Abstract
The defective human survival motor neuron 1 (SMN1) gene leads to spinal muscular atrophy (SMA), the most common genetic cause of infant mortality. We previously reported that loss of SMN results in rapid differentiation of Drosophila germline stem cells and mouse embryonic stem cells (ESCs), indicating that SMN also plays important roles in germ cell development and stem cell biology. Here, we show that in healthy mice, SMN is highly expressed in the gonadal tissues, prepubertal spermatogonia, and adult spermatocytes, whereas low SMN expression is found in differentiated spermatid and sperm. In SMA-like mice, the growth of testis tissues is retarded, accompanied with gamete development abnormalities and loss of the spermatogonia-specific marker. Consistently, knockdown of Smn1 in spermatogonial stem cells (SSCs) leads to a compromised regeneration capacity in vitro and in vivo in transplantation experiments. In SMA-like mice, apoptosis and accumulation of the R-loop structure were significantly elevated, indicating that SMN plays a critical role in the survival of male germ cells. The present work demonstrates that SMN, in addition to its critical roles in neuronal development, participates in mouse germ cell and spermatogonium maintenance.
Keywords: SMN, gametogenesis, spermatogonium
1. Introduction
Recently, we and others reported that the levels of SMN protein correlate with the capacities of stem cell proliferation and differentiation in Drosophila [1] and mice [2]. In Drosophila, SMN is highly enriched in germline stem cells and a concentration gradient of SMN is observed [1,3]. In third instar larvae, overexpression of Smn in male germline stem cells results in testis growth and an increased number of spermatocytes while mutation of the Smn gene leads to defects in stem cell self-renewal and daughter cell differentiation. In mouse embryonic stem cells (ESCs), delayed cell growth and increased differentiation signals took place after knockdown of Smn1 [2]. These findings suggest that SMN plays important roles in germ cell development and pluripotent stem cell maintenance, in addition to its well-known roles during neuronal development.
SMN is a major assembler of Sm proteins, PRMT5, and gemin proteins for the formation of the small nuclear ribonucleoprotein (snRNP) complex, which carries out pre-mRNA splicing and 3′ end processing [4,5,6]. SmB and SmD3, members of the Sm family, regulate the splicing event and are crucial for germ cell development in Drosophila [7]. PRMT5, the arginine methyltransferase that participates in the snRNP processing, is reportedly important for germline specification in both Drosophila and mouse [8,9]. SMN, as well as senataxin (SETX), interact with each other and are required for resolving R-loops, RNA:DNA hybrids that form over transcription pause sites, created by RNA polymerase II in transcription termination regions [10]. It has been reported that Setx knockout mice are defective in spermatogenesis, meiotic recombination, and meiotic sex chromosome inactivation [11]. These findings also suggest that SMN plays important roles in germ cell development and stem cell biology.
In the present work, we first documented the expression patterns of SMN in gonadal tissues in young and adult mice. The defects upon SMN deficiency in gametogenesis were further analyzed in an SMA-like mouse model (Smn1−/−;SMN2+), shRNA-based knockdown, and germ cell transplantation experiments. Our data show that SMN is not only involved in the development of germ cells but also participates in spermatogonia maintenance, providing new evidence for SMN’s participation in mammalian germ cell development.
2. Results
2.1. SMN Expression during Spermatogenesis
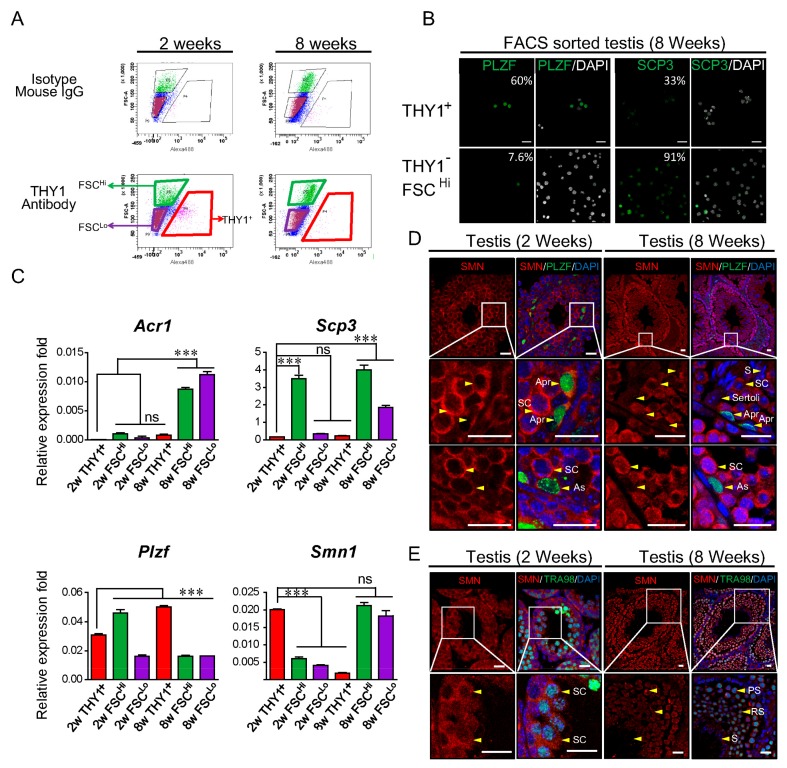
The protein levels of SMN in various adult mouse tissues were compared. Abundant SMN protein was expressed in the ovary, testis, and brain, compared to moderate levels in the spinal cord and low levels in the kidney (Supplementary Figure S1). The SMN expression profile was further examined in male germ cells. The testis of 2- and 8-week-old male mice were used to isolate spermatogonia and spermatocyte based on the level of Thy-1 Cell Surface Antigen (THY1) expression by fluorescence-activated cell sorting (FACS). Three populations were identified: Spermatogonial stem cells (SSCs, THY1+), spermatocyte (THY1−FSCHi), and sperm/spermatids/spermatocyte (THY1−FSCLo) (Figure 1A). The cell specificity of FACS was confirmed by immunofluorescent staining. The spermatogonia marker PLZF (promyelocytic leukemia zinc-finger, also called zinc finger and BTB domain-containing 16, ZBTB16) in 8-week-old mice was expressed with a high percentage in sorted THY1+ spermatogonia cells (60%); whereas THY1−FSCHi cells showed a low percentage of PLZF signal (7.6%) and high percentage of meiotic marker SCP3 (synaptonemal complex protein 3) expression (91%) (Figure 1B) [12,13,14]. The expression levels of the transcripts of Smn1 and germ cell markers were also determined in sorted populations by real-time PCR (Figure 1C). As expected, the spermatid-to-sperm stage marker acrosin prepropeptide variant 1 (Acr1) increased in 8-week-old adult THY1−FSCLo cells and the meiotic marker Scp3 showed the highest expression level in THY1−FSCHi spermatocytes. In the THY1+ SSC population, Smn1 showed a significantly high expression level in 2-week-old cells but was low in 8-week-old cells. In the THY1−FSCHi spermatocyte population, Smn1 decreased in 2-week-old cells but increased in 8-week-old cells (Figure 1C). The different expression levels of Smn1 transcripts between SSCs from 2- and 8-week-old mice are shown in Figure 1C, and the expression profile of SMN in spermatogonia was further examined with double staining of SMN and PLZF antibodies. For most paired spermatogonia (A-paired, Apr), which were connected to each other by a cytoplasmic bridge and represented the proliferating status [15,16], they expressed a high level of SMN and were compatible with the spermatocytes (Figure 1D, upper two panels). However, in A-single spermatogonia (As), which harbor potency for maintaining the spermatogonial stem cell population owing a relatively quiescent status [15,16], SMN decreased in the cytoplasm, in the testis of both prepubertal and adult males (Figure 1D, lower panels). No background signals were detected in the isotype control (Supplementary Figure S2). On the other hand, the high expression level of SMN in proliferating spermatogonia could be correlated with the higher level of Smn1 transcripts in THY1+ SSC from 2-week-old mice, as shown in Figure 1C, because most of the spermatogonia are still proliferating at this stage. Interestingly, SMN diminished in the elongated spermatid and sperm (Figure 1E). These results imply that SMN might be correlated with the differentiation and propagation of spermatogonia.
Figure 1.
The distribution of survival motor neuron (SMN) in young and adult mouse testes. (A) Fluorescence-activated cell sorting (FACS) was used to characterize and sort testicular cells from 2- and 8-week-old mice. Based on the staining intensity of Thy-1 Cell Surface Antigen (THY1) conjugated with Alexa-488 fluorescent dye, three populations were identified: SSC (THY1+), spermatocyte (THY1−FSCHi), and sperm/spermatids/spermatocyte (THY1−FSCLo). Rat Immunoglobulin G (IgG) subsequently conjugated with 488 was used as the isotype control antibody. (B) Sorted THY1+ cells from 8-week-old mice express a high percentage (60%) of promyelocytic leukemia zinc-finger (PLZF) (green, left panel), whereas THY1−FSCHi cells shows a low percentage of PLZF signal (7.6%). The meiotic marker synaptonemal complex protein 3 (SCP3) was used to characterize the THY1−FSCHi population, which mostly contains spermatocyte. The percentages of markers in different populations are indicated. (C) Determination of the expression level of Smn1 and germ cell markers in the sorted population. In the THY1+ SSC population, Smn1 showed a significantly higher expression level in 2-week-old cells (2w THY1+) but decreased to a lower level in the THY1−FSCHi spermatocyte population (2w THY1−FSCHi). In 8-week-old THY1+ SSC cells (8w THY1+), the expression level of Smn1 showed no difference compared to the spermatocyte population of 2-week-old cells but increased significantly in the THY1−FSCHi spermatocyte population (8w THY1−FSCHi). The transcript expression level of the spermatogonia-specific marker Plzf, sperm marker Acr1, and meiotic marker Scp3 showed a cell-type-dependent manner. The error bar indicates technical repeats. The expression level of each group was compared with the 2w THY1+ group. *Indicates significance, p < 0.05; **, p < 0.005. (D) Putative proliferating A-paired spermatogonia (Apr, arrowhead, upper, and middle panel) expresses the abundant amount of SMN in the testis of both 2- and 8-week-old mice, whereas undifferentiated A-single spermatogonia (As, arrowhead, lower panels) expresses PLZF (green) and possesses a lower level of SMN (red) compared with spermatocyte (SC). Sertoli cells (arrow indicated) also express a lower amount of SMN protein. (E) Validation of germ cell marker TRA98 expression in spermatocyte. SMN expressed abundantly in pachytene (PS)-stage spermatocytes colocalized with TRA98 (green); the expression decreases in round spermatocyte (RS), elongated spermatocyte (ES), and is the lowest in spermatozoa (S) in 8-week-old testis. The isotype negative control used for the double staining of antibodies is shown in Supplementary Figure S2. The scale bar in this figure represents 20 μm.
2.2. Developmental Delay in the Gonadal Tissues of SMA-Like Mice
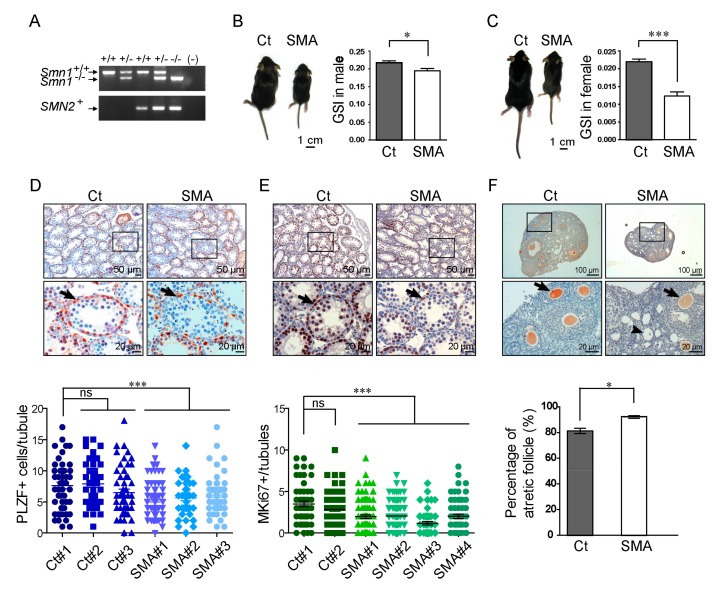
A SMN-defective mouse model that mimics human SMA pathology has made great contributions to our understanding of SMN biology and SMA pathophysiology [17,18]. Here, we used an SMA-like mouse model [18] to study whether reduced SMN causes developmental defects in the germ cells of gonadal tissues. The mice used in this study approximately represent the intermediate severity type II SMA, with a survival time about 2 weeks, which was genotyped as Smn1−/−;SMN2+ (SMA). Non-affected littermates (Smn1+/−;SMN2+, Ct) were used as control animals (Figure 2A). SMA-like mice exhibited a smaller body size, lower activity, and significantly decreased gonadosomatic index (GSI) of the testes/ovaries compared to the controls (Figure 2B,C). Immunohistochemistry (IHC) and quantitative assays of the testis revealed that fewer numbers of PLZF-positive spermatogonia cells present in the seminiferous tubules of SMA-like mice compared to those in controls (Figure 2D). Furthermore, fewer MKi67-positive spermatogonia-like cells surrounding the basement membrane of the seminiferous tubules decreased in SMA-like mice, indicating a reduced number of proliferating spermatogonia (Figure 2E). In females, a lower SMN signal (Figure 2F, upper panel) and higher frequency of atretic follicles were detected in SMA-like mice compared to control females (Figure 2F, lower panel). The decreased PLZF signal might link to the failure of spermatogonia maintenance in SMA-like mice. Thus, the function of spermatogonia was further examined by a transplant experiment.
Figure 2.
Testes and ovaries in SMA-like mice are abnormal. (A) Genotype of wildtype Smn1+/+;SMN2−, Smn1+/−;SMN2+ (littermate control, Ct), and Smn1−/−;SMN2+ (SMA-like) mice are confirmed by PCR. (-) indicates the negative control without a template. (B) Male SMA-like mice (indicated as SMA) with a phenotype has a smaller body size and significantly lower GSI than their heterozygote littermates (Smn1+/−SMN2+, indicated as Ct) at the age of 2 weeks (n = 4 Ct and 7 SMA mice). (C) The body size of female SMA-like mice (left panel), and the GSI of ovaries at 8 weeks after birth (right panel, n = 3 Ct and 4 SMA mice). (D) Reduction of PLZF-expressing germ cells (red color, arrows in larger magnification field) is shown in SMA-like mice but not in their heterozygote littermates after IHC analysis. Lower panel indicates its quantification. Each spot indicates the number of PLZF-expressing cells in each seminiferous tubule. Seminiferous tubules ranging from 100 to 150 μm are used for counting (n = 3 mice per genotype). (E) The expression of the proliferation marker, MKi67 (red color, arrow), is weaker in surrounding spermatogonia cells on the basal membrane of SMA-like mice than heterozygote controls. Hematoxylene was used as a counterstain (blue) (n = 2 for Ct and 4 for SMA). The negative control of IHC is shown in Supplementary Figure S3. (F) IHC staining demonstrates low SMN protein levels (red color, arrow) in the immature oocytes in SMA-like mice. The percentage of atretic follicles (atresia) was quantified (lower panel). * Indicates a statistically significant difference, p < 0.05. ***, p < 0.001.
2.3. Decreased Ability of Self-Renewal in SMN-Depleted SSCs
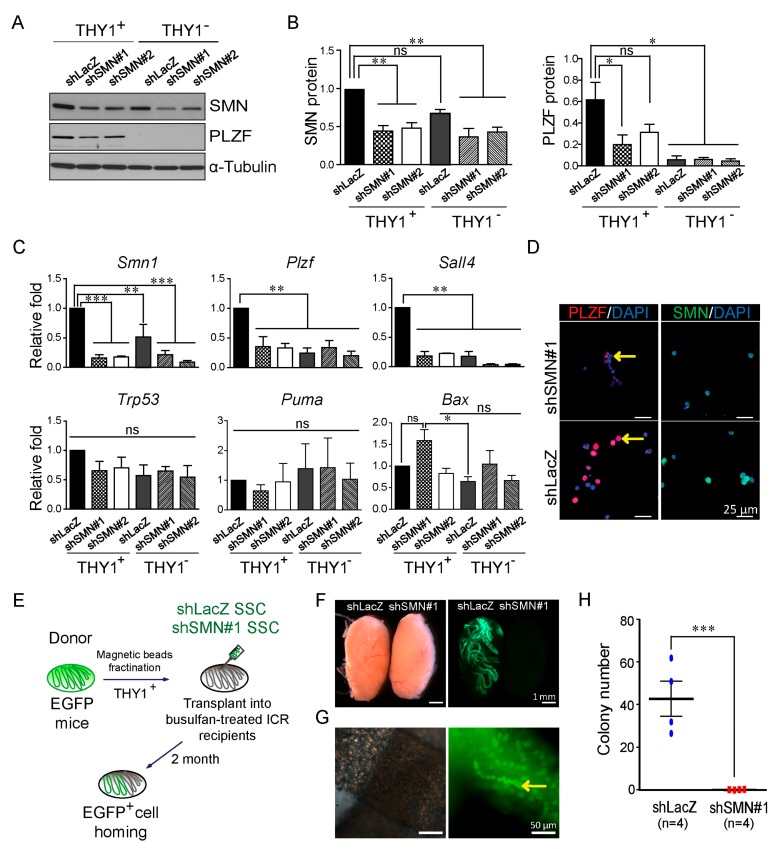
Considering the convenience of obtaining SMN-deficient spermatogonium, we performed knockdown of Smn1 in magnetic-activated cell sorting (MACS) isolated spermatogonial stem cells (SSCs) of C57/BL6 mice to evaluate the effects of SMN depletion. Antibody-enriched THY1+ SSCs were used for lentiviral-based knockdown experiments. shSMN lentivirus-transduced SSCs displayed lower expression levels of SMN and PLZF protein in the Western blotting analysis (Figure 3A,B), consistent with the result shown in the SMA-like mouse model in Figure 2. THY1− cells, which contain spermatocyte and somatic cells, did not express PLZF and showed lower expression after SMN knockdown (Figure 3A,B). The RNA levels of Smn1, sal-like protein 4 (Sall4), and Plzf genes were also decreased following SMN knockdown in THY1+ cells (Figure 3C), which was consistent with the Western blotting analysis and immunofluorescent staining results (Figure 3D). Of note, apoptotic markers, such as transformation-related protein 53 (Trp53), BCL2-binding component 3 (Bbc3, also known as Puma), and BCL2-associated X protein (Bax), had similar levels in the Smn1 knockdown and control groups (Figure 3C).
Figure 3.
Spermatogonial stem cells (SSCs) show decreased PLZF expression and lost the ability of self-renewal after SMN knockdown. (A) Western blot analysis and its quantification (B) demonstrated decreased PLZF protein following lentiviral shSMN transduction in SSCs enriched by MACS isolated from 6–7 days postpartum (dpp) testes of C57BL/6JNarl mice (shSMN#1 and shSMN#2). Compared with the control shLacZ group, a significant difference is shown. α-Tubulin is used as the internal control of the Western blot. The original Western blot is shown in Supplementary Figure S4 (n = 2). (C) Real-time PCR analysis of SMN-depleted SSCs. Smn1, spermatogonia makers, Plzf and Sall4, and apoptotic markers Trp53, Puma, and Bax were analyzed (n = 3). (D) Immunofluorescent staining demonstrates SSCs express less PLZF in the SMN knockdown group (shSMN#1, red color, arrow indicated) compared to the control shLacZ group. 4′,6-diamidino-2-phenylindole (DAPI) is used as the counterstain (blue). (E) Schematic illustration of the process of SSC transplantation. (F) SSCs carrying enhanced green fluorescent protein (EGFP) transplanted into a recipient testis shows colonization in the shLacZ group (left testis) but not in SMN-depleted SSCs (shSMN#1, right testis). (G) Aligned EGFP-positive cells in the shLacZ control group represent the homing ability and normal proliferation (arrow). (H) Quantification of the SSC homing efficiency. Each spot indicates one replicate of the transplanted mouse. *Indicates significance, p < 0.05; **, p < 0.005; ***, p < 0.001; ns indicates no significance.
Transplantation of SSCs into the seminiferous tubules of an infertile recipient animal is considered the gold standard assay to determine the germline competence of SSCs [19,20,21]. To investigate whether SMN contributes to SSC maintenance, we transplanted SMN-depleted SSCs isolated from β-Actin promoter-driven enhanced green fluorescent protein (EGFP) mice to the seminiferous tubules of a busulfan-treated recipient ICR male mice (Figure 4d for the transplant procedure). SMN deficiency completely abolished the homing ability of EGFP-SSCs transduced with shSMN, whereas control EGFP-SSCs transduced with shLacZ control shRNA could still survive, proliferate, and differentiate in the host seminiferous tubules (Figure 3E,F). The quantification of the control EGFP-SSC colony formation rate was close to a previous report [22] (Figure 3G). These results indicate that a reduction of SMN leads to defects in male germline development and spermatogonia maintenance.
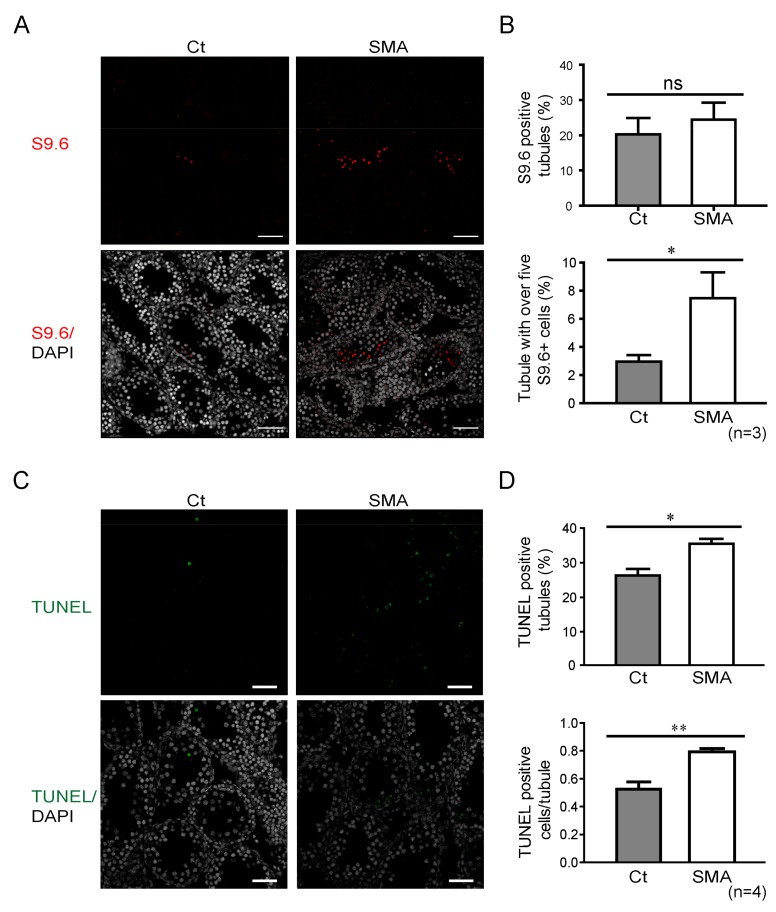
Figure 4.
Detection of RNA:DNA hybrid structure in SMA-like mice. (A) Staining result of the RNA:DNA hybrid structure, R-loop, by the detecting antibody S9.6 (red) and DAPI (white) in SMA-like mice (SMA) and littermate controls (Ct) (scale bar = 50 μm). (B) Upper panel: Percentage of S9.6-positive seminiferous tubule of SMA-like mice (SMA, n = 3) and control littermates (Ct, n = 3 mice per genotype). Lower panel: Quantification of tubules containing over five S9.6-positive cells. (C) TUNEL assay of SMA-like mice (SMA) showing a higher amount of positive signal (green). Nuclei were stained with DAPI (white) (scale bar = 50 μm). (D) Percentage of seminiferous tubules with at least one TUNEL-positive cell (upper panel) and average number of TUNEL-positive cells per seminiferous tubule (lower panel). Over 90 tubules/mouse were counted for S9.6 and TUNEL. *Indicates significance, p < 0.05; **, p < 0.005.
2.4. Increased R-loop Signal Following SMN Depletion
Accumulation of the RNA:DNA hybrid structure, R-loop, was a recently identified phenomenon in SMN-deficient cells due to the increased transcription termination, which is concomitant with the induction of the p53 pathway and DNA damage response, manifesting as γ-H2A.X activation and U12-mediated intron retention [10,23,24,25]. Because SMN is a key component of R-loop resolving, which affects transcription termination and may further contribute to cell proliferation [10,26], we tested whether SMN depletion leads to an increase of the R-loop in SMA-like mice. The IHC staining (Figure 4A) against the R-loop structural marker, S9.6, showed no significant differences between SMA-like mice and littermate controls in the total number of S9.6-positive tubules (Figure 4B, upper panel). However, a significantly higher amount of the R-loop structure appeared in the testis of SMA-like mice, which contained over five S9.6-positive cells per tubule (Figure 4B, lower panel). Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining, which detects DNA fragmentation, which is usually associated with apoptosis, was analyzed in the testis of SMA-like mice. We observed a significant increase in the percentage and average number of TUNEL-positive cells per tubule (Figure 4C,D). These data demonstrated that a deficiency of SMN would form abundant R-loop structures, which may contribute to the disruption of RNA transcription, and lead to massive cell death in testicular cells.
3. Discussion
It is well established that SMN plays key roles in neuron cell development. Genetic defects in the SMN gene is causative of the devastating neuromuscular disease SMA. In recent years, we and others have investigated the roles of SMN beyond the central neuron system. Accumulating evidence supports the notion that SMN participates in germline cell development from Drosophila to mammalian, as well as in the maintenance of pluripotent stem cells [1,2,27]. The present work strengthens this notion by demonstrating that SMN deficiency leads to retarded gonad development, and that loss of SMN causes defective SSC maintenance, in vitro and in vivo.
The present study documented the expression profiles of SMN in the testis and SSCs in mice. Higher levels of SMN were found in spermatocytes than in elongated spermatids or sperm, and surrounding sertoli cells (Figure 1). SMN protein forms a concentration gradient in the testis that is inversely proportional to the state of differentiation, which implies the possible function of SMN in germ cell development of regulating transcription activity or mRNA processing following the gradual termination of transcription during spermatogenesis [28]. This is further supported by the IHC result of the testis of young mice, where SMN colocalized with PLZF as an uneven pattern (Figure 1).
The decreased gonad mass and the reduction of MKi67 spermatogonia in the testis of SMA-like mice (Figure 2) suggest that SMN might participate in controlling cell proliferation in germ cells, which is consistent with previous studies in Drosophila and a different SMA-like mouse model (SmnC/C) with milder symptoms [1,27]. In the present work, we showed that PLZF, a DNA-binding transcriptional repressor that is crucial for regulating spermatogonia quiescence and proliferation [13,29], was decreased in the testis of SMA-like mice, indicating a loss of the spermatogia population (Figure 2) [30,31,32]. Interestingly, in the seminiferous tubules, the immunofluorescent experiments showed that spermatocytes express higher levels of SMN protein than spermatogonia, which warrants further research into the SMN biology in spermatocytes.
SALL4 is a major regulator in SSC and sustains the SSC pool by cooperation with PLZF [33,34,35]. Not surprisingly, levels of SALL4 were found to be decreased in SMN knockdown SSCs, similar to the changes of PLZF. Our results indicate that SMN deficiency leads to decreased levels of PLZF and SALL4, which directly affects male germ cell development (Figure 3). Importantly, SSCs with SMN depletion were not able to reform the population for spermatogenesis in the transplant experiments (Figure 3), suggesting that SMN is highly correlated with SSC proliferation and repopulation.
At the molecule level, our data indicate that the deficiency of SMN in germ cells results in elevated levels of unresolved RNA:DNA hybrids, the so-called R-loop, a structure that forms over transcription pause sites, contributes to genome instability, and induces repressive marks on chromatin [25,36]. SMN is the key player, which interacts with the RNA helicase SETX, affects transcription termination, and regulates polymerase II activity by resolving the R-loop structure [10,37]. Unresolved RNA:DNA hybrids, induced by SMN depletion, lead to transcriptional termination and consequently cell cycle arrest (Figure 4) [10,11]. It has also been reported that the unresolved R-loop structure due to SMN deficiency in neurons, and oxidative stress in muscles or cardiomyocytes lead to apoptosis [18,24,38,39,40,41]. Here, we provided additional evidence by showing that a loss of SMN in germ cells also induces programmed cell death in male testis and in developing oocytes in female SMA-like mice (Figure 2 and Figure 4). A similar phenomenon is also found in Setx knockout mice, including increased apoptosis and R-loop accumulation, which further supports the correlation of SMN and the R-loop structure [11].
We noticed that the gene/protein expression profiles revealed by different methods utilized in the present work, including FACS, real-time PCR, and IHC, did not always correlate with each other. This may be due to the differential sensitivities of the assays, but perhaps, more likely, it is caused by the heterogeneous nature of the cell populations. More accurate separation/characterization methods may improve this situation.
Taken together, our work demonstrates that SMN plays key roles in germ cell development in vitro and in vivo. These new roles, in addition to its well-known roles in neuronal development, may shed light on our understanding of this important protein.
4. Methods
4.1. Availability of Data and Materials
All animal maintenance, care, and procedures described within were reviewed and approved by the Institutional Animal Care and Use Committee of National Taiwan University (NTU) according to the protocol number (NTU-105-EL-68, 6th January 2016 approved, and NTU-107-EL-154, 11th January 2018 approved). All methods in the manuscript were performed in accordance with the relevant guidelines and regulations of NTU. Graphics and tables in the manuscript were prepared by the first and corresponding authors.
4.2. Animals and Tissue Sampling
Two-week-old and sexually mature 8- to 12-week-old ICR mice (BioLASCO, Taipei, Taiwan) were used for providing tissues, including ovary, testis, brain, spinal cord, and kidney, for Western blot analysis in Supplementary Figure S1. Tissues were homogenized with a grinder, and proteins were extracted by RIPA lysis buffer (20-188, Merck Millipore, Darmstadt, Germany). Transgenic Smn1-deficient mice (SMA-like) were generated from Hsieh-Li’s laboratory [18] and bred from the Rodent Model Resource Center [C57BL/6/Tg(SMN2) Hung Smn1tm1 Hung] (National Laboratory Animal Center, NLAC, Taipei, Taiwan). Testes and ovaries of SMA-like mice were collected and fixed by 4% paraformaldehyde (PFA, 30525-89, Alfa Aesar, Ward Hill, MA, USA) at different ages. Gonadosomatic index (GSI) = (testis or ovary weight divided by body weight) × 100.
4.3. Fluorescence-Activated Cell Sorting (FACS)
Testis from 2- and 8-week-old C57BL/6JNarl mice (NLAC) were digested by Collagenase IV and Trypsin, incubated with rat anti-CD90.2 (THY1) antibody (553011, BD Biosciences, San Jose, CA, USA) or isotype rat IgG (559072, BD) control, and then labeled with anti-rat 488 secondary antibody (A21208). Cells were placed into an FACS tube for isolation with three populations, including THY1+ SSCs, THY1−FSCHi spermatocyte, and THY1−FSCLo sperm/spermatids/spermatocyte, by an FACS Aria III Sorter (BD). Dead cells stained with propidium iodide (PI, 1 μg/mL) signals were excluded.
4.4. Immunohistochemistry (IHC)
The testis and ovary tissues of mice were dissected and immersed in 4% PFA overnight at 4 °C, and then embedded into wax. Ovary and testis sections were stained by mouse monoclonal anti-SMN antibody (2.5 μg/mL, 610646, BD) overnight at 4 °C. The germ cell markers, rat anti-TRA98 (6 μg/mL, ab82527, Abcam, Boston, MA, USA), rabbit anti-PLZF (1 μg/mL, sc-22839, Santa Cruz Biotechnology, Inc., CA, USA), proliferation marker MKi67 (4 μg/mL, ab16667, Abcam), and R-loop detecting antibody S9.6 (2 μg/mL, ENH001, Kerafast, Inc. Boston, MA, USA) were stained in testis sections. Isotype IgG, including mouse (554121, BD), rabbit (550875, BD) and rat (559072, BD), were used as the negative control. Secondary antibodies (4 μg/mL) purchased from Thermo (Thermo Fisher Scientific, Waltham, MA, USA) were used as following: Alexa Fluor 488 goat anti-mouse IgG (A11029), Alexa Fluor 647 goat anti-mouse IgG (A21235), Alexa Fluor 488 donkey anti-Rat IgG (A21208), and Alexa Fluor 555 goat anti-rabbit IgG (A21429). For the PLZF- and MKi67-positive cell counting in the seminiferous tubules, the same staining procedure was performed and used the standard VECTASTAIN ABC system (PK6100, Vector Laboratories, Burlingame, CA, USA), and the AEC solution (00-1122, Thermo, Waltham, MA, USA) was used for conjugated horseradish peroxidase (HRP) antibody detection to visualize the staining outcome. Hematoxylin was used for the counter stain. The confirmation of the antibody specificity is shown in the Supplementary Figures S2 and S3. For measurement of the spermatogonia in SMA-like mice, two to three non-serial sections containing 60–90 oval-shaped tubules ranging from 100 to 150 μm in diameter were counted for spermatogonia (PLZF-positive), surrounding proliferating spermatogonia-like cells (MKi67-positive), and R-loop structure (S9.6 positive). TUNEL assay was performed following the manufacturer’s instructions (In Situ Cell Death Detection Kit, 11684795910, Roche Applied Science, Mannhein, Germany) and TUNEL-positive tubules were quantified for around 90 oval-shaped tubules with a diameter ranging from 100 to 150 μm. Two to four mice were used for each genotype of mice for each assay.
4.5. Immunofluorescent Staining and Confocal Microscopy
Fixed cells were washed in D-PBS and permeabilized in 0.5% Triton X-100, followed by incubation in 2% BSA blocking solution. Samples were incubated overnight at 4 °C with primary antibodies, mouse-anti-SMN (0.8 μg/mL), anti-SCP3 (2.5 μg/mL, ab15093, Abcam), and rabbit anti-PLZF (1 μg/mL), and then washed with 0.25% PBS-Tween-20 and incubated with a secondary antibody conjugated to a fluorescence dye (Alexa Fluor 488 goat anti-mouse IgG, or Alexa Fluor 555 goat anti-rabbit IgG, A11029, and A21429, 4 μg/mL, Thermo) for 1 h. The cell nucleus was stained with 4′,6-diamidino-2-phenylindole (DAPI, 100 ng/mL, D9564, Sigma, St. Louis, MO, USA). Finally, fluorescent samples were mounted and observed with a confocal microscope (TCS SP5 II, Leica, Wetzlar, Germany).
4.6. Western Blotting
Cell lysates from tissues (30 μg/lane) were run on a 12% polyacrylamide gel and then transferred to a PVDF membrane. Following 1 h of incubation to block the non-specific binding with 5% non-fat dried milk in TBS (Bio Basic Inc., Markham, Ontario Canada), the first antibody was incubated overnight at 4 °C, including mouse anti-SMN antibody (0.03 μg/mL) and rabbit-anti-PLZF antibody (0.2 μg/mL). α-Tubulin (0.3 μg/mL, T5168, Sigma, St. Louis, MO, USA) was used as an internal control. After several washes containing 0.1% Tween-20 in TBS, the blot was incubated for 1 h with an HRP-conjugated goat anti-mouse and rabbit IgG secondary antibody (31430 and 31460, Thermo). The bound antibody was detected by a chemiluminescence detection system.
4.7. Spermatogonial Stem Cells’ (SSCs) Isolation and shRNA Knockdown
Six to seven days postpartum (dpp), the testes of C57BL/6JNarl mice were collected and dissected following the previous published study for immune-fluorescent staining, Western blotting analysis, and real-time PCR [42]. The birth date of mice was defined as 0 dpp. Briefly, Collagenase IV- and Trypsin-digested cells were incubated with anti-CD90.2 (THY1) antibody and then labeled with anti-biotin microbeads (130-090-485, Miltenyi Sigma Biotec, Bergisch Gladbach, Germany). Cells were placed into a collection tube of magnetic-activated cell sorting (MACS) to isolate THY1+ SSCs. Isolated THY1+ SSCs were cultured in the BSA medium following the recipe published previously [43] and THY1− fraction containing somatic cells was cultured in 10% FBS/DMEM medium and both fractions were assessed for shRNA knockdown on the collecting day. Control shLacZ plasmid and shRN targeting the mouse Smn1 gene were obtained from the RNAi core facility (shLacZ: TRCN0000072224, shSMN#1: TRCN0000072018, shSMN#2: TRCN0000072022). Plasmids were transfected into 293T cells to produce lentivirus for transduction into target SSCs and somatic cells. The RNA and protein of shRNA transduced SSCs and somatic cells were collected at the third day from the collecting day.
4.8. Germ Cell Transplantation
β-Actin promoter-driven EGFP (EGFP) 6–7 dpp ICR mice backcrossed with C57BL/6JNarl for 20 generations and were used for donor SSCs in the germ cell transplantation experiment [42,44]. The transplant procedure is briefly illustrated in Figure 4d. Six-week-old recipient ICR male mice were treated with 40 mg/kg busulfan (B2635, Sigma) and kept for 1 month to eliminate endogenous germ cells [45,46]. Following previously described procedures [42], the testicle THY1+ SSC cell suspension from 6–7 dpp EGFP mice (concentration of 2.5 × 106 cells/mL) was mixed with Trypan Blue dye (T8154, Sigma), and approximately 20 μL of the cell suspension was injected through the efferent duct into the rete testis of the recipients. Two months later, the testes of the recipient animals were dissected for the detection of EGFP+ colonies. Colonies were defined as groups of transplanted cells that occupied at least 0.1 mm of the tubules.
4.9. Genotyping
Mouse tails were lysed in 1X PCR buffer supplemented with proteinase K (Novagen, Darmstadt, Germany) at 55 °C. Tail lysates were heat inactivated and PCR was performed using primers specific for mouse SMN and transgenic human SMN2 [18]. Amplified PCR products were separated out by gel electrophoresis and detected by Safeview DNA staining dye (GeneMark, Taipei, Taiwan).
4.10. Gene Expression
Cell samples were collected, and total RNA was extracted by TRIzol® reagent (15596-026, Thermo) and treated with RNase-free DNase I (M6101, Promega, Madison, WI, USA). Reverse transcription was performed using the SuperScript III First-Strand Synthesis System (18080-051, Thermo) by random hexamer primers. SYBR Green PCR master mix (KK4603, Kapa Biosystems, Inc., Woburn, MA, USA) and 200 nM of the forward/reverse primers in a final volume of 10 μL were mixed with 25 ng of template cDNA. Real-time PCR was performed using the Roche LightCycler (LC480, Roche, Mannhein, Germany), with the Ct value of GAPDH serving as the internal control. Primer sequences are listed in the Supplementary Table S1.
4.11. Statistics
All data were presented as means ± standard error of the mean (SEM) and unpaired comparison was analyzed using the Student’s t-test or one-way ANOVA with Dunnett’s multiple comparison test (GraphPad Software Inc., La Jolla, CA, USA). Significance was assumed at a p-value of 0.05.
5. Conclusions
In the present work, we demonstrated the expression profile of SMN in the gonadal tissues of young and adult mice. SMN deficiency impeded gametogenesis in an SMA-like mouse model (Smn1−/−;SMN2+), especially affecting spermatogonia maintenance based on shRNA-based knockdown and germ cell transplantation experiments. SMN loss also induced programmed cell death and the accumulation of the R-loop structure in the male testis of SMA-like mice. These data show that SMN not only participates in germline development but also sustains the survival of spermatogonium in mice.
Acknowledgments
We thank Shau-Ping Lin kindly provides the EGFP mice and Hung-Fu Liao provided the details for the methodology for the transplant experiment. Thank Sheng-Wen Wang assisted for the transplant experiments. The shRNAs were generated and obtained from the National RNAi Core Facility in the Institute of Molecular Biology, Genomic Research Center, Academia Sinica. The Joint Center for Instruments and Researches, College of Bioresources and Agriculture in National Taiwan University provided the service for the laser-scanning confocal microscopy.
Supplementary Materials
Supplementary Materials can be found at https://www.mdpi.com/1422-0067/21/3/794/s1.
Author Contributions
W.-F.C. designed and performed experiments, collected and analyzed data, and wrote the first draft of the manuscript. J.X. performed interpretation and manuscript writing. T.-Y.L. and J.H. performed experiments. H.-M.H.-L. provided the SMA-like animal used in this study. Y.-M.H. and J.-L.L. provided expert advice. L.-Y.S. and C.-H.L. conceived, supervised, supported the study, and finalized the manuscript. All authors approved the final manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by grant from Ministry of Science and Technology, Taiwan (Grant Number 106-2313-B-002-039-MY3 to L. S., 106-2314-B-195-006-MY3 to C. L. and 108-2811-B-002-582 to W. C.).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Grice S.J., Liu J.L. Survival motor neuron protein regulates stem cell division, proliferation, and differentiation in Drosophila. PLoS Genet. 2011;7:e1002030. doi: 10.1371/journal.pgen.1002030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang W.F., Xu J., Chang C.C., Yang S.H., Li H.Y., Hsieh-Li H.M., Tsai M.H., Wu S.C., Cheng W.T.K., Liu J.L., et al. SMN is required for the maintenance of embryonic stem cells and neuronal differentiation in mice. Brain Struct. Funct. 2015;220:1539–1553. doi: 10.1007/s00429-014-0743-7. [DOI] [PubMed] [Google Scholar]
- 3.Lee L., Davies S.E., Liu J.L. The spinal muscular atrophy protein SMN affects Drosophila germline nuclear organization through the U body-P body pathway. Dev. Biol. 2009;332:142–155. doi: 10.1016/j.ydbio.2009.05.553. [DOI] [PubMed] [Google Scholar]
- 4.Kaida D., Berg M.G., Younis I., Kasim M., Singh L.N., Wan L., Dreyfuss G. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–668. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pillai R.S., Grimmler M., Meister G., Will C.L., Luhrmann R., Fischer U., Schumperli D. Unique Sm core structure of U7 snRNPs: Assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev. 2003;17:2321–2333. doi: 10.1101/gad.274403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matera A.G., Wang Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014;15:108–121. doi: 10.1038/nrm3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonsalvez G.B., Rajendra T.K., Wen Y., Praveen K., Matera A.G. Sm proteins specify germ cell fate by facilitating oskar mRNA localization. Development. 2010;137:2341–2351. doi: 10.1242/dev.042721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonsalvez G.B., Tian L., Ospina J.K., Boisvert F.M., Lamond A.I., Matera A.G. Two distinct arginine methyltransferases are required for biogenesis of Sm-class ribonucleoproteins. J. Cell. Biol. 2007;178:733–740. doi: 10.1083/jcb.200702147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y., Zhu T., Li Q., Liu C., Han F., Chen M., Zhang L., Cui X., Qin Y., Bao S., et al. Prmt5 is required for germ cell survival during spermatogenesis in mice. Sci. Rep. 2015;5:11031. doi: 10.1038/srep11031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao D.Y., Gish G., Braunschweig U., Li Y., Ni Z., Schmitges F.W., Zhong G., Liu K., Li W., Moffat J., et al. SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination. Nature. 2016;529:48–53. doi: 10.1038/nature16469. [DOI] [PubMed] [Google Scholar]
- 11.Becherel O.J., Yeo A.J., Stellati A., Heng E.Y., Luff J., Suraweera A.M., Woods R., Fleming J., Carrie D., McKinney K., et al. Senataxin plays an essential role with DNA damage response proteins in meiotic recombination and gene silencing. PLoS Genet. 2013;9:e1003435. doi: 10.1371/journal.pgen.1003435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan L., Liu J.G., Zhao J., Brundell E., Daneholt B., Hoog C. The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol. Cell. 2000;5:73–83. doi: 10.1016/S1097-2765(00)80404-9. [DOI] [PubMed] [Google Scholar]
- 13.Buaas F.W., Kirsh A.L., Sharma M., McLean D.J., Morris J.L., Griswold M.D., de Rooij D.G., Braun R.E. Plzf is required in adult male germ cells for stem cell self-renewal. Nat. Genet. 2004;36:647–652. doi: 10.1038/ng1366. [DOI] [PubMed] [Google Scholar]
- 14.Costoya J.A., Hobbs R.M., Barna M., Cattoretti G., Manova K., Sukhwani M., Orwig K.E., Wolgemuth D.J., Pandolfi P.P. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat. Genet. 2004;36:653–659. doi: 10.1038/ng1367. [DOI] [PubMed] [Google Scholar]
- 15.Fayomi A.P., Orwig K.E. Spermatogonial stem cells and spermatogenesis in mice, monkeys and men. Stem Cell Res. 2018;29:207–214. doi: 10.1016/j.scr.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oakberg E.F. Spermatogonial stem-cell renewal in the mouse. Anat. Rec. 1971;169:515–531. doi: 10.1002/ar.1091690305. [DOI] [PubMed] [Google Scholar]
- 17.Bebee T.W., Dominguez C.E., Chandler D.S. Mouse models of SMA: Tools for disease characterization and therapeutic development. Hum. Genet. 2012;131:1277–1293. doi: 10.1007/s00439-012-1171-5. [DOI] [PubMed] [Google Scholar]
- 18.Hsieh-Li H.M., Chang J.G., Jong Y.J., Wu M.H., Wang N.M., Tsai C.H., Li H. A mouse model for spinal muscular atrophy. Nat. Genet. 2000;24:66–70. doi: 10.1038/71709. [DOI] [PubMed] [Google Scholar]
- 19.Hermann B.P., Sukhwani M., Winkler F., Pascarella J.N., Peters K.A., Sheng Y., Valli H., Rodriguez M., Ezzelarab M., Dargo G., et al. Spermatogonial stem cell transplantation into rhesus testes regenerates spermatogenesis producing functional sperm. Cell Stem Cell. 2012;11:715–726. doi: 10.1016/j.stem.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brinster R.L., Avarbock M.R. Germline transmission of donor haplotype following spermatogonial transplantation. Proc. Natl. Acad Sci. USA. 1994;91:11303–11307. doi: 10.1073/pnas.91.24.11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brinster R.L., Zimmermann J.W. Spermatogenesis following male germ-cell transplantation. Proc. Natl. Acad. Sci. USA. 1994;91:11298–11302. doi: 10.1073/pnas.91.24.11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagano M., Avarbock M.R., Brinster R.L. Pattern and kinetics of mouse donor spermatogonial stem cell colonization in recipient testes. Biol. Reprod. 1999;60:1429–1436. doi: 10.1095/biolreprod60.6.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jangi M., Fleet C., Cullen P., Gupta S.V., Mekhoubad S., Chiao E., Allaire N., Bennett C.F., Rigo F., Krainer A.R., et al. SMN deficiency in severe models of spinal muscular atrophy causes widespread intron retention and DNA damage. Proc. Natl. Acad Sci. USA. 2017;114:E2347–E2356. doi: 10.1073/pnas.1613181114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lotti F., Imlach W.L., Saieva L., Beck E.S., Hao le T., Li D.K., Jiao W., Mentis G.Z., Beattie C.E., McCabe B.D., et al. An SMN-dependent U12 splicing event essential for motor circuit function. Cell. 2012;151:440–454. doi: 10.1016/j.cell.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skourti-Stathaki K., Kamieniarz-Gdula K., Proudfoot N.J. R-loops induce repressive chromatin marks over mammalian gene terminators. Nature. 2014;516:436–439. doi: 10.1038/nature13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chernikova S.B., Brown J.M. R-loops and genomic instability in Bre1 (RNF20/40)-deficient cells. Cell Cycle. 2012;11:2980–2984. doi: 10.4161/cc.21090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ottesen E.W., Howell M.D., Singh N.N., Seo J., Whitley E.M., Singh R.N. Severe impairment of male reproductive organ development in a low SMN expressing mouse model of spinal muscular atrophy. Sci. Rep. 2016;6:20193. doi: 10.1038/srep20193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren X., Chen X., Wang Z., Wang D. Is transcription in sperm stationary or dynamic? J. Reprod. Dev. 2017;63:439–443. doi: 10.1262/jrd.2016-093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barna M., Merghoub T., Costoya J.A., Ruggero D., Branford M., Bergia A., Samori B., Pandolfi P.P. Plzf mediates transcriptional repression of HoxD gene expression through chromatin remodeling. Dev. Cell. 2002;3:499–510. doi: 10.1016/S1534-5807(02)00289-7. [DOI] [PubMed] [Google Scholar]
- 30.Yeyati P.L., Shaknovich R., Boterashvili S., Li J., Ball H.J., Waxman S., Nason-Burchenal K., Dmitrovsky E., Zelent A., Licht J.D. Leukemia translocation protein PLZF inhibits cell growth and expression of cyclin A. Oncogene. 1999;18:925–934. doi: 10.1038/sj.onc.1202375. [DOI] [PubMed] [Google Scholar]
- 31.McConnell M.J., Chevallier N., Berkofsky-Fessler W., Giltnane J.M., Malani R.B., Staudt L.M., Licht J.D. Growth suppression by acute promyelocytic leukemia-associated protein PLZF is mediated by repression of c-myc expression. Mol. Cell Biol. 2003;23:9375–9388. doi: 10.1128/MCB.23.24.9375-9388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J.Y., English M.A., Ball H.J., Yeyati P.L., Waxman S., Licht J.D. Sequence-specific DNA binding and transcriptional regulation by the promyelocytic leukemia zinc finger protein. J. Biol. Chem. 1997;272:22447–22455. doi: 10.1074/jbc.272.36.22447. [DOI] [PubMed] [Google Scholar]
- 33.Lovelace D.L., Gao Z., Mutoji K., Song Y.C., Ruan J., Hermann B.P. The regulatory repertoire of PLZF and SALL4 in undifferentiated spermatogonia. Development. 2016;143:1893–1906. doi: 10.1242/dev.132761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phillips B.T., Gassei K., Orwig K.E. Spermatogonial stem cell regulation and spermatogenesis. Philos. Trans. R Soc. Lond. B Biol. Sci. 2010;365:1663–1678. doi: 10.1098/rstb.2010.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamaguchi Y.L., Tanaka S.S., Kumagai M., Fujimoto Y., Terabayashi T., Matsui Y., Nishinakamura R. Sall4 is essential for mouse primordial germ cell specification by suppressing somatic cell program genes. Stem Cells. 2015;33:289–300. doi: 10.1002/stem.1853. [DOI] [PubMed] [Google Scholar]
- 36.Santos-Pereira J.M., Aguilera A. R loops: New modulators of genome dynamics and function. Nat. Rev. Genet. 2015;16:583–597. doi: 10.1038/nrg3961. [DOI] [PubMed] [Google Scholar]
- 37.Nussbacher J.K., Tabet R., Yeo G.W., Lagier-Tourenne C. Disruption of RNA Metabolism in Neurological Diseases and Emerging Therapeutic Interventions. Neuron. 2019;102:294–320. doi: 10.1016/j.neuron.2019.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parker G.C., Li X., Anguelov R.A., Toth G., Cristescu A., Acsadi G. Survival motor neuron protein regulates apoptosis in an in vitro model of spinal muscular atrophy. Neurotox Res. 2008;13:39–48. doi: 10.1007/BF03033366. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Z., Lotti F., Dittmar K., Younis I., Wan L., Kasim M., Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fuller H.R., Gillingwater T.H., Wishart T.M. Commonality amid diversity: Multi-study proteomic identification of conserved disease mechanisms in spinal muscular atrophy. Neuromuscul Disord. 2016;26:560–569. doi: 10.1016/j.nmd.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 41.Maxwell G.K., Szunyogova E., Shorrock H.K., Gillingwater T.H., Parson S.H. Developmental and degenerative cardiac defects in the Taiwanese mouse model of severe spinal muscular atrophy. J. Anat. 2018;232:965–978. doi: 10.1111/joa.12793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liao H.F., Chen W.S., Chen Y.H., Kao T.H., Tseng Y.T., Lee C.Y., Chiu Y.C., Lee P.L., Lin Q.J., Ching Y.H., et al. DNMT3L promotes quiescence in postnatal spermatogonial progenitor cells. Development. 2014;141:2402–2413. doi: 10.1242/dev.105130. [DOI] [PubMed] [Google Scholar]
- 43.Kubota H., Avarbock M.R., Brinster R.L. Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proc. Natl. Acad. Sci. USA. 2004;101:16489–16494. doi: 10.1073/pnas.0407063101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsiao F.S., Cheng C.C., Peng S.Y., Huang H.Y., Lian W.S., Jan M.L., Fang Y.T., Cheng E.C., Lee K.H., Cheng W.T., et al. Isolation of therapeutically functional mouse bone marrow mesenchymal stem cells within 3 h by an effective single-step plastic-adherent method. Cell Prolif. 2010;43:235–248. doi: 10.1111/j.1365-2184.2010.00674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogawa T., Arechaga J.M., Avarbock M.R., Brinster R.L. Transplantation of testis germinal cells into mouse seminiferous tubules. Int. J. Dev. Biol. 1997;41:111–122. [PubMed] [Google Scholar]
- 46.Sato T., Katagiri K., Kubota Y., Ogawa T. In vitro sperm production from mouse spermatogonial stem cell lines using an organ culture method. Nat. Protoc. 2013;8:2098–2104. doi: 10.1038/nprot.2013.138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All animal maintenance, care, and procedures described within were reviewed and approved by the Institutional Animal Care and Use Committee of National Taiwan University (NTU) according to the protocol number (NTU-105-EL-68, 6th January 2016 approved, and NTU-107-EL-154, 11th January 2018 approved). All methods in the manuscript were performed in accordance with the relevant guidelines and regulations of NTU. Graphics and tables in the manuscript were prepared by the first and corresponding authors.