Abstract
Neurological diseases (NDs) are progressive disorders, the progression of which can be significantly affected by a range of common diseases that present as comorbidities. Clinical studies, including epidemiological and neuropathological analyses, indicate that patients with type 2 diabetes (T2D) have worse progression of NDs, suggesting pathogenic links between NDs and T2D. However, finding causal or predisposing factors that link T2D and NDs remains challenging. To address these problems, we developed a high-throughput network-based quantitative pipeline using agnostic approaches to identify genes expressed abnormally in both T2D and NDs, to identify some of the shared molecular pathways that may underpin T2D and ND interaction. We employed gene expression transcriptomic datasets from control and disease-affected individuals and identified differentially expressed genes (DEGs) in tissues of patients with T2D and ND when compared to unaffected control individuals. One hundred and ninety seven DEGs (99 up-regulated and 98 down-regulated in affected individuals) that were common to both the T2D and the ND datasets were identified. Functional annotation of these identified DEGs revealed the involvement of significant cell signaling associated molecular pathways. The overlapping DEGs (i.e., seen in both T2D and ND datasets) were then used to extract the most significant GO terms. We performed validation of these results with gold benchmark databases and literature searching, which identified which genes and pathways had been previously linked to NDs or T2D and which are novel. Hub proteins in the pathways were identified (including DNM2, DNM1, MYH14, PACSIN2, TFRC, PDE4D, ENTPD1, PLK4, CDC20B, and CDC14A) using protein-protein interaction analysis which have not previously been described as playing a role in these diseases. To reveal the transcriptional and post-transcriptional regulators of the DEGs we used transcription factor (TF) interactions analysis and DEG-microRNAs (miRNAs) interaction analysis, respectively. We thus identified the following TFs as important in driving expression of our T2D/ND common genes: FOXC1, GATA2, FOXL1, YY1, E2F1, NFIC, NFYA, USF2, HINFP, MEF2A, SRF, NFKB1, USF2, HINFP, MEF2A, SRF, NFKB1, PDE4D, CREB1, SP1, HOXA5, SREBF1, TFAP2A, STAT3, POU2F2, TP53, PPARG, and JUN. MicroRNAs that affect expression of these genes include mir-335-5p, mir-16-5p, mir-93-5p, mir-17-5p, mir-124-3p. Thus, our transcriptomic data analysis identifies novel potential links between NDs and T2D pathologies that may underlie comorbidity interactions, links that may include potential targets for therapeutic intervention. In sum, our neighborhood-based benchmarking and multilayer network topology methods identified novel putative biomarkers that indicate how type 2 diabetes (T2D) and these neurological diseases interact and pathways that, in the future, may be targeted for treatment.
Keywords: bioinformatics, computational biology, gene ontology, protein, pathways, type 2 diabetes, neurological disease
1. Introduction
Type 2 diabetes (T2D) is a global health burden that affects hundreds of millions of people [1]. It is characterized by glucose dyshomeostasis, hyperglycaemia and insulin resistance, with predisposing factors that include obesity, poor quality diet, insufficient physical activity and genetic factors [2,3]. These factors interact to cause failure of circulating glucose level regulation which can result in an inability to supply sufficient insulin and eventual beta-cell loss that exacerbates the condition [4,5]. Glucotoxicity caused by chronic hyperglycemia induces cell injury of many cell types but hepatocytes and pancreatic cells in particular [6]. Hyperglycaemia classically causes vascular disease, damaging blood vessels and leading to a range of cardiovascular diseases. In addition, hyperglycemia has a number of long term effects that exacerbate impairments of central nervous system (CNS) function and cognitive function [7]. Metabolic changes seen in T2D patients lead to chronic CNS inflammation that contribute to neurodegeneration [8]. Other T2D associated metabolic disturbances are associated with atrophy in several regions of the brain (e.g., hippocampal) that in turn are associated with cognitive impairment [9]. The brain is a very insulin-sensitive organ, so insulin resistance itself can affect memory and learning [10]. Indeed, glucose levels affect neuronal maintenance, neurogenesis, neurotransmitter regulation, cell survival and synaptic plasticity [11]. Moreover, it is notable that neurodegenerative diseases are accompanied by high production of inflammatory mediators, oxidative stress, Deoxyribonuclic acid (DNA) damage, and mitochondrial dysfunction which in turn also contribute to the degenerative cascade and exacerbate insulin resistance [12]. T2D is also associated with excessive immune system activation [13].
While the detailed mechanisms remains unclear, epidemiological, cognitive, and neuropathological evidence shows associations between T2D and neurodegenerative diseases such as Alzheimer’s disease (AD) [14], amyotrophic lateral sclerosis (ALS) [15], cerebral palsy (CP) [16], epilepsy disease (ED) [17], Huntington’s disease (HD) [18], multiple sclerosis (MS) [19], and Parkinson’s disease (PD) [20]. Interestingly, neuroimaging studies of the CNS of individuals with T2D also show structural alterations that resemble those in neurological disease patients [21]. The high incidence of T2D thus raises issues around its interaction with other diseases occurring in the same individuals.
Epidemiological studies show a particularly strong association between T2D and AD [14]. AD is characterized by the accumulation of -amyloid (A) into neuritic plaques and the presence of intracellular aggregates of tau protein in neurofibrillary tangles, amylin deposition as well as synaptic loss, neuroinflammation, and neuronal death [14]. Although its etiology still remains unclear, genetic predisposition and aging are strong risk factors for AD [22]. Moreover, the presence of (A) and tau in the pancreas and insulin-sensitive tissues and their roles in inducing peripheral insulin resistance or disruptions in insulin secretion indicate that this may contribute to the incidence of AD [14].
A wealth of evidence indicates a link between T2D and ALS [15]. ALS is a disorder characterized by progressive muscular atrophy, cognitive impairment, and pyramidal deficit, due to the degeneration of upper and lower motor neurons [23]. Motor neuron loss is associated with mutations in the Cu/Zn superoxide dismutase (SOD1) gene in this disease. ALS patients have altered lipid and glucose metabolism with increased energy consumption [24] and hypermetabolism [25]. Nevertheless, the biological mechanisms linking T2D to ALS are yet unclear even though there are clearly important risk factors in common, including environmental factors, higher body mass index (BMI), elevated cholesterol level and hyperlipidemia [26].
T2D also shows associations with CP, a neurodevelopmental disorder characterized by permanent movement-related disabilities, evident in early life, due to abnormalities in the brain centres that control movement and balance [16]. There are developmental issues in the fetus that lead to CP may have a link to maternal T2D incidence [27], and as maternal obesity and T2D might raise the risk of CP occurring [16]. Notably, children with CP may develop T2D as an adult. The etiology of CP is unclear but the role of perinatal factors such as chorioamnionitis hypoxic-ischemic encephalopathy as well as brain injury occurring during the perinatal and postnatal periods may contribute to CP [28].
T2D is also linked to ED [17]. This is a group of neurological diseases characterized by epileptic seizures. The precise relationship between T2D and epilepsy remains unclear. It is known that epilepsy or seizures are associated with autoimmune or inflammatory disorders and in the pro-inflammatory processes [29]. Additionally, hyperglycemia may exert adverse effects on the central nervous system which leads to ED [30]. Some known risk factors including degenerative brain disorders and head injuries, stroke and dementia are considered as predisposing factors to ED [31].
HD is a neurodegenerative disorder caused by an expanded CAG repeat in exon 1 of the huntingtin gene (HTT) encoding huntingtin protein and characterized by progressive disturbances of mood and motor function, and by cognitive dysfunction [32]. The pathogenetic mechanisms behind HD include misfolding and aggregation of the huntingtin protein, oxidative stress, impaired mitochondrial metabolism, excitotoxicity in affected brain regions, and impairment of the ubiquitin-proteasome system [32]. Although Huntington’s disease (HD) is primarily considered a rare neurodegenerative disorder, insulin resistance and impaired glucose metabolism contribute to its development [33].
MS is a chronic inflammatory and progressive immune-mediated disease of the central nervous system (CNS), characterized by a selective and coordinated inflammatory destruction of the myelin sheath, with damage to the axon [34]. Inflammation, demyelination, and axonal degeneration are associated with MS. Moreover, insulin resistance may induce inflammatory responses, oxidative stress and could exacerbate cognitive impairments in individuals with MS [19]. Though the links between T2D and risk of MS incidence is unclear, there are common genetic and environmental factors that contribute to the MS [34].
PD is clinically characterized by severe motor symptoms that include postural instability, resting tremor, muscular rigidity, and slowness of movements and pathologically characterized by the preferential loss of dopaminergic neurons [20]. PD features the presence of intracellular inclusions, known as Lewy bodies, rich in fibrillar -Synuclein (aS), a protein suggested being involved in synaptic vesicle recycling and docking [35]. Several epidemiological studies have demonstrated that PD patients have impaired insulin signaling and insulin resistance, and hyperglycaemia which play a role to suppress dopaminergic neuronal activity and that decreasing dopamine turnover that contributes to the possible progression of PD [36].
In sum, there is good evidence that there are pathologically and clinically significant relationships between T2D and many NDs but the association has not been widely examined. As the etiology of T2D and NDs are quite complex and their risk factors somewhat tend to overlap, their biological basis and the molecular mechanisms that underlie this link are still not well understood. Finding interactions between T2D and NDs is very difficult, but is of great interest in medical endocrinology. T2D and NDs are very complex diseases in terms of their clinical presentations, and because of this they are hard to study by conventional hypothesis-driven endocrinology research, despite the high clinical importance. Moreover, there is still a lack of bioinformatics studies addressing the relationship between T2D and NDs. The aim of this study was to identify such links between T2D and NDs, since understanding the nature of these links could bring important insights into the mechanisms that underlie these diseases. This led us to employ a bioinformatics system pipeline, to analyze gene expression data from studies of disease-affected tissues for clues to the nature of the relationship between T2D and NDs.
Here, we focus on finding ND-associated differentially expressed genes (DEGs), molecular pathways and putative discriminative biomarkers that are common to both T2D and NDs. We subsequently performed the validation of the results with gold benchmark experimentally validated databases that include dbGaP, OMIM and OMIM Expanded, and literature.
2. Materials and Methods
2.1. Datasets Employed in This Study
We query datasets from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) [37]. Queries for each disease returns a number of datasets but most of them were discarded for having a sample size below our selected cutoff sample size of at least 7, having no two conditions such as control vs case or control vs treated, replicate datasets, having undesirable formatting or irrelevant experimental focus, RNAseq datasets, and datasets from non-human organisms. Here, we used datasets by seeking those that would minimise any bias and noise for this type of analysis. This process identified 8 datasets that are highly relevant to T2D, AD, ALS, CP, ED, HD, MS, and PD are appropriate for our study.
We analyzed different human gene expression datasets with accession numbers GSE23343 [38], GSE28146 [39], GSE833 [40], GSE31243 [41], GSE22779 [42], GSE1751 [43], GSE38010 [44]. and GSE19587 [45], having control and disease affected individuals for our study. The T2D dataset (GSE23343) contained gene expression data obtained from liver biopsies of 10 T2D hyperglycemic patients and 7 normoglycemic controls using an Affymetrix Human Genome U133 Plus 2.0 arrays. The AD dataset (GSE28146) was microarray data (also Affymetrix U133 Plus 2.0 arrays) on RNA from snap-frozen brain tissue where white matter tissue was extracted by laser capture methods to collect only CA1 hippocampal gray matter. The ALS dataset (GSE833) employing Affymetrix HuGeneFL Hu6800 arrays, was a study of postmortem spinal cord gray matter samples from 7 ALS patients and from 4 control. The CP (GSE31243) dataset was generated from 40 Affymetrix human genome U133A 2.0 microarray studies of hamstring muscle samples from 20 controls (taken during tissue reconstruction) and 20 CP patients. The ED dataset (GSE22779) was a gene expression profile of peripheral blood mononuclear cells from 12 healthy control and individuals 4 with epilepsy in a study of in vivo glucocorticoid treatment; only data from blood cells extracted before glucocorticoid treatment were used. The HD dataset (GSE1751) was taken from peripheral blood cells from 14 healthy control and 12 symptomatic HD-affected individuals along with 5 presymptomatic HD patients; this also used U133A arrays. The MS dataset (GSE38010) using U133A array data from 5 MS patient brain lesions (identified histologically) compared with 2 brain tissue samples from control individuals. The PD dataset (GSE19587) was an analysis taken from affected brain areas of 12 postmortem brains of PD patients and 10 control samples of unaffected brain tissue using Affymetrix U133A Plus 2.0 arrays.
2.2. Preprocessing and Identification of Differentially Expressed Genes
We acquired gene expression microarray datasets from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO). All these datasets were generated by comparing diseased tissue against controls to identify differentially expressed genes (DEGs) associated with their respective pathology. To make uniform the mRNA expression data from different platforms and to avoid the problems of experimental systems, we normalized the gene expression data comprising disease state and control data by using the Z-score transformation () for each NDs gene expression profile using the following equation:
| (1) |
where implies standard deviation and indicates gene expression magnitude i in sample j. Such a transformation allows direct comparisons of gene expression values across samples and diseases. The datasets were normalized using the Robust Multi-Array Average expression measure (version 1.30.1) as implemented in the “affy” package (version 1.56.0) of the Bioconductor platform (version Rx64 3.3.0) in R. We performed the analysis of the microarray data using Linear Models for Microarray Data (Limma) [46]. Unpaired t-test statistic was used to identify genes differentially expressed in patients compared to normal samples. Moreover, to determine the statistical significance between groups, a two-way analysis of variance (ANOVA) with the false discovery rate (FDR) test was performed. Based on standard statistical criteria, a threshold of at least 1 log2 fold change (logFC) and t-tests giving a p-value of 0.01 were chosen. p-value < 0.01 and logFC > 1 for up-regulated genes and p-value < 0.01 and logFC < −1 for down-regulated genes were used. Genes with significantly different expression were thus selected. Gene symbols and names extracted for each disease. Gene symbol records with null or missing data were discarded for each disease. We identified both unique genes that were both over and under-expressed in NDs and T2D. We then pairwise compared the DEGs from T2D datasets with that of our AD, ALS, CP, ED, HD, MS, and PD datasets to find DEGs common to T2D and the NDs. Genes with the greatest magnitude up and down-regulation were selected from those common to the individual disease and T2D.
We then applied neighborhood benchmarking and topological methods to show the associations between genes and diseases. A gene-disease network in short GDN was built to identify gene-disease connections, in which nodes can be either diseases or genes; such a network is represented as a bipartite graph where T2D is the center of this network using Cytoscape V 3.6.1. [47]. Diseases are associated when sharing at least one significantly dysregulated gene. For gene-disease association, we consider a set of human diseases, denoted by D and a set of human genes, denoted by G to find whether gene is associated with disease . Moreover, we consider that if and is the sets of genes with significantly up and down-regulated that were associated with diseases and , respectively, then the number of shared dysregulated genes associated with both disease and is defined as follows [48]:
| (2) |
The co-occurrence is the number of common genes between two diseases in the GDN and common neighbors identified employing Jaccard Coefficient methods [48], where edge predictions score (association score) for the node pair is:
| (3) |
where G is the set of nodes and E is the set of edges. We also applied R software packages comoR [49], and POGO [50] to cross-check disease comorbidity associations.
2.3. Identification of Molecular Pathway and Gene Ontology
To obtain further insights into the molecular pathways and gene ontology (GO) of T2D that overlap with AD, ALS, CP, ED, HD, MS, and PD, we performed gene set enrichment analysis to identify pathways and GO of the overlapping DEGs with EnrichR [51]. Pathways are central to organism responses to stimuli, and pathway-based analysis is a recently developed approach to understand how complex diseases may be related to each other through their underlying molecular mechanisms [52]. GO is a conceptual model for the representation of gene functions and their relationship to gene regulation [53]. We considered 7 pathway databases: KEGG [54], Reactome [55], NCI-Nature [56], WiKi [57], BioCarta [58], Panther [59] and HumanCyc pathway database [60] and Gene ontology (GO) domain: Biological Process (BP).
2.4. Protein-Protein Interactions Analysis
Protein-protein interaction networks (PPIs) represent the physical contacts between two or more proteins molecules and are essential to all cell processes [61]. We also generated protein-protein interaction (PPI) networks based on the physical interaction of the proteins of DEGs by using information from the STRING database [62] via Network Analyst using the confidence score 900, where proteins are represented by nodes and protein interactions represented by edges. Using topological parameters, for example, a degree greater than 15°, highly interacting proteins were identified from PPI analysis.
2.5. Transcription Factors-microRNA Interactions Analysis
We studied the DEGs-Transcription Factors (TFs) and DEGs-microRNAs (miRNAs) to identify the regulatory biomolecules (i.e., TFs and miRNAs) that regulate DEGs of interest at the transcriptional and post-transcriptional level. We utilized the JASPAR database to analyze the DEGs-TFs interaction [63]. We employed miRNA-DEGs interactions from TarBase [64] and miRTarBase [65]. The topological analysis was performed via Network Analyzer in Cytoscape [47] and Network Analyst [66]. The TFs were screened out based on the degree (≥20) from the DEGs-TFs network. The miRNAs were selected based on the degree (≥15) from the DEGs-miRNAs network.
2.6. An Overview of the Analytical Approach
Our network-based systematical and quantitative pipeline to evaluate gene expression in human disease comorbidities is summarized as shown in Figure 1. The integrated pipeline of used here was implemented using the R language, code which is available on request. We developed the proposed pipeline using GEOquery [67] for downloading GEO datasets and expression set class transformation; limma [46] for differentially expressed gene identification from microarray data; genefilter [68] for filtering genes. The version of the used software and packages was R version 3.5.1, R Studio 1.0.136, Bioconductor 3.8, GEOquery 2.50.5, Affy 1.56.0, limma 3.38.3, genefilter 1.64.0.
Figure 1.
An overview of the network-based systematic and quantitative approach. Neurological diseases that were investigated comprised of Alzheimer’s disease (AD), amyotrophic lateral sclerosis (ALS), cerebral palsy (CP), epilepsy disease (ED), Huntington’s disease (HD), multiple sclerosis (MS), and Parkinson’s disease (PD).
Our approach employs gene expression analyses, disease gene association networks, signaling pathway mechanisms, gene ontology (GO) data, protein-protein interactions (PPIs) network, DEGs-Transcription Factors (TFs) interaction analysis, and DEGs-MicroRNAs (miRNAs) interaction analysis to identify putative discriminatory biomarkers between T2D and NDs. Furthermore, we also incorporated three gold benchmark verified datasets, OMIM (www.omim.org), OMIM Expanded, and dbGaP (www.ncbi.nlm.nih.gov/gap) to retrieve genes associated with known diseases and relevant disorders for validating the proof of principle of our network-based approach.
3. Results
3.1. Gene Expression Analysis
To identify and investigate the gene expression effects of T2D that may influence the progression of NDs, we analyzed the gene expression microarray data collected from the National Center for Biotechnology Information (NCBI). Based on significant p-values, we found 1320 DEGs for T2D with false discovery rate (FDR) of 0.01 and absolute logFC of 1 using R Bioconductor packages (Limma). Similarly, we identified the most significant DEGs for each ND after statistical analysis. We identified 1606 DEGs in AD, 2901 in ALS, 588 in CP, 1887 in ED, 1338 in HD, 7463 in MS and 1558 in PD which is shown in Table 1.
Table 1.
The differentially expressed gene (DEG) for all employed Datasets in the present study.
| Disease Name | GEO Platform | Tissues/Cells | GEO Accession | Raw Genes | Case Samples | Control Samples | UP Reg. Genes | Down Reg. Genes |
|---|---|---|---|---|---|---|---|---|
| Type 2 Diabetes (T2D) | Affymetrix Human Genome U133 Plus 2.0 Array | Liver | GSE23343 | 54613 | 10 | 7 | 622 | 698 |
| Alzheimer’s disease | Affymetrix Human Genome U133 Plus 2.0 Array | CA1 tissue | GSE28146 | 54675 | 22 | 8 | 847 | 759 |
| Amyotrophic lateral sclerosis | Affymetrix Human Full Length HuGeneFL Array | Spinal cord | GSE833 | 22277 | 7 | 5 | 735 | 2166 |
| Cerebral palsy | Affymetrix Human Genome U133 Plus 2.0 Array | Muscle | GSE31243 | 22277 | 20 | 20 | 243 | 345 |
| Epilepsy disease | Affymetrix Human Genome U133 Plus 2.0 Array | Peripheral Blood | GSE22779 | 54675 | 12 | 4 | 882 | 1007 |
| Huntington’s disease | Affymetrix Human Genome U133 Plus 2.0 Array | Whole Blood | GSE1751 | 22283 | 17 | 14 | 365 | 973 |
| Multiple sclerosis | Affymetrix Human Genome U133 Plus 2.0 Array | Brain | GSE38010 | 33398 | 5 | 2 | 3987 | 3476 |
| Parkinson’s disease | Affymetrix Human Genome U133A 2.0 Array | Brain | GSE19587 | 22277 | 12 | 10 | 1167 | 422 |
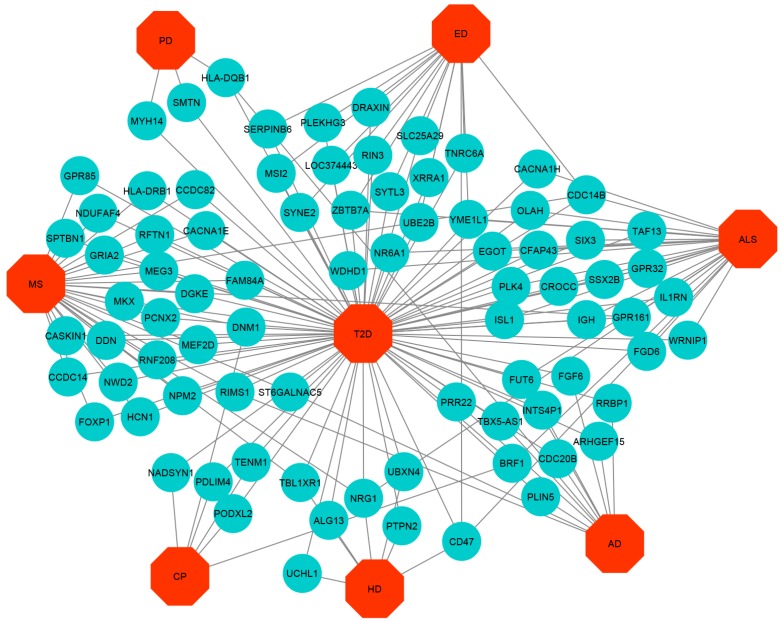
The cross-comparison analysis also identified common DEGs between T2D and each ND. We found that T2D shares 5, 5, 11, 15, 7, 35 and 21 significantly up-regulated genes whereas 12, 25, 6, 16, 7, 29 and 3 significant down-regulated genes for the AD, ALS, CP, ED, HD, MS, and PD respectively. To get statistically significant associations between T2D and the NDs, we built up- and down-regulated diseasome relationships network centered on the T2D and a link indicated between a disease and a gene when mutations in that gene are known to lead to the specific disease, as shown in Figure 2 and Figure 3 whereas two diseases are comorbid, if they share associated genes.
Figure 2.
Up-regulated Gene-Disease-network (GDN) between type 2 diabetes (T2D) and neurological diseases (NDs) comprising of commonly up-regulated genes node and different categories of diseases node represented by round shaped robin’s egg blue colour and octagon-shaped red colour.
Figure 3.
Down-regulated Gene-Disease-network (GDN) between T2D and NDs comprising of commonly down-regulated genes node and different categories of diseases node represented by round shaped robin’s egg blue colour and octagon-shaped red colour.
The most important up-regulated overlapping genes are as follows: (a) FLI1, PACSIN2, BICD1, TCP11L2, and ENTPD1 among T2D, ED, and MS, (b) PEG10 and EFCAB14 among T2D, PD, and MS, (c) ITGB8 among T2D, HD, PD, and MS, (d) FBLN1 among T2D, ALS, and MS, (e) IGFBP5 among T2D, CP, and MS, (f) SGCB among T2D, HD, and MS, (g) SLC25A30 among T2D, PD, and CP. The significant down-regulated overlapping genes are as follows: (a) ST6GALNAC5 and RIMS1 among T2D, AD, and MS, (b) ZBTB7A and YME1L1 among T2D, ALS, and ED, (c) FUT6 among T2D, AD, and ALS, (d) BRF1 among T2D, ALS, and CP, (e) CDC14B among T2D, ALS, ED, and MS, (f) CD47 among T2D, ALS, HD, and ED, (g) NRG1 among T2D, ALS, HD, and MS, (h) DNM1 among T2D, CP, and MS, (i) TLB1XR1 among T2D, HD, and MS, and (j) GPR161 among T2D, ALS, and MS.
3.2. Pathway and Functional Association Analysis
We performed pathways analysis to identify how complex diseases are interrelated with other diseases by the underlying molecular mechanisms. We performed gene set enrichment analysis to identify pathways using a bioinformatics resource: EnrichR [51] and considered 7 pathways databases to carry out tests using DEGs common between T2D and each ND. We also performed the regulatory analysis to get more insights into the molecular pathways involved in these comorbidities. We pinpointed overrepresented pathways among DEGs common to T2D and NDs and classified them into functional categories. Pathways deemed significantly enriched in the common DEG sets were reduced by manual curation to include only those pathways which have a p-value of below 0.05. We retrieved significant pathways by EnrichR which are significantly connected with DEGs of T2D and NDs as shown in Table 2.
Table 2.
Pathways common to T2D and the NDs revealed by the commonly expressed genes. These include significant pathways common to (a) T2D and AD (b) T2D and ALS (c) T2D and CP (d) T2D and ED (e) T2D and HD (f) T2D and MS and (g) T2D and PD.
| (a) Common significant pathway common between T2D and AD | ||
| Pathway Name | p -Value | Source |
| Neuroregulin receptor degredation protein-1 Controls ErbB3 receptor recycling | 3.78 | Biocarta |
| Cytokine-cytokine receptor interaction | 1.17 | KEGG |
| Toll-like receptor signaling pathway | 2.92 | KEGG |
| Glycosphingolipid biosynthesis | 3.36 | KEGG |
| IL1-mediated signaling events | 1.89 | NCI-Nature |
| TCR signaling in naive CD8+ T cells | 4.53 | NCI-Nature |
| Ubiquitin proteasome pathway | 3.09 | Panther |
| Ionotropic glutamate receptor pathway | 3.36 | Panther |
| Glutamate Neurotransmitter Release Cycle | 1.02 | Reactome |
| Signaling by Interleukins | 1.32 | Reactome |
| Cytokine Signaling in the Immune system | 1.83 | Reactome |
| Oxidative Stress-Induced Senescence | 2.07 | Reactome |
| Neurotransmitter Release Cycle | 4.23 | Reactome |
| Transmission across Chemical Synapses | 4.68 | Reactome |
| Apoptosis Modulation and Signaling | 2.97 | Wiki |
| (b) Common significant pathway common between T2D and ALS | ||
| Pathway Name | p -Value | Source |
| Neuroregulin receptor degredation protein-1 Controls ErbB3 receptor recycling | 1.48 | Biocarta |
| Cytokine-cytokine receptor interaction | 3.10 | KEGG |
| Glycosphingolipid biosynthesis | 1.05 | KEGG |
| Ubiquitin mediated proteolysis | 1.28 | KEGG |
| Glutamatergic synapse | 2.85 | KEGG |
| Immune System | 3.74 | Reactome |
| Innate Immune System | 7.98 | Reactome |
| Insulin receptor signaling cascade | 8.94 | Reactome |
| Cytokine Signaling in the Immune system | 1.05 | Reactome |
| Neuronal System | 1.17 | Reactome |
| Neurotransmitter Receptor Binding And Downstream Transmission in The Postsynaptic Cell | 1.47 | Reactome |
| Transmission across Chemical Synapses | 2.05 | Reactome |
| Adaptive Immune System | 2.30 | Reactome |
| NO/cGMP/PKG mediated Neuroprotection | 1.19 | Wiki |
| Toll-like Receptor Signaling | 3.90 | Wiki |
| (c) Common significant pathway common between T2D and CP | ||
| Pathway Name | p -Value | Source |
| Focal adhesion | 4.47 | KEGG |
| Dopaminergic synapse | 9.70 | KEGG |
| Cell adhesion molecules (CAMs) | 1.28 | KEGG |
| Rapid glucocorticoid signaling | 2.65 | NCI-Nature |
| Inflammation mediated by chemokine and cytokine signaling pathway | 4.19 | Panther |
| Adaptive Immune System | 7.50 | Reactome |
| Immune System | 1.47 | Reactome |
| Neurotransmitter Receptor Binding And Downstream Transmission In The Postsynaptic Cell | 1.21 | Reactome |
| Electric Transmission Across Gap Junctions | 1.66 | Reactome |
| Transmission across Electrical Synapses | 1.66 | Reactome |
| Neuronal System | 1.84 | Reactome |
| Neurofascin interactions | 2.32 | Reactome |
| Transmission across Chemical Synapses | 3.39 | Reactome |
| Inflammatory Response Pathway | 4.53 | Wiki |
| Insulin signaling in human adipocytes | 2.65 | Wiki |
| Toll-like Receptor Signaling Pathway | 4.68 | Wiki |
| (d) Common significant pathway common between T2D and ED | ||
| Pathway Name | p -Value | Source |
| Cell adhesion molecules (CAMs) | 1.14 | KEGG |
| Ubiquitin mediated proteolysis | 4.95 | KEGG |
| TCR signaling in naive CD8+ T cells | 4.03 | NCI-Nature |
| Rapid glucocorticoid signaling | 4.70 | NCI-Nature |
| Apoptosis signaling pathway | 2.35 | Panther |
| Ubiquitin proteasome pathway | 2.75 | Panther |
| Adaptive Immune System | 6.15 | Reactome |
| Oxidative Stress-Induced Senescence | 1.75 | Reactome |
| Immune System | 2.26 | Reactome |
| Deposition of new CENPA-containing nucleosomes at the centromere | 3.90 | Reactome |
| Spinal Cord Injury | 3.42 | Wiki |
| (e) Common significant pathway common between T2D and HD | ||
| Pathway Name | p -Value | Source |
| Neuroregulin receptor degredation protein-1 Controls ErbB3 receptor recycling | 2.59 | Biocarta |
| Glycosphingolipid biosynthesis | 1.54 | KEGG |
| Cell adhesion molecules (CAMs) | 2.32 | KEGG |
| Cytokine-cytokine receptor interaction | 3.53 | KEGG |
| Neurotrophic factor-mediated Trk receptor signaling | 2.72 | NCI-Nature |
| Ubiquitin proteasome pathway | 1.41 | Panther |
| Cholesterol biosynthesis | 4.53 | Panther |
| Immune System | 1.82 | Reactome |
| Cytokine Signaling in the Immune system | 1.54 | Reactome |
| Innate Immune System | 2.00 | Reactome |
| Adaptive Immune System | 4.10 | Reactome |
| Toll-Like Receptors Cascades | 2.12 | Reactome |
| NO/cGMP/PKG mediated Neuroprotection | 1.67 | Wiki |
| Apoptosis | 4.87 | Wiki |
| Tryptophan metabolism | 1.60 | Wiki |
| (f) Common significant pathway common between T2D and MS | ||
| Pathway Name | p -Value | Source |
| Neurotrophin signaling pathway | 1.17 | KEGG |
| Glycosphingolipid biosynthesis | 1.36 | KEGG |
| Adipocytokine signaling pathway | 1.39 | KEGG |
| Autophagy | 1.69 | KEGG |
| Glutamatergic synapse | 3.07 | KEGG |
| GABAergic synapse | 3.68 | KEGG |
| Neurotrophic factor-mediated Trk receptor signaling | 8.37 | NCI-Nature |
| Insulin/IGF pathway-protein kinase B signaling cascade | 5.05 | Panther |
| Apoptosis signaling pathway | 5.17 | Panther |
| Ubiquitin proteasome pathway | 1.16 | Panther |
| Ionotropic glutamate receptor pathway | 1.36 | Panther |
| Transmission across Chemical Synapses | 8.34 | Reactome |
| Neuronal System | 1.71 | Reactome |
| Neurotransmitter Receptor Binding And Downstream Transmission In The Postsynaptic Cell | 1.77 | Reactome |
| Insulin receptor signaling cascade | 2.65 | Reactome |
| Oxidative Stress-Induced Senescence | 1.13 | Reactome |
| Brain-Derived Neurotrophic Factor (BDNF) signaling pathway | 2.98 | Wiki |
| (g) Common significant pathway common between T2D and PD | ||
| Pathway Name | p -Value | Source |
| Neuroregulin receptor degredation protein-1 Controls ErbB3 receptor recycling | 4.04 | Biocarta |
| Toll-like receptor signaling pathway | 2.21 | KEGG |
| Cell adhesion molecules (CAMs) | 7.20 | KEGG |
| Allograft rejection | 1.70 | KEGG |
| Graft-versus-host disease | 1.96 | KEGG |
| Intestinal immune network for IgA production | 2.63 | KEGG |
| Innate Immune System | 1.07 | Reactome |
| Insulin receptor signaling cascade | 4.07 | Reactome |
| Immune System | 1.50 | Reactome |
| Chemokine receptors bind chemokines | 3.50 | Reactome |
| Adaptive Immune System | 4.73 | Reactome |
| Transmission across Electrical Synapses | 2.60 | Reactome |
Among the identified pathways, we found that cytokine-cytokine receptor interaction pathway associated with adaptive inflammatory host defenses, cell growth, differentiation, cell death [54]. Glycosphingolipid biosynthesis pathway is associated with abundant amphipathic lipids expression in the nervous system [54]; Ubiquitin proteasome pathway associated with immune response and inflammation, neural and muscular degeneration, morphogenesis of neural networks and response to stress and extracellular modulators [59]; Ionotropic glutamate receptor pathway associated with the mediatation of the majority of excitatory synaptic transmission throughout the central nervous system and synaptic plasticity [59]; Glutamate neurotransmitter release cycle maintains the neurotransmitter glutamate in the central nervous system [55]; Transmission across chemical synapses pathway associated with the communication between neurons, muscle or gland cells [55]; Glutamatergic synapse pathway associated with the regulation of several neuronal functions, such as neuronal migration, excitability, plasticity, long-term potentiation (LTP) and long-term depression (LTD) [57]; Neuronal system pathway comprised of at least 100 billion neurons are associated with the communication among astronomical number of elements with functional connection between neurons [55]; Cell adhesion molecules (CAMs) pathway associated with a vital role in the development and maintenance of the nervous system [54]; Electric transmission across gap junctions pathway associated with the function of communicating neurons in the nervous systems [55]; Transmission across electrical synapses pathway associated with the mechanical conductive link between two neighboring neurons [55]; Spinal cord injury pathway associated with the loss of muscle function, sensation, or autonomic function in the parts of the body [57]; Neurotrophic factor-mediated Trk receptor signaling pathway associated with neuronal differentiation, survival and growth [56]; Neurotrophin signaling pathway associated with differentiation and survival of neural cells [54]; Adipocytokine signaling pathway associated with the process of inflammation, coagulation, fibrinolysis, insulin resistance, diabetes and atherosclerosis [54]; Brain-derived neurotrophic factor (BDNF) signaling pathway associated with growth, differentiation, plasticity, and survival of neurons. BDNF is also implicated in various neuronal disorders such as Alzheimer’s disease, Huntington’s disease [57].
To obtain further insights into the molecular roles and biological significance, enriched common DEGs sets were processed by GO methods using Enrichr, which identifies related biological processes (BP) in order to group them in functional categories. The list of processes and terms was then curated to include those terms with a p-value below 0.05. The cell processes thus identified are summarized in Table 3.
Table 3.
Gene ontology identification of biological processes common to (a) T2D and AD (b) T2D and ALS (c) T2D and CP (d) T2D and ED (e) T2D and HD (f) T2D and MS and (g) T2D and PD.
| (a) Common significant GOs of T2D and AD | ||
| GO ID | Pathway | p -Value |
| GO:0032000 | positive regulation of fatty acid beta-oxidation | 5.99 |
| GO:0016064 | immunoglobulin mediated immune response | 6.73 |
| GO:2000269 | regulation of fibroblast apoptotic process | 6.73 |
| GO:0031998 | regulation of fatty acid beta-oxidation | 8.22 |
| GO:0051588 | regulation of neurotransmitter transport | 1.05 |
| GO:0007498 | mesoderm development | 1.64 |
| GO:0051961 | negative regulation of nervous system development | 1.71 |
| GO:0046928 | regulation of neurotransmitter secretion | 2.08 |
| (b) Common significant GOs of T2D and ALS | ||
| GO ID | Pathway | p -Value |
| GO:0006352 | DNA-templated transcription, initiation | 2.46 |
| GO:0050870 | positive regulation of T cell activation | 4.48 |
| GO:0048935 | peripheral nervous system neuron development | 1.01 |
| GO:0021522 | spinal cord motor neuron differentiation | 1.01 |
| GO:0021559 | trigeminal nerve development | 1.01 |
| GO:2000145 | regulation of cell motility | 1.10 |
| GO:0048665 | neuron fate specification | 1.15 |
| GO:1902692 | regulation of neuroblast proliferation | 1.58 |
| GO:0048663 | neuron fate commitment | 2.01 |
| GO:2000177 | regulation of neural precursor cell proliferation | 2.15 |
| GO:0014033 | neural crest cell differentiation | 2.30 |
| GO:0045597 | positive regulation of cell differentiation | 3.23 |
| GO:0051961 | negative regulation of nervous system development | 3.28 |
| GO:0019228 | neuronal action potential | 3.42 |
| GO:0021953 | central nervous system neuron differentiation | 4.68 |
| GO:0050768 | negative regulation of neurogenesis | 4.82 |
| (c) Common significant GOs of T2D and CP | ||
| GO ID | Pathway | p -Value |
| GO:0071363 | cellular response to growth factor stimulus | 5.47 |
| GO:0048681 | negative regulation of axon regeneration | 6.38 |
| GO:0070571 | negative regulation of neuron projection regeneration | 7.18 |
| GO:0099590 | neurotransmitter receptor internalization | 8.77 |
| GO:0048679 | regulation of axon regeneration | 8.77 |
| GO:0021952 | central nervous system projection neuron axonogenesis | 8.77 |
| GO:0008045 | motor neuron axon guidance | 8.77 |
| GO:0106030 | neuron projection fasciculation | 8.77 |
| GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway | 1.67 |
| GO:0071875 | adrenergic receptor signaling pathway | 1.67 |
| GO:1990090 | cellular response to nerve growth factor stimulus | 1.82 |
| GO:0010977 | negative regulation of neuron projection development | 4.08 |
| GO:0016192 | vesicle-mediated transport | 4.18 |
| GO:0001934 | positive regulation of protein phosphorylation | 4.22 |
| (d) Common significant GOs of T2D and ED | ||
| GO ID | Pathway | p -Value |
| GO:0006897 | endocytosis | 6.45 |
| GO:0007599 | hemostasis | 1.15 |
| GO:0090286 | cytoskeletal anchoring at the nuclear membrane | 1.30 |
| GO:0034214 | protein hexamerization | 1.44 |
| GO:0034063 | stress granule assembly | 1.73 |
| GO:0097205 | renal filtration | 1.73 |
| GO:0035278 | miRNA mediated inhibition of translation | 1.73 |
| GO:0071470 | cellular response to osmotic stress | 2.15 |
| GO:0034656 | nucleobase-containing small molecule catabolic process | 2.30 |
| GO:0021953 | central nervous system neuron differentiation | 4.68 |
| (e) Common significant GOs of T2D and HD | ||
| GO ID | Pathway | p -Value |
| GO:0042127 | regulation of cell proliferation | 6.92 |
| GO:0022409 | positive regulation of cell-cell adhesion | 2.16 |
| GO:0048665 | neuron fate specification | 5.19 |
| GO:0048663 | neuron fate commitment | 9.06 |
| GO:0014033 | neural crest cell differentiation | 1.04 |
| GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 1.49 |
| GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 2.65 |
| GO:0007399 | nervous system development | 3.43 |
| (f) Common significant GOs of T2D and MS | ||
| GO ID | Pathway | p -Value |
| GO:0070229 | negative regulation of lymphocyte apoptotic process | 4.32 |
| GO:0099590 | neurotransmitter receptor internalization | 5.27 |
| GO:0051588 | regulation of neurotransmitter transport | 8.67 |
| GO:0050804 | modulation of chemical synaptic transmission | 2.29 |
| GO:0046928 | regulation of neurotransmitter secretion | 3.50 |
| GO:2000146 | negative regulation of cell motility | 3.66 |
| GO:0090181 | regulation of cholesterol metabolic process | 7.75 |
| GO:0072657 | protein localization to membrane | 1.43 |
| GO:0007005 | mitochondrion organization | 1.60 |
| GO:0010595 | positive regulation of endothelial cell migration | 2.11 |
| GO:0010646 | regulation of cell communication | 2.17 |
| GO:0050658 | RNA transport | 2.70 |
| GO:0010628 | positive regulation of gene expression | 3.43 |
| GO:2000145 | regulation of cell motility | 4.71 |
| GO:0014033 | neural crest cell differentiation | 4.92 |
| (g) Common significant GOs of T2D and PD | ||
| GO ID | Pathway | p -Value |
| GO:0070486 | leukocyte aggregation | 9.16 |
| GO:0070584 | mitochondrion morphogenesis | 1.14 |
| GO:0090128 | regulation of synapse maturation | 1.26 |
| GO:0045624 | positive regulation of T-helper cell differentiation | 1.37 |
| GO:0048854 | brain morphogenesis | 1.37 |
| GO:0019228 | neuronal action potential | 2.73 |
| GO:0097061 | dendritic spine organization | 2.84 |
| GO:0030199 | collagen fibril organization | 3.40 |
| GO:0061448 | connective tissue development | 3.51 |
| GO:0048813 | dendrite morphogenesis | 3.73 |
| GO:0099601 | regulation of neurotransmitter receptor activity | 3.84 |
3.3. Protein-Protein Interactions (PPIs) Analysis
Using our enriched common disease genesets, we constructed putative PPI networks with web-based visualization resource STRING via Network Analyst using the confidence score 900 by the distinct 159 DEGs, as shown in Figure 4. The PPIs make up the so-called interactomics of the organism where anomalous PPIs cause multiple diseases. Two diseases are known to be related where one or more commonly associated protein subnetworks are shared. Using topological parameters, for example, the degree greater than 15°, highly interacting proteins were identified from PPI analysis.
Figure 4.
The protein-protein interactions (PPIs) network is built with the significantly dysregulated genes common to type 2 diabetes (T2D) and neurological diseases (NDs).
The simplified PPI networks were generated with the Cyto-Hubba plugin [69] to identify the most significant hub proteins as shown in Table 4 and the topological parameters were determined by Network Analyzer in Cytoscape [47] as shown in Figure 5.
Table 4.
Summary of hub proteins identified by protein-protein interaction analysis encoded by DEGs that are common to type 2 diabetes (T2D) and neurological diseases (NDs).
| Protein Symbol | Degree | Description | Feature |
|---|---|---|---|
| DNM1 | 65 | Dynamin 1 | GTP binding |
| DNM2 | 84 | Dynamin 2 | GTP binding and GTPase activity |
| MYH14 | 82 | Myosin Heavy Chain 14 | Calmodulin binding and motor activity |
| PACSIN2 | 60 | Protein Kinase C And Casein Kinase Substrate In Neurons | Identical protein binding and lipid binding |
| TFRC | 10 | Transferrin Receptor | Double-stranded RNA binding |
| PDE4D | 39 | Phosphodiesterase 4D | Enzyme binding and protein domain specific binding |
| ENTPD1 | 32 | Ectonucleoside Triphosphate Diphosphohydrolase 1 | Hydrolase activity and nucleoside-diphosphatase activity |
| PLK4 | 45 | Polo Like Kinase 4 | Identical protein binding and protein kinase activity |
| CDC20B | 20 | Cell Division Cycle 20B | Pathways related to DNA damage response |
| CDC14A | 36 | Cell Division Cycle 14A | Phosphatase activity and phosphoprotein phosphatase activity |
Figure 5.
The simplified PPIs network is built with the significantly dysregulated genes common to type 2 diabetes (T2D) and neurological diseases (NDs) to identify the 10 most significant hub proteins marked as red, orange and yellow colour.
This data provides evidence that PPI subnetwork exists in our enriched genesets and confirms the inclusion of relevant functional pathways. These identified hub proteins could be useful for therapeutic targets although further characterization is needed for their roles. The summary of hub protein is shown in Table 4.
3.4. Identification of Transcriptional and Post-Transcriptional Regulators of the Differentially Expressed Genes
TFs are proteins that regulate transcriptional and gene expression in all living organisms. TFs play a vital role in all cellular processes [70]. miRNAs are short RNA species involved in the post-transcriptional regulation of gene expression. The miRNAs are important biological regulators, for instance, neuronal differentiation, neurogenesis, and synaptic plasticity and they play vital roles in neurodegenerative diseases [71].
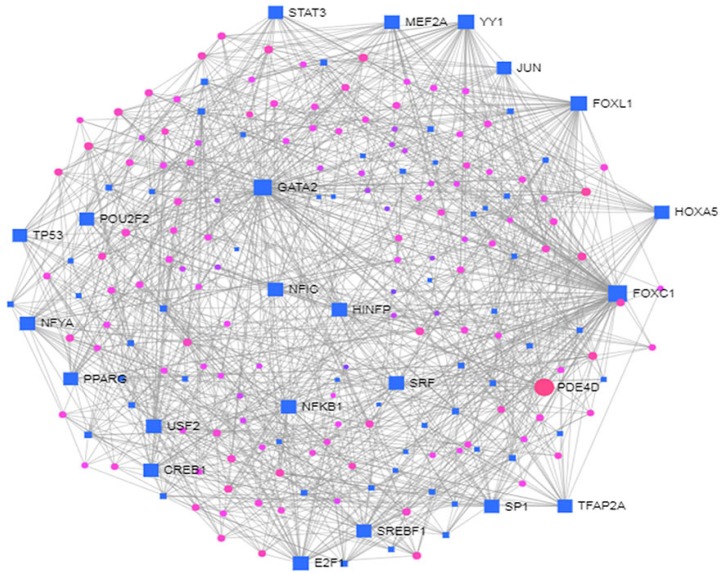
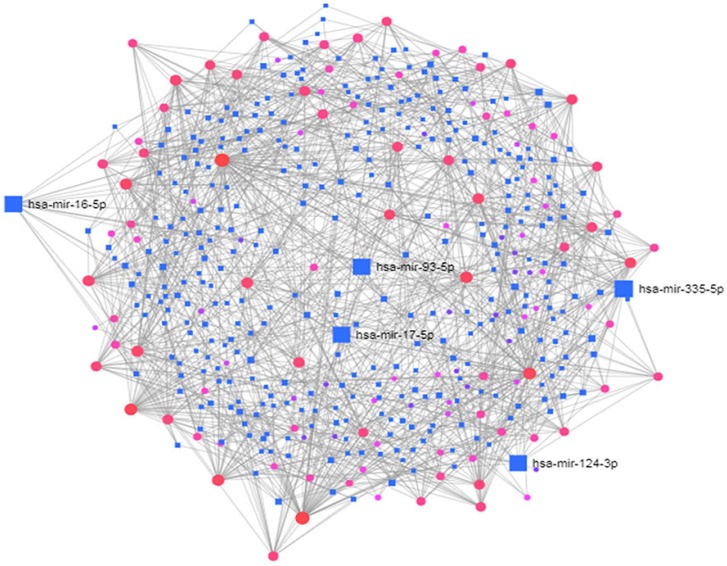
To identify the transcriptional and/or -post-transcriptional regulators of the DEGs, we performed the interaction of the DEGs-TFs analysis as shown in Figure 6 and DEGs-miRNAs interaction analysis as shown in Figure 7.
Figure 6.
The DEGs-TF interactions regulating the differentially expressed genes common to type 2 diabetes (T2D) and neurological diseases (NDs).
Figure 7.
The DEGs-miRNAs interactions regulating the differentially expressed genes common between type 2 diabetes (T2D) and neurological diseases (NDs).
The biomolecules (i.e., TFs and miRNAs) are summarized in Table 5. The FOXC1, GATA2, FOXL1, YY1, E2F1, NFIC, NFYA, USF2, HINFP, MEF2A, SRF, NFKB1, USF2, HINFP, MEF2A, SRF, NFKB1, PDE4D, CREB1, SP1, HOXA5, SREBF1, TFAP2A, STAT3, POU2F2, TP53, PPARG, JUN were identified as the key regulators of the identified DEGs.
Table 5.
Summary of transcriptional and/or post-transcriptional post regulatory biomolecules of differentially expressed genes overlapped between type 2 diabetes (T2D) and neurological diseases (NDs) that includes (a) Regulatory Transcription Factors and (b) Regulatory microRNAs.
| (a) Regulatory Transcription Factors | ||
| Symbol | Description | Feature |
| FOXC1 | Forkhead Box C1 | NA-binding transcription factor activity and transcription factor binding |
| GATA2 | GATA binding protein 2 | DNA-binding transcription factor activity and chromatin binding |
| FOXL1 | Forkhead Box L1 | NA-binding transcription factor activity and DNA-binding transcription factor activity, RNA polymerase II-specific |
| YY1 | YY1 Transcription Factor | NA-binding transcription factor activity and transcription coactivator activity |
| E2F1 | E2F transcription factor 1 | DNA-binding transcription factor activity and transcription factor binding. |
| NFIC | Nuclear Factor I C | DNA-binding transcription factor activity and proximal promoter DNA-binding transcription activator activity, RNA polymerase II-specific |
| NFYA | Nuclear Transcription Factor Y Subunit Alpha | DNA-binding transcription factor activity and transcription regulatory region DNA binding |
| USF2 | Upstream Transcription Factor 2, C-Fos Interacting | DNA-binding transcription factor activity and sequence-specific DNA binding |
| HINFP | Histone H4 Transcription Factor | DNA-binding transcription factor activity and enzyme binding |
| MEF2A | Myocyte Enhancer Factor 2A | DNA-binding transcription factor activity and protein heterodimerization activity |
| SRF | Serum Response Factor | DNA-binding transcription factor activity and sequence-specific DNA binding |
| NFKB1 | Nuclear Factor Kappa B Subunit 1 | DNA-binding transcription factor activity and sequence-specific DNA binding |
| PDE4D | Phosphodiesterase 4D | enzyme binding and protein domain specific binding |
| CREB1 | CAMP Responsive Element Binding Protein 1 | DNA-binding transcription factor activity and enzyme binding |
| SP1 | Sp1 Transcription Factor | DNA-binding transcription factor activity and sequence-specific DNA binding |
| HOXA5 | Homeobox A5 | DNA-binding transcription factor activity and RNA polymerase II proximal promoter sequence-specific DNA binding |
| SREBF1 | Sterol Regulatory Element Binding Transcription Factor 1 | DNA-binding transcription factor activity and chromatin binding |
| TFAP2A | Transcription Factor AP-2 Alpha | DNA-binding transcription factor activity and sequence-specific DNA binding |
| STAT3 | Signal Transducer And Activator Of Transcription 3 | DNA-binding transcription factor activity and sequence-specific DNA binding |
| POU2F2 | POU Class 2 Homeobox 2 | DNA-binding transcription factor activity and protein domain specific binding |
| TP53 | Tumor Protein P53 | NA-binding transcription factor activity and protein heterodimerization activity |
| PPARG | Peroxisome Proliferator Activated Receptor Gamma | DNA-binding transcription factor activity and chromatin binding |
| JUN | Jun Proto-Oncogene, AP-1 Transcription Factor Subunit | sequence-specific DNA binding |
| (b) Regulatory microRNAs | ||
| Symbol | Description | Feature |
| mir-335-5p | MicroRNA 335 | Afflicted with Alzheimer’s disease |
| mir-16-5p | MicroRNA 16 | Afflicted with apoptosis of neural cells |
| mir-93-5p | MicroRNA 93 | Involved in DNA damage pathways |
| mir-17-5p | MicroRNA 17 | Act as oncogene or tumour suppressor gene depending on the cellular context |
| mir-124-3p | MicroRNA 124 | Abundant in the brain and involved in neurodegenerative disease |
The miRNAs (mir-335-5p, mir-16-5p, mir-93-5p, mir-17-5p, mir-124-3p) were identified to provide an in-depth understanding of the DEGs at post-transcriptional regulators. The summary of transcriptional and/or post-transcriptional regulatory biomolecules of differentially expressed genes that are common to T2D and NDs is shown in Table 5.
3.5. Validating Potential Targets Using Gold Benchmark Databases and Literatures
First of all, to validate our identified potential targets, we used OMIM, OMIM Expanded, and dbGaP datasets; these datasets collect curated and validated genes that indicate disease association data from the literature. In the validation, we presented a combined relation of OMIM, OMIM Expanded, and dbGaP databases. For evaluating the validity of our work, we provided statistically significant DEGs (genes common to T2D and neurological diseases (NDs)) to the online tool EnrichR [51] and collected enriched genes and their corresponding neurological disease names from OMIM, OMIM Expanded, and dbGaP databases. To find significant NDs, manual curation is applied considering a p-value of 0.05. Then, several diseases such as cancer, infectious diseases are removed from this list because they are not of interest in this study.
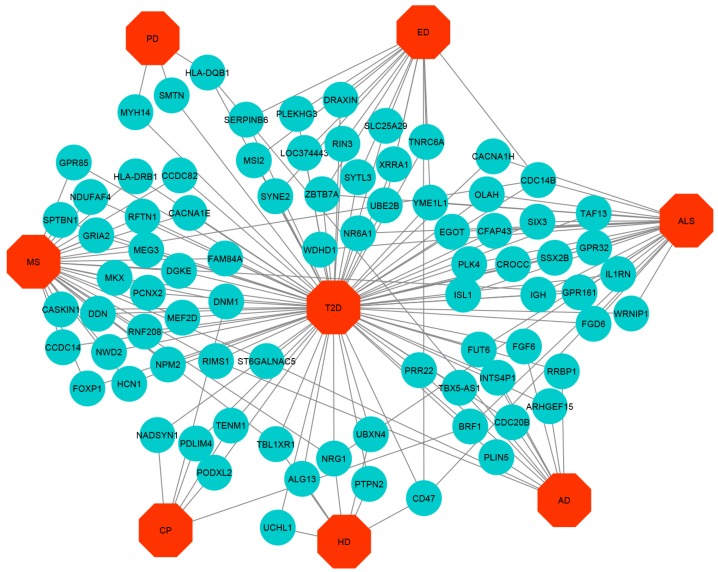
We also validated our identified potential targets by checking the biomedical literature to find genes clinically used as biomarkers for any of the NDs. We found that Van Cauwenberghe et al. [72] identified the MS4A2 gene associated with AD and Munshi et al. [73] identified the CR1 gene associated with AD. Eykens et al. [74] identified the APOE gene associated with ALS. Fahey et al. [75] identified the TENM1 gene associated with CP. UBE3A and CHD2 genes are associated with ED [76,77]. Arning et al. identified the UCHL1 [78] gene associated with HD. Baranzini et al. [79] identified the HLA-DRB1 gene to be associated with MS. Redenšek et al. identified [80] the HLA-DQB1 gene as associated with PD. This indicates that our analyses of significant genes in NDs match with existing records. We then constructed a Gene-Disease Network (GDN) based on genes and their associated neurological diseases from gold benchmark databases and literature using Cytoscape. This network showed gene-disease associations whereby if a gene mutation is known to lead to a specific disease, a link is indicated between disease and gene; this is shown in Figure 8.
Figure 8.
Disease network of type 2 diabetes (T2D) with neurological diseases (NDs) where robin’s egg blue colour round shaped nodes represent genes and octagon shaped red colour nodes represent T2D and NDs.
4. Discussion
T2D and NDs are complex diseases but we have attempted here to take advantage of this complexity by looking at pathway and looking at interactions between type 2 diabetes (T2D) and neurological diseases, due to their great clinical importance. T2D is known to affect neurological diseases but how it does this is generally unclear (though some vascular based mechanisms are usually considered) and it is very hard to study by hypothesis-driven biochical or endocrinological research. This is why we employed well-established bioinformatics methods and analytical approaches that examine functional disease overlaps in genes and pathways, and provide an important but agnostic tool to identify new factors that play a part in these comorbidity interactions and which, by implication, may be important pathogenic mechanisms for these and other related diseases. We studied the microarray gene expression datasets from publicly available repositories employing a network-based bioinformatics pipeline. We identified DEGs common to T2D and NDs and constructed diseasome networks to provide insights into the interactions of these comorbidities using the diesease-common DEGs. These DEGs enabled identification of associated dysregulated molecular pathways and related GO terms. One particular technical point is that a large number of pathways and GO categories were reduced by manual curation after filtering using a p-value threshold of 0.05. We identified different pathways by investigating cell proteins (i.e., gene products) and their interactions considering seven pathway databases.
In addition to the pathways and GO terms we investigated interactors of the products of our DEGs of interest using protein-protein interaction (PPI) analysis and hub protein identification as well as DEGs-TFs, and DEGs-miRNAs interaction that has not been previously studied for these diseases. These studies’ provide information about the molecules that could be the key drivers of pathogenesis for these comorbidities. The STRING database contains known protein-protein interactions which we used to identify the PPI for products of our genes of interest. For our purpose, we considered only experimentally verified PPI data, not predicted PPIs. We reconstructed the PPI based on the identified DEGs common to T2D and NDs and identified the central hub proteins using topological parameters. Among the hub proteins we identified, dynamin (DNM1) has been implicated in central nervous systems [81] and DNM2 is involved in Charcot-Marie-Tooth neuropathy [82]. However, a role for MYH14 in diabetes or neurodegenerative diseases is not reported. PACSIN2 expression has been found to be upregulated in diabetic kidney disease [83]. Borie et al studied the polymorphism of the TFRC gene involved in Parkinson’s disease [84]. Rahman et al. identified PDE4D commonly expressed in blood cell and brain tissues of AD [85]. A mutation of ENTPD1 has been identified in Spastic paraplegia type 64 in individuals diagnosed with suspected neurodegenerative disease patients [86]. To date, no role for PLK4, CDC20B, and CDC14A in NDs has been reported.
Transcription factors (TFs) are critical determinants of transcription of their various target genes, so their levels can identify potential biomarkers for neurodegenerative diseases. In this study, we identified relevant TFs as the regulator of the DEGs through TF-mRNA interaction networks that are relevant to the pathogenesis of T2D and NDs. TF-association has also been used by Rahman et al. in a network-based method to profile gene expression of DEGs associated with AD; they identified a number of AD-associated TFs, including JUN, YY1, E2F1, FOXC1, GATA2, SRF, USF2, PPARG, FOXL1, and NFIC [87], consistent with the present study. In contrast to TFs, microRNAs (∼ act post-transcriptionally to regulate expression. These are single nucleotide RNA which bind target mRNA, leading to the target cleavage and reduced expression. These miRNAs have many advantages of non-invasive biomarkers and can be detected in body fluids such as urine, saliva and makes them potentially attractive as biomarkers. Indeed, there are miRNAs that show good potential as biomarkers for neurodegenerative diseases [88]. Thus, we studied DEGs-miRNA interaction networks to identify relevant miRNAs as potential targets for NDs. Among the identified miRNAs, the miRNA-335 was particularly associated with AD [71]. In addition, miRNA-16 was reported to be involved in apoptosis in neural cells [89]. Furthermore, the down-expression of this miRNA is involved in the accumulation of amyloid protein precursor (APP) protein in AD [90]. The miRNA-124 is found abundantly in neural cells and as known involvement in NDs. The reduced expression is miRNA associated with AD, PD, and HD [88]. There is no evidence for ND association with miR-17-5p although it has pathogenic actions both enhancing and suppressing tumour development depending on the cellular contexts [91].
The above indicates that our approach has the potential to reveal some of the important mechanisms that underlie disease pathogenesis and provide novel hypotheses of disease mechanisms and may identify new biomarkers. Such genetic data analyses will be a key element in the development of predictive medicine and elucidating the underlying mechanisms that connect T2D and NDs and may indicate possible new drug targets. Nevertheless, our data show some limitations. It should be noted that no clinical confirmation of the roles of proteins generated from our identified genes of interest. Furthermore, the low number of samples for some diseases analyzed which may not fully sample the disease-associated genes that we used to determine the common DEGs. Thus, further experimentation is needed to properly evaluate the biological significance of the identified potential targets candidates in this study.
5. Conclusions
The present study analyzed transcriptomics datasets of the T2D and neurodegenerative diseases employing a multi-omics approach to decode the overlapped genes that were expressed between T2D and NDs. The gene set enrichment analysis revealed significantly enriched dysregulated pathways. Integration of the overlapped DEGs with different biomolecular interaction networks yielded 10 hub proteins form protein-protein interaction, regulatory TFs from DEG-TF interactions analysis, and miRNAs from DEGs-miRNAs interactions analysis. All of these hub genes and pathways are novel, that is, they have not previously been shown to be important in these diseases or in the disease interactions and most of the TFs and miRNAs identified are also novel. In this way, the present study presented molecular signatures at proteins level (i.e., hub proteins and TFs), and RNA levels (i.e., mRNAs, miRNAs), pathway and GO level but further studies are needed to establish them as biomarkers. These results indicate differentially expressed genes of T2D that may be key to the progression of NDs and may give new insights into these diseases. It also points the way to identifying mechanistic links between the T2D and various ND and that explains why their association with T2D. This study also suggests that T2D shares several common multifactorial degenerative biological processes that contribute to neuronal death, which may, in turn, lead to functional impairment. Additionally, we believe that our high-throughput transcript analysis of tissues using rigorous agnostic approaches would allow the discovery of disease-modifying therapeutic targets. Treatments aimed at attenuating the identified dysregulated pathways have the potential to ameliorate neurological dysfunctions in the T2D patient.
Acknowledgments
Thanks are due to Dou Hao, Institute of Automation, Chinese Academy of Sciences, Beijing, China who offered advice for the improvement of this work. Thanks to CAS-TWAS Presidents Fellowship for supporting Md Habibur Rahman (First Author) as a doctoral student (Fellowship No. 2016CTF014).
Author Contributions
The author contribution is as follows: conceptualization, M.H.R. and M.A.M.; methodology, M.H.R.; software, M.H.R.; validation, M.H.R., M.R.R.; formal analysis, M.H.R.; investigation, M.H.R.; resources, M.H.R.; data curation, C.C.; writing—original draft preparation, M.H.R.; writing—review and editing, S.U., X.H., J.M.W.Q., S.P.; visualization, M.R.R.; supervision, S.P.; project administration, M.A.M.; funding acquisition, X.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China(Grant no 61571438).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.DeFronzo R.A., Ferrannini E., Groop L., Henry R.R., Herman W.H., Holst J.J., Hu F.B., Kahn C.R., Raz I., Shulman G.I., et al. Type 2 diabetes mellitus. Nat. Rev. Dis. Primers. 2008;1:15019. doi: 10.1038/nrdp.2015.19. [DOI] [PubMed] [Google Scholar]
- 2.Mallorquí-Bagué N., Lozano-Madrid M., Toledo E., Corella D., Salas-Salvadó J., Cuenca-Royo A., Vioque J., Romaguera D., Martínez J.A., Wärnberg J., et al. Type 2 diabetes and cognitive impairment in an older population with overweight or obesity and metabolic syndrome: Baseline cross-sectional analysis of the predimed-plus study. Sci. Rep. 2018;8:16128. doi: 10.1038/s41598-018-33843-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu L., Huang X., Ma J., Huang J., Fan Y., Li H., Qiu J., Zhang H., Huang W. Value of three-dimensional strain parameters for predicting left ventricular remodeling after ST-elevation myocardial infarction. Int. J. Cardiovasc. Imaging. 2017;33:663–673. doi: 10.1007/s10554-016-1053-3. [DOI] [PubMed] [Google Scholar]
- 4.American Diabetes Association Diagnosis and classification of diabetes mellitus. Diabetes Care. 2004;27:S5eS10. doi: 10.2337/diacare.27.2007.s5. [DOI] [PubMed] [Google Scholar]
- 5.Xu L., Zhao H., Qiu J., Zhu W., Lei H., Cai Z., Lin W.H., Huang W., Zhang H., Zhang Y.T. The different effects of BMI and WC on organ damage in patients from a cardiac rehabilitation program after acute coronary syndrome. BioMed Res. Int. 2015;2015:942695. doi: 10.1155/2015/942695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mota M., Banini B.A., Cazanave S.C., Sanyal A.J. Molecular mechanisms of lipotoxicity and glucotoxicity in nonalcoholic fatty liver disease. Metabolism. 2016;65:1049–1061. doi: 10.1016/j.metabol.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dolan C., Glynn R., Griffin S., Conroy C., Loftus C., Wiehe P.C., Healy M.L., Lawlor B. Brain complications of diabetes mellitus: A cross-sectional study of awareness among individuals with diabetes and the general population in Ireland. Diabet. Med. 2018;35:871–879. doi: 10.1111/dme.13639. [DOI] [PubMed] [Google Scholar]
- 8.Mushtaq G., Khan J.A., Kumosani T.A., Kamal M.A. Alzheimer’s disease and type 2 diabetes via chronic inflammatory mechanisms. Saudi J. Biol. Sci. 2015;22:4–13. doi: 10.1016/j.sjbs.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verdile G., Fuller S.J., Martins R.N. The role of type 2 diabetes in neurodegeneration. Neurobiol. Dis. 2015;84:22–38. doi: 10.1016/j.nbd.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 10.Bharadwaj P., Wijesekara N., Liyanapathirana M., Newsholme P., Ittner L., Fraser P., Verdile G. The link between type 2 diabetes and neurodegeneration: Roles for amyloid-β, amylin, and tau proteins. J. Alzheimer’s Dis. 2017;59:421–432. doi: 10.3233/JAD-161192. [DOI] [PubMed] [Google Scholar]
- 11.Porte D., Jr., Baskin D.G., Schwartz M.W. Insulin signaling in the central nervous system: A critical role in metabolic homeostasis and disease from C. elegans to humans. Diabetes. 2005;54:1264–1276. doi: 10.2337/diabetes.54.5.1264. [DOI] [PubMed] [Google Scholar]
- 12.Morsi M., Kobeissy F., Magdeldin S., Maher A., Aboelmagd O., Johar D., Bernstein L. A shared comparison of diabetes mellitus and neurodegenerative disorders. J. Cell. Biochem. 2019;120:4318–14325. doi: 10.1002/jcb.28094. [DOI] [PubMed] [Google Scholar]
- 13.Chatterjee S., Mudher A. Alzheimer’s disease and type 2 diabetes: A critical assessment of the shared pathological traits. Front. Neurosci. 2018;12:383. doi: 10.3389/fnins.2018.00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez-Valbuena I., Valenti-Azcarate R., Amat-Villegas I., Riverol M., Marcilla I., de Andrea C.E., Sánchez-Arias J.A., del Mar Carmona-Abellan M., Marti G., Erro M.E., et al. Amylin as a potential link between type 2 diabetes and alzheimer disease. Ann. Neurol. 2019;86:539–551. doi: 10.1002/ana.25570. [DOI] [PubMed] [Google Scholar]
- 15.D’Ovidio F., d’Errico A., Carnà P., Calvo A., Costa G., Chiò A. The role of pre-morbid diabetes on developing amyotrophic lateral sclerosis. Eur. J. Neurol. 2018;25:164–170. doi: 10.1111/ene.13465. [DOI] [PubMed] [Google Scholar]
- 16.Crisham Janik M.D., Newman T.B., Cheng Y.W., Xing G., Gilbert W.M., Wu Y.W. Maternal diagnosis of obesity and risk of cerebral palsy in the child. J. Pediatr. 2013;163:1307–1312. doi: 10.1016/j.jpeds.2013.06.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu C.L., Chang Y.H., Sun Y., Li C.Y. A population-based study of epilepsy incidence in association with type 2 diabetes and severe hypoglycaemia. Diabetes Res. Clin. Pract. 2018;140:97–106. doi: 10.1016/j.diabres.2018.03.020. [DOI] [PubMed] [Google Scholar]
- 18.Montojo M.T., Aganzo M., González N. Huntington’s disease and diabetes: Chronological sequence of its association. J. Huntington’s Dis. 2017;6:179–188. doi: 10.3233/JHD-170253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruiz-Argüelles A., Méndez-Huerta M.A., Lozano C.D., Ruiz-Argüelles G.J. Metabolomic profile of insulin resistance in patients with multiple sclerosis is associated to the severity of the disease. Mult. Scler. Relat. Disord. 2018;25:316–321. doi: 10.1016/j.msard.2018.08.014. [DOI] [PubMed] [Google Scholar]
- 20.Shaw K. Type 2 diabetes and Parkinson’s disease. Pract. Diabetes. 2019;36:115–116. doi: 10.1002/pdi.2227. [DOI] [Google Scholar]
- 21.Schmitz T.W., Nathan Spreng R., Alzheimer’s Disease Neuroimaging Initiative Basal forebrain degeneration precedes and predicts the cortical spread of Alzheimer’s pathology. Nat. Commun. 2016;7:13249. doi: 10.1038/ncomms13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livingston G., Sommerlad A., Orgeta V., Costafreda S.G., Huntley J., Ames D., Ballard C., Banerjee S., Burns A., Cohen-Mansfield J., et al. Dementia prevention, intervention, and care. Lancet. 2017;390:2673–2734. doi: 10.1016/S0140-6736(17)31363-6. [DOI] [PubMed] [Google Scholar]
- 23.Al-Chalabi A., Hardiman O. The epidemiology of ALS: A conspiracy of genes, environment and time. Nat. Rev. Neurol. 2013;9:617–628. doi: 10.1038/nrneurol.2013.203. [DOI] [PubMed] [Google Scholar]
- 24.Dupuis L., Pradat P.F., Ludolph A.C., Loeffler J.P. Energy metabolism in amyotrophic lateral sclerosis. Lancet Neurol. 2011;10:75–82. doi: 10.1016/S1474-4422(10)70224-6. [DOI] [PubMed] [Google Scholar]
- 25.Desport J.C., Torny F., Lacoste M., Preux P.M., Couratier P. Hypermetabolism in ALS: Correlations with clinical and paraclinical parameters. Neurodegener. Dis. 2005;2:202–207. doi: 10.1159/000089626. [DOI] [PubMed] [Google Scholar]
- 26.Kioumourtzoglou M.A., Rotem R.S., Seals R.M., Gredal O., Hansen J., Weisskopf M.G. Diabetes mellitus, obesity, and diagnosis of amyotrophic lateral sclerosis: A population-based study. JAMA Neurol. 2015;72:905–911. doi: 10.1001/jamaneurol.2015.0910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cerebral Palsy Guidance Cerebral Palsy and Diabetes. [(accessed on 11 April 2019)];2019 Available online: https://www.cerebralpalsyguidance.com/cerebral-palsy/associated-disorders/diabetes/
- 28.Schendel D.E., Schuchat A., Thorsen P. Public health issues related to infection in pregnancy and cerebral palsy. Ment. Retard. Dev. Disabil. Res. Rev. 2002;8:39–45. doi: 10.1002/mrdd.10011. [DOI] [PubMed] [Google Scholar]
- 29.Marcovecchio M.L., Petrosino M.I., Chiarelli F. Diabetes and epilepsy in children and adolescents. Curr. Diabetes Rep. 2015;15:21. doi: 10.1007/s11892-015-0588-3. [DOI] [PubMed] [Google Scholar]
- 30.Soltesz G., Acsadi G. Association between diabetes, severe hypoglycemia, and electroencephalographic abnormalities. Arch. Dis. Child. 1989;64:992–996. doi: 10.1136/adc.64.7.992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferlazzo E., Gasparini S., Beghi E., Sueri C., Russo E., Leo A., Labate A., Gambardella A., Belcastro V., Striano P., et al. Epilepsy in cerebrovascular diseases: Review of experimental and clinical data with meta-analysis of risk factors. Epilepsia. 2016;57:1205–1214. doi: 10.1111/epi.13448. [DOI] [PubMed] [Google Scholar]
- 32.Schönberger S.J., Jezdic D., Faull R.L., Cooper G.J. Proteomic analysis of the human brain in Huntington’s Disease indicates pathogenesis by molecular processes linked to other neurodegenerative diseases and to type-2 diabetes. J. Huntington’s Dis. 2013;2:89–99. doi: 10.3233/JHD-120044. [DOI] [PubMed] [Google Scholar]
- 33.Lalić N.M., Marić J., Svetel M., Jotić A., Stefanova E., Lalić K., Dragašević N., Miličić T., Lukić L., Kostić V.S. Glucose homeostasis in Huntington disease: Abnormalities in insulin sensitivity and early-phase insulin secretion. Arch. Neurol. 2008;65:476–480. doi: 10.1001/archneur.65.4.476. [DOI] [PubMed] [Google Scholar]
- 34.Hou W.H., Li C.Y., Chang H.H., Sun Y., Tsai C.C. A population-based cohort study suggests an increased risk of multiple sclerosis incidence in patients with type 2 diabetes mellitus. J. Epidemiol. 2017;27:235–241. doi: 10.1016/j.je.2016.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biosa A., Outeiro T.F., Bubacco L., Bisaglia M. Diabetes Mellitus as a Risk Factor for Parkinson’s Disease: A Molecular Point of View. Mol. Neurobiol. 2018;55:8754–8763. doi: 10.1007/s12035-018-1025-9. [DOI] [PubMed] [Google Scholar]
- 36.Green H., Tsitsi P., Markaki I., Aarsland D., Svenningsson P. Novel Treatment Opportunities Against Cognitive Impairment in Parkinson’s Disease with an Emphasis on Diabetes-Related Pathways. CNS Drugs. 2019;33:143–160. doi: 10.1007/s40263-018-0601-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M., et al. NCBI geo: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2012;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Misu H., Takamura T., Takayama H., Hayashi H., Matsuzawa-Nagata N., Kurita S., Ishikura K., Ando H., Takeshita Y., Ota T., et al. A liver-derived secretory protein, selenoprotein p, causes insulin resistance. Cell Metab. 2010;12:483–495. doi: 10.1016/j.cmet.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 39.Blalock E.M., Buechel H.M., Popovic J., Geddes J.W., Landfield P.W. Microarray analyses of laser-captured hippocampus reveal distinct gray and white matter signatures associated with incipient alzheimer’s disease. J. Chem. Neuroanat. 2011;42:118–126. doi: 10.1016/j.jchemneu.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dangond F., Hwang D., Camelo S., Pasinelli P., Frosch M.P., Stephanopoulos G., Stephanopoulos G., Brown R.H., Jr., Gullans S.R. Molecular signature of late-stage human als revealed by expression profiling of postmortem spinal cord gray matter. Physiol. Genom. 2004;16:229–239. doi: 10.1152/physiolgenomics.00087.2001. [DOI] [PubMed] [Google Scholar]
- 41.Smith L.R., Chambers H.G., Subramaniam S., Lieber R.L. Transcriptional abnormalities of hamstring muscle contractures in children with cerebral palsy. PLoS ONE. 2012;7:e40686. doi: 10.1371/journal.pone.0040686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carlet M., Janjetovic K., Rainer J., Schmidt S., Panzer-Grümayer R., Mann G., Prelog M., Meister B., Ploner C., Kofler R. Expression, regulation and function of phosphofructo-kinase/fructose-biphosphatases (pfkfbs) in glucocorticoid-induced apoptosis of acute lymphoblastic leukemia cells. BMC Cancer. 2010;10:638. doi: 10.1186/1471-2407-10-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Borovecki F., Lovrecic L., Zhou J., Jeong H., Then F., Rosas H.D., Hersch S.M., Hogarth P., Bouzou B., Jensen R.V., et al. Genome-wide expression profiling of human blood reveals biomarkers for huntington’s disease. Proc. Natl. Acad. Sci. USA. 2005;102:11023–11028. doi: 10.1073/pnas.0504921102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han M.H., Lundgren D.H., Jaiswal S., Chao M., Graham K.L., Garris C.S., Axtell R.C., Ho P.P., Lock C.B., Woodard J.I., et al. Janus-like opposing roles of cd47 in autoimmune brain inflammation in humans and mice. J. Exp. Med. 2012;209:1325–1334. doi: 10.1084/jem.20101974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lewandowski N.M., Ju S., Verbitsky M., Ross B., Geddie M.L., Rockenstein E., Adame A., Muhammad A., Vonsattel J.P., Ringe D., et al. Polyamine pathway contributes to the pathogenesis of parkinson disease. Proc. Natl. Acad. Sci. USA. 2010;107:16970–16975. doi: 10.1073/pnas.1011751107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for rna-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moni M.A., Liò P. Genetic profiling and comorbidities of zika infection. J. Infect. Dis. 2017;216:703–712. doi: 10.1093/infdis/jix327. [DOI] [PubMed] [Google Scholar]
- 49.Moni M.A., Liò P. comor: A software for disease comorbidity risk assessment. J. Clin. Bioinform. 2014;4:1. doi: 10.1186/2043-9113-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moni M.A., Liò P. How to build personalized multi-omics comorbidity profiles. Front. Cell Dev. Biol. 2015;3:28. doi: 10.3389/fcell.2015.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jin L., Zuo X.-Y., Su W.-Y., Zhao X.-L., Yuan M.-Q., Han L.-Z., Zhao X., Chen Y.-D., Rao S.-Q. Pathway-based analysis tools for complex diseases: A review. Genom. Proteomics Bioinform. 2014;12:210–220. doi: 10.1016/j.gpb.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gene Ontology Consortium Gene ontology consortium: Going forward. Nucleic Acids Res. 2014;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. Kegg for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2011;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Croft D., O’kelly G., Wu G., Haw R., Gillespie M., Matthews L., Caudy M., Garapati P., Gopinath G., Jassal B., et al. Reactome: A database of reactions, pathways and biological processes. Nucleic Acids Res. 2010;39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krupa S., Anthony K., Buchoff J.R., Day M., Hannay T., Schaefer C.F. The nci-nature pathway interaction database: A cell signaling resource. Nat. Preced. 2007;2:1. doi: 10.1038/npre.2007.1311.1. [DOI] [Google Scholar]
- 57.Slenter D.N., Kutmon M., Hanspers K., Riutta A., Windsor J., Nunes N., Mélius J., Cirillo E., Coort S.L., Digles D., et al. Wikipathways: A multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 2017;46:D661–D667. doi: 10.1093/nar/gkx1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.BioCarta N.D. Biotech Software & Internet Report. RG J. 2001;2:117–120. doi: 10.1089/152791601750294344. [DOI] [Google Scholar]
- 59.Mi H., Thomas P. PANTHER pathway: An ontology-based pathway database coupled with data analysis tools. Methods Mol. Biol. 2009;563:123–140. doi: 10.1007/978-1-60761-175-2_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trupp M., Altman T., Fulcher C.A., Caspi R., Krummenacker M., Paley S., Karp P.D. Beyond the genome (BTG) is a (PGDB) pathway genome database: HumanCyc. Genome Biol. 2010;11:O12. doi: 10.1186/gb-2010-11-s1-o12. [DOI] [Google Scholar]
- 61.De Las Rivas J., Fontanillo C. Protein–protein interactions essentials: Key concepts to building and analyzing interactome networks. PLoS Comput. Biol. 2010;6:e1000807. doi: 10.1371/journal.pcbi.1000807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., et al. String v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2014;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Khan A., Fornes O., Stigliani A., Gheorghe M., Castro-Mondragon J.A., van der Lee R., Bessy A., Cheneby J., Kulkarni S.R., Tan G., et al. Jaspar 2018: Update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2017;46:D260–D266. doi: 10.1093/nar/gkx1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sethupathy P., Corda B., Hatzigeorgiou A.G. Tarbase: A comprehensive database of experimentally supported animal microrna targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hsu S.-D., Lin F.-M., Wu W.-Y., Liang C., Huang W.-C., Chan W.-L., Tsai W.-T., Chen G.-Z., Lee C.-J., Chiu C.M., et al. mirtarbase: A database curates experimentally validated microrna–target interactions. Nucleic Acids Res. 2010;39(Suppl. 1):D163–D169. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xia J., Gill E.E., Hancock R.E.W. Networkanalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat. Protoc. 2015;10:823. doi: 10.1038/nprot.2015.052. [DOI] [PubMed] [Google Scholar]
- 67.Davis S., Meltzer P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 68.Gentleman R., Carey V., Huber W., Hahne F. Genefilter: Methods for Filtering Genes from High-Throughput Experiments. [(accessed on 5 February 2020)];2015 Bioconductor. Available online: https://bioconductor.riken.jp/packages/3.0/bioc/html/genefilter.html.
- 69.Chen S.-H., Chin C.-H., Wu H.-H., Ho C.-W., Ko M.-T., Lin C.-Y. cyto-hubba: A cytoscape plug-in for hub object analysis in network biology; Proceedings of the 20th International Conference on Genome Informatics; Yokohama, Japan. 14–16 December 2009. [Google Scholar]
- 70.Cheng C., Alexander R., Min R., Leng J., Yip K.Y., Rozowsky J., Yan K.K., Dong X., Djebali S., Ruan Y., et al. Understanding transcriptional regulation by integrative analysis of transcription factor binding data. Genome Res. 2012;22:1658–1667. doi: 10.1101/gr.136838.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moradifard S., Hoseinbeyki M., Ganji S.M., Minuchehr Z. Analysis of microRNA and gene expression profiles in Alzheimer’s disease: A meta-analysis approach. Sci. Rep. 2018;8:4767. doi: 10.1038/s41598-018-20959-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Van Cauwenberghe C., Van Broeckhoven C., Sleegers K. The genetic landscape of Alzheimer disease: Clinical implications and perspectives. Genet. Med. 2016;18:421–430. doi: 10.1038/gim.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Anjana M., Ahuja Y.R. Genes associated with Alzheimer Disease. Neurol. Asia. 2010;15:109–118. [Google Scholar]
- 74.Eykens C., Robberecht W. The Genetic basis of amyotrophic lateral sclerosis: Recent breakthroughs. Adv. Genom. Genet. 2015;5:327. [Google Scholar]
- 75.Fahey M.C., Maclennan A.H., Kretzschmar D., Gecz J., Kruer M.C. The genetic basis of cerebral palsy. Dev. Med. Child Neurol. 2017;59:462–469. doi: 10.1111/dmcn.13363. [DOI] [PubMed] [Google Scholar]
- 76.GeneDx Genetic Testing for Epilepsy: A Guide for Patients. [(accessed on 26 October 2019)]; Available online: https://www.genedx.com/wpcontent/uploads/crm_docs/91040_Epilepsy-Patient-Guide.pdf.
- 77.Myers C.T., Mefford H.C. Advancing epilepsy genetics in the genomic era. Genome Med. 2015;7:1–11. doi: 10.1186/s13073-015-0214-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Arning L., Epplen J.T. Genetic modifiers of Huntington’s disease: Beyond CAG. Future Neurol. 2012;7:93–109. doi: 10.2217/fnl.11.65. [DOI] [Google Scholar]
- 79.Baranzini S.E. Revealing the genetic basis of multiple sclerosis: Are we there yet? Curr. Opin. Genet. Dev. 2011;21:317–324. doi: 10.1016/j.gde.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Redenšek S., Trošt M., Dolžan V. Genetic determinants of Parkinson’s disease: Can they help to stratify the patients based on the underlying molecular defect? Front. Aging Neurosci. 2017;9:20. doi: 10.3389/fnagi.2017.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Antoni Romeu and Lluís Arola Classical dynamin dnm1 and dnm3 genes attain maximum expression in the normal human central nervous system. BMC Res. Notes. 2014;7:188. doi: 10.1186/1756-0500-7-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sidiropoulos P.N.M., Miehe M., Bock T., Tinelli E., Oertli C.I., Kuner R., Meijer D., Wollscheid B., Niemann A., Suter U. Dynamin 2 mutations in charcot–marie–tooth neuropathy highlight the importance of clathrin-mediated endocytosis in myelination. Brain. 2012;135:1395–1411. doi: 10.1093/brain/aws061. [DOI] [PubMed] [Google Scholar]
- 83.Dumont V., Tolvanen T.A., Kuusela S., Wang H., Nyman T.A., Lindfors S., Tienari J., Nisen H., Suetsugu S., Plomann M., et al. Pacsin2 accelerates nephrin trafficking and is up-regulated in diabetic kidney disease. FASEB J. 2017;31:3978–3990. doi: 10.1096/fj.201601265R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Borie C., Gasparini F., Verpillat P., Bonnet A.-M., Agid Y., Hetet G., Brice A., Dürr A., Grandchamp B., French Parkinson’s Disease Genetic Study Group et al. Association study between iron-related genes polymorphisms and parkinson’s disease. J. Neurol. 2002;249:801–804. doi: 10.1007/s00415-002-0704-6. [DOI] [PubMed] [Google Scholar]
- 85.Rahman M., Islam T., Shahjaman M., Zaman T., Faruquee H.M., Jamal M.A.H.M., Huq F., Quinn J.M.W., Moni M.A. Discovering biomarkers and pathways shared by alzheimer’s disease and ischemic stroke to identify novel therapeutic targets. Medicina. 2019;55:191. doi: 10.3390/medicina55050191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mamelona J., Crapoulet N., Marrero A. A new case of spastic paraplegia type 64 due to a missense mutation in the entpd1 gene. Hum. Genome Var. 2019;6:5. doi: 10.1038/s41439-018-0036-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rahman M.R., Islam T., Turanli B., Zaman T., Faruquee H.M., Rahman M.M., Mollah M.N.H., Nanda R.K., Arga K.Y., Gov E., et al. Network-based approach to identify molecular signatures and therapeutic agents in alzheimer’s disease. Comput. Biol. Chem. 2019;78:431–439. doi: 10.1016/j.compbiolchem.2018.12.011. [DOI] [PubMed] [Google Scholar]
- 88.Godlewski J., Lenart J., Salinska E. Microrna in brain pathology: Neurodegeneration the other side of the brain cancer. Non-Coding RNA. 2019;5:20. doi: 10.3390/ncrna5010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Persengiev S.P., Kondova I.I., Bontrop R.E. The impact of micrornas on brain aging and neurodegeneration. Curr. Gerontol. Geriatr. Res. 2012;2012:359369. doi: 10.1155/2012/359369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu W., Liu C., Zhu J., Shu P., Yin B., Gong Y., Qiang B., Yuan J., Peng X. Microrna-16 targets amyloid precursor protein to potentially modulate alzheimer’s-associated pathogenesis in samp8 mice. Neurobiol. Aging. 2012;33:522–534. doi: 10.1016/j.neurobiolaging.2010.04.034. [DOI] [PubMed] [Google Scholar]
- 91.Cloonan N., Brown M.K., Steptoe A.L., Wani S., Chan W.L., Forrest A.R.R., Kolle G., Gabrielli B., Grimmond S.M. The mir-17-5p microrna is a key regulator of the g1/s phase cell cycle transition. Genome Biol. 2008;9:R127. doi: 10.1186/gb-2008-9-8-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]