Abstract
Biology is well-known for its ability to communicate through (i) molecularly-specific signaling modalities and (ii) a globally-acting electrical modality associated with ion flow across biological membranes. Emerging research suggests that biology uses a third type of communication modality associated with a flow of electrons through reduction/oxidation (redox) reactions. This redox signaling modality appears to act globally and has features of both molecular and electrical modalities: since free electrons do not exist in aqueous solution, the electrons must flow through molecular intermediates that can be switched between two states - with electrons (reduced) or without electrons (oxidized). Importantly, this global redox modality is easily accessible through its electrical features using convenient electrochemical instrumentation. In this review, we explain this redox modality, describe our electrochemical measurements, and provide four examples demonstrating that redox enables communication between biology and electronics. The first two examples illustrate how redox probing can acquire biologically relevant information. The last two examples illustrate how redox inputs can transduce biologically-relevant transitions for patterning and the induction of a synbio transceiver for two-hop molecular communication. In summary, we believe redox provides a unique ability to bridge bio-device communication because simple electrochemical methods enable global access to biologically meaningful information. Further, we envision that redox may facilitate the application of information theory to the biological sciences.
Keywords: bioelectronics, electrobiofabrication, molecular communication, signal processing, reverse engineering
I. Introduction
INFORMATION Age transformed our lives and the application of information theory to enable communication with our biological world could offer similarly transformative possibilities. One vision is illustrated in Fig. 1a which shows a micro/nano device (e.g., a vesicle) “talking” to biology (e.g., a cell) through the native molecular modalities used for cell-cell signaling[1], [2], [11]–[15], [3]–[10] Such bio-device communication could enable us: (i) to acquire information to understand how a complex biological system functions and to detect dysfunctions (e.g., pathological states); and (ii) to transduce biology by providing the cues to induce molecular and cellular transitions (e.g., to induce changes in gene expression). Ultimately, such bio-device communication could connect biology to the Internet of Things (or Bio-Nano-Things). [10]
Fig. 1.
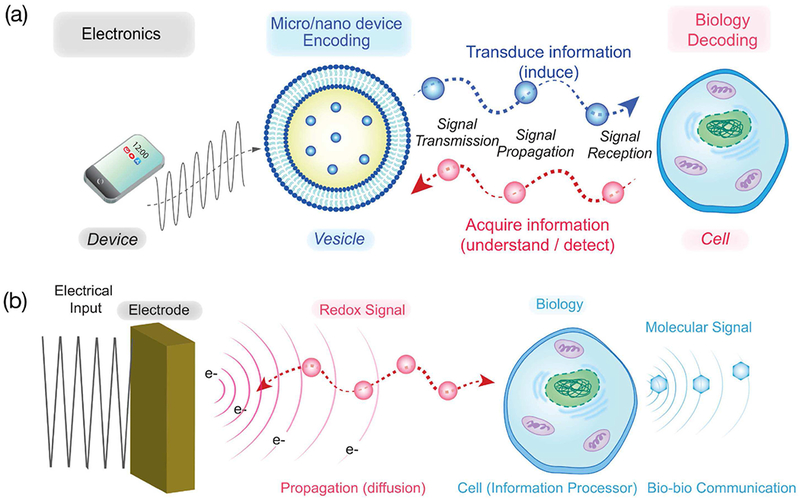
Molecular communication vision. (a) A broad vision of molecular communication involves nanoscale-devices “talking” to biology through the native molecular modalities of biology. (b) A simpler vision of communicating with biology through a global device-compatible modality.
From an experimentalist’s perspective, a key challenge to this molecular communication vision is the “hardware” needed to transmit and receive specific molecular signals. Biology uses highly evolved “components” (e.g., enzymes and receptors) to transmit and detect molecular signals with the necessary levels of sensitivity and selectivity. But these biological transmitter/receiver components are not stable. From a biological perspective, cells are constantly “turning-over” (degrading and re-synthesizing) their molecular machinery to enable them to respond to dynamically-changing environments. From a technological perspective, the short lifetime (hours to days) of bio-based transmitter/receiver components presents a substantial hurdle to creating devices capable of communicating through molecularly-specific modalities.
A related issue is how to convert the molecular information of a biological signal into a modality that allows easy communication to an outside observer. Information technology largely transmits/receives information through an electromagnetic modality but biology rarely uses electromagnetic radiation for communication. Of course, biological scientists routinely engineer cells (i) to report biologically-relevant information through such convenient optical modalities (e.g., through the expression of green fluorescent protein; GFP), or (ii) to receive optical inputs to induce desired biological responses (e.g., by opto-genetics). While successfully connecting the outside observer to the internal workings of complex biological systems, these optical modalities are generally “non-native” to the biological system being probed. Rather, cells must be purposefully engineered to allow measurement through optical outputs or to induce responses from optical inputs. Thus, although these optical approaches represent an important advance, they are intrinsically limited in their capabilities to observe the rich molecular context of a complex biological system or to participate-in the existing communication networks of life.
Fig. 1b illustrates that we are focused on a somewhat simpler problem of using a device to directly “connect” to biology without an intervening nano/micro-scale device. Yet the challenge remains of bridging biological and technological communication using a modality that offers both technological convenience of modern electronics and the biological relevance of molecular modalities. The specific question is: what modalities allow us to engage biology in meaningful (even multi-hop) communication while simultaneously providing access to convenient instrumentation for transmitting and receiving information?
The most obvious example of a modality that enables simple yet meaningful communication between technology and biology is the electrical modality associated with ionic currents across cellular membranes. Fig. 2a illustrates that comparatively simple electrode-based measurements enable electrophysiologists to discern how nerve cells use this electrical modality for communication and allow clinicians to characterize the functioning of a complex biological system (i.e., the cardiovascular system). In addition to using electrode measurements to acquire information, Fig. 2b also shows that electrodes can also be used to provide the cues to transduce a biological system by correcting life-threatening cardiac dysrhythmias.
Fig. 2.
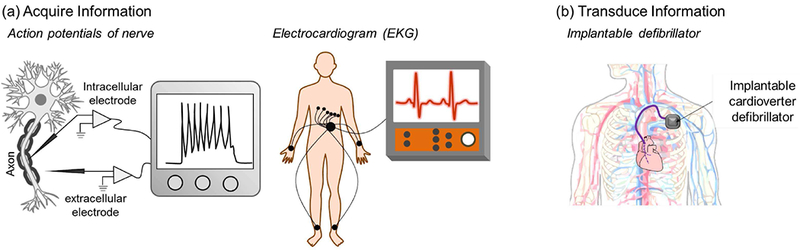
Biology’s global electrical modality associated ion flow across membranes provides easy technological access. (a) Electrode measurements that lack molecular specificity allow neurophysiologists to acquire biological information of the communication of nerve cells and allow clinicians to monitor the health of a complex organ system. (b) Electrode inputs can actuate biology to preserve life.
Thus, a critical challenge for realizing the exciting vision of molecular communication is identifying a communication modality that is technologically convenient yet biologically relevant. The electrical modality associated with ion flow across cell membranes provides an example of such a modality. But while this electrical modality enables convenient communication with the neural, cardiovascular and neuromuscular systems, it is not a particularly relevant modality for communicating with other important biological systems that range from the immune system to the biosphere (e.g., plant-microbe communication in soil). In Section II we describe the redox modality that is based on reduction and oxidation reactions. We illustrate the importance of redox reactions in biology and explain why we believe redox is a suitable modality for bio-device communication. We then describe four case studies. The first two case studies are described in Section III and focus on the use of redox for acquiring biologically-relevant information. In Section IV, we describe two case studies that illustrate how redox-based inputs can be used for transducing transitions in biomacromolecular structure and biological function. In the final section, we provide an experimentalists perspective of the importance of integrating information theory into life science applications.
II. Redox as a Communication Modality
In the above, we explained that biology sometimes communicates using molecularly-specific modalities and sometimes using a global electrical modality associated with the flow of ions across membranes. From a technology standpoint, fabricating transmitters and receivers for molecularly-specific communication will be challenging while communication through the electrical modality has been comparatively easy. Here, we suggest redox is another relevant communication modality that may also permit comparatively simple bio-device “interfacing”.
A. Redox in Biology
Reduction and oxidation (redox) reactions are ubiquitous in biology. Fig. 3a shows that cells harvest energy from their chemical fuel through a redox-pathway (respiration) in which individual electrons are transferred via membrane-bound carriers to an ultimate electron acceptor (i.e., oxygen). Fig. 3b illustrates that biosynthesis often involves the transfer of electrons using diffusible electron carriers (e.g., NADPH) that enable the generation of more-reduced cellular components (e.g., lipids). During an inflammatory response, Fig. 3c shows immune cells use enzymes to transfer electrons to oxygen to generate partially-reduced reactive oxygen species (ROS) that can defend against pathogen invasion (e.g., at a wound site). The damaging effects of high levels of ROS are responsible for their antimicrobial activities but increasing evidence suggests prolonged inflammation may be linked to oxidative stress and various human diseases[16]–[20] In fact, biology uses specific antioxidants (e.g., ascorbic acid and glutathione) to protect itself from such oxidative stresses. In contrast, low levels of ROS (especially H2O2) are believed to serve as diffusible signaling molecules that can cross cell membranes and be recognized through specialized signal transduction mechanisms (e.g., sulfur-switching) to induce diverse cellular responses (Fig. 3d). [21]–[24]
Fig. 3.
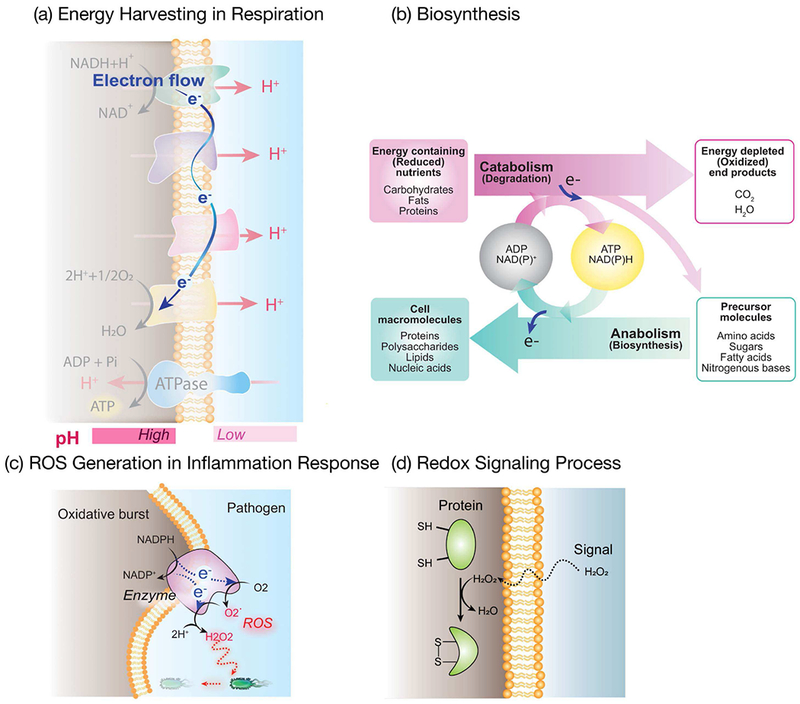
Redox is a critical modality in biology. (a) Energy harvesting in respiration occurs through a redox pathway in which individual electrons are transferred to membrane components that can be switched between reduced (with electrons) and oxidized (without electrons) states. (b) Biosynthesis often involves the transfer of electrons via diffusible electron carriers (NAD(P)H) to generate more-reduced cell components. (c) In the inflammatory response, damaging reaction oxygen species (ROS) are generated by enzymatic electron transfer to partially-reduce O2. (d) Diffusible redox-signaling molecule (H2O2) can engage cellular signal transduction mechanisms.
Because of the importance of redox in biology, there have been efforts to develop a quantitative measurement that characterizes the oxidative-reductive state much as a measurement of pH provides a quantitative measurement of acid-base context. Fig. 4 shows that acid-base and redox reactions have considerable similarities. In acid-base reactions, protons (H+) are donated or accepted, the thermodynamics is characterized by an equilibrium constant (Ka or pKa), and the Hendersen-Hasselbalch equation relates the experimentally measurable pH to one thermodynamic term and one term that relates how the ratio of concentrations adjusts to this environmental pH-context. Analogously, Fig. 4 shows that in redox reactions, electrons (e−) are donated or accepted, the thermodynamic equilibrium is characterized by a standard state redox potential (E0) and the Nernst equation also shows two terms: one thermodynamic term that describes reaction equilibrium (E0) and the other term describing the ratio of concentrations.
Fig. 4.
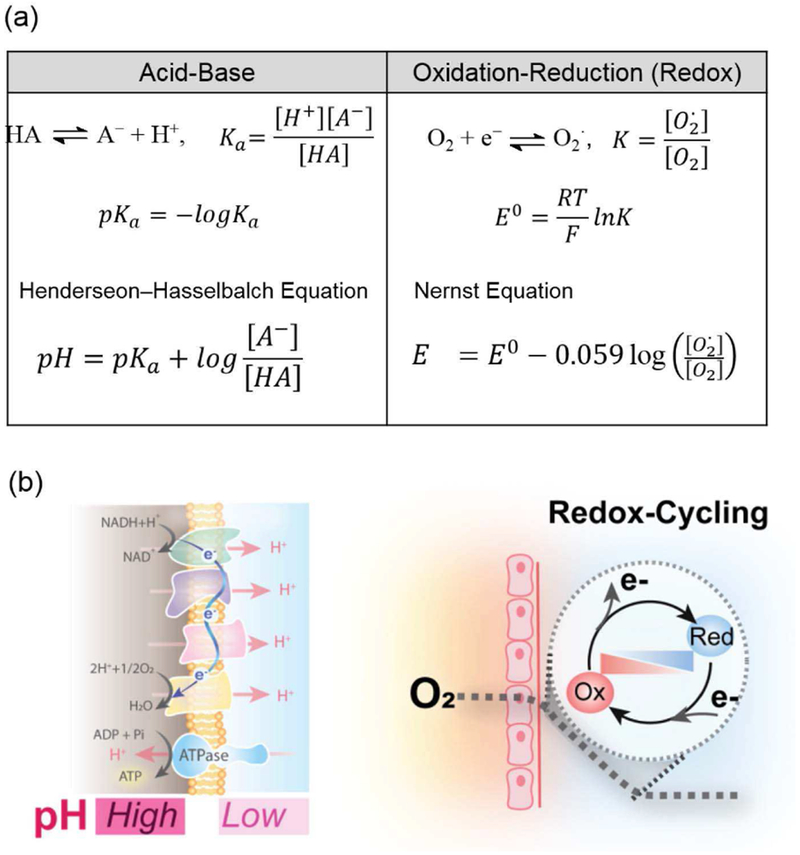
Analogy between redox and acid-base reactions. (a) Quantitative analysis of equilibrium is similar although many redox reactions do not spontaneously achieve equilibrium. (b) The importance of pH gradients across membranes is well known while redox gradients resulting from O2 gradients can lead to redox-cycling.
A critical difference between acid-base and redox reactions is the kinetics. Acid-base reactions are generally believed to be in dynamic equilibrium such that a measurement of pH provides an unambiguous indicator of the acid-base context (i.e., Hendersen-Hasselbalch equation is generally believed to be valid at all times). In contrast, redox reactions can have significant kinetic barriers such that a redox reaction that may be highly favored thermodynamically may not occur in the absence of an enzyme that catalyzes the reaction (i.e., the Nernst equation describes the theoretically possible state but not necessarily the actual state). For instance, some types of redox-active molecules can readily exchange electrons and their redox state rapidly adjusts to the environmental context (as predicted by the Nernst equation), while other types are kinetically stable and resist such changes in state. Thus unlike pH, it is not clear how to measure the redox potential (E) nor to interpret redox measurements that claim to reflect E [25] Despite this uncertainty and because of the importance of redox potential, there have been efforts to use imperfect measurements of E as a global state indicator[26]–[28] Further, it has been reported that different compartments within a cell can exist at different E values (e.g., the mitochondria is more reducing while the cytoplasm is more oxidizing),[29] and that cells progress from proliferation to differentiation to death at increasingly oxidative E values. [30] In our opinion, E measurements are imperfect but because of the importance of redox in biology, an imperfect measurement is better than no measurement.
Also illustrated in Fig. 4b, is that there can be significant and persistent gradients in pH and redox potential. For instance, electron transfer in the respiratory chain results in a pH gradient across the mitochondrial membrane and this gradient serves as the driving force for the cells to generate ATP (the energy currency of the cell). Analogous gradients in ions across membranes (Na+, Ca2+, K+ and Cl−) provide the driving force for the action potentials in Fig. 2a: for instance, a voltage-gated ion channel embedded within the cell membrane may open to allow the flow of ions that generates the currents responsible for electrical communication. From a thermodynamics perspective, these gradients in pH and ions can be considered to be stable within the constraint that the cell membrane is semi-permeable and precludes a passive diffusion of charged species (i.e., the gradient will persist in the absence of a mechanism that allows dissipation of the gradient).
Gradients in redox biology can occur due to dynamics. Specifically, steep O2 gradients occur in tissue with high metabolic activity (e.g., the gut and brain) because the rapid consumption of this oxidant results in complete depletion over short distances (~200 μm) between the O2 source (e.g., the capillaries) and the O2 sink (tissue). [31]–[38] While the exact quantitative relationship between gradients in O2 and E may be uncertain, one consequence is that these gradients provide conditions for redox-cycling. Specifically, a redox-active molecule that can readily exchange electrons only needs to diffuse small distances in a steep gradient to be exposed to a reductive environment (where it can accept electrons) and an oxidative context (where it can donate electrons).
Redox-cycling is best recognized in pathological examples. For instance, the bacterial metabolite pyocyanin is characterized as a virulence factor because of its ability to engage the host tissue in redox-cycling that is believed to induce oxidative stress that is linked to disease[39]–[42] The toxicity of the agricultural chemical paraquat is believed to result from its ability to redox-cycle and induce oxidative stress[43], [44] Finally, some anticancer drugs are purposefully designed to engage cancer cells in damaging redox-cycling reactions[45], [46] While these examples illustrate the potential damaging consequences of redox-cycling, there have also been reports suggesting that redox-state (oxidized or reduced) is a form of biologically-relevant information and redox-cycling is a mechanism for “reading” this information. For instance, the redox-cycling bacterial metabolite, pyocyanin, has been suggested to perform redox-state-dependent quorum sensing signaling function[47]
In summary, redox reactions are integral to biological processes that range from energy harvesting and biosynthesis, to immune defense and intercellular communication (i.e., redox signaling). Relevant measurements of redox could provide important insights of biological state or detect dynamic activities in complex biological systems[48] Despite this motivation, measurements of redox (e.g., of E) remain controversial and are not well accepted. Thus, the development of methods that yield meaningful redox measurements could provide an important characterization tool for redox biology. As will be discussed, we are developing an interactive methodology to measure redox features and this measurement approach enables us not only to observe but also to perturb (i.e., actuate) biological systems.
B. Electrochemical Access to Redox Information
In the above, we explained that biology uses redox to perform various functions including communication (i.e., redox signaling)[29], [49] This redox communication modality is independent from the traditional molecularly-specific modalities and the globally-acting ion-based electrical modality. From a molecular communication standpoint, our unique insight is that this redox modality has both molecular features and electrical features[50], [51] The electrical features reflect the fact that redox reactions represent a “flow” of electrons. The molecular features reflect the fact that since free electrons do not exist in aqueous solution they must be “carried” by molecular intermediates (e.g., Fig. 3b shows biology’s use of the diffusible chemical species NADPH to mediate the “flow” of electrons between degradative and biosynthetic pathways). Our overarching hypothesis is that the electrical features of this redox modality should enable relatively simple access by electrochemical measurements.
Electrochemistry is a particularly versatile methodology that starts by immersing an electrode in a solution. This electrode is our “device” in Fig. 1b. (Experimentally, the electrode of interest is termed the working electrode and it is typically immersed in the solution with two other electrodes, a counter electrode and a reference electrode). The voltage to this electrode (i.e., E) can be precisely set to have reducing (more negative) values or oxidizing (more positive) values as illustrated in Fig. 5a (note the inverted scale). This voltage serves as the thermodynamic driving force for electrons to be transferred across the electrode-solution interface. As mentioned, since free electrons do not normally exist in solution the transfer of electrons from the electrode into the solution must be accompanied by the acceptance of an electron by a solution-phase molecule. This gain of electron serves to reduce the solution-phase molecular species. In contrast, when the imposed potential is more oxidative (more positive) then electrons can be transferred from a solution phase chemical species into the electrode, in which case the solution phase chemical is oxidized. It is important to note that not all chemical species are redox-active and capable of donating or accepting an electron at an electrode (in an analogous manner, not all molecules are acids or bases and capable of donating or accepting H+). Further, if a molecule is redox-active, then its intrinsic ability to donate and accept an electron is characterized by E0 (just as an acid’s ability to donate H+ is characterized by its pKa). It is also important to note that a redox-active molecule may be unable to donate or accept an electron even in the presence of a strong thermodynamic driving force if significant kinetic barriers limit electron exchange (such barriers are an intrinsic chemical characteristic of the molecule).
Fig. 5.
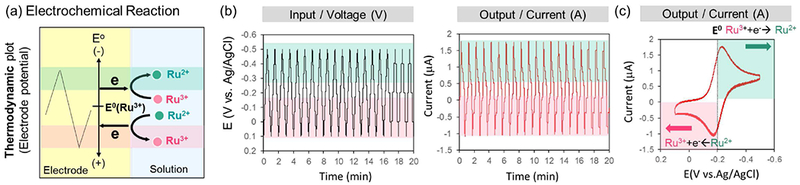
Electrochemical instrumentation can access redox information. (a) Electrochemical reactions involve the transfer of electrons at an electrode-solution interface: from the electrode perspective, electron transfer results in a readily measurable current; from the solution perspective, electron transfer switches a chemical species between its reduced (with electrons) and oxidized (without electrons) states. (b) User-defined and arbitrarily complex voltage inputs can be imposed at the electrode: cyclic inputs can generate steady (time-invarying) output currents as the mediator (Ru3+) is repeatedly switched between oxidized and reduced states. (c) An alternative representation of such data is a cyclic voltammogram (CV) which plots the current output vs. voltage input: note the arrows illustrate the direction of change of the cyclic voltage, while the change in current between oxidative (negative) and reductive (positive) occur near the mediator’s E0.
A unique feature of our electrochemical methodology is that we purposefully add diffusible redox-active chemical species that can easily exchange electrons with an electrode (i.e., they have small or no kinetic barriers to electron exchange). Fig. 5a shows one such chemical species is Ru(NH3)6Cl3 (Ru3+) which has an E0 = −0.2 V (vs a Ag/AgCl reference electrode). When the imposed voltage at the electrode is more reducing (more negative) than −0.2 V, then electrons are driven to “flow” across the electrode-solution interface to reduce Ru3+ to Ru2+. This flow of electrons is measured as a cathodic current. When the electrode voltage is set to be more oxidative (more positive) than −0.2 V, then there will be a driving force for electrons to be transferred in the opposite direction. In this case, electrons will flow from Ru2+ into the electrode, the Ru2+ will be oxidized back to Ru3+, and an anodic current will be observed. Importantly, the electrochemical reactions must occur at the electrode-solution interface and the redox-active species (e.g., Ru3+/2+ in this case) can diffuse away from the electrode surface into the bulk solution. In fact, electrochemical reactions can often be limited by diffusion of the redox-active chemical species to the electrode surface.
One desirable feature of electrochemistry is that the voltage imposed at the electrode can be varied through a user-defined and arbitrarily complex sequence of voltages. For instance, electrochemists often impose cyclic-input voltages as illustrated in Fig. 5b. In these cases, a readily reversible redox-active chemical species in the solution phase could be sequentially oxidized and reduced as the voltage cycles around its E0. This is illustrated by an example in which we added the readily-reversible redox-active chemical species, Ru3+ (E0 = −0.2 V) to a buffered solution and imposed cyclic voltage inputs. The output response from this experiment is shown in two representations; current-time curve (Fig. 5b), and current-voltage curve (Fig. 5c). The current-time representation shows the output is essentially “steady” (i.e., time-invariant) indicating the output current sequence of (Ru3+-reduction)-(Ru2+-oxidation) is repeated in response to the imposed cyclic input voltage. The current-voltage representation shows the switching of the direction of the current (oxidative vs reductive) occurs around the E0 value of Ru3+.
Conventional wisdom is that electrochemistry has significant strengths but also significant weaknesses. The strengths are that electrochemistry uses relatively inexpensive and potentially-portable instrumentation, while the measurements are simple, rapid and sensitive. There are two important limitations. First, electrochemical measurements require direct contact since electron transfer must occur at the electrode surface. As a result, electrochemical measurements can be invasive and only probe the local environment near the electrode (detection requires chemical species to diffuse to the electrode surface). A second limitation is that electrochemical measurements are not molecularly-specific in that numerous chemical species may be capable of exchanging electrons at a given imposed potential. As a result, the measured current will reflect the cumulative rates of electron transfer and it is generally impossible to discern the contributions from individual chemical species.
An important feature of electrochemistry that we believe is underutilized is its ability to be coupled with information processing. Specifically, electrochemical methods allow: arbitrarily complex input voltages to be imposed (e.g., for transmitter coding); the output currents to be sensitively measured (e.g., for receiver decoding); the electrical outputs can be measured at the same time as measurements from additional sensors (e.g., optical) to enable a fusion of sensor information; the data can be collected instantaneously in a format appropriate for real-time data analysis; and the input sequence could be adjusted on-the-fly for autonomous operation (e.g., to enlist machine learning to maximize mutual information). Our goal is to enlist these technological capabilities to enhance our ability to engage in redox-based communication with biology.
In summary, electrochemistry is an exciting measurement tool that can access biologically-relevant information by focusing on the electrical features of the redox modality[50], [51] While there are limitations to electrochemical measurements, we believe there are under-appreciated opportunities to couple electrochemical measurements with information processing to enable redox-based bio-device communication. To exploit these opportunities, we extensively enlist the use of redox-active chemical species (e.g., Ru3+) as mediators to shuttle electrons between an electrode and a local environment. Importantly, the extensive use of such diffusible mediators is uncommon in electrochemistry but, as will be seen, enables electrochemistry to be used for bio-device communication.
III. Electrochemical Redox-Probing to Acquire Information
In our opinion, the first step in developing redox as a molecular communication modality is to develop the capabilities to measure molecularly-based redox information. In this section we explain our electrochemical measurement approach and provide two examples demonstrating the acquisition of biologically-relevant redox information.
A. The Sonar Analogy
Our use of mediated electrochemical probing to search a local environment for redox-based chemical information is approximately analogous to sonar’s probing to detect/identify a physical object[51], [52] As illustrated in Fig. 6a, sonar transmits a pressure wave that propagates through a medium in search of an object that is detected by its generation of a reflected wave that can be interpreted to determine the object’s size, shape and motion. Fig. 6b illustrates that in our interactive redox-probing approach we encode information using an electrical (i.e., voltage) input sequence. The electrode serves as a transmitter that converts the electrical input into a redox signal. Specially, redox reactions at the electrode can switch the redox states of the mediators and diffusion of these mediators serve as the propagating redox signal (Note: the mediators serve as diffusible bi-stable redox switches that can be in either oxidized or reduced states). As these mediators diffuse (i.e., propagate) into the local environment, they can encounter and exchange electrons with redox-active components of this local environment. Such electron exchanges (i.e., redox reactions) serve to acquire redox information of the local environment by switching the redox-state of the mediators and this redox-based chemical information is “received” when the mediators return to the electrode and their redox-state is measured (note: the electrode serves as a co-located transmitter and receiver).
Fig. 6.
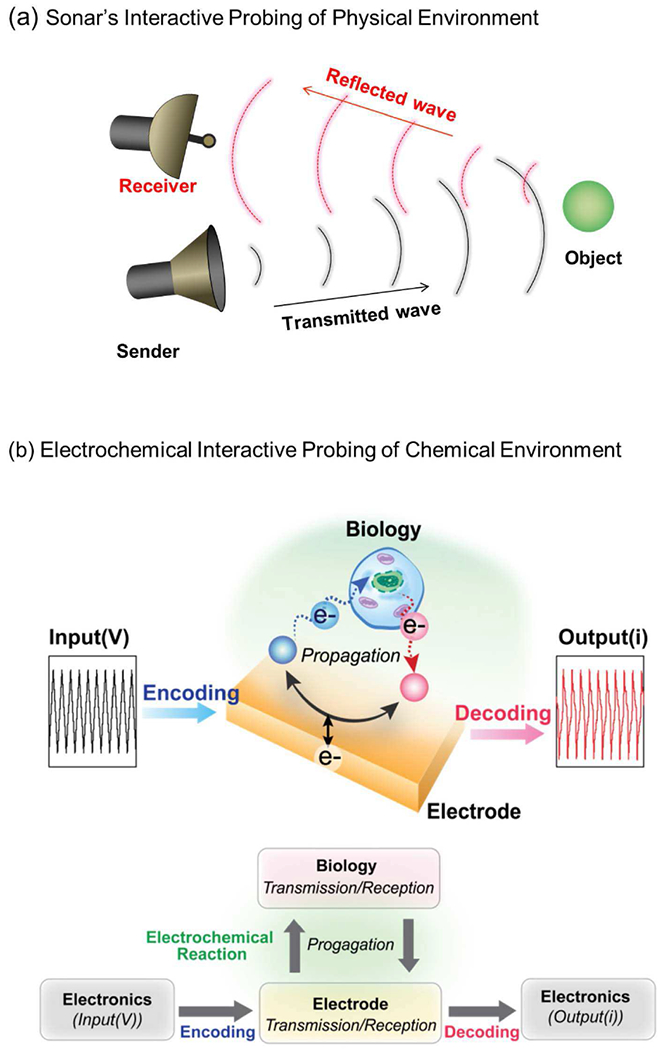
Acquiring redox-based information. (a) Sonar serves an analogy for our redox-probing method. (b) Interactive redox probing uses an electrode to provide input voltages that code and transmit redox signals that propagate by diffusion into the local environment to probe for redox activities after which the mediator diffuses back to the electrode which receives and decodes the information in the observed current.
Like sonar, we hypothesize that analysis of the input and output electrical signals will enable us to detect and characterize important redox-based signatures of the local environment. In some cases, these redox-signatures can be understood using chemical and biological information (analogous to sonar’s capabilities to identify an object from physical models of how objects reflect such pressure waves). However, in contrast to sonar, mediated electrochemical redox-probing is a new approach both conceptually and experimentally, and there are not (yet) relevant information theories (e.g., channel models) to guide interpretation or to optimize the transmitted signal. At this stage of the research, our goal is to experimentally demonstrate (somewhat empirically) that diffusion of redox-mediators can serve as a propagating signal that can access biologically meaningful redox-based information.
B. Discovering Redox-Based Signatures of Oxidative Stress
Oxidative stress refers to an imbalance between pro- and anti-oxidant activities in a biological system (e.g., your body). Oxidative stress is linked to various diseases [16], [17] which explains why we are often advised to consume foods and beverages rich in protective antioxidants. While the underlying biological mechanisms of oxidative stress are incompletely understood, emerging evidence suggests that it may be linked to inflammation[16], [19], [20], [53]–[58] As mentioned in Fig. 3c, inflammation involves the generation of damaging reactive chemical species (e.g., reactive oxygen species, ROS) that can oxidize various molecular species (e.g., proteins) and cellular components. Because of the link to diseases, there is currently intense interest in discovering biomarkers for oxidative stress to assist medical researchers and potentially even clinicians. However, it is generally unclear what should be measured, and various chemical species are being investigated as candidate molecular biomarkers. Such candidate biomarkers include: the signaling molecules that trigger inflammatory responses (e.g., inflammatory cytokines) [40], [59]–[63], the reactive species responsible for the damage [17], [64]–[66], the physiological antioxidant molecules (ascorbate and glutathione) [61], [67]–[75], the protective defense mechanisms (e.g., upregulation of ROS-degrading defense enzymes)[17], [18], [20], [53], [76], or the damaged molecules or cells [40], [77]–[79].
Conventional approaches to search for molecular biomarkers rely on advanced methods from analytical chemistry that have two limitations. First, measurement of a single (or even several) molecular species at a single time point will access only limited information of oxidative stress and such granular molecular detail may be insufficient to identify the dynamic phenomena that are acting at a global systems level[80], [81] Second, searching for detailed molecular features is complex typically requiring specialized instrumentation (mass spectrometry or immunoassays), trained personnel and long delays (hours to days).
Our approach is different, we are enlisting advances in information science (not analytical chemistry) to discover characteristic signatures of oxidative stress. Specifically, we are using mediated electrochemical probing to access chemical information at a global level (not at a detailed molecular level) and we are using information processing approaches to discern signatures of oxidative stress. In essence, our approach uses simple rapid electrochemical measurements to generate an “information-rich data stream” that can then be interpreted to identify molecular signatures of oxidative stress. Using the sonar analogy, our challenge is discovering what transmitted redox signal can best reveal signatures of oxidative stress even though: (i) there is incomplete biological information of what we are searching for, and (ii) there is no appropriate channel model for redox-probing.
Our specific clinical example is schizophrenia. Schizophrenia is a serious mental health disorder that is poorly understood and relies on subjective measures from patient interviews for diagnosis and management: there are no established blood tests or biomarkers to assist either researchers in understanding the disease or clinicians for managing the disease. Emerging research indicates that schizophrenia is linked to inflammation, redox dysregulation and oxidative stress, [18], [20], [60], [63], [76], [82]–[85] however the linkage is uncertain in part because of the absence of an accepted measurement of oxidative stress. By teaming with engineers, chemists, biostatisticians and clinicians, we are examining how the coupling of mediated electrochemical probing and signal processing can be used to discover signatures of oxidative stress from serum samples[86]
To identify redox-based signatures of oxidative stress, we probed diluted serum samples using the chemical mechanism illustrated in Fig. 7a [87] We start by adding an iridium mediator (K3IrCl6) which is initially in its inactive, colorless reduced state (Ir3+ which for convenience is designated IrRED). To initiate probing we impose an oxidative voltage that converts the mediator into its oxidized, yellow-colored state (Ir4+ which is designated IrOX). IrOX is a moderately strong oxidant (yet within the physiologically relevant range) that can exchange electrons with a wide range of molecular species (e.g., with proteins and cellular antioxidants such as ascorbate and glutathione). Importantly, IrOX is particularly sensitive to thiols (e.g., the cysteine amino acids in proteins) which are common sites of oxidative damage. [88], [89] As illustrated in Fig. 7b, the IrOX generated at the electrode propagates into the sample by diffusion where it “probes for” electron-rich molecular species from which it can extract electrons and switch it from its yellow-colored IrOX state back to its colorless state. Fig. 7b also shows that the regenerated IrRED can propagate back to the electrode by diffusion and be reoxidized.
Fig. 7.
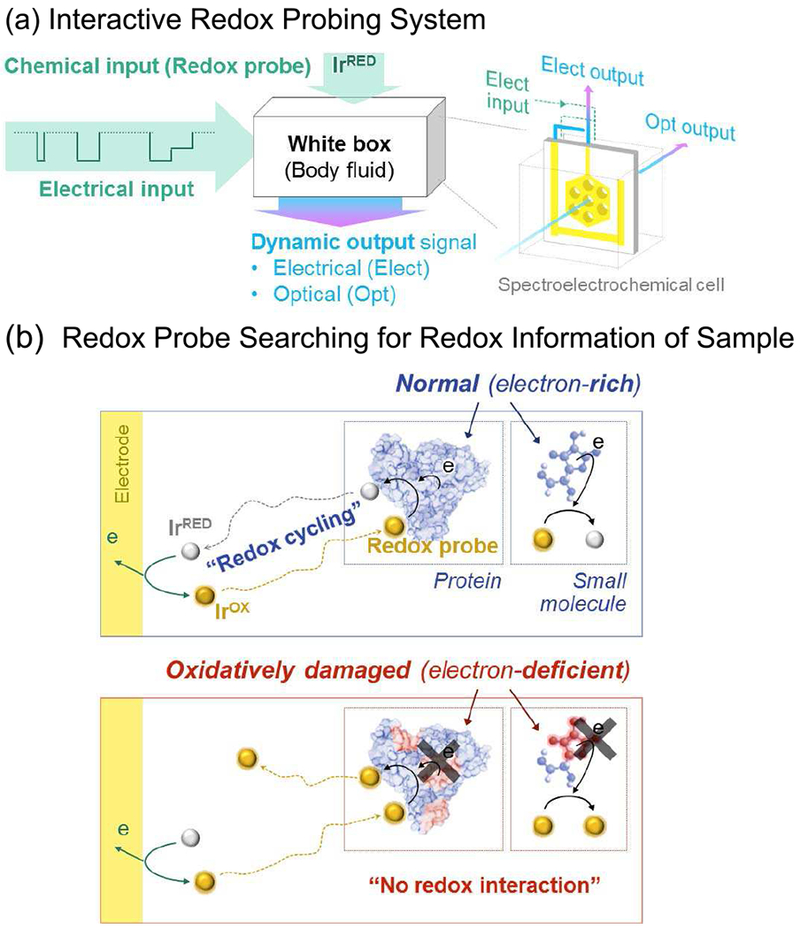
Discovering signatures of oxidative stress. (a) To discover signatures, we treated serum samples as a “white” box, used a single iridium (Ir) mediator and a 3-pulse electrical input sequence to generate electrical and optical outputs that could be analyzed to discern distinctive features. (b) The Ir mediator probes by extracting electrons from undamaged serum components to yield an amplification in electrical outputs (due to redox-cycling) and an attenuation of optical outputs (due to reaction of the yellow-colored IrOX). Reproduced with permission from Elsevier[178].
From a signal processing perspective, redox-probing involves transmitting an IrOX redox signal at the electrode (i.e., transmitter), this redox signal propagates into the local sample environment (by diffusion) where it detects redox-activity by being switched from its IrOX state to its IrRED state, and this IrRED propagates back to the electrode (i.e., receiver) where it is measured by re-oxidation of IrRED back to IrOX. From a chemistry perspective, this sequence of events is an oxidative redox-cycling mechanism and such mechanisms lead to amplified electrochemical oxidation currents. In addition to measuring the electrical current (a measure of the electron transfer rate at the electrode), Fig. 7 shows that a specialized spectroelectrochemical cell allows the simultaneous measurement of the optical absorbance associated with the yellow color and this is related to the local IrOX concentration. When IrOX undergoes oxidative redox-cycling, this optical signal is attenuated.
To illustrate the measurements, we show results from two control experiments involving well-defined buffered solutions[87] The first control experiment examined buffered solutions of varying concentrations of the physiological antioxidant glutathione (GSH). IrRED (0.5 mM final concentration) was added to these GSH-containing solutions and redox-probing was initiated by imposing a 1 min oxidative input pulse (+0.65 V vs. Ag/AgCl) to convert IrRED to IrOX (Fig. 8a). During this pulse, the electrical output was quantified as the oxidative charge transferred (Q = ∫ idt, i: current). As expected, the electrical output: (i) increases over time during this on-pulse, (ii) is amplified when GSH is present in the solution (presumably due to Ir-based redox-cycling), and (iii) is amplified to a greater extent for solutions containing higher GSH levels (the upper plot at the right in Fig. 8a). During the oxidative input pulse, we simultaneously measured the yellow color associated with the IrOX. As expected, the optical output: (i) increases over time during the oxidative on-pulse, (ii) is attenuated when GSH is present, and (iii) is attenuated to a greater extent for solutions containing higher GSH levels (lower plot at the right in Fig. 8a). The far right plot in Fig. 8a shows a close correlation between the amplified electrical output and the attenuated optical output. These results demonstrate that Ir-based redox probing is sensitive to concentration-dependent chemical information.
Fig. 8.
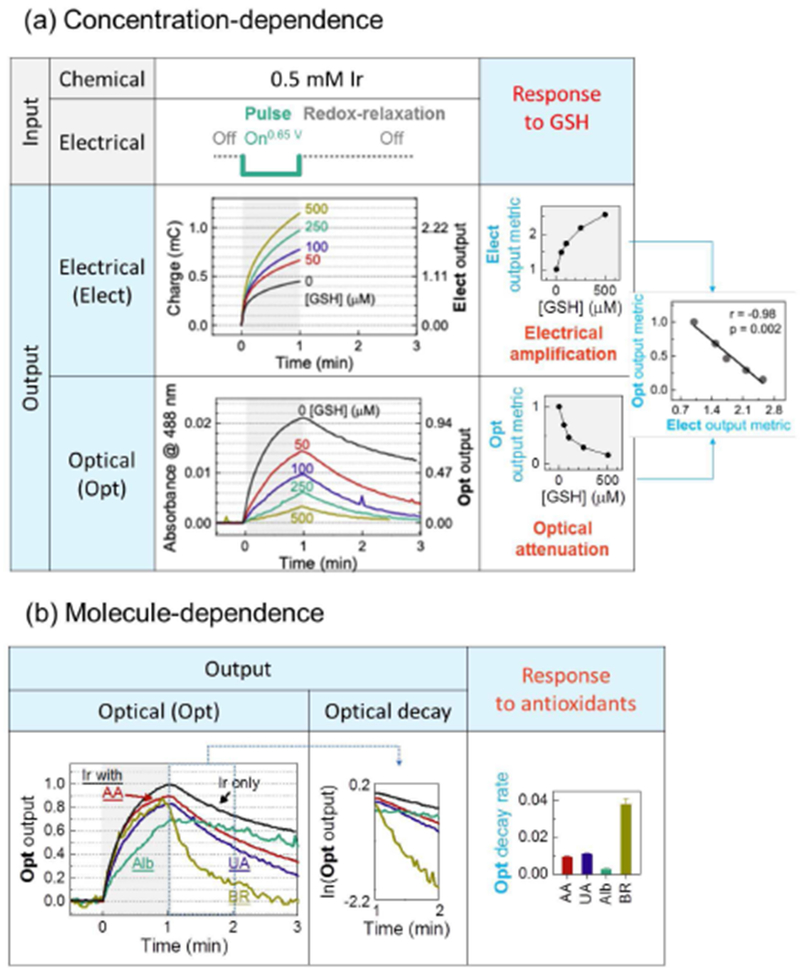
Redox-probing of control solutions illustrate the Ir-probing method. (a) Buffered solutions containing varying concentrations of the physiological antioxidant GSH were probed with Ir and a single oxidative pulse. The response characteristics show an amplified electrical output, an attenuated optical output, and a correlation between the two outputs to yield 3 signal metrics. (b) Buffered solutions of individual serum components (AA: ascorbic acid; UA: uric acid; Alb: albumin; BR: bilirubin) show more complex response characteristics (only optical outputs are shown) and a fourth signal metric based on decay (relaxation) of the optical response. Reproduced with permission from Elsevier[178].
In the second control experiment, we probed buffered solutions containing individual serum components that are expected to serve as physiological antioxidants (note these components were tested at concentrations that were 10-fold diluted compared to serum levels). Fig. 8b shows a more complex pattern of outputs emerge (only optical outputs are shown) for these different components and these differences presumably reflect differences in their chemical reactivities. Such differences in reactivities are apparent from the decay in the optical signal after the 1-min oxidative voltage has been turned off: we refer to this decay as “redox relaxation”. During this redox-relaxation period, the optical signal decays because the yellow-colored IrOX is both diffusing out of the optical window and being re-reduced to IrRED by accepting electrons from chemical components in the solution. For instance, bilirubin appears to be highly reactive with a rapid decay in the optical signal observed during the redox-relaxation period of the measurement. This decay was quantified using a first order rate model and the rate constant was used as a signal metric. The differences in this decay metric observed in bottom plot of Fig. 8b illustrates that Ir-based redox-probing can access molecule-dependent information.
The results in Fig. 8 illustrate that Ir-based redox-probing with a single 1-min oxidative pulse followed by a redox relaxation period could access concentration- and molecule-dependent information using 4 quantitative signal metrics: amplification of the electrical signal, attenuation of the optical signal, a cross-correlation of optical and electrical signals, and the optical decay during the relaxation period. For analysis of the more complex serum samples, we developed a 3 pulse-relaxation sequence that yielded a total of 14 signal metrics and we tested this sequence using clinical serum samples obtained from 10 persons diagnosed with schizophrenia and 5 healthy controls (see reference for details of the pulse sequence and signal metrics)[87] Using a Wilcoxon non-parametric test and receiver operating characteristic (ROC) analysis, Fig. 9 shows that 2 signal metrics show statistically significant capabilities to discriminate the schizophrenia and control groups based on their levels of oxidative stress. Combining information from these two metrics using multiple logistic regression analysis, we observed even greater discriminating abilities[87]
Fig. 9.
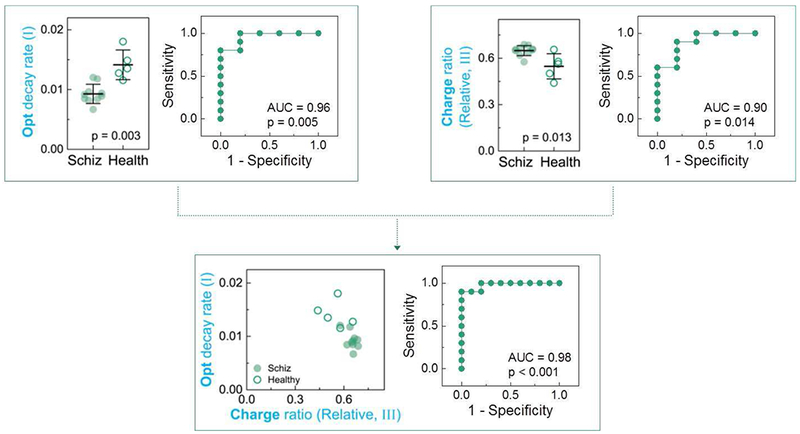
Ir-based redox-probing can discern signature differences between persons diagnosed with schizophrenia (N=10) and healthy controls (N=5). Samples were tested with a three pulse-relaxation sequence that generated 14 signal metrics. Two signal metrics showed statistically significant differences and combining these two metrics using multiple logistic regression analysis yielded even greater discriminating abilities. Reproduced with permission from Elsevier[178].
In summary, the results in Fig. 9 indicate that Ir-based redox-probing of serum samples accesses redox-based information related to oxidative stress. This measurement method is simple and rapid (the 3-pulse sequence took 40 minutes), and provides data in a convenient format for information processing. Importantly, this method accesses redox-information at a global, systems level and the biological-relevance of this information is apparent from its ability to correlate to subjective clinical measures of disease (i.e., a diagnosis of schizophrenia). Importantly, these results can be viewed as an initial validation of the methodology with considerable room for improvement by using different redox-mediators or optimizing the electrical input signal to maximize “information harvesting”.
C. Characterizing the Redox Properties of the Melanin Pigments
Melanins represent a diverse class of natural biopolymers that are present in organisms that range from bacteria to vertebrates including human and squid. In humans, the dark eumelanin pigments are believed to protect the skin and eye from radiation, free radicals and oxidative insults[90]–[93] In contrast, the lighter-colored pheomelanins that are found in persons with red-hair and fair complexions have been suggested to have deleterious photosensitizing[94] and pro-oxidant activities[95]–[98] Understanding the differences between eumelanin and pheomelanin has been challenging because these pigments are insoluble which has made it difficult to study their structures and properties. Fig. 10a shows that eumelanin is believed to be synthesized from the amino acid tyrosine, while pheomelanin is believed to be synthesized from the two amino acids tyrosine and cysteine. [99]–[101] In our studies we used mediated electrochemical probing as a reverse engineering approach to discern functionally-important differences between eumelanin and pheomelanin[102], [103]
Fig. 10.
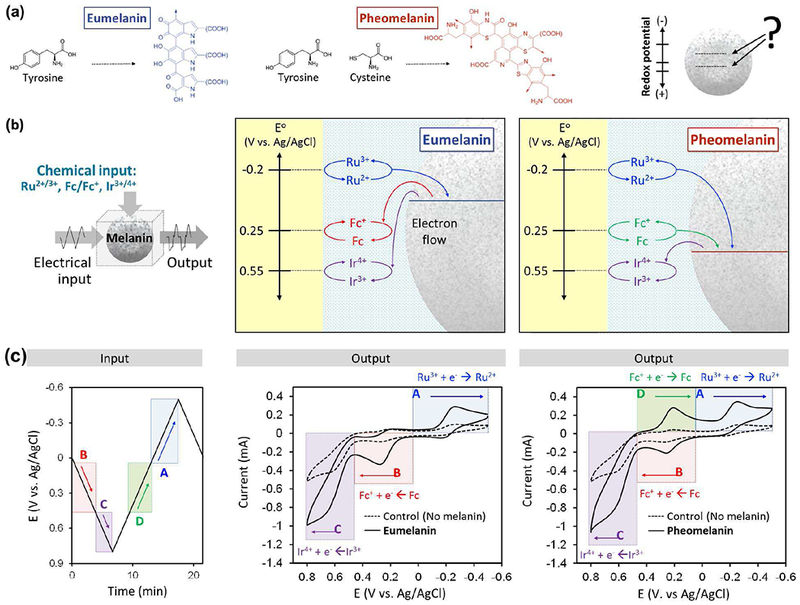
Electrochemical reverse engineering to characterize the biological pigment melanin. (a) The biosynthesis, structure and functional properties of eumelanin and pheomelanin have important differences. (b) Reverse engineering results indicate pheomelanin has a more oxidative redox potential that could explain its pro-oxidant properties. (c) Illustrative example shows that redox-probing with a cyclic electrical input sequence in the presence of 3 mediators revealed a distinct signature peak “D” for pheomelanin[104]
Experimentally, we started with synthetic models and human samples of eumelanin and pheomelanin, and we entrapped these samples in a hydrogel film that was coated onto an electrode. This hydrogel film coating serves to localize the insoluble melanin particles adjacent to the electrode.Importantly, this hydrogel film coating is permeable such that redox mediators can diffuse throughout the film to probe the redox-activities of the insoluble melanin particles. We next immersed these film-coated electrodes in solutions containing three mediators each with different redox potentials: Ru3+ (E0 = −0.2 V); Fc (E0 = +0.25 V); and Ir3+ (E0 = +0.55 V). Redox-probing was initiated using various imposed input voltage sequences while we observed the response characteristics to discern differences between these melanins[104]
We probed these samples using a variety of electrical input sequences and used chemical intuition to assist in interpreting the output response characteristics to lead to the conclusions illustrated Fig. 10b. Specifically, we observed eumelanin can accept electrons from Ru2+ and donate electrons to both Fc+ and Ir4+, while pheomelanin can accept electrons from both Ru2+ and Fc and donate electrons to Ir4+[104]
Fig. 10c shows results from one input sequence that was used to support the conclusions in Fig. 10b. In this probing, a cyclic voltage was imposed in the presence of the three mediators. The voltage was initially cycled through oxidative voltages, where Fc was first oxidized followed by the oxidation of Ir3+. When a synthetically-prepared model of eumelanin was probed, the output curve shows an oxidative current peak for Fc (peak “B”) and for Ir3+ (peak “C”). Importantly, these peak currents are amplified relative to the currents observed for a control film that lacked melanin. As noted earlier, this current amplification is a signature of oxidative redox-cycling which indicates that eumelanin is donating electrons to both the Fc+ and Ir4+ mediators which are serving to shuttle the electrons from the eumelanin to the electrode. When the input voltage was cycled to reducing potentials, then electrons could be transferred in the opposite direction to reduce Fc+ and then Ru3+ mediators. For the case of the synthetic eumelanin, an amplified Ru3+ reduction peak was observed (peak “A”) indicating that this mediator can undergo reductive redox-cycling with eumelanin. No obvious current amplification was observed for Fc+ reduction. This response characteristic indicates that eumelanin is redox-active and has a redox potential (E0) between those for Fc and Ru3+. [104]
The output response characteristics for films containing the synthetic model pheomelanin are shown at the right in Fig. 10c. For the case of pheomelanin, an amplification is observed for Ir-oxidation (peak “C”) while considerably less amplification is observed for Fc-oxidation (peak “B”). When the voltage was cycled to reducing potentials, the pheomelanin-containing film shows amplification for both Fc+-reduction (peak “D”) and Ru3+-reduction (peak “A”). This result indicates that both Fc and Ru3+ can engage pheomelanin in reductive redox-cycling. Importantly, peak “D” was unique for pheomelanin and not observed when the eumelanin-containing film was probed. These response characteristics indicate that pheomelanin has a redox potential between those for Fc and Ir3+ which would be more oxidative (more positive) than that for eumelanin[104]
From a biological perspective, the results from this study reveal features of melanin that had previously been unknown or at least under-appreciated:[103] melanins are redox-active, they can be repeatedly switched between oxidized and reduced states, and pheomelanin has a more oxidative redox potential than eumelanin. Potentially, this difference in redox potential could explain pheomelanin’s pro-oxidant activities[104] Modifications of this redox-probing methodology have also shown that melanins can undergo redox-cycling interactions with drugs and exogenous chemicals which may provide insights on the modes-of-action or side effects of drugs[105] or the mechanisms-of-action of environmental toxins[106] Further, modified redox-probing has shown subtle features of melanin’s protective radical scavenging activities[107]
More broadly, the use of a signal processing-based reverse engineering approach to characterize materials is fundamentally different from traditional materials science approaches to materials characterization. Traditionally, materials are characterized using a somewhat reductionist structural approach: the chemical structure is characterized over hierarchical length scales (e.g., from the covalent bonds to the crystal structure) and this structural knowledge is used to guide experimentation/modeling to understand and predict functionally-important properties. Mediated redox probing characterizes materials at a systems-level and especially focuses on functional properties. We believe such a systems-level approach is well-suited for materials such as melanin that: (i) are complex and not easily characterized by conventional structural approaches; and (ii) possess context-dependent functionalities that are difficult to discern from static chemical analyses (i.e., some of melanin’s functional properties depend on whether it exists in an oxidizing or reducing environment)[102], [107]
IV. Electrochemical Transmissions to Transduce Biology
To develop redox as a molecular communication modality, we believe it is necessary to demonstrate that redox-based information can be both received (i.e., measured) and transmitted. In the previous section we demonstrated the use of mediated electrochemical probing to measure (i.e., acquire) redox-based molecular information. Here we describe initial studies that demonstrate the use of electrochemically-generated molecular transmissions to induce two specific biologically-relevant responses: the patterning of biological structure and the induction of gene expression.
A. Molecularly-Induced Biological Patterning
Biology is well-known for its use of diffusible molecular signals to cue transitions in structure and function. For instance, Fig. 11a illustrates that morphogenesis is cued by gradients in signaling molecules (i.e., morphogens) that induce spatially selective differentiation of the various cells. In some cases, these molecular cues are provided in an oscillatory fashion to yield more complex segmented tissue patterns as illustrated in Fig. 11b for the emergence of vertebrae in the developing embryo. Quantitative analysis of such biological patterning often employs reaction-diffusion models[108]–[111] such as the clock-and-wavefront model used to explain the emergence of the segmented pattern of Fig. 11b[112]–[118]
Fig. 11.
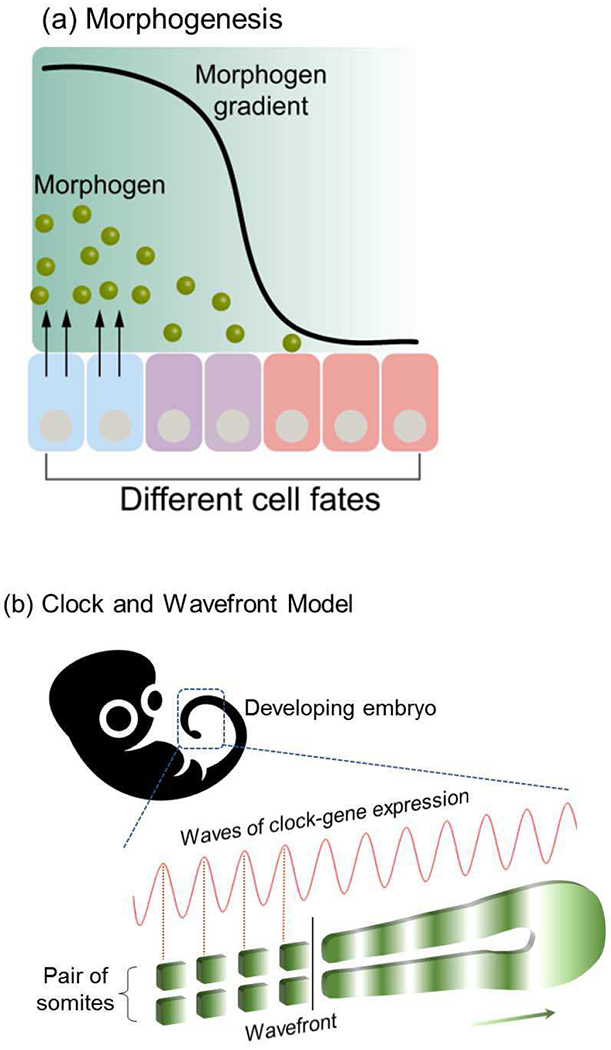
Molecular transmission that actuates biological pattern formation. (a) Morphogen diffusion is believed to cue the pattern formation in the developing embryo. (b) Oscillating molecular cues are believed to be responsible for the segmented structure of vertebrae in the developing spine. Reprinted with permission from [145]. Copyright (2018) American Chemical Society.
Recent efforts to generalize such morphogen patterning phenomena within the context of information theories[119]–[121] are enlisting a recent concept of a “pattern language” that involves Dynamic Patterning Modules. [122]–[124] For instance, this concept considers embryonic tissue as chemically excitable media that can be cued to self-organize into complex patterns in response to diffusible cues (i.e., morphogens)[124] Most relevant for the present discussion is the observation that many of the biological materials that undergo pattern formation are somewhat generic in biology.In particular, a common set of self-organizing proteins (e.g., collagen) and polysaccharides (e.g., cellulose and chitin) are a recurrent morphological motif used in biology to create complex extracellular structures[124] Thus, according to this concept, information coded into the diffusible molecular signals provide the cues that direct generic excitable materials to self-organize into unique patterns (note: the information could be encoded in the timing and/or concentrations of morphogens).
B. Electrochemically Generated Cues to Induce Self-Organization
Several groups independently reported that various stimuli-responsive self-assembling proteins (e.g., collagen[125]–[134] and silk [135]–[144]) and polysaccharides (e.g., chitosan [145], [146] and alginate[147], [148]) could be cued to self-assemble in response to imposed electrochemical inputs. Chitosan is likely the best-studied example, and its electrode-position occurs through a cathodic neutralization mechanism illustrated in Fig. 12a. As illustrated, chitosan is a pH-responsive self-assembling aminopolysaccharide. At low pH, the glucosamine residues are protonated making the chitosan polymer positively charged and water-soluble. At high pH, the glucosamine residues are deprotonated, the chains lose their charge and can undergo polymer-polymer self-associations that lead to a 3-dimensional hydrogel network. These associations result in crystalline network junctions that serve as non-covalent physical crosslinks[149]
Fig. 12.
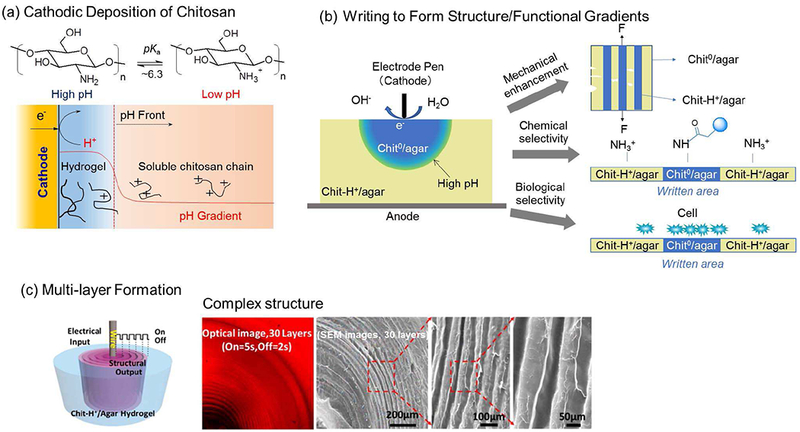
Transduction of electrochemical inputs into biomacromolecular structural outputs. (a) The pH-responsive aminopolysaccharide chitosan responds to electrode-induced pH increases by undergoing reversible structural changes (i.e., by self-assembling into a hydrogel network). (b) Blending chitosan with a second polysaccharide, agarose, yields an “excitable” medium that can respond to surface imposed electrical inputs (i.e., writing by an electrode pen) and these structural changes also result in functional changes in mechanical, chemical and biological properties. (c). Chitosan gel with multi-layer structure can be created by an oscillating electrical on/off input. Reprinted with permission from[179]. Copyright (2016) American Chemical Society.
Experimentally, electrodeposition occurs when an electrode is immersed in a slightly acidic chitosan solution (pH « 5 to 6), and a sufficiently reductive voltage (< −1.8 V vs. Ag/AgCl) is applied to promote water electrolysis. As illustrated in Fig. 12a, this electrolysis results in the localized generation of OH− and a localized region of high pH adjacent to the electrode. As the OH− diffuses away from the electrode, it can react to neutralize chitosan chains and induce their self-assembly (i.e., this self-assembly is essentially a sol-gel transition). Over time the chitosan hydrogel film grows from the electrode and this growing gelation front is co-localized with a pH front as illustrated in Fig. 12a. In essence, OH− is serving as an electrically-generated “morphogen” [150]–[152] that cues the formation of hydrogel structure as it diffuses and reacts in the medium. From an information transduction perspective, electrodeposition can be viewed as the transduction of electrical inputs into a diffusible molecular signal that induces self-assembly and the generation of hierarchical structure. We illustrate this electrical-structural transition by 2 recent examples.
The first example illustrates the electrical writing of structural patterns onto a dual-responsive, “excitable” hydrogel medium. This medium was prepared by blending two polysaccharides that can self-assemble in response to orthogonal cues. Specifically, the pH-responsive chitosan was blended with a second polysaccharide, agarose, that is thermally responsive, can be dissolved at moderately high temperatures but forms a hydrogel upon cooling below about 37 °C. A chitosan-agarose blend was prepared at low pH (5.5) and warm temperatures (85 °C) and this blend could be poured into a mold and cooled to allow the agarose hydrogel to form. This medium remains excitable (i.e., responsive) because the chitosan chains remain positively charged (designated Chit-H+) and dis-associated, but can be cued to self-assemble by providing appropriate high pH cues to deprotonate the chains to induce their self-association (designated Chit0). Fig. 12b shows that these high pH cues were supplied to localized regions using an electrode “pen” (a stainless steel acupuncture needle). Importantly, this electrode writing “excited” the chitosan-agarose medium to form stable gradients in structure and these gradients in structure were shown to be accompanied by gradients in mechanical properties, chemical reactivities and cell adhesion[153]
The second example illustrates that complex electrical inputs can be used to cue an “excitable” medium to generate complex structural patterns. As described above, a dual-responsive chitosan-agarose medium was prepared, but in this case, a wire electrode was embedded within the medium. Fig. 12c shows that the Chit-H+ chains in this excitable medium were cued to self-assemble using an oscillating imposed voltage input that repeatedly interrupted the self-assembly (gelation) process[154] Such interruptions have been observed to yield segmented structures (sometimes referred to as “onion structures” or “multilayers”) through incompletely understood mechanisms[145], [155], [156] The optical and electron microscope images in Fig. 12c show the resulting segmented structures that emerged. For instance, the optical image in Fig. 12c shows the creation of a hydrogel with 30 segments while the SEM images further illustrate the segmented structure. Although the underlying molecular mechanisms that link the diffusible input signal to the structural response are incompletely understood, the phenomenon is reproducible. Specifically, the segments grow during the “on” step while boundary regions emerge during the “off” steps, and the width and number of segments is controlled by the “on” and “off” cycles.
In summary, the above results indicate that the transmission of a time-varying electrical input signal provides a propagating signal that can induce the formation of arbitrarily complex spatially-varying structures in a responsive (i.e., excitable) medium. The propagating chemical signal in this example was the electrode-generated base (OH−), although there are several emerging examples in which different redox cues are being used to cue self-assembly[152], [157]–[159]
There are two interesting additional features to note. First, there appears to be two components to the input signal in Fig. 12c: the chemical component (OH−) that triggers self-assembly and can be quantified by current density and the charge transferred (Q = ∫ idt, i: current); and an electrical component (electric field) that can provide a force to induce motion of the charged polymer and also orient the dipoles to align the polymer chains[133], [135], [141], [145] Second, there is an interesting fundamental difference in the energetics of pattern generation between the developing embryo and the electrochemically-induced segmented hydrogel of Fig. 12c. Specifically, the diffusing morphogens in biology mobilize local cellular energy resources for pattern formation,[123] while the electrode in Fig. 12c must provide both the diffusing molecular signal and the energy required for pattern formation.
More broadly, there is considerable interest in applying concepts from information science to biology. Such efforts recognize the importance of macromolecular structure to biological information storage (e.g., genetic information in stored in DNA) and information processing (e.g., molecular recognition events are often transduced through protein conformational changes). If we extend this concept, then tissue could be considered to be a dynamic living medium where information is exchanged among cells through various modalities (e.g., molecular, mechanical and electrical)[123], [160] and this information provides the cues that guide key biological decisions such as cell adhesion, migration and differentiation. Thus, information theory may provide a framework for regenerative medicine (e.g., tissue engineering) that integrates the three, seemingly independent components (cells, material scaffolds and molecular cues) by considering the interactions (i.e., communication) among these components. Specifically, information theory may provide a framework to characterize the dynamic information flow in tissue and may assist in the development of biofabrication methods that can recapitulate such complex communication networks (as opposed to focusing on recapitulating the structural “networks”). We envision that the use of electrical inputs to generate structure from excitable biological materials (i.e., “electrobiofabrication”) could provide enabling capabilities for creating such complex communication networks for a range of life science applications.
C. Biology as a Redox Transceiver
As noted in Fig. 3c a common immune response to the detection of a bacterial pathogen is to initiate a chemically-based counter-attack by locally generating damaging reactive oxygen species (ROS). From the perspective of the bacterial pathogen, a successful defense against this ROS-based chemical counter-attack may mean the difference between life and death. Thus, bacteria have developed various mechanisms to detect such oxidative stresses and destroy the associated ROS[161]
One ROS-based oxidative stress response mechanism is the soxRSregulon[162], [163] that is illustrated in Fig. 13a and is believed to protect against one ROS, superoxide[161], [164], [165] SoxR is a redox-responsive protein with an iron–sulfur cluster that can be oxidized by superoxide and undergo a conformation change that activates expression of the superoxide-degrading enzyme superoxide dismutase (SOD). Importantly, the SoxR [2Fe-2S]3+ clusters can be re-reduced to turn off gene expression ofsoxRS regulon[164], [165] Interestingly, it has been observed that this regulon is not specific to superoxide but has been observed to be turned on by chemicals such as pyocyanin and paraquat that can undergo redox-cycling within the cell[166], [167]
Fig. 13.
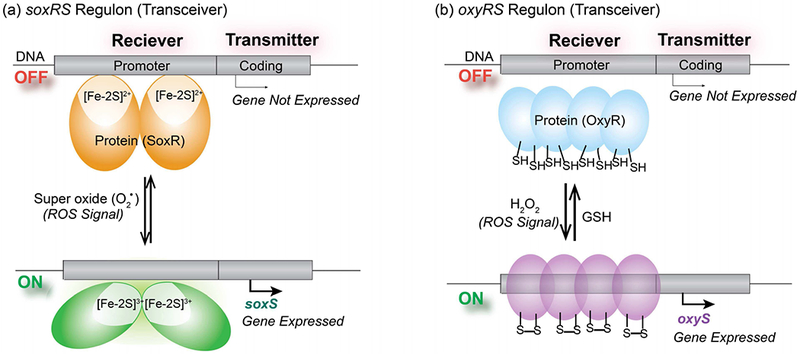
Redox-responsive elements used by E. coli serve as a biological “transceiver” to detect information of ROS-based oxidative stresses and respond by upregulating appropriate defense genes. (a) When exposed to superoxide (one specific ROS), the SoxR protein can be oxidized to undergo a conformational change that alters its binding to the DNA promoter region to enable the expression of genes to defend against this oxidative stress. This process can be reversed under reducing conditions. (b) When exposed to H2O2 (another specific ROS), the “sulfur switches” of the OxyR protein can be oxidized to undergo the conformational change that up-regulates expression of different oxidative stress response genes. These sulfur switches can be re-set by reduction to deactivate this stress response.
A second mechanism bacteria use to protect against ROS-based oxidative stresses is the oxyRS regulon that is shown Fig. 13b[168] In this case, a different ROS (H2O2) is detected by the sulfur-switching transcription factor OxyR: thiol residues of this OxyR protein are oxidized which initiates an up-regulation of the genes to produce the H2O2-degrading enzyme catalase[161], [168] Importantly, this sulfur switching mechanism is reversible in that the oxidized disulfide bond can be switched back to the reduced thiol state by intracellular reductants such glutathione[169] Thus, this sulfur-switching mechanism enables the bacteria to detect ROS-based information of oxidative stress and respond appropriately.
From a communications standpoint, these regulons serve as redox-based transceivers – they can recognize redox-based input signals (e.g., ROS or redox-cycling molecules that serve to impose oxidative stresses) and process this chemical information (by regulating gene expression) to generate appropriate output responses. Synthetic biology provides the means to alter these transceivers to fuse the receiver capabilities to alternative signal-generators (i.e., to generate alternative transmissions). currently we are developing the synthetic biology redox-toolkit to enable the design of such living transceivers. For instance, we recently re-wired E. coli by fusing the oxyRS[170] and soxRS[171] receivers to response elements that upregulate genes for chemotaxis: these synbio constructs direct their motility toward gradients in redox-active molecules.
D. Re-wiring Redox Transceiver for Two-hop Molecular Communication
A long-term vision for our research is to couple molecular communication and synthetic biology (synbio) to observe and adjust the gut microbiome to improve human health. The first step along this path (Fig. 14a) is to engineer a synbio construct to recognize and respond to device-imposed transmission. As illustrated, information that is coded in the electrode’s electrical input is converted into a redox transmission (by oxidation of a mediator). This redox transmission propagates through the solution (by diffusion of the mediator). A synbio construct (an engineered E. coli cell) receives this redox signal, perceives it as an oxidative stress, and is re-wired to decode this message by inducing gene expression. Briefly, we: selected a mediator that is known to extract electrons from bacteria to impose an oxidative stress;[172]–[174] used pyocyanin as an inducer as it has been shown to interact with the soxRS regulon (e.g., it mimics superoxide); and engineered the E. coli synbio construct to express a readily observable fluorescent protein. [175]
Fig. 14.
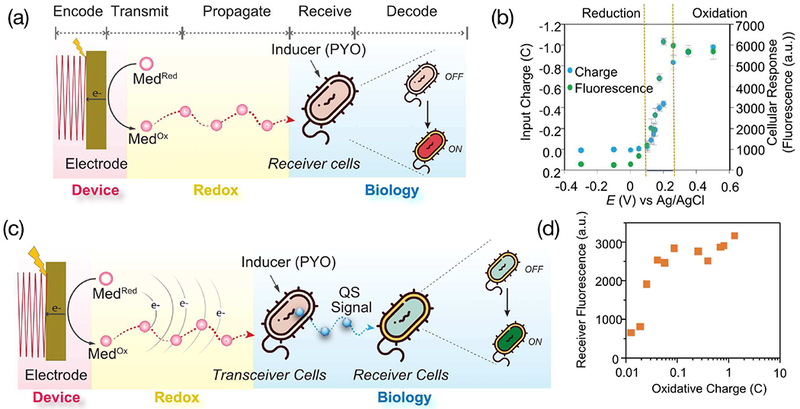
Redox-initiated actuation of biology and two-hop communication. (a) Initial test with a synbio redox “transceiver” engineered to receive redox-inputs through the soxRS regulon and re-wired to respond by upregulating expression of a gene coding for a fluorescent protein. (b) Voltage-dependent response of this synbio transceiver. (c) Schematic of two-hop molecular communication involving transceiver (or relay) cells that receive a redox input and transmit a quorum sensing (QS) signaling molecule, and a “receiver” cell that receives the QS molecular input and generates a fluorescence output response. (d) Output fluorescence vs input electrical charge for this two-hop communication. Reproduced with permission from [180].
Fig. 14b shows three types of responses. In the first region, where the imposed voltage was more reducing than the E0 for the mediator (+0.2 V vs Ag/AgCl), only small electrical currents were observed indicating little redox communication between the electrode and bacteria. Consistent with this interpretation is the observation that the cells did not appear to be oxidatively-stressed as indicated by minimal generation of the fluorescent protein.
In the second region, more oxidative voltages were imposed (between 0.1 and 0.25 V) and significant oxidative currents were observed indicating that these electrical inputs were inducing significant mediator-induced oxidative stresses on the bacteria. Consistent with this electrical measurement is the observation of increasing biological response (i.e., expression of fluorescent protein). In the third region, the imposed electrical inputs were more oxidative (greater than 0.25 V), and both the electrical and biological responses appeared to be saturated. Interestingly, Fig. 14b suggests that the biological response to the electrical input is “gated” by the mediator as the biological response is induced near this mediator’s redox potential (E0 = +0.2 V).
A second step toward coupling molecular communication and synthetic biology (synbio) for a healthy gut microbiome is to engineer the synbio construct to respond to device-imposed transmission by generating a molecular signal that can communicate with other bacteria in the microbiome. In our initial demonstration, Fig. 14c shows that we engineered an E. coli synbio transceiver (e.g., a relay cell) to receive the mediator-based redox input, process this input by upregulating genes to synthesize and transmit a bacterial quorum sensing (QS) molecule. This QS molecular signal can be recognized by a target cell population (also referred to as receiver cells). In our demonstration, we engineered a model receiver cell that recognizes the QS molecule and is re-wired to respond by expressing a readily measurable fluorescent protein[175]
Experimentally, the relay and receiver cells were co-cultured in the presence of the inducer (pyocyanin) and mediator, the electrical input was imposed by the electrode while the biological response was observed by measuring fluorescence. Fig. 14d shows the results from this two-hop communication demonstration. We should note that the electrical input (x-axis in Fig. 14d) is the charge transferred from the electrode (Q = ∫ idt) which is a quantitative measure of the extent of redox-based communication between the electrode and biology[175]
In summary, the above results demonstrate redox-mediated communication between an electrode device and a synbio construct as envisioned in Fig. 1. Previous studies (not discussed here) showed redox-based communication from biology to the electrode device to perform sensing functions[176][177] Fig. 14 shows communication in the opposite direction – from the electrode to the biology to actuate a response. Specifically, this synbio relay cell used a redox-responsive regulon to “receive” a redox-input but was “re-wired” to respond by generating a molecular QS signal capable of altering the behavior of a neighboring receiver cell population. In a broader perspective, these results illustrate the possibility for coupling the powers of electronic and biological information processing.
E. Conclusions and Perspectives
Biology uses redox to perform various functions including communication (i.e., redox signaling). As summarized in Table 1, we suggest that redox is a modality that offers the potential to bridge bio-device communication and to realize a molecular communication vision. It is important to note that the progress described here has emerged from experimentalists who are studying biological questions. To our knowledge, there has been no communication engineering analysis of this redox modality. Thus, the research on this redox modality aims to answer questions specific to biological information. In particular, biology often stores information in chemical structure (e.g., DNA) and changes in structure are integral to biological information processing (e.g., changes in protein-nucleic acid interactions are integral to regulating gene expression). In many ways, the current redox-based molecular communication research resembles biochemistry and materials science focusing on questions such as: how can structure be characterized?, what stimuli are responsible for inducing structural changes?, what properties and functions emerge from structure? We envision that these questions could be reframed from a focus on structure to a focus on information, and we imagine such a re-framing will reveal the role of redox as a modality for information processing.
TABLE 1.
Redox as a molecular communication modality.
| Features of Redox
Modality • Electrical: electron “flow” through reduction-oxidation (redox) reactions • Molecular: electrons “carried” by molecular species • Short range: requires molecular diffusion or mixing |
| Biological Relevance of Redox
Modality • Energy harvesting, biosynthesis, immune defense • Inflammation, oxidative stress • Signaling (redox signaling) |
| Measurement of Redox by
Electrodes • Simple, rapid, sensitive and convenient • Data available in real-time electronic format for analysis and transmission • Global (not molecularly-specific) • “Invasive”: requires direct electron transfer (i.e., direct contact) |
| Current
‘State-of-the-Art” • Experimental demonstrations: acquire information and actuate responses • Communication theoretical analysis: minimal |
There are two broad themes of this review. First, redox is a useful modality for bio-device communication. Redox reactions are integral to life and redox measurements provide a “portal” into important biological activities. Redox may be an especially relevant modality for observing the immune system as redox-based reactions are integral to defense and signaling. Importantly, electrochemistry provides comparatively easy access to the electrical features of redox, and electrochemistry also allows real-time access to the power of information processing. Especially important for our applications is the coupling of electrochemistry with redox-mediators that are tailored to exert specific influences on the surrounding local environmental. For instance, a sequence of oxidative input pulses with a single mediator (iridium) could generate an information-rich-data-stream that upon analysis could discern differences in the serum from persons diagnosed with schizophrenia compared to serum from healthy controls (Fig. 9). Further, the use of three mediators could “tune” probing to specific redox windows and this could reveal differences between pheomelanin and eumelanin that may be integral to specific pathologies (Fig. 10). In addition to using mediators to acquire information, our last two examples illustrated the use of mediators for transduction. Fig. 12 illustrates that imposed electrical inputs could generate diffusible signals (e.g., OH−) that could provide the cues for an “excitable” responsive medium to self-organize into complex patterns. Finally, Fig. 14 illustrates that redox provides the opportunity to connect electrochemistry and cells to access the orthogonal information processing capabilities of both electronics and biology. Thus, we believe these results provide experimental support for the exciting vision of molecular communication.
A second theme embedded throughout this review is that information sciences provide unique and complementary capabilities to the biological sciences. Traditionally, the biological sciences have focused on details because details are often essential for understanding mechanisms and designing interventions. However, it has become clear that details alone can’t usually explain systems-level biological behavior (knowledge of the genes does not generally enable predictions of phenotype). Information theory characterizes systems-level operation and uses rather abstract ideas, defining information in terms of uncertainty and evaluating the usefulness of a measurement in terms of mutual information. While such abstractions can be unsatisfying in terms of detailed mechanisms, it has the benefit of imposing a broader perspective on thinking. For instance, such abstract thinking may impose a different metric on discovery efforts in biology: is it more efficient to invest in analyzing a small number of samples to generate a detailed archival list and then apply machine learning to discern what information is useful, or could resources be better invested in a larger number of less-expensive but less-perfect measurements? Further, could information theory provide a means to understand how a cell may integrate information arriving through different modalities (molecular, mechanical and electrical) to make decisions of cell fate? Or more broadly, could information theory provide new insights for understanding and managing our biosphere: how vulnerable does our food supply, public health and infrastructure become when we plant a mono-culture crop, apply a pesticide to disrupt the existing web of life, or divert water supplies for alternative uses? Our hope is that by identifying a simple, potentially portable but powerful measurement approach (mediated electrochemical probing), we will bridge communication between biology and electronics and catalyze conversations between experimentalists and information scientists.
Acknowledgments
This work was supported with funds from the National Science Foundation (CBET #1805274 and DMREF #1435957), the National Institutes of Health (R21EB024102) and Defense Threat Reduction Agency (HDTRA1-13-0037)
Biographies

Eunkyoung Kim received the B.S. degree in Chemistry from Kyunghee University, Seoul, Korea, in 1997 and the M.S. and Ph.D. degrees in Chemistry (Electrochemistry) from Korea Advanced Institute of Science and Technology (KAIST), Daejeon, Korea, in 2004.
From 2007 to 2008, she was a Postdoctoral Fellow with the University of Pittsburgh. Since 2008, she has been a Research Associate with the Institute for Bioscience & Biotechnology Research (IBBR) and Fischell Department of Bioengineering at University of Maryland. She is the author of 60 articles and one invention. Her research interests include the integration of electrochemistry with biology to transform the biological information into the electrical information and the development of the electrochemical clinical device to measure the oxidative stress or bacterial infection.
Dr. Kim was a recipient of the Best Poster Award of Asian Conference on Electrochemistry in 2002 and the Travel Grant Award of Biological Engineering Conference on Electrofuels in 2011.

Jinyang Li is currently pursuing the Ph.D. degree in Bioengineering at University of Maryland, College Park, MD, USA. She received her bachelor’s and master’s degrees in Polymer Science from East China University of Science and Technology, Shanghai, China in 2015.
Since 2015, she has been a Graduate Assistant with the Institute for Bioscience & Biotechnology Research (IBBR) and Fischell Department of Bioengineering at University often Maryland. Her main research interest is to bridge biology with electronics via redox modality. She is currently working on electrofabrication of functional biomaterials and reverse engineering of redox biology.
Contributor Information
Eunkyoung Kim, Institute for Bioscience & Biotechnology Research, University of Maryland, College Park, MD 20742, USA.
Jinyang Li, Institute for Bioscience & Biotechnology Research, Fischell Department of Bioengineering University of Maryland, College Park, MD 20742, USA.
Mijeong Kang, Institute for Bioscience & Biotechnology Research, University of Maryland, College Park, MD 20742, USA.
Deanna L. Kelly, Maryland Psychiatric Research Center, University of Maryland School of Medicine, Baltimore, MD 21228, USA
Shuo Chen, Maryland Psychiatric Research Center, University of Maryland School of Medicine, Baltimore, MD 21228, USA.
Alessandra Napolitano, Department of Chemical Sciences, University of Naples Federico II, Via Cintia 4, I-80126 Naples, Italy.
Lucia Panzella, Department of Chemical Sciences, University of Naples Federico II, Via Cintia 4, I-80126 Naples, Italy.
Xiaowen Shi, School of Resource and Environmental Science, Hubei Biomass-Resource Chemistry, Environmental Biotechnology Key Laboratory, Wuhan University, Wuhan 430079, China.
Kun Yan, School of Resource and Environmental Science, Hubei Biomass-Resource Chemistry, Environmental Biotechnology Key Laboratory, Wuhan University, Wuhan 430079, China.
Si Wu, School of Resource and Environmental Science, Hubei Biomass-Resource Chemistry, Environmental Biotechnology Key Laboratory, Wuhan University, Wuhan 430079, China.
Jana Shen, Department of Pharmaceutical Sciences, University of Maryland School of Pharmacy, Baltimore, MD 21201, USA.
William E. Bentley, Institute for Bioscience & Biotechnology Research, Fischell Department of Bioengineering University of Maryland, College Park, MD 20742, USA
Gregory F. Payne, Institute for Bioscience & Biotechnology Research, University of Maryland, College Park, MD 20742, USA
References
- [1].Nakano T, Moore MJ, Wei F, V Vasilakos A, and Shuai J, “Molecular communication and networking: opportunities and challenges.,” IEEE Trans. Nanobioscience, vol. 11, no. 2, pp. 135–48, June 2012. [DOI] [PubMed] [Google Scholar]
- [2].Pierobon M and Akyildiz I, “A physical end-to-end model for molecular communication in nanonetworks,” IEEE J. Sel. Areas Commun, vol. 28, no. 4, pp. 602–611, May 2010. [Google Scholar]
- [3].Unluturk BD, Bicen AO, and Akyildiz IF, “Genetically Engineered Bacteria-Based BioTransceivers for Molecular Communication,” IEEE Trans. Commun, vol. 63, no. 4, pp. 1271–1281, April 2015. [Google Scholar]
- [4].Felicetti L, Femminella M, Reali G, and Liò P, “Applications of molecular communications to medicine: A survey,” Nano Communication Networks, vol. 7 pp. 27–45, March 2016. [Google Scholar]
- [5].Liu Y, Wu HC, Chhuan M, Terrell JL, Tsao CY, Bentley WE, and Payne GF, “Functionalizing Soft Matter for Molecular Communication,” ACS Biomater. Sci. Eng, vol. 1, no. 5, pp. 320–328, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Farsad N, Eckford AW, Hiyama S, and Moritani Y, “On-Chip Molecular Communication: Analysis and Design,” IEEE Trans. Nanobioscience, vol. 11, no. 3, pp. 304–314, Sep. 2012. [DOI] [PubMed] [Google Scholar]
- [7].Thomas PJ, “Every Bit Counts,” Science (80-. ), vol. 334, no. 6054, pp. 321–322, October 2011. [DOI] [PubMed] [Google Scholar]
- [8].Chou CT, “A Markovian Approach to the Optimal Demodulation of Diffusion-Based Molecular Communication Networks,” IEEE Trans. Commun, vol. 63, no. 10, pp. 3728–3743, October 2015. [Google Scholar]
- [9].Pierobon M and Akyildiz IF, “Capacity of a diffusion-based molecular communication system with channel memory and molecular noise,” IEEE Trans. Inf. Theory, vol. 59, no. 2, pp. 942–954, 2013. [Google Scholar]
- [10].Akyildiz I, Pierobon M, Balasubramaniam S, and Koucheryavy Y, “The internet of Bio-Nano things,” IEEE Commun. Mag, vol. 53, no. 3, pp. 32–40, March 2015. [Google Scholar]
- [11].Nakano T, Suda T, Okaie Y, Moore MJ, and Vasilakos AV, “Molecular Communication Among Biological Nanomachines: A Layered Architecture and Research Issues,” IEEE Trans. Nanobioscience, vol. 13, no. 3, pp. 169–197, September 2014. [DOI] [PubMed] [Google Scholar]
- [12].Nakano T, “Molecular Communication: A 10 Year Retrospective,” IEEE Trans. Mol. Biol. Multi-Scale Commun, vol. 3, no. 2, pp. 71–78, June 2017. [Google Scholar]
- [13].Gohari A, Mirmohseni M, and Nasiri-Kenari M, “Information theory of molecular communication: directions and challenges,” IEEE Trans. Mol. Biol. Multi-Scale Commun, vol. 2, no. 2, pp. 120–142, December 2016. [Google Scholar]
- [14].Hiyama S and Moritani Y, “Molecular communication: Harnessing biochemical materials to engineer biomimetic communication systems,” Nano Commun. Netw, vol. 1, no. 1, pp. 20–30, March 2010. [Google Scholar]
- [15].Akyildiz IF, Brunetti F, and Blázquez C, “Nanonetworks: A new communication paradigm,” Comput. Networks, vol. 52, no. 12, pp. 2260–2279, Aug. 2008. [Google Scholar]
- [16].Reuter S, Gupta SC, Chaturvedi MM, and Aggarwal BB, “Oxidative stress, inflammation, and cancer: How are they linked?,” Free Radic. Biol. Med, vol. 49, pp. 1603–1616, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Dalle-Donne I, Rossi R, Colombo R, Giustarini D, and Milzani A, “Biomarkers of oxidative damage in human disease,” Clinical Chemistry, vol. 52, no. 4 pp. 601–623, 2006. [DOI] [PubMed] [Google Scholar]
- [18].Leza JC, Bueno B, Bioque M, Arango C, Parellada M, Do K, O’Donnell P, and Bernardo M, “Inflammation in schizophrenia: A question of balance,” Neuroscience and Biobehavioral Reviews, vol. 55 pp. 612–626, 2015. [DOI] [PubMed] [Google Scholar]
- [19].Sawa A and Sedlak TW, “Oxidative stress and inflammation in schizophrenia,” Schizophr. Res, vol. 176, no. 1, pp. 1–2, 2016. [DOI] [PubMed] [Google Scholar]
- [20].Schiavone S and Trabace L, “Inflammation, stress response, and redox dysregulation biomarkers: Clinical outcomes and pharmacological implications for psychosis,” Front. Psychiatry, vol. 8, no. OCT, pp. 1–10, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Go Y-MM, Chandler JD, and Jones DP, “The cysteine proteome,” Free Radic. Biol. Med, vol. 84, pp. 227–245, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].García-Santamarina S, Boronat S, Hidalgo E, Garc S, Boronat S, and Hidalgo E, “Reversible cysteine oxidation in hydrogen peroxide sensing and signal transduction,” Biochemistry, vol. 53, no. 16, pp. 2560–2580, 2014. [DOI] [PubMed] [Google Scholar]
- [23].Schieber M and Chandel NS, “ROS function in redox signaling and oxidative stress,” Curr. Biol, vol. 24, no. 10, pp. R453–R462, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wang Y, Yang J, and Yi J, “Redox sensing by proteins: oxidative modifications on cysteines and the consequent events.,” Antioxid. Redox Signal, vol. 16, no. 7, pp. 649–57, 2012. [DOI] [PubMed] [Google Scholar]
- [25].Peiffer S, Klemm O, Pecher K, and Hollerung R, “Redox measurements in aqueous solutions - A theoretical approach to data interpretation, based on electrode kinetics,” J. Contam. Hydrol, 1992. [Google Scholar]
- [26].Reese AT, Cho EH, Klitzman B, Nichols SP, Wisniewski NA, Villa MM, Durand HK, Jiang S, Midani FS, Nimmagadda SN, O’Connell TM, Wright JP, Deshusses MA, and David LA, “Antibiotic-induced changes in the microbiota disrupt redox dynamics in the gut,” Elife, vol. 7, p. e35987, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hunting ER and van der Geest HG, “Predictability of bacterial activity and denitrification in aquatic sediments with continuous measurements of redox potential,” Int. J. Environ. Sci. Technol, 2011. [Google Scholar]
- [28].Jamieson LE, Jaworska A, Jiang J, Baranska M, Harrison DJ, and Campbell CJ, “Simultaneous intracellular redox potential and pH measurements in live cells using SERS nanosensors,” Analyst, 2015. [DOI] [PubMed] [Google Scholar]
- [29].Kemp M, Go Y-M, and Jones DP, “Nonequilibrium thermodynamics of thiol/disulfide redox systems: A perspective on redox systems biology,” Free Radic. Biol. Med, vol. 44, no. 6, pp. 921–937, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bigarella CL, Liang R, and Ghaffari S, “Stem cells and the impact of ROS signaling.,” Development, vol. 141, no. 22, pp. 4206–18, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Galler M, Moritz S, Liebsch G, Woertgen C, Brawanski A, and Warnat J, “Radial oxygen gradients over rat cortex arterioles,” Acta Neurochir. (Wien), 2010. [DOI] [PubMed] [Google Scholar]
- [32].Ndubuizu O and LaManna JC, “Brain Tissue Oxygen Concentration Measurements,” Antioxid. Redox Signal. , 2007. [DOI] [PubMed] [Google Scholar]
- [33].Mintun MA, Lundstrom BN, Snyder AZ, Vlassenko AG, Shulman GL, and Raichle ME, “Blood flow and oxygen delivery to human brain during functional activity: Theoretical modeling and experimental data,” Proc. Natl. Acad. Sci, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Christen T, Bolar DS, and Zaharchuk G, “Imaging brain oxygenation with MRI using blood oxygenation approaches: Methods, validation, and clinical applications,” American Journal of Neuroradiology. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Espey MG, “Role of oxygen gradients in shaping redox relationships between the human intestine and its microbiota,” Free Radic. Biol. Med, vol. 55, pp. 130–140, 2013. [DOI] [PubMed] [Google Scholar]
- [36].Pereira FC and Berry D, “Microbial nutrient niches in the gut,” Environmental Microbiology. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Albenberg L, Esipova TV, Judge CP, Bittinger K, Chen J, Laughlin A, Grunberg S, Baldassano RN, Lewis JD, Li H, Thom SR, Bushman FD, Vinogradov SA, and Wu GD, “Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota,” Gastroenterology, vol. 147, no. 5, p. 1055–1063.e8, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Secomb TW, Hsu R, Dewhirst MW, Klitzman B, and Gross JF, “Analysis of oxygen transport to tumor tissue by microvascular networks,” Int. J. Radiat. Oncol. Biol. Phys, 1993. [DOI] [PubMed] [Google Scholar]
- [39].Sies H, Biochemistry of Oxidative Stress, vol. 25, no. 12 Wiley-Blackwell, 1986, pp. 1058–1071. [Google Scholar]
- [40].Kohen R and Nyska A, “Toxicologic Pathology,” Toxicol. Pathol, vol. 30, no. 6, pp. 620–650, 2002. [DOI] [PubMed] [Google Scholar]
- [41].Hayyan M, Ali Hashim M, and AlNashef IM, “Superoxide Ion: Generation and Chemical Implications,” 2016. [DOI] [PubMed] [Google Scholar]
- [42].Sies H, “On the history of oxidative stress: Concept and some aspects of current development,” Current Opinion in Toxicology, vol. 7 Elsevier, pp. 122–126, February 2018. [Google Scholar]
- [43].Fussell KC, Udasin RG, Gray JP, Mishin V, Smith PJS, Heck DE, and Laskin JD, “Redox cycling and increased oxygen utilization contribute to diquat-induced oxidative stress and cytotoxicity in Chinese hamster ovary cells overexpressing NADPH-cytochrome P450 reductase,” Free Radic. Biol. Med, vol. 50, no. 7, pp. 874–882, April 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Bonneh-Barkay D, Reaney SH, Langston WJ, and Di Monte DA, “Redox cycling of the herbicide paraquat in microglial cultures,” Mol. Brain Res, vol. 134, no. 1, pp. 52–56, March 2005. [DOI] [PubMed] [Google Scholar]
- [45].Fang J, Seki T, and Maeda H, “Therapeutic strategies by modulating oxygen stress in cancer and inflammation,” Adv. Drug Deliv. Rev, vol. 61, no. 4, pp. 290–302, Apr. 2009. [DOI] [PubMed] [Google Scholar]
- [46].Gorrini C, Harris IS, and Mak TW, “Modulation of oxidative stress as an anticancer strategy,” Nat. Rev. Drug Discov, vol. 12, no. 12, pp. 931–947, December 2013. [DOI] [PubMed] [Google Scholar]
- [47].Koley D, Ramsey MM, Bard AJ, and Whiteley M, “Discovery of a biofilm electrocline using real-time 3D metabolite analysis,” Proc. Natl. Acad. Sci, vol. 108, no. 50, pp. 19996–20001, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Li J, Liu Y, Kim E, March JC, Bentley WE, and Payne GF, “Electrochemical Reverse Engineering: A Systems-Level Tool to Probe the Redox-Based Molecular Communication of Biology,” Free Radic. Biol. Med, vol. 105, no. October, pp. 1–22, 2017. [DOI] [PubMed] [Google Scholar]
- [49].Go Y-M and Jones DP, “The redox proteome.,” J. Biol. Chem, vol. 288, no. 37, pp. 26512–20, September 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Liu Y, Kim E, Li J, Kang M, Bentley WE, and Payne GF, “Electrochemistry for bio-device molecular communication: The potential to characterize, analyze and actuate biological systems,” Nano Commun. Netw, vol. 11, pp. 76–89, 2017. [Google Scholar]
- [51].Liu Y, Li J, Tschirhart T, Terrell JL, Kim E, Tsao C-YCY, Kelly DL, Bentley WE, and Payne GF, “Connecting Biology to Electronics: Molecular Communication via Redox Modality,” Adv. Healthc. Mater, vol. 6, no. 24, p. 1700789, 2017. [DOI] [PubMed] [Google Scholar]
- [52].Liu Y, Kim E, Lee ME, Zhang B, Elabd YA, Wang Q, White IM, Bentley WE, and Payne GF, “Enzymatic writing to soft films: Potential to filter, store, and analyze biologically relevant chemical information,” Adv. Funct. Mater, vol. 24, no. 4, pp. 480–491, 2014. [Google Scholar]
- [53].Khanna P, Ong C, Bay B, and Baeg G, “Nanotoxicity: An Interplay of Oxidative Stress, Inflammation and Cell Death,” Nanomaterials, vol. 5, no. 3, pp. 1163–1180, June 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Elmarakby AA and Sullivan JC, “Relationship between Oxidative Stress and Inflammatory Cytokines in Diabetic Nephropathy,” Cardiovasc. Ther, vol. 30, no. 1, pp. 49–59, 2012. [DOI] [PubMed] [Google Scholar]
- [55].Kohen R and Nyska A, “Invited Review: Oxidation of Biological Systems: Oxidative Stress Phenomena, Antioxidants, Redox Reactions, and Methods for Their Quantification,” Toxicol. Pathol, vol. 30, no. 6, pp. 620–650, 2002. [DOI] [PubMed] [Google Scholar]
- [56].Vaziri ND and Rodríguez-Iturbe B, “Mechanisms of Disease: oxidative stress and inflammation in the pathogenesis of hypertension,” Nat. Clin. Pract. Nephrol, vol. 2, no. 10, pp. 582–593, October 2006. [DOI] [PubMed] [Google Scholar]
- [57].Pisoschi AM and Pop A, “The role of antioxidants in the chemistry of oxidative stress: A review,” Eur. J. Med. Chem, vol. 97, pp. 55–74, 2015. [DOI] [PubMed] [Google Scholar]
- [58].Koga M, Serritella AV, Sawa A, and Sedlak TW, “Implications for reactive oxygen species in schizophrenia pathogenesis,” Schizophr. Res, vol. 176, no. 1, pp. 52–71, 2016. [DOI] [PubMed] [Google Scholar]
- [59].Chauhan A and Chauhan V, “Oxidative stress in autism,” Pathophysiology. 2006. [DOI] [PubMed] [Google Scholar]
- [60].Davison J, O’Gorman A, Brennan L, and Cotter DR, “A systematic review of metabolite biomarkers of schizophrenia,” Schizophrenia Research, vol. 195 Elsevier, pp. 32–50, May-2018. [DOI] [PubMed] [Google Scholar]
- [61].O’donnell P, Do KQ, and Arango C, “Oxidative/Nitrosative Stress in Psychiatric Disorders: Are We There Yet?,” Schizophr. Bull, vol. 40, no. 5, pp. 960–962, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Cascella NG, Kryszak D, Bhatti B, Gregory P, Kelly DL, Mc Evoy JP, Fasano A, and Eaton WW, “Prevalence of celiac disease and gluten sensitivity in the United States clinical antipsychotic trials of intervention effectiveness study population,” Schizophr. Bull, vol. 37, no. 1, pp. 94–100, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Tomasik J, Rahmoune H, Guest PC, and Bahn S, “Neuroimmune biomarkers in schizophrenia,” Schizophr. Res, vol. 176, no. 1, pp. 3–13, 2016. [DOI] [PubMed] [Google Scholar]
- [64].Ho E, Karimi Galougahi K, Liu C-C, Bhindi R, and Figtree GA, “Biological markers of oxidative stress: Applications to cardiovascular research and practice.,” Redox Biol, vol. 1, no. 1, pp. 483–91, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Valko M, Leibfritz D, Moncol J, Cronin MTD, Mazur M, and Telser J, “Free radicals and antioxidants in normal physiological functions and human disease,” Int. J. Biochem. Cell Biol, vol. 39, no. 39, pp. 44–84, 2007. [DOI] [PubMed] [Google Scholar]
- [66].Frustaci A, Neri M, Cesario A, Adams JB, Domenici E, Dalla Bernardina B, and Bonassi S, “Oxidative stress-related biomarkers in autism: Systematic review and meta-analyses,” Free Radic. Biol. Med, vol. 52, no. 10, pp. 2128–2141, 2012. [DOI] [PubMed] [Google Scholar]
- [67].Do KQ, Cabungcal JH, Frank A, Steullet P, and Cuenod M, “Redox dysregulation, neurodevelopment, and schizophrenia,” Curr. Opin. Neurobiol, vol. 19, no. 2, pp. 220–230, April 2009. [DOI] [PubMed] [Google Scholar]
- [68].Hardingham GE and Do KQ, “Linking early-life NMDAR hypo-function and oxidative stress in schizophrenia pathogenesis,” Nat. Rev. Neurosci, vol. 17, no. 2, pp. 125–134, February 2016. [DOI] [PubMed] [Google Scholar]
- [69].Conus P, Seidman LJ, Fournier M, Xin L, Cleusix M, Baumann PS, Ferrari C, Cousins A, Alameda L, Gholam-Rezaee M, Golay P, Jenni R, Woo TUW, Keshavan MS, Eap CB, Wojcik J, Cuenod M, Buclin T, Gruetter R, and Do KQ, “N-acetylcysteine in a double-blind randomized placebo-controlled trial: Toward biomarker-guided treatment in early psychosis,” Schizophr. Bull, vol. 44, no. 2, pp. 317–327, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Monin A, Baumann PS, Griffa A, Xin L, Mekle R, Fournier M, Butticaz C, Klaey M, Cabungcal JH, Steullet P, Ferrari C, Cuenod M, Gruetter R, Thiran JP, Hagmann P, Conus P, and Do KQ, “Glutathione deficit impairs myelin maturation: relevance for white matter integrity in schizophrenia patients,” Mol. Psychiatry, vol. 20, no. 7, pp. 827–838, July 2014. [DOI] [PubMed] [Google Scholar]
- [71].Zhang XY, Chen da C, Xiu MH, Tang W, Zhang F, Liu L, Chen Y, Liu J, Yao JK, Kosten TARTR, Chen DC, Xiu MH, Tang W, Zhang F, Liu L, Chen Y, Liu J, Yao JK, Kosten TARTR, and Kosten TARTR, “Plasma total antioxidant status and cognitive impairments in schizophrenia,” Schizophr. Res, vol. 139, no. 1–3, pp. 66–72, August 2012. [DOI] [PubMed] [Google Scholar]
- [72].Yao JK, Reddy R, and van Kammen DP, “Reduced level of plasma antioxidant uric acid in schizophrenia,” Psychiatry Res, vol. 80, no. 1, pp. 29–39, 1998. [DOI] [PubMed] [Google Scholar]
- [73].Yao JK, Leonard S, and Reddy R, “Altered Glutathione Redox State in Schizophrenia,” Dis. Markers, vol. 22, no. 1–2, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Yao JK, Reddy R, McElhinny LG, and van Kammen DP, “Reduced status of plasma total antioxidant capacity in schizophrenia,” Schizophr. Res, vol. 32, no. 1, pp. 1–8, 1998. [DOI] [PubMed] [Google Scholar]
- [75].Reddy R, Keshavan M, and Yao JK, “Reduced plasma antioxidants in first-episode patients with schizophrenia,” Schizophr. Res, vol. 62, no. 3, pp. 205–212, 2003. [DOI] [PubMed] [Google Scholar]
- [76].Fraguas D, ; Covadonga, Díaz-Caneja M, Rodríguez-Quiroga A, and Arango C, “Oxidative Stress and Inflammation in Early Onset First Episode Psychosis: A Systematic Review and Meta-Analysis,” Int. J. Neuropsychopharmacol, vol. 20, no. 6, pp. 435–444, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Stadtman ER, “Protein Oxidation in Aging and Age-Related Diseases,” Ann. N. Y. Acad. Sci, vol. 928, no. 1, pp. 22–38, Jan. 2006. [DOI] [PubMed] [Google Scholar]
- [78].Davies MJ, “Protein oxidation and peroxidation.,” Biochem. J, vol. 473, no. 7, pp. 805–25, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Finkel T and Holbrook NJ, “Oxidants, oxidative stress and the biology of ageing,” Nature, vol. 408, no. 6809 pp. 239–247, 2000. [DOI] [PubMed] [Google Scholar]
- [80].McDermott JE, Wang J, Mitchell H, Webb-Robertson B-J, Hafen R, Ramey J, and Rodland KD, “Challenges in Biomarker Discovery: Combining Expert Insights with Statistical Analysis of Complex Omics Data.,” Expert Opin. Med. Diagn, vol. 7, no. 1, pp. 37–51, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Vafaee F, Diakos C, Kirschner MB, Reid G, Michael MZ, Horvath LG, Alinejad-Rokny H, Cheng ZJ, Kuncic Z, and Clarke S, “A data-driven, knowledge-based approach to biomarker discovery: application to circulating microRNA markers of colorectal cancer prognosis,” npj Syst. Biol. Appl, vol. 4, no. 1, p. 20, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Chan MK, Krebs MO, Cox D, Guest PC, Yolken RH, Rahmoune H, Rothermundt M, Steiner J, Leweke FM, Van Beveren NJM, Niebuhr DW, Weber NS, Cowan DN, Suarez-Pinilla P, Crespo-Facorro B, Mam-Lam-Fook C, Bourgin J, Wenstrup RJ, Kaldate RR, Cooper JD, and Bahn S, “Development of a blood-based molecular biomarker test for identification of schizophrenia before disease onset,” Transl. Psychiatry, vol. 5, no. 7, pp. 1–10, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Yang J, Chen T, Sun L, Zhao Z, Qi X, Zhou K, Cao Y, Wang X, Qiu Y, Su M, Zhao A, Wang P, Yang P, Wu J, Feng G, He L, Jia W, and Wan C, “Potential metabolite markers of schizophrenia,” Mol. Psychiatry, vol. 18, no. 1, pp. 67–78, Jan. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Flatow J, Buckley P, and Miller BJ, “Meta-Analysis of Oxidative Stress in Schizophrenia,” Biol. Psychiatry, vol. 74, no. 6, pp. 400–409, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Lai C-Y, Scarr E, Udawela M, Everall I, Chen WJ, and Dean B, “Biomarkers in schizophrenia: A focus on blood based diagnostics and theranostics,” World J. Psychiatry, vol. 6, no. 1, p. 102, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Kim E, Winkler TE, Kitchen C, Kang M, Banis G, Bentley WE, Kelly DL, Ghodssi R, and Payne GF, “Redox Probing for Chemical Information of Oxidative Stress,” Anal. Chem, vol. 89, no. 3, pp. 1583–1592, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Kang M, Kim E, Chen S, Bentley WE, Kelly DL, and Payne GF, “Signal processing approach to probe chemical space for discriminating redox signatures,” Biosens. Bioelectron, vol. 112, no. April, pp. 127–135, 2018. [DOI] [PubMed] [Google Scholar]
- [88].Stadtman ER and Levine RL, Free radical-mediated oxidation of free amino acids and amino acid residues in proteins, vol. 25, no. 3–4 Springer-Verlag, 2003, pp. 207–218. [DOI] [PubMed] [Google Scholar]
- [89].Stadtman ER, “Oxidation of Free Amino Acids and Amino Acid Residues in Proteins by Radiolysis and by Metal-Catalyzed Reactions,” Annu. Rev. Biochem, vol. 62, no. 1, pp. 797–821, 1993. [DOI] [PubMed] [Google Scholar]
- [90].Wang A, Marino AR, Gasyna Z, Gasyna E, and Norris J, “Photoprotection by porcine eumelanin against singlet oxygen production,” Photochem. Photobiol, 2008. [DOI] [PubMed] [Google Scholar]
- [91].Rózanowska M, Sarna T, Land EJ, and Truscott TG, “Free radical scavenging properties of melanin interaction of eu- and pheo-melanin models with reducing and oxidising radicals,” Free Radic. Biol. Med, 1999. [DOI] [PubMed] [Google Scholar]
- [92].Herrling T, Jung K, and Fuchs J, “The role of melanin as protector against free radicals in skin and its role as free radical indicator in hair,” Spectrochim. Acta - Part A Mol. Biomol. Spectrosc, 2008. [DOI] [PubMed] [Google Scholar]
- [93].Nofsinger JB, Liu Y, and Simon JD, “Aggregation of eumelanin mitigates photogeneration of reactive oxygen species,” Free Radic. Biol. Med, 2002. [DOI] [PubMed] [Google Scholar]
- [94].Panzella L, Szewczyk G, D’Ischia M, Napolitano A, and Sarna T, “Zinc-induced Structural Effects Enhance Oxygen Consumption and Superoxide Generation in Synthetic Pheomelanins on UVA/Visible Light Irradiationt,” Photochem. Photobiol, vol. 86, no. 4, pp. 757–764, July 2010. [DOI] [PubMed] [Google Scholar]
- [95].Panzella L, Leone L, Greco G, Vitiello G, D’Errico G, Napolitano A, and d’Ischia M, “Red human hair pheomelanin is a potent pro-oxidant mediating UV-independent contributory mechanisms of melanomagenesis,” Pigment Cell Melanoma Res, vol. 27, no. 2, pp. 244–252, 2014. [DOI] [PubMed] [Google Scholar]
- [96].Mitra D, Luo X, Morgan A, Wang J, Hoang MP, Lo J, Guerrero CR, Lennerz JK, Mihm MC, Wargo JA, Robinson KC, Devi SP, Vanover JC, D’Orazio JA, McMahon M, Bosenberg MW, Haigis KM, Haber DA, Wang Y, Fisher DE, D/’Orazio JA, McMahon M, Bosenberg MW, Haigis KM, Haber DA, Wang Y, and Fisher DE, “An ultraviolet-radiation-independent pathway to melanoma carcinogenesis in the red hair/fair skin background,” Nature, vol. 491, no. 7424, pp. 449–453, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Hill HZ and Hill GJ, “UVA, Pheomelanin and the Carcinogenesis of Melanoma,” Pigment Cell Res, vol. 13, pp. 140–144, 2000. [DOI] [PubMed] [Google Scholar]
- [98].Wenczl E, Smit NPM, Pavel S, Schothorst AA, Van der Schans GP, Timmerman AJ, Roza L, and Kolb RM, “(Pheo)Melanin Photosensitizes UVA-Induced DNA Damage in Cultured Human Melanocytes11This work was presented in part at the Annual Meeting of the European Society for Photobiology, Stresa, September 8–13 1997 and entirely at the Annual Meeting of the Europ,” J. Invest. Dermatol, vol. 111, no. 4, pp. 678–682, 1998. [DOI] [PubMed] [Google Scholar]
- [99].Ito S and Wakamatsu K, “Quantitative analysis of eumelanin and pheomelanin in humans, mice, and other animals: a comparative review.,” Pigment cell Res, vol. 16, no. 5, pp. 523–31, October 2003. [DOI] [PubMed] [Google Scholar]
- [100].Di Donato P and Napolitano A, “1,4-benzothiazines as key intermediates in the biosynthesis of red hair pigment pheomelanins.,” Pigment cell Res, vol. 16, no. 5, pp. 532–9, October 2003. [DOI] [PubMed] [Google Scholar]
- [101].Ito S and IFPCS, “The IFPCS presidential lecture: a chemist’s view of melanogenesis.,” Pigment cell Res, vol. 16, no. 3, pp. 230–6, June 2003. [DOI] [PubMed] [Google Scholar]
- [102].Kim E, Liu Y, Leverage WT, Yin J-JJ, White IM, Bentley WE, and Payne GF, “Context-Dependent Redox Properties of Natural Phenolic Materials,” Biomacromolecules, vol. 15, no. 5, pp. 1653–1662, 2014. [DOI] [PubMed] [Google Scholar]
- [103].Kang M, Kim E, Temoçin Z, Li J, Dadachova E, Wang Z, Panzella L, Napolitano A, Bentley WE, and Payne GF, “Reverse Engineering To Characterize Redox Properties: Revealing Melanin’s Redox Activity through Mediated Electrochemical Probing,” Chem. Mater, vol. 30, no. 17, pp. 5814–5826, September 2018. [Google Scholar]
- [104].Kim E, Panzella L, Micillo R, Bentley WE, Napolitano A, and Payne GF, “Reverse Engineering Applied to Red Human Hair Pheomelanin Reveals Redox-Buffering as a Pro-Oxidant Mechanism,” Sci. Rep, vol. 5, no. November, p. 18447, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Temoçin Z, Kim E, Li J, Panzella L, Alfieri MLML, Napolitano A, Kelly DLDL, Bentley WEWE, and Payne GFGF, “The Analgesic Acetaminophen and the Antipsychotic Clozapine Can Each Redox-Cycle with Melanin,” ACS Chem. Neurosci, vol. 8, no. 12, pp. 2766–2777, December 2017. [DOI] [PubMed] [Google Scholar]
- [106].Kim E, Leverage WT, Liu Y, Panzella L, Alfieri ML, Napolitano A, Bentley WE, and Payne GF, “Paraquat-Melanin Redox-Cycling: Evidence from Electrochemical Reverse Engineering,” ACS Chem. Neurosci, vol. 7, no. 8, pp. 1057–1067, 2016. [DOI] [PubMed] [Google Scholar]
- [107].Kim E, Kang M, Tschirhart T, Malo M, Dadachova E, Cao G, Yin JJ, Bentley WE, Wang Z, and Payne GF, “Spectroelectrochemical Reverse Engineering DemonstratesThat Melanin’s Redox and Radical Scavenging Activities Are Linked,” Biomacromolecules, vol. 18, no. 12, pp. 4084–4098, 2017. [DOI] [PubMed] [Google Scholar]
- [108].Turing AM, “The Chemical Basis of Morphogenesis,” Philos. Trans. R. Soc. B Biol. Sci, vol. 237, no. 641, pp. 37–72, August 1952. [Google Scholar]
- [109].Tompkins N, Li N, Girabawe C, Heymann M, Ermentrout GB, Epstein IR, and Fraden S, “Testing Turing’s theory of morphogenesis in chemical cells,” Proc. Natl. Acad. Sci, vol. 111, no. 12, pp. 4397–4402, March 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Kondo S and Miura T, “Reaction-Diffusion Model as a Framework for Understanding Biological Pattern Formation,” Science (80-. ), vol. 329, no. 5999, pp. 1616–1620, September 2010. [DOI] [PubMed] [Google Scholar]
- [111].Vonk FJ and Richardson MK, “Serpent clocks tick faster,” Nature, vol. 454, no. 7202, pp. 282–283, July 2008. [DOI] [PubMed] [Google Scholar]
- [112].Yabe T and Takada S, “Molecular mechanism for cyclic generation of somites: Lessons from mice and zebrafish,” Dev. Growth Differ, vol. 58, no. 1, pp. 31–42, January 2016. [DOI] [PubMed] [Google Scholar]
- [113].Signon L, Nowakowski B, and Lemarchand A, “Modeling somite scaling in small embryos in the framework of Turing patterns,” Phys. Rev. E, vol. 93, no. 4, p. 042402, April 2016. [DOI] [PubMed] [Google Scholar]
- [114].Dziekan P, Nowakowski B, and Lemarchand A, “Reaction-diffusion scheme for the clock and wavefront mechanism of pattern formation,” Eur. Phys. J. B, vol. 87, no. 4, p. 77, April 2014. [Google Scholar]
- [115].Murray PJ, Maini PK, and Baker RE, “The clock and wavefront model revisited,” J. Theor. Biol, vol. 283, no. 1, pp. 227–238, August 2011. [DOI] [PubMed] [Google Scholar]
- [116].Gomez C, Özbudak EM, Wunderlich J, Baumann D, Lewis J, and Pourquié O, “Control of segment number in vertebrate embryos,” Nature, vol. 454, no. 7202, pp. 335–339, July 2008. [DOI] [PubMed] [Google Scholar]
- [117].Dias AS, de Almeida I, Belmonte JM, Glazier JA, and Stern CD, “Somites Without a Clock,” Science (80-. ), vol. 343, no. 6172, pp. 791–795, February 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Cooke J and Zeeman EC, “A clock and wavefront model for control of the number of repeated structures during animal morphogenesis.,” J. Theor. Biol, vol. 58, no. 2, pp. 455–76, May 1976. [DOI] [PubMed] [Google Scholar]
- [119].Rose C and Mian IS, “Inscribed Matter Communication: Part I,” IEEE Trans. Mol. Biol. Multi-Scale Commun, vol. 2, no. 2, pp. 209–227, December 2016. [Google Scholar]
- [120].Rose C and Mian IS, “Inscribed Matter Communication: Part II,” IEEE Trans. Mol. Biol. Multi-Scale Commun, vol. 2, no. 2, pp. 228–239, December 2016. [Google Scholar]
- [121].Mian IS and Rose C, “Communication theory and multicellular biology,” Integr. Biol, vol. 3, no. 4, p. 350, 2011. [DOI] [PubMed] [Google Scholar]
- [122].Newman SA and Bhat R, “Dynamical patterning modules: A ‘pattern language’ for development and evolution of multicellular form,” International Journal of Developmental Biology, vol. 53, no. 5–6 pp. 693–705, 2009. [DOI] [PubMed] [Google Scholar]
- [123].Benítez M, Hernández-Hernández V, Newman SA, and Niklas KJ, “Dynamical Patterning Modules, Biogeneric Materials, and the Evolution of Multicellular Plants,” Front. Plant Sci, vol. 9, p. 871, July 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Newman SA, “Physico-Genetic Determinants in the Evolution of Development,” Science (80-. ), vol. 338, no. 6104, pp. 217–219, October 2012. [DOI] [PubMed] [Google Scholar]
- [125].Kang L, Liu X, Yue Z, Chen Z, Baker C, Winberg P, and Wallace G, “Fabrication and In Vitro Characterization of Electrochemically Compacted Collagen/Sulfated Xylorhamnoglycuronan Matrix for Wound Healing Applications,” Polymers (Basel), vol. 10, no. 4, p. 415, April 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Baker HR, Merschrod S EF, and Poduska KM, “Electrochemically Controlled Growth and Positioning of Suspended Collagen Membranes,” Langmuir, vol. 24, no. 7, pp. 2970–2972, April 2008. [DOI] [PubMed] [Google Scholar]
- [127].Ramesh Kumar M, Merschrod S EF., and Poduska KM, “Correlating mechanical properties with aggregation processes in electrochemically fabricated collagen membranes,” Biomacromolecules, vol. 10, no. 7, pp. 1970–1975, July 2009. [DOI] [PubMed] [Google Scholar]
- [128].Zhuang J, Lin S, Dong L, Cheng K, and Weng W, “Magnetically Assisted Electrodeposition of Aligned Collagen Coatings,” ACS Biomater. Sci. Eng, vol. 4, no. 5, pp. 1528–1535, 2018. [DOI] [PubMed] [Google Scholar]
- [129].Uquillas JA and Akkus O, “Modeling the electromobility of type-i collagen molecules in the electrochemical fabrication of dense and aligned tissue constructs,” Ann. Biomed. Eng, vol. 40, no. 8, pp. 1641–1653, August 2012. [DOI] [PubMed] [Google Scholar]
- [130].Younesi M, Islam A, Kishore V, Panit S, and Akkus O, “Fabrication of compositionally and topographically complex robust tissue forms by 3D-electrochemical compaction of collagen,” Biofabrication, vol. 7, no. 3, p. 035001, June 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Kishore V, Iyer R, Frandsen A, and Nguyen T-U, “In vitro characterization of electrochemically compacted collagen matrices for corneal applications,” Biomed. Mater, vol. 11, no. 5, p. 055008, October 2016. [DOI] [PubMed] [Google Scholar]
- [132].Ling T, Lin J, Tu J, Liu S, Weng W, Cheng K, Wang H, Du P, and Han G, “Mineralized collagen coatings formed by electrochemical deposition,” J. Mater. Sci. Mater. Med, vol. 24, no. 12, pp. 2709–2718, 2013. [DOI] [PubMed] [Google Scholar]
- [133].Cheng X, Gurkan UA, Dehen CJ, Tate MP, Hillhouse HW, Simpson GJ, and Akkus O, “An electrochemical fabrication process for the assembly of anisotropically oriented collagen bundles,” Biomaterials, vol. 29, no. 22, pp. 3278–3288, August 2008. [DOI] [PubMed] [Google Scholar]
- [134].Nguyen T-U, Bashur CA, and Kishore V, “Impact of elastin incorporation into electrochemically aligned collagen fibers on mechanical properties and smooth muscle cell phenotype,” Biomed. Mater, vol. 11, no. 2, p. 025008, March 2016. [DOI] [PubMed] [Google Scholar]
- [135].Maniglio D, Bonani W, Bortoluzzi G, Servoli E, Motta A, and Migliaresi C, “Electrodeposition of Silk Fibroin on Metal Substrates,” J. Bioact. Compat. Polym, vol. 25, no. 5, pp. 441–454, September 2010. [Google Scholar]
- [136].Lin Y, Xia X, Shang K, Elia R, Huang W, Cebe P, Leisk G, Omenetto F, and Kaplan DL, “Tuning chemical and physical crosslinks in silk electrogels for morphological analysis and mechanical reinforcement,” Biomacromolecules, vol. 14, no. 8, pp. 2629–2635, August 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Kojic N, Panzer MJ, Leisk GG, Raja WK, Kojic M, and Kaplan DL, “Ion electrodiffusion governs silk electrogelation,” Soft Matter, vol. 8, no. 26, p. 6897, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Li M, Xiong P, Mo M, Cheng Y, and Zheng Y, “Electrophoretic-deposited novel ternary silk fibroin/graphene oxide/hydroxyapatite nanocomposite coatings on titanium substrate for orthopedic applications,” Front. Mater. Sci, vol. 10, no. 3, pp. 270–280, September 2016. [Google Scholar]
- [139].Leisk GG, Lo TJ, Yucel T, Lu Q, and Kaplan DL, “Electrogelation for protein adhesives,” Adv. Mater, vol. 22, no. 6, pp. 711–715, 2010. [DOI] [PubMed] [Google Scholar]
- [140].Elia R, Michelson CD, Perera AL, Harsono M, Leisk GG, Kugel G, and Kaplan DL, “Silk electrogel coatings for titanium dental implants,” J. Biomater. Appl, vol. 29, no. 9, pp. 1247–1255, April 2015. [DOI] [PubMed] [Google Scholar]
- [141].Lu Q, Huang Y, Li M, Zuo B, Lu S, Wang J, Zhu H, and Kaplan DL, “Silk fibroin electrogelation mechanisms,” Acta Biomater, vol. 7, no. 6, pp. 2394–2400, June 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Bressner JE, Marelli B, Qin G, Klinker LE, Zhang Y, Kaplan DL, and Omenetto FG, “Rapid fabrication of silk films with controlled architectures via electrogelation,” J. Mater. Chem. B, vol. 2, no. 31, p. 4983, June 2014. [DOI] [PubMed] [Google Scholar]
- [143].Han C, Yao Y, Cheng X, Luo J, Luo P, Wang Q, Yang F, Wei Q, and Zhang Z, “Electrophoretic Deposition of Gentamicin-Loaded Silk Fibroin Coatings on 3D-Printed Porous Cobalt-Chromium-Molybdenum Bone Substitutes to Prevent Orthopedic Implant Infections,” Biomacromolecules, vol. 18, no. 11, pp. 3776–3787, 2017. [DOI] [PubMed] [Google Scholar]
- [144].Lin Y, An B, Bagheri M, Wang Q, Harden JL, and Kaplan DL, “Electrochemically Directed Assembly of Designer Coiled-Coil Telechelic Proteins,” ACS Biomater. Sci. Eng, vol. 3, no. 12, pp. 3195–3206, 2017. [DOI] [PubMed] [Google Scholar]
- [145].Yan K, Liu Y, Zhang J, Correa SO, Shang W, Tsai CC, Bentley WE, Shen J, Scarcelli G, Raub CB, Shi XW, and Payne GF, “Electrical Programming of Soft Matter: Using Temporally Varying Electrical Inputs to Spatially Control Self Assembly,” Biomacromolecules, vol. 19, no. 2, pp. 364–373, 2018. [DOI] [PubMed] [Google Scholar]
- [146].Redepenning J, Venkataraman G, Chen J, and Stafford N, “Electrochemical preparation of chitosan/hydroxyapatite composite coatings on titanium substrates,” J. Biomed. Mater Res, vol. 66A, no. 2, pp. 411–416, Aug. 2003. [DOI] [PubMed] [Google Scholar]
- [147].Cheng Y, Luo X, Betz J, Payne GF, Bentley WE, and Rubloff GW, “Mechanism of anodic electrodeposition of calcium alginate,” Soft Matter, vol. 7, p. 5677, 2011. [Google Scholar]
- [148].Cheong M and Zhitomirsky I, “Electrodeposition of alginic acid and composite films,” Colloids Surfaces A Physicochem. Eng. Asp, vol. 328, no. 1–3, pp. 73–78, 2008. [Google Scholar]
- [149].Morrow BH, Payne GF, and Shen J, “pH-responsive self-assembly of polysaccharide through a rugged energy landscape,” J. Am. Chem. Soc, vol. 137, no. 40, pp. 13024–30, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Rydzek G, Jierry L, Parat A, Thomann J-S, Voegel J-C, Senger B, Hemmerlé J, Ponche A, Frisch B, Schaaf P, and Boulmedais F, “Electrochemically Triggered Assembly of Films: A One-Pot Morphogen-Driven Buildup,” Angew. Chemie Int. Ed, vol. 50, no. 19, pp. 4374–4377, May 2011. [DOI] [PubMed] [Google Scholar]
- [151].Maerten C, Garnier T, Lupattelli P, Chau NTT, Schaaf P, Jierry L, and Boulmedais F, “Morphogen Electrochemically Triggered Self-Construction of Polymeric Films Based on Mussel-Inspired Chemistry,” Langmuir, vol. 31, no. 49, pp. 13385–13393, 2015. [DOI] [PubMed] [Google Scholar]
- [152].Maerten C, Jierry L, Schaaf P, and Boulmedais F, “Review of Electrochemically Triggered Macromolecular Film Buildup Processes and Their Biomedical Applications,” ACS Appl Mater Interfaces, vol. 9, no. 34, pp. 28117–28138, 2017. [DOI] [PubMed] [Google Scholar]
- [153].Wu S, Yan K, Zhao Y, Tsai C-C, Shen J, Bentley WE, Chen Y, Deng H, Du Y, Payne GF, and Shi X, “Electrical Writing onto a Dynamically Responsive Polysaccharide Medium: Patterning Structure and Function into a Reconfigurable Medium,” Adv. Funct. Mater, p. 1803139, August 2018. [Google Scholar]
- [154].Yan K, Xiong Y, Wu S, Bentley WE, Deng H, Du Y, Payne GF, and Shi XW, “Electro-molecular Assembly: Electrical Writing of Information into an Erasable Polysaccharide Medium,” ACS Appl. Mater. Interfaces, vol. 8, no. 30, pp. 19780–19786, 2016. [DOI] [PubMed] [Google Scholar]
- [155].Yan K, Ding F, Bentley WE, Deng H, Du Y, Payne GF, and Shi X-W, “Coding for hydrogel organization through signal guided self-assembly,” Soft Matter, vol. 10, no. 3, p. 465, 2014. [DOI] [PubMed] [Google Scholar]
- [156].Ladet S, David L, and Domard A, “Multi-membrane hydrogels,” Nature, vol. 452, no. 7183, pp. 76–79, 2008. [DOI] [PubMed] [Google Scholar]
- [157].Mergel O, Kühn PT, Schneider S, Simon U, and Plamper FA, “Influence of Polymer Architecture on the Electrochemical Deposition of Polyelectrolytes,” Electrochim. Acta, 2017. [Google Scholar]
- [158].Hernández-Burgos K, Barton ZJ, and Rodríguez-López J, “Finding Harmony between Ions and Electrons: New Tools and Concepts for Emerging Energy Storage Materials,” Chem. Mater, vol. 29, no. 21, pp. 8918–8931, 2017. [Google Scholar]
- [159].El-Maiss J, Cuccarese M, Maerten C, Lupattelli P, Chiummiento L, Funicello M, Schaaf P, Jierry L, and Boulmedais F, “Mussel-Inspired Electro-Cross-Linking of Enzymes for the Development of Biosensors,” ACS Appl. Mater. Interfaces, vol. 10, no. 22, pp. 18574–18584, 2018. [DOI] [PubMed] [Google Scholar]
- [160].Green S and Batterman R, “Biology meets physics: Reductionism and multi-scale modeling of morphogenesis,” Stud. Hist. Philos. Sci. Part C Stud. Hist. Philos. Biol. Biomed. Sci, vol. 61, pp. 20–34, February 2017. [DOI] [PubMed] [Google Scholar]
- [161].Green J and Paget MS, “Bacterial redox sensors,” Nat. Rev. Microbiol, vol. 2, no. 12, pp. 954–966, 2004. [DOI] [PubMed] [Google Scholar]
- [162].Cabiscol E, Tamarit J, and Ros J, “Oxidative stress in bacteria and protein damage by reactive oxygen species,” Int. Microbiol, 2000. [PubMed] [Google Scholar]
- [163].Farr SB and Kogoma T, “Oxidative stress responses in Escherichia coli and Salmonella typhimurium.,” Microbiol. Rev, 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Fujikawa M, Kobayashi K, and Kozawa T, “Direct Oxidation of the [2Fe-2S] Cluster in SoxR Protein by Superoxide,” J. Biol. Chem, vol. 287, no. 42, pp. 35702–35708, October 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Blanchard JL, Wholey WY, Conlon EM, and Pomposiello PJ, “Rapid changes in gene expression dynamics in response to superoxide reveal SoxRS-dependent and independent transcriptional networks,” PLoS One, vol. 2, no. 11, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Major TTCR, “The SoxRS response of Escherichia coli is directly activated by redox-cycling drugs rather than by superoxide,” Biophys. Chem, vol. 34, no. 1, pp. 13–23, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Dietrich LEP and Kiley PJ, “A shared mechanism of SoxR activation by redox-cycling compounds,” Mol. Microbiol, vol. 79, no. January, pp. 1119–1122, 2011. [DOI] [PubMed] [Google Scholar]
- [168].Åslund F, Zheng M, Beckwith J, and Storz G, “Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thioldisulfide status.,” Proc. Natl. Acad. Sci. U. S. A, vol. 96, no. 11, pp. 6161–6165, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].García-Santamarina S, Boronat S, and Hidalgo E, “Reversible cysteine oxidation in hydrogen peroxide sensing and signal transduction,” Biochemistry, vol. 53, no. 16, pp. 2560–2580, April 2014. [DOI] [PubMed] [Google Scholar]
- [170].Virgile C, Hauk P, Wu H-CC, Shang W, Tsao C-YY, Payne GF, and Bentley WE, “Engineering bacterial motility towards hydrogen-peroxide,” PLoS One, vol. 13, no. 5, p. e0196999, May 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].McKay R, Hauk P, Wu HC, Pottash AE, Shang W, Terrell J, Payne GF, and Bentley WE, “Controlling localization of Escherichia coli populations using a two-part synthetic motility circuit: An accelerator and brake,” Biotechnol. Bioeng, vol. 114, no. 12, pp. 2883–2895, 2017. [DOI] [PubMed] [Google Scholar]
- [172].Pasco N, Baronian K, Jeffries C, and Hay J, “Biochemical mediator demand - a novel rapid alternative for measuring biochemical oxygen demand,” Appl. Microbiol. Biotechnol, vol. 53, no. 5, pp. 613–618, May 2000. [DOI] [PubMed] [Google Scholar]
- [173].Morris K, Zhao H, and John R, “Ferricyanide-Mediated Microbial Reactions for Environmental Monitoring,” Aust. J. Chem, vol. 58, no. 4, p. 237, 2005. [Google Scholar]
- [174].Catterall K, Robertson D, Hudson S, Teasdale PR, Welsh DT, and John R, “A sensitive, rapid ferricyanide-mediated toxicity bioassay developed using Escherichia coli,” Talanta, vol. 82, no. 2, pp. 751–757, Jul. 2010. [DOI] [PubMed] [Google Scholar]
- [175].Tschirhart T, Kim E, Mckay R, Ueda H, Wu HC, Pottash AE, Zargar A, Negrete A, Shiloach J, Payne GF, and Bentley WE, “Electronic control of gene expression and cell behaviour in Escherichia coli through redox signalling,” Nat. Commun, vol. 8, pp. 1–11, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Tschirhart T, Zhou XY, Ueda H, Tsao C-Y, Kim E, Payne GF, and Bentley WE, “Electrochemical Measurement of the β-Galactosidase Reporter from Live Cells: A Comparison to the Miller Assay,” ACS Synth. Biol, vol. 5, no. 1, pp. 28–35, January 2016. [DOI] [PubMed] [Google Scholar]
- [177].Liu Y, Tsao C-Y, Kim E, Tschirhart T, Terrell JL, Bentley WE, and Payne GF, “Using a Redox Modality to Connect Synthetic Biology to Electronics: Hydrogel-Based Chemo-Electro Signal Transduction for Molecular Communication,” Adv. Healthc. Mater, vol. 6, no. 1, p. 1600908, January 2017. [DOI] [PubMed] [Google Scholar]
- [178].Kang M, Kim E, Chen S, Bentley WE, Kelly DL, and Payne GF, “Signal processing approach to probe chemical space for discriminating redox signatures,” Biosens. Bioelectron, vol. 112, pp. 127–135, July 2018. [DOI] [PubMed] [Google Scholar]
- [179].Yan K, Xiong Y, Wu S, Bentley WE, Deng H, Du Y, Payne GF, and Shi X-W, “Electro-molecular Assembly: Electrical Writing of Information into an Erasable Polysaccharide Medium,” ACS Appl. Mater. Interfaces, vol. 8, no. 30, pp. 19780–19786, August 2016. [DOI] [PubMed] [Google Scholar]
- [180].Tschirhart T, Kim E, McKay R, Ueda H, Wu H-C, Pottash AE, Zargar A, Negrete A, Shiloach J, Payne GF, and Bentley WE, “Electronic control of gene expression and cell behaviour in Escherichia coli through redox signalling,” Nat. Commun, vol. 8, p. 14030, January 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]


