Abstract
Objective
Our purpose was to identify up-regulated long noncoding RNA ENST00000512916 in ameloblastoma (AB) and explore its role in the progression of AB.
Methods
We analyzed lncRNA microarray expression profile between six paired AB and normal oral mucosa (NOM) tissues. An up-regulated lncRNA, ENST00000512916 was identified and validated by real-time qPCR. Cell proliferation, migration and cell cycle were detected by CCK-8 assay, transwell chamber and flow cytometry, respectively. Western blotting analysis was used to measure the expression of cell-cycle-related proteins including CyclinD1 and Cyclin-dependent kinase (CDK) 2/4/6. In addition, Xenograft tumor model was constructed to investigate tumor growth.
Results
Real-time qPCR confirmed that lncRNA ENST00000512916 was up-regulated in AB tissues. ENST00000512916 knockdown significantly inhibited cell proliferation, migration and the expression of CDK2/4/6 in AM-1 cells. Moreover, ENST00000512916 knockdown suppressed tumor growth in vivo. We also found that ENST00000512916 overexpression significantly promoted the expression of HOXC13 in AM-1 cells. Overexpression of ENST00000512916 promoted cell cycle progression in AM-1 cells, which was reversed by HOXC13 knockdown.
Conclusion
Our findings reveal that lncRNA ENST00000512916 promotes cell proliferation, migration and cell cycle progression of AB.
Keywords: long noncoding RNA, ENST00000512916, ameloblastoma, HOXC13, proliferation
Introduction
Ameloblastoma (AB) is a common benign tumor of an odontogenic epithelial origin, with slow growth and local invasiveness.1,2 In the latest WHO Classification of Head and Neck Tumors, AB was classified into AB, unicystic, and extraosseous/peripheral types. Among them, AB is the most common type, which accounts for 91%.3 Although AB can be surgically removed, it can recur following incomplete excision, which induces AB to malignant or distant metastasis to the distal lymph nodes, lungs and kidneys, and even to the bones.4,5 Therefore, the treatment strategy of AB remains controversial. It is an urgent need to clarify the underlying molecular mechanisms associated with its biological behavior.6 Despite a large number of histological and biological studies, the underlying mechanisms of AB remain poorly understood.7,8
Abnormal non-coding RNA expression occurs extensively in cancers.9 LncRNA is a class of RNA molecule with a transcription length greater than 200 nucleotides.10 LncRNA does not encode proteins but is capable of regulating gene expression at epigenetic and transcriptional levels.11 Studies have shown that lncRNA plays an important role in the pathogenesis of tumors, which is widely involved in biological processes such as tumor cell proliferation, differentiation and apoptosis.12–14 LncRNA is involved in the regulation of tumor cell growth and proliferation via regulating the process of tumor cell cycle and cell apoptosis. As an example, lncRNA FOXC2-AS1 promotes tumor cell proliferation and tumor growth via the miR-1253/EZH2 axis.15
However, there are currently few studies involving the mechanism of lncRNA in AB.16,17 In our study, the expression profiles of lncRNAs in AB and NOM were obtained from six AB patients through microarray analysis. By screening and analyzing lncRNAs related to the development of AB, lncRNA ENST00000512916 was identified for in-depth research. Our findings reveal that lncRNA ENST00000512916 could possess potential value as a novel therapeutic target against AB.
Materials and Methods
Tissue Samples
From January 2014 to December 2015, 26 pairs of AB tumor tissues and their corresponding NOM tissues and 16 dental follicles were collected from the Department of Maxillofacial Surgery, School of Stomatology, China Medical University. The patients’ clinical information is shown in Table 1. The follow-up period was 6–30 months. All specimens were frozen in liquid nitrogen and then stored in a −80°C refrigerator. Furthermore, we collected 16 dental follicles from healthy patients submitted to extractions of impacted third molars. Histopathological diagnosis of all cases were independently achieved by two oral pathologists according to current WHO guidelines. All patients provided written informed consent. The study was approved by the Ethics Committee of School of Stomatology, China Medical University (2016–13).
Table 1.
The Clinical Information of 26 AB Patients
| Clinicopathological Characteristics | Number of Case (n = 26) | Percent (%) |
|---|---|---|
| Age (years) | ||
| <20 | 3 | 11.54% |
| 20–40 | 11 | 42.31% |
| >40 | 12 | 46.15% |
| Gender | ||
| Male | 16 | 61.54% |
| Female | 10 | 38.46% |
| Location | ||
| Maxilla | 14 | 53.85% |
| Mandible | 12 | 46.15% |
| Pathological type | ||
| Ameloblastoma | 26 | 100% |
| Unicystic | – | – |
| Extraosseous/peripheral | – | – |
| Recurrence | ||
| Yes | 11 | 42.31% |
| No | 15 | 57.69% |
Microarray Analysis
From 26 pairs of tissues, six AB tissues and their corresponding NOM tissues were used for lncRNA microarray analysis. Total RNA was extracted from the above tissues. RNA samples were assessed for RNA quantity and quality using the NanoDrop ND-1000 (Thermo Scientific). RNA integrity was evaluated by standard denaturing gel electrophoresis. The RNA was amplified and transcribed into cRNA, which was subsequently labeled with the Arrayztar RNA Flash Labeling Kit. The labeled cRNA was then hybridized to Human lncRNA Array v3.0 (8 × 60 K, Array Star, Rockville, MD, USA) via Agilent Sure Hyb. After washing, it was scanned using an Agilent DNA Microarray Scanner. Raw data were extracted and analyzed using Agilent Feature Extraction software v11.0.1.1 (Agilent Technologies), normalized by Agilent Genespring GX v12.1 software (Agilent Technologies). Differentially expressed lncRNAs with fold change>2.0 and p-value<0.05 were identified and visualized into volcano and scatter plots.
Cell Culture
Primary AM-1 cells were gifted from Iwate University of Japan. Frozen AM-1 cells were taken out of liquid nitrogen, and were cultured in K-FSM medium (GBICO, Shanghai, China) containing 1% 100× streptomycin double-antibody solution and 10% fetal bovine serum. Subculture was carried out in a 5% CO2 and 37°C cell culture incubator.
RNA Isolation and Real-Time qPCR
Total RNA was extracted from tissues or cells using TRIZOL (Invitrogen, USA). Then, extracted RNA was reverse transcribed into cDNA using M-MLV reverse transcriptase. The cDNA was mixed with SybGreen qPCR master mix reagent, primers and 50×ROX Reference Dye. Real-time qPCR results were analyzed using Rotorgene Real-Time Analysis Software. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as an internal control. The relative expression levels were calculated using 2−ΔΔCT method.18 The primers of target genes are shown in Table 2.
Table 2.
Primer Information for Real-Time qPCR
| Target Gene | Primer Information |
|---|---|
| ENST00000512916 | 5′-GACACGCGTTCTTACTTTGGCTCCTAG-3′ (F) 5′-CTGCTCGAGTAGGCCTGTCCTCAAGAC-3′ (R) |
| HOXC13 | 5′-GGAGTTCGCCTTCTACC-3′ (F) 5′-GACTGTCCCAGCCATTG-3′ (R) |
| CyclinD1 | 5′-CTTCGTTGCCCTCTGTG-3′ (F) 5′-TTGGGGTCCATGTTCTG-3′ (R) |
| CDK2 | 5′-GAAACAAGTTGACGGGA-3′ (F) 5′-ATGAGGGGAAGAGGAAT-3′ (R) |
| CDK4 | 5′-TTTGACCTGATTGGGCT-3′ (F) 5′-AGAGATTCGCTTGTGTG-3′ (R) |
| CDK6 | 5′-TCAGGTTGTTTGATGTG-3′ (F) 5′-AGCGAGTTTTATTTGTC-3′ (R) |
| GAPDH | 5′-ATGACATCAAGAAGGTGGTGAAGCAGG-3′ (F) 5′-GCGTCAAAGGTGGAGGAGTGGGT-3′ (R) |
Plasmid Transfection
The full length of ENST00000512916 was cloned into the pcDNA3.1 vector (TAKARA, Beijing, China) to overexpression ENST00000512916. Furthermore, the siRNAs of ENST00000512916 and HOXC13 were synthesized by Guangzhou Ruibo Biotechnology Co., Ltd. (Guangdong, China). The sequences of siRNAs were as follows: siRNA-ENST00000512916, 5’-GUAUAGGUAGUGAGUGGUA-3’; siRNA-HOXC13, 5’-GAAGAGGUAUUGAAUGCUA-3’. The control siRNAs (si-controls) that did not target ENST00000512916 or HOXC13 were used as the negative control. The si-ENST00000512916, si-HOXC13 or si-controls was inserted into the pcDNA3.1 vector (TAKARA, Beijing, China) to silence ENST00000512916 or HOXC13, respectively. AM-1 cells were transfected with the plasmids via Lipofectamine 3000 transfection reagent (Thermo, Shanghai, China).
Western Blot
AM-1 cells were lysed with 6×RIPA lysis plus PMSF on ice for 30 mins. The extracted proteins were separated by SDS-PAGE and transferred onto PVDF membranes (Millipore, NY, USA), and then blocked in 5% skimmed milk for 1 hr at room temperature. After that, the membranes were incubated with primary antibodies including CyclinD1 (Wuhan Sanying Biotechnology Co., Ltd., China; 1:200), CDK2 (Wuhan Sanying Biotechnology Co., Ltd., China; 1:200), CDK4 (Wuhan Sanying Biotechnology Co., Ltd., China; 1:200) and CDK6 (Wuhan Sanying Biotechnology Co., Ltd., China; 1:200) at 4°C overnight, followed by second antibodies including goat anti-rabbit (1:5000) and goat anti-rat (1:5000) for 1 hr at room temperature. Afterwards, the proteins were visualized with Super ECL Plus (Beijing Leigen Biotechnology Co., Ltd., Beijing, China). Tubulin was used as an internal control.
Cell Counting Kit-8 (CCK-8) Assay
AM-1 cells were seeded in 96-well plates. CCK-8 reagent (Beyotime, Shanghai, China) was added into each well at 24 hrs, 48 hrs and 72 hrs after transfection.19 Finally, absorbance was measured at 450 nm under a microplate reader.
Colony Formation Assay
Colony formation assay was used to measure cell proliferation. The cultured AM-1 cells were seeded in 12-well plates and cultured in an incubator for 2 weeks. The cells were fixed with 4% paraformaldehyde for 15 mins and then stained with 1 mL crystal violet for 15 mins. Finally, the number of cloned cells was counted. Colony formation rate = (the number of colony cells/the total number of cells) × 100%.
Flow Cytometry Assay
For cell apoptosis, transfected AM-1 cells were seeded in 12-well plates, separately. After adding 5 μL of Annexin V/FITC, transfected cells were incubated at room temperature for 5 mins in the dark, followed by 10μL of 20 μg/μL propidium iodide (PI) solution. Finally, the cell apoptosis was analyzed by flow cytometry. For cell cycle, cultured cells were only added 0.4 mL PI solution and incubated at 37°C for 30 mins in the dark. Finally, the cell cycle was analyzed by flow cytometry.
Transwell Chamber Assay
Cell migration ability was determined by transwell chamber assay. 4×104 AM-1 cells were seeded in the upper chamber (Millicell, Zhejiang, China) supplemented with 100 μL serum-free medium, while medium plus 10% FBS was added to the lower chamber. After incubation at 37°C for 24 hrs, the cells were fixed with 4% paraformaldehyde for 1 hr at room temperature, followed by dyeing with 0.1% crystal violet solution for 30 mins (Sigma, NY, USA). Finally, the migrated cells were counted in five different areas under a microscope.
Xenograft Tumor
Sixteen BALB/c female nude mice (4-week-old) were randomly divided into two groups. 2×106 AM-1 cells transfected with siRNA control or siRNA-ENST00000512916 were subcutaneously inoculated into the right axilla of the nude mice. The length (L) and width (W) of tumor were measured every four day. The tumor volume (V) was calculated as follows: V=1/2×L×W2. After 40 days, nude mice were sacrificed. Finally, the tumors were removed and weighed. Our study was approved by the Ethics Committee of School of Stomatology, China Medical University (2016–13). This study was performed following the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The animal use protocol was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of the China Medical University.
HOXC13 RNAscope Assay
The RNAscope assay was performed on formalin-fixed paraffin-embedded (FFPE) AB tissues using RNAscope 2.5 HD Reagent Kit (3222300; Advanced Cell Diagnostics (ACD), Hayward, CA).20 Firstly, AB tissue section was deparaffinized with xylene plus 100% ethanol, followed by incubation with pretreat solution series. Afterwards, the tissue section was hybridized with a probe Hs-HOXC13, positive control probe Hs-PPIB, negative control probe DapB in the HybEZ oven (ACD) at 40°C for 2 hrs. To ensure RNA quality, housekeeping gene PPIB served as a positive control. Then, section was subjected to signal amplification using HD 2.5 detection Kit. Furthermore, a mixture of Fast-RED solutions A and B (1:60) was used to detect hybridization signal. Followed by counterstaining with Gill’s hematoxylin, sections were dried in a 60°C dry oven for 15 mins and mounted with Ecomount. All sections were scanned using the whole-slide morphometric analysis scanning platform Aperio Scanscope CS (Leica Biosystems, Nussloch, Germany). The section was scanned at the maximum available magnification (40×), which was stored as digital high-resolution images. According to the PPIB evaluation, digital section was inspected with Aperio ImageScope v.11 (Leica Biosystems, Nussloch, Germany; 20×). Moreover, ten fields with an equal area were selected for the analysis (40×).
Statistical Analysis
Statistical analyses were performed using Graphpad Prism 7.0 (Graph Pad Software, Inc., San Diego, CA, USA). Each experiment was independently repeated at least three times. The experimental data for each group are expressed as mean ± standard deviation (SD). Two group comparisons were determined using student’s t test, while multiple group comparisons were performed using one-way ANOVA analysis. P<0.05 was considered statistically significant.
Results
LncRNA ENST00000512916 Is Up-Regulated in AB Tissues
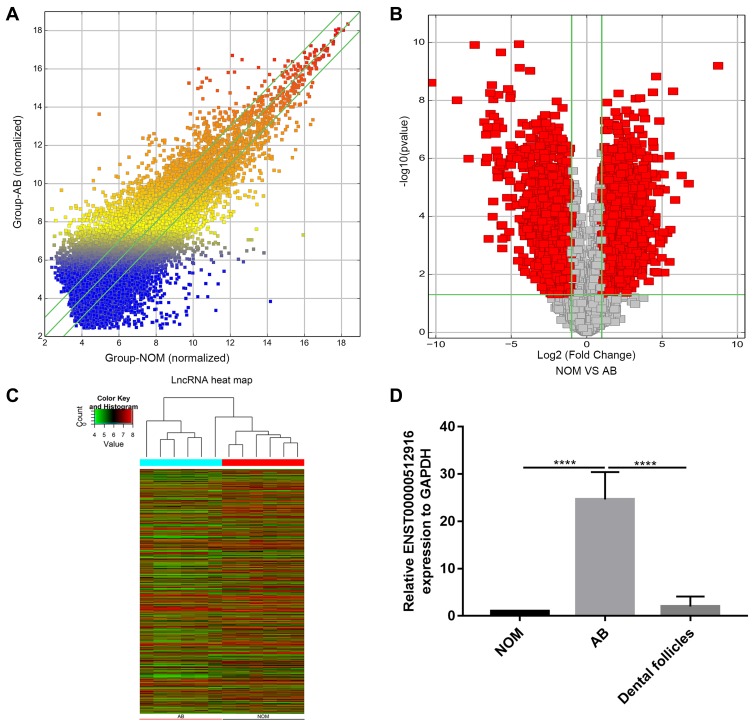
To identify differentially expressed lncRNAs in AB tissues, microarray analysis was used to perform lncRNA expression profile between AB tissues and NOM tissues (six paired samples). The scatter plot demonstrated the variation of lncRNA expression between AB tissues and NOM tissues (Figure 1A). As depicted in volcano plot, all differentially expressed lncRNAs between the two groups were identified with fold change>2 and p-value<0.05 (Figure 1B). Differentially expressed lncRNAs were further analyzed by hierarchical clustering analysis. Figure 1C shows the differences in the expression patterns of these differentially expressed lncRNAs between AB tissues and NOM tissues. Among all differentially expressed lncRNAs, ENST00000512916 (also known HOXC13-AS; 44.961201-fold change; chr12), a novel up-regulated lncRNA in AB tissues, was selected for further analysis. To validate the microarray results of ENST00000512916, real-time qPCR was performed. The results suggested that the expression levels of ENST00000512916 in AB tissues were all higher than that in NOM tissues, which were consistent with the microarray analysis results (Figure 1D). Furthermore, we also found that ENST00000512916 had the higher expression levels in AB tissues than dental follicles (Figure 1D).
Figure 1.
LncRNA ENST00000512916 is up-regulated in AB tissues. (A) The scatter plot shows the changes in lncRNA expression. LncRNAs above the top green line and below the bottom green line suggest more than a 1.5-fold change between AB group and NOM group. (B) The volcano plot demonstrates the expression patterns of all lncRNAs between AB and NOM tissues. X-axis represents log2 (fold change) and y-axis stands for –log10 (p-value). (C) The hierarchical clustering analysis suggests the differences in lncRNA expression profiling between AB and NOM tissues. (D) The expression level of ENST00000512916 was validated in 26 AB tissues using real-time qPCR. ****p-value<0.0001.
Abbreviations: AB, ameloblastoma; NOM, normal oral mucosa.
Up-Regulated lncRNA ENST00000512916 Promotes Cell Proliferation and Inhibits Apoptosis for AM-1 Cells
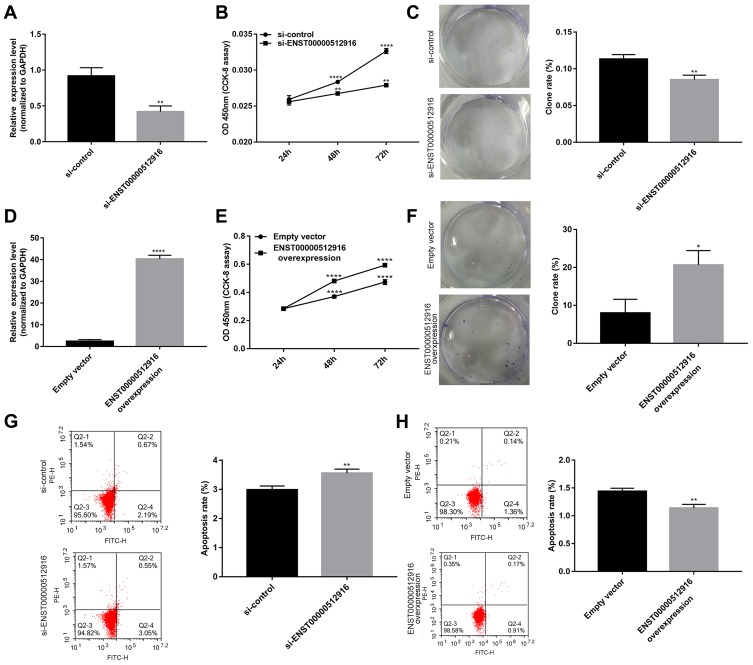
The specific siRNA was used to silence lncRNA ENST00000512916 in AM-1 cells. After 48 hrs of transfection, lncRNA ENST00000512916 expression was significantly lower in AM-1 cells compared with control group, suggesting that ENST00000512916 was successfully inhibited (Figure 2A). The cell proliferation ability was evaluated by CCK-8 and colony formation assays. After transfection of siRNA-ENST00000512916, cell viability was significantly inhibited compared to control group according to CCK-8 assay (Figure 2B). Furthermore, colony formation assay results showed that cell proliferation was significantly suppressed in siRNA-ENST00000512916 group compared to control group (Figure 2C). As shown in Figure 2D, ENST00000512916 was successfully overexpressed. We found that ENST00000512916 overexpression significantly promoted AM-1 cell viability compared to empty vector group (Figure 2E). Moreover, colony formation assay results suggested that ENST00000512916 overexpression significantly induced AM-1 cell proliferation (Figure 2F). Flow cytometry assay was used to assess the cell apoptosis. The results showed that AM-1 cell apoptosis rate was significantly higher in si-ENST00000512916 group compared to control group (Figure 2G). Furthermore, after overexpression of ENST00000512916, AM-1 cell apoptosis was significantly decreased (Figure 2H). Above results reveal that up-regulated ENST00000512916 promotes cell proliferation and inhibits apoptosis for AM-1 cells.
Figure 2.
Up-regulated lncRNA ENST00000512916 promotes cell proliferation and inhibits apoptosis for AM-1 cells. (A) Real-time qPCR results showed that ENST00000512916 was successfully silenced after transfection of siRNA-ENST00000512916 in AM-1 cells. (B) CCK8 assay was used to assess the AM-1 cell viability at 24 hrs, 48 hrs and 72 hrs after transfection with si-ENST00000512916. (C) Colony formation assay was used to detect the cell proliferation ability of AM-1 cells after transfection with si-ENST00000512916. (D) Real-time qPCR results showed that ENST00000512916 was successfully overexpressed in AM-1 cells. (E) The AM-1 cell viability was evaluated at 24 hrs, 48 hrs and 72 hrs after overexpression of ENST00000512916 using CCK-8 assay. (F) The AM-1 cell proliferation was detected after overexpression of ENST00000512916 using colony formation assay. (G) Flow cytometry assay was used to detect the AM-1 cell apoptosis after silencing ENST00000512916. (H) The AM-1 cell apoptosis was assessed after overexpression of ENST00000512916 using flow cytometry assay. *p-value<0.05; **p-value<0.01; ****p-value<0.0001.
Up-Regulated lncRNA ENST00000512916 Promotes AM-1 Cell Cycle Progression and Cell Cycle Related Protein Expression
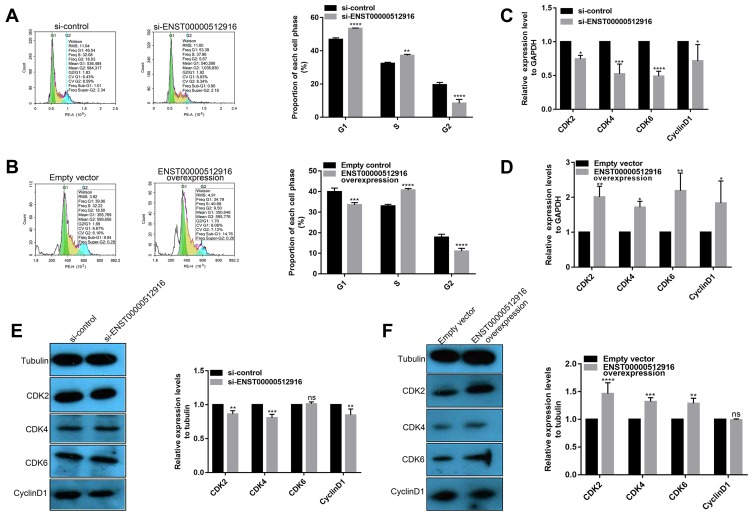
We further explored whether ENST00000512916 affected AM-1 cell cycle. Flow cytometry results showed that ENST00000512916 knockdown significantly increased G1/G2 phase (Figure 3A), while ENST00000512916 overexpression increased S phase (Figure 3B). Because the G1/S phase checkpoint plays an important role in the cell cycle, therefore, we detected cell-cycle-related proteins including CyclinD1, CDK2, CDK4 and CDK6. Real-time qPCR results showed that ENST00000512916 knockdown significantly suppressed the mRNA expression levels of CyclinD1, CDK2, CDK4 and CDK6 (Figure 3C). Moreover, ENST00000512916 overexpression significantly promoted the expression of CyclinD1, CDK2, CDK4 and CDK6 at the mRNA levels (Figure 3D). After transfection of si-ENST00000512916 in AB cells, the expression levels of CDK2, CDK4 and CyclinD1 significantly decreased, however, the expression of CDK6 did not significantly change (Figure 3E). Moreover, after overexpression of ENST00000512916, the expression levels of CDK2, CDK4 and CDK6 were significantly elevated (Figure 3F). Therefore, these results suggest that up-regulated lncRNA ENST00000512916 promotes AM-1 cell cycle progression and cell-cycle-related protein expression.
Figure 3.
Up-regulated lncRNA ENST00000512916 promotes AM-1 cell cycle progression and cell-cycle-related protein expression. Flow cytometry assay results showing the significant increased G1/G2 phase in AM-1 cells transfected with si-ENST00000512916 (A), and the significant increased S phase in AM-1cells transfected with ENST00000512916 overexpression (B). Real-time qPCR results showed the mRNA expression levels of cell-cycle-related proteins including CyclinD1, CDK2, CDK4 and CDK6 were significantly inhibited in AM-1 cells transfected with si-ENST00000512916 (C) or were significantly promoted in AM-1 cells transfected with ENST00000512916 overexpression (D). (E) Western blotting analysis showed that the expression levels of CDK2, CDK4 and CyclinD1 were significantly inhibited in AM-1 cells transfected with si-ENST00000512916. (F) Western blotting analysis showed that the expression levels of CDK2, CDK4 and CDK6 were significantly elevated in AM-1 cells transfected with ENST00000512916 overexpression. *p-value<0.05; **p-value<0.01; ***p-value<0.001; ****p-value<0.0001 and ns indicates no statistical significance.
Up-Regulated lncRNA ENST00000512916 Promotes AM-1 Cell Migration
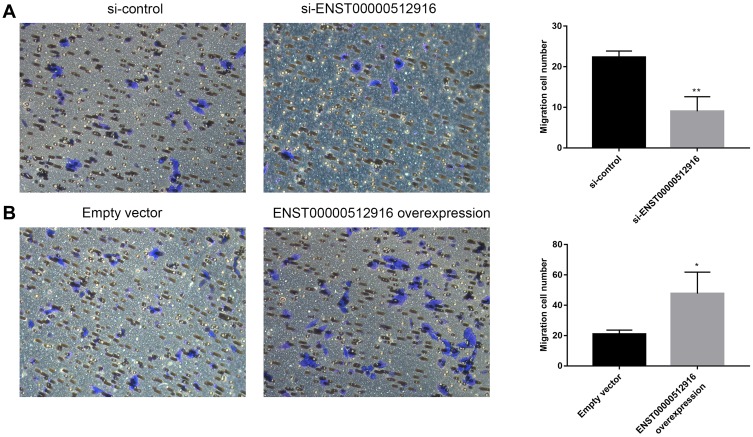
We investigated whether lncRNA ENST00000512916 could regulate AM-1 cell migration using transwell chamber assay. We found that AM-1 cell migration was significantly inhibited after transfection with si-ENST00000512916 compared with control group (Figure 4A). Furthermore, the cell migration ability of AM-1 cells was significantly promoted after ENST00000512916 overexpression (Figure 4B). Above results suggest that up-regulated lncRNA ENST00000512916 promotes AM-1 cell migration.
Figure 4.
Up-regulated lncRNA ENST00000512916 promotes AM-1 cell migration. Cell migration ability was analyzed using transwell chamber assay after transfection with si-ENST00000512916 (A) or ENST00000512916 overexpression (B). *p-value<0.05; **p-value<0.01.
Up-Regulated lncRNA ENST00000512916 Promotes AB Tumor Growth
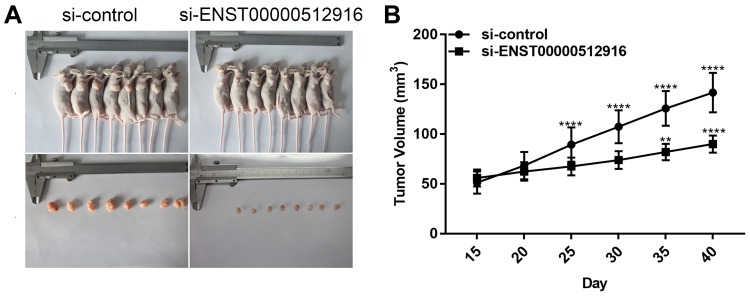
The nude mouse xenograft models were constructed to investigate the effects of ENST00000512916 on tumor growth in vivo. AM-1 cells transfected with si-ENST00000512916 or siRNA control were injected into the right side of axillary of nude mice. 40 days after injection, we found that the tumor volume was obviously smaller compared with control group (Figure 5A). Tumor growth curve depicted that after injection AM-1 cells transfected with si-ENST00000512916, the tumor growth rate was significantly slowed down (Figure 5B). Therefore, up-regulated lncRNA ENST00000512916 promotes AB tumor growth, which could become a potential therapeutic target.
Figure 5.
Up-regulated lncRNA ENST00000512916 promotes AB tumor growth. (A) Representative images of the tumors removed from nude mice after injection with AM-1 cells transfected with si-ENST00000512916 or siRNA control. (B) Tumor growth curves are shown, and the results are presented as the mean tumor weight ± SD. **p-value<0.01; ****p-value<0.0001.
HOXC13 Is Up-Regulated in AB Tissues
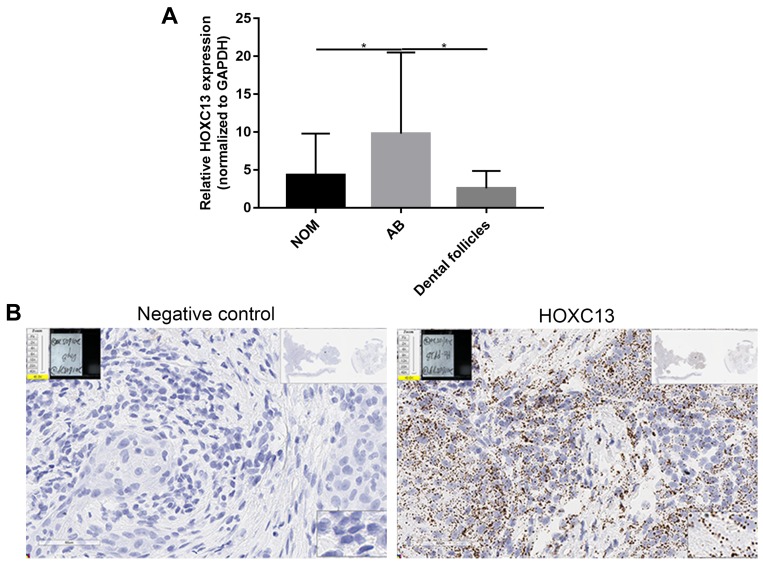
Our previous study examined the expression of HOXC13 in AB tissues.21 In this study, we found that the mRNA expression levels of HOXC13 were significantly higher in AB tissues compared to NOM tissues or dental follicles (Figure 6A). Furthermore, the expression of HOXC13 in AB tissues was confirmed using RNAscope assay (Figure 6B).
Figure 6.
HOXC13 is up-regulated in AB tissues. (A) Real-time qPCR results showed the relative expression levels of HOXC13 in 26 pairs of AB tissues compared to NOM tissues or dental follicles. (B) RNAscope assay was used to detect the expression of HOXC13 in AB tissues. *p-value<0.05.
HOXC13 Is Mediated by lncRNA ENST00000512916 in AM-1 Cells
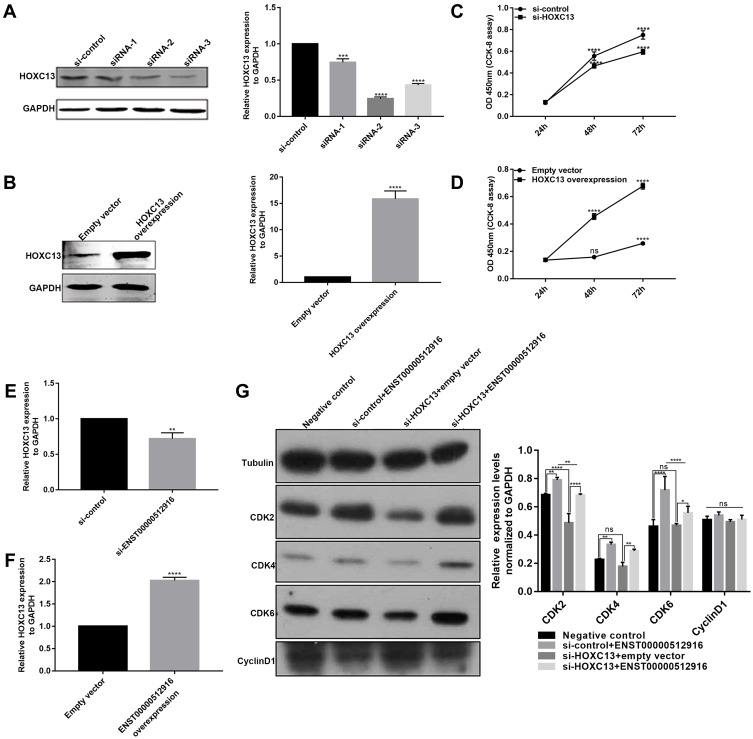
To further investigate whether HOXC13 could regulate cell proliferation, we detected the cell proliferation after AM-1 cells transfected by si-HOXC13 or HOXC13 overexpression. The effect of AM-1 cells transfected by si-HOXC13 was evaluated by Western blotting analysis. HOXC13 had the lowest protein expression levels after AM-1 cells transfected with siRNA-2; therefore, siRNA-2 was chosen for further analysis (Figure 7A). We constructed a pcDNA3.1-HOXC13 recombinant plasmid to overexpress HOXC13. As shown in Figure 7B, HOXC13 was successfully overexpressed (Figure 7B). We found that si-HOXC13 significantly inhibited AM-1 cell viability compared to control group (Figure 7C). However, overexpression of HOXC13 significantly promoted the cell viability (Figure 7D). Intriguingly, we found that si-ENST00000512916 significantly inhibited the expression of HOXC13 using real-time qPCR (Figure 7E), while ENST00000512916 overexpression significantly promoted the expression of HOXC13 (Figure 7F). Western blotting analysis results showed that si-HOXC13 significantly inhibited CDK2 expression compared to control group; however, ENST00000512916 overexpression ameliorated the inhibitory effect (Figure 7G). Above results indicated that HOXC13 could be mediated by lncRNA ENST00000512916 in AM-1 cells.
Figure 7.
HOXC13 is mediated by lncRNA ENST00000512916 in AM-1 cells. Western blotting analysis results showed that the expression levels of HOXC13 were significantly inhibited in AM-1 cells transfected with si-HOXC13 (A) or were significantly promoted after HOXC13 overexpression (B). CCK8 assay results showed that the cell viability was significantly inhibited in AM-1 cells transfected by si-HOXC13 (C) or was significantly promoted in AM-1 cells transfected by HOXC13 overexpression (D). Real-time qPCR results showed the HOXC13 expression was significantly inhibited in AM-1 cells transfected with si-ENST00000512916 (E) or was significantly elevated in AM-1 cells transfected with ENST00000512916 overexpression (F). (G) Western blotting analysis showed ENST00000512916 overexpression significantly promoted the expression levels of CDK2 and CDK4 and CDK6, which were reversed by si-HOXC13. *p-value<0.05; **p-value<0.01; ***p-value<0.001; ****p-value<0.0001 and ns indicates no statistical significance.
Discussion
AB is a common type in odontogenic tumors, which has important clinical significance.22 Therefore, it is necessary to better understand the molecular mechanisms of AB.23,24 The development of tumor is often associated with the activation of proto-oncogenes and the inactivation of tumor suppressor genes. LncRNA usually regulates the development of tumor via affecting important oncogenes and tumor suppressor genes, which participates in tumorigenesis and malignant transformation.25 In the present study, all differentially expressed lncRNAs between AB and NOM from 6 AB patients were identified via microarray analysis, among them, a novel lncRNA ENST00000512916 was obviously up-regulated.26 Furthermore, lncRNA ENST00000512916 was validated to be up-regulated in 26 AB tissues.
After AM-1 cells transfected by si-ENST00000512916, AM-1 cell proliferation ability was significantly suppressed. Cell proliferation and apoptosis are basic events in developmental and tissue homeostasis.27 The rate of cell proliferation may lead to local invasiveness of AB.28 Therefore, lncRNA ENST00000512916 could regulate AB cell proliferation and apoptosis. Identification of proliferative activity in tumors can be used to predict their biological behavior. Previous studies have found that recurrent AB has high proliferative activity.29 Our study found that inhibiting ENST00000512916 could suppress cell proliferation, indicating that ENST00000512916 possesses potential value as a therapeutic target. Then we further explored how ENST00000512916 regulated cell proliferation. We found that the expression of cyclin-dependent kinase including CDK2/4/6 was significantly decreased after inhibiting lncRNA ENST00000512916, while the expression of CDK2/4/6 was elevated after overexpressing lncRNA ENST00000512916. CDK is a serine/threonine family protein kinase that regulates the mammalian cell cycle. Cell cycle regulation disorders are closely related to the abnormal expression of CDK.30 Activation of CDK2/4/6 leads to proliferation of malignant tumor cells.31 Furthermore, CDK2/4/6 promotes cell cycle progression from G1 into S phase.32,33 We found that silencing ENST00000512916 significantly increased G1/G2 phase, while S phase was increased after overexpressed ENST00000512916. Therefore, lncRNA ENST00000512916 regulates the expression of CDK2/4/6, thereby promoting the cell cycle progression. Furthermore, we found that lncRNA ENST00000512916 could play an important role in AB metastasis using transwell chamber assay.
Our study confirmed that lncRNA ENST00000512916 could regulate the proliferation of AB cells by regulating cell cycle progression in vitro. However, the regulation of lncRNA ENST00000512916 on AB cells is not affected by other signaling pathways and transcriptional regulators. The development and progression of tumors in vivo is a long-term complex and phased process. The same lncRNA is involved in the regulation of different signaling pathways in different tumor cells. Therefore, to further confirm the regulation of lncRNA ENST00000512916 on AB proliferation and cell cycle progression, we constructed the nude mice xenograft models. After transfected AM-1 cells were injected into the right side of the nude mouse, the experimental results showed that lncRNA ENST00000512916 promoted tumor growth in vivo. Therefore, lncRNA ENST00000512916 regulates the proliferation process of AB in vivo, which could become a potential therapeutic target.
HOXC13 gene is composed of two exons spanning 7.829 kbs of genomic DNA, which encodes a conserved protein of 330 amino acids. It contains a cluster of 61 amino acids (amino acids 258–318) forming the DNA binding homeodomain.34 Increasing evidence suggests that HOXC13 regulates cell proliferation in several tumors such as lung adenocarcinoma, esophageal squamous cell carcinoma and nasopharyngeal carcinoma.35–37 However, it remains unclear whether HOXC13 regulates the proliferation of AB cells. In the present study, HOXC13 was up-regulated in 26 AB tissues. Overexpressed HOXC13 promoted cell proliferation according to CCK8 assay. Conversely, the cell proliferation ability was inhibited after HOXC13 knockdown. Therefore, up-regulated HOXC13 promotes cell proliferation in AM-1 cells.
We further confirmed whether HOXC13 was regulated by lncRNA ENST00000512916. ENST00000512916 overexpression in AB significantly promoted HOXC13 expression. We found that HOXC13 knockdown significantly decreased the expression of cell-cycle-related protein CDK2, which was ameliorated by ENST00000512916 overexpression. Therefore, HOXC13 may be mediated by lncRNA ENST00000512916 in AM-1 cells. Our future research focuses on the specific mechanism by which lncRNA ENST00000512916 regulates HOXC13 expression in AB. Furthermore, we detected the expression of ENST00000512916 in maxillary and mandibular of AB. The lesions in the two locations have different biological behaviors such as BRAF and SMO mutations and aggressiveness.38,39 Thus, in future studies, we will further explore the differences in expression and role of ENST00000512916 in different lesions of AB.
Conclusion
In our study, we identified a novel lncRNA ENST00000512916 in AB by microarray analysis. We found that the lncRNA was up-regulated in 26 AB tissues by real-time qPCR. Further research showed that ENST00000512916 may promote cell proliferation, migration and cell cycle progression in AM-1 cells. Furthermore, ENST00000512916 facilitates tumor growth in vivo. Intriguingly, we found that HOXC13 is mediated by lncRNA ENST00000512916 in AM-1 cells. Therefore, lncRNA ENST00000512916 could be considered as a potential therapeutic target against AB.
Acknowledgments
Thanks to the primary AM-1 cells gifted by Iwate University of Japan.
Funding Statement
This work was funded by the National Natural Science Foundation of China (81072197 and 81470758).
Abbreviations
LncRNAs, long noncoding RNAs; AB, ameloblastoma; NOM, normal oral mucosa; siRNA, small interfering RNA; CDK, cyclin-dependent kinase; CCK-8, cell counting kit-8.
Availability of Data and Material
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
The study was approved by the Ethics Committee of School of Stomatology, China Medical University (2016-13). All subjected provided informed consent for this study.
Author Contributions
Ming Zhong conceived and designed the study. Yan Sun, Xing Niu conducted most of the experiments and data analysis, and wrote the manuscript. Guannan Wang, Xue Qiao and Lijie Chen participated in collecting data and helped to draft the manuscript. All authors reviewed and approved the manuscript. All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no conflicts of interest in this work.
References
- 1.Yang Z, Li K, Liang Q, et al. Elevated hydrostatic pressure promotes ameloblastoma cell invasion through upregulation of MMP-2 and MMP-9 expression via Wnt/beta-catenin signalling. J Oral Pathol Med. 2018;47:836–846. doi: 10.1111/jop.12761 [DOI] [PubMed] [Google Scholar]
- 2.Yang Z, Liang Q, Yang L, et al. Marsupialization of mandibular cystic ameloblastoma: retrospective study of 7 years. Head Neck. 2018;40:2172–2180. doi: 10.1002/hed.25212 [DOI] [PubMed] [Google Scholar]
- 3.Wright JM, Vered M. Update from the 4th Edition of the World Health Organization classification of head and neck tumours: odontogenic and maxillofacial bone tumors. Head Neck Pathol. 2017;11:68–77. doi: 10.1007/s12105-017-0794-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sweeney RT, McClary AC, Myers BR, et al. Identification of recurrent SMO and BRAF mutations in ameloblastomas. Nat Genet. 2014;46:722–725. doi: 10.1038/ng.2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heikinheimo K, Huhtala JM, Thiel A, et al. The mutational profile of unicystic ameloblastoma. J Dent Res. 2019;98:54–60. doi: 10.1177/0022034518798810 [DOI] [PubMed] [Google Scholar]
- 6.Diniz MG, Franca JA, Vilas-Boas FAS, et al. The long noncoding RNA KIAA0125 is upregulated in ameloblastomas. Pathol Res Pract. 2019;215:466–469. doi: 10.1016/j.prp.2018.12.030 [DOI] [PubMed] [Google Scholar]
- 7.Kanda S, Mitsuyasu T, Nakao Y, et al. Anti-apoptotic role of the sonic hedgehog signaling pathway in the proliferation of ameloblastoma. Int J Oncol. 2013;43:695–702. doi: 10.3892/ijo.2013.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakao Y, Mitsuyasu T, Kawano S, Nakamura N, Kanda S, Nakamura S. Fibroblast growth factors 7 and 10 are involved in ameloblastoma proliferation via the mitogen-activated protein kinase pathway. Int J Oncol. 2013;43:1377–1384. doi: 10.3892/ijo.2013.2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lorenzi L, Avila Cobos F, Decock A, et al. Long noncoding RNA expression profiling in cancer: challenges and opportunities. Genes Chromosomes Cancer. 2019;58:191–199. doi: 10.1002/gcc.22709 [DOI] [PubMed] [Google Scholar]
- 10.Li CH, Chen Y. Insight into the role of long noncoding RNA in cancer development and progression. Int Rev Cell Mol Biol. 2016;326:33–65. [DOI] [PubMed] [Google Scholar]
- 11.Yoon JH, You BH, Park CH, Kim YJ, Nam JW, Lee SK. The long noncoding RNA LUCAT1 promotes tumorigenesis by controlling ubiquitination and stability of DNA methyltransferase 1 in esophageal squamous cell carcinoma. Cancer Lett. 2018;417:47–57. doi: 10.1016/j.canlet.2017.12.016 [DOI] [PubMed] [Google Scholar]
- 12.Saeinasab M, Bahrami AR, Gonzalez J, et al. SNHG15 is a bifunctional MYC-regulated noncoding locus encoding a lncRNA that promotes cell proliferation, invasion and drug resistance in colorectal cancer by interacting with AIF. J Exp Clin Cancer Res. 2019;38:172. doi: 10.1186/s13046-019-1169-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu S, Yan G, Zhang J, Yu L. Knockdown of long noncoding RNA (lncRNA) metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) inhibits proliferation, migration, and invasion and promotes apoptosis by targeting miR-124 in retinoblastoma. Oncol Res. 2018;26:581–591. doi: 10.3727/096504017X14953948675403 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Jing H, Qu X, Liu L, Xia H. A novel long noncoding RNA (lncRNA), LL22NC03-N64E9.1, promotes the proliferation of lung cancer cells and is a potential prognostic molecular biomarker for lung cancer. Med Sci Monit. 2018;24:4317–4323. doi: 10.12659/MSM.908359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y, Gu M, Liu C, et al. Long noncoding RNA FOXC2-AS1 facilitates the proliferation and progression of prostate cancer via targeting miR-1253/EZH2. Gene. 2019;686:37–42. doi: 10.1016/j.gene.2018.10.085 [DOI] [PubMed] [Google Scholar]
- 16.Gao X, Wang G, Zhang YK, Zhong M. Expression and mechanism of regulation of PP2A/Pr65 in ameloblastoma. Surgeon. 2014;12:129–133. doi: 10.1016/j.surge.2013.07.003 [DOI] [PubMed] [Google Scholar]
- 17.Wang GN, Zhong M, Chen Y, Ji J, Gao XQ, Wang TF. Expression of WNT1 in ameloblastoma and its significance. Oncol Lett. 2018;16:1507–1512. doi: 10.3892/ol.2018.8820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 19.Zhu X, Niu X, Ge C. Inhibition of LINC00994 represses malignant behaviors of pancreatic cancer cells: interacting with miR-765-3p/RUNX2 axis. Cancer Biol Ther. 2019;20:799–811. doi: 10.1080/15384047.2018.1564566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang F, Flanagan J, Su N, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong M, Wang J, Gong YB, Li JC, Zhang B, Hou L. [Expression of HOXC13 in ameloblastoma]. Zhonghua Kou Qiang Yi Xue Za Zhi. 2007;42:43–46. [PubMed] [Google Scholar]
- 22.Fonseca FP, Monteiro Benites B, Soares CD, et al. Prognostic importance of FGF2 and FGFR1 expression for patients affected by ameloblastoma. J Oral Pathol Med. 2018;47:417–424. doi: 10.1111/jop.2018.47.issue-4 [DOI] [PubMed] [Google Scholar]
- 23.Heikinheimo K, Kurppa KJ, Laiho A, et al. Early dental epithelial transcription factors distinguish ameloblastoma from keratocystic odontogenic tumor. J Dent Res. 2015;94:101–111. doi: 10.1177/0022034514556815 [DOI] [PubMed] [Google Scholar]
- 24.Neves-Silva R, Fonseca FP, de Jesus AS, et al. Tissue microarray use for immunohistochemical study of ameloblastoma. J Oral Pathol Med. 2016;45:704–711. doi: 10.1111/jop.12428 [DOI] [PubMed] [Google Scholar]
- 25.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gomes CC, Duarte AP, Diniz MG, Gomez RS. Review article: current concepts of ameloblastoma pathogenesis. J Oral Pathol Med. 2010;39:585–591. doi: 10.1111/j.1600-0714.2010.00908.x [DOI] [PubMed] [Google Scholar]
- 27.Metgud R, Gupta K. Expression of cell cycle and apoptosis-related proteins in ameloblastoma and keratocystic odontogenic tumor. Ann Diagn Pathol. 2013;17:518–521. doi: 10.1016/j.anndiagpath.2013.06.006 [DOI] [PubMed] [Google Scholar]
- 28.Cecim RL, Carmo HA, Kataoka MS, Freitas VM. de Melo Alves Junior S, Pedreira EN, Jaeger RG, Pinheiro JJ. Expression of molecules related to AKT pathway as putative regulators of ameloblastoma local invasiveness. J Oral Pathol Med. 2014;43:143–147. [DOI] [PubMed] [Google Scholar]
- 29.Abdel-Aziz A, Amin MM. EGFR, CD10 and proliferation marker Ki67 expression in ameloblastoma: possible role in local recurrence. Diagn Pathol. 2012;7:14. doi: 10.1186/1746-1596-7-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J, Pang L, Wang W, Wang L, Zhang JZH, Zhu T. Decoding molecular mechanism of inhibitor bindings to CDK2 using molecular dynamics simulations and binding free energy calculations. J Biomol Struct Dyn. 2019;1–12. [DOI] [PubMed] [Google Scholar]
- 31.Liu FY, Wang LP, Wang Q, et al. miR-302b regulates cell cycles by targeting CDK2 via ERK signaling pathway in gastric cancer. Cancer Med. 2016;5:2302–2313. doi: 10.1002/cam4.818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu G, Li JY. CDK4, CDK6, cyclin D1, p16(INK4a) and EGFR expression in glioblastoma with a primitive neuronal component. J Neurooncol. 2018;136:445–452. doi: 10.1007/s11060-017-2674-7 [DOI] [PubMed] [Google Scholar]
- 33.van Ommen-nijhof A, Konings IR, van Zeijl CJJ, et al. Selecting the optimal position of CDK4/6 inhibitors in hormone receptor-positive advanced breast cancer – the SONIA study: study protocol for a randomized controlled trial. BMC Cancer. 2018;18:1146. doi: 10.1186/s12885-018-4978-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang S, Luo Z, Zhang Y, Yuan D, Ge W, Wang X. The inconsistent regulation of HOXC13 on different keratins and the regulation mechanism on HOXC13 in cashmere goat (Capra hircus). BMC Genomics. 2018;19:630. doi: 10.1186/s12864-018-5011-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao C, Lu W, Lou W, Wang L, Xu Q. Long noncoding RNA HOXC13-AS positively affects cell proliferation and invasion in nasopharyngeal carcinoma via modulating miR-383-3p/HMGA2 axis. J Cell Physiol. 2019;234:12809–12820. doi: 10.1002/jcp.v234.8 [DOI] [PubMed] [Google Scholar]
- 36.Luo J, Wang Z, Huang J, et al. HOXC13 promotes proliferation of esophageal squamous cell carcinoma via repressing transcription of CASP3. Cancer Sci. 2018;109:317–329. doi: 10.1111/cas.13453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao Y, Luo J, Sun Q, et al. HOXC13 promotes proliferation of lung adenocarcinoma via modulation of CCND1 and CCNE1. Am J Cancer Res. 2017;7:1820–1834. [PMC free article] [PubMed] [Google Scholar]
- 38.Brown NA, Rolland D, McHugh JB, et al. Activating FGFR2-RAS-BRAF mutations in ameloblastoma. Clin Cancer Res. 2014;20:5517–5526. doi: 10.1158/1078-0432.CCR-14-1069 [DOI] [PubMed] [Google Scholar]
- 39.Fregnani ER, Perez DE, Paes de Almeida O, et al. BRAF-V600E expression correlates with ameloblastoma aggressiveness. Histopathology. 2017;70:473–484. doi: 10.1111/his.13095 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request.