Abstract
Interactions between DNA and DNA-binding proteins play an important role in many essential cellular processes. A key function of the DNA-binding protein p53 is to search for and bind to target sites incorporated in genomic DNA, which triggers transcriptional regulation. How do p53 molecules achieve “rapid” and “accurate” target search in living cells? The search dynamics of p53 were expected to include 3D diffusion in solution, 1D diffusion along DNA, and intersegmental transfer between two different DNA strands. Single-molecule fluorescence microscopy enabled the tracking of p53 molecules on DNA and the characterization of these dynamics quantitatively. Recent intensive single-molecule studies of p53 succeeded in revealing each of these search dynamics. Here, we review these studies and discuss the target search mechanisms of p53.
Keywords: p53, DNA, single molecule fluorescence microscopy, sliding, diffusion, intersegmental transfer, target search
1. Introduction
Genome editing method has been used to investigate and engineer gene functions in life science. The genome encompasses DNA sequences that encode genes, and gene editing is the genetic engineering of a specific DNA sequence, including insertion, deletion, modification, and replacement. The main player in genome editing is a type of protein that can bind to DNA, known as DNA-binding proteins. DNA-binding proteins include enzymes, which can cut DNA or ligate two DNA molecules, and transcription factors, which can activate or deactivate gene expression. These proteins are classified into DNA sequence-specific and nonspecific binders.
The sequence-specific DNA-binding proteins have a common mechanism to search for and bind to their target DNA sites encoded in a genome. Successful target binding by transcription factors can trigger the regulation of the cellular functions, whereas the failure of the target search and binding is known to cause various diseases including cancers. In addition, the target binding is delicately regulated depending on genome functioning. Accordingly, the target search and binding by DNA-binding protein are one of the indispensable reactions in life. The bacterial and human genomes comprise approximately 106 and 109 bp of DNA, respectively. In contrast, the DNA sequence that is bound by the proteins is just typically 5–30 bp. For example, assuming that a single protein searches for the target coded in 109 bp of genome by repeatedly associating and dissociating with random sites in the genome (3D diffusion), the search time is estimated to be much longer than the physiological time which is estimated from several minutes to a half of an hour in cells. However, the natural proteins complete the target search exactly within the physiological search time. This gap is called the “target search problem.” How do DNA-binding proteins solve this problem and function on the target?
To solve this problem, three target search dynamics are currently proposed: 3D diffusion; 1D diffusion, in which a protein moves along DNA; and intersegmental transfer, in which a protein transfers between two different DNA strands without dissociating [1,2,3]. Theoretical studies demonstrate that a combination of at least two dynamics can facilitate the target search [1,2,4,5], known as facilitated diffusion.
2. p53 As a Model System for Target Search Studies
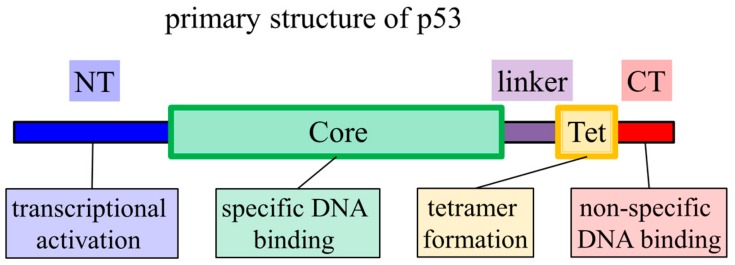
The transcription factor p53 can induce multiple tumor suppression functions, such as cell cycle arrest, DNA repair, and apoptosis. Furthermore, p53 is known as the guardian of the genome, and it contains an N-terminal (NT) domain, core domain, linker, tetramerization (Tet) domain, and C-terminal (CT) domain (Figure 1). The core and Tet domains have specific folded structures, while the others are intrinsically disordered regions [6,7,8]. The core and CT domains are involved in binding to DNA sequences in a specific and nonspecific manner, respectively [9]. The Tet domain enables the formation of a homo-tetramer [8]. In target search study, p53 is used as a model protein because it has common features that are frequently observed in DNA-binding proteins, such as oligomerization, a disordered region, and multiple DNA-binding domains [10]. A defect in target binding by p53 causes tumorigenesis [11], indicating the importance of successful target searching and binding.
Figure 1.
Domain organization and function of p53. Thin and thick boxes represent disordered and folded regions, respectively. NT, core, Tet, and CT function as transcriptional regulation, target DNA sequence binding, dimer or tetramer formation, and non-target DNA sequence binding, respectively.
3. Single-Molecule Fluorescence Microscopy
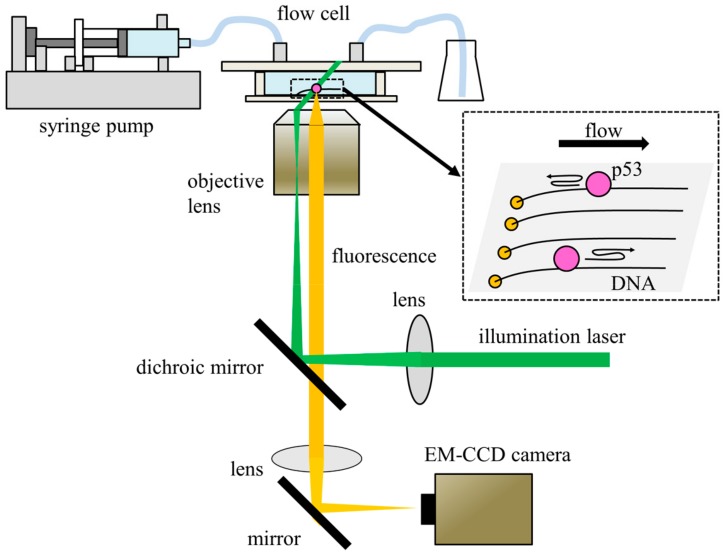
To solve the target search problem, it is necessary to differentiate and characterize each search dynamic. Single-molecule fluorescence microscopy can track a single molecule moving along DNA and/or transferring between two DNA molecules [12,13,14,15,16,17,18,19,20,21,22,23,24]. In general, the system contains a fluorescence microscope and a flow cell (Figure 2). Fluorescent dye-labeled p53 is introduced into the flow cell using a syringe pump. At least one end of each DNA molecule is tethered to the inner glass surface of the flow cell, and the tethered DNA is stretched by the flow pressure. The molecules, which are bound to the DNA, are detected as fluorescent spots using an electron-multiplying charge-coupled device (EM-CCD) camera of a fluorescence microscope. Total internal reflection fluorescence (TIRF) or highly inclined and laminated optical sheet (HILO) is used to illuminate DNA-bound molecules selectively and to reduce the unexpected background fluorescence. The positions of individual molecules are tracked using an appropriate analysis program.
Figure 2.
Schematic diagram of representative single-molecule fluorescence microscopy. The system comprises a fluorescence microscope and a flow cell. Fluorescently labeled p53 is introduced into the flow cell using a syringe pump. DNA is tethered on one end to the MPC-polymer-coated glass surface of the flow cell in a line using the DNA garden method. The tethered DNA molecules are stretched by flow pressure. TIRF or HILO is used to illuminate DNA-bound p53 molecules. Fluorescence from the p53 molecules is detected by EM-CCD. The figure is adapted from ref. [25] with some modifications.
Several elegant methods for producing DNA arrays were developed in order to acquire data on many molecules simultaneously [26,27,28]. A simple method is the “DNA garden,” in which neutravidin molecules are printed in a line on coverslips using polydimethylsiloxane (PDMS) stamps, and then biotinylated DNAs are tethered to the stamped neutravidin [28] (Figure 2). In addition, 2-methacryloyloxyethyl phosphorylcholine (MPC) polymer coating minimizes the adsorption of p53 and DNA to the flow cell, which distorts measurements. The DNA array can reduce the overlap of two adjacent DNA molecules and simplify analysis.
4. 1D Diffusion of p53 along DNA
In 2008, Tafvizi et al. succeeded in observing the movement of p53 tetramer along a non-target DNA sequence using single-molecule fluorescence microscopy [29]. A thermostable mutant of human p53 was used as pseudo-wild type to prevent irreversible aggregation observed in genotypic wild type in some cases. Also, we consider only the tetrameric form of full-length p53 throughout the manuscript because of the main oligomeric state for the target search (see in detail in Section 6). The single-molecule tracking revealed that p53 diffuses forward and backward along DNA randomly. The 1D diffusion is caused only by the thermal energy, but not the chemical energy of ATP. In other words, p53 converts the thermal energy to drive 1D search dynamics along DNA by maintaining weak interactions between p53 and DNA. Before this study, McKinney et al. indirectly observed p53 sliding off from short DNAs in ensemble gel shift assay, thereby suggesting 1D diffusion by p53 [30]. The 1D diffusion of p53 was later confirmed using similar single-molecule fluorescence microscopy by our group [31] as well as course-grained molecular dynamic (MD) simulations by two groups [32,33]. Accordingly, single-molecule tracking provided direct evidence of 1D diffusion by p53 along DNA.
Tafvizi et al. further revealed that 1D diffusion of p53 along the non-target DNA sequence does not depend on a monovalent salt concentration; this suggested “1D sliding,” in which p53 moves along DNA while maintaining contact with the DNA [2,29]. Based on similar measurements of p53 mutants with deletions in either of the DNA-binding domains, it was proposed that p53 searches for the target using restricted hopping of the core domains during 1D sliding [34]. This was later supported by a course-grained MD simulation [33]. Leith et al. also found that 1D diffusion of p53 slowed on DNA sequences that were definitely non-target sequences but had higher similarity to the target sequence [35]. This finding suggests that p53 that diffuses along DNA can read the DNA sequence through interactions between its core domains and DNA.
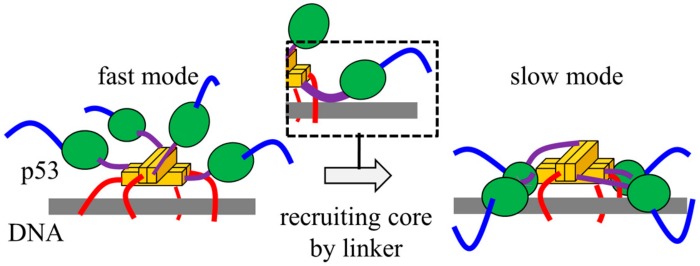
We clarified that p53 possesses two 1D diffusion modes with different diffusion coefficients by altering the structure of p53 on DNA [31,36] (Figure 3). The fast 1D diffusion mode enables an extension of the search distance by maintaining weak contacts between the CT domains and DNA. In the slow diffusion mode, p53 has a similar conformation that can recognize and bind to the target DNA sequence on non-target DNA sequence. In particular, the core domains as well as CT domains participate in p53 binding to DNA. Furthermore, the disordered linker connecting the core and Tet domains can trigger switching between the two 1D diffusion modes via electrostatic interactions between the linker and DNA [37] (Figure 3). Overall, p53 performs the target search using 1D diffusion coupled with flexible conformational changes on DNA.
Figure 3.
Schematic diagram of the p53-DNA complex structure in 1D diffusion along a non-target DNA sequence. The linker (purple) recruits the core domain (green) to DNA (grey), triggering the conformational switch between fast and slow 1D diffusion modes. The color of p53 domains is the same in Figure 1.
If p53 reaches the target DNA sequence using 1D diffusion, it will recognize and bind to the target. Considering the fast target search by DNA-binding proteins, one might predict a high efficiency of target recognition and binding [38]. Contrary to this prediction, we revealed that only ~10% of p53 molecules succeed in binding to the target DNA sequence [39]. Surprisingly, ~90% of molecules pass over the target sequence without recognition. This low probability of target recognition might be explained by a large conformational change by p53 on the target DNA sequence, as predicted in course-grained MD simulations [40]: p53 was unable to achieve the target-binding conformation before diffusing away from the target. Accordingly, the target search by p53 is decelerated due to the low target recognition probability, although 1D diffusion contributes to facilitating the target search. On the other hand, p53 function can be regulated by altering the target recognition probability in response to the cell condition [39]. Interestingly, evolution selects regulation of the target recognition probability rather than target search facilitation.
5. Ultrafast Intersegmental Transfer of p53
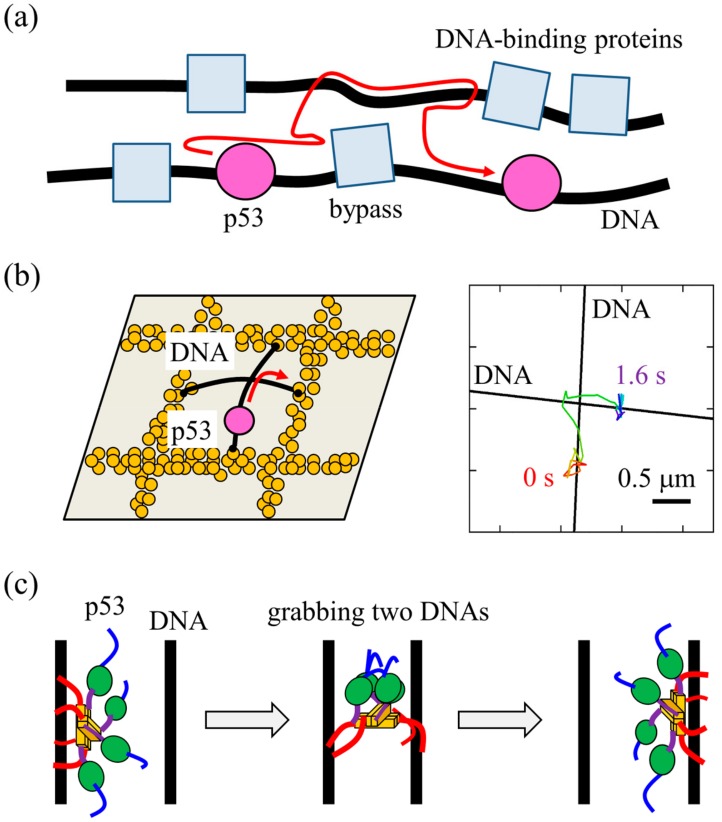
In the cell, many DNA-binding proteins bind to and cover DNA, which could block the 1D diffusion of p53 along DNA and hinder its target search. In the nucleus, genomic DNA is stored at high concentrations. In these conditions, it may be possible that p53 possesses a special mechanism to bypass the obstacles on DNA by transferring from one DNA strand to another (Figure 4a).
Figure 4.
(a) Schematic diagram of target search by p53 in the cell. p53 (pink spheres) may bypass other DNA-binding proteins (light blue squares) bound to DNA (black lines) using intersegmental transfer. (b) Single-molecule verification of intersegmental transfer of p53 between crisscrossing DNAs (left). The typical data of p53 demonstrating intersegmental transfer (right). (c) Schematic diagram of intersegmental transfer of p53. p53 binds to the first DNA mainly using the CT domain (Left), grabs the second DNA (Middle), and then releases the former while maintaining contact with the latter DNA (Right). The panels (a) and (c) are adapted from ref. [25] with some modifications, and the panel (b) is adapted from ref. [41] with some modifications.
We conducted a kinetic ensemble measurement to monitor the transfer process of p53 from fluorescent to non-fluorescent DNA [41]. Fluorescence anisotropy was used to differentiate between the p53-DNA complex and free DNA by monitoring rotational DNA diffusion. After p53 bound to the fluorescent DNA was mixed with a large quantity of non-fluorescent DNA, p53 transfer to the non-fluorescent DNA was observed. The data demonstrated that p53 transfers between two non-target DNA strands at nearly the diffusion limit (~108 M−1s−1). This suggests that even if p53 encounters DNA-bound obstacles in the cell, it can bypass these via ultrafast transfer. In contrast, p53 does not transfer from the target DNA sequence. Accordingly, once p53 binds to the target DNA sequence, it is not repelled from the target DNA by nearby DNA in the cell.
To directly examine p53 transfer between DNA segments, we prepared crisscrossing DNA in the flow cell using the modified DNA garden method and tested whether p53 moves between DNA segments at the crisscross points [41] (Figure 4b). The crisscrossing DNA was prepared according to the method developed by Greene’s group [42]. Notably, p53 changed the direction of 1D diffusion at an ~90 degree angle at a crisscross point, which gave direct evidence of intersegmental transfer. Further analysis of the mutants lacking either of the DNA-binding domains demonstrated that CT domains are required for intersegmental transfer. Therefore, during intersegmental transfer, p53 bound to the first DNA should grab the second DNA using at least two of four CT domains, after which it releases the first and transfers to the second DNA (Figure 4c). These experimental data are consistent with the preceding results based on course-grained MD simulations by the Levy and Takada groups [10,43].
6. Target Search by p53 May Be Well Designed by Biological Requirements
Considering the in vitro data described above, p53 is presumed to solve the target search problem by utilizing 3D diffusion, 1D diffusion along DNA, and intersegmental transfer between two DNAs in the cell. In 3D search, p53 can associate to non-target DNA near the diffusion limit [39]. The association rate constant of p53 is 1–10000-fold faster than those of other DNA-binding proteins including transcription factors and a nucleoid protein [44,45,46,47,48,49,50,51,52,53,54,55,56,57,58]. This ultrafast association enables a reduction in the time taken for 3D diffusion in which p53 cannot search the target in solution. The 1D diffusion enables p53 to search ~200 bp of DNA before dissociating [36] and contributes to ~10-fold elongation of the search distance compared to that in the absence of 1D diffusion. The intersegmental transfer of p53 occurs 10–10,000-fold faster than that of other DNA-binding proteins [59,60,61,62,63,64], with reactions near the diffusion limit [41]. Using such ultrafast intersegmental transfer, p53 could bypass DNA-bound obstacles in the cell, thereby facilitating the target search. In contrast, as described above, the target recognition efficiency of p53 was surprisingly low. These findings indicate that not all target searches by p53 are necessarily optimized toward facilitation.
Considering the biological functions of p53, one may wonder why the target search by p53 is not optimized in terms of facilitation. Under various cellular stresses such as DNA damage and tumorigenesis of the cell, p53 turns the function “ON” and induces DNA repair and apoptosis. In contrast, p53 should keep the function “OFF” under normal conditions. It would be dangerous if p53 were to accidentally bind to the target DNA sequences and induce apoptosis in normal conditions. To prevent this malfunction, p53 may keep target recognition low under normal conditions at the expense of target search speed in stress conditions.
To regulate multiple functional pathways as well as the discrete ON/OFF switch, p53 has three different strategies. One strategy is to increase the p53 copy number under stressed conditions [65]. The increased copy number itself can promote target searching and binding, thus triggering its function. In addition, further p53 expression enables lower affinity binding to the target DNA sequences, thereby switching the functional pathways [66]. Concomitant with the increase in copy number, the oligomeric state of p53 is altered [67,68,69], as the major oligomeric state changes from dimer to tetramer upon cell stress [70]. The affinity of the p53 dimer to the target DNA sequence is known to be much weaker than that of the tetramer [71]. Accordingly, the tetramer is the main oligomeric state involved in the target search and can regulate function. The other functional regulator is posttranslational modification [72,73,74,75,76,77]. Phosphorylation of the CT domain enhances target recognition, thereby enhancing the transcriptional activity [38,78]. Acetylation of the CT domain modulates target binding in vitro and in the cell [79,80]. Overall, these multiple strategies may be required to activate only the appropriate functional pathway according to cell conditions.
7. Target Search of p53 in Live Cells
The target search dynamics of p53 in vivo have been investigated by single-molecule fluorescence imaging. For example, Mazza et al. succeeded in tracking p53 molecules one by one in live cells using HILO illumination [81]. They showed that 20% of p53 molecules bound to chromatin and the other molecules diffused freely in the nucleus. Furthermore, Morisaki et al. found that the p53 molecules have two residence times (0.33 s and 3.5 s) for binding to transcription domains of cells, whereas the long residence time was not detected outside of the transcription domains [82]. Based on mutational analysis, they identified that the long residence time corresponds to the binding to the target sites. The percentage of the successful target binding was estimated to be 5.5% within the transcription domains. In contrast, the short residence time represents a mixture of slowly diffusing and/or transiently bound molecules. During the transient binding to genomic DNA, p53 could search for the target sites by sliding along DNA and/or using intersegmental transfer. Considering that p53 molecules search for the target sites out of the transcription domains, the percentage of the successful target binding in vivo would be quite low. Interestingly, the residence times of p53 on target and non-target DNAs in vivo are much longer than those obtained by in vitro measurements [39]. The molecular crowding effect or proteins bound to p53 may stabilize the p53/DNA complex in cells. In addition, the post-translational modifications to p53 can modulate the residence time on target sites, which enhances the transcriptional activity [80]. Further investigations are required to clarify the behavior of p53 in cells.
8. Common Target Search in Biology
Target search is not limited to DNA-binding proteins, such as p53. Various processes can be considered as target search problems, including biomolecules searching for binding partner molecules or animals seeking food. Interestingly, a common mechanism known as intermittent search is widely used in biology [38]. Intermittent search involves two different modes: a recognition mode, in which molecules can recognize their objectives, and a search mode, in which molecules can cover long distances without target recognition. In the case of p53, the search and recognition modes correspond to 3D diffusion (and/or intersegmental transfer) and 1D diffusion along DNA, respectively. Furthermore, 1D diffusion also comprises the two similar modes, thereby facilitating the target search. Accordingly, p53 utilizes the common intermittent search. It is interesting that biomolecules and living things on the micro-to-macro scale select the common search strategy in time scales from seconds to hours.
9. Summary and Future Perspectives
Single-molecule fluorescence microscopy has clarified that p53 solves the target search problem by utilizing 3D diffusion, 1D diffusion along DNA, and intersegmental transfer between two DNAs. As predicted by theoretical studies, target search by p53 can be facilitated by combining these three strategies. In particular, considering the ultrafast association to DNA, 1D diffusion along DNA, and ultrafast intersegmental transfer, target searching by p53 is somehow optimized toward facilitation. In contrast, the low target recognition by p53 decelerates target searching. Furthermore, p53 may prevent malfunction under normal conditions at the expense of the target search speed in stress conditions. Accordingly, the target search strategy of p53 may be well designed to satisfy biological requirements.
Further single-molecule investigations could deepen the understanding of target search by p53. In the cell, histone proteins bind to and condense DNA, which may affect target search by p53 [83]. Single-molecule measurements with the use of histone-bound DNA [84], an in vivo-mimicking system used in vitro would offer further interpretation of the in vivo single-molecule data [80,81,82]. Also, single-molecule microscopy could be used to validate important DNA binding regions as well as local DNA structures of p53 [85,86,87,88,89,90] and to clarify the molecular mechanism for functional regulations of truncated and/or posttranslational modified forms of p53 [91,92,93,94,95]. Additionally, p53 may search for the target using hops and short jumps along DNA, as observed in EcoRV [96]. To resolve these dynamics, the spatial and/or temporal resolutions of the single-molecule microscope must be improved. Recently, we succeeded in improving such temporal resolution from 33 ms to 0.5 ms and found that p53 hops and jumps along DNA (Subekti et al., submitted). Furthermore, cryo-electron microscopy could determine heterogeneous structures of p53 at the single-molecule level [97,98,99]. In addition to the single-molecule measurements, the regulation of the target search dynamics as well as target binding by an artificially designed peptide may be useful for medical applications [100].
Since many DNA-binding proteins exist in the cell and have different structures and functions, it is interesting how such DNA-binding proteins solve the target search problem. For example, if certain DNA-binding proteins differ in structure and function, their target search mechanisms may each differ as well [101,102,103,104,105]. In the near future, the target search mechanism of various DNA-binding proteins will be revealed by single-molecule fluorescence microscopy. Comparisons between p53 and other proteins will allow the characterization of the special action of p53 in performing target search. Furthermore, such detailed characterization could give us suggestions for engineering of gene-editing DNA-binding proteins toward higher editing efficiency and lower off-target editing.
Acknowledgments
All authors thank the corroborators regarding our studies of target search by p53 for their helpful discussion. This article received no external funding.
Abbreviations
| NT | N-terminal |
| Tet | tetramerization |
| CT | C-terminal |
| TIRF | total internal reflection fluorescence |
| HILO | highly inclined and laminated optical sheet |
| PDMS | polydimethylsiloxane |
| MPC | 2-methacryloyloxyethyl phosphorylcholine |
| MD | molecular dynamics |
Author Contributions
All authors wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Halford S.E., Marko J.F. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tafvizi A., Mirny L.A., van Oijen A.M. Dancing on DNA: Kinetic aspects of search processes on DNA. Chemphyschem. 2011;12:1481–1489. doi: 10.1002/cphc.201100112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamagata K., Murata A., Itoh Y., Takahashi S. Characterization of facilitated diffusion of tumor suppressor p53 along DNA using single-molecule fluorescence imaging. J. Photochem. Photobiol. C Photochem. Reviews. 2017;30:36–50. doi: 10.1016/j.jphotochemrev.2017.01.004. [DOI] [Google Scholar]
- 4.Slutsky M., Mirny L.A. Kinetics of protein-DNA interaction: Facilitated target location in sequence-dependent potential. Biophys. J. 2004;87:4021–4035. doi: 10.1529/biophysj.104.050765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt H.G., Sewitz S., Andrews S.S., Lipkow K. An integrated model of transcription factor diffusion shows the importance of intersegmental transfer and quaternary protein structure for target site finding. PLoS ONE. 2014;9:e108575. doi: 10.1371/journal.pone.0108575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joerger A.C., Fersht A.R. The tumor suppressor p53: From structures to drug discovery. Cold Spring Harb. Perspect. Biol. 2010;2:a000919. doi: 10.1101/cshperspect.a000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laptenko O., Tong D.R., Manfredi J., Prives C. The Tail That Wags the Dog: How the Disordered C-Terminal Domain Controls the Transcriptional Activities of the p53 Tumor-Suppressor Protein. Trends Biochem. Sci. 2016;41:1022–1034. doi: 10.1016/j.tibs.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamada R., Toguchi Y., Nomura T., Imagawa T., Sakaguchi K. Tetramer formation of tumor suppressor protein p53: Structure, function, and applications. Biopolymers. 2016;106:598–612. doi: 10.1002/bip.22772. [DOI] [PubMed] [Google Scholar]
- 9.Anderson M.E., Woelker B., Reed M., Wang P., Tegtmeyer P. Reciprocal interference between the sequence-specific core and nonspecific C-terminal DNA binding domains of p53: Implications for regulation. Mol. Cell. Biol. 1997;17:6255–6264. doi: 10.1128/MCB.17.11.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vuzman D., Levy Y. Intrinsically disordered regions as affinity tuners in protein-DNA interactions. Mol. Biosyst. 2012;8:47–57. doi: 10.1039/C1MB05273J. [DOI] [PubMed] [Google Scholar]
- 11.Hainaut P., Hollstein M. p53 and human cancer: The first ten thousand mutations. Adv. Cancer Res. 2000;77:81–137. doi: 10.1016/s0065-230x(08)60785-x. [DOI] [PubMed] [Google Scholar]
- 12.Kabata H., Kurosawa O., Arai I., Washizu M., Margarson S.A., Glass R.E., Shimamoto N. Visualization of single molecules of RNA-polymerase sliding along DNA. Science. 1993;262:1561–1563. doi: 10.1126/science.8248804. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y.M., Austin R.H., Cox E.C. Single molecule measurements of repressor protein 1D diffusion on DNA. Phys. Rev. Lett. 2006;97:048302. doi: 10.1103/PhysRevLett.97.048302. [DOI] [PubMed] [Google Scholar]
- 14.Gorman J., Chowdhury A., Surtees J.A., Shimada J., Reichman D.R., Alani E., Greene E.C. Dynamic basis for one-dimensional DNA scanning by the mismatch repair complex Msh2-Msh6. Mol. Cell. 2007;28:359–370. doi: 10.1016/j.molcel.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komazin-Meredith G., Mirchev R., Golan D.E., van Oijen A.M., Coen D.M. Hopping of a processivity factor on DNA revealed by single-molecule assays of diffusion. Proc. Natl. Acad. Sci. USA. 2008;105:10721–10726. doi: 10.1073/pnas.0802676105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blainey P.C., Luo G., Kou S.C., Mangel W.F., Verdine G.L., Bagchi B., Xie X.S. Nonspecifically bound proteins spin while diffusing along DNA. Nat. Struct. Mol. Biol. 2009;16:1224–1229. doi: 10.1038/nsmb.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biebricher A., Wende W., Escude C., Pingoud A., Desbiolles P. Tracking of single quantum dot labeled EcoRV sliding along DNA manipulated by double optical tweezers. Biophys. J. 2009;96:L50–L52. doi: 10.1016/j.bpj.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorman J., Plys A.J., Visnapuu M.L., Alani E., Greene E.C. Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice. Nat. Struct. Mol. Biol. 2010;17:932–938. doi: 10.1038/nsmb.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunn A.R., Kad N.M., Nelson S.R., Warshaw D.M., Wallace S.S. Single Qdot-labeled glycosylase molecules use a wedge amino acid to probe for lesions while scanning along DNA. Nucleic Acids Res. 2011;39:7487–7498. doi: 10.1093/nar/gkr459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forget A.L., Dombrowski C.C., Amitani I., Kowalczykowski S.C. Exploring protein-DNA interactions in 3D using in situ construction, manipulation and visualization of individual DNA dumbbells with optical traps, microfluidics and fluorescence microscopy. Nat. Protoc. 2013;8:525–538. doi: 10.1038/nprot.2013.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee J.B., Cho W.K., Park J., Jeon Y., Kim D., Lee S.H., Fishel R. Single-molecule views of MutS on mismatched DNA. DNA Repair. 2014;20:82–93. doi: 10.1016/j.dnarep.2014.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin J., Countryman P., Buncher N., Kaur P., E L., Zhang Y., Gibson G., You C., Watkins S.C., Piehler J., et al. TRF1 and TRF2 use different mechanisms to find telomeric DNA but share a novel mechanism to search for protein partners at telomeres. Nucleic Acids Res. 2014;42:2493–2504. doi: 10.1093/nar/gkt1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones N.D., Lopez M.A., Jr., Hanne J., Peake M.B., Lee J.B., Fishel R., Yoder K.E. Retroviral intasomes search for a target DNA by 1D diffusion which rarely results in integration. Nat. Commun. 2016;7:11409. doi: 10.1038/ncomms11409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kostiuk G., Dikic J., Schwarz F.W., Sasnauskas G., Seidel R., Siksnys V. The dynamics of the monomeric restriction endonuclease BcnI during its interaction with DNA. Nucleic Acids Res. 2017;45:5968–5979. doi: 10.1093/nar/gkx294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamagata K., Itoh Y., Subekti D.R.G. How does tumor suppressor protein p53 solve the target DNA search problem? (Japanese) BUTSURI. 2019;74:472–475. [Google Scholar]
- 26.Yamamoto T., Kurosawa O., Kabata H., Shimamoto N., Washizu M. Molecular surgery of DNA based on electrostatic micromanipulation. Ieee Trans. Ind. Appl. 2000;36:1010–1017. doi: 10.1109/28.855954. [DOI] [Google Scholar]
- 27.Fazio T., Visnapuu M.L., Wind S., Greene E.C. DNA curtains and nanoscale curtain rods: High-throughput tools for single molecule imaging. Langmuir. 2008;24:10524–10531. doi: 10.1021/la801762h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Igarashi C., Murata A., Itoh Y., Subekti D.R.G., Takahashi S., Kamagata K. DNA garden: A simple method for producing arrays of stretchable DNA for single-molecule fluorescence imaging of DNA binding proteins. Bull. Chem. Soc. Jpn. 2017;90:34–43. doi: 10.1246/bcsj.20160298. [DOI] [Google Scholar]
- 29.Tafvizi A., Huang F., Leith J.S., Fersht A.R., Mirny L.A., van Oijen A.M. Tumor suppressor p53 slides on DNA with low friction and high stability. Biophys. J. 2008;95:L01–L03. doi: 10.1529/biophysj.108.134122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKinney K., Mattia M., Gottifredi V., Prives C. p53 linear diffusion along DNA requires its C terminus. Mol. Cell. 2004;16:413–424. doi: 10.1016/j.molcel.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 31.Murata A., Ito Y., Kashima R., Kanbayashi S., Nanatani K., Igarashi C., Okumura M., Inaba K., Tokino T., Takahashi S., et al. One-dimensional sliding of p53 along DNA is accelerated in the presence of Ca(2+) or Mg(2+) at millimolar concentrations. J. Mol. Biol. 2015;427:2663–2678. doi: 10.1016/j.jmb.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 32.Khazanov N., Levy Y. Sliding of p53 along DNA can be modulated by its oligomeric state and by cross-talks between its constituent domains. J. Mol. Biol. 2011;408:335–355. doi: 10.1016/j.jmb.2011.01.059. [DOI] [PubMed] [Google Scholar]
- 33.Terakawa T., Kenzaki H., Takada S. p53 searches on DNA by rotation-uncoupled sliding at C-terminal tails and restricted hopping of core domains. J. Am. Chem. Soc. 2012;134:14555–14562. doi: 10.1021/ja305369u. [DOI] [PubMed] [Google Scholar]
- 34.Tafvizi A., Huang F., Fersht A.R., Mirny L.A., van Oijen A.M. A single-molecule characterization of p53 search on DNA. Proc. Natl. Acad. Sci. USA. 2011;108:563–568. doi: 10.1073/pnas.1016020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leith J.S., Tafvizi A., Huang F., Uspal W.E., Doyle P.S., Fersht A.R., Mirny L.A., van Oijen A.M. Sequence-dependent sliding kinetics of p53. Proc. Natl. Acad. Sci. USA. 2012;109:16552–16557. doi: 10.1073/pnas.1120452109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murata A., Itoh Y., Mano E., Kanbayashi S., Igarashi C., Takahashi H., Takahashi S., Kamagata K. One-dimensional search dynamics of tumor suppressor p53 regulated by a disordered C-terminal domain. Biophys. J. 2017;112:2301–2314. doi: 10.1016/j.bpj.2017.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subekti D.R.G., Murata A., Itoh Y., Fukuchi S., Takahashi H., Kanbayashi S., Takahashi S., Kamagata K. The disordered linker in p53 participates in nonspecific binding to and one-dimensional sliding along DNA revealed by single-molecule fluorescence measurements. Biochemistry. 2017;56:4134–4144. doi: 10.1021/acs.biochem.7b00292. [DOI] [PubMed] [Google Scholar]
- 38.Benichou O., Loverdo C., Moreau M., Voituriez R. Intermittent search strategies. Rev. Modern Phys. 2011;83:81–129. doi: 10.1103/RevModPhys.83.81. [DOI] [PubMed] [Google Scholar]
- 39.Itoh Y., Murata A., Sakamoto S., Nanatani K., Wada T., Takahashi S., Kamagata K. Activation of p53 facilitates the target search in DNA by enhancing the target recognition probability. J. Mol. Biol. 2016;428:2916–2930. doi: 10.1016/j.jmb.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 40.Terakawa T., Takada S. p53 dynamics upon response element recognition explored by molecular simulations. Sci. Rep. 2015;5:17107. doi: 10.1038/srep17107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Itoh Y., Murata A., Takahashi S., Kamagata K. Intrinsically disordered domain of tumor suppressor p53 facilitates target search by ultrafast transfer between different DNA strands. Nucleic Acids Res. 2018;46:7261–7269. doi: 10.1093/nar/gky586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gorman J., Wang F., Redding S., Plys A.J., Fazio T., Wind S., Alani E.E., Greene E.C. Single-molecule imaging reveals target-search mechanisms during DNA mismatch repair. Proc. Natl. Acad. Sci. USA. 2012;109:E3074–E3083. doi: 10.1073/pnas.1211364109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takada S., Kanada R., Tan C., Terakawa T., Li W., Kenzaki H. Modeling Structural Dynamics of Biomolecular Complexes by Coarse-Grained Molecular Simulations. Acc. Chem. Res. 2015;48:3026–3035. doi: 10.1021/acs.accounts.5b00338. [DOI] [PubMed] [Google Scholar]
- 44.Neylon C., Brown S.E., Kralicek A.V., Miles C.S., Love C.A., Dixon N.E. Interaction of the Escherichia coli replication terminator protein (Tus) with DNA: A model derived from DNA-binding studies of mutant proteins by surface plasmon resonance. Biochemistry. 2000;39:11989–11999. doi: 10.1021/bi001174w. [DOI] [PubMed] [Google Scholar]
- 45.Takayama Y., Clore G.M. Impact of protein/protein interactions on global intermolecular translocation rates of the transcription factors Sox2 and Oct1 between DNA cognate sites analyzed by z-exchange NMR spectroscopy. J. Biol. Chem. 2012;287:26962–26970. doi: 10.1074/jbc.M112.382960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spinner D.S., Liu S., Wang S.W., Schmidt J. Interaction of the myogenic determination factor myogenin with E12 and a DNA target: Mechanism and kinetics. J. Mol. Biol. 2002;317:431–445. doi: 10.1006/jmbi.2002.5440. [DOI] [PubMed] [Google Scholar]
- 47.Kim J.G., Takeda Y., Matthews B.W., Anderson W.F. Kinetic studies on Cro repressor-operator DNA interaction. J. Mol. Biol. 1987;196:149–158. doi: 10.1016/0022-2836(87)90517-1. [DOI] [PubMed] [Google Scholar]
- 48.Luo Y., North J.A., Rose S.D., Poirier M.G. Nucleosomes accelerate transcription factor dissociation. Nucleic Acids Res. 2014;42:3017–3027. doi: 10.1093/nar/gkt1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Esadze A., Iwahara J. Stopped-flow fluorescence kinetic study of protein sliding and intersegment transfer in the target DNA search process. J. Mol. Biol. 2014;426:230–244. doi: 10.1016/j.jmb.2013.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carlsson B., Haggblad J. Quantitative determination of DNA-binding parameters for the human estrogen receptor in a solid-phase, nonseparation assay. Anal. Biochem. 1995;232:172–179. doi: 10.1006/abio.1995.0004. [DOI] [PubMed] [Google Scholar]
- 51.Cho S., Wensink P.C. DNA binding by the male and female doublesex proteins of Drosophila melanogaster. J. Biol. Chem. 1997;272:3185–3189. doi: 10.1074/jbc.272.6.3185. [DOI] [PubMed] [Google Scholar]
- 52.Golebiowski F.M., Gorecki A., Bonarek P., Rapala-Kozik M., Kozik A., Dziedzicka-Wasylewska M. An investigation of the affinities, specificity and kinetics involved in the interaction between the Yin Yang 1 transcription factor and DNA. FEBS J. 2012;279:3147–3158. doi: 10.1111/j.1742-4658.2012.08693.x. [DOI] [PubMed] [Google Scholar]
- 53.Saitoh F., Kawamura S., Yamasaki N., Tanaka I., Kimura M. Arginine-55 in the beta-arm is essential for the activity of DNA-binding protein HU from Bacillus stearothermophilus. Biosci. Biotechnol. Biochem. 1999;63:2232–2235. doi: 10.1271/bbb.63.2232. [DOI] [PubMed] [Google Scholar]
- 54.Nakadai T., Shimada M., Shima D., Handa H., Tamura T.A. Specific interaction with transcription factor IIA and localization of the mammalian TATA-binding protein-like protein (TLP/TRF2/TLF) J. Biol. Chem. 2004;279:7447–7455. doi: 10.1074/jbc.M305412200. [DOI] [PubMed] [Google Scholar]
- 55.Wanandi I., Waldschmidt R., Seifart K.H. Mammalian transcription factor PBP. Characterization of its binding properties to the proximal sequence element of U6 genes. J. Biol. Chem. 1993;268:6629–6640. [PubMed] [Google Scholar]
- 56.Kwon H., Park S., Lee S., Lee D.K., Yang C.H. Determination of binding constant of transcription factor AP-1 and DNA. Application of inhibitors. Eur. J. Biochem. 2001;268:565–572. doi: 10.1046/j.1432-1327.2001.01897.x. [DOI] [PubMed] [Google Scholar]
- 57.Okahata Y., Niikura K., Sugiura Y., Sawada M., Morii T. Kinetic studies of sequence-specific binding of GCN4-bZIP peptides to DNA strands immobilized on a 27-MHz quartz-crystal microbalance. Biochemistry. 1998;37:5666–5672. doi: 10.1021/bi980037k. [DOI] [PubMed] [Google Scholar]
- 58.Sugo N., Morimatsu M., Arai Y., Kousoku Y., Ohkuni A., Nomura T., Yanagida T., Yamamoto N. Single-Molecule Imaging Reveals Dynamics of CREB Transcription Factor Bound to Its Target Sequence. Sci. Rep. 2015;5:10662. doi: 10.1038/srep10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Iwahara J., Zweckstetter M., Clore G.M. NMR structural and kinetic characterization of a homeodomain diffusing and hopping on nonspecific DNA. Proc. Natl. Acad. Sci. USA. 2006;103:15062–15067. doi: 10.1073/pnas.0605868103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Iwahara J., Clore G.M. Direct observation of enhanced translocation of a homeodomain between DNA cognate sites by NMR exchange spectroscopy. J. Am. Chem. Soc. 2006;128:404–405. doi: 10.1021/ja056786o. [DOI] [PubMed] [Google Scholar]
- 61.Takayama Y., Sahu D., Iwahara J. NMR studies of translocation of the Zif268 protein between its target DNA Sites. Biochemistry. 2010;49:7998–8005. doi: 10.1021/bi100962h. [DOI] [PubMed] [Google Scholar]
- 62.Takayama Y., Clore G.M. Interplay between minor and major groove-binding transcription factors Sox2 and Oct1 in translocation on DNA studied by paramagnetic and diamagnetic NMR. J. Biol. Chem. 2012;287:14349–14363. doi: 10.1074/jbc.M112.352864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Esadze A., Kemme C.A., Kolomeisky A.B., Iwahara J. Positive and negative impacts of nonspecific sites during target location by a sequence-specific DNA-binding protein: Origin of the optimal search at physiological ionic strength. Nucleic Acids Res. 2014;42:7039–7046. doi: 10.1093/nar/gku418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Giuntoli R.D., Linzer N.B., Banigan E.J., Sing C.E., de la Cruz M.O., Graham J.S., Johnson R.C., Marko J.F. DNA-segment-facilitated Dissociation of Fis and NHP6A from DNA detected via single-molecule mechanical response. J. Mol. Biol. 2015;427:3123–3136. doi: 10.1016/j.jmb.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Y.V., Wade M., Wong E., Li Y.C., Rodewald L.W., Wahl G.M. Quantitative analyses reveal the importance of regulated Hdmx degradation for P53 activation. Proc. Natl. Acad. Sci. USA. 2007;104:12365–12370. doi: 10.1073/pnas.0701497104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu M., Ye H., Tang Z., Shao C., Lu G., Chen B., Yang Y., Wang G., Hao H. p53 dynamics orchestrates with binding affinity to target genes for cell fate decision. Cell Death Dis. 2017;8:e3130. doi: 10.1038/cddis.2017.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kawaguchi T., Kato S., Otsuka K., Watanabe G., Kumabe T., Tominaga T., Yoshimoto T., Ishioka C. The relationship among p53 oligomer formation, structure and transcriptional activity using a comprehensive missense mutation library. Oncogene. 2005;24:6976–6981. doi: 10.1038/sj.onc.1208839. [DOI] [PubMed] [Google Scholar]
- 68.Rajagopalan S., Huang F., Fersht A.R. Single-Molecule characterization of oligomerization kinetics and equilibria of the tumor suppressor p53. Nucleic Acids Res. 2011;39:2294–2303. doi: 10.1093/nar/gkq800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fischer N.W., Prodeus A., Malkin D., Gariepy J. p53 oligomerization status modulates cell fate decisions between growth, arrest and apoptosis. Cell Cycle. 2016;15:3210–3219. doi: 10.1080/15384101.2016.1241917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gaglia G., Guan Y., Shah J.V., Lahav G. Activation and control of p53 tetramerization in individual living cells. Proc. Natl. Acad. Sci. USA. 2013;110:15497–15501. doi: 10.1073/pnas.1311126110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weinberg R.L., Veprintsev D.B., Fersht A.R. Cooperative binding of tetrameric p53 to DNA. J. Mol. Biol. 2004;341:1145–1159. doi: 10.1016/j.jmb.2004.06.071. [DOI] [PubMed] [Google Scholar]
- 72.Beckerman R., Prives C. Transcriptional regulation by p53. Cold Spring Harb. Perspect. Biol. 2010;2:a000935. doi: 10.1101/cshperspect.a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bieging K.T., Mello S.S., Attardi L.D. Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Canc. 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hamard P.J., Lukin D.J., Manfredi J.J. p53 basic C terminus regulates p53 functions through DNA binding modulation of subset of target genes. J. Biol. Chem. 2012;287:22397–22407. doi: 10.1074/jbc.M111.331298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hamard P.J., Barthelery N., Hogstad B., Mungamuri S.K., Tonnessen C.A., Carvajal L.A., Senturk E., Gillespie V., Aaronson S.A., Merad M., et al. The C terminus of p53 regulates gene expression by multiple mechanisms in a target- and tissue-specific manner in vivo. Genes Dev. 2013;27:1868–1885. doi: 10.1101/gad.224386.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marouco D., Garabadgiu A.V., Melino G., Barlev N.A. Lysine-specific modifications of p53: A matter of life and death? Oncotarget. 2013;4:1556–1571. doi: 10.18632/oncotarget.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Laptenko O., Shiff I., Freed-Pastor W., Zupnick A., Mattia M., Freulich E., Shamir I., Kadouri N., Kahan T., Manfredi J., et al. The p53 C terminus controls site-specific DNA binding and promotes structural changes within the central DNA binding domain. Mol. Cell. 2015;57:1034–1346. doi: 10.1016/j.molcel.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Retzlaff M., Rohrberg J., Kupper N.J., Lagleder S., Bepperling A., Manzenrieder F., Peschek J., Kessler H., Buchner J. The regulatory domain stabilizes the p53 tetramer by intersubunit contacts with the DNA binding domain. J. Mol. Biol. 2013;425:144–155. doi: 10.1016/j.jmb.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 79.Friedler A., Veprintsev D.B., Freund S.M., von Glos K.I., Fersht A.R. Modulation of binding of DNA to the C-terminal domain of p53 by acetylation. Structure. 2005;13:629–636. doi: 10.1016/j.str.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 80.Loffreda A., Jacchetti E., Antunes S., Rainone P., Daniele T., Morisaki T., Bianchi M.E., Tacchetti C., Mazza D. Live-cell p53 single-molecule binding is modulated by C-terminal acetylation and correlates with transcriptional activity. Nat. Commun. 2017;8:313. doi: 10.1038/s41467-017-00398-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mazza D., Abernathy A., Golob N., Morisaki T., McNally J.G. A benchmark for chromatin binding measurements in live cells. Nucleic Acids Res. 2012;40:e119. doi: 10.1093/nar/gks701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Morisaki T., Muller W.G., Golob N., Mazza D., McNally J.G. Single-molecule analysis of transcription factor binding at transcription sites in live cells. Nat. Commun. 2014;5:4456. doi: 10.1038/ncomms5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kanada R., Terakawa T., Kenzaki H., Takada S. Nucleosome Crowding in Chromatin Slows the Diffusion but Can Promote Target Search of Proteins. Biophys. J. 2019;116:2285–2295. doi: 10.1016/j.bpj.2019.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Visnapuu M.L., Greene E.C. Single-molecule imaging of DNA curtains reveals intrinsic energy landscapes for nucleosome deposition. Nat. Struct. Mol. Biol. 2009;16:1056–1062. doi: 10.1038/nsmb.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jagelska E.B., Brazda V., Pecinka P., Palecek E., Fojta M. DNA topology influences p53 sequence-specific DNA binding through structural transitions within the target sites. Biochem. J. 2008;412:57–63. doi: 10.1042/BJ20071648. [DOI] [PubMed] [Google Scholar]
- 86.Jagelska E.B., Pivonkova H., Fojta M., Brazda V. The potential of the cruciform structure formation as an important factor influencing p53 sequence-specific binding to natural DNA targets. Biochem. Biophys. Res. Commun. 2010;391:1409–1414. doi: 10.1016/j.bbrc.2009.12.076. [DOI] [PubMed] [Google Scholar]
- 87.Coufal J., Jagelska E.B., Liao J.C., Brazda V. Preferential binding of p53 tumor suppressor to p21 promoter sites that contain inverted repeats capable of forming cruciform structure. Biochem. Biophys. Res. Commun. 2013;441:83–88. doi: 10.1016/j.bbrc.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 88.Brazda V., Cechova J., Battistin M., Coufal J., Jagelska E.B., Raimondi I., Inga A. The structure formed by inverted repeats in p53 response elements determines the transactivation activity of p53 protein. Biochem. Biophys. Res. Commun. 2017;483:516–521. doi: 10.1016/j.bbrc.2016.12.113. [DOI] [PubMed] [Google Scholar]
- 89.Brazda V., Coufal J. Recognition of Local DNA Structures by p53 Protein. Int. J. Mol. Sci. 2017;18:375. doi: 10.3390/ijms18020375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brazda V., Fojta M. The Rich World of p53 DNA Binding Targets: The Role of DNA Structure. Int. J. Mol. Sci. 2019;20:5605. doi: 10.3390/ijms20225605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Garcia-Alai M.M., Tidow H., Natan E., Townsley F.M., Veprintsev D.B., Fersht A.R. The novel p53 isoform “delta p53” is a misfolded protein and does not bind the p21 promoter site. Protein Sci. 2008;17:1671–1678. doi: 10.1110/ps.036996.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sauer M., Bretz A.C., Beinoraviciute-Kellner R., Beitzinger M., Burek C., Rosenwald A., Harms G.S., Stiewe T. C-terminal diversity within the p53 family accounts for differences in DNA binding and transcriptional activity. Nucleic Acids Res. 2008;36:1900–1912. doi: 10.1093/nar/gkn044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Meek D.W., Anderson C.W. Posttranslational modification of p53: Cooperative integrators of function. Cold Spring Harb. Perspect. Biol. 2009;1:a000950. doi: 10.1101/cshperspect.a000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Botchkarev V.A., Flores E.R. p53/p63/p73 in the epidermis in health and disease. Cold Spring Harb. Perspect. Med. 2014;4:a015248. doi: 10.1101/cshperspect.a015248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nguyen T.A., Menendez D., Resnick M.A., Anderson C.W. Mutant TP53 posttranslational modifications: Challenges and opportunities. Hum. Mutat. 2014;35:738–755. doi: 10.1002/humu.22506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bonnet I., Biebricher A., Porte P.-L., Loverdo C., Benichou O., Voituriez R., Escude C., Wende W., Pingoud A., Desbiolles P. Sliding and jumping of single EcoRV restriction enzymes on non-cognate DNA. Nucleic Acids Res. 2008;36:4118–4127. doi: 10.1093/nar/gkn376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Aramayo R., Sherman M.B., Brownless K., Lurz R., Okorokov A.L., Orlova E.V. Quaternary structure of the specific p53-DNA complex reveals the mechanism of p53 mutant dominance. Nucleic Acids Res. 2011;39:8960–8971. doi: 10.1093/nar/gkr386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kearns S., Lurz R., Orlova E.V., Okorokov A.L. Two p53 tetramers bind one consensus DNA response element. Nucleic Acids Res. 2016;44:6185–6199. doi: 10.1093/nar/gkw215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Martin T.G., Bharat T.A., Joerger A.C., Bai X.C., Praetorius F., Fersht A.R., Dietz H., Scheres S.H. Design of a molecular support for cryo-EM structure determination. Proc. Natl. Acad. Sci. USA. 2016;113:E7456–E7463. doi: 10.1073/pnas.1612720113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kamagata K., Mano E., Itoh Y., Wakamoto T., Kitahara R., Kanbayashi S., Takahashi H., Murata A., Kameda T. Rational design using sequence information only produces a peptide that binds to the intrinsically disordered region of p53. Sci. Rep. 2019;9:8584. doi: 10.1038/s41598-019-44688-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang F., Redding S., Finkelstein I.J., Gorman J., Reichman D.R., Greene E.C. The promoter-search mechanism of Escherichia coli RNA polymerase is dominated by three-dimensional diffusion. Nat. Struct. Mol. Biol. 2013;20:174–181. doi: 10.1038/nsmb.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sternberg S.H., Redding S., Jinek M., Greene E.C., Doudna J.A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nelson S.R., Dunn A.R., Kathe S.D., Warshaw D.M., Wallace S.S. Two glycosylase families diffusively scan DNA using a wedge residue to probe for and identify oxidatively damaged bases. Proc. Natl. Acad. Sci. USA. 2014;111:E2091–E2099. doi: 10.1073/pnas.1400386111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cuculis L., Abil Z., Zhao H., Schroeder C.M. TALE proteins search DNA using a rotationally decoupled mechanism. Nat. Chem. Biol. 2016;12:831–837. doi: 10.1038/nchembio.2152. [DOI] [PubMed] [Google Scholar]
- 105.Kamagata K., Mano E., Ouchi K., Kanbayashi S., Johnson R.C. High Free-Energy Barrier of 1D Diffusion Along DNA by Architectural DNA-Binding Proteins. J. Mol. Biol. 2018;430:655–667. doi: 10.1016/j.jmb.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]