AP-1 transcription factor is recruited to the majority of chromatin remodeling sites during T cell activation. The activity of AP-1 is induced by co-stimulation and is required for chromatin opening. The sites of remodeling overlap with risk loci of immunological diseases.
Abstract
Activation of T cells is dependent on the organized and timely opening and closing of chromatin. Herein, we identify AP-1 as the transcription factor that directs most of this remodeling. Chromatin accessibility profiling showed quick opening of closed chromatin in naive T cells within 5 h of activation. These newly opened regions were strongly enriched for the AP-1 motif, and indeed, ChIP-seq demonstrated AP-1 binding at >70% of them. Broad inhibition of AP-1 activity prevented chromatin opening at AP-1 sites and reduced the expression of nearby genes. Similarly, induction of anergy in the absence of co-stimulation during activation was associated with reduced induction of AP-1 and a failure of proper chromatin remodeling. The translational relevance of these findings was highlighted by the substantial overlap of AP-1–dependent elements with risk loci for multiple immune diseases, including multiple sclerosis, inflammatory bowel disease, and allergic disease. Our findings define AP-1 as the key link between T cell activation and chromatin remodeling.
Introduction
Upon encountering an antigen, naive T helper cells are activated and differentiate over several days into various effector lineages that contribute to immune responses (O’Shea and Paul, 2010; Russ et al., 2013). These differentiated effector cells secrete different sets of cytokines and have specific functions in orchestrating immune responses against pathogens. In the contraction phase of the response, most effector cells die, but a few survive and become long-lived memory cells (Youngblood et al., 2017). We and others have demonstrated that epigenetic states induced during T cell activation, differentiation, and memory formation are associated with T cell lineage stability and plasticity, cytokine production in effector cells, and rapid recall response in the memory cells (Vahedi et al., 2012; Barski et al., 2009; Komori et al., 2015; Smith et al., 2009; Hawkins et al., 2013; Mukasa et al., 2010; Mazzoni et al., 2015; Sekimata et al., 2009; Wei et al., 2009; Ohkura et al., 2012). An outstanding question in the field is how the epigenetic changes are induced and targeted to specific loci during primary activation of T cells.
The differentiation of T cells is a multistep process starting with T cell activation. The activation is accomplished through simultaneous stimulation of the TCR and costimulatory receptors such as CD28. Downstream NFATs, AP-1 (a heterodimer of FOS and JUN proteins), and NF-κB are activated via Ca2+-calcineurin, MAPK, and PI3K/PKC pathways (Fathman and Lineberry, 2007; Crabtree and Olson, 2002; Zhu and Paul, 2010; Jain et al., 1994; Rochman et al., 2015). Concurrently with activation signals, differentiation signals provided by the cytokine milieu lead to the activation of JAK–STAT pathways, induction of lineage-specific transcription factors (TFs), and eventually lineage-specific cytokine gene expression (Zhu et al., 2010). The Il2 locus has previously been used as a model to study activation-induced transcriptional regulation. The Il2 promoter has several AP-1 and NFAT binding sites that are conserved between human and mouse (Rooney et al., 1995; Macián et al., 2001). The binding sites are adjacent, and AP-1 and NFAT form a heteromer (Jain et al., 1994; Chen et al., 1998) and synergize to induce Il2 expression (Walters et al., 2013; Nguyen et al., 2010). Mutation of these binding sites prevents Il2 expression (Walters et al., 2013). NF-κB and several other TFs also participate in Il2 regulation during T cell activation via their binding sites near the promoter (Thaker et al., 2015; Skerka et al., 1995). However, the mechanisms of transcriptional regulation during T cell activation are not common for all genes. For example, IL2 expression is dependent on new protein synthesis, but IL10, IFNG, and TNF are not (Sareneva et al., 1998).
Herein, we profiled chromatin accessibility during the early stages of T cell activation in human primary naive CD4 T cells. We were struck by the massive number of regions undergoing remodeling within 5 h of activation and the considerable enrichment of AP-1 motifs. Chromatin immunoprecipitation sequencing (ChIP-seq) demonstrated AP-1 binding at the majority of these regions, often together with its partner, NFAT1. AP-1 was also strongly present at superenhancer (SE) elements formed during activation. Whereas prior studies have focused on genetic disruption of individual AP-1 members, herein we broadly blocked the AP-1 family in human naive T cells by electroporating a dominant-negative protein (A-FOS); this resulted in loss of chromatin remodeling and T cell activation. Conversely, AP-1–associated chromatin changes were absent during induction of T cell anergy. The translational significance of these findings to clinical medicine was supported by the overlap of activation-specific enhancers and AP-1 binding sites with single-nucleotide polymorphisms (SNPs) associated with increased risk for a variety of diseases, most substantially found for multiple sclerosis.
Results
Characterizing open chromatin regions
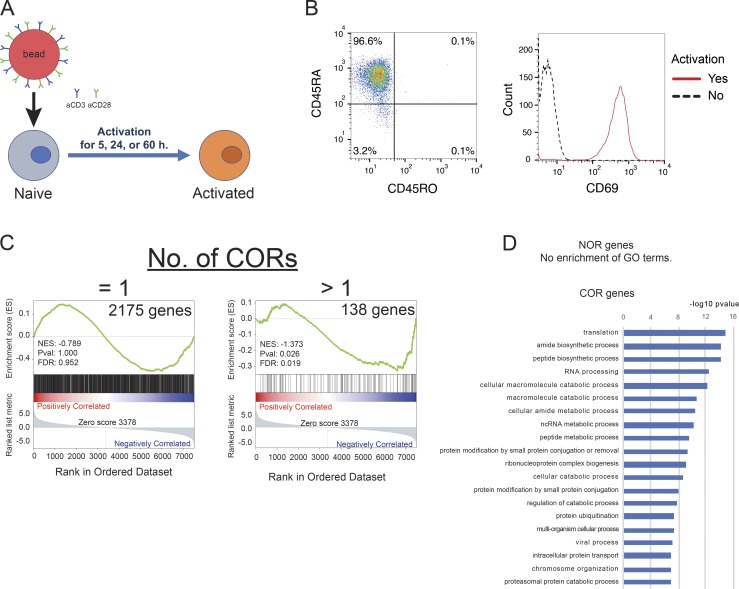
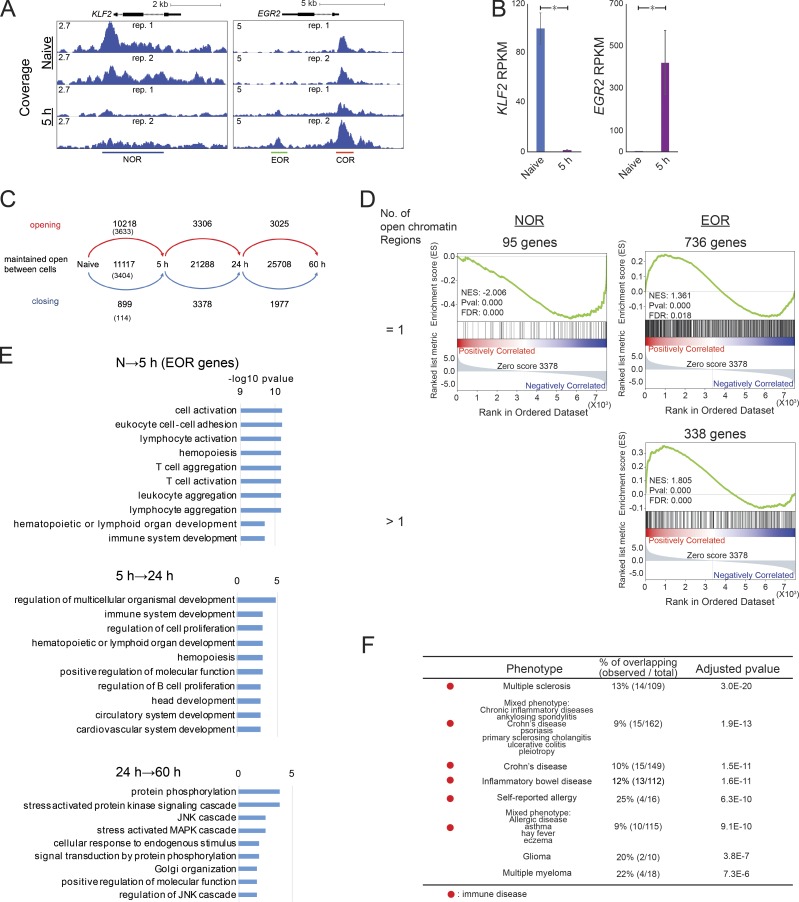
Human naive CD4 T cells isolated from the blood of healthy donors were activated with anti-CD3/CD28 beads for 5, 24, and 60 h (Fig. S1, A and B). Open chromatin in resting and activated cells was profiled by assay for transposase-accessible chromatin (ATAC-seq), and differentially accessible regions were identified (Fig. 1 A). Herein, we will refer to loci that are accessible only in naive cells or only in activated cells as naive open regions (NORs) or effector open regions (EOR), respectively, and to those loci maintained in both naive and activated cells as common open regions (CORs). For example, KLF2, encoding a quiescence-related TF (Kuo et al., 1997) down-regulated upon activation, had a NOR, whereas EGR2, an early growth response TF that is induced during activation, has both a COR and an EOR (Fig. 1, A and B). Overall, genome-wide analysis indicates that the chromatin remodeling that occurs at the early stages of T cell activation is dramatic (Fig. 1 C): in addition to the 11,117 open regions present in resting T cells (NORs plus CORs), 10,218 regions became accessible after 5 h of activation (EORs). Only 899 of open regions closed. We observed fewer changes at later time points, indicating that chromatin remodeling is more active immediately after activation. For the 5-h time point, we also identified a high-confidence set of regions that were consistently identified as NOR, COR, or EOR in T cells from two separate donors and were used for further analysis (Fig. 1 C). As expected, chromatin opening was associated with gene expression changes; genes nearest to EORs were enriched among genes up-regulated during T cell activation (false discovery rate [FDR] = 0.018 for 736 genes with one EOR and FDR < 0.001 for 338 genes with more than one EOR; Fig. 1 D). In contrast to EOR genes, genes possessing NORs were down-regulated during T cell activation (FDR < 0.001 for 95 genes with one NOR; Fig. S1 C). Furthermore, we found that the set of genes in the vicinity of 5-h EORs (EOR genes) were enriched for genes related to T cell function and activation (Gene Ontology [GO] terms “lymphocyte activation” and “T cell activation”) by GO analysis, whereas those genes that became accessible at later time points (5–24 h and 24–60 h) were involved in cell migration, proliferation, and metabolism (Fig. 1 E). We also found that NOR and COR gene sets were not enriched with genes related to T cell function and activation (Fig. S1 D). Further, we intersected the EORs with the genetic variants that related to various human conditions using the regulatory element locus intersection (RELI) approach (Harley et al., 2018). The results show that the risk SNPs for autoimmune or allergic diseases (e.g., multiple sclerosis, inflammatory bowel disease, Crohn disease, self-reported allergy, and combined allergic disease SNPs [asthma, hay fever, or eczema]) have significant overlap with EORs (Fig. 1 F and Table S1; all significant overlaps for NOR, COR, and EOR), whereas the risk SNPs for diseases such as coronary heart disease, Alzheimer's disease, and breast cancer do not (data not shown). Risk loci for glioma and multiple myeloma also significantly overlapped with EORs, probably due to cell cycle regulator (CDKN1A/B) involvement in both tumorigenesis and T cell activation. Thus, the formation of open chromatin at early time points during T cell activation is likely to play an important role in T cell activation, and its disruption may play a key role in immune disease processes.
Figure S1.
Experimental scheme and characterization of NORs and CORs. (A) Experimental scheme: activation of primary human naive CD4 T cells using beads covered with anti-CD3 and anti-CD28 antibodies. (B) Flow cytometric analysis of isolated human naive CD4 T cells. Left: Counts for CD45RA and CD45RO. Right: Counts for CD69 with or without 5 h of activation. (C) GSEA compares the gene list ranked by expression fold change during activation with the sets of genes that are located next to one or more COR. NES, normalized enrichment score. (D) GO analysis of genes adjacent to NORs or CORs. Top GO biological processes terms and −log10 P values are shown. Representative data were pooled for A and B from three independent experiments using a total of three donors. Open chromatin was defined in C and D from two independent experiments.
Figure 1.
Characterization of open chromatin during T cell activation. (A) The UCSC genome browser screenshots show open chromatin at the KLF2 and EGR2 loci in naive and activated T cells at 5 h. The y axis shows ATAC-seq coverage by estimated fragments normalized to the number of mapped reads. (B) The bar plots show expression of KLF2 and EGR2 genes by RNA-seq. Mean and standard error are shown. *, FDR < 0.01 from DEseq2. (C) The flow diagram shows the dynamics of open chromatin regions identified in naive T cells and T cells activated for the indicated time period. The numbers indicate the number of newly opened, maintained open, or newly closed chromatin regions. The numbers in parentheses indicate the number of highly reliable regions used for further analyses. (D) GSEA compares the gene list ranked by expression fold change during activation with the sets of genes that are located next to one or more than one NOR or EOR. NES, normalized enrichment score. (E) GO analysis of genes adjacent to chromatin regions that open during T cell activation. Top GO biological processes terms and −log10 P values are shown. N, naive. (F) Overlap between disease risk SNPs and EORs. Significance of overlap between disease risk SNPs and EORs as calculated by the RELI approach. Only the top 10 GWAS terms are shown. The full list for NORs, CORs, and EORs is available in Table S1. Representative data were pooled for A and B from two independent experiments using a total of three donors. Open chromatin was defined in C–F from the two independent experiments.
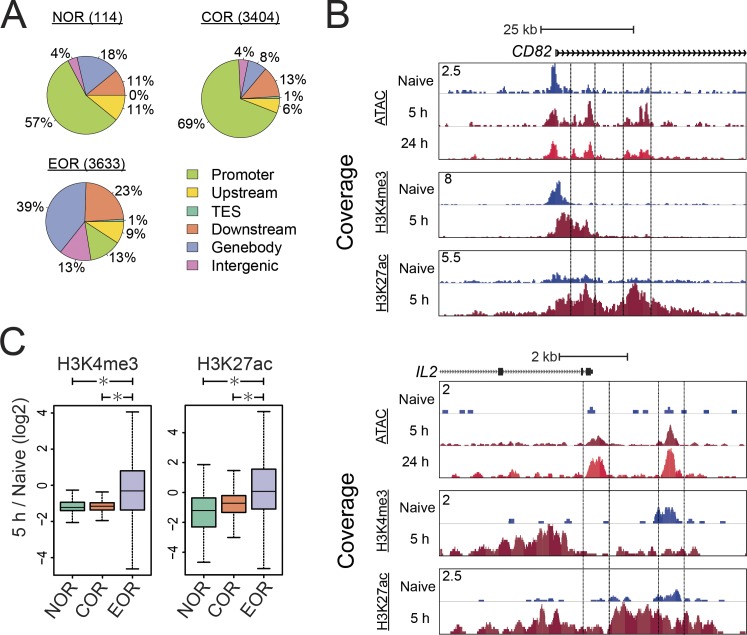
Interestingly, the majority of NOR and COR loci were found in promoter regions, whereas the majority of EOR loci were not (Fig. 2 A). These findings suggest that these newly accessible sites might serve as regulatory elements. To investigate their function, we examined changes of histone modifications at these open chromatin regions (Fig. 2, B and C). We observed the gain of positive chromatin modifications, H3K27ac and H3K4me3, at the EORs in the activation-inducible CD82 and IL2 loci, suggesting that the EOR elements may function as enhancers. Genome-wide analysis showed that the levels of H3K27ac and H3K4me3 at the EOR were significantly increased in the transition from naive to activated cells at 5 h (Fig. 2 C). Though H3K27ac is considered to be a hallmark of enhancers, H3K4me3 is often also present at these elements in addition to promoters (Ernst et al., 2011; Wang et al., 2008). In conjunction with the EOR association with transcriptional up-regulation, these findings suggest that EORs are likely to serve as transcriptional enhancers.
Figure 2.
EORs function as transcriptional enhancers during T cell activation. (A) The pie charts show the number and distribution of open chromatin areas relative to types of gene locations. TES, transcription end site. (B) The UCSC genome browser screenshot of ATAC-seq and ChIP-seq for H3K4me3 and H3K27ac at the CD82 and IL2 loci. The y axis shows the ATAC/ChIP-seq coverage by estimated fragments normalized to the number of mapped reads. (C) Changes in the H3K4me3 and H3K27ac levels at the open chromatin regions during T cell activation. The y axis shows the log2 of the normalized ratio of ChIP-seq signals between naive and 5-h activated T cells (5 h/naive). *, P < 0.01 (Wilcoxon rank-sum test). In the boxplot, lower whisker, lower hinge, line inside the box, upper hinge, and upper whisker show 5th,25th,50th,75th,and 95th percentile, respectively. Representative data were pooled for A–C from an experiment using a total of three donors.
The TFs AP-1 and NFAT1 bind effector open chromatin regions
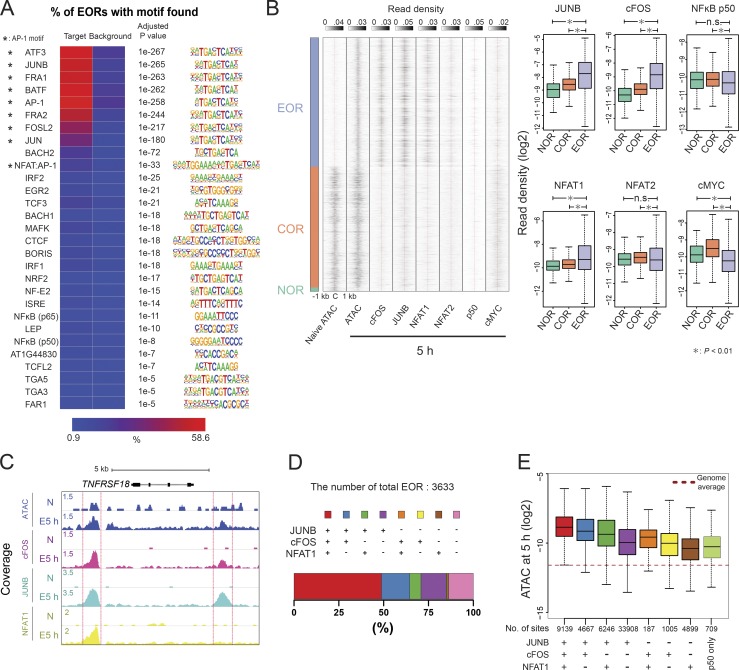
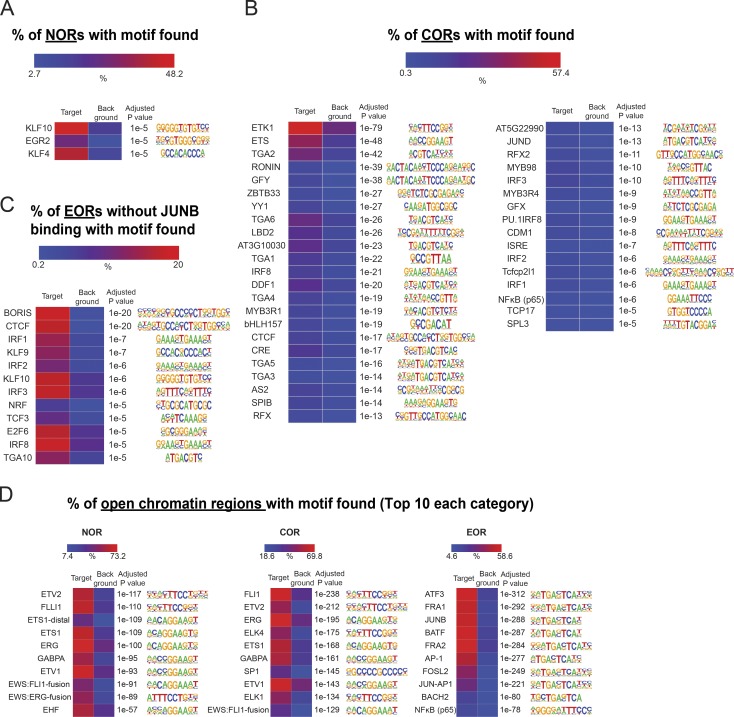
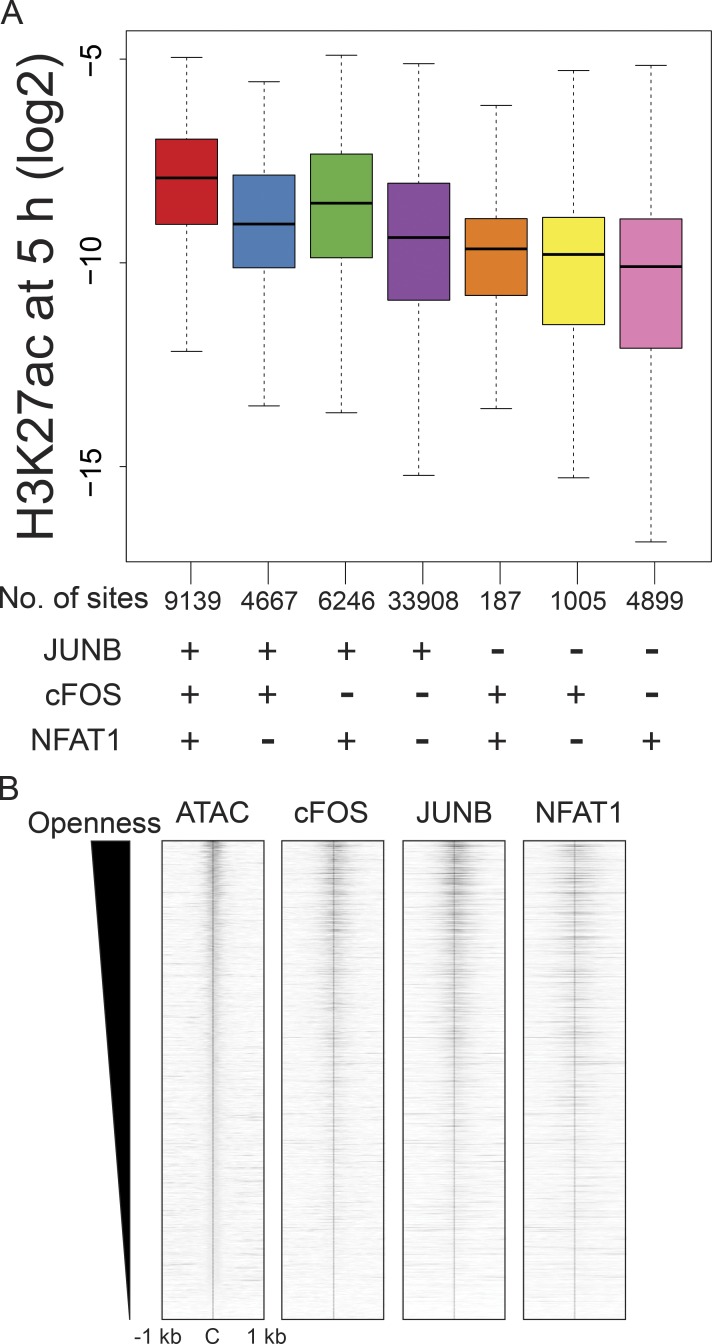
To identify TFs that may play a role in chromatin remodeling during activation, we used HOMER software (Heinz et al., 2010) to identify TF-binding sites overrepresented within the differentially accessible regions (Fig. 3 A and Fig. S2, A and B). Analysis showed that DNA motifs of AP-1 or the AP-1/NFAT composite element were enriched in EORs, but not in NORs and CORs. Interestingly, the NFAT motif alone was not enriched as strongly. BORIS and CTCF motifs were prominent at EOR sites not bound by AP-1 (Fig. S2 C), suggesting that nuclear organization also may be affected by activation-induced chromatin opening. In contrast to EORs, NORs were enriched in motifs for EGR and KLF, which have been reported to be negative regulators of T cell activation (Safford et al., 2005) and maintain the naive state in T cells (Yamada et al., 2009). For CORs, motifs of ETS, which is important in T cell development and activation (Panagoulias et al., 2016; Muthusamy et al., 1995), were prominent. Indeed, enrichment of JUNB motifs and binding at the sites of chromatin remodeling were previously reported in mouse T cells (Bevington et al., 2016), and our own analysis of motif enrichment in published mouse T cell ATAC data (Miraldi et al., 2019) produced similar results (Fig. S2 D). To test whether AP-1 and NFAT TFs actually bind EORs in human T cells, we performed ChIP-seq for cFOS and JUNB (components of AP-1), NFAT1, NFAT2, NF-κB, and cMYC, which are known to be important for T cell activation and proliferation (Liu et al., 2016; Trushin et al., 1999; Wong et al., 1999; Chou et al., 2014; Wang et al., 2011). cFOS, JUNB, and NFAT1 were present in EORs more than in NORs and CORs, whereas NF-κB and cMYC were not enriched in EORs (Fig. 3 B). For example, the TNFRSF18 gene (GITR) locus had two EOR regions (Fig. 3 C). One of them was bound by AP-1 and NFAT1, but the other was only bound by AP-1. Previously, it was reported that AP-1 and NFAT formed a heteromer on the IL2 promoter (Chen et al., 1998), leading us to examine combinatorial effects of the TFs on the opening of chromatin genome-wide (Fig. 3 D). Remarkably, >70% of EORs were bound by AP-1 (cFOS/JUNB) alone or AP-1 and NFAT1 together. Further, almost all NFAT1-bound EORs were also bound by AP-1 (cFOS or JUNB). Interestingly, the triple combination of cFOS, JUNB, and NFAT1 resulted in the highest accessibility, followed by the cFOS/JUNB dimer, whereas NFAT1 sites not bound by AP-1 showed minimal opening (Fig. 3 E), in agreement with the NFAT-only motif not being enriched in EORs. Moreover, the H3K27ac level was also the highest in regions with the triple combination (Fig. S3 A). Furthermore, the level of chromatin openness was correlated with the strength of cFOS, JUNB, and NFAT1 binding (Fig. S3 B). These data suggest either a role for AP-1 in chromatin remodeling during T cell activation or passive binding of AP-1 complexes to enhancers opened by other TFs.
Figure 3.
NFAT1 and AP-1 bind to EORs. (A) DNA-binding motifs enriched in EORs. The heatmap shows the percentage of EORs with motifs. The overrepresented motifs were identified by HOMER analysis and selected with an adjusted P ≤ 10−5 and target/background >2. Asterisk indicates AP-1 motifs. (B) NFAT1 and AP-1 bind to EORs. Left: The fragment density heatmaps show read density of the ATAC-seq and TF ChIP-seq signal at open chromatin regions. C, center of open chromatin regions. Right: The boxplots show the TF ChIP-seq read density at open chromatin regions in activated T cells at 5 h. *, P < 0.01 (Wilcoxon rank-sum test). (C) The UCSC genome browser screenshot shows ChIP-seq for AP-1 and NFAT1 at the TNFRSF18/GITR locus. The regions marked with red lines are EORs. The TF ChIP-seq was normalized between naive and 5-h–activated samples using spike-in of Drosophila genomic DNA (see Materials and methods). N, naive cells. (D) The stacked bar plot shows the percentage of EORs overlapping with significant TF ChIP-seq peaks. (E) The boxplot shows the ATAC-seq signal in activated T cells at 5 h in the regions bound by a given combination of TFs. The genome average indicates the average ATAC-seq signal for the whole genome. The y axis indicates the ATAC-seq tag density over peaks. Representative data were pooled for A–E from one to three independent experiments using a total of three donors. n.s., not significant. In the boxplots, lower whisker, lower hinge, line inside the box, upper hinge, and upper whisker show 5th, 25th, 50th, 75th, and 95th percentile, respectively.
Figure S2.
DNA-binding motifs enriched in open chromatin regions. (A and B) Enriched DNA motifs in NORs and CORs. The heatmap shows the percentage of regions with motifs. The overrepresented motifs were identified by HOMER analysis and selected with an adjusted P value ≤ 10−5 and target/background >2. (C) Shown are the same analyses as in A and B but for EORs not bound by JUNB. (D) Shown are the enriched DNA motifs in ATAC-seq data (naive vs. activated for 6 h) of mouse CD4 T cells from Miraldi et al. (2019).
Figure S3.
Combinatorial effects of AP-1 and NFAT1 to H3K27ac and openness. (A) Boxplot showing the H3K27ac ChIP-seq signal in activated cells at 5 h in the regions bound by a given combination of TFs. The y axis shows the H3K27Ac ChIP-seq tag density for the TF ChIP-seq peaks. (B) Correlation of openness with TF enrichments in EORs. The heatmap shows the normalized fragment density of ATAC-seq and TF ChIP-seq in a 1-kb radius of the center of the EOR. C, center of open regions. Representative data were pooled for A and B from one or two independent experiments using a total of three donors. In the boxplot, lower whisker, lower hinge, line inside the box, upper hinge, and upper whisker show 5th, 25th, 50th, 75th, and 95th percentile, respectively.
Formation of SEs involves chromatin opening
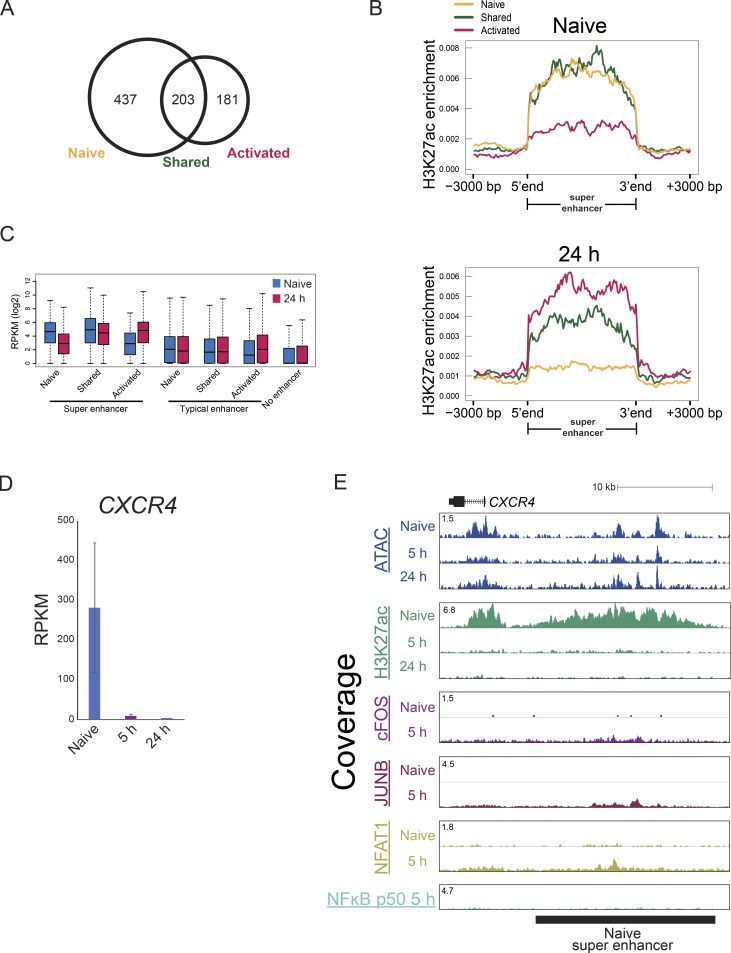
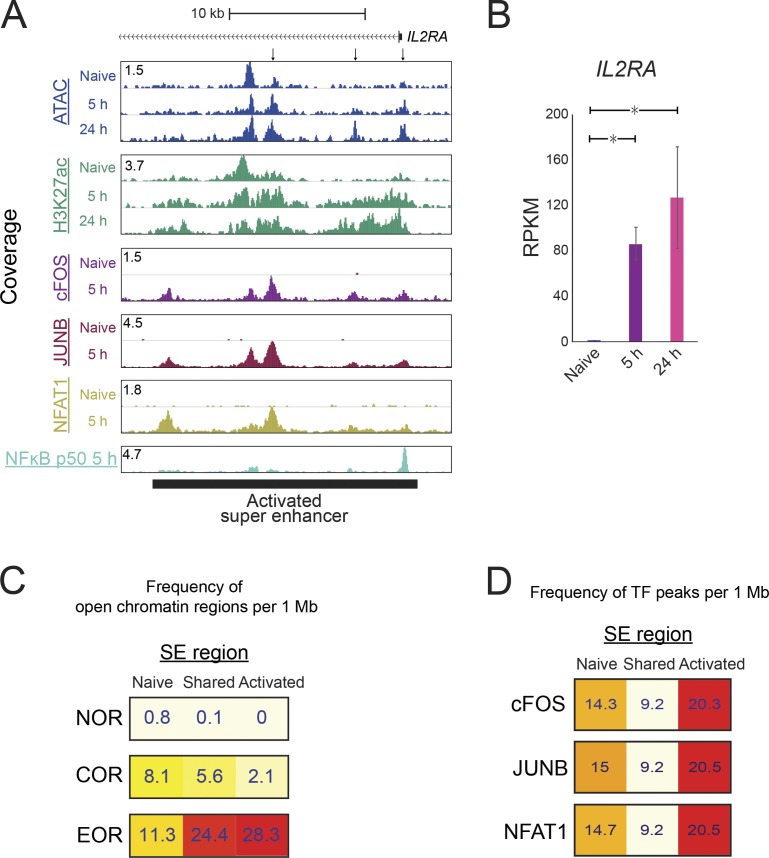
Combinations of several nearby enhancer elements with unusually high total level of H3K27 acetylation and/or binding of mediator or BRD4 proteins are known as SEs and are believed to induce expression of cell type–specific genes (Whyte et al., 2013; Lovén et al., 2013). Therefore, we examined whether EORs were involved in the assembly of SEs. We identified 640 and 384 SEs in naive and activated T cells, respectively, and 203 of them were shared between the cell states (Fig. S4, A and B). As expected, the presence of SEs resulted in high expression of nearby genes (Fig. S4 C). SE formation during T cell activation was associated with chromatin remodeling. For example, the IL2RA SE formation was accompanied by open chromatin formation after activation (Fig. 4, A and B). In addition to the EOR frequency being higher in activated SEs (28.3 EOR/1 Mb), the frequency of EORs also was higher in shared SE (24.4 frequency EOR/1 Mb; Fig. 4 C). SEs in naive cells seem to rely on NOR and COR in contrast to SE in activated cells (Fig. 4 C and Fig. S4, D and E). Furthermore, AP-1 and NFAT1 were bound to activated and shared SEs at high frequency in contrast to naive SEs (Fig. 4 D). Collectively, these results suggest that AP-1 and NFAT1 likely play a role in the formation of SEs during T cell activation.
Figure S4.
Detection and characterization of SEs in naive and activated T cells. (A) Venn diagram showing the overlap of SEs in naive and 24-h–activated T cells. (B) Average tag density profile of H3K27ac within SE regions in naive and activated T cells at 24 h. Naive indicates naive SE, shared indicates a shared SE; and activated indicates an activated SE. (C) The boxplot shows expression of genes with naive, shared, or activated SEs or without SEs (“No”) in naive and 24-h–activated T cells. (D) The bar plots show expression of CXCR4 by RNA-seq. The mean and standard error of the mean are shown. (E) UCSC genome browser screenshot showing the ATAC signal and H3K27ac and TF ChIP-seq at a naive SE in the CXCR4 locus. Representative data were pooled for A–E from one to three independent experiments using a total of three donors. In the boxplot, lower whisker, lower hinge, line inside the box, upper hinge, and upper whisker show 5th, 25th, 50th, 75th, and 95th percentile, respectively.
Figure 4.
SE formation is associated with open chromatin formation. The SEs were identified using H3K27ac ChIP-seq. (A) The UCSC genome browser screenshot shows the ATAC-seq and ChIP-seq signal for H3K27ac and TFs around the SE located in the IL2RA locus. The TF ChIP-seq was normalized between naive and 5-h–activated samples using spike-in of Drosophila genomic DNA (see Materials and methods). (B) The bar plots show the expression of IL2RA by RNA-seq. The mean and standard error of the mean are shown; n = 2. *, FDR < 0.01 from DEseq2. (C) The heatmap shows the frequency of open chromatin regions per 1 Mb of an SE. The frequency was calculated for SEs formed only in naive cells, only in activated cells, or in both cell types (shared). (D) The heatmap shows the frequency of cFOS, JUNB, and NFAT1 peaks in the SE regions. Representative data were pooled for A, C, and D from one to three independent experiments using a total of three donors. RPKM, reads per kilobase per million mapped to transcriptome.
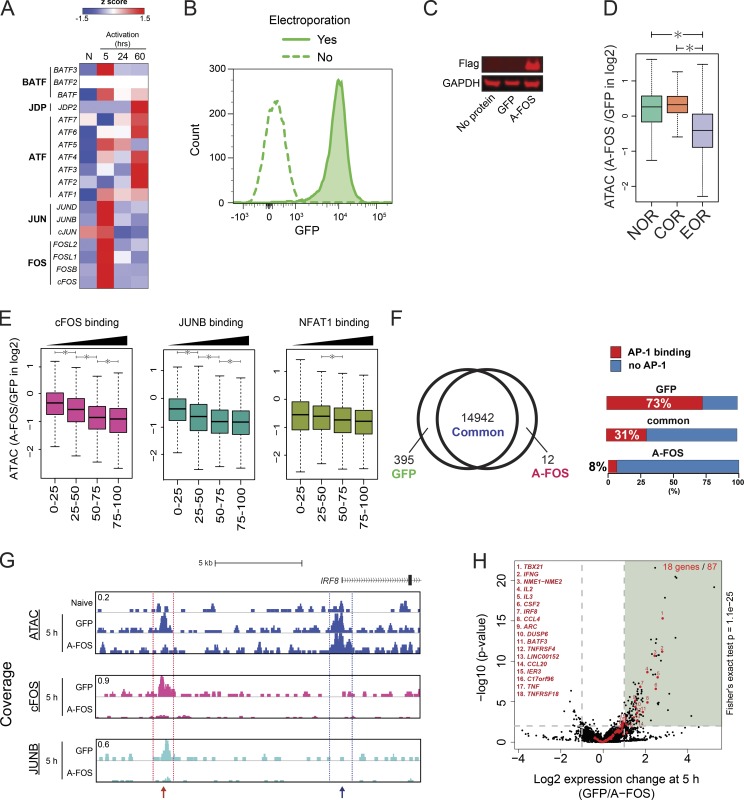
AP-1 activity is required for open chromatin formation during T cell activation
To examine the role of AP-1 in the formation of open chromatin, we sought to inhibit AP-1 activity during T cell activation. Unfortunately, due to coexpression of multiple JUN and FOS family members in T cells and their common up-regulation during activation (Fig. 5 A; Jain et al., 1994), knockout or knock-down strategies are unlikely to be successful. For this reason, we instead used the AP-1 dominant-negative protein A-FOS (Biddie et al., 2011). A-FOS can sequester JUN isoforms and prevent FOS–JUN complex formation and DNA binding. Bacterially expressed A-FOS or GFP control proteins were purified and electroporated into naive T cells before activation. Electroporation efficiency was close to 100% (Fig. 5 B), A-FOS protein remained in T cells for ≥5 h after electroporation (Fig. 5 C), and electroporation did not affect T cell activation (data not shown). We observed that upon A-FOS electroporation, chromatin opening was decreased at EORs, but not at NORs and CORs (Fig. 5 D). This blocking effect was more significant in EORs strongly bound by AP-1 (Fig. 5 E). Indeed, among 395 regions that were accessible in GFP-electroporated cells, but not in the cells receiving A-FOS, 73% were bound by both FOS and JUNB (Fig. 5 F). For example, the IRF8 gene has two EORs; the distal one is bound by AP-1, whereas the proximal one is not. As expected, the EOR bound by AP-1 was lost, whereas an EOR not bound by AP-1 was not affected. (Fig. 5 G). The specific effect of A-FOS protein transduction provides reassurance that the electroporation procedure was functional and well received by the T cells. Inhibiting AP-1 resulted in down-regulation of activation-inducible genes such as IRF8, TBX21, IFNG, and CSF2 (Fig. 5 H) but did not affect down-regulated genes during T cell activation (0 of 41 genes). Furthermore, decreased chromatin opening was observed at 8 of the 18 AP-1 activation-inducible genes down-regulated by A-FOS. These results indicate that AP-1 binding is required for opening chromatin during T cell activation at many genomic loci.
Figure 5.
A-FOS, a dominant-negative regulator of AP-1, inhibits EOR formation. (A) The heatmap shows the changes in mRNA expression level of AP-1 (FOS, JUN, ATF, and JDP) and BATF family proteins during T cell activation by RNA-seq. The color shows the z-score. N, naive T cells. (B) Flow cytometry shows the high efficiency of protein electroporation. GFP was added to cells, and the mixture was either subjected to electroporation or not. (C) Western blot shows the A-FOS level in electroporated cells. No protein indicates cells electroporated without protein, GFP indicates cells electroporated with GFP, and A-FOS indicates cells electroporated with Flag-tagged A-FOS. Anti-FLAG antibody was used to detect A-FOS. (D) The boxplot shows the ratio of ATAC-seq signal in open chromatin regions between T cells electroporated with GFP and A-FOS after 5 h of activation. *, P < 0.01 (Wilcoxon rank-sum test). (E) EORs were separated into quartiles on the basis of TFs’ ChIP-seq read density within peaks. The boxplot shows the ratio of ATAC-seq signal in open chromatin regions between T cells electroporated with GFP and A-FOS after 5-h activation. *, P < 0.01 (Wilcoxon rank-sum test). (F) Left: Venn diagram showing the overlap of open chromatin regions between GFP- and A-FOS–electroporated T cells. Right: Stacked bar plot showing the percentage of differential open chromatin regions overlapping with significant AP-1 (both FOS and JUNB) ChIP-seq peaks. (G) UCSC genome browser screenshot showing the ATAC signal at the IRF8 locus in naive cells and activated cells with GFP or A-FOS. The red arrow indicates an EOR dependent on AP-1. The blue arrow indicates an EOR independent of AP-1. ChIP-seq for AP-1 proteins was normalized between GFP and A-FOS samples using spike-in of Drosophila genomic DNA (see Materials and methods). (H) The volcano plot showing the changes in gene expression between GFP- and A-FOS–electroporated T cells by RNA-seq. The x axis shows the ratio of expression between GFP and A-FOS T cells (log2), and the y axis shows the log10 DEseq2 P value for differential expression. Red dots represent activated genes (naive → 5-h activation). Black dots represent other genes. The vertical gray lines are at −1 and 1 on the x axis. The horizontal gray line is at 2 on the y axis. The P value for overlap between genes induced during T cell activation and suppressed by A-FOS electroporation was calculated using the Fisher's exact test. Representative data were pooled for A–H from one to three independent experiments using a total of three donors. In the boxplots, lower whisker, lower hinge, line inside the box, upper hinge, and upper whisker show 5th, 25th, 50th, 75th, and 95th percentile, respectively.
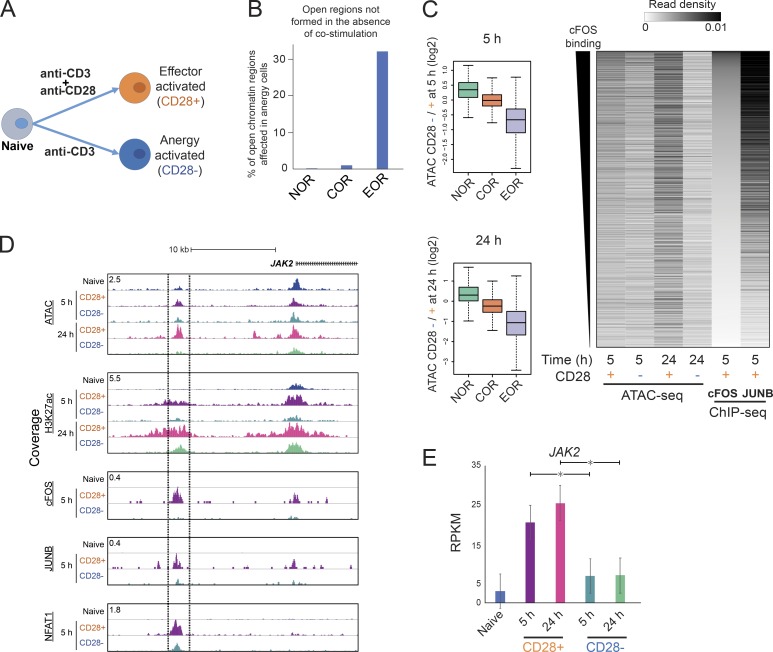
Co-stimulation is required for open chromatin formation
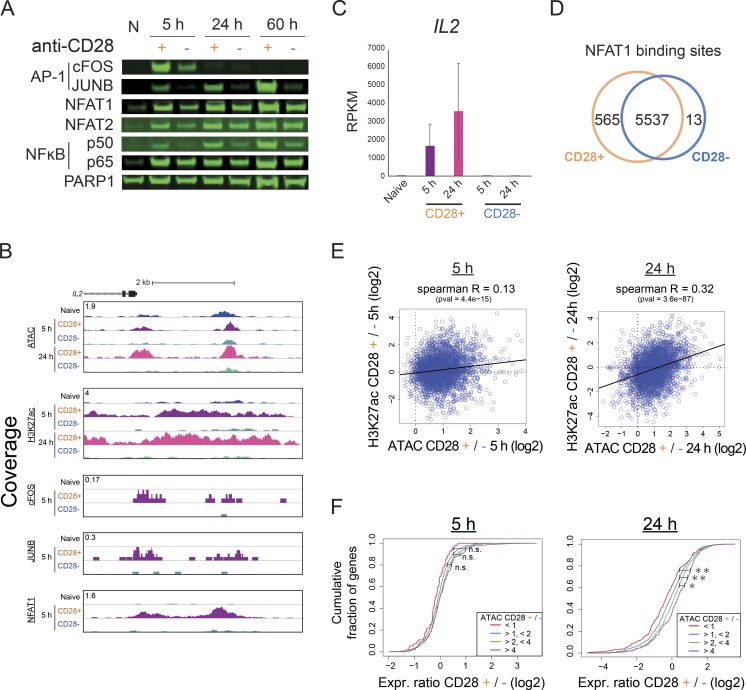
To better understand the role of AP-1–induced chromatin remodeling in the greater context of the immune response, we next performed T cell activation in the absence of CD28 co-stimulation (Fig. 6 A). Previous studies have shown that the lack of co-stimulation reduces activation of AP-1 and eventually leads to the induction of anergy (Macián et al., 2002; Kriegel et al., 2009; Rochman et al., 2015). Indeed, cells not receiving CD28 co-stimulation (anergy) showed dramatic reduction of nuclear translocation of AP-1 and, to a smaller degree, NF-κB p50, whereas nuclear levels of NFATs and NF-κB p65 were reduced only slightly (Fig. S5 A). Genome-wide, the reduction of AP-1 activity was accompanied by decreased chromatin opening at EORs (Fig. 6, B and C). For example, in the absence of CD28 signaling, the AP-1–bound EOR in the vicinity of the JAK2 and IL2 genes was not remodeled and did not become acetylated on H3K27 (Figs. 6 D and S5 B); consequently, both JAK2 and IL2 demonstrated reduced gene expression (Figs. 6 E and S5 C). The promoter-associated COR, also bound by AP-1, remained but with decreased H3K27ac levels. Interestingly, NFAT binding was still detectable at the majority of sites, including those that lost the ATAC signal in the absence of co-stimulation, suggesting that NFAT is not sufficient for opening chromatin (Figs. 6 D and S5 D), with IL2 being one of the exceptions. Failure of EOR formation in co-stimulation-deficient cells was especially prominent in regions that have higher openness among EORs in effector-activated cells at 24 h (Fig. 6 C). We tested whether failure of chromatin remodeling due to the lack of CD28/AP-1 affected the histone modifications and transcriptome in cells that did not receive co-stimulation. Indeed, failure of open chromatin formation led to reduced H3K27ac level and gene expression at nearby genes (5 h, Spearman R = 0.13, P = 4.4e-15; 24 h, Spearman R = 0.32, P = 3.6e-87; Fig. S5, E and F). These results suggest that the induction of anergy by activation in the absence of co-stimulation is associated with the failure of AP-1–dependent chromatin remodeling.
Figure 6.
Incomplete open chromatin formation during induction of T cell anergy in the absence of co-stimulation. (A) Experimental approach. Naive cells were activated with beads covered with anti-CD3 and anti-CD28 or only anti-CD3 antibodies. (B) Barplot shows the percentage of open chromatin regions not formed at 5 h in cells that did not receive co-stimulation. (C) Chromatin fails to open in T cells that lack co-stimulation. Left: Boxplot showing the ratio of ATAC signal between cells activated without and with CD28 co-stimulation at 5 and 24 h. Right: Heatmap showing ATAC-seq and TF ChIP-seq read density in EORs. EORs were sorted on the basis of cFOS signal. (D) UCSC genome browser screenshot showing the ATAC-seq and ChIP-seq for H3K27ac, AP-1, and NFAT1 in the JAK2 locus. The region bordered by lines is open in fully activated cells, but not in cells lacking co-stimulation. ChIP-seq for TF proteins was normalized between CD28+/− samples using spike-in of Drosophila genomic DNA (see Materials and methods). (E) Barplot showing expression of JAK2 by RNA-seq. The mean and standard error of the mean are shown; n = 2. *, FDR < 0.01 from DEseq2. Representative data were pooled for A–E from one to three independent experiments using a total of three donors.
Figure S5.
Expression and chromatin changes during anergy induction in the absence of co-stimulation. (A) Western blot showing the nuclear levels of TFs during T cell activation with or without co-stimulation. N, naive T cells. (B) UCSC genome browser screenshot showing the ATAC-seq and ChIP-seq in the IL2 locus. (C) Barplot showing the expression of IL2 by RNA-seq. The mean and standard error of the mean are shown. (D) Venn diagram showing the overlap between the NFAT1 sites detected in the cells stimulated with or without co-stimulation. (E) Correlation between the change in chromatin opening (ATAC) and H3K27ac (ChIP-seq) at the EOR at 5 h (left) and 24 h (right). pval, P value. (F) Cumulative distribution of the fold changes between effector (CD28+) and anergic (CD28−) conditions for genes that have the indicated ratios of ATAC value in EORs between activated and anergic T cells at 5 h (left) and 24 h (right). The x axis shows the ratio of expression between effector and anergic conditions (log2), and the y axis shows the cumulative fraction of genes. Each line indicates the gene groups by the ratio of the ATAC value (for all EOR peaks that are nearby; effector/anergic: red, <1; blue, >1 and <2; green, >2 and <4; and purple, >4). *, P < 0.05; **, P < 0.01 (Kolmogorov–Smirnov test). Representative data were pooled for A–F from one to three independent experiments using a total of three donors. n.s., not significant.
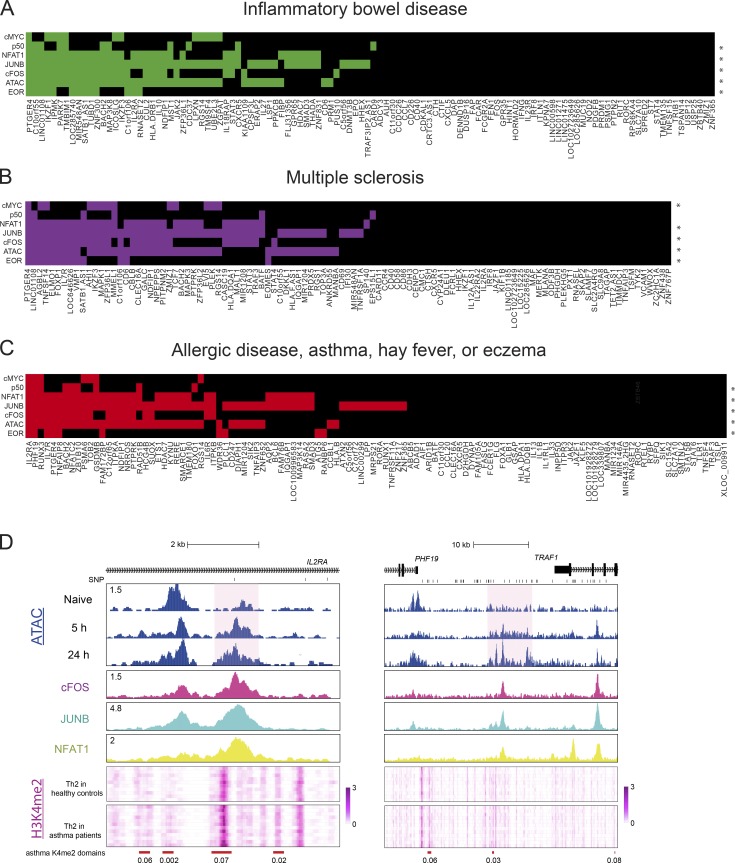
AP-1 sites overlap risk loci for immunological diseases
CD4 T cells control functions of other immune cells and are the key regulators of immune response and homeostasis. Impairment of T cell activation may lead to persistent infections and cancer, and overactivation of T cells may cause autoimmune or atopic disease. Aside from elucidating the epigenetic mechanism of T cell activation, our data demonstrate a significant overlap between putative regulatory elements employed in T cell activation (EORs) with previously unexplained risk loci for several autoimmune diseases, such as Crohn disease, multiple sclerosis, and inflammatory bowel disease (Fig. 1 F; Soderquest et al., 2017). Previously AP-1 sites were found to be enriched near SNPs for autoimmune diseases (Farh et al., 2015). To gain further clues into potential disease mechanisms, we repeated the RELI analysis with TF ChIP-seq data in order to identify the candidate TFs that may bind risk loci. This analysis revealed that risk loci for multiple immune diseases were enriched for AP-1 and NFAT1 (Fig. 7, A–C). We next endeavored to check whether the EORs overlapping disease risk loci are differentially open (or modified) in T cells from patients. Although we could not find such data for our top hits (multiple sclerosis and inflammatory bowel disease), a recent study (Seumois et al., 2014) reported H3K4me2 profiles for Th2 cells obtained from patients with asthma and healthy controls. We found that two allergic disease risk loci (CD25 and PHF19) that significantly overlap with EORs (Fig. 7 C) showed a significant increase in the level of H3K4me2 in patients with asthma compared with healthy controls (FDR < 0.1; Fig. 7 D). These findings indicate the importance of proper chromatin remodeling for immune homeostasis.
Figure 7.
Intersection of immune disease risk loci, open chromatin regions, and TF-binding interactions with the genome. (A–C) Heatmaps for inflammatory bowel disease (A), multiple sclerosis (B), and allergic disease, asthma, hay fever, or eczema (C) risk loci are shown. The x axis displays disease-associated loci. An asterisk means that the genomic coordinates of the ChIP-seq peaks measuring open chromatin regions or TF binding (at 5 h) significantly intersect with the disease risk loci at a RELI-corrected P value threshold of 10−6. A colored box indicates that the given locus contains at least one disease-associated variant located within an open chromatin region or a ChIP-seq peak for the given TF. (D) The genome browser screenshots shows the ATAC-seq in naive and activated T cells, ChIP-seq (cFOS, JUNB, and NFAT1 at 5 h activation), and heatmap of the H3K4me2 level in Th2 cells of patients with asthma and healthy controls in the IL2RA and PHF19 loci. Red bars indicate differentially enriched loci (FDR is shown under the bars), and a pink background indicates EORs. Representative data were pooled for A–C from one to three independent experiments using a total of three donors.
Discussion
T cell activation is associated with dramatic chromatin decondensation that is essential for T cell differentiation and function (Rawlings et al., 2011; Lee et al., 2015). Genome-wide regulatory elements are bound by TFs that recruit chromatin-remodeling complexes and chromatin-modifying enzymes, leading to expression of activation-related genes. We mapped putative regulatory elements in human naive and activated CD4 T cells by ATAC-seq and identified the regions that undergo remodeling at early time points during T cell activation. Many of them stay open in the resting memory cells. These regions also gained “positive” histone modification, such as H3K4me3 and H3K27ac, whereas the nearby genes tended to demonstrate increased expression. Interestingly, these regulatory elements showed significant overlap with SNPs associated with a number of immune diseases, underscoring their importance for the regulation of the immune system. Next, we identified the TF-binding motifs that are present at the sites of remodeling and thus may control chromatin remodeling at these elements. To confirm the results of the computational analysis, we monitored actual binding of NFAT1 and AP-1, as well as NF-κB and cMYC, during activation by ChIP-seq. Comparing TF-binding and chromatin-opening data suggested that AP-1 is the factor responsible for opening chromatin at regulatory elements during T cell activation. Indeed, delivery of the AP-1 dominant-negative protein A-FOS into naive T cells resulted in the T cells’ failure to remodel chromatin. Finally, we found that a lack of AP-1 activation during the induction of anergy resulted in a failure of T cell chromatin remodeling.
It has long been known that activation of T cells requires two signals. Work from the Rao laboratory and others has shown that insufficient induction of AP-1 in the absence of co-stimulation fails to induce Il2 gene expression and eventually leads to anergy (Macián et al., 2002; Kriegel et al., 2009; Rochman et al., 2015). However, murine cFOS knockout T cells demonstrated normal response to activation, suggesting either that cFOS is not necessary (Jain et al., 1994) or that the other FOS family members, such as FOSB, FRA-1, and FRA-2, could substitute for cFOS (Fleischmann et al., 2000; Gruda et al., 1996). Indeed, all AP-1 family genes are dramatically up-regulated upon T cell activation (Fig. 5 A). Given that the AP-1 family includes 18 genes, generating a complete deletion was impractical. Furthermore, manipulating expression in resting naive T cells is difficult, because successful viral transduction requires T cell proliferation, and although plasmid electroporation is possible, the process impairs cell activation (data not shown). We overcame this dual problem by (i) using the A-FOS dominant-negative protein that sequesters JUN family members into unproductive A-FOS–JUN complexes (Biddie et al., 2011) and (ii) electroporating the protein into resting naive T cells, which (unlike plasmid electroporation) did not affect T cell activation. In the presence of A-FOS, T cells failed to remodel their chromatin at multiple AP-1–bound sites. This experiment explains the role of AP-1 in T cell activation: establishing chromatin remodeling. Without CD28-driven AP-1 induction, naive CD4 T cells could not establish open chromatin regions to act as enhancer elements for inducing gene expression in T cell activation. Our results further confirmed the hypothesis (Jain et al., 1994) that although individual FOS family proteins are redundant, induction of the AP-1 family is critical for T cell activation.
How does binding of AP-1 lead to chromatin remodeling? Previous studies have shown that AP-1 proteins can recruit components of the BAF chromatin-remodeling complex. In particular, SMARCD1 was shown to be an AP-1–interacting partner by a yeast two-hybrid system screen (Ito et al., 2001). ATF3 was shown to physically interact with SMARCA4 (BRG1-ATPase; Xu et al., 2011), and a more recent study showed the interaction of FOS with 10 out of 15 BAF members and the ability of other FOS family members (FOSD and FOSL1/2) to recruit both SMARCB1 and SMARCD1 (Vierbuchen et al., 2017).
Other TFs are involved in regulating the epigenome during T cell activation. Recent studies discussed the roles of STATs, GATA3, and T-BET (Vahedi et al., 2012; Durant et al., 2010; Wan et al., 2015; Pham et al., 2013; Kanhere et al., 2012). Moreover, although AP-1 is required for chromatin remodeling at the regulatory elements for T cell activation, our study does not completely answer the question of specificity; many cell types express AP-1 and NFATs but do not induce T cell genes such as IL2 (Armstrong and Bischoff, 2004; Turner et al., 1998; Ranger et al., 2000; Jauliac et al., 2002). Thus, the specificity would need to be established either by a preexisting epigenetic state or by interaction with additional TFs. For this reason, AP-1 cannot be considered to be a pioneer factor (Zaret and Mango, 2016) by itself. RUNX family proteins, which are broadly expressed in lymphocytes and regulate expression of IL2 (Djuretic et al., 2007; Ono et al., 2007), and ETS family members (Bevington et al., 2016) may play a role, but involvement of additional TFs is also possible.
Materials and methods
Isolating, culturing, and activating human CD4 T cells
Blood filters containing cells from deidentified donors were supplied by Hoxworth Blood Center at the University of Cincinnati under exemption granted Cincinnati Children’s Hospital Medical Center Institutional Review Board. Peripheral blood mononuclear cells were isolated by Ficoll density gradient centrifugation. Isolated peripheral blood mononuclear cells were processed with the EasySep Human Naive CD4+ T Cell Isolation Kit (#19555; STEMCELL) to negatively isolate human CD4+/CD45RO− cells (naive T cells), and the purity was confirmed by flow cytometry (Fig. S1 B). Isolated naive T cells were cultured in RPMI media with L-glutamine (#SH30027.01; HyClone) supplemented with 10% FBS and 25 µM 2-ME before activation. For bead activation, 100 µl anti-mouse pluriBeads (#31-GTaMS-10; pluriSelect) were incubated with 24 ng anti-human CD3 antibody (#BE0001-2; Bio X Cell) and 52 ng of either anti-human CD28 antibody (#BE0248; Bio X Cell) or mouse IgG (#12-371; Millipore) with rotation for 3–4 h at room temperature (RT). The beads were centrifuged and washed with PBS twice. Antibody-bound pluriBeads were directly added to the cultures of human CD4+ T cells, and the cultures were shaken for 4 h and swirled every 10 min until the cells were bound to the beads.
Flow cytometry assay
Cells were stained with PE-Cy7 mouse anti-human CD45RO (#560608; BioLegend), FITC mouse anti-human CD45RA (#130-092-247; Miltenyi Biotec), and FITC anti-human CD69 (#310904; BioLegend) antibodies. Flow cytometry was performed on the BD FACS Canto II using the BD FACSDIVA.
Isolating nuclear proteins and Western blotting
T cells were collected and lysed with nuclear isolation buffer (10 mM Hepes-KOH, pH 8.0, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM dithiothreitol, and 1× protease inhibitors) for 10 min with shaking every 3 min. The nuclear pellet was collected by centrifugation and was suspended and washed with nuclear isolation buffer twice. The nuclear pellet was directly lysed in SDS sample buffer with 8 M urea. SDS-PAGE was performed in the Bolt Bis-Tris system (Thermo Fisher Scientific). Transfer was performed with 120 V for 90 min to a polyvinylidene difluoride membrane. Blocking of the membrane was performed with Odyssey Blocking Buffer (#927-50000; LI-COR) for 60 min at RT with shaking. The antibody incubation was performed at 4°C overnight with an orbital shaker. Antibodies to cFOS (#2250), JUNB (#3753), NFAT1 (#5861), NFAT2 (#8032), NF-κB p50 (#12540), and PARP1 (#9532) from Cell Signaling; NF-κB p65 (c15310256) from Diagenode; and GAPDH (NB600-502) from Novus were used as primary antibodies. Membranes were washed with Tris-buffered saline, pH 7.5, 0.1% Tween for 5 min three times and were incubated with IRDye secondary antibodies in 1/15,000 dilution in blocking buffer for 60 min at RT with shaking. Membranes were washed with TBS-Tween for 5 min three times again and with TBS once. Images were taken with an Odyssey Fluorescent Imaging system (LI-COR).
ATAC-seq
50,000 naive or activated T cells were collected and processed in a transposase reaction, and library preparation was as described by Buenrostro et al. (2013).
ChIP and library preparation by ChIPmentation
Fixing solution (50 mM Hepes-KOH, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, and 8.8% formaldehyde) was added directly to T cell culture at a final formaldehyde concentration of 0.88% for fixation. After the incubation for 4 min on ice, 2 M glycine solution was added to a final concentration of 125 mM and incubated at room temperature for 10 min to stop fixation. Cells were transferred to tubes and washed with cold PBS twice. All buffers used in the following procedures were supplemented with 1× protease inhibitor solution (#P8340; Sigma) before use. Fixed cells were resuspended in L1 buffer (50 mM Hepes-KOH, pH 8.0, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, and 1× protease inhibitors) and incubated on ice for 10 min with shaking every 3 min. Isolated nuclei were incubated with L2 buffer (10 mM Tris-HCl, pH 8.0, 200 mM NaCl, 1 mM EDTA, and 0.5 mM EGTA) with rotation for 10 min at RT. Then, isolated nuclei were carefully washed and resuspended in Tris- EDTA + 0.1% SDS solution. Cells were sonicated (peak power 105.0, duty factor 10.0, and cycle/burst 200) for 45 s in microtubes at 4°C using a S220 focused ultrasonicator (Covaris) to obtain the 200- to 500-bp fragments of chromatin. The chromatin solution was centrifuged, and the supernatant chromatin was collected. Triton X-100, glycerol, NaCl, and sodium deoxycholate were added into the chromatin solution to final concentrations of 1%, 5%, 150 mM, and 0.1%, respectively. Protease inhibitors (#P8340; Sigma) were also added.
ChIPs were performed in an IP-Star Compact automation system (Diagenode). In brief, ∼5–10 µg chromatin solution corresponding to 1–2 million cells and 5–10 µl Protein A or G Dynabeads (Thermo Fisher Scientific) were used per reaction. Antibodies against cFOS (sc-7202; Santa Cruz Biotechnology), cMyc (sc-764X; Santa Cruz Biotechnology), H3K27ac (pAb-196-050; Diagenode), or H3K4me3 (#17-614; Millipore) were used. The other antibodies were the same as used in Western blotting. The Dynabeads were sequentially washed with wash buffer 1 (RIPA 150 mM NaCl: 10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 0.1% sodium deoxycholate, and 1% Triton X-100), wash buffer 2 (RIPA 250 mM NaCl), wash buffer 3 (50 mM Tris-HCl, pH 8.0, 2 mM EDTA, and 0.2% N-Lauroylsarcosine sodium salt), and wash buffer 4 (TE + 0.2% Triton X-100) for 15 min each. Washed Dynabeads were processed with transposase per ChIPmentation protocol to accomplish tagmentation of ChIPed DNA (Schmidl et al., 2015). Tagmented DNA was incubated with proteinase K in elution buffer (TE with 250 mM NaCl and 0.3% SDS) for 4 h at 65°C and purified from beads with the Qiagen MinElute DNA kit. PCR amplification and purification were performed in the same way as for ATAC-seq.
RNA isolation and RNA sequencing (RNA-seq)
Total RNA was isolated with the Aurum Total RNA Mini Kit (#7326820; Bio-Rad) including on-column DNaseI digest. PolyA selection and fragmentation of RNA were performed according to the TruSeq Stranded polyA RNA Sample Preparation Guide from Illumina. The fragmented RNA was subjected to first-strand cDNA synthesis. For second-strand cDNA synthesis and adapter ligation, the IP-Star Compact automation system was used. After that, library construction was performed according to the Illumina protocol.
Sequencing, read mapping, peak calling, peak comparison, and calculation enrichment
DNA libraries for ATAC-seq, ChIP-seq, and RNA-seq were sequenced on HiSeq2500/4000 (Illumina) at the DNA Sequencing and Genotyping Core at Cincinnati Children’s Hospital Medical Center. Sequencing data were deposited to the Gene Expression Omnibus under accession no. GSE116698. Data analysis was performed in BioWardrobe (Kartashov and Barski, 2015). Briefly, 75-bp reads were aligned to the human genome (hg19) using Bowtie (Langmead et al., 2009; for ATAC-seq and ChIP-seq) or STAR (Dobin et al., 2013; for RNA-seq). STAR was supplied with RefSeq annotation. For peak calling in ATAC-seq and ChIP-seq, MACS2 (Zhang et al., 2008) software was used.
Browser screenshots
Browser screenshots show the coverage by estimated fragments; reads were extended to estimated (by MACS2) fragment length, normalized to total mapped read number, and displayed as coverage on a mirror of the University of California, Santa Cruz (UCSC) genome browser.
Peak comparison
MAnorm analysis was performed in BioWardrobe using peaks identified with MACS2 (Shao et al., 2012). To define NORs and EORs, differential fold enrichment >2 and FDR ≤0.05 thresholds were used. For identifying “common” open chromatin regions (CORs), merged common regions from MAnorm analysis with FDR >0.05 were used.
Boxplots and tag density
Uniquely mapped reads in ATAC-seq and ChIP-seq samples were extended to MACS2 estimated fragment length. At each nucleotide position, the number of reads mapping there per million of total uniquely mapped reads was calculated and converted into average density over the peak. The density was used for boxplots, heatmaps, density profiles, and scatter plots.
ChIP-seq normalization by Drosophila melanogaster genomic DNA
Because ChIP-seq is a relative method (Lovén et al., 2012), in the situations where global change in protein levels occurs (e.g., AP-1 in GFP-electroporated vs. A-FOS–electroporated T cells), an external normalization is required. To compare ChIP enrichment for AP-1 and NFAT1 between GFP- and A-FOS–electroporated cells (Fig. 5 G) or cells with or without CD28 co-stimulation (Fig. 6 D and S5 B), normalization was performed by spiking in Drosophila genomic DNA. Immunoprecipitated DNA was mixed with 10 pg Drosophila genomic DNA, which was tagmented by Nextera Tn5 enzyme, before amplification. After library amplification of the DNA, the reads were aligned to the dm3 Drosophila genome and the hg19 human genome. The mapping rate to dm3 is ∼2%, and the number of mapped reads was used for normalizing tag density between the samples.
Differential expression analysis
Differential expression in RNA-seq between samples was analyzed by DEseq2 software (Love et al., 2014).
Motif enrichment analysis
Isolated genomic coordinates of open chromatin regions were used for identifying TF motifs by HOMER software (Heinz et al., 2010). We ran HOMER with the empirical datasets together with the control datasets, which comprised the same length of DNA sequence from the flanking regions within 20,000 bp of each region.
GO enrichment analysis and GSEA
Isolated genes nearby open chromatin regions were processed with the ToppFun function in the ToppGene Suite (Chen et al., 2007) website (https://toppgene.cchmc.org/enrichment.jsp) for ontology enrichment analysis. Gene set enrichment analysis (GSEA) was performed with manually defined gene sets located next to the open chromatin regions (Mootha et al., 2003; Subramanian et al., 2005).
Identifying SEs
For identifying SEs, ChIP-seq of H3K27ac and the same criteria and software as described in Whyte et al. (2013) and Lovén et al. (2013) were used.
RELI
Briefly, SNPs that are in linkage disequilibrium with index risk SNPs for various diseases from the National Institutes of Health genome-wide association study (GWAS) catalog were identified. RELI software was used to calculate their overlap with ChIP-seq and ATAC-seq peaks with risk loci (Harley et al., 2018).
Analysis of H3K4me2 data in patients with asthma
H3K4me2 data in Th2 cells from patient donors with asthma and healthy controls were collected from GSE53646 (Seumois et al., 2014) and initially processed in BioWardrobe as described above. H3K4me2 data are shown in the WUSTL genome browser within BioWardrobe (Zhou et al., 2011). The H3K4me2 differential enrichment domains between patients with asthma and healthy controls were identified by using rgt-THOR software (Allhoff et al., 2016).
A-FOS and GFP protein preparation
A-FOS and GFP cDNA fragments were amplified from CMV500 A-FOS (#33353; Addgene) and pmaxGFP (Lonza) vectors by PCR, and a 6xHis-GST DNA fragment was also amplified from pGEX vector. A-FOS, GFP, and 6xHis-GST were individually mixed with digested plasmid DNA (GE Healthcare Life Sciences) with NcoI restriction enzyme in an InFusion cloning reaction (TakaraBio). The resulting plasmid was transformed into the BL21 bacterial strain. A single colony was picked and transferred to 10 ml TB-SB (Terrific Broth–Super Broth) medium (TB/SB 1:1) with Kanamycin, cultured overnight at 37°C, transferred to 500 ml TB-SB medium, and cultured until the optical density at 600 nm reached 0.8. For protein expression, isopropyl β-D-1-thiogalactopyranoside was added into the culture to a final concentration of 0.5 mM, and bacteria were cultured for another 6–8 h at 34°C. The bacteria were spun down, washed with PBS twice, and stored at −80°C. The bacterial pellet was resuspended in 20 ml lysis buffer (50 mM Na2HPO4, pH 8, 300 mM NaCl, 10% glycerol, 0.1% Triton X-100, and 20 mM imidazole with the addition of protease inhibitors [#04693159001; Roche], lysozyme [100 µg/ml], DNaseI [0.2 μ/ml], and β-mercaptoethanol to 1 mM), sonicated with a probe sonicator, and then frozen, thawed, and sonicated again. The lysate was cleared by centrifugation and loaded on a 5-ml HisTrap HP column (#17524801; GE Healthcare Life Sciences) in Buffer A (50 mM Na2HPO4, pH 8, 300 mM NaCl, and 20 mM imidazole) using AKTA Start (GE Healthcare Life Sciences). The bound proteins were eluted by the gradient of Buffer B (50 mM Na2HPO4, pH 8, 300 mM NaCl, and 500 mM imidazole). The fractions containing protein were combined and subjected to dialysis against PBS with 1 mM β-mercaptoethanol overnight. After dialysis, Triton X-100 was added to the protein solution, and the His6-GST tag was cut with His-tagged HRV3C protease (#SAE0045; Sigma) at RT for 5 h. To remove the tags and the protease, imidazole was added to 20 mM, the solution was again passed through a 5-ml HisTrap HP column, and the flow-through was collected. The purity was confirmed by SDS-PAGE (data not shown). The Endotoxin Removal Beads (#130-093-657; Miltenyi Biotec) were used to remove endotoxins.
Protein electroporation
A-FOS and GFP proteins were electroporated using Neon Nucleofector (Invitrogen) and the Neon Transfection System 100 µl Kit (#MPK10025; Invitrogen). Isolated naive cells (3.5 million cells) were mixed with 25 µg A-FOS or GFP in T buffer and pulsed (voltage, 2,500 V; width, 15 ms; one pulse). For activation, the cells were stimulated 2 h after electroporation as described above.
Online supplemental material
Fig. S1 shows an experimental scheme and characterization of NORs and CORs. Fig. S2 shows DNA-binding motifs enriched in open chromatin regions. Fig. S3 shows combinatorial effects of AP-1 and NFAT1 to H3K27ac and openness. Fig. S4 shows detection and characterization of SEs in naive and activated T cells. Fig. S5 shows expression and chromatin changes during anergy induction in the absence of co-stimulation. Table S1 lists significant GWAS datasets overlapping NORs, CORs, or EORs.
Supplementary Material
List of significant GWAS datasets overlapping NORs, CORs, or EORs
Acknowledgments
We thank the Cincinnati Children’s Hospital Medical Center DNA Sequencing Core for the next-generation sequencing; R. Giulitto, C. Habel, and all blood donors at the University of Cincinnati Hoxworth Blood Center for blood donations; and S. Hottinger for editorial assistance.
This research was supported in part by the American Association of Immunologists Careers in Immunology Fellowship to M. Yukawa; the National Institutes of Health Director’s New Innovator Award (DP2 GM119134) to A. Barski; National Institutes of Health P30 grant DK078392; and National Institute of Neurological Disorders and Stroke grant R01 NS099068, the Lupus Research Alliance (“Novel Approaches”), the Cincinnati Children’s Research Foundation (Endowed Scholar), the Cincinnati Children’s Hospital Medical Center Center for Pediatric Genomics Pilot Study, and Cincinnati Children’s Hospital Medical Center Trustee Awards to M.T. Weirauch.
A.V. Kartashov and A. Barski are co-founders of Datirium, LLC. Datirium provides installation and support services for the BioWardrobe platform used in this paper. The remaining authors declare no competing financial interests.
Author contributions: M. Yukawa conducted experiments with support from S. Jagannathan, S. Vallabh, and A. Barski and performed data analysis with support from A.V. Kartashov, X. Chen, M.T. Weirauch, and A. Barski; M. Yukawa and A. Barski conceived the project and wrote the manuscript; and all authors read the paper.
References
- Allhoff M., Seré K., F Pires J., Zenke M., and G Costa I.. 2016. Differential peak calling of ChIP-seq signals with replicates with THOR. Nucleic Acids Res. 44:e153 10.1093/nar/gkw680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong E.J., and Bischoff J.. 2004. Heart valve development: endothelial cell signaling and differentiation. Circ. Res. 95:459–470. 10.1161/01.RES.0000141146.95728.da [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A., Jothi R., Cuddapah S., Cui K., Roh T.Y., Schones D.E., and Zhao K.. 2009. Chromatin poises miRNA- and protein-coding genes for expression. Genome Res. 19:1742–1751. 10.1101/gr.090951.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevington S.L., Cauchy P., Piper J., Bertrand E., Lalli N., Jarvis R.C., Gilding L.N., Ott S., Bonifer C., and Cockerill P.N.. 2016. Inducible chromatin priming is associated with the establishment of immunological memory in T cells. EMBO J. 35:515–535. 10.15252/embj.201592534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddie S.C., John S., Sabo P.J., Thurman R.E., Johnson T.A., Schiltz R.L., Miranda T.B., Sung M.-H., Trump S., Lightman S.L., et al. . 2011. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol. Cell. 43:145–155. 10.1016/j.molcel.2011.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., and Greenleaf W.J.. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 10:1213–1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Glover J.N., Hogan P.G., Rao A., and Harrison S.C.. 1998. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature. 392:42–48. 10.1038/32100 [DOI] [PubMed] [Google Scholar]
- Chen J., Xu H., Aronow B.J., and Jegga A.G.. 2007. Improved human disease candidate gene prioritization using mouse phenotype. BMC Bioinformatics. 8:392 10.1186/1471-2105-8-392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C., Pinto A.K., Curtis J.D., Persaud S.P., Cella M., Lin C.-C., Edelson B.T., Allen P.M., Colonna M., Pearce E.L., et al. . 2014. c-Myc-induced transcription factor AP4 is required for host protection mediated by CD8+ T cells. Nat. Immunol. 15:884–893. 10.1038/ni.2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree G.R., and Olson E.N.. 2002. NFAT signaling: choreographing the social lives of cells. Cell. 109(2, Suppl):S67–S79. 10.1016/S0092-8674(02)00699-2 [DOI] [PubMed] [Google Scholar]
- Djuretic I.M., Levanon D., Negreanu V., Groner Y., Rao A., and Ansel K.M.. 2007. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat. Immunol. 8:145–153. 10.1038/ni1424 [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant L., Watford W.T., Ramos H.L., Laurence A., Vahedi G., Wei L., Takahashi H., Sun H.-W., Kanno Y., Powrie F., and O’Shea J.J.. 2010. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 32:605–615. 10.1016/j.immuni.2010.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J., Kheradpour P., Mikkelsen T.S., Shoresh N., Ward L.D., Epstein C.B., Zhang X., Wang L., Issner R., Coyne M., et al. . 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 473:43–49. 10.1038/nature09906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh K.K.-H., Marson A., Zhu J., Kleinewietfeld M., Housley W.J., Beik S., Shoresh N., Whitton H., Ryan R.J.H., Shishkin A.A., et al. . 2015. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 518:337–343. 10.1038/nature13835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fathman C.G., and Lineberry N.B.. 2007. Molecular mechanisms of CD4+ T-cell anergy. Nat. Rev. Immunol. 7:599–609. 10.1038/nri2131 [DOI] [PubMed] [Google Scholar]
- Fleischmann A., Hafezi F., Elliott C., Remé C.E., Rüther U., and Wagner E.F.. 2000. Fra-1 replaces c-Fos-dependent functions in mice. Genes Dev. 14:2695–2700. 10.1101/gad.187900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruda M.C., van Amsterdam J., Rizzo C.A., Durham S.K., Lira S., and Bravo R.. 1996. Expression of FosB during mouse development: normal development of FosB knockout mice. Oncogene. 12:2177–2185. [PubMed] [Google Scholar]
- Harley J.B., Chen X., Pujato M., Miller D., Maddox A., Forney C., Magnusen A.F., Lynch A., Chetal K., Yukawa M., et al. . 2018. Transcription factors operate across disease loci, with EBNA2 implicated in autoimmunity. Nat. Genet. 50:699–707. 10.1038/s41588-018-0102-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins R.D., Larjo A., Tripathi S.K., Wagner U., Luu Y., Lönnberg T., Raghav S.K., Lee L.K., Lund R., Ren B., et al. . 2013. Global chromatin state analysis reveals lineage-specific enhancers during the initiation of human T helper 1 and T helper 2 cell polarization. Immunity. 38:1271–1284. 10.1016/j.immuni.2013.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., and Glass C.K.. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38:576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Yamauchi M., Nishina M., Yamamichi N., Mizutani T., Ui M., Murakami M., and Iba H.. 2001. Identification of SWI.SNF complex subunit BAF60a as a determinant of the transactivation potential of Fos/Jun dimers. J. Biol. Chem. 276:2852–2857. 10.1074/jbc.M009633200 [DOI] [PubMed] [Google Scholar]
- Jain J., Nalefski E.A., McCaffrey P.G., Johnson R.S., Spiegelman B.M., Papaioannou V., and Rao A.. 1994. Normal peripheral T-cell function in c-Fos-deficient mice. Mol. Cell. Biol. 14:1566–1574. 10.1128/MCB.14.3.1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauliac S., López-Rodriguez C., Shaw L.M., Brown L.F., Rao A., and Toker A.. 2002. The role of NFAT transcription factors in integrin-mediated carcinoma invasion. Nat. Cell Biol. 4:540–544. 10.1038/ncb816 [DOI] [PubMed] [Google Scholar]
- Kanhere A., Hertweck A., Bhatia U., Gökmen M.R., Perucha E., Jackson I., Lord G.M., and Jenner R.G.. 2012. T-bet and GATA3 orchestrate Th1 and Th2 differentiation through lineage-specific targeting of distal regulatory elements. Nat. Commun. 3:1268 10.1038/ncomms2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartashov A.V., and Barski A.. 2015. BioWardrobe: an integrated platform for analysis of epigenomics and transcriptomics data. Genome Biol. 16:158 10.1186/s13059-015-0720-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori H.K., Hart T., LaMere S.A., Chew P.V., and Salomon D.R.. 2015. Defining CD4 T cell memory by the epigenetic landscape of CpG DNA methylation. J. Immunol. 194:1565–1579. 10.4049/jimmunol.1401162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegel M.A., Rathinam C., and Flavell R.A.. 2009. E3 ubiquitin ligase GRAIL controls primary T cell activation and oral tolerance. Proc. Natl. Acad. Sci. USA. 106:16770–16775. 10.1073/pnas.0908957106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C.T., Veselits M.L., and Leiden J.M.. 1997. LKLF: A transcriptional regulator of single-positive T cell quiescence and survival. Science. 277:1986–1990. 10.1126/science.277.5334.1986 [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., and Salzberg S.L.. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.D., Bingham K.N., Mitchell T.Y., Meredith J.L., and Rawlings J.S.. 2015. Calcium mobilization is both required and sufficient for initiating chromatin decondensation during activation of peripheral T-cells. Mol. Immunol. 63:540–549. 10.1016/j.molimm.2014.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Berry C.T., Ruthel G., Madara J.J., MacGillivray K., Gray C.M., Madge L.A., McCorkell K.A., Beiting D.P., Hershberg U., et al. . 2016. T cell receptor-induced nuclear factor κB (NF-κB) signaling and transcriptional activation are regulated by STIM1- and Orai1-mediated calcium entry. J. Biol. Chem. 291:8440–8452. 10.1074/jbc.M115.713008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovén J., Orlando D.A., Sigova A.A., Lin C.Y., Rahl P.B., Burge C.B., Levens D.L., Lee T.I., and Young R.A.. 2012. Revisiting global gene expression analysis. Cell. 151:476–482. 10.1016/j.cell.2012.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovén J., Hoke H.A., Lin C.Y., Lau A., Orlando D.A., Vakoc C.R., Bradner J.E., Lee T.I., and Young R.A.. 2013. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 153:320–334. 10.1016/j.cell.2013.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macián F., López-Rodríguez C., and Rao A.. 2001. Partners in transcription: NFAT and AP-1. Oncogene. 20:2476–2489. 10.1038/sj.onc.1204386 [DOI] [PubMed] [Google Scholar]
- Macián F., García-Cózar F., Im S.-H., Horton H.F., Byrne M.C., and Rao A.. 2002. Transcriptional mechanisms underlying lymphocyte tolerance. Cell. 109:719–731. 10.1016/S0092-8674(02)00767-5 [DOI] [PubMed] [Google Scholar]
- Mazzoni A., Santarlasci V., Maggi L., Capone M., Rossi M.C., Querci V., De Palma R., Chang H.D., Thiel A., Cimaz R., et al. . 2015. Demethylation of the RORC2 and IL17A in human CD4+ T lymphocytes defines Th17 origin of nonclassic Th1 cells. J. Immunol. 194:3116–3126. 10.4049/jimmunol.1401303 [DOI] [PubMed] [Google Scholar]
- Miraldi E.R., Pokrovskii M., Watters A., Castro D.M., De Veaux N., Hall J.A., Lee J.-Y., Ciofani M., Madar A., Carriero N., et al. . 2019. Leveraging chromatin accessibility for transcriptional regulatory network inference in T Helper 17 Cells. Genome Res. 29:449–463. 10.1101/gr.238253.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha V.K., Lindgren C.M., Eriksson K.-F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., et al. . 2003. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34:267–273. 10.1038/ng1180 [DOI] [PubMed] [Google Scholar]
- Mukasa R., Balasubramani A., Lee Y.K., Whitley S.K., Weaver B.T., Shibata Y., Crawford G.E., Hatton R.D., and Weaver C.T.. 2010. Epigenetic instability of cytokine and transcription factor gene loci underlies plasticity of the T helper 17 cell lineage. Immunity. 32:616–627. 10.1016/j.immuni.2010.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthusamy N., Barton K., and Leiden J.M.. 1995. Defective activation and survival of T cells lacking the Ets-1 transcription factor. Nature. 377:639–642. 10.1038/377639a0 [DOI] [PubMed] [Google Scholar]
- Nguyen T.N., Kim L.J., Walters R.D., Drullinger L.F., Lively T.N., Kugel J.F., and Goodrich J.A.. 2010. The C-terminal region of human NFATc2 binds cJun to synergistically activate interleukin-2 transcription. Mol. Immunol. 47:2314–2322. 10.1016/j.molimm.2010.05.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea J.J., and Paul W.E.. 2010. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 327:1098–1102. 10.1126/science.1178334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura N., Hamaguchi M., Morikawa H., Sugimura K., Tanaka A., Ito Y., Osaki M., Tanaka Y., Yamashita R., Nakano N., et al. . 2012. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity. 37:785–799. 10.1016/j.immuni.2012.09.010 [DOI] [PubMed] [Google Scholar]
- Ono M., Yaguchi H., Ohkura N., Kitabayashi I., Nagamura Y., Nomura T., Miyachi Y., Tsukada T., and Sakaguchi S.. 2007. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature. 446:685–689. 10.1038/nature05673 [DOI] [PubMed] [Google Scholar]
- Panagoulias I., Georgakopoulos T., Aggeletopoulou I., Agelopoulos M., Thanos D., and Mouzaki A.. 2016. Transcription factor Ets-2 acts as a preinduction repressor of interleukin-2 (IL-2) transcription in naive T helper lymphocytes. J. Biol. Chem. 291:26707–26721. 10.1074/jbc.M116.762179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham D., Yu Q., Walline C.C., Muthukrishnan R., Blum J.S., and Kaplan M.H.. 2013. Opposing roles of STAT4 and Dnmt3a in Th1 gene regulation. J. Immunol. 191:902–911. 10.4049/jimmunol.1203229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranger A.M., Gerstenfeld L.C., Wang J., Kon T., Bae H., Gravallese E.M., Glimcher M.J., and Glimcher L.H.. 2000. The nuclear factor of activated T cells (NFAT) transcription factor NFATp (NFATc2) is a repressor of chondrogenesis. J. Exp. Med. 191:9–22. 10.1084/jem.191.1.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings J.S., Gatzka M., Thomas P.G., and Ihle J.N.. 2011. Chromatin condensation via the condensin II complex is required for peripheral T-cell quiescence. EMBO J. 30:263–276. 10.1038/emboj.2010.314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochman Y., Yukawa M., Kartashov A.V., and Barski A.. 2015. Functional characterization of human T cell hyporesponsiveness induced by CTLA4-Ig. PLoS One. 10:e0122198–e18. 10.1371/journal.pone.0122198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney J.W., Sun Y.L., Glimcher L.H., and Hoey T.. 1995. Novel NFAT sites that mediate activation of the interleukin-2 promoter in response to T-cell receptor stimulation. Mol. Cell. Biol. 15:6299–6310. 10.1128/MCB.15.11.6299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ B.E., Prier J.E., Rao S., and Turner S.J.. 2013. T cell immunity as a tool for studying epigenetic regulation of cellular differentiation. Front. Genet. 4:218 10.3389/fgene.2013.00218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safford M., Collins S., Lutz M.A., Allen A., Huang C.-T., Kowalski J., Blackford A., Horton M.R., Drake C., Schwartz R.H., and Powell J.D.. 2005. Egr-2 and Egr-3 are negative regulators of T cell activation. Nat. Immunol. 6:472–480. 10.1038/ni1193 [DOI] [PubMed] [Google Scholar]
- Sareneva T., Matikainen S., Vanhatalo J., Melén K., Pelkonen J., and Julkunen I.. 1998. Kinetics of cytokine and NFAT gene expression in human interleukin-2-dependent T lymphoblasts stimulated via T-cell receptor. Immunology. 93:350–357. 10.1046/j.1365-2567.1998.00440.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidl C., Rendeiro A.F., Sheffield N.C., and Bock C.. 2015. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat. Methods. 12:963–965. 10.1038/nmeth.3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimata M., Pérez-Melgosa M., Miller S.A., Weinmann A.S., Sabo P.J., Sandstrom R., Dorschner M.O., Stamatoyannopoulos J.A., and Wilson C.B.. 2009. CCCTC-binding factor and the transcription factor T-bet orchestrate T helper 1 cell-specific structure and function at the interferon-gamma locus. Immunity. 31:551–564. 10.1016/j.immuni.2009.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seumois G., Chavez L., Gerasimova A., Lienhard M., Omran N., Kalinke L., Vedanayagam M., Ganesan A.P.V., Chawla A., Djukanović R., et al. . 2014. Epigenomic analysis of primary human T cells reveals enhancers associated with TH2 memory cell differentiation and asthma susceptibility. Nat. Immunol. 15:777–788. 10.1038/ni.2937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z., Zhang Y., Yuan G.-C., Orkin S.H., and Waxman D.J.. 2012. MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets. Genome Biol. 13:R16 10.1186/gb-2012-13-3-r16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerka C., Decker E.L., and Zipfel P.F.. 1995. A regulatory element in the human interleukin 2 gene promoter is a binding site for the zinc finger proteins Sp1 and EGR-1. J. Biol. Chem. 270:22500–22506. 10.1074/jbc.270.38.22500 [DOI] [PubMed] [Google Scholar]
- Smith A.E., Chronis C., Christodoulakis M., Orr S.J., Lea N.C., Twine N.A., Bhinge A., Mufti G.J., and Thomas N.S.B.. 2009. Epigenetics of human T cells during the G0-->G1 transition. Genome Res. 19:1325–1337. 10.1101/gr.085530.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderquest K., Hertweck A., Giambartolomei C., Henderson S., Mohamed R., Goldberg R., Perucha E., Franke L., Herrero J., Plagnol V., et al. . 2017. Genetic variants alter T-bet binding and gene expression in mucosal inflammatory disease. PLoS Genet. 13:e1006587–e23. 10.1371/journal.pgen.1006587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaker Y.R., Schneider H., and Rudd C.E.. 2015. TCR and CD28 activate the transcription factor NF-κB in T-cells via distinct adaptor signaling complexes. Immunol. Lett. 163:113–119. 10.1016/j.imlet.2014.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trushin S.A., Pennington K.N., Algeciras-Schimnich A., and Paya C.V.. 1999. Protein kinase C and calcineurin synergize to activate IkappaB kinase and NF-kappaB in T lymphocytes. J. Biol. Chem. 274:22923–22931. 10.1074/jbc.274.33.22923 [DOI] [PubMed] [Google Scholar]
- Turner H., Gomez M., McKenzie E., Kirchem A., Lennard A., and Cantrell D.A.. 1998. Rac-1 regulates nuclear factor of activated T cells (NFAT) C1 nuclear translocation in response to Fcepsilon receptor type 1 stimulation of mast cells. J. Exp. Med. 188:527–537. 10.1084/jem.188.3.527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahedi G., Takahashi H., Nakayamada S., Sun H.-W., Sartorelli V., Kanno Y., and O’Shea J.J.. 2012. STATs shape the active enhancer landscape of T cell populations. Cell. 151:981–993. 10.1016/j.cell.2012.09.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T., Ling E., Cowley C.J., Couch C.H., Wang X., Harmin D.A., Roberts C.W.M., and Greenberg M.E.. 2017. AP-1 transcription factors and the BAF complex mediate signal-dependent enhancer selection. Mol. Cell. 68:1067–1082.e12. 10.1016/j.molcel.2017.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters R.D., Drullinger L.F., Kugel J.F., and Goodrich J.A.. 2013. NFATc2 recruits cJun homodimers to an NFAT site to synergistically activate interleukin-2 transcription. Mol. Immunol. 56:48–56. 10.1016/j.molimm.2013.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan C.-K., Andraski A.B., Spolski R., Li P., Kazemian M., Oh J., Samsel L., Swanson P.A. II, McGavern D.B., Sampaio E.P., et al. . 2015. Opposing roles of STAT1 and STAT3 in IL-21 function in CD4+ T cells. Proc. Natl. Acad. Sci. USA. 112:9394–9399. 10.1073/pnas.1511711112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Zang C., Rosenfeld J.A., Schones D.E., Barski A., Cuddapah S., Cui K., Roh T.-Y., Peng W., Zhang M.Q., and Zhao K.. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 40:897–903. 10.1038/ng.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Dillon C.P., Shi L.Z., Milasta S., Carter R., Finkelstein D., McCormick L.L., Fitzgerald P., Chi H., Munger J., and Green D.R.. 2011. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 35:871–882. 10.1016/j.immuni.2011.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G., Wei L., Zhu J., Zang C., Hu-Li J., Yao Z., Cui K., Kanno Y., Roh T.-Y., Watford W.T., et al. . 2009. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 30:155–167. 10.1016/j.immuni.2008.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., and Young R.A.. 2013. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 153:307–319. 10.1016/j.cell.2013.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H.K., Kammer G.M., Dennis G., and Tsokos G.C.. 1999. Abnormal NF-kappa B activity in T lymphocytes from patients with systemic lupus erythematosus is associated with decreased p65-RelA protein expression. J. Immunol. 163:1682–1689. [PubMed] [Google Scholar]
- Xu Y.Z., Thuraisingam T., Marino R., and Radzioch D.. 2011. Recruitment of SWI/SNF complex is required for transcriptional activation of the SLC11A1 gene during macrophage differentiation of HL-60 cells. J. Biol. Chem. 286:12839–12849. 10.1074/jbc.M110.185637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T., Park C.S., Mamonkin M., and Lacorazza H.D.. 2009. Transcription factor ELF4 controls the proliferation and homing of CD8+ T cells via the Krüppel-like factors KLF4 and KLF2. Nat. Immunol. 10:618–626. 10.1038/ni.1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngblood B., Hale J.S., Kissick H.T., Ahn E., Xu X., Wieland A., Araki K., West E.E., Ghoneim H.E., Fan Y., et al. . 2017. Effector CD8 T cells dedifferentiate into long-lived memory cells. Nature. 552:404–409. 10.1038/nature25144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K.S., and Mango S.E.. 2016. Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr. Opin. Genet. Dev. 37:76–81. 10.1016/j.gde.2015.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., and Liu X.S.. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Maricque B., Xie M., Li D., Sundaram V., Martin E.A., Koebbe B.C., Nielsen C., Hirst M., Farnham P., et al. . 2011. The human epigenome browser at Washington University. Nat. Methods. 8:989–990. 10.1038/nmeth.1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., and Paul W.E.. 2010. Peripheral CD4+ T-cell differentiation regulated by networks of cytokines and transcription factors. Immunol. Rev. 238:247–262. 10.1111/j.1600-065X.2010.00951.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Yamane H., and Paul W.E.. 2010. Differentiation of effector CD4 T cell populations (*). Annu. Rev. Immunol. 28:445–489. 10.1146/annurev-immunol-030409-101212 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of significant GWAS datasets overlapping NORs, CORs, or EORs