Abstract
The TRPV1 cation nonselective ion channel plays an essential role in thermosensation and perception of other noxious stimuli. TRPV1 can be activated by low extracellular pH, high temperature, or naturally occurring pungent molecules such as allicin, capsaicin, or resiniferatoxin. Its noxious thermal sensitivity makes it an important participant as a thermal sensor in mammals. However, details of the mechanism of channel activation by increases in temperature remain unclear. Here, we used a combination of approaches to try to understand the role of the ankyrin repeat domain (ARD) in channel behavior. First, a computational modeling approach by coarse-grained molecular dynamics simulation of the whole TRPV1 embedded in a phosphatidylcholine and phosphatidylethanolamine membrane provides insight into the dynamics of this channel domain. Global analysis of the structural ensemble shows that the ARD is a region that sustains high fluctuations during dynamics at different temperatures. We then performed biochemical and thermal stability studies of the purified ARD by the means of circular dichroism and tryptophan fluorescence and demonstrate that this region undergoes structural changes at similar temperatures that lead to TRPV1 activation. Our data suggest that the ARD is a dynamic module and that it may participate in controlling the temperature sensitivity of TRPV1.
Significance
This work demonstrates that the temperature-dependent dynamics of the ankyrin repeat domain of TRPV1 channels, as probed by coarse-grained molecular dynamics, correspond to the experimentally determined dynamics of an isolated ankyrin repeat domain domain. These results show that this region of TRPV1 channels undergoes significant conformational change as a function of increased temperature and suggest that it participates in the temperature-dependent structural changes that lead to the channel opening.
Introduction
TRPV1 is a nonspecific cation channel implicated in nociception by chemicals, temperature, and pH (1, 2, 3). This channel is one of the chemosensors involved in the sensation of pain and thermal stimuli, and it participates in a diverse range of cellular processes (4,5). The latter has been evidenced from studies in which the deletion of TRPV1 in mice alters noxious and mild temperature sensation (6,7), whereas knockout of other thermo-TRPs such as TRPV2, TRPV3, and TRPV4 shows little effect in sensory transduction in rodents (6,8,9). Moreover, whereas deletion of TRPV1 in rodents does not affect corporal temperature, blockage of TRPV1 in vivo triggers hyperthermia (10).
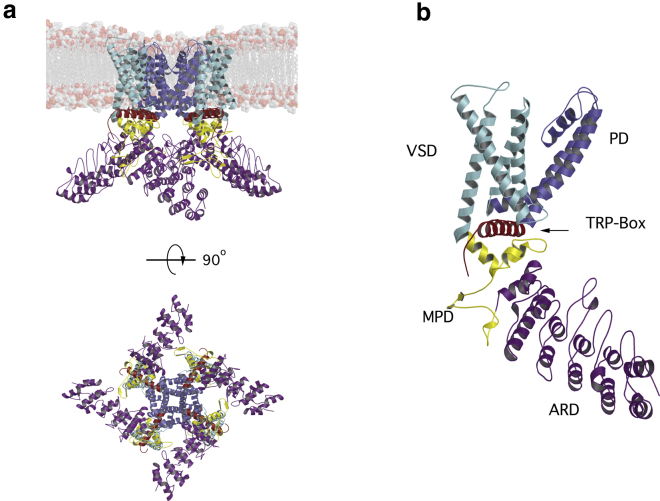
The rat TRPV1 structure is a tetramer (Fig. 1 a), with every monomer consisting of 838 amino acids (Fig. 1 c). The structure solved by cryo-EM is from a minimal-functional TRPV1 that lacks 100 amino acids from the N-termini and 80 amino acids in the C-termini and which is also missing a longer S5-pore extracellular loop named the turret. This minimal-functional 586 amino acid construct provides a model for the full-length channel, although without unfolded loops. In TRPV1, the structure of the membrane-embedded domains is canonical with other ion channels like voltage-gated potassium, sodium, and calcium ion channels. A tetramer is formed by a voltage sensor-like domain (VSD)-like domain, surrounding a pore formed by the contribution of the four pore domains (PD) of each subunit.
Figure 1.
Depiction of the all-atom model of TRPV1, derived from the PDB: 3J5P structure. (a, top) Shown is a side view of the membrane-TRPV1 system. The coloring of the channel domains is as follows: ARD, violet; MPD, yellow; VSD, cyan; PD, blue; and TRP-box, red. The lipid headgroups of the membrane bilayer are represented as spheres. (a, bottom) The intracellular view of the channel is represented with the same coloring code. Inter-subunit interactions are seen between MPD and ARD. (b) Shown is a depiction of a single subunit of TRPV1; VSD and pore domain (PD) are membrane-embedded domains, and ARD is fully cytosolic. The membrane proximal domain (MPD) has complex interactions with ARD, VSD, and TRP-box. The TRP-box is sandwiched between the MPD and VSD. The repaired loops are not shown. To see this figure in color, go online.
The C-terminus of TRPV1 has been shown to be functionally important. Several modulating lipids, such as phosphatidylinositol 4,5-bisphosphate and lysophosphatidic acid (LPA), bind in this region (11,12). In the cryo-EM structures, the α helix after the S6 transmembrane segment is well defined and is known as the TRP-box; it is a conserved domain in almost all the members of the TRP family. In all the solved structures for TRPV1 (13, 14, 15), TRPV2 (16), TRPV3 (17), TRPV4 (18), TRPV5 (19), and TRPV6 (20), the TRP-box is folded in the same manner. In the case of TRPV1, cryo-EM experiments show that the TRP-box displays a conformational change when the channel is bound to capsaicin (13) or resiniferatoxin and double-knot toxin together (13), similar to what happens in TRPV2 sensitized by 2-aminoethoxydiphenyl borate (16). Until now, the structure of the distal part of the C-terminus remains unknown, but there is some evidence that it is located near the ankyrin repeat domain (ARD) and that these two structures may undergo intradomain interactions (21).
The ARD and the membrane proximal domain (MPD), a structure located between the ARD and the first transmembrane segment, have a noncanonical interaction only seen in TRP channels. A “finger” loop from ANK4 touches the MPD at a recognition site in a neighboring subunit (Figs. 1 a and S3; (15,20)). Although some of the functions of these domains are still not known, others have been well characterized and include the direct interaction of the ARD with calmodulin (19,22) and the possible participation of the MPD in heat activation (23). Thus, the ARD and MPD are functional regulatory domains, and it should be noted that, before the first cryo-EM structures for TRP channels were obtained, the intracellular domains were visualized as individual and not interconnected domains. Thanks to structural analysis, today, we know that the intracellular domains of TRPV ion channels form tetramers and that coupling between these domains regulates channel function (20,24). In this context, the overall nature of ARD interactions is poorly understood, but some evidences correlate with the notion that they play a leading role in the functional mechanism of the channel (22,25,26). For example, there is some evidence of the ARD contributing to gating by selective oxidation of intracellular cysteines present in the ARD (3,27). Additionally, a complex formed by TRPV5 and calmodulin was solved recently (19). This interaction can be of interest because it also exists in TRPV1 and other members of the family, and it is located between C-termini and the ARD.
To further study the role of the ARD in TRPV1, we studied the integral relationships between ARD function and channel gating. For this purpose, we employed coarse-grained molecular dynamics of the whole TRPV1 channel embedded in a POPC/POPE membrane and also performed experiments with the purified ARD in solution, making use of the thermal shift unfolding, CD, and tryptophan fluorescence measurements. We find that the ARD is a dynamical domain and that these dynamics are modulated by temperature in a way that might be relevant to activation by temperature.
Methods
Model preparation
The cryo-electron microscopy (cryo-EM) structure of a minimal-functional rat TRPV1, solved at 3.4 Å resolution, was used as the initial coordinates to perform coarse-grained molecular dynamics (Protein Data Bank, PDB: 3J5P) (15). The missing loops and side chains were built in the Swiss Modeler web server (28,29). To generate our model, the server used the whole channel model (15) and the transmembrane cryo-EM structure solved in nanodisks (14). The repaired TRPV1 structure was prepared for coarse graining by means of the Martinize.py program (30). A 20 nm per side, square POPC/POPE (coarse-grained) membrane was prepared in CHARMM-GUI (31), and the channel model was embedded into the membrane using Pymol scripts written in-house. The TRPV1-POPC/POPE structure was prepared for molecular dynamics (MD) simulation in the Gromacs 4.5 suite (32,33). The model was minimized and solvated with coarse-grained water and surrounded by 30 mM NaCl ions. A 200 ns NVT equilibration dynamics with a Berendsen thermostat was performed before the production run (30,32).
Molecular dynamics simulations
We used coarse-grained molecular dynamics (CGMD) simulations to characterize the dynamics of the ion channel in the membrane. We simulated the initial repaired model of the APO state (PDB: 3J5P) at various temperatures varying from 290 to 350 K (17–77°C), sampling every 5 K. The structure of TRPV1 was embedded in a POPC/POPE bilayer with a MARTINI force field; several equilibrating simulations with temperature coupling were performed, and then, an equilibrating simulation with pressure and temperature was performed until the root mean-square deviation (RMSD) was stable at 298 K (30). A production run at 290 K was sampled as an initial structure. From a stable point in a 290 K run, five replicas were prepared. The RMSD corresponds to the mean of the whole protein main chain over the whole simulation time, and the root mean-square fluctuation (RMSF) corresponds to the mean of a single amino acid position over time. Both were calculated with Gromacs 4.5. RMSF values for each domain (the average over the four monomers on each simulated replica at the same temperature) were calculated employing the gmx RMSF function, fitting independently each domain through the whole analyzed trajectories (last 500 ns). RMSFs were calculated removing the roto-translations of each domain (ARD, VSD, MPD, PD, TRP-box) by fitting the trajectory and calculating its RMSF values independently for each domain; only the last 500 ns of each trajectory were used to perform these calculations. All simulated replicas (five replicas for each of the different simulated temperatures) started from the same equilibrated structural model. On each replica, the initial velocities were reassigned following its proper Maxwell-Boltzmann distribution.
The output trajectories were used as input for principal component analysis (PCA) of the atomic displacements around the average. This analysis was performed in Bio3D for the four subunits (34). Production runs were performed in the “Miztli” HP Cluster Platform 3000SL supercomputer at the Supercomputing Facility of the National Autonomous University of México (UNAM) (Mexico City, Mexico).
ARD expression and purification
The DNA sequence encoding for the ARD of rat TRPV1 (residues 100–262) was cloned into a pET28c vector (MilliporeSigma, Burlington, MA). For expression, the Escherichia coli BL21 (DE3) strain (New England BioLabs, Ipswich, MA) was transformed with this vector. Batch cultures of transformed cells were grown in Luria Bertani broth (Sigma-Aldrich, St. Louis, MO) to an OD of 0.7–0.8, and protein expression was then induced with isopropyl-β-D-1-thiogalactopyranoside 0.1 mM (GoldBio, St. Louis, MO) for 12 h at room temperature. The cell pellet was lysed in Tris 50 mM (pH 8), dithiothreitol 0.1 mM, and NaCl 300 mM (Sigma-Aldrich). The lysate was purified by Histrap FF columns (GE Healthcare, Chicago, IL) with an imidazole gradient from 0 to 0.5 M. The eluted protein was concentrated and desalted in a PD-10 column (GE Healthcare) against a Tris 50 mM (pH 8) and dithiothreitol 0.1 mM buffer. Then, the protein was injected into a cation exclusion chromatography column (MonoQ) and eluted against Tris 50 mM (pH 8) and dithiothreitol 0.1 mM buffer with an increased linear gradient from 0 to 1 M of NaCl. The final step was a size exclusion chromatography in a handmade 275 mL Superdex 75 Column (Dimensions 2.50 × 60 cm; GE Healthcare). ARD concentration was determined using a theoretical extinction coefficient of 1.21 L/mg·cm obtained using the ProtParam Expasy WebServer (35) and by bicinchoninic acid assays with reduction compatible reagents (Pierce Chemical, Dallas, TX).
Thermal shift unfolding
Assays were prepared in a 96-well plate for real-time PCR; each well was prepared with 1 μL of SYPRO Dye 1X (Sigma-Aldrich), 1 μL of 1 mg/mL solution of purified ARD, and 1 μL of solubility and stability II kit (Hampton Research, Aliso Viejo, CA). Water was added to fill 10 μL per well. A real-time thermocycler QuantStudio 3 (Thermo Fisher Scientific, Waltham, MA) was used for collecting data from 20 to 80°C, sampling every 1°C. The data were exported and analyzed in IgorPro 6 (WaveMetrics).
Size exclusion chromatography-multiple angle light scattering
The size exclusion chromatography-multiple angle light scattering analysis was performed on a DAWN HELEOS multiangle light scattering detector, with 18 angle detectors and a 658.9 nm laser beam (Wyatt Technology, Santa Barbara, CA) and an Optilab T-rEX refractometer (Wyatt Technology) in line with a Superdex 75 10/300 GL (GE Healthcare Life Sciences, Chicago, IL) size exclusion chromatography analytical column. Experiments were performed using an isocratic pump (Agilent Technologies, Santa Clara, CA) with a flow of 0.5 mL/min at room temperature (25°C). Data collection was performed with ASTRA 6.1 software (Wyatt Technology). For the experiments, 300 μL at 1 mg/mL protein were loaded on the columns with a running buffer of 20 mM glycine (pH 9.5) and 200 mM NaCl. The molecular weight and the ratio of gyration were calculated by the ASTRA software.
CD spectroscopy
Circular dichroism (CD) spectroscopy was carried out in a JASCO J-715 spectropolarimeter (JASCO, Oklahoma City, OK) with a 1-mm path length cuvette; the sample was prepared at 250 μg/mL in 5 mM sodium phosphate buffer. Samples were heated at a 0.5 K min−1 rate with a Peltier device (JASCO). A thermal unfolding curve was generated by measuring a spectrum for every step. CD data were analyzed in custom-made scripts and plotted with Igor Pro 6 (WaveMetrics). Normalized ellipticity data obtained at 208 nm were fitted to the following sigmoid function to calculate the Tm:
| (1) |
Tryptophan fluorescence spectroscopy
Intrinsic fluorescence spectra were obtained in a PC1 spectrofluorometer (ISS, Copenhagen, Denmark) configured with a 10 W halogen lamp as the excitation source. All samples were prepared at 50 μg/L. Tryptophan fluorescence spectra were obtained using an excitation wavelength of 295 nm; emission spectra were collected from 310 to 400 nm, every 4 nm, using excitation and emission slits of 0.25 and 1 mm, respectively. The temperature was controlled with a Peltier device (Quantum Northwest, Liberty Lake, WA). Normalized intensity at 330 nm were fitted to Eq. 1.
Results
Cytosolic domains of TRPV1 fluctuate more at higher temperatures
It is generally assumed that TRPV1 is formed by a cluster of functional domains (Fig. 1 c), each with a set of specific interactions. The ARD, MPD, and TRP-box domains are intracellular. Several groups have found that the minimal-functional TRPV1 structure needs the N-termini from the ANK1 motif upwards (15,21,36), and no more than 100 amino acids from the C-terminus can be deleted without rendering the channel nonfunctional (36).
As an initial point in our research to understand the behavior of TRPV1 as a function of temperature, we carried out coarse-grained molecular dynamics simulations at different temperatures using a full-length model of TRPV1 embedded in a phosphatidylcholine (POPC)/phosphatidylethanolamine (POPE) lipid membrane (Fig. 1 a). Even though using a coarse-grained force field at a temperature different to 300 K does not guarantee sampling a canonical ensemble, our simulated systems may give us a clue about the TRPV1 dynamics at higher temperatures. Our simulations were performed without an elastic network model constraint. The simulated systems did not show high amounts of structural drifts from our initial TRPV1 model, as shown in Fig. S1.
The initial model for simulation was derived from the closed TRPV1 structure (PDB: 3J5P) solved by cryo-EM (15). The model was simulated for 200 ns in temperature-controlled conditions (NVT) for stabilization and then 200 ns of pressure and temperature-controlled conditions (NPT) with Berendsen thermostat and pressure coupling (see Methods). The simulations were equilibrated until the channel was in a stable RMSD state; from that point on, five replicas were simulated at 290, 295, 300, 305, 310, 315, 320, 325, 330, 335, and 340 K. At these temperatures, independent runs of 1000 ns were simulated to sample for an adequate time to observe critical large-scale structural transitions.
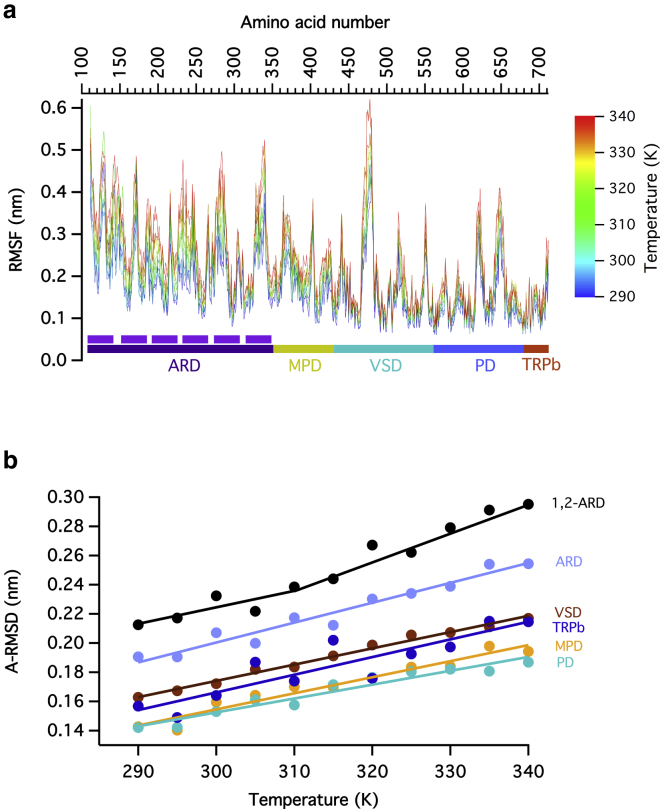
In Fig. 2 a, we show the RMSF calculated for all four subunits of the whole TRPV1 tetramer and over all simulated replicas. Temperatures from 290 to 340 K were sampled, and each point is the average of RMSF of a particular residue for all the temperatures. The RMSF deviation, which correlates the fluctuation per amino acid during the trajectory of the dynamics, shows that there are some regions that fluctuate more than others as the temperature is increased (Fig. S2). The regions with a well-defined secondary structure have less movement, and the loops and less structured chains move more (Fig. 2 a). The transmembrane region of the channel has smaller magnitude movements in relation to the cytosolic part; this is likely due to the two-dimensional restriction of the lipid membrane. The data shows that the mean RMSF greatly increases in the N- and C-termini, even though these are regions with well-defined secondary structure. Interestingly, the loop between S1 and S2 transmembrane segments also has a high value of RMSF and of RMSF deviation.
Figure 2.
Analysis of the structural fluctuations induced by temperature. (a) RMSF of the beads that correspond to the main chain of each amino acid in coarse-grained molecular dynamics, calculated over different subunits and different replicas for each simulated temperature. The ANKs are indicated by purple bars. The ANKs that have the greatest RMSF are ANK1 and ANK2. (b) The calculated average value of the RMSD (A-RMSD) of each functional domain during the last 500 ns of each simulated replica and each of the four subunits at a given temperature is shown as a function of the simulation temperature. The data is fit to a linear function. Notice that the ARD 1–2 data has an increased slope at 310 K. To see this figure in color, go online.
All of the N-terminus (ARD + MPD) shows a significant increase in the main movement as the temperature rises. The ankyrin motifs (ANK) are represented in Fig. 1 a. ANK1 and ANK2 are not solved in the cryo-EM structure (Fig. S3 c), and in our analysis, these repeats presented a large average RMSF. Although the C-terminus also has a high value of temperature-dependent RMSF, for the purpose of this study, it will not be discussed.
The RMSD for all the subunits increases in proportion to temperature. When the average RMSD (A-RMSD) of each region is calculated and plotted separately as a function of temperature, it can be seen that all regions fluctuate more as the temperature increases. If we increase the simulation temperature, we can expect that the A-RMSD value will increase at the same rate in each domain, but because a heat-sensitive domain will absorb more thermal energy, it would also have a higher increase rate and overall higher A-RMSD values compared to less sensitive domains.
As can be seen in Fig. 2 b, the ARD domain, and particularly repeats 1 and 2, show overall higher A-RMSD values. Interestingly, at 310 K, the increase rate of A-RMSD with the temperature also increases. This measure will indicate which domains absorb more energy in the form of increased vibration.
Protein dynamics and inter-subunit interactions of TRPV1
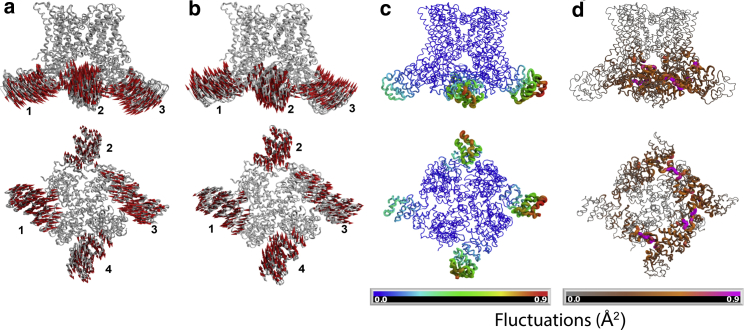
To study the coarse-grained dynamics of TRPV1 and provide a quantitative interpretation of the simulations, we performed PCA (37,38). We identified a component in which the ARDs undergo alternate up-and-down movements. Fig. 3, a and b shows the structure at two different stages of the oscillation. The arrows indicate the direction of the movement of each of the coarse-grained beads of only the ARD. The bottom panels show a view from the intracellular region of the channel. These fluctuations represent more than 70% of the movements of each trajectory, and this is seen at all the simulated temperatures.
Figure 3.
PCA of TRPV1 movements at 290 K. (a and b) The first component shows an alternating upwards and downwards movement of adjacent ARDs. The red arrows indicate the direction of movement of the ARD domain only. The subunits are numbered for clarity. (a) Domains 1 and 3 moved downwards, whereas 2 and 4 are upwards. (b) Opposed domains moved upwards (1,3) and downwards (2,4). The bottom panels show the intracellular view of the channel after a 90° rotation. (c) Shown are fluctuations calculated by RMSF per bead relative to main chain. ARDs show the highest values of fluctuations. Only the movement of ARDs is represented. Bottom panel, view of a 90° rotation of the channel is shown. The scale represents the value of the calculated fluctuation. (d) Shown are the deformation values of motion relative to neighbors. The MPDs show the highest correlated movement with the ARD (pink color). The values were calculated with Bio3D (see Methods) (33,36). The scale represents the calculated deformation values. The proximal ARD (ANK3 to MPD) was highly correlated. To see this figure in color, go online.
Fluctuation analyses were performed from the trajectories by means of the greater principal component (PC) eigenvectors (34). Fig. 3 c shows the fluctuation analysis of the 290 K trajectory taken from the same mode of vibration from Fig. 3, a and b. It can be seen that the highest values of the fluctuations occur at the ankyrin repeats one and two, and the fluctuations extend into the whole ARD.
A deformation displacement analysis was performed from the PCA; this is obtained by the averaged values of each individual component and by measuring the contribution of each backbone bead to the energy density as a function of position (37). This analysis provides information on the correlation between regions with high deformation and regions that are most affected by this deformation. Fig. 3 d shows that the regions with higher deformation are the ARD and that the regions with highest correlation with the deformation of the ARD is the MPD. This is a reflection of the physical interactions between these two domains but also means that the conformational changes in one domain are transmitted to the other in the interaction pair.
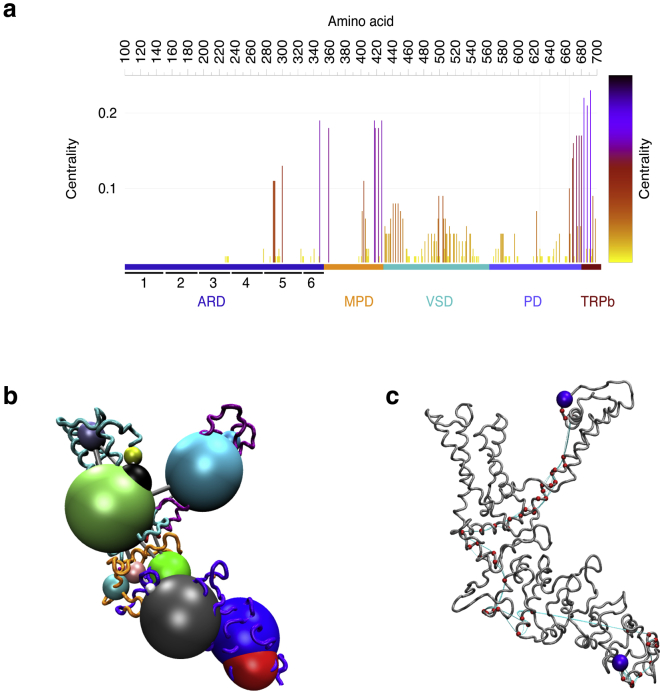
To further explore the allosteric pathway of conformational change, we calculated the eigenvector centrality for each node (Fig. 4 a); this centrality value measures the influence of each residue on the correlation network, and nodes with many edges indicate residues with a high number of connections, yielding insights into the internal dynamic coordination of the TRPV1 domains. The centrality values shown in Fig. 4 a indicate the importance of the MPD domain to connect the motion between the ARD domain and the PD domain that contains the selectivity filter. Additionally, we also performed a correlation network analysis in our 290 K trajectories to identify protein segments with correlated motions and the potential dynamic coordination of between distal sites (ARD) and other regions of the channel. The correlation matrices showed strong couplings between the ARD domain and the pore gate through the MPD. These couplings are seen as groups of amino acid residues, which move in a coordinated fashion. The size of the spheres in Fig. 4 b indicates the size of these coupled groups of residues.
Figure 4.
Correlation network analysis. (a) Shown are network centrality values per amino acid, color coded by its value. The vertical lines highlight residues Gly643 and Val679. (b) Shown is three-dimensional molecular structure mapping the consensus network communities and residue couplings, and the community network is depicted with colored spheres whose relative radius indicates the number of residues in a particular community. Different colors are used to indicate different groups of amino acid residues. The linking gray lines show their relative intercommunity coupling. (c) The most likely path of allosteric communication linking the 1-ARD and 2-ARD domains and residues Gly643 and Ile679. The green spheres show the position of the 1-ARD and 2-ARD and the residues at the gate (Ile679) and the selectivity filter (Gly643). To see this figure in color, go online.
To further characterize the dynamic linkage of the ARD and the PD and identify the most probable residues involved in the allosteric signaling, we carried out an optimal path analysis with the Bio3D software. The resulting calculated path (Fig. 3 c) describes the most favorable way in which dynamic communication could flow among the 1-ARD, 2-ARD, and the PD domain. This methodology involves the calculation of the shortest path through all network edges that represent a residue correlation between our selected node pairs, ARD and PD domains. This result is consistent with our suggestion that the ARD domain may allosterically modulate gating through the MPD domain.
The conformation of the ARD shows a thermal dependency at physiological temperatures
The simulation data illustrate that the intracellular domains of TRPV1 undergo conformational changes at temperatures that are relevant to the physiological activation of the channel. The region that shows the largest mobility is the ARD, a 260 amino acid region in the N-terminus of TRPV channels. In TRPV1, the ARD is implicated in adenosine triphosphate binding, Ca2+-calmodulin modulation, and regulation by cysteine oxidation (3,22,26,27,39). In the whole channel, the ARD has several complex interactions with the pre-S1 region (MPD, 90 amino acids) in the same subunit and a characteristic ARD-MPD interaction with the adjacent subunit (Fig. 1, a and c; (15)). The ARD also has an interaction with the distal C-terminus; this region is not well solved in TRPV1 cryo-EM structure, but in TRPV2 and TRPV3, it is (16,17). The functional significance of these interactions remains unknown.
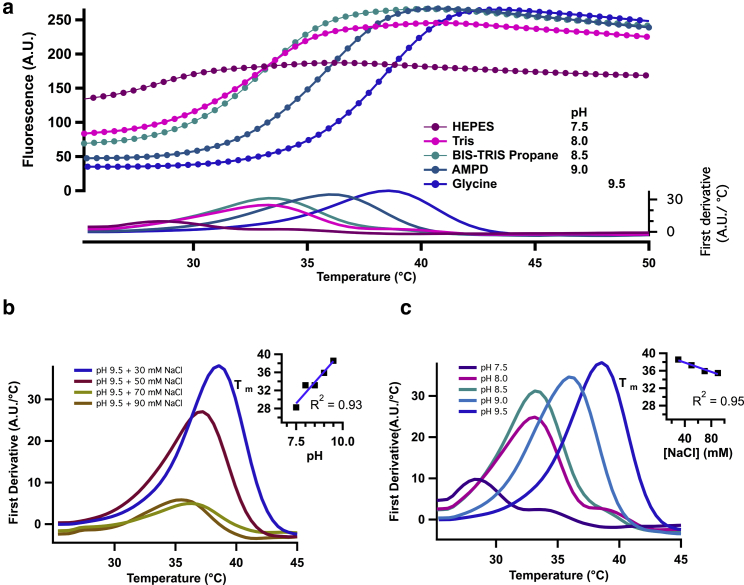
As a first approximation to study the biophysical properties of the N-terminus, we purified an ARD from rat TRPV1 and screened its stability by a thermal shift assay (Fig. 5), utilizing several pH and salt concentration conditions. Fig. 5 a shows the fluorescence signal from the dye SYPRO orange as a reporter of the thermal unfolding process. SYPRO emits a fluorescence signal by binding to hydrophobic regions exposed during unfolding. The fluorescence signals show a characteristic sigmoid dependence on temperature, which is a feature of a two-state unfolding process in soluble proteins. To characterize the cooperativity of the change in fluorescence, the first derivative was calculated for every trace (Fig. 5, a–c).
Figure 5.
Thermal shift unfolding of ARD. (a) Representative raw SYPRO fluorescence signals obtained in a real-time thermocycler are plotted as a function of temperature. The lower panel shows the first derivative of each signal. (b) First derivative (FD) plot (A.U./°C) of similar data as in (a) obtained at different pH values. The melting temperature, Tm, was calculated as the temperature in which the maximum occurs. Inset shows the Tm of the ARD at different pH values. (c) Shown is the melting temperature Tm of ARD in glycine buffer (pH 9.5) at different sodium chloride concentration. The inset shows the dependence of Tm on the NaCl concentration. To see this figure in color, go online.
The data show that the first derivative of the fluorescence increases linearly as a function of pH. The data in Fig. 5 a (inset) indicate that the cooperativity of the unfolding of the ARD is highest at basic pH because the value of the derivative increases as pH increases. The inset in Fig. 5 b shows that, additionally to the increased cooperativity, the domain becomes more stable because the Tm value, defined as the temperature at which the derivative is maximal, shifts to higher temperatures. Next, the dependence on salt concentration was analyzed for the pH 9.5 condition. In Fig. 5 c, we show the data for glycine buffer (pH 9.5) and several NaCl concentrations. The steepness of the fluorescence change is higher at low salt concentrations, and the Tm decreases with increasing salt concentration, with a slope of −0.05 and a change in Tm from 36 to 38°C. The behavior of ARD in glycine buffer (pH 9.5) was also studied by size exclusion chromatography-multiple angle light scattering to control for its possible oligomeric assembly. The experiment shows that the ARD is monodisperse in these conditions (Fig. S5).
Significantly, these series of experiments indicate that the thermal transitions of the isolated ARD occur in a range of temperatures that is rather narrow (Tm = 36–38°C for the most stable condition) and very close to the physiological temperature of mammals.
Stability of the ARD in solution
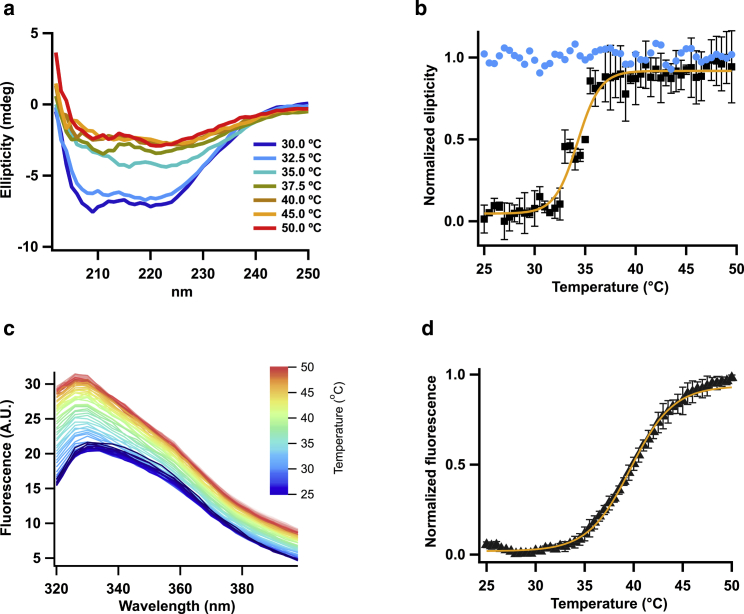
Because SYPRO thermal shifts are mostly qualitative, we compared the thermal sensitivity of the ARD by means of tryptophan fluorescence and CD experiments. Fig. 6 a shows far-ultraviolet CD spectra (between 200 and 240 nm) of ARD obtained at increasing temperatures (see Methods for details). The CD spectrum at 25°C is consistent with the high α-helical content of the ARD structure. The shape and ellipticity of individual spectra changes as temperature is increased, indicating the presence of thermal unfolding of the ARD.
Figure 6.
Thermal stability of ARD. (a) Shown are CD spectra of the ARD at the temperatures indicated in the inset. (b) The normalized ellipticity change at 208 nm is plotted as a function of temperature. Data are mean ± SE for n = 3 experiments (black circles). The blue circles are the data from one experiment at an attempt at refolding of the ARD by returning the temperature to 30°C and indicate an irreversible unfolding transition. The continuous curve represents a fit to a sigmoid function (slope 1.13 ± 0.14). The value of Tm from the fit is 34.2 ± 0.2°C. (c) Shown are tryptophan fluorescence spectra of the ARD at the temperatures indicated by the color scale. (d) The normalized fluorescence intensity increase for n = 3 experiments is plotted as the mean ± SE. The continuous curve represents a fit to a sigmoid function (slope 2.17 ± 0.10). The value of Tm from the fit of intensity data is 39.7 ± 0.1°C. To see this figure in color, go online.
Because the main CD signal is due to a macro dipole formed by the sum of dipoles of all the secondary structures, the decrease of ellipticity is characteristic of a random array of secondary structures, even when a single α helix does not have zero signal. In the case of the ARD, the loss of signal can be associated with an increase of mobility between motifs. To characterize the change in CD, we normalized the signal at 208 nm and plotted it as a function of temperature (Fig. 6 b). The unfolding occurs with a very steep dependence on temperature, with a Tm of ∼34°C, which is in agreement with the Tm observed in the thermal shift experiments. We attempted to measure the reversibility of the unfolding reaction; this result is plotted as blue circles in Fig. 6 b and shows that thermal unfolding of the ARD is irreversible. In comparison, the closest ARD previously studied is from the human TRPV4, which has a Tm of 37.1°C measured by CD, but its reversibility was not tested (39).
Thermal unfolding was also measured by tryptophan fluorescence experiments. The ARD has an exposed Trp at position 272 of the ANK4 repeat. Excitation of this Trp at 295 nm and measurement of the fluorescence at 330 nm as a function of increasing temperature shows an increase in the fluorescence counts (Fig. 6 c). The fluorescence increases (dequenching) in a temperature-dependent manner, and a sigmoid function was fitted with a Tm of 39.7°C (Fig. 6 d). Interestingly, the fluorescence of this Trp remains blue shifted (334 nm at 20°C to 327 nm at 50°C) as if it did not get exposed to solvent, and a temperature-dependent conformational change reduces quenching, perhaps separating Trp272 from a quenching residue.
Discussion
All TRPV channels of the thermo-TRP category are characterized by the presence of several ankyrin repeats in the N-terminus. Elucidation of their functional role has been a main objective of TRP channel structure-function studies. Other ARDs from different proteins show a variety of functions, from replication/translation regulation to protein-protein interaction and ligand binding (40, 41, 42).
TRPV1 is one of the best-studied members of the thermo-TRP channel subfamily. Notwithstanding the wealth of available information, the mechanisms of heat sensitivity and heat-dependent activation remain poorly understood. Several mechanisms of thermal sensing have been proposed. The transmembrane regions could contribute with a significant heat capacity and serve as thermal sensor domains (43,44), the PD could act as a temperature sensing domain (45), and the intracellular and transmembrane domains could act cooperatively to contribute to thermal sensation through a mechanism involving a linker domain (23). In TRPA1 channels, the ARD, although much bigger than the TRPV1 ARDs, have been implicated in temperature sensation (46,47).
A few TRPV1 structures determined by cryo-EM exist and are useful in interpreting experiments and proposing new ones, but the quality of these structures is, in general, poor. All of them are missing electronic density that should correspond to the initial ankyrin repeats of the ARD. Possible reasons for this are as follows: high density of the side chain of the amino acids, the presence of salt bridges, or the loss of density near Asp and Glu residues (48). Another possibility is higher mobility of this whole region of the protein structure. Our coarse-grained molecular dynamics simulations with a repaired model show that indeed the amino terminus and, in particular, the ARD are highly dynamic regions. An important result stemming from these simulations is that increased temperatures also modulate the high mobility of ARDs. Experimentally, the deletion of the ARD results in nonfunctional TRPV1 channels (15,36), a result that has hindered an understanding of the role of ARs in heat sensitivity and other forms of gating in this channel. The ARDs are responsible for autorecognition and probably an automodulation mechanism by self-interaction.
Our approach identifies the dynamics of the ARD of TRPV1 as important for the overall function of the ion channel. The low frequency movement represented by the first normal mode identified in this study suggests that the ARD is a dynamical module that might contribute to the response of TRPV1 to temperature. We propose that these fluctuations might be important in controlling temperature-dependent gating of TRPV1. At the very least, our results highlight the importance of the ARD and its interactions with other parts of the channel in regulating channel gating.
The structure of the ARD of TRPV1 was previously solved by crystallography (26). These authors also showed that basic pH is better for ARD purification.
Our data from CGMD simulations support a highly dynamical conformation of the ARD and show the breaking of some contacts between the second and third ankyrin repeat motifs at higher temperatures. This kind of perturbation may decrease the stability of the ARD as illustrated by the unfolding experiments in designed ARDs (41,49) and might contribute to a conformational change coupled to channel opening.
A previous study using coarse-grained molecular dynamics also identified a possible allosteric pathway of activation that initiated at intracellular domains (50). Our simulations are consistent with this result and provide further dissection of the specific role of the ARD. Fully atomistic simulations have identified specific residues involved in the opening transition at the PD (51,52).
Our biochemical data provide experimental support for the fluctuations observed in the simulations. Importantly, we observe conformational changes in the structure of the isolated ARD that occur at physiologically relevant temperatures. It is entirely possible that the Tms of these ARDs measured in isolation would not correlate with the actual range of activation of the parent channel because factors such as the solutions employed for experiments or the absence of a membrane context are entirely different. Also, an important factor that will almost certainly determine this correlation is the coupling between heat-sensitive transitions and actual channel opening.
Our results indicate that the structure of the ARD undergoes important changes over a range of ∼25°C. The CD data indicates that at 50°C, the structure is highly altered but still contains a significant amount of α-helical structure. This result might indicate that the ARD is not completely unfolded into a random coil. This is also indicated by the fact that the tryptophan fluorescence emission spectrum, which measures a local conformational change, remains blue shifted through the heating process, indicating that the local structure around it is preserved. An important observation is the fact that this structural change is not reversible. In accordance with this, we have recently shown that temperature-dependent gating of full-length TRPV1 is also irreversible, and this irreversibility is reflected in a large hysteresis during the activation processes (53). It is thus possible that the behavior of the ARD observed here could be related to this irreversible gating process.
Some naturally occurring splicing variants of TRPV1 have been described. The ARD and MPD are part of two different exons, and the splicing variants ARD-less and MPD-less express ion channels that are not sensitive to temperature (54). In our simulations and structural analysis, it is observed that the distal ARD (ANK1–2) approaches the MPD domain. This interaction is hard to measure by molecular dynamics because it is known that, in this region, a C-terminal peptide forms a third β strand of the MPD finger loop. Our analysis of the correlations of fluctuation during a simulated trajectory also highlights this interaction because the highest correlated movements happen precisely between the proximal ARD and the MPD.
The importance of the ARD-MPD interaction in TRPV1 function is also highlighted by experiments by Yao et al., in which several TRPV1-MPD chimeras were generated between rat TRPV2, human TRPV2, murine TRPV3, and murine TRPV4. All the chimeras increased the sensitivity to temperature. These results are consistent with the ARD being in direct contact with the adjacent MPD in such a way that its ability to interact with a viable MPD might play a role in the capacity of the TRPV channels to being thermally sensitive.
In conclusion, structure-function studies and natural occurring variants of the N-terminus (ARD + MPD) support its fundamental role for TRPV1 function. Our study contributes to an understanding of the dynamics of ARD and suggests that it participates in the regulation of thermal sensing, although its function as a thermal sensor or an element in temperature coupling remains to be elucidated.
Author Contributions
E.L.-d.-G. designed experiments, performed experiments and simulations, analyzed data, and wrote the article. L.D. performed simulations, analyzed data, and read the article. G.E.R.-Y. contributed materials, performed molecular biology, and read the article. D.A.F.-V. helped with CD and fluorescence experiments, read the article, and analyzed data. A.T.-L. helped with SECS-MALS and thermal shift experiments, read the article, and analyzed data. T.R. designed experiments and wrote the article. L.D.I. designed experiments, analyzed data, and wrote the article.
Acknowledgments
E.L.-d.-G. is grateful to Consejo Nacional de Ciencia y Tecnología for a Ph.D. scholarship (grant 297659). We acknowledge support from DGAPA-PAPIIT (Direccion General de Asuntos del Personal Academico-Programa de Apoyo a Proyectos de Investigacion e Inovacion Tecnologica) grant IN209515 to L.D.I. and IN200717 to T.R. We also acknowledge support from Consejo Nacional de Ciencia y Tecnología-Ciencia Básica grant 252644 to L.D.I. and A1-S-8760 to T.R. We are especially grateful to Dirección General de Cómputo y de Tecnologías de Información y Comunicación-UNAM Laboratorio Nacional de Cómputo de Alto Desempeño-UNAM-Dirección General de Cómputo y de Tecnologías de Información y Comunicación project No. 290 for use of the Miztli Supercomputer. This article is dedicated to the memory of E.L.-d.-G.
Editor: Baron Chanda.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.10.041.
Supporting Material
References
- 1.Aneiros E., Cao L., Grimm C. The biophysical and molecular basis of TRPV1 proton gating. EMBO J. 2011;30:994–1002. doi: 10.1038/emboj.2011.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caterina M.J., Schumacher M.A., Julius D. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature. 1997;389:816–824. doi: 10.1038/39807. [DOI] [PubMed] [Google Scholar]
- 3.Salazar H., Llorente I., Rosenbaum T. A single N-terminal cysteine in TRPV1 determines activation by pungent compounds from onion and garlic. Nat. Neurosci. 2008;11:255–261. doi: 10.1038/nn2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caterina M.J., Julius D. The vanilloid receptor: a molecular gateway to the pain pathway. Annu. Rev. Neurosci. 2001;24:487–517. doi: 10.1146/annurev.neuro.24.1.487. [DOI] [PubMed] [Google Scholar]
- 5.Pingle S.C., Matta J.A., Ahern G.P. Capsaicin receptor: TRPV1 a promiscuous TRP channel. Handb. Exp. Pharmacol. 2007;179:155–171. doi: 10.1007/978-3-540-34891-7_9. [DOI] [PubMed] [Google Scholar]
- 6.Caterina M.J., Leffler A., Julius D. Impaired nociception and pain sensation in mice lacking the capsaicin receptor. Science. 2000;288:306–313. doi: 10.1126/science.288.5464.306. [DOI] [PubMed] [Google Scholar]
- 7.Yarmolinsky D.A., Peng Y., Zuker C.S. Coding and plasticity in the mammalian thermosensory system. Neuron. 2016;92:1079–1092. doi: 10.1016/j.neuron.2016.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang S.M., Li X., Caterina M.J. TRPV3 and TRPV4 ion channels are not major contributors to mouse heat sensation. Mol. Pain. 2011;7:37. doi: 10.1186/1744-8069-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park U., Vastani N., Caterina M.J. TRP vanilloid 2 knock-out mice are susceptible to perinatal lethality but display normal thermal and mechanical nociception. J. Neurosci. 2011;31:11425–11436. doi: 10.1523/JNEUROSCI.1384-09.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iida T., Shimizu I., Caterina M. Attenuated fever response in mice lacking TRPV1. Neurosci. Lett. 2005;378:28–33. doi: 10.1016/j.neulet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 11.Nieto-Posadas A., Picazo-Juárez G., Rosenbaum T. Lysophosphatidic acid directly activates TRPV1 through a C-terminal binding site. Nat. Chem. Biol. 2011;8:78–85. doi: 10.1038/nchembio.712. [DOI] [PubMed] [Google Scholar]
- 12.Ufret-Vincenty C.A., Klein R.M., Gordon S.E. Mechanism for phosphoinositide selectivity and activation of TRPV1 ion channels. J. Gen. Physiol. 2015;145:431–442. doi: 10.1085/jgp.201511354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao E., Liao M., Julius D. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature. 2013;504:113–118. doi: 10.1038/nature12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao Y., Cao E., Cheng Y. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature. 2016;534:347–351. doi: 10.1038/nature17964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao M., Cao E., Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zubcevic L., Herzik M.A., Jr., Lee S.Y. Cryo-electron microscopy structure of the TRPV2 ion channel. Nat. Struct. Mol. Biol. 2016;23:180–186. doi: 10.1038/nsmb.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zubcevic L., Herzik M.A., Jr., Lee S.Y. Conformational ensemble of the human TRPV3 ion channel. Nat. Commun. 2018;9:4773. doi: 10.1038/s41467-018-07117-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng Z., Paknejad N., Yuan P. Cryo-EM and X-ray structures of TRPV4 reveal insight into ion permeation and gating mechanisms. Nat. Struct. Mol. Biol. 2018;25:252–260. doi: 10.1038/s41594-018-0037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes T.E.T., Pumroy R.A., Moiseenkova-Bell V.Y. Structural insights on TRPV5 gating by endogenous modulators. Nat. Commun. 2018;9:4198. doi: 10.1038/s41467-018-06753-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGoldrick L.L., Singh A.K., Sobolevsky A.I. Opening of the human epithelial calcium channel TRPV6. Nature. 2018;553:233–237. doi: 10.1038/nature25182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De-la-Rosa V., Rangel-Yescas G.E., Islas L.D. Coarse architecture of the transient receptor potential vanilloid 1 (TRPV1) ion channel determined by fluorescence resonance energy transfer. J. Biol. Chem. 2013;288:29506–29517. doi: 10.1074/jbc.M113.479618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenbaum T., Gordon-Shaag A., Gordon S.E. Ca2+/calmodulin modulates TRPV1 activation by capsaicin. J. Gen. Physiol. 2004;123:53–62. doi: 10.1085/jgp.200308906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao J., Liu B., Qin F. Modular thermal sensors in temperature-gated transient receptor potential (TRP) channels. Proc. Natl. Acad. Sci. USA. 2011;108:11109–11114. doi: 10.1073/pnas.1105196108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh A.K., McGoldrick L.L., Sobolevsky A.I. Structure and gating mechanism of the transient receptor potential channel TRPV3. Nat. Struct. Mol. Biol. 2018;25:805–813. doi: 10.1038/s41594-018-0108-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kasimova M.A., Yazici A., Carnevale V. Ion channel sensing: are fluctuations the crux of the matter? J. Phys. Chem. Lett. 2018;9:1260–1264. doi: 10.1021/acs.jpclett.7b03396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lishko P.V., Procko E., Gaudet R. The ankyrin repeats of TRPV1 bind multiple ligands and modulate channel sensitivity. Neuron. 2007;54:905–918. doi: 10.1016/j.neuron.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 27.Chuang H.H., Lin S. Oxidative challenges sensitize the capsaicin receptor by covalent cysteine modification. Proc. Natl. Acad. Sci. USA. 2009;106:20097–20102. doi: 10.1073/pnas.0902675106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bordoli L., Schwede T. Automated protein structure modeling with SWISS-MODEL workspace and the protein model portal. Methods Mol. Biol. 2012;857:107–136. doi: 10.1007/978-1-61779-588-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waterhouse A., Bertoni M., Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Jong D.H., Singh G., Marrink S.J. Improved parameters for the Martini coarse-grained protein force field. J. Chem. Theory Comput. 2013;9:687–697. doi: 10.1021/ct300646g. [DOI] [PubMed] [Google Scholar]
- 31.Hsu P.C., Bruininks B.M.H., Im W. CHARMM-GUI Martini Maker for modeling and simulation of complex bacterial membranes with lipopolysaccharides. J. Comput. Chem. 2017;38:2354–2363. doi: 10.1002/jcc.24895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pronk S., Páll S., Lindahl E. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Der Spoel D., Lindahl E., Berendsen H.J. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 34.Grant B.J., Rodrigues A.P., Caves L.S. Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics. 2006;22:2695–2696. doi: 10.1093/bioinformatics/btl461. [DOI] [PubMed] [Google Scholar]
- 35.Sippl M.J., Wiederstein M. Detection of spatial correlations in protein structures and molecular complexes. Structure. 2012;20:718–728. doi: 10.1016/j.str.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hellwig N., Albrecht N., Schaefer M. Homo- and heteromeric assembly of TRPV channel subunits. J. Cell Sci. 2005;118:917–928. doi: 10.1242/jcs.01675. [DOI] [PubMed] [Google Scholar]
- 37.Hinsen K. Analysis of domain motions by approximate normal mode calculations. Proteins. 1998;33:417–429. doi: 10.1002/(sici)1097-0134(19981115)33:3<417::aid-prot10>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 38.Skjaerven L., Martinez A., Reuter N. Principal component and normal mode analysis of proteins; a quantitative comparison using the GroEL subunit. Proteins. 2011;79:232–243. doi: 10.1002/prot.22875. [DOI] [PubMed] [Google Scholar]
- 39.Inada H., Procko E., Gaudet R. Structural and biochemical consequences of disease-causing mutations in the ankyrin repeat domain of the human TRPV4 channel. Biochemistry. 2012;51:6195–6206. doi: 10.1021/bi300279b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barrick D., Ferreiro D.U., Komives E.A. Folding landscapes of ankyrin repeat proteins: experiments meet theory. Curr. Opin. Struct. Biol. 2008;18:27–34. doi: 10.1016/j.sbi.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferreiro D.U., Cervantes C.F., Komives E.A. Stabilizing IkappaBalpha by “consensus” design. J. Mol. Biol. 2007;365:1201–1216. doi: 10.1016/j.jmb.2006.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mosavi L.K., Cammett T.J., Peng Z.Y. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13:1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chowdhury S., Jarecki B.W., Chanda B. A molecular framework for temperature-dependent gating of ion channels. Cell. 2014;158:1148–1158. doi: 10.1016/j.cell.2014.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma L., Yang F., Zheng J. Exploring functional roles of TRPV1 intracellular domains with unstructured peptide-insertion screening. Sci. Rep. 2016;6:33827. doi: 10.1038/srep33827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang F., Jara-Oseguera A., Swartz K.J. Heat activation is intrinsic to the pore domain of TRPV1. Proc. Natl. Acad. Sci. USA. 2018;115:E317–E324. doi: 10.1073/pnas.1717192115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cordero-Morales J.F., Gracheva E.O., Julius D. Cytoplasmic ankyrin repeats of transient receptor potential A1 (TRPA1) dictate sensitivity to thermal and chemical stimuli. Proc. Natl. Acad. Sci. USA. 2011;108:E1184–E1191. doi: 10.1073/pnas.1114124108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jabba S., Goyal R., Grandl J. Directionality of temperature activation in mouse TRPA1 ion channel can be inverted by single-point mutations in ankyrin repeat six. Neuron. 2014;82:1017–1031. doi: 10.1016/j.neuron.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hryc C.F., Chen D.H., Chiu W. Accurate model annotation of a near-atomic resolution cryo-EM map. Proc. Natl. Acad. Sci. USA. 2017;114:3103–3108. doi: 10.1073/pnas.1621152114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parra R.G., Espada R., Ferreiro D.U. Structural and energetic characterization of the ankyrin repeat protein family. PLoS Comput. Biol. 2015;11:e1004659. doi: 10.1371/journal.pcbi.1004659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng W., Qin F. A combined coarse-grained and all-atom simulation of TRPV1 channel gating and heat activation. J. Gen. Physiol. 2015;145:443–456. doi: 10.1085/jgp.201411335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Darré L., Furini S., Domene C. Permeation and dynamics of an open-activated TRPV1 channel. J. Mol. Biol. 2015;427:537–549. doi: 10.1016/j.jmb.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 52.Kasimova M.A., Yazici A.T., Carnevale V. A hypothetical molecular mechanism for TRPV1 activation that invokes rotation of an S6 asparagine. J. Gen. Physiol. 2018;150:1554–1566. doi: 10.1085/jgp.201812124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sánchez-Moreno A., Guevara-Hernández E., Islas L.D. Irreversible temperature gating in trpv1 sheds light on channel activation. eLife. 2018;7:e36372. doi: 10.7554/eLife.36372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schumacher M.A., Moff I., Levine J.D. Molecular cloning of an N-terminal splice variant of the capsaicin receptor. Loss of N-terminal domain suggests functional divergence among capsaicin receptor subtypes. J. Biol. Chem. 2000;275:2756–2762. doi: 10.1074/jbc.275.4.2756. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.