Abstract
Background
Lung cancer (LC) has become the top cause responsible for cancer-related deaths. Cell division cycle-associated (CDCA) genes exert an important role in the life process. Dysregulation in the process of cell division may lead to malignancy.
Methods
Transcriptional data on CDCA gene family and patient survival data were examined for lung cancer (LC) patients from the GEPIA, Oncomine, cBioPortal, and Kaplan–Meier Plotter databases.
Results
CDCA1/2/3/4/5/7/8 expression levels were higher in lung adenocarcinoma tissues, and the CDCA1/2/3/4/5/6/7/8 expression levels were increased in squamous cell LC tissues compared with those in noncarcinoma lung tissues. The expression levels of CDCA1/2/3/4/5/8 showed correlation with tumor classification. The Kaplan–Meier Plotter database was employed to carry out survival analysis, indicating that increased CDCA1/2/3/4/5/6/7/8 expression levels were increased in squamous cell LC tissues compared with those in noncarcinoma lung tissues. The expression levels of P < 0.05). Only LC patients with increased CDCA3/4/5/8 expression were significantly related to lower post-progression survival (PPS) (P < 0.05). Only LC patients with increased CDCA gene family and patient survival data were examined for lung cancer (LC) patients from the GEPIA, Oncomine, cBioPortal, and Kaplan–Meier Plotter databases. CDCA8, INCENP, AURKB, and BIRC5); CORUM: 127: NDC80 kinetochore complex; M129: the PID PLK1 pathway; and GO: 0007080: mitotic metaphase plate congression, all of which were remarkably modulated since the alterations affected CDCA gene family and patient survival data were examined for lung cancer (LC) patients from the GEPIA, Oncomine, cBioPortal, and Kaplan–Meier Plotter databases.
Conclusions
Upregulated CDCA genes' expression levels in LC tissues probably play a crucial part in LC oncogenesis. The upregulated CDCA genes' expression levels are used as the potential prognostic markers to improve patient survival and the LC prognostic accuracy. CDCA genes probably exert their functions in tumorigenesis through the PLK1 pathway.CDCA gene family and patient survival data were examined for lung cancer (LC) patients from the GEPIA, Oncomine, cBioPortal, and Kaplan–Meier Plotter databases. CDCA gene family and patient survival data were examined for lung cancer (LC) patients from the GEPIA, Oncomine, cBioPortal, and Kaplan–Meier Plotter databases. CDCA gene family and patient survival data were examined for lung cancer (LC) patients from the GEPIA, Oncomine, cBioPortal, and Kaplan–Meier Plotter databases.
1. Introduction
In the United States, lung cancer (LC) has turned into the top cause responsible for cancer-related deaths. According to estimation, there are over 200 thousand new LC cases and over 100 thousand deaths in 2019 [1]. LC can be classified as small-cell lung cancer (SCLC) as well as non-small-cell lung cancer (NSCLC). Among them, squamous cell carcinoma (SCC) and adenocarcinoma represent the two major NSCLC types. Nowadays, some studies have found that the platinum-based chemotherapy regimens generate a plateau, and the median overall survival (OS) is 8–14 months [2, 3]. Great progress has been made in gene-targeted therapies and immunotherapies in treating NSCLC patients, and metastatic LC patients treated with these therapies can survive for a longer period than before (over 2 years) [2, 3]. Mutations in epidermal growth factor receptor (EGFR), as well as rearrangement of ROS1 and anaplastic lymphoma kinase (ALK), are suggested as the first-line treatment for metastatic LC, which contribute a lot to cancer patient OS. [2, 3]. Besides, remarkable progress has been made in a new gene study, which has been recommended in the clinical guidelines, like neurotrophic tyrosine kinase receptor (NTRK) gene fusion. Larotrectinib has been added as the treatment option for metastatic NSCLC patients, which is sensitive to the NTRK gene fusion [4].
There are 8 respective members in the cell division cycle-associated (CDCA) gene and protein families, namely, CDCA1-8. Cell division takes an important role in the life process. It has been suggested in numerous reports that any dysregulation in the process of cell division may lead to malignancy [5–7]. CDCA2 plays a role in modulating the response of DNA injury in the cell cycle, which is achieved through binding onto protein phosphatase 1 γ (PP1γ) [8, 9]. CDCA3 functions modulate the progression of the cell cycle, and the expression level is regulated via protein degradation and transcription at the G1 phase in the cell cycle [10]. Moreover, CDCA4 can regulate the cell cycle, which is associated with the transition of the G1/S phase [11] and regulates the expression of p53 [12]. CDCA5 serves as a primary regulatory factor for the sister chromatid separation and cohesion [13]. In the undifferentiated hematopoietic populations, CDCA7 can be triggered in the precursors of hematopoietic stem cells in the murine embryo and is maintained afterwards. Additionally, CDCA8 plays an essential role in regulating mitosis [14].
This study aimed to evaluate systematically the association of CDCAs mRNA expression with LC patient survival. The CDCAs mRNA expression was detected in both normal and LC tissues. Then, the significance of all CDCA family members in predicting the prognosis for LC was analyzed based on the Kaplan–Meier Plotter database, and later the gene–gene interaction network of CDCAs was established to examine the underlying mechanisms of action. This study explored the CDCAs clinical value, so as to provide a certain theoretical foundation for making an early diagnosis, prognosis evaluation, and specific treatment for LC.
2. Materials and Methods
Each dataset used in the current work was searched based on the published literature. Gene Expression Omnibus (GEO) datasets and The Cancer Genome Atlas (TCGA) dataset were used for the analysis in the Oncomine dataset, the Gene Expression Profiling Interactive Analysis (GEPIA) dataset, and the Kaplan–Meier Plotter dataset. Additionally, the informed consent of participated subjects has been submitted by the researchers, which could be searched in the TCGA database and GEO datasets.
2.1. Oncomine Analyses
The transcription levels of CDCAs among various cancer types were examined based on the online cancer microarray database, namely, the Oncomine gene expression array dataset (www.oncomine.org). Moreover, CDCAs mRNA expression was compared between the clinical tumor samples and normal specimens. The P value was generated by Student's t-test. The threshold fold change and P value were set at 2 and 0.01, respectively.
2.2. The Gene Expression Profiling Interactive Analysis (GEPIA) Dataset
As the latest designed interactive web server, GEPIA was used to analyze RNA sequencing materials based on the GTEx and TCGA projects with the normalized processing pipeline. GEPIA allows us to offer the differential expression analyses on normal and tumor tissues, as well as the access to the profiling of cancer type and pathologic stage, analysis of patient survival, detection of a similar gene, and dimensionality reduction and correlation analyses.
2.3. The Kaplan–Meier Plotter
Kaplan–Meier Plotter (http://www.kmplot.com), the online database, was used to evaluate the prognostic significance of CDCAs mRNA expression, which offered the data on LC patient survival and gene expression. To examine the postprogression survival (PPS), progression-free survival (PFS), and overall survival (OS) of LC cases, all patient specimens were divided into two groups (namely, high and low expression groups) according to the median expression. Afterwards, the Kaplan–Meier survival plot was used for the evaluation on the basis of hazard ratio (HR) and the corresponding 95% confidence intervals (CI), as well as the log-rank Pvalue. The Kaplan–Meier plots were obtained through the CDCAs Jetset best probe set alone, where the number at risk was suggested under the major plot.
2.4. Bioinformatic Analysis and Functional Enrichment
The online database Metascape (http://metascape.org) has integrated more than 40 bioinformatic knowledge bases, which enables us to extract rich annotations, identify the enriched pathways, and construct the protein-protein interaction (PPI) network based on the lists of protein and gene identifiers. The CDCA genes were analyzed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) approaches of Metascape, so as to search for linked genes with the highest alteration frequency.
3. Results
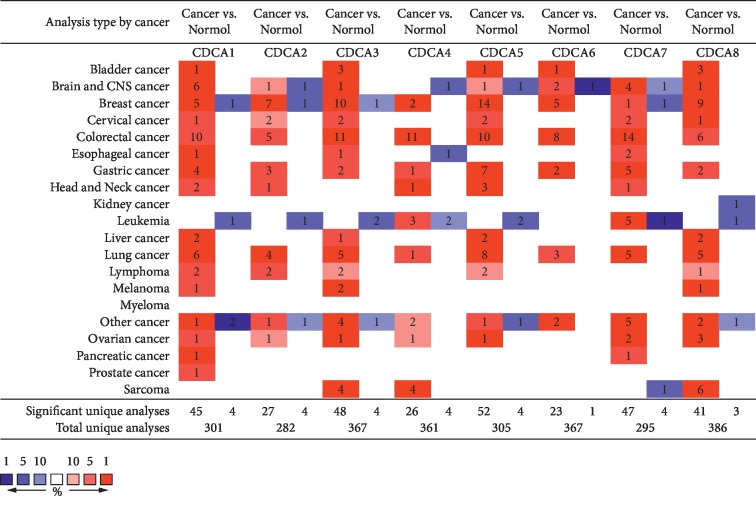
Eight CDCA factors are recognized in mammalian cells. In the present study, the Oncomine databases were used to compare CDCAs transcriptional levels between cancer tissues and normal specimens (Figure 1). According to our results, the mRNA expression of CDCAs was remarkably upregulated in LC patients of many databases. In terms of the Garber dataset, CDCA1 overexpression was detected in SCLC and SCC tissues, with the fold changes of 13.086 and 9.240, respectively [15]. In Hou et al.'s dataset, CDCA1 was overexpressed in SCC, large-cell LC, and adenocarcinoma, and the fold changes were 10.202, 13.352, and 5.248, respectively [16]. According to Okayama's dataset, CDCA1 overexpression was detected in lung adenocarcinoma, and the fold change was 3.267 [17]. For CDCA2, Hou et al.'s dataset showed that the fold changes in lung adenocarcinoma, SCC, and large-cell LC were 2.752, 4.844, and 5.076, separately [16]. Okayama et al.'s dataset also indicated CDCA2 overexpression in lung adenocarcinoma, and the fold change was 2.511 [17]. CDCA3 overexpression was found in lung adenocarcinoma, and the fold change was suggested to be 4.143 by Su et al.'s dataset [18], 2.828 by Okayama et al.'s dataset [17], and 3.551 by Hou's dataset. In Hou's dataset, CDCA3 was also expressed, and the fold change in SCC was 7.717 and that in large-cell LC was 4.431 [16]. CDCA4 was found to be overexpressed in Hou's dataset, and the fold change in SCC was 3.354 [16]. For CDCA5, the fold changes in Garber Lung's dataset were shown to be 7.928, 5.343, and 3.557 in large-cell LC, SCC, and lung adenocarcinoma in comparison with the common tissues, respectively [15]. Hou's dataset demonstrated the fold changes of 5.533, 6.249, and 2.853 in SCC, large-cell LC, and lung adenocarcinoma, respectively [16]. In addition, the CDCA5 fold changes in lung adenocarcinoma were 3.324 and 2.291 in Selamat et al.'s [19] and Okayama et al.'s datasets [17], respectively. For CDCA6, the fold changes presented in Hou's dataset were 5.371, 3.744, and 2.267 in large-cell LC, SCC, and lung adenocarcinoma compared with common tissues, respectively [16]. For CDCA7, the fold changes displayed in Hou's dataset were 5.997, 9.075, and 7.392 in lung adenocarcinoma, SCC, and large-cell LC, respectively [16]. Okayama's dataset showed that the fold change was 6.000 in lung adenocarcinoma [17]. Besides, Selamat's dataset indicated that the fold change was 2.935 in lung adenocarcinoma. For CDCA8, in Hou's dataset, the fold changes in lung adenocarcinoma, SCC, and large-cell LC were 2.935, 3.743, and 4.913, respectively, compared with normal tissues [16]. Selamat et al.'s dataset showed a fold change of 2.000 in lung adenocarcinoma [19], while Okayama et al.'s dataset presented a fold change of 5.763 in lung adenocarcinoma [17] (Table 1).
Figure 1.
CDCA expression at transcription level among various cancer types (the ONCOMINE).
Table 1.
The significant changes of CDCA expression in transcription level between types of lung cancer and normal lung tissues (Oncomine Database).
| Type of lung cancer versus normal lung tissue | Fold change | P value | t-test | Source and/or reference | |
|---|---|---|---|---|---|
| CDCA1 | Small cell lung carcinoma | 13.086 | 1.21E – 5 | 8.683 | Garber et al. [15] |
| Squamous cell lung carcinoma | 9.240 | 4.71E – 6 | 6.905 | Garber et al. [15] | |
| Squamous cell lung carcinoma | 10.202 | 1.55E – 19 | 18.306 | Hou et al. [16] | |
| Lung adenocarcinoma | 5.248 | 7.31E – 15 | 10.550 | Hou et al. [16] | |
| Large-cell lung carcinoma | 13.352 | 2.73E – 8 | 8.647 | Hou et al. [16] | |
| Lung adenocarcinoma | 3.267 | 2.26E – 12 | 10.264 | Okayama et al. [17] | |
|
| |||||
| CDCA2 | Lung adenocarcinoma | 2.752 | 3.07E – 15 | 10.285 | Hou et al. [16] |
| Squamous cell lung carcinoma | 4.844 | 1.20E – 13 | 12.093 | Hou et al. [16] | |
| Large-cell lung carcinoma | 5.076 | 1.34E – 6 | 6.586 | Hou et al. [16] | |
| Lung adenocarcinoma | 2.511 | 1.03E – 12 | 10.242 | Okayama et al. [17] | |
|
| |||||
| CDCA3 | Lung adenocarcinoma | 4.143 | 2.60E – 11 | 8.366 | Su et al. [18] |
| Squamous cell lung carcinoma | 7.717 | 5.79E – 26 | 21.275 | Hou et al. [16] | |
| Lung adenocarcinoma | 3.551 | 9.34E – 16 | 10.511 | Hou et al. [16] | |
| Large-cell lung carcinoma | 4.431 | 1.08E – 8 | 9.131 | Hou et al. [16] | |
| Lung adenocarcinoma | 2.828 | 3.60E – 12 | 10.001 | Okayama et al. [17] | |
| CDCA4 | Squamous cell lung carcinoma | 3.354 | 1.66E – 13 | 12.179 | Hou et al. [16] |
|
| |||||
| CDCA5 | Large-cell lung carcinoma | 7.928 | 2.49E – 6 | 10.744 | Garber et al. [15] |
| Squamous cell lung carcinoma | 5.343 | 8.05E – 7 | 8.173 | Garber et al. [15] | |
| Lung adenocarcinoma | 3.557 | 3.03E – 5 | 7.382 | Garber et al. [15] | |
| Squamous cell lung carcinoma | 5.533 | 1.77E – 23 | 21.214 | Hou et al. [16] | |
| Large-cell lung carcinoma | 6.249 | 1.74E – 8 | 8.843 | Hou et al. [16] | |
| Lung adenocarcinoma | 2.853 | 8.10E – 14 | 9.704 | Hou et al. [16] | |
| Lung adenocarcinoma | 3.324 | 9.19E – 20 | 13.055 | Selamat et al. [19] | |
| Lung adenocarcinoma | 2.291 | 2.02E – 9 | 8.518 | Okayama et al. [17] | |
|
| |||||
| CDCA6 | Large-cell lung carcinoma | 5.371 | 7.64E – 7 | 6.902 | Hou et al. [16] |
| Squamous cell lung carcinoma | 3.744 | 5.28E – 10 | 8.850 | Hou et al. [16] | |
| Lung adenocarcinoma | 2.267 | 1.35E – 8 | 6.564 | Hou et al. [16] | |
|
| |||||
| CDCA7 | Lung adenocarcinoma | 5.997 | 9.23E – 17 | 6.009 | Hou et al. [16] |
| Squamous cell lung carcinoma | 9.075 | 1.91E – 19 | 15.046 | Hou et al. [16] | |
| Large-cell lung carcinoma | 7.392 | 6.18E – 6 | 5.779 | Hou et al. [16] | |
| Lung adenocarcinoma | 6.000 | 1.26E – 16 | 15.108 | Okayama et al. [17] | |
| Lung adenocarcinoma | 2.935 | 2.00E – 14 | 9.214 | Selamat et al. [19] | |
|
| |||||
| CDCA8 | Lung adenocarcinoma | 2.935 | 3.18E – 15 | 10.818 | Hou et al. [16] |
| Squamous cell lung carcinoma | 3.743 | 3.32E – 17 | 15.713 | Hou et al. [16] | |
| Large-cell lung carcinoma | 4.913 | 1.07E – 8 | 9.151 | Hou et al. [16] | |
| Lung adenocarcinoma | 2.000 | 4.40E – 17 | 11.529 | Selamat et al. [19] | |
| Lung adenocarcinoma | 5.763 | 5.05E – 10 | 9.625 | Okayama et al. [17] | |
3.1. Associations of CDCAs mRNA Expression with Clinicopathological Variables in LC Patients
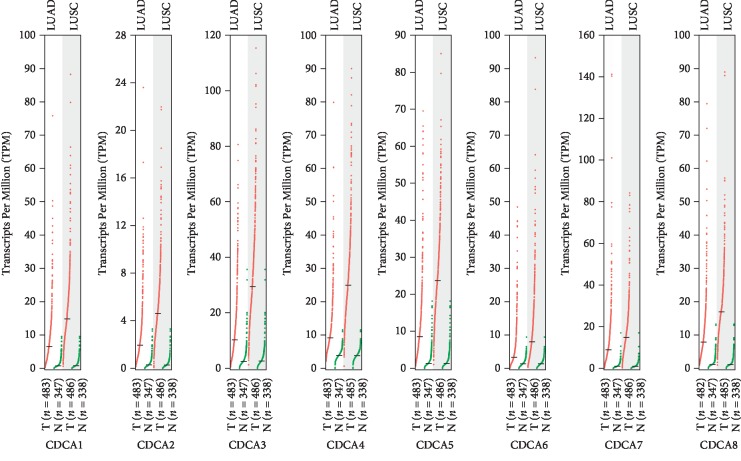
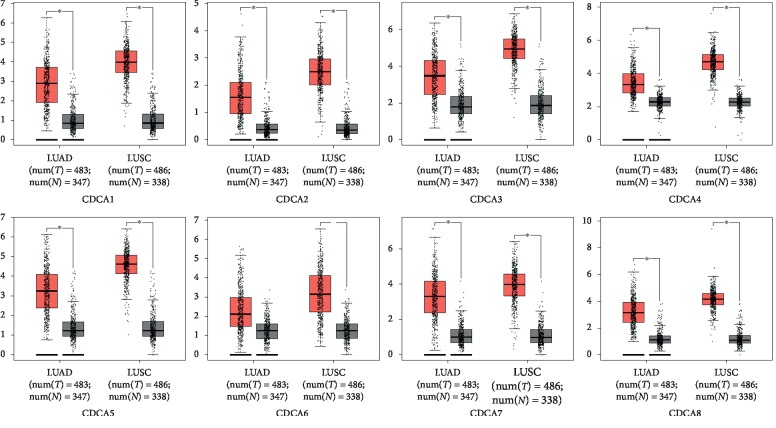
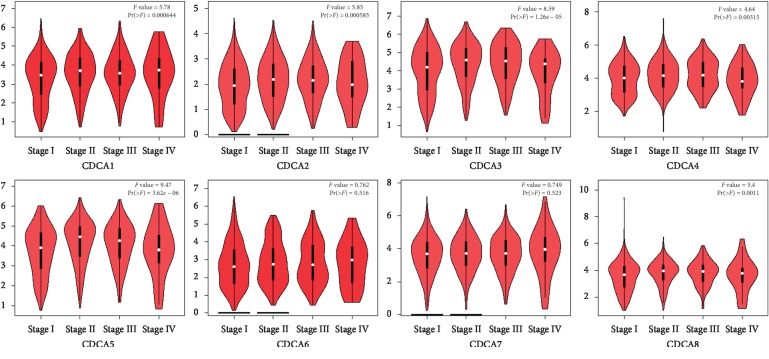
The GEPIA dataset (http://gepia.cancer-pku.cn/) was performed to compare the mRNA expression of CDCAs in LC tissues with that in normal lung tissues. According to our findings, the CDCA1/2/3/4/5/6/7/8 expression levels were upregulated in LC tissues relative to that in noncarcinoma ones (Figures 2 and 3). Additionally, the association of the expression of CDCA genes with the LC stage was analyzed. There were significant differences in CDCA1/2/3/4/5/8 expression (Figure 4).
Figure 2.
CDCA expression in LC (GEPIA).
Figure 3.
CDCA expression in LC presented in the form of a boxplot (GEPIA).
Figure 4.
Correlation of the CDCA expression with tumor stage among LC cases (GEPIA).
3.2. Relationship between Elevated CDCA 2/3/4/5/7/8 mRNA Expression and Dismal Prognosis for LC Cases
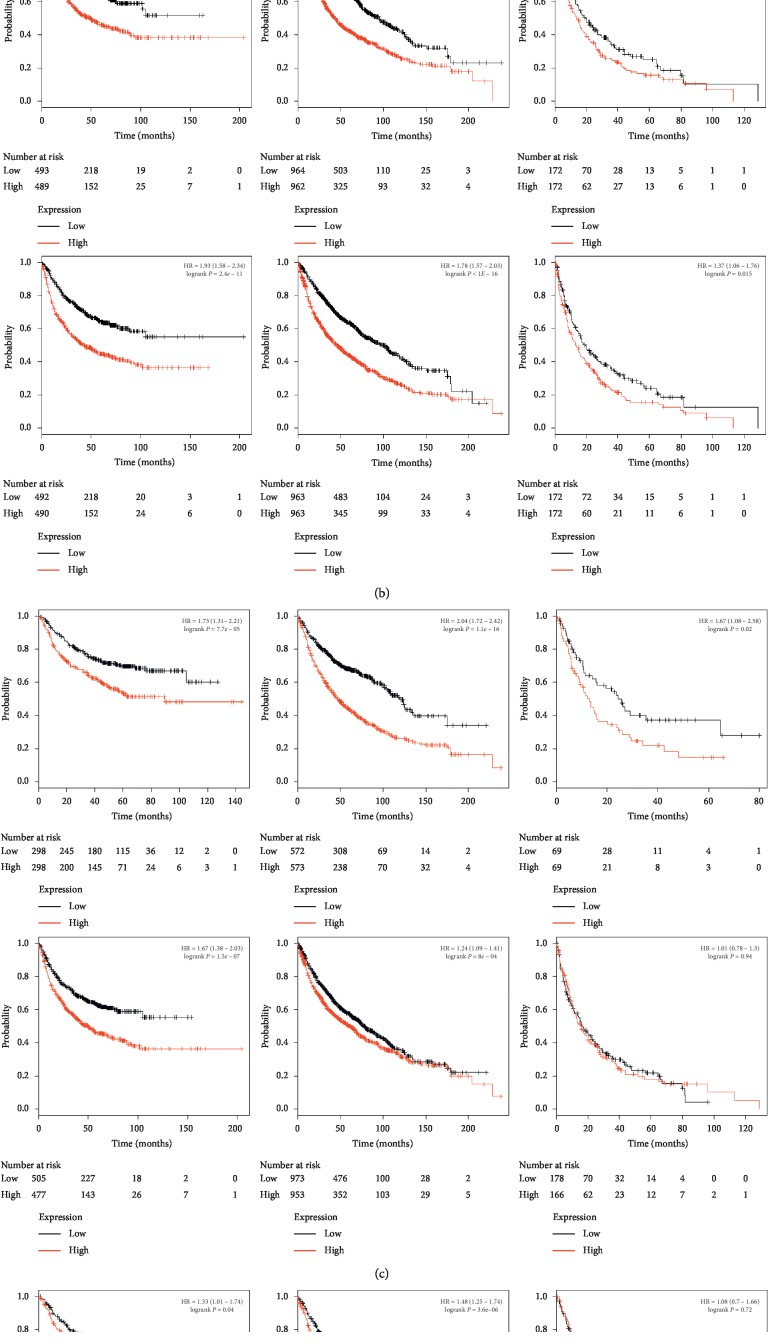
The crucial CDCAs efficiency in LC patient survival was also found. The Kaplan–Meier Plotter approach was utilized to examine the relationship of mRNA expression of CDCAs with LC patient survival based on the public datasets. Our results suggested that increased CDCA 1–8 showed a significant relationship with poorer OS and PFS (P < 0.05). Only LC patients with upregulated CDCA3/4/5/8 expression were significantly correlated with the lower PPS (P < 0.05) (Figure 5).
Figure 5.
Significance of the CDCA mRNA expression in predicting the prognosis for LC cases (Kaplan–Meier plotter).
3.3. Genetic Alteration and Correlation
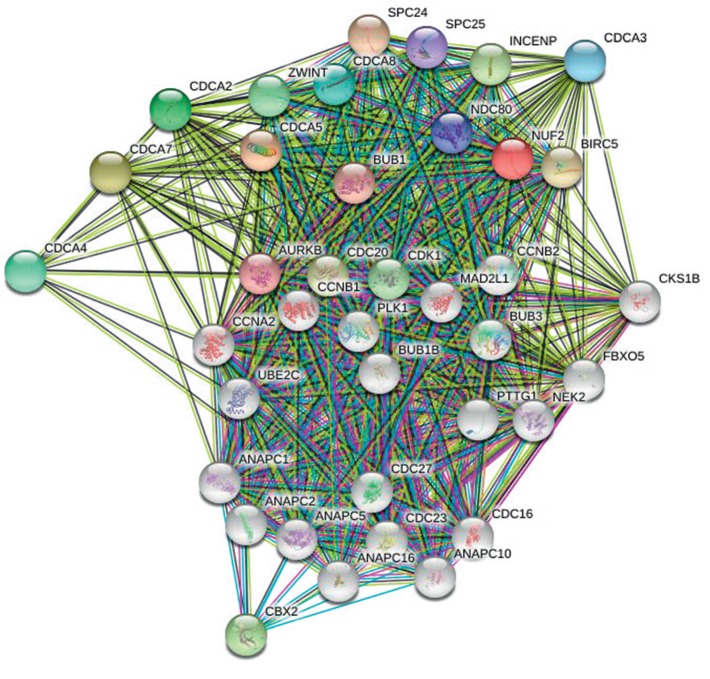
3.3.1. Pathway Enrichment Analyses and Predicted Functions of CDCA Genes among LC Cases
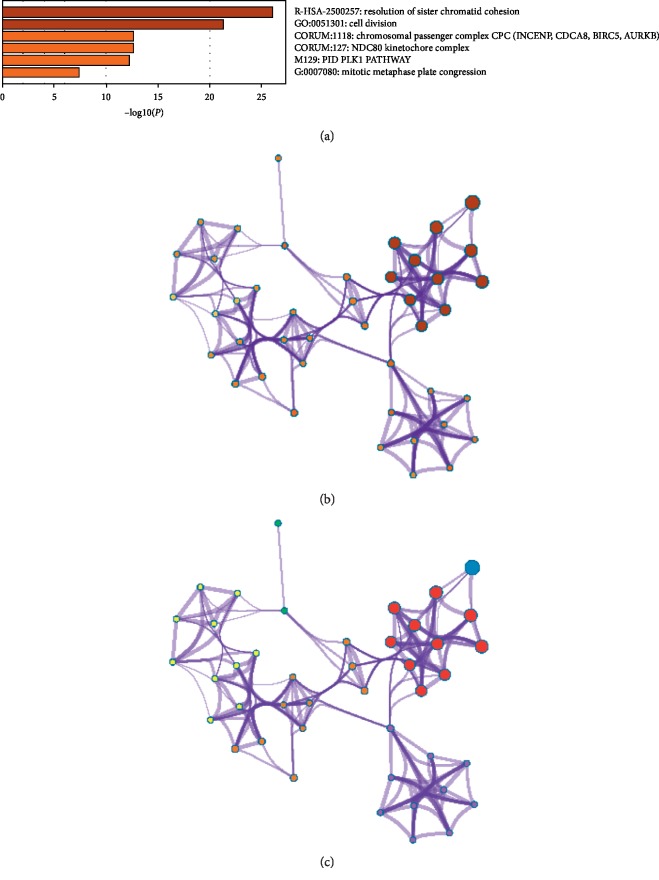
Genes showing coexpression with CDCA genes would be examined using the String and Functional protein association networks. NUF2, CDCA2, CDCA3, CDCA4, CDCA5, CDCA, CDCA7, CDCA8, CDC20, AURKB, CBX2, CDK1, ZWINT, BUB1, NDC80, SPC24, SPC25, BIRC5, and INCENP were discovered in our results (Figure 6). Then, the lists of all the CDCA genes expressed, together with linked genes displaying the highest alteration frequency, were compiled before they were analyzed by the KEGG and GO approaches in Metascape (Figure 7). According to our results, the processes below were subjected to the influence of CDCA gene alteration: R-HAS-2500257: resolution of sister chromatid cohesion; GO:0051301: cell division; CORUM: 1118: Chromosomal passenger complex (CPC, including CDCA8, INCENP, AURKB, and BIRC5); CORUM: 127: NDC80 kinetochore complex; M129: PID PLK1 pathway; and GO: 0007080: mitotic metaphase plate congression.
Figure 6.
Gene coexpression among LC cases (STRING).
Figure 7.
Functions of CDCA genes as well as those showing significant correlation with CDCA gene alterations.
4. Discussion
CDCA1, one of the Ndc80 complex members, plays a role in regulating mitosis [20], which is coexpressed with the known cell cycle genes [21] (such as cyclin and topoisomerase II). Some studies demonstrate that CDCA1 overexpression is related to the dismal prognosis for patients with colorectal cancer (CRC) [22, 23]. Moreover, the study conducted by Hayama, et al. [21] showed that CDCA1 knockdown using small interfering RNA remarkably suppressed the growth of NSCLC cells. Furthermore, CDCA1 has been used as the vaccination for patients with advanced biliary tract cancer and prostate cancer, and well toleration is achieved in these phase I clinical trials [24, 25]. The current study suggested that The Cancer Genome Atlas and the Oncomine datasets revealed higher CDCA1 expression in LC tissues than in noncarcinoma tissues. A high CDCA1 level revealed a significant correlation with worse OS in all LC patients.
CDCA2 acts as the PP1γ expression regulator, which inhibits the activation of DNA damage response [8, 9]. Recent research results demonstrate that CDCA2 methylation in HeLa cells promotes cell proliferation and suppresses apoptosis [26]. Additionally, CDCA2 overexpression promotes the proliferation of CRC cells and oral squamous cell carcinoma (OSCC) cells [27, 28]. Furthermore, a study on lung adenocarcinoma suggests that CDCA2 proliferates lung adenocarcinoma cells and predicts the poor prognosis for these patients [29]. Our results indicated that CDCA2 expression level in LC tissues was upregulated relative to that in noncarcinoma tissues. The expression of CDCA2 showed a correlation with the LC stage. High CDCA2 expression level displayed a significant correlation with the improved OS for all LC patients.
CDCA3 controls the G1 phase [30], which acts as one of the prognostic genes for hepatocellular carcinoma (HCC) [31] and is also involved in LC cell proliferation, migration, invasion, and apoptosis [30], as well as CRC cell proliferation [32]. Moreover, it has been reported that CDCA3 expression is related to prognosis for bladder cancer cases [33] and luminal A breast cancer [34]. Current studies show that overexpression of CDCA3 frequently occurs in the process of oral carcinogenesis [35]. It was discovered that CDCA3 expression was upregulated among LC tissues compared with that in noncarcinoma counterparts, but not with the LC stage. Additionally, the upregulated CDCA3 expression showed a significant correlation with the improved PFS, OS, and PPS among all LC patients.
CDCA4 protein expression is found in some human cells, which can be induced when cells enter the G1/S phase in the cell cycle [11]. In a previous study, Hayashi et al. showed that CDCA4 participated in cell proliferation [11]. Moreover, CDCA4 is involved in the triple-negative breast cancer (TNBC) cells [36], and it is shown that RNA interference of CDCA4 markedly increases cell apoptotic rate. In addition, one recent study suggests that CDCA4 enhances human BC cell proliferation and reduces their apoptosis [37]. In this study, we found that CDCA4 expression was increased in human LC tissues relative to that in noncarcinoma tissues, and such expression showed a correlation with the LC stage. The upregulated CDCA4 expression showed a marked correlation with the improved PFS and OS of all LC patients.
A recent study shows that CDCA5 probably serves as a biomarker for the prognosis, treatment, and diagnosis for HCC [38–40]. It also exerts a vital part in the proliferation of HCC cells [41, 42], OSCC [41, 42], and bladder cancer [43]. For digestive system cancer, CDCA5 is found to play crucial roles in the proliferation of gastric cancer cells [44]. Moreover, CDCA5 is also differentially expressed in patients with localized and locally advanced prostate cancer [45]. Regarding LC, the transactivation of CDCA5 and its phosphorylation exert vital parts in the proliferation of LC cells [13]. Wu et al. [46] also indicated that CDCA5 acted as a novel promising target for NSCLC diagnosis and treatment. In this study, the CDCA5 expression level was downregulated in LC tissues compared with that in noncarcinoma counterparts. Besides, such expression showed an association with the LC stage. Obviously, the high CDCA5 expression displayed a significant correlation with the improved OS for all LC patients.
CDCA7 has been recognized as an MYC-target gene [47]. A recent study shows that CDCA7 is overexpressed in lymphoid tumors, and CDCA7 knockdown decreases the growth rate of the lymphoid tumor, without inhibiting the proliferation of normal cells [48]. In this study, the CDCA7 expression level was upregulated in human LC tissues compared with that in noncarcinoma counterparts, and such expression showed no correlation with the LC stage. Obviously, the high CDCA7 expression displayed a remarkable correlation with the improved PFS and OS in all LC patients.
CDCA8 protein has been identified as an integral part of the vertebrate chromosomal passenger complex (cPc) [49]. The expression of CDCA8 is closely associated with tumor progression, N stage, T stage, and grade of bladder cancer [50]. CDCA8 is related to the distant metastasis risk of breast cancer [51, 52]. With regard to renal cancer, CDCA8 has also certain prognostic value [53]. CDCA8 promotes the malignant progression of cutaneous melanoma [54]. Furthermore, CDCA8 also exerts a vital part during lung carcinogenesis [55]. In this study, the CDCA8 expression level was upregulated in LC tissues relative to that in noncarcinoma counterparts, and such expression exerted no correlation with the LC stage. Obviously, the high CDCA8 expression showed a close association with the improved PFS and OS of all LC patients.
Besides, KEGG and GO analyses were also carried out to find the correlations between CDCA genes' expression and linked genes of the highest alteration frequency and the prognosis for LC. According to our results, attention should be paid to some pathways including R-HAS-2500257: resolution of sister chromatid cohesion; GO:0051301: cell division; CORUM: 1118: chromosomal passenger complex (CPC, including CDCA8, INCENP, AURKB, and BIRC5); CORUM: 127: NDC80 kinetochore complex; M129: PID PLK1 pathway; and GO: 0007080: mitotic metaphase plate congression. Previous studies show that the Polo-like kinase 1(PLK1) is highly expressed in LC, which predicts the poor survival in metastatic LC patients [56, 57]. In addition, the PLK1 pathway plays a certain role in the progression of HCC [58], glioma [59], and lung adenocarcinoma [60].
The current research systemically examines the expression of CDCA genes and its prognostic significance in LC, which sheds more light on the complexity and heterogeneity of LC biological properties at the molecular level. Based on our results, CDCAs upregulation in LC tissues probably exerts a crucial part during LC oncogenesis. Besides, CDCAs upregulation can serve as a potential prognostic marker to improve the survival and prognostic accuracy for LC. Moreover, CDCA genes probably exert their functions in tumorigenesis through the PLK1 pathway.
Acknowledgments
This work was supported by the grant from the Medical Research Fund of Guangdong Province (no. A2018237).
Contributor Information
Jielan Lai, Email: laijl@sysucc.org.cn.
Huan Li, Email: lihuan@sysucc.org.cn.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
All authors declare no conflicts of interest.
Authors' Contributions
HL and LJL designed the research protocol, analyzed data, and revised the manuscript. CXC and SLC searched and analyzed the data. CXC and LLP wrote the manuscript and participated in analyzing data. HHY and ML participated in searching the data. QYZ made a lot of work in reviewing and revising the manuscript. All authors read and approved the final manuscript. Chongxiang Chen and Siliang Chen contributed equally to this work.
References
- 1.Siegel R. L., Miller K. D., Jemal A. Cancer statistics, 2019. CA: A Cancer Journal for Clinicians. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Reck M., Rodriguez-Abreu D., Robinson A. G., et al. Updated analysis of KEYNOTE-024: pembrolizumab versus platinum-based chemotherapy for advanced non-small-cell lung cancer with PD-L1 tumor proportion score of 50% or greater. Journal of Clinical Oncology. 2019;37(7):537–546. doi: 10.1200/jco.18.00149. [DOI] [PubMed] [Google Scholar]
- 3.Zhao D., Chen X., Qin N., et al. The prognostic role of EGFR-TKIs for patients with advanced non-small cell lung cancer. Scientific Reports. 2017;7 doi: 10.1038/srep40374.40374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drilon A., Laetsch T. W., Kummar S., et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. New England Journal of Medicine. 2018;378(8):731–739. doi: 10.1056/nejmoa1714448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Preston-Martin S., Pike M. C., Ross R. K., Jones P. A., Henderson B. E. Increased cell division as a cause of human cancer. Cancer Research. 1990;50(23):7415–7421. [PubMed] [Google Scholar]
- 6.Vader G., Lens S. M. The Aurora kinase family in cell division and cancer. Biochim Biophys Acta. 2008;1786(1):60–72. doi: 10.1016/j.bbcan.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Collins I., Garrett M. Targeting the cell division cycle in cancer: CDK and cell cycle checkpoint kinase inhibitors. Current Opinion In Pharmacology. 2005;5(4):366–373. doi: 10.1016/j.coph.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 8.Peng A., Lewellyn A. L., Schiemann W. P., Maller J. L. Repo-man controls a protein phosphatase 1-dependent threshold for DNA damage checkpoint activation. Current Biology. 2010;20(5):387–396. doi: 10.1016/j.cub.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vagnarelli P. Repo-man at the intersection of chromatin remodelling, DNA repair, nuclear envelope organization, and cancer progression. Cancer Biology and the Nuclear Envelope. 2014;773:401–414. doi: 10.1007/978-1-4899-8032-8_18. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida K. Cell-cycle-dependent regulation of the human and mouse Tome-1 promoters. FEBS Letters. 2005;579(6):1488–1492. doi: 10.1016/j.febslet.2005.01.055. [DOI] [PubMed] [Google Scholar]
- 11.Hayashi R., Goto Y., Ikeda R., Yokoyama K. K., Yoshida K. CDCA4 is an E2F transcription factor family-induced nuclear factor that regulates E2F-dependent transcriptional activation and cell proliferation. Journal of Biological Chemistry. 2006;281(47):35633–35648. doi: 10.1074/jbc.m603800200. [DOI] [PubMed] [Google Scholar]
- 12.Tategu M., Nakagawa H., Hayashi R., Yoshida K. Transcriptional co-factor CDCA4 participates in the regulation of JUN oncogene expression. Biochimie. 2008;90(10):1515–1522. doi: 10.1016/j.biochi.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 13.Nguyen M.-H., Koinuma J., Ueda K., et al. Phosphorylation and activation of cell division cycle associated 5 by mitogen-activated protein kinase play a crucial role in human lung carcinogenesis. Cancer Research. 2010;70(13):5337–5347. doi: 10.1158/0008-5472.can-09-4372. [DOI] [PubMed] [Google Scholar]
- 14.Higuchi T., Uhlmann F. Passenger acrobatics. Nature. 2003;426(6968):780–781. doi: 10.1038/426780a. [DOI] [PubMed] [Google Scholar]
- 15.Garber M. E., Troyanskaya O. G., Schluens K., et al. Diversity of gene expression in adenocarcinoma of the lung. Proceedings of the National Academy of Sciences. 2001;98(24):13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hou J., Aerts J., den Hamer B., et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5(4) doi: 10.1371/journal.pone.0010312.e10312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okayama H., Kohno T., Ishii Y., et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Research. 2012;72(1):100–111. doi: 10.1158/0008-5472.can-11-1403. [DOI] [PubMed] [Google Scholar]
- 18.Su L.-J., Chang C.-W., Wu Y.-C., et al. Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics. 2007;8(1):p. 140. doi: 10.1186/1471-2164-8-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selamat S. A., Chung B. S., Girard L., et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Research. 2012;22(7):1197–1211. doi: 10.1101/gr.132662.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeLuca J. G., Howell B. J., Canman J. C., Hickey J. M., Fang G., Salmon E. D. Nuf2 and Hec1 are required for retention of the checkpoint proteins Mad1 and Mad2 to kinetochores. Current Biology. 2003;13(23):2103–2109. doi: 10.1016/j.cub.2003.10.056. [DOI] [PubMed] [Google Scholar]
- 21.Hayama S., Daigo Y., Kato T., et al. Activation of CDCA1-KNTC2, members of centromere protein complex, involved in pulmonary carcinogenesis. Cancer Research. 2006;66(21):10339–10348. doi: 10.1158/0008-5472.can-06-2137. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi Y., Takano A., Miyagi Y., et al. Cell division cycle-associated protein 1 overexpression is essential for the malignant potential of colorectal cancers. International Journal of Oncology. 2014;44(1):69–77. doi: 10.3892/ijo.2013.2177. [DOI] [PubMed] [Google Scholar]
- 23.Miyata Y., Kumagai K., Nagaoka T., et al. Clinicopathological significance and prognostic value of Wilms’ tumor gene expression in colorectal cancer. Cancer Biomarkers. 2015;15(6):789–797. doi: 10.3233/cbm-150521. [DOI] [PubMed] [Google Scholar]
- 24.Aruga A., Takeshita N., Kotera Y., et al. Phase I clinical trial of multiple-peptide vaccination for patients with advanced biliary tract cancer. Journal of Translational Medicine. 2014;12(1):p. 61. doi: 10.1186/1479-5876-12-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Obara W., Sato F., Takeda K., et al. Phase I clinical trial of cell division associated 1 (CDCA1) peptide vaccination for castration resistant prostate cancer. Cancer Science. 2017;108(7):1452–1457. doi: 10.1111/cas.13278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li C.-W., Chen B.-S. Investigating core genetic-and-epigenetic cell cycle networks for stemness and carcinogenic mechanisms, and cancer drug design using big database mining and genome-wide next-generation sequencing data. Cell Cycle. 2016;15(19):2593–2607. doi: 10.1080/15384101.2016.1198862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng Y., Qian W., Zhang Y., et al. CDCA2 promotes the proliferation of colorectal cancer cells by activating the AKT/CCND1 pathway in vitro and in vivo. BMC Cancer. 2019;19(1):p. 576. doi: 10.1186/s12885-019-5793-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uchida F., Uzawa K., Kasamatsu A., et al. Overexpression of CDCA2 in human squamous cell carcinoma: correlation with prevention of G1 phase arrest and apoptosis. PLoS One. 2013;8(2) doi: 10.1371/journal.pone.0056381.e56381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi R., Zhang C., Wu Y., et al. CDCA2 promotes lung adenocarcinoma cell proliferation and predicts poor survival in lung adenocarcinoma patients. Oncotarget. 2017;8(12):19768–19779. doi: 10.18632/oncotarget.15519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu Q., Fu J., Luo B., et al. OY-TES-1 may regulate the malignant behavior of liver cancer via NANOG, CD9, CCND2 and CDCA3: a bioinformatic analysis combine with RNAi and oligonucleotide microarray. Oncology Reports. 2015;33(4):1965–1975. doi: 10.3892/or.2015.3792. [DOI] [PubMed] [Google Scholar]
- 31.Guan L., Luo Q., Liang N., Liu H. A prognostic prediction system for hepatocellular carcinoma based on gene co-expression network. Experimental and Therapeutic Medicine. 2019;17(6):4506–4516. doi: 10.3892/etm.2019.7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qian W., Zhang Z., Peng W., et al. CDCA3 mediates p21-dependent proliferation by regulating E2F1 expression in colorectal cancer. International Journal of Oncology. 2018;53(5):2021–2033. doi: 10.3892/ijo.2018.4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li S., Liu X., Liu T., et al. Identification of biomarkers correlated with the TNM staging and overall survival of patients with bladder cancer. Frontiers in Physiology. 2017;8:p. 947. doi: 10.3389/fphys.2017.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perez-Pena J., Alcaraz-Sanabria A., Nieto-Jimenez C., et al. Mitotic read-out genes confer poor outcome in luminal A breast cancer tumors. Oncotarget. 2017;8(13):21733–21740. doi: 10.18632/oncotarget.15562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uchida F., Uzawa K., Kasamatsu A., et al. Overexpression of cell cycle regulator CDCA3 promotes oral cancer progression by enhancing cell proliferation with prevention of G1 phase arrest. BMC Cancer. 2012;12:p. 321. doi: 10.1186/1471-2407-12-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pang S., Xu Y., Chen J., Li G., Huang J., Wu X. Knockdown of cell division cycle-associated protein 4 expression inhibits proliferation of triple negative breast cancer MDA-MB-231 cells in vitro and in vivo. Oncology Letters. 2019;17(5):4393–4400. doi: 10.3892/ol.2019.10077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu Y., Wu X., Li F., Huang D., Zhu W. CDCA4, a downstream gene of the Nrf2 signaling pathway, regulates cell proliferation and apoptosis in the MCF7/ADM human breast cancer cell line. Molecular Medicine Reports. 2017;17(1):1507–1512. doi: 10.3892/mmr.2017.8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cai C., Wang W., Tu Z. Aberrantly DNA methylated-differentially expressed genes and pathways in hepatocellular carcinoma. Journal of Cancer. 2019;10(2):355–366. doi: 10.7150/jca.27832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tian Y., Wu J., Chagas C., et al. CDCA5 overexpression is an Indicator of poor prognosis in patients with hepatocellular carcinoma (HCC) BMC Cancer. 2018;18(1):p. 1187. doi: 10.1186/s12885-018-5072-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J., Xia C., Pu M., et al. Silencing of CDCA5 inhibits cancer progression and serves as a prognostic biomarker for hepatocellular carcinoma. Oncology Reports. 2018;40(4):1875–1884. doi: 10.3892/or.2018.6579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen H., Chen J., Zhao L., et al. CDCA5, transcribed by E2F1, promotes oncogenesis by enhancing cell proliferation and inhibiting apoptosis via the AKT pathway in hepatocellular carcinoma. Journal of Cancer. 2019;10(8):1846–1854. doi: 10.7150/jca.28809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tokuzen N., Nakashiro K., Tanaka H., Iwamoto K., Hamakawa H. Therapeutic potential of targeting cell division cycle associated 5 for oral squamous cell carcinoma. Oncotarget. 2016;7(3):2343–2353. doi: 10.18632/oncotarget.6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pan S., Zhan Y., Chen X., Wu B., Liu B. Identification of biomarkers for controlling cancer stem cell characteristics in bladder cancer by network analysis of transcriptome data stemness indices. Frontiers in Oncology. 2019;9:p. 613. doi: 10.3389/fonc.2019.00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen T., Huang Z., Tian Y., et al. Role of triosephosphate isomerase and downstream functional genes on gastric cancer. Oncology Reports. 2017;38(3):1822–1832. doi: 10.3892/or.2017.5846. [DOI] [PubMed] [Google Scholar]
- 45.Tolkach Y., Merseburger A., Herrmann T., Kuczyk M., Serth J., Imkamp F. Signatures of adverse pathological features, androgen insensitivity and metastatic potential in prostate cancer. Anticancer Research. 2015;35(10):5443–5451. [PubMed] [Google Scholar]
- 46.Wu Q., Zhang B., Sun Y., et al. Identification of novel biomarkers and candidate small molecule drugs in non-small-cell lung cancer by integrated microarray analysis. OncoTargets and Therapy. 2019;12:3545–3563. doi: 10.2147/ott.s198621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prescott J. E., Osthus R. C., Lee L. A., et al. A novel c-Myc-responsive Gene, JPO1, participates in neoplastic transformation. Journal Of Biological Chemistry. 2001;276(51):48276–48284. doi: 10.1074/jbc.m107357200. [DOI] [PubMed] [Google Scholar]
- 48.Martin-Cortazar C., Chiodo Y., Jimenez R. P., et al. CDCA7 finely tunes cytoskeleton dynamics to promote lymphoma migration and invasion. Haematologica. 2019;105(1) doi: 10.3324/haematol.2018.215459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Phan N. N., Wang C. Y., Li K. L., et al. Distinct expression of CDCA3, CDCA5, and CDCA8 leads to shorter relapse free survival in breast cancer patient. Oncotarget. 2018;9(6):6977–6992. doi: 10.18632/oncotarget.24059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bi Y., Chen S., Jiang J., et al. CDCA8 expression and its clinical relevance in patients with bladder cancer. Medicine. 2018;97(34) doi: 10.1097/md.0000000000011899.e11899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cai Y., Mei J., Xiao Z., et al. Identification of five hub genes as monitoring biomarkers for breast cancer metastasis in silico. Hereditas. 2019;156:p. 20. doi: 10.1186/s41065-019-0096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kabisch M., Lorenzo B. J., Dunnebier T., et al. Inherited variants in the inner centromere protein (INCENP) gene of the chromosomal passenger complex contribute to the susceptibility of ER-negative breast cancer. Carcinogenesis. 2015;36(2):256–271. doi: 10.1093/carcin/bgu326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gu Y., Lu L., Wu L., Chen H., Zhu W., He Y. Identification of prognostic genes in kidney renal clear cell carcinoma by RNA-seq data analysis. Molecular Medicine Reports. 2017;15(4):1661–1667. doi: 10.3892/mmr.2017.6194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ci C., Tang B., Lyu D., et al. Overexpression of CDCA8 promotes the malignant progression of cutaneous melanoma and leads to poor prognosis. International Journal of Molecular Medicine. 2018;43(1):404–412. doi: 10.3892/ijmm.2018.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hayama S., Daigo Y., Yamabuki T., et al. Phosphorylation and activation of cell division cycle associated 8 by aurora kinase B plays a significant role in human lung carcinogenesis. Cancer Research. 2007;67(9):4113–4122. doi: 10.1158/0008-5472.can-06-4705. [DOI] [PubMed] [Google Scholar]
- 56.Shin S. B., Jang H. R., Xu R., Won J. Y., Yim H. Active PLK1-driven metastasis is amplified by TGF-beta signaling that forms a positive feedback loop in non-small cell lung cancer. Oncogene. 2019;39(4):767–785. doi: 10.1038/s41388-019-1023-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liao Y., Yin G., Wang X., Zhong P., Fan X., Huang C. Identification of candidate genes associated with the pathogenesis of small cell lung cancer via integrated bioinformatics analysis. Oncology Letters. 2019;18(4):3723–3733. doi: 10.3892/ol.2019.10685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li R., Jiang X., Zhang Y., et al. Cyclin B2 overexpression in human hepatocellular carcinoma is associated with poor prognosis. Archives Of Medical Research. 2019;50(1):10–17. doi: 10.1016/j.arcmed.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 59.Zhang R., Wei R. L., Du W., et al. Long noncoding RNA ENST00000413528 sponges microRNA-593-5p to modulate human glioma growth via polo-like kinase 1. CNS Neuroscience & Therapeutics. 2019;25(8):842–854. doi: 10.1111/cns.13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dalvi P. S., Macheleidt I. F., Lim S.-Y., et al. LSD1 inhibition attenuates tumor growth by disrupting PLK1 mitotic pathway. Molecular Cancer Research. 2019;17(6):1326–1337. doi: 10.1158/1541-7786.mcr-18-0971. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.