performed comprehensive integrative clinical sequencing (ICS), including paired tumor/normal DNA sequencing and RNA-seq, for pediatric patients who presented at our institution over a period of five years with acute lymphoblastic or myelogenous leukemia (ALL and AML; n=43) and high-risk clinical features (high white blood cell count, extramedullary disease, or refractory and/or relapsed disease).
Results
We found that RAS- and Ras-pathway aberrations, including N-RAS, NF1 and PTPN11, are frequent somatic mutations and, importantly, associated with decreased event free and overall survival (OS) (P=0.04, median event free survival 22.8 vs. 5.6 months; P=0.04, median OS 124 vs. 22.5 months).
Conclusions
We thus propose that hyperactive Ras signaling confers inferior survival in high-risk pediatric acute leukemia and that Ras pathways should be molecularly characterized to inform clinical decision making and to identify patients for experimental clinical trials and RAS-targeted therapy.
Keywords: Integrative clinical sequencing (ICS), pediatric leukemia, Ras
Introduction
With advances in genomic sequencing technology, the hope of precision medicine is being harnessed to both better understand disease biology and to provide targeted therapies to minimize toxicity. While innovative laboratory techniques have improved sequencing depth and sensitivity, translating these findings into meaningful diagnostic, prognostic, and therapeutic information for patients remains a major clinical challenge and likely requires large clinical volumes to generate this data. For example, while early phase 1/2 data showed ~40% response to IDH inhibition in adult AML, next-generation sequencing (NGS) identified that non-responders in larger clinical trials had concurrent Ras mutations (1,2). These studies highlight how comprehensive sequencing can yield insightful information that better informs treatment plans. However, with rare exceptions, the majority of these studies have been conducted in adult patients, with mean ages typically in the 60s (3), and where mutational burden is higher and the molecular landscape quite divergent from pediatric malignancies (4-8). Illustratively, recent studies confirm that pediatric malignancies possess fewer and different mutations as compared to their adult counterparts and therefore likely represent biologically distinct disease processes. Together, this accumulating evidence supports that further studies are needed to define the unique genetic signatures and their clinical implications specific to pediatric diseases.
In this vein, large genomic datasets in pediatric cancers have started to become available, but the correlative clinical data is not yet widely accessible. To address this limitation, we completed comprehensive ICS on pediatric and young adult patients over a period of five years who presented with high-risk clinical features at our institution and analyzed this data in the context of their clinical course. Across the spectrum of high risk acute leukemia, our analysis demonstrates that patients who harbor mutations predictive of hyperactive Ras signaling experience shorter event-free (EFS) and overall survival (OS). Notably, this data complements and expands upon studies in specific cohorts of ALL, including relapsed T-cell ALL (9), pre-B cell ALL (10), early T-cell precursor ALL (11), hypodiploid ALL (12), and MLL-rearranged infant ALL (13) and AML (14), by examining pediatric and young adult patients across a spectrum of presentations of acute leukemia. These analyses have uncovered an association of Ras aberrancy with inferior survival, early relapse of ALL, more frequent central nervous system (CNS) involvement and, importantly, sensitivity to MEK inhibition (15). Together, these data highlight the clinical impact of Ras mutational status and the importance of interrogating Ras-mediated leukemic transformation to uncover novel biology and strategically design targeted treatment regimens for high-risk pediatric leukemia patients.
Methods
High-risk or relapsed/refractory pediatric and young adult patients with hematologic malignancies were consented to undergo ICS through the PEDS-MIONCOSEQ study at the C. S. Mott Children’s Hospital from 2012–2017 (UM IRBMED: HUM00056496). Patients with chronic myeloid leukemia, juvenile myelomonocytic leukemia, and mixed phenotype and mixed lineage acute leukemia were excluded from the analysis. Specific methods of sequencing procedures have been described previously (8). Briefly, nucleic acid preparation and high-throughput sequencing were performed using standard protocols, adhering to Clinical Laboratory Improvement Amendments (CLIA). Our cohort was sequenced using two panels: prior to January 2016, we used whole-exome sequencing and, since that time, a 1,700 gene panel replaced WES. Paired-end whole-exome/OncoSeq 1,700 libraries and transcriptome libraries from bone marrow or peripheral blood samples that were matched with normal DNA were prepared and sequenced. Sequences were analyzed to detect putative somatic mutations, insertions and deletions, copy-number alterations, gene fusions, and gene expression. Statistical analysis was performed using Prism GraphPad software (San Diego, CA, USA). Differences between groups (Ras-aberrant vs. non-Ras aberrant) for clinical data were calculated using a Chi-Square test or unpaired, two-tailed student’s t-test, depending on the analysis. EFS and OS were calculated from date of initial diagnosis using Kaplan-Meier analyses to either first event using Gehan-Breslow-Wilcoxon test (relapse/death for EFS) or log-rank test for death (OS).
Results
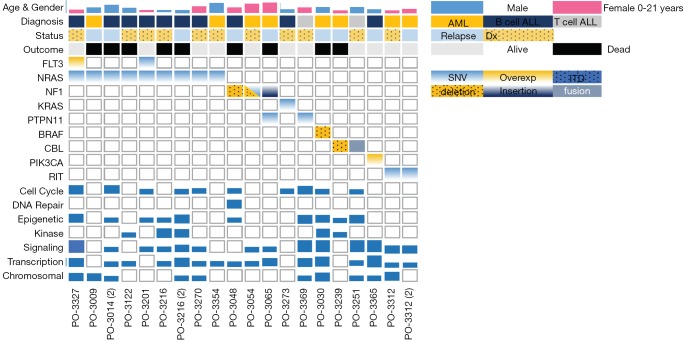
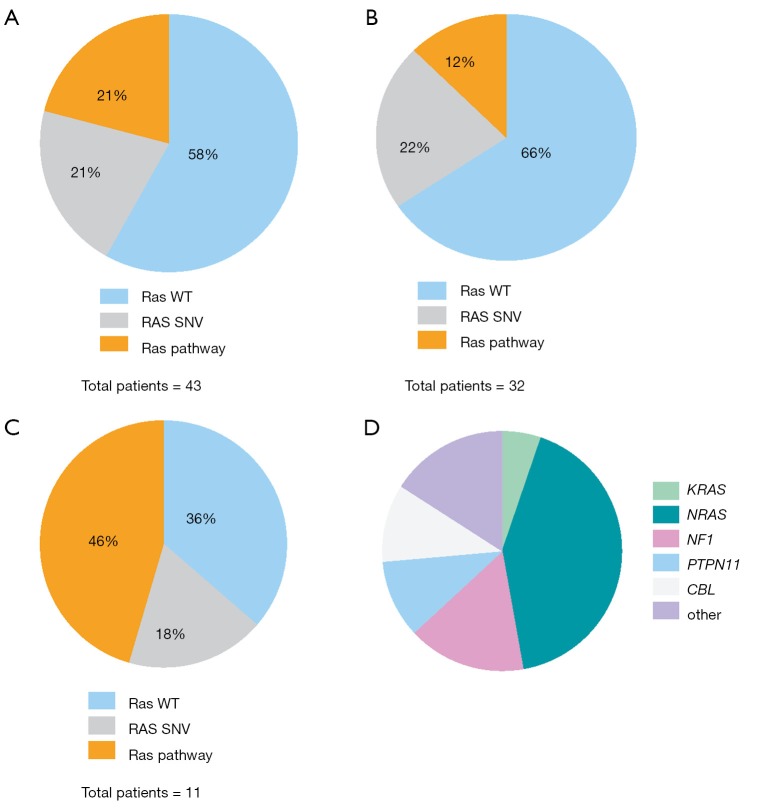
Among the acute leukemia patients sequenced, 32 (74%) had acute lymphoblastic leukemia (ALL) and 11 patients (26%) had acute myeloid leukemia (AML). Of these, 22 (51%) were sequenced at the patient’s initial diagnosis (n=18/22 with ALL, n=4/22 with AML), while the remaining 21 cases (49%) were patients with either relapsed or refractory disease (14/21 =ALL, 7/21 =AML; Table 1). Our initial analysis revealed that the most prevalent activating mutations were for Ras-signaling pathways (Figure 1, Table S1). In this group, we include all NRAS, KRAS or HRAS mutations, along with well-characterized Ras-pathway aberrations (NF1, PTPN11, PI3K, BRAF). Importantly, while FLT3 mutations also activate Ras signaling (16), patients with FLT3 mutations (12/55 patients sequenced during this analysis) were excluded from this study given its established role conferring inferior survival in leukemia (17). It is noteworthy, however, that concurrent Ras mutations have recently been shown to promote chemoresistance in FLT3 mutant leukemia and co-mutational burden may, in fact, synergize to confer inferior survival (18). Nonetheless, patients in our cohort had a much higher percentage, 42% (n=18/43), of RAS and Ras-pathway aberrations than would be anticipated based on previously published pediatric (14,19-21) and adult leukemia series (22) but in-line with recent reports of large cohort studies and of high-risk and relapsed leukemia patients (4,5,15,23-25). Of the 42% of mutations detected, approximately half were Ras-pathway aberrations (21%), while the remainder (21%) were point mutations in the Ras family of oncogenes (Figure 2A,B,C, Table 2), with single nucleotide variants in NRAS being more common than KRAS (Figure 2D). We next compared the frequency of these mutations by disease type and clinical status at sample collection. Surprisingly, pediatric patients with myeloid neoplasms did not have a significantly higher prevalence of RAS and Ras-pathway aberrations (7/11; 64%) compared to pediatric ALL patients (11/32; 34%) (P=0.09; Figure 2B,C; Table 1). To understand if mutations conferring hyperactive Ras signaling were enriched at disease recurrence, we compared the frequency of these mutations in unmatched diagnostic and relapsed/refractory samples, but observed no difference, as 11 of 22 (50%) diagnostic samples contained RAS or Ras-pathway aberrations, while 7 of 21 (33%) relapsed samples harbored these mutations (P=0.27; Table 1). These data suggest that a considerable number of high-risk pediatric leukemia patients, whether at diagnosis or relapse, harbor mutations predictive of hyperactive Ras signaling.
Table 1. Characteristics of Patients with and without Ras-aberrant mutations.
| Patient characteristic | Non-Ras aberrant (N=25) | Ras-aberrant (N=18) | P |
|---|---|---|---|
| Demographics | |||
| Age (mean ± SEM) | 0.2–22.4 (9.3±1.5) | 0.5–17.9 (8.6±1.4) | 0.76 |
| Male (%) | 15 [60] | 8 [44] | 0.313 |
| Clinical information | |||
| WBC (mean ± SEM) k/μL | 66.8±31 | 40.4±22 | 0.49 |
| CNS involvement (%) | 8 [32] | 6 [33] | 0.93 |
| ICU admission (%) | 3 [12] | 4 [22] | 0.37 |
| Sample type (%) | |||
| ALL | 21 [84] | 11 [61] | |
| AML | 4 [16] | 7 [39] | |
| Diagnostic (%) | 11 [44] | 11 [61] | |
| ALL | 10 [40] | 8 [44] | |
| AML | 1 [4] | 3 [17] | |
| Relapse/refractory (%) | 14 [56] | 7 [39] | |
| ALL | 11 [44] | 3 [17] | |
| AML | 3 [12] | 4 [22] | |
| Treatment | |||
| Allogeneic BMT (%) | 14 [56] | 6 [33] | 0.14 |
| Number of treatment regimens received (mean ± SEM) | 1–11 (3.4±0.5) | 1–5 (2.6±0.38) | 0.27 |
| Mutational landscape | |||
| Range of mutations detected (mean ± SEM) | 1–20 (7.4±1.1) | 2–25 (8.2±1.6) | 0.69 |
| KMT2A present (%) | 6 [24] | 6 [33] | 0.5 |
| Chromosomal abnormalities detected (mean ± SEM) | 0.52±0.15 | 0.89±0.25 | 0.22 |
Data are shown as mean ± SEM or n [%]. SEM, standard error of mean; WBC, white blood cell; CNS, central nervous system; ICU, intensive care unit; ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; BMT, bone marrow transplant.
Figure 1.
Overview of genetic landscape in sequenced pediatric patients with Ras aberrancies. Prevalence of RAS and Ras-pathway aberrations in hematologic malignancies broken down by gender, age, disease type, disease status at sequencing and clinical outcome with concurrent mutational burden.
Figure 2.
Frequency of RAS and Ras-pathway mutations in pediatric hematologic malignancies. (A) RAS and Ras-pathway mutations in all patients; (B) RAS and Ras-pathway mutations in ALL; (C) percentage of RAS and Ras pathway aberrations in AML; (D) frequency of genetic mutations identified to predict hyperactive Ras signaling in sequenced patients. ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia.
Table 2. Detected mutations in RAS or Ras pathway genes [n].
| Gene [total mutation No.] | Mutations detected by gene |
|---|---|
| NRas [10] | All single nucleotide variants: G12D [2]; G12S [2]; G12A [2]; G13D [1]; G13R [1]; Q61H [2] |
| KRas [1] | G12D [1] |
| PTPN11 [3] | All single nucleotide variants: E76Q [1]; A461G [1]; G483V [1] |
| NF1 [4] | Single nucleotide variant Y333I [1]; homozygous deletion [1]; frameshift deletion [2] |
| CBL [2] | Deletion [1]; MLL-CBL fusion [1] |
| Other | BRAF loss [1]; RIT1 (p.F82C); PIK3CA |
MLL, mixed lineage leukemia; CBL, Casitas B-Lineage Lymphoma Proto-oncogene.
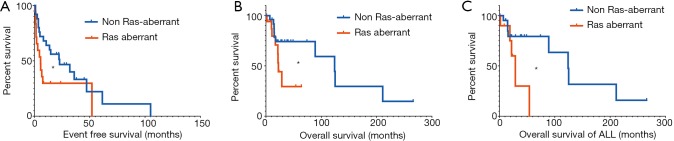
Given these findings, we next sought to understand if RAS and Ras-pathway aberrations had any impact on clinical outcomes and should therefore be considered in medical management. Importantly, in adult leukemia, RAS mutations have not been prognostic to clinical outcomes (26,27). To understand this in pediatric leukemias, we compared EFS in Ras-aberrant patients to that of non-Ras-aberrant patients and observed a significantly shorter EFS in the Ras-aberrant patients (median EFS 5.6 vs. 22.8 months; P=0.04) (Figure 3A). In an attempt to uncover if hyperactive Ras signaling contributed only to lower EFS and perhaps early relapse but bore no ultimate effect on OS, we compared OS of Ras-aberrant patients to non-Ras aberrant patients and again observed significantly shorter OS (median OS 22.5 vs. 124 months; P=0.04) (Figure 3B). Given that myeloid and lymphoid malignancies are distinct diseases with overall different expected survival, we attempted to independently analyze clinical outcomes in each disease cohort. Due to low patient numbers with AML, we were unable to make meaningful conclusions for AML. However, we were able to identify inferior OS in ALL patients with genetic lesions in Ras pathway genes compared to those without (P=0.03; median survival 28.7 vs. 124 months) (Figure 3C), similar to other studies (25). Notably, the OS for both Ras-aberrant and non-Ras aberrant patients is much lower than reported for standard-risk pediatric leukemia patients and highlights the high-risk disease features represented by our cohort (28).
Figure 3.
Survival of sequenced patients. (A) Event free survival comparing Ras aberrant (red) to non-Ras aberrant (blue); (B) overall survival of Ras aberrant (red) and non-Ras aberrant (blue) patients; (C) overall survival of ALL patients with mutations predictive of Ras aberrancy (red) or non-Ras aberrant (blue). *, P<0.05. ALL, acute lymphoblastic leukemia.
Given the inferior survival of patients with mutations predictive of Ras aberrancy, we examined if these patients are more likely to present with high-risk clinical features, and could therefore account for higher mortality. Notably, there was no difference in CNS involvement between Ras-aberrant and non-Ras aberrant patients (33% vs. 32%, P=0.93) (Table 1), total white blood cell count at diagnosis/relapse (40.4±22 vs. 66.8±31; P=0.49) or initial ICU admission (22% vs. 12%, P=0.37) (Table 1). This data suggests that hyperactive Ras signaling confers inferior survival without initially evident high-risk clinical features. Our observations are supported by the association of NRAS mutations with aggressive clinical behavior including early relapse (15,29) and resistance to chemotherapy, including vincristine and MTX (30), and IDH inhibition (1).
Next, we wanted to dissect if Ras-aberrant mutations could have a direct role in inferior survival. To do this, we examined if Ras-aberrant mutations occurred in older pediatric patients, where mutational frequency is likely higher and therefore the accumulation of secondary mutations more likely and could result in chemotherapy resistance and/or further genomic instability. However, we saw no difference in the mean age between Ras-aberrant versus Ras non-aberrant patients (8.6±1.4 vs. 9.3±1.5 years; P=0.76; Table 1). Consistent with this, we also observed no difference in the mean number of mutations detected in Ras-aberrant versus non Ras-aberrant samples (8.2±1.6 vs. 7.4±1.1; P=0.69). Furthermore, we observed no difference between patient cohorts in the frequency of additional high-risk genetic features, including patients with KMT2A fusions (6/18, 33% and 6/25, 24%; P=0.5; Table 1) or the mean number of chromosomal abnormalities detected (0.89±0.25; 0.52±0.15; P=0.22). Together, these data suggest that mutations predictive of Ras aberrancy are associated with inferior survival without requiring the accumulation of further genetic lesions that contribute to treatment resistance and genomic instability.
Discussion
Improved efficiency in sequencing techniques have allowed us to define genetic alterations for pediatric leukemia in real-time. However, with these results arises a great need to ascribe their function and clinical significance. Here, using ICS, we show across a spectrum of high risk acute pediatric leukemia that RAS and Ras-pathway aberrations are associated with inferior EFS and OS. These findings are not dependent on other high-risk genetic features, such as KMT2A fusions, or on chromosomal or mutational burden. Rather, we propose Ras aberrancy is an independent risk factor for aggressive clinical behavior and that the downstream consequences of hyperactive Ras signaling directly contribute to inferior survival and, potentially, therapy resistance. Notably, while oncogenic Ras has not been validated as a leukemia-initiating event in adult disease, its biologic significance in pediatric leukemia has yet to be defined. Hence, murine models that activate Ras signaling may best recapitulate the steps of pediatric leukemogenesis, where mutational burden is lower and therefore driven by fewer, distinct, and perhaps more fully penetrant genetic lesions. These models may provide insightful evidence regarding the potential role of Ras aberrancy in disease development and treatment resistance.
Importantly, large pediatric cohort studies have now identified mutations in RAS and Ras pathways as frequent lesions in many pediatric cancers (4,5,24), specifically in the relapsed or refractory setting (6,15,31). This finding, when combined with inferior survival, early relapse and chemotherapy resistance, illuminate two fundamental questions regarding the precise role of hyperactive Ras signaling in both leukemogenesis and treatment refractoriness. In murine models, it is known that oncogenic NRasG12D/+ induces hematopoietic stem cell (HSC) dysregulation, where hyperactive Ras signaling drives both pre-leukemic proliferation and clonal expansion but also HSC self-renewal (32). Importantly, complementary studies have revealed that quiescent pre-LSCs persist despite negative minimal residual disease testing (MRD), are therefore resistant to treatment, and can hence form the basis of relapse (33,34). Combined, murine models support that hyperactive Ras signaling drives leukemogenesis through HSC dysregulation but accumulating clinical data suggests that this may ultimately lead to treatment refractoriness and relapse. Understanding these paradigms is of critical importance in the research laboratory and the clinic if we are to better define disease-initiating cell populations and more effectively target disease-propagating events that may be specific to pediatric diseases. Ultimately, these results suggest that real-time ICS looking to identify RAS mutations and known Ras-pathway aberrations holds great promise in realizing the power of precision oncology in pediatrics. If our findings are validated by larger cohorts, we propose these alterations should be part of expanded disease stratification when tailoring treatment regimens based on patient genomics, such as the addition of MEK inhibitors (15), in order to improve outcomes.
Table S1. Characteristics of patients with RAS or Ras pathway mutations.
| Age (years) | Disease | Presenting white blood cell count (k/μL) | CNS | Ras pathway mutation | Mutation frequency | Diagnosis or relapse | Co-occurring mutations | Total somatic lesions |
|---|---|---|---|---|---|---|---|---|
| 4 | AML | 0.6 | Negative | NRAS (p.G13R) | 0.46 | Relapse | Copy loss of 6q and 20q | 14 |
| 14.3 | Pre B ALL | 5.6 | Positive | NRAS (p.Q61H) | 0.38 | Relapse | p53, MLL2 | 7 |
| 13.6 | AML | 49.8 | Negative | BRAF | Refractory | CSF3R, EIF4A2-MECOM, Chr7q, Chr3q loss,Chr21 gain | 6 | |
| 8.5 | Pre B ALL | 6.1 | Negative | NF1 | Deletion | Relapse | p53, MAX, KDM6A | 5 |
| 17.9 | AML | 0.7 | Positive | NF1 (p.Y333I) | 0.23 | Diagnosis | TSC2, CBFB-MYH11 fusion | 3 |
| 17.9 | AML | 3.7 | Negative | PTPN11 (E76Q) | 0.12 | Relapse | NF1, WT1, BIRC-CEBPZ, EDF1-NOTCH | 5 |
| 5.2 | ALL | 39.5 | Negative | NRAS (p.G12S) | 0.31 | Diagnosis | ETV6-RUNX1, BCR-ABL1 | 3 |
| 0.9 | Pre B ALL | 33 | Positive |
NRAS (p.G12D) (p.G12S) |
0.02 | Diagnosis | KMT2D, FLT3, CDKN2A | 9 |
| 0.5 | Pre B ALL | 30.5 | Positive | NRAS (p.G12A) | 0.32 to 0.38 | Diagnosis + relapse | PIK3R1, MLL | 4 |
| 1.8 | AML | 0.4 | Negative | CBL | Refractory | MLL | 3 | |
| 11.5 | T ALL | 1.1 | Negative | CBL | Diagnosis | NOTCH1, IL7R, SH2B3 | 4 | |
| 13 | Pre B ALL | 2.3 | Positive | NRAS (p.G12A) | 0.03 | Diagnosis | PAX5 | 4 |
| 1.8 | Pre B ALL | 7.9 | Negative | KRAS (p.G12D) | 0.03 | Diagnosis | CCNB3 | 2 |
| 4 | AML | 12.7 | Negative | RIT1 (p.F82C) | 0.41 | Diagnosis | KIT | 8 |
| 6 | ALL | 3.1 | Negative | NRAS (p.G13D) | 0.01 | Diagnosis | DOT1L, RUNX1 | 21 |
| 18 | AML | 89 | Positive | NRAS | Diagnosis | HNRNPH1-ERG | 3 | |
| (p.G12D) | 0.01 | |||||||
| (p.Q61H) | 0.08 | |||||||
| 6 | ALL | 35 | Negative | PIK3CA | Relapse | XBP1, BCL2L14, ETV6 | 18 | |
| 11 | T cell ALL | 407 | Negative | PTPN11 | Diagnosis | JAK1, JAK3, NOTCH1 | 25 | |
| (p.A461G) | 0.46 | |||||||
| (p.G483V) | 0.45 |
AML, acute myeloid leukemia; ALL, acute lymphoblastic leukemia; CNS, central nervous system.
Acknowledgments
We would like to thank our patients and families for participating in this study and advancing our knowledge and molecular understanding of pediatric oncology.
Funding: GM Ney was funded by an NIH T32 in Molecular Hematology (5-T32-HL007622) and is a St. Baldrick’s Foundation Fellow with Tough Like Ike Support. Q Li is supported through NIH/NHLBI R01HL132392 and is an American Cancer Society Research Scholar recipient. R Mody is supported by Hyundai Hope on Wheels Scholar award. Clinical sequencing was supported by grant 1UM1HG006508 from the National Institutes of Health Clinical Sequencing Exploratory Research Award (PI: AM Chinnaiyan).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the University of Michigan Institutional Review Board (approval number: HUM00056495).
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Amatangelo MD, Quek L, Shih A, et al. Enasidenib induces acute myeloid leukemia cell differentiation to promote clinical response. Blood 2017;130:732-41. 10.1182/blood-2017-04-779447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stein EM, DiNardo CD, Pollyea DA, et al. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood 2017;130:722-31. 10.1182/blood-2017-04-779405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu J, Lichtenberg T, Hoadley KA, et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018;173:400-16.e11. 10.1016/j.cell.2018.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gröbner SN, Worst BC, Weischenfeldt J, et al. The landscape of genomic alterations across childhood cancers. Nature 2018;555:321-7. 10.1038/nature25480 [DOI] [PubMed] [Google Scholar]
- 5.Ma X, Liu Y, Liu Y, et al. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature 2018;555:371-6. 10.1038/nature25795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eleveld TF, Oldridge DA, Bernard V, et al. Relapsed neuroblastomas show frequent RAS-MAPK pathway mutations. Nat Genet 2015;47:864-71. 10.1038/ng.3333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stieglitz E, Taylor-Weiner AN, Chang TY, et al. The genomic landscape of juvenile myelomonocytic leukemia. Nat Genet 2015;47:1326-33. 10.1038/ng.3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mody RJ, Wu YM, Lonigro RJ, et al. Integrative Clinical Sequencing in the Management of Refractory or Relapsed Cancer in Youth. JAMA 2015;314:913-25. 10.1001/jama.2015.10080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richter-Pechańska P, Kunz JB, Hof J, et al. Identification of a genetically defined ultra-high-risk group in relapsed pediatric T-lymphoblastic leukemia. Blood Cancer J 2017;7:e523. 10.1038/bcj.2017.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang DC, Chen SH, Liu HC, et al. Mutational status of NRAS, KRAS, and PTPN11 genes is associated with genetic/cytogenetic features in children with B-precursor acute lymphoblastic leukemia. Pediatr Blood Cancer 2018;65:e26786. 10.1002/pbc.26786 [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, Ding L, Holmfeldt L, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 2012;481:157-63. 10.1038/nature10725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmfeldt L, Wei L, Diaz-Flores E, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet 2013;45:242-52. 10.1038/ng.2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andersson AK, Ma J, Wang J, et al. The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat Genet 2015;47:330-7. 10.1038/ng.3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meshinchi S, Stirewalt DL, Alonzo TA, et al. Activating mutations of RTK/ras signal transduction pathway in pediatric acute myeloid leukemia. Blood 2003;102:1474-9. 10.1182/blood-2003-01-0137 [DOI] [PubMed] [Google Scholar]
- 15.Irving J, Matheson E, Minto L, et al. Ras pathway mutations are prevalent in relapsed childhood acute lymphoblastic leukemia and confer sensitivity to MEK inhibition. Blood 2014;124:3420-30. 10.1182/blood-2014-04-531871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilliland DG, Griffin JD. The roles of FLT3 in hematopoiesis and leukemia. Blood 2002;100:1532-42. 10.1182/blood-2002-02-0492 [DOI] [PubMed] [Google Scholar]
- 17.Kiyoi H, Naoe T, Nakano Y, et al. Prognostic implication of FLT3 and N-RAS gene mutations in acute myeloid leukemia. Blood 1999;93:3074-80. [PubMed] [Google Scholar]
- 18.McMahon CM, Ferng T, Canaani J, et al. Clonal Selection with RAS Pathway Activation Mediates Secondary Clinical Resistance to Selective FLT3 Inhibition in Acute Myeloid Leukemia. Cancer Discov 2019;9:1050-63. 10.1158/2159-8290.CD-18-1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunger SP, Mullighan CG. Redefining ALL classification: toward detecting high-risk ALL and implementing precision medicine. Blood 2015;125:3977-87. 10.1182/blood-2015-02-580043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts KG, Mullighan CG. Genomics in acute lymphoblastic leukaemia: insights and treatment implications. Nat Rev Clin Oncol 2015;12:344-57. 10.1038/nrclinonc.2015.38 [DOI] [PubMed] [Google Scholar]
- 21.Iacobucci I, Mullighan CG. Genetic Basis of Acute Lymphoblastic Leukemia. J Clin Oncol 2017;35:975-83. 10.1200/JCO.2016.70.7836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen Y, Zhu YM, Fan X, et al. Gene mutation patterns and their prognostic impact in a cohort of 1185 patients with acute myeloid leukemia. Blood 2011;118:5593-603. 10.1182/blood-2011-03-343988 [DOI] [PubMed] [Google Scholar]
- 23.Faber ZJ, Chen X, Gedman AL, et al. The genomic landscape of core-binding factor acute myeloid leukemias. Nat Genet 2016;48:1551-6. 10.1038/ng.3709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolouri H, Farrar JE, Triche T, et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat Med 2018;24:103-12. 10.1038/nm.4439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jerchel IS, Hoogkamer AQ, Aries IM, et al. RAS pathway mutations as a predictive biomarker for treatment adaptation in pediatric B-cell precursor acute lymphoblastic leukemia. Leukemia 2018;32:931-40. 10.1038/leu.2017.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowen DT, Frew ME, Hills R, et al. RAS mutation in acute myeloid leukemia is associated with distinct cytogenetic subgroups but does not influence outcome in patients younger than 60 years. Blood 2005;106:2113-9. 10.1182/blood-2005-03-0867 [DOI] [PubMed] [Google Scholar]
- 27.Metzeler KH, Herold T, Rothenberg-Thurley M, et al. Spectrum and prognostic relevance of driver gene mutations in acute myeloid leukemia. Blood 2016;128:686-98. 10.1182/blood-2016-01-693879 [DOI] [PubMed] [Google Scholar]
- 28.Matloub Y, Bostrom BC, Hunger SP, et al. Escalating intravenous methotrexate improves event-free survival in children with standard-risk acute lymphoblastic leukemia: a report from the Children's Oncology Group. Blood 2011;118:243-51. 10.1182/blood-2010-12-322909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindsley RC, Saber W, Mar BG, et al. Prognostic Mutations in Myelodysplastic Syndrome after Stem-Cell Transplantation. N Engl J Med 2017;376:536-47. 10.1056/NEJMoa1611604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oshima K, Khiabanian H, da Silva-Almeida AC, et al. Mutational landscape, clonal evolution patterns, and role of RAS mutations in relapsed acute lymphoblastic leukemia. Proc Natl Acad Sci U S A 2016;113:11306-11. 10.1073/pnas.1608420113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X, Stewart E, Shelat AA, et al. Targeting oxidative stress in embryonal rhabdomyosarcoma. Cancer Cell 2013;24:710-24. 10.1016/j.ccr.2013.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Q, Bohin N, Wen T, et al. Oncogenic Nras has bimodal effects on stem cells that sustainably increase competitiveness. Nature 2013;504:143-7. 10.1038/nature12830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shlush LI, Zandi S, Mitchell A, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 2014;506:328-33. 10.1038/nature13038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corces-Zimmerman MR, Hong WJ, Weissman IL, et al. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc Natl Acad Sci U S A 2014;111:2548-53. 10.1073/pnas.1324297111 [DOI] [PMC free article] [PubMed] [Google Scholar]