Abstract
MicroRNA (miRNA) and mitofusin-2 (Mfn2) are important in the development of cardiac hypertrophy, but the target relationship and mechanism associated with Ca2+ handling between SR and mitochondria under hypertrophic condition is not established. Mfn2 expression, Mfn2-mediated interorganelle Ca2+ cross-talk, and target regulation by miRNA-20b (miR-20b) were evaluated using animal/cellular hypertrophic models with state-of-the-art techniques. The results demonstrated that Mfn2 was downregulated and miR-20b was upregulated upon the target binding profile under hypertrophic condition. Our data showed that miR-20b induced cardiac hypertrophy that was reversed by recombinant adeno-associated virus vector 9 (rAAV9)-anti-miR-20b or miR-20b antisense inhibitor (AMO-20b). The deleterious action of miR-20b on Mfn2 expression/function and mitochondrial ATP synthesis was observed and reversed by rAAV9-anti-miR-20b or AMO-20b. The targeted regulation of miR-20b on Mfn2 was confirmed by luciferase reporter and miRNA-masking. Importantly, the facts that mitochondrial calcium uniporter (MCU) activation by Spermine increased the cytosolic Ca2+ into mitochondria, manifested as enhanced histamine-mediated Ca2+ release from mitochondrial, suggesting that Ca2+ reuptake/buffering capability of mitochondria to cytosolic Ca2+ is injured by miR-20b-mediated Mfn2 signaling, by which leads cytosolic Ca2+ overload and cardiac hypertrophy through Ca2+ signaling pathway. In conclusion, pro-hypertonic miR-20b plays crucial roles in cardiac hypertrophy through downregulation of Mfn2 and cytosolic Ca2+ overload by weakening the buffering capability of mitochondria.
Introduction
Cardiac hypertrophy is associated closely with mitochondrial dysfunction.1 Morphological and functional integrity of mitochondria are regulated by the highly dynamic processes of fusion and fission events,2 in which mitofusin 2 (Mfn2) is a key player in these events.3 Additionally, Mfn2 participates in various cellular processes including mitochondrial metabolism and energy supply, mitochondrial membrane potential, mitophagy, and mtDNA stability.4,5 More importantly, Mfn2 plays an important role in maintaining or buffering the cytosolic Ca2+ through uptaking Ca2+ into mitochondria.6,7 Mfn2 is enriched at the interface of mitochondria and mitochondria-associated sarcoplasmic reticulum (SR) in cardiomyocytes,6,8 highly suggesting the critical role of Mfn2 in the interorganelle Ca2+ cross-talk between SR and mitochondria; however, its exact function of Mfn2 remains a matter of intense debate.2,6 Although Mfn2 is likely a putative inhibitor of cardiac hypertrophy,9,10 the evidence is limited, especially the underlying mechanism of functional expression of Mfn2 in the development of myocardial hypertrophy, and we have strong reason to believe that Mfn2-mediated interorganelle communication of Ca2+ plays a crucial role in pathophysiology of cardiac hypertrophy. MicroRNAs (miRNAs) are important gene modulators at post-transcriptional level11,12 and promising novel therapeutic targets for cardiovascular diseases. Determining whether miRNAs share the same seed site with Mfn2 in cardiac hypertrophy is another goal of the current study. Intriguingly, our present data have shown that miRNA-20b (miR-20b) was substantially upregulated in the hypertrophic heart. However, current data regarding miR-20b only focus on a variety of cancers as a tumor suppressor13, 14, 15, 16 and no such research has been done so far in cardiac hypertrophy.
On the basis of these facts, we hypothesized that miR-20b is likely to directly and negatively regulate Mfn2 expression leading to interorganelle Ca2+ communication between SR and mitochondria. Interestingly, by using the state-of-the-art techniques, the current study has demonstrated for the first time that the direct target relationship between miR-20b and Mfn2 that has been confirmed and upregulated miR-20b during hypertrophy impairs Mfn2-mediated Ca2+ reuptake/buffering capability of mitochondria, consequently leading to the cardiac hypertrophy. These novel findings extend our current understanding and shed new light for prophylactic and therapeutic strategies of clinical management of cardiac hypertrophy-related cardiovascular diseases.
Results
Upregulated miR-20b in In Vivo and In Vitro Models of Myocardial Hypertrophy
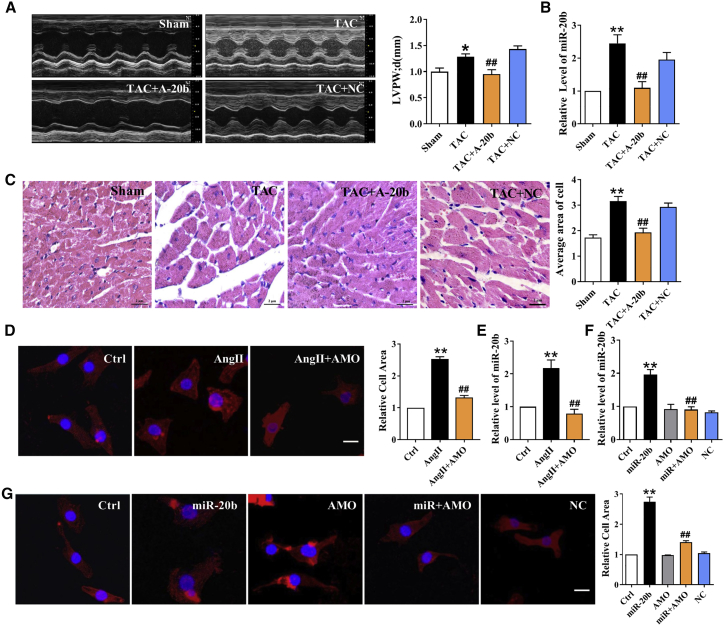
Transverse aortic constriction (TAC) in mice, a widely accepted in vivo hypertrophic model, was used in the current study, which was confirmed by echocardiographic analysis (Figure 1A; Table S1), the ratios of heart/body weight and heart weight/tibia length (Figures S1A–S1C), and the biomarkers including atrial natriuretic peptide (ANP), brain natriuretic peptide (BNP), and β-myosin heavy chain (β-MHC) (Figure S1D). By using this in vivo model, Mfn2 was downregulated markedly (Figure 1B). To confirm this result, we selected an in vitro hypertrophic model of neonatal rat ventricular cardiomyocytes (NRVCs) treated with angiotensin II (Ang II) as well (Figure S1E) and the expression profile of Mfn2 was consistently downregulated (Figure 1C). Interestingly, under exact experimental conditions, the miR-20b expression was significantly upregulated among those miRNAs showing binding profiles with Mfn2 (Figures 1D and 1E), suggesting the potential involvement in cardiac hypertrophy via targeted regulation of Mfn2.
Figure 1.
miR-20b Is Upregulated in TAC Mice and Hypertrophic Cardiomyocytes
(A) Exemplary picture of echocardiography and statistical analysis of left ventricular posterior wall at end-diastole (LVPW;d). LVPW;d of TAC mice was thicker compared with that of sham mice. n = 11–16. (B) Decreased expression of Mfn2 in TAC mice. n = 8. (C) Expression of Mfn2 in Ang II-incubated cardiomyocytes was downregulated. n = 6. (D) miRNAs expression levels in TAC and sham mice. miR-20b level in myocardium was upregulated in TAC mice. n = 12–15. (E) The expression levels of miRNAs in NRVCs treatment with or without Ang II. n = 10–12. Averaged data were presented as mean ± SEM, *p < 0.05 and **p < 0.01 versus Sham or Ctrl (Control).
Confirmation of miR-20b-Mediated Induction of Myocardial Hypertrophy
To test the effect of miR-20b, we introduced recombinant adeno-associated virus vector 9 (rAAV9)-anti-miR-20b into TAC mice to knockdown endogenous miR-20b. The wall thickness of left ventricle was reduced in TAC mice treated with rAAV9-anti-miR-20b as compared with those treated with rAAV9-negative control (NC) (Figure 2A; Table S2). The successful uptake of rAAV9-anti-miR-20b or scramble rAAV9-NC was identified by quantitative real-time PCR (Figure 2B). Similar trends in changing the ratios of heart/body weight and heart weight/tibia length and the mRNA levels of hypertrophic biomarkers were also confirmed (Figures S2A–S2D). This observation was further supported by the notion of histological sectioning with H&E staining showing a significant increase in the cross-sectional area of ventricular tissues after TAC and a restoration by administration of rAAV9-anti-miR-20b (Figure 2C). To confirm these in vivo observations, we transfected miR-20b antisense inhibitor (AMO-20b) into Ang II-treated NRVCs and the result showed that increased cell surface area induced by upregulation of endogenous miR-20b was abolished significantly (Figure 2D). The successful transfection of AMO-20b was verified by quantitative real-time PCR (Figure 2E). To further assess this finding, we transfected miR-20b mimic, AMO-20b, or NC miRNA into NRVCs and verified the efficiency (Figure 2F). Notably, miR-20b mimic increased the cell surface area and that was reversed by co-transfection of miR-20b mimic and AMO-20b, as well as no change with NC (Figure 2G). Consistently, similar trends were also verified at the mRNA levels of ANP, BNP, and β-MHC (Figures S2E and S2F). These results strongly imply that gain- and loss-of-function of miR-20b may play a crucial role in cardiac hypertrophy, while knockdown of miR-20b may exert protection against hypertrophy.
Figure 2.
miR-20b Induces Cardiomyocyte Hypertrophy
(A) TAC-induced increases of LVPW;d were abrogated by A-20b. A-20b, rAAV9-anti-miR-20b; NC, negative control. n = 8–12. (B) The successful uptake of A-20b or scramble NC was verified. n = 7–9. (C) A-20b reversed the increase of cross-sectional area of ventricular tissues after TAC in H&E staining. Magnification ×400; scale bar represents 2 μm. n = 6 different sections. (D) Representative images and statistical analysis showing changes of the cell surface area in Ang II-treated cells with or without AMO-20b. Magnification of 200×; scale bar represents 5 μm. n = 6 different sections. (E) The successful transfection of AMO-20b was verified. n = 9. (F) The successful transfection of miR-20b was verified. n = 6. (G) Representative images and statistical analysis showing that miR-20b-induced increase of cell surface area was reversed by co-transfection with AMO-20b. n = 6. Averaged data were presented as mean ± SEM, *p < 0.05 and **p < 0.01 versus Sham or Ctrl, #p < 0.05 and ##p < 0.01 versus TAC, Ang II, or miR-20b.
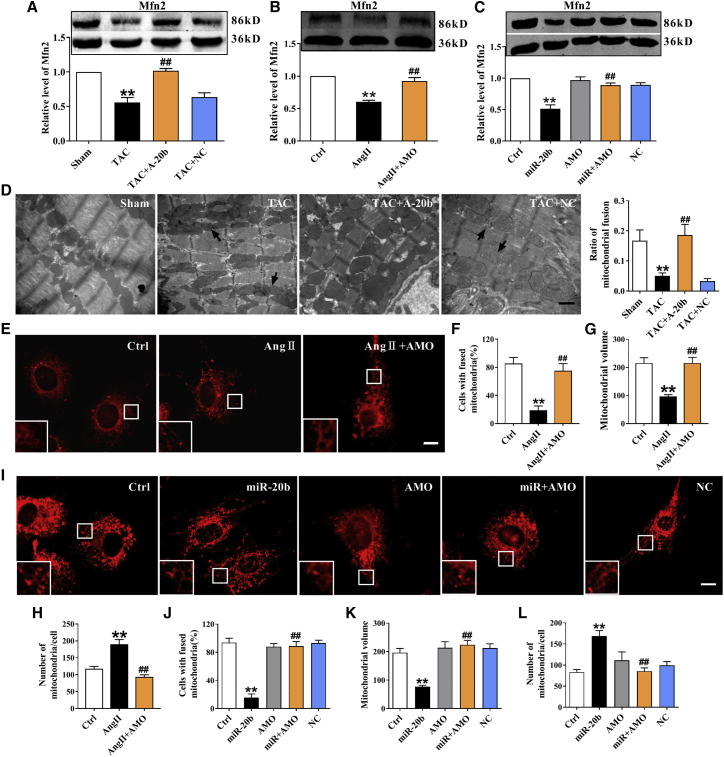
miR-20b-Mediated Negative Regulation of Mfn2 in Cardiac Hypertrophic Models
Western blot results showed that the expression of Mfn2 was significantly repressed in TAC mice and restored by rAAV9-anti-miR-20b, rather than scramble NC (Figure 3A). Significant downregulation of Mfn2 was also observed in the in vitro study. In stark contrast, administration of AMO-20b markedly elevated the protein level of Mfn2 (Figures 3B and 3C). Additionally, differential expression profiles of Mfn2 at mRNA and protein levels (Figures S3A–S3C) led us to speculate a possible post-transcriptional regulation. Because Mfn2 is associated with mitochondrial dynamics, the mitochondrial fusion was evaluated by transmission electron microscopy (TEM) and the result showed that the ratio of mitochondrial fusion (numbers of fused mitochondria/total mitochondria) was reduced by TAC procedure and restored by rAAV9-anti-miR-20b (Figure 3D). Similar and quantitative alterations were also detected in NRVCs using mitotracker, a fluorescent indicator of mitochondria. Apparently, Ang II treatment induced mitochondrial fragmentation and significant decrease in percentage of cells with fused mitochondria from 80% down to 20% (Figures 3E and 3F). Importantly, mitochondrial network integrity was also tested and the result showed that Ang II decreased the mean volume of individual mitochondrion and increased total number of mitochondria per cell (Figures 3G and 3H), which was reversed by AMO-20b (Figures 3E–3H). Similar results were confirmed in the model of overexpression of miR-20b (Figures 3I–3L). Moreover, an inverse change in ATP synthesis would not be surprised in hypertrophic models and in the presence of rAAV9-anti-miR-20b or AMO-20b (Figures S3D–S3F). These data implicate a direct target and negative regulation of miR-20b to Mfn2 under hypertrophic conditions.
Figure 3.
The Effect of miR-20b on Mfn2 in Both In Vivo and In Vitro Hypertrophic Models
(A) TAC-induced decrease of Mfn2 at protein levels in the mouse heart were improved by A-20b. n = 11. (B) Ang II-induced downregulation of Mfn2 at protein levels in NRVCs were abolished by AMO-20b transfection. n = 9. (C) miR-20b overexpression-induced reduction of Mfn2 at protein levels were reversed by co-transfection of miR-20b mimic and AMO-20b. n = 9. (D) A-20b reversed the decrease in the ratio of mitochondrial fusion induced by TAC in TEM examination. Magnification ×20,000; scale bar represents 1 μm. n = 6–13 different sections. (E) Representative images of mitotracker fluorescence staining showing that Ang II-induced mitochondrial fragmentation was abrogated by AMO-20b. Magnification of 400×; scale bar represents 5 μm. (F) Statistical analysis of percentage of cells with fused mitochondria in the model of Ang II treatment. n = 4 different times. (G and H) AMO-20b transfection reversed Ang II-induced the decrease of the mean volume of individual mitochondrion and the increase of total number of mitochondria per cell. n = 17–25 different cells. (I) Representative images of mitotracker fluorescence showing that miR-20b-induced decrease of mitochondrial fusion was restored by co-transfection with AMO-20b. (J–L) Statistical analysis of percentage of cells with fused mitochondria, mean volume of individual mitochondrion, and total number of mitochondria per cell in overexpression model of miR-20b. n = 4 different times, and 17–23 different cells. Averaged data were presented as mean ± SEM, *p < 0.05 and **p < 0.01 versus Sham or Ctrl, #p < 0.05 and ##p < 0.01 versus TAC, Ang II, or miR-20b.
Confirmation of Targeted Relationship between miR-20b and Mfn2
As expected, a complementary binding site between miR-20b and Mfn2 was predicted (Figure 4A) using the database of miRNA targets (http://www.targetscan.org/) and the database of the University of California Santa Cruz (UCSC). To validate this finding, we constructed luciferase reporter vectors containing a segment of the 3′ UTR of Mfn2 (MFN2) or a mutated 3′ untranslated region (3′ UTR) of Mfn2 (Mut-MFN2). By co-transfection of constructs with miR-20b mimic into HEK293 cells (Figure 4B), the luciferase activity was markedly inhibited and the silencing effects were recognized by co-transfection of constructs with AMO-20b, whereas no effect was observed with the corresponding mutant construct. To further confirm that the observed changes of Mfn2 were the direct results of miR-20b, we employed the miRNA-masking antisense oligodeoxynucleotides (ODN) techniques (miR-mask) as previously developed.17 Instead of binding to the target miRNA like an AMO, the miR-20b-mask was designed upon the base pair of miR-20b binding sites in the 3′ UTRs of Mfn2 genes labeled MFN2-ODN. As expected, the miR-mask (MFN2-ODN), unlike AMO-20b, failed to affect miR-20b while co-transfection (Figure 4C) blocked the repressive effects of miR-20b on Mfn2 (Figure 4D), implying that Mfn2 is the target gene for miR-20b. Moreover, this Mfn2-specific mask and related attenuation of cell hypertrophy by the detection of the cell surface area (Figures 4E and 4G), mitotracker (Figures 4F and 4H–4J), and hypertrophic biomarkers (Figure S3G) would really strengthen our conclusion.
Figure 4.
Validation of Mfn2 as a Target Gene of miR-20b
(A) The binding (seed) site for miR-20b in the 3′ UTR of the Mfn2 gene, as predicted by the TargetScan algorithm. The mutant sequences were equivalent to the wild-type with the exception of mutations at the 3′ end of the target site. (B) Luciferase reporter gene assay for measuring interactions between miR-20b and its binding sites in the 3′ UTRs of Mfn2 mRNAs in HEK293T cells. Cells were transfected with luciferase-target motif chimeric vector alone (Ctrl) or with miR-20b or NC using lipofectamine 2000. n = 3. (C) Lack of effect of gene-specific MFN2-ODN on miR-20b expression in NRVCs by quantitative real-time PCR. n = 6. (D) Derepression of target genes of miR-20b by MFN2-ODN, determined by western blot analysis. n = 6. (E and G) Representative images and statistical analysis showing that miR-20b-induced increase of cell surface area was abrogated by MFN2-ODN. Magnification ×200; scale bar represents 5 μm. n = 6 different sections. (F and H–J) Representative images and statistical analysis of mitotracker fluorescence showing that miR-20b-induced decrease of mitochondrial fusion was reversed by MFN2-ODN. Magnification ×400; scale bar represents 5 μm. n = 4 different times, and 19–23 different cells. Averaged data were presented as mean ± SEM, *p < 0.05 and **p < 0.01 versus Ctrl, #p < 0.05 and ##p < 0.01 versus miR-20b.
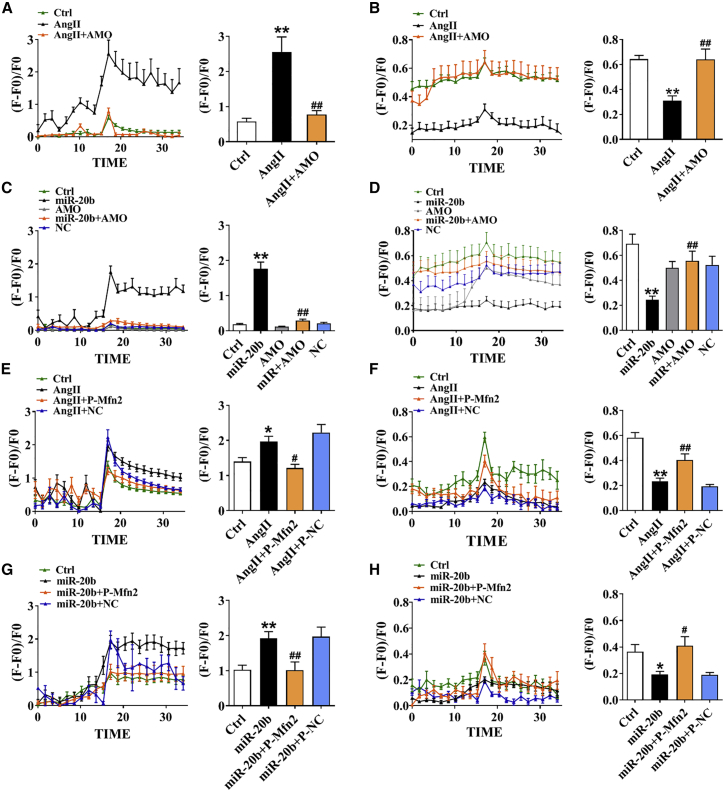
miR-20b-Facilitated Cardiac Hypertrophy via Dysregulated Ca2+ Interaction of Mfn2 between SR and Mitochondria
Mfn2 plays an important role in SR-mitochondria tethering and interorganelle Ca2+ communication, whereas the development of hypertrophy is directly associated with the increase in cytosolic Ca2+ mainly released from SR calcium pool. Our current results showed that 10 mM caffeine-mediated intracellular Ca2+ release from SR as control was further increased by Ang II and reversed by AMO-20b (Figure 5A). By contrast, mitochondrial Ca2+ level induced by 100 μM histamine was attenuated by Ang II and mitigated by AMO-20b (Figure 5B). Similar results were obtained in the model of overexpression of miR-20b (Figures 5C and 5D), suggesting that upregulation of miR-20b induced by hypertrophy promotes cytosolic Ca2+ increase in cardiomyocytes and mitochondrial Ca2+ decrease. To verify the role of Mfn2 in interorganelle Ca2+ communication, we silenced or overexpressed Mfn2 (screened the optimal Mfn2 small interfering RNA [siRNA]: si-Mfn2 [si2], 0.05 nmol/mL, and Mfn2 overexpression plasmid: P-Mfn2 [P5], 5 μg/mL; Figures S4A–S4D). Detectable results in the presence of caffeine or histamine indicated that transfection of si-Mfn2 significantly elevated the cytoplasmic Ca2+ from SR but attenuated that from mitochondria (Figures S4E and S4F), while these Ca2+ events were reversed by P-Mfn2 (Figures S4G and S4H). In the hypertrophic models induced by Ang II or overexpression of miR-20b, increased cytosolic and decreased mitochondrial Ca2+ could be abolished by P-Mfn2, rather than P-NC (Figures 5E–5H). These results strongly demonstrate that the direct impact of miR-20b on SR-mitochondrial Ca2+ interaction is very likely to be mediated via Mfn2.
Figure 5.
The Effect of miR-20b on Ca2+ Mobilization between SR and Mitochondria
(A and B) In Ang II model (NRVCs), Ca2+ transients elicited by 10 mM caffeine (n = 6–11) or 100 μM histamine (n = 12–14) were recorded in SR and mitochondria; 10 averaged time points before and after the peak were included (left panel) and averaged peaks of Ca2+ concentration were presented (right panel). (C and D) In miR-20b overexpression model, Ca2+ transients of elicited by caffeine (n = 13–15) or histamine (n = 10–15) were recorded in SR and mitochondria. (E and F) P-Mfn2 reversed Ang II-induced SR and mitochondrial Ca2+ transients elicited by caffeine (n = 8–16) or histamine (n = 11–16). (G and H) P-Mfn2 reversed miR-20b-induced SR and mitochondrial Ca2+ transients elicited by caffeine (n = 6–15) or histamine (n = 10–16). Averaged data were presented as mean ± SEM, *p < 0.05 and **p < 0.01 versus Ctrl, #p < 0.05 and ##p < 0.01 versus Ang II or miR-20b.
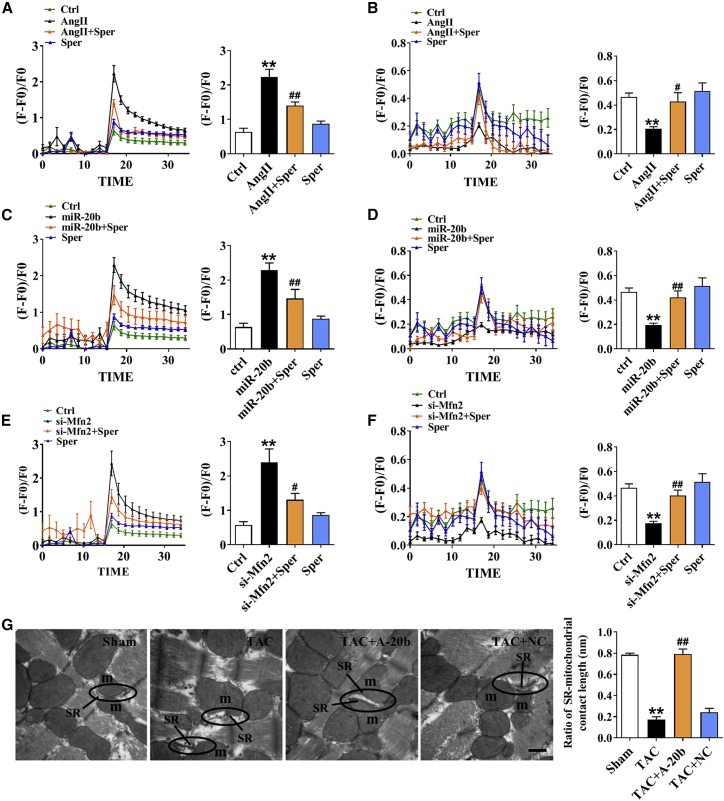
Direct Impact of Attenuated Buffering of Mitochondria on Augmented Cytosolic Ca2+
Mitochondrial calcium uniporter (MCU) is the key channel of mitochondrial Ca2+ uptake.18 It is essential to answer whether the Ca2+ events occurred in cytosolic Ca2+ and mitochondria under hypertrophic condition can be modified by MCU activation. Our data showed that the increase in cytosolic Ca2+ and the decrease in mitochondrial Ca2+ in Ang II-treated model were reversed in the presence of Spermine, a MCU activator (Figures 6A and 6B). These effects of Spermine were also confirmed in the model of miR-20b overexpression (Figures 6C and 6D) and direct silence of Mfn2 (Figures 6E and 6F). These phenomena suggest that miR-20b lead cardiomyocyte hypertrophy by inhibition of Mfn2 and thereby the reduction of buffering capability of SR-derived Ca2+ into the mitochondria through MCU. Notably, this was further supported the notion of physical connection of SR and mitochondria juxtaposition by measuring the ratio of SR-mitochondrial contact length (contact length of SR-mitochondria/total length of SR) using TEM. Our result showed that the contact length between SR and mitochondria was significantly reduced in TAC mice and restored completely by rAAV9-anti-miR-20b (Figure 6G).
Figure 6.
The Effect of miR-20b on Buffering Capability of Mitochondria
(A and B) Ang II induced inversed Ca2+ transients elicited by caffeine (n = 8–16) or histamine (n = 8–15) between SR and mitochondria, which were rescued by 20 μM Spermine (pretreatment for 2 h before staining). (C and D) Similar changes in miR-20b overexpression model. n = 8–16 for caffeine, n = 11–16 for histamine. (E and F) Similar changes in si-Mfn2 model. n = 6–16 for caffeine, n = 11–18 for histamine. (G) A-20b reversed the decrease in the contact length between SR and mitochondria induced by TAC in TEM examination. Images were viewed at a magnification of 40,000×; scale bar represents 500 nm. n = 4–8. Averaged data were presented as mean ± SEM, *p < 0.05 and **p < 0.01 versus Ctrl or Sham, #p < 0.05 and ##p < 0.01 versus Ang II, miR-20b, si-Mfn2, or TAC.
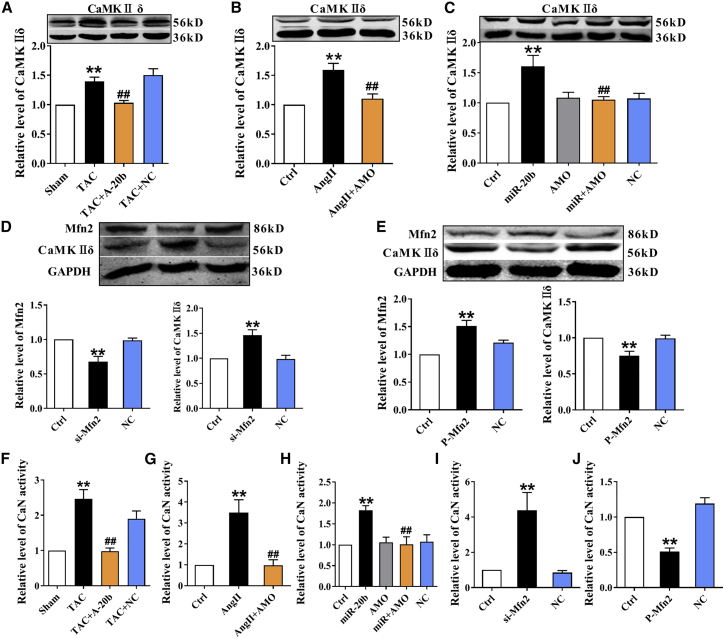
Verification of Ca2+-Dependent Pro-hypertrophic Signaling Pathway
The Ca2+-mediated signaling pathway is a key downstream player of miR-20b/Mfn2 modification under hypertrophic conditions; it is no doubt that the key pro-hypertrophic proteins Ca2+/calmodulin-dependent protein kinase II (CaMKIIδ) (the predominant myocardial isoform) and calcineurin (CaN) were investigated using in vivo (TAC) and in vitro (Ang II-treated and miR-20b overexpression) hypertrophic models. Apparently, in stark contrast to Mfn2, CaMKIIδ expression was significantly increased in all models and reversed by rAAV9-anti-miR-20b or AMO-20b (Figures 7A–7C), suggesting that miR-20b activates CaMKIIδ-mediated hypertrophic pathway. To verify the involvement of Mfn2 in these findings, we also tested CaMKIIδ expression by si-Mfn2 and P-Mfn2 and the results showed that si-Mfn2 and P-Mfn2 individually and inversely altered the expressions of Mfn2 and CaMKIIδ (Figures 7D and 7E), demonstrating that the pro-hypertrophic effect of miR-20b on Ca2+ signaling is due at least partially to Mfn2. Similar and quantitative alterations were also observed in CaN activity (Figures 7F–7J), suggesting that miR-20b promotes Mfn2/Ca2+/CaN-mediated hypertrophic signaling pathway.
Figure 7.
The Effect of miR-20b on Ca2+-Dependent Pro-Hypertrophic Signaling Pathways
(A) TAC-induced increase of the expression of CaMKIIδ in the mouse heart was abolished by A-20b. n = 9. (B) Ang II-induced upregulation of the expression of CaMKIIδ in NRVCs was abrogated by AMO-20b transfection. n = 5. (C) miR-20b overexpression-induced elevation of the expression of CaMKIIδ was reversed by co-transfection of miR-20b mimic and AMO-20b. n = 9. (D) Si-Mfn2 inversely altered the expressions of Mfn2 and CaMKIIδ. n = 8. (E) P-Mfn2 inversely altered the expressions of Mfn2 and CaMKIIδ. n = 8. (F) TAC-induced increase of CaN activity in the mouse heart was abolished by A-20b. n = 17. (G and H) CaN activity in model of Ang II administration and overexpression model of miR-20b in NRCVs. n = 8–10 and 8–11. (I and J) Si-Mfn2 and P-Mfn2 inversely altered CaN activity. n = 6–7 and 6–9. Averaged data were presented as mean ± SEM, *p < 0.05 and **p < 0.01 versus Sham or Ctrl, #p < 0.05 and ##p < 0.01 versus TAC, Ang II, miR-20b, si-Mfn2, or P-Mfn2.
Discussion
Major Findings
Here we identify miR-20b as a new pro-hypertrophic miRNA in the heart by targeting Mfn2. Overexpression of miR-20b induces the hypertrophic responses, and knockdown of it is able to mitigate the pathological phenotypes. Downregulation of Mfn2 by miR-20b decreases the tethering of SR to mitochondria, prevents mitochondrial Ca2+ uptake as a buffering sponge, and further increases cytoplasmic Ca2+ level, which then activates Ca2+ signaling transcription pathways and finally induces cardiac hypertrophy. To the best of our knowledge, our study is the first to identify the pathophysiological role of miR-20b in the progress of cardiac hypertrophy and the protective role and mechanism of Mfn2 in hypertrophic injury. These findings may provide new insights into the mechanism of cardiac hypertrophy and open new perspectives of therapeutic targets for cardiac hypertrophy, as well as the basis for a prophylaxis and effective clinical treatment against heart failure.
Identification of miR-20b as Target miRNA and Targeted Regulation of Mfn2
By binding to the 3′ UTR of Mfn2 gene with miRNAs, we found that miR-15a and miR-20b substantially elevated with hypertrophic state. In view of relatively low expression of miR-15a in our model, we only focused on the targeted regulation of miR-20b on Mfn2. It has been well documented that miR-20b functions as a tumor suppressor and demonstrates a wide range of biological activity in the development of various cancers.13, 14, 15, 16,19, 20, 21 miR-20b has also been implicated in infectious diseases.22, 23, 24, 25, 26 In addition, circulating miR-20b is related to type 2 diabetes27 and diabetic retinopathy.28 However, the research regarding miR-20b on the heart is very limited even if miR-20b is important for apoptosis, differentiation, and mitochondrial function in the P19 cell model of cardiac differentiation in vitro.29 Another study has also shown that miR-20b suppresses the expression of transcription factor ZFP-148 and promotes cardiomyocytes’ survival in viral myocarditis.30 Some circulating miRNA levels including miR-20b are significantly increased in response to hypertension-induced heart failure.31 Additionally, modulation of miR-20b with resveratrol and longevinex is linked with their anti-angiogenic cardioprotection in ischemia/reperfusion injury.32 Even so, there is no research regarding the effect of miR-20b on cardiac hypertrophy. Therefore, the current study is designed to examine this notion and underlying mechanisms using a mouse model of TAC and a cellular model of Ang II. And we first demonstrate that hypertrophic stimuli induce pronounced upregulation of miR-20b along with the changes of hypertrophic indicators. In stark contrast, knockdown of miR-20b by rAAV9-anti-miR-20b or AMO-20b offers robust protective effects against hypertrophic injury. The current evidence clearly tells us that miR-20b is a key player in the pathophysiology of cardiac hypertrophy.
It has been well documented that a large number of miRNAs are associated with the progress of cardiac hypertrophy and heart failure by targeting to hypertrophic genes,33,34 whereas the involvement of mitochondrial dynamics remains far from complete.1 So far, the downregulated Mfn2, a key functional protein in mitochondrial fusion, in myocardial hypertrophy is soly published information available9,10 without known mechanism, and little supporting evidence implying that the importance of Mfn2 in cardiac hypertrophy comes from the finding that MFN2 gene can be targeted modulation by miRNAs in the heart, which include miR-106a, miR-214, and miR-20a.35, 36, 37 Here, we present evidence that miR-20b, a newly identified miRNA, plays a pro-hypertrophic role via directly targeting Mfn2. Our current data not only identify that miR-20b downregulates Mfn2 along with the evidences showing damaged mitochondrial fusion but also confirms the direct regulatory relationship by luciferase reporter assay and miR-mask techniques. Hence our present work definitely enriches the network mechanisms of miRNAs with Mfn2. And to the best of our knowledge, this is the first time the critical role of miR-20b in cardiac hypertrophy has been demonstrated, and we suggest that miR-20b/Mfn2 axis may be beneficial for repairing hypertrophic injury. It is known that each single miRNA has the potential to regulate a particular cellular process by targeting multiple genes. It is conceivable that miR-20b controls cardiac hypertrophy by diverse mechanisms that merit future studies to exploit these possibilities.
Mfn2-Mediated Interaction of SR to Mitochondria as the Mechanism of Mfn2-Induced Hypertrophy
As mentioned above, miRNAs could target inhibition of Mfn2 in cardiac hypertrophy. However, the details about the molecular mechanism by which Mfn2 causes hypertrophy are largely unknown. Cytoplasmic Ca2+ overload is accepted for the common mechanism of cardiac hypertrophy, but understanding the source of elevated Ca2+ under miR-20b-induced cardiac hypertrophy is another key question that needs to be answered. SR is the main release channels of cytoplasmic Ca2+ in the heart, and two major proteins are involved: ryanodine receptor type 2 (RYR2)18,38 and inositol 1,4,5 triphosphate receptors (IP3Rs).18,38, 39, 40 Ca2+ uptake from cytosol back to SR is controlled by SR Ca2+ ATPase 2a (SERCA2a).18 While mitochondrial Ca2+ uptake is a two-step process, cytosolic Ca2+ is initially taken up into the mitochondrial outer membrane by voltage-dependent anion-selective channel protein-1 (VDAC1) and then transported from the inner membrane into the matrix through mitochondrial calcium uniporter (MCU).18 The most recently identified MCU protein complex includes MCU, mitochondrial Ca2+ uptake 1 (MICU1), and MICU2.41 As the pore-forming component, MCU is a highly selective Ca2+ channel that takes up immense levels of Ca2+ across the mitochondrial inner membrane.42,43 MICU1 and MICU2, functioning as “gatekeepers,” sense MCU Ca2+ levels and maintain mitochondrial Ca2+ homeostasis.44,45 Research shows that mitochondria readily sense the cyclic changes in free cytosolic Ca2+ released from SR and buffer such an increase by re-uptake Ca2+ into the matrix to prevent excessive elevation.18 Therefore, the SR and mitochondria are the most important organelles to efficiently maintain intracellular Ca2+ homeostasis.46 Furthermore, Mfn2 dimers serve as molecular bridges between SR to mitochondria;7 therefore, Mfn2 is critical for cytosolic Ca2+ homeostasis by a normal interorganelle Ca2+ signaling.
The interorganelle communication mediated by Mfn2 has been strongly implicated in mitochondrial calcium overload and apoptosis. Herein, we questioned whether Mfn2 offers the same cellular protection during hypertrophy. Excitingly, we found that downregulation of Mfn2 by transfection of si-Mfn2 increased cytoplasmic Ca2+ from SR but attenuated mitochondrial Ca2+, whereas these phenomena could be completely reversed by Mfn2 overexpression through P-Mfn2 transfection in both physiological and hypertrophic conditions (Ang II-treated or miR-20b overexpression), indicating that the effect of miR-20b on cytoplasmic and mitochondrial Ca2+ communication is linked by Mfn2. Numerous studies make it undeniable that SR proteins such as RYRs, IP3Rs, and SERCA2a are all involved and play important roles in intracellular Ca2+ regulation.18,38,47,48 However, the underlying mechanism is complicated and difficult to be investigated thoroughly, which is a limitation for our current observation. We could still focus on the Ca2+ uptake of mitochondria from cytosolic by activation of MCU using Spermine to verify that the pro-hypertrophic effect of miR-20b is due largely to the reduction in Ca2+ shuttling from SR to mitochondria by impairing the buffering capacity of mitochondria. As hypothesized, our observations confirm that Mfn2 links SR and mitochondria to regulate interorganelle Ca2+ delivery and maintain cytoplasmic Ca2+ level during pathogenesis of cardiac hypertrophy. From a morphological point of view, the proper contact between SR and mitochondria is another key to restrict mitochondrial import of SR-derived Ca2+.46 The existing morphological evidence with TEM confirmed that miR-20b loosened the physical contacts between these two compartments, the one of plausible explanation of the weakened Ca2+ shuttling from SR to mitochondria mediated by Mfn2, followed by the disruption of mitochondrial buffering, and eventual cytosolic Ca2+ overload. Moreover, the changes in mitochondrial Ca2+ levels impact on the metabolic regulation of Ca2+-activated matrix dehydrogenase and thus modulate ATP production.49 This may well explain why miR-20b inhibits ATP synthesis and aggravates cardiac hypertrophy under this experimental condition.
Ca2+-mediated signaling transductions participate in cardiac hypertrophy by two transcription mechanisms. One is CaN-mediated activation of nuclear factor of activated T cells (NFAT) and zinc-finger transcription factor GATA4.50 Elevated cytoplasmic Ca2+ binds to CaM and activates CaN, which then induces dephosphorylated NFAT translocation into the nucleus and interacts with GATA4 to implicate hypertrophic genes expression. The other is Ca2+/CaMKII-mediated activation of myocyte enhancer factor-2 (MEF-2).51 Ca2+/CaM activate CaMKIIδ to increase nuclear pre-hypertrophic signaling by phosphorylation of histone deacetylase (HDAC) and derepression of MEF-2. In the present study, both Ca2+-dependent pro-hypertrophic signaling pathways are believed to be activated by miR-20b/Mfn2 axis.
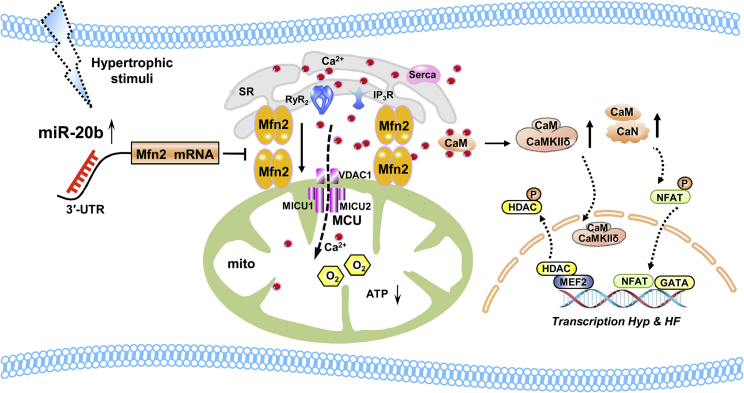
Our novel finding taken together with existing evidence, we outline the potential signaling pathway that aberrant upregulation of miR-20b in response to hypertrophic injury is likely to impair Mfn2 function by attenuating the physical and functional Ca2+ cross-talk between SR-mitochondria resulting in the decrease in the uptaking/buffering capability of mitochondria to Ca2+, and such a reduction may be presumable reason to lead a subsequent cytosolic Ca2+ elevation, trigger cytoplasmic Ca2+ signaling, and finally result in cardiac hypertrophy (Figure 8). This study has demonstrated for the first time to the best of our knowledge that the regulation of miR-20b on Mfn2 in cardiac hypertrophy and the miRNA-based prophylactic and therapeutic actions will shed a new light on clinical significance for cardiac hypertrophy and heart failure. Our results provide an interesting lead to protect cardiomyocytes from hypertrophic injury. SR-mitochondria communication can be expected to favor the development of more specific therapeutic strategies to selectively improve mitochondrial function in hypertrophy. Definitely, the present study cannot exclude other potential pathways to contribute to SR-mitochondrial cross-talk like Mfn2 in cardiac hypertrophy such as FUN14 domain containing 1 (FUNDC1)40 and macromolecular complex composed of VDAC1, Grp75, and IP3R1.38 So, it is very much anticipated to illustrate the diverse mechanism of Mfn2-mediated interorganelle interplay. Further studies are warranted to exploit these possibilities and gain more molecular and functional insights.
Figure 8.
Schematic Illustrations Explaining the Possible Targeting and Signaling Mechanisms by which miR-20b Causes Myocardial Hypertrophy
Overexpression of miR-20b induced by cardiac hypertrophy produces the repression of Mfn2 and interferes with Mfn2-mediated Ca2+ interaction of SR to mitochondria, which attenuates mitochondrial Ca2+ buffering capacity and increases cytosolic Ca2+ level. These changes trigger the activation of cytoplasmic Ca2+-dependent pro-hypertrophic signaling pathways and finally promote cardiac hypertrophy and even heart failure. Mfn2, mitofusin 2; RyR2, Ryanodine Receptor 2; IP3R, Inositol 1,4,5 Triphosphate Receptors; SERCA2a, SR Ca2+ ATPase 2a; VDAC1, voltage-dependent anion-selective channel protein 1; MCU, mitochondrial calcium uniporter; MICU1, mitochondrial Ca2+ uptake 1; MICU2, mitochondrial Ca2+ uptake 2; CaM, calmodulin; CaN, calcineurin; CaMKII, calcium/calmodulin-dependent protein kinase II; NFAT, nuclear factor of activated T cells; MEF-2, myocyte enhancer factor-2; HDAC, histone deacetylase; HF, heart failure.
Materials and Methods
Animals
Adult male Kunming mice (weight 22–25 g) were obtained from the Animal Centre of the Second Affiliated Hospital of Harbin Medical University and housed at 23°C ± 1°C with 55% ± 5% of humidity and maintained on a 12 h dark-light artificial cycle (lights on at 07:00 A.M.) with food and water available ad libitum. Before surgery procedures, mice were anesthetized with Avertin (0.2 g/kg, intraperitoneally [i.p.], T48402, Sigma-Aldrich, USA). All protocols about animals were pre-approved by Institutional Animal Care and Use Committee of Harbin Medical University, which are in accordance with the recommendations of the Panel on Euthanasia of the American Veterinary Medical Association and the National Institutes of Health publication Guide for the Care and Use of Laboratory Animals (https://www.nap.edu/topic/).
Pressure-Overload Cardiac Hypertrophy
TAC was applied for pressure-overload cardiac hypertrophy. Mouse was placed in a supine position and the chest was opened to identify the thoracic aorta. The aortic arch with 26-gauge blunt needle was tied with 6/0 silk suture, and the chest was then closed after the needle was removed. The sham group was received TAC procedures without aorta tied. After 4 weeks of TAC, echocardiography was performed and the heart was quickly excised, cleaned in PBS, and weighted. For comparison for varying weights of the hearts, the length of tibia was measured correspondingly.
Synthesis and Administration of rAAV9-anti-miR-20b
rAAV was used as carrier, among which rAAV9 is the most efficient vector for myocardial transduction. For this regard, rAAV9-anti-miR-20b or rAAV9-NC was produced (PackGene Biotech, Guangzhou, China) and the sequences were delivered into mouse, respectively, through tail vein injection at 1 × 1011 vg (viral genomes) per animal at 5 weeks before TAC. Animals were divided into four groups: sham mice, TAC mice, TAC mice received rAAV9-anti-miR-20b, and TAC mice received rAAV9-NC (n = 10–20 for each group).
Culture of NRVCs and Preparation of Cellular Model of Hypertrophy
The enzymatic dispersion techniques were used to isolate single ventricular myocytes from neonatal SD rats. Ventricular tissues from 1- to 3-day-old rats were dissected, minced in Dulbecco’s modified Eagle’s medium (DMEM, SH30022.01, HyClone, USA) and cells were dissociated with 0.25% Trypsin–EDTA Solution (C0201, Beyotime, China). After the centrifugation, the collected isolated cells were plated onto 25 cm2 cell culture flask at incubator for 100 min. The cells were then seeded in a 6-well plate (2 × 105/well) in DMEM containing 10% fetal bovine serum (FBS, SV30087.03, HyClone, USA) for further experiments. This procedure yielded cultures with 90% ± 5% of myocytes with beating observed under the light microscope. NRVCs were incubated with 0.1 μM of Ang II for 24 h to establish the cellular model of myocardial hypertrophy.
Synthesis of Various Oligonucleotides and Transfection Procedures
miR-20b mimic, NC miRNA, and AMO-20b were synthesized by RIBOBIO (Guangzhou, China). miRNA-masking antisense oligodeoxynucleotides (ODNs), Mfn2 siRNA, and Mfn2 overexpression plasmid were also synthesized (RIBOBIO or PackGene Biotech, Guangzhou, China). For transfection, NRVCs were washed with serum-free medium once and then incubated with 2 mL fresh FBS-free medium in 6-well plates. Then, NRVCs (2 × 105/well) were transfected with 0.2 nmol miR-20b, AMO-20b, ODNs, or NC miRNAs, or 0.05 nmol/mL Mfn2 siRNAs or 5 μg/mL overexpression plasmids, respectively, for 48 h with X-tremeGENE siRNA Transfection Reagent (Cat.# 04476093001, Roche, Switzerland).
Echocardiography
Anesthetized mice were placed on a platform. Cardiac anatomical and functional parameters were evaluated by 2-dimensional transthoracic echocardiography using a Visual Sonic Ultrasound system (Vevo2100, VisualSonics, Canada). The heart was imaged in a parasternal short-axis view at the level of the papillary muscles with M-mode to determine wall thickness, end-diastolic, and end-systolic dimensions. Left ventricular (LV) wall thickness was used as an index of cardiac hypertrophy.
TEM Detection
The mouse heart was removed and immersed in stationary liquid (pH 7.3) containing 3% glutaraldehyde in 0.1 mM sodium phosphate buffer and 0.45 mM Ca2+. Tissue samples were then fixed in 2% osmic acid (OsO4) in phosphate-buffered solution with 1.5% potassium ferricyanide. After dehydration with a concentration gradient of alcohol solutions, tissues were then embedded in epon with propylene oxide as an intermediary solvent. Ultrathin sections were obtained, mounted onto formvar-coated slot grids, and stained with uranyl acetate and lead citrate. Images were examined by an electron microscope (H-7650 Hitachi, Tokyo, Japan).
Histological Analysis
Cardiac tissue was fixed with 4% paraformaldehyde and embedded in paraffin. 5 μm-thick sections were stained with H&E according to standard protocols. The slices were visualized under a microscope (Axio Scope A1, Zeiss, Germany). The area of cardiomyocyte was measured using ImageJ and expressed as averaged area calculated from the ratio of total area to nuclear number from each of six independent experiments.
Quantitative Real-Time PCR for mRNAs
Total RNA samples were isolated from mouse heart or NRVCs using phenol/chloroform and complementary DNA synthesis was performed with high-capacity cDNA reverse transcription reagent kits (FSQ-101, Toyobo, Japan). The SYBR Green PCR Master Mix Kit (04913914001, Roche, Switzerland) was used in quantitative real-time PCR with LightCycler96 Real-Time PCR System (Roche, Switzerland) to quantify target genes. U6 or GAPDH served as an internal control. The primers for quantitative real-time PCR were commercially designed (Table S3).
Western Blot Analysis
Protein samples extracted from mouse hearts or NRVCs were used for immunoblotting analysis. Protein concentration was determined by the BCA Protein Assay Kit (P0010, Beyotime, China) with bovine serum albumin (B2064, Sigma-Aldrich, USA) as the standard. Equal amounts of protein (80 μg) were loaded on a 10% SDS-PAGE gel. The lysate was resolved by electrophoresis (70 V for 30 min and 110 V for 1.5 h) and transferred onto nitrocellulose membranes. After blocking in 5% nonfat milk for 2 h at room temperature, the membranes were treated with anti-Mfn2 (1:500, ab56889, Abcam, UK) or CaMKIIδ (1:1,000, ab181052, Abcam, UK) at 4°C overnight. The membranes were then incubated with secondary antibody (1:10,000) for 1 h at room temperature. Blots were detected with the Odyssey infrared imaging system (Li-Cor, USA). Protein loading was confirmed by immunoblotting for GAPDH (1:10,000, AC002, ABclonal, USA) as an internal control. Western blot bands were quantified using Quantity One software to measure band intensity (area × OD) for each group and normalized to actin band intensity. The final results were expressed as fold changes compared with the control values.
Measurement of Cell Surface Area
Cardiomyocytes were fixed with 4% paraformaldehyde for 30 min, penetrated by 0.4% Triton X-100 for 1 h, and then blocked by goat serum for 1 h. The cells were first incubated with anti-sarcomeric alpha actinin antibody (ab9465, Abcam, UK) at 4°C overnight and subsequently with Alexa Fluor 594 donkey anti-mouse antibody (ab150108, Abcam, UK) for 1 h at room temperature. Then the cells were incubated with DAPI (C1005, Beyotime, China) for 10 min. Immunofluorescence was visualized under a fluorescence microscope (Axio Scope A1, Zeiss, Germany). Cell surface area was measured by Carl Zeiss Software. The results were calculated as the averaged area of cell and expressed as relative level by normalizing the data to control values from each of six independent experiments.
Mitochondrial Dynamics Analysis
Cells were incubated for 30 min with MitoTrackerR Red (400 nm; M7512, Invitrogen, USA) and maintained in PBS solution. The images were captured with a confocal laser scanning microscope (FluoView FV1000, Olympus, Germany). The number and individual volume of each mitochondrium were quantified with ImageJ. Each experiment was done at least three times, and 17–25 cells per condition were quantified. An increase in mitochondrial volume and a decrease in the number of mitochondria were considered as fusion criteria. The percentage of cells (<10% punctiform) with a fusion pattern was also determined.
ATP Detection
ATP production was measured using ATP detection kit (S0026, Beyotime, China) according to the manufacturer’s instruction. The standard curve was obtained with defined ATP concentrations, from which the rate of mitochondrial ATP production was calculated.
Luciferase Reporter Assay
To generate reporter vectors bearing miRNA-binding sites, we synthesized the 3′ UTR of Mfn2 and their mutant variant (PackGene Biotech, Guangzhou, China). The construct was inserted into the multiple cloning sites downstream of the luciferase gene (Xhol and Not I sites) in the psiCHECK-2 luciferase reporter vector (Promega, USA). For luciferase assay, 0.1 μg of luciferase reporters containing 3′ UTR was co-transfected with NC, miR-20b, or AMO-20b and 10 ng of psiCHECK-2, respectively, into HEK293 cells using lipofectamine 2000 transfection reagent (11668027, Invitrogen, USA) according to the manufacturer’s instructions. Luciferase activity was measured 48 h after transfection with a dual luciferase reporter assay kit (Promega, USA).
Measurement of Ca2+ Transient
Cells were stained with 5 μL Fluo-3 AM (F1242, Thermo Fisher Scientific, USA) at 37°C for 50 min and the fluorescence was determined by confocal. After 20 s of baseline recording, 10 mM caffeine (C0750, Sigma-Aldrich, USA) was added to monitor SR Ca2+ release. In order to monitor mitochondrial Ca2+ release, cells were stained with 2.5 μL Rhod-2 AM (R1244, Thermo Fisher Scientific, USA) at 37°C for 50 min and 100 mM histamine (MB0303, Meilunbio, China) was added after 20 s of baseline recording. 20 μM Spermine (HY-B1777, MedChemExpress, USA) was preincubated for 2 h before staining to observe the effect of MCU activation. Confocal images were recorded every 1.7 s (FluoView FV1000, Olympus, Germany) at 488 nm and 591 nm excitation using a 40× objective. The images were analyzed and quantified using Olympus-700.
CaN Phosphatase Activity Assay
CaN phosphatase activity was determined by CaN phosphatase activity detection kit (A068, Nanjing JianCheng, China) according to the manufacturer’s instructions, and it was calculated by formula that was described in the manufacturer’s instructions.
Statistical Analysis
Averaged data were presented as mean ± SEM. The two-tailed Student’s t test was applied for comparisons between the two groups. Multi-group’s comparisons were performed by one-way ANOVA. SPSS19.0 software was used for all statistical analyses and p <0.05 was considered significant difference.
Author Contributions
Y.Q. performed experiments and analyzed and interpreted the data. R.C. analyzed the data and wrote and edited the manuscript. C.L., Y.Y., W.Z., and J.Z. performed experiments and analyzed data. M.Z. and B.L. wrote and edited the manuscript and supplied funding. C.X. performed study design, drafted the manuscript, supplied funding, and supervised the work. R.Z. performed conceptualization, study design, data interpretation, and drafting the manuscript and provided project funding.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
Research funding was provided by the National Natural Science Foundation of China (81470523, 81670207, 81703510, and 81573431) and Natural Science Foundation of Heilongjiang Province (LH2019H008).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.01.017.
Contributor Information
Chaoqian Xu, Email: chaoqx68@126.com.
Rong Zhang, Email: rongzhang77@163.com.
Supplemental Information
References
- 1.Zhao Y., Ponnusamy M., Liu C., Tian J., Dong Y., Gao J., Wang C., Zhang Y., Zhang L., Wang K., Li P. MiR-485-5p modulates mitochondrial fission through targeting mitochondrial anchored protein ligase in cardiac hypertrophy. Biochim Biophys Acta Mol Basis Dis. 2017;1863:2871–2881. doi: 10.1016/j.bbadis.2017.07.034. [DOI] [PubMed] [Google Scholar]
- 2.Filadi R., Pendin D., Pizzo P. Mitofusin 2: from functions to disease. Cell Death Dis. 2018;9:330. doi: 10.1038/s41419-017-0023-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papanicolaou K.N., Kikuchi R., Ngoh G.A., Coughlan K.A., Dominguez I., Stanley W.C., Walsh K. Mitofusins 1 and 2 are essential for postnatal metabolic remodeling in heart. Circ. Res. 2012;111:1012–1026. doi: 10.1161/CIRCRESAHA.112.274142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng M., Xiao R.P. Role of mitofusin 2 in cardiovascular oxidative injury. J. Mol. Med. (Berl.) 2010;88:987–991. doi: 10.1007/s00109-010-0675-5. [DOI] [PubMed] [Google Scholar]
- 5.Zhou Y., Carmona S., Muhammad A.K.M.G., Bell S., Landeros J., Vazquez M., Ho R., Franco A., Lu B., Dorn G.W., 2nd Restoring mitofusin balance prevents axonal degeneration in a Charcot-Marie-Tooth type 2A model. J. Clin. Invest. 2019;130:1756–1771. doi: 10.1172/JCI124194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Brito O.M., Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y., Csordás G., Jowdy C., Schneider T.G., Csordás N., Wang W., Liu Y., Kohlhaas M., Meiser M., Bergem S. Mitofusin 2-containing mitochondrial-reticular microdomains direct rapid cardiomyocyte bioenergetic responses via interorganelle Ca(2+) crosstalk. Circ. Res. 2012;111:863–875. doi: 10.1161/CIRCRESAHA.112.266585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandhok G., Lazarou M., Neumann B. Structure, function, and regulation of mitofusin-2 in health and disease. Biol. Rev. Camb. Philos. Soc. 2018;93:933–949. doi: 10.1111/brv.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang L., Moore X.L., Gao X.M., Dart A.M., Lim Y.L., Du X.J. Down-regulation of mitofusin-2 expression in cardiac hypertrophy in vitro and in vivo. Life Sci. 2007;80:2154–2160. doi: 10.1016/j.lfs.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Yu H., Guo Y., Mi L., Wang X., Li L., Gao W. Mitofusin 2 inhibits angiotensin II-induced myocardial hypertrophy. J. Cardiovasc. Pharmacol. Ther. 2011;16:205–211. doi: 10.1177/1074248410385683. [DOI] [PubMed] [Google Scholar]
- 11.Piubelli C., Meraviglia V., Pompilio G., D’Alessandra Y., Colombo G.I., Rossini A. microRNAs and Cardiac Cell Fate. Cells. 2014;3:802–823. doi: 10.3390/cells3030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Islas J.F., Moreno-Cuevas J.E. A MicroRNA Perspective on Cardiovascular Development and Diseases: An Update. Int. J. Mol. Sci. 2018;19:19. doi: 10.3390/ijms19072075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang D., Yang Z., Long F., Luo L., Yang B., Zhu R., Sang X., Cao G., Wang K. Long noncoding RNA MALAT1 mediates stem cell-like properties in human colorectal cancer cells by regulating miR-20b-5p/Oct4 axis. J. Cell. Physiol. 2019;234:20816–20828. doi: 10.1002/jcp.28687. [DOI] [PubMed] [Google Scholar]
- 14.Braicu C., Raduly L., Morar-Bolba G., Cojocneanu R., Jurj A., Pop L.A., Pileczki V., Ciocan C., Moldovan A., Irimie A. Aberrant miRNAs expressed in HER-2 negative breast cancers patient. J. Exp. Clin. Cancer Res. 2018;37:257. doi: 10.1186/s13046-018-0920-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo Y., He J., Tao X., Wang H., Fang Q., Guo S., Song C. miR-20b negatively regulates VEGF expression by targeting STAT3 in H22 hepatocellular carcinoma cells. Oncol. Rep. 2018;40:2806–2813. doi: 10.3892/or.2018.6651. [DOI] [PubMed] [Google Scholar]
- 16.Hong S., Yu S., Li J., Yin Y., Liu Y., Zhang Q., Guan H., Li Y., Xiao H. MiR-20b displays tumor-suppressor functions in papillary thyroid carcinoma by regulating the MAPK/ERK signaling pathway. Thyroid. 2016;26:1733–1743. doi: 10.1089/thy.2015.0578. [DOI] [PubMed] [Google Scholar]
- 17.Hong H., Tao T., Chen S., Liang C., Qiu Y., Zhou Y., Zhang R. MicroRNA-143 promotes cardiac ischemia-mediated mitochondrial impairment by the inhibition of protein kinase Cepsilon. Basic Res. Cardiol. 2017;112:60. doi: 10.1007/s00395-017-0649-7. [DOI] [PubMed] [Google Scholar]
- 18.Lopez-Crisosto C., Pennanen C., Vasquez-Trincado C., Morales P.E., Bravo-Sagua R., Quest A.F.G., Chiong M., Lavandero S. Sarcoplasmic reticulum-mitochondria communication in cardiovascular pathophysiology. Nat. Rev. Cardiol. 2017;14:342–360. doi: 10.1038/nrcardio.2017.23. [DOI] [PubMed] [Google Scholar]
- 19.Huang T., Alvarez A.A., Pangeni R.P., Horbinski C.M., Lu S., Kim S.H., James C.D., J Raizer J., A Kessler J., Brenann C.W. A regulatory circuit of miR-125b/miR-20b and Wnt signalling controls glioblastoma phenotypes through FZD6-modulated pathways. Nat. Commun. 2016;7:12885. doi: 10.1038/ncomms12885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu M., Wang D., Li N. MicroRNA-20b downregulates HIF-1alpha and inhibits the proliferation and invasion of osteosarcoma cells. Oncol. Res. 2016;23:257–266. doi: 10.3727/096504016X14562725373752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silva J., García V., Zaballos Á., Provencio M., Lombardía L., Almonacid L., García J.M., Domínguez G., Peña C., Diaz R. Vesicle-related microRNAs in plasma of nonsmall cell lung cancer patients and correlation with survival. Eur. Respir. J. 2011;37:617–623. doi: 10.1183/09031936.00029610. [DOI] [PubMed] [Google Scholar]
- 22.Biswas S., Haleyurgirisetty M., Lee S., Hewlett I., Devadas K. Development and validation of plasma miRNA biomarker signature panel for the detection of early HIV-1 infection. EBioMedicine. 2019;43:307–316. doi: 10.1016/j.ebiom.2019.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu X., Liao S., Bai H., Wu L., Wang M., Wu Q., Zhou J., Jiao L., Chen X., Zhou Y. Integrating exosomal microRNAs and electronic health data improved tuberculosis diagnosis. EBioMedicine. 2019;40:564–573. doi: 10.1016/j.ebiom.2019.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qian K., Zhang L., Shi K. Triptolide prevents osteoarthritis via inhibiting hsa-miR-20b. Inflammopharmacology. 2019;27:109–119. doi: 10.1007/s10787-018-0509-6. [DOI] [PubMed] [Google Scholar]
- 25.Singh A.K., Rooge S.B., Varshney A., Vasudevan M., Bhardwaj A., Venugopal S.K., Trehanpati N., Kumar M., Geffers R., Kumar V., Sarin S.K. Global microRNA expression profiling in the liver biopsies of hepatitis B virus-infected patients suggests specific microRNA signatures for viral persistence and hepatocellular injury. Hepatology. 2018;67:1695–1709. doi: 10.1002/hep.29690. [DOI] [PubMed] [Google Scholar]
- 26.Coskun M., Bjerrum J.T., Seidelin J.B., Troelsen J.T., Olsen J., Nielsen O.H. miR-20b, miR-98, miR-125b-1*, and let-7e* as new potential diagnostic biomarkers in ulcerative colitis. World J. Gastroenterol. 2013;19:4289–4299. doi: 10.3748/wjg.v19.i27.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katayama M., Wiklander O.P.B., Fritz T., Caidahl K., El-Andaloussi S., Zierath J.R., Krook A. Circulating exosomal miR-20b-5p is elevated in type 2 diabetes and could impair insulin action in human skeletal muscle. Diabetes. 2019;68:515–526. doi: 10.2337/db18-0470. [DOI] [PubMed] [Google Scholar]
- 28.Platania C.B.M., Maisto R., Trotta M.C., D’Amico M., Rossi S., Gesualdo C., D’Amico G., Balta C., Herman H., Hermenean A. Retinal and circulating miRNA expression patterns in diabetic retinopathy: An in silico and in vivo approach. Br. J. Pharmacol. 2019;176:2179–2194. doi: 10.1111/bph.14665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu S., Hu X., Yu Z., Peng Y., Zhu J., Liu X., Li M., Han S., Zhu C. Effect of miR-20b on apoptosis, differentiation, the BMP signaling pathway and mitochondrial function in the P19 cell model of cardiac differentiation in vitro. PLoS ONE. 2015;10:e0123519. doi: 10.1371/journal.pone.0123519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu H.F., Gao X.T., Lin J.Y., Xu X.H., Hu J., Ding Y.J., Zhu S.H. MicroRNA-20b suppresses the expression of ZFP-148 in viral myocarditis. Mol. Cell. Biochem. 2017;429:199–210. doi: 10.1007/s11010-017-2947-7. [DOI] [PubMed] [Google Scholar]
- 31.Dickinson B.A., Semus H.M., Montgomery R.L., Stack C., Latimer P.A., Lewton S.M., Lynch J.M., Hullinger T.G., Seto A.G., van Rooij E. Plasma microRNAs serve as biomarkers of therapeutic efficacy and disease progression in hypertension-induced heart failure. Eur. J. Heart Fail. 2013;15:650–659. doi: 10.1093/eurjhf/hft018. [DOI] [PubMed] [Google Scholar]
- 32.Mukhopadhyay P., Das S., Ahsan M.K., Otani H., Das D.K. Modulation of microRNA 20b with resveratrol and longevinex is linked with their potent anti-angiogenic action in the ischaemic myocardium and synergestic effects of resveratrol and γ-tocotrienol. J. Cell. Mol. Med. 2012;16:2504–2517. doi: 10.1111/j.1582-4934.2011.01480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sayed D., Hong C., Chen I.Y., Lypowy J., Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ. Res. 2007;100:416–424. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- 34.Da Costa Martins P.A., De Windt L.J. MicroRNAs in control of cardiac hypertrophy. Cardiovasc. Res. 2012;93:563–572. doi: 10.1093/cvr/cvs013. [DOI] [PubMed] [Google Scholar]
- 35.Guan X., Wang L., Liu Z., Guo X., Jiang Y., Lu Y., Peng Y., Liu T., Yang B., Shan H. miR-106a promotes cardiac hypertrophy by targeting mitofusin 2. J. Mol. Cell. Cardiol. 2016;99:207–217. doi: 10.1016/j.yjmcc.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 36.Lv L., Li T., Li X., Xu C., Liu Q., Jiang H., Li Y., Liu Y., Yan H., Huang Q. The lncRNA Plscr4 controls cardiac hypertrophy by regulating miR-214. Mol. Ther. Nucleic Acids. 2018;10:387–397. doi: 10.1016/j.omtn.2017.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun D., Li C., Liu J., Wang Z., Liu Y., Luo C., Chen Y., Wen S. Expression profile of micrornas in hypertrophic cardiomyopathy and effects of microrna-20 in inducing cardiomyocyte hypertrophy through regulating gene mfn2. DNA Cell Biol. 2019;38:796–807. doi: 10.1089/dna.2019.4731. [DOI] [PubMed] [Google Scholar]
- 38.Paillard M., Tubbs E., Thiebaut P.A., Gomez L., Fauconnier J., Da Silva C.C., Teixeira G., Mewton N., Belaidi E., Durand A. Depressing mitochondria-reticulum interactions protects cardiomyocytes from lethal hypoxia-reoxygenation injury. Circulation. 2013;128:1555–1565. doi: 10.1161/CIRCULATIONAHA.113.001225. [DOI] [PubMed] [Google Scholar]
- 39.Nakayama H., Bodi I., Maillet M., DeSantiago J., Domeier T.L., Mikoshiba K., Lorenz J.N., Blatter L.A., Bers D.M., Molkentin J.D. The IP3 receptor regulates cardiac hypertrophy in response to select stimuli. Circ. Res. 2010;107:659–666. doi: 10.1161/CIRCRESAHA.110.220038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bertero E., Maack C. Calcium signaling and reactive oxygen species in mitochondria. Circ. Res. 2018;122:1460–1478. doi: 10.1161/CIRCRESAHA.118.310082. [DOI] [PubMed] [Google Scholar]
- 41.Wang W., Xie Q., Zhou X., Yao J., Zhu X., Huang P., Zhang L., Wei J., Xie H., Zhou L., Zheng S. Mitofusin-2 triggers mitochondria Ca2+ influx from the endoplasmic reticulum to induce apoptosis in hepatocellular carcinoma cells. Cancer Lett. 2015;358:47–58. doi: 10.1016/j.canlet.2014.12.025. [DOI] [PubMed] [Google Scholar]
- 42.Kirichok Y., Krapivinsky G., Clapham D.E. The mitochondrial calcium uniporter is a highly selective ion channel. Nature. 2004;427:360–364. doi: 10.1038/nature02246. [DOI] [PubMed] [Google Scholar]
- 43.Baughman J.M., Perocchi F., Girgis H.S., Plovanich M., Belcher-Timme C.A., Sancak Y., Bao X.R., Strittmatter L., Goldberger O., Bogorad R.L. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 2011;476:341–345. doi: 10.1038/nature10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Csordás G., Golenár T., Seifert E.L., Kamer K.J., Sancak Y., Perocchi F., Moffat C., Weaver D., de la Fuente Perez S., Bogorad R. MICU1 controls both the threshold and cooperative activation of the mitochondrial Ca2+ uniporter. Cell Metab. 2013;17:976–987. doi: 10.1016/j.cmet.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ahuja M., Muallem S. The gatekeepers of mitochondrial calcium influx: MICU1 and MICU2. EMBO Rep. 2014;15:205–206. doi: 10.1002/embr.201438446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dorn G.W., 2nd, Song M., Walsh K. Functional implications of mitofusin 2-mediated mitochondrial-SR tethering. J. Mol. Cell. Cardiol. 2015;78:123–128. doi: 10.1016/j.yjmcc.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garrido-Moreno V., Díaz-Vegas A., López-Crisosto C., Troncoso M.F., Navarro-Marquez M., García L., Estrada M., Cifuentes M., Lavandero S. GDF-11 prevents cardiomyocyte hypertrophy by maintaining the sarcoplasmic reticulum-mitochondria communication. Pharmacol. Res. 2019;146:104273. doi: 10.1016/j.phrs.2019.104273. [DOI] [PubMed] [Google Scholar]
- 48.Garcia M.I., Boehning D. Cardiac inositol 1,4,5-trisphosphate receptors. Biochim Biophys Acta Mol Cell Res. 2017;1864:907–914. doi: 10.1016/j.bbamcr.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leal N.S., Schreiner B., Pinho C.M., Filadi R., Wiehager B., Karlström H., Pizzo P., Ankarcrona M. Mitofusin-2 knockdown increases ER-mitochondria contact and decreases amyloid β-peptide production. J. Cell. Mol. Med. 2016;20:1686–1695. doi: 10.1111/jcmm.12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilkins B.J., Molkentin J.D. Calcium-calcineurin signaling in the regulation of cardiac hypertrophy. Biochem. Biophys. Res. Commun. 2004;322:1178–1191. doi: 10.1016/j.bbrc.2004.07.121. [DOI] [PubMed] [Google Scholar]
- 51.Anderson M.E., Brown J.H., Bers D.M. CaMKII in myocardial hypertrophy and heart failure. J. Mol. Cell. Cardiol. 2011;51:468–473. doi: 10.1016/j.yjmcc.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.