Highlights
-
•
Found increased subcortical asymmetry associated with autism.
-
•
Utilized a new measure of shape asymmetry for analysis of structural differences.
-
•
Observed significantly increased shape asymmetry of the hippocampus.
-
•
Observed significantly increased volumetric asymmetry in the lateral ventricles.
-
•
Focalized abnormalities may result in detectable shape (but not volume) differences.
Keywords: Autism, Asymmetry, MRI, Amygdala, Hippocampus, Ventricles
Abstract
Autism spectrum disorder (ASD) is a prevalent and fast-growing pervasive neurodevelopmental disorder worldwide. Despite the increasing prevalence of ASD and the breadth of research conducted on the disorder, a conclusive etiology has yet to be established and controversy still exists surrounding the anatomical abnormalities in ASD. In particular, structural asymmetries have seldom been investigated in ASD, especially in subcortical regions. Additionally, the majority of studies for identifying structural biomarkers associated with ASD have focused on small sample sizes. Therefore, the present study utilizes a large-scale, multi-site database to investigate asymmetries in the amygdala, hippocampus, and lateral ventricles, given the potential involvement of these regions in ASD. Contrary to prior work, we are not only computing volumetric asymmetries, but also shape asymmetries, using a new measure of asymmetry based on spectral shape descriptors. This measure represents the magnitude of the asymmetry and therefore captures both directional and undirectional asymmetry. The asymmetry analysis is conducted on 437 individuals with ASD and 511 healthy controls using T1-weighted MRI scans from the Autism Brain Imaging Data Exchange (ABIDE) database. Results reveal significant asymmetries in the hippocampus and the ventricles, but not in the amygdala, in individuals with ASD. We observe a significant increase in shape asymmetry in the hippocampus, as well as increased volumetric asymmetry in the lateral ventricles in individuals with ASD. Asymmetries in these regions have not previously been reported, likely due to the different characterization of neuroanatomical asymmetry and smaller sample sizes used in previous studies. Given that these results were demonstrated in a large cohort, such asymmetries may be worthy of consideration in the development of neurodiagnostic classification tools for ASD.
1. Introduction
Autism spectrum disorder (ASD) is a pervasive neurodevelopmental disorder that begins in early childhood and persists throughout life. ASD is characterized by impairments in communication and social interaction, along with the exhibition of stereotyped repetitive patterns of behavior and restricted interests (DSM-V, American Psychiatric Association, 2013). Prevalence estimates continue to suggest that ASD is one of the most prevalent developmental disorders (CDC 2014; Brownsell et al., 2011; Elsabbagh et al., 2012; Rice et al., 2012), estimated to occur in approximately one in every 160 individuals worldwide (World Health Organization, 2013). Despite the prevalence of the disorder, however, research has not yet been able to determine a conclusive etiology (Hughes, 2008; Pendergrass et al., 2014).
In recent years, extensive research utilizing magnetic resonance imaging (MRI) has been conducted to investigate structural and functional differences found in ASD, and various neuroanatomical differences have been proposed as potential biomarkers for the disorder (for a recent review, see Li et al., 2017). Although MRI has enabled researchers to search for neuroanatomical biomarkers and attempt to develop MRI-based classification algorithms and computer-aided diagnostic systems to facilitate diagnosis (Dekhil et al., 2017; Ecker et al., 2010; Ismail, 2016; Mateos-Perez et al., 2018; Mostapha et al., 2015; Nielsen et al., 2012; Plitt et al., 2014), findings of potential structural biomarkers for ASD have been mixed and partly inconclusive.
The amygdala has consistently been a structure of interest in the search for a neuropathological classification of autism, given its important role in emotional and social functions that are impaired in individuals with ASD (Baron-Cohen et al., 2000; Brothers, 1990; Howard et al., 2000). The hippocampus is frequently investigated in tandem with the amygdala, due to its connection to the amygdala within the limbic system and its involvement in core functions in the “social brain” (Crespi and Badcock, 2008; Goodman et al., 2014; Groen et al., 2010; Hughes, 2008). Despite that both structures have been studied in individuals with ASD, research has demonstrated inconsistent evidence for structural differences in these regions. While many researchers have found increased volume in the amygdala (Groen et al., 2010; Howard et al., 2000; Kim et al., 2010; Munson et al., 2006; Murphy et al., 2012; Nordahl et al., 2012; Sparks et al., 2002; Schumann et al., 2004) others have found no evidence for this increase (Haar et al., 2014) and one study even found significantly decreased volume within the amygdala (Aylward et al., 1999). Similarly, while some studies have found increased volume in the hippocampus (Groen et al., 2010; Murphy et al., 2012, Rojas et al., 2006), others have found no significant differences in hippocampal volume (Aylward et al., 1999; Piven et al., 1998) and some have found significantly decreased volume within the hippocampus (Eilam-Stock et al., 2016; Nicolson et al., 2006; Schumann et al., 2004; Sparks et al., 2002).
Ventricles have also gained much attention in ASD research, given that morphometric differences such as lesions and hypertrophy of neighboring brain volume (Carper et al., 2002; DeLong and Bauman, 1987; Herbert, 2005; Vidal et al., 2008) have been found in ASD, and such issues of hemispheric development are likely to exert influence on the shape and size of the lateral ventricles (Geschwind and Galaburda, 1985). The majority of studies have found greater ventricular volumes associated with ASD (Filipek et al., 1992; Howard et al., 2000; Lange et al., 2015; McAlonan et al., 2002; Movsas et al., 2013; Palmen et al., 2005; Piven et al., 1995; Turner et al., 2016; Wolfe et al., 2015) and longitudinal research has also recently identified ventricular enlargement as a risk factor for ASD in a low birth weight population (Movsas et al., 2013). However, some studies have not found evidence for increased ventricular volumes (Creasey et al., 1986; Garber et al., 1989; Hardan et al., 2001; Jacobson et al., 1988), and one study even found reductions in ventricular volume in the right and left frontal and occipital horns (Vidal et al., 2008). Although several studies have investigated volumetric differences in the ventricles, little attention has been given to asymmetry of the lateral ventricles and the neighboring subcortical structures in ASD.
Although recent research has linked asymmetric brain alterations to several psychiatric disorders, such as anorexia nervosa (Titova et al., 2013) and schizophrenia (Oertel-Knochel et al., 2012), clear asymmetry patterns for ASD have not yet been established. While Herbert et al. (2005) found ASD-related asymmetry differences in the higher-order association cortex and Haznedar et al. (2006) found a reversal of typical hemispheric asymmetry in ASD patients, these results were observed in small samples (N = 16 and N = 15, respectively) and have yet to be replicated in larger samples. Atypical diffusion tensor hemispheric asymmetry has been observed within the superior temporal gyrus and temporal stem of individuals with ASD (Lange et al., 2010), however, these results have also yet to be replicated in a large sample. Dougherty et al. (2016) recently found evidence for abnormal asymmetry of the fusiform gyrus associated with symptom severity in a large sample of males with ASD, however, aside from these studies, asymmetry research on structures thought to underlie impaired processes in ASD is quite sparse. Recently, Monterrey et al. (2017) reported incidental findings of hippocampal asymmetry in twin pairs with ASD (N = 15), as well as ventricular abnormalities (i.e. either enlargement or asymmetry), which occurred more frequently in twin pairs with ASD (N = 18 pairs) than in control twin pairs (N = 10 pairs), however, these findings have not yet been replicated in larger samples. While relationships have been found between other psychiatric disorders and abnormal asymmetries of subcortical structures, such as the amygdala in schizophrenia (Niu et al., 2004), the hippocampus in depression (Xia et al., 2004), and both the hippocampus and the amygdala in Alzheimer's disease (Wachinger et al., 2016), asymmetry patterns within these subcortical structures have not yet been established for ASD.
It is important to note that the majority of studies conducted on structural abnormalities in ASD, especially studies investigating subcortical structures, have relied on small sample sizes. Although the recent advent of the Autism Brain Imaging Data Exchange (ABIDE) database has permitted research using a larger number of subjects, few studies have utilized the ABIDE sample to investigate potential ASD-related differences in subcortical regions (Cerliani et al., 2015; Plitt et al., 2014; Turner et al., 2016). The ABIDE dataset has received some criticism due to its breadth across various sites utilizing different scanners, given the inherent confounding effects of scanner variations (Auzias et al., 2015), however, research investigating such confounding scanner effects has demonstrated that robust results may still be obtained across images obtained from different scanners in multisite analyses (Noble et al., 2017).
Additionally, ABIDE presents the unique opportunity to include a large sample of females within analyses. Research including females with ASD is limited, likely in part due to the differential prevalence rates among males and females (Fombonne, 2009), but also due to the fact that several studies on ASD have excluded females from analyses altogether. Therefore, the present study will include females in the analyses, given that a large sample is available in the ABIDE dataset.
Given the scarcity of asymmetry research and the problems inherent to findings in small ASD samples, we therefore aim to investigate ASD-related asymmetry within subcortical structures, namely, the amygdala, hippocampus, and lateral ventricles within a large sample of ASD patients and healthy controls. The development of hemispheric asymmetry is noted to underlie important functions, such as visuospatial processing (Zhen et al., 2017) and language (Vigneau et al. 2006), and although many asymmetries are known to emerge throughout normal development, others emerge as a result of disordered hemispheric development, such as disturbed neuronal migration or the emergence of atrophic lesions (Geschwind and Galaburda, 1985). Such disturbances in neuronal development have been hypothesized to underlie various learning disabilities and psychiatric conditions (Geschwind and Galaburda, 1985). Although asymmetries within subcortical structures have previously been linked to an increased susceptibility to cognitive and psychiatric disorders (for a detailed review and large-scale asymmetry study, see Guadalupe et al., 2016), little is known about the relationship between subcortical asymmetries and ASD. Thus, we analyze asymmetries of contralateral brain structures that have been previously implicated in ASD, as they present a unique, subject-specific reference element for comparison and can potentially serve as a personalized marker of the disorder. Concurrently to our work, Postema et al. (2019) have also investigated subcortical volume asymmetries in ASD.
While most existing asymmetry analyses utilize purely volumetric measures of asymmetry, we aim to analyze both volumetric and shape asymmetries in the present study, given that clear asymmetry patterns for these structures have not yet been established for ASD. In this article, we will work with a new measure of structural brain asymmetry that has previously been used to study Alzheimer's disease (Wachinger et al., 2016; Wachinger et al., 2018). This method operates on spectral shape descriptors in the BrainPrint (Wachinger et al., 2015), which describe the geometry of a brain structure with a high-dimensional vector. Hence, it has the potential to be more sensitive to anatomical variations than commonly used volume measurements. This shape asymmetry measure captures the magnitude of the asymmetry and therefore combines directional and undirectional asymmetry. Directional asymmetry refers to hemispheric differences that systematically show a stronger effect on one of the hemispheres, e.g., larger changes on the right than on the left. In contrast, undirectional asymmetry does not have a consistent hemispheric effect. Alternative approaches, such as voxel-wise techniques or statistical shape models compute statistics across the population and are well suited for measuring directional asymmetry, but they cannot detect undirectional asymmetry. In addition, these techniques only allow for a qualitative assessment of shape asymmetry, but do not provide quantitative measure of shape asymmetry. The sensitive representation of geometry together with ability to identify undirectional asymmetry are particularly promising for ASD, given the complex and multi-faceted nature of the disorder, which may be accompanied by subtle focalized differences in morphology that are difficult to detect using purely volumetric measures.
2. Method
2.1. MR image acquisition
Structural MRI data were obtained from the ABIDE 1 database of preprocessed MR images (Craddock et al., 2013; http://preprocessed-connectomes-project.org/abide/). The ABIDE database contains MRI data for 539 individuals with ASD and 573 healthy controls, aged 6–65 years, collected at 17 international sites. All sites acquired MRI data using 3-Tesla scanners with T1-weighted scans with isotropic voxels (1 × 1 × 1-mm resolution). All data were obtained with informed consent, approved by institutional review boards (IRBs) at their respective collection sites, and were fully anonymized in accordance with HIPAA regulations. Data were also visually inspected and assigned quality ratings from three independent raters prior to online distribution. Further information on data acquisition and site-specific details (i.e., protocols, test batteries used, and scanning parameters) is available at http://fcon_1000.projects.nitrc.org/indi/abide/abide_I.html
2.2. Sample selection
Following inspection of the demographics of the ABIDE dataset, subjects older than 35 (N = 21) were removed both due to the relatively low number of subjects aged older than 35 within the dataset, and to lessen the potential confounding effects of normal brain aging experienced during mid- and late-life stages on our results. Only individuals with full-scale IQ (FSIQ) scores within two standard deviations of the overall ABIDE sample mean were included in the final sample to reduce the potential influence of outlying FSIQ scores. Finally, quality ratings provided by ABIDE were then used to remove subjects with scans rated as “poor” quality from the dataset. These quality ratings were obtained following manual inspection of the raw images in the ABIDE dataset by three independent reviewers. In cases where any rater disagreement occurred or where low ratings were given, images were visually inspected by the authors to ensure that segmentation was performed successfully before including these scans in analyses. Visual inspection resulted in the removal of two participants with ASD and two control participants with poor quality scans, which prevented the calculation of intracranial volume. This yielded data for a total of 948 individuals (ASD = 437, Controls = 511) from 17 different sites.
2.3. Subjects
The present study used a sample of 437 ASD patients (382 males), with ages ranging from seven to 34.6 (M = 15.95, SD=6.17), and 511 healthy controls (419 males), with ages ranging from 6.47 to 34.1 (M = 16.13, SD = 6.20). The two groups did not differ significantly in age (t = 0.43, p = 0.66) nor in variances in age (F = 0.99, p = 0.45). Our ASD sample is comprised of individuals diagnosed with all formerly used DSM-IV-TR subtypes for ASD (now classified together under ASD in the DSM-V; APA 2013), please see Table 1 for further detailed demographic information and summary statistics.
Table 1.
Demographic information and summary statistics by diagnosis.
| Autism spectrum disorder | Healthy controls | |
|---|---|---|
| N | 437 | 511 |
| Sex (M/F) | 382/55 | 419/92 |
| % (M/F) | 87.4/12.5 | 82.0/18.0 |
| Age (years) | 15.95 ± 6.17 | 16.12 ± 6.20 |
| FSIQ Mean | 106.38 ± 14.18 | 111.07 ± 11.74 |
| Intracranial Volume (mm3) | 1336,921 ± 272,115.55 | 1322,270 ± 254,188.55 |
| Volumetric Amygdala Asymmetry | 185.21 ± 164.95 | 168.9 ± 141.56 |
| Amygdala Shape Asymmetry | 2.13 ± 1.29 | 2.07 ± 1.14 |
| Volumetric Hippocampal Asymmetry | 285.05 ± 398.59 | 259.35 ± 351.15 |
| Hippocampal Shape Asymmetry | 2.15 ± 1.64 | 1.91 ± 1.14 |
| Volumetric Ventricular Asymmetry | 1637.52 ± 1843.34 | 1300.51 ± 1403.56 |
| Ventricular Shape Asymmetry | 3.76 ± 2.29 | 3.87 ± 2.34 |
Demographic information and summary statistics by diagnosis. Note: the ASD group is comprised of individuals diagnosed with all formerly used DSM-IV-TR subtypes, including 289 (66.1%) individuals diagnosed with Autism, 73 (16.7%) individuals diagnosed with Asperger's, 25 (5.7%) individuals diagnosed with Pervasive Development Disorder, and three (0.7%) individuals diagnosed as ASD-Not Otherwise Specified. DSM-IV-TR subtype information was not provided for 47 (11%) of ASD patients. Asymmetry values are non-directional.
All ASD patients and healthy controls had mean full-scale IQ (FSIQ) scores ≥79. Patients FSIQ scores ranged from 79 to 137 (M = 106.38, SD = 14.18). Healthy controls FSIQ scores ranged from 79 to 138 (M = 111.07, SD = 11.74). Despite that the two subject groups had similar FSIQ ranges, the variances of the two groups were significantly different (F = 1.46, p < 0.0001), and control subjects’ FSIQ scores were significantly higher than those of ASD patients, t(848) = 5.23, p < 0.0001. No significant difference in intracranial volume was found between ASD patients and controls.
2.4. Image analysis
A graphical overview of the image analysis steps for computing the brain asymmetry with the BrainPrint is presented in Fig. 1. The BrainPrint (Wachinger et al., 2015) description is based on the automated segmentation of anatomical brain structures with FreeSurfer (Dale and Sereno 1993; Dale et al., 1999; Fischl et al., 1999a, Fischl et al., 1999b; Fischl et al., 2002). We accessed files from the FreeSurfer v5.1 output of the ABIDE Preprocessed initiative (http://preprocessed-connectomes-project.org/abide/index.html). After image segmentation, geometric representations (surface and volumetric meshes) are extracted for the cortical and subcortical structures via the marching cubes algorithm. Marching cubes is a standard algorithm for extracting meshes from voxel maps, where we used the implementation from the FreeSurfer package. Each segmented brain structure is then represented by a mesh, and based on that mesh, a shape descriptor is computed. The shape descriptor transforms the complex geometric representation in a vector, which is easier to work with in the follow-up analyses. It is important that the descriptor captures all the relevant shape information. We use shapeDNA (Reuter et al., 2006) as shape descriptor, which performed among the best in a comparison of methods for non-rigid 3D shape retrieval (Lian et al., 2013). shapeDNA is based on the eigenvalues of the Laplace-Beltrami operator and, therefore, is isometry invariant (including rigid motion and reflections). Prior applications of shapeDNA and the Laplace-Beltrami operator in medical image analysis are discussed in (Wachinger et al., 2015). Eigenvalues of the Laplace-Beltrami operator Δ can be computed via finite element analysis by solving the Laplacian eigenvalue problem (Helmholtz equation) on the given shape:
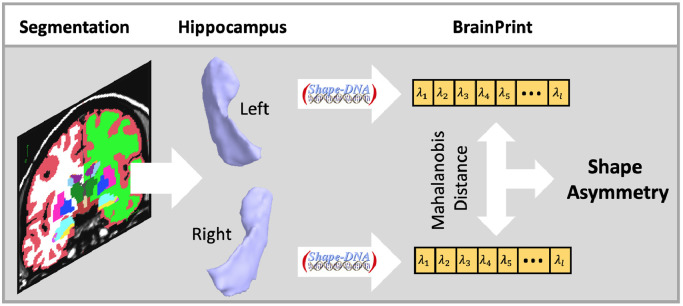
Fig. 1.
Graphical overview of steps for computing the brain asymmetry with the BrainPrint. MRI scans are segmented with FreeSurfer and meshes for the lateralized structure of interest, here hippocampus, are created. The computation of the shapeDNA results in the spectral shape descriptor in BrainPrint. The Mahalanobis distance between shape vectors yields measure of shape asymmetry.
The solution consists of eigenvalue and eigenfunction fi pairs, sorted by eigenvalues, (a positive diverging sequence). The first l non-zero eigenvalues are computed using the finite element methods and form the shapeDNA: , where we set in this study. To achieve scale independence, we normalize the eigenvalues:
where vol is the Riemannian volume of the D-dimensional manifold (Reuter et al., 2006, Wolfe et al., 2015), i.e., the surface area for 2D manifolds, or the volume for 3D solids.
A key property of the eigenvalues is their isometry invariance, i.e., length-preserving deformations will not change the spectrum. Isometry invariance includes rigid body motion as well as reflections, and, therefore, permits the comparison of shapes across individuals or hemispheres by directly comparing the shapeDNA without any complex and potentially error-prone image or geometry registration. The BrainPrint consists of the spectra for subcortical structures on the 2D boundary surfaces (triangle meshes) and for cortical structures on the full 3D solid (Wachinger et al., 2015). The BrainPrint is very stable with respect to re-meshing and mesh density, as long as the meshes are dense enough to represent the underlying geometry, since the Laplace-Beltrami operator is designed to be independent of the mesh, as opposed to graph Laplace operators (Reuter et al., 2006).
2.5. Brain asymmetry from BrainPrint
Based on the BrainPrint, we measure the asymmetry of the amygdala, hippocampus, and lateral ventricles. Since shapeDNA is invariant to reflections, we can directly compute the Mahalanobis distance between the descriptors of a lateralized brain structure, s
where we use a diagonal covariance matrix Σ with the ith element , for 1 ≤ i ≤ l, to reduce the impact of higher eigenvalues on the distance (Wachinger et al., 2015). The asymmetry measure is computed independently per subject and therefore presents a within-subject measure; it represents directional and undirectional asymmetry, but does not differentiate between the two types of asymmetry. The approach completely avoids lateral processing bias as it works on both hemispheres independently. Due to the pose invariance of spectral shape descriptors we can directly measure shape asymmetry by computing the distance in a symmetric fashion. Guaranteeing symmetric processing can be rather involved for most other shape representations that first require the construction of local correspondences, where choosing a target hemisphere for registration can potentially bias subsequent analyses. To the best of our knowledge, there is no alternative measure of shape asymmetry.
2.6. Statistical analyses
Data were analyzed using linear regression models. Amygdalar, hippocampal, and ventricular asymmetry values were investigated as dependent variables in separate models. Diagnosis group and sex were set as independent variables. Binary classification was used for diagnostic grouping, such that all subjects with a diagnosis of ASD were included together in analyses, regardless of their DSM-IV-TR subtypes. Age, scanning site, intracranial volume, and FSIQ were included as covariates. We evaluated the inclusion of a quadratic age term to the model to account for non-linear aging related effects. However, the likelihood ratio test between linear and quadratic models was not significant, so that we used a linear age term in the final model. All analyses were conducted using R version 3.2.2 for 64-bit Windows (R Core Team, 2015) and RStudio version 0.99.486 for Windows (Rstudio, 2015).
3. Results
Multiple regression analyses were calculated to predict volumetric and shape asymmetries in the amygdala, hippocampus, and lateral ventricles based on diagnosis and sex, while controlling for age, scanning site, intracranial volume, and FSIQ. A summary of regression results and the predictive values (β) of diagnosis for each of the structural regions can be seen in Table 2. We use Bonferroni correction to account for multiple comparisons and to adjust the significance threshold accordingly. A graphical illustration of regions investigated in the present study, depicted by their significance level of their differences in shape asymmetry, can be seen in Fig. 2.
Table 2.
Regression results and predictive values of diagnosis, sex, age, ICV, and FIQ on asymmetry.
| Region (Asymmetry Type) | Model | Diagnosis | Sex | Age | ICV | FIQ | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R2 | F | p | t | p | t | p | t | p | t | p | t | p | ||||||
| Amygdala (volumetric) | 0.05 | 3.39 | <0.0001 | −19.25 | −1.93 | 0.05 | −19.84 | −1.40 | 0.16 | 0.44 | 0.41 | 0.68 | −0.01 | −1.61 | 0.11 | 0.38 | 0.99 | 0.33 |
| Amygdala (shape) | 0.16 | 8.72 | <0.0001 | −0.02 | −0.27 | 0.79 | −0.29 | −2.72 | 0.006 | −0.01 | −0.33 | 0.74 | −0.01 | −5.01 | <0.0001 | −0.01 | −2.12 | 0.03 |
| Hippocampus (volumetric) | 0.04 | 2.59 | <0.0001 | −17.95 | −0.73 | 0.47 | −115.80 | −3.32 | 0.0008 | −0.09 | −0.03 | 0.97 | 0.01 | −2.13 | 0.03 | −0.33 | −0.35 | 0.73 |
| Hippocampus (shape) | 0.10 | 5.48 | <0.0001 | −0.23 | −2.66 | 0.008 | −0.41 | −3.22 | 0.001 | 0.01 | −0.011 | 0.99 | 0.01 | −4.71 | <0.0001 | 0.01 | −0.16 | 0.87 |
| Ventricles (volumetric) | 0.06 | 3.66 | <0.0001 | −321.41 | −3.04 | 0.002 | 139.12 | 0.93 | 0.36 | 25.57 | 2.28 | 0.02 | 0.01 | 6.63 | <0.0001 | 0.36 | 0.09 | 0.93 |
| Ventricles (shape) | 0.03 | 1.13 | 0.30 | 0.05 | 0.34 | 0.73 | −0.13 | −0.88 | 0.38 | 0.02 | 1.38 | 0.17 | −0.01 | −1.66 | 0.09 | 0.01 | 1.21 | 0.23 |
Regression results and predictive values of diagnosis, sex, age, ICV, and FIQ on asymmetry. Estimated regression coefficients reflect differences in ASD relative to controls. For diagnosis, negative beta values indicate that ASD>Controls, positive values indicate that ASD<Controls. For sex, negative beta values indicate that males>females, positive values indicate that males<females. All F(df1, df2) = 25, 922. Bold values indicate significance after Bonferroni correction.
Fig. 2.
Illustration of brain structures investigated in the present study. Coloring is according to significance of shape asymmetry with respect to diagnosis, where red indicates significant and blue indicates no significant group-related differences in asymmetry. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
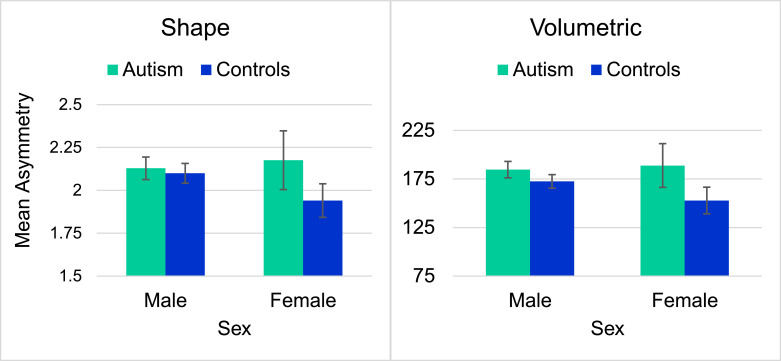
For the amygdala, diagnosis had no significant effect on shape or volumetric asymmetry, however, sex was a significant predictor of shape asymmetry. Irrespective of diagnosis, we observed significantly greater amygdalar shape asymmetry for males compared to females. Plots for the volumetric and shape asymmetry of the amygdala can be seen in Fig. 3. Although visual inspection of Fig. 3 appears to suggest that a diagnosis by sex interaction could be present, Welch's Two Sample T-Tests did not show any significant group differences in the amygdala for either measure of asymmetry.
Fig. 3.
Mean amygdala asymmetry values by diagnosis and sex.
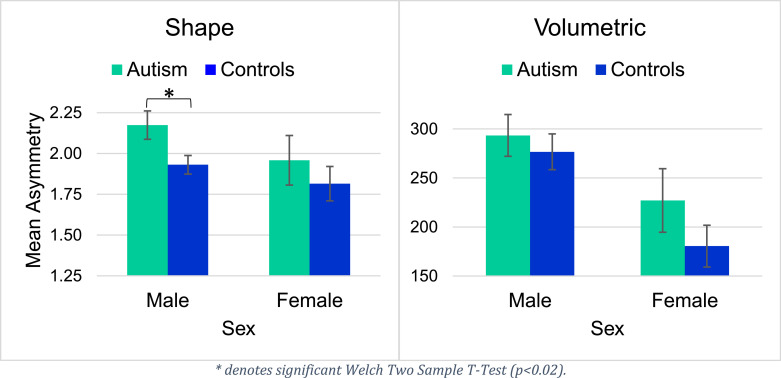
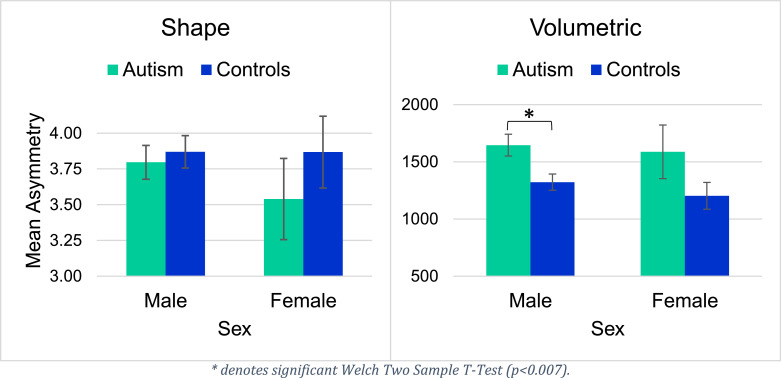
For the hippocampus, diagnosis significantly predicted shape asymmetry, but did not explain volumetric hippocampal asymmetry. Sex was found to be a significant predictor for both volumetric and shape asymmetries in the hippocampus. We observed significantly greater shape asymmetry of the hippocampus in ASD compared to healthy controls, as well as a significantly greater shape and volumetric asymmetry of the hippocampus for males compared to females across both participant groups. In Supplementary Figure 1, we illustrate a scatter plot between the predicted and the measured asymmetry values. Welch's Two-Sample T-Tests revealed significant group differences in hippocampal shape asymmetry in males (p < 0.02), but not in females. Plots for volumetric and shape asymmetry of the hippocampus can be seen in Fig. 4.
Fig. 4.
Mean hippocampal asymmetry values by diagnosis and sex.
Notably, none of the regression factors explained ventricular shape asymmetry, but diagnosis did significantly predict volumetric ventricular asymmetry. We observed significantly greater volumetric ventricular asymmetry in individuals with ASD than in healthy controls. In Supplementary Figure 2, we illustrate a scatter plot between the predicted and the measured asymmetry values. Welch's Two-Sample T-Tests also revealed significant group differences in volumetric ventricular asymmetry in males (p < 0.007), though not in females. Fig. 5 displays mean ventricular asymmetry values.
Fig. 5.
Mean ventricular asymmetry values by diagnosis and sex.
Additionally, age was not associated with either type of asymmetry in the amygdala or the hippocampus, but age was a significant predictor for volumetric ventricular asymmetry (p = 0.03). As age increased, asymmetry within the ventricles increased, for both ASD patients and controls. The interaction between age and diagnosis was not significant for any of the three regions, however. Furthermore, the interaction between sex and age was not significant for any of the three regions.
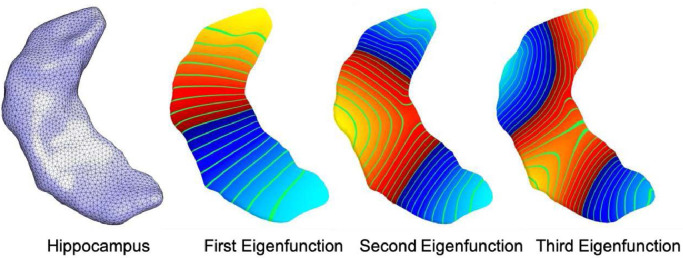
As there are significant associations between hippocampal shape asymmetry and Autism, we illustrate the hippocampus mesh together with three non-constant eigenfunctions in Fig. 6. The eigenfunctions demonstrate natural vibrations of the shape when oscillating at a frequency specified by the square root of the eigenvalue. The main variation of the first eigenfunction is from top to bottom and the second one from left to right. The third one already shows a more complex pattern.
Fig. 6.
Visualization of the hippocampus mesh and the first three non-constant eigenfunctions of the Laplace-Beltrami operator calculated on the surface. Increasing positive values of the eigenfunctions are shown in the color gradient from red to yellow and decreasing negative values are shown from dark blue to light blue. Level sets are shown in green. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
The present study is the first to report volumetric and shape asymmetry results for the amygdala, hippocampus, and ventricles in a large cohort of individuals with ASD. We examined these differences in asymmetry in a large sample of ASD patients and typically developing healthy control subjects using linear regression analyses and a new measure of asymmetry based on spectral shape descriptors, introduced by Wachinger et al. (2016). While we did not find significant differences in shape or volume asymmetry of the amygdala in ASD patients, we did observe significantly increased shape asymmetry in the hippocampus, as well as increased volumetric asymmetry in the lateral ventricles in ASD patients.
Although we anticipated significant results for the amygdala, given its important role in impaired functions in ASD, diagnosis did not explain either measure of asymmetry in the amygdala. However, patients with ASD did have greater mean amygdalar asymmetry values than controls, for both measures of amygdalar asymmetry. Sex was found to be a significant predictor for shape asymmetry of the amygdala, but not for volumetric asymmetry, although males were found to have greater asymmetry than females for both measures of asymmetry in the amygdala. The absence of significant sex differences in volumetric amygdalar asymmetry is consistent with the recent report from Guadalupe et al. (2016), who found no significant differences in volumetric asymmetry of the amygdala between the sexes in more than 14,000 subjects, the largest sample to date.
Diagnosis was found to be a significant predictor for hippocampal shape asymmetry, however, diagnosis did not explain volumetric hippocampal asymmetry, despite that ASD patients did have higher mean volumetric hippocampal asymmetry values than controls. Sex was a significant predictor for both shape and volumetric asymmetries in the hippocampus, such that males had greater asymmetry than females, which is consistent with sex effects on volumetric hippocampal asymmetry previously described by Guadalupe et al. (2016). The opposite pattern of regression results was found for the ventricles—diagnosis explained volumetric asymmetry, but not shape asymmetry, and sex did not explain either measure of ventricular asymmetry. We argue that our differential findings for the hippocampus and lateral ventricles could simply be a result of structural differences of these two regions. Given the heterogeneous nature of the hippocampus and its substructures, compared to the more homogenous nature of the ventricles, this may account for the differential results of the volumetric and shape asymmetry analyses.
Since normalized eigenvalues were used for the shape descriptors, where the volume information is removed, it is possible that the shape descriptor does not detect purely volumetric differences that do not exert any influence on the shape of the structure. For the ventricles, which are fluid-filled structures and therefore intrinsically more homogeneous, it is reasonable to assume that significant changes in volume are more likely to occur than changes in the shape of the ventricles in the course of the disorder. In contrast, considering the heterogeneous composition of the hippocampus and its subfields, it is likely that focalized changes may occur within these subfields, which could result in detectable shape differences but not volumetric differences. Our hypothesis is consistent with previous findings of hippocampal shape abnormalities observed in ASD (Dager et al., 2007). An asymmetry in texture features of the hippocampus has been reported by Chaddad et al. (2017).
While Dager et al. (2007) did not specifically investigate asymmetry, they did find differences in hippocampal shape which distinguished children with ASD from typically developing children. They observed a pattern of upward bending of both the head and tail of the hippocampus, as well as an inward deformation of the subiculum in individuals with ASD, which is in line with our argument that focalized changes may occur in ASD, and that shape descriptors are sensitive enough to detect such focalized changes in subfields of the hippocampal formation. Given that our results showed hippocampal shape differences within a large cohort, hippocampal shape alterations may be worthy of consideration when developing MR-based classification algorithms and computer-aided diagnostic systems for ASD. Moreover, the supplementary role of shape analyses is certainly worthy of consideration, not only in the context of research on ASD, but for research on anatomical variations in general, given that such analyses permit the detection of focalized abnormalities that could otherwise remain undetected using purely volumetric measures.
It is important to note the methodological limitations of this study. Firstly, although FSIQ was controlled for, the ASD and control samples had an uneven number of subjects and were not matched with each other based on FSIQ, and FSIQ did differ significantly between the two groups. Additionally, given that the ASD sample was limited to relatively high-functioning individuals with ASD (FSIQ mean = 106.58, SD = 14.23), this may have had a significant influence on the results. Lower-functioning individuals with ASD could potentially exhibit significant structural abnormalities that differ from those found in high-functioning individuals with ASD. Thus, the generalizability of the current findings to the whole ASD spectrum is limited.
Second, several researchers have raised valid concerns and criticisms regarding the ABIDE dataset. Some of the most prevalent of these concerns include the confounding effects of scanner variations across ABIDE data collection sites (Auzias et al., 2015; Sato et al., 2016), the age range of subjects included in the dataset (Kazeminejad and Sotero, 2019), and significant differences in sample characterization variables, such as age and IQ across sites (Sato et al., 2016). However, although MRI data processing parameters as well as scanner parameters (e.g. scanner field strength, differing pule sequences) can increase variability and impact reliability of results (Han et al., 2006), some studies have demonstrated that site and scanner effects observed in multisite analyses may be minimal and outweighed by significant subject effects (Noble et al., 2017). Therefore, we believe that the use of ABIDE is still valid, however, future studies may benefit from analyzing these structures within additional datasets and employing cross-validation techniques.
Finally, our study also succumbs to the causal inference problem that often occurs with investigations of this nature. Although we observed differences in asymmetry among individuals with ASD and healthy controls, it is not clear whether ASD or the asymmetry causes the other. Longitudinal studies would provide stronger evidence in this regard, given that they would provide the opportunity to investigate the developmental trajectory of the disorder and its associated morphometric alterations. Future studies investigating longitudinal structural differences in individuals with ASD at varying levels of functioning and symptom severity are certainly warranted in order to better understand the relationship between symptom severity of ASD and structural patterns that emerge over time.
In summary, this study investigated volumetric and shape asymmetry in Autism Spectrum Disorder. We analyzed data from a large multi-site MRI dataset and found increased shape asymmetry of the hippocampus in individuals with ASD relative to controls. In addition, we observed increased volumetric asymmetry of the lateral ventricles in ASD. We believe that contralateral brain structures present a unique, within-patient reference element for disease progression. Therefore, asymmetries of contralateral structures could provide a personalized measure of the accumulation of past disease and disordered processes. The potential for these asymmetries and other previously reported structural abnormalities to serve as biomarkers for ASD warrants further studies utilizing large cohorts, especially longitudinal analyses and classification experiments, if this information is to be incorporated in a neurodiagnostic tool for ASD.
Disclosures
No commercial support was received for the preparation of this manuscript and the authors have no conflicts of interest to report.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the original data sources on which this study is based.
CRediT authorship contribution statement
Rose Richards: Conceptualization, Data curation, Formal analysis, Investigation, Resources, Writing - original draft, Writing - review & editing, Visualization, Project administration. Ellen Greimel: Conceptualization, Resources, Writing - review & editing. Dorit Kliemann: Writing - review & editing. Inga K. Koerte: Writing - review & editing. Gerd Schulte-Körne: Writing - review & editing, Supervision. Martin Reuter: Methodology, Resources, Writing - review & editing. Christian Wachinger: Conceptualization, Methodology, Software, Formal analysis, Investigation, Resources, Data curation, Writing - original draft, Writing - review & editing, Visualization, Supervision, Project administration, Funding acquisition.
Acknowledgements
This work was supported in part by the Faculty of Medicine at LMU (FöFoLe) and the Bavarian State Ministry of Science and the Arts in the framework of the Centre Digitisation.Bavaria (ZD.B).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.nicl.2020.102207.
Contributor Information
Rose Richards, Email: roserm@umich.edu.
Christian Wachinger, Email: Christian.wachinger@med.uni-muenchen.de.
Appendix. Supplementary materials
References
- American Psychiatric Association . fifth ed. Author; Washington, DC: 2013. Diagnostic and Statistical Manual of Mental Disorders. [Google Scholar]
- Auzias G., Takerkart S., Deruelle C. On the influence of confounding factors in multisite brain morphometry studies of developmental pathologies: application to autism spectrum disorder. IEEE J. Biomed. Health Inform. 2015;20(3):810–817. doi: 10.1109/JBHI.2015.2460012. [DOI] [PubMed] [Google Scholar]
- Aylward E.H., Minshew N.J., Goldstein G., Honeycutt N.A., Augustine A.M., Yates K.O., Pearlson G.D. MRI volumes of amygdala and hippocampus in non–mentally retarded autistic adolescents and adults. Neurology. 1999;53(9) doi: 10.1212/wnl.53.9.2145. ... 2145-2145. [DOI] [PubMed] [Google Scholar]
- Baron-Cohen S., Ring H.A., Bullmore E.T., Wheelwright S., Ashwin C., Williams S.C.R. The amygdala theory of autism. Neurosci. Biobehav. Rev. 2000;24(3):355–364. doi: 10.1016/s0149-7634(00)00011-7. [DOI] [PubMed] [Google Scholar]
- Brownsell S., Bradley D., Cardinaux F., Hawley M. IEEE International Conference on Healthcare Informatics, Imaging and Systems Biology (HISB) 2011. Autism: a systems biology disease; pp. 359–366. [Google Scholar]
- Brothers L. The neural basis of primate social communication. Motiv. Emot. 1990;14(2):81–91. [Google Scholar]
- Carper A.R., Moses P., Tigue D.Z., Courchesne E. Cerebral lobes in autism: early hyperplasia and abnormal age effects. Neuroimage. 2002;16(4):1038–1051. doi: 10.1006/nimg.2002.1099. [DOI] [PubMed] [Google Scholar]
- Cerliani L., Mennes M., Thomas R.M., Martino D., A. T., Keysers C. Increased functional connectivity between subcortical and cortical resting-state networks in autism spectrum disorder. JAMA Psychiatry. 2015;72(8):767. doi: 10.1001/jamapsychiatry.2015.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention Prevalence of autism spectrum disorder among children aged 8 years. Morbid. Mortal. Week. Rep. 2014;63(2):1–24. [Google Scholar]
- Chaddad A., Desrosiers C., Toews M. Multi-scale radiomic analysis of sub-cortical regions in MRI related to autism, gender and age. Sci. Rep. 2017;7:45639. doi: 10.1038/srep45639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craddock C., Benhajali Y., Chu C., Chouinard F., Evans A., Jakab A., Khundrakpam B.S., Lewis J.D., Li Q., Milham M., Yan C., Bellec P. The neuro bureau preprocessing initiative: open sharing of preprocessed neuroimaging data and derivatives. Neuroinformatics 2013; Stockholm, Sweden; 2013. [Google Scholar]
- Creasey H., Rumsey J.M., Schwartz M., Duara R., Rapoport J.L., Rapoport S.I. Brain morphometry in autistic men as measured by volumetric computed tomography. Arch. Neurol. 1986;43(7):669–672. doi: 10.1001/archneur.1986.00520070027012. [DOI] [PubMed] [Google Scholar]
- Crespi B., Badcock C. Psychosis and autism as diametrical disorders of the social brain. Behav. Brain Sci. 2008;31(3):241–261. doi: 10.1017/S0140525X08004214. [DOI] [PubMed] [Google Scholar]
- Dager S.R., Wang L., Friedman S.D., Shaw D.W., Constantino J.N., Artru A.A., Csernansky J.G. Shape mapping of the hippocampus in young children with autism spectrum disorder. Am. J. Neuroradiol. 2007;28(4):672–677. ... [PMC free article] [PubMed] [Google Scholar]
- Dale A.M., Fischl B., Sereno M.I. Cortical surface-based analysis: I. Segmentation and surface reconstruction. Neuroimage. 1999;9(2):179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Dale A.M., Sereno M.I. Improved localization of cortical activity by combining eeg and meg with mri cortical surface reconstruction: a linear approach. J. Cognit. Neurosci. 1993;5(2):162–176. doi: 10.1162/jocn.1993.5.2.162. [DOI] [PubMed] [Google Scholar]
- Dekhil O., Ismail M., Shalaby A., Switala A., Elmaghraby A., Keynton R., El-Baz A. Biomedical Imaging (ISBI 2017), 2017 IEEE 14th International Symposium on. IEEE; 2017. A novel CAD system for autism diagnosis using structural and functional MRI; pp. 995–998. ... [Google Scholar]
- DeLong G.R., Bauman M.L. Brain lesions in autism. In: Schopler E., Mesibov G.B., editors. Neurobiological Issues in Autism. Current Issues in Autism. Springer; Boston, MA: 1987. [Google Scholar]
- Dougherty C.C., Evans D.W., Katuwal G.J., Michael A.M. Asymmetry of fusiform structure in autism spectrum disorder: trajectory and association with symptom severity. Mol. Autism. 2016;7:28. doi: 10.1186/s13229-016-0089-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker C., Marquand A., Mourão-Miranda J., Johnston P., Daly E.M., Brammer M.J., Murphy D.G. Describing the brain in autism in five dimensions—magnetic resonance imaging-assisted diagnosis of autism spectrum disorder using a multiparameter classification approach. J. Neurosci. 2010;30(32):10612–10623. doi: 10.1523/JNEUROSCI.5413-09.2010. ... [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilam-Stock T., Wu T., Spagna A., Egan L.J., Fan J., Agarwal V., Subramanyam A.A. Neuroanatomical alterations in high-functioning adults with autism spectrum disorder. Front. Neurosci. 2016;10:237. doi: 10.3389/fnins.2016.00237. doi: 10.3389/fnins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsabbagh M., Divan G., Koh Y.J., Kim Y.S., Kauchali S., Marcı´n C. Global prevalence of autism and other pervasive developmental disorders. Autism Res. 2012;5:160–179. doi: 10.1002/aur.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipek P.A., Richelme C., Kennedy D.N., Rademacher J., Pitcher D.A., Zidel S., Caviness V.S. Morphometric analysis of the brain in developmental language disorders and autism. Ann. Neurol. 1992;32(475):76–149. [Google Scholar]
- Fischl B., Salat D.H., Busa E., Albert M., Dieterich M., Haselgrove C., Montillo A. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–355. doi: 10.1016/s0896-6273(02)00569-x. ... [DOI] [PubMed] [Google Scholar]
- Fischl B., Sereno M.I., Dale A.M. Cortical surface-based analysis: II: inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999;9(2):195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Fischl B., Sereno M.I., Tootell R.B., Dale A.M. High-resolution intersubject averaging and a coordinate system for the cortical surface. Hum. Brain Mapp. 1999;8(4):272–284. doi: 10.1002/(SICI)1097-0193(1999)8:4<272::AID-HBM10>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fombonne E. Epidemiology of pervasive developmental disorders. Pediatr. Res. 2009;65(6):591–598. doi: 10.1203/PDR.0b013e31819e7203. [DOI] [PubMed] [Google Scholar]
- Garber H.J., Ritvo E.R., Chiu L.C., Griswold V.J., Kashanian A. A magnetic resonance imaging study of autism: normal fourth ventricle size and absence of pathology. Am. J. Psychiatry. 1989;146(4):532. doi: 10.1176/ajp.146.4.532. [DOI] [PubMed] [Google Scholar]
- Geschwind N., Galaburda A. Cerebral lateralization. biological mechanisms, associations, and pathology: I. A hypothesis and a program for research. Arch. Neurol. 1985;42:428–459. doi: 10.1001/archneur.1985.04060050026008. [DOI] [PubMed] [Google Scholar]
- Goodman J., Marsh R., Peterson B.S., Packard M.G. Annual research review: the neurobehavioral development of multiple memory systems–implications for childhood and adolescent psychiatric disorders. J. Child Psychol. Psychiatry. 2014;55(6):582–610. doi: 10.1111/jcpp.12169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen W., Teluij M., Buitelaar J., Tendolkar I. Amygdala and hippocampus enlargement during adolescence in autism. J. Am. Acad. Child Adolesc. Psychiatry. 2010;49(6):552–560. doi: 10.1016/j.jaac.2009.12.023. [DOI] [PubMed] [Google Scholar]
- Guadalupe T., Mathias S.R., Theo G.M., Whelan C.D., Zwiers M.P., Abe Y., Aribisala B.S. Human subcortical brain asymmetries in 15,847 people worldwide reveal effects of age and sex. Brain Imaging Behav. 2016:1–18. doi: 10.1007/s11682-016-9629-z. ... [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haar S., Berman S., Behrmann M., Dinstein I. Anatomical abnormalities in autism? Cereb. Cortex. 2014 doi: 10.1093/cercor/bhu242. [DOI] [PubMed] [Google Scholar]
- Han X., Jovicich J., Salat D., van der Kouwe A., Quinn B., Czanner S., Maguire P. Reliability of MRI-derived measurements of human cerebral cortical thickness: the effects of field strength, scanner upgrade and manufacturer. Neuroimage. 2006;32(1):180–194. doi: 10.1016/j.neuroimage.2006.02.051. ... [DOI] [PubMed] [Google Scholar]
- Hardan A.Y., Minshew N.J., Mallikarjuhn M., Keshavan M.S. Brain volume in autism. J. Child Neurol. 2001;16(6):421. doi: 10.1177/088307380101600607. [DOI] [PubMed] [Google Scholar]
- Haznedar M.M., Buchsbaum M.S., Hazlett E.A., LiCalzi E.M., Cartwright C., Hollander E. Volumetric analysis and three-dimensional glucose metabolic mapping of the striatum and thalamus in patients with autism spectrum disorders. Am. J. Psychiatry. 2006;163(7):1252–1263. doi: 10.1176/ajp.2006.163.7.1252. [DOI] [PubMed] [Google Scholar]
- Herbert R.M. Large brains in autism: the challenge of pervasive abnormality. The Neuroscientist. 2005;11(5):417–440. doi: 10.1177/0091270005278866. [DOI] [PubMed] [Google Scholar]
- Herbert M.R., Ziegler D.A., Deutsch C.K., O'brien L.M., Kennedy D.N., Filipek P.A., Caviness V.S. Brain asymmetries in autism and developmental language disorder: a nested whole-brain analysis. Brain. 2005;128(1):213–226. doi: 10.1093/brain/awh330. ... [DOI] [PubMed] [Google Scholar]
- Howard M.A., Cowell P.E., Boucher J., Broks P., Mayes A., Farrant A., Roberts N. Convergent neuroanatomical and behavioural evidence of an amygdala hypothesis of autism. Neuroreport. 2000;11(13):2931–2935. doi: 10.1097/00001756-200009110-00020. [DOI] [PubMed] [Google Scholar]
- Hughes J.R. A review of recent reports on autism: 1000 studies published in 2007. Epilepsy Behav. 2008;13(3):425–437. doi: 10.1016/j.yebeh.2008.06.015. [DOI] [PubMed] [Google Scholar]
- Ismail M.M.T. Univeristy of Louisville's Institutional Repository of Electronic Theses and Dissertations. Paper 2599. 2016. A CAD system for early diagnosis of autism using different imaging modalities. [DOI] [Google Scholar]
- Jacobson R., Le Couteur A., Howlin P., Rutter M. Selective subcortical abnormalities in autism. Psychol. Med. 1988;18(1):39–48. doi: 10.1017/s0033291700001860. [DOI] [PubMed] [Google Scholar]
- Kazeminejad A., Sotero R.C. Topological properties of resting-state fMRI functional networks improve machine learning-based autism classification. Front. Neurosci. 2019;12:1018. doi: 10.3389/fnins.2018.01018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.E., Lyoo I.K., Estes A.M., Renshaw P.F., Shaw D.W., Friedman S.D., Dager S.R. Laterobasal amygdalar enlargement in 6-to 7-year-old children with autism spectrum disorder. Arch. Gen. Psychiatry. 2010;67(11):1187–1197. doi: 10.1001/archgenpsychiatry.2010.148. ... [DOI] [PubMed] [Google Scholar]
- Lange N., DuBray M.B., Lee J.E., Froimowitz M.P., Froehlich A., Adluru N., Alexander A.L. Atypical diffusion tensor hemispheric asymmetry in autism. Autism Res. 2010;3(6):350–358. doi: 10.1002/aur.162. ... [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange N., Travers B.G., Bigler E.D., Prigge M.B.D., Froehlich A.L., Nielsen J.A. Longitudinal volumetric brain changes in autism spectrum disorder ages 6–35 years. Autism Res. 2015;8:82–93. doi: 10.1002/aur.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Karnath H.O., Xu X. Candidate biomarkers in children with autism spectrum disorder: a review of MRI studies. Neurosci. Bull. 2017:1–19. doi: 10.1007/s12264-017-0118-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian Z., Godil A., Bustos B., Daoudi M., Hermans J., Kawamura S., Ohkita Y. A comparison of methods for non-rigid 3D shape retrieval. Pattern Recognit. 2013;46(1):449–461. ... [Google Scholar]
- Mateos-Perez J.M., Dadar M., Lacalle-Aurioles M., Iturria-Medina Y., Zeighami Y., Evans A.C. Structural neuroimaging as clinical predictor: a review of machine learning applications. NeuroImage. 2018 doi: 10.1016/j.nicl.2018.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlonan G.M., Daly E., Kumari V., Critchley H.D., van Amelsvoort T., Suckling J., Schmitz N. Brain anatomy and sensorimotor gating in asperger’s syndrome. Brain. 2002;125(7):1594–1606. doi: 10.1093/brain/awf150. ... [DOI] [PubMed] [Google Scholar]
- Monterrey J.C., Philips J., Cleveland S., Tanaka S., Barnes P., Hallmayer J.F., Hardan A.Y. Incidental brain MRI findings in an autism twin study. Autism Res. 2017;10(1):113–120. doi: 10.1002/aur.1720. ... [DOI] [PubMed] [Google Scholar]
- Mostapha M., Casanova M.F., Gimel'farb G., El-Baz A. Towards non-invasive image-based early diagnosis of autism. In: Navab N., Hornegger J., Wells W., Frangi A., editors. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015. Vol. 9350. Springer; Cham: 2015. (Lecture Notes in Computer Science). [Google Scholar]
- Movsas T.Z., Pinto-Martin J.A., Whitaker A.H., Feldman J.F., Lorenz J.M., Korzeniewski S.J., Paneth N. Autism spectrum disorder is associated with ventricular enlargement in a low birth weight population. J. Pediatr. 2013;163(1):73–78. doi: 10.1016/j.jpeds.2012.12.084. ... [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson J., Dawson G., Abbott R., Faja S., Webb S.J., Friedman S.D., Shaw D., Artru A., Dager S.R. Amygdalar volume and behavioral development in autism. Arch. Gen. Psychiatry. 2006;63(6):686–693. doi: 10.1001/archpsyc.63.6.686. [DOI] [PubMed] [Google Scholar]
- Murphy C.M., Deeley Q., Daly E.M., Ecker C., O’Brien F.M., Hallahan B., Robertson D.M. Anatomy and aging of the amygdala and hippocampus in autism spectrum disorder: an in vivo magnetic resonance imaging study of asperger syndrome. Autism Res. 2012;5(1):3–12. doi: 10.1002/aur.227. ... [DOI] [PubMed] [Google Scholar]
- Nicolson R., DeVito T.J., Vidal C.N., Sui Y., Hayashi K.M., Drost D.J., Thompson P.M. Detection and mapping of hippocampal abnormalities in autism. Psychiatry Res. 2006;148(1):11–21. doi: 10.1016/j.pscychresns.2006.02.005. ... [DOI] [PubMed] [Google Scholar]
- Nielsen J.A., Zielinski B.A., Fletcher P.T., Alexander A.L., Lange N., Bigler E.D., Anderson J.S. Multisite functional connectivity MRI classification of autism: abide results. Front. Hum. Neurosci. 2012;7 doi: 10.3389/fnhum.2013.00599. ... 599-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu L., Matsui M., Zhou S.Y., Hagino H., Takahashi T., Yoneyama E., Kawasaki Y., Suzuki M., Seto H., Ono T. Volume reduction of the amygdala in patients with schizophrenia: a magnetic resonance imaging study. Psychiatry Res. 2004;132(1):41–51. doi: 10.1016/j.pscychresns.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Noble S., Scheinost D., Finn E.S., Shen X., Papademetris X., McEwen S.C., Mirzakhanian H. Multisite reliability of MR-based functional connectivity. Neuroimage. 2017;146:959–970. doi: 10.1016/j.neuroimage.2016.10.020. ... [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordahl C.W., Scholz R., Yang X., Buonocore M.H., Simon T., Rogers S., Amaral D.G. Increased rate of amygdala growth in children aged 2 to 4 years with autism spectrum disorders: a longitudinal study. Arch. Gen. Psychiatry. 2012;69(1):53–61. doi: 10.1001/archgenpsychiatry.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oertel-Knochel V., Knochel C., Stablein M., Linden D.E. Abnormal functional and structural asymmetry as biomarker for schizophrenia. Curr. Top. Med. Chem. 2012;12(21):2434–2451. doi: 10.2174/156802612805289926. [DOI] [PubMed] [Google Scholar]
- Palmen S.J., Pol H.E.H., Kemner C., Schnack H.G., Durston S., Lahuis B.E., Van Engeland H. Increased gray-matter volume in medication-naive high-functioning children with autism spectrum disorder. Psychol. Med. 2005;35(4):561–570. doi: 10.1017/s0033291704003496. ... [DOI] [PubMed] [Google Scholar]
- Pendergrass S., Girirajan S., Selleck S. Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing. 2014. Uncovering the etiology of autism spectrum disorders: genomics, bioinformatics, environment, data collection and exploration, and future possibilities; pp. 422–426. [PMC free article] [PubMed] [Google Scholar]
- Piven J., Arndt S., Bailey J., Havercamp S., Andreasen N.C., Palmer P. An MRI study of brain size in autism. Am. J. Psychiatry. 1995;152(8):1145. doi: 10.1176/ajp.152.8.1145. [DOI] [PubMed] [Google Scholar]
- Piven J., Bailey J., Ranson B.J., Arndt S. No difference in hippocampus volume detected on magnetic resonance imaging in autistic individuals. J. Autism Dev. Disord. 1998;28(2):105–110. doi: 10.1023/a:1026084430649. [DOI] [PubMed] [Google Scholar]
- Plitt M., Barnes K.A., Martin A. Functional connectivity classification of autism identifies highly predictive brain features but falls short of biomarker standards. NeuroImage. 2014;7:359–366. doi: 10.1016/j.nicl.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postema M.C., Van Rooij D., Anagnostou E., Arango C., Auzias G., Behrmann M., Filho B., G. C., S. C., R. D., Deruelle C. Altered structural brain asymmetry in autism spectrum disorder in a study of 54 datasets. Nat. Commun. 2019;10(1):1–12. doi: 10.1038/s41467-019-13005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2015. R: A Language and Environment for Statistical Computing (Version 3.2.2)http://www.R-project.org Vienna, Austria. URL: [Google Scholar]
- Reuter M., Wolter F.E., Peinecke N. Laplace–Beltrami spectra as ‘Shape-DNA’ of surfaces and solids. Comput.-Aided Des. 2006;38(4):342–366. [Google Scholar]
- Rice C.E., Rosanoff M., Dawson G., Durkin M.S., Croen L.A., Singer A., Yeargin-Allsopp M. Evaluating changes in the prevalence of the autism spectrum disorders (ASDs) Public Health Rev. 2012;34(2):1–22. doi: 10.1007/BF03391685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas D.C., Peterson E., Winterrowd E., Reite M.L., Rogers S.J., Tregellas J.R. Regional gray matter volumetric changes in autism associated with social and repetitive behavior symptoms. BMC Psychiatry. 2006;6(1):56. doi: 10.1186/1471-244X-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RStudio . 2015. RStudio: Integrated Development Environment for R (Version 0.99.486) [Computer software] Boston, MA, United States. [Google Scholar]
- Sato J.R., Balardin J., Vidal M.C., Fujita A. Identification of segregated regions in the functional brain connectome of autistic patients by a combination of fuzzy spectral clustering and entropy analysis. J. Psychiatry Neurosc. 2016;41(2):124–132. doi: 10.1503/jpn.140364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann C.M., Hamstra J., Goodlin-Jones B.L., Lotspeich L.J., Kwon H., Buonocore M.H., Amaral D.G. The amygdala is enlarged in children but not adolescents with autism; the hippocampus is enlarged at all ages. J. Neurosci. 2004;24(28):6392–6401. doi: 10.1523/JNEUROSCI.1297-04.2004. ... [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks B.F., Friedman S.D., Shaw D.W., Aylward E.H., Echelard D., Artru A.A. Brain structural abnormalities in young children with autism spectrum disorder. Neurology. 2002;59:184–192. doi: 10.1212/wnl.59.2.184. [DOI] [PubMed] [Google Scholar]
- Titova O.E., Hjorth O.C., Schioth H.B., Brooks S.J. Anorexia nervosa is linked to reduced brain structure in reward and somatosensory regions: a meta-analysis of vbm studies. BMC Psychiatry. 2013;13:110. doi: 10.1186/1471-244X-13-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A.H., Greenspan K.S., van Erp T.G. Pallidum and lateral ventricle volume enlargement in autism spectrum disorder. Psychiatry Res. 2016;252:40–45. doi: 10.1016/j.pscychresns.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal C.N., Nicolson R., Boire J.Y., Barra V., DeVito T.J., Hayashi K.M., Toga A.W. Three-dimensional mapping of the lateral ventricles in autism. Psychiatry Res. 2008;163(2):106–115. doi: 10.1016/j.pscychresns.2007.11.002. ... [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneau M. Meta-analyzing left hemisphere language areas: phonology, semantics, and sentence processing. Neuroimage. 2006;30:1414–1432. doi: 10.1016/j.neuroimage.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Wachinger C., Golland P., Kremen W., Fischl B., Reuter M. BrainPrint: a discriminative characterization of brain morphology. Neuroimage. 2015;109:232–248. doi: 10.1016/j.neuroimage.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachinger C., Salat D.H., Weiner M., Reuter M. Whole-brain analysis reveals increased neuroanatomical asymmetries in dementia for hippocampus and amygdala. Brain. 2016;139(12):3253–3266. doi: 10.1093/brain/aww243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachinger C., Nho K., Saykin A.J., Reuter M., Rieckmann A. A longitudinal imaging genetics study of neuroanatomical asymmetry in alzheimer’s disease. Biol. Psychiatry. 2018 doi: 10.1016/j.biopsych.2018.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe F.H., Auzias G., Deruelle C., Chaminade T. Focal atrophy of the hypothalamus associated with third ventricle enlargement in autism spectrum disorder. Neuroreport. 2015;26(17):1017–1022. doi: 10.1097/WNR.0000000000000461. [DOI] [PubMed] [Google Scholar]
- Organization World Health. Author; Geneva, Switzerland: 2013. Autism Spectrum Disorders & Other Developmental disorders: from Raising Awareness to Building Capacity. [Google Scholar]
- Xia J., Chen J., Zhou Y., Zhang J., Yang B., Xia L., Wang C. Volumetric MRI analysis of the amygdala and hippocampus in subjects with major depression. J. Huazhong Univ. Sci. Technol. 2004;24(5) doi: 10.1007/BF02831120. 500–2–50506. [DOI] [PubMed] [Google Scholar]
- Zhen Z. Quantifying the variability of scene-selective regions: interindividual, interhemispheric, and sex differences. Hum. Brain Mapp. 2017;38:2260–2275. doi: 10.1002/hbm.23519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.