Abstract
The cancer-targeting gene virotherapy might be a useful strategy for the treatment of cancer, because it could combine the advantages of both gene therapy and virotherapy. This study aimed to construct a triple-regulated oncolytic adenovirus, Ad-RGD-Survivin-ZD55-miR-143, carrying the therapeutic gene miR-143 and evaluate its possible antitumor effect in colorectal cancer. We observed that miR-143 was lowly expressed in patients with colorectal cancer. The upregulation of miR-143 could inhibit cell proliferation and induce cell apoptosis by targeting KRAS in colorectal cancer cells. Then, Ad-RGD-Survivin-ZD55-miR-143 was successfully constructed in this study. Cells infected with Ad-RGD-Survivin-ZD55-miR-143 could inhibit cell proliferation, suppress cell migration and invasion, arrest cells at the G1 phase, and induce cellular apoptosis. At the same time, Ad-RGD-Survivin-ZD55-miR-143 decreased the expression of PARP-1 and KRAS protein in vitro. In a HCT116 xenograft model, intratumoral injection of Ad-RGD-Survivin-ZD55-miR-143 resulted in reduced tumor growth. Furthermore, Ad-RGD-Survivin-ZD55-miR-143 induced apoptosis and decreased the expression level of KRAS in HCT116 xenograft cells. Our results suggested that Ad-RGD-Survivin-ZD55-miR-143 produced a strong antitumor effect by targeting KRAS and that this strategy could broaden the therapeutic options for treating colorectal cancer.
Keywords: colorectal cancer, oncolytic adenovirus, microRNA-143, KRAS
Introduction
Colorectal cancer (CRC) is the third most common type of cancer all over the world. In 2018, over 1.8 million new CRC cases were diagnosed, and 881,000 deaths were estimated to occur worldwide.1 The survival of CRC patients is related to the stage at diagnosis, with a 5-year survival rate of 90% at early diagnosis and less than 10% in patients with distant metastases.2 The treatments for primary and metastatic CRC include laparoscopic surgery for primary disease; resection for some forms of metastatic disease; and radiotherapy and neo-adjuvant and palliative chemotherapy for quite advanced disease. Despite advances in surgical and medical therapies, cure rates and long-term survival have changed little in the past several decades.3,4 Especially, the prognosis and overall survival for metastatic CRC is still poor.5 Therefore, it is essential to develop more effective therapeutic strategies for metastatic CRC.
MicroRNAs (miRNAs) are short endogenous non-coding RNAs that inhibit protein translation or messenger RNA (mRNA) decay by interacting with the 3′ untranslated regions (3′ UTRs) of the mRNA of target genes.6 miRNAs presumably play an important role in the development and progression of human malignancy because they can affect the translation and stability of targeted oncogenes or tumor suppressors.7 MiR-143 which is downregulated in CRC tissues, are involved in the regulation of several cellular processes including proliferation, migration and chemoresistance.8, 9, 10 Furthermore, Guo et al.8 found that the expression of miR-143 has significantly decreased in liver metastatic CRC tissues compared with primary CRC tissues and that overexpression of miR-143 showed the inhibitory effect on the migration and invasion potential of CRC cells in vivo. The overexpression of miR-143 in experimental cancer models appeared to be beneficial as a potential cancer therapy. Therefore, miR-143 could act as a tumor suppressor gene for CRC.
Adenovirus (or Ad) vectors are the most commonly used vectors in gene therapeutic applications, because they have high transduction efficiency, broad cell tropism, high gene expression, and mature production technology.11 The adenovirus vectors can be genetically modified to selectively replicate in malignant cells, causing cancer cell lysis and inflammation, which, in turn, can stimulate host immune responses to cancer cells.12 The new modified adenoviruses are called oncolytic adenoviruses.13 Recently, oncolytic adenoviruses have been commonly used in clinical trials for cancer gene therapy.14,15 Among them, the cancer-targeting gene virotherapy (CTGVT) strategy16 might be a useful strategy for the treatment of advanced or metastatic cancer. For example, inserting the vascular endothelial cell growth inhibitor (VEGI) into a selectively replicating adenovirus with an E1B-55kDa gene deletion (ZD55) to construct ZD55-VEGI-251 leads to a much more severe cytopathic effect than control viruses on human cancer cell line HeLa, hepatoma cell line SMMC-7721, and CRC cell line SW620.17 Thus, the CTGVT strategy has broad spectrum antitumor properties, because it could combine the advantages of both gene therapy and virotherapy.
In our previous study, we constructed a conditionally replicative adenovirus, Ad-ZD55-miR-143, which exhibited specific antitumor effects by targeting KRAS in vitro.18 However, the antitumor effect of Ad-ZD55-miR-143 was not ideal in follow-up experiments, because it did not increase the chance of vector particles to reach its target cells. Therefore, we constructed a new triple-regulated oncolytic adenovirus, Ad-RGD (Arg-Gly-Asp)-Survivin-ZD55-miR-143, and evaluated its possible antitumor effects on CRC in this study.
Results
Expression of miR-143 Was Decreased in CRC Specimens, and KRAS Was a Target Gene of miR-143
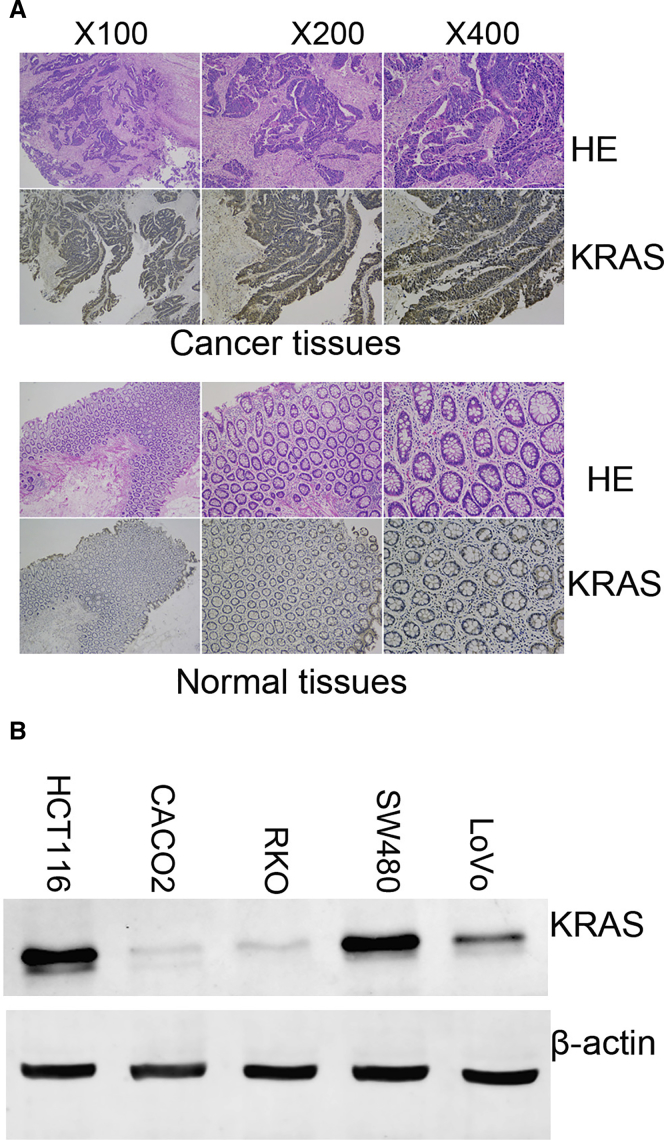
The expression levels of miR-143 in CRC specimens and adjacent normal tissues were measured by qRT-PCR. As shown in Figure S1A, miR-143 expression was significantly decreased in CRC specimens compared with adjacent normal tissues. Then, we found two potential binding sites for miR-143 that were located 1,720–1,728 and 3,772–3,780 bp downstream of the 5′ end of the KRAS 3′ UTR (Figure S1B). The putative binding sites were cloned into a luciferase reporter vector, and the luciferase activity was detected by dual-luciferase reporter assays. As depicted in Figure S1C, the relative luciferase activity was evaluated by normalizing the renilla luciferase (RL) activity with firefly luciferase (FL) activity. We found that the luciferase activity of co-transfection of psiCHECK-2/KRAS 3′ UTR and miR-143 was lower than that of co-transfection with psiCHECK-2/KRAS 3′ UTR and negative control (NC) groups. This is because the RL mRNA was degraded by miR-143 interacting with KRAS 3′ UTR fragments of the psiCHECK-2 reporter plasmid. Subsequently, we measured the expression levels of KRAS in CRC specimens and several CRC cell lines. The expression level of KRAS in cancer tissues was higher than that in normal tissues, and it was mainly expressed in the cytoplasm of cancer cells (Figure 1A). Compared with HCT116 and SW480 cells, CACO2, RKO, and LoVo cells showed low expression of KRAS (Figure 1B). The aforementioned results indicated that the expression of miR-143 was significantly decreased in human CRC specimens, and KRAS was a direct target of miR-143.
Figure 1.
The Expression of KRAS in CRC Tissues and Cells
(A) Representative images of immunohistochemical (IHC) staining of KRAS in CRC specimens and normal tissues. (B) The expression levels of KRAS in CRC cell lines.
Overexpression of miR-143 Inhibited Cell Proliferation and Induced Cell Apoptosis
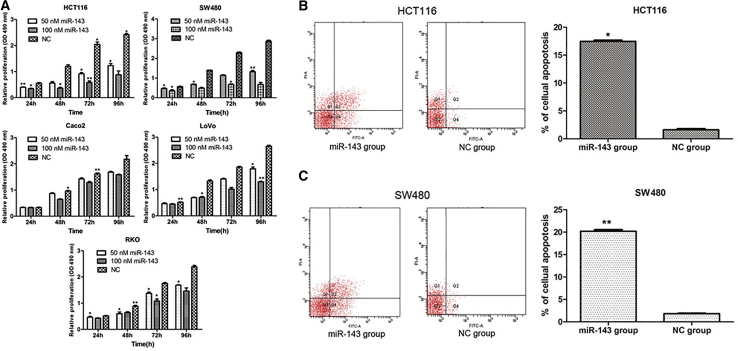
CRC cells were transfected with either 50 or 100 nM miR-143. Then, the viability of cells was measured by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium) assays. We found that the viability of miR-143 groups was consistently significantly lower than that of NC groups in HCT116, SW480, LoVo, and RKO cells. However, there was little change in Caco2 cells (Figure 2A). Furthermore, we also found that the inhibition of miR-143 in HCT116, SW480, and LoVo cells showed a time- and dose-dependent manner. Referring to the results of Figure 1B, HCT116 and SW480 cell lines were selected for use in the subsequent experiments.
Figure 2.
Overexpression of miR-143 Inhibited Cell Proliferation and Induced Cell Apoptosis
(A) Upregulation of miR-143 in CRC cells significantly repressed cell proliferation. OD, optical density. (B and C) MiR-143 overexpression induced early and late apoptosis of (B) HCT116 and (C) SW480 cells. *p < 0.05; **p < 0.01.
HCT116 and SW480 cells were transfected with miR-143 and NC mimics (100 nM) for 72 h. The proportion of apoptotic cells was evaluated using flow-cytometric analysis of annexin V-fluorescein isothiocyanate/propidium iodide (FITC/PI) staining. As indicated in Figures 2B and 2C, the percentage of the early and late apoptotic HCT116 and SW480 cells were higher in the miR-143 group compared with the NC group.
Recombinant Adenovirus Ad-RGD-Survivin-ZD55-miR-143 Was Successfully Constructed and Selectively Targeted to CRC Cells
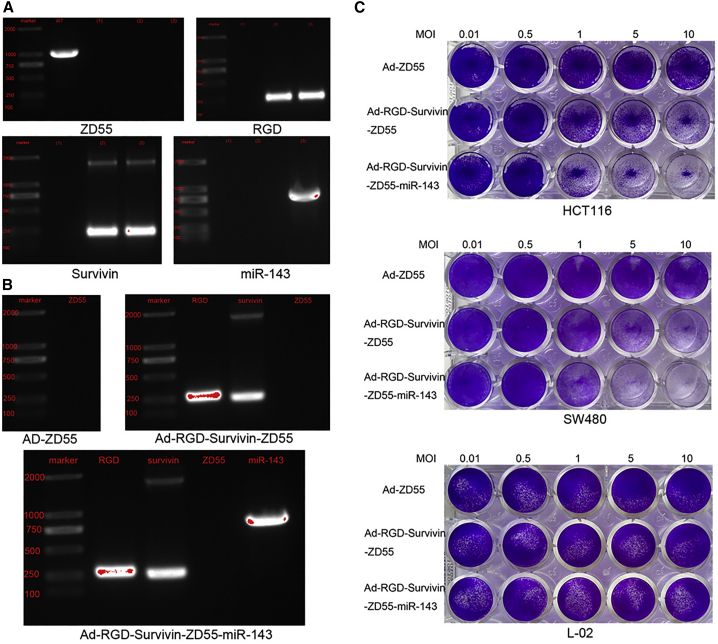
The viral DNA was extracted and acted as a template for the PCR. ZD55, RGD, Survivin, and miR-143 were detected by the agarose gel electrophoresis assays and DNA sequencing. As shown in Figure 3A, about 1,000-bp gene fragments were obtained from the wild-type (WT) adenovirus line, while Ad-ZD55, Ad-RGD-Survivin-ZD55, and Ad-RGD-Survivin-ZD55-miR-143 lines were not observed. The results signified that three new recombinant viruses were not contaminated by WT adenovirus. Using primers for RGD and Survivin, about 200-bp and 269-bp fragments were, respectively, amplified in Ad-RGD-Survivin-ZD55 and Ad-RGD-Survivin-ZD55-miR-143 lines. About 960 bp was only observed in the Ad-RGD-Survivin-ZD55-miR-143 line when using miR-143 primers. The information in Figure 3B indicates that the recombinant virus Ad-RGD-Survivin-ZD55-miR-143 contained RGD and Survivin promoter, deleted the E1B-55kDa region, and successfully carried the therapeutic gene miR-143, which revealed that Ad-RGD-Survivin-ZD55-miR-143 was successfully constructed.
Figure 3.
Recombinant Adenoviruses Were Successfully Constructed and Could Selectively Target to CRC Cells
(A) The viral DNA was amplified by PCR using ZD55, RGD, Survivin, and miR-143 primers. The numbers (1), (2), and (3) represent Ad-ZD55, Ad-RGD-Survivin-ZD55, and Ad-RGD-Survivin-ZD55-miR-143, respectively. (B) Verification of Ad-ZD55, Ad-RGD-Survivin-ZD55, and Ad-RGD-Survivin-ZD55-miR-143 by agarose gel electrophoresis. (C) HCT116, SW480, and L-02 cells were infected with recombinant adenoviruses at different MOIs. HCT116 and SW480 cells were sensitive to Ad-RGD-Survivin-ZD55-miR-143. However, the viability of L-02 cells remained almost unchanged.
HCT116, SW480, and L-02 cells were infected with different recombinant adenoviruses at multiplicities of infection (MOIs) of 0.01, 0.5, 1, 5, and 10. After 5 days, crystal violet staining was observed with the camera. As shown in Figure 3C, the dye of the Ad-RGD-Survivin-ZD55 and Ad-RGD-Survivin-ZD55-miR-143 groups was gradually lightened from an MOI of 1 in HCT116 and SW480 cells. Furthermore, the cytopathic effect of Ad-RGD-Survivin-ZD55-miR-143 was higher than that of Ad-RGD-Survivin-ZD55. The cytological effect of three recombinant viruses was not obvious in L-02 cells. These results indicated that Ad-RGD-Survivin-ZD55-miR-143 could selectively target to HCT116 and SW480 cells.
Recombinant Adenovirus Ad-RGD-Survivin-ZD55-miR-143 Has the Antitumor Effect In Vitro
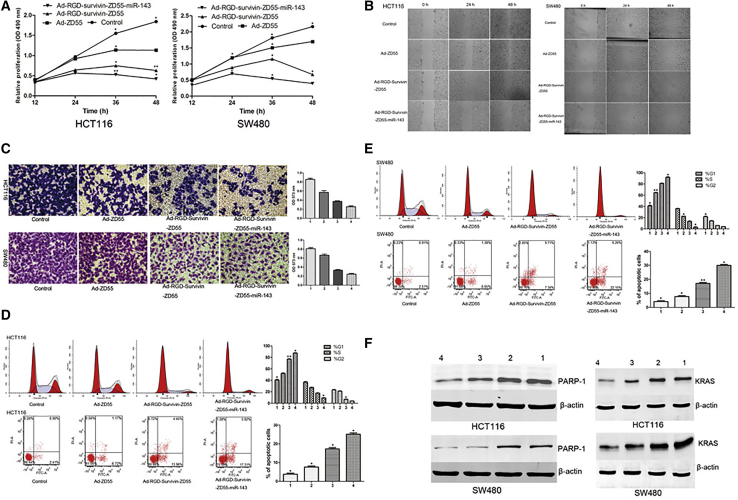
HCT116 and SW480 cells were infected with the recombinant adenoviruses at an MOI of 5. The uninfected cells acted as controls. Results of MTT assays showed that all recombinant viruses significantly inhibited cell proliferation of HCT116 and SW480 cells, compared with the control groups. Among them, Ad-RGD-Survivin-ZD55-miR-143 has the strongest ability to inhibit cell proliferation, followed by Ad-RGD-Survivin-ZD55. However, Ad-ZD55 has a relatively weak ability to inhibit cell proliferation, which might be related to a lack of ability to target CRC cells and the absence of therapeutic genes (Figure 4A).
Figure 4.
The Antitumor Effect of Recombinant Adenoviruses In Vitro
(A) Effect of recombinant adenoviruses on cell proliferation in HCT116 and SW480 cells by MTT assay. (B) Migration ability of cells was determined by wound-healing assay at the 0-h, 24-h, and 48-h time points (image magnification, 100×). (C) The invasion ability of cells was detected by Transwell assays. Images of crystal violet-stained invaded cells were obtained on an inverted microscope with 200× magnification. (D and E) The cell cycle and cell apoptosis of HCT116 (D) and SW480 (E) were evaluated by flow cytometry. The statistical figures represented the distribution of cell cycle and the proportion of early and late apoptotic cells. (F) The expression levels of PARP-1 and KRAS protein of the Ad-RGD-Survivin-ZD55-miR-143 group were lower than Ad-ZD55, Ad-RGD-Survivin-ZD55, and control groups. The numbers 1, 2, 3, and 4 represent control, Ad-ZD55, Ad-RGD-Survivin-ZD55, and Ad-RGD-Survivin-ZD55-miR-143, respectively. *p < 0.05; **p < 0.01.
To study how recombinant adenoviruses affected cell migration and invasion, we performed wound-healing assays and Transwell assays with HCT116 and SW480 cells infected with the recombinant adenoviruses at an MOI of 5. As shown in Figure 4B, the cell-free gaps of the control, Ad-ZD55, Ad-RGD-Survivin-ZD55, and Ad-RGD-Survivin-ZD55-miR-143 groups of HCT116 cells were 46.52 μm, 52.38 μm, 189.73 μm, and 278.85 μm, respectively, at 48 h. In SW480 cells, the cell-free area of the Ad-RGD-Survivin-ZD55 group was significantly wider than all other groups at 48 h. Thus, Ad-RGD-Survivin-ZD55-miR-143 could hamper the extent of closure of HCT116 and SW480 cells. The results of Figure 4C showed that the number of HCT116 and SW480 cells in the Ad-RGD-Survivin-ZD55-miR-143 groups was significantly lower than those in the control, Ad-ZD55, and Ad-RGD-Survivin-ZD55 groups. Ad-RGD-Survivin-ZD55-miR-143 could affect cell invasion of HCT116 and SW480 cells, which could also be illustrated by the optical density (OD) value of the eluent. The OD value of the Ad-RGD-Survivin-ZD55-miR-143 group was the lowest in all of the groups, confirming that the number of cells through the Matrigel was the lowest.
HCT116 cells were infected with different recombinant viruses at an MOI of 5. 36 h after infection, the cell-cycle distributions and the apoptotic rates were evaluated by flow cytometry. As shown in Figure 4D, Ad-RGD-Survivin-ZD55-miR-143 and Ad-RGD-Survivin-ZD55 caused more cells to stay in G1 phase. The statistical results also showed that the proportion of G1 cells in the Ad-RGD-Survivin-ZD55-miR-143 group was higher than in other groups. We also found that the proportion of early and late apoptosis of HCT116 cells induced by the Ad-RGD-Survivin-ZD55-miR-143 group was higher than in other groups. In SW480 cells, the proportion of S-phase and G2-phase cells of Ad-RGD-Survivin-ZD55-miR-143 was significantly reduced compared with other groups. The proportion of early and late apoptotic cells induced by Ad-RGD-Survivin-ZD55-miR-143 in SW480 cells was also significantly higher than in other groups (Figure 4E).
The aforementioned results indicated that Ad-RGD-Survivin-ZD55-miR-143 could target and kill CRC cells. In this case, CRC cells could survive by initiating their own repair mechanism. In eukaryotic cells, PARP-1 was involved in the repair of DNA damage, and its expression level was increased in a variety of tumors. HCT116 and SW480 cells were infected with different recombinant viruses at an MOI of 5. 72 h after infection, the expression of PARP-1 in the recombinant viruses was lower than in the control group, and the downregulated expression of PARP-1 was the most obvious in the Ad-RGD-Survivin-ZD55-miR-143 group (Figure 4F). Furthermore, the expression of KRAS protein in the Ad-RGD-Survivin-ZD55-miR-143 group was lower than that in the Ad-ZD55, Ad-RGD-Survivin-ZD55, and control groups, which indicated that the therapeutic gene miR-143 could be effectively expressed in the recombinant adenovirus and exerted an anti-tumor effect by targeting KRAS in HCT116 and SW480 cells.
Recombinant Adenovirus Ad-RGD-Survivin-ZD55-miR-143 Has the Antitumor Effect In Vivo
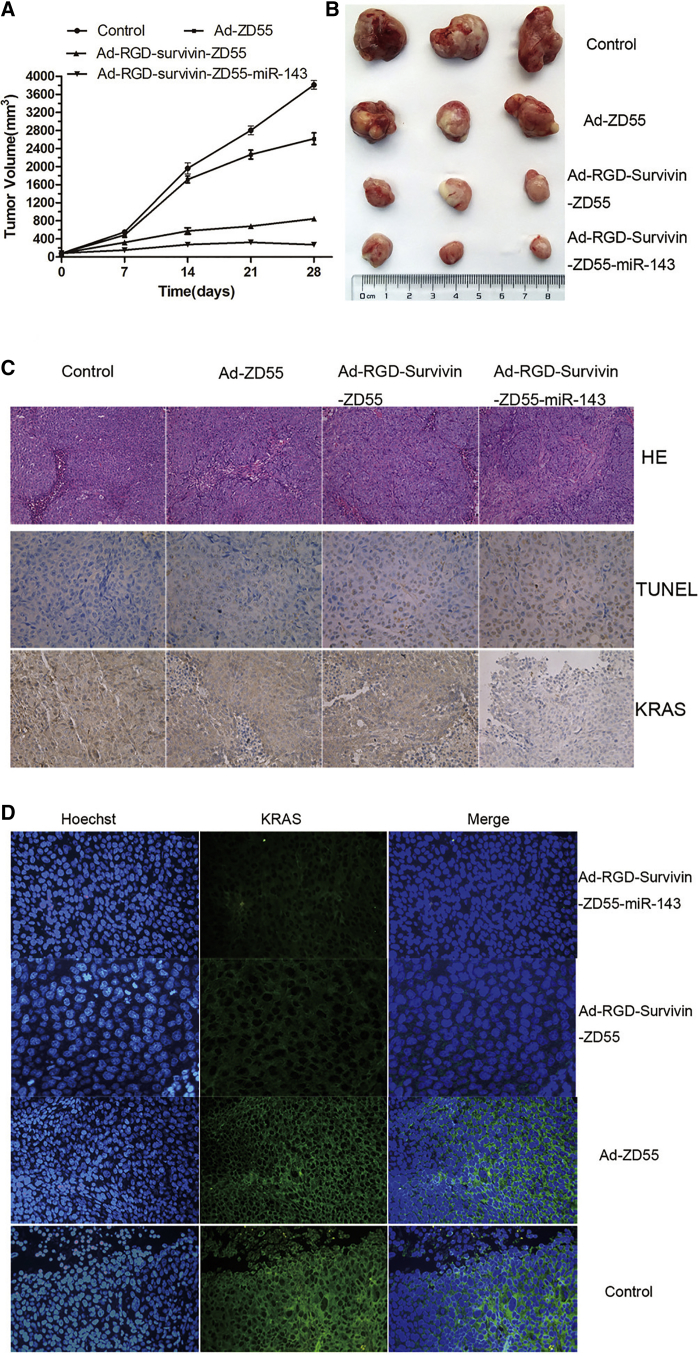
To investigate the antitumor efficacy of Ad-RGD-Survivin-ZD55-miR-143 in vivo, nude mouse tumor xenografts were randomly divided into 4 groups. Tumors treated with recombinant adenoviruses exhibited more growth inhibition compared with those treated with saline alone, and the Ad-RGD-Survivin-ZD55-miR-143 group achieved a significantly greater therapeutic effect than the Ad-ZD55 and Ad-RGD-Survivin-ZD55 groups (Figure 5A). After a 28-day post-treatment interval, all tumors were removed from the nude mice (Figure 5B). The volume of each tumor was evaluated: the tumor volumes of nude mice treated with Ad-ZD55 and control group were 2,618.52 ± 34.77 mm3 and 3,813.60 ± 49.36 mm3, respectively. Those treated with Ad-RGD-Survivin-ZD55-miR-143 and Ad-RGD-Survivin-ZD55 were 272.34 ± 12.28 mm3 and 786.58 ± 15.73 mm3, respectively. In H&E staining, cell necrosis obviously appeared in the Ad-RGD-Survivin-ZD55-miR-143 group, while cell growth was intact in the control group. In terminal deoxynu-cleotidyl transferase (TdT)-mediated dUTP-biotin nick end-labeling (TUNEL) staining, the Ad-RGD-Survivin-ZD55-miR-143 group caused more apoptosis than the Ad-ZD55, Ad-RGD-Survivin-ZD55, and control groups. In immunohistochemical (IHC) staining, the expression of KRAS in the Ad-RGD-Survivin-ZD55-miR-143 group was significantly weaker than in other groups (Figure 5C). Subsequently, the results of immunofluorescence (IF) staining (Figure 5D) also showed that the expression level of KRAS in the cell cytoplasm of recombinant virus groups was lower than in the control group, while only sporadic expression of KRAS was observed in the Ad-RGD-Survivin-ZD55-miR-143 group.
Figure 5.
The Antitumor Effect of Recombinant Adenoviruses In Vivo
(A) Ad-RGD-Survivin-ZD55-miR-143 inhibited the growth of the HCT116 xenograft tumors. Tumor growth curves were plotted, and the xenograft tumor volumes of recombinant virus groups were compared to those of the control group (*p < 0.05; **p < 0.01). (B) All tumors of recombinant viruses and control groups were removed from the nude mice, and images were captured. (C) Representative images of H&E staining (200× magnification), TUNEL staining (400× magnification), and IHC staining of KRAS (400× magnification) were obtained by an inverted microscope. (D) The expression level of KRAS in cell cytoplasm of recombinant virus groups was lower than that in the control group (400× magnification).
Discussion
The disadvantages of traditional gene therapy include low transfection, no specificity, and poor efficacy. The CTGVT strategy could enhance the therapy efficacy, in which the expression of transgene might be increased markedly along with the preferable replication of viruses in tumor cells but not in normal cells.16 Selecting the appropriate therapeutic gene and delivering the gene to target tumor cells is crucial for the success of the CTGVT strategy.
miRNAs might play either as oncogenes or as suppressor genes in the development and progression of human malignancy. Substantial breakthroughs have been published on the diagnosis and prognosis of tumors with miRNAs.19,20 However, there are few clinical trials that have been reported on the treatment of cancer with miRNAs. miR-143 is known to act as a tumor suppressor in several cancers, such as CRC,8, 9, 10,18,21, 22, 23 gastric cancer,24 and bladder cancer.25 Several published papers also indicated that miR-143 acts as a biomarker for the prognosis of CRC, and a low expression of it was associated with better 5-year disease-free survival of CRC.21, 22, 23 In this study, we detected the expression level of miR-143 in CRC tissues and validated the targeted gene of miR-143, and then we explored the impact of miR-143 on the proliferation and cell apoptosis of CRC cells. We found that the expression of miRNA-143 in CRC tissues was lower than in adjacent normal tissues, which was in agreement with previous reports.8, 9, 10 In MTT assays, the proliferative ability of HCT116, RKO, SW480, and LoVo cells was greater inhibited by overexpression of miR-143. The results of flow-cytometric analysis revealed that overexpression of miR-143 in HCT116 and SW480 could induce cellular apoptosis. The results of luciferase assays indicated that miR-143 could bind with its putative target site in the 3′ UTR of KRAS, suggesting that KRAS was a direct target of miR-143. We detected the expression of KRAS in CRC tissues as well as in cells. The expression level of KRAS in cancer tissues was higher than that in normal tissues in IHC assays. The protein levels of KRAS in HCT116 and SW480 were significantly higher than those in Caco2, RKO, and LoVo cells. Approximately 40% of sporadic CRC patients had KRAS mutation26 and were precluded from receiving anti-EGFR targeted therapies. Therefore, targeting KRAS is a promising treatment strategy for CRC. All of the findings confirmed that miR-143 might be an appropriate therapeutic gene for the CTGVT strategy.
Adenovirus vectors based on human serotype 5 of species C are widely used as gene delivery vehicles that enabled high-titer production and highly efficient gene transfer into targeted cells.27 The oncolytic adenovirus carrying therapeutic genes has shown to exert promising antitumor effects on different types of cancers.28, 29, 30 In our study, we used the AdEasy system31 to construct new recombinant adenovirus Ad-RGD-Survivin-ZD55-miR-143: replacing the original E1A promoter with tumor-specific Survivin promoter could transcriptionally control viral replication; deleting the E1B-55kDa region could lose the ability to replicate in normal cells; and inserting an RGD motif into the HI loop of the adenovirus knob could elevate the infection efficiency. All genes of Ad-RGD-Survivin-ZD55-miR-143 were detected to confirm that the recombinant adenovirus was successfully constructed without WT adenovirus contamination. The results of crystal violet staining assays indicated that a cytopathic effect was observed in HCT116 and SW480 cells compared with L-02 cells. L-02 cells acted as a control because a majority of medicines needed to be metabolized through the liver. These findings suggest that the recombinant adenovirus Ad-RGD-Survivin-ZD55-miR-143 is safe and could selectively target CRC cells.
Finally, we investigated the antitumor effect of recombinant adenoviruses in vitro and in vivo. We found that Ad-RGD-Survivin-ZD55-miR-143 inhibited cell growth, suppressed cell migration and invasion, arrested cells at the G1 phase, and induced cell apoptosis in HCT116 and SW480 cells. Ad-RGD-Survivin-ZD55-miR-143 inhibited the protein levels of PARP-1 and KRAS in HCT116 and SW480 cells. The expression of PARP-1 was increased in CRC32 and worked as a survival factor when DNA damage was occurring. Cogoi et al.33 provided compelling evidence that PARP-1 was an activator of the KRAS promoter. Ad-RGD-Survivin-ZD55-miR-143 inhibited the expression of PARP-1 and prevented the survival of KRAS-positive cells, but the specific mechanism needs further analysis. In animal experiments, we found that xenograft tumor growth in nude mice was significantly inhibited by Ad-RGD-Survivin-ZD55-miR-143. Additionally, H&E staining of xenograft tumor tissues confirmed that more patches of necrosis were observed in the Ad-RGD-Survivin-ZD55-miR-143 group compared to other groups. TUNEL showed that Ad-RGD-Survivin-ZD55-miR-143 induced more severe apoptosis in cancer cells than other groups. IF and IHC assays showed that the expression level of KRAS was decreased in Ad-RGD-Survivin-ZD55-miR-143 groups. Hence, these results validated that the tumor suppressor role of Ad-RGD-Survivin-ZD55-miR-143 in CRC tumorigenesis was achieved by targeting KRAS.
Conclusions
In conclusion, a triple-regulated oncolytic adenoviral vector carrying the miR-143 gene was successfully constructed. This new virus could selectively replicate in CRC cells and exhibited specific antitumor effects in vitro and in vivo. It might provide an alternative safe and effective therapeutic method for the future treatment of CRC.
Materials and Methods
Specimens
Thirty-six paired CRC specimens and adjacent normal tissues were collected from the Department of General Surgery of the Shanghai Tenth People’s Hospital. The pathological information of specimens was confirmed by the Department of Pathology of the Shanghai Tenth People’s Hospital, and the specimens were preserved in liquid nitrogen. None of the patients received chemotherapy prior to surgery. The specimens were only collected after patients provided informed consent, and the usage of specimens was approved by the ethics committee of the Shanghai Tenth People’s Hospital.
Cells and Transfection
Human normal liver cell line L-02, human CRC cell line HCT116, Caco2, SW480, RKO, and LoVo were purchased from the Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China). The HEK293A and HEK293T cells were stored in our laboratory. All cells were cultured according to the manufacturer’s instructions.
Cells (2 × 105) were seeded in a 6-well culture plate and cultured with Dulbecco’s modified Eagle’s medium (DMEM) (GIBCO, USA) without serum or antibiotics. Once cell density reached about 40%, they were transfected with miR-143 (GenePharma, Shanghai, China) or NC mimics using Lipofectamine transfection reagents (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. After 5–6 h of incubation, DMEM was replaced by DMEM with 10% fetal bovine serum (FBS) (GIBCO, Waltham, MA, USA).
PCR
Total RNAs were extracted from 36 paired CRC specimens and adjacent normal tissues by using the miRcute microRNA Isolation Kit (Tiangen, Beijing, China). The miR-143 stem-loop primer, U6 primer, and EzOmics SYBR qPCR kit were all purchased from Biomics Biotechnology (Jiangsu, China). Expression levels of miR-143 were analyzed by using a one-step qRT-PCR kit according to the manufacturer’s instructions. Real-time PCR was performed on a 7900HT Fast Real-Time PCR instrument (Applied Biosystems, Singapore, Singapore) using the following primers: miR-143, 5′-CCGCGCGTGAGATGAAGCACTG-3′ (sense) and 5′-ATCCAGTGCAGGGTCCGAGG-3′ (antisense); U6, 5′-TGCGGGTG-CTCGCTTCGCAGC-3′ (sense) and 5′-CCAGTGC-AGGGTCCGAGGT-3′ (antisense). The stem-loop primer is 5′-GTCCTATCCAGTGCAGGGTCCGAGGTGCACTGGATACG-ACAAAATATGGAAC-3′. One-step PCR parameters for miRNA quantification were as follows: 37°C for 60 min for reverse transcription, 10 min at 95°C, followed by 40 cycles of 20 s at 95°C, 30 s at 62°C, and 30 s at 72°C. Ct values were collected at the end of the PCR. Each sample was tested in triplicate, and the relative quantification equation was used to calculate the relative expression.
Dual-Luciferase Reporter Assay
HEK293T cells grown in the 24-well plate were co-transfected with miR-143 (100 nmol/L) and psiCHECK-2/KRAS 3′ UTR reporter plasmids (100 ng) by using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA). The cells were harvested 36 h after transfection, and the dual-luciferase reporter system (Promega, Madison, WI, USA) was used to detect the change in relative luciferase activity. FL and RL activity were measured, and FL was co-transfected as an internal control.
Western Blot Analysis
Cells were harvested and lysed with the RIPA lysis buffer (Beyotime, Jiangsu, China). The protein concentrations were determined using the BCA Protein Assay Kit (Beyotime). Equal amounts of proteins were separated by 10% SDS-PAGE and transferred onto 0.45-μm nitrocellulose membranes (Beyotime). The membranes were blocked with 5% skim milk for 1 h and then were incubated with primary antibodies and the appropriate secondary antibodies. Immunoreactive protein bands were detected using the Odyssey Scanning System (Li-Cor, Lincoln, NE, USA).
Construction and Identification of Recombinant Adenoviruses
pZD55 and pshuttle-Survivin-ZD55 were constructed as described previously.18,34 The expression cassette of miR-143 was inserted into pshuttle-Survivin-ZD55 to produce the pshuttle-Survivin-ZD55-miR-143 plasmid. The oncolytic adenoviral plasmid pAdeasy-RGD-Survivin-ZD55-miR-143 was generated using homologous recombination of the shuttle vector and adenoviral backbone plasmid pAdeasy-RGD in the Escherichia coli strain BJ5183. Recombinant adenovirus Ad-RGD-Survivin-ZD55-miR-143 was packaged and amplified in HEK293A cells followed by centrifugation in a graded CsCl solution for purification. Viral genomes for identification were extracted using the TIANamp Genomic DNA Kit (Tiangen, Beijing, China) according to the manufacturer’s instructions. The existence of the RGD peptide-coding sequence, the Survivin promoter, the E1B-55kDa deletion, miR-143, and WT contaminants were shown using PCR and sequencing with the appropriate primers (Table 1).
Table 1.
Primers of Viral Genomes
| Name and Primer | 5′–3′ Sequence |
|---|---|
| RGD | |
| FOR | CCGCGCGTGAGATGAAGCACTG |
| RE | TGCGGGTGCTCGCTTCGCAGC |
| Survivin | |
| FOR | CCGCTCGAGCGCGTTCTTTGAAAGC |
| RE | ACGCGTCGACTGCCGCCGCCG |
| ZD55 | |
| FOR | CACAAGAATCGCCTGCTA |
| RE | GCTGGCTCCGTGAATGGT |
| miR-143 | |
| FOR | TCGTCTAGCATCGAAGATCTTAGTTATTAATAGTAATCAATTACGGGG |
| RE | CCTCAGCACCTTCCAGATCTGGAAACACTCTGTCCTTCCTG |
Cytopathic Effect Assay
The cytopathic effect procedure was observed by crystal violet staining. Different cells were dispensed into 24-well plates and infected with different recombinant adenoviruses at various MOIs. After 4 days, the culture medium was removed and washed twice with phosphate-buffered saline (PBS). Then, 500 μL 2% crystal violet solution was added to each well for 15 min at an ambient temperature. The 24-well plates were washed with distilled water and documented as photographs.
Cell Proliferation Assay
Cell proliferation was assessed by MTT (Sigma, St. Louis, MO, USA) assays. Cells in 96-well plates were transfected with miR-143 mimics or infected with different recombinant adenoviruses at various MOIs. 20 μL MTT (5 mg/mL) solution was added in each well, and incubation was continued at 37°C for 4 h. The culture medium was discarded, and 150 μL dimethyl sulfoxide (DMSO) was added. After 10 min of low-speed (100 rpm) shaking for dissolving the formazan production, absorbance was measured at 490 nm with a microplate spectrophotometer.
Cell Migration Assay
Cell migration was detected by scratch wound-healing assays. Cells were seeded in 6-well plates and grown to full confluence. A denuded zone of constant width was made through each well using a sterile pipette tip. Each well was rinsed slowly with PBS three times to remove the detached cells. After that, DMEM with serum-free medium was added into each well, and cells of each well were infected with different recombinant adenoviruses at a MOI of 5. Cell migration to the wounded region was observed using an inverted microscope and photographed (100× magnification) at the same position. Images were captured at 0, 24, and 48 h to monitor the wound-healing process.
Cell Invasion Assay
Transwell invasion assays were performed to evaluate invasion ability by using a precoated cell invasion kit (Corning, Corning, NY, USA) and Matrigel (BD Biosciences, Billerica, MA, USA) in Transwell chambers. The chamber was filled with CRC cells infected with adenoviruses and added into a 24-well plate containing DMEM with 20% FBS. Cells were allowed to invade from the Transwell chamber to the bottom wells. After 48 h of incubation, the invaded cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Images of the stained cells were obtained using an inverted microscope (200× magnification). The crystal violet was completely dissolved by 33% acetic acid, and then absorbance was measured at 570 nm with a microplate spectrophotometer.
Cell Cycle and Apoptosis Assay
Cells were infected with different recombinant adenoviruses at an MOI of 5. After 36 h, cell-culture supernatants and adherent cells were harvested and washed three times with cold PBS. Then, the cells were fixed with 70% ethanol at 4°C overnight. The cells were stained with PI staining solution (BD Biosciences). After incubation for 15 min at room temperature in the dark, the cells were analyzed with a flow cytometer (FACSCanto II, BD Biosciences, Billerica, MA, USA). For the determination of apoptotic cell death, the cells were stained using the Annexin V-FITC/PI Apoptosis Detection Kit (Beyotime, Jiangsu, China) according to the manufacturer’s instructions. Briefly, the cells were incubated in the dark for 15 min at room temperature after adding annexin V-FITC. Subsequently, PI was added, and the cells were incubated in dark for 5 min. Last, all samples were processed by flow cytometry (BD Biosciences).
Tumor Xenografts in Nude Mice
The animal experiments were approved by the ethics review board of the Shanghai Tenth People’s Hospital. Four-week-old female BALB/c nude mice were purchased from the Animal Core Facility (Shanghai, China). 5 × 106 HCT116 cells were mixed with Matrigel (BD Biosciences) at a ratio of 2:1 and injected subcutaneously into the right flanks of mice for the tumorigenicity assay. The growth of xenograft was monitored regularly using Vernier calipers to measure the tumor size, and the volume of xenograft was calculated using the following equation: volume = length × width2 × 0.5. When the tumors reached about 100 mm3 in size, the mice were randomly divided into four equal groups (6 mice per group): control, Ad-ZD55, Ad-RGD-Survivin-ZD55, and Ad-RGD-Survivin-ZD55-miR-143. Mice were treated with multisite intratumor injections of the various recombinant adenoviruses at 1 × 109 plaque forming units (PFUs) per mouse in 100 μL PBS. Mice in the control groups received 100 μL saline alone. The injections were administered once every other day, through 5 injections. The growth of xenograft curves was then plotted using the aforementioned equation.
Histological Analysis and TUNEL Staining
The xenograft specimens were fixed by formalin and then paraffin embedded, sliced into 4-μm sections, and stained using H&E. TUNEL staining was used to detect apoptotic cells by using an in situ cell death detection kit (Roche, Palo Alto, CA, USA). The positive indices were counted from five randomly selected high-power fields and expressed as the percentage of total cells counted.
IHC and IF Assay
The sections were dewaxed in xylene and rehydrated in graded concentrations of ethyl alcohol. Then, the slides were incubated in 3% H2O2 for 10 min to inhibit the endogenous peroxidase activity. Next the slides were placed in sodium citrate buffer for antigen retrieval, a high voltage was applied for 3 min (pH 6.0), and the slides were placed in FBS as blocking antibody for 10 min. The slides were incubated with a human polyclonal antibody against KRAS (1:150, Abcam) at 4°C overnight. After washing with PBS, the sections were incubated with a second antibody for 30 min. Finally, the sections were visualized with diaminobenzidine solution and counterstained with hematoxylin. The percentage of cells with KRAS staining and the staining intensity were scored as follows: 0, negative; 1+, <10% positive cells; 2+, 10%–50% positive cells; 3+, >50% positive cells. The positive KRAS staining was for sections with 2+ or 3+ immunostaining. For the IF assay, KRAS was applied and incubated overnight at 4°C. After fluorescent labeling, the second antibody was applied. Finally, the slides were stained with Hoechst for nuclear staining at room temperature for 5 min. Representative figures were captured with a fluorescence microscope.
Statistical Analysis
Data from at least three separate experiments were presented as mean ± standard error of the mean (SEM). Data were evaluated by Student’s t test (for two-group comparison) or one-way ANOVA with Bonferroni’s post hoc test (for multiple-group comparison). Differences were considered statistically significant only when the p value was less than 0.05 or 0.01.
Author Contributions
L.F. and B.X. designed and directed this study. Q.L. and H.G. constructed recombinant adenoviruses. X.D. and J.L. performed cell proliferation, migration, and invasion assays. W.J. and J.Z. conducted western blotting assays. X.Z. performed cell-cycle and apoptosis assays. H.S. and Q.L. performed dual-luciferase reporter assays, tumor xenografts, TUNEL staining, and the IHC and IF assays. Q.L. and S.B. drafted the manuscript. T.D. edited the manuscript. All authors read and approved the final manuscript.
Acknowledgments
This work was supported by grants from the Scientific Research Foundation for the Returned Overseas Chinese Scholars, Chinese Ministry of Education (no. 020114001).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2020.01.005.
Contributor Information
Bin Xu, Email: pfdbsxubin@163.com.
Lin Fang, Email: fangzhuren301@163.com.
Supplemental Information
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Benson A.B., 3rd, Venook A.P., Bekaii-Saab T., Chan E., Chen Y.J., Cooper H.S., Engstrom P.F., Enzinger P.C., Fenton M.J., Fuchs C.S. Rectal cancer, version 2.2015. J. Natl. Compr. Canc. Netw. 2015;13:719–728, quiz 728. doi: 10.6004/jnccn.2015.0087. [DOI] [PubMed] [Google Scholar]
- 3.Creasy J.M., Sadot E., Koerkamp B.G., Chou J.F., Gonen M., Kemeny N.E., Saltz L.B., Balachandran V.P., Kingham T.P., DeMatteo R.P. The impact of primary tumor location on long-term survival in patients undergoing hepatic resection for metastatic colon cancer. Ann. Surg. Oncol. 2018;25:431–438. doi: 10.1245/s10434-017-6264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lujan H.J., Plasencia G., Jacobs M., Viamonte M., 3rd, Hartmann R.F. Long-term survival after laparoscopic colon resection for cancer: complete five-year follow-up. Dis. Colon Rectum. 2002;45:491–501. doi: 10.1007/s10350-004-6227-8. [DOI] [PubMed] [Google Scholar]
- 5.Franko J., Shi Q., Meyers J.P., Maughan T.S., Adams R.A., Seymour M.T., Saltz L., Punt C.J.A., Koopman M., Tournigand C. Prognosis of patients with peritoneal metastatic colorectal cancer given systemic therapy: an analysis of individual patient data from prospective randomised trials from the Analysis and Research in Cancers of the Digestive System (ARCAD) database. Lancet Oncol. 2016;17:1709–1719. doi: 10.1016/S1470-2045(16)30500-9. [DOI] [PubMed] [Google Scholar]
- 6.Makeyev E.V., Maniatis T. Multilevel regulation of gene expression by microRNAs. Science. 2008;319:1789–1790. doi: 10.1126/science.1152326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 8.Guo L., Fu J., Sun S., Zhu M., Zhang L., Niu H., Chen Z., Zhang Y., Guo L., Wang S. MicroRNA-143-3p inhibits colorectal cancer metastases by targeting ITGA6 and ASAP3. Cancer Sci. 2019;110:805–816. doi: 10.1111/cas.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pekow J., Meckel K., Dougherty U., Butun F., Mustafi R., Lim J., Crofton C., Chen X., Joseph L., Bissonnette M. Tumor suppressors miR-143 and miR-145 and predicted target proteins API5, ERK5, K-RAS, and IRS-1 are differentially expressed in proximal and distal colon. Am. J. Physiol. Gastrointest. Liver Physiol. 2015;308:G179–G187. doi: 10.1152/ajpgi.00208.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen X., Guo X., Zhang H., Xiang Y., Chen J., Yin Y., Cai X., Wang K., Wang G., Ba Y. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;28:1385–1392. doi: 10.1038/onc.2008.474. [DOI] [PubMed] [Google Scholar]
- 11.Crystal R.G. Adenovirus: the first effective in vivo gene delivery vector. Hum. Gene Ther. 2014;25:3–11. doi: 10.1089/hum.2013.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mizuguchi H., Kay M.A., Hayakawa T. Approaches for generating recombinant adenovirus vectors. Adv. Drug Deliv. Rev. 2001;52:165–176. doi: 10.1016/s0169-409x(01)00215-0. [DOI] [PubMed] [Google Scholar]
- 13.Lichty B.D., Breitbach C.J., Stojdl D.F., Bell J.C. Going viral with cancer immunotherapy. Nat. Rev. Cancer. 2014;14:559–567. doi: 10.1038/nrc3770. [DOI] [PubMed] [Google Scholar]
- 14.Taguchi S., Fukuhara H., Homma Y., Todo T. Current status of clinical trials assessing oncolytic virus therapy for urological cancers. Int. J. Urol. 2017;24:342–351. doi: 10.1111/iju.13325. [DOI] [PubMed] [Google Scholar]
- 15.Eissa I.R., Bustos-Villalobos I., Ichinose T., Matsumura S., Naoe Y., Miyajima N., Morimoto D., Mukoyama N., Zhiwen W., Tanaka M. The current status and future prospects of oncolytic viruses in clinical trials against melanoma, glioma, pancreatic, and breast cancers. Cancers (Basel) 2018;10:E356. doi: 10.3390/cancers10100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X.Y. Targeting gene-virotherapy of cancer and its prosperity. Cell Res. 2006;16:879–886. doi: 10.1038/sj.cr.7310108. [DOI] [PubMed] [Google Scholar]
- 17.Xiao T., Fan J.K., Huang H.L., Gu J.F., Li L.Y., Liu X.Y. VEGI-armed oncolytic adenovirus inhibits tumor neovascularization and directly induces mitochondria-mediated cancer cell apoptosis. Cell Res. 2010;20:367–378. doi: 10.1038/cr.2009.126. [DOI] [PubMed] [Google Scholar]
- 18.Luo Q., Basnet S., Dai Z., Li S., Zhang Z., Ge H. A novel E1B55kDa-deleted oncolytic adenovirus carrying microRNA-143 exerts specific antitumor efficacy on colorectal cancer cells. Am. J. Transl. Res. 2016;8:3822–3830. [PMC free article] [PubMed] [Google Scholar]
- 19.Adams B.D., Parsons C., Walker L., Zhang W.C., Slack F.J. Targeting noncoding RNAs in disease. J. Clin. Invest. 2017;127:761–771. doi: 10.1172/JCI84424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bertoli G., Cava C., Castiglioni I. MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 2015;5:1122–1143. doi: 10.7150/thno.11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caritg O., Navarro A., Moreno I., Martínez-Rodenas F., Cordeiro A., Muñoz C., Ruiz-Martinez M., Santasusagna S., Castellano J.J., Monzó M. Identifying high-risk stage II colon cancer patients: a three-microRNA-based score as a prognostic biomarker. Clin. Colorectal Cancer. 2016;15:e175–e182. doi: 10.1016/j.clcc.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 22.Jacob H., Stanisavljevic L., Storli K.E., Hestetun K.E., Dahl O., Myklebust M.P. Identification of a sixteen-microRNA signature as prognostic biomarker for stage II and III colon cancer. Oncotarget. 2017;8:87837–87847. doi: 10.18632/oncotarget.21237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J.X., Song W., Chen Z.H., Wei J.H., Liao Y.J., Lei J., Hu M., Chen G.Z., Liao B., Lu J. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol. 2013;14:1295–1306. doi: 10.1016/S1470-2045(13)70491-1. [DOI] [PubMed] [Google Scholar]
- 24.Lei C., Du F., Sun L., Li T., Li T., Min Y., Nie A., Wang X., Geng L., Lu Y. miR-143 and miR-145 inhibit gastric cancer cell migration and metastasis by suppressing MYO6. Cell Death Dis. 2017;8:e3101. doi: 10.1038/cddis.2017.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Avgeris M., Mavridis K., Tokas T., Stravodimos K., Fragoulis E.G., Scorilas A. Uncovering the clinical utility of miR-143, miR-145 and miR-224 for predicting the survival of bladder cancer patients following treatment. Carcinogenesis. 2015;36:528–537. doi: 10.1093/carcin/bgv024. [DOI] [PubMed] [Google Scholar]
- 26.Phipps A.I., Buchanan D.D., Makar K.W., Win A.K., Baron J.A., Lindor N.M., Potter J.D., Newcomb P.A. KRAS-mutation status in relation to colorectal cancer survival: the joint impact of correlated tumour markers. Br. J. Cancer. 2013;108:1757–1764. doi: 10.1038/bjc.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agrawal B., Gupta N., Vedi S., Singh S., Li W., Garg S., Li J., Kumar R. Heterologous immunity between adenoviruses and hepatitis C virus (HCV): recombinant adenovirus vaccine vectors containing antigens from unrelated pathogens induce cross-reactive immunity against HCV antigens. Cells. 2019;8:E507. doi: 10.3390/cells8050507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rahal A., Musher B. Oncolytic viral therapy for pancreatic cancer. J. Surg. Oncol. 2017;116:94–103. doi: 10.1002/jso.24626. [DOI] [PubMed] [Google Scholar]
- 29.Foreman P.M., Friedman G.K., Cassady K.A., Markert J.M. Oncolytic virotherapy for the treatment of malignant glioma. Neurotherapeutics. 2017;14:333–344. doi: 10.1007/s13311-017-0516-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fajardo C.A., Guedan S., Rojas L.A., Moreno R., Arias-Badia M., de Sostoa J., June C.H., Alemany R. Oncolytic adenoviral delivery of an EGFR-targeting T-cell engager improves antitumor efficacy. Cancer Res. 2017;77:2052–2063. doi: 10.1158/0008-5472.CAN-16-1708. [DOI] [PubMed] [Google Scholar]
- 31.Luo J., Deng Z.L., Luo X., Tang N., Song W.X., Chen J., Sharff K.A., Luu H.H., Haydon R.C., Kinzler K.W. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat. Protoc. 2007;2:1236–1247. doi: 10.1038/nprot.2007.135. [DOI] [PubMed] [Google Scholar]
- 32.Sulzyc-Bielicka V., Domagala P., Hybiak J., Majewicz-Broda A., Safranow K., Domagala W. Colorectal cancers differ in respect of PARP-1 protein expression. Pol. J. Pathol. 2012;63:87–92. [PubMed] [Google Scholar]
- 33.Cogoi S., Paramasivam M., Membrino A., Yokoyama K.K., Xodo L.E. The KRAS promoter responds to Myc-associated zinc finger and poly(ADP-ribose) polymerase 1 proteins, which recognize a critical quadruplex-forming GA-element. J. Biol. Chem. 2010;285:22003–22016. doi: 10.1074/jbc.M110.101923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu B., Zheng W.Y., Jin D.Y., Wang D.S., Liu X.Y., Qin X.Y. Treatment of pancreatic cancer using an oncolytic virus harboring the lipocalin-2 gene. Cancer. 2012;118:5217–5226. doi: 10.1002/cncr.27535. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.