Abstract
In the era of opioid abuse epidemics, there is an increased demand for understanding how opioid receptors can be allosterically modulated to guide the development of more effective and safer opioid therapies. Among the modulators of the μ-opioid (MOP) receptor, which is the pharmacological target for the majority of clinically used opioid drugs, are monovalent and divalent cations. Specifically, the monovalent sodium cation (Na+) has been known for decades to affect MOP receptor signaling by reducing agonist binding, whereas the divalent magnesium cation (Mg2+) has been shown to have the opposite effect, notwithstanding the presence of sodium chloride. Although ultra-high-resolution opioid receptor crystal structures have revealed a specific Na+ binding site and molecular dynamics (MD) simulation studies have supported the idea that this monovalent ion reduces agonist binding by stabilizing the receptor inactive state, the putative binding site of Mg2+ on the MOP receptor, as well as the molecular determinants responsible for its positive allosteric modulation of the receptor, are unknown. In this work, we carried out tens of microseconds of all-atom MD simulations to investigate the simultaneous binding of Mg2+ and Na+ cations to inactive and active crystal structures of the MOP receptor embedded in an explicit lipid-water environment and confirmed adequate sampling of Mg2+ ion binding with a grand canonical Monte Carlo MD method. Analyses of these simulations shed light on 1) the preferred binding sites of Mg2+ on the MOP receptor, 2) details of the competition between Mg2+ and Na+ cations for specific sites, 3) estimates of binding affinities, and 4) testable hypotheses of the molecular mechanism underlying the positive allosteric modulation of the MOP receptor by the Mg2+ cation.
Significance
Overprescription of opioid drugs in the late 1990s, followed by abuse of both prescription and illicit opioid drugs, has led to what is nowadays called the “opioid overdose crisis” or “opioid epidemic,” with more than 130 Americans dying daily from opioid overdose at the time of writing. To reduce opioid doses, and thereby prevent or treat overdose and opioid use disorders, attention has recently shifted to the use of coanalgesics. Understanding how opioid receptor targets can be allosterically modulated by these elements, including cations, is key to the development of improved therapeutics. Here, we provide an atomic-level understanding of the mechanism by which magnesium binds to the μ-opioid receptor and enhances opioid drug efficacy by stabilizing the receptor activated state.
Introduction
Opioid analgesics such as morphine remain the “gold standard” for the treatment of acute (e.g., postoperative) or chronic (e.g., cancer) pain notwithstanding the multitude of side effects they cause, including respiratory depression, tolerance, addiction, dependence, constipation, nausea, vomiting, dizziness, fatigue, and itching (1). Among all side effects, respiratory depression is certainly the most feared by physicians given the almost 400,000 casualties from overdosing on prescription or illicit opioids during 1999–2017 in the United States alone (2).
Almost all clinically used opioid drugs act through the μ-opioid (MOP) receptor (3), a member of the G protein-coupled receptor (GPCR) superfamily. Like other GPCRs, this receptor exists as an ensemble of multiple inactive, active, and intermediate conformations, and this conformational plasticity forms the basis for the receptor signaling diversity achieved through receptor interaction with different intracellular proteins, such as G proteins (typically Gαi/o subtypes) or arrestins (typically β-arrestin-2) (4). Depending on the MOP receptor conformation stabilized by the drug and the intracellular protein recruited by the receptor, biological functions that are either clinically desirable or detrimental can ensue. Indeed, although studies using MOP receptor knockout mice confirmed that the MOP receptor is responsible for the antinociceptive action of morphine (3), β-arrestin recruitment by the MOP receptor was shown to contribute to some of the side effects of this classical opioid drug (5, 6, 7), supporting the hypothesis that biasing the conformational equilibrium of MOP receptor toward the G protein pathway may lead the way to develop safer opioid therapies.
In principle, there are several ways one could possibly bias the conformational equilibrium of the MOP receptor toward a specific conformational state, whether using a small molecule, a peptide, a protein, or other allosteric modulators, including cations. Indeed, it has been known for more than 40 years that the MOP receptor can be differentially modulated by cations (8). Whereas the monovalent Na+ cation can decrease agonist affinity at the MOP receptor (8), most likely through stabilization of the inactive conformational state of the receptor (e.g., see (9,10)), the divalent Mg2+ cation has the opposite effect (e.g., see (11)), suggesting it stabilizes an active-like conformation of the receptor. Notably, similar conclusions were drawn for other GPCRs based on inferences from biochemical and pharmacological studies (12, 13, 14, 15, 16, 17, 18).
Although various high-resolution x-ray crystal structures of inactive forms of GPCRs have revealed the atomic details of Na+ binding (19, 20, 21, 22) and several molecular dynamics (MD) simulation studies have supported the stabilization of inactive conformations of the receptor by this monovalent cation (e.g., see (9,10,23,24)), limited information exists to date about the preferred binding site(s) of Mg2+ and the molecular mechanism underlying the positive allosteric effect of this divalent cation. A recent interdisciplinary study combining the results of 19F-NMR and MD simulations on another prototypic GPCR, the adenosine A2A receptor (18), provided further support to the negative allosteric modulation of receptors by Na+ and their positive allosteric modulation by Ca2+ and Mg2+. Not only did this work allow quantification of the effects of these cations on the conformational ensemble of the adenosine A2A receptor, but it also suggested important molecular determinants involved in the allosteric activation of this receptor in the presence of physiological cations. Notable results of these studies were the unexpected finding that Na+ also stabilized an intermediate state that had previously been associated with partial agonism, whereas Mg2+ cations drove G protein-binding cleft opening, and consequent receptor activation, upon bridging specific acidic residues on the extracellular region of transmembrane (TM) helices 5 and 6 (TM5 and TM6) of the receptor (18).
In our work, we studied the free binding of Mg2+ to the MOP receptor in the presence of physiological concentrations of Na+ by all-atom standard MD simulations of the receptor embedded in an explicit 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC)/cholesterol environment and supplemented them with a grand canonical Monte Carlo MD (GCMC-MD) method (25, 26, 27) to confirm thorough exploration of the Mg2+ bound states. The goal of this study was not only to predict energetically favorable binding sites of Mg2+ at the MOP receptor but also to provide atomic details of the molecular mechanism regulating the positive allosteric modulation of the receptor induced by this divalent cation. This information is important considering that magnesium is known to enhance opioid-induced analgesia in different types of surgical procedures without increasing opioid side effects at therapeutic doses, thus suggesting that its use in combination with opioid analgesics may be useful in clinical practice (28).
Methods
MD simulations and system setup
Three different simulation setups were built for the ligand-free murine MOP receptor (residues 65–347), using the CHARMM-GUI web server (29). Whereas one of them was based on the x-ray crystal structure of the inactive MOP receptor (Protein Data Bank (PDB): 4DKL (30)), the other two system setups used the crystal structure of the active MOP receptor (PDB: 5C1M (31)) and differed in the protonation state of the highly conserved D1142.50 residue. The missing loop between TM5 and TM6 in 4DKL (residues M264–K269) was added using the Modeller software (32). Both ligand-free inactive and active MOP receptor models were embedded in a lipid bilayer with a POPC/cholesterol ≈ 9:1 ratio and an area of 7.5 × 7.5 nm2. A POPC/10% cholesterol lipid bilayer is a typical membrane model used in MD simulations of GPCRs considering 1) the abundance of phosphatidylcholine lipids in cell membranes (33), 2) a 10% sterol/lipids ratio representing a mean value across membranes of different cell compartments, and 3) the suggested impact of cholesterol on the receptor conformational dynamics (34).
The membrane and protein were then solvated with explicit TIP3P water molecules and 0.15 M concentrations of both Na+ and Mg2+ cations and neutralized with Cl− ions. All ions were placed randomly in the solvent volume, at least 10 Å away from the protein and membrane. Each full simulation system contained ∼70,000 atoms and had an initial volume of 7.5 × 7.5 × 11.9 nm3. The CHARMM36 force field (35) was used to model protein, lipids, and ions, and all-atom MD simulations were carried out using the GROMACS software package version 2018.1 (36). After a first energy minimization step, six short equilibrations runs with gradually decreasing harmonic constraints on lipid and protein heavy atoms were carried out, following the CHARMM-GUI membrane builder equilibration protocol. After these short equilibration runs, the three systems were further equilibrated for 100 ns in the NPT ensemble without constraints using the Nosé-Hoover thermostat (37) at 310 K (coupling constant ps) and the Berendsen barostat (38) at 1 atm (coupling constant ps). A final equilibration run of 20 ns was carried out under the production simulation conditions in which the Berendsen barostat was replaced by the Parrinello-Rahman barostat (39) (coupling constant of ps at 1 atm). Different production runs ranging between ∼0.7 and ∼2.4 μs, for a total simulation time of over 20 μs, followed, beginning from equidistant frames selected from the previous 20 ns equilibration (see Table S1 for details). Long-range electrostatic interactions (for interparticle distances beyond 1.2 nm) were taken into account using the particle mesh Ewald algorithm (40). The van der Waals interactions were switched off gradually between 1.0 and 1.2 nm. Periodic boundary conditions were applied to the simulation boxes, and an integration time step of 2 fs was used for all simulations.
GCMC-MD simulations
Despite extensive unbiased MD simulations, Mg2+ coordination complexes with direct interactions to acidic residues were not observed, possibly because of the large desolvation free energy for Mg2+ (41,42) and in agreement with the results of MD simulations carried out on another prototypic GPCR (18). To enable sampling of Mg2+ direct coordination complexes and check whether other important coordination sites had been missed by standard MD simulations, we applied a grand canonical Monte Carlo method combined with MD (GCMC-MD) (25, 26, 27) that successfully reproduced Mg2+coordination on RNA molecules (27). This method combines a Monte Carlo insertion/deletion protocol using an oscillating chemical potential with short MD trajectories to sample potential ion coordination complexes. Specifically, we utilized this method to insert and delete water molecules and Mg2+ ions into a defined GCMC region around the MOP receptor with a target concentration of water molecules and Mg2+ ions that matched their respective simulation concentrations. We used the previously described excess chemical potentials of −5.6 kcal/mol for water molecules and −437.38 kcal/mol for Mg2+ ions (25,43). This GCMC region was defined separately for different simulation systems as a cuboid with a volume of ∼52 × 52 × 94 Å3 centered on the geometrical center of each simulation box. We applied this protocol on representative conformations of the most populated Mg2+ macrosites extracted from our unbiased MD simulations (see below). For each protein conformation, we performed two initial iterations of GCMC to populate the GCMC simulation region with the target water molecules and Mg2+ ions. These were followed by 100 iterations of GCMC-MD sampling, each of which featured 200,000 steps of GCMC, a brief 5000-step steepest descent minimization, a 100 ps constant pressure equilibration, and finally a 1 ns constant volume run. The MD steps were performed with Gromacs version 2018.6. The MD trajectories of the GCMC-MD protocol were then analyzed to identify Mg2+ microsites and to calculate their bound population as described below.
Identification of cation binding sites
We used a two-step approach to identify the preferred binding sites for each cation on each of the simulated MOP receptor systems. First, so-called cation binding microsites were detected by the presence of either Na+ or Mg2+ cations within the closest minimum distance from heavy atoms of protein residues. The value of for the two cations was selected after inspection of the radial distribution functions of the Na+-protein and Mg2+-protein distances, respectively, calculated from combined MD and GCMC-MD trajectories of all simulated active and inactive MOP receptor systems. Based on the radial distribution functions shown in Fig. S1, values of Å or Å were chosen to indicate direct Na+ or Mg2+ coordination to protein residues, respectively. However, unlike Na+, the majority of Mg2+ coordination complexes were water mediated and we therefore chose to select a second closest minimum distance for Mg2+ cations ( Å) to characterize microsites containing these coordination states. Given the large number of microsites obtained using the aforementioned criteria, for each simulated MOP receptor system, we aggregated microsites into macrosites of cation binding based on the location of coordinating residues within said microsites. For instance, microsites involving cation contacts with D1473.32 were labeled as “orthosteric site,” whereas microsites involving coordination with D1142.50 or N1503.35 were labeled as “allosteric site.” Similarly, sites in the extracellular loop (ECL) region were labeled ECL1, ECL2, or ECL3 if containing G131–G136, A206–H223, or L305–T311 residues, respectively.
Calculation of cation binding affinity at macrosites
The cation binding affinity for the th macrosite was estimated based on its marginal occupancy probability pi, defined as the fraction of MD simulation frames in which the macrosite is occupied. This analysis was performed separately for frames sampled during the GCMC-MD protocol to assess the affinity of Mg2+ to equivalent macrosites identified during said protocol. Before running these calculations, we verified that the number of cation binding events at each macrosite, estimated in terms of the number of contacts formed by a cation at each macrosite across the multiple MD simulation trajectories, was sufficient to obtain reliable estimates of occupation probabilities from population counts. The results we obtained from analysis of the MD simulation runs, ranging from ∼40 to ∼1000 binding events for Mg2+ and 20 to ∼500 binding events for Na+ (see Table S2), confirmed that occupation probabilities could be calculated reliably. Assuming that binding to the protein does not change the ion concentration, the binding affinity can be estimated via the relation as follows:
where C is the molar concentration of each cation species present in the simulation box.
Allosteric coupling between macrosites of cation binding
We assessed the effect of each cation species on the binding of the other by calculating cooperativity coefficients between pairs of binding macrosites derived from standard MD simulations. Specifically, we considered the joint probability distribution of the cation species x occupying the macrosite i and the cation species y occupying the microsite j, marginalizing the occupancy of all the other macrosites on the receptor. Indicating with the affinity of the cation species x at macrosite i when macrosite j is not occupied, and correspondingly, with the affinity of the cation species y at macrosite j when macrosite i is not occupied, the presence of a cation at macrosite j changes the affinity of the cation species x at macrosite i to , whereas the closure of the thermodynamic cycle ensures that when macrosite i is occupied, the cation binding affinity at macrosite j becomes . We report these cation binding affinity changes as , with negative values implying negative modulation (i.e., a reduction in the cation binding affinity at a given macrosite induced by the presence of another cation species). We estimate the values of from the ratios as follows:
Quantification of cation effect on extracellular loop gating
To elucidate the effect of Mg2+ binding on the conformational plasticity of the extracellular loop 2 and 3 (ECL2/3) region of the receptor, which has often been linked to the allosteric modulation of ligand binding, we outlined a simple activation model of the MOP receptor and used our MD simulations to derive its thermodynamic properties. Frames sampled from GCMC-MD simulations were not included in this analysis because of a limited sampling of protein motions in this method. Specifically, our simple model only considers agonist binding and defines the active state of the receptor (with either neutral or charged D2.50) as the ligand-bound state, whereas the inactive receptor conformation is classified as a ligand-unbound state. We consider two different states of the ECL2/3 region of the MOP receptor, which we labeled open (O) or closed (C), depending on whether the minimum distance between the heavy atoms of residue pairs E310ECL3−R211ECL2 and Q212ECL2−D216ECL2 sampled during simulation was larger or smaller than 10 Å, respectively. Indicating with and the ligand binding affinity for the ECL2/3 region open and closed states, respectively, and with and the equilibrium constants for ECL2/3 region opening in the bound and unbound receptors, we have that , and by applying the mass-action law, we can write the ligand-bound fraction of the receptor at ligand concentration as follows:
where
is a coefficient that measures the allosteric coupling between ligand binding and loop dynamics.
If the dynamics of the ECL2/3 region do not depend on the presence of the ligand, then and . In this case, the ligand binding thermodynamics are not affected by ECL2/3 region gating, and the ligand binding affinity is , that is, the same as to the open receptor. If, however, the closed state of the ECL2/3 region is more stable in the ligand-bound complex, and . In this case, (i.e., the affinity of the ligand) and the stability of the active state are increased by ECL2/3 region closure. Accordingly, this model can capture the basic mechanism by which ECL2/3 region closure increases the binding affinity of a ligand.
To quantify the effect of Na+ and Mg2+ cations on ECL2/3 region gating, we assessed the coupling between the ECL2/3 loop region conformational dynamics and cation binding. For a given Mg2+ concentration [M], we characterized the dynamics between the Mg2+-bound state of the open ECL2/3 region and the Mg2+-free state of the open ECL2/3 region (O) as where is the equilibrium constant reflecting the affinity of Mg2+ for the open state, and indicates the MOP receptor state (unbound/inactive, bound/active). Similarly, for the closed state of the ECL2/3 region, . Indicating with the equilibrium constant between the open and closed states of the ECL2/3 region in the absence of Mg2+, at magnesium concentration , we have that the equilibrium between the ECL2/3 region open and closed states is as follows:
Using this expression for and , we obtain the dependence of the allosteric coefficient ρ on the Mg2+ concentration. Specifically, in the absence of magnesium, , whereas for high-magnesium concentrations , the allosteric coefficient is as follows:
The values of and (see Table S3) can be estimated from the occupancy of the extracellular binding sites in the closed and open states of the ECL2/3 region in the inactive MOP receptor trajectories, whereas the value of was obtained from the probability of the ECL2/3 region being in an open state in the same system. The values of , , and (see Table S3) were estimated from the corresponding fractions in the inactive and active MOP receptor trajectories. Values of and are reported in Table S4.
Pocket surface area and mouth of the pocket area estimations
To quantify the accessibility of the orthosteric binding site through open or closed states of the ECL2/3 region in the three simulated MOP receptors, we calculated two commonly used geometrical descriptors of binding sites, specifically their pocket surface area and mouth of the pocket area, using the Computed Atlas of Surface Topography of proteins (CASTp) web server (44). The server uses the atomic van der Waals radii of the atoms provided in a molecular topology to detect concave surfaces in a structure, which are labeled as pockets. The mouth of each pocket is described as the constriction plane between the concave surface and the exterior convex surface of the molecule. The areas of both the mouth and concave pocket surface were calculated here using the α-shape method (45) and a probe radius of 2.0 Å. Table S5 lists these calculated quantities for representative protein conformations of the three most populated macrosites for each of the three simulated MOP receptors.
Calculation of coinformation
To elucidate the molecular determinants of the positive allosteric modulation of the MOP receptor induced by the Mg2+ cation binding, we calculated the coinformation between receptor residue pairs and Mg2+ occupancy of macrosites identified in MD simulations within the ECL2/3 region of the MOP receptor. Results of GCMC-MD runs were not included because of limited protein sampling in the protocol as implemented. Specifically, we describe residue dynamics using the Cartesian coordinates of the center of mass of the residue heavy atoms , whereas Mg2+ binding was described using a binary variable representing the occupancy state of any of the Mg2+ macrosites in the ELC2/3 region, . The mutual information is a common quantity used to describe the long-range allosteric coupling between the dynamics of different regions of the receptor. In this work, we assessed the modulatory effect of Mg2+ binding by calculating the so-called coinformation quantity:
where is the mutual information between the two residues, and is the conditional mutual information between the residues, conditioned upon Mg2+ occupancy. This coinformation is the difference between the information shared by Ri and Rj for a given value of and the information shared by Ri and Rj irrespective of and thus measures the effect of on the amount of information shared between Ri and Rj. We note that negative values of the coinformation indicate that explains (some of) the observed correlation between Ri and Rj. Table S6 lists the normalized coinformation values derived from the following:
To reduce calculation time, trajectories were analyzed with a time step of 0.2 ns, and the mutual information and conditional mutual information values needed to calculate were estimated using the MDEntropy python library (46) and the k-nearest neighbor algorithm with five bins per Cartesian dimension.
Results and Discussion
To elucidate the atomic details of the mechanism by which Mg2+ binds and allosterically modulates the MOP receptor in the presence of Na+ ions, we carried out MD simulations of experimentally determined inactive and active structures of the receptor embedded in a lipid-water environment and in the presence of both cations. Unlike the inactive MOP receptor state, in which the highly conserved D2.50 residue is likely to be charged and bound to a Na+ cation, from previous simulation studies by us and others, we know that the D2.50 does not bind Na+ in the receptor’s active state, and its protonation state depends on its proximity to cations (23,47). Thus, we carried out MD simulations of one ligand-free inactive MOP receptor crystal structure corresponding to PDB: 4DKL (30) and two forms of the ligand-free active MOP receptor crystal structure corresponding to PDB: 5C1M (31), differing only in the protonation state of the D2.50 residue.
MD simulations were run at equal concentrations (0.15 M) of Na+ and Mg2+. Whereas the Na+ concentration is close to extracellular physiological conditions, the Mg2+ concentration is significantly higher than its normal value under the same conditions, but this choice was necessary to observe binding events of this divalent cation by standard MD simulations. Notably, the positive allosteric modulation of ligand-free receptors by Mg2+ cations can only be observed experimentally at high Mg2+ concentrations (0.1–0.5 M) as recently demonstrated for the adenosine A2A receptor (18).
To assess whether the reported MD simulations had missed important bound states of Mg2+ ions because of their slow desolvation kinetics, we supplemented these simulations with a GCMC-MD procedure that overcomes desolvation barriers by sampling Mg2+ insertion around the protein. As reported below, the Mg2+ coordination complexes identified by GCMC-MD mostly matched those identified by MD and exhibited similar affinities (see Table 1), instilling confidence in the MD simulation results. A notable difference is the observed binding of Mg2+ to the allosteric and orthosteric binding sites of the inactive MOP receptor in the GCMC-MD simulations, but not in the MD simulations. Another difference concerns Mg2+ binding to the orthosteric binding site of the active MOP receptor with neutral D2.50 in the GCMC-MD runs, an event that is also not seen in the MD simulations. A possible reason for this lack of agreement is the absence of Na+ ions in the GCMC-MD runs and their preferred occupancy of the orthosteric and allosteric sites in the MD simulations. However, one should not forget that interactions between metal ions and protein residues are notoriously hard to describe by MD simulations. Despite intense efforts (see (48) for a recent review), the reliability of these models is still under debate as evidence emerges that polarizable models or quantum calculations might be required to properly describe ion-binding events (49).
Table 1.
Magnesium Binding to the MOP Receptor
| Kd (MD) | Kd (GCMC) | |
|---|---|---|
| Inactive MOP Receptor | ||
| E310ECL3/D216ECL2 | 0.73 | 0.37 |
| D216ECL2/G1312.67 | 4.85 | 7.35 |
| D216/C217ECL2 | 0.73 | 1.00 |
| Allosteric site | – | 0.10 |
| Orthosteric site | – | 1.35 |
| Active MOP Receptor with Charged D2.50 | ||
| E310ECL3/D216ECL2 | 0.64 | 1.99 |
| D216ECL2/S214ECL2 | 0.85 | 0.21 |
| Orthosteric site | 0.56 | 0.39 |
| Allosteric site | – | 0.07 |
| Active MOP Receptor with Neutral D2.50 | ||
| E310ECL3/D216ECL2 | 4.85 | 1.35 |
| D216ECL2/G1312.67 | 0.85 | 0.79 |
| Q212ECL2/D216ECL2 | 0.92 | 1.21 |
| Orthosteric site | – | 0.22 |
| Allosteric site | – | 1.73 |
Estimates of Mg2+ binding affinity (in Molar units) to sites sampled by MD and GCMD-MD.
Mg2+ binds predominantly to the MOP receptor extracellular region and exhibits higher binding affinity for the active receptor conformation
To characterize likely binding sites of Mg2+ on the inactive or active MOP receptor (so-called macrosites) at physiological concentrations of Na+, we first identified microsites of single or multiple residues that were found to be forming direct interactions with Na+ and direct or water-mediated interactions with Mg2+ cations (see Methods for details). Given the large number of microsites obtained by this detailed classification, we grouped them into macrosites of cation binding at different regions of the receptor. The analysis of the occupancy probability of these macrosites (see Methods) revealed a small number of binding regions with significant marginal occupancy probability (>3%).
Figs. 1, 2, and 3 report the molecular and energetic details of all macrosites identified with cation binding affinity less than 5 M in MD simulations of ligand-free inactive MOP receptor, ligand-free active MOP receptor with charged D2.50, and ligand-free active MOP receptor with neutral D2.50, respectively. Values of the Na+ and Mg2+ binding affinities for these macrosites derived from occupancy probabilities in the MD or GCMC-MD simulation trajectories (see Methods for details) are listed in Tables 1 and 2 for Mg2+ and Na+ ions, respectively.
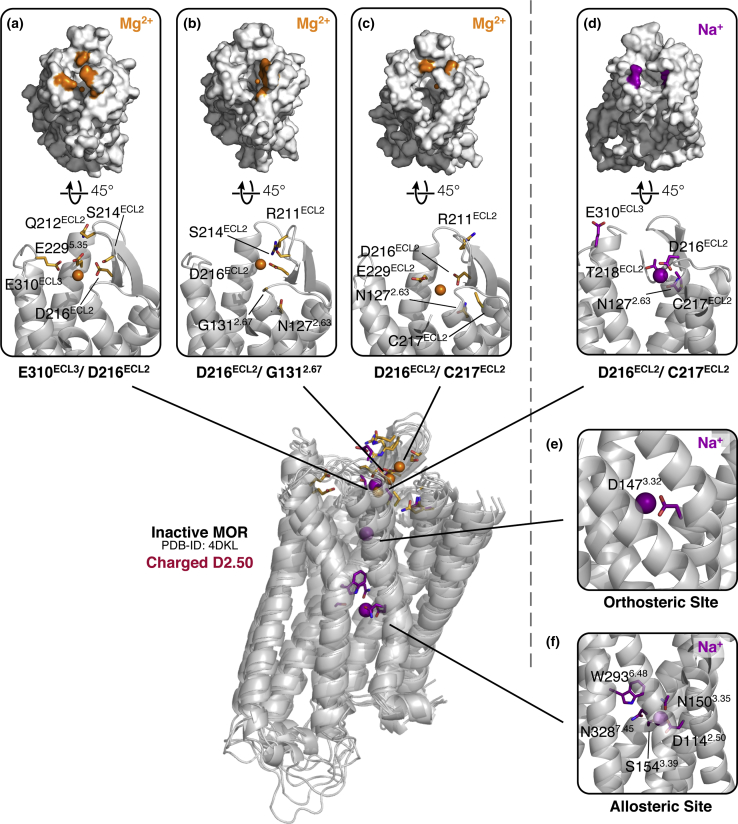
Figure 1.
Molecular and energetic details of all macrosites identified with cation binding affinity less than 8 M in simulations of ligand-free inactive MOP receptor. (a)–(c) and (d)–(f )show predicted binding sites for Mg2+ and Na+ ions, respectively. Representative residues of each cation binding macrosite (i.e., residues that are most frequently involved in Na+ and Mg2+ binding in microsites) are indicated in orange and purple colors, respectively, on the surface representations of the inactive MOP receptor. In the insets, the same residues are shown as sticks, whereas purple and orange spheres refer to Na+ and Mg2+ cations, respectively. Macrosites in the extracellular region of the MOP receptor are labeled by the residues that most frequently bind Na+ and Mg2+ cations in microsites. To see this figure in color, go online.
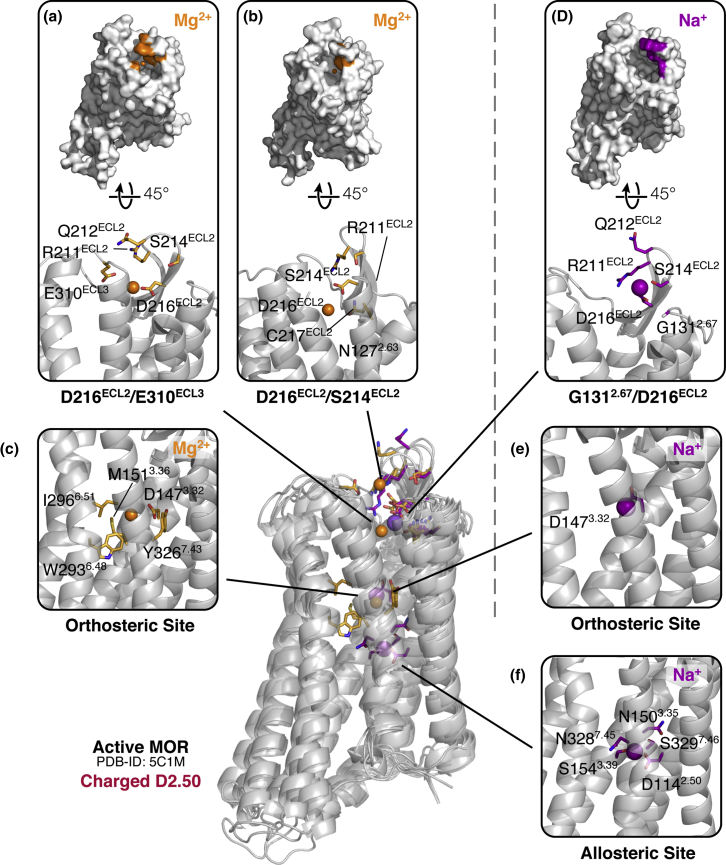
Figure 2.
Molecular and energetic details of all macrosites identified with cation binding affinity less than 8 M in simulations of the ligand-free active MOP receptor with a charged D2.50 residue. (a)–(c) and (d)–(f )show predicted binding sites for Mg2+ and Na+ ions, respectively. See the caption of Fig. 1 for details. To see this figure in color, go online.
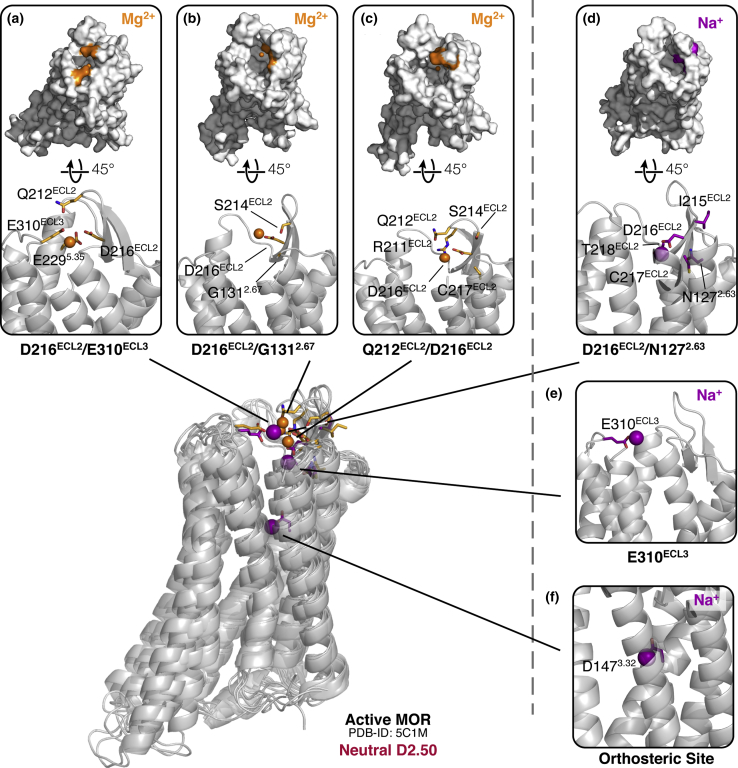
Figure 3.
Molecular and energetic details of all macrosites identified with cation binding affinity less than 8 M in simulations of the ligand-free active MOP receptor with a neutral D2.50 residue. (a)–(c) and (d)–(f )show predicted binding sites for Mg2+ and Na+ ions, respectively. See the caption of Fig. 1 for details. To see this figure in color, go online.
Table 2.
Sodium Binding to the MOP Receptor
| Kd (MD) | |
|---|---|
| Inactive | |
| D216ECL2/C127ECL2 | 0.50 |
| Orthosteric site | 0.60 |
| Allosteric site | 0.01 |
| Active Charged D2.50 | |
| G1312.67/D216ECL2 | 1.18 |
| Orthosteric site | 0.36 |
| Allosteric site | 1.29 |
| Active Neutral D2.50 | |
| D216ECL2/N1272.63 | 0.62 |
| E310ECL3 | 1.79 |
| Orthosteric site | 3.36 |
Estimates of Na+ binding affinities (in Molar units) to sites sampled by MD.
As expected, in the ligand-free inactive MOP receptor system (Fig. 1), the Na+ cation has the highest affinity for the allosteric binding site (in particular, residues D1142.50, N1503.35, S1543.39, N3287.45, and W2936.48) revealed by high-resolution crystal structures of various GPCRs. We calculate Kd = 0.01 M for Na+ at this site (Fig. 1 f), which agrees with our previously published calculations from simulations of the MOP receptor in the presence of Na+ only and their experimental validation (9). In the absence of ligands, sodium ions can also bind to the orthosteric binding site, establishing direct interactions with the conserved D1473.32 residue (Fig. 1 e). In addition to these binding sites within the transmembrane bundle, the Na+ cation was also found to bind at an extracellular site (specifically, residues N1272.63, D216ECL2, C217ECL2, T218ECL2, and E310ECL3; see Fig. 1 d) in the inactive MOP receptor system, albeit with the significantly lower affinity of 0.5 M. Unlike Na+, Mg2+ ions bound primarily at three slightly different sites on the extracellular region of the inactive MOP receptor system, which all had the D216ECL2 residue coordinating the cation (Fig. 1, a–c). The Mg2+ cation binding affinity for these sites was equally low, ranging from 0.7 to ∼5.0 M. As expected, in the presence of sodium ions, no binding of Mg2+ to the orthosteric and allosteric sites was observed. In GCMC simulations, which were carried out in the absence of Na+ ions, binding of Mg2+ was also observed to the orthosteric and allosteric binding sites of the inactive MOP receptor, where the ion established direct chelation with D1473.32 or D1142.50 residues. We note that in all GCMC-MD trajectories, Mg2+ never formed direct binding with residues in the extracellular region, except for a marginal fraction (<2% of frames) in the inactive MOP receptor, where direct contacts were established with E310ECL2. Not shown in Fig. 1 is a Mg2+ binding site observed in the vicinity of helix 8, which is likely an artifact of having run MD simulations with a zwitterionic C-terminus.
Although Na+ had a much higher affinity than Mg2+ for the inactive MOP receptor, this difference was reduced in the simulations of the active MOP receptor systems (see Figs. 2 and 3). For instance, the affinity of Na+ for the crystallographic allosteric site decreased by two orders of magnitude from ∼0.01 to 1.3 M in the ligand-free active MOP receptor system with a charged D2.50 (Fig. 2 f). This is consistent with the knowledge that this site is partially collapsed in active GPCR structures and not suitable to the same type of cation binding (19). Interestingly, the Na+ affinity for the extracellular region decreases, albeit less dramatically, from 0.5 to 1.2 M (Fig. 2 c). On the other hand, not only did the Mg2+ binding affinity for the extracellular loop regions slightly increase in the ligand-free active MOP receptor system with a charged D2.50 compared to the simulated inactive receptor system (from 0.7–5.0 M to 0.6–0.8 M; compare Fig. 1, a–c with Fig. 2, a and c), but the Mg2+ cation was also found to bind at the orthosteric ligand binding site defined by residue D1473.32 with a of 0.6 M, the highest affinity value for a macrosite for any ion in this system (see Fig. 2 c). Notably, although the orthosteric ligand binding site was equally accessible in the ligand-free inactive MOP receptor system (see Table S5), the Mg2+ cation did not bind at this site during MD simulation of the ligand-free inactive MOP receptor system. In contrast, GCMD-MD simulations carried out in the absence of Na+ did reveal binding of Mg2+ to both the orthosteric and the allosteric sites on the receptor. Notably, the Monte Carlo sampling revealed (data not shown) that the binding of Mg2+ to D1473.32 at the orthosteric site is not via direct interaction, which was the case for the inactive receptor, but through water-mediated contacts, in agreement with the MD simulation results.
Not surprisingly, the change of the D2.50 protonation state to neutral in the simulated active MOP receptor system had a significant impact on the binding affinity of both Na+ and Mg2+ cations for the ligand-free MOP receptor (Fig. 3) at macrosites within the TM bundle, therefore implying a relationship between cation binding and local electrostatics. In this system, both the Na+ affinity for the allosteric binding site defined by D2.50 and the Mg2+ affinity for the orthosteric ligand binding site defined by D1473.32 fell below the cutoff value of 5 M chosen for significance. GCMC-MD simulations of the active MOP receptor system with a neutral D2.50 did not yield binding of Mg2+ to the allosteric site, and the Mg2+ binding to the orthosteric site was reduced with respect to that observed in the active receptor with charged D2.50. Notably, the affinity values of Mg2+ for the extracellular loop region (ranging from 0.8 to 5.0 M; see Fig. 3, a–c) were closer to those recorded for the ligand-free inactive MOP receptor system than for the active receptor system with a charged D2.50, whereas Na+ binding affinity for the extracellular region dropped to 1.8 M (Fig. 3, d and e).
Mg2+ binding to the extracellular region of the MOP receptor promotes occlusion of the ligand-binding pocket
A unique feature of Mg2+ binding to the MOP receptor extracellular region, particularly negatively charged residues D216ECL2 and E310ECL3, is that it can produce a transient closure of the orthosteric ligand-binding pocket by bringing ECL 2 and ECL 3 closer together. These two extracellular loops effectively act as a “gate,” and when closer together, they can hinder access of the ligand to the pocket as well as its departure from it. This phenomenon appears to be a unique feature of the divalent Mg2+ cation because it was not observed when the monovalent Na+ cation was bound at this site. The surface representations of the MOP receptor systems in Figs. 1, 2, and 3 and the mouth and pocket surface area quantifications reported in Table S5 clearly show the different degree of accessibility of the MOP receptor orthosteric ligand-binding pocket when Na+ or Mg2+ is bound at the extracellular region of the receptor. Notably, visual inspection of the trajectories, as well as smaller values of the equilibrium constant HM (see Table S3), reveal that the Mg2+-induced ECL2/3 loop closure occurs with higher frequency in the simulated active MOP receptor conformations than in the inactive receptor system, suggesting a coupling between activation and ECL2/3 closure.
To quantify the effect of Mg2+-induced ECL2/3 closure on ligand binding and receptor activation, we calculated a coupling coefficient, as normally done to measure the allosteric modulation between different receptor degrees of freedom. Specifically, we considered a first approximation in which the agonist binding affinity is expressed as , where is the agonist binding affinity to the receptor when the ECL2/3 gate is “open,” and is an “allosteric coefficient” that takes into account the Mg2+occupancy of the extracellular binding sites in the closed and open states of the ECL2/3 region, thus capturing the effect of cation binding on ECL2/3 closure. The same coefficient ρ was used to express the constitutive activity ratio between active and inactive MOP receptors as R/ρ, where R is the active/inactive receptor ratio in the absence of ECL2/3 gating. Values of the ρ coefficient calculated for different Mg2+ concentrations are shown in Table S4 (see Table S3 for values of its components and Methods for equations). In the absence of allosteric modulation by the cation, ρ is expected to be equal to 1. On the other hand, if ρ < 1, the agonist binding affinity is predicted to increase, as is the active fraction of the receptor. Comparison between the inactive MOP receptor system and the active MOP receptor with neutral D2.50 yields , which suggests a 4-fold increase in agonist binding affinity in the presence of magnesium as well as a fourfold increase of the constitutively active fraction of the receptor (see Table S3). Similarly, comparing the inactive MOP receptor and the active MOP receptor with charged D2.50 yields a value of , corresponding to a fivefold change (see Table S4).
Na+ and Mg2+ compete for binding at ECL2 sites
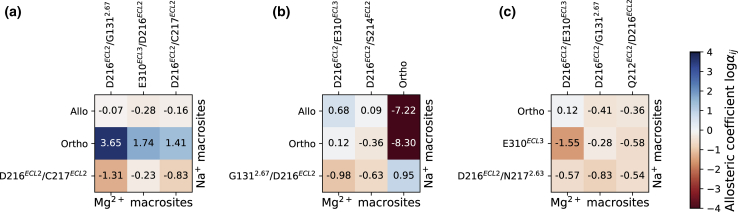
To address possible binding competition between Na+ and Mg2+ cations for specific sites on the MOP receptor, we calculated the allosteric modulation coefficient for the cation binding affinity for each of the three simulated MOP receptor systems. Negative values of this coefficient in the plots of Fig. 4 (red color) indicate a negative cooperativity, namely, that the binding of Na+ at site decreases the affinity of Mg2+ at site (and vice versa), whereas positive values (Fig. 4, blue) would suggest that binding of one ion increases the affinity of the other. As expected, most of the coupling coefficients in the plots of Fig. 4 are negative, suggesting that there is active competition for the available binding sites in all simulated MOP receptor systems. Specifically, in the inactive MOP receptor system (Fig. 4 a), Mg2+ competes (negative values of ) with Na+ for any of the extracellular binding sites. Notably, the coupling between the Na+ allosteric binding site and the Mg2+ E310ELC3/D216ECL2 site is also negative, albeit not very pronounced , suggesting a reduction of the Na+ affinity for its allosteric binding site in the presence of Mg2+ at the E310ELC3/D216ECL2 site in the inactive MOP receptor. Conversely, this analysis reveals a strong positive cooperativity (up to ) between the presence of Na+ at the orthosteric site and magnesium binding in the loop region.
Figure 4.
Allosteric coefficients for cooperative binding at Na+ and Mg2+ binding sites in the simulated MOP receptor systems. (a)–(c) refer to inactive, active with charged D2.50, and active with neutral D2.50 MOP receptor, respectively. The coefficients are reported as , with negative values (in red) corresponding to negative cooperativity and positive values (blue) corresponding to positive cooperativity of the cation binding affinity. To see this figure in color, go online.
In the active MOP receptor (Fig. 4, b and c), a negative coupling is also observed between all pairs of binding sites in the extracellular region of the receptor. As expected from simple electrostatic considerations and from the proximity of the orthosteric and allosteric sites, we observe a strong negative modulation of the binding of Mg2+ to the orthosteric ligand-binding pocket induced by Na+ binding at the D2.50 allosteric binding site in the active receptor with a charged D2.50 (Fig. 4 b). This high negative cooperativity appears to be the reason for the lack of Mg2+ binding at the orthosteric ligand binding site when an Na+ cation is present at its allosteric binding site and vice versa.
Cation binding modulates the information flow across the receptor
To further elucidate the molecular mechanism underlying Mg2+ positive allosteric modulation of the MOP receptor, we analyzed the impact of Mg2+ binding on the information flow across the receptor. Reasoning that receptor allosteric modulation can be captured by the mutual information between residues, we characterized the residues mediating the modulatory effect of the cation by calculating the normalized coinformation between pairs of receptor residues (i, j) and Mg2+ occupancy of macrosites within the ECL2/3 region of the MOP receptor (see Methods).
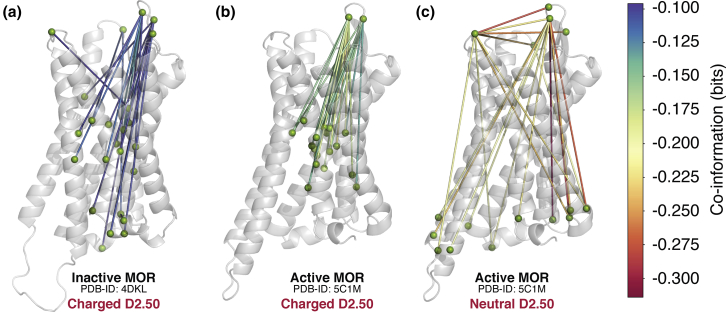
The receptor residue pairs whose correlation is maximally affected by Mg2+ binding (i.e., the top 20 pairs with the highest coinformation values) are listed in Table S6, and their location is shown in Fig. 5 for the three simulated MOP receptor systems. Accordingly, in the inactive MOP receptor system (Fig. 5 a), Mg2+ binding to the ECL2/3 region most dramatically affects the mutual information between specific residues in ECL2 (i.e., Y210, R211, I215, and S222), ECL3 (i.e., P309), TM2 (i.e., A1022.38, N1092.45, A1132.49), TM3 (D1473.32, Y1493.34, N1503.35, M1513.36, and T1533.38), intracellular loop (ICL) 2 (i.e., L176), TM5 (i.e., C2355.41), TM6 (i.e., V2886.43 and C2926.47), the intracellular end of TM7, flanking the NPxxY motif (L3247.41, C3307.47, L3317.48, L3397.56), ICL4 (i.e., E341), and H8 (i.e., N342 and K344).
Figure 5.
Location of the MOP receptor residues pairs whose correlation is maximally affected by Mg2+ binding in the three simulated receptor systems. (a)–(c) refer to inactive, active with charged D2.50, and active with neutral D2.50 MOP receptor, respectively. Lines between residues are colored according to their coinformation value, with the most negative values shown in red. To see this figure in color, go online.
Notably, coinformation values are much more negative in the simulated active MOP receptor systems and especially in the system with a neutral D2.50 (see Fig. 5 c; Table S6), reflecting a tighter coupling between magnesium occupancy of macrosites within the ECL2/3 loop region and the receptor dynamics in the active conformational state. In the active MOP receptor with a charged D2.50 residue, the most affected regions of the receptor by Mg2+ binding share many similarities with those of the inactive MOP receptor (compare Fig. 5, a and b), involving specific residues within ECL2 (Y210ECL2 and R211ECL2), TM2 (N1092.45), TM3 (N1503.35, M1513.36, and T1533.38), TM6 (C2926.47), and TM7 (L3247.41 and L3317.48). On the other hand, the coinformation observed in the active receptor with neutral D2.50 is strikingly different from that of the MOP receptor with charged D2.50 (compare Fig. 5, b and c). Notably, among the most affected receptor regions by cation binding in this system are ECL2/3 residue pairs, specifically P309ECL3 with K209ECL2, R211ECL2, Q212ECL2, or S222ECL2 as well as the intra-ECL2 pair R211ECL2-S222ECL2. As expected by a positive allosteric modulator, magnesium binding also modulates the information transfer from ECL2/3 residues (i.e., R211ECL2, S222ECL2, and P309ECL3) to the intracellular G protein-binding region of the MOP receptor, including the intracellular ends of TM1 (I931.57 and V941.58), TM3 (C1703.55), TM4 (P1814.39), TM5 (L2575.63 and K2605.66), and TM6 (D2726.27 and R2736.28) as well as ICL1 (M99), ICL2 (V173), and ICL4 (D340).
Taken together, these results support a mechanism by which Mg2+ binding to the MOP receptor active state with neutral D2.50 (i.e., the expected most probable protonation state for an active receptor) has a direct effect on the dynamics of the ECL2/3 region, which in turn is strongly coupled with the dynamics of the intracellular region of the receptor, whereas this coupling is less effective in the inactive receptor. Notably, a similar mechanism was recently proposed for a different GPCR subtype (18), suggesting a possible molecular paradigm of GPCR allosteric modulation by cations.
Conclusions
Our simulations provide atomic details of the molecular mechanism by which magnesium cations preferentially bind to the extracellular region of the MOP receptor in the presence of sodium and allosterically modulate the G protein-binding region of the receptor. Specifically, the results reported here support a mechanism by which Mg2+ cations, unlike Na+ cations, promote states of the active MOP receptor with ECL2 and ECL3 folding back over the extracellular opening of the receptor TM bundle, thus impairing agonist unbinding and possibly enhancing agonist binding affinity. Mutations of residues involved in Mg2+ binding to the ECL2/3 region (e.g., the acidic residues E310ELC3 and D216ECL2) as well as mutations of residues identified as mediators of the cation allosteric effect (e.g., R211ECL2, Q212ECL2, and P309ECL3) are predicted to affect its positive allosteric modulation of the MOP receptor.
Although Na+ and Mg2+ cations actively compete for binding to extracellular sites of the MOP receptor, a larger number of binding sites are available to Mg2+ in this region. Thus, despite the low (Molar) range of affinities for these sites, we would predict that Mg2+ cations bind to the extracellular region of the MOP receptor even at lower concentrations. Notably, in the presence of sodium, Mg2+ was never observed to bind to the Na+ allosteric site, most likely because of its double positive charge and an unfavorable balance between desolvation and binding. Mg2+ could, however, bind at the orthosteric ligand binding site with a significantly higher affinity than sodium. Furthermore, we observed a strong negative cooperativity between Mg2+ binding at the orthosteric site and Na+ binding at the allosteric site, suggesting that Mg2+ binding could reduce the fraction of bound Na+, thus reducing its inhibitory effect.
Author Contributions
X.H. and S.R. carried out the MD and GCMC-MD simulations, respectively. All authors analyzed the results and wrote the manuscript.
Acknowledgments
The authors thank Dr. Alexander D. MacKerell Jr. and members of his research team (in particular, Drs. Sunhwan Jo, Abhishek Kognole, and Himanshu Goel) at the University of Maryland for generously providing the software used in this work to perform GCMC-MD sampling and for assistance in its use.
This work was supported by National Institutes of Health grant DA045473. Computations were run on resources available through the Scientific Computing facility at the Icahn School of Medicine at Mount Sinai and the Extreme Science and Engineering Discovery Environment under MCB080077, which is supported by National Science Foundation grant number ACI-1053575.
Editor: Vasanthi Jayaraman.
Footnotes
Xiaohu Hu and Davide Provasi contributed equally to this work.
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.10.007.
Supporting Material
References
- 1.Pasternak G.W. Opiate pharmacology and relief of pain. J. Clin. Oncol. 2014;32:1655–1661. doi: 10.1200/JCO.2013.53.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scholl L., Seth P., Baldwin G. Drug and opioid-involved overdose deaths – United States, 2013–2017. MMWR Morb. Mortal Wkly. Rep. 2018;67:1419–1427. doi: 10.15585/mmwr.mm675152e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kieffer B.L. Opioids: first lessons from knockout mice. Trends Pharmacol. Sci. 1999;20:19–26. doi: 10.1016/s0165-6147(98)01279-6. [DOI] [PubMed] [Google Scholar]
- 4.Latorraca N.R., Venkatakrishnan A.J., Dror R.O. GPCR dynamics: structures in motion. Chem. Rev. 2017;117:139–155. doi: 10.1021/acs.chemrev.6b00177. [DOI] [PubMed] [Google Scholar]
- 5.Bohn L.M., Lefkowitz R.J., Lin F.T. Enhanced morphine analgesia in mice lacking beta-arrestin 2. Science. 1999;286:2495–2498. doi: 10.1126/science.286.5449.2495. [DOI] [PubMed] [Google Scholar]
- 6.Maguma H.T., Dewey W.L., Akbarali H.I. Differences in the characteristics of tolerance to μ-opioid receptor agonists in the colon from wild type and β-arrestin2 knockout mice. Eur. J. Pharmacol. 2012;685:133–140. doi: 10.1016/j.ejphar.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raehal K.M., Walker J.K., Bohn L.M. Morphine side effects in beta-arrestin 2 knockout mice. J. Pharmacol. Exp. Ther. 2005;314:1195–1201. doi: 10.1124/jpet.105.087254. [DOI] [PubMed] [Google Scholar]
- 8.Pert C.B., Pasternak G., Snyder S.H. Opiate agonists and antagonists discriminated by receptor binding in brain. Science. 1973;182:1359–1361. doi: 10.1126/science.182.4119.1359. [DOI] [PubMed] [Google Scholar]
- 9.Hu X., Wang Y., Filizola M. Kinetic and thermodynamic insights into sodium ion translocation through the μ-opioid receptor from molecular dynamics and machine learning analysis. PLoS Comput. Biol. 2019;15:e1006689. doi: 10.1371/journal.pcbi.1006689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shang Y., LeRouzic V., Filizola M. Mechanistic insights into the allosteric modulation of opioid receptors by sodium ions. Biochemistry. 2014;53:5140–5149. doi: 10.1021/bi5006915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez F.D., Bardaji E., Traynor J.R. Differential effects of Mg2+ and other divalent cations on the binding of tritiated opioid ligands. J. Neurochem. 1992;59:467–472. doi: 10.1111/j.1471-4159.1992.tb09393.x. [DOI] [PubMed] [Google Scholar]
- 12.Burgmer U., Schulz U., Mohr K. Interaction of Mg2+ with the allosteric site of muscarinic M2 receptors. Naunyn Schmiedebergs Arch. Pharmacol. 1998;357:363–370. doi: 10.1007/pl00005180. [DOI] [PubMed] [Google Scholar]
- 13.Johansson B., Parkinson F.E., Fredholm B.B. Effects of mono- and divalent ions on the binding of the adenosine analogue CGS 21680 to adenosine A2 receptors in rat striatum. Biochem. Pharmacol. 1992;44:2365–2370. doi: 10.1016/0006-2952(92)90681-8. [DOI] [PubMed] [Google Scholar]
- 14.Mazzoni M.R., Martini C., Lucacchini A. Regulation of agonist binding to A2A adenosine receptors: effects of guanine nucleotides (GDP[S] and GTP[S]) and Mg2+ ion. Biochim. Biophys. Acta. 1993;1220:76–84. doi: 10.1016/0167-4889(93)90100-4. [DOI] [PubMed] [Google Scholar]
- 15.Pi M., Faber P., Quarles L.D. Identification of a novel extracellular cation-sensing G-protein-coupled receptor. J. Biol. Chem. 2005;280:40201–40209. doi: 10.1074/jbc.M505186200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silve C., Petrel C., Ruat M. Delineating a Ca2+ binding pocket within the venus flytrap module of the human calcium-sensing receptor. J. Biol. Chem. 2005;280:37917–37923. doi: 10.1074/jbc.M506263200. [DOI] [PubMed] [Google Scholar]
- 17.Urwyler S. Allosteric modulation of family C G-protein-coupled receptors: from molecular insights to therapeutic perspectives. Pharmacol. Rev. 2011;63:59–126. doi: 10.1124/pr.109.002501. [DOI] [PubMed] [Google Scholar]
- 18.Ye L., Neale C., Prosser R.S. Mechanistic insights into allosteric regulation of the A2A adenosine G protein-coupled receptor by physiological cations. Nat. Commun. 2018;9:1372. doi: 10.1038/s41467-018-03314-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu W., Chun E., Stevens R.C. Structural basis for allosteric regulation of GPCRs by sodium ions. Science. 2012;337:232–236. doi: 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller-Gallacher J.L., Nehmé R., Tate C.G. The 2.1 Å resolution structure of cyanopindolol-bound β1-adrenoceptor identifies an intramembrane Na+ ion that stabilises the ligand-free receptor. PLoS One. 2014;9:e92727. doi: 10.1371/journal.pone.0092727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang C., Srinivasan Y., Kobilka B.K. High-resolution crystal structure of human protease-activated receptor 1. Nature. 2012;492:387–392. doi: 10.1038/nature11701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fenalti G., Giguere P.M., Stevens R.C. Molecular control of δ-opioid receptor signalling. Nature. 2014;506:191–196. doi: 10.1038/nature12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vickery O.N., Carvalheda C.A., Zachariae U. Intracellular transfer of Na+ in an active-state G-protein-coupled receptor. Structure. 2018;26:171–180.e2. doi: 10.1016/j.str.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vickery O.N., Machtens J.P., Zachariae U. Structural mechanisms of voltage sensing in G protein-coupled receptors. Structure. 2016;24:997–1007. doi: 10.1016/j.str.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lakkaraju S.K., Raman E.P., MacKerell A.D., Jr. Sampling of organic solutes in aqueous and heterogeneous environments using oscillating excess chemical potentials in grand canonical-like Monte Carlo-molecular dynamics simulations. J. Chem. Theory Comput. 2014;10:2281–2290. doi: 10.1021/ct500201y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lakkaraju S.K., Yu W., MacKerell A.D., Jr. Mapping functional group free energy patterns at protein occluded sites: nuclear receptors and G-protein coupled receptors. J. Chem. Inf. Model. 2015;55:700–708. doi: 10.1021/ci500729k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lemkul J.A., Lakkaraju S.K., MacKerell A.D., Jr. Characterization of Mg2+ distributions around RNA in solution. ACS Omega. 2016;1:680–688. doi: 10.1021/acsomega.6b00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bujalska-Zadrożny M., Tatarkiewicz J., Naruszewicz M. Magnesium enhances opioid-induced analgesia - what we have learnt in the past decades? Eur. J. Pharm. Sci. 2017;99:113–127. doi: 10.1016/j.ejps.2016.11.020. [DOI] [PubMed] [Google Scholar]
- 29.Jo S., Kim T., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 30.Manglik A., Kruse A.C., Granier S. Crystal structure of the μ-opioid receptor bound to a morphinan antagonist. Nature. 2012;485:321–326. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang W., Manglik A., Kobilka B.K. Structural insights into μ-opioid receptor activation. Nature. 2015;524:315–321. doi: 10.1038/nature14886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Šali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 33.van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sengupta D., Prasanna X., Chattopadhyay A. Exploring GPCR-lipid interactions by molecular dynamics simulations: excitements, challenges, and the way forward. J. Phys. Chem. B. 2018;122:5727–5737. doi: 10.1021/acs.jpcb.8b01657. [DOI] [PubMed] [Google Scholar]
- 35.Best R.B., Zhu X., Mackerell A.D., Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abraham M.J., Murtola T., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 37.Nosé S. A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 1984;52:255–268. [Google Scholar]
- 38.Berendsen H.J.C., Postma J.P.M., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 39.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 40.Darden T., York D., Pedersen L. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 41.Allnér O., Nilsson L., Villa A. Magnesium ion-water coordination and exchange in biomolecular simulations. J. Chem. Theory Comput. 2012;8:1493–1502. doi: 10.1021/ct3000734. [DOI] [PubMed] [Google Scholar]
- 42.Bleuzen A., Pittet P.A., Merbach A.E. Water exchange on magnesium(II) in aqueous solution: a variable temperature and pressure 17O NMR study. Magn. Reson. Chem. 1997;35:765–773. [Google Scholar]
- 43.Sun D., Lakkaraju S.K., MacKerell A.D., Jr. Determination of ionic hydration free energies with grand canonical Monte Carlo/molecular dynamics simulations in explicit water. J. Chem. Theory Comput. 2018;14:5290–5302. doi: 10.1021/acs.jctc.8b00604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tian W., Chen C., Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 2018;46:W363–W367. doi: 10.1093/nar/gky473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edelsbrunner H., Mücke E.P. Three-dimensional alpha shapes. ACM Trans. Graph. 1994;13:43–72. [Google Scholar]
- 46.Hernández C.X., Pande V.S. MDEntropy: information-theoretic analyses for molecular dynamics. J. Open Source Softw. 2017;2:427. [Google Scholar]
- 47.Ranganathan A., Dror R.O., Carlsson J. Insights into the role of Asp79(2.50) in β2 adrenergic receptor activation from molecular dynamics simulations. Biochemistry. 2014;53:7283–7296. doi: 10.1021/bi5008723. [DOI] [PubMed] [Google Scholar]
- 48.Li P., Merz K.M., Jr. Metal ion modeling using classical mechanics. Chem. Rev. 2017;117:1564–1686. doi: 10.1021/acs.chemrev.6b00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jing Z., Liu C., Ren P. Many-body effect determines the selectivity for Ca2+ and Mg2+ in proteins. Proc. Natl. Acad. Sci. USA. 2018;115:E7495–E7501. doi: 10.1073/pnas.1805049115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.