Abstract
Defective nitrate signaling in plants causes disorder in nitrogen metabolism, and it negatively affects nitrate transport systems, which toggle between high- and low-affinity modes in variable soil nitrate conditions. Recent discovery of a plasma membrane nitrate transceptor protein NRT1.1—a transporter cum sensor—provides a clue on this toggling mechanism. However, the general mechanistic description still remains poorly understood. Here, we illustrate adaptive responses and regulation of NRT1.1-mediated nitrate signaling in a wide range of extracellular nitrate concentrations. The results show that the homodimeric structure of NRT1.1 and its dimeric switch play an important role in eliciting specific cytosolic calcium waves sensed by the calcineurin-B-like calcium sensor CBL9, which activates the kinase CIPK23, in low nitrate concentration that is, however, impeded in high nitrate concentration. Nitrate binding at the high-affinity unit initiates NRT1.1 dimer decoupling and priming of the Thr101 site for phosphorylation by CIPK23. This phosphorylation stabilizes the NRT1.1 monomeric state, acting as a high-affinity nitrate transceptor. However, nitrate binding in both monomers, retaining the unmodified NRT1.1 state through dimerization, attenuates CIPK23 activity and thereby maintains the low-affinity mode of nitrate signaling and transport. This phosphorylation-led modulation of NRT1.1 activity shows bistable behavior controlled by an incoherent feedforward loop, which integrates nitrate-induced positive and negative regulatory effects on CIPK23. These results, therefore, advance our molecular understanding of adaptation in fluctuating nutrient availability and are a way forward for improving plant nitrogen use efficiency.
Significance
NRT1.1, an important plasma protein residing in plant root cells, plays dual roles as nitrate sensor and transporter. This NRT1.1-mediated nitrate signaling provides a key to improving plants’ nitrogen use efficiency while reducing nitrogen fertilization. To facilitate this effort, we use structural and biochemical modeling to reveal bistable control of NRT1.1-mediated nitrate signaling by its upstream components in response to a wide range of variation of extracellular nitrate concentrations.
Introduction
Nitrate signaling is essential for a variety of critical responses of plant cells, including the repression of key nitrate transporter NRT2.1 expression, stimulation of ANR1-regulated lateral root, inhibition of L-glutamate-induced root architecture modulation, and stimulation of seed germination (1, 2, 3, 4, 5). These large palettes of nitrate-induced responses are seen to be regulated adaptively depending on extracellular concentrations of nitrate and evidently depend on the nitrate transporter NRT1.1 (6). Besides nitrate transportation, NRT1.1, a plasma membrane protein, modulates these responses by triggering independent signaling pathways irrespective of its transport function in Arabidopsis thaliana (7). A short-term, nitrate-induced transcriptional response of nitrogen assimilatory and transporter genes, known as primary nitrate response (PNR), is negatively regulated by the phosphorylation of NRT1.1 at threonine 101 residue (8,9). This PNR has, however, remained unaffected by P492L substitution, which suppresses NRT1.1 transport activity. It indicates that a nonphosphorylated form of NRT1.1 is predominant in nitrate signaling and is also independent of its transport activity (7). Phosphomimetic (T101D) and phosphodefective (T101A) point mutations in Arabidopsis showed different NRT1.1-dependent signaling responses (7). Although T101 phosphorylation is responsible for switching between low- and high-affinity modes of nitrate transportation, it has also remained important for maintaining biphasic PNR in low (<1 mM) and high (>1 mM) nitrate concentrations (10). Furthermore, the same phosphorylation at the T101 site directs the action of NRT1.1 toward the activation of specific signaling responses for lateral root development and feedback regulation by high nitrogen (1,11). Thus, the dynamic processes of phosphorylation/dephosphorylation of NRT1.1 at the T101 site, which depend on soil nitrate availability, play significant roles in nitrate signaling and modulate several physiological and developmental responses of plant cell. In this work, we have shown how the homodimeric structure of NRT1.1 and the nitrate-induced dynamic interactions with the immediate downstream effectors affect and regulate the NRT1.1 phosphorylation.
The nitrate transporter NRT1.1 is a 590-amino-acid homodimer, consisting of two monomers in an asymmetric unit. Each monomer contains 12 transmembrane (TM)-spanning α-helices, with amino (TM1–TM6)- and carboxyl (TM7–TM12)-terminal bundles (Fig. 1 D). These two bundles are linked by the lateral helix, consisting of 84 amino acids spanning horizontally in the outward direction from the transporter domain (12). A recent study reports large local conformational differences between the two monomers, which is further associated with differential nitrate-binding affinity (13). One of the monomers has almost five times higher affinity than the other, suggesting their differential roles for biphasic regulation of NRT1.1 (13). The phosphorylation site, Thr101, is located at the bottom of TM3 on the cytosolic side and is surrounded by hydrophobic residues from TM2 and TM4. In the apo state, the Thr101 residue is in a deep hydrophobic pocket demarcated by the hydrophobic residues Leu 96, Leu 100, and Ile 104 from the dimer interface. Upon nitrate binding in the high-affinity monomer, Thr101 is shifted to the interface and primed for phosphorylation. In contrast, such conformational changes and priming have not been observed in the low-affinity monomer (13). A recent study suggests a potential structural significance for this phosphorylation (12). Although the unphosphorylated NRT1.1 forms a structurally coupled dimer and functions as a low-affinity transporter, the phosphorylation of the Thr101 site of NRT1.1 triggers dimer decoupling and allows adopting a high-affinity transport mode (14). However, an alternative hypothesis suggests that phosphorylation increases structural flexibility and changes in packing in the N-terminal bundles, resulting in increased nitrate transport (12). In this study, using extensive mathematical methods and computational techniques, we have reconciled these two hypotheses and illustrated the role of dimerization in NRT1.1 mediated nitrate signaling.
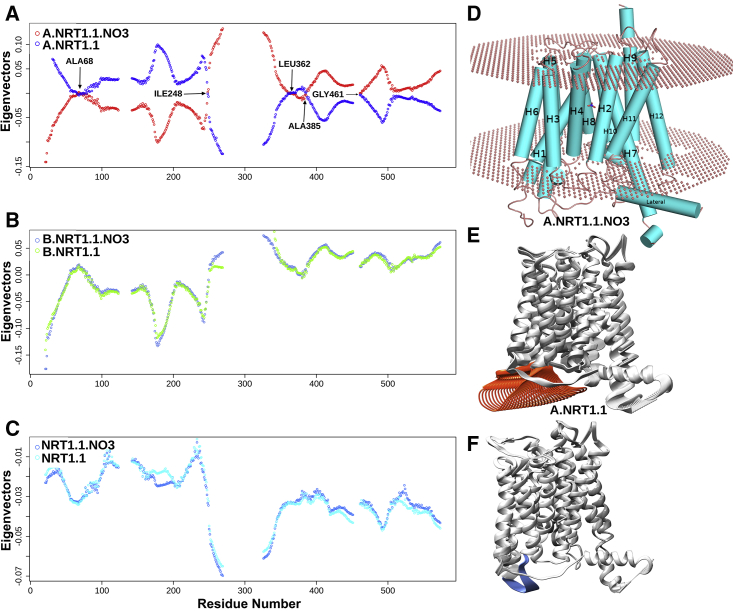
Figure 1.
Collective motion of NRT1.1 molecule shows large differences in the high-affinity state. (A) The monomeric nitrate-bound high-affinity unit shows antiphase dynamics relative to its nitrate-unbound monomer A. Several antiphase tipping residues, indicated by arrows, are responsible for maintaining this antiphase asynchronous collective motion. (B) and (C) show in-phase, synchronous motions for NRT1.1 dimer and the monomer B, with and without nitrate binding. (D) NRT1.1 monomer containing 12 transmembrane (TM)-spanning α-helices with amino-(TM1–TM6) and carboxy-terminal (TM7–TM12) bundles. (E) Nitrate-bound high-affinity monomer A shows a large-amplitude motion of the loop (orange color) connecting the C-terminal portion of helix H4 to the N-terminal of helix H5 and playing an important role for releasing the nitrate in the cytosol. (F) Without nitrate binding, such motions of the dynamic loop are absent (blue color). To see this figure in color, go online.
The NRT1.1 protein acts as a toggle switch through the phosphorylation/dephosphorylation of Thr101. At low nitrate concentration, nitrate-bound NRT1.1 triggers specific calcium waves through the action of an unknown phospholipase C, and blocking these waves severely affects several nitrate-induced responses (15, 16, 17). These calcium waves have also been reported to occur in response to abiotic stress and in the regulation of ion homeostasis (18,19). Nitrate binding to NRT1.1 activates the downstream effector CBL9-interacting kinase, CIPK23, at low nitrate concentration. The calcium-dependent interaction of CBL9 with the self-inhibitory NAF motif relieves the self-inhibition and activates the kinase CIPK23 (20). This nitrate-induced calcium-dependent regulation of the activity of the system ensures colocalization of the CIPK23.CBL9 interacting pairs toward the NRT1.1 at the plasma membrane (21). Here, we build a molecular model of the CIPK23.NRT1.1 complex, and, using this complex, we characterized this dynamic interaction. To demonstrate the biphasic change of NRT1.1 depending on extracellular nitrate concentrations, we constructed a mathematical model that includes these interacting components of nitrate signaling. The model is formed based on a three-node network topology in which two nodes (NRT1.1 and CIPK23) are activated by nitrate binding. These two nodes then act on the third node (NRT1.1 phosphorylated state) with opposite effects, in which one activates and the other inhibits the phosphorylation depending on the nitrate concentration. This network topology, known as incoherent feedforward control structure, leads to a transient response shaping the adaptive regulation of NRT1.1. A similar adaptive control through an incoherent feedforward network was observed in the social amoeba chemotaxis signaling pathway (22).
Materials and Methods
Construction of lipid bilayer for NRT1.1
To perform normal mode analysis (NMA) in a membrane environment, the membrane-bound dimeric NRT1.1 in both apo- and nitrate-bound forms were obtained from the OPM database (23). OPM uses the PPM 2.0 server to calculate orientation of proteins in membranes (23). OPM is a curated web resource that provides spatial positions of membrane-bound peptides and proteins of known three-dimensional structure in the lipid bilayer. The PPM 2.0 server uses a generic version of the 1,2-dioleoyl-sn-glycero-3-phosphocholine bilayer and combines an all-atom representation of a solute, an anisotropic solvent representation of the lipid bilayer, and a universal solvation model (23).
NMA for NRT1.1
To assess a broad range of potential large conformational changes in NRT1.1, the DynOmics ENM Server was used to perform NMA of the membrane-bound NRT1.1 (24). NMA is a tool for exploring functional motions of proteins. It uses two elastic network models (ENMs) — the Gaussian network model (GNM) and the anisotropic network model—to evaluate the dynamics of structurally resolved systems from individual molecules to large complexes and assemblies in the context of their physiological environment.
In the GNM, network nodes are the C-α atoms and the elastic springs represented the interactions. We used the GNM with an interaction cutoff distance of 7.3 Å and spring constant scaling factor cutoff of 1 Å for the calculation of the elastic network model. The fluctuation between any two residues i and j is given by
where Fij is the distance between residues i, j at any given instant of time and Fij0 denotes distance at their equilibrium positions.
For an N-node (residues) network, the GNM potential in terms of ΔXi, ΔYi, and ΔZi components of ΔFi can be written as
where γ is the uniform force constant for all springs and Γij is the ijth element of the Kirchhoff matrix of inter-residue interactions defined by
Expressing the X, Y, Z components of the fluctuation vectors ΔFi as three N-dimensional vectors ΔX, ΔY, and ΔZ, the total potential reduces to the following simplified form:
The correlations between the fluctuation of residues i, j are finally found as a statistical mechanical average, and then it reduces to the following equation:
In the GNM, the determinant of the Kirchhoff matrix is zero, and Γ−1 is therefore calculated from the eigenvalue decomposition Γ = ABAT, where A is the orthogonal matrix whose kth column vector ak is the kth eigenvector of Γ and B is the diagonal matrix of eigenvalues λk. The first eigenvalue is identically zero so that each of the remaining N − 1 eigenvalues define the frequency of the N − 1 modes.
We calculated the first 20 slowest modes of NRT1.1. The eigenvectors of these modes represent the global motions, and the constrained residues help in identifying critical regions such as hinge-bending regions, thereby giving an idea of domain motions around these regions. We plotted the first slowest mode of NRT1.1 in different conditions, which showed a significant difference in motions.
Nitrate-bound monomer A is docked with the nitrate-unbound monomer B
To analyze the consequences of high-affinity nitrate binding on monomer B, nitrate-bound monomer A was docked to nitrate-unbound monomer B using the PRISM web server (25). Given two protein structures, PRISM predicts the complex by checking interface matching between the structures and also accounts for flexible structural similarity and evolutionary interface conservation. The predictions are scored according to binding energies using the CHARMM22 force field.
Modeling the ATP.CIPK23 complex
250 poses of ATP docked to CIPK23 were generated and divided into 37 clusters using SwissDock (26). Out of 250 poses, five modes bound ATP at the already-known position (Table S2). The ATP.CIPK23 complex was formed out of the pose having the lowest ΔG-value.
Modeling the NRT1.1.CIPK23 complex
X-ray crystallographic data of NRT1.1 (Protein Data Bank (PDB): 5A2N and 5A2O) (12) and CIPK23 (PDB: 4CZT) (20) were obtained from the PDB repository. The overall structure of kinase CIPK23 is the same as cAMP-dependent protein kinase (PDB: 1ATP), which usually serves as a prototype for the entire kinome (27). Superimposition of the two structures shows 1.5 Å RMSD in the activation loop region. ATP docking to CIPK23 at the known position was guided by the kinase 1 ATP using SwissDock. Interaction of ATP-bound active CIPK23 with NRT1.1 was modeled using HADDOCK version 2.2 (28). The two structures were docked rigidly, and their interfaces were refined with both the backbone and side-chain flexibility. At the rigid-body stage, 10,000 models were generated, and the 400 best scoring models, ranked by the HADDOCK scoring function, were further refined in the semiflexible and solvated protocols. The final 400 models were clustered based on the fraction of common contacts with the threshold of 0.75, and each cluster was scored on the average score of its best four models (29). The desolvation energy term was turned off for this final scoring step. The docking calculations were driven by using the biochemical data of Ho et al. (10).
Mathematical model
NI = /25, where , the nitrate concentration gradient, ranges from 0.1 to 25 mM.
System stability analysis and biphase quantification using Lyapunov exponent
Because the system is nonlinear, we used a numerical method to calculate the steady-state solutions of the above system. The stability of the steady-state solutions is determined by the eigenvalues of the Jacobian matrix, J, evaluated at the steady states. The system was solved at 100 linearly spaced values of NI ranging from 0.1 to 25 so as to get 100 eigenvalues (also called local Lyapunov exponents). The absolute values of these Lyapunov exponents plotted against the increasing nitrate gradient indicated two distinct phases of NRT1.1P apart from the transient range. Determinant (J)ss > 0 and Trace (J)ss < 0 for all steady states confirmed the stability of the system.
Results and Discussion
Nitrate-induced priming of the phosphorylation site T101 depends on intermonomer dynamics
The homodimer, NRT1.1, consists of two monomers in an asymmetric unit. It has dual-affinity modes of nitrate transport that switch depending on extracellular nitrate concentrations. Nitrate-binding affinity significantly differs between the monomers, which further affects intermonomer interactions (13). While looking into the interface contacts between the monomers, we observed that the polar interface residues B.Thr111 and A.Thr111 establish H-bonds with A.Val229 (2.65 Å), B.Val229 (2.96 Å), and B.Ser233 (3.31 Å), respectively, in the absence of nitrate binding. Also, the hydrophobic residue B.Leu96 at the interface forms an H-bond with A.Arg21 (2.98 Å) (Fig. S1). Although both monomers retain 4.5% of the total surface area as interface solvent-accessible area, they lose all the direct H-bond contacts between the interface residues after nitrate binding. However, they still remain intact because of strong hydrophobic interactions. Additionally, the composition of polar and charged residues at the interface remains almost unaltered, which implies the role of nitrate-driven specific allostery involving the dimer interface residues. A recent study reported that nitrate binding triggers more changes in chemical interactions in monomer A, leading to redistribution of rigid clusters of atoms and formation of a largest rigid cluster with residues 30–94, an interlinking nitrate-binding pocket, and the phosphorylation site T101. In contrast, this rigidity-based allostery is relatively weak in monomer B, which negatively affects the priming of B.T101 for potential phosphorylation in low nitrate concentration (13).
Phosphorylation site Thr101, located on the cytosolic side of the bottom of helix H3, is surrounded by residues from helices H2 and H4. In the apo state, Thr101 is in the deep hydrophobic pocket demarcated by the hydrophobic residues Leu 96, Leu 100, and Ile 104 from the dimer interface side (Fig. S1). Upon nitrate binding in the high-affinity monomer A, Thr101 is shifted to the interface and primed for phosphorylation at low nitrate concentration. In contrast, such conformational changes and priming have not been observed in the low-affinity monomer. A recent study suggests a potential structural significance of this phosphorylation (12). Although the unmodified NRT1.1 forms a structurally coupled dimer and functions as a low-affinity transporter, phosphorylation of Thr101 triggers dimer decoupling, and the protein adopts a high-affinity transport mode (14).
To probe the connections between dimer coupling/decoupling and the relative intermonomer dynamics, NMA has been performed with ENMs that integrate the GNM and the anisotropic network model. This allows us to assess the magnitude and direction of atomic fluctuations and to predict the potential collective motions of the biomolecule. The lowest-frequency normal modes that provide a high degree of collectivity distinguish between the monomeric and dimeric motion of nitrate-bound and unbound NRT1.1. Interestingly, the collective motions of the dimeric NRT1.1 show in-phase synchronous dynamics irrespective of the nitrate binding (Fig. 1, B and C). This corroborates the experimental results of Sun et al. (14), who reported structurally coupled NRT1.1 dimer as an “in-phase” homodimeric transporter. In contrast to the dimeric NRT1.1, the monomeric high-affinity unit shows antiphase asynchronous dynamics relative to its nitrate-unbound monomer A (Fig. 1 A). Several antiphase tipping residues (Ala 68, Leu 362) are responsible for maintaining this antiphase asynchronous collective motion. TM portions of helices H1, H7, H2, H8, H4, H10, H5, and H11, forming the nitrate transport tunnels in the inward-facing open configuration, have mostly remained invariant by maintaining their pairwise angles in both apo and nitrate-bound states. However, NMA shows substantial changes in the volumes of nitrate transport tunnels between the positive and negative motions. In particular, high-affinity nitrate-bound monomer A shows significant changes in the tunnel volume (∼2-fold), with the largest ΔGtransfer = −71.6 kcal/mol among all the monomers in both nitrate-bound and unbound states (Tables S1 and S2). This difference is positively correlated with the large-amplitude motion of the loop (residues 172–182) connecting the C-terminal portion of helix H4 to the N-terminal of helix H5. Together, these indicate the important role of the dynamic loop for releasing the nitrate in the cytosol in the high-affinity nitrate-binding state (Fig. 1, E and F).
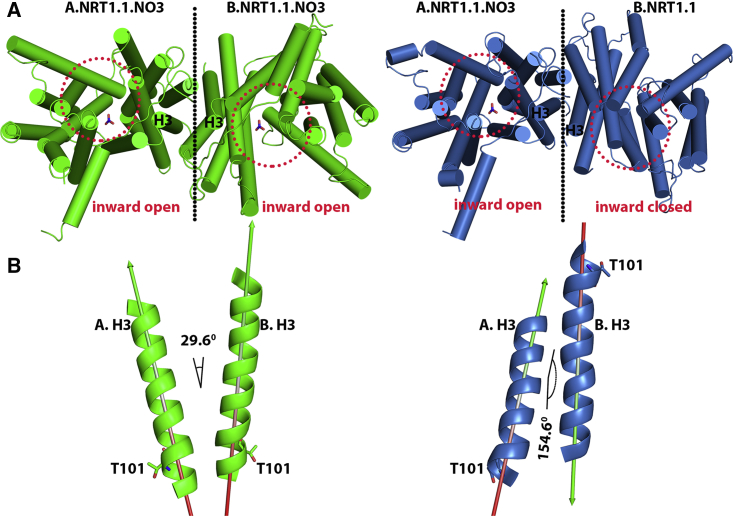
To evaluate the effects of nitrate binding in the high-affinity monomer A on the low-affinity monomer B, nitrate-bound monomer A is docked to nitrate-unbound monomer B (Fig. 2 A). In all the predicted poses, monomer B is reoriented with the reorganization of all the TM helices that buffer nitrate transport activity through the monomer B (Fig. S2). In particular, the lowest-energy pose (i.e., −64.7 kcal/mol) shows rotational conformation of the monomer B relative to the monomer A. The interface helix H3 of monomers A and B, on which the phosphorylation site Thr101 is located, makes an angle of ∼29.6° in the nitrate-bound dimeric structure (Fig. 2 A). However, these helices form an angle of ∼154.6° with an almost opposite orientation of the H3 helix of nitrate-unbound monomer B in the docked dimeric structure (Fig. 2 B). The docked dimer also shows significant loss of the interface area, ∼262.6 Å2, between the monomers. These results, therefore, show that nitrate binding disrupts the in-phase, synchronous motions only in high-affinity monomer A, which results in loss of interface area and primes dimer decoupling. The resultant conformational dynamics also reorients the helices blocking nitrate transport activity through monomer B.
Figure 2.
In low nitrate concentration, nitrate-induced intermonomer rotational dynamics cause dimer decoupling and attenuation of nitrate transport through the monomer B. (A) Both the monomers are nitrate-bound and show an inward-open nitrate transport tunnel. In this case, the interface helices A.H3 and B.H3, in which the phosphorylation site T101 is located at the bottom, made the angle 29.6° with the same orientation. (B) In contrast, whereas nitrate is bound only on monomer A, there is large rotational dynamics of monomer B, in which the interface helices A.H3 and B.H3 show the opposite orientation with an angle of 154.6°. To see this figure in color, go online.
Dynamic loop stabilization is attained through CIPK23 binding
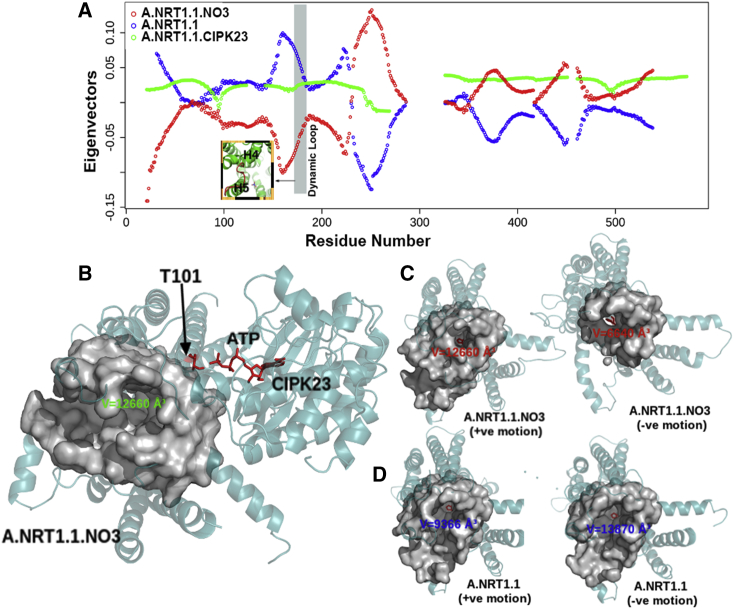
The dynamics of nitrate-bound monomer A shows high-amplitude domain motions in the dynamic loop (residues 172–182) that buffers the nitrate transport (Fig. 1 E (orange); Fig. 3 A). Moreover, the volume of the nitrate tunnel largely varies between the positive and negative motions in the nitrate-bound monomeric state, relative to modest variation in the nitrate-bound dimeric and apo states (Fig. 3, C and D; Fig. S3). This variation is unfavorable for stable transportation and release of nitrate in the cytosol. It is observed that the interactions of NRT1.1 with the active kinase CIPK23 increases the variational entropy by ∼2-fold. Moreover, the collective atomic motions of NRT1.1 and hence of the dynamic loop (Fig. 3 A) are stabilized, resulting in stable high-affinity transport of nitrate through monomer A (Fig. 3 B).
Figure 3.
Interaction of NRT1.1 with the active kinase CIPK23 increases the vibrational entropy (∼2-fold) and has stabilizing effects on the collective motions. (A) Eigenvectors represent a relatively stable state of the A.NRT1.1.CIPK23 complex and the loss of loop dynamics that allow maintaining stable nitrate transport. (B) Formation of relatively stable large nitrate transport tunnel in the presence of CIPK23, with a large volume of 12,660 Å3. (C) and (D) show a large change in the volume of nitrate transport tunnel in the absence of CIPK23 interactions with monomer A. To see this figure in color, go online.
CIPK23 interacts only with the high-affinity nitrate-bound monomer
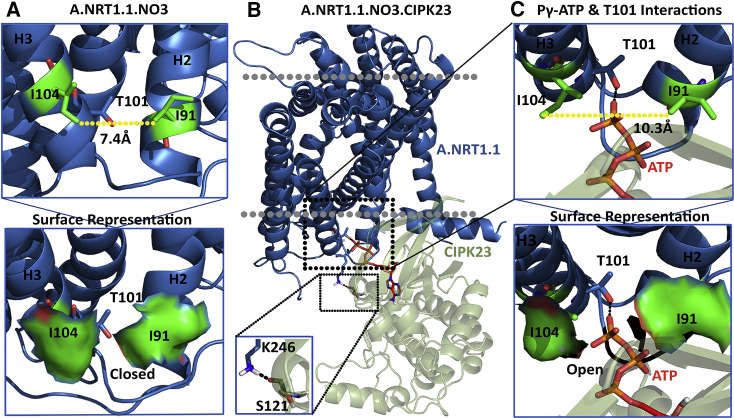
The biochemical data provide valuable experimental constraints for modeling. We used HADDOCK 2.2 (28) to build molecular models of CIPK23.NRT1.1 complex by imposing the structural constraints from the available biochemical data of Ho et al. (10). The kinase domain of CIPK23 interacted with the nitrate-bound monomer A of NRT1.1 and formed a stable complex (Fig. 4 B). Interestingly, Pγ of ATP-bound CIPK23 interacted with the P-site Thr101, apart from other polar interactions between CIPK23 and NRT1.1 that increase the stability of the complex and are necessary for efficient phosphorylation (Fig. 4 A). The distance between Pγ of ATP and the hydroxyl oxygen of the P-site Thr101 is significantly low (2.0 Å). Nitrate-unbound monomers A and B and nitrate-bound monomer B did not form such a complex with CIPK23, and Pγ of ATP did not even come within the 5 Å neighborhood of P-site Thr101 in either of these complexes (Fig. 4 C; Fig. S4). While looking into the NRT1.1.CIPK23 complexes, we also found that in the nitrate-bound monomer A.CIPK23 complex, the interface area is ∼100 Å2 greater and the ΔG-value is ∼2-fold less than in the CIPK23 complex with nitrate-unbound monomers A and B and nitrate-bound monomer B (Table 1). These observations indicate that CIPK23 can specifically interact with and phosphorylate only the high-affinity monomer A of NRT1.1.
Figure 4.
CIPK23 interacts only with the high-affinity nitrate-bound monomer. (A) The hydrophobic pocket within which the phosphorylation site T101 is located inside is closed by the local conformational state of the interface residues I101 and I91. (B) Active CIPK23 closely interacts with the inward-open part of the NRT1.1monomer A in the presence of nitrate. (C) Opening of the hydrophobic pocket and subsequent priming of T101 for phosphorylation by the active CIPK23. To see this figure in color, go online.
Table 1.
Relative Effects of Nitrate Binding and CIPK23 Interactions with NRT1.1
| Monomer A | Monomer B | |||
|---|---|---|---|---|
| NRT1.1.NO3 | dist. (I91, I104) = 7.4 A | I104 (ϕ, ψ) = (−75.6, −45.7) | dist. (I91, I104) = 8.0 A | I104 (ϕ, ψ) = (−71.2, −53.9) |
| NRT1.1.NO3.CIPK23 | dist. (IL91, I104) = 10.3 A | I104 (ϕ, ψ) = (−59.3, −56.7) | dist. (I91, I104) = 8.1 A | I104 (ϕ, ψ) = (−70.2, −56.4) |
| interface area = 556.9 A2 | ΔG = −8.2 kcal/mol | interface area = 439.0 A2 | ΔG = −3.3 kcal/mol | |
| NRT1.1 | dist. (I91, I104) = 7.1 A | I104 (ϕ, ψ) = (−88.2, −46.1) | dist. (I91, I104) = 7.8 A | I104 (ϕ, ψ) = (−67.4, −44.7) |
| NRT1.1.CIPK23 | dist. (I91, I104) = 8.5 A | I104 (phi, psi) = (−67.7, −54.3) | dist. (I91, I104) = 8.3 A | I104 (ϕ, ψ) = (−63.8, −53.8) |
| interface area = 450.9 A2 | ΔG = −4.9 kcal/mol | interface area = 441.5 A2 | ΔG = −3.6 kcal/mol | |
The P-site Thr101, harbored on the H3 helix, lies in a deep pocket surrounded by hydrophobic residues ILE91, PHE95, LEU96, LEU100, and ILE104. Although ILE91 belongs to the H2 helix, LEU100 and ILE104 belong to the H3 helix, and PHE95 and LEU96 form the connecting loop between the H2 and H3 helices. A recent study showed that nitrate binding induces conformational changes that prime the Thr101 site for phosphorylation (13). Though all the neighboring residues seem to occupy strategic positions mounting over P-site Thr101, side chains of ILE91 and ILE104 are still positioned such that they form a gate hindering the accessibility of ATP to T101. Interestingly, our analyses of the complex formation reveal that in monomer A of NRT1.1, the ILE104 side chain rotates away from the cavity upon complex formation with CIPK23, resulting in increased distance between gate residues by 3 Å. This opens the hydrophobic cavity, thereby allowing kinase to deliver Pγ of ATP to the hydroxyl oxygen of the P-site Thr101. Surprisingly, no such conformational positioning in the side chains of the gate residues in nitrate-unbound monomers A and B and nitrate-bound monomer B upon complex formation is reported, and hence, the opening to the T101 site remains closed (Fig. S4). These analyses suggest that the joint effect of high-affinity nitrate binding and CIPK23 interaction results in significant conformational changes around the P-site Thr101 pocket that enhance its full opening toward the surface, resulting in phosphorylation. The detailed comparison between the monomers before and after complex formation is shown in the table (Table 1).
Activation of calcium-dependent signals regulates the bistable states of NRT1.1
At low nitrate concentration (<1 mM), nitrate ions bind only in the high-affinity monomer A, leading to high-amplitude domain motions and subsequent decoupling of the dimer NRT1.1. Nitrate treatments of NRT1.1 also affect the downstream nitrate signaling components by activating phospholipase C, which increases cytoplasmic Ca2+ concentration (15). The calcineurin-B-like calcium sensor CBL9 senses these specific changes in [Ca2+]cyt and binds to the self-inhibitory motif NAF of the kinase CIPK23, which eventually relieves the self-inhibition and activates the kinase CIPK23. At high nitrate concentration, nitrate ions bind to both the monomers of the dimer NRT1.1, causing synchronous in-phase domain motions that couple the dimer and negatively affect the specific calcium waves that activate CBL9. As a result, it indirectly inhibits CIPK23 by maintaining the activity of the self-inhibitory motif NAF. These two phases of NRT1.1 are noted at the extreme of nitrate continuum along which extracellular nitrate varies between the low and high concentrations, ranging from ∼0.1–25 mM. At the lower extreme, NRT1.1 and CBL9.CIPK23 will have close interactions and produce the highest levels of phosphorylated NRT1.1. In contrast, at the high extreme of the nitrate concentrations, CIPK23 will be completely inactive, and the phosphorylated NRT1.1 concentration will be zero. In between these extremes, dynamic interactions between these positive and negative calcium-dependent signals, SX and SY, determine the level of phosphorylated NRT1.1.
To illustrate these dynamic interactions, we find an incoherent feedforward loop (iFFL), which is a specific biochemical control circuit with three-node network topology (30). Upon receiving the two input signals SX and SY—associated with the low and high soil nitrate concentrations, respectively—the disparate responses of NRT1.1 have distinct regulatory effects on the dynamics of phosphorylation: one that negatively regulates the activity of CBL9.CIPK23 complex (X), and the other one that positively regulates its activity for phosphorylating T101 site (Fig. 5 A). These two dynamic regulatory effects are integrated by a Boolean gate function that modulates the phosphorylated state of NRT1.1. The gate function is designed based on the toggling character of the phosphorylation switch as reported by Ho et al. (10). We have then constructed a mathematical model for illustrating the abovementioned dynamic regulation and for the establishment of biphasic stable states along the increasing gradient of nitrate concentrations (Fig. 5 B). This model consists of an incoherent feedforward regulatory circuit in which the nitrate-activated NRT1.1 (X1) modulates its phosphorylated state (X3) either directly, by nitrate binding at the high-affinity site and activating the CBL9.CIPK23 complex (X2), or indirectly, by binding at both the sites (high- and low-affinity sites) and inactivating X2. These two direct and indirect effects have opposite signs and are integrated by a gate function GX3. Changes of their concentrations are described by the ordinary differential equations as stated in Fig. 5. The first equation describes the immediate response of NRT1.1 to nitrate availabilities by the Hill function with the Hill coefficient h1 and dissociation constant k1. In the second equation, the phosphorylation and dephosphorylation are quantified as Michaelis-Menten kinetics with the Michaelis constant k11 for phosphorylation and k12 for the dephosphorylation (31,32). The phosphorylation rate is repressed by the negative regulatory function, X = 1/. The third equation describes the joint effects of negative and positive regulatory functions, X and Y = on the phosphorylated state x3, which are integrated by a gate function .The model parameterization (Table S3) has been carried out to establish a biphasic state along the nitrate gradient. The fixed parameter values keep model dynamics in the positive quadrant throughout and eventually allow for attending a feasible steady state with a given nitrate concentration in the range 0.1–25 mM. Sensitivities of model parameters are tested by increasing/decreasing their numerical values up to twofold from the fixed levels. It has been found that the parameters a1, a3, b3, k1, k12, k2, and h2 are nonsensitive because their changes have no effects on the biphasic stable states. In contrast, the changes of a2, b2, k11, and h1 have distorted the biphasic pattern and thus remain sensitive model parameters.
Figure 5.
An iFFL regulates the bistable states of NRT1.1. (A) Decoupling and coupling of the NRT1.1 dimer generates specific calcium waves that active the kinase CIPK23, having positive and negative effects on NRT1.1P, the phosphorylation state. (B) An incoherent feedforward circuit that integrates calcium-dependent negative (X) and positive (Y) effects on NRT1.1P. (C) The model predicts bistability. At low nitrate concentration, it establishes the high-affinity state, and at the high nitrate concentration, it establishes the low-affinity state. (D) At low nitrate concentration, the NRT1.1P attains its saturation level exponentially, whereas at the high nitrate concentration, the NRT1.1P displays a unimodal response with the peak and then continuously slides down to a lower stable state. (E) The model predicts the continuous sliding from the high-concentration phosphorylated state to the lowest level along the increasing gradient of nitrate concentrations. (F) The calculated Lyapunov exponent at the steady states along the increasing nitrate gradient from 0.1 to 25 mM: it is indicating the biphasic changes of NRT1.1P with increasing soil nitrate concentrations. This allows us to predict the low, the high, and the transient ranges of soil nitrate concentrations that were categorized with respect to the chosen parameter values. To see this figure in color, go online.
The model predicts two different response patterns: at low nitrate concentration, the concentration of the phosphorylated state NRT1.1P (mM) increases exponentially over the time and eventually reaches a plateau (i.e., 0.6 mM) with saturation characteristics (Fig. 5 D). In contrast, at high nitrate concentrations, NRT1.1P displays unimodal characteristics with a peak before skidding to a lower steady state (i.e., 0.001 mM), which is almost 600 times lower than the saturated state at low nitrate concentration. As the nitrate concentration gradually reduces from its highest level of 25 mM, NRT1.1P quickly reaches its lower peak value and then decreases to its lowest stable state in a shorter period of time. Time-dependent changes in the ratio of dimeric NRT1.1/monomeric NRT1.1-P show two distinct trends. Although the ratio increases steadily after a short time period at high nitrate concentration, the ratio quickly decreases at the lowest level at low nitrate levels (Fig. S5).
Both the low- and high-nitrate-associated NRT1.1P states are observed to be stable, as the changes of nitrate concentrations within the ranges of 0.1–1 and 3.0–25 mM do not affect the steady-state concentration much (standard deviation of 0.004 for the low nitrate range; standard deviation of 0.03 for the high nitrate range), and the eigenvalues of the corresponding Jacobian matrix are shown to be negative (Fig. 5 C). There is an intermediate range of nitrate concentrations that corresponds to transient NRT1.1P states. In silico experiments showed that with randomly chosen nitrate concentrations from the uniform distribution between 1.0 and 3.0 mM (transient nitrate level), the model predicts a trajectory connecting the one NRT1.1-P steady state with the NRT1.1 unphosphorylated state, indicating continuous sliding from the high-concentration phosphorylated state to a lowest level along the increasing gradient of nitrate concentrations (Fig. 5 E).
To quantify and establish the bistable states of NRT1.1 (i.e., phosphorylated and unphosphorylated states) in parallel to the experimental results of Ho et al. (10), we have suitably chosen the parameters and calculated the associated Lyapunov exponent to establish a stable biphase (Fig. 5 F). We have used the standard method, in which the maximal eigenvalues of the Jacobian matrix at each steady state for specific nitrate inputs have been calculated. It allows us to predict the low, high, and transient ranges of soil nitrate concentrations that are categorized with respect to the chosen parameter values.
Conclusions
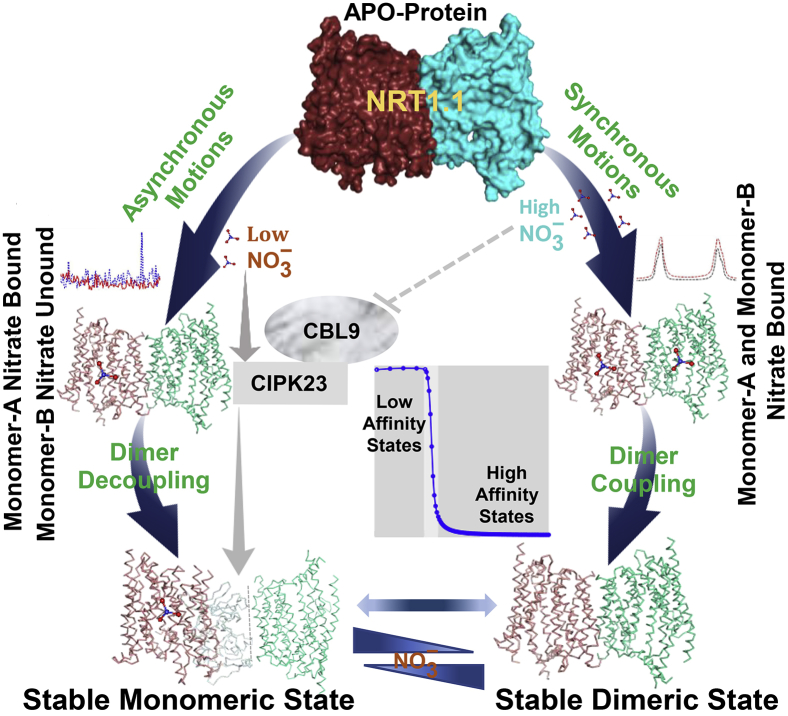
Nitrate-induced dynamic interactions between NRT1.1 and the kinase CIPK23 are examined depending on the extracellular nitrate concentrations. This study shows that nitrate-dependent modulation of this interaction establishes NRT1.1 bistable states that are positively correlated to the biphasic nature of the PNR. The results show that the homodimeric structure of NRT1.1 plays an important role in eliciting specific cytosolic calcium waves sensed by the calcineurin-B-like calcium sensor CBL9, which activates the kinase CIPK23, in low nitrate concentration that is impeded in high nitrate concentration. The dimeric structure of NRT1.1 is well-maintained by nitrate binding to both the monomers that have differential nitrate-binding affinity, with concurrent inhibition of CIPK23 activity. However, at low nitrate concentration, nitrate binds only at the high-affinity monomer, inducing significant changes in collective atomic motions that are transferred allosterically to the low-affinity monomer. It causes the loss of interface interactions and reorientation of the low-affinity unit through conformational reorientation of nitrate-channeling helices, eventually resulting in dimer decoupling and inhibition of nitrate binding at the low-affinity unit. Furthermore, nitrate binding at the high-affinity monomer initiates dimer decoupling and primes the T101 site for the phosphorylation by CIPK23. It stably maintains the monomeric phosphorylated state of NRT1.1, thereby regulating high-affinity nitrate transport and signaling.
We see a coherence between the seemingly different hypotheses: 1) “NRT1.1 monophasic high-affinity switching due to dimer-decoupling” reported by Sun et al. (14) and 2) “the increase in transport rate due to packing/repacking in the N-bundle” reported by Parker et al. (12). In support of the dimer decoupling hypothesis, the analyses show that high-affinity nitrate binding results in loss of crucial contacts at the interface between the monomers, apart from losing a significant percentage of interface area. This is further supported by the antiphase asynchronous dynamics of the two monomers, in contrast to the dimeric NRT1.1 showing in-phase synchronous motions. In line with the concept of structural flexibility of the N-bundle and the enhancement of the transport rate, we show that the flexibility of the transport tunnel increases with enhanced tunnel volume by almost onefold. Furthermore, it is noted that the high-affinity nitrate binding causes a large-amplitude motion of an identified loop between the N and C bundle that could disrupt the transport. But, these loop motions are stabilized by the CIPK23-led phosphorylation of NRT1.1 T101 residue, which allows stable high-affinity transport.
The dynamical responses of NRT1.1 to uniform changes in nitrate concentrations show bistability. Whereas stable phosphorylated states are maintained at low nitrate concentration (0.1–1 mM), the dephosphorylated states prevail in a large range (3.0–25 mM) of nitrate variation (Fig. 6). The transition between these states is not abrupt, but it changes continuously along the gradient of nitrate concentrations. This bistability is achieved through the dynamic interactions between two calcium-dependent signals elicited by the dimerization switch: one that activates the kinase CIPK23 and the other that impedes the CIPK23 activity by its self-inhibitory allosteric mechanism. An incoherent feedforward network involving these dynamic interactions accurately describes this bistability. This analysis reveals that the nitrate transceptor NRT1.1 is transiently regulated by an iFFL that combines both the dynamic effects on CIPK23. The results, therefore, suggest that adaptive responses of nitrate signaling are modulated by the upstream components of the nitrate signaling integrated into an iFFL.
Figure 6.
Graphical representation of NRT1.1-mediated bistable control of nitrate signaling: at low nitrate concentration, high-affinity monomer A is bound by nitrate, which induces asynchronous motions that initiate dimer decoupling and then eventually establishes a stable monomeric state through the interactions with CBL9-activated kinase CIPK23. At high nitrate concentrations, both the monomers are bound by nitrate, which maintains synchronous motions with stable dimer coupling by indirectly inhibiting the kinase CIPK23 activity. To see this figure in color, go online.
Author Contributions
M.R., S.B., and A.C. designed the study and conducted computational experiments and analytical analysis. A.C., M.R., G.-Q.S., A.B.M., and M.B. have reviewed and written the article.
Acknowledgments
Our thanks to project students Ayushi Bhutra, Govind Swami, Neha Bhookar, and Kartik Sharma of the School of Mathematics, Statistics and Computational Sciences, Central University of Rajasthan for their support while completing a part of computational study of this work. We also thank Swadesh Pal, IIT Kanpur and Vijay Pal Bajiya, Central University of Rajasthan for their valuable efforts in mathematical calculations. We highly appreciate Prof. Yi-Fang Tsay, Institute of Molecular Biology, Academia Sinica, Taiwan, for her critical comments on the first version of this manuscript.
This research was supported by SERB-EMR grant (file no. EMR/2015/001671) and by the Central University of Rajasthan; M.R. and S.B. received their Ph.D. fellowships from the Central University of Rajasthan.
Editor: Andrew Plested.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.10.018.
Contributor Information
Gui-Quan Sun, Email: gquansun@126.com.
Amit Chakraborty, Email: amitc.maths@curaj.ac.in.
Supporting Material
References
- 1.Muños S., Cazettes C., Gojon A. Transcript profiling in the chl1-5 mutant of Arabidopsis reveals a role of the nitrate transporter NRT1.1 in the regulation of another nitrate transporter, NRT2.1. Plant Cell. 2004;16:2433–2447. doi: 10.1105/tpc.104.024380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Remans T., Nacry P., Gojon A. The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc. Natl. Acad. Sci. USA. 2006;103:19206–19211. doi: 10.1073/pnas.0605275103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nacry P., Bouguyon E., Gojon A. Nitrogen acquisition by roots: physiological and developmental mechanisms ensuring plant adaptation to a fluctuating resource. Plant Soil. 2013;370:1–29. [Google Scholar]
- 4.O’Brien J.A., Vega A., Gutiérrez R.A. Nitrate transport, sensing, and responses in plants. Mol. Plant. 2016;9:837–856. doi: 10.1016/j.molp.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 5.Lin Y.L., Tsay Y.F. Influence of differing nitrate and nitrogen availability on flowering control in Arabidopsis. J. Exp. Bot. 2017;68:2603–2609. doi: 10.1093/jxb/erx053. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y.Y., Cheng Y.H., Tsay Y.F. Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol. 2018;69:85–122. doi: 10.1146/annurev-arplant-042817-040056. [DOI] [PubMed] [Google Scholar]
- 7.Bouguyon E., Brun F., Gojon A. Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nat. Plants. 2015;1:15015. doi: 10.1038/nplants.2015.15. [DOI] [PubMed] [Google Scholar]
- 8.Wang R., Tischner R., Crawford N.M. Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol. 2004;136:2512–2522. doi: 10.1104/pp.104.044610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu H.C., Wang Y.Y., Tsay Y.F. AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J. 2009;57:264–278. doi: 10.1111/j.1365-313X.2008.03685.x. [DOI] [PubMed] [Google Scholar]
- 10.Ho C.H., Lin S.H., Tsay Y.F. CHL1 functions as a nitrate sensor in plants. Cell. 2009;138:1184–1194. doi: 10.1016/j.cell.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 11.Krouk G., Tillard P., Gojon A. Regulation of the high-affinity NO3- uptake system by NRT1.1-mediated NO3- demand signaling in Arabidopsis. Plant Physiol. 2006;142:1075–1086. doi: 10.1104/pp.106.087510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parker J.L., Newstead S. Molecular basis of nitrate uptake by the plant nitrate transporter NRT1.1. Nature. 2014;507:68–72. doi: 10.1038/nature13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rashid M., Bera S., Chakraborty A. Adaptive regulation of nitrate transceptor NRT1.1 in fluctuating soil nitrate conditions. iScience. 2018;2:41–50. doi: 10.1016/j.isci.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun J., Bankston J.R., Zheng N. Crystal structure of the plant dual-affinity nitrate transporter NRT1.1. Nature. 2014;507:73–77. doi: 10.1038/nature13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riveras E., Alvarez J.M., Gutiérrez R.A. The calcium ion is a second messenger in the nitrate signaling pathway of Arabidopsis. Plant Physiol. 2015;169:1397–1404. doi: 10.1104/pp.15.00961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krouk G. Nitrate signalling: calcium bridges the nitrate gap. Nat. Plants. 2017;3:17095. doi: 10.1038/nplants.2017.95. [DOI] [PubMed] [Google Scholar]
- 17.Kudla J., Becker D., Schumacher K. Advances and current challenges in calcium signaling. New Phytol. 2018;218:414–431. doi: 10.1111/nph.14966. [DOI] [PubMed] [Google Scholar]
- 18.Choi W.G., Toyota M., Gilroy S. Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc. Natl. Acad. Sci. USA. 2014;111:6497–6502. doi: 10.1073/pnas.1319955111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manishankar P., Wang N., Kudla J. Calcium signaling during salt stress and in the regulation of ion homeostasis. J. Exp. Bot. 2018;69:4215–4226. doi: 10.1093/jxb/ery201. [DOI] [PubMed] [Google Scholar]
- 20.Chaves-Sanjuan A., Sanchez-Barrena M.J., Albert A. Structural basis of the regulatory mechanism of the plant CIPK family of protein kinases controlling ion homeostasis and abiotic stress. Proc. Natl. Acad. Sci. USA. 2014;111:E4532–E4541. doi: 10.1073/pnas.1407610111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hashimoto K., Eckert C., Kudla J. Phosphorylation of calcineurin B-like (CBL) calcium sensor proteins by their CBL-interacting protein kinases (CIPKs) is required for full activity of CBL-CIPK complexes toward their target proteins. J. Biol. Chem. 2012;287:7956–7968. doi: 10.1074/jbc.M111.279331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takeda K., Shao D., Firtel R.A. Incoherent feedforward control governs adaptation of activated ras in a eukaryotic chemotaxis pathway. Sci. Signal. 2012;5:ra2. doi: 10.1126/scisignal.2002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lomize M.A., Pogozheva I.D., Lomize A.L. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 2012;40:D370–D376. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H., Chang Y.Y., Yang L.W. DynOmics: dynamics of structural proteome and beyond. Nucleic Acids Res. 2017;45:W374–W380. doi: 10.1093/nar/gkx385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baspinar A., Cukuroglu E., Gursoy A. PRISM: a web server and repository for prediction of protein-protein interactions and modeling their 3D complexes. Nucleic Acids Res. 2014;42:W285–W289. doi: 10.1093/nar/gku397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grosdidier A., Zoete V., Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011;39:W270–W277. doi: 10.1093/nar/gkr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng J., Trafny E.A., Sowadski J.M. 2.2 A refined crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MnATP and a peptide inhibitor. Acta Crystallogr. D Biol. Crystallogr. 1993;49:362–365. doi: 10.1107/S0907444993000423. [DOI] [PubMed] [Google Scholar]
- 28.van Zundert G.C.P., Rodrigues J.P.G.L.M., Bonvin A.M.J.J. The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016;428:720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 29.Rodrigues J.P., Trellet M., Bonvin A.M. Clustering biomolecular complexes by residue contacts similarity. Proteins. 2012;80:1810–1817. doi: 10.1002/prot.24078. [DOI] [PubMed] [Google Scholar]
- 30.Mangan S., Alon U. Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. USA. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tyson J.J., Chen K.C., Novak B. Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Curr. Opin. Cell Biol. 2003;15:221–231. doi: 10.1016/s0955-0674(03)00017-6. [DOI] [PubMed] [Google Scholar]
- 32.Ferrell J.E., Jr., Ha S.H. Ultrasensitivity part III: cascades, bistable switches, and oscillators. Trends Biochem. Sci. 2014;39:612–618. doi: 10.1016/j.tibs.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.