Abstract
G protein-coupled receptors (GPCRs) are important membrane proteins in higher eukaryotes that carry out a vast array of cellular signaling and act as major drug targets. The serotonin1A receptor is a prototypical member of the GPCR family and is implicated in neuropsychiatric disorders such as anxiety and depression, besides serving as an important drug target. With an overall goal of exploring the functional consequence of altered receptor dynamics, in this work, we probed the role of the actin cytoskeleton in the dynamics, ligand binding, and signaling of the serotonin1A receptor. We monitored receptor dynamics utilizing single particle tracking, which provides information on relative distribution of receptors in various diffusion modes in addition to diffusion coefficient. We show here that the short-term diffusion coefficient of the receptor increases upon actin destabilization by cytochalasin D. In addition, analysis of individual trajectories shows that there are changes in relative populations of receptors undergoing various types of diffusion upon actin destabilization. The release of dynamic constraint was evident by an increase in the radius of confinement of the receptor upon actin destabilization. The functional implication of such actin destabilization was manifested as an increase in specific agonist binding and downstream signaling, monitored by measuring reduction in cellular cAMP levels. These results bring out the interdependence of GPCR dynamics with cellular signaling.
Significance
Biological membranes carry many cellular functions, mediated by membrane proteins. An important function is signaling, which enables the cellular exterior to cross talk with the cellular interior. Although signaling is routinely measured (based on specific readouts), analysis of physical factors contributing to signaling is scant. We report here that receptor lateral diffusion is a major factor in signaling. We measured lateral diffusion of an important G protein-coupled receptor, the serotonin1A receptor, which acts as a signaling hub, using single particle tracking under control and actin-destabilized conditions. Our analysis shows that actin destabilization leads to change in receptor diffusion, which manifests as an increase in functional readouts (ligand binding and cAMP signaling). Our results demonstrate the interdependence of membrane protein dynamics with signaling.
Introduction
G protein-coupled receptors (GPCRs) are the largest and most diverse membrane protein family in eukaryotes. They are involved in a gamut of signal transduction processes across the plasma membrane (1, 2, 3). A common mechanism of signal transduction by GPCRs requires their activation by extracellular ligands followed by relay of signals to the cellular interior through coordinated structural changes in their transmembrane (or extramembranous) regions (4, 5). GPCRs regulate several crucial physiological processes that include cellular metabolism, neurotransmission, growth, immune responses, and cellular differentiation. As a result, GPCRs have emerged as major drug targets (6, 7) and account for ∼40% of current drug targets across all clinical areas (8). However, only a small fraction (∼15%) of all GPCRs are current drug targets (9). This opens up novel avenues for future drug development for diseases that are currently not treatable by available drugs. The serotonin1A receptor is an important neurotransmitter GPCR, is extensively studied among the 16 serotonin receptors, and mediates a multitude of neurological, behavioral, and cognitive functions (10, 11, 12, 13, 14). The serotonin1A receptor plays a crucial role in human physiology and has emerged as an important drug target in developing therapeutics against neuropsychiatric disorders such as anxiety, depression, and even cancer (15, 16).
Membrane proteins carry out the majority of functions in biological membranes. Although ∼40% of genes present in humans code for membrane proteins (17, 18), the progress in structural biology of membrane proteins in general (19), and GPCRs in particular (20, 21) has been rather slow. In addition, it is becoming increasingly clear that we need to appreciate and understand characteristic dynamics and conformational plasticity of membrane proteins for deciphering their function (22, 23). Since membrane proteins are embedded in membrane lipids, knowledge of membrane lipid and protein dynamics is crucial for understanding cellular function. It is in this overall context that measurement of live cell membrane dynamics, with a goal of correlating it with receptor function, assumes relevance in contemporary membrane biology. Consequently, receptor dynamics (lateral diffusion) in membranes determines the overall efficacy of the signal transduction process (24, 25, 26).
Lateral diffusion in the plasma membrane constitutes a fundamental biophysical process that dictates the dynamics of lipid-protein and protein-protein interactions (27). Confined lateral diffusion of lipids and proteins is often associated with compositional heterogeneity (domain) in cell membranes over various spatiotemporal scales (28). A major source of confined lateral diffusion in membranes is the presence of an intricate network of actin cytoskeleton below the membrane (29, 30, 31). Actin is one of the most ubiquitous cytosolic proteins in eukaryotic cells and is maintained in a dynamic equilibrium between polymeric (filamentous or F-actin) and monomeric (globular or G-actin) forms (32). The dynamic equilibrium between actin polymerization and depolymerization is coordinated by various actin binding proteins implicated in a wide range of physiological processes (33, 34). Such tunable equilibrium between F- and G-actin serves as a common mechanism for various signal transduction processes (35). Although evidence of direct interaction of actin cytoskeleton with GPCRs is lacking (36), lateral dynamics of membrane proteins and lipids is shown to be affected by the actin cytoskeleton (26, 31, 37, 38). The destabilization of the actin cytoskeleton could therefore provide a useful handle to explore the cytoskeleton-dependent lateral diffusion of membrane components.
Measurement of membrane dynamics could be challenging because of inherent noise in cellular systems. Nonetheless, significant progress has been made in recent years toward the measurement of membrane dynamics that allows the detection, identification, and tracking of individual membrane-bound molecules with high spatiotemporal resolution under physiological conditions (39, 40, 41, 42). A major advantage of imaging single-molecule diffusion is that it circumvents ensemble averaging from multiple molecules. As a result, single-molecule based approaches enable observation of heterogeneous behavior (and transient phenomena) within subpopulations of molecules (43). Single-molecule techniques such as single particle tracking (SPT) have emerged as a powerful approach to explore the organization and lateral diffusion of membrane-bound molecules (31, 42, 44, 45). Although SPT has been used to probe dynamics of membrane-bound molecules (39, 46), the application of SPT to GPCRs is relatively rare (47, 48). In this work, we explored the organization and dynamics of the serotonin1A receptor in live cells utilizing an SPT approach under conditions of actin cytoskeleton destabilization. By analysis of diffusion modes, we further demonstrated how actin cytoskeleton could influence diffusion behavior of the serotonin1A receptor. Importantly, our results show that such actin-cytoskeleton-dependent dynamics could play a major role in GPCR function and regulate downstream signaling.
Materials and Methods
Materials
Bovine serum albumin (BSA), CaCl2, cytochalasin D (CD), dimethyl sulfoxide (DMSO), doxycycline, D-glucose, EDTA, forskolin, gentamycin sulfate, 3-isobutyl-1-methylxanthine (IBMX), MgCl2, MnCl2, NaHCO3, penicillin, (4-(2′-methoxy)-phenyl-1-[2′-(N-2″-pyridinyl)-p-fluorobenzamido]ethyl-piperazine (p-MPPF), serotonin, streptomycin, Triton X-100, and Tris were purchased from Sigma-Aldrich (St. Louis, MO). 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) was obtained from Calbiochem (San Diego, CA). Alexa Fluor 546 phalloidin and Qdot 655 streptavidin conjugate were from Molecular Probes (Eugene, OR). Anti-myc tag antibody C-terminal (biotin) was purchased from Abcam (Cambridge, UK). Anti-myc antibody Alexa Fluor 488 conjugate was purchased from Millipore (Bedford, MA). Dulbecco’s modified Eagle’s medium (DMEF): nutrient mixture F-12 (Ham) (1:1), fetal calf serum (FCS), and hygromycin B were obtained from Invitrogen Life Technologies (Carlsbad, CA). [3H]8-hydroxy-2(di-N-propylamino)tetralin ([3H]8-OH-DPAT), specific activity 141 Ci/mmol and [3H]p-MPPF, specific activity 74.2 Ci/mmol were obtained from MP Biomedicals (Santa Ana, CA). GF/B glass microfiber filters were from Whatman International (Kent, UK). Homogeneous time-resolved fluorescence (HTRF) cAMP-Gi assay kit was purchased from CisBio Bioassays (Codolet, France). Purified water through a Millipore Milli-Q system was used for all experiments.
Cells and cell culture
Human embryonic kidney (HEK)-293 cells stably expressing the N-terminal myc-tagged serotonin1A receptor were generated as described previously (49). Cells were maintained in DMEM/F-12 (1:1) supplemented with 10% FCS, 60 μg/mL penicillin, 50 μg/mL streptomycin, 50 μg/mL gentamycin sulfate, and 250 μg/mL hygromycin B (complete DMEM/F-12) in a humidified incubator with 5% CO2 at 37°C. The cell culture medium was supplemented with 1 μg/mL doxycycline 24 h before experiments for inducing receptor expression.
CD treatment of cells
Actin cytoskeleton destabilization in HEK-293 cells expressing the myc-tagged serotonin1A receptor was performed utilizing CD as described earlier (26, 50, 51) with few modifications. Briefly, 2 mM CD stock solution was prepared in DMSO and working stock was made upon diluting this stock in serum-free DMEM/F-12 medium. Cells were grown in complete DMEM/F-12 media for 3 days and subsequently incubated in serum-free DMEM/F-12 media for 3 h. Actin cytoskeleton was destabilized by treatment of HEK-293 cells with 5 μM CD in serum-free DMEM/F-12 medium for 30 min at 37°C. After CD treatment, the media was discarded, and cells were gently washed using phosphate-buffered saline (PBS) to remove excess CD.
MTT viability assay
Viability of HEK-293 cells under actin destabilization condition was assessed utilizing MTT assay as described earlier (51).
F-actin labeling of cells
F-actin labeling in cells was performed as described previously (35). Briefly, HEK-293 cells expressing the myc-tagged serotonin1A receptor were plated at a density of ∼104 cells on 22 mm glass coverslips, grown in complete DMEM/F-12 medium for 3 days, and subsequently incubated in serum-free DMEM/F-12 medium for 3 h. Following this, cells were treated with 5 μM CD as described above. After CD treatment, cells were washed with buffer A (PBS containing 0.5 mM MgCl2 and 1 mM CaCl2) and fixed with 3.5% (v/v) formaldehyde for 10 min. The fixed cells were permeabilized in buffer A containing 0.5% Triton X-100 (v/v) for 6 min. Cells were washed with buffer A, stained with Alexa Fluor 546 phalloidin for 60 min at room temperature, and subsequently mounted on glass slides.
Fluorescence microscopy and F-actin quantitation
F-actin was imaged on an inverted Zeiss LSM 510 Meta confocal microscope (Jena, Germany). Imaging of F-actin cytoskeleton was carried out by exciting Alexa Fluor 546 phalloidin using a 561 nm diode pumped solid state (DPSS) laser, and emission was collected using a bandpass filter of 575–630 nm. The level of F-actin was estimated using a quantitative high-resolution confocal microscopic technique previously developed by us (35). Briefly, z-section images were acquired using a 63×/1.4 NA oil immersion objective with a fixed z-step size of 0.32 μm under 1 airy condition. Maximal intensity projections of 11 z-sections (corresponding to ∼3.5 μm from the base into the cell) were obtained, and the area of the projected images was calculated using the analysis software provided with the microscope. Iso-surfaces (defined as voxel contours of equal fluorescence intensity) were generated from the z-sections using Imaris 6.0.0 (Bitplane, Zurich, Switzerland) after thresholding the fluorescence intensity of z-sections, followed by application of a Gaussian filter. The estimated volumes of actin cytoskeleton enclosed by isosurfaces were normalized to the projected total area of cells for a given field.
Confocal microscopic imaging of receptor localization
To obtain representative confocal microscopic images for localization of the receptor, HEK-293 cells expressing the myc-tagged serotonin1A receptor were plated at a density of ∼104 cells on 22 mm glass coverslips and grown in complete DMEM/F-12 medium for 3 days. Cells were washed with PBS, fixed with 3.5% (v/v) formaldehyde for 10 min, and subsequently permeabilized with 0.5% Triton X-100 (v/v) for 6 min. Cells expressing the serotonin1A receptor with a myc-tag were then stained with anti-myc antibody Alexa Fluor 488 conjugate (1:100 dilution) in PBS for 1 h, washed with PBS, and mounted using Vectashield antifade mounting medium containing 4',6-diamidino-2-phenylindole (DAPI). Imaging was carried out on an inverted Zeiss LSM 880 confocal microscope. Serotonin1A receptors with an N-terminal myc-tag were imaged by exciting anti-myc antibody Alexa Fluor 488 conjugate using the 488 nm line of an argon laser and the emission was collected from 500 to 560 nm. DAPI was excited at 405 nm, and emission was collected from 410 to 460 nm. Midplane images of cells were acquired with a 63×/1.4 NA oil immersion objective under 1 airy condition.
Flow cytometric analysis of receptor expression level
HEK-293 cells expressing the serotonin1A receptor were collected in cold PBS on ice. Cells were fixed with 3.5% (v/v) formaldehyde for 30 min on ice and centrifuged at 800 × g for 3 min at 4°C. Cells were washed with PBS, and the plasma membrane associated myc-tagged serotonin1A receptors were stained with anti-myc antibody Alexa Fluor 488 conjugate (1:100 dilution) in PBS containing 2% FCS for 1 h on ice. Subsequently, cells were washed in cold PBS and then resuspended in PBS containing 2% FCS. The receptor population associated with membranes was quantified using a MoFlo flow cytometer (Dako Cytomation, Fort Collins, CO), and data were acquired and analyzed using Summit 5.4 analysis software (Beckman Coulter, Fullerton, CA). Alexa Fluor 488 was excited at 488 nm, and emission was collected using a 525/40 bandpass filter. Mode count values were obtained by flow cytometric analysis of 10,000 cells for each condition.
Radioligand binding assay in live cells
HEK-293 cells expressing the serotonin1A receptor were grown for 72 h in complete DMEM/F-12 media and were detached from culture flasks in PBS containing 0.25 mM EDTA. Cells were spun at 500 × g for 5 min and resuspended in serum-free DMEM/F-12 medium for counting using a hemocytometer. Cells (∼106) in serum-free DMEM/F-12 medium were incubated at ∼25°C for 15 min in presence of 1 nM [3H]8-OH-DPAT (specific agonist) or 1 nM [3H]p-MPPF (specific antagonist). Nonspecific binding was obtained by performing the assay in presence of 10 μM unlabeled serotonin (for [3H]8-OH-DPAT) or 10 μM unlabeled p-MPPF (for [3H]p-MPPF). Ligand binding was terminated by rapid filtration under vacuum through Millipore multiport filtration apparatus using Whatman GF/B glass microfiber filters (1 μm pore size) that were presoaked in 0.3% (w/v) polyethylenimine for 3 h (52). After washing the filters for three times with 5 ml of cold water (∼4°C) and subsequent drying, the remaining radioactivity was measured using ∼5 ml scintillation fluid in a Packard Tri-Carb 2900 liquid scintillation counter (Perkin Elmer, Waltham, MA).
Saturation binding assay
Saturation binding assays were carried out to estimate binding parameters in control and actin cytoskeleton destabilized cells. Assays were carried out with increasing concentrations of the radiolabeled agonist [3H]8-OH-DPAT (0.1–7.5 nM) in intact cells. Nonspecific binding was determined by performing the assay in presence of 10 μM unlabeled serotonin. Ligand-binding assays were carried out at ∼25°C with ∼106 cells for each ligand concentration. The concentration of bound ligand (RLbound) was determined from (49)
| (1) |
where B is the bound radioactivity in disintegrations per minute, V is the assay volume in ml, and SA is the specific activity of the radioligand. The saturation binding data could be fitted best to a one-site ligand binding equation. The binding parameters, the dissociation constant (Kd) and the number of maximal binding sites (Bmax) were calculated by nonlinear regression analysis of the binding data using GraphPad Prism software, version 4.0 (San Diego, CA).
Cellular signaling assay
HEK-293 cells stably expressing serotonin1A receptors were plated at a density of ∼105 cells in poly-l-lysine-coated six-well plates and grown in complete medium in a humidified incubator with 5% CO2 at 37°C for 48 h. After treatment with CD, cells were treated with 50 μM IBMX (basal), 50 μM IBMX/10 μM forskolin (forskolin-stimulated), or 50 μM IBMX/10 μM forskolin/10 μM serotonin (agonist treatment) and incubated for 30 min at 37°C. After discarding media, cells were washed once with PBS. Cells were lifted using a cell scraper, counted using a hemocytometer, and added at 6000 cells/well to a low-volume HTRF 96-well plate (CisBio Bioassays). The ability of the serotonin1A receptor to inhibit the forskolin-stimulated increase in cAMP levels was assessed using the fluorescence resonance energy transfer (FRET)-based HTRF cAMP-Gi assay kit (CisBio Bioassays). Fluorescence was measured at 620 nm (cAMP-cryptate donor emission) and 655 nm (anti-cAMP-d2 acceptor emission) upon excitation of the donor at 320 nm using an EnSpire Multimode Plate Reader (Perkin Elmer). cAMP levels were calculated as a ratio of the acceptor/donor emission. Values for serotonin-induced cAMP reduction were normalized and expressed as a percentage to the values obtained for only IBMX-treated cells.
SPT experiments
HEK-293 cells expressing the N-terminal myc-tagged serotonin1A receptor were plated on coverslips 72 h before the experiments. Streptavidin-conjugated quantum dots (QDs) were precoupled to a biotinylated anti-c-myc antibody (at a molar ratio of 10:1, respectively) by incubating for 15 min in PBS with 1% BSA (w/v) and 2% D-glucose (w/v) (buffer B). Before microscopic observations, the coverslips were mounted in a chamber (25 mm inner diameter), and myc-tagged serotonin1A receptors were labeled with 0.03 nM antibody-conjugated QDs in 100 μL buffer B at room temperature (∼22°C) for 30 min. Subsequently, cells were washed three times with 3 mL buffer B to remove any unbound QDs. Trajectories of QDs at the cell surface were tracked with a Cascade II 512 EM-CCD camera (Roper Scientific, Tucson, AZ) operating at 25 Hz acquisition frequency on a Zeiss Axio-observer A1 microscope at room temperature (∼22°C). QDs were illuminated with an X-CITE 120 light source with a metal halide vapor short arc lamp and observed through a Fluar 100×/1.3 NA oil immersion ultraviolet objective associated to a 1.6× multiplier tube lens connected to the camera. The duration of the recordings was set to 80 s.
The trajectories of all QDs in a video sequence were analyzed using the Multiple Target Tracing program developed by Sergé et al. (53). Trajectories of more than 20 s were further analyzed with a home-made program written in Visual Basic (VBA Excel, Microsoft) computing the mean-square displacement as a function of time interval, MSD(nδt), using the equation:
| (2) |
where n is the number of time intervals, δt is the time interval between two successive frames (40 ms), N is the total number of frames (in our case, N ≤ 2000 frames), and x(t) and y(t) are the QD coordinates at time t. For obtaining accurate and precise values of the diffusion coefficient, short-term diffusion coefficient (D1–2) was estimated from the first two MSD(nδt) points D1–2 = [MSD(2δt) − MSD(δt)]/(4δt) independent of the diffusion mode of the particles (54, 55). To identify transient or continuous confinement events, a confinement index Λ(t), as established by Meilhac et al. (56), was calculated on sliding intervals using the equation
| (3) |
where Δr2 is the variance of the trajectory segment of duration Δt under study. A value of Λ(t) > 4 for a period longer than Δt is characteristic of confined diffusion. The size of the domains and the diffusion coefficient inside the confined trajectory segments were determined by fitting MSD(t) with its theoretical expression for confined diffusion (56). The unconfined trajectories, partial or total, were analyzed using standard procedures (57) and classified as having either random or directed diffusion.
Results
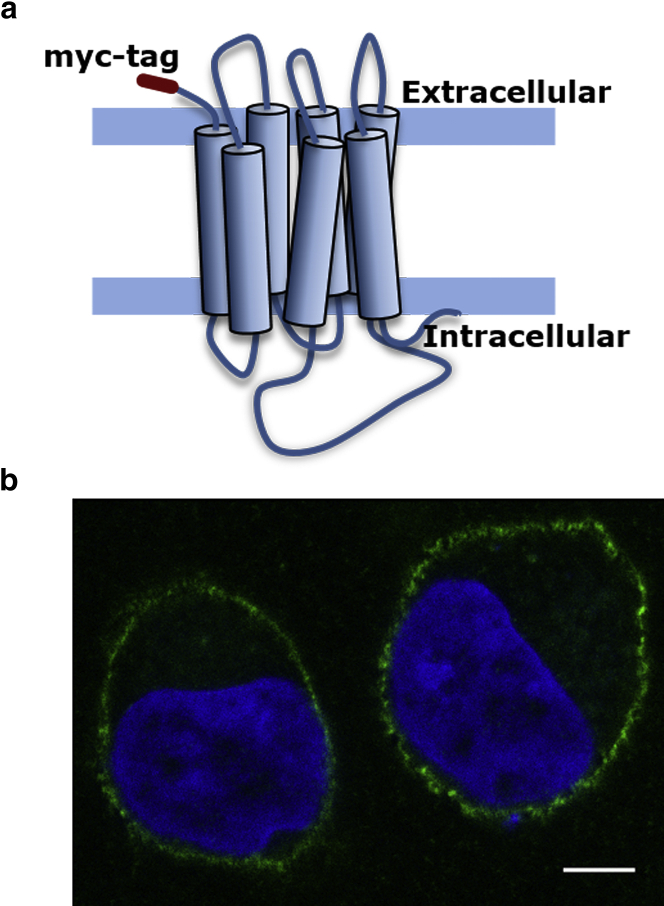
We utilized HEK-293 cells stably expressing serotonin1A receptors (generated by transfection with a construct encoding the receptor with a N-terminal myc-tag; see Fig. 1 a) for measuring receptor dynamics, ligand binding, and cellular signaling. This stable cell line allows us to fluorescently label the serotonin1A receptor with streptavidin-conjugated QDs precoupled to a biotinylated anti-c-myc antibody and track the trajectory of the receptor by SPT. Confocal microscopic imaging at a midplane section of HEK-293 cells showed a characteristic plasma membrane localization of the serotonin1A receptor (Fig. 1 b). Importantly, we recently showed that the serotonin1A receptor expressed in HEK-293 cells with N-terminal myc-tag retains all characteristics of the native receptor in terms of ligand binding, G-protein coupling, and receptor trafficking (49).
Figure 1.
(a) A schematic representation of topological features of the human serotonin1A receptor with a myc-tag (maroon) at its N-terminal stably expressed in HEK-293 cells. The boundaries of the membrane are shown as blue horizontal bars. (b) A representative confocal microscopic image of HEK-293 cells showing the distribution of the myc-tagged serotonin1A receptor labeled with anti-myc antibody conjugated to Alexa Fluor 488 (green). The nucleus is labeled with DAPI (blue). The scale bar represents 10 μm. To see this figure in color, go online.
Treatment with cytochalasin D leads to loss of F-actin in HEK-293 cells
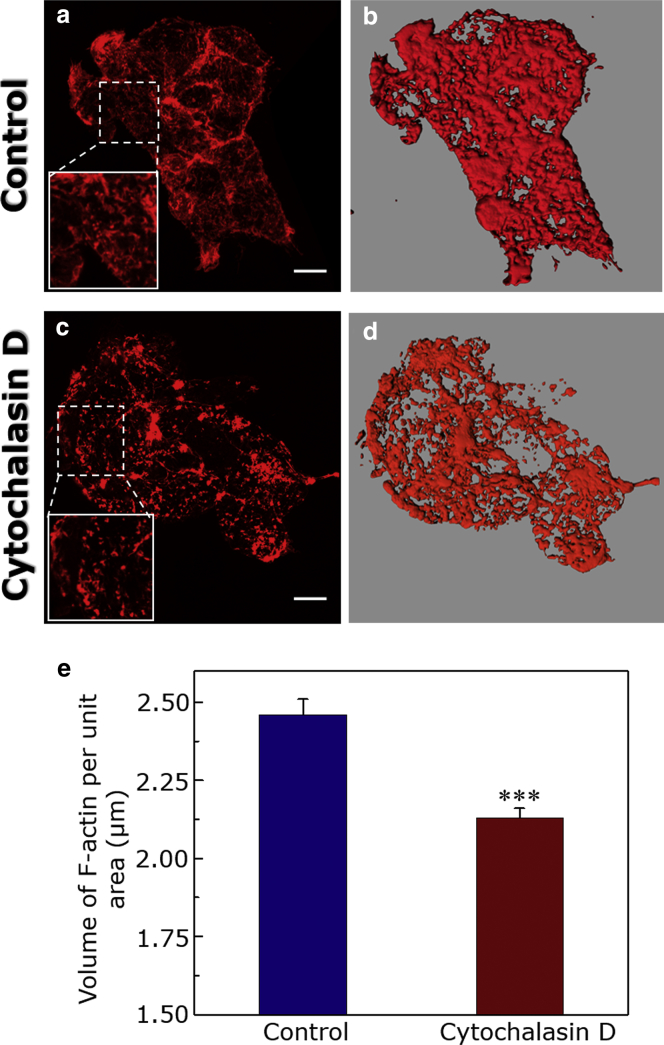
Cytochalasin D (CD), a potent inhibitor of actin polymerization, is commonly used to destabilize F-actin in live cells (26, 50). In vitro studies have shown that CD shifts the equilibrium toward F-actin depolymerization upon binding to the barbed (fast-growing) end of F-actin filaments (58). In vivo action of CD has been attributed to the above effect that is observed in vitro and a secondary cellular response (59). To analyze lateral diffusion of the receptor as a consequence of reorganization of the actin cytoskeleton, it is crucial to quantitatively measure the amount of F-actin in any given condition. Unfortunately, intensity-based analysis for quantitating the reorganization of actin cytoskeleton is often complicated by the fact that CD treatment leads to fragmentation of F-actin into smaller aggregates, which appear brighter under a fluorescence microscope. To circumvent this problem, we previously developed a quantitative high-resolution microscopy-based approach to measure F-actin content using an image reconstruction method (35). To quantitatively estimate the extent of F-actin depolymerization, cells were treated with 5 μM CD, and F-actin was labeled with Alexa Fluor 546 phalloidin. As shown in Fig. 2, treatment of HEK-293 cells with CD resulted in considerable fragmentation of F-actin (Fig. 2 b). The figure shows confocal images of maximal intensity projections of the actin cytoskeleton (stained with Alexa Fluor 546 phalloidin) of HEK-293 cells treated with CD. We found that treatment with higher concentrations of CD in HEK-293 cells changes the overall cellular morphology within few minutes of incubation. We therefore chose an optimal concentration (5 μM) and time period (30 min) of CD treatment to retain the cellular morphology during subsequent experiments. To assess the effect of actin destabilization on viability of HEK-293 cells, we measured cell viability using MTT viability assay after 5 μM CD treatment. Cell viability was not affected at 5 μM CD, which was used during subsequent experiments (see Fig. S1 a). Importantly, actin-destabilized cells remain viable for at least 30 min, even after removal of CD from the culture medium (Fig. S1 a), and retained healthy morphology after actin destabilization (Fig. S1 b). This ensures the reliability of SPT data from actin cytoskeleton destabilized cells.
Figure 2.
Reorganization of actin cytoskeleton in HEK-293 cells expressing the serotonin1A receptor. The actin cytoskeleton of HEK-293 cells was stained with Alexa Fluor 546 phalloidin. Imaging of F-actin cytoskeleton was carried out by exciting Alexa Fluor 546 phalloidin using a 561 nm DPSS laser, and emission was collected using a bandpass filter of 575–630 nm. The level of F-actin was estimated using a quantitative high-resolution confocal microscopic technique previously developed by us (35). z-section images were acquired using a 63×/1.4 NA oil immersion objective with a fixed z-step size of 0.32 μm under 1 airy condition. (a) and (c) show maximal intensity projections of 11 sections from the base of the coverslip (∼3.5 μm from the base into the cell) of the actin cytoskeleton in control and 5 μM CD-treated cells, respectively. Loss of F-actin filaments and formation of F-actin aggregates can be observed upon treatment with 5 μM of CD. The scale bars represent 10 μm. Iso-surfaces (defined as voxel contours of equal fluorescence intensity) were generated from the z-sections using Imaris 6.0.0 (Bitplane) after thresholding the fluorescence intensity of z-sections, followed by application of a Gaussian filter, and are shown in (b) and (d). To quantitate F-actin, the volume enclosed by isosurfaces was normalized to the projected area of cells obtained using the software provided with the LSM 510 Meta confocal microscope. Values obtained upon quantitation of F-actin in control and CD-treated cells are shown in (e). Data represent means ± standard error (SE) of at least 11 independent measurements from three independent experiments (∗∗∗ corresponds to significant (p < 0.0001) difference in F-actin content in cells treated with CD relative to control). See Materials and Methods for other details. To see this figure in color, go online.
Fig. 2, c and d shows isosurface images (by joining voxels of equal intensity) corresponding to projected images shown in Fig. 2, a and b. To obtain a quantitative estimate of F-actin levels, the volume enclosed by the isosurfaces was normalized to the respective projected area of cells in all cases. The F-actin content upon actin destabilization quantitated using this approach is shown in Fig. 2 e. As shown in the figure, CD treatment resulted in ∼15% reduction in cellular F-actin level. We further explored the modulation of receptor dynamics (lateral diffusion) as a consequence of actin destabilization.
Modulation of receptor diffusion upon actin destabilization monitored by SPT
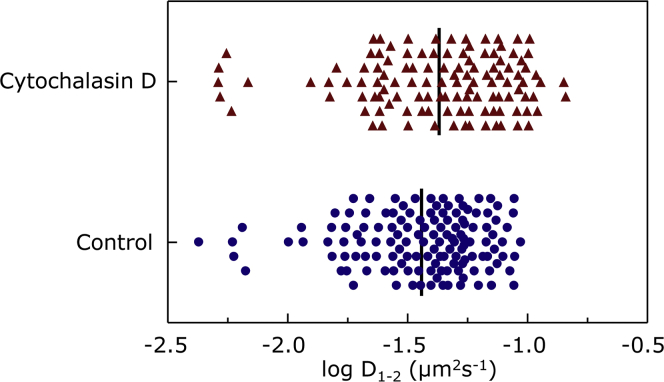
Before systematic acquisition of video sequences, the experimental conditions were optimized to ensure reliability of results. We first checked the specificity of the QDs for labeling the serotonin1A receptor. When cells were incubated with streptavidin-conjugated QDs coupled to biotinylated anti-c-myc antibody, we observed ∼20 QDs per cell, that remained bound even after washing the cells several times. In case of QDs alone (without prior coupling to biotinylated anti-c-myc antibody), we observed only two to three QDs on each cell. We recorded a series of 80 s sequences for each cell to compute the short-term diffusion coefficient (D1-2) for varying antibody/QD ratios. In agreement with previous literature (55), we chose the antibody/QD ratio of 1:10, which gives maximal D1–2, because this was used for optimal functionalization of the QDs with minimal perturbation of receptor diffusion. Although we found a reasonable density of QDs at the surface of the cells, we could obtain analyzable tracks only from those QDs that were present on top of the cells. We carried out data acquisition in control condition and after treating the cells with 5 μM CD. As a first step, we performed a global analysis of all trajectories collected for each condition. The distributions of the short-term diffusion coefficient D1–2 observed for each condition are shown in Fig. 3. We observed a small shift in the distribution of D1–2 toward higher values for 5 μM CD-treated cells. The average D1–2-value for control cells was 4.2 ± 2.1 × 10−2 μm2 s−1 and for cells treated with 5 μM CD was 5.3 ± 3.1 × 10−2 μm2 s−1. Destabilization of the actin cytoskeleton, therefore, has negligible averaged effects on diffusion coefficient.
Figure 3.
Distribution of log D1–2 calculated from the trajectories of serotonin1A receptors obtained by SPT on HEK-293 cells in the absence (control, blue circles, n = 133) and in the presence (red triangles, n = 117) of 5 μM CD. Trajectories of QDs at the cell surface were tracked with a Cascade II 512 EM-CCD camera (Roper Scientific) operating at 25 Hz acquisition frequency on a Zeiss Axio-observer A1 microscope at room temperature (∼22°C). The duration of the recordings was set to 80 s. The trajectories of all QDs in a video sequence were analyzed using the Multiple Target Tracing program developed by Sergé et al. (53). Trajectories of more than 20 s were further analyzed with a home-made program written in Visual Basic (VBA Excel, Microsoft). The black vertical lines correspond to the mean of the distribution of log D1–2. See text for more details. To see this figure in color, go online.
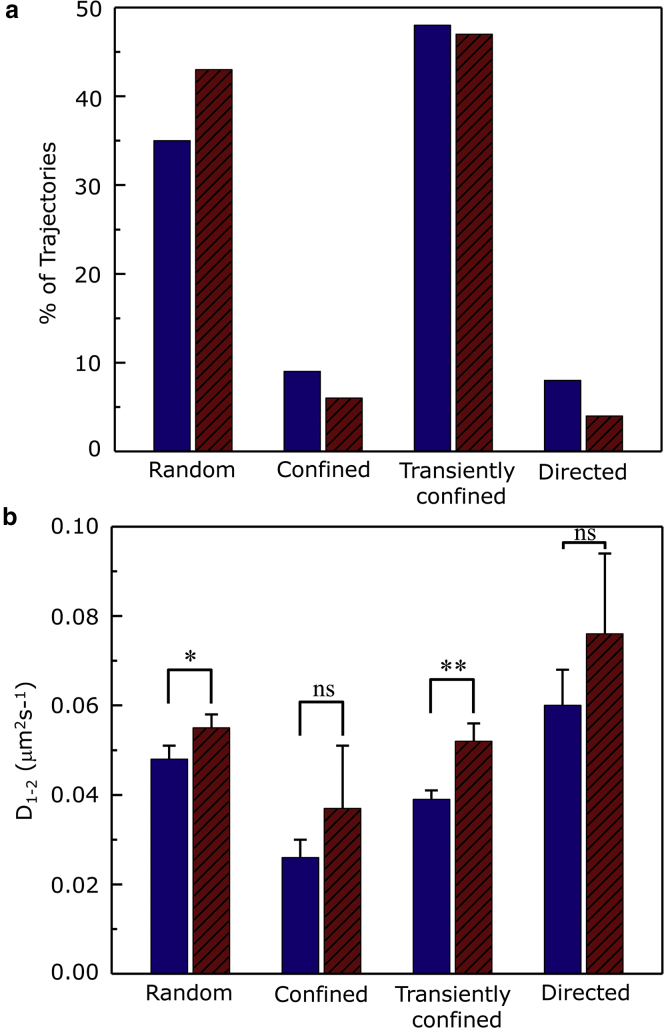
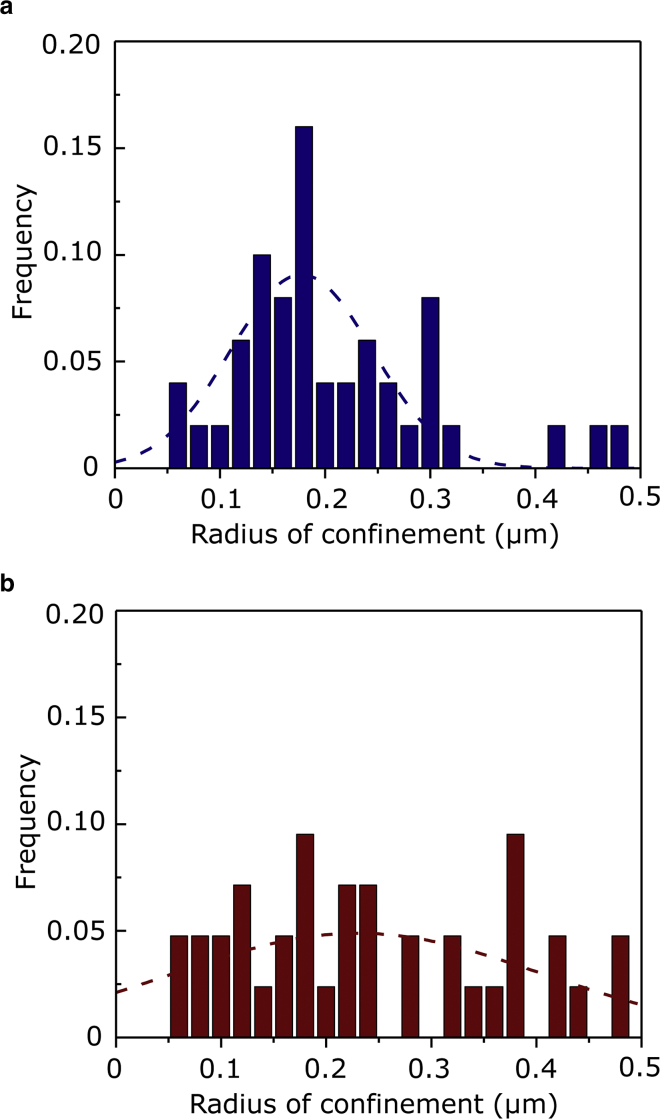
A unique feature of SPT is that it is possible to analyze individual trajectories and determine their distribution among distinct diffusion modes. We identified four types of diffusion by analysis of trajectories: random, confined, transiently confined, and directed diffusion (Fig. 4). Interestingly, after actin destabilization after CD treatment, the fraction of receptors characterized by random diffusion mode increases, whereas those of the confined and directed diffusion modes exhibit a reduction. On the other hand, the fraction of receptors with transiently confined diffusion mode remains invariant (see Fig. 4 a). This observation is consistent with the proposition that the actin cytoskeleton compartmentalizes the plasma membrane (60) and is implicated in the internalization of receptors (61). Destabilization of the actin cytoskeleton appears to release dynamic constraints that could promote random diffusion, along with a small increase in D1–2 for each diffusion mode (Fig. 4 b). The effect of release of dynamic constraints upon actin destabilization gets further manifested on the serotonin1A receptor diffusion, as is evident from the overall flattening of the distribution of the radius of confinement (R) and an overall shift toward larger values of the confinement radius (see Fig. 5; Table 1). For control cells, in case of receptors undergoing confined diffusion, the average radius of confinement was ∼212 nm (Table 1). The corresponding value upon actin destabilization using 5 μM CD resulted in a significant increase (∼25%) in the average radius of confinement, thereby suggesting release of dynamic constraints of receptors from cytoskeleton-induced domains.
Figure 4.
(a) The relative distribution of SPT trajectories of serotonin1A receptors in HEK-293 cells among the four diffusion modes observed in the absence (blue bars, n = 133) and presence (red hatched bars, n = 117) of 5 μM CD. (b) Average short-term diffusion coefficient (D1–2 (μm2 s−1)) of the serotonin1A receptor measured by SPT on HEK-293 cells is shown for each diffusion mode in the absence (blue bars) and presence (red hatched bars) of 5 μM CD. Data represent means ± SE of at least four independent experiments with at least eight cells for each experiment (∗, ∗∗ correspond to significant (p < 0.05), (p < 0.01) differences in short-term diffusion coefficient (D1–2 (μm2 s−1)) for cells treated with CD relative to control). The number of trajectories analyzed in each condition is given in Table 1. See text for more details. To see this figure in color, go online.
Figure 5.
A histogram distribution of domain radii of confinement in the confined trajectories measured by SPT for serotonin1A receptors (a) in the absence (blue bars, n = 64) and (b) presence (red bars, n = 55) of 5 μM CD. To identify transient or continuous confinement events, a confinement index Λ(t), as established by Meilhac et al. (56), was calculated, and a value of Λ(t) > 4 for a period longer than Δt is characteristic of confined diffusion. The size of the domains and the diffusion coefficient inside the confined trajectory segments were determined by fitting MSD(t) with its theoretical expression for confined diffusion (56). See text for other details. To see this figure in color, go online.
Table 1.
Diffusion Parameters of the Serotonin1A Receptor for Different Diffusion Modes
| Diffusion Modes | |||||
|---|---|---|---|---|---|
| Random | Confined | Directed | Transiently Confined |
||
| Random | Confined | ||||
| (a) Control | |||||
| D1–2 (10−2μm2/s) | 4.8 ± 0.3 | 2.6 ± 0.4 | 6.0 ± 0.8 | 3.9 ± 0.2 | 3.2 ± 0.3 |
| # of trajectories | 47 | 12 | 10 | 64 | |
| R (nm) | – | 212 ± 40 | – | – | 218 ± 27 |
| (b) CD-treateda | |||||
| D1–2 (10−2μm2/s) | 5.5 ± 0.3 | 3.7 ± 1.4 | 7.6 ± 1.8 | 5.2 ± 0.4 | 4.4 ± 0.5 |
| # of trajectories | 50 | 7 | 5 | 55 | |
| R (nm) | – | 264 ± 54 | – | – | 288 ± 22 |
The concentration of CD was 5 μM.
Ligand binding to the serotonin1A receptor upon actin cytoskeleton destabilization
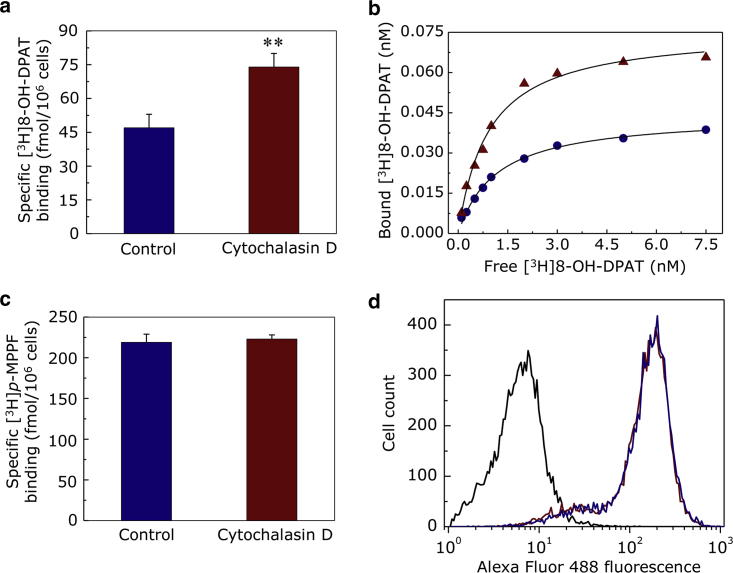
Ligand binding to GPCRs is the first step toward initiation of signaling in response to an extracellular stimulus. To monitor changes in ligand binding to the serotonin1A receptor upon cytoskeletal destabilization, we carried out whole-cell ligand binding of the serotonin1A receptor upon CD treatment in cells. Previous work from our laboratory has established that the agonist preferentially binds to receptors that are exclusively coupled to G-proteins, whereas the antagonist binding is insensitive to the G-protein coupling state of the receptor (62, 63). Fig. 6 a shows the increase in specific agonist ([3H]8-OH-DPAT) binding to the serotonin1A receptor upon destabilization of actin cytoskeleton. The figure shows that the specific agonist binding in CD-treated cells increases by ∼57% relative to control cells. The increase in specific agonist binding upon actin destabilization could possibly be due to an increase in the population of the G-protein-coupled pool of serotonin1A receptors because agonist binding requires G-protein coupling. Actin destabilization, therefore, appears to result in increased probability of interaction of GPCRs with G-proteins. Another possibility could be that there is an increase in agonist-binding affinity of the serotonin1A receptor due to subtle conformational changes upon actin destabilization.
Figure 6.
(a) Specific binding of the agonist [3H]8-OH-DPAT to serotonin1A receptors in intact HEK-293 cells for control and 5 μM CD treatment. Cells (∼106) in serum-free DMEM/F-12 medium were incubated at ∼25°C for 15 min in presence of 1 nM [3H]8-OH-DPAT (specific agonist). Nonspecific binding was obtained by performing the assay in presence of 10 μM unlabeled serotonin. Data represent means ± SE from at least three independent experiments (∗∗ corresponds to significant (p < 0.001) difference in specific [3H]8-OH-DPAT binding in cells treated with CD relative to control). (b) Saturation binding analysis of specific [3H]8-OH-DPAT binding to the serotonin1A receptor in intact cells is shown. The concentration of [3H]8-OH-DPAT ranged from 0.1 to 7.5 nM. Nonspecific binding was determined by performing the assay in presence of 10 μM unlabeled serotonin. Ligand-binding assays were carried out at ∼25°C with ∼106 cells for each ligand concentration. The saturation binding data could be fitted best to a one-site ligand binding equation. Representative binding plots are shown for control (blue circles) and 5 μM CD-treated (red triangles) cells. The curves are nonlinear regression fits to the experimental data. See Table 2 for more details. (c) Specific binding of the antagonist [3H]p-MPPF to serotonin1A receptors in intact HEK-293 cells for control and 5 μM CD treatment is shown. Cells (∼106) in serum-free DMEM/F-12 medium were incubated at ∼25°C for 15 min in presence of 1 nM [3H]p-MPPF (specific antagonist). Nonspecific binding was obtained by performing the assay in presence of 10 μM unlabeled p-MPPF. Values are expressed as percentages of specific radioligand binding in control cells. Data represent means ± SE from at least three independent experiments (d) An overlay of representative flow cytometric histograms indicating the population of serotonin1A receptors on the cell surface corresponding to control (blue) and CD-treated cells (red). The plasma membrane associated receptor population was labeled with anti-myc antibody Alexa Fluor 488 conjugate. Alexa Fluor 488 was excited at 488 nm, and emission was collected using a 525/40 bandpass filter. Mode count values were obtained by flow cytometric analysis of 10,000 cells for each condition. Fluorescence from unstained cells are represented as a black histogram toward lower value on the fluorescence axis. See Materials and Methods for other details. To see this figure in color, go online.
To gain further insight on the increase in agonist binding upon actin destabilization, we performed saturation binding assays, and representative binding plots are shown in Fig. 6 b. Our analysis of saturation binding of the specific agonist [3H]8-OH-DPAT in control and CD-treated condition showed that there was no significant difference in the agonist-binding affinity (Kd), as is apparent from Table 2. Instead, we observed a significant increase in the number of binding sites (Bmax) upon destabilization of actin cytoskeleton, as shown in Table 2. These results indicate that the increase in specific agonist binding upon CD treatment is possibly due to an increase in the G-protein-coupled state of the serotonin1A receptor. These results are supported by the observation that specific antagonist ([3H]p-MPPF) binding to the serotonin1A receptor remained invariant upon actin destabilization (Fig. 6 c). Taken together, these results indicate that the total cell surface receptor numbers do not change during actin destabilization. This conclusion is further reinforced by using a quantitative flow cytometric assay (Fig. 6 d), in which we measured cell surface population of the serotonin1A receptor by exclusively labeling the membrane-associated population of receptors with anti-myc antibody Alexa Fluor 488 conjugate. Fig. 6 d shows a representative flow cytometric histogram with overlapping histogram of fluorescence intensity, indicating invariance of total serotonin1A receptor population (irrespective of G-protein coupling) on the cell surface upon actin cytoskeleton destabilization.
Table 2.
Binding Parameters of the Agonist [3H]8-OH-DPAT Binding to Serotonin1A Receptor Expressed in HEK-293 Cells
| Parameters | Control | CD-treated |
|---|---|---|
| Kd | 1.1 ± 0.1 nM | 1.0 ± 0.1 nM |
| Bmax | 49 ± 3 fmol/106 cells | 76 ± 3 fmol/106 cells |
The binding parameters shown represent means ± SE from three independent experiments, whereas saturation binding data shown in Fig. 6b is from a representative experiment. The concentration of CD was 5 μM.
Actin cytoskeleton dependent signaling by the serotonin1A receptor
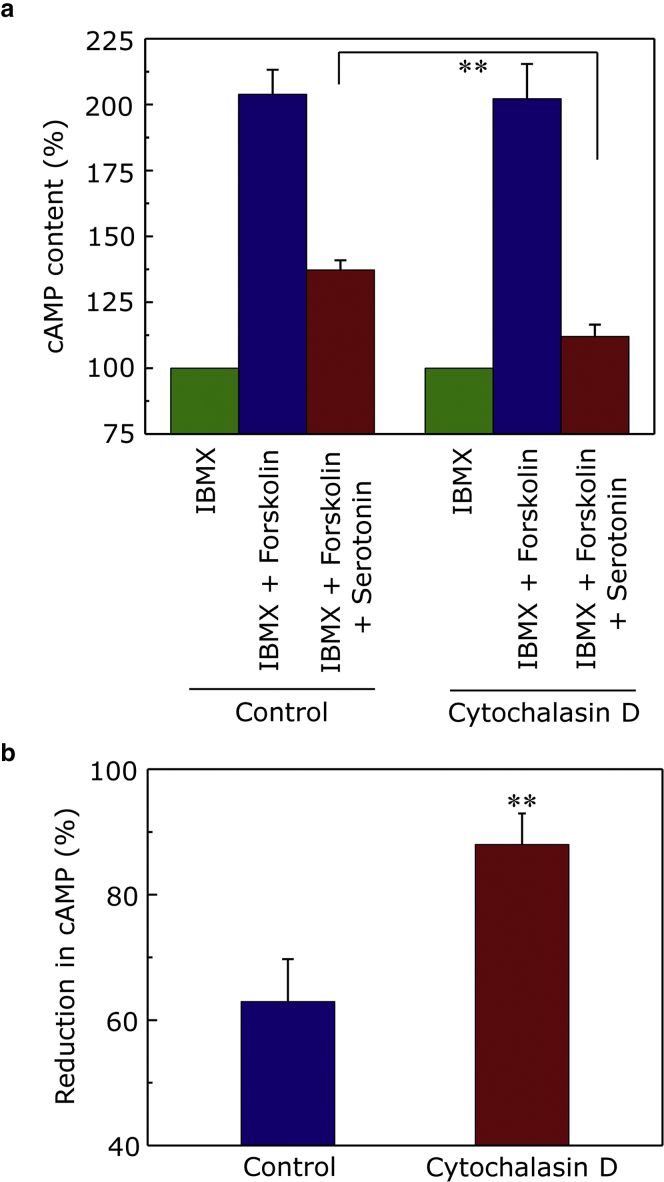
As a downstream effect of agonist stimulation, the serotonin1A receptor was shown to activate the Gi/Go class of G-proteins in HEK-293 cells (64, 65). This results in the inhibition of adenylyl cyclase activity which leads to reduction in cellular cAMP levels (66). To assess the effect of receptor dynamics (upon actin destabilization) on its signaling efficacy, we monitored serotonin (agonist)-stimulated reduction of cAMP levels by the serotonin1A receptor under these conditions. Because the basal levels of cAMP are low (difficult to measure), the cellular level of cAMP first needs to be increased using agents such as forskolin to assess Gi-mediated reduction in cAMP levels. We utilized a FRET-based assay (67, 68) that measures serotonin-induced reduction in forskolin-stimulated cAMP levels. We previously showed that forskolin-stimulated increase in intracellular levels of cAMP could induce significant actin cytoskeleton reorganization and lead to differential mobility of the serotonin1A receptor (26). Fortunately, we found that pretreatment of cells with CD considerably reduces the effect of forskolin treatment on the organization of actin cytoskeleton and could be used for cAMP measurements (26). The relative cAMP level, normalized to that found in basal condition (in presence of only IBMX, the phosphodiesterase inhibitor), is shown in Fig. 7. As shown in Fig. 7, forskolin stimulation resulted in ∼100% increase in cellular cAMP levels. Importantly, we observed that treatment with serotonin suppressed the forskolin-stimulated enhancement of cAMP levels in control as well as CD-treated cells (albeit to different extents). The relative reduction in cAMP levels mediated by the serotonin1A receptor upon treatment with serotonin, normalized to that found upon forskolin stimulation alone, is shown in Fig. 7 b. The figure shows that actin destabilization leads to further reduction in serotonin-stimulated cAMP levels relative to control cells. Interestingly, we observed that even at a 10-fold lower ligand concertation (1 μM), actin destabilization led to further reduction in serotonin-stimulated cAMP levels relative to control cells, as observed in the case of 10 μM serotonin (Fig. S2). However, such effects on cAMP levels were absent when cells were treated with 100 nM serotonin (Fig. S2). Taken together, these results suggest that the signaling efficiency of the serotonin1A receptor is enhanced upon actin destabilization. Importantly, we ruled out the possibility that actin destabilization alone could regulate the activity of adenylyl cyclase by measuring the level of basal and forskolin-stimulated cAMP levels in cells pretreated with CD (see Fig. S3). No significant difference was observed in the basal as well as forskolin-stimulated cAMP levels in control and CD-treated cells, thereby suggesting that CD treatment alone does not stimulate or inhibit adenylyl cyclase activity. These results therefore suggest that the release of constraints by actin destabilization favors signaling through enhanced dynamic interaction between the receptor and G-protein. In other words, the actin cytoskeleton induced spatial confinement plays an important role in regulation of signal transduction by GPCRs.
Figure 7.
Estimation of cellular cAMP levels in HEK-293 cells stably expressing serotonin1A receptors. (a) The ability of the serotonin1A receptor to inhibit forskolin-stimulated increase in cAMP levels upon treatment with 10 μM serotonin was assessed under control and CD-treated conditions. After treatment with CD, cells were treated with 50 μM IBMX (basal), 50 μM IBMX/10 μM forskolin (forskolin-stimulated), or 50 μM IBMX/10 μM forskolin/10 μM serotonin (agonist treatment) and incubated for 30 min at 37°C. The phosphodiesterase inhibitor IBMX (50 μM) was present during all treatments to prevent breakdown of cAMP. The ability of the serotonin1A receptor to inhibit the forskolin-stimulated increase in cAMP levels was assessed using the FRET-based HTRF cAMP-Gi assay kit (CisBio Bioassays). Fluorescence was measured at 620 nm (cAMP-cryptate donor emission) and 655 nm (anti-cAMP-d2 acceptor emission) upon excitation of the donor at 320 nm. cAMP levels were calculated as a ratio of the acceptor/donor emission. Data are normalized to cAMP levels in the presence of IBMX for each condition. Data represent means ± SE of at least three independent experiments (∗∗ corresponds to significant (p < 0.001) difference in cAMP content under serotonin-treated condition for cells treated with CD relative to control). (b) The relative reduction in forskolin-stimulated levels of cAMP in HEK-293 expressing the serotonin1A receptor upon activation by 10 μM serotonin is shown. Data are normalized to cAMP levels in the presence of 50 μM IBMX/10 μM forskolin (forskolin-stimulated) for each condition. Data represent means ± SE of at least three independent experiments (∗∗ corresponds to significant (p < 0.001) difference in cAMP content under serotonin-treated condition for cells treated with CD relative to control). See Materials and Methods for other details. To see this figure in color, go online.
Discussion
In this work, we explored the role of the actin cytoskeleton on the dynamics, ligand binding, and signaling of the serotonin1A receptor stably expressed in HEK-293 cells using SPT, which can provide information on relative distribution of receptors among diffusion modes. Our results show that the short-term diffusion coefficient of the serotonin1A receptor exhibits an increase upon actin destabilization due to release of dynamic constraints. Analysis of individual trajectories revealed that the population of receptors undergoing random diffusion increases, whereas those of the confined and directed diffusion modes decrease. The fraction of transiently confined receptors remains invariant. Analysis of the radius of confinement of the receptor revealed that there was an overall shift toward larger values of the confinement radius upon actin destabilization using 5 μM CD, which corresponds to ∼15% reduction in F-actin level. The functional implication of such actin destabilization was apparent from the increase in specific agonist binding and cAMP signaling.
The short-term diffusion coefficient of the serotonin1A receptor by SPT was found to be ∼0.05 μm2 s−1 and is similar to the diffusion coefficient of other class A GPCRs such as the μ-opioid receptor (47) and neurokinin-1 receptor (48), obtained using SPT. To the best of our knowledge, our previous results (26, 50, 69), along with these results, represent the first report of lateral diffusion coefficient of a functional GPCR (the serotonin1A receptor) monitored utilizing three independent approaches differing in spatiotemporal resolutions. However, the diffusion coefficient obtained by SPT for the serotonin1A receptor appears to be lower than that probed by fluorescence recovery after photobleaching (FRAP) (∼0.14 μm2 s−1) (26, 69). A possible reason for this discrepancy could be that the temperature at which FRAP measurements were performed previously (∼37°C) (69) is significantly higher than the temperature at which measurements were carried out in our work (∼22°C). Interestingly, the diffusion coefficient values obtained from our SPT measurements are much lower than the corresponding value obtained using z-scanning fluorescence correlation spectroscopy (∼4 μm2 s−1) (50). The difference in the magnitude of diffusion coefficients of the serotonin1A receptor obtained by these techniques could be attributed to the varying spatiotemporal resolutions associated with these approaches (42, 70, 71). SPT and FCS represent two different approaches for observing diffusion behavior. Single-molecule techniques such as SPT track single-fluorescent molecules at low concentrations and connect single localizations into trajectories. On the other hand, FCS is an ensemble method and is based on the calculation of autocorrelation functions from intensity fluctuations due to diffusing particles through an observation volume defined by a focused laser. SPT provides specific information on single particles and is best suited for the examination of heterogeneous systems, whereas FCS yields averaged information and spatial information. The characteristic timescales that govern the length scales that are probed by the techniques merit discussion. SPT is limited to a time resolution of ∼10 ms relative to the timescale of FCS (which could be tens of microseconds). It is possible that at such short times, the environment is less crowded, and this leads to larger values of the diffusion coefficient. There are other interesting differences between these two approaches. For example, FCS (but not SPT) can provide precise concentration of the fluorophore, and FCS is also more sensitive of the two methods. The big advantage with SPT lies in the fact that it allows for distinguishing subpopulations undergoing various modes of diffusion, which is very useful in a microheterogeneous environment such as biological membranes.
The plasma membrane acts as the first portal where the cellular response to various extracellular stimuli is initiated. Because molecules in the plasma membrane are dynamic, functional association between them would depend on the probability of their interaction with signaling partners (downstream effectors). When viewed from this perspective, signaling mediated by membrane proteins could be proposed to be a consequence of differential mobility of various interacting molecules (72, 73, 74). This constitutes the basis of the “mobile receptor” hypothesis that proposes that receptor-effector interactions at the plasma membrane are determined by lateral diffusion of the interacting components. Lateral diffusion of membrane components is modulated by the actin cytoskeleton. For example, the actin cytoskeleton was shown to be an important regulator of the lateral mobility of GPCRs such as the metabotropic glutamate receptor 5 (75, 76). Previous SPT measurements have reinforced the proposal of actin cytoskeleton dependent dynamics of membrane molecules (37). Actin destabilization could therefore offer a convenient handle to modulate the lateral diffusion of membrane proteins and receptors. As our present results show, this could lead to enhanced downstream signaling. Interestingly, we previously showed using FRAP that activation of the serotonin1A receptor with ligands or activators of G-proteins leads to an increase in diffusion (77). In a complimentary fashion, our results using SPT show that release of dynamic constraints by actin destabilization results in increased signaling. We conclude that measurements of receptor dynamics could provide novel insight toward developing a conceptual framework for cellular signaling.
Author Contributions
S.S., P.S., and P.P. performed experiments and analyzed data. S.S., P.S., P.P., L.S., and A.C. designed experiments. S.S., P.S., L.S. and A.C. wrote the manuscript.
Acknowledgments
We thank members of the Chattopadhyay laboratory for critically reading the manuscript.
This work was supported by Indo-French Centre for the Promotion of Advanced Research (project number 4603-2) to A.C. and L.S. A.C. gratefully acknowledges support from SERB Distinguished Fellowship (Department of Science and Technology, Govt. of India). P.S. thanks the Council of Scientific and Industrial Research for the award of a Shyama Prasad Mukherjee Fellowship. A.C. is a Distinguished Visiting Professor at the Indian Institute of Technology Bombay (Mumbai), and Adjunct Professor at the RMIT University (Melbourne, Australia), Tata Institute of Fundamental Research (Mumbai) and Indian Institute of Science Education and Research (Kolkata), and an Honorary Professor at the Jawaharlal Nehru Centre for Advanced Scientific Research (Bengaluru).
Editor: Andrew Plested.
Footnotes
Sandeep Shrivastava and Parijat Sarkar contributed equally to this work.
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.08.034.
Contributor Information
Laurence Salomé, Email: salome@ipbs.fr.
Amitabha Chattopadhyay, Email: amit@ccmb.res.in.
Supporting Material
References
- 1.Katritch V., Cherezov V., Stevens R.C. Structure-function of the G protein-coupled receptor superfamily. Annu. Rev. Pharmacol. Toxicol. 2013;53:531–556. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chattopadhyay A. GPCRs: lipid-dependent membrane receptors that act as drug targets. Adv. Biol. 2014;2014:143023. [Google Scholar]
- 3.Sakmar T.P. Introduction: G-protein coupled receptors. Chem. Rev. 2017;117:1–3. doi: 10.1021/acs.chemrev.6b00686. [DOI] [PubMed] [Google Scholar]
- 4.Weis W.I., Kobilka B.K. The molecular basis of G protein-coupled receptor activation. Annu. Rev. Biochem. 2018;87:897–919. doi: 10.1146/annurev-biochem-060614-033910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erlandson S.C., McMahon C., Kruse A.C. Structural basis for G protein-coupled receptor signaling. Annu. Rev. Biophys. 2018;47:9.1–9.18. doi: 10.1146/annurev-biophys-070317-032931. [DOI] [PubMed] [Google Scholar]
- 6.Jacobson K.A. New paradigms in GPCR drug discovery. Biochem. Pharmacol. 2015;98:541–555. doi: 10.1016/j.bcp.2015.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sriram K., Insel P.A. G protein-coupled receptors as targets for approved drugs: how many targets and how many drugs? Mol. Pharmacol. 2018;93:251–258. doi: 10.1124/mol.117.111062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan H.C.S., Li Y., Yuan S. New binding sites, new opportunities for GPCR drug discovery. Trends Biochem. Sci. 2019;44:312–330. doi: 10.1016/j.tibs.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 9.Insel P.A., Sriram K., Chinn A.M. GPCRomics: an approach to discover GPCR drug targets. Trends Pharmacol. Sci. 2019;40:378–387. doi: 10.1016/j.tips.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pucadyil T.J., Kalipatnapu S., Chattopadhyay A. The serotonin1A receptor: a representative member of the serotonin receptor family. Cell. Mol. Neurobiol. 2005;25:553–580. doi: 10.1007/s10571-005-3969-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalipatnapu S., Chattopadhyay A. Membrane organization and function of the serotonin(1A) receptor. Cell. Mol. Neurobiol. 2007;27:1097–1116. doi: 10.1007/s10571-007-9189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Müller C.P., Carey R.J., De Souza Silva M.A. Serotonin and psychostimulant addiction: focus on 5-HT1A-receptors. Prog. Neurobiol. 2007;81:133–178. doi: 10.1016/j.pneurobio.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 13.Gordon J.A., Hen R. The serotonergic system and anxiety. Neuromolecular Med. 2004;5:27–40. doi: 10.1385/NMM:5:1:027. [DOI] [PubMed] [Google Scholar]
- 14.Glikmann-Johnston Y., Saling M.M., Stout J.C. Hippocampal 5-HT1A receptor and spatial learning and memory. Front. Pharmacol. 2015;6:289. doi: 10.3389/fphar.2015.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lacivita E., Leopoldo M., Perrone R. 5-HT1A receptor, an old target for new therapeutic agents. Curr. Top. Med. Chem. 2008;8:1024–1034. doi: 10.2174/156802608785161385. [DOI] [PubMed] [Google Scholar]
- 16.Fiorino F., Severino B., Perissutti E. 5-HT(1A) receptor: an old target as a new attractive tool in drug discovery from central nervous system to cancer. J. Med. Chem. 2014;57:4407–4426. doi: 10.1021/jm400533t. [DOI] [PubMed] [Google Scholar]
- 17.Ahram M., Litou Z.I., Al-Tawallbeh G. Estimation of membrane proteins in the human proteome. In Silico Biol. (Gedrukt) 2006;6:379–386. [PubMed] [Google Scholar]
- 18.Almén M.S., Nordström K.J., Schiöth H.B. Mapping the human membrane proteome: a majority of the human membrane proteins can be classified according to function and evolutionary origin. BMC Biol. 2009;7:50. doi: 10.1186/1741-7007-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lacapère J.J., Pebay-Peyroula E., Etchebest C. Determining membrane protein structures: still a challenge! Trends Biochem. Sci. 2007;32:259–270. doi: 10.1016/j.tibs.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 20.Hanson M.A., Stevens R.C. Discovery of new GPCR biology: one receptor structure at a time. Structure. 2009;17:8–14. doi: 10.1016/j.str.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venkatakrishnan A.J. Structure and activation mechanism of GPCRs. In: Lebon G., editor. Structure and Function of GPCRs. Topics in Medicinal Chemistry. vol. 30. Springer, Cham; 2019. pp. 53–64. [Google Scholar]
- 22.Latorraca N.R., Venkatakrishnan A.J., Dror R.O. GPCR dynamics: structures in motion. Chem. Rev. 2017;117:139–155. doi: 10.1021/acs.chemrev.6b00177. [DOI] [PubMed] [Google Scholar]
- 23.Alhadeff R., Vorobyov I., Warshel A. Exploring the free-energy landscape of GPCR activation. Proc. Natl. Acad. Sci. USA. 2018;115:10327–10332. doi: 10.1073/pnas.1810316115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jans D.A., Peters R., Fahrenholz F. Vasopressin V2-receptor mobile fraction and ligand-dependent adenylate cyclase activity are directly correlated in LLC-PK1 renal epithelial cells. J. Cell Biol. 1991;114:53–60. doi: 10.1083/jcb.114.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calvert P.D., Govardovskii V.I., Makino C.L. Membrane protein diffusion sets the speed of rod phototransduction. Nature. 2001;411:90–94. doi: 10.1038/35075083. [DOI] [PubMed] [Google Scholar]
- 26.Ganguly S., Pucadyil T.J., Chattopadhyay A. Actin cytoskeleton-dependent dynamics of the human serotonin1A receptor correlates with receptor signaling. Biophys. J. 2008;95:451–463. doi: 10.1529/biophysj.107.125732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edidin M. Getting there is only half the fun. Curr. Top. Membr. 1996;43:1–13. [Google Scholar]
- 28.Jacobson K., Mouritsen O.G., Anderson R.G. Lipid rafts: at a crossroad between cell biology and physics. Nat. Cell Biol. 2007;9:7–14. doi: 10.1038/ncb0107-7. [DOI] [PubMed] [Google Scholar]
- 29.Kwik J., Boyle S., Edidin M. Membrane cholesterol, lateral mobility, and the phosphatidylinositol 4,5-bisphosphate-dependent organization of cell actin. Proc. Natl. Acad. Sci. USA. 2003;100:13964–13969. doi: 10.1073/pnas.2336102100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kusumi A., Fujiwara T.K., Suzuki K.G. Dynamic organizing principles of the plasma membrane that regulate signal transduction: commemorating the fortieth anniversary of Singer and Nicolson’s fluid-mosaic model. Annu. Rev. Cell Dev. Biol. 2012;28:215–250. doi: 10.1146/annurev-cellbio-100809-151736. [DOI] [PubMed] [Google Scholar]
- 31.Kusumi A., Nakada C., Fujiwara T. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophys. Biomol. Struct. 2005;34:351–378. doi: 10.1146/annurev.biophys.34.040204.144637. [DOI] [PubMed] [Google Scholar]
- 32.Davidson A.J., Wood W. Unravelling the actin cytoskeleton: a new competitive edge? Trends Cell Biol. 2016;26:569–576. doi: 10.1016/j.tcb.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.dos Remedios C.G., Chhabra D., Nosworthy N.J. Actin binding proteins: regulation of cytoskeletal microfilaments. Physiol. Rev. 2003;83:433–473. doi: 10.1152/physrev.00026.2002. [DOI] [PubMed] [Google Scholar]
- 34.Pollard T.D. Actin and actin-binding proteins. Cold Spring Harb. Perspect. Biol. 2016;8:a018226. doi: 10.1101/cshperspect.a018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ganguly S., Saxena R., Chattopadhyay A. Reorganization of the actin cytoskeleton upon G-protein coupled receptor signaling. Biochim. Biophys. Acta. 2011;1808:1921–1929. doi: 10.1016/j.bbamem.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 36.Vázquez-Victorio G., González-Espinosa C., Macías-Silva M. GPCRs and actin-cytoskeleton dynamics. Methods Cell Biol. 2016;132:165–188. doi: 10.1016/bs.mcb.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 37.Fujiwara T., Ritchie K., Kusumi A. Phospholipids undergo hop diffusion in compartmentalized cell membrane. J. Cell Biol. 2002;157:1071–1081. doi: 10.1083/jcb.200202050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Calebiro D., Rieken F., Lohse M.J. Single-molecule analysis of fluorescently labeled G-protein-coupled receptors reveals complexes with distinct dynamics and organization. Proc. Natl. Acad. Sci. USA. 2013;110:743–748. doi: 10.1073/pnas.1205798110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lagerholm B.C., Weinreb G.E., Thompson N.L. Detecting microdomains in intact cell membranes. Annu. Rev. Phys. Chem. 2005;56:309–336. doi: 10.1146/annurev.physchem.56.092503.141211. [DOI] [PubMed] [Google Scholar]
- 40.Kasai R.S., Kusumi A. Single-molecule imaging revealed dynamic GPCR dimerization. Curr. Opin. Cell Biol. 2014;27:78–86. doi: 10.1016/j.ceb.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 41.Yu J. Single-molecule studies in live cells. Annu. Rev. Phys. Chem. 2016;67:565–585. doi: 10.1146/annurev-physchem-040215-112451. [DOI] [PubMed] [Google Scholar]
- 42.Marguet D., Salomé L. Lateral diffusion in heterogeneous cell membranes. In: Bassereau P., Sens P., editors. Physics of Biological Membranes. Springer Nature; 2018. pp. 169–189. [Google Scholar]
- 43.Destainville N., Dumas F., Salomé L. What do diffusion measurements tell us about membrane compartmentalisation? Emergence of the role of interprotein interactions. J. Chem. Biol. 2008;1:37–48. doi: 10.1007/s12154-008-0005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen H., Tauzin L.J., Landes C.F. Single particle tracking: from theory to biophysical applications. Chem. Rev. 2017;117:7331–7376. doi: 10.1021/acs.chemrev.6b00815. [DOI] [PubMed] [Google Scholar]
- 45.Haanappel E., Salomé L. G-protein-coupled receptors: membrane diffusion and organization matter. In: Chattopadhyay A., editor. Membrane Organization and Dynamics, Springer Series in Biophysics. Volume 20. Springer International; 2017. pp. 243–258. [Google Scholar]
- 46.Wieser S., Schütz G.J. Tracking single molecules in the live cell plasma membrane-Do’s and Don’t’s. Methods. 2008;46:131–140. doi: 10.1016/j.ymeth.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 47.Daumas F., Destainville N., Salomé L. Confined diffusion without fences of a g-protein-coupled receptor as revealed by single particle tracking. Biophys. J. 2003;84:356–366. doi: 10.1016/S0006-3495(03)74856-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Veya L., Piguet J., Vogel H. Single molecule imaging deciphers the relation between mobility and signaling of a prototypical G protein-coupled receptor in living cells. J. Biol. Chem. 2015;290:27723–27735. doi: 10.1074/jbc.M115.666677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar G.A., Sarkar P., Chattopadhyay A. Exploring endocytosis and intracellular trafficking of the human serotonin1A receptor. Biochemistry. 2019;58:2628–2641. doi: 10.1021/acs.biochem.9b00033. [DOI] [PubMed] [Google Scholar]
- 50.Ganguly S., Chattopadhyay A. Cholesterol depletion mimics the effect of cytoskeletal destabilization on membrane dynamics of the serotonin1A receptor: a zFCS study. Biophys. J. 2010;99:1397–1407. doi: 10.1016/j.bpj.2010.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roy S., Kumar G.A., Chattopadhyay A. Integrity of the actin cytoskeleton of host macrophages is essential for Leishmania donovani infection. Biochim. Biophys. Acta. 2014;1838:2011–2018. doi: 10.1016/j.bbamem.2014.04.017. [DOI] [PubMed] [Google Scholar]
- 52.Bruns R.F., Lawson-Wendling K., Pugsley T.A. A rapid filtration assay for soluble receptors using polyethylenimine-treated filters. Anal. Biochem. 1983;132:74–81. doi: 10.1016/0003-2697(83)90427-x. [DOI] [PubMed] [Google Scholar]
- 53.Sergé A., Bertaux N., Marguet D. Dynamic multiple-target tracing to probe spatiotemporal cartography of cell membranes. Nat. Methods. 2008;5:687–694. doi: 10.1038/nmeth.1233. [DOI] [PubMed] [Google Scholar]
- 54.Michalet X. Mean square displacement analysis of single-particle trajectories with localization error: Brownian motion in an isotropic medium. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2010;82:041914. doi: 10.1103/PhysRevE.82.041914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mascalchi P., Haanappel E., Salomé L. Probing the influence of the particle in single particle tracking measurements of lipid diffusion. Soft Matter. 2012;8:4462–4470. [Google Scholar]
- 56.Meilhac N., Le Guyader L., Destainville N. Detection of confinement and jumps in single-molecule membrane trajectories. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2006;73:011915. doi: 10.1103/PhysRevE.73.011915. [DOI] [PubMed] [Google Scholar]
- 57.Saxton M.J., Jacobson K. Single-particle tracking: applications to membrane dynamics. Annu. Rev. Biophys. Biomol. Struct. 1997;26:373–399. doi: 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- 58.Sampath P., Pollard T.D. Effects of cytochalasin, phalloidin, and pH on the elongation of actin filaments. Biochemistry. 1991;30:1973–1980. doi: 10.1021/bi00221a034. [DOI] [PubMed] [Google Scholar]
- 59.Schliwa M. Action of cytochalasin D on cytoskeletal networks. J. Cell Biol. 1982;92:79–91. doi: 10.1083/jcb.92.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jacobson K., Liu P., Lagerholm B.C. The lateral organization and mobility of plasma membrane components. Cell. 2019;177:806–819. doi: 10.1016/j.cell.2019.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wolfe B.L., Trejo J. Clathrin-dependent mechanisms of G protein-coupled receptor endocytosis. Traffic. 2007;8:462–470. doi: 10.1111/j.1600-0854.2007.00551.x. [DOI] [PubMed] [Google Scholar]
- 62.Harikumar K.G., Chattopadhyay A. Differential discrimination of G-protein coupling of serotonin(1A) receptors from bovine hippocampus by an agonist and an antagonist. FEBS Lett. 1999;457:389–392. doi: 10.1016/s0014-5793(99)01082-0. [DOI] [PubMed] [Google Scholar]
- 63.Javadekar-Subhedar V., Chattopadhyay A. Temperature-dependent interaction of the bovine hippocampal serotonin(1A) receptor with G-proteins. Mol. Membr. Biol. 2004;21:119–123. doi: 10.1080/09687680310001058335. [DOI] [PubMed] [Google Scholar]
- 64.Malmberg A., Strange P.G. Site-directed mutations in the third intracellular loop of the serotonin 5-HT(1A) receptor alter G protein coupling from G(i) to G(s) in a ligand-dependent manner. J. Neurochem. 2000;75:1283–1293. doi: 10.1046/j.1471-4159.2000.751283.x. [DOI] [PubMed] [Google Scholar]
- 65.Giulietti M., Vivenzio V., Nardi B. How much do we know about the coupling of G-proteins to serotonin receptors? Mol. Brain. 2014;7:49. doi: 10.1186/s13041-014-0049-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Raymond J.R., Olsen C.L., Gettys T.W. Cell-specific physical and functional coupling of human 5-HT1A receptors to inhibitory G protein α-subunits and lack of coupling to Gsα. Biochemistry. 1993;32:11064–11073. doi: 10.1021/bi00092a016. [DOI] [PubMed] [Google Scholar]
- 67.Tardieu J.-L. Selecting a cyclic AMP kit for assaying GPCR target activation. Nat. Methods. 2008;5 iii–iv. [Google Scholar]
- 68.Ward R.J., Pediani J.D., Milligan G. Spatial intensity distribution analysis quantifies the extent and regulation of homodimerization of the secretin receptor. Biochem. J. 2017;474:1879–1895. doi: 10.1042/BCJ20170184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pucadyil T.J., Chattopadhyay A. Cholesterol depletion induces dynamic confinement of the G-protein coupled serotonin(1A) receptor in the plasma membrane of living cells. Biochim. Biophys. Acta. 2007;1768:655–668. doi: 10.1016/j.bbamem.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 70.Baker A., Saulière A., Salomé L. Functional membrane diffusion of G-protein coupled receptors. Eur. Biophys. J. 2007;36:849–860. doi: 10.1007/s00249-007-0214-7. [DOI] [PubMed] [Google Scholar]
- 71.Sarkar P., Chattopadhyay A. Exploring membrane organization at varying spatiotemporal resolutions utilizing fluorescence-based approaches: implications in membrane biology. Phys. Chem. Chem. Phys. 2019;21:11554–11563. doi: 10.1039/c9cp02087j. [DOI] [PubMed] [Google Scholar]
- 72.Cuatrecasas P. Membrane receptors. Annu. Rev. Biochem. 1974;43:169–214. doi: 10.1146/annurev.bi.43.070174.001125. [DOI] [PubMed] [Google Scholar]
- 73.Kahn C.R. Membrane receptors for hormones and neurotransmitters. J. Cell Biol. 1976;70:261–286. doi: 10.1083/jcb.70.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peters R. Lateral mobility of proteins and lipids in the red cell membrane and the activation of adenylate cyclase by β-adrenergic receptors. FEBS Lett. 1988;234:1–7. doi: 10.1016/0014-5793(88)81290-0. [DOI] [PubMed] [Google Scholar]
- 75.Sergé A., Fourgeaud L., Choquet D. Receptor activation and homer differentially control the lateral mobility of metabotropic glutamate receptor 5 in the neuronal membrane. J. Neurosci. 2002;22:3910–3920. doi: 10.1523/JNEUROSCI.22-10-03910.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sergé A., Fourgeaud L., Choquet D. Active surface transport of metabotropic glutamate receptors through binding to microtubules and actin flow. J. Cell Sci. 2003;116:5015–5022. doi: 10.1242/jcs.00822. [DOI] [PubMed] [Google Scholar]
- 77.Pucadyil T.J., Kalipatnapu S., Chattopadhyay A. G-protein-dependent cell surface dynamics of the human serotonin1A receptor tagged to yellow fluorescent protein. Biochemistry. 2004;43:15852–15862. doi: 10.1021/bi0480887. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.