Abstract
Cysteine palmitoylation, a form of S-acylation, is a key posttranslational modification in cellular signaling. This type of reversible lipidation occurs in both plasma and organellar membranes, and is catalyzed by a family of integral membrane proteins known as DHHC acyltransferases. The first step in the S-acylation process is the recognition of free acyl coenzyme A (acyl-CoA) from the lipid bilayer. The DHHC enzyme then becomes autoacylated at a site defined by a conserved Asp-His-His-Cys motif. This reaction entails ionization of the catalytic Cys. Intriguingly, in known DHHC structures, this catalytic Cys appears to be exposed to the hydrophobic interior of the lipid membrane, which would be highly unfavorable for a negatively charged nucleophile, thus hindering autoacylation. Here, we use biochemical and computational methods to reconcile these seemingly contradictory facts. First, we experimentally demonstrate that human DHHC20 is active when reconstituted in POPC nanodiscs. Microsecond-long all-atom molecular dynamics simulations are then calculated for human DHHC20 and for different acyl-CoA forms, also in a POPC membrane. Strikingly, we observe that human DHHC20 induces a drastic deformation in the membrane, particularly on the cytoplasmic side, where autoacylation occurs. As a result, the catalytic Cys becomes hydrated and optimally positioned to encounter the cleavage site in acyl-CoA. In summary, we hypothesize that DHHC enzymes locally reshape the membrane to foster a morphology that is specifically adapted for acyl-CoA recognition and autoacylation.
Significance
Palmitoylation, the most common form of S-acylation and the only reversible type of protein lipidation, is ubiquitous in eukaryotic cells. In humans, for example, it has been estimated that as much as ∼10% of the proteome becomes palmitoylated, i.e., thousands of proteins. Accordingly, protein palmitoylation touches every important aspect of human physiology, both in health and in disease. Despite its biological and biomedical importance, little is known about the molecular mechanism of the enzymes that catalyze this posttranslational modification, known as DHHC acyltransferases. Here, we leverage the recently determined atomic-resolution structure of human DHHC20 to gain insights into the mechanism of this class of enzymes using both experimental and computational approaches.
Introduction
DHHC acyltransferases are integral membrane proteins that catalyze cysteine acylation (S-acylation), one of the most common posttranslational modifications in eukaryotic cells (1,2). (The term DHHC reflects a consensus Asp-His-His-Cys catalytic motif.) Indeed, it has been estimated that as much as ∼10% of the human proteome becomes palmitoylated, including over 40% of all known synaptic proteins (3). Accordingly, palmitoylation touches every important aspect of human cellular physiology, both in health and in disease. Human DHHC proteins have been specifically implicated in the development of multiple cancer types, as well as Huntington’s disease, schizophrenia, and X-linked mental retardation (2,4).
S-acylation is unique in that it is the only form of lipidation that can be biochemically reversed, in particular through the action of thioesterases (5,6). Reversible posttranslational modifications are used by the cell to regulate the activity and colocalization (or segregation) of proteins; S-acylation is thus specifically important in membrane signaling (2). S-acylation proceeds in two stages (7). First, a DHHC enzyme embedded in the lipid bilayer binds fatty acyl coenzyme A (acyl-CoA), and the active-site cysteine becomes autoacylated. This process involves the formation of a thioester bond between the fatty acyl chain (hereafter referred to as “acyl chain”) and the cysteine; CoA is cleaved off as a result. In the second stage, the acyl chain covalently linked to DHHC is transferred to a protein substrate (2).
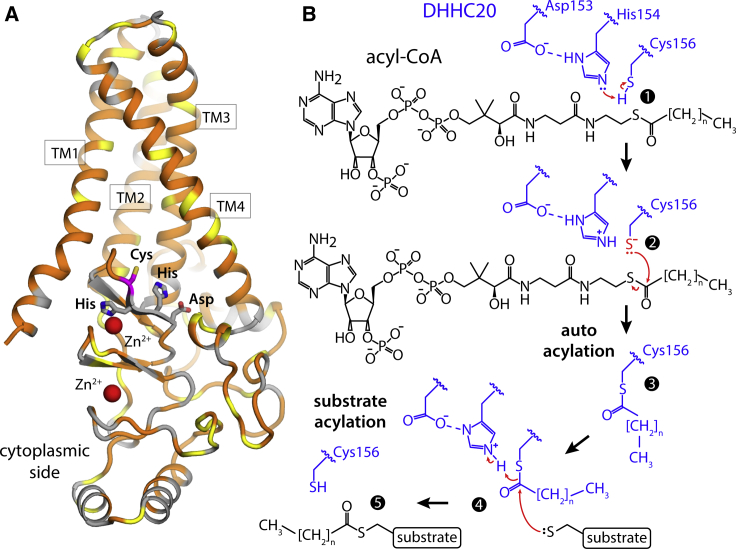
The only human DHHC protein of known structure is DHHC20 (hDHHC20) (8). hDHHC20 is known to be selective for C16:0 acyl chains (8), i.e., it specializes in palmitoylation, which is the most prevalent form of S-acylation (9). Acylation occurs with other chain lengths, but suboptimally (8). hDHHC20 consists of four transmembrane helices and a cytoplasmic domain (Fig. 1 A). The available structure (8) does not fully clarify the mode of acyl-CoA recognition; however, it does reveal the location of the catalytic Asp-His-His-Cys motif and what appears to be the binding surface for the acyl chain in the transmembrane span. Based on these insights, Rana et al. have proposed a catalytic mechanism whereby His154, polarized by Asp153, acts as a base and causes the deprotonation of Cys156. Ionized Cys156 then acts as a thiolate nucleophile and attacks the acyl-CoA carbonyl to create a thioester bond (Fig. 1 B; (10)).
Figure 1.
Structure of human DHHC20 and proposed catalytic mechanism. (A) The existing crystal structure of hDHHC20 (PDB: 6BMN) is shown. The overall fold of the structure is represented with cartoons, indicating the four transmembrane spans (TM1–TM4). Hydrophobic and/or aromatic residues are colored in orange, serine and threonine residues in yellow, and others in gray. The side chains that define the catalytic site (Asp153-His154-His155-Cys156) are highlighted, as well as two structural Zn2+ ions. (B) Scheme for the hypothetical two-stage S-acylation reaction mediated by DHHC20: steps 1–3 make up the autoacylation stage, whereby an acyl-chain is linked to Cys156; in steps 4–5, this acyl-chain is transferred to a substrate. Red arrows indicate the proposed reaction pathway. His213, in TM4, lies within 5 Å of both Asp153 and His154, and might also be involved in this reaction (data not shown for clarity); its role might be to capture a proton initially bound to His154 before acyl-CoA recognition and at the onset of the autoacylation reaction described in (B). To see this figure in color, go online.
It has been puzzling, however, that the catalytic Cys appears to be well within the hydrophobic, transmembrane region of the protein. Shielded from water, the proposed deprotonation of this side chain would be energetically costly and hinder the autoacylation reaction. An equally important question is whether this side chain is accessible to the reactive thioester group in free acyl-CoA. Here, we seek to resolve these questions by combining experimental and computational approaches. First, we use biochemical assays to evaluate the functionality of purified hDHHC20 reconstituted in a simple phospholipid bilayer. Using the same type of lipid bilayer, atomically detailed molecular dynamics (MD) simulations are then calculated, both for hDHHC20 and for three acyl-CoA variants, to gain insights into this recognition process as it pertains to the membrane.
Materials and Methods
Protein expression and purification
Expression and purification of hDHHC20 from Pichia were carried out as described previously (8) with the following modifications: the hDHHC20 expression construct was cloned into a modified pPICZ-N vector with a His10 tag, followed by an mVenus coding sequence and, finally, by a PreScission cleavage site at the N-terminus of the hDHHC20-encoding DNA sequence (11). hDHHC20 was purified with an intact mVenus tag and quantified using nanodrop (εmax = 105,000 M−1). MSPE3D1 was produced as described previously (12) with the following modifications: the TEV cleavage site in the original pET28a MSPE3D1 construct (#20066; Addgene, Cambridge, MA) was mutated to a PreScission cleavage site. The N-terminal His6 tag of MSPE3D1 was removed using PreScission.
Reconstitution in POPC nanodiscs
The hDHHC20 nanodisc reconstitution was carried out in the following condition: 6 μM hDHHC20 mVenus, 18 μM MSPE3D1, 1.44 mM POPC, 20 mM sodium cholate in 25 mM Tris, 137 mM NaCl, 27 mM KCl (pH 7.4) buffer. The reaction mixture was left agitating gently at 4°C for 2 h, and freshly washed amberlite beads (∼0.8–1 g per reaction volume) were added to capture the residual detergent. The reaction was left agitating gently overnight at 4°C. The supernatant was filtered through a 0.45-μm PVDF filter before gel filtration on Superdex 200 Increase column (GE Healthcare, Chicago, IL) using 25 mM Tris, 137 mM NaCl, 27 mM KCl (pH 7.4) buffer. The fractions containing hDHHC20 nanodiscs were collected and concentrated using Millipore 50-kDa concentrators and quantified (εmax = 105,000 M−1).
Autoacylation assay in lipid nanodiscs
The autoacylation assay for DHHC enzymes has been described in detail elsewhere (13). In brief, reaction mixture 1 containing 50 mM HEPES, 50 mM NaCl, 2 mM 2-oxoglutarate, 0.5 mM NAD+, and 0.4 mM thiamine pyrophospate (TPP) was made in 4°C, with and without hDHHC20 nanodiscs. Mixture 2 was prepared containing 40 μM palmitoyl-CoA, 50 mM HEPES, 50 mM NaCl, and 0.2 mUnits of 2-oxoglutarate dehydrogenase and stored in 4°C. In 96-well CORNING (Corning, NY) black-well solid-bottom plates (catalog #: 12-566-09), 30 μL of reaction mixture 1 with and without hDHHC20 nanodiscs was plated, dithiothreitol (DTT) was added (varying concentrations from 50 to 0.01 mM), and NADH fluorescence kinetics were measured after adding 30 μL of mixture 2 containing palmitoyl-CoA using a fluorescence plate reader. The fluorescence of the wells containing hDHHC20 nanodiscs was normalized in reference to that of the wells lacking the enzyme, and the normalized fluorescence was converted to [NADH, μM] using the following equation derived from an NADH calibration curve: RFU = 88.4 × [NADH, μM] + 22.
2-bromopalmitate inhibition assay in nanodiscs
The 2-bromopalmitate (2-BP) inhibition of hDHHC20 autoacylation was monitored by varying concentrations of 2-BP in reaction mixture 1 containing hDHHC20 nanodiscs for a final concentration range of 65.7 μM to 2.0 nM. Dimethyl sulfoxide (DMSO) vehicle controls of reaction mixture 1, both with and without hDHHC20 nanodiscs, were used to normalize the inhibitor-treated wells, and the resulting data were plotted and analyzed with GraphPad (nonlinear regression, log(inhibitor) versus response − variable slope).
Molecular dynamics simulations
MD simulations of hDHHC20 were based on the existing apo-state crystal structure (Protein Data Bank, PDB: 6BMN) (8). Protonation probabilities for each of the ionizable residues in the protein in the configuration observed in the crystal structure were evaluated using a Monte Carlo algorithm and electrostatic energy calculations, as described elsewhere (14). These calculations showed that His154 is more likely to be ionized than not, at pH 7 (∼75%), owing to its proximity with Asp153 (Fig. 1 A). His231, which is also conserved and within 5 Å of both Asp153 and His154, showed a protonation probability of ∼10%. His154 was therefore set in the protonated state, whereas His231 was set as neutral. The protein structure with two Zn2+ ions bound was embedded in a preequilibrated POPC bilayer using GRIFFIN (15). The internal geometry of the Zn2+ binding sites was maintained throughout the study through a network of ion-protein and protein-protein distance restraints involving the four coordinating residues in each case (i.e., a total of 10 distance restraints per ion). The resulting simulation system includes 222 POPC lipids, 21,633 water molecules, 31 Na+, and 43 Cl− (100 mM NaCl plus counterions of the protein net charge) in an orthorhombic box of ∼89 × 89 × 119 Å. The preparation of the simulations entailed a multistage equilibration phase whereby a series of (primarily) internal-coordinate restraints acting on different portions of the protein structure are gradually weakened and ultimately removed over 100 ns. A trajectory of 1 μs was then calculated for each of the two equilibrated configurations, with no conformational restraints (aside from those preserving the internal geometry of the Zn2+ binding sites).
Simulations were also carried for caprylyl-CoA (C8:0), palmitoyl-CoA (C16:0), and behenyl-CoA (C22:0) in a POPC bilayer. These simulation systems consist of 240 POPC lipids, 17,887 water (16) molecules, 34 Na+, and 30 Cl− (100 mM NaCl plus counterions of the acyl-CoA net charge) in an orthorhombic box of ∼87 × 87 × 109 Å. The palmitoyl-CoA simulation was prepared by placing the lipid outside the membrane in the water layer. Palmitoyl-CoA inserted spontaneously in the bilayer within ∼200 ns and remained therein for the duration of trajectory, which was 1 μs long. The simulations of caprylyl-CoA and behenyl-CoA were prepared by elongating or shortening the acyl chain in palmitoyl-CoA once inserted in the membrane.
All MD simulations were carried out using NAMD version 2.9 (17) and the CHARMM36 force field (18, 19, 20) at constant temperature (298 K) and constant pressure (1 atm) with periodic boundary conditions. Electrostatic interactions were calculated using PME, with a real-space cutoff of 12 Å. A cutoff of 12 Å was also used for computing van der Waals interactions, with a smooth switching function taking effect at 10 Å. The integration time step was 2 fs.
Force field parameters for palmitoyl-CoA (C16:0), caprylyl-CoA (C8:0), and behenyl-CoA (C22:0) were derived from existing CHARMM27 parameters for acetyl-CoA (16). Adaptations were made by incorporating the relevant CHARMM36 parameters for protein and ADP into the acyl-CoA parameter set. All three force fields are included as Data S1.
The shape of each of the leaflets in the POPC membrane was evaluated by analyzing the z-coordinate of the ester oxygen layer across the xy plane, averaged over all simulation snaphots. To do so, the xy plane was discretized on a two-dimensional lattice of points spaced by 1 Å. The characteristic value of z for each lattice point (x, y) was quantified by averaging the z-coordinate of the nearest ester oxygen atoms; the contribution of each of these neighboring atoms, identified for each individual snapshot, was weighted according to its proximity to the point considered—that is,
where the index f denotes a specific simulation snapshot, and the index r denotes a specific ester atom in one of the two layers. The weight w for a given atom r, a given snapshot f, and a given lattice point (x, y) was read from a Gaussian function of d, where d is the distance (in the xy plane) between the atom and the lattice point considered. The standard deviation of this Gaussian function was 0.76 Å (i.e., half the atomic radius). Weights were considered only for atoms up to four standard deviations away from a given lattice point.
Results
hDHHC20 is active in POPC nanodiscs
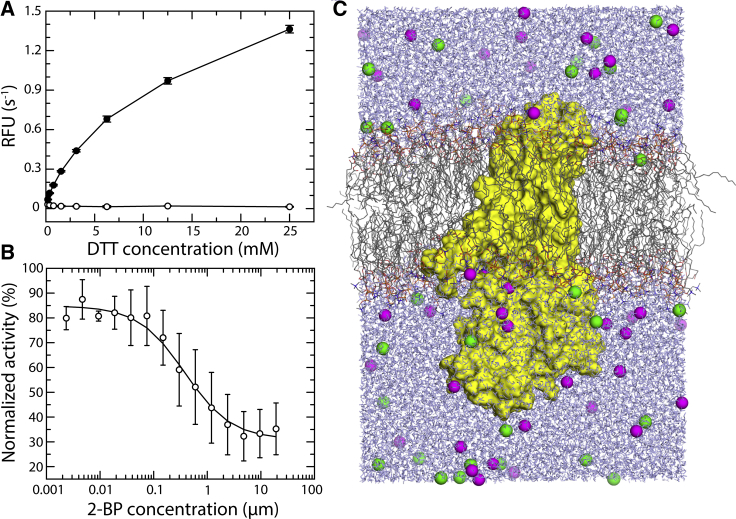
We first sought to identify a type of lipid bilayer that is biologically representative and yet simple enough to permit an unequivocal interpretation of molecular simulation data. Given that phosphatidylcholine lipids are the most abundant in eukaryotic membranes (21), POPC seemed the natural choice. To evaluate whether POPC membranes are a plausible model system for mechanistic studies, we sought to demonstrate the catalytic activity of purified hDHHC20 reconstituted into MSPE3D1 nanodiscs consisting of POPC lipids. To our knowledge, there is no prior report of any DHHC enzyme functionally reconstituted in lipid nanodiscs. Upon addition of acyl-CoA, we observed robust autoacylation activity in samples containing hDHHC20 but no detectable activity for protein-less nanodiscs (Fig. 2 A). Addition of the DHHC inhibitor 2-BP resulted in the expected concentration-dependent inhibitory effect (Fig. 2 B). These results demonstrate that POPC nanodiscs are a valid model system to examine the mechanism of hDHHC20.
Figure 2.
Functional reconstitution of hDHHC20 in a POPC bilayer. (A) An experimental assay of the autoacylation activity of hDHHC20 in POPC nanodiscs, either in the presence of acyl-CoA (closed circles) or in its absence (open circles), is shown. Autoacylation of hDHHC20 is spontaneous; addition of DTT breaks the thioester linkage between Cys156 and acyl-CoA, thereby regenerating the enzyme from the autoacylated form and enabling multiple rounds of catalysis. (B) Impairment of the autoacylation reaction assayed in (A) by addition of 2-BP, a competitive inhibitor of hDHHC20, is shown. (C) An all-atom molecular dynamics simulation system, including hDHHC20 (yellow surface), a POPC bilayer (gray/blue/orange/red lines), water (blue lines), and 100 mM NaCl (green/magenta spheres), is shown. The figures show the final snapshot of one of the two calculated trajectories, each 1 μs long. To see this figure in color, go online.
hDHHC20 deforms the lipid bilayer
In view of these experimental results, we prepared a simulation system for hDHHC20 in a POPC bilayer, with both the protein and its environment represented in atomic detail (Fig. 2 C). We then calculated two independent MD trajectories, each 1 μs long. These trajectories revealed no substantial changes in the architecture of protein relative to the starting condition. For example, relative to the crystal structure, the RMS difference in the backbone of the transmembrane domain is, on average, 0.8 ± 0.1 Å for one simulation and 0.9 ± 0.1 Å for the other. When all secondary-structure elements in the protein are considered, these differences are 2.2 ± 0.4 and 2.4 ± 0.6 Å.
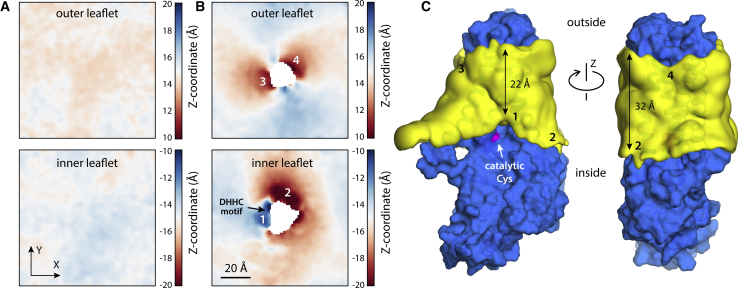
By contrast, we observed that the structure of the lipid bilayer changed significantly in the course of both simulations. In the absence of a protein, a simulated bilayer is essentially flat, on average (Fig. 3 A). Only small deviations are observed, due to thermal fluctuations that have not been completely averaged out. hDHHC20, however, induces a pronounced deformation in the surrounding membrane (Fig. 3 B). This deformation is most prominent on the cytoplasmic side, where the membrane bends toward its center, right in front of the catalytic site; elsewhere along the protein perimeter, the membrane is elevated instead (Fig. 3 B). The magnitude of these perturbations is clearly significant; the displacement of the cytoplasmic ester layer in either direction is ∼5 Å, i.e., ∼1/3 of the leaflet width. Accordingly, these deformations translate into a clear change in membrane width; near the catalytic site, the bilayer is ∼8–10 Å thinner than elsewhere in the protein perimeter (Fig. 3 C).
Figure 3.
Human DHHC20 deforms the lipid bilayer to expose the catalytic cysteine. (A) MD simulations of a pure POPC bilayer are shown. Having set the bilayer midplane as the xy plane (i.e., z = 0), the plots quantify the mean z-coordinate of the ester layer of POPC for different values of x and y, either for the outer leaflet or the inner leaflet. The data are based on a trajectory of 330 ns. The data show that the bilayer is essentially flat, aside from minor thermal fluctuations that have not yet averaged out. (B) MD simulations of the same POPC bilayer with hDHHC20 embedded are shown. The plots again quantify the mean z-coordinate of the two ester layers in the membrane. The data are based on two trajectories of 1 μs each. Regions showing significant deformations are numbered. Regions 1, 3, and 4 reflect depressions, i.e., the ester layer in either leaflet bends toward the membrane center; region 2 is an elevation. (C) Shown are 3D density maps for the POPC alkyl-chain core (yellow surface) near the protein surface (blue) calculated from the MD trajectories. The catalytic cysteine (Cys156) is highlighted (magenta). The four regions where the membrane is deformed are again numbered, as in (B). The approximate minimal and maximal widths of the alkyl-chain core of the bilayer are also indicated. To see this figure in color, go online.
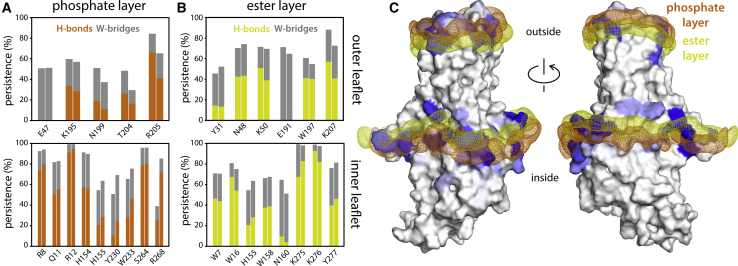
The deformation of the bilayer observed in our simulations is explained by the amino acid composition of the protein surface. Analysis of the frequency of hydrogen-bonding interactions between residues on the hDHHC20 surface and lipid molecules reveals two layers of ionized or polar side chains that persistently associate with either the ester or phosphate layers in the POPC membrane (Fig. 4). The shape of this double layer of polar interactions correlates with the shape of the bilayer around the protein perimeter (Fig. 4), suggesting these interactions contribute to sustain the energetic cost of the observed membrane deformation.
Figure 4.
Polar interactions between hDHHC20 side chains and surrounding lipids. (A) The plot quantifies the persistence of observed interactions between protein side chains and POPC phosphate groups, i.e., the fraction of the simulated time in which the interaction was detected. The data are based on two independent trajectories of 1 μs each. Two types of interactions are described: direct hydrogen bonds (H-bonds) and hydrogen bonds bridged by one water molecule (W-bridge). A direct H-bond was recorded when the distance between a protein donor and a POPC acceptor is 3.2 Å or less (POPC does not contain any H-bond donors). A W-bridge was recorded when a water molecule is found to be at 3.2 Å or less of both an H-bond acceptor in POPC and either an H-bond donor or acceptor in the protein. (B) shows the same as (A) for interactions between protein side chains and POPC ester groups. (C) Shown are 3D density maps for the phosphate and ester layers of POPC (orange and yellow mesh, respectively) near the protein surface, calculated from the MD trajectories. Protein residues that interact with either layer are colored in shades of blue; greater intensity denotes more persistent interactions, as quantified in (A) and (B). To see this figure in color, go online.
Membrane perturbation exposes catalytic Cys to water and to reactive group in acyl-CoA
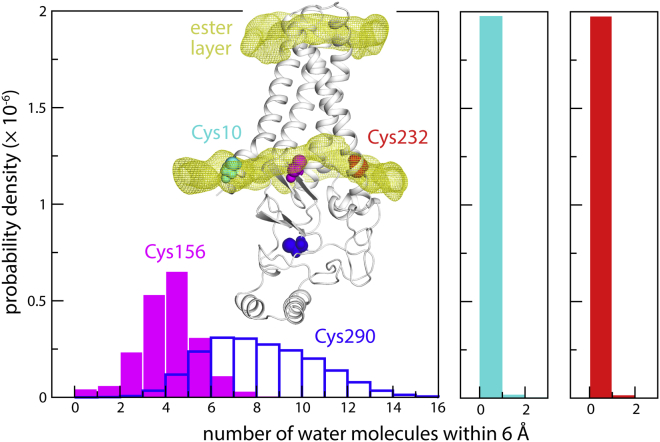
As mentioned, the structure of hDHHC20 shows Cys156 well within the hydrophobic, transmembrane span of the protein, seemingly at odds with a putative reaction intermediate in which this side chain is transiently ionized. The membrane perturbations revealed in our MD simulations resolve this apparent contradiction. As shown in Fig. 5, the localized depression of the bilayer near the catalytic site exposes Cys156 to water to a significant degree. This result is most evident by comparison with Cys10 and Cys232, which are at the same level in the transmembrane domain but are almost entirely dehydrated. Indeed, the range of hydration numbers we observe for Cys156 overlaps signigicantly with that of Cys290, which is in the cytosolic domain and is fully exposed to solvent (Fig. 5).
Figure 5.
Hydration of the catalytic cysteine. The number of water molecules near Cys156 is quantified in terms of a probability distribution derived from two MD trajectories. Analogous data are shown for a surface-exposed cysteine in the cytoplasmic domain (Cys290) and for two cysteines near the inner ester layer (Cys10, Cys232). The inset indicates the location of the four cysteines; calculated density maps for the two ester layers are also shown. To see this figure in color, go online.
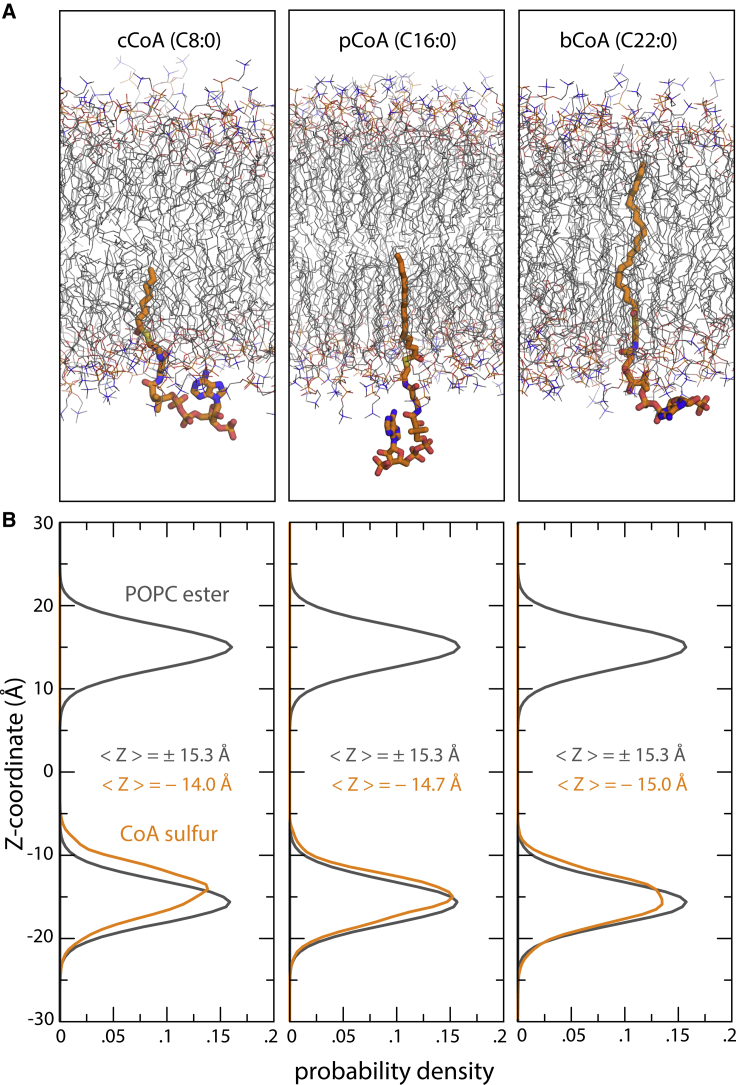
The depression of the membrane near the catalytic site also happens to position Cys156 at the level of the ester layer of the cytoplasmic leaflet (Fig. 5). We wondered whether the reactive thioester group in acyl-CoA might also tend to reside in this layer; despite the ubiquitous importance in biology of fatty acyl-CoA, we could not find a previous study that addresses this question. Thus, we calculated 1-μs trajectories for molecular systems consisting of one acyl-CoA molecule in a POPC lipid bilayer without a protein (Fig. 6 A); we considered three acyl-chain lengths—namely, C8:0 (caprylyl-CoA), C16:0 (palmitoyl-CoA), and C22:0 (behenyl-CoA). All three are substrates of hDHHC20, but as mentioned, palmitoyl-CoA is preferred (10). Caprylyl-CoA and behenyl-CoA are the shortest and longest acyl-chain lengths for which hDHHC20 autoacylation is detectable, albeit at a much slower rate (10). Interestingly, analysis of our trajectory data shows, for all three lengths, that the reactive site in acyl-CoA is indeed aligned with the ester layer of the POPC membrane (Fig. 6 B). The position of acyl-CoA therefore seems to be largely determined by the headgroup rather than by the length of the hydrophobic chain. Thus, by deforming the membrane and aligning Cys156 to the ester layer, hDHHC20 appears to promote encounters between the reactive sites in the protein and substrate.
Figure 6.
MD simulations of acyl-CoA in POPC. (A) Snapshots of three forms of acyl-CoA differing in the length of the acyl chain from MD simulations are shown. From left to right, these are caprylyl-CoA (cCoA), palmitoyl-CoA (pCoA), and behenyl-CoA (bCoA). Surrounding lipid molecules are also shown (thin lines). Water, ions, and hydrogens are omitted for clarity. (B) For each form of acyl-CoA, the plots quantify the most probable location of the sulfur atom in which acyl-CoA is cleaved off (orange) relative to the membrane center and to the two ester layers in POPC. The distributions shown derive from three 1-μs MD trajectories (one for each form of acyl-CoA). To see this figure in color, go online.
The binding site for the acyl-CoA chain
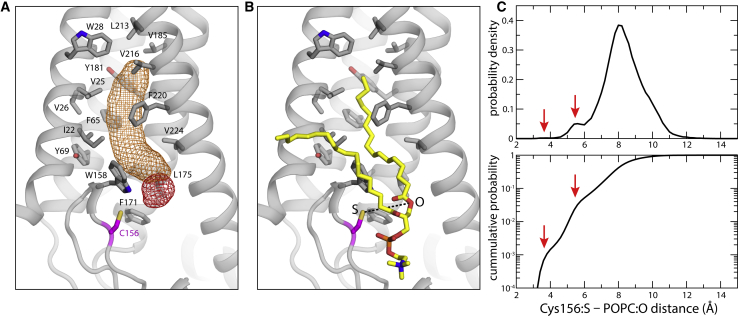
Although no structure of DHHC bound to acyl-CoA has been reported yet, the structure of hDHHC20 inhibited by 2-BP has been determined. 2-BP also reacts with Cys156 to form a covalent linkage (8). In this structure, the palmitate chain of 2-BP occupies a hydrophobic tunnel formed within the protein on the cytoplasmic half of the transmembrane span. Because reactivity with acyl-CoA or 2-BP necessarily begins with noncovalent recognition of these ligands, we wondered whether we would observe POPC lipids bound to this site in our simulations. Indeed, analysis of the simulation data did in fact reveal that this tunnel is persistently occupied by one alkyl chain of a POPC molecule (Fig. 7, A and B). The site is empty at the beginning of the two MD trajectories, but two different POPC lipids move to occupy the cavity early on. Interestingly, in both cases, it is the C16:0 alkyl chain that enters the hydrophobic pocket. We next asked whether the range of configurations explored by this bound lipid during the simulations might be conducive to autoacylation if translated to palmitoyl-CoA. Indeed, we find that in ∼5% of the configurations the distance between the reactive sites in the protein and the lipid is only 6 Å or less (Fig. 7 C). Although this is a small population it is worth noting that the thioester bond formed upon DHHC autoacylation does not break spontaneously (i.e., in the absence of a thiol acceptor, in either a small molecule or a protein); this irreversibility implies that autoacylation will progress steadily even if the likelihood of reactive configurations is small. Taken together with the structure of the DHHC20-2-BP complex, these observations indicate that this cavity is indeed the site of recognition of the hydrophobic chain of acyl-CoA before autoacylation. These simulations, however, do not offer clues on the mode of recognition of the acyl-CoA headgroup because it is chemically very different from that of POPC.
Figure 7.
Occupancy of the putative acyl-CoA binding site by POPC. (A) Shown is the 3D density map for the POPC alkyl chain (orange mesh) that occupies the putative binding site for acyl-CoA throughout both of the simulated trajectories. A density map for the corresponding ester group is also shown (red mesh). The hydrophobic and/or aromatic side chains lining the binding pocket are highlighted, as is the catalytic cysteine Cys156. The site is empty at the beginning of both simulations. Two different POPC lipids are observed to bind to the protein early on in the equilibration phase. In both cases, it is the C16:0 alkyl chain that enters the hydrophobic pocket. (B) Shown is the snapshot of the bound POPC lipid at the end of one of the two 1-μs MD trajectories (also shown in A). In this snapshot, the distance between the sulfur atom of Cys156 and the oxygen atom in POPC that is analogous to the reactive sulfur in pCoA (dashed lines) is 8.1 Å. (C) The top panel shows the probability distribution for the S-O distance described in (B), calculated from all trajectory data; the lower panel shows the integral of this probability distribution. Red arrows indicate low-population states that would likely foster the formation of a thioester bond if acyl-CoA was bound as POPC. The peaks at 5.5 and 8 Å are populated in both simulations. The small peak at 3.5 Å is, however, only observed in one of the 1-μs trajectories. To see this figure in color, go online.
Discussion
Despite the physiological importance of protein palmitoylation, little is known about the molecular mechanism of the enzymes that catalyze this posttranslational modification, known as DHHC acyltransferases. In a breakthrough, the atomic structure of human DHHC20 was recently determined, both for an apo state and in complex with the inhibitor 2-BP (8). These structures not only provided the necessary framework to generate atomically detailed mechanistic hypotheses but also opened up a range of new questions. Particularly pertinent among them are the nature of the interactions between DHHC enzymes and the surrounding membrane and how the membrane, in turn, defines the chemical environment around the active site. This study builds upon that discovery and seeks to begin to provide specific insights into these questions.
More specifically, we first aimed to establish an experimental in vitro system to evaluate the activity of the enzyme in the context of a lipid membrane, rather than in detergent, which had been the condition of previous functional assays (8). This development was essential because S-acylation requires that DHHC proteins first recognize acyl-CoA molecules residing in the membrane. The fatty acyl group then becomes covalently linked to a membrane-proximal cysteine and is ultimately transferred to a substrate protein. The membrane thus appears to be an integral element of this process. Although lipid nanodiscs are imperfect models, it has been argued that they might be more realistic than liposomes, for example, in terms of lipid diffusion rates or lateral pressure profile, in that the morphological restraints imposed by the scaffold protein mimic the crowded environment of biological membranes (22,23). We thus believe that the nanodisc system developed here will enable us to examine a range of mechanistic aspects, such as the molecular basis for the varying substrate specificities of different DHHC enzymes, or how the lipid composition of the membrane defines catalytic rates. Another area of interest is of course the interaction of the enzyme with potential drugs; indeed, two human DHHC enzymes have been proposed as targets for novel therapeutic interventions against cancer (36,37).
Our second aim was to begin to understand the interplay between DHHC enzymes and the membrane in atomic detail. To this end, we resorted to MD simulations, which had not been previously employed to study this class of enzymes. The simulations enabled us to clarify a puzzling observation in the crystal structure of hDHHC20—namely, that the catalytic cysteine (Cys156) is well within the hydrophobic transmembrane span of the protein. Shielded from water, the transient ionization of Cys156, which must precede the formation of the thioester bond with the chain of acyl-CoA, would be highly disfavored. (Low-dielectric environments such as the interior of the membrane or a protein tend to shift the pKa of ionizable residues in favor of their neutral form; in the case of Cys, whose solution pKa is 8.6, the shift would be upwards.) The answer to this puzzle is that hDHHC20 induces a pronounced depression of the membrane near the catalytic site precisely to expose Cys156 to water. Interestingly, our simulations show that this perturbation of the membrane also aligns Cys156 with the ester layer, which is precisely where the cleavage site in acyl-CoA resides. These insights underscore the notion that the interplay between membrane and protein is an integral element of the mechanism of this class of enzymes. These results also provide a solid foundation for future simulation studies; for example, a direct examination of the process of recognition of acyl-CoA ought to reveal important clues into the determinants of the chain-length specificity of hDHHC20.
Whether the position of the active site relative to the membrane center is a conserved feature across the DHHC family remains to be clarified. Therefore, it is also unclear whether other DHHC proteins deform the membrane as DHHC20 does, and whether this deformation is functionally crucial or provides some kind of specialization. It is conceivable that the catalytic site is shifted in other DHHC proteins either toward the cytoplasm or to the membrane center, reflecting variations in other structural elements. If so, it would be of interest to correlate this aspect with the functional characteristics of each protein. For example, the precise positioning of the catalytic domain and the degree to which it perturbs the surrounding bilayer might impact the chain-length selectivity of the enzyme.
The ligands of many membrane-integral enzymes and transporters are hydrophobic (or at least amphipathic), implying these ligands are most likely recognized (and released) laterally, from (and to) the membrane instead of through the aqueous phase. In particular, membrane-integral enzymes with lipid-like ligands include many transferases (24, 25, 26, 27, 28), as well as some translocases (29), synthases (30), polymerases (31), and proteases (32). Little is known about how most of these proteins interact with the membrane, but the increasing availability of structural data will surely help advance our understanding. For example, in the structure of the isoprenylcysteine carboxyl methyltransferase, the arrangement of two of the transmembrane helices appears to indicate a depression in the lipid membrane near the active site to help accommodate the substrates (24). Similar membrane perturbations have been noted for the rhomboid protease GlpG based on pioneering MD simulations (33,34), later corroborated by EPR spectroscopy (35). Taken together with our observations for DHHC20, the paradigm that seems to be emerging for these types of membrane proteins is that they have evolved highly specific ways to perturb the morphology of the surrounding lipid bilayer, precisely to facilitate the recognition of substances and substrates that reside therein.
Author Contributions
R.S. performed the computational work and analyzed the data. J.S. performed the experimental work and analyzed the data. A.B. designed and supervised the experimental work and data analysis. J.D.F.-G. designed and supervised the computational work and data analysis. All authors contributed to writing the manuscript and preparing figures.
Acknowledgments
This work was funded by the Divisions of Intramural Research of the National Heart, Lung and Blood Institute (R.S. and J.D.F.-G.) and of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (J.S. and A.B.), National Institutes of Health. Computational resources were in part provided by the National Institutes of Health High-Performance Computing facility Biowulf.
Editor: Michael Grabe.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.11.003.
Contributor Information
Anirban Banerjee, Email: anirban.banerjee@nih.gov.
José D. Faraldo-Gómez, Email: jose.faraldo@nih.gov.
Supporting Material
. CHARMM Forcefield Parameters for Palmitoyl-CoA, Caprylic-CoA, and Behenic-CoA
References
- 1.Khoury G.A., Baliban R.C., Floudas C.A. Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Sci. Rep. 2011;1:90. doi: 10.1038/srep00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang H., Zhang X., Lin H. Protein lipidation: occurrence, mechanisms, biological functions, and enabling technologies. Chem. Rev. 2018;118:919–988. doi: 10.1021/acs.chemrev.6b00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanders S.S., Martin D.D., Hayden M.R. Curation of the mammalian palmitoylome indicates a pivotal role for palmitoylation in diseases and disorders of the nervous system and cancers. PLoS Comput. Biol. 2015;11:e1004405. doi: 10.1371/journal.pcbi.1004405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Korycka J., Łach A., Sikorski A.F. Human DHHC proteins: a spotlight on the hidden player of palmitoylation. Eur. J. Cell Biol. 2012;91:107–117. doi: 10.1016/j.ejcb.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 5.Zaballa M.E., van der Goot F.G. The molecular era of protein S-acylation: spotlight on structure, mechanisms, and dynamics. Crit. Rev. Biochem. Mol. Biol. 2018;53:420–451. doi: 10.1080/10409238.2018.1488804. [DOI] [PubMed] [Google Scholar]
- 6.Blaskovic S., Blanc M., van der Goot F.G. What does S-palmitoylation do to membrane proteins? FEBS J. 2013;280:2766–2774. doi: 10.1111/febs.12263. [DOI] [PubMed] [Google Scholar]
- 7.Gottlieb C.D., Linder M.E. Structure and function of DHHC protein S-acyltransferases. Biochem. Soc. Trans. 2017;45:923–928. doi: 10.1042/BST20160304. [DOI] [PubMed] [Google Scholar]
- 8.Rana M.S., Kumar P., Banerjee A. Fatty acyl recognition and transfer by an integral membrane S-acyltransferase. Science. 2018;359:eaao6326. doi: 10.1126/science.aao6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchell D.A., Vasudevan A., Deschenes R.J. Protein palmitoylation by a family of DHHC protein S-acyltransferases. J. Lipid Res. 2006;47:1118–1127. doi: 10.1194/jlr.R600007-JLR200. [DOI] [PubMed] [Google Scholar]
- 10.Rana M.S., Lee C.J., Banerjee A. The molecular mechanism of DHHC protein acyltransferases. Biochem. Soc. Trans. 2019;47:157–167. doi: 10.1042/BST20180429. [DOI] [PubMed] [Google Scholar]
- 11.Rana M.S., Wang X., Banerjee A. An improved strategy for fluorescent tagging of membrane proteins for overexpression and purification in mammalian cells. Biochemistry. 2018;57:6741–6751. doi: 10.1021/acs.biochem.8b01070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ritchie T.K., Grinkova Y.V., Sligar S.G. Chapter 11 - reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 2009;464:211–231. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamel L.D., Deschenes R.J., Mitchell D.A. A fluorescence-based assay to monitor autopalmitoylation of zDHHC proteins applicable to high-throughput screening. Anal. Biochem. 2014;460:1–8. doi: 10.1016/j.ab.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eicher T., Seeger M.A., Pos K.M. Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB. eLife. 2014;3:03145. doi: 10.7554/eLife.03145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Staritzbichler R., Anselmi C., Faraldo-Gómez J.D. GRIFFIN: a versatile methodology for optimization of protein-lipid interfaces for membrane protein simulations. J. Chem. Theory Comput. 2011;7:1167–1176. doi: 10.1021/ct100576m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aleksandrov A., Field M. Efficient solvent boundary potential for hybrid potential simulations. Phys. Chem. Chem. Phys. 2011;13:10503–10509. doi: 10.1039/c0cp02828b. [DOI] [PubMed] [Google Scholar]
- 17.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klauda J.B., Venable R.M., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Best R.B., Zhu X., Mackerell A.D., Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vanommeslaeghe K., Hatcher E., Mackerell A.D., Jr. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mörs K., Roos C., Glaubitz C. Modified lipid and protein dynamics in nanodiscs. Biochim. Biophys. Acta. 2013;1828:1222–1229. doi: 10.1016/j.bbamem.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 23.Martinez D., Decossas M., Loquet A. Lipid internal dynamics probed in nanodiscs. ChemPhysChem. 2017;18:2651–2657. doi: 10.1002/cphc.201700450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diver M.M., Pedi L., Long S.B. Atomic structure of the eukaryotic intramembrane RAS methyltransferase ICMT. Nature. 2018;553:526–529. doi: 10.1038/nature25439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mao G., Zhao Y., Zhang X.C. Crystal structure of E. coli lipoprotein diacylglyceryl transferase. Nat. Commun. 2016;7:10198. doi: 10.1038/ncomms10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu G., Xu Y., Zhang X.C. Crystal structure of E. coli apolipoprotein N-acyl transferase. Nat. Commun. 2017;8:15948. doi: 10.1038/ncomms15948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wiktor M., Weichert D., Caffrey M. Structural insights into the mechanism of the membrane integral N-acyltransferase step in bacterial lipoprotein synthesis. Nat. Commun. 2017;8:15952. doi: 10.1038/ncomms15952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anandan A., Evans G.L., Vrielink A. Structure of a lipid A phosphoethanolamine transferase suggests how conformational changes govern substrate binding. Proc. Natl. Acad. Sci. USA. 2017;114:2218–2223. doi: 10.1073/pnas.1612927114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung B.C., Zhao J., Lee S.Y. Crystal structure of MraY, an essential membrane enzyme for bacterial cell wall synthesis. Science. 2013;341:1012–1016. doi: 10.1126/science.1236501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X., Yin Y., Liu Z. Structure and mechanism of an intramembrane liponucleotide synthetase central for phospholipid biosynthesis. Nat. Commun. 2014;5:4244. doi: 10.1038/ncomms5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sjodt M., Brock K., Kruse A.C. Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis. Nature. 2018;556:118–121. doi: 10.1038/nature25985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y., Zhang Y., Ha Y. Crystal structure of a rhomboid family intramembrane protease. Nature. 2006;444:179–180. doi: 10.1038/nature05255. [DOI] [PubMed] [Google Scholar]
- 33.Bondar A.N., del Val C., White S.H. Rhomboid protease dynamics and lipid interactions. Structure. 2009;17:395–405. doi: 10.1016/j.str.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bondar A.N. Mechanisms by which lipids influence conformational dynamics of the GlpG intramembrane protease. J. Phys. Chem. B. 2019;123:4159–4172. doi: 10.1021/acs.jpcb.8b11291. [DOI] [PubMed] [Google Scholar]
- 35.Kreutzberger A.J.B., Ji M., Urban S. Rhomboid distorts lipids to break the viscosity-imposed speed limit of membrane diffusion. Science. 2019;363::eaao0076. doi: 10.1126/science.aao0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Runkle K.B., Kharbanda A., Witze E.S. Runkle, A. Kharbanda, et al., E.S. Witze, Inhibition of DHHC20-Mediated EGFR Palmitoylation Creates a Dependence on EGFR Signaling. Mol. Cell. 2016;62:385–396. doi: 10.1016/j.molcel.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao H., Lan J., Xu J. Inhibiting PD-L1 palmitoylation enhances T-cell immune responses against tumours. Nat. Biomed. Eng. 2019;3:306–317. doi: 10.1038/s41551-019-0375-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
. CHARMM Forcefield Parameters for Palmitoyl-CoA, Caprylic-CoA, and Behenic-CoA