Regulation of Bcl6 expression during follicular helper T cell differentiation remains incompletely understood. Here, Long et al. show that T cell activation induces H3K36me2 methyltransferase Nsd2, in a CD28- and ICOS-dependent manner, to promote Bcl6 expression and Tfh differentiation.
Abstract
Follicular helper T (Tfh) cells provide essential help for humoral immune response. Transcriptional factor Bcl6 is the master regulator for Tfh generation and is induced very early after T cell activation in a CD28-dependent manner, but how CD28 signal promotes Bcl6 early expression remains unknown. Here we found that CD28 signal quickly induces expression of the H3K36me2 methytransferase Nsd2, which is required for Bcl6 expression as early as the first cell division after T cell activation. Nsd2 deficiency in T cells leads to decreased Bcl6 expression, impaired Tfh generation, compromised germinal center response, and delayed virus clearance. Ectopic Bcl6 expression rescues the Tfh defect of Nsd2 KO cells. ICOS signal is dispensable for early Nsd2 induction but required for sustained Nsd2 expression, which is critical for Tfh maintenance. Overexpression of Nsd2 increases Bcl6 expression and enhances Tfh generation; 4-mo-old mice even develop spontaneous Tfh. Overall, our study reveals Nsd2 as a critical epigenetic regulator for Tfh differentiation.
Introduction
Upon antigen stimulation, naive T cells differentiate into functionally distinct subsets. Follicular helper T (Tfh) cells are the specialized subset providing essential help for B cells in germinal centers (GCs) and thus are predominantly required for efficient humoral immune response (Liu et al., 2013; Qi, 2016; Vinuesa et al., 2016). By facilitating high-affinity antibody generation, Tfh cells have an important role in protective immunity against pathogen infections. In addition, aberrant Tfh differentiation is implicated in antibody-mediated autoimmune diseases, such as systemic lupus erythematosus (Crotty, 2014; Ueno et al., 2015).
The transcription factor Bcl6 is selectively expressed in Tfh cells and has been demonstrated to be the master regulator for Tfh differentiation (Johnston et al., 2009; Nurieva et al., 2009; Yu et al., 2009), although other transcription factors such as ASCL2, IRF4, BATF, TCF1, and Maf are also required (Bauquet et al., 2009; Betz et al., 2010; Bollig et al., 2012; Choi et al., 2015; Liu et al., 2014). Bcl6 promotes Tfh generation by multiple mechanisms, including repressing expression of key transcription factors for helper T cell 1 (Th1), Th2, and Th17 differentiation; inhibiting Blimp1 expression; and down-regulating T cell egress molecules, thus facilitating Tfh migration toward the B cell follicle (Johnston et al., 2009; Nurieva et al., 2009; Yu et al., 2009). In contrast to our profound understanding of the mechanisms by which Bcl6 promotes Tfh differentiation, it remains much less clear how Bcl6 expression itself is regulated. Previous studies revealed that Bcl6 was induced as early as the first cell division after T cell activation, and that CD28 costimulatory signal from dendritic cell–priming was critically required for such Bcl6 early expression (Baumjohann et al., 2011; Weber et al., 2015). But how CD28 signal induces Bcl6 early expression is unknown. Another costimulatory signal, inducible T cell costimulator (ICOS), is not required for Bcl6 early induction (Weber et al., 2015) but could promote Bcl6 expression by inactivating Foxo1 and helping to maintain Tfh (Choi et al., 2011; Stone et al., 2015; Weber et al., 2015).
Epigenetic regulations including histone modifications could respond quickly to external stimuli and combine various signals, and thus play essential roles in cell differentiation and plasticity events (Allis and Jenuwein, 2016). During CD4+ T cell differentiation, specific epigenetic modifications, especially histone methylation, correlate with distinct lineages and play critical roles (Nakayamada et al., 2012; Wilson et al., 2009). In terms of Tfh differentiation, H3K27me3 modification has been revealed to be critically involved (Wei et al., 2009). UTX (ubiquitously transcribed tetratricopeptide repeat, X chromosome; KDM6A)-mediated H3K27me3 demethylation enforces Tfh-related gene expression such as IL6ra to sustain Tfh lineage, and deficiency in UTX leads to impaired GC B cell response and antibody production against chronic virus infection (Cook et al., 2015). In addition, the H3K27me3 methyltransferase Ezh2 is required for Tfh generation (Chen et al., 2019; Li et al., 2018). H3K36 methylation is another important histone modification and has gained attention in recent years (Wagner and Carpenter, 2012). Unlike H3K27me3 modification, which occurs mainly near the gene promoter region, H3K36 methylation, especially H3K36me2/3, decorates the gene body region and associates with active transcription (Wagner and Carpenter, 2012). Our previous study revealed that Nsd2 expressed in B cells is required for GC B cell selection by regulating adhesive interaction between GC B cells and follicular dendritic cells (Chen et al., 2018). However, the role of H3K36me in T cell differentiation has not been investigated yet.
Here we found that H3K36me2 modification mediated by methyltransferase Nsd2 is required for Bcl6 expression and Tfh differentiation. Nsd2 expression is quickly induced upon T cell activation in a CD28-dependent manner, and ICOS signal is required for sustained Nsd2 expression. Nsd2 deficiency leads to impaired Tfh generation and maintenance, whereas its overexpression increases Tfh differentiation and GC response.
Results
CD28-dependent Nsd2 is required for Bcl6 early expression in T cells
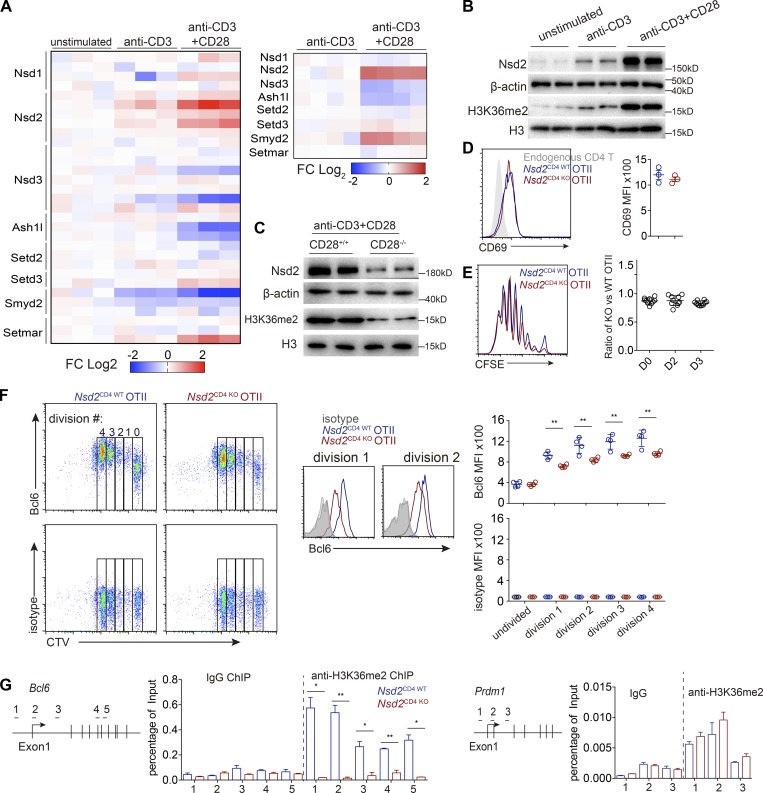
To explore how CD28 signal promotes Bcl6 early expression, we initially analyzed published gene expression profiles of anti-CD3 plus anti-CD28 stimulated T cells (DuPage et al., 2015). When we focused on the H3K36 methyltransferases, we found that only Nsd2 is significantly up-regulated by CD28 signal in both human and mouse T cells (Fig. 1 A). Nsd2 is a key methyltransferase for H3K36 dimethylation (H3K36me2), which is a permissive marker associated with transcriptional elongation (Kuo et al., 2011). At the protein level, CD3 plus CD28 stimulation also promoted Nsd2 expression much more strongly than CD3 stimulation alone (Fig. 1 B). The necessity of CD28 signal for efficient Nsd2 expression was also confirmed by stimulation experiments with CD28-deficient T cells (Fig. 1 C).
Figure 1.
CD28-dependent Nsd2 promotes Bcl6 early expression in activated T cells. (A) Heatmap depicting relative mRNA expression of eight known H3K36 methyltransferases in T cells stimulated as indicated (previously published data, Gene Expression Omnibus: GSE58998). Left, microarray analysis of human CD4+ T cells; right, RNA-seq analysis of mouse CD4+ T cells. (B) Immunoblotting analysis of Nsd2 protein in mouse CD4+ T cells after 24-h stimulation as indicated. (C) CD4+ T cells from WT or CD28-deficient mice were stimulated with anti-CD3 and anti-CD28 for 24 h and analyzed for Nsd2 expression. (D) Flow cytometry analysis of CD69 expression on WT and Nsd2-deficient OT II in transfer recipient spleens 2 d after OVA immunization. (E) Proliferation of cotransferred carboxyfluorescein succinimidyl ester (CFSE)–labeled OT II T cells at different time points after OVA immunization. FACS plot shows the representative CFSE dilution at day 3. (F) CTV-labeled OT II T cells were adoptively transferred into WT recipients, followed by subcutaneous immunization with OVA/Alum, and then stained for Bcl6 or isotype control 48 h later (left). Histograms show overlay of Bcl6 in WT and Nsd2 KO T cells in first and second cell division (middle). Mean fluorescence intensity (MFI) is summarized on right. (G) WT and Nsd2-deficient OT II T cells were transferred into recipients, followed by OVA/Alum immunization. OT II T cells were then sorted out at day 3 for ChIP-qPCR analysis of H3K36me2 modification on the Bcl6 and Prdm1 gene loci. Data are representative of three (D–F) or two (B, C, and G) experiments. For bar graphs, bars represent means, and dots represent individual mice. Statistical analysis was done with Student’s t test. Error bars show means ± SD. *, P < 0.05; **, P < 0.01.
We then generated T cell–specific Nsd2-deficient mice by crossing CD4Cre to Nsd2 floxed mice and obtained CD4Cre+Nsd2fl/fl mice (Nsd2CD4 KO mice). Compared with control Nsd2CD4 WT mice (either CD4Cre+Nsd2+/+ or CD4Cre-Nsd2fl/fl), deletion of Nsd2 in T cells did not lead to significant T cell development defects (Fig. S1, A–C). We crossed the mice with an OT II TCR transgenic background and performed adoptive transfer experiments to examine Bcl6 early expression during cell division, as previously reported (Baumjohann et al., 2011). Although T cell activation and proliferation were not affected by Nsd2 ablation (Fig. 1, D and E), we found that Bcl6 expression was dramatically impaired as early as the first cell division after T cell activation in the absence of Nsd2 (Fig. 1 F). Chromatin immunoprecipitation (ChIP) analysis showed that Nsd2 deletion severely reduced H3K36me2 modification across the Bcl6 gene body, suggesting that Nsd2 potentially directly regulates Bcl6 expression (Fig. 1 G). As a control, the H3K36me2 modifications on other Tfh related transcription factors, such as Prdm1, Ascl2, and Maf, were not affected by Nsd2 deletion (Figs. 1 G and S4). Overall, these data revealed that, during T cell activation, Nsd2 expression is up-regulated in a CD28-dependent manner, which then promotes Bcl6 early induction.
Figure S1.
Nsd2 is not required for T cell development. (A) Thymocyte development analysis by flow cytometry. (B) T cell cellularity in spleen and lymph nodes. (C) T reg cell analysis in spleen. In A–C, bars represent means, and dots represent individual mice. Error bars show means ± SD. Statistical analysis was done with Student’s t test. ns, not significant; DP, CD4+CD8+ double positive; pLN, peripheral lymph node.
Figure S4.
ChIP-qPCR for H3K36me2 modifications on Tfh-related genes. OT II T cells from Fig. 1 G were used for experiments. Data were all representative of two experiments. Error bars show means ± SD.
Nsd2 is specifically required for Tfh differentiation and GC response
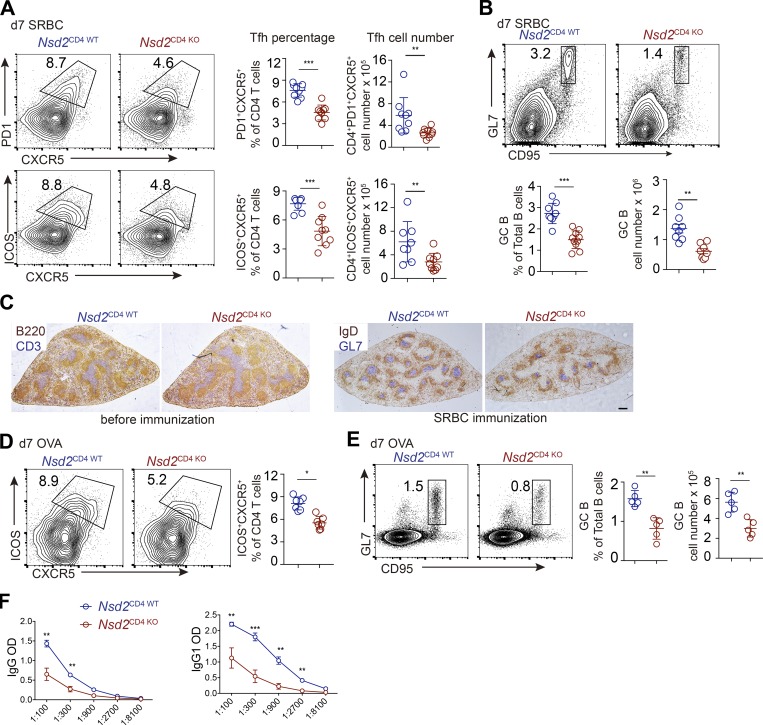
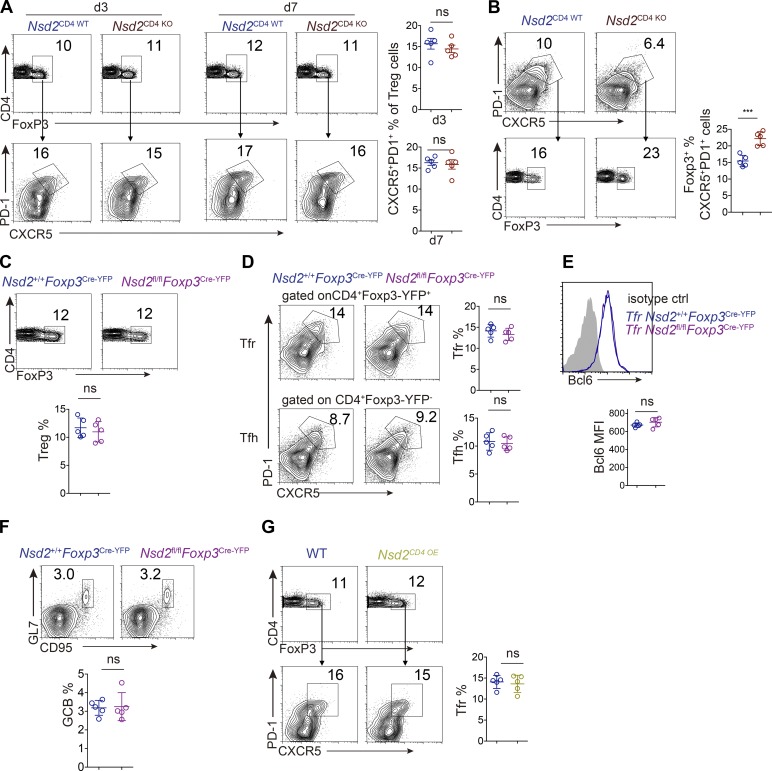
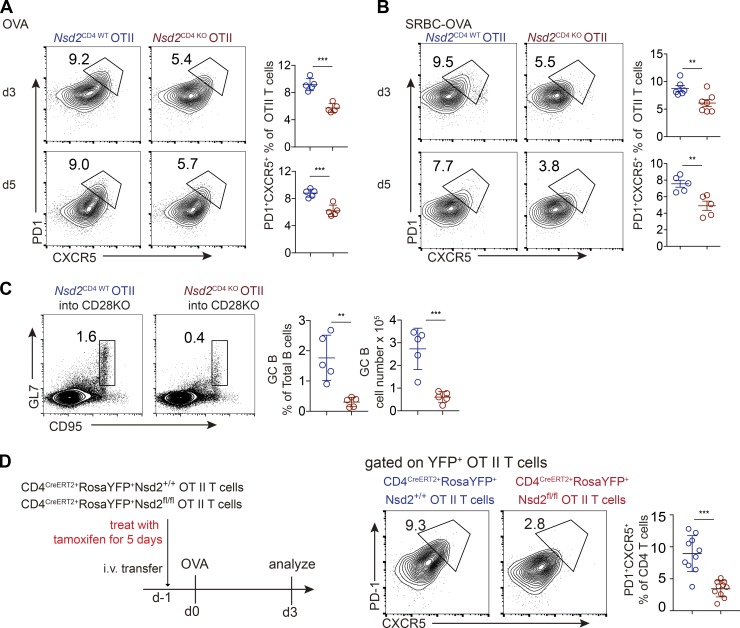
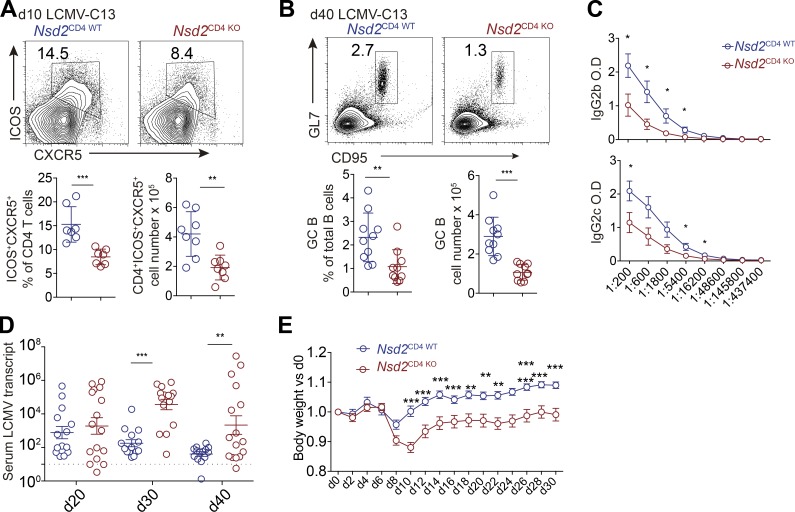
Since Nsd2 promotes Bcl6 early expression, we assessed the role of Nsd2 in Tfh differentiation. Upon particulate antigen sheep red blood cell (SRBC) immunization, Tfh generation was significantly decreased in Nsd2CD4 KO mice compared with Nsd2CD4 WT mice, by either CXCR5+PD1+ or CXCR5+ICOS+ staining (Fig. 2 A). Accordingly, the GC B cell response was also strongly reduced in Nsd2CD4 KO mice (Fig. 2 B). The reduced GC response was also revealed by immunohistological staining. T cell–specific deletion of Nsd2 did not affect the splenic structure before immunization, whereas it led to severely decreased GC on sections (Fig. 2 C). When the mice were subjected to soluble antigen OVA immunization, the Tfh differentiation and GC response were also significantly reduced compared with Nsd2CD4 WT mice (Fig. 2, D and E). Consistent with decreased GC response, the OVA specific antibody in serum from OVA immunized Nsd2CD4 KO mice was dramatically reduced (Fig. 2 F). Follicular regulatory T cells (Tfr) are a subset of regulatory T cells (T reg cells), which also express Bcl6 (Chung et al., 2011; Linterman et al., 2011). Although both localize in B cell follicles, Tfh and Tfr cells have opposite functions and derive from distinct progenitors (Sage and Sharpe, 2016). We analyzed Tfr differentiation by staining CXCR5/PD1 among Foxp3+ cells in Nsd2CD4 KO mice and, interestingly, found that Tfr differentiation was not affected by Nsd2 ablation (Fig. S2 A). When we first gated on the reduced CXCR5+PD1+ fraction and then applied the Foxp3 gate, we observed increased Foxp3+ percentages (Fig. S2 B), suggesting that Nsd2 is specifically required for Tfh differentiation, but not for Tfr differentiation. To further validate whether Nsd2 regulates Tfr cells, we crossed Nsd2fl/fl mice with Foxp3Cre-YFP mice to obtain T reg cell– and Tfr-specific conditional KO mice. In these mice, we found that T reg, Tfr, Tfh, and GC B cell responses were all normal (Fig. S2, C–F). Therefore, we concluded that Nsd2 is specifically required for Tfh differentiation but not for Tfr cells. Our data suggest that the Bcl6 regulation mechanism might be different in Tfh and Tfr cells. In fact, a previous study revealed distinct Bcl6 expression levels in Tfh and Tfr cells, with the latter expressing lower amounts, and Tfr cells even express a fair amount of Blimp1, which is devoid in Tfh (Linterman et al., 2011), also suggesting potentially different mechanisms for Bcl6 regulation in these two subset cells.
Figure 2.
Nsd2 loss impaired Tfh differentiation and GC B cell response. (A and B) Nsd2CD4WT and Nsd2CD4KO mice were immunized with SRBCs and analyzed by flow cytometry for Tfh (A) or GC B cells (GCB; B) at day 7. Data are representative of three independent experiments. (C) Immunohistological analysis of splenic cryosections before and after SRBC immunization. B220 stains for B cells; CD3 stains for T cells; IgD stains for naive B cells; and GL7 stains for GCB cells. Scale bar: 100 mm. (D and E) Nsd2CD4WT and Nsd2CD4KO mice were immunized with OVA and analyzed by flow cytometry for Tfh (D) or GCB (E) at day 7. Data are representative of three independent experiments. (F) Anti-OVA specific antibody in the serum from OVA immunized mice in D and E was analyzed by ELISA. In A, B, D, and E, bars represent means, and dots represent individual mice. Statistical analysis was done with Student’s t test; error bars show means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Figure S2.
Tfr cell differentiation analysis. (A and B) Tfr analysis by flow cytometry with splenocytes from SRBC immunized Nsd2CD4WT and NSD2CD4KO mice at day 7. (C–F) Analysis of T reg cell (C), Tfr and Tfh (D), and Bcl6 expression (E) and GC B cell response (F) in Nsd2+/+ Foxp3Cre-YFP, and Nsd2fl/fl Foxp3Cre-YFP mice immunized with SRBC for 7 d. (G) Analysis of Tfr cells in Nsd2 OE mice at day 7 after SRBC immunization. Data are all representative of more than three experiments. In all bar graphs, bars represent means, and dots represent individual mice. Error bars show means ± SD. Statistical analysis was done with Student’s t test. ns, not significant; ***, P < 0.001.
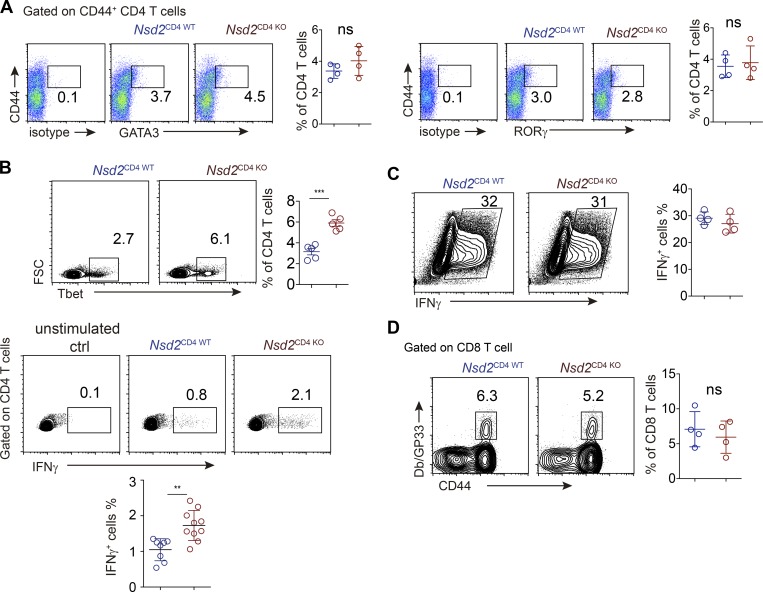
We also analyzed cell differentiation of other T cell lineages and found that Th2 and Th17 differentiation were unaffected in the absence of Nsd2 (Fig. S3 A). Interestingly, Nsd2 deficiency led to increased Th1 differentiation, as revealed by both T-bet staining and cytokine staining (Fig. S3 B). Given that Nsd2 regulates Bcl6 expression as early as the first cell division after T cell activation, and previous studies reported that during early T cell fate determination, a bifurcation between Tfh and Th1 occurs (Choi et al., 2011; Shen et al., 2018), we reasoned that the increased Th1 lineage could be due to skewed Tfh differentiation under conditions permissive for both Tfh and Th1 differentiation. To test this hypothesis, we analyzed T cell differentiation under Th1-prone conditions by in vitro differentiation experiments. The data showed that Nsd2 KO and WT T cells differentiated into Th1 at comparable levels (Fig. S3 C), suggesting that Nsd2 ablation has no effect on the bona fide capacity of Th1 lineage decision. Overall, these observations revealed a specific role of Nsd2 for Tfh lineage differentiation, but not for other T cell lineages.
Figure S3.
Th cell differentiation analysis. (A) Th2 and Th17 analysis of CD4 T cells from Nsd2CD4WT and NSD2CD4KO mice immunized with NP-OVA/Alum/LPS. (B) Th1 cell differentiation analysis by transcriptional factor staining (upper) or cytokine staining (lower) in splenic CD4+ T cells from NP-OVA/Alum/LPS immunized mice. For cytokine analysis, cells were restimulated with PMA/ionomycin for 4 h. (C) In vitro Th1 differentiation analysis. Sorted naive CD4+ T cells of indicated genotypes were stimulated with anti-CD3 and anti-CD28 in the presence of IL12 (10 ng/ml), IFNγ (10 ng/ml), and anti-IL4 (5 µg/ml) for 4 d. Cells were restimulated with PMA/ionomycin/GolgiPlug for 4 h before FACS staining for INFγ. Data are all representative of more than two experiments. (D) GP33-tetramer staining for splenic LCMV-specific CD8 T cells in mice infected with LCMV-cl13 10 d earlier. In all bar graphs, bars represent means, and dots represent individual mice. Error bars show means ± SD. Statistical analysis was done with Student’s t test. ns, not significant; **, P < 0.01; ***, P < 0.001. FSC, forward scatter.
Nsd2 promotes Tfh differentiation in a cell-intrinsic manner
Using the T cells from OT II transgenic Nsd2CD4 KO mice, we performed adoptive transfer experiments to analyze whether the role of Nsd2 in Tfh differentiation is cell intrinsic. Nsd2CD4 WT and Nsd2CD4 KO OT II T cells were transferred into congenically distinct recipient mice, and the mice were then immunized with either soluble OVA or particulate SRBC-OVA. We found that Tfh differentiation at day 3 or 5 was significantly reduced in Nsd2 KO T cells compared with WT T cells upon both forms of immunization (Fig. 3, A and B). To assess the functional consequence of such Tfh defects under OT II background, we transferred OT II T cells of each genotype into CD28 KO recipients (the endogenous CD28-deficient T cells cannot generate Tfh for B cell help), immunized the mice with OVA conjugated to 4-hydroxy-3-nitrophenylacetyl (NP-OVA), and analyze GC response 7 d later. The data showed dramatically reduced GC response in CD28 KO mice transferred with Nsd2-deficient T cells compared with WT T cells (Fig. 3 C).
Figure 3.
Nsd2 is required for Tfh differentiation in a cell-intrinsic manner. (A and B) Nsd2CD4WT and Nsd2CD4KO OT II T cells were adoptively transferred into congenically distinct receipt mice, immunized with OVA (A) or SRBC-OVA (B), and analyzed at days 3 and 5. Data are representative of four independent experiments. (C) Nsd2CD4WT and NSD2CD4KO OT II T cells were adoptively transferred into CD28−/− mice, immunized with NP-OVA, and analyzed for GC B cells at day 7. Data are representative of three independent experiments. (D) CD4CreERT2+RosaYFP+Nsd2+/+ and CD4CreERT2+RosaYFP+Nsd2fl/fl mice on OT II TCR transgenic background were treated with tamoxifen for 5 d consecutively, and OT II T cells from these mice were adoptively transferred into recipient mice, which were subjected to OVA immunization and analyzed for Tfh at day 3. n = 10. Data were pooled from three experiments. In all bar graphs, bars represent means, and dots represent individual mice. Error bars show means ± SD. Statistical analysis was done with Student’s t test. **, P < 0.01; ***, P < 0.001.
Although the T cell numbers in Nsd2CD4 KO mice did not show significant alteration compared with Nsd2CD4 WT mice, there seemed to be a slight trend of reduction (Fig. S1 B). In addition, the observation of unaltered cell number cannot rule out the possibility that T cell function might be affected during thymocyte development, since CD4Cre starts to express early in thymus. Therefore, to more precisely assess the role of Nsd2 in Tfh differentiation, we adopted an inducible KO system by crossing CD4CreERT2 mice (expressing a fusion protein with a tamoxifen-sensitive estrogen receptor variant and Cre) with OT II TCR transgenic Nsd2fl/fl mice. The mice also carry loxP-STOP-loxP-YFP at Rosa26 locus (RosaYFP+), serving as a reporter for Cre activity. We treated the adult mice with tamoxifen to acutely ablate Nsd2 in mature T cells and then immediately adoptively transferred the cells into recipient mice for Tfh generation. We found that under this setting, acute Nsd2 ablation in mature T cells led to a striking reduction of Tfh differentiation (Fig. 3 D). Overall, these data revealed that Nsd2 is critically required for Tfh differentiation in a cell-intrinsic manner.
ICOS signal–dependent sustained Nsd2 expression is required for Tfh maintenance
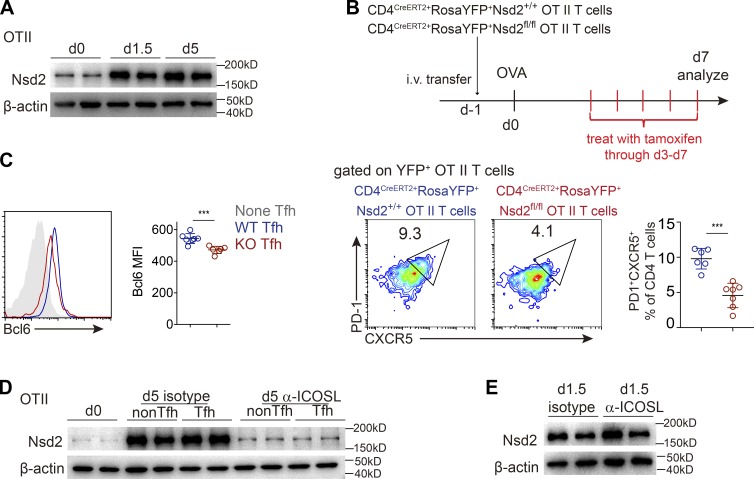
When we analyzed the Nsd2 expression dynamics in vivo, we found that Nsd2 was sustained at high levels. Once up-regulated after T cell activation and 5 d after activation, Nsd2 was still abundantly expressed (Fig. 4 A), leading us to consider whether Nsd2 is also important for Tfh maintenance. Using the CD4CreERT2 inducible KO system in Fig. 3 D, we found that ablating Nsd2 in transferred OT II T cells starting from day 3 after immunization, the time point when Tfh had already been generated, resulted in significantly reduced Tfh at day 7 (Fig. 4 B). Similar to the role of Nsd2 in Bcl6 early expression, inducible ablation of Nsd2 after Tfh generation also decreased Bcl6 at late time points (Fig. 4 C). These data suggested that Nsd2 not only is required for initial Bcl6 expression to promote Tfh early differentiation, but also is important for Tfh maintenance via Bcl6 regulation.
Figure 4.
ICOS signal–dependent sustained Nsd2 expression is required for Tfh maintenance. (A) Recipient mice were adoptively transferred with WT OT II T cells and immunized with OVA on the next day (day 0). OT II T cells were then sorted out at time points indicated for Western blot analysis of Nsd2 expression. Data are representative of three independent experiments. (B) OT II T cells were transferred into recipient mice, which were subjected to OVA immunization and tamoxifen treatment as shown on the upper panel. On day 7 after immunization, Tfh differentiation of YFP+ OT II T cells was analyzed by flow cytometry. Data are representative of three independent experiments. (C) Tfh cells of each genotype from B were analyzed for Bcl6 expression. Data are representative of three independent experiments. (D and E) Adoptive transfer of OT II T cells and OVA immunization were done as in A. Mice were treated with anti-ICOSL blockade antibody or isotype control daily starting from day 3 (D) or day 0 (E). CXCR5−PD1− and CXCR5+PD1+ OT II T cells at day 5 (D) or total OT II T cells at day 1.5 (E) after immunization were sorted out for Western blot. For bar graphs in B and C, bars represent means, and dots represent individual mice. Error bars show means ± SD. Statistical analysis was done with Student’s t test. ***, P < 0.001.
Previous studies reported that ICOS signal is required for Tfh maintenance and late-stage differentiation (Crotty, 2014; Weber et al., 2015). We therefore tested whether ICOS signal mediated the sustained high level of Nsd2. Indeed, we found that 2-d ICOS ligand (ICOSL) blockade led to severe reduction of Nsd2 in both activated non-Tfh cells and Tfh cells at day 5 (Fig. 4 D). Notably, ICOSL blockade did not affect Nsd2 expression at early time points of T cell activation (Fig. 4 E), suggesting that ICOS signal is required for sustaining Nsd2 expression at late time points of T cell activation, but not for early Nsd2 expression.
Bcl6 overexpression rescues the defect of Tfh differentiation in the absence of Nsd2
We sorted CXCR5+PD1+ Tfh cells from Nsd2CD4 WT and Nsd2CD4 KO mice for mRNA sequencing (RNA-seq) and identified 310 differentially expressed genes (DEGs) between these Nsd2 WT and KO Tfh cells. When we focused on the known Tfh-relevant genes, only expression of Bcl6 and CXCR5 was reduced in KO cells (Fig. 5 A). There was no significant alteration for other Tfh-relevant genes, including Maf, BATF, Ascl2, Irf4, Slamf1, Tcf7, Lef1, Icos, Cd40, etc. (Liu et al., 2013). Although Cxcr5 mRNA is reduced in Nsd2CD4 KO cells, the H3K36me2 mark on its locus was unaffected, suggesting that it is not a target of Nsd2 (Fig. S4). We also performed ChIP–quantitative PCR (qPCR) for other Tfh-related genes and did not find that any of them lost H3K36me2 modifications (Fig. S4).
Figure 5.
Ectopic Bcl6 expression fully rescues the Tfh differentiation defect in the absence of Nsd2. (A) RNA-seq analysis of Nsd2 WT and KO CXCR5+PD1+ Tfh from SRBC immunized mice. DEGs (Padjust < 0.05 and log2[fold change] greater than 0.5 or less than −0.5) are shown in black, and non–DEGs are shown in gray. Tfh related genes are highlighted in red dots (DEGs) or blue dots (non-DEGs). (B) Flow cytometry analysis of Bcl6 expression in gated Tfh. Data are representative of three independent experiments. (C) Nsd2 WT and KO OT II T cells were retrovirally transduced with Bcl6 or empty vector control (Ctrl) and adoptively transferred into recipients, which were then subjected to OVA immunization. Tfh differentiation of the transduced cells was analyzed by flow cytometry 3 d later. Data are representative of three independent experiments. (D and E) OT II T cells were transduced as in C and transferred into CD28KO receipts, which were immunized with NP-OVA/Alum for 7 d, and GC B cell response (D) and anti-NP antibody (E) were measured. Data are representative of two independent experiments. In B–D, bars represent means, and dots represent individual mice. Error bars show means ± SD. Statistical analysis was done with Student’s t test. n.s., not significant; **, P < 0.01; ***, P < 0.001. MFI, mean fluorescence intensity.
Flow cytometry analysis confirmed the reduced Bcl6 protein in Nsd2 KO Tfh (Fig. 5 B). To validate whether Bcl6 expression regulation is the predominant mechanism by which Nsd2 promotes Tfh differentiation, we performed rescue experiments by retrovirally overexpressing Bcl6 in Nsd2-deficient T cells. In the empty vector control transduction group, Nsd2 KO T cells differentiated less to Tfh compared with WT cells. However, in the Bcl6 transduction group, Tfh differentiation efficiency was comparable between Nsd2 WT and KO cells (Fig. 5 C), meaning that the Tfh differentiation defect in the absence of Nsd2 is fully rescued by Bcl6 overexpression. Overexpression of Bcl6 also rescued GC B cell responses and antibody titers (Fig. 5, D and E). These data suggest that Nsd2 promotes Tfh differentiation predominantly via up-regulating Bcl6 expression, although we cannot completely rule out whether there are other mechanisms involved.
T cell–expressed Nsd2 is required for clearance of chronic lymphocytic choriomeningitis virus (LCMV) infection
Tfh generation and humoral immune response are indispensable for control of chronic virus infections such as LCMV-cl13 (Bergthorsdottir et al., 2001; Chou et al., 2016; Cook et al., 2015; Harker et al., 2011). When we infected Nsd2CD4 WT and Nsd2CD4 KO mice with LCMV-cl13, we found that both Tfh differentiation and GC response were significantly impaired in Nsd2CD4 KO mice (Fig. 6, A and B). LCMV specific antibodies in mouse serum were also reduced in Nsd2 T cell–deficient mice (Fig. 6 C). Accordingly, we observed that the virus clearance in Nsd2CD4 KO mice was delayed (Fig. 6 D). LCMV-cl13 infection led to body weight loss in both Nsd2CD4 WT and Nsd2CD4 KO mice. However, compared with Nsd2CD4 WT mice, body weight of Nsd2CD4 KO mice recovered more slowly (Fig. 6 E). CD8 T cells are also important for virus clearance; however, it seemed that LCMV-specific CD8 T cell frequency was not affected by Nsd2 deficiency (Fig. S3 D), although we cannot completely exclude the role of Nsd2 for CD8 T cell function. Overall, our data suggest that Nsd2 protects mice from chronic LCMV infection by promoting Tfh generation and antibody response.
Figure 6.
T cell–expressed Nsd2 is required for clearance of chronic LCMV virus infection. Nsd2CD4WT and Nsd2CD4KO mice were infected with LCMV-cl13 and analyzed at indicated time points. (A and B) Tfh (A) and GC B cell (GCB; B) frequency and number were analyzed by flow cytometry at day 10 or 40. n = 8 for Tfh; n = 10 for GCB. Data were pooled from three independent experiments. (C) LCMV specific antibodies in serum were measured by ELISA at day 40 after infection. Data are representative of two independent experiments. (D) Virus load in the serum of mice after LCMV cl13 infection. n = 16. Data were pooled from three experiments. (E) Body weight before and after infection. Results are presented relative to original body weight (day 0). Data were pooled from three experiments. In A, B, and D, bars represent means, and dots represent individual mice. Statistical analysis was done with Student’s t test. Error bars show means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Nsd2 overexpression in T cells leads to increased Tfh and GC response
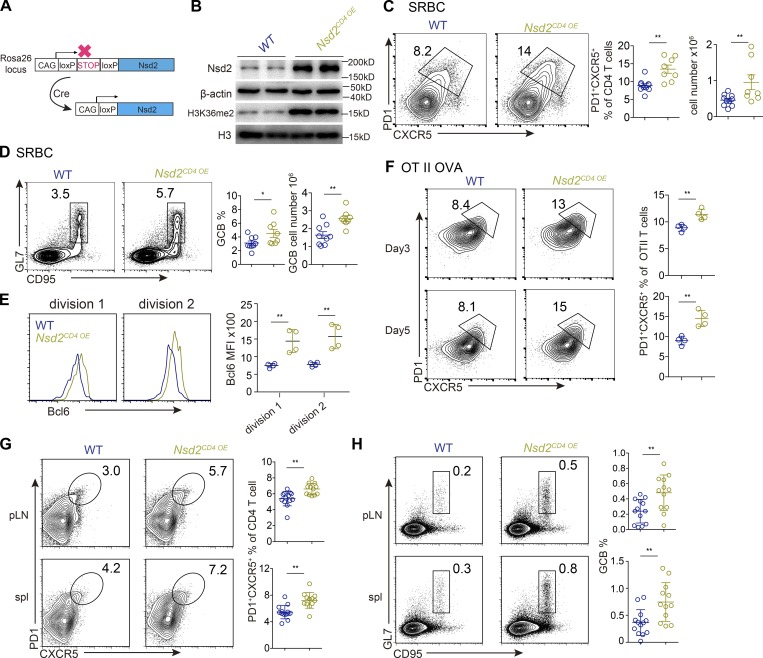
We further tested the role of Nsd2 in Tfh differentiation by gain-of-function approaches. Mice harboring loxP-STOP-loxP cassette and Nsd2 cDNA at Rosa26 locus (Nsd2OE/+) were crossed with CD4Cre mice to obtain T cell–specific Nsd2 overexpression mice (Nsd2CD4 OE; Fig. 7 A). In this mouse strain, H3K36me2 modification was also increased (Fig. 7 B). For control mice (WT), we used either CD4Cre-Nsd2OE/+ or CD4Cre+Nsd2+/+ littermate mice. Upon SRBC immunization, we observed stronger Tfh differentiation and GC B cell response in Nsd2-overexpressing mice compared with WT mice (Fig. 7, C and D). The Tfr cells were not affected by Nsd2 overexpression (Fig. S2 G). We then crossed the Nsd2CD4 OE mice to OT II TCR transgenic mice to analyze the effect of Nsd2 overexpression on the early Bcl6 expression. As shown in Fig. 7 E, the Nsd2-overexpressed T cells showed significantly increased Bcl6 levels at first cell division after OVA immunization. Consistently, we also observed increased Tfh differentiation for Nsd2CD4 OE OT II T cells (Fig. 7 F). Notably, in the 4-mo-old Nsd2CD4 OE mice, we observed spontaneous Tfh generation and GC response (Fig. 7, G and H), strongly demonstrating that Nsd2 is a critical regulator of Tfh differentiation.
Figure 7.
Nsd2 overexpression in T cells leads to increased Tfh and GC response. (A) Scheme of Nsd2 overexpression in mice. (B) Western blot of Nsd2 and H3K36me2 in CD4+ T cells from indicated mice. Data are representative of two independent experiments. (C and D) Mice of indicated genotype were immunized with SRBCs and analyzed by flow cytometry for Tfh (C) and GC B cell (GCB; D) response at day 7. Data are representative of three independent experiments. (E) CTV-labeled OT II T cells were transferred into recipient mice and immunized with OVA for 2 d. Histogram shows Bcl6 expression of cells in first and second cell division based on CTV dilution. Data are representative of three independent experiments. (F) OT II T cells were adoptively transferred into congenically distinct recipient mice, immunized with OVA, and analyzed for Tfh differentiation at days 3 and 5. Data are representative of three independent experiments. (G and H) Unimmunized WT and Nsd2CD4 OE littermate mice were analyzed at age 4 mo by flow cytometry for Tfh (G) and GCB (H). Data were pooled from four experiments. In C–H, bars represent means, and dots represent individual mice. Error bars show means ± SD. Statistical analysis was done with Student’s t test. *, P < 0.05; **, P < 0.01. MFI, mean fluorescence intensity; pLN, peripheral lymph node; spl, spleen.
Discussion
Bcl6 is the most essential transcription factor for Tfh fate determination and is expressed early after antigen-specific stimulation in a CD28-dependent manner. However, little is known regarding the epigenetic mechanism that promotes Bcl6 expression during early Tfh differentiation. In this study, we report that H3K36me2 methyltransferase Nsd2 is robustly induced by CD28 costimulatory signaling to promote early Bcl6 expression during T cell activation. The other costimulatory signal, ICOS signaling, is not involved in the early induction of Nsd2, but is required for the maintenance of Nsd2 at later time points. Deficiency of Nsd2 in T cells lead to impaired Tfh differentiation and maintenance due to reduced Bcl6 expression. Ectopic Nsd2 expression results in enhanced Tfh generation and GC B cell responses, and 4-mo-old Nsd2 overexpression mice even develop spontaneous Tfh cells and GCs. These data demonstrate that the histone methyltransferase Nsd2 acts as a critical epigenetic regulator of Bcl6 expression and Tfh differentiation.
CD28 and ICOS are distinct coreceptors for T cell differentiation, with CD28 expressed on both naive and active T cells, whereas ICOS is expressed only after T cell activation (Smith-Garvin et al., 2009), and these two signals are both required for Tfh differentiation and maintenance (Barnett et al., 2014; Vinuesa et al., 2016). Despite the in-depth studies about the role and mechanism of ICOS signaling in Tfh development, much less is known regarding how CD28 signal induces Bcl6. Our data thus provide a valuable mechanistic explanation for this question. We found that CD28 signaling rapidly induces Nsd2 to promote Bcl6 expression and Tfh differentiation. ICOS is essential for Tfh development and is believed to be able to promote Bcl6 expression (Akiba et al., 2005; Bossaller et al., 2006; Choi et al., 2011; Gigoux et al., 2009), although it is not required for early Bcl6 expression (Weber et al., 2015). By repressing KLF2, ICOS signal critically maintains the Tfh phenotype (Weber et al., 2015). Moreover, ICOS signal inactivates FOXO1 to indirectly up-regulate Bcl6 expression (Stone et al., 2015). At the posttranslation level, ICOS signal helps to maintain Bcl6 protein levels in an osteopontin-dependent manner (Leavenworth et al., 2015). In addition, ICOS signal derived from bystander B cells could promote T cell migration into B cell follicles (Xu et al., 2013). Our study provides another level of mechanistic insight into how ICOS promotes Tfh differentiation. We found that ICOS signal promotes Nsd2 expression at late but not early stages of Tfh differentiation, and that the sustained high level of Nsd2 at late stages is required for Tfh maintenance. Therefore, both CD28 and ICOS signaling are required for Nsd2 expression, although at different time points, to promote Tfh differentiation.
Tfh cells provide critical help for B cell–mediated humoral response and thus are essential for protection against pathogen infections. Here we showed that Tfh defects in the absence of Nsd2 lead to impaired clearance of chronic virus infection. In addition to their role in pathogen infection, Tfh cells have also been implicated in other pathological situations, such as autoimmune diseases, especially the autoantibody-mediated autoimmune diseases (Crotty, 2014; Ueno et al., 2015). Given that Nsd2 is an epigenetic enzyme, the activity of which could be easily targeted by small molecules in principle, our findings may help to design better vaccines and develop therapeutic strategies for autoimmune diseases.
Materials and methods
Mice
Nsd2fl/fl mice and Nsd2OE mice were previously described (Li et al., 2017). CD4Cre transgenic mice, CD4CreERT2 transgenic mice, Rosa26loxP-Stop-loxP-YFP reporter mice, OT II transgenic mice, CD28−/− mice, and B6 (C57BL/6J) and B6-CD45.1 (Ptprca Pepcb/BoyJ) mice were obtained from the Jackson Laboratory. All mice were maintained on C57BL/6J background. All control mice were littermate controls. 6–12-wk-old male and female mice were used for experiments. 4-mo-old mice were used for spontaneous Tfh analysis. Mice were maintained in a specific pathogen–free environment, and animal protocols were reviewed and approved by the Institutional Animal Care and Use Committee of Nanjing Medical University and the Institutional Biomedical Research Ethics Committee of the Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences.
Flow cytometry and cell sorting
For surface staining, cells were isolated from spleen or lymph nodes, incubated with indicated antibodies on ice for 30 min, and washed with staining buffer (2% FBS in PBS with 0.1% NaN3 and 50 mM EDTA). For intracellular staining, cells were fixed with 1% paraformaldehyde in PBS and permeabilized with cold methanol as previously described (Chen et al., 2018). Data were acquired on a Beckman Cytoflex and analyzed with FlowJo 10 software. Cell sorting was performed with a BD Aria II. Sorting efficiency was tested, and only cells with high purity (>98%) were used for the experiments.
Mouse immunization and treatment
For induction of Tfh and GC response, mice were immunized with 2 × 108 SRBCs (i.p.) or 100 µg NP-OVA (or OVA) precipitated with aluminum hydroxide gel adjuvant (Alum) plus 1 µg LPS (i.p. or s.c.). For adoptive transfers, 106 OT II T cells were injected i.v. into recipient mice. Acute ablation of Nsd2 was done as previously described (Wang et al., 2014). Briefly, CD4CreERT2+RosaYFP+ OT II mice were treated daily for 5 d with tamoxifen (Sigma-Aldrich) at a dose of 1 mg per mouse. 106 OT-II T cells from the treated mice were then injected i.v. into recipient mice. For in vivo blockade of ICOSL, mice were treated with 500 µg monoclonal antibody to ICOSL (HK5.3; Bio-X-Cell) by i.p. injection daily as previously described (Wang et al., 2014; Weber et al., 2015).
Early Bcl6 expression analysis during CD4+ T cell activation
Cell Trace Violet (CTV)–labeled OT-II cells (5 × 105) were adoptively transferred into recipients, followed by s.c. immunization with 100 µg OVA in Alum plus 1 µg LPS. Peripheral lymph nodes were dissected 48 h later and analyzed by flow cytometry (Baumjohann et al., 2011).
RNA-seq
CXCR5+PD1+ Tfh from SRBC immunized mice were sorted out at day 7 for RNA extraction and library construction at RiboBio Co., Ltd. Two biological replicates (two Nsd2CD4 WT samples and two Nsd2CD4 KO samples) were performed, and each sample combined a mixture of RNA from at least three mice. Sequencing was done on an Illumina Hiseq 2500 platform and SE50 single-end module. Sequence reads were mapped to mm10 reference genome using Hisat2 (Kim et al., 2015), and differential gene expression analysis was detected using Bioconductor DESeq2 package with a threshold of Padjust < 0.05 and |log2(fold change)| >0.5 (Love et al., 2014). Genes with fragments per kilobase of transcript per million mapped reads <1 were not included for analysis. Raw data files and processed files have been uploaded to Gene Expression Omnibus public database under accession no. GSE130550.
Immunoblot analysis
For immunoblot analysis, 106 sorted CD4+ T cells were lysed in 100 µl SDS loading buffer. Nonspecific binding to membranes was blocked by 5% milk in PBS with 0.05% Tween20 for 1 h at room temperature, then membranes were incubated with primary antibodies diluted in 5% BSA in PBS overnight at 4°C with constant rotation. The membranes were incubated with secondary antibodies diluted in PBST for 1 h at room temperature. Chemiluminescence was detected using Immobilon Western Chemiluminescent HRP Substrate (Millipore).
T cell in vitro stimulation
CD4+CD25− T cells were activated with anti-CD3 (5 µg/ml; 145-2C11; Bio-X-Cell) or anti-CD3 (5 µg/ml) plus anti-CD28 (5 µg/ml; 37.51; Bio-X-Cell) at a concentration of 106 cells/ml in RPMI supplemented with 10% FBS, nonessential amino acids, sodium pyruvate, l-glutamine, Hepes, and β-mercaptoethanol.
Retroviral transduction and cell transfer
Transduction was performed as previously described (Chen et al., 2018; Wang et al., 2014). Briefly, virus was packaged in the Plat-E cell line (Cell Biolabs) by transfecting MSCV-GFP-Bcl6 plasmid, and the viral supernatants were harvested at day 2 after transfection. CD4+ T cells were stimulated for 24 h with anti-CD3 plus anti-CD28 and spin infected with viral supernatant. The culture medium was then replaced with complete T cell medium supplemented with 100 U/ml recombinant human IL-2, incubated for 2 d, and adoptively transferred into recipient mice, which were then immunized with OVA.
ChIP-qPCR assay
Briefly, CD4+ T cells were cross-linked in 1% formaldehyde for 5 min at room temperature, quenched with 0.125 M glycine for 5 min at room temperature, and lysed in 0.2% SDS with a protease-inhibitor tablet for 10 min on ice. Genomic DNA were then fragmented by sonicating. For immunoprecipitation, anti-dimethyl histone H3 (07-274; Millipore) and Whsc1/Nsd2 (ab75395; Abcam) were used. DNA purification was performed using the ChIP DNA Clean & Concentrator (ZYMO Research) according to the manufacturer’s instructions.
LCMV infection
For LCMV chronic infection, age-matched mice were infected intravenously with LCMV cl13 (106 plaque-forming units). After infection, mice were weighed daily, and body weight was calculated as a percentage of initial weight on day 0. For detection of serum viral load, RNA was extracted from 10 µl of serum from infected mice using ZR Viral RNA Kit (ZYMO Research). RT-qPCR was done as described (McCausland and Crotty, 2008).
ELISA
OVA specific antibodies were detected using plates coated with OVA (albumin from chicken egg white; Sigma-Aldrich), goat anti-mouse IgG (Southern Biotech), and goat anti-mouse IgG1 HRP (Southern Biotech). LCMV antibody titers were measured with coated cell lysates from LCMV-infected BHK-21 cells, goat anti-mouse IgG2c HRP (Southern Biotech), and goat anti-mouse IgG2b HRP (Southern Biotech).
Statistical analyses
Student’s t tests were used for bioinformatics analysis, except where ANOVA was used as indicated in figure legends. Unless otherwise indicated, the data in figures are displayed as the mean ± SD. P values are denoted in figures by ns, not significant; *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
Online supplemental material
Fig. S1 shows analysis of T cell development. Fig. S2 shows Tfr differentiation analysis. Fig. S3 shows Th and CD8 T cell differentiation analysis. Fig. S4 shows anti-H3K36me2 ChIP-qPCR analysis.
Acknowledgments
We thank all the laboratory members for helpful discussions.
This work was supported by National Key R&D Program of China (2018YFC1003900), National Natural Science Foundation of China grants 31570881(to X. Wang) and 81771670 (to X. Wang), Postgraduate Research & Practice Innovation Program of Jiangsu Province grant KYCX18_1448 (to J. Chen), and Key Development Program of Nanjing Medical University grant NMUD201809 (to X. Wang).
The authors declare no competing financial interests.
Author contributions: X. Long and X. Wang conceptualized the project, designed the experimental approaches, analyzed the data, and prepared the manuscript as senior authors. X. Long, L. Zhang, Y. Zhang, M. Min, B. Lin, J. Chen, X. Ma, S. Zhai, Y. Lu, Z. Cai, and Y. Liu performed most of the experiments. N. Che, W. Tan, and J. Qin provided resources and supervised a specific subset of experiments and analysis.
References
- Akiba H., Takeda K., Kojima Y., Usui Y., Harada N., Yamazaki T., Ma J., Tezuka K., Yagita H., and Okumura K.. 2005. The role of ICOS in the CXCR5+ follicular B helper T cell maintenance in vivo. J. Immunol. 175:2340–2348. 10.4049/jimmunol.175.4.2340 [DOI] [PubMed] [Google Scholar]
- Allis C.D., and Jenuwein T.. 2016. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17:487–500. 10.1038/nrg.2016.59 [DOI] [PubMed] [Google Scholar]
- Barnett L.G., Simkins H.M.A., Barnett B.E., Korn L.L., Johnson A.L., Wherry E.J., Wu G.F., and Laufer T.M.. 2014. B cell antigen presentation in the initiation of follicular helper T cell and germinal center differentiation. J. Immunol. 192:3607–3617. 10.4049/jimmunol.1301284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumjohann D., Okada T., and Ansel K.M.. 2011. Cutting Edge: Distinct waves of BCL6 expression during T follicular helper cell development. J. Immunol. 187:2089–2092. 10.4049/jimmunol.1101393 [DOI] [PubMed] [Google Scholar]
- Bauquet A.T., Jin H., Paterson A.M., Mitsdoerffer M., Ho I.C., Sharpe A.H., and Kuchroo V.K.. 2009. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nat. Immunol. 10:167–175. 10.1038/ni.1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergthorsdottir S., Gallagher A., Jainandunsing S., Cockayne D., Sutton J., Leanderson T., and Gray D.. 2001. Signals that initiate somatic hypermutation of B cells in vitro. J. Immunol. 166:2228–2234. 10.4049/jimmunol.166.4.2228 [DOI] [PubMed] [Google Scholar]
- Betz B.C., Jordan-Williams K.L., Wang C., Kang S.G., Liao J., Logan M.R., Kim C.H., and Taparowsky E.J.. 2010. Batf coordinates multiple aspects of B and T cell function required for normal antibody responses. J. Exp. Med. 207:933–942. 10.1084/jem.20091548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollig N., Brüstle A., Kellner K., Ackermann W., Abass E., Raifer H., Camara B., Brendel C., Giel G., Bothur E., et al. 2012. Transcription factor IRF4 determines germinal center formation through follicular T-helper cell differentiation. Proc. Natl. Acad. Sci. USA. 109:8664–8669. 10.1073/pnas.1205834109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossaller L., Burger J., Draeger R., Grimbacher B., Knoth R., Plebani A., Durandy A., Baumann U., Schlesier M., Welcher A.A., et al. 2006. ICOS deficiency is associated with a severe reduction of CXCR5+CD4 germinal center Th cells. J. Immunol. 177:4927–4932. 10.4049/jimmunol.177.7.4927 [DOI] [PubMed] [Google Scholar]
- Chen C., Zhai S., Zhang L., Chen J., Long X., Qin J., Li J., Huo R., and Wang X.. 2018. Uhrf1 regulates germinal center B cell expansion and affinity maturation to control viral infection. J. Exp. Med. 215:1437–1448. 10.1084/jem.20171815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Cao G., Wu J., Wang X., Pan Z., Gao J., Tian Q., Xu L., Li Z., Hao Y., et al. 2019. The histone methyltransferase EZH2 primes the early differentiation of follicular helper T cells during acute viral infection. Cell. Mol. Immunol. 10.1038/s41423-019-0219-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.S., Gullicksrud J.A., Xing S., Zeng Z., Shan Q., Li F., Love P.E., Peng W., Xue H.-H., and Crotty S.. 2015. LEF-1 and TCF-1 orchestrate T(FH) differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nat. Immunol. 16:980–990. 10.1038/ni.3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.S., Kageyama R., Eto D., Escobar T.C., Johnston R.J., Monticelli L., Lao C., and Crotty S.. 2011. ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity. 34:932–946. 10.1016/j.immuni.2011.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C., Verbaro D.J., Tonc E., Holmgren M., Cella M., Colonna M., Bhattacharya D., and Egawa T.. 2016. The transcription factor AP4 mediates resolution of chronic viral infection through amplification of germinal center B cell responses. Immunity. 45:570–582. 10.1016/j.immuni.2016.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y., Tanaka S., Chu F., Nurieva R.I., Martinez G.J., Rawal S., Wang Y.-H., Lim H., Reynolds J.M., Zhou X.H., et al. 2011. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat. Med. 17:983–988. 10.1038/nm.2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook K.D., Shpargel K.B., Starmer J., Whitfield-Larry F., Conley B., Allard D.E., Rager J.E., Fry R.C., Davenport M.L., Magnuson T., et al. 2015. T follicular helper cell-dependent clearance of a persistent virus infection requires T cell expression of the histone demethylase UTX. Immunity. 43:703–714. 10.1016/j.immuni.2015.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. 2014. T follicular helper cell differentiation, function, and roles in disease. Immunity. 41:529–542. 10.1016/j.immuni.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuPage M., Chopra G., Quiros J., Rosenthal W.L., Morar M.M., Holohan D., Zhang R., Turka L., Marson A., and Bluestone J.A.. 2015. The chromatin-modifying enzyme Ezh2 is critical for the maintenance of regulatory T cell identity after activation. Immunity. 42:227–238. 10.1016/j.immuni.2015.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigoux M., Shang J., Pak Y., Xu M., Choe J., Mak T.W., and Suh W.-K.. 2009. Inducible costimulator promotes helper T-cell differentiation through phosphoinositide 3-kinase. Proc. Natl. Acad. Sci. USA. 106:20371–20376. 10.1073/pnas.0911573106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harker J.A., Lewis G.M., Mack L., and Zuniga E.I.. 2011. Late interleukin-6 escalates T follicular helper cell responses and controls a chronic viral infection. Science. 334:825–829. 10.1126/science.1208421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R.J., Poholek A.C., DiToro D., Yusuf I., Eto D., Barnett B., Dent A.L., Craft J., and Crotty S.. 2009. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 325:1006–1010. 10.1126/science.1175870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., and Salzberg S.L.. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 12:357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo A.J., Cheung P., Chen K., Zee B.M., Kioi M., Lauring J., Xi Y., Park B.H., Shi X., Garcia B.A., et al. 2011. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol. Cell. 44:609–620. 10.1016/j.molcel.2011.08.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavenworth J.W., Verbinnen B., Yin J., Huang H., and Cantor H.. 2015. A p85α-osteopontin axis couples the receptor ICOS to sustained Bcl-6 expression by follicular helper and regulatory T cells. Nat. Immunol. 16:96–106. 10.1038/ni.3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Zeng Z., Xing S., Gullicksrud J.A., Shan Q., Choi J., Badovinac V.P., Crotty S., Peng W., and Xue H.-H.. 2018. Ezh2 programs TFH differentiation by integrating phosphorylation-dependent activation of Bcl6 and polycomb-dependent repression of p19Arf. Nat. Commun. 9:5452 10.1038/s41467-018-07853-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Xue W., Yuan H., Dong B., Ding Y., Liu Y., Jiang M., Kan S., Sun T., Ren J., et al. 2017. AKT-mediated stabilization of histone methyltransferase WHSC1 promotes prostate cancer metastasis. J. Clin. Invest. 127:1284–1302. 10.1172/JCI91144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linterman M.A., Pierson W., Lee S.K., Kallies A., Kawamoto S., Rayner T.F., Srivastava M., Divekar D.P., Beaton L., Hogan J.J., et al. 2011. Foxp3+ follicular regulatory T cells control the germinal center response. Nat. Med. 17:975–982. 10.1038/nm.2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Chen X., Zhong B., Wang A., Wang X., Chu F., Nurieva R.I., Yan X., Chen P., van der Flier L.G., et al. 2014. Transcription factor achaete-scute homologue 2 initiates follicular T-helper-cell development. Nature. 507:513–518. 10.1038/nature12910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Nurieva R.I., and Dong C.. 2013. Transcriptional regulation of follicular T-helper (Tfh) cells. Immunol. Rev. 252:139–145. 10.1111/imr.12040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCausland M.M., and Crotty S.. 2008. Quantitative PCR technique for detecting lymphocytic choriomeningitis virus in vivo. J. Virol. Methods. 147:167–176. 10.1016/j.jviromet.2007.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayamada S., Takahashi H., Kanno Y., and O’Shea J.J.. 2012. Helper T cell diversity and plasticity. Curr. Opin. Immunol. 24:297–302. 10.1016/j.coi.2012.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva R.I., Chung Y., Martinez G.J., Yang X.O., Tanaka S., Matskevitch T.D., Wang Y.-H., and Dong C.. 2009. Bcl6 mediates the development of T follicular helper cells. Science. 325:1001–1005. 10.1126/science.1176676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H. 2016. T follicular helper cells in space-time. Nat. Rev. Immunol. 16:612–625. 10.1038/nri.2016.94 [DOI] [PubMed] [Google Scholar]
- Sage P.T., and Sharpe A.H.. 2016. T follicular regulatory cells. Immunol. Rev. 271:246–259. 10.1111/imr.12411 [DOI] [PubMed] [Google Scholar]
- Shen E., Wang Q., Rabe H., Liu W., Cantor H., and Leavenworth J.W.. 2018. Chromatin remodeling by the NuRD complex regulates development of follicular helper and regulatory T cells. Proc. Natl. Acad. Sci. USA. 115:6780–6785. 10.1073/pnas.1805239115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Garvin J.E., Koretzky G.A., and Jordan M.S.. 2009. T cell activation. Annu. Rev. Immunol. 27:591–619. 10.1146/annurev.immunol.021908.132706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone E.L., Pepper M., Katayama C.D., Kerdiles Y.M., Lai C.-Y., Emslie E., Lin Y.C., Yang E., Goldrath A.W., Li M.O., et al. 2015. ICOS coreceptor signaling inactivates the transcription factor FOXO1 to promote Tfh cell differentiation. Immunity. 42:239–251. 10.1016/j.immuni.2015.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno H., Banchereau J., and Vinuesa C.G.. 2015. Pathophysiology of T follicular helper cells in humans and mice. Nat. Immunol. 16:142–152. 10.1038/ni.3054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinuesa C.G., Linterman M.A., Yu D., and MacLennan I.C.M.. 2016. Follicular helper T cells. Annu. Rev. Immunol. 34:335–368. 10.1146/annurev-immunol-041015-055605 [DOI] [PubMed] [Google Scholar]
- Wagner E.J., and Carpenter P.B.. 2012. Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 13:115–126. 10.1038/nrm3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Geng J., Wen X., Bi E., Kossenkov A.V., Wolf A.I., Tas J., Choi Y.S., Takata H., Day T.J., et al. 2014. The transcription factor Foxp1 is a critical negative regulator of the differentiation of follicular helper T cells. Nat. Immunol. 15:667–675. 10.1038/ni.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J.P., Fuhrmann F., Feist R.K., Lahmann A., Al Baz M.S., Gentz L.-J., Vu Van D., Mages H.W., Haftmann C., Riedel R., et al. 2015. ICOS maintains the T follicular helper cell phenotype by down-regulating Krüppel-like factor 2. J. Exp. Med. 212:217–233. 10.1084/jem.20141432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G., Wei L., Zhu J., Zang C., Hu-Li J., Yao Z., Cui K., Kanno Y., Roh T.-Y., Watford W.T., et al. 2009. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 30:155–167. 10.1016/j.immuni.2008.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C.B., Rowell E., and Sekimata M.. 2009. Epigenetic control of T-helper-cell differentiation. Nat. Rev. Immunol. 9:91–105. 10.1038/nri2487 [DOI] [PubMed] [Google Scholar]
- Xu H., Li X., Liu D., Li J., Zhang X., Chen X., Hou S., Peng L., Xu C., Liu W., et al. 2013. Follicular T-helper cell recruitment governed by bystander B cells and ICOS-driven motility. Nature. 496:523–527. 10.1038/nature12058 [DOI] [PubMed] [Google Scholar]
- Yu D., Rao S., Tsai L.M., Lee S.K., He Y., Sutcliffe E.L., Srivastava M., Linterman M., Zheng L., Simpson N., et al. 2009. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 31:457–468. 10.1016/j.immuni.2009.07.002 [DOI] [PubMed] [Google Scholar]