Using a polyclonal Treg cell replacement system, Pratama et al. show that naive Tconv cells have the capacity to convert into both RORγ+ and Helios+ Treg cells in the colon. The developmental maturity (or cell states) of the starting Tconv cells determines the phenotype of the resulting pTreg cells.
Abstract
RORγ+ and Helios+ Treg cells in the colon are phenotypically and functionally distinct, but their origins and relationships are poorly understood. In monocolonized and normal mice, single-cell RNA-seq revealed sharing of TCR clonotypes between these Treg cell populations, potentially denoting a common progenitor. In a polyclonal Treg cell replacement system, naive conventional CD4+ (Tconv) cells, but not pre-existing tTregs, could differentiate into RORγ+ pTregs upon interaction with gut microbiota. A smaller proportion of Tconv cells converted into Helios+ pTreg cells, but these dominated when the Tconv cells originated from preweaning mice. T cells from infant mice were predominantly immature, insensitive to RORγ-inducing bacterial cues and to IL6, and showed evidence of higher TCR-transmitted signals, which are also characteristics of recent thymic emigrants (RTEs). Correspondingly, transfer of adult RTEs or Nur77high Tconv cells mainly yielded Helios+ pTreg cells, recapitulating the infant/adult difference. Thus, CD4+ Tconv cells can differentiate into both RORγ+ and Helios+ pTreg cells, providing a physiological adaptation of colonic Treg cells as a function of the age of the cell or of the individual.
Introduction
Regulatory T (Treg) cells that express the transcription factor (TF) FoxP3 are important players in maintaining immunological homeostasis in the intestines (Sharma and Rudra, 2018; Russler-Germain et al., 2017; Tanoue et al., 2016). They can be divided into two major subsets based on their expression of additional TFs. The first expresses the nuclear hormone receptor RORγ and the TF c-Maf (Ohnmacht et al., 2015; Sefik et al., 2015; Yang et al., 2016; Yissachar et al., 2017; Xu et al., 2018; Neumann et al., 2019; Wheaton et al., 2017), which are also key regulators for Th17 cells and group 3 innate lymphoid cells (Sawa et al., 2010; Spits and Cupedo, 2012; Ivanov et al., 2006). RORγ+ Treg cells predominate in the colon, and their induction is highly dependent on commensal bacteria through molecular mediators that remain uncertain but may involve cross-talk with the enteric nervous system (Yissachar et al., 2017). The second subset expresses Helios and Gata3 and predominates in the small intestine (Wohlfert et al., 2011; Schiering et al., 2014; Sefik et al., 2015; Ohnmacht et al., 2015). Accumulation of Helios+ Treg cells does not require the microbiota. Rather, they express the receptor for IL33 (also known as ST2), expand in response to this cytokine (Schiering et al., 2014; He et al., 2017), and are hence connected to IL33-inducing stress pathways (Peine et al., 2016; Molofsky et al., 2015). RORγ+ and Helios+ Treg cells have nonredundant functions, as genetic inactivation of RORγ+ Treg cells results in increased proinflammatory cytokine production at baseline and in greater susceptibility in colitis models (Sefik et al., 2015; Ohnmacht et al., 2015; Neumann et al., 2019).
The origins of, and the relationship between, RORγ+ and Helios+ Treg cells are still incompletely understood. Helios is often considered to be a marker for Treg cells generated in the thymus (tTreg cells; Thornton et al., 2010). Although this relation is known to have exceptions (Akimova et al., 2011; Gottschalk et al., 2012), it suggests that colonic Helios+ Treg cells are tTreg cells, similar to those found in lymphoid organs. In contrast, the lack of Helios in RORγ+ Treg cells, their induction by gut bacteria, and their delayed appearance in the gut only after colonization by an adult microbiota led to the initial suggestion that this population was peripherally generated Treg (pTreg) cells. Indeed, experimental conversion of FoxP3− conventional CD4+ T cells (Tconv cells), in vitro and in vivo, supported this notion (Nutsch et al., 2016; Solomon and Hsieh, 2016; Yang et al., 2018). The two Treg cell subsets should then be quite distinct in terms of their differentiation pathways, and hence of their TCRs. This dichotomy was in line with earlier studies showing that microbe-responsive Treg cells were not positively selected with any efficiency in the thymus, but appeared only in the periphery (Lathrop et al., 2011; Geuking et al., 2011; Atarashi et al., 2011). However, several lines of evidence later suggested more intricate relationships between Helios+ and RORγ+ Treg cells. First, RORγ could be induced in tTreg cells by TCR-mediated activation in vitro in the presence of IL6 (Kim et al., 2017; Yang et al., 2018), which is of potential relevance because RORγ+ Treg cells depend on IL6 in vivo (Ohnmacht et al., 2015; Yissachar et al., 2017). Second, using a transgenic mouse model expressing a TCR reactive to an antigen of microbial origin, Hsieh and colleagues showed that Tconv cells could be efficiently converted in vitro and in vivo by exposure to cognate microbial antigen, mostly to RORγ+ Treg cells via a FoxP3+RORγ− intermediate (Nutsch et al., 2016; Solomon and Hsieh, 2016), thus suggesting that colonic Treg cells of Helios+ and RORγ+ phenotypes might be interconnected.
Here, we revisited the relationships between colonic Treg cell and Tconv populations by exploiting the potential of single-cell RNA sequencing (scRNA-seq) to provide detailed phenotypic and specificity information on colonic T cells from normal mice, and by using a Treg cell replacement strategy to elucidate the relationships between RORγ+ Treg cells, Helios+ Treg cells, and Tconv cells. There was substantial sharing of TCRs between Helios+ Treg cells and RORγ+ Treg cells, hinting at a common precursor. We observed robust conversion from Tconv cells to both Helios+ and RORγ+ pTreg cells, the balance between the two depending on the age and maturity of the starting naive Tconv cells. This dichotomy conditioned the sequential colonization by the two colonic Treg cell populations in developing mice, which may thus play temporally complementary roles in immune homeostasis in the colon.
Results
Shared TCR specificities between Tconv, RORγ+, and Helios+ Treg cells
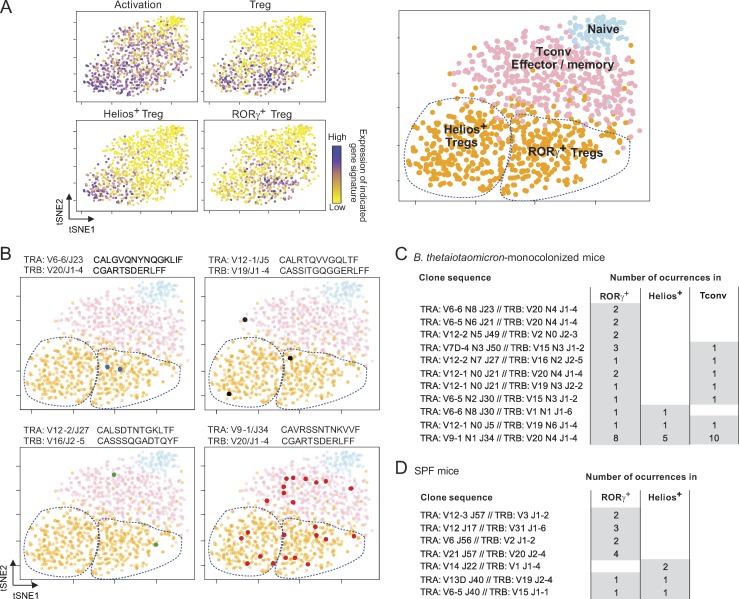
In general, the TCR repertoires of tTreg cell and Tconv cells are nonoverlapping (Hsieh et al., 2004; Pacholczyk et al., 2006; Wong et al., 2007; Liu et al., 2009). Therefore, according to the hypothesis that RORγ+ Treg cells are pTreg cells and that Helios+ Treg cells are tTreg cells, one would expect some overlap between the repertoires of Tconv cells and RORγ+ Treg cells, but not with Helios+ Treg cells. Consistent with this notion, Solomon and Hsieh (2016) reported mostly distinct TCRα repertoires in RORγ+ versus other Treg cells in mice carrying a single-chain TCRβ transgene with reactivity to a bacterial peptide (Solomon and Hsieh, 2016), although in fact ≤20% of CDR3 motifs were found in both Treg cell populations. To re-examine this question in the context of polyclonal repertoires, we leveraged our previous adaptation of the InDrop protocol for scRNA-seq (Zilionis et al., 2017; Zemmour et al., 2018) to obtain both the transcriptome and the TCRα and β variable region sequences from individual cells. We used the particular context of germ-free (GF) mice monocolonized with an RORγ+ Treg cell–inducing microbe (Sefik et al., 2015; Geva-Zatorsky et al., 2017) to facilitate the TCR repertoire analysis by restricting the potential antigenic diversity. We thus obtained data that met the usual quality-control criteria (Fig. S1, A and B) for 1,102 CD4+ T cells from the colon lamina propria (LP) 2 wk after monocolonization with Bacteroides thetaiotaomicron. The transcriptomes were used to parse these cells into either the Treg cell or Tconv cell subsets (Fig. 1 A) using the integrated expression of gene sets identified in a recent study (DiSpirito et al., 2018; Table S1). Of the 142 colonic T cells for which both TCRα and TCRβ sequences could be determined unambiguously, we found 14 clonotypes shared by two or more cells (repetition defined here as identical TCRα and TCRβ V and joining nucleotide sequences). The CDR3 sequences of these repeated clonotypes included N-region diversity, indicating a common origin rather than a recurring recombination-driven rearrangement (Komori et al., 1993). Some of these repeated clonotypes were found only in RORγ+ Treg cells (Fig. 1 B, top left), but many were shared between two cell types, most often RORγ+ Treg cells and Tconv cells (Fig. 1, B and C; and Table S1). One particularly frequent clonotype was found in all three subsets (Fig. 1 B, bottom right). These results support the notion that RORγ+ Treg cells can convert peripherally from Tconv cells when the inducing microbe is introduced into GF mice, and also suggest that RORγ+ and Helios+ Treg cells might be more closely related developmentally than previously thought. We also analyzed single Treg cells from colonic LP of normal specific pathogen–free (SPF) mice (Zemmour et al., 2018). Although repeated clonotypes were less frequent than in monocolonized mice, as expected, shared clonotypes were found in RORγ+ and Helios+ Treg cells (Fig. 1 D and Table S1), generalizing the above observations.
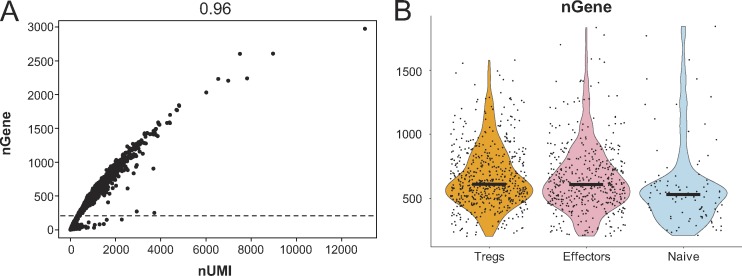
Figure S1.
scRNA-seq quality control analysis and sequences of repeated TCR clonotypes. (A) Scatter plot showing the number of unique genes and unique molecular identifiers (UMIs) detected in each cell. Each dot represents a cell. Cells with <200 detected genes (below the dotted line) were excluded from further analysis. The r2 value of the correlation is shown above. (B) Violin plots showing the number of unique genes detected in cells from different clusters (Treg cells, effectors, and naive). Black bars indicate median values.
Figure 1.
Shared TCR clonotypes between Tconv, RORγ+, and Helios+ Treg cells. (A) scRNA-seq was performed to determine both transcriptome and TCRα and β sequences of CD4+ T cells from the colon LP of B. thetaiotaomicron–monocolonized mice. Each cell is shown as a dot on the dimensionality reduction plot (tSNE), color-coded according to its relative expression of the indicated gene signatures. Based on these profiles, four cell clusters were identified (right). (B) Cells expressing one of four recurrent TCRαβ clonotypes; colored dots show their position on the same tSNE plots as in A. V/J alleles and CDR3 sequences are indicated above each plot. (C and D) Repeated TCR clonotypes shared by various T cell subsets in B. thetaiotaomicron–monocolonized mice (C) or SPF B6 mice (D). Data in C are pooled from two independent experiments.
Conventional naive CD4+ T cells differentiate mainly, but not exclusively, into RORγ+ Treg cells in the colon
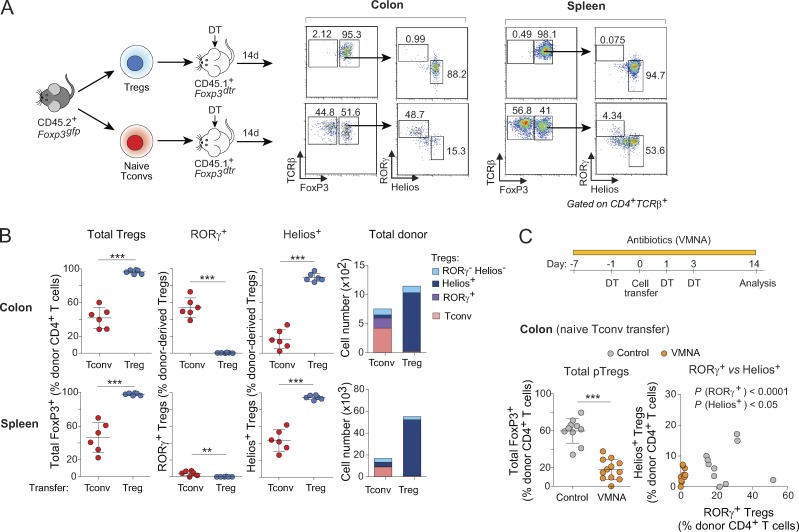
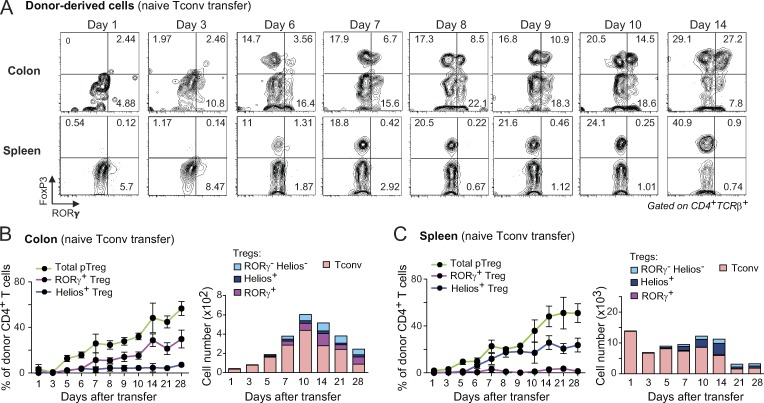
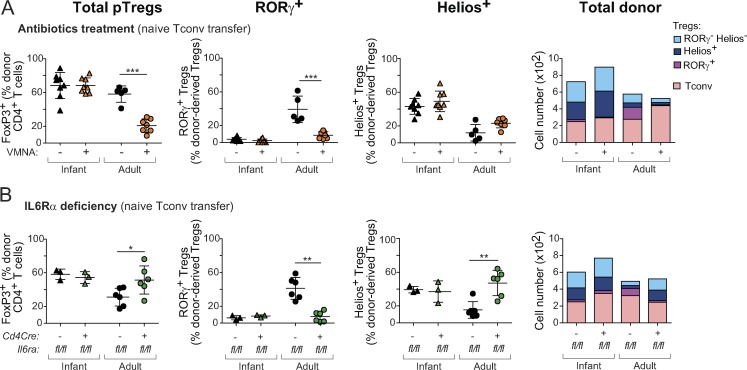
To directly analyze the relationships between the various T cell populations in the colon, we used a novel cell-transfer strategy, opting for several characteristics. First, cell donors were nontransgenic mice, for a broad perspective. Second, we did not use RAG-deficient hosts, in which immunological organs are poorly formed and Treg cells are unstable (indeed, Nutsch et al. [2016] have demonstrated that alymphoid hosts do not support pTreg cell formation); we also wished to avoid irradiated hosts, in which damage to the radio-sensitive colonic epithelium would confound the results. Rather, we used a “Treg cell replacement” protocol, wherein Treg cells or Tconv cells originating from wild-type mice were injected into a Foxp3dtrhost, at the same time as endogenous Treg cells were being eliminated by treatment with diphtheria toxin (DT), i.e., before severe autoinflammatory symptoms of Treg cell deficiency. In practice, we sorted and transferred 106 naive Tconv cells or 105 Treg cells from pooled spleen and LNs of Foxp3gfp reporter mice into congenically marked Foxp3dtr hosts (Kim et al., 2007) immediately after onset of DT treatment (Fig. 2 A; note that treatment with DT eliminated both RORγ+ and Helios+ Treg cells from the colon of these hosts [Fig. S2 A]). 2 wk after transfer, all donor Treg cells remained FoxP3+ (Fig. 2 A, top) in both the spleen and colon (tabulated in Fig. 2 B). In the colon, these cells maintained an RORγ− Helios+ phenotype, indicating that Helios+ Treg cells do not readily become RORγ+ in spite of a competent microbiota in the gut of these hosts (Fig. S2 A). In contrast, 30–40% of donor Tconv cells converted into pTreg cells (Fig. 2, A and B). In the spleen, almost all of these pTreg cells were RORγ− Helios+; in the colon, most (∼60%) donor-derived pTreg cells up-regulated RORγ, although 10–20% expressed Helios (Fig. 2, A and B; these also expressed Gata3 as expected). Unlike previous studies (Nutsch et al., 2016; Solomon and Hsieh, 2016), we did not observe donor-derived RORγ+ Treg cells in the mesenteric LNs (Fig. S2 B), suggesting that conversion probably occurs directly in the colonic LP. Donor T cells needed to be naive for conversion to occur effectively in this system, as CD4+ T cells with CD44hiCD62Llo phenotypes yielded few pTreg cells (Fig. S2 C). We assessed the impact of the intestinal microbiota on these conversion events by also treating recipient mice with a broad-spectrum antibiotic combination of vancomycin, metronidazole, neomycin, and ampicillin (VMNA). The numbers of converted RORγ+ pTreg cells were severely curtailed by VMNA pretreatment of the hosts, and there was also a modest but significant reduction in the numbers of Helios+ pTreg cells (Fig. 2 C). These results support the notion that RORγ+ and Helios+ pTreg cells can arise from Tconv cells under the influence of microbes, which is compatible with the TCR sequencing results.
Figure 2.
Tconv cells differentiate mainly, but not exclusively, into RORγ+ Treg cells in the colon. (A) Schematic diagram (left) and flow cytometric analyses (right) of a transfer experiment where either 106 naive Tconv or 105 Treg cells from pooled LNs of CD45.2+ Foxp3gfp mice were transferred into CD45.1+ Foxp3dtr hosts. Hosts were treated with DT on days −1, 1, and 3 after transfer and sacrificed at day 14. (B) Frequencies and average numbers of donor-derived Treg cells in the whole colon (top) and spleen (bottom) of hosts as described in A (n = 6). (C) In a Tconv cell transfer as in A, hosts were given either antibiotic-supplemented (VMNA) or normal water. Frequencies of donor-derived Treg cells in the colon are shown (bottom; n ≥ 10). Summary plots show data pooled from three (A and B) and four (C) independent experiments. Means ± SD. **, P < 0.01; ***, P < 0.001 using Student’s t test.
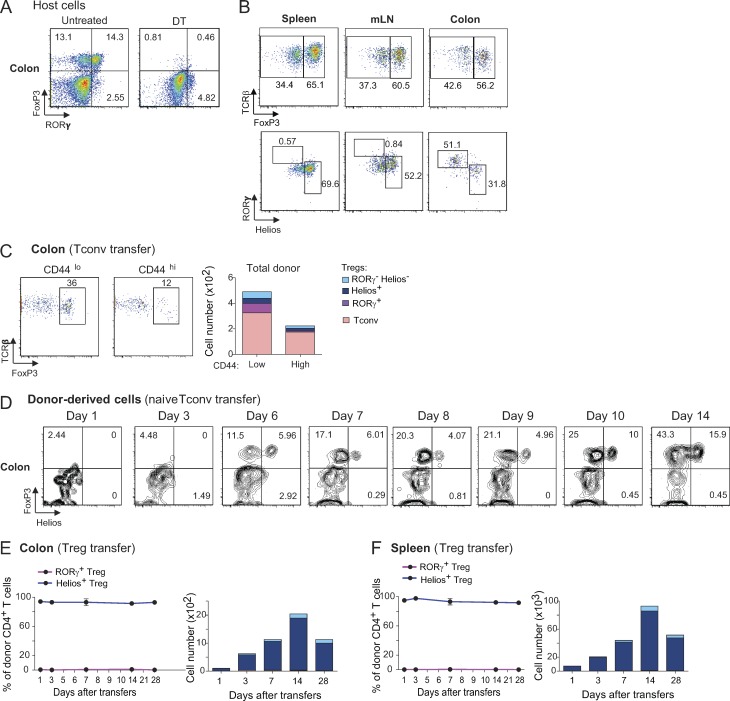
Figure S2.
Naive Tconv cells differentiate mostly into RORγ+ Treg cells while splenic Treg cells maintain their Helios+ phenotypes. (A) Representative dot plots of CD4+ T cells in the colon of Foxp3dtr mice before (left) and 30 h after (right) DT injection. Frequencies of cells in each quadrant are shown. (B) Representative dot plots of donor CD4+ T cells (top) and donor-derived Treg cells (bottom) in the spleen, mesenteric LNs (mLN), and colon of Foxp3dtr recipients. Data are representative of two independent experiments. (C) Flow cytometric analysis of a transfer experiment where 0.25 × 106 CD44lo or CD44hi CD4+ Tconv cells were transferred into Foxp3dtr hosts (left). Average numbers of donor-derived cells are shown (right; n ≥ 8). (D) Representative dot plots of donor CD4+ T cells in the colon of Foxp3dtr recipients at different days after transfer. Frequencies of cells in each quadrant are shown. (E and F) LN Treg cells were transferred into Foxp3dtr recipients. Frequencies (left) and average numbers (right) of donor-derived cells at different days after transfer in the colon (E) and spleen (F) of recipients (n ≥ 3 for each time point). Summary plots show data pooled from three independent experiments. Means ± SD.
A previous analysis of conversion of microbe-specific T cells showed that a FoxP3+ Treg cell population appeared 3 d before RORγ+ Treg cells, suggesting that RORγ+ Treg cells may differentiate from a FoxP3+RORγ− intermediate (Solomon and Hsieh, 2016). To better trace the relationship between Treg cell subsets, we evaluated the kinetics of conversion in our Treg cell replacement system. The bulk of donor Tconv cells resided in the spleen during the first few days after transfer, but distinct pTreg cell populations started to accumulate after day 6 (Fig. 3, A–C; and Fig. S2 D), largely synchronously in the colon and the spleen (Fig. 3, B and C). This timeframe was consistent with results in a transgenic model (Solomon and Hsieh, 2016). The appearance of RORγ+ Treg cells was only marginally delayed relative to that of total FoxP3+ pTreg cells overall, and continued to increase in parallel to a maximum around day 14, after which donor-derived pools started to be outcompeted by host-derived cells (Fig. 3, B and C). As expected from the results above, transferred splenic Treg cells accumulated while maintaining their Helios+ phenotype throughout the course of the experiment (Fig. S2, E and F). Hence, pre-existing Helios+ Treg cells that entered and/or expanded in the colon do not turn on RORγ expression. In addition, the conversion of Tconv cells into RORγ+ or Helios+ pTreg cells in the colon seems to follow parallel kinetics. Together, these observations suggest that RORγ does not appear secondarily in preformed FoxP3+ cells, but occurs only in the colon at the time when Tconv cells are converting into pTreg cells, or immediately thereafter.
Figure 3.
Donor Tconv cells up-regulate FoxP3 and RORγ at around the same time in the colon. (A) Representative dot plots of donor CD4+ T cells in the colon (top) and spleen (bottom) of Foxp3dtr recipients at different days following naive T cell transfer, per Fig. 2 A. (B and C) Frequencies (left) and average numbers (right) of donor-derived Treg cells in the colon (B) and spleen (C) of recipients described in A (n ≥ 3 for each time point). Summary plots show data pooled from three independent experiments. Means ± SD.
Variation in donor Tconv cells affects the phenotypes of converted pTreg cells
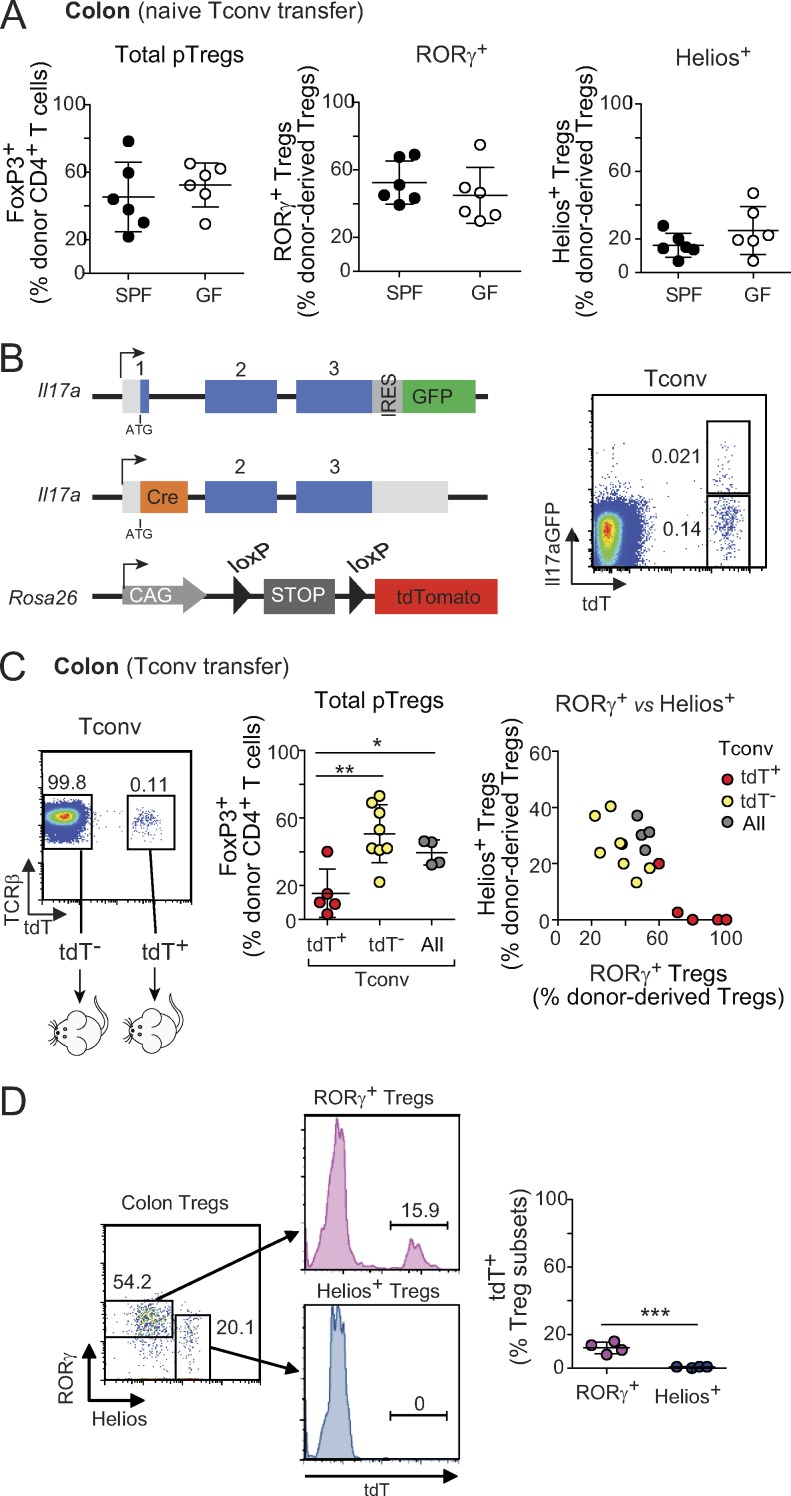
Since pTreg cells generated in the colon seemed to adopt two possible fates, we asked what parameters might affect the balance of outcomes. Continuous commensal antigen stimulation is essential for maintenance of the RORγ+ Treg cell pool (Sefik et al., 2015; Ohnmacht et al., 2015), so one might hypothesize that prior exposure of Tconv cells to microbial antigens is required for efficient RORγ+ pTreg cell conversion. To this end, we transferred naive Tconv cells from animals raised in either a GF or an SPF facility. There was no difference in total conversion to FoxP3+ pTreg cells, or in RORγ+ vs Helios+ pTreg cell frequencies (Fig. 4 A). Thus, donor cells do not need memory from pre-exposure to microbes, and microbial presence in the hosts is sufficient to drive RORγ+ pTreg cell conversion.
Figure 4.
Variation in donor Tconv cells affects the phenotypes of converted pTreg cells. (A) Frequencies of donor-derived Treg cells in the colon of Foxp3dtr hosts that received naive T cells from SPF or GF mice (n = 6). (B and C) Il17 fate-mapping mice were used as donors in the Treg cell replacement experiment. (B) Simplified diagram of Il17a and Rosa26 loci in these mice, and representative dot plot of Tconv cells showing tdT and GFP expression. (C) Transfer strategy and frequencies of donor-derived Treg cells in Foxp3dtr hosts that received 2.5 × 105 tdT−, tdT+, or total Tconv cells (n ≥ 4). (D) TdT expression among RORγ+ and Helios+ colonic Treg cells from Il17a fate-mapping mice (n = 4). Summary plots show data pooled from two (A and D) and four (C) independent experiments. Means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001 using Student’s t test. IRES, internal ribosome entry site; loxP, locus of X-over P1; CAG, hybrid construct consisting of the cytomegalovirus enhancer fused to the chicken beta-actin promoter.
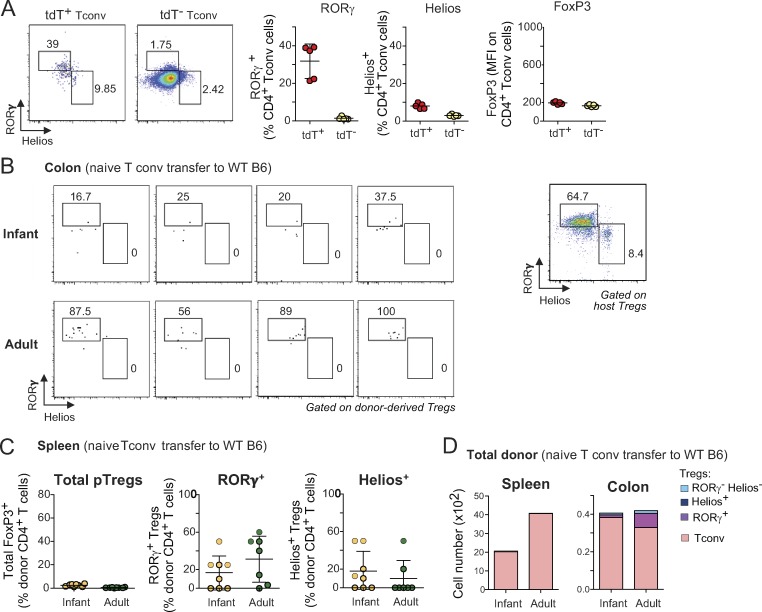
Is there, however, a predisposition of some naive Tconv cells to convert to RORγ+ pTreg cells? RORγ+ Treg cells and Il17-expressing Tconv cells (Th17) share some characteristics (foremost the expression of RORγ), and there have been reports that their fates might be inter-related (i.e., that Th17 cells could transdifferentiate to adopt an anti-inflammatory Treg cell phenotype [Gagliani et al., 2015], or that Treg cells could secrete Il17 under inflammatory conditions in vitro and in vivo [Komatsu et al., 2014; Yang et al., 2008; Osorio et al., 2008; Xu et al., 2007]). Thus, one possible explanation for the alternative fates adopted by pTreg cells was that RORγ+ Treg cells represent “exTh17” cells. We tested this hypothesis using Il17 fate-mapping mice (Il17aCregfp × Rosa26tdTomato; Hirota et al., 2011), which have tdTomato+ (tdT+) cells in which the Il17a locus was active at some time in their past, and a smaller fraction of tdT+GFP+ Th17 cells in which it is currently active (Fig. 4 B). TdT+ and tdT− Tconv cells from these mice were introduced into the Treg cell replacement system as above. Interestingly, tdT+ Tconv cells converted with lower efficiency than did their tdT− counterparts (Fig. 4 C, left), but their progeny included a markedly higher proportion of RORγ+ Treg cells (Fig. 4 C, right), likely resulting from their pre-existing expression of RORγ (Fig. S3 A). Importantly, however, tdT− Tconv cells still gave rise to high proportions of RORγ+ Treg cells. Thus, Tconv cells do not need to be ex-Th17 to convert into RORγ+ Treg cells. To analyze the relationship further, we examined colonic Treg cells isolated directly from the Il17a fate-reporter mice (Fig. 4 D). All of the Helios+ Treg cells were tdT−, suggesting an origin independent of Th17 cells. Some of the RORγ+ Treg cells were tdT+, in line with the transfer data, but these cells were clearly a small minority. Thus, while prior activity of the Il17a locus, and most likely of Rorc, can predispose Tconv cells to conversion to RORγ+ Treg cells, this is not a prerequisite for most cells.
Figure S3.
Pre-existing expression of RORγ on Tconv cells and availability of a Treg cell niche contribute to the differentiation of RORγ+ and Helios+ pTreg cells. (A) Representative dot plots (left) and expression of RORγ, Helios, and FoxP3 (right) on tdT+ and tdT− LN Tconv cells (n = 5). (B) Flow cytometric analyses of donor-derived Treg cells in the colon of unmanipulated B6 hosts that received naive T cells from infant (top left) or adult (bottom left) donors. Representative dot plot of host colonic Treg cells in the same experiment is shown (right). (C) Frequencies of donor-derived Treg cells in the spleen of unmanipulated B6 hosts that received naive cells from adult or infant donors (n = 8). Frequencies of RORγ+ and Helios+ Treg cells are not calculated in mice where no pTreg cells were observed. (D) Average numbers of donor-derived CD4+ T cells in the transfers shown in B and C. Data are pooled from two (A) and three (B–D) independent experiments. Means ± SD.
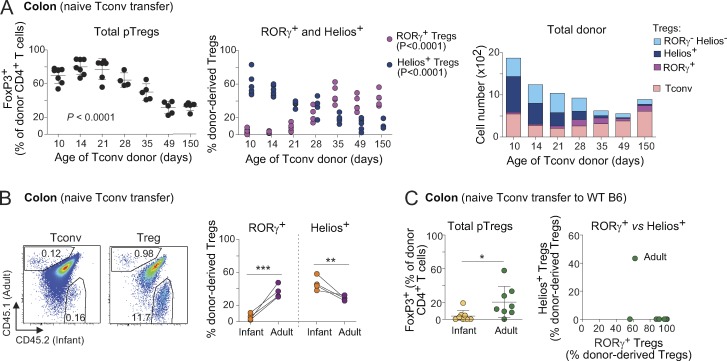
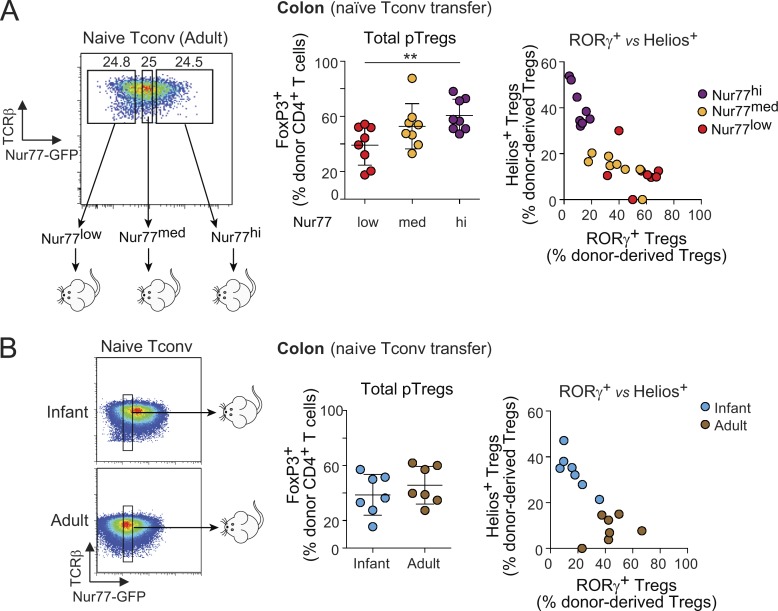
We and others have shown that the proportions of RORγ+ and Helios+ Treg cells in the colon evolve with age. Virtually all colonic Treg cells are Helios+ in neonatal and infant mice, as RORγ+ Treg cells start to appear only at ∼15 d of age in SPF conditions (Sefik et al., 2015; Nutsch et al., 2016). It was assumed that this switch is tied to the profound changes in the composition of the gut microbiota that occur around weaning, and there was evidence for this interpretation (Nutsch et al., 2016). Nevertheless, we asked whether Tconv cells from young mice might also be more inclined than adult Tconv cells to convert into Helios+ pTreg cells. Naive Tconv cells from mice of different ages were transferred into adult Foxp3dtr recipients, DT-treated as above. Strikingly, naive CD4+ T cells from infant mice (defined hereafter as 10–14 d of age), while converting into Treg cells more efficiently overall than those of adults (Fig. 5 A, left), mostly turned into Helios+ pTreg cells and were largely refractory to RORγ expression (Fig. 5 A, middle). In contrast, naive T cells from adult mice (35–150 d old) exhibited less Treg cell conversion in the same experiments, but predominantly to RORγ+ Treg cells. Interestingly, this transition occurred progressively over time, rather than abruptly around the weaning period (Fig. 5 A).
Figure 5.
Naive T cells from infant mice are predisposed to convert into Helios+ Treg cells in Treg cell–depleted hosts. (A) Frequencies and average numbers of donor-derived Treg cells in the whole colon of Foxp3dtr hosts that received naive T cells from mice of different ages (n ≥ 4). (B) A 1:1 mixture of naive T cells from CD45.1+ adult and CD45.2+ infant donors were transferred into CD45.1/2+ Foxp3dtr hosts. Representative dot plots of colonic Tconv (left) and Treg cells (middle) and frequencies of donor-derived Treg cells (right) are shown. Linked data points are from the same recipients (n = 4). (C) Frequencies of donor-derived Treg cells in the colon of unmanipulated B6 hosts (left) that received naive Tconv cells from adult or infant donors 7 d after transfer (n = 8). Due to low pTreg cell numbers after transfer of infant Tconv into normal hosts, only frequencies of RORγ+ and Helios+ pTreg cells from adult Tconv cells are shown (right). Summary plots show data pooled from two (B) and four (A and C) independent experiments. Means ± SD. P < 0.0001 using one-way ANOVA (A); *, P < 0.05; **, P < 0.01; ***, P < 0.001 using Student’s t test (B and C).
To determine whether this difference was Tconv cell–intrinsic, or resulted from intercrine effects, we transferred a 1:1 mix of congenically marked naive Tconv cells from adult and infant mice into Foxp3dtr hosts. The same distinction between infant and adult Tconv cell fates was observed after mixed transfer: in the same mouse, Tconv cells of infant origin converted more efficiently and mostly into Helios+ pTreg cells, while cells of adult origin mainly turned into RORγ+ pTreg cells (Fig. 5 B). Thus, even when being in the same host and in the presence of the same adult microbiota, there is a strong cell-intrinsic effect controlling the fate of infant or adult Tconv cells.
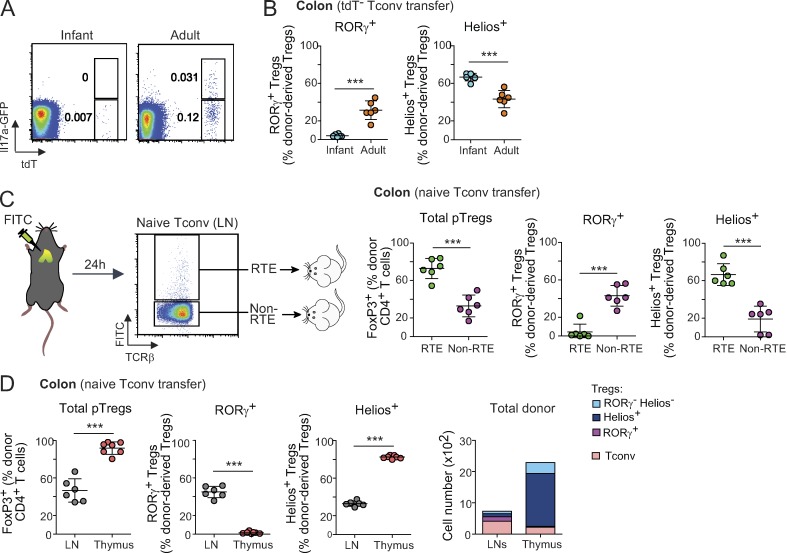
The Treg cell replacement system was designed to test the fate of polyclonal T cell pools in a nonirradiated host with normal lymphoid structures. One caveat, however, is that this is a setting in which conversion to pTreg cells is stimulated by the strong homeostatic drive to restore Treg cell pools (Kim et al., 2007; Feuerer et al., 2009), which might influence the outcome. We thus sought to validate our findings using unmanipulated B6 mice as hosts. As expected (Yang et al., 2018; Solomon and Hsieh, 2016), the extent of pTreg cell conversion was greatly diminished compared with that of Foxp3dtr hosts, presumably owing to limitations in niche size (Fig. 5 C and Fig. S3, B–D), which was particularly true after transferring naive Tconv cells from infant donors. Naive T cells from adult donors again gave rise predominantly to RORγ+ Treg cells, with no Helios+ Treg cells whatsoever (Fig. 5 C). Due to the very low cell numbers, the phenotype of pTreg cells derived from Tconv cells of infant mice was difficult to reliably evaluate (Fig. 5 C and Fig. S3 B), but most appeared to be RORγ−. These data confirm, in a Treg cell–replete environment, the differential ability of infant and adult Tconv cells to become RORγ+ pTreg cells.
Mechanism of differential infant versus adult conversion to pTreg cells
Having found that pTreg cells originating from adult or infant precursors adopted different phenotypes, we asked what molecular parameters might distinguish them. First, we assessed whether differentiation of Tconv cells from infant mice into Helios+ Treg cells is microbe-dependent, as are adult RORγ+ pTreg cells. Naive Tconv cells from adult or infant mice were transferred into Foxp3dtr recipients, half of which were treated with VMNA as above (Fig. 6 A). In contrast to adult Tconv cells, for which both overall conversion and RORγ+ pTreg cell proportions were strongly inhibited by antibiotic treatment as expected, conversion from infant Tconv cells was largely unaltered and resulted in Helios+ pTreg cells as in control mice. This difference confirms that infant Tconv cells are insensitive to the microbe-derived cues that induce Rorc expression in adult-derived Tconv cells. The observation also implies that the Helios+ phenotype may be a default pathway adopted in the absence of, or in the inability to sense, RORγ-inducing cues.
Figure 6.
Several checkpoints distinguish infant and adult pTreg cell conversion. (A and B) Frequencies and average numbers of donor-derived Treg cells in the colon of Foxp3dtr hosts that received naive T cells from infant or adult origin. (A) Recipients were treated, or not, with broad-spectrum antibiotics (per Fig. 2 C; n ≥ 5; data for adult donors are the same as in Fig. 2 C). (B) Donor Tconv cells were from Il6rafl/flxCd4Cre (deficient in the IL6 receptor) or control Il6rafl/fl littermates (n ≥ 3). Data are pooled from two (B) and three (A) independent experiments. Means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001 using Student’s t test.
IL6 is important for the generation or maintenance of RORγ+ Treg cells, with a twofold reduction in its absence (Ohnmacht et al., 2015; Yissachar et al., 2017), an effect that can be reproduced in vitro (Kim et al., 2017; Yang et al., 2018). We compared the role of IL6 signals in Treg cell conversion from infant or adult naive T cells using Il6rafl/flxCd4Cre mice. IL6R deficiency in naive Tconv cells of adult donors slightly increased donor-derived pTreg cell numbers in the colon, consistent with reports of pTreg cell inhibition by IL6 (Bettelli et al., 2006; Mangan et al., 2006), but with a significantly lower proportion of RORγ+ Treg cells (and correspondingly more Helios+ Treg cells; Fig. 6 B). These trends were not observed when young donor cells were used, with no difference between IL6R-deficient mice and their proficient littermates. Therefore, IL6-mediated signaling is important for conversion of Tconv cells from adult mice into RORγ+ Treg cells, but not for those from infant mice, possibly because IL6 signaling pathways are not yet active at this stage. Overall, the differential sensitivity to microbes and to IL6 suggests that pTreg cell conversion from adult and infant Tconv cells is regulated by distinct molecular checkpoints.
Why can’t Tconv cells from infant mice give rise to RORγ+ Treg cells?
The striking change over time in naive CD4+ T cells’ ability to convert to RORγ+ Treg cells had significant implication for setting the immunoregulatory tone in the colon, so it was important to decipher the underlying mechanisms. First, we asked whether the difference might be due simply to the higher proportion of “Il17a-experienced” cells in adult mice, which was readily evidenced in the Il17a fate-reporter mice (Fig. 7 A). However, when this difference was factored out by sorting only naive tdT− Tconv cells from both adult and infant mice, higher proportions of RORγ+ pTreg cells still emerged from Tconv cells from adults relative to infants (Fig. 7 B). Hence the skewed conversion into RORγ+ Treg cells from adult Tconv cells is not simply due to more abundant Il17a-experienced cells in adult mice.
Figure 7.
Dominance of RTE phenotypes in Tconv cells from infant mice promotes Helios+ pTreg cell conversion. (A) Representative dot plots of LN Tconv cells from infant or adult Il17a fate-mapping mice. Frequencies of tdT+ and GFP+tdT+ cells are indicated. (B) Frequencies of donor-derived Treg cells in the colon of Foxp3dtr hosts that received 106 naive tdT− Tconv cells from infant or adult Il17a fate-mapping mice (n = 6). (C) 2.5 × 105 RTEs and non-RTEs from adult mice were transferred to Foxp3dtr hosts. Sort strategy (left), and frequencies of donor-derived Treg cells (right) in the colon of hosts (n = 6). (D) LN naive T cells or CD4+ CD8− FoxP3− thymocytes from adult donors were transferred to Foxp3dtr hosts. Frequencies and average numbers of donor-derived Treg cells (n ≥ 6). Summary plots show data pooled from two independent experiments. Means ± SD. ***, P < 0.001 using Student’s t test.
Young animals naturally possess a high proportion of T cells that have recently emerged from the thymus, known as recent thymic emigrants (RTEs; Fink, 2013; Cunningham et al., 2018). RTEs have a distinct phenotype compared with mature naive T cells; for example, CD4+ RTEs produced less IL2 and IFNγ than their mature naive counterparts and had a bias toward T helper (Th) type 2 cell responses (Hendricks and Fink, 2011; Bhaumik et al., 2013; Friesen et al., 2016). Pertinent to this study, RTEs had an increased tendency to convert into Treg cells, especially when the Treg cell niche was empty (Bhaumik et al., 2013; Paiva et al., 2013), in part due to lower sensitivity to inhibitory cytokines like IL6 (Paiva et al., 2013) or IFNγ (Bhaumik et al., 2013). Hence, a plausible hypothesis was that the higher number of RTEs in young donors caused the difference in RORγ+ versus Helios+ pTreg cells. We labeled RTEs in adult mice by intrathymic FITC injection (Scollay et al., 1980) and sorted FITC-labeled naive Tconv cells and corresponding FITC− bulk cells from LNs 24 h later (Fig. 7 C) for Treg cell replacement in DT-treated Foxp3dtr hosts. 2 wk later, RTEs had converted into Treg cells more effectively than non-RTEs, and they up-regulated Helios instead of RORγ (Fig. 7 C). Similarly, CD4 single-positive thymocytes showed a high propensity for pTreg cell differentiation, predominantly to a Helios+ phenotype (Fig. 7 D). Although all donor cells here were from adult mice, the outcome recapitulated the infant/adult dichotomy, suggesting that the cell-autonomous decision between Helios+ or RORγ+ pTreg cell fate largely depends on the developmental maturity of the precursor Tconv cells.
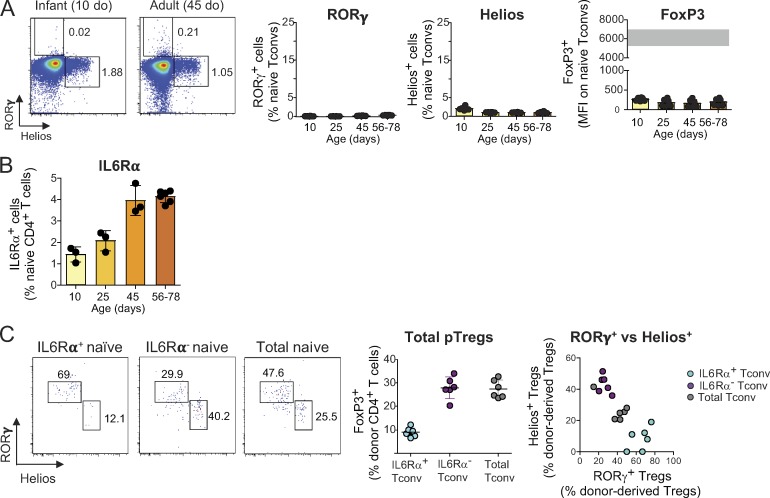
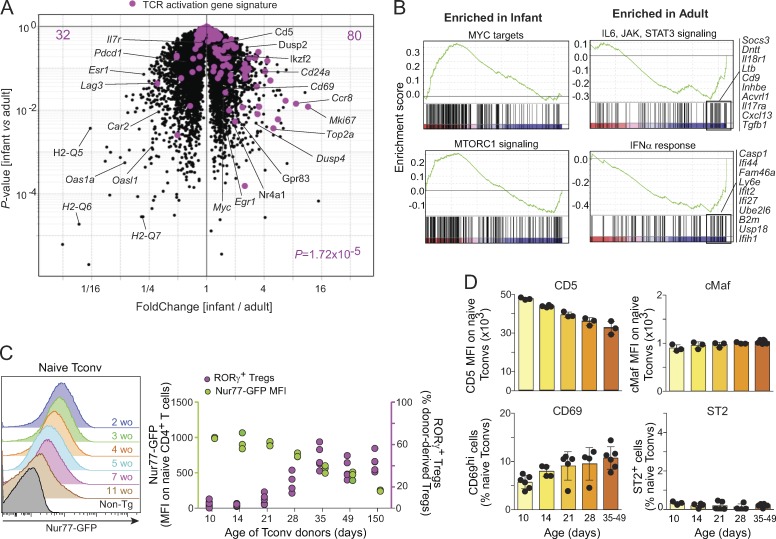
In an attempt to elucidate the molecular mechanisms that drive immature and mature Tconv cells to different pTreg cell fates, we first measured the expression of FoxP3, RORγ, and Helios on naive Tcells from mice of different ages. There was no age-dependent difference in any of these molecules (Fig. S4 A). For a broader approach, we compared the transcriptomes of naive CD4+ T cells from infant and adult mice by low-input RNA-seq (in biological triplicates). The differences (Fig. 8 A) were surprisingly extensive (212 and 277 transcripts up- or down-regulated at an arbitrary FoldChange of 2 and false discovery rate of 0.1). As expected (Boursalian et al., 2004; Houston and Fink, 2009), these included differences in maturation markers (Cd24a and H2-Q7). We noted an overexpression in infant Tconv cells of several transcripts indicative of activation through the TCR (Nr4a1, Cd5, Cd69, Dusp2, Dusp4, Myc, and Egr1). This was further evidenced as a shift of a generic T cell activation signature (Wakamatsu et al., 2013; highlighted in Fig. 8 A), and gene set enrichment analysis (GSEA) indicated a higher expression in infant Tconv cells of genes related to mTORC1 signaling and of c-Myc targets (Fig. 8, A and B). Since the expression of Nr4a1 (encodes Nur77; Moran et al., 2011; Baldwin and Hogquist, 2007) and Cd5 (Azzam et al., 1998) reflects the strength of TCR signals, this difference might suggest that Tconv cells from infant mice perceive trophic TCR signals more strongly than their adult counterparts.
Figure S4.
IL6 signaling contributes to driving the conversion of adult Tconv cells into RORγ+ Treg cells. (A) Representative dot plots (left), frequencies of RORγ+ and Helios+ cells, and mean fluorescence intensity (MFI) of FoxP3 on naive CD4+ T cells from mice of different ages (n = 6). The gray bar represents the range of FoxP3 mean fluorescence intensity of Treg cells. (B) Frequencies of IL6Rα+ cells among naive CD4+ T cells from mice of different ages (n ≥ 3). (C) Representative dot plots (left) and frequencies and average numbers of donor-derived Treg cells in the colon of Foxp3dtr hosts that received IL6Rα+ naive T cells, IL6Rα− naive T cells, or unfractionated naive T cells from adult donors (n = 6). Summary plots show data pooled from two independent experiments. Means ± SD. do, days old.
Figure 8.
Molecular differences between infant and adult Tconv cells. (A and B) Low-input RNA-seq was performed on 1,000 LN naive Tconv cells from infant and adult mice. (A) Differential expression shown as a volcano plot (fold change versus P value). TCR activation signature genes are highlighted in purple. Number of genes and χ2-test P value are indicated. (B) GSEA showing differentially enriched hallmark gene sets. Data are average values of three biological replicates. (C) Nur77-GFP expression on naive T cells from mice of different ages (left) and how they correlate with frequencies of RORγ+ pTreg cells following Tconv cell transfer from mice of different ages (n ≥ 3; RORγ+ frequency data taken from Fig. 5 A). (D) Mean fluorescence intensities of CD5 and c-Maf on naive T cells, and frequencies of CD69hi and ST2+ among naive T cells from mice of different ages (n ≥ 3). Data are pooled from three independent experiments. Means ± SD. MFI, mean fluorescence intensity; wo, weeks old; Tg, transgenic.
To follow this lead from the transcriptome data, we used Nur77gfp knock-in mice, in which GFP levels reflect the strength of TCR stimulation (Moran et al., 2011). Nur77GFP expression was highest in naive T cells from infant mice and decreased gradually with age (Fig. 8 C), with a time course that evoked the transitions in pTreg cell conversion seen in Fig. 5 A. CD5 expression followed a similar trend (Fig. 8 D). On the other hand, the early activation gene CD69 seemed to increase slightly with age, suggesting that it is not the immediate engagement of TCR that underlies the difference, but rather the downstream integration reflected by Nr4a1 and Cd5 levels.
Additionally, GSEA showed higher expression in adult Tconv cells of genes belonging to the IL6-JAK-STAT3 pathway (Fig. 8 B), consistent with the requirement for IL6Rα in adult Tconv cell conversion (Fig. 6 B) and the higher expression of IL6Rα in adult naive Tconv cells (Fig. S4 B). Naive CD4+ T cells expressing IL6Rα were predisposed to convert into RORγ+ Treg cells following transfer into Foxp3dtr mice (Fig. S4 C). In contrast, the expression of c-Maf, which has recently been shown to be important for RORγ+ Treg cell differentiation (Xu et al., 2018; Neumann et al., 2019; Wheaton et al., 2017), did not change with age (Fig. 8 D). Similarly, the level of ST2 (the receptor for IL33), which is preferentially expressed by Helios+ Treg cells and important for their expansion and stability (Schiering et al., 2014), remained low in all age groups analyzed (Fig. 8 D). These observations underscore the contribution of IL6 signaling to the age-dependent difference in pTreg cell phenotypes.
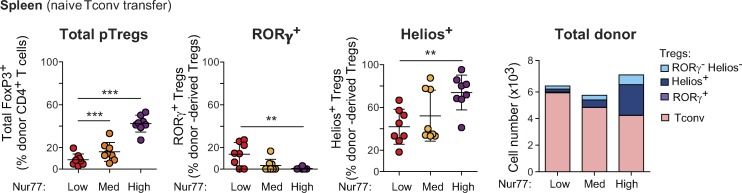
To test the significance of these observations and validate the relationship between Tconv cell activation status and the resulting pTreg cell phenotypes, we introduced naive Tconv cells from adult Nur77gfp mice into our Treg cell replacement system after sorting into three bins according to their intensity of GFP expression (Fig. 9 A). The extent of Treg cell conversion was proportional to the expression of GFP in donor cells, with infant-like Nur77GFPhi Tconv cells converting most efficiently to FoxP3+ Treg cells overall (Fig. 9 A, left) and yielding mostly Helios+ pTreg cells (Fig. 9 A). Conversely, adult-like Nur77GFPlo cells preferentially converted to RORγ+ pTreg cells. Similar trends were observed in the spleen of these mice, but only Nur77GFPhi cells gave a substantial extent of pTreg cell conversion there (Fig. S5). Hence, the status of TCR signaling pathways in the naive Tconv cell progenitors determines the conversion into either RORγ+ or Helios+ pTreg cells.
Figure 9.
Nur77 expression on donor Tconv cells correlates with the phenotypes of converted pTreg cells. (A) Naive Tconv cells from adult Nur77gfp reporter mice were sorted into three bins before transfer into DT-treated Foxp3dtr hosts (n = 8), yielding the colon pTreg cells shown at right. (B) Naive T cells with an identical window of Nur77GFP expression were sorted from adult or infant mice (n = 7). Summary plots show data pooled from two (B) and three (A) independent experiments. Means ± SD. **, P < 0.01 using Student’s t test.
Figure S5.
Tconv cells expressing a high amount of Nur77 preferentially convert to Helios+ pTreg cells in the spleen of recipients. Frequencies (left) and average numbers (right) of donor-derived Treg cells in the spleen of Foxp3dtr hosts that received Nur77low, Nur77med, or Nur77high naive Tconv cells from adult mice (n = 8). Summary plots show data pooled from three independent experiments. Means ± SD. **, P < 0.01; ***, P < 0.001 using Student’s t test.
In the experiments reported in Fig. 9 A, Nur77 may simply be an indicator of the activation status of the cells, but one could also hypothesize that it directly plays a role in determining the difference in fate of Tconv cells upon conversion to pTreg cells, which is plausible given its sizeable transcriptional footprint in T cells (Fassett et al., 2012; Liu et al., 2019; Chen et al., 2019). To distinguish between these scenarios, and to ask whether Nur77 can account for the difference between infant and adult Tconv cells, we sorted naive Tconv cells from infant or adult Nur77gfp mice in a matched window of GFP fluorescence (Fig. 9 B, left). If Nur77 was indeed the causal factor, equalizing its expression should eliminate the difference in pTreg cell subsets arising from infant and adult donors. In fact, while the percentage of total donor-derived FoxP3+ Treg cells became comparable for infant and adult donors, the relative proportion of RORγ+ pTreg cells remained higher for progeny from adult Tconv than from infant Tconv cells (Fig. 9 B). Hence, it is not Nur77 itself, but rather the cell activation state that it reflects, that predetermines the probability of converting Tconv cells to adopt RORγ+ or Helios+ pTreg cell phenotypes.
Discussion
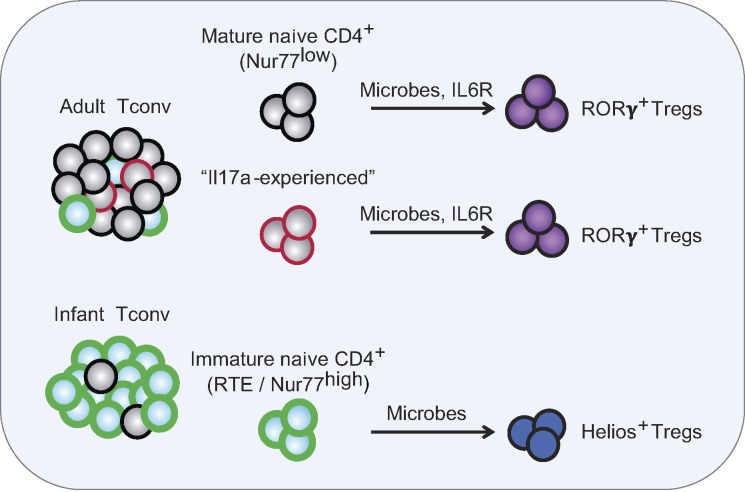
Using a combination of TCR-based lineage tracing and direct Treg cell replacement experiments, we have revisited the generation of pTreg cells in the colon, focusing in particular on the relationships between the RORγ+ and Helios+ Treg cell subsets. The key conclusions are that naive Tconv cells have the capacity to convert into both RORγ+ and Helios+ Treg cells in the colon, and that the developmental maturity (or cell states) of the starting Tconv cells determines the phenotype of the resulting pTreg cells. This study provides a new T cell–intrinsic perspective into the ontogeny and regulation of the two main Treg cell subsets in the colon (Fig. 10).
Figure 10.
Integrated model. Adult and infant naive Tconv cells differ in their composition: mostly immature RTEs in infants, and mostly mature naive T cells in adults, which also contain more Il17a-experienced CD4+ T cells. Both mature naive (Nur77low) and Il17a-experienced T cells preferentially convert to RORγ+ pTreg cells in the colon. This process is dependent on the presence of microbes and IL6R-mediated signals. In contrast, RTEs (Nur77high) that dominate in infant mice are skewed to convert to Helios+ Treg cells, with a lesser dependence on gut microbes and IL6R.
Since the gut microbiota is important for RORγ+ Treg cell differentiation, it was widely assumed that the appearance of colonic RORγ+ Treg cells around weaning (Nutsch et al., 2016; Sefik et al., 2015) was due solely to the profound changes in microbiota that result from the introduction of solid food. We now show that there are also T cell–intrinsic differences between pTreg cell precursors in infants and adults, and these likely contribute as well to the developmental timing of RORγ+ Treg cells. These results are not mutually exclusive with those from the Hsieh and Eberl groups, who showed that the environment of neonatal and infant mice is not conducive to differentiation of RORγ+ pTreg cells (Nutsch et al., 2016; Al Nabhani et al., 2019). Rather, they imply that the temporally delayed appearance of RORγ+ Treg cells in the colon has two roots, cell intrinsic (maturation stage of the CD4+ T cells, as reflected by the RTE transcriptome) and cell extrinsic (microbial influences). A caveat worth mentioning is that one cannot, in the quantitative comparison between the outcomes of such transfer experiments, distinguish differences in rates of conversion from differences in proliferative expansion and/or competitive fitness of the cells that result from conversion. Some of the influences may be affecting either or both aspects.
For the most part, the temporal shift could be ascribed to the proportion of RTEs among Tconv cell pools, since the biases could be recapitulated by purifying RTEs from adult mice. RTEs dominate the Tconv cell pool in infant mice since they take ∼3 wk to fully mature (Berzins et al., 1998; Boursalian et al., 2004; Cunningham et al., 2018). What, then, are the differences in Tconv cells that underlie the shift in potential? Several cellular characteristics of RTEs and/or of Tconv cells from infant mice might contrive to yield Helios+ pTreg cells. First, RTEs are prone to differentiate along the Th2 pathway (Hendricks and Fink, 2011), and Helios+ pTreg cells that developed from RTEs or infant Tconv cells co-expressed Gata3, the key transcriptional regulator of Th2 cells (Wan, 2014). Second, RTEs exhibit enhanced proliferation in a lymphopenic environment but compete less effectively with mature naive counterparts in lympho-replete conditions (Houston et al., 2011), which corresponds well to the present observations: greater responsiveness to conversion-inducing cues (IL2?) in the Treg cell–depleted environment (Fig. 2) and in lymphopenic conditions (Paiva et al., 2013), and far lower conversion in a lympho-replete environment (Fig. 5). This sensitivity seemed related to their greater activation status and to Nur77 levels. The “matched Nur77” experiment in Fig. 9 demonstrates that Nur77 is not the causal factor by itself, and that other elements also distinguish infant and adult Tconv cells. Third, RTEs seem less attuned to bacterial-derived inducers of RORγ+ Treg cell differentiation, and to the influence of IL6, consistent with a previous report that documented a lower sensitivity of RTEs to IL6 (Paiva et al., 2013), possibly linked to lower expression of IL6Rα. The greater reliance of adult/non-RTE cells on IL6 signals is consistent with their heightened expression of the IL6R–STAT3 pathway (Fig. 8).
Our profiling and transfer data point to differences in cell state as responsible for the varying fates of infant and adult Tconv cells. It is also possible that these reflect a gradual change in TCR repertoire, as young cells (infant/RTEs) capable of differentiation into Helios+ pTreg cells are depleted from the matured pool. A plausible scenario is that higher-affinity TCRs (reflected by high Nur77 expression) preferentially induce Helios+ pTreg cells, since strong TCR signals have been linked to Helios expression in other T cell contexts (Daley et al., 2013; Gottschalk et al., 2012). However, the quasi-exclusive conversion into Helios+ pTreg cells of RTEs, not all of which would be expected to express high-affinity TCRs, suggests that other elements of the cells’ signaling apparatus are at play. It may rather be the downstream integration of these TCR-driven signals (e.g., via differential sensitivity to IL7 or IL6) that determines the outcome. In addition, Helios expression was already slightly higher in Tconv cells from infant than adult mice. As Helios is one of the transcriptional cofactors of FoxP3 that is important for its activity (Kwon et al., 2017), Helios expression in the starting Tconv cells might positively reinforce the Helios+ pTreg cell phenotype and dampen the expression of RORγ.
In addition, Il17a-experienced Tconv cells that have expressed Il17a, and perhaps also Rorc, at some point in their past preferentially converted to RORγ+ pTreg cells. This is consistent with the notion that reciprocal exchanges can occur between Treg cell and Th17 populations (Gagliani et al., 2015; Komatsu et al., 2014; Yang et al., 2008), and one might speculate that it corresponds to a more open status of the chromatin at the Rorc locus in Il17a-experienced Tconv cells. However, this pathway seemed to play a minor contribution to the RORγ+ pTreg cell compartment overall.
This time-controlled development of the two populations of colonic Treg cells may denote an evolutionary advantage to a two-stage process: in early life, Helios+ Treg cells with amphiregulin-mediated barrier-fortifying function (Schiering et al., 2014) are the dominant subset, since there are only a few bacterial species to contend with (most of them Lactobacillus commensals from breast milk that are controlled by maternal IgA). A contribution to sterile tissue homeostasis may be particularly valuable at a time of rapid tissue growth. Later, RORγ+ pTreg cells become dominant when nutritional input changes radically and brings a far more diverse microbiota, and maintaining immunological tolerance to commensals becomes paramount. In adults, the potential of colonic Tconv cells to generate both Helios+ and RORγ+ pTreg cells enables a continued renewal of these pools, as needed in the face of novel microbial challenges or of situations that deplete Treg cell pools. Finally, niche competition between the two pTreg cell types may balance their relative proportions.
In conclusion, this study provides a better understanding of the origins of and relationships between RORγ+ and Helios+ Treg cells in the colon, and highlights the influence of Tconv cell states in determining the balance of these two pTreg cell fates, in addition to changes in environmental modulators. The ability to manipulate these states may prove clinically beneficial in inducing a particular pTreg cell subset to restore gut homeostasis.
Materials and methods
Mice
B6.CD45.1+, B6.CD45.2+, Foxp3ires-gfp/B6 (or Foxp3gfp; Bettelli et al., 2006), Foxp3tm3(DTR/GFP)Ayr/J (or Foxp3dtr; Kim et al., 2007), Foxp3tm10.1(Casp9,-Thy1)Ayr (or Foxp3thy1.1; Liston et al., 2008), Il17atm1.1(icre)Stck/J (or Il17aCre; Hirota et al., 2011), Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J (or Rosa26tdTomato; Madisen et al., 2010), Il17atm1Bcgen/J (or Il17agfp), Tg(Cd4-cre)1Cwi/BfluJ (or CD4Cre; Lee et al., 2001), Il6ratm1.1Drew/J (or Il6rafl; McFarland-Mancini et al., 2010), and Tg(Nr4a1-EGFP/cre)820Khog/J (or Nur77gfp; Moran et al., 2011) mice were obtained from the Jackson Laboratory. Il17a fate-reporter mice were generated by crossing Il17aCre mice to Rosa26tdTomato, and then to Il17agfp mice. All mice were backcrossed and maintained on the B6 background in our SPF facility at Harvard Medical School. A GF C57BL/6J breeding nucleus was obtained from L. Bry (Brigham and Women's Hospital, Harvard Medical School. Boston, MA) and maintained in GF isolators. Gnotobiotic mice were generated by gavaging 4-wk-old GF mice once with 108 CFU bacteria and housing them in sterile isolators for 2 wk before analysis. Adult (7–10 wk old) and infant (10–14 d old) male and female littermates were used, unless indicated otherwise. All experimentation was performed following animal protocols approved by the Harvard Medical School Institutional Animal Use and Care Committee (protocol IS00001257).
Bacteria
B. thetaiotaomicron (Bthet.ATCC29741) culture was obtained from the American Type Culture Collection.
Antibiotic treatment
For antibiotic treatment, a mixture of 1 g/liter of ampicillin sodium salt (Sigma-Aldrich), 1 g/liter of metronidazole (Sigma-Aldrich), 0.5 g/liter vancomycin hydrochloride (Research Products International), and 1 g/liter neomycin sulfate (Thermo Fisher Scientific) plus 2.5 g/liter of the sweetener Equal were used.
T cell transfers
For Treg cell or Tconv cell transfer, 105 Treg cells or 106 naive CD4+ Tconv cells were sorted from pooled (inguinal, mesenteric, brachial, axillary, and cervical) LNs of adult (7–10 wk old) or infant (10–14 d old) CD45.2+ Foxp3gfp mice using MoFlo (Beckman Coulter) and were i.v. injected into gender-matched 6–8-wk-old CD45.1+ Foxp3dtr or B6 mice. Treg cells were sorted as DAPI− CD4+ TCRβ+ Foxp3GFP+, whereas naive CD4+ Tconv cells were gated as DAPI− CD4+ TCRβ+ CD44lo Foxp3GFP−. In certain experiments, recipients were given antibiotic-supplemented water from 7 d before transfer until the day of analysis. When GF mice or Il6rafl/fl CD4Cre mice were used as donors, naive CD4+ Tconv cells were sorted as DAPI− CD4+ TCRβ+ CD44lo CD25−. For CD4+ CD44hi Tconv cell transfer: 0.25 × 106 CD44lo or CD44hi Tconv cells were sorted from pooled spleen and LNs of adult male CD45.2+ Foxp3gfp mice using MoFlo and were i.v.-injected into male 6–8-wk-old CD45.1+ Foxp3dtr mice. For Il17aCregfp Rosa26tdTomato cell transfer: 0.25 × 106 tdT+ or tdT− Tconv cells were sorted from pooled spleen and LNs of adult or infant male CD45.2+ Il17a fate-mapping mice using MoFlo and were i.v.-injected into male 6–8-wk-old CD45.1+ Foxp3dtr mice. For RTE transfer: 0.25 × 106 FITC+ or FITC− naive Tconv cells were sorted from pooled LNs of adult male CD45.2+ Foxp3thy1.1 mice using MoFlo and were i.v.-injected into male 6–8-wk-old CD45.1+ Foxp3dtr mice. For CD4 single-positive thymocytes transfer: 106 T cells were sorted from either thymi or LNs of adult male CD45.2+ Foxp3gfp mice using MoFlo and were i.v.-injected into male 6–8-wk-old CD45.1+ Foxp3dtr mice. Thymocytes were gated as DAPI− CD4+ CD8− TCRβ+ Foxp3GFP−, while LN Tconv cells were sorted as DAPI− CD4+ TCRβ+ CD44lo Foxp3GFP−. Treg cell depletion was achieved by injecting Foxp3dtr mice i.p. with DT (Sigma-Aldrich) in PBS at 20 ng/g of body weight. DT was injected 1 d prior, and 1 and 3 d after, transfer. 1 (for transfer into unmanipulated B6 hosts) or 2 wk (for transfer into Foxp3dtr recipients) after transfer, frequencies of donor-derived cells in spleen and colon of recipient mice were analyzed by flow cytometry.
Cell isolation and flow cytometry
A single-cell suspension of mouse splenocytes, thymocytes, or LNs was obtained by physical dissociation with a 40-μm cell strainer (Falcon). For splenocytes, lysis of RBCs was performed with 1 ml of ACK lysing buffer (Lonza) for 2 min on ice. Lymphocytes from colonic LP were isolated through a procedure outlined previously (Sefik et al., 2015). In brief, the entire colon was incubated in RPMI (Gibco) containing 1 mM dithiothreitol, 20 mM EDTA, and 2% FCS at 37°C for 15 min to remove epithelial cells. The colon was then minced and dissociated in RPMI containing 1.5 mg/ml collagenase II (Gibco), 0.5 mg/ml Dispase (Gibco), and 1% FCS, at 37°C for 45 min with constant stirring. The digested materials were washed and filtered through a 40-μm cell strainer at least twice to obtain a single-cell suspension. Antibody staining was performed in ice-cold buffer (RPMI with 2% FCS) for 30 min at a dilution of 1/100 with antibodies to CD5 (53–7.3), CD8 (53–6.7), CD25 (PC61), CD44 (IM7), CD45 (30-F11), CD45.1 (A20), CD45.2 (104), CD69 (H1.2F3), CD90.1/Thy1.1 (OX-7), and TCRβ (H57-597; all from BioLegend), IL6Rα (15A7; homemade), and CD4 (RM4-5; Invitrogen). For analysis of TFs, cells were fixed, permeabilized, and intracellularly stained for Foxp3 (FJK-16s), Gata3 (TWAJ), RORγ (AFKJS-9; all from Invitrogen), c-Maf (sym0F1; eBioscience), and Helios (22F6; BioLegend) according to the manufacturer’s (eBioscience’s) instructions. Cells were acquired with an LSRII flow cytometer (BD Biosciences) or a MoFlo cell sorter (Beckman Coulter), and data were analyzed using FlowJo software.
RTE labeling in adult mice
Intrathymic injection of FITC has been described in detail previously (Scollay et al., 1980). Briefly, thymic lobes were injected with ∼10 μl of 350 μg/ml FITC (Invitrogen), randomly labeling 30–60% of thymocytes. Mice were euthanized 24 h later, and lymphocytes from LNs were analyzed by FACS.
RNA-seq
Biological triplicates of 103 naive CD4+ T cells (DAPI− CD4+ TCRβ+ CD44lo Foxp3GFP−) from pooled LNs of 10-d-old or 7-wk-old Foxp3gfp mice were double-sorted using MoFlo into 5 μl buffer TCL (Qiagen) containing 1% 2-mercaptoethanol (Sigma-Aldrich). Smart-Seq2 libraries were prepared by the Broad Technology Labs and sequenced using the Broad Genomics Platform (Picelli et al., 2014). In brief, total RNA was captured and purified on RNAClean XP beads (Beckman Coulter). Polyadenylated mRNA was selected using an anchored oligo(dT) primer and reverse-transcribed to cDNA. First-strand cDNA was subjected to limited PCR amplification followed by transposon-based fragmentation using the Nextera XT DNA Library Preparation Kit (Illumina). Samples were then PCR-amplified using barcoded primers such that each sample carried a specific combination of Illumina P5 and P7 barcodes and were pooled before sequencing. Paired-end sequencing was performed on an Illumina NextSeq500 using 2 × 25 bp reads. Reads were aligned to the mouse genome (Gencode GRCm38 primary assembly; https://www.gencodegenes.org/mouse/). Transcripts were quantified by the Broad Technology Labs computational pipeline with Cuffquant version 2.2.1 (Trapnell et al., 2012). Raw read count tables were normalized by the median of ratios method with the DESeq2 package from Bioconductor and then converted to GCT and CLS format. Poor-quality samples with <3 million uniquely mapped reads were automatically excluded from normalization. Normalized reads were further filtered by minimal expression and analyzed by Multiplot Studio in the GenePattern software package. Pathway enrichment analysis was performed by querying hallmark gene sets in the Molecular Signatures Database (v6.2; Subramanian et al., 2005) using the GSEA tool.
scRNA-seq library preparation and data analysis
scRNA-seq was performed using the InDrop protocol that has been described in detail elsewhere (Zilionis et al., 2017; Zemmour et al., 2018). In brief, 3 × 104 CD4+ T cells (DAPI− CD4+ TCRβ+) from the colon of 6-wk-old B. thetaiotaomicron–monocolonized mice were sorted using MoFlo into RPMI medium containing 2% FCS. Cells were then pelleted by centrifugation at 500 g for 5 min and resuspended in PBS containing 15% OptiPrep Density Gradient Medium (Sigma-Aldrich) at a concentration of 80,000 cells/ml. Around 2,000 single cells per sample were then encapsulated in droplets of 3–4 nl containing a primer hydrogel bead and the SuperScript III RT buffer (Invitrogen). RT was performed immediately after encapsulation. After purification of the DNA/RNA duplex with 1.2× AMPure XP beads (Beckman Coulter), second-strand cDNA synthesis (NEB) was performed per the manufacturer’s instructions. The library was then amplified by in vitro transcription using HiScribe T7 High Yield RNA Synthesis Kit (NEB). After purification, half of the amplified RNA was used for further processing, while the rest was saved for TCRαβ sequencing. For transcriptome analysis, the amplified RNA was fragmented using the magnesium RNA fragmentation kit (Ambion) and purified using AMPure Beads (1.2×). RT with random hexamers was then performed using PrimeScript RT (Takara Clontech) per the manufacturer’s instructions. A final PCR using Kapa HiFi HotStart PCR mix (Kapa Biosystems) was performed to amplify the library and to add the P5-P7 and Illumina index primers. Library size was measured with a High Sensitivity D1000 ScreenTape (Agilent Technologies), quantified by quantitative PCR, and sequenced using NextSeq500.
Single-cell demultiplexing was performed against the possible barcode space, and only reads mapping unambiguously and with less than two mismatches were kept. For each single-cell library, reads were mapped to the mouse mm10 transcriptome using tophat2. Duplicate reads, those mapping to multiple regions, or those having a low alignment score (mapping quality < 10) were filtered out. A final matrix with genes in rows and cells in columns was then constructed. Data were then analyzed using the Seurat R toolkit (Butler et al., 2018). Briefly, cells were first filtered based on the number of unique genes. Cells with <200 unique genes were excluded from further analysis. Gene expression values for each cell were then normalized by the total expression, multiplied by an arbitrary scale factor of 10,000, and log-transformed. A principle component analysis was performed on the top 200 most variable genes that were expressed in >1% of the cells. The number of statistically significant principal components was determined by comparison with principle component analysis over a randomized matrix as described previously (Klein et al., 2015). The data were then visualized using the t-distributed stochastic neighbor embedding (tSNE) dimensionality reduction algorithm (van der Maaten and Hinton, 2008), on the significant PCs. The activated T cell, Treg cell, RORγ+ Treg cell, or Helios+ Treg cell gene signature score for each single cell was calculated by summing the counts for the genes identified previously to be up-regulated in that particular subset (DiSpirito et al., 2018; list of genes provided in Table S1).
Paired single-cell TCRαβ sequencing
A detailed version of this protocol has been described previously (Zemmour et al., 2018). The material used for this process was the same as for the whole-transcriptome library construction, by taking half of the amplified RNA immediately before the fragmentation step. In brief, the RNA library was amplified by RT using T cell receptor alpha variable region (TRAV) or T cell receptor beta variable region (TRBV) external primers (Table S1). After purification with 0.5× AMPure Beads (Beckman Coulter), cDNA was further amplified by PCR using TRAV or TRBV internal primers containing the Illumina PE1 sequence. A second PCR incorporating the P5 Illumina sequence was then performed. The PCR product was again purified and size-selected twice with 0.5× AMPure beads. Library size was assessed using a High sensitivity D5000 ScreenTape (Agilent Technologies). The TCRα and TCRβ libraries had an expected size of 1,470 and 1,230 bp respectively. Library concentration was determined by quantitative PCR, and sequencing was done using paired-end Nano MiSeq.
Similar to the transcriptome analysis, single-cell demultiplexing was performed against the possible barcode space, and only reads mapping unambiguously and with fewer than two mismatches were kept. TCRα and TCRβ alignment (V, D, J alignment and CDR3 identification) was done individually for each single cell against the mouse IMGT database (http://www.imgt.org/) using the MiXCR 1.8.1 software (Bolotin et al., 2015). TCRα and TCRβ sequences and transcriptome data were matched to the same single-cell barcode unambiguously with fewer than two mismatches. Only single-cell barcodes with both TCRα and TCRβ sequences (with MiXCR score > 100) and for which the transcriptome was available were kept for further analysis. Single cells were grouped in clonotypes when sharing the same TCRα and TCRβ nucleotide sequences.
Data availability
RNA-seq data comparing infant versus adult mice and scRNA-seq data of colon T cells have been deposited to the Gene Expression Omnibus database under accession nos. GSE132255 and GSE132573, respectively.
Statistical analyses
Data were presented as means ± SD. Statistical significance, indicated by asterisks, was determined by Student’s t test (two-tailed, unpaired) or one-way ANOVA using GraphPad Prism 5.0. P values <0.05 were considered significant: *, P < 0.05; **, P < 0.01; ***, P < 0.001. χ2 test was used to determine the enrichment of certain gene signatures in RNA-seq datasets.
Online supplemental material
Fig. S1 shows the scRNA-seq quality control analysis. Fig. S2 compares the fates of naive, effector Tconv cells, and tTreg cells following adoptive transfer into Foxp3dtr recipients. Fig. S3 shows that pre-existing expression of RORγ on Tconv cells and availability of a Treg cell niche contribute to the differentiation of RORγ+ pTreg cells. Fig. S4 details the contribution of IL6R signaling to RORγ+ pTreg cell conversion. Fig. S5 shows that high expression of Nur77 on Tconv cells correlates with Helios+ pTreg cells conversion in the spleen. Table S1 contains additional information on the scRNA-seq experiment.
Supplementary Material
C. Lists of primers for TCR sequencing
Acknowledgments
We thank Drs. V. Kuchroo (Il17aCre × Rosa26tdTomato × Il17agfp mice; Evergrande Center for Immunological Diseases, Harvard Medical School and Brigham and Women’s Hospital, Boston, MA), A. Rudensky (Foxp3thy1.1 mice; Memorial Sloan Kettering Cancer Center, New York, NY), and D. Zemmour for insightful discussions, data, and mouse lines, and K. Hattori, G. Buruzula, A. Wood, C. Araneo, K. Seddu, B. Vijaykumar, and L. Yang for help with mice, cell sorting, profiling, and computational biology. Cell sorting was at Joslin Flow Core (CF-0003-11-04 and S10OD021740).
This work was supported by grants from the National Institutes of Health (RO1-AI51530 and RO1-AI125603) and by the Evergrande Center for Immunological Diseases.
The authors declare no competing financial interests.
Author contributions: A. Pratama, D. Mathis, and C. Benoist designed the research. A. Pratama and A. Schnell performed the research. A. Pratama, A. Schnell, D. Mathis, and C. Benoist analyzed the data. A. Pratama and C. Benoist wrote the manuscript.
References
- Akimova T., Beier U.H., Wang L., Levine M.H., and Hancock W.W.. 2011. Helios expression is a marker of T cell activation and proliferation. PLoS One. 6:e24226 10.1371/journal.pone.0024226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Nabhani Z., Dulauroy S., Marques R., Cousu C., Al Bounny S., Déjardin F., Sparwasser T., Bérard M., Cerf-Bensussan N., and Eberl G.. 2019. A weaning reaction to microbiota is required for resistance to immunopathologies in the adult. Immunity. 50:1276–1288.e5. 10.1016/j.immuni.2019.02.014 [DOI] [PubMed] [Google Scholar]
- Atarashi K., Tanoue T., Shima T., Imaoka A., Kuwahara T., Momose Y., Cheng G., Yamasaki S., Saito T., Ohba Y., et al. 2011. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 331:337–341. 10.1126/science.1198469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzam H.S., Grinberg A., Lui K., Shen H., Shores E.W., and Love P.E.. 1998. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J. Exp. Med. 188:2301–2311. 10.1084/jem.188.12.2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin T.A., and Hogquist K.A.. 2007. Transcriptional analysis of clonal deletion in vivo. J. Immunol. 179:837–844. 10.4049/jimmunol.179.2.837 [DOI] [PubMed] [Google Scholar]
- Berzins S.P., Boyd R.L., and Miller J.F.. 1998. The role of the thymus and recent thymic migrants in the maintenance of the adult peripheral lymphocyte pool. J. Exp. Med. 187:1839–1848. 10.1084/jem.187.11.1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettelli E., Carrier Y., Gao W., Korn T., Strom T.B., Oukka M., Weiner H.L., and Kuchroo V.K.. 2006. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 441:235–238. 10.1038/nature04753 [DOI] [PubMed] [Google Scholar]
- Bhaumik S., Giffon T., Bolinger D., Kirkman R., Lewis D.B., Weaver C.T., and Randolph D.A.. 2013. Retinoic acid hypersensitivity promotes peripheral tolerance in recent thymic emigrants. J. Immunol. 190:2603–2613. 10.4049/jimmunol.1200852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin D.A., Poslavsky S., Mitrophanov I., Shugay M., Mamedov I.Z., Putintseva E.V., and Chudakov D.M.. 2015. MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods. 12:380–381. 10.1038/nmeth.3364 [DOI] [PubMed] [Google Scholar]
- Boursalian T.E., Golob J., Soper D.M., Cooper C.J., and Fink P.J.. 2004. Continued maturation of thymic emigrants in the periphery. Nat. Immunol. 5:418–425. 10.1038/ni1049 [DOI] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., and Satija R.. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36:411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., López-Moyado I.F., Seo H., Lio C.J., Hempleman L.J., Sekiya T., Yoshimura A., Scott-Browne J.P., and Rao A.. 2019. NR4A transcription factors limit CAR T cell function in solid tumours. Nature. 567:530–534. 10.1038/s41586-019-0985-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham C.A., Helm E.Y., and Fink P.J.. 2018. Reinterpreting recent thymic emigrant function: defective or adaptive? Curr. Opin. Immunol. 51:1–6. 10.1016/j.coi.2017.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley S.R., Hu D.Y., and Goodnow C.C.. 2013. Helios marks strongly autoreactive CD4+ T cells in two major waves of thymic deletion distinguished by induction of PD-1 or NF-κB. J. Exp. Med. 210:269–285. 10.1084/jem.20121458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiSpirito J.R., Zemmour D., Ramanan D., Cho J., Zilionis R., Klein A.M., Benoist C., and Mathis D.. 2018. Molecular diversification of regulatory T cells in nonlymphoid tissues. Sci. Immunol. 3:eaat5861 10.1126/sciimmunol.aat5861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassett M.S., Jiang W., D’Alise A.M., Mathis D., and Benoist C.. 2012. Nuclear receptor Nr4a1 modulates both regulatory T-cell (Treg) differentiation and clonal deletion. Proc. Natl. Acad. Sci. USA. 109:3891–3896. 10.1073/pnas.1200090109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerer M., Shen Y., Littman D.R., Benoist C., and Mathis D.. 2009. How punctual ablation of regulatory T cells unleashes an autoimmune lesion within the pancreatic islets. Immunity. 31:654–664. 10.1016/j.immuni.2009.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink P.J. 2013. The biology of recent thymic emigrants. Annu. Rev. Immunol. 31:31–50. 10.1146/annurev-immunol-032712-100010 [DOI] [PubMed] [Google Scholar]
- Friesen T.J., Ji Q., and Fink P.J.. 2016. Recent thymic emigrants are tolerized in the absence of inflammation. J. Exp. Med. 213:913–920. 10.1084/jem.20151990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliani N., Amezcua Vesely M.C., Iseppon A., Brockmann L., Xu H., Palm N.W., de Zoete M.R., Licona-Limón P., Paiva R.S., Ching T., et al. 2015. Th17 cells transdifferentiate into regulatory T cells during resolution of inflammation. Nature. 523:221–225. 10.1038/nature14452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuking M.B., Cahenzli J., Lawson M.A., Ng D.C., Slack E., Hapfelmeier S., McCoy K.D., and Macpherson A.J.. 2011. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 34:794–806. 10.1016/j.immuni.2011.03.021 [DOI] [PubMed] [Google Scholar]
- Geva-Zatorsky N., Sefik E., Kua L., Pasman L., Tan T.G., Ortiz-Lopez A., Yanortsang T.B., Yang L., Jupp R., Mathis D., et al. 2017. Mining the human gut microbiota for immunomodulatory organisms. Cell. 168:928–943.e11. 10.1016/j.cell.2017.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk R.A., Corse E., and Allison J.P.. 2012. Expression of Helios in peripherally induced Foxp3+ regulatory T cells. J. Immunol. 188:976–980. 10.4049/jimmunol.1102964 [DOI] [PubMed] [Google Scholar]
- He Z., Chen L., Souto F.O., Canasto-Chibuque C., Bongers G., Deshpande M., Harpaz N., Ko H.M., Kelley K., Furtado G.C., and Lira S.A.. 2017. Epithelial-derived IL-33 promotes intestinal tumorigenesis in Apc Min/+ mice. Sci. Rep. 7:5520 10.1038/s41598-017-05716-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks D.W., and Fink P.J.. 2011. Recent thymic emigrants are biased against the T-helper type 1 and toward the T-helper type 2 effector lineage. Blood. 117:1239–1249. 10.1182/blood-2010-07-299263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K., Duarte J.H., Veldhoen M., Hornsby E., Li Y., Cua D.J., Ahlfors H., Wilhelm C., Tolaini M., Menzel U., et al. 2011. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat. Immunol. 12:255–263. 10.1038/ni.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston E.G. Jr., and Fink P.J.. 2009. MHC drives TCR repertoire shaping, but not maturation, in recent thymic emigrants. J. Immunol. 183:7244–7249. 10.4049/jimmunol.0902313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston E.G. Jr., Higdon L.E., and Fink P.J.. 2011. Recent thymic emigrants are preferentially incorporated only into the depleted T-cell pool. Proc. Natl. Acad. Sci. USA. 108:5366–5371. 10.1073/pnas.1015286108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.S., Liang Y., Tyznik A.J., Self S.G., Liggitt D., and Rudensky A.Y.. 2004. Recognition of the peripheral self by naturally arising CD25+ CD4+ T cell receptors. Immunity. 21:267–277. 10.1016/j.immuni.2004.07.009 [DOI] [PubMed] [Google Scholar]
- Ivanov I.I., McKenzie B.S., Zhou L., Tadokoro C.E., Lepelley A., Lafaille J.J., Cua D.J., and Littman D.R.. 2006. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 126:1121–1133. 10.1016/j.cell.2006.07.035 [DOI] [PubMed] [Google Scholar]
- Kim J.M., Rasmussen J.P., and Rudensky A.Y.. 2007. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 8:191–197. 10.1038/ni1428 [DOI] [PubMed] [Google Scholar]
- Kim B.S., Lu H., Ichiyama K., Chen X., Zhang Y.B., Mistry N.A., Tanaka K., Lee Y.H., Nurieva R., Zhang L., et al. 2017. Generation of RORγt+ antigen-specific T regulatory 17 cells from Foxp3+ precursors in autoimmunity. Cell Reports. 21:195–207. 10.1016/j.celrep.2017.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A.M., Mazutis L., Akartuna I., Tallapragada N., Veres A., Li V., Peshkin L., Weitz D.A., and Kirschner M.W.. 2015. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 161:1187–1201. 10.1016/j.cell.2015.04.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu N., Okamoto K., Sawa S., Nakashima T., Oh-hora M., Kodama T., Tanaka S., Bluestone J.A., and Takayanagi H.. 2014. Pathogenic conversion of Foxp3+ T cells into TH17 cells in autoimmune arthritis. Nat. Med. 20:62–68. 10.1038/nm.3432 [DOI] [PubMed] [Google Scholar]
- Komori T., Okada A., Stewart V., and Alt F.W.. 1993. Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science. 261:1171–1175. 10.1126/science.8356451 [DOI] [PubMed] [Google Scholar]
- Kwon H.K., Chen H.M., Mathis D., and Benoist C.. 2017. Different molecular complexes that mediate transcriptional induction and repression by FoxP3. Nat. Immunol. 18:1238–1248. 10.1038/ni.3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop S.K., Bloom S.M., Rao S.M., Nutsch K., Lio C.W., Santacruz N., Peterson D.A., Stappenbeck T.S., and Hsieh C.S.. 2011. Peripheral education of the immune system by colonic commensal microbiota. Nature. 478:250–254. 10.1038/nature10434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.P., Fitzpatrick D.R., Beard C., Jessup H.K., Lehar S., Makar K.W., Pérez-Melgosa M., Sweetser M.T., Schlissel M.S., Nguyen S., et al. 2001. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 15:763–774. 10.1016/S1074-7613(01)00227-8 [DOI] [PubMed] [Google Scholar]
- Liston A., Nutsch K.M., Farr A.G., Lund J.M., Rasmussen J.P., Koni P.A., and Rudensky A.Y.. 2008. Differentiation of regulatory Foxp3+ T cells in the thymic cortex. Proc. Natl. Acad. Sci. USA. 105:11903–11908. 10.1073/pnas.0801506105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Nguyen P., Liu W., Cheng C., Steeves M., Obenauer J.C., Ma J., and Geiger T.L.. 2009. T cell receptor CDR3 sequence but not recognition characteristics distinguish autoreactive effector and Foxp3(+) regulatory T cells. Immunity. 31:909–920. 10.1016/j.immuni.2009.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Wang Y., Lu H., Li J., Yan X., Xiao M., Hao J., Alekseev A., Khong H., Chen T., et al. 2019. Genome-wide analysis identifies NR4A1 as a key mediator of T cell dysfunction. Nature. 567:525–529. 10.1038/s41586-019-0979-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R., et al. 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13:133–140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan P.R., Harrington L.E., O’Quinn D.B., Helms W.S., Bullard D.C., Elson C.O., Hatton R.D., Wahl S.M., Schoeb T.R., and Weaver C.T.. 2006. Transforming growth factor-β induces development of the T(H)17 lineage. Nature. 441:231–234. 10.1038/nature04754 [DOI] [PubMed] [Google Scholar]
- McFarland-Mancini M.M., Funk H.M., Paluch A.M., Zhou M., Giridhar P.V., Mercer C.A., Kozma S.C., and Drew A.F.. 2010. Differences in wound healing in mice with deficiency of IL-6 versus IL-6 receptor. J. Immunol. 184:7219–7228. 10.4049/jimmunol.0901929 [DOI] [PubMed] [Google Scholar]
- Molofsky A.B., Savage A.K., and Locksley R.M.. 2015. Interleukin-33 in tissue homeostasis, injury, and inflammation. Immunity. 42:1005–1019. 10.1016/j.immuni.2015.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran A.E., Holzapfel K.L., Xing Y., Cunningham N.R., Maltzman J.S., Punt J., and Hogquist K.A.. 2011. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J. Exp. Med. 208:1279–1289. 10.1084/jem.20110308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann C., Blume J., Roy U., Teh P.P., Vasanthakumar A., Beller A., Liao Y., Heinrich F., Arenzana T.L., Hackney J.A., et al. 2019. c-Maf-dependent Treg cell control of intestinal TH17 cells and IgA establishes host-microbiota homeostasis. Nat. Immunol. 20:471–481. 10.1038/s41590-019-0316-2 [DOI] [PubMed] [Google Scholar]
- Nutsch K., Chai J.N., Ai T.L., Russler-Germain E., Feehley T., Nagler C.R., and Hsieh C.S.. 2016. Rapid and efficient generation of regulatory T cells to commensal antigens in the periphery. Cell Reports. 17:206–220. 10.1016/j.celrep.2016.08.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnmacht C., Park J.H., Cording S., Wing J.B., Atarashi K., Obata Y., Gaboriau-Routhiau V., Marques R., Dulauroy S., Fedoseeva M., et al. 2015. MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORγt+ T cells. Science. 349:989–993. 10.1126/science.aac4263 [DOI] [PubMed] [Google Scholar]
- Osorio F., LeibundGut-Landmann S., Lochner M., Lahl K., Sparwasser T., Eberl G., and Reis e Sousa C.. 2008. DC activated via dectin-1 convert Treg into IL-17 producers. Eur. J. Immunol. 38:3274–3281. 10.1002/eji.200838950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacholczyk R., Ignatowicz H., Kraj P., and Ignatowicz L.. 2006. Origin and T cell receptor diversity of Foxp3+CD4+CD25+ T cells. Immunity. 25:249–259. 10.1016/j.immuni.2006.05.016 [DOI] [PubMed] [Google Scholar]
- Paiva R.S., Lino A.C., Bergman M.L., Caramalho I., Sousa A.E., Zelenay S., and Demengeot J.. 2013. Recent thymic emigrants are the preferential precursors of regulatory T cells differentiated in the periphery. Proc. Natl. Acad. Sci. USA. 110:6494–6499. 10.1073/pnas.1221955110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peine M., Marek R.M., and Löhning M.. 2016. IL-33 in T cell differentiation, function, and immune homeostasis. Trends Immunol. 37:321–333. 10.1016/j.it.2016.03.007 [DOI] [PubMed] [Google Scholar]
- Picelli S., Faridani O.R., Björklund A.K., Winberg G., Sagasser S., and Sandberg R.. 2014. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9:171–181. 10.1038/nprot.2014.006 [DOI] [PubMed] [Google Scholar]
- Russler-Germain E.V., Rengarajan S., and Hsieh C.S.. 2017. Antigen-specific regulatory T-cell responses to intestinal microbiota. Mucosal Immunol. 10:1375–1386. 10.1038/mi.2017.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa S., Cherrier M., Lochner M., Satoh-Takayama N., Fehling H.J., Langa F., Di Santo J.P., and Eberl G.. 2010. Lineage relationship analysis of RORgammat+ innate lymphoid cells. Science. 330:665–669. 10.1126/science.1194597 [DOI] [PubMed] [Google Scholar]
- Schiering C., Krausgruber T., Chomka A., Fröhlich A., Adelmann K., Wohlfert E.A., Pott J., Griseri T., Bollrath J., Hegazy A.N., et al. 2014. The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature. 513:564–568. 10.1038/nature13577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scollay R.G., Butcher E.C., and Weissman I.L.. 1980. Thymus cell migration. Quantitative aspects of cellular traffic from the thymus to the periphery in mice. Eur. J. Immunol. 10:210–218. 10.1002/eji.1830100310 [DOI] [PubMed] [Google Scholar]
- Sefik E., Geva-Zatorsky N., Oh S., Konnikova L., Zemmour D., McGuire A.M., Burzyn D., Ortiz-Lopez A., Lobera M., Yang J., et al. 2015. MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORγ+ regulatory T cells. Science. 349:993–997. 10.1126/science.aaa9420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., and Rudra D.. 2018. Emerging functions of regulatory T cells in tissue homeostasis. Front. Immunol. 9:883 10.3389/fimmu.2018.00883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon B.D., and Hsieh C.S.. 2016. Antigen-specific development of mucosal Foxp3+RORγt+ T cells from regulatory T cell precursors. J. Immunol. 197:3512–3519. 10.4049/jimmunol.1601217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spits H., and Cupedo T.. 2012. Innate lymphoid cells: emerging insights in development, lineage relationships, and function. Annu. Rev. Immunol. 30:647–675. 10.1146/annurev-immunol-020711-075053 [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanoue T., Atarashi K., and Honda K.. 2016. Development and maintenance of intestinal regulatory T cells. Nat. Rev. Immunol. 16:295–309. 10.1038/nri.2016.36 [DOI] [PubMed] [Google Scholar]
- Thornton A.M., Korty P.E., Tran D.Q., Wohlfert E.A., Murray P.E., Belkaid Y., and Shevach E.M.. 2010. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J. Immunol. 184:3433–3441. 10.4049/jimmunol.0904028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., and Pachter L.. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7:562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L., and Hinton G.. 2008. Visualizing high-dimensional data using t-SNE. J. Mach. Learn. Res. 9:2579–2605. [Google Scholar]
- Wakamatsu E., Mathis D., and Benoist C.. 2013. Convergent and divergent effects of costimulatory molecules in conventional and regulatory CD4+ T cells. Proc. Natl. Acad. Sci. USA. 110:1023–1028. 10.1073/pnas.1220688110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y.Y. 2014. GATA3: a master of many trades in immune regulation. Trends Immunol. 35:233–242. 10.1016/j.it.2014.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheaton J.D., Yeh C.H., and Ciofani M.. 2017. Cutting edge: c-Maf is required for regulatory T cells to adopt RORγt+ and follicular phenotypes. J. Immunol. 199:3931–3936. 10.4049/jimmunol.1701134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlfert E.A., Grainger J.R., Bouladoux N., Konkel J.E., Oldenhove G., Ribeiro C.H., Hall J.A., Yagi R., Naik S., Bhairavabhotla R., et al. 2011. GATA3 controls Foxp3+ regulatory T cell fate during inflammation in mice. J. Clin. Invest. 121:4503–4515. 10.1172/JCI57456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J., Obst R., Correia-Neves M., Losyev G., Mathis D., and Benoist C.. 2007. Adaptation of TCR repertoires to self-peptides in regulatory and nonregulatory CD4+ T cells. J. Immunol. 178:7032–7041. 10.4049/jimmunol.178.11.7032 [DOI] [PubMed] [Google Scholar]
- Xu L., Kitani A., Fuss I., and Strober W.. 2007. Cutting edge: regulatory T cells induce CD4+CD25-Foxp3- T cells or are self-induced to become Th17 cells in the absence of exogenous TGF-β. J. Immunol. 178:6725–6729. 10.4049/jimmunol.178.11.6725 [DOI] [PubMed] [Google Scholar]
- Xu M., Pokrovskii M., Ding Y., Yi R., Au C., Harrison O.J., Galan C., Belkaid Y., Bonneau R., and Littman D.R.. 2018. c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature. 554:373–377. 10.1038/nature25500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.O., Nurieva R., Martinez G.J., Kang H.S., Chung Y., Pappu B.P., Shah B., Chang S.H., Schluns K.S., Watowich S.S., et al. 2008. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 29:44–56. 10.1016/j.immuni.2008.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B.H., Hagemann S., Mamareli P., Lauer U., Hoffmann U., Beckstette M., Föhse L., Prinz I., Pezoldt J., Suerbaum S., et al. 2016. Foxp3(+) T cells expressing RORγt represent a stable regulatory T-cell effector lineage with enhanced suppressive capacity during intestinal inflammation. Mucosal Immunol. 9:444–457. 10.1038/mi.2015.74 [DOI] [PubMed] [Google Scholar]
- Yang J., Zou M., Pezoldt J., Zhou X., and Huehn J.. 2018. Thymus-derived Foxp3+ regulatory T cells upregulate RORγt expression under inflammatory conditions. J. Mol. Med. (Berl.). 96:1387–1394. 10.1007/s00109-018-1706-x [DOI] [PubMed] [Google Scholar]
- Yissachar N., Zhou Y., Ung L., Lai N.Y., Mohan J.F., Ehrlicher A., Weitz D.A., Kasper D.L., Chiu I.M., Mathis D., and Benoist C.. 2017. An intestinal organ culture system uncovers a role for the nervous system in microbe-immune crosstalk. Cell. 168:1135–1148.e12. 10.1016/j.cell.2017.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemmour D., Zilionis R., Kiner E., Klein A.M., Mathis D., and Benoist C.. 2018. Single-cell gene expression reveals a landscape of regulatory T cell phenotypes shaped by the TCR. Nat. Immunol. 19:291–301. 10.1038/s41590-018-0051-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilionis R., Nainys J., Veres A., Savova V., Zemmour D., Klein A.M., and Mazutis L.. 2017. Single-cell barcoding and sequencing using droplet microfluidics. Nat. Protoc. 12:44–73. 10.1038/nprot.2016.154 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
C. Lists of primers for TCR sequencing
Data Availability Statement
RNA-seq data comparing infant versus adult mice and scRNA-seq data of colon T cells have been deposited to the Gene Expression Omnibus database under accession nos. GSE132255 and GSE132573, respectively.