Abstract
Expression systems for highly toxic protein genes must be conditional and suppress leakage expression to almost zero because even faint leakage expression may kill host cells, inhibit host growth, and cause loss of plasmids containing the toxic protein genes. The most widely used conditional expression systems are controlled only at the transcriptional level, and complete suppression of leakage expression is challenging. Recent progress on translational control has enabled construction of dual transcriptional-translational control systems in which leakage expression is strongly suppressed. This review summarizes the principles, features, and practical examples of dual transcriptional-translational control systems in bacteria, and provides future perspectives on these systems.
Keywords: tight control of gene expression, unnatural amino acid incorporation, riboswitch, ribozyme, STAR, antisense RNA, translational regulation, transcriptional regulation
1. Introduction
Heterologous gene expression is a fundamental technique for the production of recombinant proteins. A variety of bacteria are used as hosts for heterologous gene expression [1,2,3,4]. A problem with this technique is that about 50% of artificially expressed proteins are toxic to the host bacteria, even in self-gene overexpression [5,6]. This toxicity inhibits host bacterial growth and often kills host bacteria. In addition, loss of plasmids containing the toxic protein genes frequently occurs. To overcome this problem, conditional expression systems, in which toxic proteins are produced only in the presence of inducers, are widely used [7]. In the proliferative stage of host bacteria, the growth of bacteria is not affected because the production of toxic proteins is suppressed in the absence of inducers. After sufficient bacterial growth, the toxic proteins can be produced in the presence of inducers. However, the switchability of most “biological” conditional expression systems is not as perfectly tight as that of electronic switches. This means a small, but problematic, number of proteins are produced, even in the absence of inducers (leakage expression) [8]. Highly toxic proteins affect the growth rate and viability of host bacteria, even with faint leakage expression. Therefore, complete suppression of leakage expression is essential for production of active highly toxic proteins. However, this is a challenging problem.
Dual transcriptional-translational control, in which heterologous gene expression is regulated at the transcriptional and translational levels, has recently been developed to solve this problem in bacteria. In this review, the principles, features, and recent practical examples of dual transcriptional-translational control systems are described, together with a discussion of future perspectives on these systems.
2. Advantages of Dual Transcriptional-Translational Control
The most widely-used inducible expression systems are regulated at the transcriptional level only. In these systems, transcription is activated by an inducer, such as a specific chemical or physical stimulus. The operator/repressor system derived from the lactose operon in Escherichia coli (lacO/I), which is activated by lactose or its persistent analog, isopropyl β-D-1-thiogalactopyranoside (IPTG), is a classic example [9,10]. LacO/I continues to be used widely, but the level of leakage expression is greatly affected by promoter selection. Leakage expression of the native lactose promoter (Plac) and lacO/I pair is about 0.1% in an optimal setting, but reaches 2% in combination with Ptac, a stronger derivative of Plac, indicating insufficient tightness for expression of highly toxic genes [11]. The operator/repressor system derived from the arabinose operon (araO/C) in Escherichia coli is a widely used expression system that can tightly regulate transcription of a target gene [12]. Leakage expression of araO/C is <0.1%. However, a moderately strong promoter derived from the araBAD operon (PBAD) and an araO/C-regulated expression construct for the colicin E3 enzymatic domain (ColE3e), of which a few molecules can kill E. coli by cleaving 16S rRNA, cannot be maintained in a multicopy plasmid, indicating that the leakage level is too high [13,14,15]. Complete suppression of leakage transcription is still challenging despite the effort put into the development of tight transcriptional regulatory systems that permit functional expression of highly toxic proteins. Several conditional expression systems that work at the translational level have been reported, as described below, but complete suppression of leakage translation is also challenging in these systems.
Dual transcription-translation control provides a novel scaffold on which to construct an extremely tight expression system in which leakage expression is suppressed to the minimum. In conventional systems that regulate expression only at the transcriptional level, all leakage-expressed mRNAs are translated to toxic proteins; production of these mRNAs therefore must be zero to suppress the toxic protein production completely. In contrast, leakage mRNAs are faintly translated with additional translational suppression, and production of toxic proteins can be completely suppressed, even if transcription of the toxic protein gene is not perfectly suppressed. In other words, dual transcriptional-translational control may achieve extreme suppression of leakage expression, even if the transcriptional or translational regulation is leaky. In the following section, several dual transcriptional-translational control systems are described based on the order of the publication date.
3. Site-Specific Unnatural Amino Acid Incorporation
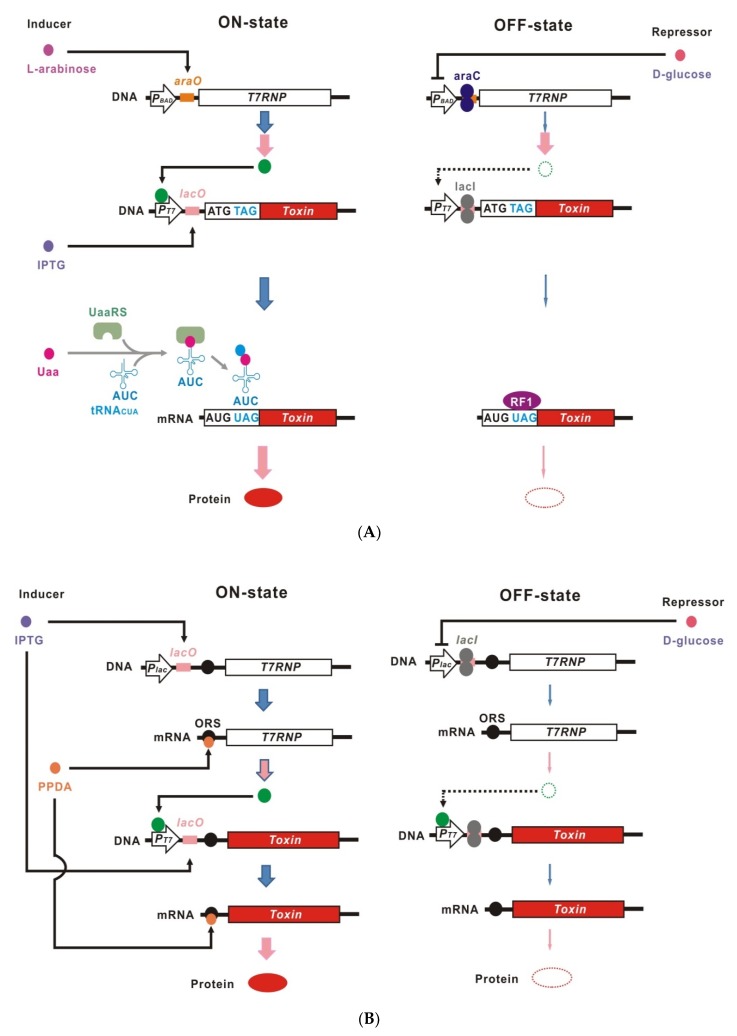
Incorporation of a site-specific unnatural amino acid (Uaa) into ribosomally synthesized proteins in vivo at a position encoded by an amber stop codon [16,17,18,19,20] was originally developed for structural analysis, labeling, chemical ligation, and functional modification of proteins through the replacement of canonical natural amino acids [16,17,18]. Site-specific Uaa incorporation can also be used to control translation of target mRNA (Figure 1A) [21,22,23,24]. Typically, amber stop codons are inserted near the translation initiation site of the coding region of target genes. Genes encoding the UAA-specific aminoacyl tRNA synthetase (UaaRS) and the cognate tRNACUA are also introduced into the host bacteria. Once the Uaa is provided in the culture medium, it is taken up into the intracellular space and then incorporated in proteins at the inserted amber stop codons, causing full-length translation of target mRNAs by amber stop codon read through (ON-state). In the absence of Uaa, translation is interrupted at the inserted amber stop codons, resulting in inhibition of functional target protein production (OFF-state).
Figure 1.
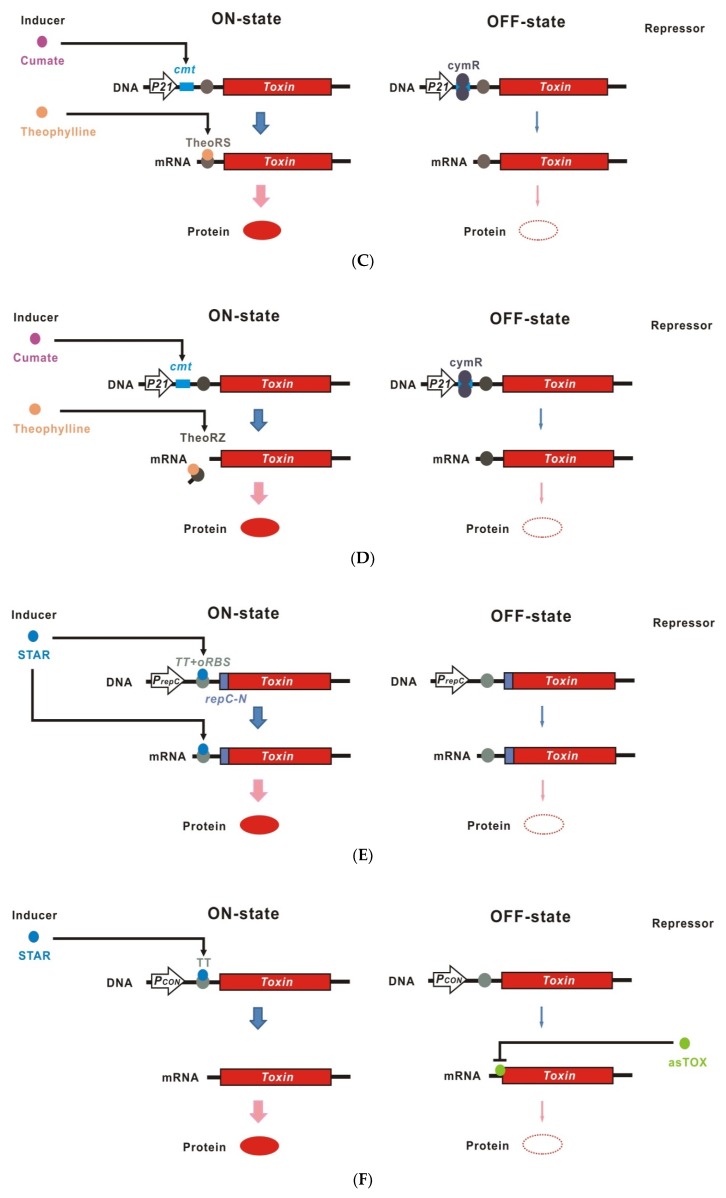
Architecture of dual transcriptional-translational control systems. Blue and pink arrows indicate transcription and translation, respectively. (A) HYZEL. RF1, peptide release factor 1. (B) RiboTite(tT/tT). (C) cymO/R and theophylline riboswitch control. P21, synthetic P21 promoter. cmt, operator sequence of the cumate degradation operon. CymR, CymR regulator. TheoRS, theophylline riboswitch. (D) cymO/R and theophylline ribozyme control. TheoRZ, theophylline ribozyme. (E) Transcriptional terminator and occluded RBS control. PrepC, promoter regulating repC. TT + oRBS, transcriptional terminator and occluded RBS. repC-N, first 12 nucleotides of repC. (F) Transcriptional terminator and anti-toxin mRNA antisense RNA control. PCON, a constitutive promoter. TT, transcriptional terminator. asTOX, antisense RNA against the RBS-start codon region of toxin gene mRNA.
Translational control using site-specific Uaa incorporation can regulate translation in an all-or-nothing manner, and also at any intermediate magnitude by adjustment of the Uaa concentration in the medium [22]. The EC90/EC10 ratio is 55 in translational control using a 3-iodo-L-tyrosine (IY) incorporation system derived from the tyrosyl-RS/tRNA pair of the archaeon Methanocaldococus jannaschii. Such intermediate-level expression is due to a uniform response of each individual cell, rather than changes in the population averages of induced and non-induced cells.
The level of leakage translation varies among Uaa incorporation systems. For instance, leakage translation is only about 1% in the Nε-benzyloxycarbonyl-L-lysine (ZK) system, which was developed from pyrrolysyl-tRNA synthetase PylS and its cognate tRNACUA PylT of the archaeon Methanosarcina mazei, whereas that of the IY system is 6%–25% [21,22,23,25,26]. Various methods and settings can suppress the level of leakage translation. Lower expression of UaaRS and tRNACUA suppresses leakage translation, but yield also decreases when expression is too low [21]. Positive-feedback regulation of UaaRS and tRNACUA was recently developed to suppress leakage translation [23], through a mechanism in which sufficient amounts of UaaRS and tRNACUA are supplied in the ON-state, whereas expression of these genes is suppressed in the OFF-state. Such positive-feedback regulation enables lower leakage translation without a severe loss of yield. The IY system equipped with positive-feedback regulation achieved a gain (expression in the ON-state/leakage expression) of 1.4 × 103 (synonymous with 0.07% leakage expression), which was 3 × 102-fold higher than that of the parent system. This gain is comparable to that with araO/C [12]. Leakage translation is also suppressed through multiplexing of inserted amber stop codons, despite a loss of yield [21]. Some other methods have been proposed to suppress leakage translation, but experimental evidence is yet to be provided [24].
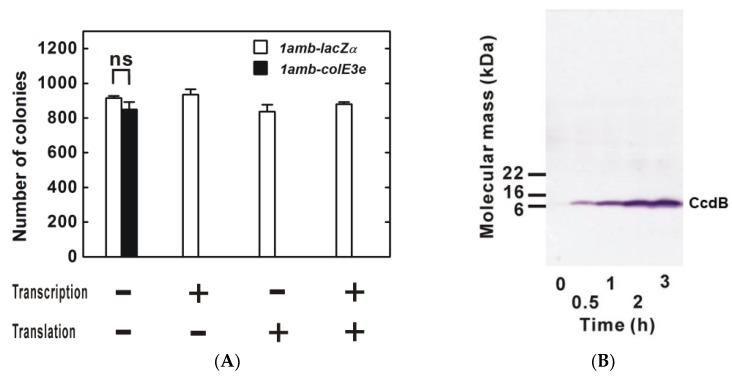
In 2014, the HYZEL (High-Yield and ZEro-Leakage) dual transcription-translation control expression system using Uaa incorporation was reported in E. coli (Figure 1A) [21]. In HYZEL, transcription of a toxic gene is controlled through a cascade under the T7 promoter (PT7) with lacO/I in the host bacterium BL21-AI, in which T7 RNA polymerase (T7RNP) is conditionally expressed under the control of PBAD and araO/C. The recombinant toxic protein is produced in the presence of L-arabinose, IPTG and Uaa (ON-state), whereas production is suppressed in the absence of these inducers and the presence of D-glucose, which causes catabolite suppression (OFF-state). HYZEL with translation of the toxic protein controlled by ZK incorporation tolerated an expression construct for ColE3e containing a single amber stop codon insertion, which suggests that leakage expression is almost zero (Kato Y, unpublished data, Figure 2A), while the yield of recombinant protein was not affected by insertion of 1 or 2 amber stop codons [21]. In addition, the DNA gyrase inhibitor, CcdB, a well-known toxic protein to E. coli, has been successfully produced using HYZEL (Kato Y, unpublished data, Figure 2B).
Figure 2.
Production of highly-toxic proteins using HYZEL. (A) Maintenance of ColE3e expression construct. The expression construct for the ColE3e gene, which contains a single amber stop codon insertion, was introduced into E. coli BL21-AI with another plasmid that constitutively expresses the specific UaaRS for ZK and its cognate tRNACUA. A V5-LacZ with a gene containing an amber stop codon insertion was used as non-toxic protein control. Leakage expression of ColE3e killed the host bacteria using single repression at the transcriptional or translational level. In contrast, the host bacteria survived in dual transcriptional-translational repression. Data are shown as mean ± s.e.m. of three biological replicates. Statistical analyses were performed using single-factor analysis of variance (ANOVA) with an α of 0.05 (ns, not significant). (B) Production of the DNA gyrase inhibitor, CcdB. CcdB with a V5 epitope tag added at the N-terminus was produced using HYZEL. An amber stop codon was inserted next to the translation start codon in the V5 epitope tag. The V5-CcdB expression construct driven by T7RNP was cotransformed into BL21-AI with a plasmid that constitutively expressed the specific UaaRS for IY and its cognate tRNACUA. Bacteria carrying these plasmids were cultured overnight in LB medium containing D-glucose for catabolite repression against PBAD-araC/O, which regulates T7RNP gene expression. V5-CcdB production was induced by changing the medium containing IY, L-arabinose and IPTG. V5-CcdB production was shown by western blot using an anti-V5 antibody. Time after the medium change is shown below the photo.
A drawback of HYZEL is that Uaa must be incorporated in the recombinant proteins, causing an unintended modification of the native amino acid sequence. This problem may be solved by using a Uaa designed to be incorporated in N-terminal tags or signal sequences that are removed after natural or artificial processing [21]. An alternative approach is use of a Uaa that can be converted into a natural amino acid by a chemical or biological process after incorporation into proteins [27,28].
4. Riboswitches
A riboswitch is a cis-regulatory element of mRNA that is usually located in the 5′ untranslated region (5′UTR) [29,30,31]. Binding of a specific ligand to the binding region of a riboswitch (aptamer domain) induces a conformational change that alters gene expression at the transcriptional or translational level. The Add-A translational ON riboswitch is an adenine-sensing riboswitch found in the bacterium Vibrio vulnificus [32]. In the absence of adenine, the ribosome-binding site (RBS) of mRNA is occluded by formation of a repressor stem in the riboswitch. In contrast, binding of adenine to the aptamer domain induces a conformational change in the riboswitch, resulting in initiation of translation by release of the RBS. A modified riboswitch, which is specifically controlled by the unnatural ligand pyrimido[4,5-d]pyrimidine-2,4-diamine (PPDA), has also been constructed and is referred to as the PPDA orthogonal riboswitch (PPDA-ORS) [33,34,35].
RiboTite is a tight conditional expression system that uses a combination of PT7-lacO/I controlled transcriptional regulation and PPDA-ORS-controlled translational regulation, as first reported in 2015 (Figure 1B) [36,37]. RiboTite is a cascade-regulatory system, similarly to HYZEL. In the tightest system among several variations of RiboTite, named tT/tT, the T7RNP gene is regulated by lacO/I at the transcriptional level and by PPDA-ORS at the translational level. A recombinant protein gene is transcribed by T7RNP and regulated by lacO/I and PPDA-ORS. tT/tT achieved low leakage expression (0.1%) and high gain (850-fold), suggesting that tightness was increased by >240-fold by addition of PPDA-ORS regulation, whereas the volumetric yield decreased by about 60%. In addition, the EC90/EC10 ratio increased from 10 in the parent system lacking PPDA-ORS regulation to 245 in tT/tT. Using tT/tT, the type II-like bacteriocin, epidermicin NI01 from Staphylococcus epidermidis, which has toxicity against E. coli in the cytoplasm, was produced at 2-fold higher than in the parent system [36,38]. Moreover, an expression construct of the toxic gene encoding levansucrase (sacB) was safely maintained in a modified tT/tT that omitted PPDA-ORS regulation of a recombinant protein gene, tT/t, in the presence of 5% sucrose, in which SucB had toxicity [36,39,40].
A dual transcriptional-translational control similar to RiboTite was independently developed in Actinobacteria and reported in 2016 (Figure 1C) [41]. This system has transcriptional regulation using cymO/R, an operator-repressor system that is derepressed by cumate (p-isopropylbenzoic acid), and translational regulation using a theophylline riboswitch, which regulates translation of mRNA by masking and releasing of the RBS, as well as an Add-A translational ON riboswitch [42,43]. The dual cymO/R-theophylline riboswitch control achieved a gain of 20- to 25-fold over that of a single control using a theophylline riboswitch only. No activity was detected in a leakage expression test using a gusA reporter, suggesting that this dual control is highly tight. Using dual cymO/R-theophylline riboswitch control, production of the macrodiolide antibiotic pamamycin, which is extremely toxic to streptomycetes, was successful in Streptomyces albus by tight control of pamJ, which encodes an essential enzyme for pamamycin biosynthesis [44,45]. Dual control using the operator-repressor system rolR/O, which is derived from the Corynebacterium glutamicum resorcinol catabolic operon, and the theophylline riboswitch has also been reported [41].
5. Ribozymes
Ribozymes are RNAs with catalytic activities [46,47], and some ribozymes are activated by binding of specific ligands. Theophylline ribozyme is a hammerhead ribozyme that recognizes and site-specifically digests a specific RNA sequence [48], and is activated in the presence of theophylline. Gene expression can be controlled at the translational level using this ribozyme. This control element, in which a complementary sequence to the RBS inhibits translation and the theophylline ribozyme, is inserted in the 5′UTR near the translation initiation site. Theophylline ribozyme removes this control element by self-digestion in the presence of theophylline, resulting in release of the RBS and translation initiation of mRNA. A dual control system using cumO/R and theophylline ribozyme was reported in 2016 (Figure 1D) [41]. No leakage expression was detected using a gusA expression construct regulated by this dual control system.
6. Antisense RNA
Antisense RNAs can regulate both transcription and translation in trans [49,50,51,52,53]. A dual transcriptional-translational control system regulated by a single antisense RNA was reported in 2017 (Figure 1E) [54]. This system was designed based on the regulatory element of the plasmid replication protein RepC in plasmid pT181 in the bacterium Staphylococcus aureus [55]. A transcriptional terminator is located in the 5′UTR near the translation initiation site of repC. Formation of the transcriptional terminator also inhibits translation of repC because the RBS is located in the terminator hairpin, similarly to the toehold switch, which suggests that terminator hairpin formation is a natural dual transcriptional-translational control element [51]. A specific controller antisense RNA, small transcription activating RNA (STAR), interrupts hairpin formation [50]. Expression of a recombinant protein gene with its coding sequence fused to the first 12 nucleotides of repC is controlled at the transcriptional and translational levels by a single regulator STAR. This dual control system achieved <0.1% leakage expression and a 923-fold gain.
A dual control system with antisense RNAs was reported in 2018, using combined STAR-regulated transcriptional control and antisense RNA-regulated translational control (Figure 1F) [56]. STAR and the antisense RNA are provided from transgenes regulated by a 3OC6-inducible PLux promoter and a tetracycline-inducible PTet promoter, respectively. In the ON-state, only expression of STAR is induced. In contrast, expression of antisense RNAs against STAR and the RBS and start codon is induced in the OFF-state, resulting in inhibition of transcription induction and translation initiation. With the optimal setting, 4% leakage expression was recorded for this system.
7. Conclusions
Dual transcriptional-translational control is a powerful approach for achieving a minimal leakage system for the expression of toxic proteins. Some applications, such as ColE3e and CcdB in HYZEL, SucB and epidermicin in RiboTite, and pamamycin in the dual cumO/R and theophylline riboswitch control system, clearly support the potency of this approach. Dual control is also promising for other purposes in which low leakage is desired, such as switching of enzyme gene expression in metabolic engineering and biocomputing in synthetic biology [57,58]. For example, precise expression control of Cre recombinase, which was achieved using the dual cumO/R and theophyllin riboswitch control system, is useful both for genome editing and for non-volatile memory in biocomputing [41]. In addition, most dual control systems contain many regulatory factors, suggesting that Boolean multi-input logic gates can be constructed for integration of environmental signals in synthetic gene circuits [21,36,41,56]. However, such multi-regulator systems require many regulatory elements for control of a single target gene expression. This is a possible drawback because the number of well-characterized regulatory elements is limited, even in E. coli. Dual transcription-translation control using a single regulatory element may be an ideal solution to this problem (Figure 1E) [56].
Other forms of post-transcriptional regulation, such as mRNA degradation and antisense RNA against protein coding regions, may also be used to construct a dual or multi-control system [59,60]. Moreover, some methods have also been used to suppress leakage expression without improving the tightness of transcription or translation, such as inhibition of specific RNA polymerase activity in the OFF-state (e.g., lysozyme inhibition of T7RNP), phage delivery of a specific RNA polymerase after sufficient bacterial growth, and an adjustable plasmid copy number [5,61,62]. Construction of further improved tight control systems is likely to be achieved using multi-layered mechanisms working at different biological levels.
In conclusion, multi-layer control, such as the use of the dual transcriptional-translational control approaches described here, is a rational method to achieve tight regulation. This is the state-of the-art for building a reliable biological regulator out of unreliable components, indicating that the fuzziness of biological elements may be overcome to achieve a finely controlled biological system that behaves like an electronic device.
Funding
This research was funded by JSPS grant number JP16K08121.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Terpe K. Overview of bacterial expression systems for heterologous protein production: From molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 2006;72:211–222. doi: 10.1007/s00253-006-0465-8. [DOI] [PubMed] [Google Scholar]
- 2.Dermain A., Vaishnav P. Production of recombinant proteins by microbes and higher organisms. Biotechnol. Adv. 2009;27:297–306. doi: 10.1016/j.biotechadv.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 3.Chen R. Bacterial expression systems for recombinant protein production. E. coli and beyond. Biotechnol. Adv. 2012;30:1102–1107. doi: 10.1016/j.biotechadv.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 4.Ferrer-Miralles N., Villaverde A. Bacterial cell factories for recombinant protein production; expanding the catalogue. Microb. Cell Fact. 2013;12:113. doi: 10.1186/1475-2859-12-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saïda F., Uzan M., Odaert B., Bontems F. Expression of highly toxic genes in E. coli: Special strategies and genetic tools. Curr. Prot. Pept. Sci. 2006;7:47–56. doi: 10.2174/138920306775474095. [DOI] [PubMed] [Google Scholar]
- 6.Kitagawa M., Ara T., Arifuzzaman M., Ioka-Nakamichi T., Inamoto E., Toyonaga H., Mori H. Complete set of ORF clones of Escherichia coli ASKA library (A Complete S et of E. coli K -12 ORF A rchive): Unique Resources for Biological Research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 7.Giacalone M.J., Gentitle A.M., Lovitt B.T., Berkly N.L., Gunderson C.W., Surber M.W. Toxic protein expression in Escherichia coli using a rhamnose-based tightly regulated and tunable promoter system. BioTechniques. 2006;40:355–364. doi: 10.2144/000112112. [DOI] [PubMed] [Google Scholar]
- 8.Ellefson J.W., Meyer A.J., Hughes R.A., Cannon J.R., Brodbelt J.S., Ellington A.D. Directed evolution of genetic parts and circuits by compartmentalized partnered replication. Nat. Biotechnol. 2014;32:97–101. doi: 10.1038/nbt.2714. [DOI] [PubMed] [Google Scholar]
- 9.Jacob F., Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961;3:318–356. doi: 10.1016/S0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 10.Du M., Kodner S., Bai L. Enhancement of LacI binding in vivo. Nucl. Acids Res. 2019;47:9609–9618. doi: 10.1093/nar/gkz698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lanzer M., Bujard H. Promoters largely determine the efficiency of repressor action. Proc. Natl. Acad. Sci. USA. 1988;85:8973–8977. doi: 10.1073/pnas.85.23.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guzman L., Belin D., Carson M.J., Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/JB.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masaki H., Ohta T. Colicin E3 and its immunity genes. J. Mol. Biol. 1985;182:217–227. doi: 10.1016/0022-2836(85)90340-7. [DOI] [PubMed] [Google Scholar]
- 14.Lazzaroni J.-C., Dubuisson J.-F., Vianney A. The Tol proteins of Escherichia coli and their involvement in the translocation of group A colicins. Biochimie. 2002;84:391–397. doi: 10.1016/S0300-9084(02)01419-0. [DOI] [PubMed] [Google Scholar]
- 15.Anthony L.C., Suzuki H., Filutowicz M. Tightly regulated vectors for the cloning and expression of toxic genes. J. Microbiol. Meth. 2004;58:243–250. doi: 10.1016/j.mimet.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Liu C.C., Schultz P.G. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 17.Xiao H., Schultz P.G. At the interface of chemical and biological synthesis: An expanded genetic code. CSH Perspect. Biol. 2016;8:a023945. doi: 10.1101/cshperspect.a023945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chin J.W. Expanding and reprogramming the genetic code. Nature. 2017;550:53–60. doi: 10.1038/nature24031. [DOI] [PubMed] [Google Scholar]
- 19.Mukai T., Lajoie M.J., Englert M., Söll D. Rewriting the genetic code. Annu. Rev. Microbiol. 2017;71:557–577. doi: 10.1146/annurev-micro-090816-093247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terasaka N., Iwane Y., Geiermann A.S., Goto Y., Suga H. Recent developments of engineered translational machineries for the incorporation of non-canonical amino acids into polypeptides. Int. J. Mol. Sci. 2015;16:6513–6531. doi: 10.3390/ijms16036513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minaba M., Kato Y. High-yield, zero-leakage expression system with a translational switch using site-specific unnatural amino acid incorporation. Appl. Environ. Microbiol. 2014;80:1718–1725. doi: 10.1128/AEM.03417-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kato Y. Tunable translational control using site-specific unnatural amino acid incorporation in Escherichia coli. PeerJ. 2015;3:e904. doi: 10.7717/peerj.904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato Y. Tight translational control using site-specific unnatural amino acid incorporation with positive feedback gene circuits. ACS Synth. Biol. 2018;7:1956–1963. doi: 10.1021/acssynbio.8b00204. [DOI] [PubMed] [Google Scholar]
- 24.Kato Y. Translational Control using an Expanded Genetic Code. Int. J. Mol. Sci. 2019;20:887. doi: 10.3390/ijms20040887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakamoto K., Murayama K., Oki K., Iraha F., KatoMurayama M., Takahashi M., Ohtake K., Kobayashi T., Kuramitsu S., Shirouzu M., et al. Genetic encoding of 3-iodo-L-tyrosine in Escherichia coli for single-wavelength anomalous dispersion phasing in protein crystallography. Structure. 2009;17:335–344. doi: 10.1016/j.str.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 26.Yanagisawa T., Ishii R., Fukunaga R., Kobayashi T., Sakamoto K., Yokoyama S. Multistep engineering of pyrrolysyl-tRNA synthetase to genetically encode Nε-(o-Azidobenzyloxycarbonyl)lysine for site-specific protein modification. Chem. Biol. 2008;15:1187–1197. doi: 10.1016/j.chembiol.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 27.Xuan W., Schultz P.G. A strategy for creating organisms dependent on noncanonical amino acids. Angew. Chem. Int. Ed. 2017;56:9170–9173. doi: 10.1002/anie.201703553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Volkwein W., Maier C., Krafczyk R., Jung K., Lassak J. A versatile toolbox for the control of protein levels using Nɛ-acetyl-L-lysine dependent amber suppression. ACS Synth. Biol. 2017;6:1892–1902. doi: 10.1021/acssynbio.7b00048. [DOI] [PubMed] [Google Scholar]
- 29.Pavlova N., Kaloudas D., Penchovsky R. Riboswitch distribution, structure, and function in bacteria. Gene. 2019;708:38–48. doi: 10.1016/j.gene.2019.05.036. [DOI] [PubMed] [Google Scholar]
- 30.McCown P.J., Corbino K.A., Stav S., Sherlock M.E., Breaker R.R. Riboswitch diversity and distribution. RNA. 2017;23:995–1011. doi: 10.1261/rna.061234.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hallberg Z.F., Su Y., Kitto R.Z., Hammond M.C. Engineering and in vivo applications of riboswitches. Annu. Rev. Biochem. 2017;86:515–539. doi: 10.1146/annurev-biochem-060815-014628. [DOI] [PubMed] [Google Scholar]
- 32.Mandal M., Breaker R.R. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 33.Dixon N., Duncan J.N., Geerlings T., Dunstan M.S., McCarthy J.E., Leys D., Micklefield J. Reengineering orthogonally selective riboswitches. Proc. Natl. Acad. Sci. USA. 2010;107:2830–2835. doi: 10.1073/pnas.0911209107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dixon N., Robinson C.J., Geerlings T., Duncan J.N., Drummond S.P., Micklefeld J. Orthogonal riboswitches for tuneable coexpression in bacteria. Angew. Chem. Int. Ed. Engl. 2012;51:3620–3624. doi: 10.1002/anie.201109106. [DOI] [PubMed] [Google Scholar]
- 35.Robinson C.J., Vincent H.A., Wu M.-C., Lowe P.T., Dunstan M.S., Leys D., Micklefield J. Modular riboswitch toolsets for synthetic genetic control in diverse bacterial species. J. Am. Chem. Soc. 2014;136:10615–10624. doi: 10.1021/ja502873j. [DOI] [PubMed] [Google Scholar]
- 36.Morra R., Shankar J., Robinson C.J., Halliwell S., Butler L., Upton M., Hay S., Micklefield J., Dixon N. Dual transcriptional-translational cascade permits cellular level tuneable expression control. Nucl. Acids Res. 2016;44:e21. doi: 10.1093/nar/gkv912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horga L.G., Halliwell S., Castiñeiras T.S., Wyre C., Matos C.F.R.O., Yovcheva D.S., Kent R., Morra R., Williams S.G., Smith D.C., et al. Tuning recombinant protein expression to match secretion capacity. Microb. Cell Fact. 2018;17:199. doi: 10.1186/s12934-018-1047-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sandiford S., Upton M. Identifcation, characterization, and recombinant expression of epidermicin NI01, a novel unmodified bacteriocin produced by Staphylococcus epidermidis that displays potent activity against Staphylococci. Antimicrob. Agents Chemother. 2012;56:1539–1547. doi: 10.1128/AAC.05397-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gay P., Le Coq D., Steinmetz M., Berkelman T., Kado C.I. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J. Bacteriol. 1985;164:918–921. doi: 10.1128/JB.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharan S.K., Thomason L.C., Kuznetsov S.G., Court D.L. Recombineering: A homologous recombination-based method of genetic engineering. Nat. Protoc. 2009;4:206–223. doi: 10.1038/nprot.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horbal L., Luzhetskyy A. Dual control system—A novel scaffolding architecture of an inducible regulatory device for the precise regulation of gene expression. Metab. Eng. 2016;37:11–23. doi: 10.1016/j.ymben.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horbal L., Fedorenko V., Luzhetskyy A. 2014. Novel and tightly regulated resorcinol and cumate-inducible expression systems for Streptomyces and other actinobacteria. Appl. Microbiol. Biotechnol. 2014;98:8641–8655. doi: 10.1007/s00253-014-5918-x. [DOI] [PubMed] [Google Scholar]
- 43.Rudolph M.M., Vockenhuber M.P., Suess B. Synthetic riboswitches for the conditional control of gene expression in Streptomyces coelicolor. Microbiology. 2013;159:1416–1422. doi: 10.1099/mic.0.067322-0. [DOI] [PubMed] [Google Scholar]
- 44.McCann P.A., Pogell B.M. Pamamycin: A new antibiotic and stimulator of aerial mycelia formation. J. Antibiot. 1979;32:673–678. doi: 10.7164/antibiotics.32.673. [DOI] [PubMed] [Google Scholar]
- 45.Rebets Y., Brötz E., Manderscheid N., Tokovenko B., Myronovskyi M., Metz P., Petzke L., Luzhetskyy A. Insights into the pamamycin biosynthesis. Angew. Chem. Int. Ed. Engl. 2015;54:2280–2284. doi: 10.1002/anie.201408901. [DOI] [PubMed] [Google Scholar]
- 46.Park S.V., Yang J.S., Jo H., Kang B., Oh S.S., Jung G.Y. Catalytic RNA, ribozyme, and its applications in synthetic biology. Biotechnol. Adv. 2019;37:107452. doi: 10.1016/j.biotechadv.2019.107452. [DOI] [PubMed] [Google Scholar]
- 47.De la Peña M., García-Robles I., Cervera A. The Hammerhead Ribozyme: A Long History for a Short RNA. Molecules. 2017;22:78. doi: 10.3390/molecules22010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wieland M., Hartig J.S. Improved aptazyme design and in vivo screening enable riboswitching in bacteria. Angew. Chem. Int. Ed. Engl. 2008;47:2604–2607. doi: 10.1002/anie.200703700. [DOI] [PubMed] [Google Scholar]
- 49.Lucks J.B., Qi L., Mutalik V.K., Wang D., Arkin A.P. Versatile RNA-sensing transcriptional regulators for engineering genetic networks. Proc. Natl. Acad. Sci. USA. 2011;108:8617–8622. doi: 10.1073/pnas.1015741108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chappell J., Takahashi M.K., Lucks J.B. Creating small transcription activating RNAs. Nat. Chem. Biol. 2015;11:214–220. doi: 10.1038/nchembio.1737. [DOI] [PubMed] [Google Scholar]
- 51.Green A.A., Silver P.A., Collins J.J., Yin P. Toehold switches: De-novo-designed regulators of gene expression. Cell. 2014;159:925–939. doi: 10.1016/j.cell.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noh M., Yoo S.M., Yang D., Lee S.Y. Broad-spectrum gene repression using scaffold engineering of synthetic sRNAs. ACS Synth. Biol. 2019;8:1452–1461. doi: 10.1021/acssynbio.9b00165. [DOI] [PubMed] [Google Scholar]
- 53.Thomason M.K., Storz G. Bacterial antisense RNAs: How many are there, and what are they doing. Annu. Rev. Genet. 2010;44:167–188. doi: 10.1146/annurev-genet-102209-163523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Westbrook A.M., Lucks J.B. Achieving large dynamic range control of gene expression with a compact RNA transcription–translation regulator. Nucleic Acids Res. 2017;45:5614–5624. doi: 10.1093/nar/gkx215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Novick R.P., Iordanescu S., Projan S.J., Kornblum J., Edelman I. pT181 plasmid replication is regulated by a countertranscript-driven transcriptional attenuator. Cell. 1989;59:395–404. doi: 10.1016/0092-8674(89)90300-0. [DOI] [PubMed] [Google Scholar]
- 56.Lee Y.J., Kim S.J., Moon T.S. Multilevel regulation of bacterial gene expression with the combined STAR and antisense RNA system. ACS Synth. Biol. 2018;7:853–865. doi: 10.1021/acssynbio.7b00322. [DOI] [PubMed] [Google Scholar]
- 57.Peralta-Yahya P.P., Zhang F., del Cardayre S.B., Keasling J.D. Microbial engineering for the production of advanced biofuels. Nature. 2012;488:320–328. doi: 10.1038/nature11478. [DOI] [PubMed] [Google Scholar]
- 58.Ho J.M.L., Bennett M.R. Improved memory devices for synthetic cells. Science. 2018;360:150–151. doi: 10.1126/science.aat3236. [DOI] [PubMed] [Google Scholar]
- 59.Carothers J.M., Goler J.A., Juminaga D., Keasling J.D. Model-driven engineering of RNA devices to quantitatively program gene expression. Science. 2011;334:1716–1719. doi: 10.1126/science.1212209. [DOI] [PubMed] [Google Scholar]
- 60.O’Connor C.D., Timmis K.N. Highly repressible expression system for cloning genes that specify potentially toxic proteins. J. Bacteriol. 1987;169:4457–4462. doi: 10.1128/JB.169.10.4457-4462.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Studier F.W. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J. Mol. Biol. 1991;219:37–44. doi: 10.1016/0022-2836(91)90855-Z. [DOI] [PubMed] [Google Scholar]
- 62.Unnithan S., Green L., Morrissey L., Binkley J., Singer B., Karam J., Gold L. Binding of the bacteriophage T4 regA protein to mRNA targets: An initiator AUG is required. Nucleic Acids Res. 1990;18:7083–7092. doi: 10.1093/nar/18.23.7083. [DOI] [PMC free article] [PubMed] [Google Scholar]