Abstract
Purpose
Ventilator-associated pneumonia caused by Pseudomonas aeruginosa (P. aeruginosa) is a major health-care problem. In this study, we explored the epidemiology of virulence determinants among multi-drug-resistant (MDR) clinical P. aeruginosa isolates from hospitalized patients with ventilator-associated pneumonia in intensive care units in Upper Egypt.
Patients and Methods
MDR P. aeruginosa isolates were screened for the presence of eight virulence factors and typed by ERIC-PCR.
Results
A total of 39 clinical MDR isolates were selected out of 173 isolated P. aeruginosa showing a combination of adhesion and cytotoxicity virulence patterns, with the detection of aprA, exoU, exoS, lasB, algD, toxA in 74.3%, 58.9%, 46.1%, 41.2%, 30.7%, 20.5% of the isolates, respectively. The MDR isolates were grouped into 13 different virulence profiles according to the pattern of virulence gene distribution. exoU genotype was more predominant among the P. aeruginosa isolates with more than 48% of the isolates harboring this gene alone, 7% harboring both exoU and exoS and 43.5% harboring exoS gene. An intermediate degree of diversity was detected by ERIC-PCR typing where the isolates were clustered in 7 major groups, indicating possible cross-infection within the hospital.
Conclusion
Our results highlight the increased frequency of virulent P. aeruginosa isolates with a shift to the more virulent cytotoxic exoU genotype. Further hospital infection-control measures are mandatory to control the hospital cross-transmission of these highly virulent isolates. This study could vastly be a help to develop efficient treatment policies against P. aeruginosa induced ventilator-associated pneumonia.
Keywords: P. aeruginosa, virulence, exoU, MDR
Introduction
Pseudomonas aeruginosa (P. aeruginosa) is one of the most common causes of healthcare-associated infections being responsible for urinary tract, respiratory and surgical site infections.1,2 This opportunistic pathogen is considered as a major health hazard, especially in immunodeficient patients.3 Furthermore, infections caused by multi-drug-resistant (MDR) P. aeruginosa isolates are associated with longer duration of hospitalization, increased costs, as well as increased morbidity and mortality rates.4
P. aeruginosa has a repertoire of virulence factors that markedly contribute to its pathogenicity5 e.g., lipopolysaccharides, adhesion factor (pili type IV), flagella, exo-proteases (alkaline protease “AprA”, elastase, staphylolysin, protease IV), phospholipase C, exotoxins “Exo A”, exoenzymes S, T, and U, as well as sialidase.5–9 In addition, P. aeruginosa has the exquisite ability to form biofilms, which render it more resistant to antimicrobials.10
Exoenzymes S, Y, T, and U are encoded by the genes exoS, exoY exoT, and exoU, respectively, with exoS being the most predominant.11 While former studies have shown that the existence of exoS is related to increased virulence in burn wounds and lung infections,12 ExoU is 100 times more cytotoxic than ExoS.5,13 LasB elastase, a zinc metalloprotease encoded by the lasB gene, attacks eukaryotic proteins such as elastin and collagen.10,14 In addition, two phospholipases C encoded by plcH and plcN genes can hydrolyze phospholipids. The high frequency of virulence factor phospholipase C gene (plcH) in MDR P. aeruginosa clinical isolates probably plays an important role in the pathogenesis of this bacterium. The gene algD encodes the GDP-mannose dehydrogenase enzyme, which is the first element in the alginate biosynthetic cluster essential for alginate biosynthesis. Alginate is a linear exopolysaccharide, which protects the bacterium from antibiotics and the host’s immune response.10
The increased invasiveness and pathogenicity as a result of harboring these virulence factors upsurge the morbidity despite the use of antibiotics.15 However, the arsenal of virulence mechanisms harbored by P. aeruginosa varies according to the settings and the environment of infection.16
In previous studies done in this region, a high incidence of MDR P. aeruginosa was observed among isolates from infected burn and surgical wounds.17,18 The identification of virulence genes’ profile is crucial for developing efficient policies against P. aeruginosa infections; the aim of this study was to evaluate the distribution of different virulence genes among MDR P. aeruginosa isolated from different Egyptian ICUs.
Materials and Methods
In this cross-sectional study, samples were collected from respiratory tract infections over a period of 15 months between 2017 and 2019 from patients admitted to the different ICUs in Minia University Hospital (a tertiary care hospital). A written informed consent was obtained from every patient or his caregiver. The study was carried out as per the Helsinki declarations and was approved by the Ethical committee of the Faculty of Medicine, Minia University. A total of 173 P. aeruginosa isolates were phenotypically identified by routine cultural and biochemical methods. MDR isolates were selected for further characterization by antibiotic sensitivity testing using the disc-diffusion method according to CLSI 2017, using P. aeruginosa ATCC 27853 as a reference strain. MDR is identified as resistance to three or more groups of antimicrobials.
DNA Extraction, Identification and Virulence Gene Detection
Genomic DNA was extracted using a boiling method as described previously; one loopful of fresh bacteria (grown overnight on Brain-Heart Infusion agar plates) was picked up and suspended in 200 µL of sterile DNase/RNase-free water and boiled for 10 min. The bacterial suspension was then centrifuged at 12,000 rpm at 4°C for 10 min, and the supernatant was collected and kept at −20°C.19
Detection of the following virulence genes was carried out by conventional PCR: GDP-mannose dehydrogenase enzyme for alginate (algD), alkaline protease (aprA), elastase (lasB), exoenzyme S and U (exoS and exoU) and exotoxin A (toxA), hemolytic phospholipase C (plcH) and non-hemolytic phospholipase C (plcN), and the used primers are listed in Table 1. The amplification was carried in a 25μL volume, containing 12.5 μL PCR Master mix (DreamTaq Green PCR master mix, Thermo scientific), 1μL of each primer (forward and reverse), 1 μL of template DNA and nuclease-free water.
Table 1.
Primers Used in This Study
| Target Gene | Sequence (5ʹ – 3ʹ) | Amplicon Size (bp) | Annealing Temperature |
|---|---|---|---|
| toxA | F: 5ʹGGTAACCAGCTCAGCCACAT 3ʹ | 352 | 48°C |
| R: 5ʹTGATGTCCAGGTCATGCTTC 3’ | |||
| plcN | F: 5ʹ GTTATCGCAACCAGCCCTAC 3ʹ | 466 | 48°C |
| R: 5ʹAGGTCGAACACCTGGAACAC | |||
| plcH | F: 5ʹGAAGCCATGGGCTACTTCAA 3ʹ | 307 | 48°C |
| R: 5ʹAGAGTGACGAGGAGCGGTAG 3’ | |||
| algD | F:5ʹ ATGCGAATCAGCATCTTTGGT 3ʹ | 1311 | 48°C |
| R:5ʹ CTACCAGCAGATGCCCTCGGC 3ʹ | |||
| lasB | F:5ʹCCAGCCCGCTGACCCACAAGCTGTA 3ʹ | 650 | 46°C |
| R: 5ʹCATTCCTTCCTGGAGTGCYRGCCG 3’ | |||
| aprA | F:5ʹ CCTGATCKGGCCGATAACTGCAAT 3ʹ | 1580 | 46°C |
| R:5ʹ GGAAGACASCTATCAATTCGAACAG 3’ | |||
| exoU | F: 5ʹ GGCACATATCTCCGGTTCCTTC 3ʹ | 761 | 55°C |
| R: 5ʹ TCAACTCAGCTGCCAACCATGC 3’ | |||
| exoS | F: 5ʹATGGCGTGTTCCGAGTCA 3ʹ | 1587 | 55°C |
| R: 5ʹ AGGTGTCGGTTCGTGACGTCT 3’ | |||
| ERIC primers | ERIC-1R, 5VCACTTAGGGGTCCTCGAATGTA-3V | ||
| ERIC-2, 5V-AAGTAAGTGACTGGGGTGAGCG-3V |
Abbreviations: toxA, exotoxin A; plcH, hemolytic phospholipase C; plcN, non-hemolytic phospholipase C; algD, GDP-mannose dehydrogenase enzyme for alginate; lasB, elastase; aprA, alkaline protease; exoU, exoenzyme U; exoS, exoenzyme S; ERIC, Enterobacterial Repetitive Intergenic Consensus-PCR.
The PCR conditions for lasB and aprA comprised denaturation at 94°C (4 mins), followed by 25 cycles of: 94°C (1 min), 46°C (40 s), 72°C (1 min) and a final extension at 72°C (2 mins). The exoS and exoU amplification conditions included denaturation at 95°C (5 mins), followed by 35 cycles of 95°C (1 min), 55°C (1 min), 72°C (1 min) followed by final extension at 72°C (10 mins).11,20 For the rest of the genes, the conditions were as follows: preliminary denaturation at 94°C (5 min), 35 cycles of denaturation at 94°C (60 s), annealing at 48°C (60 s) and extension at 72°C (90 s), with a last extension cycle at 72°C for 10 min.21 PCR products were run on 1.5% agarose gel and were afterward visualized under UV lamb.
Enterobacterial Repetitive Intergenic Consensus-PCR (ERIC-PCR Typing)
ERIC PCR was performed to produce the repetitive sequence between the two primers.22 The PCR started with an initial denaturation at 94°C for 7 min, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 53°C for 1 min and extension at 72°C for 2 min and a final extension cycle at 72°C for 15 min. Amplicons were loaded in 1.5% agarose gel, which was run at 80 V for 3 hrs. Dendrogram was generated by using GelJ software using Dice Similarity method; UPGMA linkage.23
Statistical Analysis
Statistical analysis was performed using Graph-pad Prism 6.
Results
Identification and Virulence-Profiling
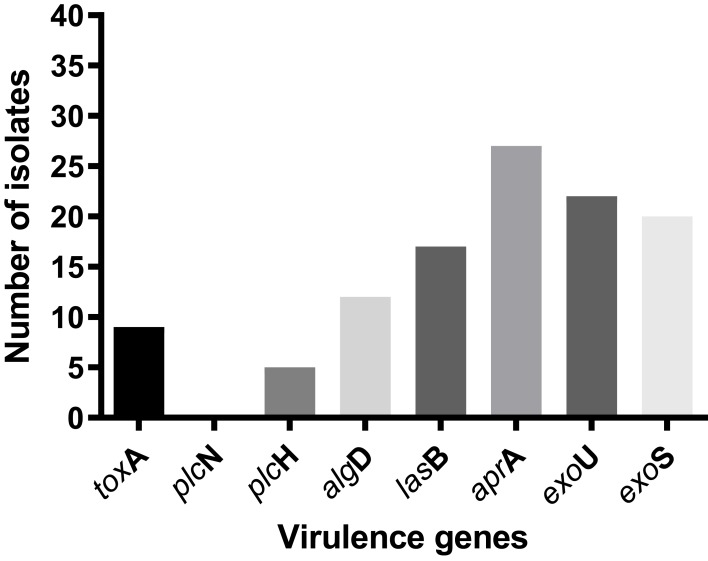
This study included 39 (22.5%) MDR isolates from a total of 173 P. aeruginosa isolates (the antimicrobial-susceptibility profile of the MDR isolates to different anti-pseudomonal drugs is illustrated in Supplementary Figure 1). The prevalence of eight virulence genes among MDR P. aeruginosa nosocomial isolates was investigated, and the frequencies of toxA, plcN; plcH; algD; lasB; aprA; exoU and exoS genes among the MDR isolates were 9 (23%); 0 (0%); 5 (12.8%); 12 (30.7%); 17 (43.5%); 27 (67.2%); 22 (50.4%) and 20 (51.3%), respectively (Figure 1). None of the isolates harbored plc N gene. Only three isolates (7.6%) showed a simultaneous presence of exoU and exoS genes (Table 2).
Figure 1.
Frequency of virulence genes in MDR P. aeruginosa.
Table 2.
Characteristics of MDR P.aeruginosa Isolates from Different ICUs in Egypt
| Isolate No. | toxA | plcN | plcH | algD | lasB | aprA | exoU | exoS | Number of Virulence Genes |
|---|---|---|---|---|---|---|---|---|---|
| PA-M1 | - | - | - | - | - | + | + | - | 2 |
| PA-M2 | - | - | - | - | - | + | + | - | 2 |
| PA-M3 | - | - | - | - | + | + | - | + | 3 |
| PA-M4 | - | - | - | - | - | - | - | + | 1 |
| PA-M5 | - | - | - | - | + | + | + | - | 3 |
| PA-M6 | - | - | - | - | - | - | + | + | 2 |
| PA-M7 | - | - | - | - | - | - | + | - | 1 |
| PA-M8 | - | - | - | - | + | + | - | + | 3 |
| PA-M9 | - | - | - | - | - | - | - | + | 1 |
| PA-M10 | - | - | + | + | - | - | + | - | 3 |
| PA-M11 | - | - | - | + | + | + | + | - | 4 |
| PA-M12 | - | - | - | - | + | + | - | + | 3 |
| PA-M13 | - | - | - | - | + | + | + | - | 3 |
| PA-M14 | - | - | - | - | + | + | + | - | 3 |
| PA-M15 | + | - | + | + | + | + | - | + | 6 |
| PA-M16 | - | - | - | - | - | + | + | - | 2 |
| PA-M17 | + | - | - | + | - | + | - | + | 4 |
| PA-M18 | + | - | - | + | - | + | + | - | 4 |
| PA-M19 | + | - | - | + | - | + | - | + | 4 |
| PA-M20 | - | - | - | - | - | + | + | - | 2 |
| PA-M21 | - | - | - | - | - | + | + | - | 2 |
| PA-M22 | - | - | - | - | + | + | - | + | 3 |
| PA-M23 | - | - | - | - | - | - | - | + | 1 |
| PA-M24 | - | - | - | - | - | - | - | + | 1 |
| PA-M25 | - | - | - | - | - | - | + | + | 2 |
| PA-M26 | - | - | - | - | - | - | + | - | 1 |
| PA-M27 | + | - | + | - | + | - | + | - | 4 |
| PA-M28 | - | - | - | - | + | + | + | - | 3 |
| PA-M29 | - | - | + | + | - | - | + | - | 3 |
| PA-M30 | - | - | - | + | - | + | + | + | 4 |
| PA-M31 | - | - | - | - | + | + | - | + | 3 |
| PA-M32 | + | - | - | + | + | - | - | + | 4 |
| PA-M33 | - | - | - | - | + | + | + | - | 3 |
| PA-M34 | + | - | + | + | + | + | - | + | 6 |
| PA-M35 | - | - | - | - | - | + | + | - | 2 |
| PA-M36 | - | - | - | - | + | + | + | - | 3 |
| PA-M37 | - | - | - | - | + | + | - | + | 3 |
| PA-M38 | + | - | - | + | - | + | - | + | 4 |
| PA-M39 | + | - | - | + | - | + | - | + | 4 |
Abbreviations: PA, P. aeruginosa; toxA, exotoxin A; plcH, hemolytic phospholipase C; plcN, non-hemolytic phospholipase C; algD, GDP-mannose dehydrogenase enzyme for alginate; lasB, elastase; aprA, alkaline protease; exoU, exoenzyme U; exoS, exoenzyme S; ERIC, Enterobacterial Repetitive Intergenic Consensus-PCR.
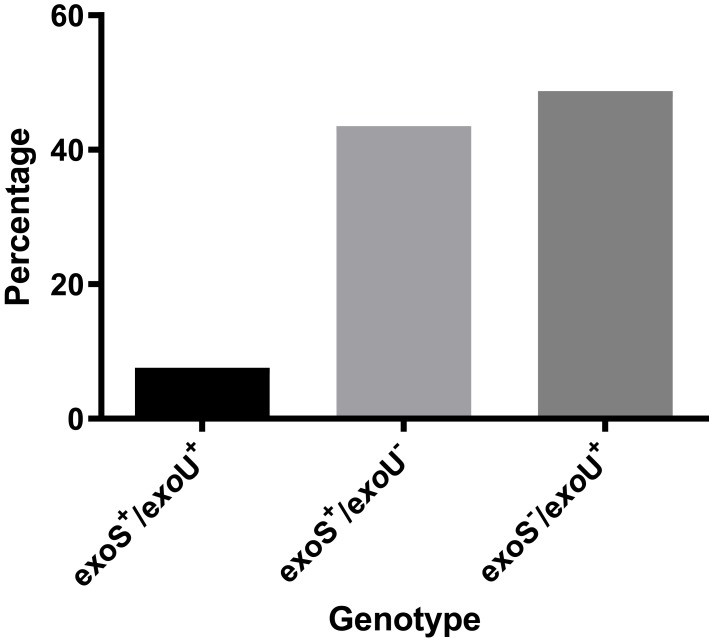
The majority of the isolates (23 isolates) had between 1 and 4 virulence genes, and only two isolates showed 6 virulence genes altogether, while no isolates possessed more than 6 virulence genes. There were 13 different virulence profiles of MDR P. aeruginosa isolates with types 1, 2 and 3 being the most common, while the rarest were types 10–13 (Table 3). Furthermore, due to the danger of exoU-harboring isolates, the isolates were classified into three categories according to the presence or absence of exoU and exoS genes (Figure 2), where exoS−/exoU+ genotype was the commonest genotype (48.7%).
Table 3.
Virulence Profiles of MDR P. aeruginosa Isolates
| Profile | Virulence Gens | Number/% |
|---|---|---|
| (Profile 1) | lasB-aprA-exoS | 6 (15%) |
| (Profile 2) | lasB-aprA-exoU | 6 (15%) |
| (Profile 3) | aprA-exoU | 6 (15%) |
| (Profile 4) | toxA-algD-aprA-exoS | 5(12.8%) |
| (Profile 5) | exoS | 4(10%) |
| (Profile 6) | exoU | 2 (5%) |
| (Profile 7) | exoU/exoS | 2(5%) |
| (Profile 8) | toxA-plcH-algD-lasB-aprA-exoS | 2(5%) |
| (Profile 9) | plcH-algD-exoU | 2(5%) |
| (Profile 10) | toxA-algD-aprA-exoU | 1(2%) |
| (Profile 11) | toxA-plcH-lasB-exoU | 1(2%) |
| (Profile 12) | algD-lasB-aprA-exoU | 1(2%) |
| (Profile 13) | algD-aprA-exoU-exoS | 1(2%) |
Abbreviations: toxA, exotoxin A; plcH, hemolytic phospholipase C; plcN, non-hemolytic phospholipase C; algD, GDP-mannose dehydrogenase enzyme for alginate; lasB, elastase; aprA, alkaline protease; exoU, exoenzyme U; exoS, exoenzyme S; ERIC, Enterobacterial Repetitive Intergenic Consensus-PCR.
Figure 2.
Distribution of exoU, exoS genotypes among MDR P. aeruginosa isolates.
Genotyping by ERIC-PCR
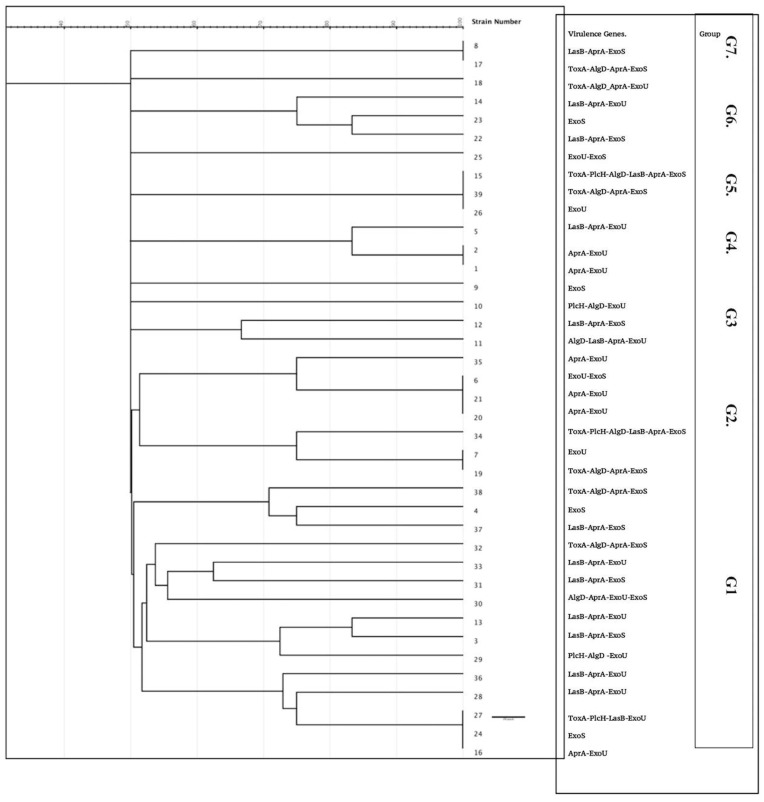
This study revealed intermediate diversity of ERIC fingerprints, with 2 major clusters (1 and 2) enclosing 22 isolates, three groups with 3 isolates and two groups with 2 closely related isolates. Four isolates showed a unique pattern (9, 10, 15, and 25), while groups (3 and 7) were composed of 2 isolates (Figure 3). The difference in the ERIC regions is not strictly related to the occurrence of virulence genes. Furthermore, isolates with shared ERIC types displayed different virulence factor arrays.
Figure 3.
Dendrogram illustrating the genetic relationship among MDR P. aeruginosa isolates.
Discussion
One of the very important pathogens causing opportunistic health-care-acquired infections is P. aeruginosa, because of its capacity to invade tissues using a myriad of enzymes and toxins.24 It is contemplated as a threat to health particularly in patients with deficient immune response: as those in intensive care units (ICU) and burn units.3 In fact, P. aeruginosa is the most commonly isolated pathogen from patients with ventilator-associated pneumonia (VAP) in ICUs.25 Epidemiological evaluation of P. aeruginosa on the molecular level is extremely important so as to comprehend the potential risk elements related to the infection process, assist in finding epidemic strain(s) in a particular population of patients, and recognize virulent strain(s).26
In the current study, MDR P.aeruginosa showed a frequency of 22.5%, which is far less than previous results found in the same region.18 These could be due to differences in the nature of infection with different associated risk factors.27 In addition to the fact that the epidemiology of MDR microorganism infections fluctuates from year to year between hospitals, wards, departments in addition to the geographical zone.28 Furthermore, a better implementation of antimicrobial stewardship in different ICUs is another reason for this decrease in MDR rate.
Identification of the P. aeruginosa armor of virulence factors is crucial to understand the pathogenesis of this opportunistic pathogen and is important in exploring new antimicrobial objectives especially in MDR strains. In this study, most of the isolates (23 isolates) had at least one virulence gene with a maximum of 6 virulence genes detected in 2 isolates only. The virulence phenotype was a mixture of adhesion and cytotoxicity, where aprA (encoding alkaline metalloproteinase) detected in 74.3% of the isolates; exoU (encoding exoenzyme U) in 58.9%; exoS (encoding exoenzyme S) in 46.1%; lasB (encoding elastase LasB) in 41.2%; algD (encoding GDP-mannose 6-dehydrogenase) in 30.7%; toxA (encoding exotoxin A) in 20.5% of the isolates and plcH (encoding hemolytic phospholipase C precursor) in 10.2% of the isolates.
The high incidence of aprA among our isolates is consistent with other studies,21,29 as this enzyme is essential for bacterial survival in tissues by diminishing the activation of Toll-like receptor 5 (TLR-5) and degrading host proteins as cytokines and complement.30
We found a high frequency of exoU in our isolates indicating that more than half of the isolates display the cytotoxic phenotype,31 which is associated with more pulmonary damage than invasive phenotypes.32 This finding is consistent with Rodulfo et al,33 who found a high frequency of exoU in MDR P. aeruginosa isolates. On the other hand, exoS was detected in a relatively similar frequency as exoU; with 17 isolates (43%) harboring exoS gene alone and 3 isolates possessing both genes (7.6%). These results are different from those obtained earlier by our team, where exoS genotype was much more frequent than exoU.34 This could be due to the difference in sample source as in the earlier study samples were from infected surgical wounds.
Elastase lasB, an important virulence attribute detected in a relatively high frequency in our isolates, is known to evade the immune response and particularly target alveolar macrophages.20,32 Our results are concurring with several studies showing a high frequency of lasB in isolates from different sources.35–37
GDP-mannose dehydrogenase enzyme encoded by algD (which contributes to bacterial adhesion) was not very frequent in our isolates. These findings are not in agreement with Al Dawodeyah et al who found a very high frequency of algD among their respiratory isolates.37 As this polysaccharide is a biofilm-related one, this might indicate the absence or lack of biofilm formation by our isolates, which needs further confirmation in future work.
Exotoxin A (a very potent and is the most toxic virulence determinant of P. aeruginosa) was detected in a smaller number of our isolates. This finding is not concurring with a study carried by Nikbin et al or Khattab et al, who found that almost all of their pulmonary isolates harbored toxA.38,39
Two phospholipases C determined by plcH and plcN genes are known to hydrolyze the phospholipids contained in pulmonary surfactants. We did not find any plcN in our isolates; however, plcH was detected in about 10% of MDR P. aeruginosa. This adds more threat, as plcH increases the virulence (causing organ injury and increased mortality) compared to plcN, which has no pathogenic effect.21 Our results are comparable to those done by Fadhil et al,24 while higher incidence of both genes was reported in different studies involving P. aeruginosa from various hospital sources.6,21
Bacterial phenotypic typing methods as phage typing and serotyping are now replaced by molecular methods as pulse-field gel electrophoresis (PFGE), ribotyping, and different PCR-based methods such as ERIC-PCR. ERIC-PCR is a technique that uses the difference in the position and number of ERIC sequences as an indicator of bacterial diversity.40 Bacterial genotyping using ERIC-PCR is of tremendous importance in epidemiological studies carried on P. aeruginosa to evaluate genetic relations, particularly in health-care-acquired infections. In the current study, we found intermediate diversity of ERIC patterns, with no close relation to the presence of different virulence genes. In addition, we found that sharing the same ERIC group did not imply having the same virulence determinant pattern. Great genetic heterogeneity in P. aeruginosa isolates was reported by several studies using ERIC-PCR, RAPD, and PFGE.3,36,41 We used ERIC-PCR as it is cheap, reliable and has a good discriminatory power for P. aeruginosa genotyping.42 Our results suggest that nosocomial transmission of the isolates in this study and its cross-infection is relatively limited due to the relative degree of diversity among the ERIC-patterns apart from isolates in the first cluster, where cross-acquisition may have occurred. P. aeruginosa cross-infection is reported in several studies done in ICUs with variable rates between different hospitals.43–45
Conclusion
In conclusion, pulmonary isolates obtained from ICUs in Upper Egypt possess a wide array of virulence determinants in addition to its MDR attributes. Furthermore, we report here a high incidence of the virulent exoU genotype, the presence of which in MDR P. aeruginosa found in our ICUs is of tremendous significance due to the harmful blend of increased cytotoxicity and limited treatment options for these patients.
Funding Statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Ethics Approval and Consent to Participate
A written consent was obtained from each patient or his/her caregiver and the study was ethically approved by the Ethical Committee of the Faculty of Medicine, Minia University.
Disclosure
The authors declare no conflicts of interest in this work.
References
- 1.Fazeli H, Nasr Esfahani B, Sattarzadeh M, Mohammadi BH. Antibiotyping and genotyping of Pseudomonas aeruginosa strains isolated from Mottahari Hospital in Tehran, Iran by ERIC-PCR. Infect Epidemiol Microbiol. 2017;3(2):41–45. [Google Scholar]
- 2.Rossolini G, Mantengoli E. Treatment and control of severe infections caused by multiresistant Pseudomonas aeruginosa. Clin Microbiol Infect. 2005;11:17–32. doi: 10.1111/j.1469-0691.2005.01161.x [DOI] [PubMed] [Google Scholar]
- 3.Nanvazadeh F, Khosravi AD, Zolfaghari MR, Parhizgari N. Genotyping of Pseudomonas aeruginosa strains isolated from burn patients by RAPD-PCR. Burns. 2013;39(7):1409–1413. doi: 10.1016/j.burns.2013.03.008 [DOI] [PubMed] [Google Scholar]
- 4.Gales AC, Castanheira M, Jones RN, Sader HS. Antimicrobial resistance among gram-negative bacilli isolated from Latin America: results from SENTRY antimicrobial surveillance program (Latin America, 2008–2010). Diag Microbiol Infect Dis. 2012;73(4):354–360. doi: 10.1016/j.diagmicrobio.2012.04.007 [DOI] [PubMed] [Google Scholar]
- 5.Benie CKD, Dadié A, Guessennd N, et al. Characterization of virulence potential of Pseudomonas aeruginosa isolated from bovine meat, fresh fish, and smoked fish. Euro J Microbiol Immunol. 2017;7(1):55–64. doi: 10.1556/1886.2016.00039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fazeli N, Momtaz H. Virulence gene profiles of multidrug-resistant Pseudomonas aeruginosa isolated from Iranian hospital infections. Iran Red Cresc Med J. 2014;16:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krall R, Schmidt G, Aktories K, Barbieri JT. Pseudomonas aeruginosa ExoT is a Rho GTPase-activating protein. Infect Immun. 2000;68(10):6066–6068. doi: 10.1128/IAI.68.10.6066-6068.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaver CM, Hauser AR. Relative contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to virulence in the lung. Infect Immun. 2004;72(12):6969–6977. doi: 10.1128/IAI.72.12.6969-6977.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bricha S, Ounine K, Oulkheir S, Haloui N, Attarassi B. Virulence factors and epidemiology related to Pseudomonas aeruginosa. Tunis J Infect Dis. 2009;2:7–14. [Google Scholar]
- 10.Wolska K, Szweda P. Genetic features of clinical Pseudomonas aeruginosa strains. Pol J Microbiol. 2009;58(3):255–260. [PubMed] [Google Scholar]
- 11.Agnello M, Wong-Beringer A, Cascales E. Differentiation in quinolone resistance by virulence genotype in Pseudomonas aeruginosa. PLoS One. 2012;7(8):e42973. doi: 10.1371/journal.pone.0042973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Almeida K, Calomino MA, Deutsch G, et al. Molecular characterization of multidrug-resistant (MDR) Pseudomonas aeruginosa isolated in a burn center. Burns. 2017;43(1):137–143. doi: 10.1016/j.burns.2016.07.002 [DOI] [PubMed] [Google Scholar]
- 13.Sawa T, Shimizu M, Moriyama K, Wiener-Kronish JP. Association between Pseudomonas aeruginosa type III secretion, antibiotic resistance, and clinical outcome: a review. Crit Care. 2014;18(6):668. doi: 10.1186/s13054-014-0668-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toder D, Ferrell S, Nezezon J, Rust L, Iglewski B. lasA and lasB genes of Pseudomonas aeruginosa: analysis of transcription and gene product activity. Infect Immun. 1994;62(4):1320–1327. doi: 10.1128/IAI.62.4.1320-1327.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makedou KG, Tsiakiri EP, Bisiklis AG, et al. Changes in antibiotic resistance of the most common gram-negative bacteria isolated in intensive care units. J Hosp Infect. 2005;60(3):245–248. doi: 10.1016/j.jhin.2005.01.013 [DOI] [PubMed] [Google Scholar]
- 16.Dubern JF, Cigana C, De Simone M, et al. Integrated whole‐genome screening for P seudomonas aeruginosa virulence genes using multiple disease models reveals that pathogenicity is host specific. Environ Microbiol. 2015;17(11):4379–4393. doi: 10.1111/1462-2920.12863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hassuna NA, Mohamed AHI, Abo-Eleuoon SM, Rizk HA. High prevalence of multidrug resistant Pseudomonas aeru. Arch Clin Microbiol. 2015;6:4. [Google Scholar]
- 18.Raouf MR, Sayed M, Rizk HA, Hassuna NA. High incidence of MBL-mediated imipenem resistance among Pseudomonas aeruginosa from surgical site infections in Egypt. J Infect Dev Countr. 2018;12(07):520–525. doi: 10.3855/jidc.9936 [DOI] [PubMed] [Google Scholar]
- 19.Gholami A, Majidpour A, Talebi-Taher M, Boustanshenas M, Adabi M. PCR-based assay for the rapid and precise distinction of Pseudomonas aeruginosa from other Pseudomonas species recovered from burns patients. J Prev Med Hyg. 2016;57(2):E81. [PMC free article] [PubMed] [Google Scholar]
- 20.Andrejko M, Zdybicka-Barabas A, Janczarek M, Cytryńska MJABP. Three Pseudomonas aeruginosa strains with different protease profiles. Acta Bioch Polo. 2013;60(1):83–90. [PubMed] [Google Scholar]
- 21.Badamchi A, Masoumi H, Javadinia S, Asgarian R, Tabatabaee A. Molecular detection of six virulence genes in Pseudomonas aeruginosa isolates detected in children with urinary tract infection. Microb Pathog. 2017;107:44–47. doi: 10.1016/j.micpath.2017.03.009 [DOI] [PubMed] [Google Scholar]
- 22.Syrmis MW, O’carroll MR, Sloots TP, et al. Rapid genotyping of Pseudomonas aeruginosa isolates harboured by adult and paediatric patients with cystic fibrosis using repetitive-element-based PCR assays. J Med Microbiol. 2004;53(11):1089–1096. doi: 10.1099/jmm.0.45611-0 [DOI] [PubMed] [Google Scholar]
- 23.Heras J, Domínguez C, Mata E, et al. GelJ–a tool for analyzing DNA fingerprint gel images. BMC Bioinform. 2015;16:270. doi: 10.1186/s12859-015-0703-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fadhil L, Al-Marzoqi AH, Al Taee ZM, Shalan AA. Molecular and phenotypic study of virulence genes in a pathogenic strain of Pseudomonas aeruginosa isolated from various clinical origins by PCR: profiles of genes and toxins. Res J Pharm Biolog Chem Sci. 2016;7(1):590–598. [Google Scholar]
- 25.Barbier F, Andremont A, Wolff M, Bouadma L. Hospital-acquired pneumonia and ventilator-associated pneumonia: recent advances in epidemiology and management. Cur Opin Pulm Med. 2013;19(3):216–228. doi: 10.1097/MCP.0b013e32835f27be [DOI] [PubMed] [Google Scholar]
- 26.Tazumi A, Maeda Y, Buckley T, et al. Molecular epidemiology of clinical isolates of Pseudomonas aeruginosa isolated from horses in Ireland. Irish Vet J. 2009;62(7):456. doi: 10.1186/2046-0481-62-7-456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bassetti M, Vena A, Croxatto A, Righi E, Guery B. How to manage Pseudomonas aeruginosa infections. Drugs in Context. 2018;7:1–18. doi: 10.7573/17404398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horcajada JP, Montero M, Oliver A, et al. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin Microbiol Rev. 2019;32(4):e00031–e000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ra’oof WA. Distribution of algD, lasB, pilB and nan1 genes among MDR clinical isolates of Pseudomonas aeruginosa in respect to site of infection. Tikrit Med J. 2011;17(2):148–160. [Google Scholar]
- 30.Bardoel BW, van Kessel KPM, van Strijp JAG, Milder FJ. Inhibition of Pseudomonas aeruginosa virulence: characterization of the AprA–AprI interface and species selectivity. J Mol Biol. 2012;415(3):573–583. doi: 10.1016/j.jmb.2011.11.039 [DOI] [PubMed] [Google Scholar]
- 31.Roy-Burman A, Savel RH, Racine S, et al. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J Infect Dis. 2001;183(12):1767–1774. doi: 10.1086/320737 [DOI] [PubMed] [Google Scholar]
- 32.Fleiszig S, Wiener-Kronish JP, Miyazaki H, et al. Pseudomonas aeruginosa-mediated cytotoxicity and invasion correlate with distinct genotypes at the loci encoding exoenzyme S. Infect Immun. 1997;65(2):579–586. doi: 10.1128/IAI.65.2.579-586.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodulfo H, Arcia A, Hernández A, et al. Virulence Factors and Integrons are Associated with MDR and XDR Phenotypes in Nosocomial Strains of Pseudomonas Aeruginosa in a Venezuelan University Hospital. Vol. 61 Revista do Instituto de Medicina Tropical de São Paulo; 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hassuna NA. Molecular Detection of the virulent ExoU genotype of Pseudomonas aeruginosa isolated from infected surgical incisions. Surg Infect (Larchmt). 2016;17(5):610–614. doi: 10.1089/sur.2016.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karatuna O, Yagci A. Analysis of quorum sensing-dependent virulence factor production and its relationship with antimicrobial susceptibility in Pseudomonas aeruginosa respiratory isolates. Clin Microbiol Infect. 2010;16(12):1770–1775. doi: 10.1111/j.1469-0691.2010.03177.x [DOI] [PubMed] [Google Scholar]
- 36.Asadpour L. Antimicrobial resistance, biofilm-forming ability and virulence potential of Pseudomonas aeruginosa isolated from burn patients in northern Iran. J Glob Antimicrob Resist. 2018;13:214–220. doi: 10.1016/j.jgar.2018.01.018 [DOI] [PubMed] [Google Scholar]
- 37.Al Dawodeyah HY, Obeidat N, Abu-Qatouseh LF, Shehabi AA. Antimicrobial resistance and putative virulence genes of Pseudomonas aeruginosa isolates from patients with respiratory tract infection. Germs. 2018;8(1):31–40. doi: 10.18683/germs.2018.1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nikbin V, Aslani MM, Sharafi Z, et al. Molecular identification and detection of virulence genes among Pseudomonas aeruginosa isolated from different infectious origins. Iran J Microbiol. 2012;4(3):118. [PMC free article] [PubMed] [Google Scholar]
- 39.Khattab M, Nour M, ElSheshtawy NJJMBT. Genetic identification of Pseudomonas aeruginosa virulence genes among different isolates. Microb Biochem Technol. 2015;7(5):274–277. [Google Scholar]
- 40.Van Belkum A, Tassios P, Dijkshoorn L, et al. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin Microbiol Infect. 2007;13:1–46. doi: 10.1111/j.1469-0691.2007.01786.x [DOI] [PubMed] [Google Scholar]
- 41.Zarei O, Shokoohizadeh L, Hossainpour H, Alikhani MY. Molecular analysis of Pseudomonas aeruginosa isolated from clinical, environmental and cockroach sources by ERIC-PCR. BMC Res Notes. 2018;11(1):668. doi: 10.1186/s13104-018-3765-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson LA, Sharp PM. Enterobacterial repetitive intergenic consensus (ERIC) sequences in Escherichia coli: evolution and implications for ERIC-PCR. Mol Biol Evol. 2006;23(6):1156–1168. doi: 10.1093/molbev/msj125 [DOI] [PubMed] [Google Scholar]
- 43.Ruimy R, Genauzeau E, Barnabe C, et al. Genetic diversity of Pseudomonas aeruginosa strains isolated from ventilated patients with nosocomial pneumonia, cancer patients with bacteremia, and environmental water. Infect Immun. 2001;69(1):584–588. doi: 10.1128/IAI.69.1.584-588.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bertrand X, Thouverez M, Talon D, et al. Endemicity, molecular diversity and colonisation routes of Pseudomonas aeruginosa in intensive care units. Int Care Med. 2001;27(8):1263–1268. doi: 10.1007/s001340100979 [DOI] [PubMed] [Google Scholar]
- 45.Di Martino P, Gagnière H, Berry H, Bret L. Antibiotic resistance and virulence properties of Pseudomonas aeruginosa strains from mechanically ventilated patients with pneumonia in intensive care units: comparison with imipenem-resistant extra-respiratory tract isolates from uninfected patients. Microb Infect. 2002;4(6):613–620. doi: 10.1016/S1286-4579(02)01579-4 [DOI] [PubMed] [Google Scholar]