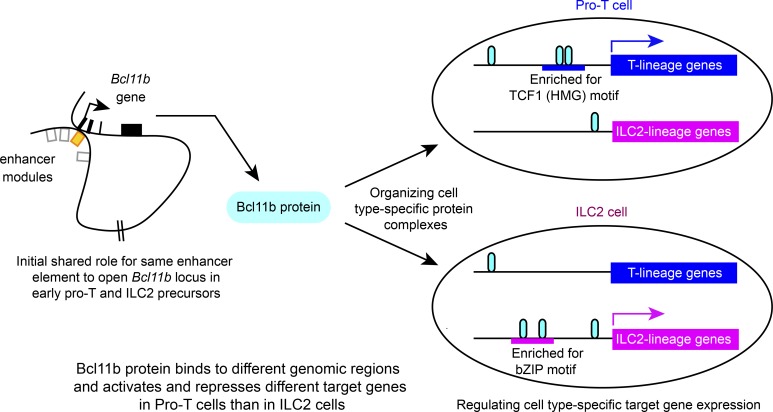
Bcl11b binds to distinctive genomic regions with different partners and regulates completely different target genes in pro-T and ILC2 cells. Despite these divergences in Bcl11b function, a shared enhancer supports initial Bcl11b locus opening in both pro-T and ILC2 lineages.
Abstract
The zinc finger transcription factor, Bcl11b, is expressed in T cells and group 2 innate lymphoid cells (ILC2s) among hematopoietic cells. In early T-lineage cells, Bcl11b directly binds and represses the gene encoding the E protein antagonist, Id2, preventing pro-T cells from adopting innate-like fates. In contrast, ILC2s co-express both Bcl11b and Id2. To address this contradiction, we have directly compared Bcl11b action mechanisms in pro-T cells and ILC2s. We found that Bcl11b binding to regions across the genome shows distinct cell type–specific motif preferences. Bcl11b occupies functionally different sites in lineage-specific patterns and controls totally different sets of target genes in these cell types. In addition, Bcl11b bears cell type–specific post-translational modifications and organizes different cell type–specific protein complexes. However, both cell types use the same distal enhancer region to control timing of Bcl11b activation. Therefore, although pro-T cells and ILC2s both need Bcl11b for optimal development and function, Bcl11b works substantially differently in these two cell types.
Graphical Abstract
Introduction
The zinc finger transcription factor Bcl11b is shared between only two known classes of hematopoietic cells in mice: T cells and the type 2 subset of innate lymphoid cells (ILCs). In both, it plays an important functional role (Avram and Califano, 2014; Califano et al., 2015; Kojo et al., 2017; Liu et al., 2010; Longabaugh et al., 2017; Walker et al., 2015; Yu et al., 2015). However, in the murine T cell lineage, a conspicuous part of its role involves blocking access to natural killer–like developmental programs (Li et al., 2010a; Li et al., 2010b) and specifically repressing the Id2 gene (Hosokawa et al., 2018a). Another Bcl11b repression target in early T cells (Hosokawa et al., 2018a), Zbtb16 (encoding PLZF), is positively required in ILC common precursors, but is declining by the time committed ILC2 precursors activate Bcl11b (Constantinides et al., 2015; Harly et al., 2018; Seillet et al., 2016; Yu et al., 2016). In contrast, Id2 is a factor with a continuing role in all ILCs, which persists, stably co-expressed with Bcl11b, in normal ILC2 cells (Seillet et al., 2016; Serafini et al., 2015; Walker et al., 2015; Wang et al., 2017; Yu et al., 2016; Zook and Kee, 2016). If Bcl11b always repressed Id2, the ILC2 phenotype would not exist. To understand how this apparent contradiction is resolved, we have directly compared the ways Bcl11b is used in early T-lineage cells and in ILC2 cells.
Here, we document the genomic sites where Bcl11b is engaged in early primary pro-T cells, in a pro-T cell line, and in an ILC2 cell line; the motif enrichments at the sites that Bcl11b occupies; the interaction partners of Bcl11b as determined by proteomics of co-immune precipitated proteins; and the functionally responsive target genes of Bcl11b, in both the T-lineage and ILC-lineage contexts. The results show that while many Bcl11b binding sites are shared, a large minority of the sites in each cell type are lineage specific, and these are associated with distinct, lineage-specific motif co-enrichment patterns and associations of Bcl11b with different interacting proteins. Bcl11b interaction partners of the Runx and GATA factor families, also shared between the cell types, are also deployed to lineage-specific sites. The functional Bcl11b target gene sets in the two cell types are even more divergent: nearly all of the genes responding acutely to Bcl11b disruption, whether Bcl11b dependent or Bcl11b repressed, are cell lineage–specific. In contrast, the regulation of the Bcl11b locus itself has similar features in T and ILC2 cells, as shown by a common role of an early-acting distal enhancer element (Li et al., 2013; Ng et al., 2018) in heritably enabling Bcl11b expression. Thus, despite being expressed in both, Bcl11b does not exert homologous functions in ILC2 cells and pro-T cells.
Results and discussion
Bcl11b binds to distinct regions across the genome in pro-T and ILC2 cells
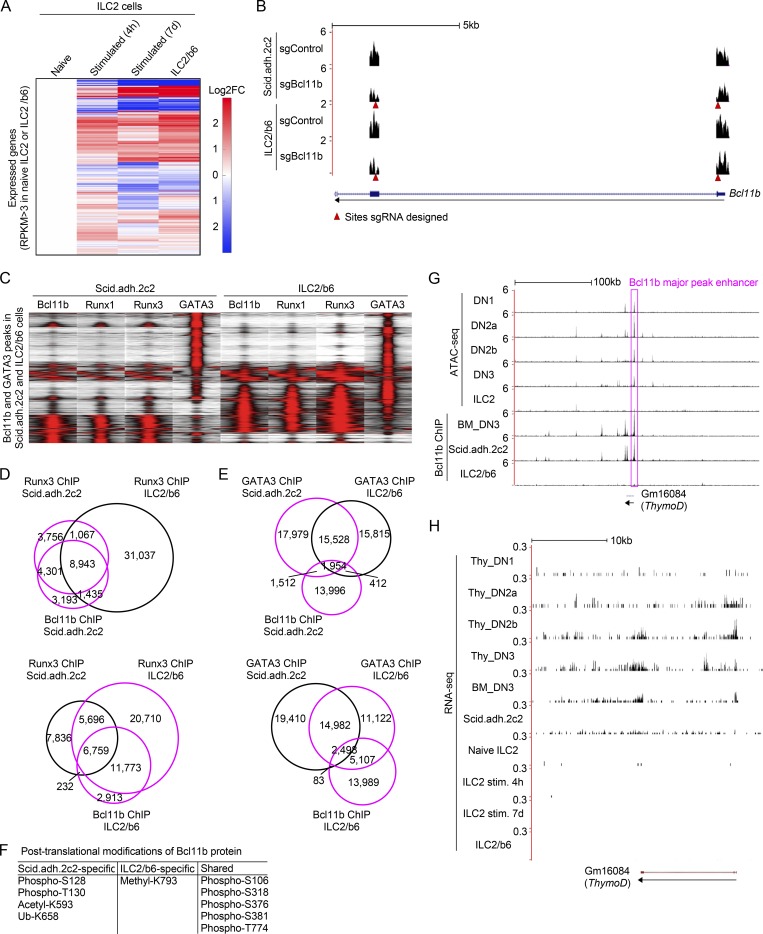
We previously reported that Bcl11b directly represses Id2 expression in pro-T cells, preventing these immature T cell precursors from adopting an innate-like fate (Hosokawa et al., 2018a). However, normal development and function of ILC2 cells depend on co-expression of both Bcl11b and Id2. To address this seeming contradiction, we tested whether Bcl11b action mechanisms might differ in early T-lineage and ILC2 cells. Bcl11b might bind to different sites in the two cell contexts. Alternatively, because Bcl11b can work either as an activator or as a repressor, it might bind to the same sites but exert different effects due to recruitment of different partner factors. To compare the molecular mechanisms through which Bcl11b controls cell type–specific gene regulation in the two contexts, we first examined the DNA binding patterns of Bcl11b across the genome in pro-T cells with those in ILC2 cells. Because of the cell numbers needed for chromatin immunoprecipitation (ChIP) followed by massively parallel DNA sequencing (ChIP-seq) and the rarity of primary ILC2 cells, we took advantage of an ILC2 cell line, ILC2/b6, which can be grown continuously in tissue culture supplemented with IL-2, IL-7, and IL-33 (Zhang et al., 2017). Fig. S1 A shows that the gene expression profile of ILC2/b6 cells was almost indistinguishable from that of primary ILC2 cells after stimulation for 4 h or 7 d (Shih et al., 2016; Yagi et al., 2014). We used these cells for Bcl11b ChIP-seq analysis, comparing the ILC2/b6 Bcl11b ChIP-seq results with those from primary double-negative (DN)2b/DN3 cells (henceforth called “DN3”) and from a DN3-like cell line, Scid.adh.2c2.
Figure S1.
Characterization of ILC2 and pro-T cell transcriptomes, Runx binding patterns, Bcl11b modifications, and activities in the Bcl11b enhancer region. (A) Heat maps show hierarchical clustering analyses of the expression of all expressed genes, which have RPKM >3 in naive ILC2 cells or an ILC2 cell line, ILC2/b6 cells, in naive ILC2, stimulated ILC2 for 4 h or 7 d (Shih et al., 2016; Yagi et al., 2014), and ILC2/b6 cells. (B) Representative RNA-seq tracks are shown for Bcl11b-deficient Scid.adh.2c2 or ILC2/b6 cells at the Bcl11b locus (around exon1 and 2). Red arrowheads show sites against which sgRNA was designed. (C) Tag count distributions for Bcl11b, Runx1, Runx3, and GATA3 in Scid.adh.2c2 and ILC2/b6 cells are shown. All Bcl11b and GATA3 binding sites identified in the DN3 and ILC2/b6 cells were included in the analysis. (D) Venn diagrams show the number of Runx3 ChIP peaks in Scid.adh.2c2 and ILC2/b6 cells with Bcl11b ChIP peaks in Scid.adh.2c2 cells (top) or ILC2/b6 cells (bottom). (E), Venn diagrams show the number of GATA3 ChIP peaks in Scid.adh.2c2 and ILC2/b6 cells with Bcl11b ChIP peaks in Scid.adh.2c2 cells (top) or ILC2/b6 cells (bottom). (F) Post-translational modifications of Bcl11b protein detected by mass spectrometry analysis are shown. (G) Representative ATAC-seq tracks for thymic DN subsets and mature small intestine ILC2 cells (Yoshida et al., 2019; downloaded from GEO accession no. GSE100738) and binding profiles of Bcl11b in DN3, Scid.adh.2c2, and ILC2/b6 cells are shown around the Bcl11b major peak enhancer (magenta rectangle) and ThymoD locus. The low activity state shown here in mature ILC2 cells contrasts with the state inferred to exist in ILC2 precursors, based on evidence in Fig. 5. (H) Representative RNA-seq tracks (mapped with STAR v2.4.0) at the ThymoD locus are shown for thymic DN subsets (Yoshida et al., 2019; GSE100738), in vitro cultured bone marrow precursor–derived DN3, Scid.adh.2c2, mature naive ILC2, mature ILC2 stimulated (stim.) for 4 h or 7 d (Shih et al., 2016; Yagi et al., 2014), and ILC2/b6 cells. Data from this study are based on two replicate RNA-seq results (A, B, and H) and are based on reproducible ChIP-seq peaks in two replicate samples (C–E and G).
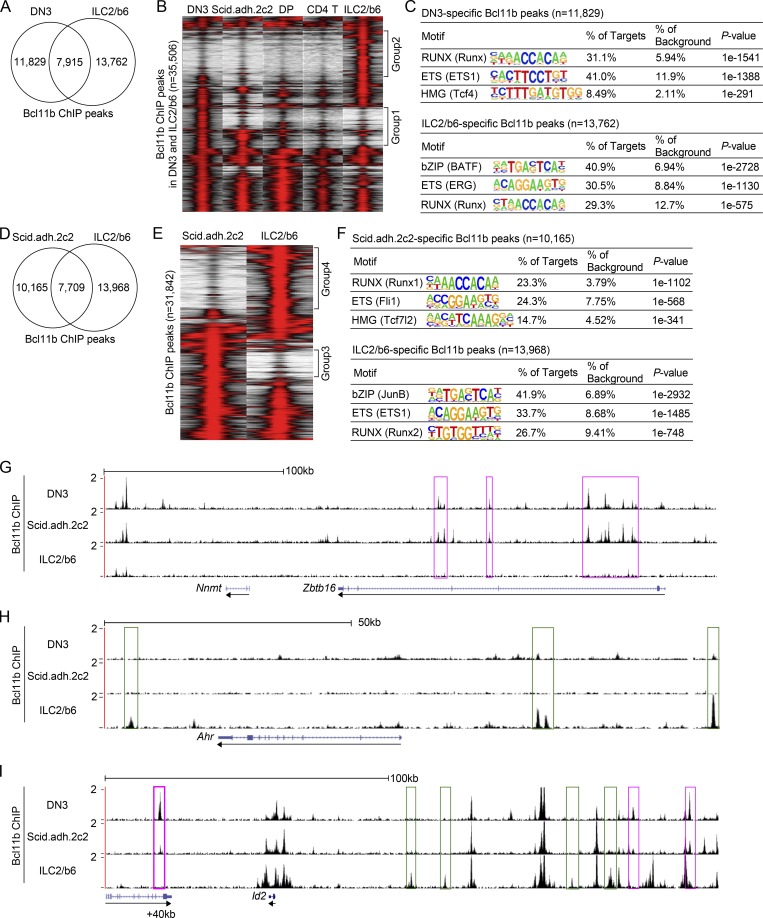
Compared with previously identified Bcl11b ChIP peaks in primary DN2b/DN3 cells (Longabaugh et al., 2017), the data showed a conspicuous divergence between ILC2 and DN3 Bcl11b binding sites. Many binding sites were shared, but there were also many DN3-specific and many ILC2-specific sites (Fig. 1, A and B; group 1 and group 2 regions, respectively). The group 1 sites included many pro-T cell specific occupancies that were less prominent in later T-lineage cells, such as later CD4+ CD8+ (double positive, DP) thymocytes and mature peripheral CD4+ T cells (Fig. 1 B; Hu et al., 2018), in agreement with the stage-specific unique roles that we have previously demonstrated for Bcl11b around the time of lineage commitment (DN2-DN3 stages; Longabaugh et al., 2017). The binding profile of Bcl11b in Scid.adh.2c2 cells was intermediate between those in DN3 and DP cells (Fig. 1 B). However, the group 2 sites were fully ILC2-lineage specific. The pro-T and ILC2 lineage-specific sites were also enriched for different motifs (Fig. 1 C). Whereas Runx and ETS (E26 transformation-specific) family motifs were by far the most enriched motifs at the DN3-specific sites (group 1), with TCF/LEF (T-cell factor/ lymphoid enhancer-binding factor; high mobility group) factor motifs a distant third, the most highly enriched motifs at ILC2-specific Bcl11b binding sites were BATF-subfamily bZIP motifs (AP-1 family “BATF” or “JunB”), beyond the frequency of the Runx and ETS-family motifs in these cells (group 2). Similar results were seen when Bcl11b binding sites in ILC2/b6 cells were directly compared with those in the Scid.adh.2c2 pro-T–like cell line (Fig. 1, D–F).
Figure 1.
Bcl11b binds distinct regions across the genome in pro-T and ILC2 cells. (A) Bcl11b ChIP-seq analyses were performed using an ILC2 cell line, ILC2/b6. Venn diagrams show the number of Bcl11b ChIP peaks in ILC2/b6 and DN3 (Longabaugh et al., 2017) cells. (B) Tag count distributions for Bcl11b binding in DN3 (Longabaugh et al., 2017), Scid.adh.2c2, DP, CD4 T (Hu et al., 2018), and ILC2/b6 cells are shown. Each lane represents the merged tag directories from two biological replicates. All Bcl11b binding sites identified as reproducible in the DN3 and ILC2/b6 cells were included in the analysis. (C) The top three enriched sequence motifs of DN3-specific (top) and ILC2/b6-specific (bottom) Bcl11b peaks are shown. TF family names are shown for motifs, with specific HOMER position weight matrix names in parentheses. (D) Venn diagrams show the number of Bcl11b ChIP peaks in Scid.adh.2c2 and ILC2/b6 cells. (E) Tag count distributions for Bcl11b binding in Scid.adh.2c2 and ILC2/b6 cells are shown for reproducible Bcl11b sites, with each lane representing merged tag directories of two biological replicates. (F) The top three enriched sequence motifs of Scid.adh.2c2-specific (top) and ILC2/b6-specific (bottom) Bcl11b peaks are shown. (G–I) Representative binding profiles of Bcl11b in DN3, Scid.adh.2c2, and ILC2/b6 cells around Zbtb16 (G), Ahr (H), and Id2 (I) loci are shown. Pro-T–specific and ILC2-specific Bcl11b binding sites are labeled with magenta and green rectangles, respectively. Data are based on ChIP-seq peaks scored as reproducible in two replicate samples (A–F). Tracks shown are representative of two independent experiments (G–I).
In some genetic regions, all Bcl11b occupancies were cell type specific, but in others only a few cell type–specific sites were interspersed with other, shared-occupancy sites, as shown in the representative genomic regions shown in Fig. 1, G–I. In primary pro-T cells and the pro-T cell line, Bcl11b bound multiple sites at the Zbtb16 locus, a repression target in this context, whereas it bound no sites at this locus in ILC2/b6 cells (Fig. 1 G, magenta rectangles; an intronic region is active in ILC precursors [Mao et al., 2017], but not in these cells). Conversely, sites linked to Ahr were strongly bound in the ILC2/b6 cells (Fig. 1 H, green rectangles), but this locus completely lacked Bcl11b binding in the pro-T cell samples. Interestingly, Bcl11b peaks were found in both cell types around the Id2 locus, which is highly expressed in ILC2 cells but repressed in pro-T cells, and these included not only shared sites but also both ILC2-specific sites (Fig. 1 I, green rectangles) and pro-T–specific sites (magenta rectangles). In pro-T cells, Bcl11b directly represses Id2, and we previously determined that one of the Bcl11b binding sites about 40 kb downstream of the Id2 locus transcriptional start site (+40k) mediates significant repressive activity in pro-T cells (Hosokawa et al., 2018a). This site was bound less strongly in the Scid.adh.2C2 cell line, where repression is leaky, than in primary pro-T cells, but it was completely unbound in ILC2/b6 cells (Fig. 1 I). Thus, Bcl11b binds to functionally different sites in lineage-specific ways.
Bcl11b controls different sets of genes in pro-T and ILC2 cells
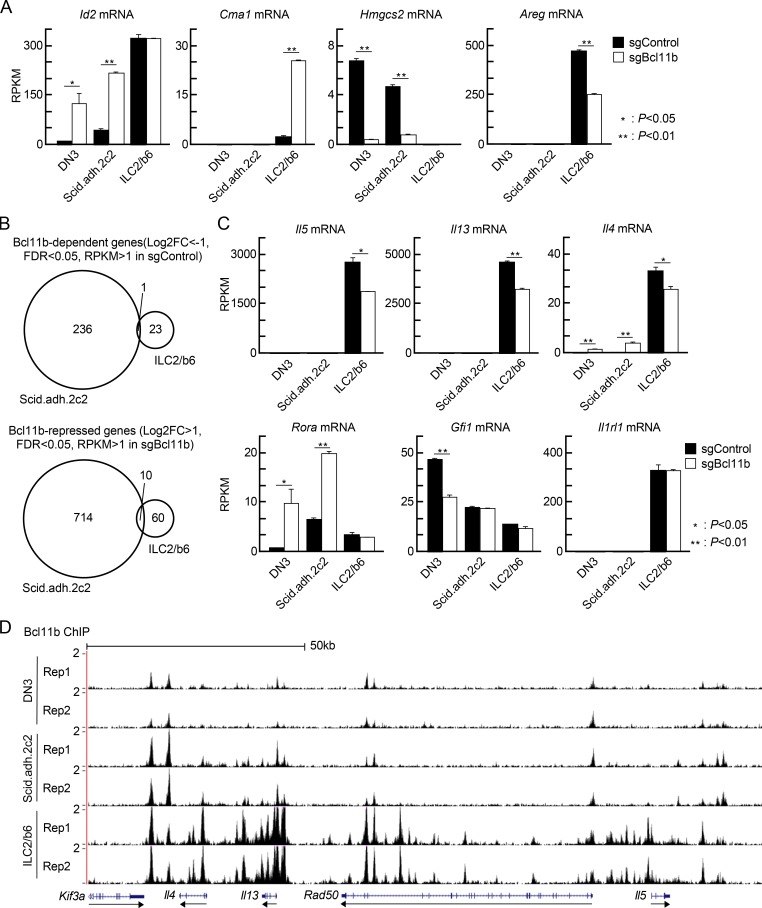
To compare the Bcl11b-regulated target genes in pro-T and ILC2 cells, we disrupted the Bcl11b gene in Scid.adh.2c2 and ILC2/b6 cells, using a multi-vector retroviral transduction strategy for CRISPR/Cas9 mutagenesis, and determined the effects on transcriptome expression. First, Cas9-GFP transduced cells were sorted, and then they were transduced with retroviral vectors with CFP (a brighter version called mTurquoise2) reporters that also encoded sgRNA against Bcl11b or an irrelevant (luciferase) sgRNA control (Hosokawa et al., 2018a). 3 d after sgRNA infection, GFP+CFP+ doubly infected cells were sorted for RNA sequencing (RNA-seq) analyses. Gaps in RNA-seq reads confirmed that the sgBcl11b transduction into the cells nicely induced biallelic deletions at the targeted sites in the Bcl11b locus in both cell types (Fig. S1 B). Expression of Id2 was up-regulated by disruption of Bcl11b in DN3 and Scid.adh.2c2 cells as expected (Hosokawa et al., 2018a; Fig. 2 A). However, Id2 expression in ILC2/b6 cells was high and was not changed by Bcl11b disruption (Fig. 2 A). In contrast, expression of another gene, Cma1, was up-regulated by loss of Bcl11b in ILC2/b6 cells only, indicating that it is an ILC2-specific target of repression by Bcl11b (Fig. 2 A). Targets of cell type–specific activation by Bcl11b were also identified: Bcl11b-dependent expression of Hmgcs2 in pro-T cells and of Areg in ILC2 cells was also cell type specific (Fig. 2 A).
Figure 2.
Bcl11b controls distinct sets of genes in pro-T and ILC2 cells. (A) Scid.adh.2c2 and ILC2/b6 cells were first infected with Cas9-GFP, and GFP+ cells were expanded; then they were transduced with sgBcl11b-CFP retroviruses. 3 d after second infection, GFP+CFP+ cells were sorted and subjected to RNA-seq analysis. RPKM values for Id2, Cma1, Hmgcs2, and Areg in control and Bcl11b KO DN3 (Hosokawa et al., 2018a), Scid.adh.2c2, and ILC2/b6 cells are shown with SD. **, P < 0.01; *, P < 0.05 by Student’s t test. (B) Venn diagrams show the number of Bcl11b-dependent (log2 FC less than −1, FDR <0.05, RPKM >1 in sgControl, top) and Bcl11b-repressed (log2 FC >1, FDR <0.05, RPKM >1 in sgBcl11b, bottom) genes in Scid.ad.2c2 and ILC2/b6 cells. (C) RPKM values for Il5, Il13, Il4, Rora, Gfi1, and Il1rl1 in control and Bcl11b KO DN3, Scid.adh.2c2, and ILC2/b6 cells are shown with SD. **, P < 0.01; *, P < 0.05 by Student’s t test. Data for A–C are based on two replicate RNA-seq samples. (D) Two replicates of binding profiles of Bcl11b in DN3, Scid.adh.2c2, and ILC2/b6 cells around Th2 cytokine loci are shown.
Globally, the functional impacts of Bcl11b on gene expression were more cell type specific than the binding site choices. We defined differentially expressed genes (DEGs) after Cas9-mediated Bcl11b deletion in Scid.adh.2c2 and ILC2/b6 cells, based on false discovery rate (FDR) <0.05, |log2 fold change (FC)| >1 (i.e., greater than two times increase or decrease in expression), and average reads per kilobase million (RPKM) >1 across samples (Table S1). Both positive and negative effects of Bcl11b deletion in Scid.adh.2c2 were more extensive than the effects in ILC2/b6 cells, but in both cell types, 2.6–3.0 times as many genes could be inferred to be Bcl11b repressed as to be Bcl11b dependent. The overwhelming majority of DEGs in both cell types were cell type specific in their regulation by Bcl11b, even though ILC2/b6-specific DEGs were frequently expressed in Scid.adh.2c2 cells (>20%), and vice versa (>55%; Table S1, “RPKM values for DEGs”). Only 1 of 237 Bcl11b-dependent genes in Scid.adh.2c2 cells was also among the 24 Bcl11b-dependent genes in ILC2/b6 cells (Fig. 2 B, top), while only 10 of 724 Bcl11b-repressed genes in Scid.adh.2c2 were also among the 70 in ILC2/b6 cells (Fig. 2 B, bottom). This overlap in DEGs was seen to be limited even though the acute deletion system used here replicated many of the changes previously reported in primary Bcl11b−/− ILC2 cells generated from in vivo steady-state knockout experiments (Califano et al., 2015). For example, expression of Il5, Il13, and Il4 was reproducibly decreased in Bcl11b-deficient ILC2/b6 cells compared with controls (Fig. 2 C). The lineage-specific Bcl11b response of the type 2 cytokine genes was in accord with the ILC2/b6-specific binding of Bcl11b to multiple sites around Il4, Il13, Rad50, and Il5 in the type 2 cytokine gene cluster (Fig. 2 D). In control and Bcl11b-deficient ILC2/b6 cells, expression levels of Il1rl1 (IL-33R) and Rora and Gfi1 were comparable (Fig. 2 C), whereas Rora was up-regulated and Gfi1 was down-regulated by loss of Bcl11b in pro-T cells. Taken together, the data show that Bcl11b binds to distinct genomic regions and controls almost completely different activation and repression target genes in pro-T cells and in ILC2 cells.
Cell type–specific binding profiles of Runx factors and GATA3 in pro-T and ILC2 cells
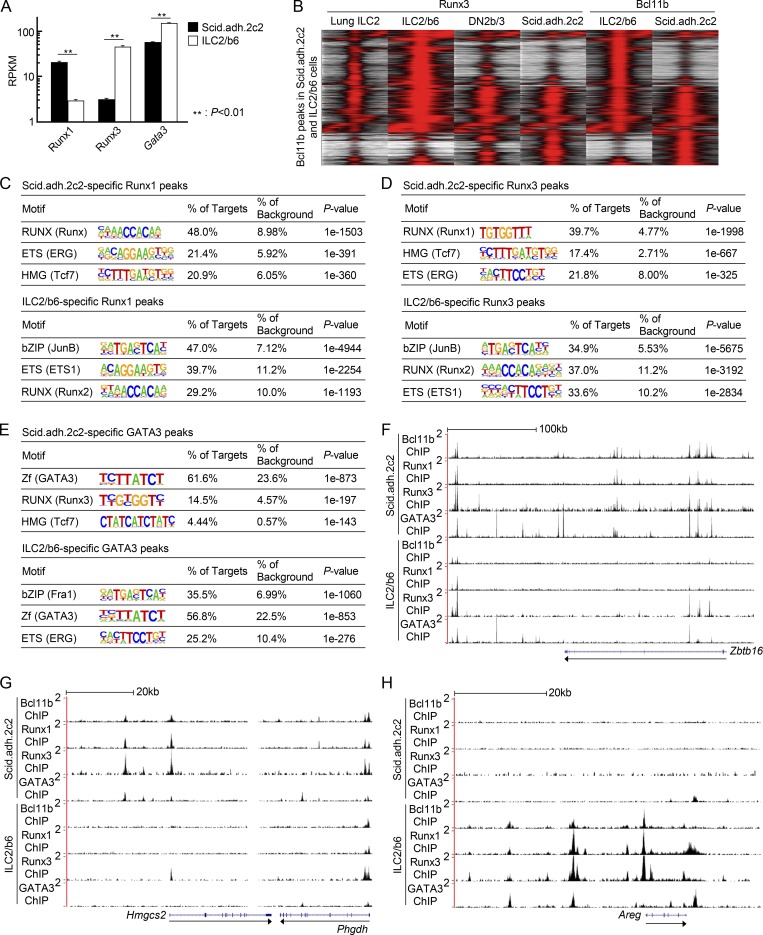
To investigate how Bcl11b could exert such different actions, we examined the binding profiles of known and likely Bcl11b interaction partners that might be differentially active in pro-T and ILC2 cells. We previously reported that Runx1 is one of the most important binding partners of Bcl11b in pro-T cells (Hosokawa et al., 2018a), and Runx factors are important in ILCs as well (Ebihara et al., 2017; Miyamoto et al., 2019). However, ILC2 cells express less Runx1 and more Runx3 than Scid.adh.2c2 (Fig. 3 A) or primary DN2b pro-T cells (not shown). If Runx3 systematically preferred different genomic sites than Runx1, then the altered Runx3/Runx1 ratio might play a role in directing Bcl11b to ILC2-specific sites. Also, GATA3 is crucial to control development and function of both pro-T and ILC2 cells, but is more highly expressed in ILC2 cells (Fig. 3 A) and might thus open up additional chromatin sites for Bcl11b. Therefore, we tested whether genomic DNA binding preferences of Runx3 versus Runx1 and GATA3 could dictate Bcl11b site choices in Scid.adh.2c2 pro-T cells and ILC2/b6 cells.
Figure 3.
Distribution of Runx1, Runx3, and GATA3 binding sites across the genome in pro-T and ILC2 cells. (A) RPKM values for Runx1, Runx3, and Gata3 in Scid.adh.2c2 and ILC2/b6 cells are shown with SD. **, P < 0.01 by Student’s t test. (B) Tag count distributions for Runx3 and Bcl11b in activated primary lung ILC2, ILC2/b6, bone marrow precursor–derived DN2b/3, and Scid.adh.2c2 cells are shown. All Bcl11b binding sites identified in the Scid.adh.2c2 and ILC2/b6 cells were included in the analysis. Note that lung ILC2 data (GEO accession no. GSE111871) were generated using a different cross-linking protocol (Miyamoto et al., 2019). (C–E) The top three enriched sequence motifs of Scid.adh.2c2-specific (top) and ILC2/b6-specific (bottom) Runx1 (C), Runx3 (D), and GATA3 (E) peaks are shown. (F–H) Representative binding profiles of Bcl11b, Runx1, Runx3, and GATA3 in Scid.adh.2c2 and ILC2/b6 cells around Zbtb16 (F), Hmgcs2 (G), and Areg (H) loci are shown. Data are based on two replicate RNA-seq results (A) and are based on reproducible ChIP-seq peaks in two replicate samples (B–E), or are representative of results with replicate samples of each type generated from two independent experiments (F–H).
In fact, in both cell lines, Runx3 had similar site binding profiles with Bcl11b and showed the same cell-type specificity as Bcl11b (Fig. 3 B). Similar to Runx1 (Hosokawa et al., 2018a), Runx3 co-bound with Bcl11b at “pro-T cell sites” in Scid.adh.2c2 cells and a highly overlapping set of sites in primary DN2b/DN3 cells, but was found at the distinct “ILC2” sites of Bcl11b binding in ILC2/b6 cells (Fig. 3 B and Fig. S1, C and D) and in recently published data from primary lung ILC2 cells activated in vitro (Miyamoto et al., 2019; Fig. 3 B, lane 1). Thus, Runx3 did not define the specific ILC2 sites for binding, but followed the cell context in its own site choice. Binding profiles of GATA3 in both cell types clearly differed from the sites for Bcl11b and Runx family factors (Fig. S1, C and E). However, GATA3 also had pro-T–specific and ILC2-specific binding sites across the genome, and the ILC2-specific GATA3 sites were enriched more than the shared GATA3 sites for overlap with the ILC2-specific sites for Bcl11b.
These lineage-specific shifts in site preferences for multiple factors were associated with distinct transcription factor binding motifs. At ILC2/b6-specific Runx binding sites as at ILC2/b6-specific Bcl11b sites, the most highly enriched motif was a bZIP motif, regardless of which Runx binding factor was precipitated (Fig. 3, C and D), contrasting with the preferred Runx, ETS family, and TCF/LEF motifs seen in pro–T cell–specific sites. Whereas the GATA3 motif itself was by far the top enriched motif in Scid.adh.2c2-specific GATA3 sites, the ILC2-specific GATA3 binding sites also had a bZIP motif most highly enriched over background (Fig. 3 E).
Thus, globally, Runx1, Runx3, and Bcl11b binding sites across the genome were often coincident within a cell type, but differed markedly between pro-T and ILC2 contexts. Examples of these distinct patterns are illustrated for Zbtb16 (pro-T cell–biased binding), Hmgcs2 (pro-T cell–specific binding), and Areg (ILC2-specific binding; Fig. 3, F–H). These results show sharp lineage-specific binding differences both for the Runx factors, usually co-recruited with Bcl11b, and for GATA3, often recruited independently of Bcl11b. They also indicate a likely role for a bZIP family member in defining ILC2-specific occupancy sites of Bcl11b, Runx factors, and GATA3.
Proteomic evidence for distinct Bcl11b protein interaction partners in pro-T and ILC2 cells
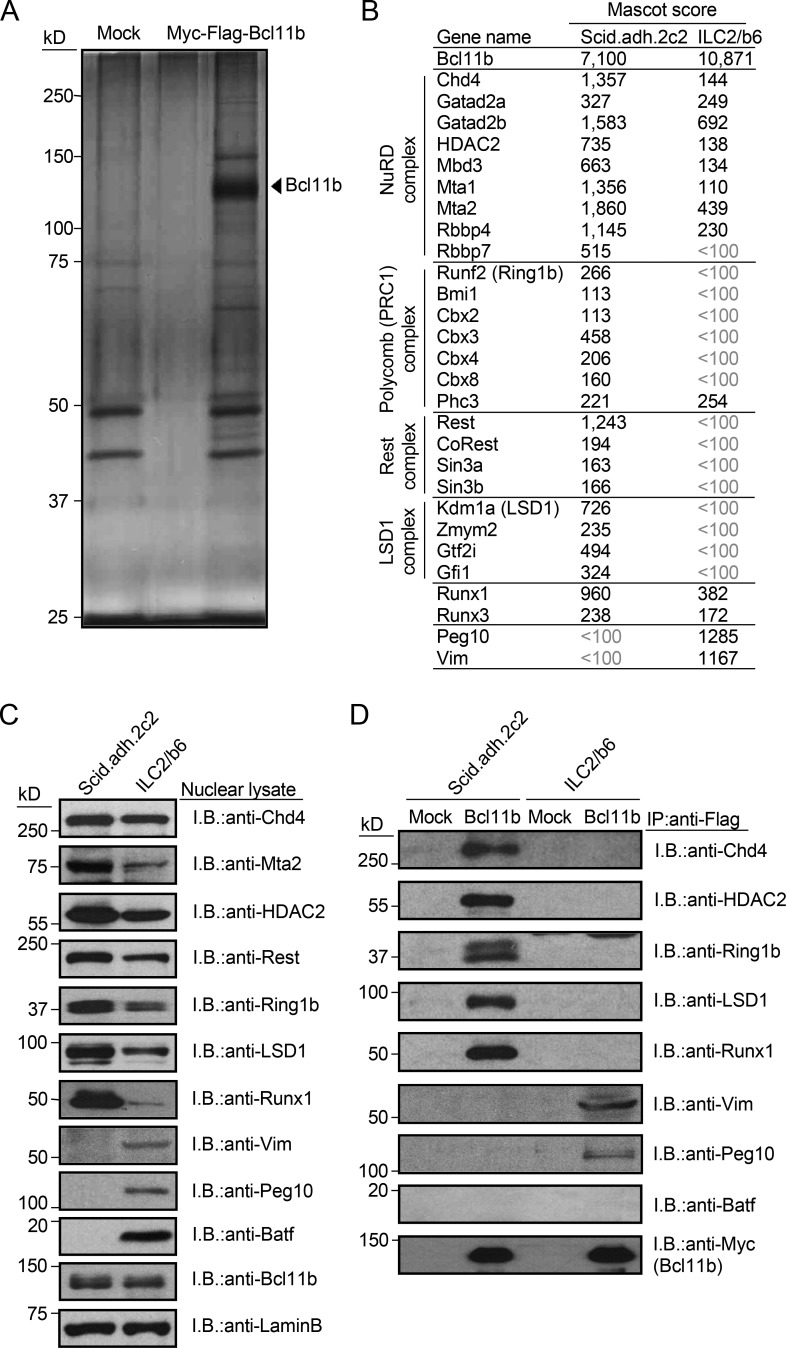
Bcl11b nucleates several distinct chromatin-binding complexes in pro-T cells, including NuRD, polycomb (PRC1), Rest, and Runx1 complexes, as we and others have shown (Cismasiu et al., 2005; Hosokawa et al., 2018a). We hypothesized that if Bcl11b organized different complexes in pro-T and ILC2 cells, this could be a reason why it binds to different genomic regions in pro-T and ILC2 cells. To identify components of Bcl11b-containing complexes, ILC2/b6 cells were transduced with Myc-Flag–tagged Bcl11b for tandem affinity purification. While forced Bcl11b expression is more inhibitory in these cells than in pro-T cells, we were able to isolate tagged Bcl11b complexes by two-step affinity purification followed by SDS/PAGE and silver staining (Fig. 4 A). Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis identified ∼90 molecules with supra-threshold enrichment from the ILC2/b6 cells, and these had only moderate overlap with the major interaction partners identified in the Scid.adh.2c2 cells (Table S2). As in Scid.adh.2c2 cells (Hosokawa et al., 2018a), proteins involved in transcriptional regulation and chromatin remodeling appeared most enriched (Table S2), but statistical confidence scores (Mascot scores; Materials and methods) showed most shared factors to have substantially lower or sub-threshold scores in ILC2 cells (Fig. 4 B). Overall protein levels of major Bcl11b interaction partners in pro-T cells were lower in ILC2 cells, especially Mta2 and Runx1 (Fig. 4 D), which are particularly important for both activation and repression of Bcl11b targets in pro-T cells (Hosokawa et al., 2018a). Some ILC2-specific associations of Bcl11b were found with other proteins, e.g., Peg10, Vim, Jak3, and Dock2 (Table S2), and co-immunoprecipitation followed by immunoblotting tests of a sample of these candidate partners supported these cell type differences (Fig. 4, C and D). Finally, the mass spectrometry detected several cell type–specific differences in post-translational modifications in Bcl11b protein from Scid.adh.2c2 and ILC2/b6 cells, which could contribute to differential association preferences even when the partners were present in both cell types (Fig. S1 F).
Figure 4.
Bcl11b-binding molecules in pro-T and ILC2 cells. (A) Total extracts from Myc-Flag-Bcl11b–expressing ILC2/b6 cells were subjected to two-step affinity purification followed by SDS-PAGE and silver staining. All of the visible bands were subjected to mass spectrometry analysis. (B) Representative Bcl11b-binding molecules in pro-T cells, identified previously (Hosokawa et al., 2018a), and ILC2/b6 are shown with Mascot scores in Scid.adh.2c2 and ILC2/b6 cells. The full list of Bcl11b-binding molecules is shown in Table S2. (C) Nuclear lysates from Scid.adh.2c2 and ILC2/b6 cells were subjected to immunoblotting (I.B.) with the indicated antibodies. (D) Total extracts from Mock- or Myc-Flag-Bcl11b–transduced Scid.adh.2c2 and ILC2/b6 cells were subjected to immunoprecipitation (IP) with anti-Flag mAb followed by immunoblotting with the indicated antibodies. Data in A, C, and D are representative of two independent experiments.
The bZIP family protein BATF was of great interest, because the enrichment of its likely target motifs (Batf or JunB) around ILC2-specific occupancy sites for Bcl11b, Runx1, Runx3, and GATA3 had appeared to be the strongest feature distinguishing the ILC2/b6 from pro-T cell transcription factor binding patterns (Fig. 1, C and F; and Fig. 3, C–E). BATF indeed had stronger expression in ILC2/b6 cells than in Scid.adh.2c2 pro-T cells (Fig. 4 C). However, we failed to detect BATF or any ILC2-specific AP-1 family members as direct interaction partners of the epitope-tagged Bcl11b protein in the mass spectrometry analysis in ILC2 cells (Table S2), or in co-immunoprecipitation and immunoblotting (Fig. 4 D). This suggests that even if Bcl11b might follow BATF to regions where it binds on the genome, it might not form a direct protein complex with it. Binding of Bcl11b across the genome in general occurs frequently at open chromatin sites with active histone marks (Hosokawa et al., 2018a; Hu et al., 2018), and such domains might commonly be established by BATF/AP-1 family proteins in these ILC2 cells.
Shared regulation of Bcl11b expression in pro-T and ILC2 cells by a distal enhancer region downstream of Bcl11b locus
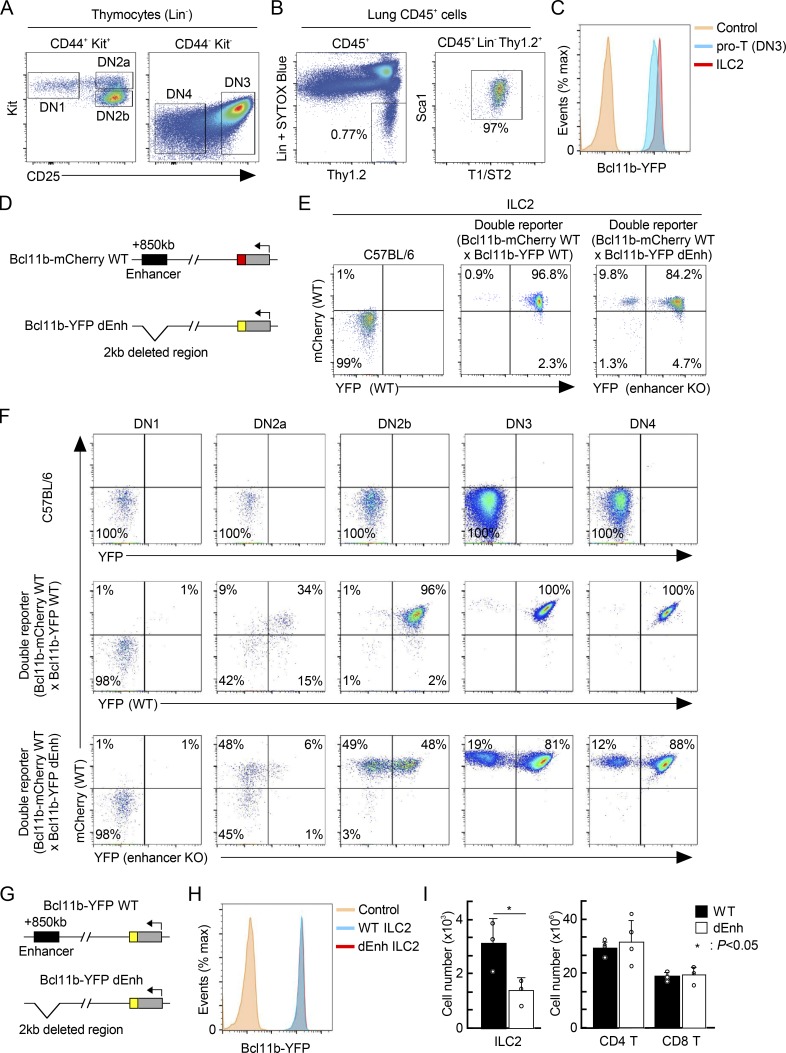
Bcl11b is expressed only in T cells and ILC2 cells among hematopoietic cells, with comparable or higher levels in lung ILC2 cells than in thymic DN3 pro-T cells (Fig. 5, A–C). Expression of Bcl11b is controlled by the hit-and-run licensing functions of Notch signaling, GATA3, and TCF1, and the continuous magnitude control of Runx1 in pro-T cells (Kueh et al., 2016). Both T and ILC2 lineages make strong developmental use of Notch signaling, GATA3, and TCF1 (Cherrier et al., 2018; De Obaldia and Bhandoola, 2015; Koga et al., 2018). However, most precursors of these cells presumably diverge before T cell precursors migrate to the thymus, long before Bcl11b is activated, and it is thus unknown whether they use the same or different genetic circuitry to activate Bcl11b in the first place. Notch signaling, GATA3, and TCF1 may act initially on a distal enhancer +850 kb downstream of the Bcl11b promoter to control the initial timing of Bcl11b activation (Li et al., 2013; Ng et al., 2018). The assay for transposase-accessible chromatin sequencing (ATAC-seq) landscapes around this enhancer appear different in early pro-T cells than in mature small intestine ILC2 (Fig. S1 G; Yoshida et al., 2019); Bcl11b itself bound to this region in pro-T but not ILC2/b6 cells; and the noncoding RNA transcript, ThymoD, which characterizes post-commitment T-lineage cells (Isoda et al., 2017), was not detectable in primary mature ILC2 cells or the ILC2/b6 cell line (Fig. S1 H). However, this need not exclude a common activation mechanism in pro-T and earlier ILC2 precursors. The distal “major peak” enhancer accelerates the earliest Bcl11b expression in pro-T cells, working in a hit-and-run way to increase the likelihood that an allele of Bcl11b will become activated at all (Ng et al., 2018). Thus, in a given cell lineage, if the percentage of cells expressing a Bcl11b allele depends on this distal enhancer, it can be a kind of time stamp for an earlier regulatory state within the cell lineage.
Figure 5.
Expression of Bcl11b in primary ILC2 cells is controlled by a distal enhancer region. (A) Flow cytometric analysis of thymocytes was performed. Representative c-Kit/CD25 profiles in Lin− thymocytes from Bcl11b-YFP mice are shown. Results are representative of four animals. (B) Flow cytometric analysis of lymphocytes from the lung was performed. Representative Thy1.2/Lin and T1/ST2/Sca1 profiles in CD45+ lymphocytes from Bcl11b-YFP mice are shown. Results are representative of four animals. (C) Representative Bcl11b-YFP profiles in pro-T (DN3) cells (A) and lung ILC2 cells (B) from Bcl11b-YFP reporter mice are shown. Results are representative of four animals. (D) Schematic of WT and dEnh two-color Bcl11b reporter mice is shown. (E) Flow cytometric analysis of lymphocytes from the lung was performed. Representative mCherry/YFP profiles in ILC2 cells from WT (left), two-color Bcl11b reporter mice with mCherry-WT/YFP-WT alleles (middle), or mCherry-WT/YFP-dEnh (right) are shown with the percentages of cells in each quadrant. Results are representative of six animals. (F) Flow cytometric analysis of thymocytes was performed. Representative c-Kit/CD25 and mCherry/YFP profiles in Lin− thymocytes from WT (top), two-color Bcl11b reporter mice with mCherry-WT/YFP-WT alleles (middle), or mCherry-WT/YFP-dEnh (bottom) are shown with the percentages of cells in each quadrant. Results are representative of six animals. (G) Schematic of WT and dEnh Bcl11b-YFP reporter alleles is shown. (H) Representative Bcl11b-YFP profiles in lung ILC2 cells from WT-YFP or dEnh-YFP reporter mice are shown. Results are representative of four animals. (I) Cell numbers of ILC2 cells (n = 3) in the lung (left) and CD4 and CD8 T cells (n = 4) in the spleen (right) from WT or dEnh mice are shown with SD. *, P < 0.05 by Student’s t test.
To determine whether the same distal enhancer region is also involved in activation of Bcl11b expression in ILC2 cells, we crossed a WT or enhancer-deleted (dEnh) allele of Bcl11b (also expressing YFP) with a WT allele (also expressing mCherry) in mice (Fig. 5 D), and compared fresh lung ILC2 cells from these animals (Ng et al., 2018; Fig. 5 E). Whereas both alleles were expressed in >96% of ILC2 cells from animals with WT/WT loci, in mice with WT/dEnh loci, ∼10% of lung ILC2 cells selectively failed to express the enhancer-disrupted allele (mCherry+YFP−; Fig. 5 E). This phenotype mirrored the percentage of T-lineage cells that ultimately failed to activate the enhancer-disrupted allele, similar to the monoallelic expression frequency in DN4 pre-T cells from the thymus (Fig. 5 F), and DP, CD4, and CD8 mature T cells from these mice (Ng et al., 2018). The monoallelic expression was due to a clear delay in activation between DN2a and DN3 pro-T stages (Fig. 5 F), and the limited permissive time window for locus activation in overall T cell development as previously described (Ng et al., 2018). The failure to activate Bcl11b from a mutant distal major peak enhancer, although not fully penetrant, also had long-term consequences for ILC2 cells. When we compared mice with homozygous delEnh Bcl11b alleles (YFP) against mice with homozygous WT alleles (YFP), as expected, ILC2 and T cells were generated in substantial numbers in both, and these cells expressed normal levels of Bcl11b-YFP per cell (Fig. 5, G and H). However, the dEnh homozygous mice had significantly fewer total ILC2 cells in the lung than WT mice (Fig. 5 I), a striking difference because numbers of mature CD4 and CD8 T cells in the spleen were normal. Thus, loss of the distal “pro-T cell” enhancer had put about half the ILC2 cells through a developmental bottleneck for which they could not compensate as well as T cells. Therefore, despite their distinct uses of Bcl11b in later mature function, both T and ILC2 cells use the same distal enhancer region stochastically to control timing and likelihood of Bcl11b activation in the developmental pathway.
Concluding remarks
The effector functional similarities between T and ILC2 cells and their shared dependence on Notch, TCF1, GATA3, Bcl11b, and Runx factors can make it appear that these lineages are basically similar except for the E protein–dependent recombination of TCR genes. Similarly, when related cell types share requisite transcription factors, it is tempting to assume that these factors are working on the same targets. This would indeed be predicted if most transcription factor activity were determined primarily by their sequence-specific abilities to “read” the genome to locate the same optimal regulatory sites in all cells. In this work, we have shown that despite likely sharing of the gene network circuitry that activates the Bcl11b locus, early T cells and ILC2 cells actually use Bcl11b protein in markedly different ways across the genome. This result joins others that emphasize the conditionality of transcription factor site choice (e.g., Chronis et al., 2017; Guertin et al., 2014; Heinz et al., 2010; Hosokawa et al., 2018b) to raise important challenges for predictive systems biology of transcriptional regulation. Here, not only Bcl11b but also the Runx factors, which it frequently accompanies, and GATA3, which binds mostly distinct sites, have shown notably different patterns of genomic site choice in the primary and immortalized pro-T cells from those in ILC2 cells. A substantial positive influence on site choice in ILC2 cells appears to be BATF or other bZIP family transcription factors that provide the predominant signature motif at ILC2-specific sites for Bcl11b, Runx factors, and GATA3 alike. However, sites like the Id2 repression site used by Bcl11b in pro-T cells are emptied in ILC2s. Thus, even in two cell types that share as many regulatory factors as pro-T cells and ILC2s, the same transcription factors can regulate substantially different target genes.
Materials and methods
Mice
C57BL/6 (referred to as B6) mice were purchased from the Jackson Laboratory. Bcl11b-mCherry WT (backcrossed to C57BL/6 mice 10 times), Bcl11b-YFP WT (backcrossed to C57BL/6 mice 10 times), and Bcl11b-YFP dEnh (backcrossed to C57BL/6 mice six times) mice were described previously (Ng et al., 2018). All animals were bred and maintained in the California Institute of Technology Laboratory Animal Facility, under specific pathogen–free conditions, and the protocol supporting animal breeding for this work was reviewed and approved by the Institute Animal Care and Use Committee of the California Institute of Technology.
Cells and cell culture
Primary DN2/3 pro-T cells were generated from bone marrow–derived precursors by differentiation on OP9-DL1 stromal cells, exactly as previously described (Hosokawa et al., 2018a).
Scid.adh.2c2 cells (Dionne et al., 2005) were cultured in RPMI 1640 (Gibco) with 10% FBS (Sigma-Aldrich), sodium pyruvate (Gibco), nonessential amino acids (Gibco), Pen-Strep-Glutamine (Gibco), and 50 μM β-mercaptoethanol (Sigma-Aldrich).
An ILC2 cell line, ILC2/b6 (Zhang et al., 2017) was cultured in OP9 medium (α-MEM, 20% FBS, 50 μM β-mercaptoethanol, and Pen-Step-Glutamine) supplemented with 10 ng/ml of IL-2, IL-7, and IL-33 (Pepro Tech Inc.).
CRISPR/Cas9-mediated deletion of Bcl11b in pro-T and ILC2/b6 cells
The method for Cas9-mediated transduction using sequential retroviral vector infections and the vectors used were described previously in detail (Hosokawa et al., 2018a). Briefly, Scid.adh.2c2 or ILC2/b6 were infected with a retroviral vector encoding Cas9-IRES-GFP. 2 d after the first infection, Cas9-introduced GFP+ cells were sorted and cultured for 7 d. Then, they were transduced with a second retrovirus, marked with a CFP reporter, to introduce sgRNA targeting a negative control (luciferase) or Bcl11b. 3 d after the second infection, GFP+CFP+ cells were sorted and subjected to RNA-seq analysis.
Flow cytometry analysis
For staining of thymocytes, surface antibodies against PECy7-CD45, BV510-CD44, APC-c-Kit, APCe780-CD25, and a biotin-conjugated lineage cocktail (CD4, CD8α, CD11b, CD11c, TER-119, NK1.1, TCRβ, and TCRγδ) were used for staining. Lymphocytes were isolated from the lung based on the protocol, previously described (Moro et al., 2015). For ILC2 cells from the lung, antibodies against PECy7-CD45, APCe780-Thy1.2, APC-Sca1, BV510-S1/ST2, and a biotin-conjugated lineage cocktail (CD3, CD4, CD5, CD8α, FcɛR1, NK1.1, F4/80, CD11c, Gr1, CD19, and TER-119) were used for staining. Prior to cell surface staining, cells were treated with 2.4G2 cell supernatant. All of the cells were analyzed using a flow cytometer, MacsQuant 10 (Miltenyi) with FlowJo software (Tree Star).
Two-step affinity purification of Bcl11b complexes from ILC2/b6 cells
ILC2/b6 cells were infected with either Myc-Flag-Bcl11b–containing retrovirus or empty vector control (pMxs-IRES-GFP) as described previously (Champhekar et al., 2015). 3 d after infection, Myc-Flag–tagged Bcl11b-infected GFP+ cells were sorted and expanded for 2 wk, then solubilized with protease inhibitor-containing immunoprecipitation buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% glycerol, 0.1% Tween, 1 mM EDTA, 10 mM NaF, 1 mM dithiothreitol, and a protease inhibitor cocktail [Roche Applied Science]), lysed on ice for 30 min with gentle shaking, and sonicated on a Misonix S-4000 sonicator (Qsonica) for three cycles, amplitude 20 for 30 s followed by 30 s of rest. The insoluble materials were removed by centrifugation, and immunoprecipitation with anti-Flag M2 agarose (Sigma-Aldrich) was performed overnight at 4°C. Immune complexes were eluted from the agarose by 3xFlag peptide (Sigma-Aldrich), and the eluted Bcl11b complexes were subjected to a second immunoprecipitation with anti-Myc gel (MBL). Immune complexes were eluted from the gel with Myc peptide (MBL) and separated by SDS-PAGE. The bands were excised from the gel and subjected to a mass spectrometric analysis to identify corresponding proteins. The gel pieces were washed twice with 100 mM bicarbonate in acetonitrile, and the proteins were digested with trypsin. After adding 0.1% formic acid to the supernatant, the peptides were analyzed by LC-MS/MS with an Advance UHPLC (Bruker) and an Orbitrap Velos Pro Mass Spectrometer (Thermo Fisher Scientific). The resulting MS/MS dataset was analyzed using the Mascot software program (Matrix Science). Mascot score is the probability that the observed match is a random event (Mascot score >100 means absolute probability <1 × 10−10).
Immunoblotting
Nuclear extracts were prepared using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific). The antibodies used for the immunoblot analyses were anti-Chd4 (A301-081A; Bethyl), anti-Mta2 (sc-9447; Santa Cruz Biotechnology), anti-HDAC2 (ab12169; Abcam), anti-Rest (12C11-1B11; Caltech Protein Expression Center), anti-Ring1b (A302-869A; Bethyl), anti-LSD1 (ab17721; Abcam), anti-Runx1 (ab23980; Abcam), anti-Batf (sc-100974; Santa Cruz Biotechnology), anti-Bcl11b (ab18465; Abcam), anti-Vim (ab20346; Abcam), anti-Peg10 (ab181249; Abcam), and anti-Lamin B (sc-6217; Santa Cruz Biotechnology).
Because of the low recovery of Runx1 in co-immunoprecipitation with Bcl11b complexes from ILC2/b6 cells, independently made antibodies against Runx1 and Runx3 from the Weizmann Institute (Levanon et al., 2014) were also tested for immunoblotting. While showing higher Runx3 in total nuclear extracts of ILC2/b6 cells, co-immunoprecipitation results for Runx1 and Runx3 (data not shown) were similar to those shown for Runx1 using ab23980 in Fig. 4 D
ChIP and ChIP-seq
107 cells were fixed with 1% formaldehyde in α-MEM for 10 min at RT (for GATA3), or with 1 mg/ml disuccinimidyl glutarate (Thermo Fisher Scientific) in PBS for 30 min at room temperature followed by an additional 10 min with addition of formaldehyde up to 1% (for Bcl11b, Runx1, and Runx3). The reaction was quenched by addition of 1/10 volume of 0.125 M glycine, and the cells were washed with HBSS (Gibco). Pelleted nuclei were dissolved in lysis buffer (0.5% SDS, 10 mM EDTA, 0.5 mM EGTA, 50 mM Tris-HCl, pH 8, and protease inhibitor cocktail) and sonicated on a Bioruptor (Diagenode) for 18 cycles of 30 s of sonication followed by 30 s of rest, with max power. 6 μg per 107 cells of anti-Bcl11b antibodies (a mixture of A300-383A [Bethyl], A300-385A [Bethyl], ab18465 [Abcam], and 12120 [CST]), anti-Runx1 antibody (ab23980), anti-GATA3 (a mixture of sc-268 [Santa Cruz Biotechnology] and MAB26051 [R&D Systems]), or anti-Runx3 antibody (Levanon et al., 2014) were each separately prebound to Dynabeads anti-Rabbit Ig, Dynabeads anti-Mouse Ig, or Dynabeads Protein A/G (Invitrogen) and then added individually to the diluted chromatin complexes in parallel aliquots. The samples were incubated overnight at 4°C and then washed and eluted for 6 h at 65°C in ChIP elution buffer (20 mM Tris-HCl, pH 7.5, 5 mM EDTA, 50 mM NaCl, 1% SDS, and 50 μg/ml proteinase K). Precipitated chromatin fragments were cleaned up using Zymo ChIP DNA Clean & Concentrator. ChIP-seq libraries were constructed using NEBNext ChIP-Seq Library Preparation Kit (E6240; NEB) and sequenced on Illumina HiSeq2500 in single-read mode with the read length of 50 nt. Analysis pipelines used are described below under ChIP-seq analysis and RNA-seq analysis. All analyses are based on results from at least two biologically separate replicates.
mRNA preparation and RNA-seq
Total RNA was isolated from samples of 1–2 × 105 cultured cells using an RNeasy Micro Kit (Qiagen). Libraries were constructed using NEBNext Ultra RNA Library Prep Kit for Illumina (E7530; NEB) from ∼1 µg of total RNA following the manufacturer’s instructions. Libraries were sequenced on Illumina HiSeq2500 in single-read mode with the read length of 50 nt. Base calls were performed with RTA 1.13.48.0 followed by conversion to FASTQ with bcl2fastq 1.8.4 and produced ∼30 million reads per sample.
ChIP-seq analysis
Base calls were performed with RTA 1.13.48.0 followed by conversion to FASTQ with bcl2fastq 1.8.4 and produced ∼30 million reads per sample. ChIP-seq data were mapped to the mouse genome build NCBI37/mm9 using Bowtie (v1.1.1; http://bowtie-bio.sourceforge.net/index.shtml) with “-v 3 -k 11 -m 10 -t --best --strata” settings, and HOMER tag directories were created with makeTagDirectory and visualized in the UCSC Genome Browser (http://genome.ucsc.edu; Speir et al., 2016). The NCBI37/mm9 assembly was chosen for ChIP-seq sample mapping in this study to ease comparisons with numerous previous data tracks from our laboratory and others. ChIP peaks were identified with findPeaks.pl against a matched control sample using the settings “-P .1 -LP .1 -poisson .1 -style factor.” The identified peaks were annotated to genes with the annotatePeaks.pl command against the mm9 genomic build in the HOMER package. Peak calls were always based on data from at least two independent biological replicates. Peak reproducibility was determined by a HOMER adaptation of the Irreproducibility Discovery Rate package (Karmel, 2014) according to ENCODE guidelines (Kundaje, 2012), as we have described previously (Ungerbäck et al., 2018). Only reproducible high-quality peaks, with a normalized peak score ≥ 15, were considered for further analysis. Motif enrichment analysis was performed with the findMotifsGenome.pl command in the HOMER package using a 200-bp window. Tag density plots and heat maps were created with annotatePeaks.pl (-hist or -hist & -ghist, respectively) in a 2,000-bp region surrounding the indicated transcription factor binding peak center, and by hierarchically clustering the tag count profiles in Cluster3 (de Hoon et al., 2004) with average linkage followed by TreeView visualization (Saldanha, 2004). Bcl11b ChIP-seq data in DN3, DP, and CD4 T cells, and Runx1 and Runx3 ChIP-seq data in in vitro stimulated lung ILC2 cells used in this study are previously published (Hu et al., 2018; Longabaugh et al., 2017; Miyamoto et al., 2019; Gene Expression Omnibus [GEO] accession nos. GSE93572, GSE93572, and GSE111871, respectively).
RNA-seq analysis
RNA-sequenced reads were mapped onto the mouse genome build NCBI37/mm9 with STAR (v2.4.0; Dobin et al., 2013) and post-processed with RSEM (v1.2.25; http://deweylab.github.io/RSEM/; Li and Dewey, 2011) according to the settings in the ENCODE (Encyclopedia of DNA Elements) Consortium long-rna-seq-pipeline (https://github.com/ENCODE-DCC/long-rna-seq-pipeline/blob/master/DAC/STAR_RSEM.sh) with the minor modifications that settings “--output-genome-bam --sampling-for-bam” was added to rsem-calculate-expression. STAR and RSEM reference libraries were created from genome build NCBI37/mm9 together with the Ensembl gene model file Mus_musculus.NCBIM37.66.gtf. The resulting bam-files were used to create HOMER (Heinz et al., 2010) tag directories (makeTagDirectory with “-keepAll” setting). For analysis of statistical significance among DEGs, the raw gene counts were derived from each tag directory with analyzeRepeats.pl with the “-noadj -condenseGenes” options followed by the getDiffExpression.pl command using EdgeR (v3.6.8; http://bioconductor.org/packages/release/bioc/html/edgeR.html; Robinson et al., 2010). For data visualization, RPKM-normalized reads were derived using the analyzeRepeats.pl command with the options “–count exons –condenseGenes –rpkm” followed by log transformation. The normalized datasets were hierarchically clustered with “average” linkage and visualized in MatLab (clustergram). RNA-seq data for naive primary ILC2 cells and stimulated ILC2 cells (4 h and 7 d) were taken from previous publications (Shih et al., 2016; Yagi et al., 2014; GEO accession nos. GSE77695 and GSE47851, respectively).
UCSC Genome Browser BigWig visualization
BigWigs were generated from the aligned SAM or BED-file formats using Samtools (Li et al., 2009), Bedtools (Quinlan and Hall, 2010), and the UCSC genomeCoverageBed and bedGraphToBigWig and normalized to 1 million reads. For visualization of RNA-seq tracks, bamToBed and genomeCoverageBed were used with the “-split” setting enabled. BigWig files were up-loaded to the UCSC Genome Browser (http://genome.ucsc.edu; Speir et al., 2016) for visualization.
DEGs were defined using EdgeR, typically with FDR <0.05, |log2 FC| >1, and RPKM >1 except where otherwise indicated, based on measurements from at least two biologically independent replicates for each sample type. The statistical significance of differences between datasets was determined by two-sided Student’s t test, Fisher’s exact test using Excel, or the R package. Statistical details of experiments can be found in the figure legends.
Data availability
The GEO accession no. for all the new deep-sequencing data reported in this paper is GSE131082
Online supplemental material
Fig. S1 shows a characterization of ILC2/b6 compared with primary ILC2-cell transcriptomes, evidence for successful Bcl11b disruption by Cas9, a comparison of Runx factor binding patterns in ILC2 and pro-T cells, a comparison of Bcl11b protein post-translational modifications in ILC2 and pro-T cells, and comparisons of ATAC accessibilities and noncoding transcription across the distal Bcl11b superenhancer (ThymoD) region in pro-T cells and mature ILC2 cells. Table S1 lists the expression values and differential expression statistics for genes scored as Bcl11b repressed or Bcl11b dependent in the ILC2 and pro-T cell lines. Table S2 lists the proteins found to interact with Bcl11b in the ILC2 and pro-T cell lines, with Mascot scores and gene-ontology (GO) term enrichments.
Supplementary Material
DEGs after Cas9-mediated disruption of Bcl11b gene in Scid.adh.2c2 and ILC2/b6 cells
Bcl11b-interacting molecules in Scid.adh.2c2 and ILC2/b6 cells identified by LC-MS/MS
Acknowledgments
We thank A. Bhandoola (National Cancer Institute, National Institutes of Health) for helpful discussions; D. Perez, J. Tijerina, and R. Diamond for cell sorting and advice; I. Soto for mouse colony care; V. Kumar for library preparation and sequencing; H. Amrhein and D. Trout for computational assistance; I. Antoshechkin for sequencing management; and members of the Rothenberg group for valuable discussion and reagents.
This work was supported by grants from the US Public Health Service to E.V. Rothenberg (R01 AI135200, R01AI083514, and R01HD076915); the Japan Society for the Promotion of Science KAKENHI grant number JP19H03692, the Mochida Memorial Foundation for Medical and Pharmaceutical Research, and the Takeda Science Foundation (to H. Hosokawa); and the Japan Society for the Promotion of Science KAKENHI grant numbers JP19H03708, JP18K08464, JP19K07635, and JP18K07439, the Takeda Science Foundation, the Naito Foundation, the SENSHIN Medical Research Foundation, and the Novartis Research Foundation (to T. Tanaka). This work was partly performed in the Cooperative Research Project Program of the Medical Institute of Bioregulation at Kyushu University and was supported by the Donated Fund of Next Generation Hormone Academy for Human Health & Longevity. This work was partly supported by the California Institute of Regenerative Medicine Bridges to Stem Cell Research Program (Pasadena City College and California Institute of Technology, to M. Romero-Wolf), the L. A. Garfinkle Memorial Laboratory Fund and the Al Sherman Foundation, special project funds from the Provost and Division of Biology & Biological Engineering of the California Institute of Technology, and the California Institute of Technology Albert Billings Ruddock Professorship (to E.V. Rothenberg).
The authors declare no competing financial interests.
Author contributions: H. Hosokawa designed the study, performed experiments, analyzed data, and wrote the manuscript. M. Romero-Wolf performed experiments and analyzed data. Q. Yang provided the ILC2 cell line (ILC2/b6) and helpful discussions and edited the manuscript. D. Levanon and Y. Groner provided unique biological reagents and valuable criticism and edited the manuscript. Y. Motomura, K. Moro, and T. Tanaka performed experiments, analyzed data, and provided helpful discussions and comments on the manuscript. E.V. Rothenberg designed and supervised the study, analyzed data, and wrote the manuscript.
References
- Avram D., and Califano D.. 2014. The multifaceted roles of Bcl11b in thymic and peripheral T cells: impact on immune diseases. J. Immunol. 1932059–2065. 10.4049/jimmunol.1400930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Califano D., Cho J.J., Uddin M.N., Lorentsen K.J., Yang Q., Bhandoola A., Li H., and Avram D.. 2015. Transcription factor Bcl11b controls identity and function of mature Type 2 innate lymphoid cells. Immunity. 43354–368. 10.1016/j.immuni.2015.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champhekar A., Damle S.S., Freedman G., Carotta S., Nutt S.L., and Rothenberg E.V.. 2015. Regulation of early T-lineage gene expression and developmental progression by the progenitor cell transcription factor PU.1. Genes Dev. 29832–848. 10.1101/gad.259879.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrier D.E., Serafini N., and Di Santo J.P.. 2018. Innate lymphoid cell development: A T cell perspective. Immunity. 481091–1103. 10.1016/j.immuni.2018.05.010 [DOI] [PubMed] [Google Scholar]
- Chronis C., Fiziev P., Papp B., Butz S., Bonora G., Sabri S., Ernst J., and Plath K.. 2017. Cooperative binding of transcription factors orchestrates reprogramming. Cell. 168442–459.e20. 10.1016/j.cell.2016.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cismasiu V.B., Adamo K., Gecewicz J., Duque J., Lin Q., and Avram D.. 2005. BCL11B functionally associates with the NuRD complex in T lymphocytes to repress targeted promoter. Oncogene. 246753–6764. 10.1038/sj.onc.1208904 [DOI] [PubMed] [Google Scholar]
- Constantinides M.G., Gudjonson H., McDonald B.D., Ishizuka I.E., Verhoef P.A., Dinner A.R., and Bendelac A.. 2015. PLZF expression maps the early stages of ILC1 lineage development. Proc. Natl. Acad. Sci. USA. 1125123–5128. 10.1073/pnas.1423244112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon M.J., Imoto S., Nolan J., and Miyano S.. 2004. Open source clustering software. Bioinformatics. 201453–1454. 10.1093/bioinformatics/bth078 [DOI] [PubMed] [Google Scholar]
- De Obaldia M.E., and Bhandoola A.. 2015. Transcriptional regulation of innate and adaptive lymphocyte lineages. Annu. Rev. Immunol. 33607–642. 10.1146/annurev-immunol-032414-112032 [DOI] [PubMed] [Google Scholar]
- Dionne C.J., Tse K.Y., Weiss A.H., Franco C.B., Wiest D.L., Anderson M.K., and Rothenberg E.V.. 2005. Subversion of T lineage commitment by PU.1 in a clonal cell line system. Dev. Biol. 280448–466. 10.1016/j.ydbio.2005.01.027 [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2915–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebihara T., Seo W., and Taniuchi I.. 2017. Roles of RUNX complexes in immune cell development. Adv. Exp. Med. Biol. 962395–413. 10.1007/978-981-10-3233-2_24 [DOI] [PubMed] [Google Scholar]
- Guertin M.J., Zhang X., Coonrod S.A., and Hager G.L.. 2014. Transient estrogen receptor binding and p300 redistribution support a squelching mechanism for estradiol-repressed genes. Mol. Endocrinol. 281522–1533. 10.1210/me.2014-1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harly C., Cam M., Kaye J., and Bhandoola A.. 2018. Development and differentiation of early innate lymphoid progenitors. J. Exp. Med. 215249–262. 10.1084/jem.20170832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., and Glass C.K.. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa H., Romero-Wolf M., Yui M.A., Ungerbäck J., Quiloan M.L.G., Matsumoto M., Nakayama K.I., Tanaka T., and Rothenberg E.V.. 2018a Bcl11b sets pro-T cell fate by site-specific cofactor recruitment and by repressing Id2 and Zbtb16. Nat. Immunol. 191427–1440. 10.1038/s41590-018-0238-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa H., Ungerbäck J., Wang X., Matsumoto M., Nakayama K.I., Cohen S.M., Tanaka T., and Rothenberg E.V.. 2018b Transcription factor PU.1 represses and activates gene expression in early T cells by redirecting partner transcription factor binding. Immunity. 481119–1134.e7. 10.1016/j.immuni.2018.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G., Cui K., Fang D., Hirose S., Wang X., Wangsa D., Jin W., Ried T., Liu P., Zhu J., et al. 2018. Transformation of accessible chromatin and 3D nucleome underlies lineage commitment of early T cells. Immunity. 48227–242.e8. 10.1016/j.immuni.2018.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isoda T., Moore A.J., He Z., Chandra V., Aida M., Denholtz M., Piet van Hamburg J., Fisch K.M., Chang A.N., Fahl S.P., et al. 2017. Non-coding transcription instructs chromatin folding and compartmentalization to dictate enhancer-promoter communication and T cell fate. Cell. 171103–119.e18. 10.1016/j.cell.2017.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmel A. 2014. HOMER-IDR. Available at: https://github.com/karmel/homer-idr. Last updated 2014-08 (accessed April 4, 2016).
- Koga S., Hozumi K., Hirano K.I., Yazawa M., Terooatea T., Minoda A., Nagasawa T., Koyasu S., and Moro K.. 2018. Peripheral PDGFRα+gp38+ mesenchymal cells support the differentiation of fetal liver-derived ILC2. J. Exp. Med. 2151609–1626. 10.1084/jem.20172310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojo S., Tanaka H., Endo T.A., Muroi S., Liu Y., Seo W., Tenno M., Kakugawa K., Naoe Y., Nair K., et al. 2017. Priming of lineage-specifying genes by Bcl11b is required for lineage choice in post-selection thymocytes. Nat. Commun. 8702 10.1038/s41467-017-00768-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueh H.Y., Yui M.A., Ng K.K.H., Pease S.S., Zhang J.A., Damle S.S., Freedman G., Siu S., Bernstein I.D., Elowitz M.B., and Rothenberg E.V.. 2016. Asynchronous combinatorial action of four regulatory factors activates Bcl11b for T cell commitment. Nat. Immunol. 17956–965. 10.1038/ni.3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundaje A. 2012. (2012) ENCODE: TF ChIP-seq peak calling using the Irreproducibility Discovery Rate (IDR) framework. Available at: https://sites.google.com/site/anshulkundaje/projects/idr (accessed April 4, 2016).
- Levanon D., Negreanu V., Lotem J., Bone K.R., Brenner O., Leshkowitz D., and Groner Y.. 2014. Transcription factor Runx3 regulates interleukin-15-dependent natural killer cell activation. Mol. Cell. Biol. 341158–1169. 10.1128/MCB.01202-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., and Dewey C.N.. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 12323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., and Durbin R.. 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 252078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Leid M., and Rothenberg E.V.. 2010a An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science. 32989–93. 10.1126/science.1188989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Burke S., Wang J., Chen X., Ortiz M., Lee S.C., Lu D., Campos L., Goulding D., Ng B.L., et al. 2010b Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science. 32985–89. 10.1126/science.1188063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhang J.A., Dose M., Kueh H.Y., Mosadeghi R., Gounari F., and Rothenberg E.V.. 2013. A far downstream enhancer for murine Bcl11b controls its T-cell specific expression. Blood. 122902–911. 10.1182/blood-2012-08-447839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Li P., and Burke S.. 2010. Critical roles of Bcl11b in T-cell development and maintenance of T-cell identity. Immunol. Rev. 238138–149. 10.1111/j.1600-065X.2010.00953.x [DOI] [PubMed] [Google Scholar]
- Longabaugh W.J.R., Zeng W., Zhang J.A., Hosokawa H., Jansen C.S., Li L., Romero-Wolf M., Liu P., Kueh H.Y., Mortazavi A., and Rothenberg E.V.. 2017. Bcl11b and combinatorial resolution of cell fate in the T-cell gene regulatory network. Proc. Natl. Acad. Sci. USA. 1145800–5807. 10.1073/pnas.1610617114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao A.P., Ishizuka I.E., Kasal D.N., Mandal M., and Bendelac A.. 2017. A shared Runx1-bound Zbtb16 enhancer directs innate and innate-like lymphoid lineage development. Nat. Commun. 8863 10.1038/s41467-017-00882-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto C., Kojo S., Yamashita M., Moro K., Lacaud G., Shiroguchi K., Taniuchi I., and Ebihara T.. 2019. Runx/Cbfβ complexes protect group 2 innate lymphoid cells from exhausted-like hyporesponsiveness during allergic airway inflammation. Nat. Commun. 10447 10.1038/s41467-019-08365-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro K., Ealey K.N., Kabata H., and Koyasu S.. 2015. Isolation and analysis of group 2 innate lymphoid cells in mice. Nat. Protoc. 10792–806. 10.1038/nprot.2015.047 [DOI] [PubMed] [Google Scholar]
- Ng K.K.H., Yui M.A., Mehta A., Siu S., Irwin B., Pease S., Hirose S., Elowitz M.B., Rothenberg E.V., and Kueh H.Y.. 2018. A stochastic epigenetic switch controls the dynamics of T-cell lineage commitment. eLife. 7e37851 10.7554/eLife.37851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., and Hall I.M.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha A.J. 2004. Java Treeview--extensible visualization of microarray data. Bioinformatics. 203246–3248. 10.1093/bioinformatics/bth349 [DOI] [PubMed] [Google Scholar]
- Seillet C., Mielke L.A., Amann-Zalcenstein D.B., Su S., Gao J., Almeida F.F., Shi W., Ritchie M.E., Naik S.H., Huntington N.D., et al. 2016. Deciphering the innate lymphoid cell transcriptional program. Cell Reports. 17436–447. 10.1016/j.celrep.2016.09.025 [DOI] [PubMed] [Google Scholar]
- Serafini N., Vosshenrich C.A., and Di Santo J.P.. 2015. Transcriptional regulation of innate lymphoid cell fate. Nat. Rev. Immunol. 15415–428. 10.1038/nri3855 [DOI] [PubMed] [Google Scholar]
- Shih H.Y., Sciumè G., Mikami Y., Guo L., Sun H.W., Brooks S.R., Urban J.F. Jr., Davis F.P., Kanno Y., and O’Shea J.J.. 2016. Developmental acquisition of regulomes underlies innate lymphoid cell functionality. Cell. 1651120–1133. 10.1016/j.cell.2016.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speir M.L., Zweig A.S., Rosenbloom K.R., Raney B.J., Paten B., Nejad P., Lee B.T., Learned K., Karolchik D., Hinrichs A.S., et al. 2016. The UCSC Genome Browser database: 2016 update. Nucleic Acids Res. 44(D1):D717–D725. 10.1093/nar/gkv1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerbäck J., Hosokawa H., Wang X., Strid T., Williams B.A., Sigvardsson M., and Rothenberg E.V.. 2018. Pioneering, chromatin remodeling, and epigenetic constraint in early T-cell gene regulation by SPI1 (PU.1). Genome Res. 281508–1519. 10.1101/gr.231423.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J.A., Oliphant C.J., Englezakis A., Yu Y., Clare S., Rodewald H.R., Belz G., Liu P., Fallon P.G., and McKenzie A.N.J.. 2015. Bcl11b is essential for group 2 innate lymphoid cell development. J. Exp. Med. 212875–882. 10.1084/jem.20142224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.C., Qian L., Zhao Y., Mengarelli J., Adrianto I., Montgomery C.G., Urban J.F. Jr., Fung K.M., and Sun X.H.. 2017. Downregulation of E protein activity augments an ILC2 differentiation program in the thymus. J. Immunol. 1983149–3156. 10.4049/jimmunol.1602009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R., Zhong C., Northrup D.L., Yu F., Bouladoux N., Spencer S., Hu G., Barron L., Sharma S., Nakayama T., et al. 2014. The transcription factor GATA3 is critical for the development of all IL-7Rα-expressing innate lymphoid cells. Immunity. 40378–388. 10.1016/j.immuni.2014.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Lareau C.A., Ramirez R.N., Rose S.A., Maier B., Wroblewska A., Desland F., Chudnovskiy A., Mortha A., Dominguez C., et al. Immunological Genome Project . 2019. The cis-regulatory atlas of the mouse immune system. Cell. 176897–912.e20. 10.1016/j.cell.2018.12.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Wang C., Clare S., Wang J., Lee S.C., Brandt C., Burke S., Lu L., He D., Jenkins N.A., et al. 2015. The transcription factor Bcl11b is specifically expressed in group 2 innate lymphoid cells and is essential for their development. J. Exp. Med. 212865–874. 10.1084/jem.20142318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Tsang J.C., Wang C., Clare S., Wang J., Chen X., Brandt C., Kane L., Campos L.S., Lu L., et al. 2016. Single-cell RNA-seq identifies a PD-1hi ILC progenitor and defines its development pathway. Nature. 539102–106. 10.1038/nature20105 [DOI] [PubMed] [Google Scholar]
- Zhang K., Xu X., Pasha M.A., Siebel C.W., Costello A., Haczku A., MacNamara K., Liang T., Zhu J., Bhandoola A., et al. 2017. Cutting Edge: Notch signaling promotes the plasticity of group-2 innate lymphoid cells. J. Immunol. 1981798–1803. 10.4049/jimmunol.1601421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zook E.C., and Kee B.L.. 2016. Development of innate lymphoid cells. Nat. Immunol. 17775–782. 10.1038/ni.3481 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DEGs after Cas9-mediated disruption of Bcl11b gene in Scid.adh.2c2 and ILC2/b6 cells
Bcl11b-interacting molecules in Scid.adh.2c2 and ILC2/b6 cells identified by LC-MS/MS
Data Availability Statement
The GEO accession no. for all the new deep-sequencing data reported in this paper is GSE131082