Abstract
The increasing prevalence of adult and adolescent obesity and its associated risk of colorectal cancer reinforces the urgent need to elucidate the underlying mechanisms contributing to the promotion of colon cancer in obese individuals. Adiponectin is an adipose tissue-derived adipokine, whose levels are reduced during obesity. Both epidemiological and preclinical data indicate that adiponectin suppresses colon tumorigenesis. We have previously demonstrated that both adiponectin and AdipoRon, a small-molecule adiponectin receptor agonist, suppress colon cancer risk in part by reducing the number of Lgr5+ stem cells in mouse colonic organoids. However, the mechanism by which the adiponectin signaling pathway attenuates colon cancer risk remains to be addressed. Here, we have hypothesized that adiponectin signaling supports colonic stem cell maintenance through modulation of the biophysical properties of the plasma membrane (PM). Specifically, we investigated the effects of adiponectin receptor activation by AdipoRon on the biophysical perturbations linked to the attenuation of Wnt-driven signaling and cell proliferation as determined by LEF luciferase reporter assay and colonic organoid proliferation, respectively. Using physicochemical sensitive dyes, Di-4-ANEPPDHQ and C-laurdan, we demonstrated that AdipoRon decreased the rigidity of the colonic cell PM. The decrease in membrane rigidity was associated with a reduction in PM free cholesterol levels and the intracellular accumulation of free cholesterol in lysosomes. These results suggest that adiponectin signaling plays a role in modulating cellular cholesterol homeostasis, PM biophysical properties, and Wnt-driven signaling. These findings are noteworthy because they may in part explain how obesity drives colon cancer progression.
Significance
The biophysical properties of the plasma membrane (PM) determined by the composition and compartmentalization of lipid-ordered domains and their associated proteins has been recently recognized to be relevant to the fields of cell biology, cancer biology, and cellular biophysics. Alterations in the organization of PM lipids through the activation of the adiponectin signaling pathway indicates a potential therapeutic regulation of aberrant Wnt signaling in the highly susceptible obese population. This study draws a novel, to our knowledge, link between AdipoRon, alteration of PM cholesterol homeostasis, and canonical Wnt signaling. The broad findings of this work support the development of plasma-membrane-targeted therapies in cancer.
Introduction
The plasma membrane (PM) is a dynamic cellular structure composed of a myriad of lipids and proteins (1). Free cholesterol is an important component of the PM that influences the dynamic lipid-driven assemblies of specialized membrane environments in the lateral dimension also referred to as nanodomains or ordered membrane domains (2, 3). These membrane components are exquisitely organized through various forms of interactions (4, 5). The nomenclature used to describe these interactions varies (6); however, what is known is that perturbations to this organization can lead to disruption of cellular events (7, 8). Thus, the PM functions as the crucial interface between cells and environmental signals influencing stemness, cell division, and intracellular membrane trafficking.
Wnt/β-catenin signaling originates at the PM via the initial binding of canonical Wnt ligands to Wnt receptors (9, 10, 11). At the base of colonic crypts, Lgr5+ stem cells are highly dependent on Wnt signaling as a key determinant of stem cell homeostasis, proliferation, and differentiation (12, 13, 14). Wnt receptor Frizzled and coreceptor low-density lipoprotein receptor-related protein 6 (LRP6) and its close relative LRP5, are distributed throughout the PM. However, binding of canonical Wnt ligands occurs in ordered PM domains (15). Specifically, phosphorylation of LRP6, which is necessary for activation of Wnt/β-catenin signaling, occurs in liquid-ordered PM domains (16, 17). In the absence of Wnt pathway activating ligands, cytoplasmic β-catenin protein is degraded by the “destruction complex” composed of the scaffolding protein Axin, the tumor suppressor adenomatous polyposis coli protein, casein kinase 1, and glycogen synthase kinase 3 (GSK3). Casein kinase 1 and GSK3 sequentially phosphorylate the amino terminal region of β-catenin, resulting in β-catenin recognition by an E3 ubiquitin ligase subunit, β-Trcp, and subsequent β-catenin ubiquitination and proteasomal degradation (18, 19). Typically, dysregulation of Wnt/β-catenin signaling leads to congenital defects/disorders and promotes cancer risk (9, 20).
Colorectal cancer is the second leading cause of cancer-related mortality in the United States (21), and up to 20% of all cancer-related deaths may be attributed to obesity (22). The relative risk of colon cancer increases proportionally to body mass index (BMI) (23). An abundant body of epidemiological evidence suggests that the risk of colon cancer is strongly associated with increasing BMI (24), whereas an increase in BMI is inversely associated with circulating adiponectin levels in men and women (25). Adiponectin is an ∼30-kDa monomeric adipokine secreted from adipose tissue into the bloodstream at circulating levels of 3–30 μg/mL (26, 27). Interestingly, human epidemiological and preclinical data suggest that low levels of adiponectin are correlated with increased risk of colon cancer (28, 29, 30, 31). Supporting evidence for the protective role of adiponectin against colon cancer has been reported both in mice and cell culture studies (31, 32, 33, 34, 35, 36, 37, 38). Thus, there is an urgent need to elucidate the link between adipose-derived adipokines and colon cancer risk.
AdipoRon is a synthetic, orally active small-molecule adiponectin receptor agonist that ameliorates obesity-related diseases such as diabetes (39) and cardiovascular complications (40). Furthermore, AdipoRon suppresses pancreatic tumor growth (41) and has been proposed as a possible colorectal cancer prevention strategy (42). Recently, we demonstrated that AdipoRon reduces the number of colonic Lgr5+ stem cells in a mouse colonic organoid model (43); however, the mechanism underlying this effect is not known. Based on these findings, we hypothesized that AdipoRon may serve as a modifier of Wnt signaling.
Here, we demonstrate that AdipoRon suppresses stem cell proliferation through attenuation of canonical Wnt and R-Spondin-1-dependent Wnt signaling. Because Wnt signaling originates from rigid domains in the PM, we probed the effects of AdipoRon on the biophysical properties of the PM. Using multiple cell membrane models, for example, isolated giant PM vesicles (GPMVs), two-dimensional cell lines and three-dimensional (3D) colonic organoids, we provide evidence that AdipoRon reduces PM rigidity. This decrease in PM rigidity is associated with an influx of extracellular cholesterol and the depletion and subsequent migration of the PM pool of free cholesterol intracellularly. The results of this influx are 1) a large increase in the number of lysosomal-like vesicles, which are enriched in free cholesterol, 2) the upregulation of cholesterol biosynthesis and esterification pathways, and 3) the suppression of Wnt signaling. Collectively, our results suggest that adiponectin signaling plays a role in modulating cellular cholesterol homeostasis, PM biophysical properties, and Wnt-driven signaling in the intestine. These findings are noteworthy because they may in part explain how obesity drives colon cancer progression.
Materials and Methods
Cell culture
Conditionally immortalized young adult mouse colonic (YAMC) epithelial cells (Research Resource Identifier: CVCL_6E40) were originally obtained from R.H. Whitehead of the Ludwig Cancer Institute (Melbourne, Australia). YAMC cells (passages 12–20) were cultured under permissive conditions; 33°C and 5% CO2 in Roswell Park Memorial Institute (RPMI) 1640 medium, no glutamine (21870076; Gibco, Carlsbad, CA), supplemented with 5% fetal bovine serum (FBS; SH300084.03; HyClone, Logan, UT), 2 mM GlutaMAX (35050061; Gibco), 5 μg/mL insulin, 5 μg/mL transferrin, 5 ng/mL selenious acid (354351; Corning, Corning, NY), and 5 IU/mL of murine interferon-γ (11276905001; Roche, Basel, Switzerland). YAMC cells were authenticated (07/24/15) by STR profiling (CellCheck Plus) by IDEXX BioResearch (Westbrook, Maine). DKOB8 human colorectal adenocarcinoma cells were acquired (09/19/03) from Dr. Patrick Casey (Duke University) who maintained them for Dr. Gideon Bollag of Onyx Pharmaceuticals (San Francisco, California) (44). DKOB8 cells (passages 7–12) were maintained at 37°C and 5% CO2 in Dulbecco’s Modified Eagle Medium (DMEM), high glucose, GlutaMAX medium (10569010; Gibco) supplemented with 5% FBS. All cell lines used tested negative for mycoplasma bacteria (05/09/18) as assessed by a universal mycoplasma detection kit (30-1012K; American Type Culture Collection, Manassas (ATCC), VA). Select cultures were treated for 24 or 48 h with 5, 10, 20, or 50 μM AdipoRon (SML0998; Sigma-Aldrich, St. Louis, MO) or 0.1% dimethylsulfoxide (DMSO, 4-X; ATCC). Select cultures were treated for 24 h with 5 or 10 μg/mL of recombinant mouse full-length adiponectin (ALX-522-059-C050; Enzo Life Sciences, Farmingdale, NY). In addition, select cultures were incubated with complete media containing 10 mM methyl-β-cyclodextrin (MβCD, C4555; Sigma-Aldrich) for 30 min to deplete cholesterol from the PM.
Wnt reporter assay
Leading Light Wnt Reporter 3T3 mouse fibroblasts were obtained (02/05/19) from Enzo Life Sciences. For culturing purposes, 3T3 cells (passages 1–20) were maintained at 37°C and 5% CO2 in DMEM without phenol red, containing 4.5 g/mL glucose, and 4 mM glutamine supplemented with Leading Light Wnt Reporter Growth Medium Concentrate (ENZ-60003-0001; Enzo Life Sciences). The luciferase activity assay was conducted with some modification to the manufacturer’s protocol. Cells were pretreated with AdipoRon or DMSO before adding Wnt activators: recombinant murine Wnt3a (315-20; PeproTech, Rocky Hill, NJ), recombinant murine R-Spondin-1 (315-32; PeproTech), LiCl (Wnt-LICL-0500; Enzo Life Sciences), or CHIR99021 (04-0004; Stemgent, Cambridge, MA). Protein concentration was measured by Pierce BCA protein assay kit (23225; Thermo Fisher Scientific, Waltham, MA). Luciferase luminescence and protein absorbance was measured on a CLARIOstar microplate reader (BMG LABTECH, Cary NC).
Organoid culture
Isolated mouse colonic crypts from wild-type or Lgr5-EGFP-IRES-creERT2 (Lgr5-GFP) mice were counted and embedded in Matrigel (356231; Corning) at three to six crypts per microliter and cultured in crypt culture medium containing Advanced DMEM/F12 (12634010; Gibco) supplemented with 2 mM GlutaMAX, 10 mM HEPES (H0887; Sigma-Aldrich), 50 ng/mL EGF (PMG8045; Life Technologies, Carlsbad, CA), 0.2 μM LDN-193189 (C5361-2s; Cellagen Technology, San Diego, CA), 10% R-spondin-1 conditioned medium, 1 μM N-acetyl-l-cysteine (A9165; Sigma-Aldrich), 1X N2 (17502048; Life Technologies), 1X B27 (17504044; Life Technologies), and 50% Wnt conditioned medium as described previously (45). Isolated intestinal crypts were centrifuged for 3 min at 500 × g, resuspended in the appropriate volume of Matrigel (30 μL) containing 10 μM Y-27632 (Y0503; Sigma-Aldrich) and 1 μM Jagged-1 (AS-61298; AnaSpec, Fremont CA) in flat-bottom 24-well plates. After Matrigel polymerization, cells were overlaid with 500 μL of crypt culture medium supplemented with 10 μM Y-27632, 1 μM Jagged-1, and 2.5 μM CHIR99021. Y-27632, Jagged-1, and CHIR99021 were withdrawn from crypt culture medium 2 days after plating. The crypt media was changed every 2 days. For AdipoRon-related treatments, 0.1% DMSO or 10 μM AdipoRon was added to cultures for 48 h. To measure organoid metabolic activity, CellTiter-Blue Cell Viability Assay (G8080; Promega, Madison, WI) was used according to manufacturer’s instructions. Fluorescence was measured on a CLARIOstar microplate reader.
Crypt isolation
Mouse colons were removed, washed with ice-cold Dulbecco’s phosphate buffered saline (DPBS; 14190144; Gibco) without calcium and magnesium, everted on a disposable mouse gavage needle (FTP-20-38; Instech Laboratories, Plymouth Meeting, PA), and incubated in 15 mM EDTA (ED4SS; Sigma-Aldrich) in DPBS at 37°C for 35 min as previously described (46). Subsequently, after transfer to chilled DPBS, crypts were mechanically separated from the connective tissue by rigorous vortexing. Crypts were embedded in Matrigel and overlaid with crypt culture media as previously described (46).
GPMV generation and isolation
GPMV generation was performed as described previously (47). In brief, plated cells were washed with the GPMV buffer (10 mM HEPES, 150 mM NaCl, 2 mM CaCl2 (pH 7.4)) twice, then GPMV buffer containing vesiculation agents (25 mM paraformaldehyde (PFA; 15713-S; Electron Microscopy Sciences, Hatfield, PA), 2 mM dithiothreitol (DTT; D0632; Sigma)) was added for at least 1 h at 37°C. The GPMV-rich solution was then transferred to a 1.7-mL tube and centrifuged at 100 × g for 10 min to pellet cell debris. After centrifugation, the GPMV supernatant was transferred to a new 1.7-mL tube. GPMVs were stored at 4°C for 1–2 days without visible degradation or used immediately for staining.
Membrane order measurement via confocal microscopy and image-based flow cytometry
Cells were stained with 5 μM Di-4-ANEPPDHQ (Di4; D36802; Invitrogen, Carlsbad, CA) for membrane order determination as previously described (48, 49, 50, 51, 52, 53). In brief, cells were seeded into an eight-chamber cell imaging cover glass (0030742036; Eppendorf, Hamburg, Germany; or C8-1.5H-N; Cellvis, Mountain View, CA), gently washed with DPBS, then Di4 in live cell imaging solution (A14291DJ; Invitrogen) was added and cells immediately imaged to avoid dye internalization. For experiments involving organoids, organoids were gently removed from the Matrigel by the addition of cold DPBS. Collected organoids were gently spun down at 100 × g for 3 min at 4°C and resuspended in cold live cell imaging solution before the addition of Di4 and transfered to an eight-chamber cell imaging cover glass. Imaging experiments were conducted using a Leica DMi8 confocal microscope (Wetzlar, Germany). YAMC and DKOB8 cells were imaged with a 1.15 NA 40× plan apochromat oil objective, whereas organoids were imaged with a 0.30 NA 10× plan apochromat air objective at room temperature. Laser light at 488 nm was used to excite Di4, and emission wavelengths were collected in two channels representing ordered (O: 500–580 nm) and disordered (D: 620–700 nm). GPMVs were stained with 5 μM C-laurdan (CL; T0001-SFC; SFCprobes, South Korea) and imaged with a 1.3 NA 40× plan apochromat oil objective at room temperature. Laser light at 405 nm was used to excite CL, and emission wavelengths were collected in two channels representing ordered (O: 415–455 nm) and disordered (D: 490–530 nm). Generalized polarization (GP) was calculated using the equation below:
Here, G is a calibration factor determined using a solution of 500 μM Di4 or 50 μM CL in DMSO following a procedure described previously (52). A reference value of −0.85 and 0.207 was used for Di4 and CL, respectively. The same laser power and detector settings were used for every experiment. Image processing was conducted using Fiji/ImageJ (National Institutes of Health) software, with a GP plugin and a custom-built macro. Briefly, images were converted to eight-bit tiffs, combined into RBG images. A threshold was applied to exclude background pixels and converted into GP images. The average GP was determined from the region of interest of cells. Additionally, GPMVs were stained with 1 μM Di4 and imaged via image-based flow cytometry via Amnis FlowSight (Luminex, Austin, TX) (53). Laser light at 488 nm was used to excite Di4, and emission wavelengths were collected in two preset channels representing ordered (O: 480–560 nm) and disordered (D: 640–745 nm). Because there is no way to acquire a calibration image, the G factor was omitted, and GP was calculated as stated above using Amnis IDEAS software.
Lysotracker staining
DKOB8 cells were grown in normal 5% FBS or in 5% lipoprotein-depleted FBS (880100; Kalen Biomedical, Germantown, Maryland) before staining with LysoTracker Red DND-99 (L7528; Invitrogen). After treatment and before fixation, 50 nM LysoTracker was utilized to stain cells for 30 min at 37°C as suggested by the manufacturer. After 30 min, cells were washed twice with DBPS and fixed. Cells were imaged on a Leica DMi8 confocal microscope.
Filipin III staining
Cells were fixed in 4% PFA for 15 min at room temperature. Subsequently, cells were washed with DPBS, and 50 μg/mL Filipin III (F4767; Sigma-Aldrich) was added for 45 min at room temperature in the dark as previously described (54). After 45 min, excess Filipin III was removed by washing with DPBS twice. GPMVs were also stained with 50 μg/mL Filipin III for 45 min at room temperature in the dark. Cells were imaged on a Leica DMi8 confocal microscope when costained with LysoTracker or on a Keyence BZ-X710 all-in-one fluorescence microscope (Itasca, IL) when imaging Filipin III alone. Quantitative image analysis was conducted using BZ-X Analyzer Hybrid Cell Count software (Keyence).
Phalloidin staining
After fixation, cells were washed twice with DPBS and permeabilized with 0.1% Triton X-100 (X-100-500ML; Sigma-Aldrich) in DPBS for 5 min. After permeabilization, cells were washed twice with DPBS and stained with 1 unit or 5 μL (stock: 200 units/mL) Alexa Fluor 488 Phalloidin (A12379; Invitrogen) in DPBS containing 1% Blocker bovine serum albumin (BSA; 37525; Thermo Fisher Scientific) per coverslip for 20 min according to the manufacturer’s instructions. After 20 min, cells were washed twice with DPBS and imaged on a Leica DMi8 confocal microscope.
Annexin V and SYTOX staining
An Annexin V-Alexa Fluor 647 conjugate assay (A23204; Invitrogen) was used to measure cell apoptosis. SYTOX Green (S7020; Invitrogen) was used to identify dead cells. Cells were seeded in a glass-like polymer coverslip bottom 24-well plate (P24-1.5P; Cellvis). 24 h after seeding, cells were treated with AdipoRon or vehicle control (0.1% DMSO). Annexin V staining was performed according to the manufacturer’s protocol. Fluorescence was measured directly from the plate on a Keyence BZ-X710 all-in-one fluorescence microscope. Quantitative image analysis was conducted using BZ-X Analyzer Hybrid Cell Count software (Keyence).
GPMV PM and actin staining
YAMC cells were seeded into an eight-chamber cell imaging cover glass. After 24 h, cells were incubated with 1:1000 CellMask Green Plasma Membrane Stain (C37608; Invitrogen) and 1 μM SiR-actin (CY-SC001; Cytoskeleton, Denver, CO) in live cell imaging solution for 30 min at 37°C. After staining, cells were rinsed with GPMV buffer and incubated with GPMV vesiculation buffer for ∼1 h before imaging with the Leica DMi8 confocal microscope.
EdU staining
Colonic mouse organoids from Lgr5-EGFP-IRES-creERT2 mice were incubated with 10 μM EdU for 1 h and harvested from Matrigel. Organoids were dissociated with 0.25% trypsin-EDTA (25200056; Gibco) and filtered through a 30-μm cell strainer (04-0042-2316; Sysmex Partec, Kobe, Japan). Subsequently, the Click-iT Plus EdU Alexa Fluor 647 Cell Proliferation Kit (Invitrogen, C10640) was utilized to determine the S-phase cell population, and nuclei were stained with Hoechst 33342 (H3570; Invitrogen) according to the manufacturer’s instructions. Cell cycle profiling was assessed by running cells through the Amnis FlowSight imaging system.
RNA-seq and ingenuity pathway analysis
DKOB8 cells were treated with 20 μM AdipoRon or vehicle control (0.1% DMSO) for 24 h before RNA was extracted using the Zymo Quick-RNA Miniprep Kit (11-327; Genesee Scientific, San Diego, CA). RNA quality was assessed using an Agilent bioanalyzer (Santa Clara, CA). Only RNA quality of RIN ≥ 8 was used. QuantSeq 3′ mRNA-seq library prep lot for Illumina from Lexogen (Vienna, Austria) was used. Sequencing was performed on a NextSeq 500 (Illumina, San Diego, CA). After the removal of all genes with counts per million values less than 1, publicly available R software, EdgeR, was used to identify differentially expressed genes (55). Ingenuity Pathway Analysis (Qiagen, Hilden, Germany) was conducted on all differently expressed genes (56). This in silico analysis is based on prior knowledge of expected effects between transcriptional regulators and their target genes stored in the Ingenuity Knowledge Base and the observed effects as seen in the differentially expressed gene data.
Amplex-based cholesterol quantification
Cells were grown in a T-175 flask (0030712129; Eppendorf), and GPMVs were isolated as described above. After pelleting cell debris, the GPMV-rich supernatant was then transferred into several 1.7-mL tubes and centrifuged at 20,000 × g for 1 h at 4°C to pellet the GPMVs. Lipids were extracted from GPMVs by the Folch (2:1, v/v, chloroform/methanol) method. Briefly, 1 mL methanol was added and transferred between tubes, then transferred to a glass vial. Subsequently, 2 mL chloroform and 0.6 mL cold KCl (0.1 M) was added to the GPMV lysate, vortexed, and centrifuged to generate two layers. The lower phase was collected and set aside, then an additional 2 mL chloroform was added to the original mixture, and the extraction was repeated. The lower phase was once again collected and added to the first collection. The combined lower phases were then dried under nitrogen and resuspended in 1 mL Folch. 600 and 300 μL of the sample was used for cholesterol and phosphate assays, respectively. Free and esterified cholesterol concentrations were determined using an Amplex Red Cholesterol Assay Kit (A12216; Invitrogen) according to the manufacturer’s instructions. The cholesterol concentration was normalized to the phospholipid concentration using an inorganic phosphate assay (57). Briefly, 300 μL of the lipid extract was mixed with 30 μL 10% Mg(NO3)2-6H2O in MeOH (w/v) in a disposable borosilicate glass tube and dried with N2. Using a Bunsen burner in a fume hood, individual tubes were flamed for ∼30 s until the solution had dried and formed a white residue. The tubes were allowed to cool at room temperature and then 100 μL of 1 N HCl was added to dissolve the precipitate. An additional 600 μL of ddH20 was added, and the sample was vortexed for 2 min to completely dissolve the precipitate. Inorganic phosphate was determined using 200 μL of this solution in triplicate using the Phosphate Colorimetric Assay Kit (K410; BioVision, Milpitas, CA), according to the manufacturer’s instructions.
Statistics
Two-tailed Student’s t-tests or one-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test were used to assess statistical significance of the differences between means across experimental treatments. All data are presented as mean ± standard error (SE), and all analyses were conducted using Prism 7 statistical software (GraphPad Software).
Results and Discussion
Lgr5+ stem cells are considered the cells of origin of intestinal cancer (58). We previously established that AdipoRon, like adiponectin, reduces the number of Lgr5+ stem cells in mouse colonic organoids (43), supporting its use as a possible colorectal cancer chemoprevention strategy (42). However, the mechanism underlying AdipoRon’s effect on stemness has not been investigated. Thus, we determined the effect of AdipoRon on canonical stem cell signaling and the downstream mechanism of action.
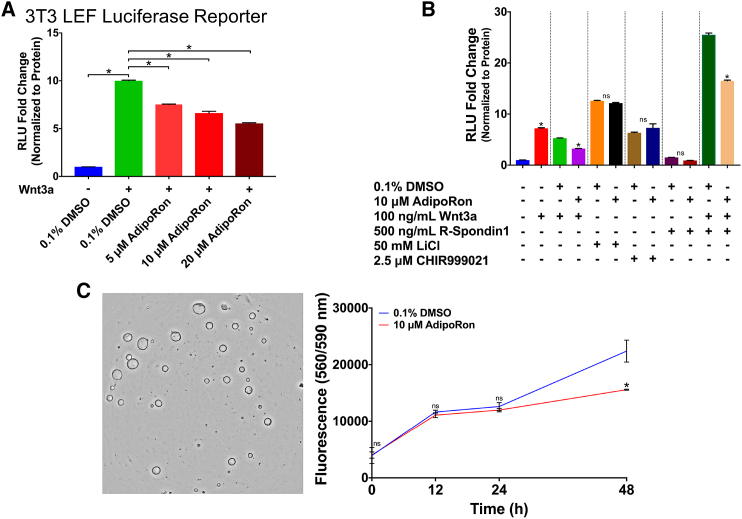
AdipoRon reduces Wnt-driven signaling
Wnt/β-catenin signaling plays a critical role in maintaining intestinal stem cells. To assess the ability of AdipoRon to modulate Wnt/β-catenin signaling, we utilized a mouse fibroblast (Leading Light 3T3) cell line stably expressing an LEF luciferase reporter vector. Cells treated with physiologically relevant levels (5–20 μM) of AdipoRon (39) for 24 h dose dependently reduced the Wnt3a-stimulated increase in reporter activity (Fig. 1 A). To investigate the causative link between AdipoRon and canonical Wnt/β-catenin signaling, we used GSK3β inhibitors, LiCl and CHIR99021, to activate Wnt/β-catenin signaling downstream of Wnt3a coreceptor complex formation at the level of cytosolic destruction complex inactivation. AdipoRon had no effect on LiCl- or CHIR99021-mediated Wnt/β-catenin signaling (Fig. 1 B). These results indicate that AdipoRon inhibits Wnt signaling upstream of the formation of the destruction complex. Because the Lgr5 receptor is essential for the binding of R-Spondin-1 and Rnf43/Znrf3 and potentiates Wnt signaling by allowing Wnt3a-Frizzled/LRP6 receptor complexes to persist on the PM (59), we subsequently determined the effect of AdipoRon on R-Spondin-1 enhanced Wnt signaling. Interestingly, cotreatment of cells with R-Spondin-1 and Wnt3a generated a threefold higher response than Wnt3a alone, whereas AdipoRon suppressed the ability of R-Spondin-1 to enhance Wnt signaling (Fig. 1 B). Furthermore, AdipoRon had no effect on downstream GSK3β-mediated modulation of Wnt signaling (Fig. 1 B). These results demonstrate that AdipoRon attenuates both canonical Wnt- and R-Spondin-1-dependent Wnt signaling, both of which originate at the PM cell surface.
Figure 1.
AdipoRon suppresses Wnt-driven signaling. (A) Quantification of luciferase reporter activity from mouse fibroblasts (Leading Light 3T3) cells stably expressing an LEF luciferase reporter vestor. Cells were pretreated with 0.1% DMSO or indicated doses of AdipoRon for 12 h before stimulation, with 100 ng/mL of recombinant murine Wnt3a for 12 h in the presence of DMSO or AdipoRon (n = 3 wells per treatment). (B) Quantification of luciferase reporter activity. Mouse fibroblasts were pretreated with 0.1% DMSO or 10 μM AdipoRon for 12 h before stimulation with Wnt pathway activators (100 ng/mL Wnt3a, 500 ng/mL R-Spondin-1, 50 mM LiCl, or 2.5 μM CHIR99021) for 12 h in the presence of DMSO or AdipoRon (n = 3 wells per treatment). (C) Murine colonic organoid metabolic activity assessed by CellTiter-Blue Cell Viability Assay (n = 3 wells per treatment per time point). Unless otherwise indicated, data are mean ± SE; statistical significance between treatments (∗p < 0.01) was determined using (A) one-way ANOVA with Dunnett’s multiple comparisons test or (B and C) an unpaired t-test. To see this figure in color, go online.
To investigate the physiological importance of AdipoRon-mediated suppression of Wnt signaling, we used mouse colonic organoids whose growth is highly dependent on Wnt signaling. After a 48 h incubation period, AdipoRon reduced organoid metabolic activity as compared to the control (Fig. 1 C). To address if AdipoRon specifically impacts stem cell proliferation, we performed an EdU-based cell proliferation assay on organoids derived from Lgr5-GFP mice. AdipoRon treatment induced a ∼33 and ∼51% reduction of proliferating total cells and stem cells, respectively (Fig. S1).
The attenuation of Wnt-dependent signaling and stem-cell-dependent proliferation by AdipoRon is in agreement with our previous studies (43) yet contradicts a previous report conducted in mouse embryonic fibroblast in which adiponectin, but not AdipoRon, reduced Wnt/β-catenin target gene expression (60). The timing of AdipoRon treatment in these studies may explain the apparent disparate results: AdipoRon treatment was conducted for a total of 24 h in our study versus 2 h in the aforementioned study. Furthermore, high-molecular-weight forms of adiponectin also bind PAQR3 (61) and T-cadherin; however, the impact of their interaction on cell signaling is unclear (62). Collectively, these results demonstrate that AdipoRon attenuates both Wnt-dependent signaling and stem-cell-dependent phenotypes.
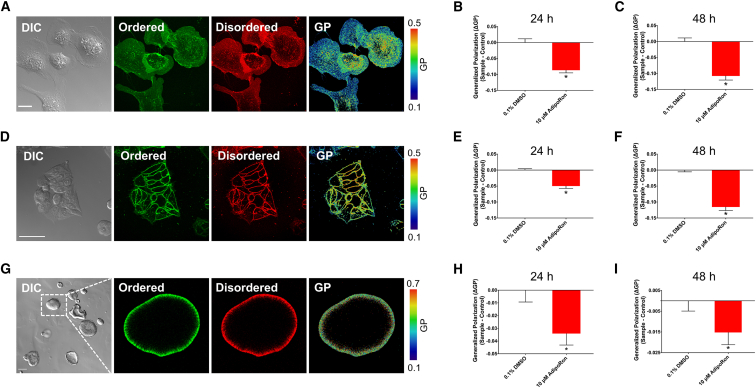
PM biophysical properties are altered by AdipoRon
The binding of Wnt3a to its coreceptors occurs in compartmentalized PM ordered domains (15). These ordered domains are referred to as “lipid rafts” and are rigid (highly ordered) as compared to fluid “nonraft” domains. To explore the effects of AdipoRon on membrane rigidity, we performed ratiometric confocal imaging of the polarity sensitive dye Di-4-ANEPPDHQ (Di4) (52, 53). Di4 was selected over other dyes, for example, Laurdan, because Di4 exhibits slower internalization kinetics in live cells and therefore provides a more representative measure of PM-specific organization (63, 64). Furthermore, we imaged the cells immediately after the addition of the dye to prevent its eventual internalization (64).
Our initial experiments were conducted with YAMC cells because this nontransformed cell model faithfully recapitulates the in vivo effects of dietary lipids (65, 66, 67) and allows for lipidomic and proteomic studies under well-controlled conditions (68). Cells treated with AdipoRon (10 μM) for 24 or 48 h exhibited a reduction in PM rigidity, as indicated by a reduction of Δ GP (Fig. 2, A–C). We verified that these biophysical effects were not specific to murine cells by performing similar experiments using DKOB8 human colorectal adenocarcinoma cells (Fig. 2, D–F). Because cells grown in a two-dimensional environment do not always recapitulate effects seen in 3D environments, we recapitulated these effects using mouse colonic 3D organoids (Fig. 2, G–I). Importantly, treatments that reduced organoid metabolic activity and proliferation (Figs. 1 C and S1) also reduced organoid PM rigidity (Fig. 2, H and I). Di4 is a leaflet-selective dye, which specifically reports on the biophysical properties of the outer leaflet when used as described above (69); therefore, it is critical to rule out the contribution of apoptosis-driven outer and inner leaflet lipid scrambling from the observations (70). Apoptotic lipid scrambling was detected using Annexin V, which binds PS exposed on the exoplasmic leaflet. We ruled out the contribution of apoptosis-driven lipid scrambling because AdipoRon only increased Annexin V staining at a high (50 μM) dose (Fig. S2). Stimulation of cells with Wnt3a causes the translocation of free cholesterol from the outer to the inner leaflet (71), which reduces the rigidity of the outer leaflet (15). Therefore, reduced activation of the Wnt pathway itself may alter cholesterol leaflet asymmetry and impact membrane order. Further work is required to determine the specific contribution of Wnt-mediated biophysical interactions to AdipoRon’s observed effects. We next performed a set of complementary experiments to verify that the observed reduction in membrane rigidity was not an artifact of stimulation of the adiponectin receptors by a synthetic ligand. In comparison to AdipoRon, physiologically relevant doses of adiponectin (26, 27), the natural ligand for the adiponectin receptors, produced similar effects on membrane rigidity (Fig. S3).
Figure 2.
Plasma membrane (PM) biophysical properties are altered by AdipoRon. (A) Representative confocal images of young adult mouse colonic (YAMC) epithelial cell stained with Di-4-ANEPPDHQ: DIC, ordered, disordered, and generalized polarization (GP). (B) Quantification of cell membrane rigidity ΔGP (sample − control) values. Cells were treated with 0.1% DMSO or 10 μM AdipoRon for 24 h (n = 8–16 fields of view per treatment). (C) ΔGP values of cells treated for 48 h (n = 8–16 fields of view per treatment). (D) DKOB8 cells, (E) 24 h (n = 5–10 fields of view per treatment), and (F) 48 h (n = 7–10 fields of view per treatment). (G) Mouse colonic organoids, (H) 24 h (n = 30–62 organoids per treatment), and (I) 48 h (n = 73–91 organoids per treatment). Unless otherwise indicated, data are mean ± SE; statistical significance between treatments (∗p < 0.01) was determined using an unpaired t-test. Scale bars, (A) 20 μm, (D) 50 μm, and (G) 100 μm. To see this figure in color, go online.
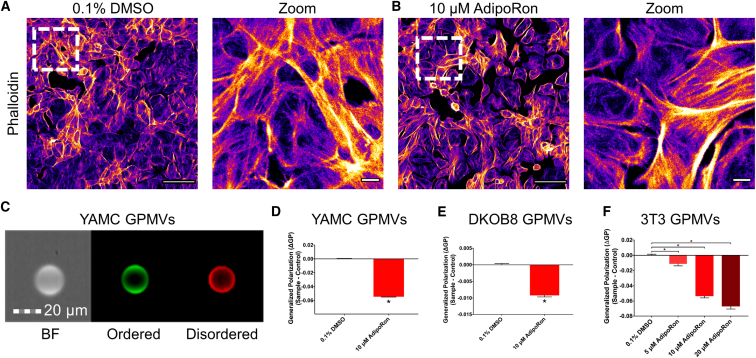
The biophysical properties of the PM are mediated by the combination of lipid-lipid and lipid-protein interactions, such as phospholipid-cholesterol and phospholipid-cytoskeleton, respectively (72, 73). Therefore, we investigated whether AdipoRon-mediated changes to PM order are direct (lipid-lipid) or indirect (protein-lipid) in nature. We initially addressed this question by performing a qualitative assessment of cytoskeletal structure using confocal imaging of cellular filamentous actin labeled with phalloidin. Treatment of YAMC cells with AdipoRon had no obvious effect on cytoskeletal structure (Fig. 3, A and B). To further assess the PM-specific effects of AdipoRon, we complemented our live cell experiments with studies using a PM model system consisting of cytoskeletal free membrane blebs termed GPMVs (47). GPMVs are microscopic (∼5–15 μm) PM spheres harvested from live cells after chemical treatment (74). This model has been used to probe PM biological processes by minimizing the number of cellular variables (e.g., cytoskeleton), thus decreasing experimental complexity, while retaining the functionality of the PM (75). The F-actin binding probe SiR-actin and a green PM stain were used to assess the cytoskeletal characteristics of GPMVs. Under basal conditions, the fluorescent signal from GPMVs generated from YAMC cells labeled with a PM stain displayed sharp outlines along the GPMV border and very little signal from SiR-actin (Fig. S4). These results indicate that GPMVs derived from YAMC cells are largely devoid of a structured cytoskeleton. GPMVs generated from YAMC, DKOB8, and 3T3 cells incubated with AdipoRon (0–20 μM) were subsequently assessed for membrane order using image-based flow cytometry (Amnis FlowSight) (53, 76). GPMVs derived from all cell types exposed to AdipoRon displayed reduced membrane order (Fig. 3, C–F). This effect was consistent with previous observations in live YAMC and DKOB8 whole cells (Fig. 2, A–F). Because the GP value determined by Di4 can be influenced by the experimental conditions including laser power, camera gain, and the choices of filters used (64), we verified that the results obtained using image-based flow cytometry on GPMVs were comparable to those produced in live cells (Fig. S5, A–C). Furthermore, to rule out probe-specific effects (63, 77), we conducted a set of experiments on GPMVs labeled with a complementary polarity sensitive probe, C-laurdan (78, 79) (Fig. S6).
Figure 3.
The effect of AdipoRon on PM rigidity is independent of cytoskeletal influences. YAMC cells were treated with (A) 0.1% DMSO or (B) 10 μM AdipoRon for 48 h, fixed in 4% PFA, permeabilized and stained for F-actin (phalloidin-Alexa Fluor 488). (C) A representative FlowSight image of GPMV generated from YAMC cells stained with Di-4-ANEPPDHQ: brightfield (BF), ordered, and disordered. (D) ΔGP values of YAMC GPMVs. Cells were treated for 24 h before GPMV generation (n = 5,797–24,448 GPMVs per treatment). (E) ΔGP values of DKOB8 GPMVs. Cells were treated for 24 h before GPMV generation (n = 5,797–24,448 GPMVs per treatment). (F) ΔGP values of 3T3 GPMVs. Cells were treated for 24 h before GPMV generation (n = 367–1,868 GPMVs per treatment). Unless otherwise indicated, data are mean ± SE; statistical significance between treatments (∗p < 0.05, ∗∗p < 0.01) was determined using an (D and E) unpaired t-test or (F) one-way ANOVA with Dunnett’s multiple comparisons test. Scale bars, (A and B) 100 μm, 10 μm (zoom), and (C) 20 μm. To see this figure in color, go online.
The reduction of membrane rigidity may explain the observed decreases in Wnt signaling because these domains are required for the formation of the Wnt signaling complex (15, 80). Future work is required to determine if intracellular trafficking (81) or the nanoscale PM localization (82, 83) of relevant Wnt receptors is impacted by AdipoRon. Furthermore, the adiponectin receptors were shown to be capable of remodeling PM composition and rigidity in that knockdown of the adiponectin receptors in a variety of mammalian cells increased the rigidity of the PM through increased lipid saturation (84). This is highly relevant because an increase in PM rigidity and lipid saturation through lipid remodeling has been linked to tumorigenesis (85, 86). Therefore, follow-up experiments are needed to determine the impact of AdipoRon on the lipid composition and unsaturation index of the PM. Overall, these findings underscore the ability of AdipoRon to induce a robust reduction of PM rigidity independent of cytoskeletal influence.
AdipoRon alters cholesterol trafficking
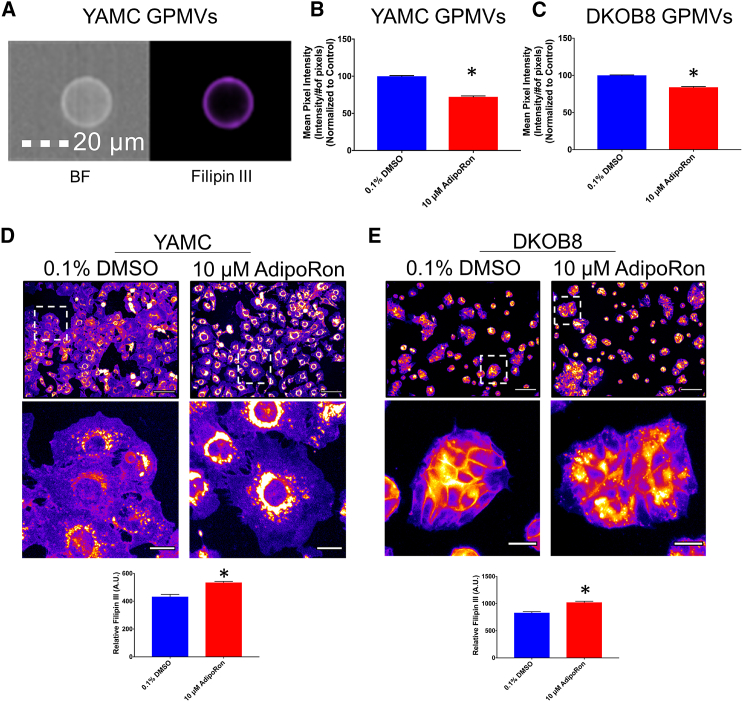
We next investigated the mechanism by which AdipoRon reduces PM order. Free cholesterol plays a major role in regulating PM rigidity (87, 88). To assess the role of free cholesterol involvement in membrane order in our model systems, we utilized the cholesterol-depleting agent MβCD (3, 89) and Filipin III, which specifically binds free cholesterol and not the esterified storage form of cholesterol (90, 91). As expected, MβCD reduced the levels of free cholesterol (up to 50%), subsequently decreasing membrane order (Fig. S7). To assess the effect of AdipoRon on PM free cholesterol, we performed image-based flow cytometry on Filipin-III-stained GPMVs (76). Filipin III intensity of GPMVs generated from AdipoRon-treated YAMC and DKOB8 cells was reduced (Fig. 4, A–C). In addition to cholesterol, Filipin III can bind other molecules (92). Therefore, we used a complementary Amplex-Red-based method to quantify the levels of free cholesterol in isolated GPMVs. Similar to our Filipin-III-based observations, AdipoRon reduced the level of free cholesterol (Fig. S8). These data support the hypothesis that AdipoRon reduces membrane rigidity by decreasing PM free cholesterol levels.
Figure 4.
AdipoRon alters cholesterol trafficking. (A) A representative image of GPMV derived from YAMC cells and stained with Filipin III is shown. Filipin III intensity values of GPMVs derived from (B) YAMC and (C) DKOB8 cells treated with 0.1% DMSO or 10 μM AdipoRon for 24 h before GPMV generation (n = 1126–7910 GPMVs per treatment) are shown. (D) YAMC and (E) DKOB8 cells were treated with 0.1% DMSO or 10 μM AdipoRon for 24 h. Cells were then fixed in 4% PFA for 15 min, followed by labeling of free cholesterol with 50 μg/mL Filipin III for 45 min at room temperature in the dark (n = 9 fields of view per treatment). Unless otherwise indicated, data are mean ± SE; statistical significance between treatments (∗p < 0.01) was determined using an unpaired t-test. Scale bars, (A) 20 μm, (D and E) 100, and 20 μm (zoom). To see this figure in color, go online.
Because cholesterol is not degraded in mammalian cells and is readily shuttled between different cellular compartments (93), the reduction of membrane rigidity could be explained by transport of cholesterol out of the PM to an intracellular compartment. Thus, we labeled whole cells with Filipin III to monitor cholesterol trafficking. Interestingly, AdipoRon treatment (10 μM) of YAMC and DKOB8 cells resulted in an overall increase in intracellular Filipin III staining (Fig. 4, D and E). The observed increase in intracellular free cholesterol was unexpected because the PM is typically its primary location (∼90%) in the cell (94, 95), whereas the remaining ∼10% associated with the endocytic pathway (96, 97). It is important to note that the accumulation of intracellular cholesterol can also be found in an esterified form and stored along with other neutral lipids (98), which cannot be detected by Filipin III (99, 100).
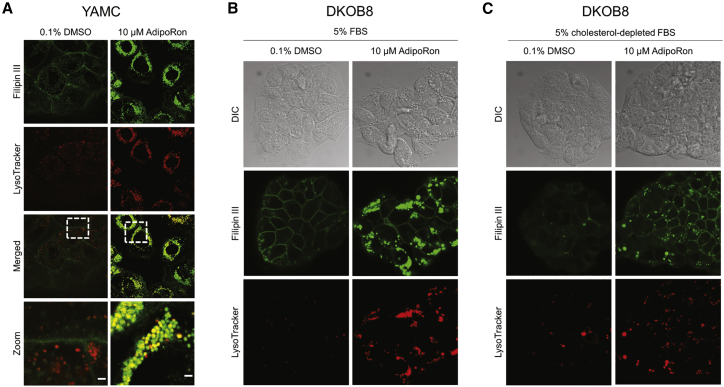
The majority of Filipin III staining, that is, free cholesterol accumulation, after AdipoRon exposure was localized to intracellular vesicle-like structures, resembling a Niemann-Pick type C (NPC) disease phenotype (101). In NPC disease, accumulated intracellular free cholesterol colocalizes with lysosomes (102, 103). To determine if AdipoRon (10 μM) produced similar effects, we performed colabeling studies with Filipin III and LysoTracker. LysoTracker intensity was dramatically increased in cells treated with AdipoRon (Fig. 5 A). Furthermore, LysoTracker signal strongly colocalized with Filipin III staining (Fig. 5 A). These data indicate the AdipoRon treatment induced the formation of free cholesterol enriched lysosomes.
Figure 5.
AdipoRon reroutes PM and extracellular cholesterol into lysosomes. (A) YAMC cells were treated with 0.1% DMSO or 10 μM AdipoRon for 48 h before staining lysosomes with 50 nM LysoTracker Red DND-99 for 30 min before fixing with 4% PFA for 15 min. After fixation, free cholesterol was stained with 50 μg/mL Filipin III for 45 min at room temperature in the dark. Zoom scale bar, 2 μm. DKOB8 cells were treated with 0.1% DMSO or 10 μM AdipoRon for 24 h in either (B) 5% FBS or (C) 5% cholesterol-depleted FBS then stained and fixed as described above. To see this figure in color, go online.
Cholesterol accumulation can be a result of upregulated synthesis or altered uptake/export. To explore which route AdipoRon influences, we conducted a series of experiments using lipoprotein-depleted FBS. Lipoprotein-depleted FBS contains 35 times less cholesterol than standard FBS according to the manufacturer. Under these conditions, cholesterol accumulation would primarily be a result of synthesis and not uptake. The use of lipoprotein-depleted FBS drastically attenuated the ability of AdipoRon to accumulate intracellular free cholesterol and reduced the amount of lysosomal vesicles versus normal FBS (Fig. 5, B and C). These data indicate that under normal growth conditions, AdipoRon increased the uptake of extracellular cholesterol and redirected the pool of PM free cholesterol into intracellular lysosomal-like vesicles. Interestingly, even under lipoprotein-depleted FBS conditions, AdipoRon treatment increased the levels of intracellular free cholesterol and lysosomes although not to the extent as normal FBS (Fig. 5, B and C). We then used RNA-seq to investigate differential gene expression in AdipoRon-treated DKOB8 cells. The Ingenuity Pathway Analysis was used to assign differentially expressed genes to functional networks. Pathways involving cholesterol biosynthesis were identified as the top hits (Fig. S9; Tables S1 and S2). Interestingly, the induction of cholesterol synthesis seems counterintuitive for a cell that is presumably overloaded with cholesterol. We interpret this as the cell losing its ability to accurately sense intracellular cholesterol. Furthermore, ACAT2, a critical mediator of cholesterol esterification, was upregulated (Table S2), which may be a response of the cell to reduce the pool of free cholesterol (Fig. S8). Further work is needed to assess the contribution of cholesterol synthesis to AdipoRon-induced intracellular free cholesterol accumulation.
Finally, the intracellular accumulation of free cholesterol has been proposed as a mechanism by which chemotherapeutic drugs, for example, itraconazole, attenuates Wnt signaling (104). Because cells treated with itraconazole show a similar cholesterol phenotype to AdipoRon and NPC1 cells, it is possible that itraconazole’s effects are mediated by a decrease in PM cholesterol. Interestingly, Wnt signaling is also attenuated in NPC1 cells (15); however, PM rigidity is increased (105). This highlights the need to determine if the effects of AdipoRon are solely mediated by a reduction in PM free cholesterol or are in part driven by intracellular free cholesterol accumulation. Collectively, these results demonstrate that AdipoRon reduces the PM-specific pool of free cholesterol while increasing the intracellular accumulation of free cholesterol into lysosomal-like vesicles.
Conclusion
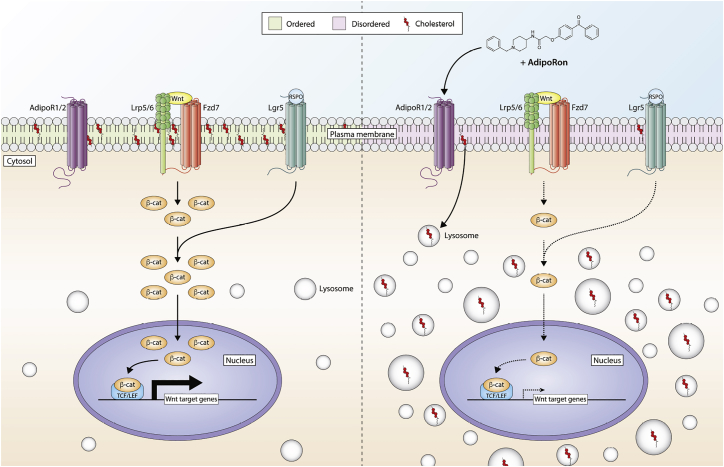
In this study, we characterized the effect of AdipoRon on Wnt signaling and PM biophysical properties. We show that AdipoRon treatment attenuates canonical and R-Spondin-1 enhanced Wnt signaling, both of which are important for maintaining stemness. Through the use of quantitative fluorescent microscopy techniques, we demonstrate that AdipoRon reduces PM rigidity in multiple cell types. The reduction in membrane order was caused by a reduction in PM levels of free cholesterol. This was the result of a redirection of PM free cholesterol and extracellular cholesterol into intracellular lysosomal-like vesicles (Fig. 6).
Figure 6.
Summary diagram highlighting the effect of AdipoRon on Wnt signaling. AdipoRon treatment attenuates canonical and R-Spondin-1 enhanced Wnt signaling by redirecting plasma membrane (PM) free cholesterol to intracellular pools, thereby decreasing PM rigidity. To see this figure in color, go online.
The complex relationship between obesity and cancer progression is not fully understood; however, our data support the hypothesis that adiponectin signaling plays an important role by influencing Wnt signaling through maintenance of membrane fluidity (31). Therefore, we postulate that obese individuals, with low levels of circulating adiponectin, will exhibit an increased cell membrane rigidity and enhanced Wnt signaling phenotype, making them susceptible to tumorigenesis. This is in part supported by observations that erythrocytes from obese individuals have more rigid membranes (106, 107), which suggests an opportunity to normalize these changes by PM-targeted countermeasures (53, 108, 109). Collectively, our novel, to our knowledge, findings pave the way for future experiments exploring the link between obesity, PM rigidity, Wnt signaling, and colon tumorigenesis.
Author Contributions
M.L.S., N.R.F., D.N.M., and R.S.C. designed the studies and wrote the manuscript. M.L.S., N.R.F., R.C., and R.C.W. performed the experimental work.
Acknowledgments
This work was supported by the Allen Endowed Chair in Nutrition & Chronic Disease Prevention and the National Institutes of Health (R35-CA197707 and P30-ES029067). N.R.F. is supported by the Texas A&M University Regulatory Science in Environmental Health and Toxicology training grant (T32-ES026568) and is a former recipient of a predoctoral fellowship in pharmacology/toxicology from the Pharmaceutical Research and Manufacturers of America Foundation. M.L.S. is a recipient of the National Science Foundation Texas A&M University System Louis Stokes Alliance for Minority Participation Bridge to the Doctorate Fellowship (HRD-1612776). Rachel Choate is supported by the National Science Foundation Research Experiences for Undergraduates Site Summer Undergraduate Research Program in Biochemistry (NSF DBI-1358941).
Editor: Ilya Levental.
Footnotes
Michael L. Salinas and Natividad R. Fuentes contributed equally to this work.
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.09.009.
Supporting Material
References
- 1.Barrera N.P., Zhou M., Robinson C.V. The role of lipids in defining membrane protein interactions: insights from mass spectrometry. Trends Cell Biol. 2013;23:1–8. doi: 10.1016/j.tcb.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 2.Simons K., Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 3.Sezgin E., Levental I., Eggeling C. The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 2017;18:361–374. doi: 10.1038/nrm.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia-Parajo M.F., Cambi A., Jacobson K. Nanoclustering as a dominant feature of plasma membrane organization. J. Cell Sci. 2014;127:4995–5005. doi: 10.1242/jcs.146340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nussinov R., Jang H., Tsai C.J. Oligomerization and nanocluster organization render specificity. Biol. Rev. Camb. Philos. Soc. 2015;90:587–598. doi: 10.1111/brv.12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sevcsik E., Schütz G.J. With or without rafts? Alternative views on cell membranes. BioEssays. 2015;38:129–139. doi: 10.1002/bies.201500150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Y., Maxwell K.N., Levental I. Bile acids modulate signaling by functional perturbation of plasma membrane domains. J. Biol. Chem. 2013;288:35660–35670. doi: 10.1074/jbc.M113.519116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim W., Fan Y.Y., Chapkin R.S. n-3 polyunsaturated fatty acids suppress the localization and activation of signaling proteins at the immunological synapse in murine CD4+ T cells by affecting lipid raft formation. J. Immunol. 2008;181:6236–6243. doi: 10.4049/jimmunol.181.9.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacDonald B.T., Tamai K., He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Driehuis E., Clevers H. WNT signalling events near the cell membrane and their pharmacological targeting for the treatment of cancer. Br. J. Pharmacol. 2017;174:4547–4563. doi: 10.1111/bph.13758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Angers S., Moon R.T. Proximal events in Wnt signal transduction. Nat. Rev. Mol. Cell Biol. 2009;10:468–477. doi: 10.1038/nrm2717. [DOI] [PubMed] [Google Scholar]
- 12.Gregorieff A., Clevers H. Wnt signaling in the intestinal epithelium: from endoderm to cancer. Genes Dev. 2005;19:877–890. doi: 10.1101/gad.1295405. [DOI] [PubMed] [Google Scholar]
- 13.Fevr T., Robine S., Huelsken J. Wnt/beta-catenin is essential for intestinal homeostasis and maintenance of intestinal stem cells. Mol. Cell. Biol. 2007;27:7551–7559. doi: 10.1128/MCB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perochon J., Carroll L.R., Cordero J.B. Wnt signalling in intestinal stem cells: lessons from mice and flies. Genes (Basel) 2018;9:138. doi: 10.3390/genes9030138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sezgin E., Azbazdar Y., Ozhan G. Binding of canonical Wnt ligands to their receptor complexes occurs in ordered plasma membrane environments. FEBS J. 2017;284:2513–2526. doi: 10.1111/febs.14139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakane H., Yamamoto H., Kikuchi A. LRP6 is internalized by Dkk1 to suppress its phosphorylation in the lipid raft and is recycled for reuse. J. Cell Sci. 2010;123:360–368. doi: 10.1242/jcs.058008. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto H., Sakane H., Kikuchi A. Wnt3a and Dkk1 regulate distinct internalization pathways of LRP6 to tune the activation of beta-catenin signaling. Dev. Cell. 2008;15:37–48. doi: 10.1016/j.devcel.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 18.He X., Semenov M., Zeng X. LDL receptor-related proteins 5 and 6 in Wnt/beta-catenin signaling: arrows point the way. Development. 2004;131:1663–1677. doi: 10.1242/dev.01117. [DOI] [PubMed] [Google Scholar]
- 19.Kimelman D., Xu W. beta-catenin destruction complex: insights and questions from a structural perspective. Oncogene. 2006;25:7482–7491. doi: 10.1038/sj.onc.1210055. [DOI] [PubMed] [Google Scholar]
- 20.Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 21.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2017. CA Cancer J. Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 22.Calle E.E., Rodriguez C., Thun M.J. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N. Engl. J. Med. 2003;348:1625–1638. doi: 10.1056/NEJMoa021423. [DOI] [PubMed] [Google Scholar]
- 23.Renehan A.G., Tyson M., Zwahlen M. Body-mass index and incidence of cancer: a systematic review and meta-analysis of prospective observational studies. Lancet. 2008;371:569–578. doi: 10.1016/S0140-6736(08)60269-X. [DOI] [PubMed] [Google Scholar]
- 24.Ning Y., Wang L., Giovannucci E.L. A quantitative analysis of body mass index and colorectal cancer: findings from 56 observational studies. Obes. Rev. 2010;11:19–30. doi: 10.1111/j.1467-789X.2009.00613.x. [DOI] [PubMed] [Google Scholar]
- 25.Arita Y., Kihara S., Matsuzawa Y. Paradoxical decrease of an adipose-specific protein, adiponectin, in obesity. Biochem. Biophys. Res. Commun. 1999;257:79–83. doi: 10.1006/bbrc.1999.0255. [DOI] [PubMed] [Google Scholar]
- 26.Fang H., Judd R.L. Comprehensive Physiology. John Wiley & Sons, Inc.; 2018. Adiponectin regulation and function; pp. 1031–1063. [Google Scholar]
- 27.Giannessi D., Maltinti M., Del Ry S. Adiponectin circulating levels: a new emerging biomarker of cardiovascular risk. Pharmacol. Res. 2007;56:459–467. doi: 10.1016/j.phrs.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 28.Wei E.K., Giovannucci E., Mantzoros C.S. Low plasma adiponectin levels and risk of colorectal cancer in men: a prospective study. J. Natl. Cancer Inst. 2005;97:1688–1694. doi: 10.1093/jnci/dji376. [DOI] [PubMed] [Google Scholar]
- 29.Kelesidis I., Kelesidis T., Mantzoros C.S. Adiponectin and cancer: a systematic review. Br. J. Cancer. 2006;94:1221–1225. doi: 10.1038/sj.bjc.6603051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamaji T., Iwasaki M., Tsugane S. Interaction between adiponectin and leptin influences the risk of colorectal adenoma. Cancer Res. 2010;70:5430–5437. doi: 10.1158/0008-5472.CAN-10-0178. [DOI] [PubMed] [Google Scholar]
- 31.Di Zazzo E., Polito R., Moncharmont B. Adiponectin as link factor between adipose tissue and cancer. Int. J. Mol. Sci. 2019;20:839. doi: 10.3390/ijms20040839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim A.Y., Lee Y.S., Kim J.B. Adiponectin represses colon cancer cell proliferation via AdipoR1- and -R2-mediated AMPK activation. Mol. Endocrinol. 2010;24:1441–1452. doi: 10.1210/me.2009-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zakikhani M., Dowling R.J.O., Pollak M.N. The effects of adiponectin and metformin on prostate and colon neoplasia involve activation of AMP-activated protein kinase. Cancer Prev. Res. (Phila.) 2008;1:369–375. doi: 10.1158/1940-6207.CAPR-08-0081. [DOI] [PubMed] [Google Scholar]
- 34.Shackelford D.B., Shaw R.J. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat. Rev. Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujisawa T., Endo H., Nakajima A. Adiponectin suppresses colorectal carcinogenesis under the high-fat diet condition. Gut. 2008;57:1531–1538. doi: 10.1136/gut.2008.159293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otani K., Kitayama J., Nagawa H. Adiponectin suppresses tumorigenesis in Apc(Min)(/+) mice. Cancer Lett. 2010;288:177–182. doi: 10.1016/j.canlet.2009.06.037. [DOI] [PubMed] [Google Scholar]
- 37.Saxena A., Chumanevich A., Fayad R. Adiponectin deficiency: role in chronic inflammation induced colon cancer. Biochim. Biophys. Acta. 2012;1822:527–536. doi: 10.1016/j.bbadis.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mutoh M., Teraoka N., Wakabayashi K. Loss of adiponectin promotes intestinal carcinogenesis in Min and wild-type mice. Gastroenterology. 2011;140:2000–2008. e1-e2. doi: 10.1053/j.gastro.2011.02.019. [DOI] [PubMed] [Google Scholar]
- 39.Okada-Iwabu M., Yamauchi T., Kadowaki T. A small-molecule AdipoR agonist for type 2 diabetes and short life in obesity. Nature. 2013;503:493–499. doi: 10.1038/nature12656. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y., Zhao J., Wang Y.J. AdipoRon, the first orally active adiponectin receptor activator, attenuates postischemic myocardial apoptosis through both AMPK-mediated and AMPK-independent signalings. Am. J. Physiol. Endocrinol. Metab. 2015;309:E275–E282. doi: 10.1152/ajpendo.00577.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akimoto M., Maruyama R., Takenaga K. Antidiabetic adiponectin receptor agonist AdipoRon suppresses tumour growth of pancreatic cancer by inducing RIPK1/ERK-dependent necroptosis. Cell Death Dis. 2018;9:804. doi: 10.1038/s41419-018-0851-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malih S., Najafi R. AdipoRon: a possible drug for colorectal cancer prevention? Tumour Biol. 2015;36:6673–6675. doi: 10.1007/s13277-015-3911-3. [DOI] [PubMed] [Google Scholar]
- 43.DeClercq V., McMurray D.N., Chapkin R.S. Obesity promotes colonic stem cell expansion during cancer initiation. Cancer Lett. 2015;369:336–343. doi: 10.1016/j.canlet.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Habets G.G., Knepper M., Bollag G. cDNA array analyses of K-ras-induced gene transcription. Methods Enzymol. 2001;332:245–260. doi: 10.1016/s0076-6879(01)32207-3. [DOI] [PubMed] [Google Scholar]
- 45.Sato T., Vries R.G., Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 46.Fan Y.Y., Davidson L.A., Chapkin R.S. Murine colonic organoid culture system and downstream assay applications. Methods Mol. Biol. 2016;1576:171–181. doi: 10.1007/7651_2016_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sezgin E., Kaiser H.J., Levental I. Elucidating membrane structure and protein behavior using giant plasma membrane vesicles. Nat. Protoc. 2012;7:1042–1051. doi: 10.1038/nprot.2012.059. [DOI] [PubMed] [Google Scholar]
- 48.Fan Y.Y., Fuentes N.R., Chapkin R.S. Remodelling of primary human CD4+ T cell plasma membrane order by n-3 PUFA. Br. J. Nutr. 2018;119:163–175. doi: 10.1017/S0007114517003385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim W., Khan N.A., Chapkin R.S. Regulatory activity of polyunsaturated fatty acids in T-cell signaling. Prog. Lipid Res. 2010;49:250–261. doi: 10.1016/j.plipres.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hou T.Y., McMurray D.N., Chapkin R.S. Omega-3 fatty acids, lipid rafts, and T cell signaling. Eur. J. Pharmacol. 2016;785:2–9. doi: 10.1016/j.ejphar.2015.03.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chapkin R.S., DeClercq V., Fan Y.Y. Mechanisms by which pleiotropic amphiphilic n-3 PUFA reduce colon cancer risk. Curr. Colorectal Cancer Rep. 2014;10:442–452. doi: 10.1007/s11888-014-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Owen D.M., Rentero C., Gaus K. Quantitative imaging of membrane lipid order in cells and organisms. Nat. Protoc. 2011;7:24–35. doi: 10.1038/nprot.2011.419. [DOI] [PubMed] [Google Scholar]
- 53.Fuentes N.R., Salinas M.L., Chapkin R.S. Emerging role of chemoprotective agents in the dynamic shaping of plasma membrane organization. Biochim Biophys Acta Biomembr. 2017;1859:1668–1678. doi: 10.1016/j.bbamem.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vanier M.T., Latour P. Laboratory diagnosis of Niemann–Pick disease type C: the filipin staining test. Methods Cell Biol. 2015;126:357–375. doi: 10.1016/bs.mcb.2014.10.028. [DOI] [PubMed] [Google Scholar]
- 55.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krämer A., Green J., Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duck-Chong C.G. A rapid sensitive method for determining phospholipid phosphorus involving digestion with magnesium nitrate. Lipids. 1979;14:492–497. [Google Scholar]
- 58.Barker N., Ridgway R.A., Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 59.de Lau W., Peng W.C., Clevers H. The R-spondin/Lgr5/Rnf43 module: regulator of Wnt signal strength. Genes Dev. 2014;28:305–316. doi: 10.1101/gad.235473.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reinke L., Lam A.P., Gottardi C.J. Adiponectin inhibits Wnt co-receptor, Lrp6, phosphorylation and β-catenin signaling. Biochem. Biophys. Res. Commun. 2016;470:606–612. doi: 10.1016/j.bbrc.2016.01.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garitaonandia I., Smith J.L., Lyons T.J. Adiponectin identified as an agonist for PAQR3/RKTG using a yeast-based assay system. J. Recept. Signal Transduct. Res. 2009;29:67–73. doi: 10.1080/10799890902729456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hug C., Wang J., Lodish H.F. T-cadherin is a receptor for hexameric and high-molecular-weight forms of Acrp30/adiponectin. Proc. Natl. Acad. Sci. USA. 2004;101:10308–10313. doi: 10.1073/pnas.0403382101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sezgin E., Sadowski T., Simons K. Measuring lipid packing of model and cellular membranes with environment sensitive probes. Langmuir. 2014;30:8160–8166. doi: 10.1021/la501226v. [DOI] [PubMed] [Google Scholar]
- 64.Sezgin E., Waithe D., Eggeling C. Spectral imaging to measure heterogeneity in membrane lipid packing. ChemPhysChem. 2015;16:1387–1394. doi: 10.1002/cphc.201402794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ma D.W., Seo J., Chapkin R.S. n-3 PUFA alter caveolae lipid composition and resident protein localization in mouse colon. FASEB J. 2004;18:1040–1042. doi: 10.1096/fj.03-1430fje. [DOI] [PubMed] [Google Scholar]
- 66.Turk H.F., Barhoumi R., Chapkin R.S. Alteration of EGFR spatiotemporal dynamics suppresses signal transduction. PLoS One. 2012;7:e39682. doi: 10.1371/journal.pone.0039682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Turk H.F., Monk J.M., Chapkin R.S. Inhibitory effects of omega-3 fatty acids on injury-induced epidermal growth factor receptor transactivation contribute to delayed wound healing. Am. J. Physiol. Cell Physiol. 2013;304:C905–C917. doi: 10.1152/ajpcell.00379.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Seo J., Barhoumi R., Chapkin R.S. Docosahexaenoic acid selectively inhibits plasma membrane targeting of lipidated proteins. FASEB J. 2006;20:770–772. doi: 10.1096/fj.05-4683fje. [DOI] [PubMed] [Google Scholar]
- 69.Jin L., Millard A.C., Loew L.M. Characterization and application of a new optical probe for membrane lipid domains. Biophys. J. 2006;90:2563–2575. doi: 10.1529/biophysj.105.072884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pyrshev K.A., Klymchenko A.S., Demchenko A.P. Apoptosis and eryptosis: Striking differences on biomembrane level. Biochim. Biophys. Acta. 2018;1860:1362–1371. doi: 10.1016/j.bbamem.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 71.Liu S.-L., Sheng R., Cho W. Orthogonal lipid sensors identify transbilayer asymmetry of plasma membrane cholesterol. Nat. Chem. Biol. 2017;13:268–274. doi: 10.1038/nchembio.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chichili G.R., Rodgers W. Cytoskeleton-membrane interactions in membrane raft structure. Cell. Mol. Life Sci. 2009;66:2319–2328. doi: 10.1007/s00018-009-0022-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gaus K., Chklovskaia E., Harder T. Condensation of the plasma membrane at the site of T lymphocyte activation. J. Cell Biol. 2005;171:121–131. doi: 10.1083/jcb.200505047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Scott R.E. Plasma membrane vesiculation: a new technique for isolation of plasma membranes. Science. 1976;194:743–745. doi: 10.1126/science.982044. [DOI] [PubMed] [Google Scholar]
- 75.Sezgin E., Levental I., Schwille P. Partitioning, diffusion, and ligand binding of raft lipid analogs in model and cellular plasma membranes. Biochim. Biophys. Acta. 2012;1818:1777–1784. doi: 10.1016/j.bbamem.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 76.Fuentes N.R., Kim E., Chapkin R.S. Omega-3 fatty acids, membrane remodeling and cancer prevention. Mol. Aspects Med. 2018;64:79–91. doi: 10.1016/j.mam.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Amaro M., Reina F., Sezgin E. Laurdan and Di-4-ANEPPDHQ probe different properties of the membrane. J. Phys. D Appl. Phys. 2017;50:134004. doi: 10.1088/1361-6463/aa5dbc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim H.M., Choo H.J., Cho B.R. A two-photon fluorescent probe for lipid raft imaging: C-laurdan. ChemBioChem. 2007;8:553–559. doi: 10.1002/cbic.200700003. [DOI] [PubMed] [Google Scholar]
- 79.Dodes Traian M.M., González Flecha F.L., Levi V. Imaging lipid lateral organization in membranes with C-laurdan in a confocal microscope. J. Lipid Res. 2012;53:609–616. doi: 10.1194/jlr.D021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Özhan G., Sezgin E., Weidinger G. Lypd6 enhances Wnt/β-catenin signaling by promoting Lrp6 phosphorylation in raft plasma membrane domains. Dev. Cell. 2013;26:331–345. doi: 10.1016/j.devcel.2013.07.020. [DOI] [PubMed] [Google Scholar]
- 81.Carmon K.S., Lin Q., Liu Q. LGR5 interacts and cointernalizes with Wnt receptors to modulate Wnt/β-catenin signaling. Mol. Cell. Biol. 2012;32:2054–2064. doi: 10.1128/MCB.00272-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim I., Pan W., Wu D. Clathrin and AP2 are required for PtdIns(4,5)P2-mediated formation of LRP6 signalosomes. J. Cell Biol. 2013;200:419–428. doi: 10.1083/jcb.201206096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Janda C.Y., Dang L.T., Garcia K.C. Surrogate Wnt agonists that phenocopy canonical Wnt and β-catenin signalling. Nature. 2017;545:234–237. doi: 10.1038/nature22306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ruiz M., Ståhlman M., Pilon M. AdipoR1 and AdipoR2 maintain membrane fluidity in most human cell types and independently of adiponectin. J. Lipid Res. 2019;60:995–1004. doi: 10.1194/jlr.M092494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang B., Rong X., Tontonoz P. Phospholipid remodeling and cholesterol availability regulate intestinal stemness and tumorigenesis. Cell Stem Cell. 2018;22:206–220.e4. doi: 10.1016/j.stem.2017.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bi J., Ichu T.A., Mischel P.S. Oncogene amplification in growth factor signaling pathways renders cancers dependent on membrane lipid remodeling. Cell Metab. 2019;30:525–538.e8. doi: 10.1016/j.cmet.2019.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Róg T., Pasenkiewicz-Gierula M., Karttunen M. Ordering effects of cholesterol and its analogues. Biochim. Biophys. Acta. 2009;1788:97–121. doi: 10.1016/j.bbamem.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 88.Krause M.R., Regen S.L. The structural role of cholesterol in cell membranes: from condensed bilayers to lipid rafts. Acc. Chem. Res. 2014;47:3512–3521. doi: 10.1021/ar500260t. [DOI] [PubMed] [Google Scholar]
- 89.Mahammad S., Parmryd I. Cholesterol depletion using methyl-β-cyclodextrin. Methods Mol. Biol. 2015;1232:91–102. doi: 10.1007/978-1-4939-1752-5_8. [DOI] [PubMed] [Google Scholar]
- 90.Schroeder F., Holland J.F., Bieber L.L. Fluorometric evidence for the binding of cholesterol to the filipin complex. J. Antibiot. (Tokyo) 1971;24:846–849. doi: 10.7164/antibiotics.24.846. [DOI] [PubMed] [Google Scholar]
- 91.Maxfield F.R., Wüstner D. Analysis of cholesterol trafficking with fluorescent probes. Methods Cell Biol. 2012;108:367–393. doi: 10.1016/B978-0-12-386487-1.00017-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Arthur J.R., Heinecke K.A., Seyfried T.N. Filipin recognizes both GM1 and cholesterol in GM1 gangliosidosis mouse brain. J. Lipid Res. 2011;52:1345–1351. doi: 10.1194/jlr.M012633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tabas I. Consequences of cellular cholesterol accumulation: basic concepts and physiological implications. J. Clin. Invest. 2002;110:905–911. doi: 10.1172/JCI16452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lange Y., Swaisgood M.H., Steck T.L. Plasma membranes contain half the phospholipid and 90% of the cholesterol and sphingomyelin in cultured human fibroblasts. J. Biol. Chem. 1989;264:3786–3793. [PubMed] [Google Scholar]
- 95.Lange Y., Ye J., Steck T. Cholesterol movement in Niemann-Pick type C cells and in cells treated with amphiphiles. J. Biol. Chem. 2000;275:17468–17475. doi: 10.1074/jbc.M000875200. [DOI] [PubMed] [Google Scholar]
- 96.Lange Y. Disposition of intracellular cholesterol in human fibroblasts. J. Lipid Res. 1991;32:329–339. [PubMed] [Google Scholar]
- 97.Lange Y., Ye J., Steck T.L. Circulation of cholesterol between lysosomes and the plasma membrane. J. Biol. Chem. 1998;273:18915–18922. doi: 10.1074/jbc.273.30.18915. [DOI] [PubMed] [Google Scholar]
- 98.Shyu P., Jr., Wong X.F.A., Thibault G. Dropping in on lipid droplets: insights into cellular stress and cancer. Biosci. Rep. 2018;38 doi: 10.1042/BSR20180764. BSR20180764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Muller C.P., Stephany D.A., Wunderlich J.R. Filipin as a flow microfluorometry probe for cellular cholesterol. Cytometry. 1984;5:42–54. doi: 10.1002/cyto.990050108. [DOI] [PubMed] [Google Scholar]
- 100.Hassall D.G., Graham A. Changes in free cholesterol content, measured by filipin fluorescence and flow cytometry, correlate with changes in cholesterol biosynthesis in THP-1 macrophages. Cytometry. 1995;21:352–362. doi: 10.1002/cyto.990210407. [DOI] [PubMed] [Google Scholar]
- 101.Pipalia N.H., Huang A., Maxfield F.R. Automated microscopy screening for compounds that partially revert cholesterol accumulation in Niemann-Pick C cells. J. Lipid Res. 2006;47:284–301. doi: 10.1194/jlr.M500388-JLR200. [DOI] [PubMed] [Google Scholar]
- 102.Karten B., Peake K.B., Vance J.E. Mechanisms and consequences of impaired lipid trafficking in Niemann-Pick type C1-deficient mammalian cells. Biochim. Biophys. Acta. 2009;1791:659–670. doi: 10.1016/j.bbalip.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 103.Sokol J., Blanchette-Mackie J., Comly M.E. Type C Niemann-Pick disease. Lysosomal accumulation and defective intracellular mobilization of low density lipoprotein cholesterol. J. Biol. Chem. 1988;263:3411–3417. [PubMed] [Google Scholar]
- 104.Miyamoto S., Narita T., Mutoh M. Novel screening system revealed that intracellular cholesterol trafficking can be a good target for colon cancer prevention. Sci. Rep. 2019;9:6192. doi: 10.1038/s41598-019-42363-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.von Einem B., Weber P., Schneckenburger H. Cholesterol-dependent energy transfer between fluorescent proteins-insights into protein proximity of APP and BACE1 in different membranes in Niemann-Pick type C disease cells. Int. J. Mol. Sci. 2012;13:15801–15812. doi: 10.3390/ijms131215801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Faloia E., Garrapa G.G., Mazzanti L. Physicochemical and functional modifications induced by obesity on human erythrocyte membranes. Eur. J. Clin. Invest. 1999;29:432–437. doi: 10.1046/j.1365-2362.1999.00458.x. [DOI] [PubMed] [Google Scholar]
- 107.Ferretti G., Curatola G., Giorgi P.L. Erythrocyte membrane fluidity and changes in plasma lipid composition: a possible relationship in childhood obesity. Biochem. Med. Metab. Biol. 1991;46:1–9. doi: 10.1016/0885-4505(91)90044-l. [DOI] [PubMed] [Google Scholar]
- 108.Fuentes N.R., Mlih M., Chapkin R.S. Long-chain n-3 fatty acids attenuate oncogenic KRas-driven proliferation by altering plasma membrane nanoscale proteolipid composition. Cancer Res. 2018;78:3899–3912. doi: 10.1158/0008-5472.CAN-18-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Erazo-Oliveras A., Fuentes N.R., Chapkin R.S. Functional link between plasma membrane spatiotemporal dynamics, cancer biology, and dietary membrane-altering agents. Cancer Metastasis Rev. 2018;37:519–544. doi: 10.1007/s10555-018-9733-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.