Rag GTPase–dependent nutrient mTORC1 signaling plays a critical role in control of T reg cell suppression of autoimmunity and tumor immunity. These findings establish nutrients as a novel class of signaling molecule.
Abstract
Foxp3+ regulatory T (T reg) cells are pivotal regulators of immune tolerance, with T cell receptor (TCR)–driven activated T reg (aT reg) cells playing a central role; yet how TCR signaling propagates to control aT reg cell responses remains poorly understood. Here we show that TCR signaling induces expression of amino acid transporters, and renders amino acid–induced activation of mTORC1 in aT reg cells. T reg cell–specific ablation of the Rag family small GTPases RagA and RagB impairs amino acid–induced mTORC1 signaling, causing defective amino acid anabolism, reduced T reg cell proliferation, and a rampant autoimmune disorder similar in severity to that triggered by T reg cell–specific TCR deficiency. Notably, T reg cells in peripheral tissues, including tumors, are more sensitive to Rag GTPase–dependent nutrient sensing. Ablation of RagA alone impairs T reg cell accumulation in the tumor, resulting in enhanced antitumor immunity. Thus, nutrient mTORC1 signaling is an essential component of TCR-initiated T reg cell reprogramming, and Rag GTPase activities may be titrated to break tumor immune tolerance.
Introduction
Thymus-derived regulatory T (T reg) cells, defined by expression of the transcription factor forkhead box P3 (Foxp3), play a central role in the control of immune tolerance to self-antigens and commensals, while excessive T reg cell activities impede immune responses to pathogens and tumors (Sakaguchi et al., 2008; Josefowicz et al., 2012; Bluestone et al., 2015; Panduro et al., 2016; Shevach, 2018). Due to an intermediate level of TCR signaling involved in T reg cell selection and Foxp3-induced TCR signal tuning, recent thymic emigrant T reg cells display a resting phenotype characterized by low expression of the T cell activation marker CD44 and high expression of the lymph node–homing molecule CD62L (Smigiel et al., 2014). Following antigen reencountering in secondary lymphoid organs, resting T reg (rT reg) cells are converted to CD44hiCD62Llo activated T reg (aT reg) cells and migrate to peripheral tissues (Huehn et al., 2004; Luo et al., 2016; Miyara et al., 2009; Sugiyama et al., 2013). T reg cell–specific ablation of the TCRα-chain depletes aT reg cells, but not rT reg cells, resulting in rampant autoimmunity, supporting an important function for TCR-driven aT reg cells in control of immunological self-tolerance (Levine et al., 2014; Vahl et al., 2014).
To define how TCR stimulation promotes aT reg cell responses is a field of active research. The mechanistic target of rapamycin complex 1 (mTORC1) kinase is a master regulator of cell growth and proliferation through induction of macromolecule biosynthesis and cell anabolism (Laplante and Sabatini, 2012). Compared with conventional CD4+ T cells, T reg cells exhibit elevated TCR-dependent mTORC1 activation (Vahl et al., 2014). T reg cell–specific ablation of the mTORC1 component regulatory-associated protein mTOR (RAPTOR) results in a lethal autoimmune disease, revealing a crucial role for mTORC1 signaling in control of T reg cell–mediated immune tolerance (Zeng et al., 2013).
Antigen stimulation can modulate mTORC1 signaling through Akt-induced inactivation of the tuberous sclerosis complex (TSC) that functions as a GTPase-activating protein for the lysosomal small GTPase Rheb, an activator of mTORC1 (Inoki et al., 2003; Tee et al., 2003). In addition, nutrients, in particular amino acids, promote mTORC1 recruitment to the lysosome to be activated by Rheb. Notably, antigen stimulation activates the glutamine transporter ASCT2 and induces expression of the System L amino acid transporter Slc7a5 via calcineurin-dependent mechanisms (Nakaya et al., 2014; Sinclair et al., 2013). Slc7a5- or ASCT2-deficient T cells exhibit defective mTORC1 signaling (Nakaya et al., 2014; Sinclair et al., 2013). In addition, a recent study showed that mice fed with an amino acid–reduced diet have decreased numbers of T reg cells associated with attenuated mTORC1 activation, which is phenocopied in mice with T reg cell–specific deletion of the amino acid transporter Slc3a2 (CD98 heavy chain; Ikeda et al., 2017). However, whether the T reg cell defects are caused by compromised amino acid metabolism or attenuated amino acid–induced mTORC1 signaling is unknown.
How nutrient availability promotes mTORC1 activation has started to be revealed and involves several endomembrane small GTPases. The lysosomal Rag family of small GTPases was the first reported to facilitate amino acid–induced mTORC1 signaling (Kim et al., 2008; Sancak et al., 2008). Mammals have four Rag proteins, RagA–D, which form obligatory heterodimers made of RagA or the highly related RagB binding to RagC or RagD that are also homologous to one another (Kim et al., 2008; Sancak et al., 2008). Upon amino acid stimulation, the active Rag complex consisting of RagA/B in the guanosine triphosphate (GTP)–bound state and RagC/D in the guanosine diphosphate–bound state promotes mTORC1 translocation to the lysosome. In addition to Rag GTPases, Arf1 can recruit mTORC1 to the lysosome following glutamine stimulation (Jewell et al., 2015), and Rab1A supports amino acid–induced mTORC1 activation by promoting its recruitment to the Golgi apparatus (Thomas et al., 2014). Nonetheless, the in vivo function of these GTPases in control of mTORC1 signaling in T reg cells has not been studied.
Results and discussion
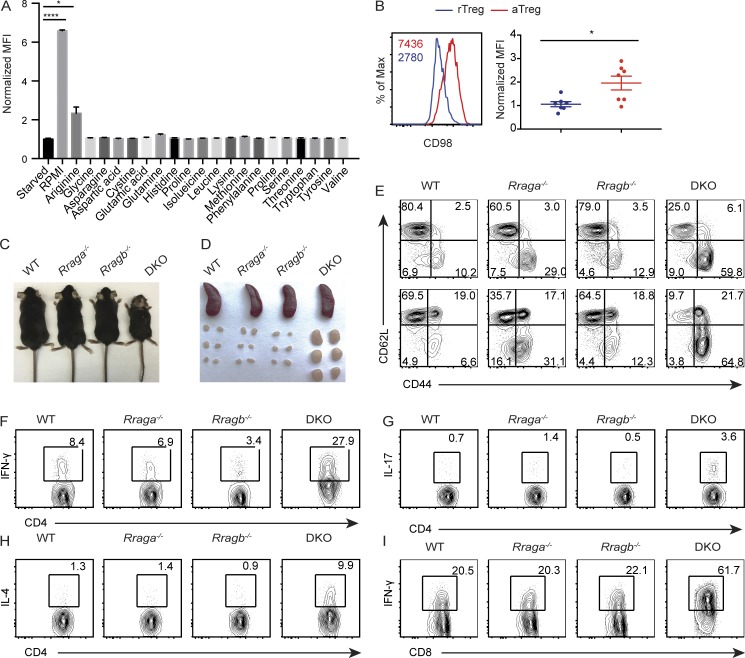
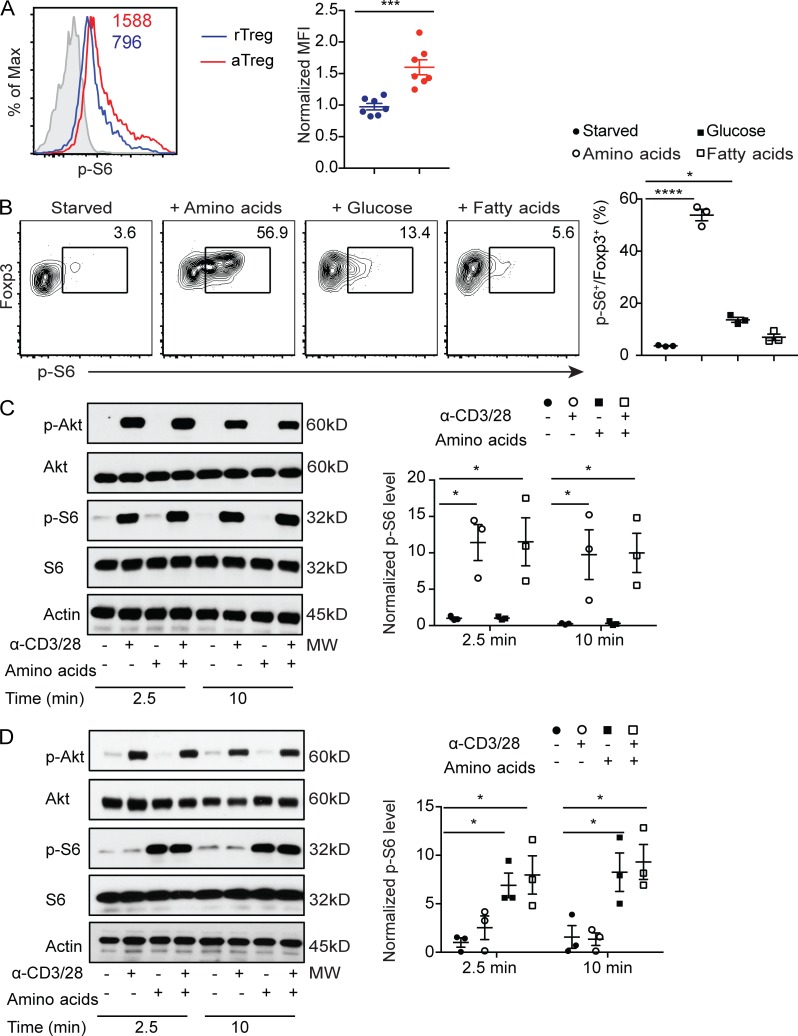
Mice with T reg cell–specific ablation of RAPTOR develop a lethal autoimmune disease with comparable kinetics to that of T reg cell–specific TCRα-deficient mice (Levine et al., 2014; Vahl et al., 2014; Zeng et al., 2013), supporting a critical role for TCR-induced mTORC1 signaling in control of T reg cell–dependent immune tolerance. Notably, phosphorylation of the mTORC1 signaling pathway marker S6 ribosomal protein (p-S6) was higher in aT reg cells than rT reg cells (Fig. S1). mTORC1 is activated by integrating signals of growth factors and nutrients (Laplante and Sabatini, 2012). Macronutrients including amino acids, glucose, and fatty acids all promote cell growth; yet amino acids most potently induced mTORC1 signaling in aT reg cells (Fig. 1 B). Specifically, arginine appeared to be the strongest inducer of mTORC1 activation, although the magnitude was much less than the combination of all amino acids (Fig. S1 A). In line with the critical role of amino acids in mTORC1 signaling, aT reg cells expressed higher levels of CD98 than rT reg cells (Fig. S1 B), implying that enhanced mTORC1 activity in aT reg cells is promoted by increased amino acid uptake.
Figure S1.
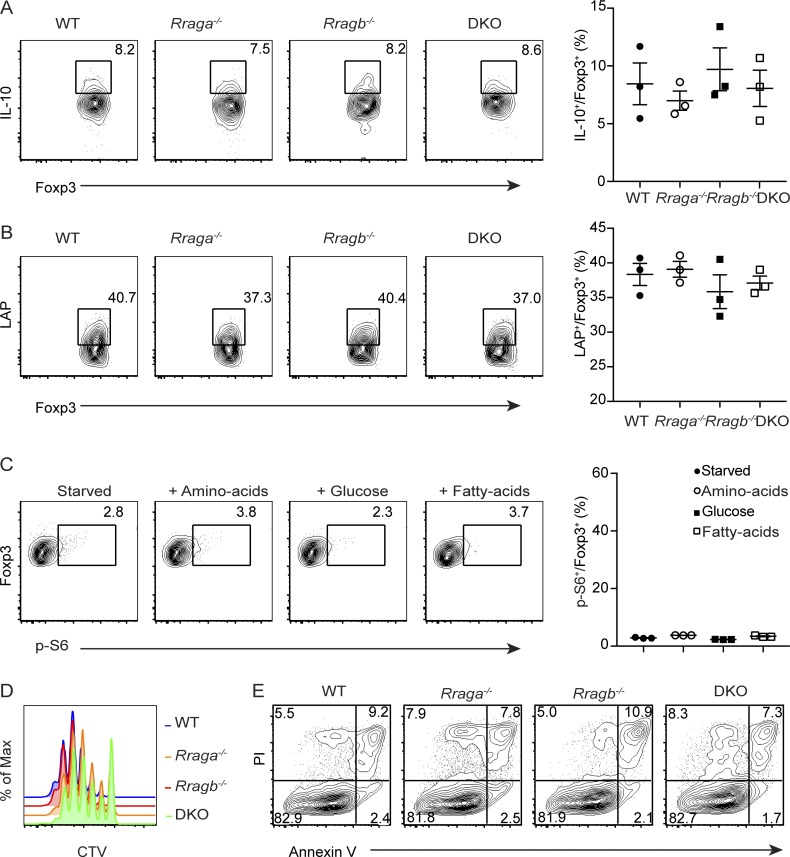
Mice lacking RagA and RagB in T reg cells develop lethal autoimmune disease. (A) Quantification of p-S6 expression in aT reg cells that were deprived of amino acids for 1 h and refed with all 20 amino acids or individual amino acid as indicated for 1 h (n = 3). Data are pooled from three independent experiments. (B) Flow cytometric analysis and quantification of CD98 expression in splenic aT reg and rT reg cells. Numbers in graphs indicate MFI (n = 7 mice each group). Data are pooled from three independent experiments. (C) Representative images of 5-wk-old WT, Rraga−/−, Rragb−/−, and DKO mice. Data are representative of more than five independent experiments. (D) Representative images of spleens and peripheral (axillary, brachial, and inguinal) lymph nodes from 5-wk-old WT, Rraga−/−, Rragb−/−, and DKO mice. Data are representative of more than five independent experiments. (E) CD44 and CD62L expression in splenic CD4+Foxp3− (top) and CD8+ (bottom) T cells from 5-wk-old WT, Rraga−/−, Rragb−/−, and DKO mice. Data are representative of more than five independent experiments. (F–I) IFN-γ, IL-4, and IL-17 expression in splenic CD4+Foxp3− T cells and IFN-γ expression in splenic CD8+ T cells from 5-wk-old WT, Rraga−/−, Rragb−/−, and DKO mice. Data are representative of more than five independent experiments. Numbers in gates or quadrants indicate percentage of cells. Data in plots indicate means ± SEM. *, P < 0.05; ****, P < 0.0001 (unpaired t test; A and B). Max, maximum.
Figure 1.
Amino acids induce sustained mTORC1 signaling in aT reg cells. (A) Flow cytometric analysis and quantification of p-S6 expression in splenic aT reg and rT reg cells. Numbers in graphs indicate mean fluorescence intensity (MFI; n = 7 mice each group). Data are pooled from three independent experiments. (B) Flow cytometric analysis and quantification of p-S6 expression in aT reg cells that were deprived of nutrient for 1 h and refed with amino acids, glucose, or fatty acids for 1 h (n = 3). Data are pooled from three independent experiments. (C and D) Immunoblot analysis of p-S6 and phosphorylated-Akt (p-Akt) levels in rT reg (C) and aT reg (D) cells rested for 1 h in amino acid–deficient media, followed by stimulation with or without α-CD3/28 cross-linking in the presence or absence of amino acids for the indicated times. p-S6 expression level normalized to total S6 protein is shown (n = 3). Data are pooled from three independent experiments. Data in plots indicate mean ± SEM. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001 (unpaired t test [A and B], one-way ANOVA with the Dunnett post hoc test [C and D]). Max, maximum; MW, molecular weight.
To further assess how T reg cell activation status affects the growth factor and nutrient arms of mTORC1 signaling, we performed in vitro culture experiments with anti-CD3 and anti-CD28 (α-CD3/28) in the absence or presence of amino acids. α-CD3/28 induced mTORC1 signaling concomitant with Akt activation in rT reg cells (Fig. 1 C), while amino acids were dispensable for mTORC1 activation (Fig. 1 C). Following 18 h stimulation by α-CD3/28 and IL-2, rT reg cells differentiated to aT reg cells characterized by low expression of CD62L and high expression of CD44 and CD98 (data not shown). Strikingly, restimulation with amino acids, but not with α-CD3/28, potently activated mTORC1 in aT reg cells (Fig. 1 D). These findings demonstrate that aT reg cells are preferentially dependent on nutrients for sustained mTORC1 signaling.
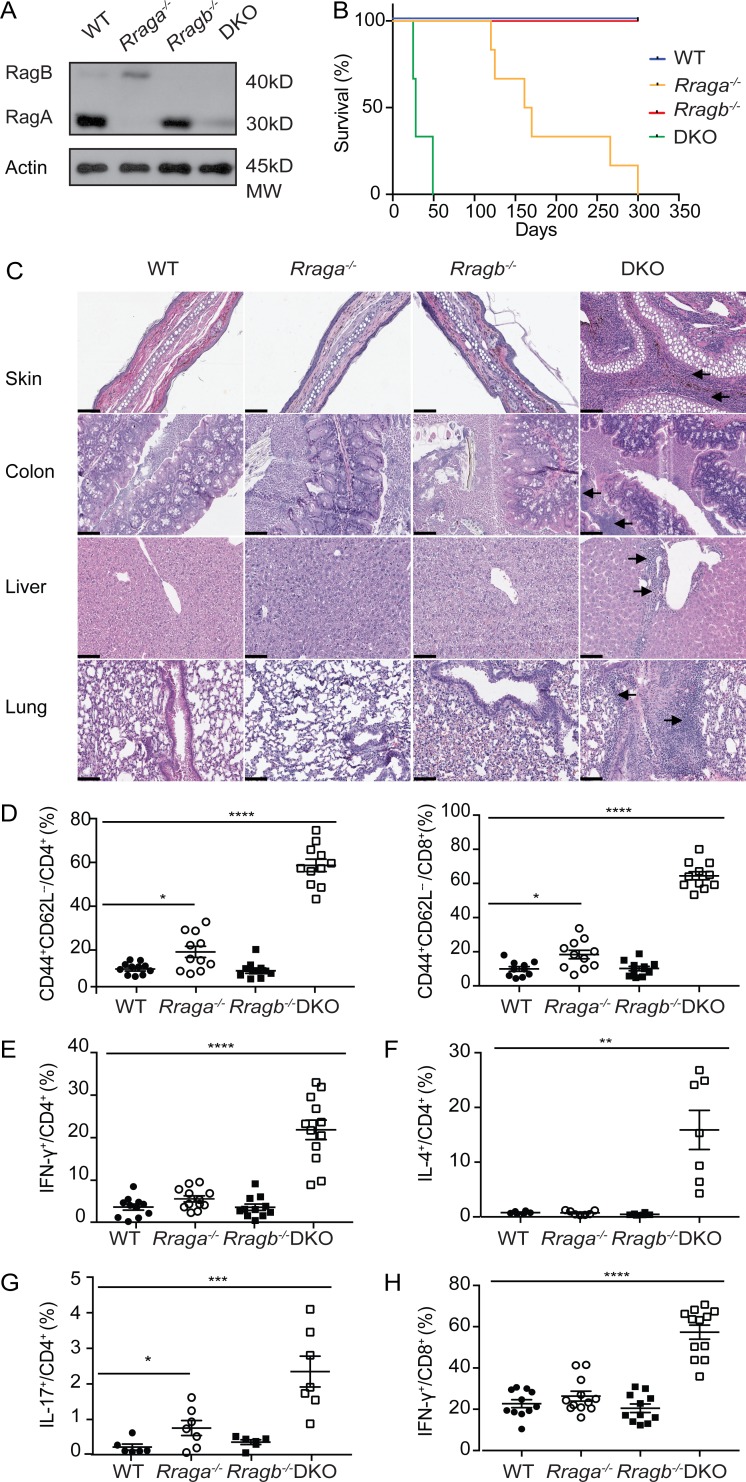
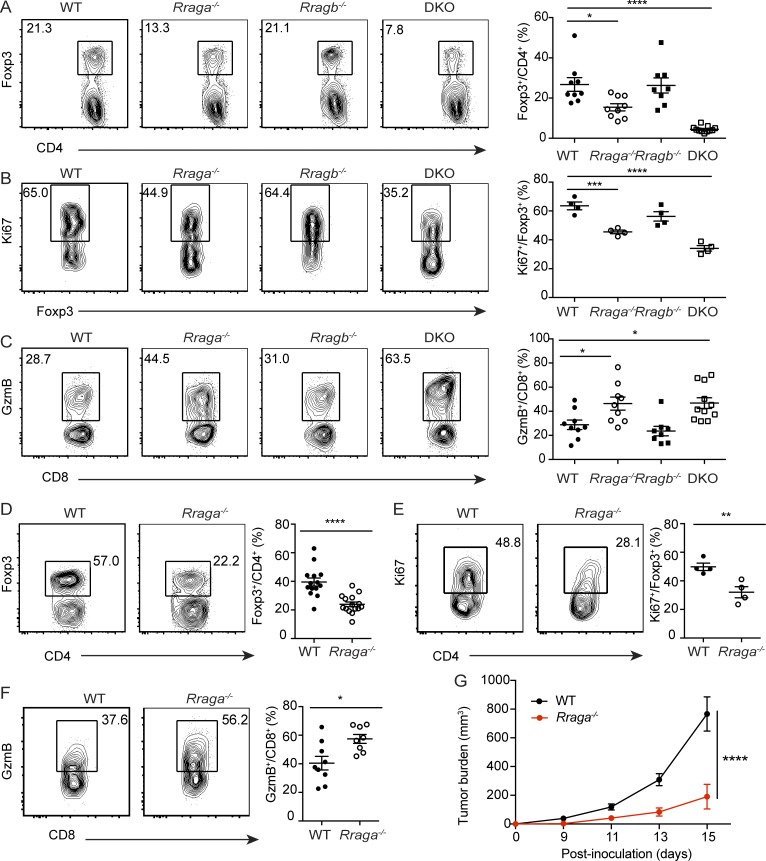
We wished to identify the molecular mechanisms of nutrient mTORC1 signaling in T reg cells. To this end, we focused on the evolutionarily conserved Rag family of small GTPases, whose activity is regulated by an elaborate network of regulatory proteins that participate in nutrient sensing (Saxton and Sabatini, 2017). Previous studies have shown that despite similar biochemical activities of RagA and RagB in nutrient signaling, RagA is more abundantly expressed than RagB in most tissues (Efeyan et al., 2014). In fact, whole-body ablation of RagA, but not RagB, results in an embryonic lethal phenotype in mice (Efeyan et al., 2014). Immunoblotting analysis revealed that RagA was indeed expressed at a higher level than RagB in T reg cells (Fig. 2 A). To investigate the in vivo function of RagA, we crossed mice carrying floxed alleles of Rraga (Rragafl/fl) with Foxp3Cre transgenic mice, herein designated as Rraga−/− mice, which depleted RagA protein specifically in T reg cells (Fig. 2 A and data not shown). Rraga−/− mice appeared healthy at young age with no signs of autoimmunity (Fig. 2, B and C; and Fig. S1, C and D), although increased mortality was observed at old age (Fig. 2 B).
Figure 2.
T reg cell–specific ablation of RagA and RagB results in lethal autoimmunity in mice. (A) Immunoblot analysis of RagA and RagB levels in T reg cells isolated from WT, Rraga−/−, Rragb−/−, and DKO mice. Data are representative of three independent experiments. (B) Survival curves of WT, Rraga−/−, Rragb−/−, and DKO mice (n = 10–12 mice per group). Data are representative of two independent experiments. (C) Hematoxylin and eosin staining of skin, colon, lung, and liver from WT, Rraga−/−, Rragb−/−, and DKO mice. Original magnification, 20×; scale bars, 100 µm. Black arrows indicate immune cell infiltration. Data are representative of three independent experiments. (D) Quantification of splenic activated CD44hiCD62Llo CD4+Foxp3− (left) and CD44hiCD62Llo CD8+ (right) T cells from 5-wk-old WT, Rraga−/−, Rragb−/−, and DKO mice (n = 11 mice per group). Data are pooled from five independent experiments. (E–H) Quantification of IFN-γ, IL-4, and IL-17 expression in splenic CD4+Foxp3− T cells and IFN-γ expression in splenic CD8+ T cells from 5-wk-old WT, Rraga−/−, Rragb−/−, and DKO mice (n = 6–12 mice per group). Data are pooled from five independent experiments. Data in plots indicate mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (one-way ANOVA with the Tukey post-hoc test; D–H).
Notably, in the absence of RagA, RagB was substantially elevated in T reg cells (Fig. 2 A). To investigate the potential compensatory function of RagB, we crossed mice carrying floxed alleles of Rragb (Rragbfl/fl) with Foxp3Cre transgenic mice, herein designated as Rragb−/− mice, which were further bred to the RagA-deficient background (Fig. 2 A). While Rragb−/− mice appeared indistinguishable from WT controls, mice with T reg cell–specific loss of both RagA and RagB (double KO [DKO]) developed a fulminant autoimmune disease with hunched posture and lack of mobility, in addition to crusting of the ears, eyelids, and tail (Fig. S1 C), which culminated in complete mortality before 7 wk of age (Fig. 2 B). Moreover, compared with WT, Rraga−/− and Rragb−/− mice, DKO mice exhibited lymphadenopathy (Fig. S1 D) and had a dense leukocyte infiltration in multiple vital organs (Fig. 2 C).
High frequencies of CD44hiCD62Llo effector/memory phenotype conventional Foxp3−CD4+ T cells and CD8+ T cells were also observed in DKO mice, while their abundance was slightly increased in the absence of RagA (Fig. 2 D and Fig. S1 E). Furthermore, conventional CD4+ T cells from DKO mice produced copious amounts of inflammatory cytokines IFN-γ, IL-4, and IL-17 (Fig. 2, E–G; and Fig. S1, F–H), while CD8+ T cells from DKO mice produced high levels of IFN-γ (Fig. 2 H and Fig. S1 I). Such an autoimmune phenotype is comparable in severity to those developed in T reg cell–specific TCRα- or RAPTOR-deficient mice (Levine et al., 2014; Zeng et al., 2013), revealing crucial, yet redundant, functions of RagA and RagB in control of T reg cell–mediated immune tolerance.
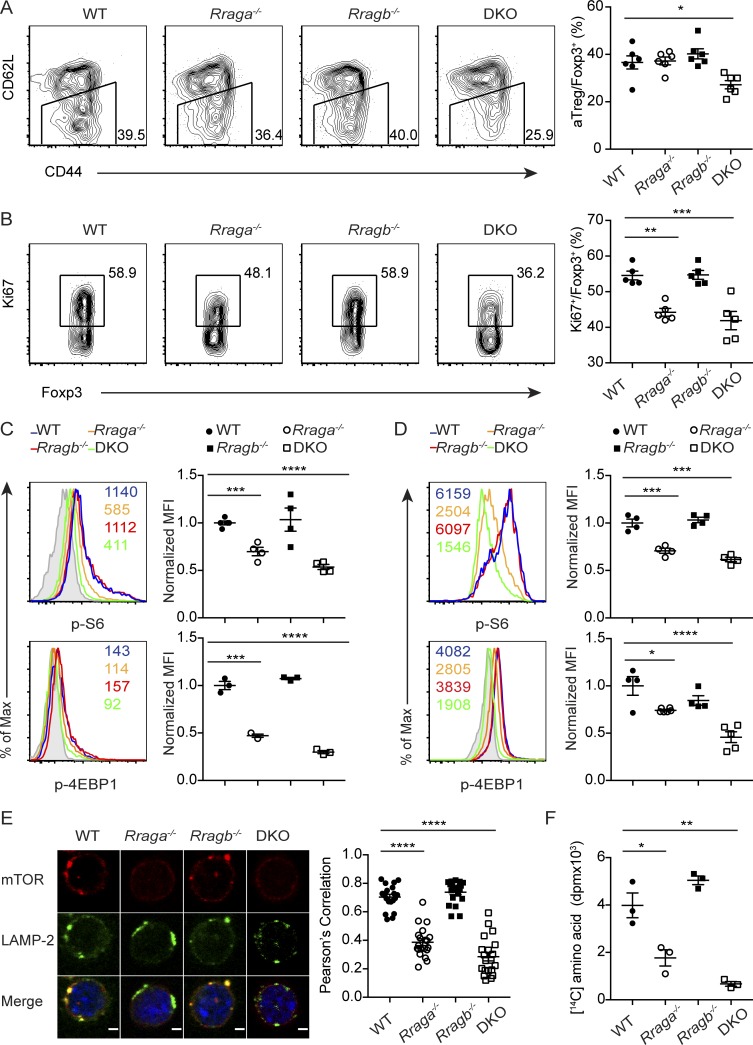
To investigate how Rag GTPase deficiency affected T reg cell responses, we characterized T reg cells in the secondary lymphoid organs of WT, Rraga−/−, Rragb−/−, and DKO mice. The frequencies of splenic and lymph node aT reg cells were substantially reduced in 9–12-d-old DKO mice before their development of immunopathology in peripheral tissues (Fig. 3 A and data not shown), although T reg cells from four genotypes produced comparable amounts of the immunoregulatory cytokines IL-10 and TGF-β1 (as shown by staining of its latency-associated peptide; Fig. S2, A and B). Reduced expression of the cell proliferation marker Ki67 in T reg cells was also observed in DKO mice, and to a lesser extent in Rraga−/− mice (Fig. 3 B), which was associated with attenuated mTORC1 signaling as reflected by decreased p-S6 and phosphorylation of the mTORC1 signaling pathway marker eukaryotic translation initiation factor 4E–binding protein 1 (p-4EBP1) staining (Fig. 3 C). Furthermore, neither amino acids nor glucose could activate mTORC1 in RagA and RagB double-deficient aT reg cells (Fig. 1 B and Fig. S2 C), revealing a crucial function for the Rag family of small GTPases in controlling nutrient-induced mTORC1 signaling. To substantiate these findings, we performed in vitro culture experiments with CellTrace Violet (CTV)–labeled rT reg cells purified from WT, Rraga−/−, Rragb−/−, and DKO mice. Following 18 h stimulation by α-CD3/28 and IL-2, reduced mTORC1 signaling was observed in RagA and RagB double-deficient as well as RagA-deficient T reg cells (Fig. 3 D). Furthermore, T reg cell proliferation was substantially inhibited in the absence of RagA, which was exaggerated with the additional depletion of RagB (Fig. S2, D and E), while T reg cell survival was comparable among the four genotypes (Fig. S2, D and E). These findings reveal a crucial function for Rag GTPases in promoting mTORC1 signaling and T reg cell proliferation.
Figure 3.
Rag GTPases promote amino acid-induced mTORC1 signaling, amino acid anabolism, and aT reg cell proliferation. (A) Flow cytometric analysis of CD44 and CD62L expression in splenic T reg cells and quantification of frequencies of aT reg cells among total T reg cells of 9–12-d-old WT, Rraga−/−, Rragb−/−, and DKO mice (n = 6 mice per group). Data are pooled from three independent experiments. (B) Flow cytometric analysis and quantification of Ki67 expression in splenic T reg cells from 9–12-d-old WT, Rraga−/−, Rragb−/−, and DKO mice (n = 5 mice per group). Data are pooled from three independent experiments. (C and D) Flow cytometric analysis and quantification of p-S6 and p-4EBP1 in ex vivo (C) or 18-h–aT reg cells (D) from WT, Rraga−/−, Rragb−/−, and DKO mice. Numbers in graphs indicate MFI (n = 3–4 mice per group). Data are pooled from three independent experiments. (E) Immunofluorescence staining of mechanistic target of rapamycin (mTOR) and LAMP-2 in 18-h–aT reg cells from WT, Rraga−/−, Rragb−/−, and DKO mice. Original magnification, 63×; zoom of 2.5; scale bar, 2 µm. Quantification of colocalization between mTOR and LAMP-2 in T reg cells from four genotypes (n = 20 cells per group). Data are pooled from two independent experiments. (F) T reg cells from WT, Rraga−/−, Rragb−/−, and DKO mice were stimulated with α-CD3/28 and IL-2 for 18 h and pulse-labeled with L-[U-14C]-amino acid for another 6 h. The amount of radioactive amino acids incorporated into protein is shown (n = 3). Data are pooled from three independent experiments. Data in plots indicate mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (one-way ANOVA with the Tukey post hoc test; A–F). Max, maximum.
Figure S2.
RagA and RagB promote T reg cell mTORC1 signaling and proliferation. (A and B) Flow cytometric analysis and quantification of IL-10 and TGF-β1 latency-associated peptide (LAP) expression in splenic T reg cells from WT, Rraga−/−, Rragb−/−, and DKO mice (n = 3 mice per group). Data are pooled from two independent experiments. (C) Flow cytometric analysis and quantification of p-S6 expression in DKO aT reg cells that were deprived of nutrient for 1 h and refed with amino acids, glucose, or fatty acids for 1 h (n = 3). Data are pooled from three independent experiments. (D and E) rT reg cells from WT, Rraga−/−, Rragb−/−, and DKO mice were labeled with CTV and stimulated with α-CD3/28 and IL-2 for 72 h. (D) T reg cell proliferation was assessed by flow cytometric analysis of CTV dilution. Data are representative of three independent experiments. (E) T reg cell death was measured by propidium iodide (PI) and Annexin V staining. Data are representative of three independent experiments. Numbers in gates or quadrants indicate percentage of cells. Data in plots indicate mean ± SEM (one-way ANOVA with the Tukey post hoc test; A–C).
Previous studies have shown that following amino acid stimulation, endomembrane small GTPases including Rag GTPases recruit mTORC1 to the outer surface of organelles, in particular, lysosomes to be activated by Rheb (Kim et al., 2008; Sancak et al., 2008). To determine the impact of Rag GTPase deficiency on mTORC1 lysosomal translocation in T reg cells, we isolated rT reg cells from WT, Rraga−/−, Rragb−/−, and DKO mice, and activated them in a nutrient-rich medium supplemented with α-CD3/28 and IL-2 for 18 h. Colocalization of mTOR and the lysosome marker LAMP-2 was assessed by confocal microscopy. Compared with WT and RagB-deficient T reg cells, RagA-deficient T reg cells exhibited partial loss of mTOR recruitment to the lysosomal surface, while RagA and RagB double-deficient T reg cells failed to recruit mTOR to the lysosome (Fig. 3 E). These findings demonstrate an essential role for the Rag GTPase–dependent nutrient-sensing pathway in promoting mTORC1 lysosomal translocation in T reg cells.
Proliferating cells uptake increased amounts of nutrients such as amino acids for their biosynthetic needs (Hosios et al., 2016). To determine whether compromised amino acid–induced mTORC1 signaling in Rag GTPase–deficient T reg cells affected amino acid anabolism, we performed pulse-labeling experiments with radioactive amino acids. While RagB-deficient T reg cells had 14C amino acid protein incorporation comparable to that of WT T reg cells, DKO and RagA-deficient T reg cells exhibited substantially diminished 14C amino acid labeling (Fig. 3 F). Thus, Rag GTPase–mediated amino acid–induced mTORC1 signaling promotes amino acid anabolism to support T reg cell proliferation.
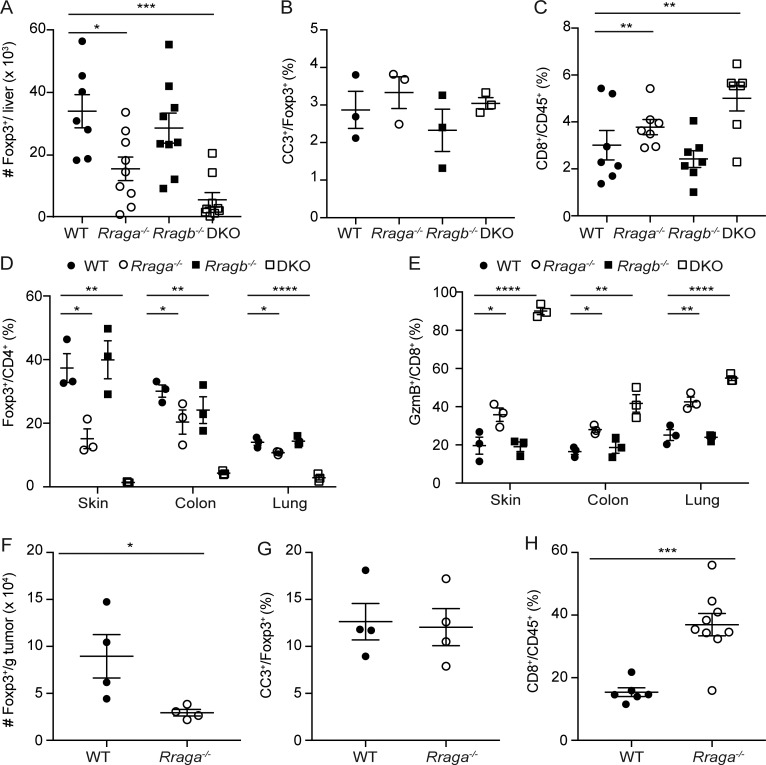
aT reg cells are present in both secondary lymphoid organs and nonlymphoid tissues (Luo et al., 2016). While splenic and lymph node aT reg cells were selectively depleted in DKO mice (Fig. 3 A and data not shown), nonlymphoid tissue T reg cells such as those in the liver were reduced in both DKO mice and Rraga−/− mice (Fig. 4 A and Fig. S3 A). Similar to T reg cells in the secondary lymphoid organs, proliferation was also partially inhibited in RagA-deficient T reg cells and further inhibited in DKO T reg cells, while there was no difference in T reg cell survival (Fig. 4 B and Fig. S3 B). Our previous findings reveal that T reg cells in tissues are not essential for the control of CD4+ T cells but are critical for limiting the activation of CD8+ T cell response (Luo et al., 2016). Recent studies suggest that T reg cells suppress CD4+ and CD8+ T cell response through different mechanisms. Specifically, the capture of IL-2 is essential to control CD8+ T cell activation while dispensable for CD4+ T cell response (Chinen et al., 2016). Therefore, it might be possible that T reg cells in tissues preferentially deplete IL-2 from the environment to control CD8+ T cell response. In line with this notion, we observed that CD8+ T cells, but not CD4+ T cells, expanded in the liver of DKO mice and Rraga−/− mice (Fig. S3 C and data not shown). Moreover, high fractions of liver CD8+ T cells from these mice expressed the cytolytic molecule granzyme B (GzmB; Fig. 4 C). Furthermore, the reduction of T reg cells and enhanced CD8+ T cell responses were observed in other peripheral tissues, including the skin, the colon, and the lung (Fig. S3, D and E). These findings reveal that T reg cells in peripheral tissues are more dependent on Rag GTPase–mediated mTORC1 signaling, with RagA deficiency alone causing impaired T reg cell accumulation, CD8+ T cell expansion, and effector differentiation.
Figure 4.
Rag GTPases promote T reg cell generation in nonlymphoid tissues and tumor. (A) Flow cytometric analysis and quantification of Foxp3 expression in liver CD4+ T cells from 9–12-d-old WT, Rraga−/−, Rragb−/−, and DKO mice (n = 8–11 mice per group). Data are pooled from five independent experiments. (B) Flow cytometric analysis and quantification of Ki67 expression in liver T reg cells from WT, Rraga−/−, Rragb−/−, and DKO mice (n = 4 mice per group). Data are pooled from two independent experiments. (C) Flow cytometric analysis and quantification of GzmB expression in liver CD8+ T cells from 9–12-d-old WT, Rraga−/−, Rragb−/−, and DKO mice (n = 8–11 mice per group). Data are pooled from five independent experiments. (D–G) 6–8-wk-old WT or Rraga−/− mice received subcutaneous injection of B16 melanoma cells. (D) Flow cytometric analysis and quantification of Foxp3 expression in tumor-infiltrating CD4+ T cells from WT and Rraga−/− mice (n = 14–15 mice per group). Data are pooled from five independent experiments. (E) Flow cytometric analysis and quantification of Ki67 expression in tumor T reg cells from WT and Rraga−/− mice (n = 4 mice per group). Data are pooled from two independent experiments. (F) Flow cytometric analysis and quantification of GzmB expression in tumor-infiltrating CD8+ T cells from WT and Rraga−/− mice (n = 8 or 9 mice per group). Data are pooled from five independent experiments. (G) Tumor growth curves of WT and Rraga−/− mice are shown (n = 11 or 12 mice per group). Data are pooled from four independent experiments. Numbers in gates indicate percentage of cells. Data in plots indicate mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (one-way ANOVA with the Tukey post hoc test [A–C], unpaired t test [D–F], and two-way ANOVA [G]).
Figure S3.
Rag GTPase depletion results in CD8+ T cell expansion in nonlymphoid tissues and tumor. (A) Quantification of absolute numbers of liver T reg cells from 9–12-d-old WT, Rraga−/−, Rragb−/−, and DKO mice (n = 7–9 mice per group). Data are pooled from more than five independent experiments. (B) Quantification of cleaved caspase 3 (CC3) expression in T reg cells from 9–12-d-old WT, Rraga−/−, Rragb−/−, and DKO mice (n = 3 mice per group). Data are pooled from two independent experiments. (C) Percentage of CD8+ T cells among total CD45+ immune cells in the liver of 9–12-d-old WT, Rraga−/−, Rragb−/−, and DKO mice (n = 5–7 mice per group). Data are pooled from more than five independent experiments. (D) Flow cytometric analysis and quantification of Foxp3 expression in skin, colon, and lung CD4+ T cells from 9–12-d-old WT, Rraga−/−, Rragb−/−, and DKO mice (n = 3 mice per group). Data are pooled from two independent experiments. (E) Flow cytometric analysis and quantification of GzmB expression in skin, colon, and lung CD8+ T cells from 9–12-d-old WT, Rraga−/−, Rragb−/−, and DKO mice (n = 3 mice per group). Data are pooled from two independent experiments. (F) Quantification of absolute numbers of tumor T reg cells from WT and Rraga−/− mice (n = 4 mice per group). Data are pooled from two independent experiments. (G) Quantification of CC3 expression in T reg cells from tumor T reg cells from WT and Rraga−/− mice (n = 4 mice per group). Data are pooled from two independent experiments. (H) Percentage of CD8+ T cells among total tumor-infiltrating CD45+ immune cells in B16-tumor-bearing WT and Rraga−/− mice (n = 6–9 mice per group). Data are pooled from more than five independent experiments. Data in plots indicate mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (one-way ANOVA with the Tukey post hoc test [A–E] and unpaired t test [B]).
T reg cells infiltrate various tumor types (Nishikawa and Sakaguchi, 2010; Quezada et al., 2011; Zou, 2006), and increased numbers of tumor-associated T reg cells have been correlated with poor cancer patient survival (Curiel et al., 2004). Our previous studies have revealed that compared with T reg cells from healthy tissues, tumor-infiltrating T reg cells display a more activated phenotype (Luo et al., 2016). In addition, we found that the strength of the Akt-Foxo1 signaling pathway in T reg cells can be titrated to selectively repress tumor T reg cell trafficking, resulting in enhanced antitumor immunity (Luo et al., 2016). To determine whether the Rag GTPase–mediated nutrient signaling could also be tuned to preferentially break tumor immune tolerance, we inoculated 6–8-wk-old WT and Rraga−/− mice with B16 melanoma cells. In line with our previous findings (Luo et al., 2016), tumor-associated T reg cells greatly expanded in WT mice, which made up to 60% tumor-infiltrating CD4+ T cells (Fig. 4 D). In the absence of RagA, the proportion of T reg cells was reduced approximately twofold (Fig. 4 D and Fig. S3 F). Tumor-associated T reg cells in Rraga−/− mice were less proliferative compared with WT T reg cells with comparable survival status (Fig. 4 E and Fig. S3 G).
The diminished T reg cell response in Rraga−/− mice was associated with substantial expansion of tumor-infiltrating CD8+ T cells, a higher proportion of which expressed GzmB (Fig. 4 F, Fig. S3 H, and data not shown). Associated with the enhanced effector CD8+ T cell response, tumor growth was substantially inhibited in Rraga−/− mice (Fig. 4 G). These findings reveal that partial repression of the nutrient-sensing pathway in T reg cells can revive antitumor immunity without triggering overt autoimmunity.
Adoptive T reg cell transfer experiments in correcting the neonatal thymectomy-triggered autoimmunity have revealed that disease-suppressing T reg cells are enriched in the draining lymph nodes of the affected organs (Samy et al., 2005). These observations together with the finding that inhibition of TCR signaling in T reg cells causes rampant autoimmunity support a model in which self-antigens are transferred to tissue-draining lymph nodes by APCs to drive the conversion and expansion of disease-suppressing aT reg cells (Li and Rudensky, 2016). Using mouse parabionts, we have shown that aT reg cells exhibit much slower turnover than rT reg cells (Luo et al., 2016). Notably, a recent study revealed that acute inhibition of mTORC1 signaling with rapamycin depletes aT reg cells (Sun et al., 2018), suggesting that prolonged mTORC1 activation is required for their maintenance. However, as APC interaction with T reg cells is dynamically regulated and short-lived, how transient TCR stimulation propagates to induce sustained mTORC1 signaling has been enigmatic. The revelation of an essential function for the Rag GTPase–dependent nutrient sensing in aT reg cell mTORC1 activation offers an explanation. In essence, although TCR stimulation can acutely activate mTORC1 via the immediate–early signaling events including Akt-mediated repression of the TSC complex, transcriptional induction of nutrient transporters such as the amino acid transporter Slc7a5 promotes long-lasting nutrient uptake to enable persistent nutrient-induced mTORC1 signaling. Yet it is currently unknown whether the Rag GTPase–mediated nutrient-sensing response represents an essential facet of TCR signaling–initiated metabolic reprograming, or simply functions to supersede the transient nature of TCR-triggered TSC inactivation. Future studies in investigating whether inactivation of the TSC complex rescues the defects of Rag GTPase–deficient T reg cells can be used to differentiate these possibilities. The revelation of a crucial function for amino acid–dependent activation of Rag GTPases in T reg cells also establishes nutrients as a novel class of signaling molecules that work in concert with antigens, costimulatory molecules and cytokines to control T cell tolerance. Manipulation of the nutrient sensing pathway might provide new strategies of T reg cell targeting to rectify faulty immune responses.
Materials and methods
Mice
Rragafl/fl and Rragbfl/fl mice described previously (Efeyan et al., 2014) were backcrossed at least six generations to the C57BL/6 background. Foxp3YFP/Cre transgenic mice were described previously (Rubtsov et al., 2008). In all experiments, littermate controls were used when possible. Both male and female mice were included. All mice were maintained under specific pathogen–free conditions, and animal experimentation was conducted in accordance with procedures approved by the Institutional Animal Care and Use Committee of Memorial Sloan Kettering Cancer Center.
Tumor model
6–8-wk-old WT or Rraga−/− mice were injected with B16.F10 melanoma (1.25 × 105 cells subcutaneously). Tumors were measured regularly with a caliper. Tumor volume was calculated using the equation (L × W2) × 0.52, where L is length and W is width. Tumor-bearing mice were sacrificed 15–18 d after inoculation.
Cell isolation
Mice were first perfused with 25 ml PBS, and lymphocytes were isolated as follows. For spleens and peripheral (axillary, brachial, and inguinal) lymph nodes, single-cell suspension was obtained by tissue disruption with glass slides. For liver and lung, tissues were chopped and digested with 1 mg/ml Collagenase D (Worthington) for 30 min at 37°C. For colon, tissues were dissected and washed in PBS. Intestines were cut in small pieces and incubated in 1 mM dithiothreitol in PBS with gentle agitation for 30 min. The supernatant that contains intraepithelial lymphocytes was discarded. The remaining tissues were further digested with RPMI plus 5% FBS and 1 mg/ml Collagenase D for 30 min at 37°C. For skin, mouse ear skin was harvested and minced finely with scissors and further digested with RPMI plus 5% FBS and 2 mg/ml collagenase XI, 0.5 mg/ml hyaluronidase, and 0.1 mg/ml DNase I for 1 h at 37°C. For tumor, tissues were prepared by mechanical disruption followed by 1 h treatment with 1 mg/ml Collagenase Type 3 (Worthington) and 4 µg/ml DNase I (Sigma-Aldrich) at 37°C. After collagenase treatment, cells isolated from the liver and tumor were filtered through a 70-µM cell strainer, layered in a 44% and 66% Percoll gradient (Sigma-Aldrich), and centrifuged at 1,900 g for 30 min without brake. Cells at the interface were collected and analyzed by flow cytometry.
Flow cytometry
Fluorochrome-conjugated, biotinylated antibodies against TCR-β (H57-595), CD4 (RM4-5), CD8 (17A2), CD44 (IM7), CD62L (MEL-14), Foxp3 (FJK-16s), IFN-γ (XMG1.2), IL-4 (11B11), CD98 (RL388), and IL-10 (JES5-16E3) were purchased from Thermo Fisher Scientific. Antibodies against IL-17a (TC11-18H10.1) and Ki67 (B56) were purchased from BD Biosciences. Anti-GzmB (GB11) was purchased from Invitrogen. Antibodies against CC3 (D3E9), p-4EBP1 (236B4), and p-S6 (S235/236) were purchased from CST. Antibody against latency-associated peptide (TW7-16B4) was purchased from BioLegend. Cell surface staining was performed by incubating cells with specific antibodies for 20 min on ice in the presence of 2.4G2 mAb to block FcγR binding. Foxp3, GzmB, IFN-γ, IL-4, IL-17a, IL-10, Ki67, CC3, p-4EBP1, and p-S6 staining was performed using the intracellular transcription factor or cytokine staining kits from Tonbo. Secondary antibodies with fluorochrome conjugation were used for the staining of unconjugated antibodies. To determine cytokine expression, isolated cells were stimulated with 50 ng/ml phorbol o-myristate 13-acetate (Sigma-Aldrich), 1 mM ionomycin (Sigma-Aldrich), and GolgiStop (BD Biosciences) for 4 h before staining. All samples were acquired and analyzed with an LSRII flow cytometer (Becton Dickinson) and FlowJo software (TreeStar).
In vitro T cell culture
rT reg cells (CD4+Foxp3+CD62LhiCD44lo) were purified from spleen and lymph nodes of Foxp3YFP/Cre mice by flow cytometry sorting (BD FACS Aria). Sorted cells were first labeled with CTV (Invitrogen) before they were used in cell proliferation experiments. T reg cells were cultured with plate-bound α-CD3 (5 µg/ml), soluble α-CD28 (2 µg/ml), and IL-2 (200 U/ml) for the indicated time periods in complete T cell media (RPMI 1640 media supplemented with 10% FBS, 1 mM sodium pyruvate, nonessential amino acids [Gibco], 10 mM Hepes, 55 µM 2-mercaptoethanol, 100 U/ml penicillin G, and 0.1 mg/ml streptomycin).
In vitro T cell restimulation
Sorted rT reg cells or 18-h–aT reg cells were rested for 1 h in amino acid–deficient media before incubation with biotinylated α-CD3/28 in the presence or absence of amino acids for the amount of time indicated. Following acute α-CD3/28 cross-linking, cells were immediately spun down, washed once in cold PBS, and lysed using 1× lysis buffer (Cell Signaling) in the presence of proteinase and phosphatase inhibitors. Total protein extracts were further processed for immunoblotting. For nutrient starved and refed experiments, 18-h–aT reg cells were rested for 1 h in HBSS or amino acid–deficient media and refed with all 20 amino acids, glucose, fatty acids (Lipid Mixture 1, Chemically Defined from Sigma-Aldrich), or individual amino acid for 1 h. Cells were immediately spun down, washed once in cold PBS, fixed, and permeabilized for p-S6 staining. Concentrations of refed glucose and individual amino acid are shown in the Table 1
Table 1. Amino acids and glucose concentrations in RPMI 1640.
| Component | Concentration (mg/liter) |
|---|---|
| Glycine | 10 |
| L-Arginine | 200 |
| L-Asparagine | 50 |
| L-Aspartic acid | 20 |
| L-Cystine | 65 |
| L-Glutamic acid | 20 |
| L-Glutamine | 300 |
| L-Histidine | 15 |
| L-Hydroxyproline | 20 |
| L-Isoleucine | 50 |
| L-Leucine | 50 |
| L-Lysine | 40 |
| L-Methionine | 15 |
| L-Phenylalanine | 15 |
| L-Proline | 20 |
| L-Serine | 30 |
| L-Threonine | 20 |
| L-Tryptophan | 5 |
| L-Tyrosine | 29 |
| L-Valine | 20 |
| D-Glucose | 2,000 |
Amino acid–labeling study
rT reg cells were activated in the presence of α-CD3/28 and IL-2 for 18 h, and L-[U-14C]-amino acid mixture (0.05 µCi/ml; American Radiolabeled Chemicals) was added for an additional 6 h of culture. After incubation, cells were collected, washed twice with PBS, and lysed in TRIzol reagent (Life Technologies). Protein was extracted and purified by bi-phasic extraction according to the manufacturer’s instructions. Purified protein was lysed in 10 or more volumes of Emulsifier-Safe liquid scintillation cocktail (Perkin Elmer), and incorporated 14C was quantified by liquid scintillation counting.
Immunoblotting
Total protein extracts were dissolved in SDS sample buffer, separated on 10% SDS-PAGE gels, and transferred to a polyvinylidene difluoride membrane (Millipore). The membranes were probed with RagA/B (D8B5), p-Akt (S473) (736E11), Akt (4691), p-S6 (S235/236; D57.2.2E), and S6 (2217) antibodies (Cell Signaling), as well as anti-Actin (AC-15; Sigma-Aldrich), and visualized with the Immobilon Western Chemiluminescent HRP Substrate (Millipore). Quantification for protein levels was done with ImageJ software.
Histopathology
Organs from euthanized animals were fixed in Safefix II (Protocol) and embedded in paraffin. 5-µm sections were stained with hematoxylin and eosin.
Immunofluorescence
rT reg cells were cultured for 18 h with plate-bound α-CD3 (5 µg/ml), soluble α-CD28 (2 µg/ml) and IL-2 (200 U/ml) and plated on coverslips. Cells were rinsed with PBS and fixed with 4% paraformaldehyde. The slides were rinsed with PBS, and cells were permeabilized with 0.1% Triton X-100 in PBS for 5 min. After rinsing with PBS, the slides were incubated with primary antibodies in 5% normal donkey serum for 3 h, washed three times with PBS, and incubated with secondary antibodies for 1 h. Slides were washed three times with PBS and mounted on glass coverslips using ProLong Gold (Invitrogen). Images were acquired on a Leica-Upright Point scanning confocal microscope with a 63× oil objective and zoom of 2.5. Colocalization analysis was done with Imaris software, and at least 10 different fields were randomly selected per sample.
Statistical analysis
All data are presented as the mean values ± SEM. Comparisons between groups were analyzed using unpaired t test, one-way ANOVA with the Tukey post hoc test unless indicated otherwise, or two-way ANOVA using Prism 7 software (GraphPad). P < 0.05 was considered significant.
Online supplemental material
Fig. S1 shows that mice lacking RagA and RagB in T reg cells develop lethal autoimmune disease. Fig. S2 shows that RagA and RagB promote T reg cell mTORC1 signaling and proliferation. Fig. S3 shows that Rag GTPase depletion results in CD8+ T cell expansion in nonlymphoid tissues and tumor.
Acknowledgments
This work was supported by the National Institutes of Health (T32 GM007739 and F30 AI129273-03 to M.H. Do and R01 AI 102888 to M.O. Li), the Howard Hughes Medical Institute (Faculty Scholar Award to M.O. Li), and the Memorial Sloan Kettering Cancer Center Support Grant/Core Grant (P30 CA08748).
The authors declare no competing financial interests.
Author contributions: M.H. Do and M.O. Li were involved in all aspects of this study, including planning and performing experiments, analysis and interpretation of data, and writing the manuscript. X. Wang, X. Zhang, C. Chou, B.G. Nixon, K.J. Capistrano, and M. Peng assisted with mouse colony management and performed experiments. A. Efeyan and D.M. Sabatini provided key mouse lines utilized in the studies.
References
- Bluestone J.A., Bour-Jordan H., Cheng M., and Anderson M.. 2015. T cells in the control of organ-specific autoimmunity. J. Clin. Invest. 1252250–2260. 10.1172/JCI78089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinen T., Kannan A.K., Levine A.G., Fan X., Klein U., Zheng Y., Gasteiger G., Feng Y., Fontenot J.D., and Rudensky A.Y.. 2016. An essential role for the IL-2 receptor in Treg cell function. Nat. Immunol. 171322–1333. 10.1038/ni.3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curiel T.J., Coukos G., Zou L., Alvarez X., Cheng P., Mottram P., Evdemon-Hogan M., Conejo-Garcia J.R., Zhang L., Burow M., et al. 2004. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat. Med. 10942–949. 10.1038/nm1093 [DOI] [PubMed] [Google Scholar]
- Efeyan A., Schweitzer L.D., Bilate A.M., Chang S., Kirak O., Lamming D.W., and Sabatini D.M.. 2014. RagA, but not RagB, is essential for embryonic development and adult mice. Dev. Cell. 29321–329. 10.1016/j.devcel.2014.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosios A.M., Hecht V.C., Danai L.V., Johnson M.O., Rathmell J.C., Steinhauser M.L., Manalis S.R., and Vander Heiden M.G.. 2016. Amino Acids Rather than Glucose Account for the Majority of Cell Mass in Proliferating Mammalian Cells. Dev. Cell. 36540–549. 10.1016/j.devcel.2016.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huehn J., Siegmund K., Lehmann J.C., Siewert C., Haubold U., Feuerer M., Debes G.F., Lauber J., Frey O., Przybylski G.K., et al. 2004. Developmental stage, phenotype, and migration distinguish naive- and effector/memory-like CD4+ regulatory T cells. J. Exp. Med. 199303–313. 10.1084/jem.20031562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K., Kinoshita M., Kayama H., Nagamori S., Kongpracha P., Umemoto E., Okumura R., Kurakawa T., Murakami M., Mikami N., et al. 2017. Slc3a2 Mediates Branched-Chain Amino-Acid-Dependent Maintenance of Regulatory T Cells. Cell Reports. 211824–1838. 10.1016/j.celrep.2017.10.082 [DOI] [PubMed] [Google Scholar]
- Inoki K., Li Y., Xu T., and Guan K.L.. 2003. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 171829–1834. 10.1101/gad.1110003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell J.L., Kim Y.C., Russell R.C., Yu F.X., Park H.W., Plouffe S.W., Tagliabracci V.S., and Guan K.L.. 2015. Metabolism. Differential regulation of mTORC1 by leucine and glutamine. Science. 347194–198. 10.1126/science.1259472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz S.Z., Lu L.F., and Rudensky A.Y.. 2012. Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 30531–564. 10.1146/annurev.immunol.25.022106.141623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E., Goraksha-Hicks P., Li L., Neufeld T.P., and Guan K.L.. 2008. Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 10935–945. 10.1038/ncb1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M., and Sabatini D.M.. 2012. mTOR signaling in growth control and disease. Cell. 149274–293. 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A.G., Arvey A., Jin W., and Rudensky A.Y.. 2014. Continuous requirement for the TCR in regulatory T cell function. Nat. Immunol. 151070–1078. 10.1038/ni.3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.O., and Rudensky A.Y.. 2016. T cell receptor signalling in the control of regulatory T cell differentiation and function. Nat. Rev. Immunol. 16220–233. 10.1038/nri.2016.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C.T., Liao W., Dadi S., Toure A., and Li M.O.. 2016. Graded Foxo1 activity in Treg cells differentiates tumour immunity from spontaneous autoimmunity. Nature. 529532–536. 10.1038/nature16486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyara M., Yoshioka Y., Kitoh A., Shima T., Wing K., Niwa A., Parizot C., Taflin C., Heike T., Valeyre D., et al. 2009. Functional delineation and differentiation dynamics of human CD4+ T cells expressing the FoxP3 transcription factor. Immunity. 30899–911. 10.1016/j.immuni.2009.03.019 [DOI] [PubMed] [Google Scholar]
- Nakaya M., Xiao Y., Zhou X., Chang J.H., Chang M., Cheng X., Blonska M., Lin X., and Sun S.C.. 2014. Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity. 40692–705. 10.1016/j.immuni.2014.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa H., and Sakaguchi S.. 2010. Regulatory T cells in tumor immunity. Int. J. Cancer. 127759–767 [DOI] [PubMed] [Google Scholar]
- Panduro M., Benoist C., and Mathis D.. 2016. Tissue Tregs. Annu. Rev. Immunol. 34609–633. 10.1146/annurev-immunol-032712-095948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quezada S.A., Peggs K.S., Simpson T.R., and Allison J.P.. 2011. Shifting the equilibrium in cancer immunoediting: from tumor tolerance to eradication. Immunol. Rev. 241104–118. 10.1111/j.1600-065X.2011.01007.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov Y.P., Rasmussen J.P., Chi E.Y., Fontenot J., Castelli L., Ye X., Treuting P., Siewe L., Roers A., Henderson W.R. Jr., et al. 2008. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 28546–558. 10.1016/j.immuni.2008.02.017 [DOI] [PubMed] [Google Scholar]
- Sakaguchi S., Yamaguchi T., Nomura T., and Ono M.. 2008. Regulatory T cells and immune tolerance. Cell. 133775–787. 10.1016/j.cell.2008.05.009 [DOI] [PubMed] [Google Scholar]
- Samy E.T., Parker L.A., Sharp C.P., and Tung K.S.. 2005. Continuous control of autoimmune disease by antigen-dependent polyclonal CD4+CD25+ regulatory T cells in the regional lymph node. J. Exp. Med. 202771–781. 10.1084/jem.20041033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y., Peterson T.R., Shaul Y.D., Lindquist R.A., Thoreen C.C., Bar-Peled L., and Sabatini D.M.. 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 3201496–1501. 10.1126/science.1157535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton R.A., and Sabatini D.M.. 2017. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 169361–371. 10.1016/j.cell.2017.03.035 [DOI] [PubMed] [Google Scholar]
- Shevach E.M. 2018. Foxp3+ T Regulatory Cells: Still Many Unanswered Questions-A Perspective After 20 Years of Study. Front. Immunol. 91048 10.3389/fimmu.2018.01048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair L.V., Rolf J., Emslie E., Shi Y.B., Taylor P.M., and Cantrell D.A.. 2013. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat. Immunol. 14500–508. 10.1038/ni.2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smigiel K.S., Richards E., Srivastava S., Thomas K.R., Dudda J.C., Klonowski K.D., and Campbell D.J.. 2014. CCR7 provides localized access to IL-2 and defines homeostatically distinct regulatory T cell subsets. J. Exp. Med. 211121–136. 10.1084/jem.20131142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama D., Nishikawa H., Maeda Y., Nishioka M., Tanemura A., Katayama I., Ezoe S., Kanakura Y., Sato E., Fukumori Y., et al. 2013. Anti-CCR4 mAb selectively depletes effector-type FoxP3+CD4+ regulatory T cells, evoking antitumor immune responses in humans. Proc. Natl. Acad. Sci. USA. 11017945–17950. 10.1073/pnas.1316796110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun I.H., Oh M.H., Zhao L., Patel C.H., Arwood M.L., Xu W., Tam A.J., Blosser R.L., Wen J., and Powell J.D.. 2018. mTOR Complex 1 Signaling Regulates the Generation and Function of Central and Effector Foxp3+ Regulatory T Cells. J. Immunol. 201481–492. 10.4049/jimmunol.1701477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee A.R., Manning B.D., Roux P.P., Cantley L.C., and Blenis J.. 2003. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 131259–1268. 10.1016/S0960-9822(03)00506-2 [DOI] [PubMed] [Google Scholar]
- Thomas J.D., Zhang Y.J., Wei Y.H., Cho J.H., Morris L.E., Wang H.Y., and Zheng X.F.. 2014. Rab1A is an mTORC1 activator and a colorectal oncogene. Cancer Cell. 26754–769. 10.1016/j.ccell.2014.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahl J.C., Drees C., Heger K., Heink S., Fischer J.C., Nedjic J., Ohkura N., Morikawa H., Poeck H., Schallenberg S., et al. 2014. Continuous T cell receptor signals maintain a functional regulatory T cell pool. Immunity. 41722–736. 10.1016/j.immuni.2014.10.012 [DOI] [PubMed] [Google Scholar]
- Zeng H., Yang K., Cloer C., Neale G., Vogel P., and Chi H.. 2013. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature. 499485–490. 10.1038/nature12297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W. 2006. Regulatory T cells, tumour immunity and immunotherapy. Nat. Rev. Immunol. 6295–307. 10.1038/nri1806 [DOI] [PubMed] [Google Scholar]