Graphical abstract
Keywords: CRISPR/Cas9, Convolutional neural network, Bidirectional gate recurrent unit network, sgRNA, On-target
Abstract
CRISPR/Cas9 is a hot genomic editing tool, but its success is limited by the widely varying target efficiencies among different single guide RNAs (sgRNAs). In this study, we proposed C-RNNCrispr, a hybrid convolutional neural networks (CNNs) and bidirectional gate recurrent unit network (BGRU) framework, to predict CRISPR/Cas9 sgRNA on-target activity. C-RNNCrispr consists of two branches: sgRNA branch and epigenetic branch. The network receives the encoded binary matrix of sgRNA sequence and four epigenetic features as inputs, and produces a regression score. We introduced a transfer learning approach by using small-size datasets to fine-tune C-RNNCrispr model that were pre-trained from benchmark dataset, leading to substantially improved predictive performance. Experiments on commonly used datasets showed C-RNNCrispr outperforms the state-of-the-art methods in terms of prediction accuracy and generalization. Source codes are available at https://github.com/Peppags/C_RNNCrispr.
1. Introduction
CRISPR/Cas9 originated from bacterial immune system has been adopted for a promising genomic editing tool in recent years [1]. It consists of two components: a nuclease activity-carrying Cas9 protein and a specificity-programming single guide RNA (sgRNA) [2], [3]. Recognition and cleavage work via complementarity of a ~20-bp sequence within the sgRNA to the genome target region flanked by a 3′ NGG protospacer adjacent motif (PAM) based on Watson-Crick base pairing [1]. The success of CRISPR/Cas9 system for genome engineering of prokaryotic hosts largely depends on sgRNA activity. However, different activities among various sgRNAs still represent a significant limitation, leading to inconsistent target efficiency [4]. Moreover, the specific features that determine sgRNA activity remain largely unexplored. Therefore, accurate prediction of sgRNA activity would facilitate the design of sgRNAs by maximizing aimed activity at the desired target site while minimizing off-target cleavage [5].
Numerous computational methods for sgRNA activity prediction have been developed based on different rules. Existing tools fall into three classes, namely alignment-based, hypothesis-driven and learning-based methods [6]. Alignment-based tools aligned the sgRNA from the given genome by locating PAM. For example, CRISPRdirect performs sgRNA selection based on investigating the entire genome for perfect matches with the candidate target sequence and their seed sequence flanking the PAM [7]. Hypothesis driven-based tools score the sgRNA mainly considering the contribution of specific genome context factors. For instance, ECRISP ranks sgRNAs by taking into account on-target specificity and the number of off-targets [8]. Machine learning-based methods predict the sgRNA cleavage efficacy based on training a model by integrating different features affecting the efficiency. For example, sgRNA Designer considers the position of the target site relative to the transcription start site and position within the protein when evaluating the efficacy of candidate sgRNAs [9]. sgRNA Scorer unravels both locus accessibility and sequence composition of the sgRNA that are important in determining sgRNA target efficacy [10]. However, the accuracy of machine learning-based tools varies widely among the constructed features [11]. Moreover, the hand-crafted features may result in redundancy, further leading to the poor prediction results. Therefore, machine learning-based methods have obvious drawbacks, e.g. requiring expert domain knowledge and showing low generalization. To learn more about the computational methods used to facilitate the process of CRISPR/Cas9 sgRNA target efficacy prediction, we refer readers to [6], [12] for a comprehensive reading.
More recently, deep learning [13] is another exciting and promising approach being applied in the genomics field. It is a variation of machine learning that uses neural networks to automatically extract novel features from input data. Deep learning has made impressive advances in areas such as computer vision [14] and natural language processing (NLP) [15]. Besides, deep learning-based methods which are mainly based on convolutional neural networks (CNNs) are attractive solutions for CRISPR sgRNA target efficacy prediction problems. Currently, several attractive strategies have been explored for this issue. To the best of our knowledge, Seq-deepCpf1 is the first published deep learning method to predict CRISPR/Cpf1 gRNA on-target activity. It used CNN to extract features from the input gRNA sequence [16]. DeepCRISPR has been successfully applied for predicting CRISPR/Cas9 sgRNA on-target knockout efficiency and whole genome off-target profiles by incorporating deep convolutionary neural network (DCDNN)-based auto-encoder as well as CNN [5]. DeepCas9 used CNN to automatically learn the sequence determinants and predict the activities of sgRNAs across multiple species genomes [17]. These three models all used CNNs to extract features from the input genomic sequence. Overall, they are superior to machine learning-based tools in prediction accuracy.
CNNs are multilayer architectures where the successive layers are designed to learn abstract features, until the last layer produces an output value. They use weight-sharing strategy to capture local patterns in data such as sequences, which is analogous to taking the position weight matrix of a motif and scanning it across the DNA sequence [18]. CNNs perform well when some spatially invariant patterns of the inputs are expected. But they are restricted to learn the local patterns. Recurrent neural networks (RNN), particularly based on long short-term memory network (LSTM) [19] and gated recurrent unit (GRU) [20], have been designed for sequential or time-series data [21]. The hidden layers of RNN are regarded as memory states which can retain information from previous sequence and be updated at each step. Several tools have been introduced in the literature to demonstrate the synergistic improvements of CNN-RNN models due to the complementary in their modeling capability. SPEID achieved competitive performance using types of epigenetic data for enhancer-promoter interaction prediction in a unified CNN-RNN model [22]. DeeperBind added a LSTM layer to learn the dependencies between sequence features identified by CNN, further improving the prediction of protein binding specificity [23]. DanQ, a CNN combined bi-directional LSTM (BLSTM) framework, has recently been introduced to quantify function of DNA sequences by incorporating the motifs and a complex regulatory grammar between the motifs [24]. Pan et al. proposed a hybrid CNN-BLSTM based iDeepS to concurrently identify the binding sequence and structure motifs from RNA sequences [25].
The previous success of CNN-RNN in bioinformatics motivated us to extend its applications to CRISPR/Cas9 sgRNA on-target activity prediction. In this work, we introduced C-RNNCrispr, a hybrid architecture combining CNN with bidirectional GRU (BGRU), to predict sgRNA cleavage efficacy. The intuition of this hybrid architecture is to use CNN for feature extraction while using the BGRU to model sequential dependencies of sgRNA features. C-RNNCrispr contains two branches: sgRNA branch and epigenetic branch, respectively being used to extract sgRNA and epigenetic features. Particularly, we first represented sgRNA sequences and its related epigenetic features by one-hot encoding, which transforms the inputs into two 4 × 23 binary matrices for subsequent convolution operations. Second, the encoded sgRNA and epigenetic matrices were respectively fed into sgRNA branch and epigenetic branch for abstract features extraction. Third, the outputs of these two branches were integrated by element-wise multiplication. Finally, the outputs of the merged layer were fed into a linear regression layer to grade sgRNA cleavage efficiency. Besides, we proposed a transfer learning strategy to address the small-size sample problem. To be specific, we first pre-trained the proposed C-RNNCrispr on the benchmark dataset. Subsequently, we fine-tuned the pre-trained C-RNNCrispr on small-size cell-line datasets to predict sgRNA on-target activity. Experiments results showed that C-RNNCrispr consistently surpasses other available state-of-the-art prediction methods.
2. Methods
2.1. Data resources
We utilized the publicly available datasets packaged by Chuai et al. [5], available at https://github.com/bm2-lab/DeepCRISPR. It contains a benchmark dataset and four cell-line independent datasets. For benchmark dataset, each observation in the data contains a 23-nt sgRNA sequence and a binary class label indicating the high-efficiency and low-efficiency sgRNAs. There are 180,512 unique sgRNA sequences in this dataset. We used this dataset to build our C-RNNCrispr model. HCT116 and HELA datasets were generated in human HCT116 and Hela cells [26], HEK293T dataset generated in human Hek293t cell was original published in [10]. HL60 dataset generated in human Hl60 cell was original published in [27]. Each observation contains a sgRNA sequence and the measured knockout efficacy. After removing the redundancy, the number of datasets HCT116, HEK293T, HELA and hl60 was 4239, 2333, 8101 and 2076, respectively.
The above datasets were processed by Chuai et al. [5]. In their study, four epigenetic features of each sgRNA sequence was obtained from ENCODE [28], including CTCF binding information obtained from ChIP-Seq assay, H3K4me3 information from ChIP-Seq assay, chromatin accessibility information from DNase-Seq assay, and DNA methylation information from RRBS assay. Each epigenetic feature was denoted by a symbolic sequence with length of 23, with notations “A” and “N” meaning the present and absent of the epigenetic feature at a particular base position of DNA regions.
The binary cleavage efficacy was obtained by converting the measured sgRNA efficacy using a log-fold change of 1 as the cutoff. The high-activity sgRNAs were denoted by 1 and the low-activity ones were represented by 0. Besides, numerical cleavage efficacy was defined by applying a collaborative filtering-based data normalization method [29]. To be specific, a matrix was formulated where each row represented the experiments and each column denoted one gRNA. Normalized numerical sgRNA cleavage efficacy value was defined as
| (1) |
where denoted the n-th sgRNA in the m-th experiments. , and represented the mean values for each row, the mean values for each column and the mean values of , respectively. For each sgRNA, the log2-fold change was calculated. Subsequently, sgRNAs within each gene were ranked by carrying out the rank-based normalization method proposed by Doench et al. [9]. The resulting normalized ranks were averaged across cell types and rescaled into 0–1. Here, 1 means the successful on-target cleavage efficacy. Each observation in the above four cell-line datasets contained the 23-nt sgRNA sequence, chromosome, start site, end site, strand, four types of symbolic epigenetic features, normalized numerical cleavage efficacy and the binary efficiency. We used these datasets to evaluate and compare the proposed C-RNNCrispr with several current deep learning and existing machine learning prediction methods.
2.2. Sequence encoding
The input sequence should be numerically encoded before being fed into deep learning models. Several methods have been proposed to represent the input sequence, such as one-hot encoding and k-mer embedding computed by word2vec [30]. For one-hot encoding representation, the input sequence is represented by a 4 × L matrix where 4 is the size of nucleotides vocabulary (A, C, G and T) and L is the length of the sequence. Each position in the sequence is related to a vector of length four with a single non-zero element corresponding to the nucleotide in that position. Specifically, the nucleotides A, C, G and T are encoded as four one-hot vectors and. Notably, one-hot encoding simply transforms DNA sequences into images with binary values rather than real continuous-values for each pixel with four channels corresponding to A, C, G and T, which may lead to restrictive effects on the performance. When using the k-mer embedding, the input sequence is split into overlapped k-mers of length k using a sliding window with stride s. Subsequently, each k-mer in the obtained sequence is mapped into a d-dimensional vector using word2vec method [30]. Word2vec is an unsupervised learning algorithm which maps k-mers from the vocabulary to vectors of real numbers in a low-dimensional space. The value of k-mer length and stride can be determined by model training. We refer readers to the original publication for details [30]. A recent study showed that using k-mer embedding to represent input sequence gained superiority of model performance than one-hot encoding [31]. However, the improvement in performance comes at the cost of the training time for sequence modeling. Overall, sequence representations denoted by one-hot encoding are sparse, high-dimensional and hardcoded, whereas k-mer embedding representations are dense, relatively low-dimensional, and learned from data.
It is noteworthy that one-hot encoding has been adapted in several previous methods on sgRNA on-target activity prediction [5], [16], [17]. For fair comparison, we chose this approach to encode sgRNA sequence with 23 nucleotides in length. Therefore, a 1-by-23 nucleotide sequence was denoted by a 4 × 23 binary matrix. Four kinds of epigenetic features (mentioned in Section 2.1) were analyzed in this work. We used a one-dimensional binary vector to encode each of epigenetic feature. The presence of the specific epigenetic feature at a particular position is represented by 1 while its absence is denoted by 0. As a consequence, four epigenetic features are denoted by a 4 (types of epigenetic features) × 23 (sequence length) binary matrix. The encoded sgRNA and epigenetic features binary matrices were subsequently fed into C-RNNCrispr-based model for training and testing.
2.3. C-RNNCrispr model
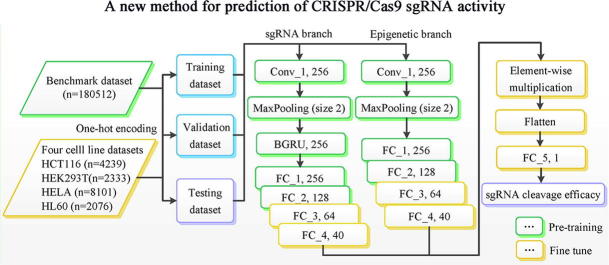
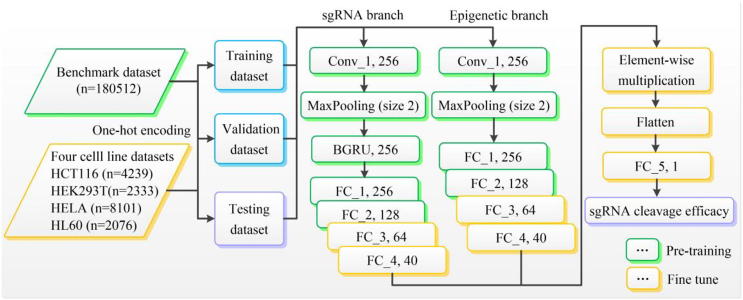
A convolutional neural network (CNN) [32] is a type of deep, feed-forward artificial neural network that can capture the hierarchical spatial representations, thus avoiding laborious manual feature engineering. Recurrent neural network (RNN) [33] is a variation of deep neural networks. Unlike CNN, RNN has an internal state that is updated as the network reads the input sequence. This internal memory allows RNN to capture interactions between the elements along the sequence, and is thus widely used in the field of NLP [34]. For details of CNN and RNN, see Supplementary Note. CNN excels at capturing local patterns in sequence data by using weight-sharing strategy but it fails at learning sequential correlations. Inversely, RNN achieves excellent performance for sequential modelling while fails to derive features in parallel. Many studies support the idea that the combination of CNN and RNN can achieve better performance [24], [35], [36]. To be specific, the convolutional modules stage scans the sequence using a series of 1D convolutional filter to capture sequence patterns. The following RNN stage is used for learning complex high-level relationships by considering the orientations and spatial relationships between the motifs. Inspired by these studies, we proposed a unified CNN-RNN framework to predict CRISPR/Cas9 sgRNA on-target activity. Fig. 1 and the following description give a summary of the basic architectural structure of C-RNNCrispr used.
Fig. 1.
An overview of C-RNNCrispr architecture. We trained the C-RNNCrispr from scratch on the benchmark dataset. Then, we fine-tuned the well-trained pre-trained C-RNNCrispr model on small-size datasets.
As shown in Fig. 1, C-RNNCrispr consists of two branches, viz. sgRNA branch and epigenetic branch. The sgRNA branch is applied to extract the abstract features of sgRNA sequences, whereas epigenetic branch is necessary to reveal the hidden knowledge of epigenetic information. Note that, the structure of epigenetic branch is similar to sgRNA branch except that a bidirectional gate recurrent unit network (BGRU, a special kind of variants for RNN) layer is absent. For the example of sgRNA branch, it receives a 4 (size of nucleotides vocabulary) × 23 (sequence length) binary matrix as an input. The first layer of the sub-network is a one-dimensional (1D) convolutional layer (conv_1), which consists of an array of 256 filters that convolves with the input sequence. Our rationale for including a convolution layer before BGRU layer is that CNNs achieve excellent performance for extracting sgRNA sequence features while keeping the number of model parameters tractable by applying convolutional operator. Rectified linear units (ReLU) [37] is subsequently used to keep only positive filter values and set the remaining to zeros, where .
The second layer is a local max-pooling layer (pool_1) with window size of 2. It connects with the outputs of previous layer by propagating only the largest output of each kernel with each stride for download sampling.
The third layer is a BGRU layer with dimension of 256. The motivation for adding a BGRU layer is that it is apt at enhancing the relevance between features of the sequences. The outputs of two parallel GRUs are concatenated to obtain our final feature representation containing both the forward and backward information of sgRNA sequence.
Next, the obtained features are followed by four fully connected layers (fc_1, fc_2, fc_3 and fc_4) with the sizes of 256, 128, 64 and 40 with ReLU activations, respectively. We used dropout for model regularization to avoid overfitting. The dropout rate will be given in Section 2.5.1.
The features of the last fully connected layer of sgRNA and epigenetic branches are integrated using element-wise multiplication operator for features merging. Finally, the outputs of the merged features are fed into a linear regression transformation to make a prediction of sgRNA on-target activity.
2.4. Experimental settings
The proposed C-RNNCrispr model was implemented using Python 3.6.4 and Keras 2.1.0 (http://keras.io) with TensorFlow (1.4.0) as the backend. All experiments were carried out on a desktop computer with Intel (R) Core (TM) i7-7800X CPU @ 3.50 GHz, Ubuntu 16.04.5 LTS and 32 GB RAM, as well as two NVIDIA GeForce GTX 1080 Ti GPUs with 11 GB of memory per GPU. We used mean square error (MSE) as the loss function for the regression task. During training process, we applied the RMSprop algorithm [38] for stochastic optimization of the objective of the loss function.
2.5. Implementation of the C-RNNCrispr model
2.5.1. Model selection and pre-training
Model selection was performed by testing the variants of C-RNNCrispr as summarized in Table 1 on the benchmark dataset. The dataset was randomly divided into a training dataset and an independent testing dataset with 80% and 20% classes. Experiments were performed under 5-fold cross-validation in the training phase. During each training test, the training data was randomly split into equal five parts. Among them, four parts were regarded as training dataset, while the remaining one part was taken as testing dataset. Supplementary Table S1 summarizes the number of the training samples, validation samples and testing samples of all the experiments in our study.
Table 1.
The variants of C-RNNCrispr models compared in this work.
| Model | Architecture |
|---|---|
| C-RNNCrispr_std | Using one convolutional layer with 256 1D filtering kernels of length 5 with dropout layer |
| C-RNNCrispr_2conv | Using two convolutional layers with 256 1D filtering kernels of length 5 |
| C-RNNCrispr_len7 | Using one convolutional layer with 256 1D filtering kernels of length 7 |
| C-RNNCrispr_avepool2 | Using average-pooling layer of window size 2 |
| C-RNNCrispr_ndrop | Using one convolutional layer with 256 1D filtering kernels of length 5 without using dropout layer |
Note: The descriptions of four C-RNNCrispr variants are relative to C-RNNCrispr_std model descripted in Section 2.3.
It is worth noting that the number of BGRU (1), neurons per layer of CNN (256) and BGRU (256) with recurrent dropout rate of 0.2 of C-RNNCrispr were manually set empirically. We constructed five C-RNNCrispr variants by various parameters: the number of convolution layers, the window size of convolution filters, the window size of pooling layer and the dropout rate. Fig. 1 depicts the standard architecture of C-RNNCrispr (C-RNNCrispr_std). Mini-batch gradient descent was performed for optimization and further reduced the gradient variance during training process. Each experiment was run for 200 epochs, and batch size was set to 256.
Bayesian optimization [39] is an automatic tuning method for optimizing the given learning algorithm via modeling the generalization performance. Hyperopt [40] and hyperas [41] (https://github.com/maxpumperla/hyperas) were used to carry out the Bayesian optimization created with Keras. We used the benchmark data to train models with different hyperparameters (i.e., dropout out rate, activation function, batch size and epoch) suggested iteratively by Bayesian optimization. The selection was applied over the following set of parameters: dropout coefficient (0.2, 0.3, 0.4, 0.5), activation function (‘ReLU’, ‘ELU’, ‘LeakyReLU’), batch size (128, 256, 512) and epoch (100, 200). The training data and test data were generated in the same way in Section 2.5.1. After enough iterations, we took the best hyperparameters that showed the minimum average validation loss as the final parameters of the model. The hyperparameters were as follows: activation function: ‘ReLU’; batch size: 256; epoch: 200. The dropout rates were 0.2 (keeping 80% of the connections) and 0.3 for specific layers of C-RNNCrispr. Specifically, the dropout rate following the layers of max_pooling layer, GBRU, and four fully connected layers of sgRNA branch was set to 0.2, 0.3, 0.3, 0.2, 0.2 and 0.2, respectively. Similarly, the dropout rate following the max_pooling layer and four fully connected layers of epigenetic branch was set to 0.3, 0.2, 0.3, 0.3 and 0.2, respectively. The dropout rate following the multiply layer was 0.2. We then carried out the final hyperparameters to pre-train C-RNNCrispr model again from scratch on benchmark dataset under 5-fold cross-validation.
2.5.2. Transfer learning
Transfer learning is the process of migration of trained model parameters to a new model to help train the new model. Previous studies in computer vision have demonstrated that deep models with better performance are learned via transfer learning from large scale datasets to other datasets of limited scales [42], [43]. Motivated by these studies, we investigated four transfer learning strategies (i.e., fine tune, frozen CNN, frozen BGRU and frozen FC) of borrowing information from benchmark dataset, and determined the optimal one for testing new cell line. For complete details see Table 2.
Table 2.
Four transfer learning strategies for C-RNNCrispr model.
| Strategy | Transfer learning procedure |
|---|---|
| Fine tune | Only the weights in the last two fully connected layers of sgRNA and epigenetic branches as well as the last fully connected layer of C-RNNCrispr are trainable |
| Frozen CNN | Freeze the weights of CNN layers |
| Frozen BGRU | Freeze the weights of BGRU layer |
| Frozen FC | Freeze the weights of fully connected layers |
We compared C-RNNCrispr with these four transfer learning strategies and determined the one which achieved the best performance in terms of Spearman correlation and area under the ROC curve (AUROC) as the final transfer learning strategy for the following analysis. Four cell line datasets were used under 5-fold cross-validation for performance evaluation. The training data and testing data for each cell line were constructed in the same way as described in Section 2.5.1. We first pre-trained C-RNNCrispr from scratch on the benchmark dataset under 5-fold cross-validation. Next, we applied the above transfer learning strategies on small-size cell-line datasets to evaluate and compare the predictive performance. Here, we provide a detailed description of fine tune strategy. Besides the last two fully connected layers of sgRNA branch and epigenetic branch as well as the element-wise multiplication and the last fully connected layers of C-RNNCrispr, all the layers of these two branches were frozen. After borrowing weights of the pre-trained C-RNNCrispr base network, we fine-tuned C-RNNCrispr to minimize the MSE loss function using the RMSprop optimizer for small-size cell lines. Through fine tune, C-RNNCrispr could effectively prevent overfitting when applying for small-size datasets. For any given cell line of interest, the training process is described as below:
-
(1)
Pre-train C-RNNCrispr from scratch on benchmark dataset for 200 epochs.
-
(2)
Freeze the convolution, BGRU, max-pooling layers, the first two fully connected layers of sgRNA branch. On the other hand, freeze the convolution, max-pooling layers and the first two fully connected layers of the epigenetic branch.
-
(3)
Train the last two fully connected layers of both the above two branches, the element-wise multiplication layer and the last fully connected layer of C-RNNCrispr with training data from cell line of interest for another 200 epochs.
-
(4)
Evaluate C-RNNCrispr model on the test data.
2.6. Settings of other methods
We ran the Python code of Seq_deepCpf1 (downloaded from Github at https://github.com/MyungjaeSong/Paired-Library) using the same data and training process. It is noteworthy that the input of Seq_deepCpf1 is a 4-by-34 binary matrix. We changed the input shape of Seq_deepCpf1 model into 4-by-23 into to match the size of the data in this work. We used the benchmark dataset to pre-train the model. For fair comparison, we only fine-tuned the weights parameters in the last two layers (1681 free parameters) for cell line-specific prediction.
The source R code of DeepCas9 can be downloaded from Github at https://github.com/lje00006/DeepCas9. We constructed DeepCas9 model following the description in [17] and trained it using the same training and validation data in Python. For fair compared with other deep learning-based methods, we also applied transfer learning for DeepCas9 (DeepCas9 plus transfer learning). Specifically, we used fine tune by only training the top two fully connected layers (80769 free parameters) of DeepCas9 architecture. The source code of DeepCRISPR were run by getting from https://github.com/bm2-lab/DeepCRISPR. The performance of sgRNA Designer and sgRNA Scorer were taken from Chuai et al. [5].
2.7. Performance measures
In order to evaluate the performance of C-RNNCrispr, we used Spearman correlation coefficient between efficiency scores and predicted scores. We used Spearman correlation because it is more robust to outliers compared with Pearson correlation coefficient [44]. In addition, it was adopted in previous sgRNA activity prediction studies [5], [9], [16], [17]. We also calculated AUROC to comprehensively quantify the overall predictive model performance of C-RNNCrispr. The value of AUROC is in [0, 1], where 1 equates to a successful performance. In this study, we applied 0.5 AUROC as the baseline.
3. Results
3.1. Performance comparisons for different architectures under 5-fold cross-validation on benchmark dataset
We compared the performance of different model architectures trained on benchmark dataset under 5-fold cross-validation in Table 3. Some interesting conclusions can be extracted: first, amongst the compared architectures, C-RNNCrispr_std achieved the best performance for predicting sgRNA on-target activity with mean Spearman correlation and AUROC values of 0.877 and 0.976, respectively. Second, CNN with one convolutional layer (C-RNNCrispr_std) surpassed that with two convolutional layers (C-RNNCrispr_2conv). Third, the performance of C-RNNCrispr_std was a little superior than C-RNNCrispr_len7 and C-RNNCrispr_avepool2. Finally, we noticed that C-RNNCrispr_std outperformed C-RNNCrispr_ndrop, which is expected since dropout regularization contributes to the prevention or mitigation of overfitting. Together, these results demonstrated that C-RNNCrispr_std achieves the best generalization performance amongst these architectures. Therefore, we chose C-RNNCrispr_std for the following experiments.
Table 3.
Performance comparisons amongst different architectures under 5-fold cross-validation on benchmark dataset.
| Model | Spearman correlation | AUROC |
|---|---|---|
| C-RNNCrispr_std | 0.8770.062 | 0.9760.002 |
| C-RNNCrispr_2conv | 0.8330.000 | 0.9720.003 |
| C-RNNCrispr_len7 | 0.8330.000 | 0.9750.004 |
| C-RNNCrispr_avepool2 | 0.8330.000 | 0.9740.005 |
| C-RNNCrispr_ndrop | 0.8330.001 | 0.9690.001 |
Note: Performance is shown as mean ± standard deviation. This representation also applies to Table 4. The best performance (as measured by each metric) across different architectures is highlighted in bold for clarification. These highlights also apply to Table 4, Table 6, Supplementary Tables 2, 3 and 4.
3.2. Efficacy of CNN and BGRU
Next, we evaluated the effectiveness of CNN and BGRU for our C-RNNCrispr on sgRNA activity prediction. First, we verified the efficacy of the convolution stage by proposing a variant deep architecture (without CNN) getting rid of the convolutional layers and max-pooling layers of sgRNA branch from full C-RNNCrispr model. Then we compared this variant architecture with C-RNNCrispr under 5-fold cross-validation on benchmark dataset. The training data and test data were generated identically to the way described in Section 2.5.1. As expected, we discovered that removing convolutional layer of sgRNA branch leads to 0.046 and 0.001 decrease on average of Spearman correlation and AUROC values, respectively. Second, we performed experiments on another variant of C-RNNCrispr by removing the BGRU layer in the sgRNA branch (without BGRU). In this case, we observed Spearman correlation and AUROC score became less compared with full C-RNNCrispr, with 0.060 and 0.034 decline on average (Table 4). For the sake of clarity, C-RNNCrispr achieved the highest Spearman correlation and AUROC values amongst these architectures.
Table 4.
Performance comparison among C-RNNCrispr and its two variant architectures (i.e., without CNN and without BGRU) on benchmark dataset under 5-fold cross-validation.
| Model | Spearman correlation | AUROC |
|---|---|---|
| C-RNNCrispr | 0.8770.062 | 0.9760.002 |
| without CNN | 0.8310.001 | 0.9710.002 |
| without BGRU | 0.8170.001 | 0.9420.004 |
In view of the above two aspects, convolutional operations were capable of extracting the abstract features of sgRNA and epigenetic sequences. Besides, BGRU stage was indispensable in our architecture for capturing the sequence dependencies of sgRNA. We conclude that combination of CNN and BGRU can boost the power of C-RNNCrispr for sgRNA activity prediction.
3.3. Effect of transfer learning on small-sample data learning
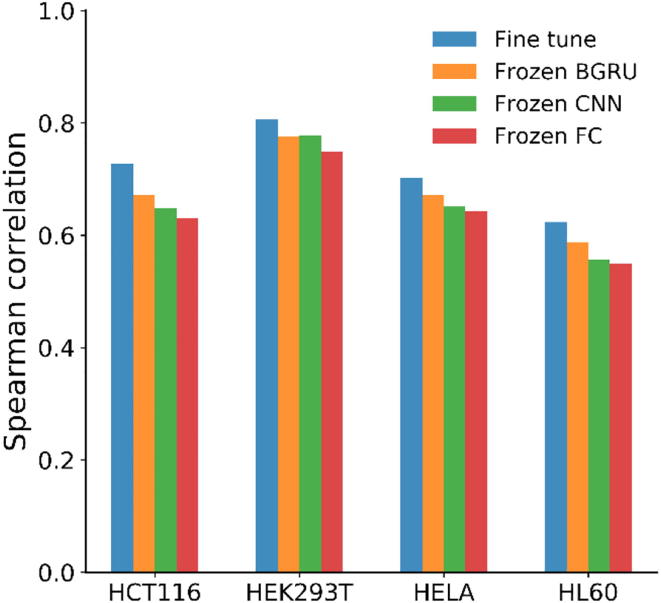
In this section, we focus on analyzing how the general feature representations from the C-RNNCrispr base network can be transferred and help the small-sample cell lines data learning for sgRNA target efficacy prediction. For this purpose, we compared the performance of the above mentioned transfer learning schemes (the basic training process was introduced in Section 2.5.2). The experiments were performed under 5-fold cross-validation on the above four small-size cell-line datasets. The training data and testing data for each cell line were constructed identically to the way described in Section 2.7. As shown in Fig. 2, it is clear that amongst the transfer learning strategies, fine tune clearly outperformed others. For example, using benchmark data to pre-train C-RNNCrispr, we obtained the Spearman correlation of 0.727, 0.648, 0.672 and 0.630 on HCT116 dataset for fine tune, frozen CNN, frozen BGRU and frozen FC, respectively. Supplementary Table S2 shows the results of AUROC values based on these transfer learning schemes. As can be seen, fine tune strategy performed as well as, or even slightly better than the other transfer learning strategies on datasets HCT116 and HEK293T, with values of 0.937 and 0.976, respectively. Therefore, we applied fine tune strategy on small-size cell line datasets to boost the predictive performance.
Fig. 2.
Performance comparison of different transfer learning strategies for sgRNA activity prediction on four cell line-specific training data under 5-fold cross-validation.
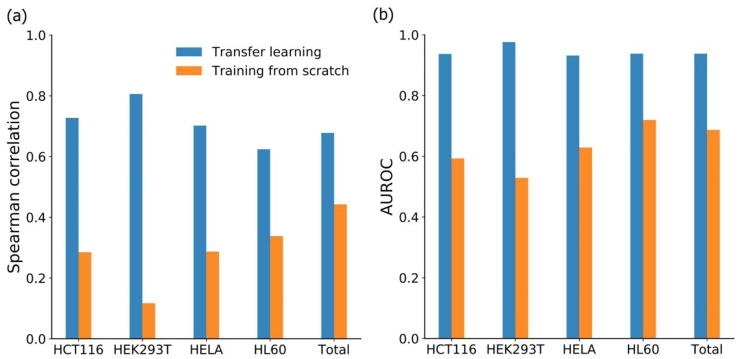
In order to further verify the advantage of transfer learning (i.e. via fine tune) with benchmark dataset on small-size cell line data, we compared the results between using cell line-specific training data to train from scratch and using fine-tune strategy. Intuitively, there was a significant improvement for each cell line in terms of these two evaluation criteria (Fig. 3). For example, the standard training method yield Spearman correlation values of 0.285, 0.117, 0.287, 0.338, and 0.443 on HCT116, HEK293T, HELA, HL60 and total datasets, much smaller than 0.727, 0.806, 0.702, 0.624, and 0.682 obtained from fine tune strategy. The superior performance of fine tune for modeling power of C-RNNCrispr is clear.
Fig. 3.
Performance comparison of C-RNNCrispr training from scratch and transfer learning via fine tune for each cell line data by 5-fold cross-validation.
3.4. Comparison with current algorithms
We next compared our C-RNNCrispr with current algorithms using sgRNA and epigenetic data. Prior to this, we briefly comment on some comparisons of C-RNNCrispr and other existing deep learning-based methods for sgRNA on-target activity prediction (see Table 5). First, both Seq_deepCpf1 and DeepCas9 performed based on 1D convolution model (1D CNN) considering only sgRNA sequence composition. DeepCRISPR and C-RNNCrispr performed by incorporating both sgRNA sequence and epigenetic data. Second, DeepCRISPR used two dimensional CNN (2D CNN) while C-RNNCrispr used hybrid 1D CNN and BGRU. Third, besides DeepCas9, all methods used transfer learning technique. Thus, we only compared with DeepCRISPR among these methods when considering both sgRNA and epigenetic data. Moreover, we also compared C-RNNCrispr with two current machine learning-based tools, including CRISPR Designer and sgRNA Scorer.
Table 5.
Existing deep learning-based methods for sgRNA on-target activity prediction.
| Model | Model | Sequence | Training mode | Reference |
|---|---|---|---|---|
| Seq_deepCpf1 | 1D CNN | sgRNA | Transfer learning | [16] |
| DeepCRISPR | 2D CNN | sgRNA + Epi | Transfer learning | [5] |
| DeepCas9 | 1D CNN | sgRNA | From scratch | [17] |
| C-RNNCrispr | 1D CNN-BGRU | sgRNA + Epi | Transfer learning | – |
Note: sgRNA + Epi, sequence features and epigenetic features.
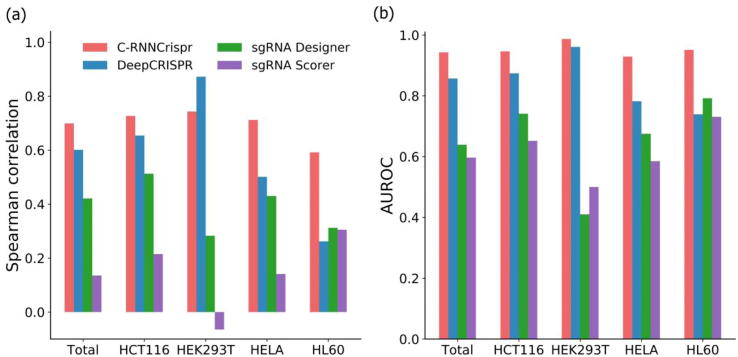
The above four cell line datasets were applied under 5-fold cross-validation for performance evaluation. The training data and testing data for each cell line were built in the same way as described in Section 2.5.1. Fig. 4a shows the Spearman correlations of compared methods and C-RNNCrispr. In general, deep learning-based model outperformed the machine learning-based tools. To be specific, our C-RNNCrispr gained superiority to other methods in three cell lines and total dataset, whereas for dataset HEK293T, it performed lower than DeepCRISPR. As depicted in Fig. 4b, it is clear that C-RNNCrispr outperformed other methods in terms of AUROC scores. Detailed Spearman correlation and AUROC scores for individual datasets are provided in Supplementary Table S3. We conclude that C-RNNCrispr is competitive against other existing methods.
Fig. 4.
Performance comparison of C-RNNCrispr and other learning-based prediction models on different testing cell line data under 5-fold cross-validation.
To further evaluate the generalization capability of the proposed method, we trained C-RNNCrispr using leave-one-cell-line-out procedure and made comparisons with the above mentioned methods. The training and testing data for each cell line were constructed followed the procedure illustrated in Section 2.5.1. In the training stage, for a cell line of interest to be predicted, we used the training data from other three cell lines. In the testing stage, we evaluated the model on the test data of the given cell line of interest. Taking leave-HCT116-out as an example, we trained the model by incorporating training data from HEK293T, HELA, and HL60 cell lines (lacking training data from HCT116), and evaluated the model on HCT116 test data. Intuitively, we observed that our C-RNNCrispr achieve the best performance (Fig. 5). Specifically, compared with the next-best DeepCRISPR, C-RNNCrispr showed the superior performance in all cell line datasets except for HCT116. C-RNNCrispr achieved mean Spearman correlation of 0.692, which was 0.286 higher than DeepCRISPR. In addition, C-RNNCrispr clearly outperformed other models in all cell line datasets in terms of AUROC. For more details, see Supplementary Table S4. These observations suggest that C-RNNCrispr gained the superiority of generalizability for sgRNA activity prediction.
Fig. 5.
Performance comparison of C-RNNCrispr and other learning-based prediction models on different testing cell line data with a leave-one-cell-out procedure.
3.5. Case studies
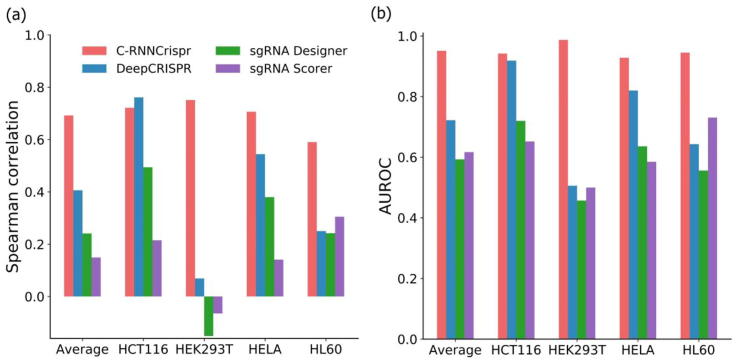
From the above investigation, we have observed that our C-RNNCrispr showed satisfactory performance for sgRNA activity prediction. Previous machine learning-based methods demonstrated that sgRNA sequence composition is critical for the cleavage efficiency [45], [46]. The first case study was conducted to investigate whether C-RNNCrispr can effectively predict sgRNA activity using sgRNA sequence composition features. For this purpose, we evaluated the prediction ability of C-RNNCrispr by incorporating only sgRNA sequence features. We only used the sgRNA branch to evaluate the sgRNA activity. We compared C-RNNCrispr with three deep learning-based models (i.e., DeepCRISPR, Seq_deepCpf1 and DeepCas9) for predicting sgRNA activity on four cell line datasets. Notably, the inputs of Seq_deepCpf1 and DeepCas9 only include the sgRNA sequence. For fair comparison, we applied DeepCRISPR using sgRNA sequence only. In addition, Seq_deepCpf1, DeepCRISPR and C-RNNCrispr applied fine-tuning of the benchmark dataset pre-trained model on cell line of interest dataset (see Table 5). More concretely, Seq_deepCpf1 only updated the weights parameters in the last two layers of a benchmark dataset pre-trained CNN model on small sample cell line of interest datasets. Analogously, DeepCRISPR used the encoder part of the DCDNN-based model as the pre-trained model. C-RNNCrispr applied the fine-tune strategy (Section 2.5.3) for improving the performance. DeepCas9 trained the model from scratch. To make a fair comparison, we retrained it by applying transfer learning, namely DeepCas9 + transfer learning. Specifically, all layers except the last two fully connected layers were fine tune on small sample cell line of interest. The training data and test data for each cell line were generated in the same way as described in Section 3.4.
We note that our C-RNNCrispr consistently outperforms the other methods in terms of Spearman correlation (see Table 6). On average, C-RNNCrispr shows a Spearman correlation value of 0.663, with 0.026 higher than the second best Seq_deepCpf1. Besides, C-RNNCrispr also presents better performance in terms of AUROC than other models except for dataset HCT116, convincing us that C-RNNCrispr is more powerful for sgRNA activity prediction. We also note that, with the help of transfer learning, DeepCas9 plus fine tune surpasses DeepCas9 training from scratch. This observation is in accordance with the conclusion of Section 3.3. Taken together, our C-RNNCrispr can effectively predict sgRNA activity using sequence composition information only.
Table 6.
Performance comparisons amongst five deep learning models based on target sequence composition on four cell line datasets under 5-fold cross-validation.
| Model | HCT116 | HEK293T | HELA | HL60 | Average |
|---|---|---|---|---|---|
| (a) Spearman correlation | |||||
| C-RNNCrispr | 0.724 | 0.665 | 0.685 | 0.577 | 0.663 |
| Seq_deepCpf1 | 0.672 | 0.665 | 0.651 | 0.558 | 0.637 |
| DeepCRISPR | 0.650 | 0.035 | 0.510 | 0.200 | 0.349 |
| DeepCas9 | 0.603 | −0.116 | 0.418 | 0.118 | 0.256 |
| DeepCas9 + TF | 0.683 | 0.572 | 0.675 | 0.495 | 0.606 |
| (b) AUROC | |||||
| C-RNNCrispr | 0.934 | 0.978 | 0.925 | 0.936 | 0.943 |
| Seq_deepCpf1 | 0.939 | 0.978 | 0.921 | 0.928 | 0.942 |
| DeepCRISPR | 0.887 | 0.474 | 0.788 | 0.584 | 0.683 |
| DeepCas9 | 0.784 | 0.470 | 0.677 | 0.535 | 0.617 |
| DeepCas9 + TF | 0.902 | 0.905 | 0.902 | 0.887 | 0.899 |
Note: The top table records Spearman correlation values while the bottom one records AUROC values. DeepCas9 + TF: DeepCas9 + transfer learning. The performance of DeepCRISPR is take from [5].
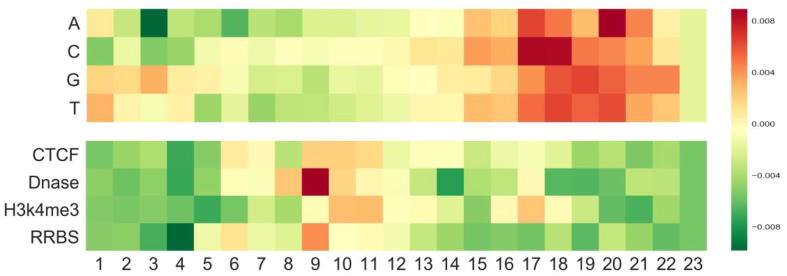
The second case study was conducted to reveal the biological insights into the sgRNA on-target activity prediction. Using the method in a previous study [47], we developed a heuristic to interpret our C-RNNCrispr network by visualizing the importance of all possible nucleotides and their corresponding epigenetic features at different locations. Briefly, we generated special sequences denoting the presence of the nucleotide and epigenetic features at a specific position and respectively fed them into the sgRNA branch and epigenetic branch of the well trained C-RNNCrispr model, subsequently took the outputs for visualization. For complete details see Supplementary Note. Fig. 6 shows the importance of all four nucleotides and epigenetic features at different positions. This allowed us to reveal general patterns of CRISPR-mediated DNA editing and make a number of observations. The positions adjacent to the PAM are more crucial than the PAM-distal region for sgRNA activity prediction. This is consistent with previous observations that perfect base-pairing with 10–12 bp immediately upstream the PAM (PAM-proximal) determines Cas9 specificity, whereas multiple PAM-distal mismatches can be tolerated [48]. In agreement with previous studies that presence of an A or T nucleotide at position 20 (1 bp adjacent to PAM) increased the proportion of indel [49], we found the presence of A or T is favored at this position. We also noted that position 17 (immediately 5′ of the cleavage site) is the most important. The presence of a C nucleotide is informative at this position since the cut site usually resides 3 bp upstream the PAM. The homopolymers (a run of two or more identical nucleotides) are found to be favored at position 17–19, which coincides with a previous finding that the presence of homopolymers adjacent to the cut site increased the proportion of deletions [49]. Most of the top epigenetic features were obtained by convolving the middle region of the input matrix. Opening-chromatin information of Dnase is found to be favored at 3 bp upstream of the PAM, which is in accord with a previous study that considering the target site accessibility can boost the predictive performance of gRNA activity [16]. It has a general preference for Dnase while relative avoidance of DNA methylation (H3K4me3) for high sgRNA efficiency. The same observation was also obtained by Chuai et al. [5].
Fig. 6.
Visualization of the importance of different nucleotides and epigenetic features at different positions for our C-RNNCrispr. The colors represent the contribution of the position-specific nucleotide and epigenetic features to determining an efficient sgRNA. The nucleotides and epigenetic features are arranged vertically, whereas the positions of the sequence are placed horizontally.
4. Discussion
In this study, we introduced C-RNNCrispr, a hybrid CNN-BGRU based model for CRISPR/Cas9 sgRNA activity prediction. An intuition of using a combination of CNN and BGRU is to use CNN for feature extraction and apply BGRU for modeling sequential dependencies of sgRNA features. Experimental results on publicly available datasets show C-RNNCrispr can adaptively learn sequence characteristics of sgRNA and epigenetic features; thereby avoiding manual feature extraction.
Numerous studies have demonstrated the synergistic improvements of unified CNN-RNN models in virtue of the complementary in their modeling capability, such as DeeperBind [23] and DanQ [24]. However, there is no consensus on the relative superiority between CNNs and RNNs for sequential data. Though RNNs are popular for natural language data with strong long-range dependencies, there are some studies reported that CNNs perform as well as RNNs [50], [51]. For instance, Zhuang et al. proposed a simple CNN for predicting enhancer-promoter interactions with DNA sequence data, which performs equally well with hybrid CNN-RNN model [52]. They used only moderately large sample sizes of the training data. It is perhaps that there is no strong long-range dependency in DNA sequence data in their study, possibly too subtle to be detected. If so, it would suggest unnecessary to use RNNs.
Previous studies in computer vision have demonstrated that CNN transfer learning from ImageNet to other datasets of limited scales contributed to better performing deep models [43], [53]. We have explored four transfer learning strategies (Section 3.3) to fine-tune the benchmark dataset pre-trained C-RNNCrispr model on small sample cell line datasets. Each model gained clearly better predictive results than the naive way. This result is expected given that there may be commonalities among cell line-specific sequence features. Therefore, fine tune is ideal for borrowing information from other cell lines for the task of predicting sgRNA activity when training data was limited.
Compared with several state-of-the-art learning-based tools, we found that our C-RNNCrispr coupled with a fine tune strategy integrating data from benchmark dataset would perform competitively against other methods. Moreover, we noticed that even given the sgRNA sequence only, C-RNNCrispr still surpassed amongst the compared deep learning-based models. This result implies that C-RNNCrispr can represent and capture nontrivial patterns or relationship between sequence information of sgRNA sequences, it is due to the proposed CNN-BGRU based model combines the advantages of CNN for capturing local patterns of sequences and BGRU for modeling sequential dependencies. Note that, the architectures that combine CNN with BGRU indeed provide performance over the CNN model. However, the improvement in accuracy comes at the expense of the increased computational cost. We ran our experiments on an Ubuntu server with two NVIDIA GeForce GTX 1080 Ti GPUs with 11 GB of memory per GPU. Typical running time of each experiment for model training was 1 h for the variant without BGRU (see Section 3.2), 9.8 h for the variant without CNN and almost 5.5 h for the C-RNNCrispr network including CNN and GBRU modules (see details in Supplementary Table S5). Because BGRU are expensive for processing long sequence, but 1D converts are cheap, it can be a good idea to use 1D convert as a preprocessing step before a BGRU, shortening the sequence and extracting useful representations for the BGRU to process. Considering that sgRNA sequence is more important than its corresponding epigenetic features, thus, we used no BGRU for the epigenetic branch to reduce the computational cost.
Although C-RNNCrispr has improved the performance for sgRNA activity prediction and become an advantageous approach, there are still several avenues of interest to investigate. Our future work will focus on three areas. One area is about exploring other deep learning-based frameworks and exploiting methods for optimal hyperparameters selection, which may yield better performance. Notably, there are certain characteristic differences between biological sequence and image data of computer vision, the technical details of optimizing parameters determination may differ. The second area is about expanding the feature space. Currently, we only use sgRNA sequence data and four epigenetic features including CTCF binding, H3K4me3, chromatin accessibility as well as DNA methylation. Other informative features such as cutting positions, physicochemical property and RNA fold score can be exploited to boost the predictive power. The third area is sgRNA off-target site prediction. The limitation of the current study is that C-RNNCrispr can only be used for sgRNA on-target activity prediction. Note that, CRISPR can tolerate mismatches in sgRNA-DNA at various positions in a sequence-dependent manner, leading to off-target mutations [54], [55]. Therefore, this issue should be critically resolved and completely avoided when applying CRISPR/Cas9 gene editing to clinical applications. Various tools have been proposed to predict off-target score by considering the positions of the mismatches to the guide sequence (MIT score [54] and CFD score [9], etc.). Subsequently, two machine learning-based methods Elevation [56] and CRISTA [57] expand the feature set, including features such as sgRNA secondary structure, genomic location for off-target prediction. Zhang et al. proposed an ensemble learning framework to predict the off-target activities. It found ensemble learning using AdaBoost outperformed other individual off-target predictive tools and adding PhyloP can enhance the predictive capabilities [58]. It was not until 2018 that deep learning-based methods have been integrated for CRISPR off-target prediction, such as DeepCRISPR and CNN_std [59]. The above two CNN-based models showed good performance in off-target activity prediction. To learn more about the computational methods used to facilitate the process of CRISPR/Cas9 sgRNA off-target activity prediction, we refer readers to [12], [60], [61] for details. Integrating the sgRNA off-target site prediction and our on-target activity prediction is worth of generating for providing more comprehensive guidance for optimal sgRNAs selection. These are interesting topics deserve to be explored in the future.
5. Conclusion
In this study, we present C-RNNCrispr, a unified CNN-BGRU architecture for CRISPR/Cas9 sgRNA on-target activity prediction. We applied a CNN to automatically learn the abstract features of sgRNA and four epigenetic features (i.e., CTCF binding, H3K4me3, chromatin accessibility and DNA methylation). On the other hand, we used BGRU to model the sequential dependencies of sgRNA features. Compared with three deep learning based models (i.e., Seq_deepCpf1, DeepCRISPR and DeepCas9) and two machine learning-based models (e.g. sgRNA Designer and sgRNA Scorer), C-RNNCrispr can effectively learn the features of sgRNA sequence and epigenetic features. We also introduced a transfer learning strategy to boost the predictive power of C-RNNCrispr in dealing with small-size datasets. Experimental results on the published datasets indicated that the effectiveness of our C-RNNCrispr for CRISPR/Cas9 sgRNA cleavage efficacy prediction.
Conflicts of interest
None declared.
Acknowledgement
This research was funded by the National Natural Science Foundation of China Grant No. 61872396, U1611265 and 61872395, and also by Pearl River Nova Program of Guangzhou, China Grant No. 201710010044.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.01.013.
Contributor Information
Zhiming Dai, Email: daizhim@mail.sysu.edu.cn.
Xianhua Dai, Email: issdxh@mail.sysu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo J., Wang T., Guan C., Liu B., Luo C., Xie Z. Improved sgRNA design in bacteria via genome-wide activity profiling. Nucleic Acids Res. 2018;46:7052–7069. doi: 10.1093/nar/gky572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moreno-Mateos M.A., Vejnar C.E., Beaudoin J.D., Fernandez J.P., Mis E.K., Khokha M.K. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods. 2015;12:982–988. doi: 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chuai G., Ma H., Yan J., Chen M., Hong N., Xue D. DeepCRISPR: optimized CRISPR guide RNA design by deep learning. Genome Biol. 2018;19:80. doi: 10.1186/s13059-018-1459-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan J., Chuai G., Zhou C., Zhu C., Yang J., Zhang C. Benchmarking CRISPR on-target sgRNA design. Brief Bioinform. 2017 doi: 10.1093/bib/bbx001. [DOI] [PubMed] [Google Scholar]
- 7.Naito Y., Hino K., Bono H., Ui-Tei K. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics. 2015;31:1120–1123. doi: 10.1093/bioinformatics/btu743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heigwer F., Kerr G., Boutros M. E-CRISP: fast CRISPR target site identification. Nat Methods. 2014;11:122. doi: 10.1038/nmeth.2812. [DOI] [PubMed] [Google Scholar]
- 9.Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chari R., Mali P., Moosburner M., Church G.M. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat Methods. 2015;12:823–826. doi: 10.1038/nmeth.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hinz J.M., Laughery M.F., Wyrick J.J. Nucleosomes inhibit Cas9 endonuclease activity in vitro. Biochemistry. 2015;54:7063–7066. doi: 10.1021/acs.biochem.5b01108. [DOI] [PubMed] [Google Scholar]
- 12.Wilson L.O.W., O'Brien A.R., Bauer D.C. The current state and future of CRISPR-Cas9 gRNA design tools. Front Pharmacol. 2018;9:749. doi: 10.3389/fphar.2018.00749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 14.Zamir AR, Sax A, Shen W, Guibas LJ, Malik J, Savarese S, editors. Taskonomy: disentangling task transfer learning. Proceedings of the IEEE conference on computer vision and pattern recognition; 2018.
- 15.Lopez MM, Kalita J. Deep learning applied to NLP. arXiv preprint arXiv:170303091 2017.
- 16.Kim H.K., Min S., Song M., Jung S., Choi J.W., Kim Y. Deep learning improves prediction of CRISPR-Cpf1 guide RNA activity. Nat Biotechnol. 2018;36:239–241. doi: 10.1038/nbt.4061. [DOI] [PubMed] [Google Scholar]
- 17.Xue L., Tang B., Chen W., Luo J. Prediction of CRISPR sgRNA activity using a deep convolutional neural network. J Chem Inf Model. 2018 doi: 10.1021/acs.jcim.8b00368. [DOI] [PubMed] [Google Scholar]
- 18.Zou J., Huss M., Abid A., Mohammadi P., Torkamani A., Telenti A. A primer on deep learning in genomics. Nat Genet. 2019;51:12–18. doi: 10.1038/s41588-018-0295-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sundermeyer M, Schlüter R, Ney H, editors. LSTM neural networks for language modeling. Thirteenth annual conference of the international speech communication association; 2012.
- 20.Chung J, Gulcehre C, Cho K, Bengio Y. Empirical evaluation of gated recurrent neural networks on sequence modeling. arXiv preprint arXiv:14123555 2014.
- 21.Goodfellow I., Bengio Y., Courville A. MIT press; 2016. Deep learning. [Google Scholar]
- 22.Singh S, Yang Y, Poczos B, Ma J. Predicting enhancer-promoter interaction from genomic sequence with deep neural networks. bioRxiv 2018;085241. [DOI] [PMC free article] [PubMed]
- 23.Hassanzadeh HR, Wang MD, editors. DeeperBind: enhancing prediction of sequence specificities of DNA binding proteins. IEEE international conference on bioinformatics & biomedicine; 2017. [DOI] [PMC free article] [PubMed]
- 24.Quang D., Xie X. DanQ: a hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences. Nucleic Acids Res. 2016;44:e107-e. doi: 10.1093/nar/gkw226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan X., Rijnbeek P., Yan J., Shen H.B. Prediction of RNA-protein sequence and structure binding preferences using deep convolutional and recurrent neural networks. BMC Genomics. 2018;19:511. doi: 10.1186/s12864-018-4889-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hart T., Chandrashekhar M., Aregger M., Steinhart Z., Brown K.R., MacLeod G. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell. 2015;163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 27.Wang T., Wei J.J., Sabatini D.M., Lander E.S. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Consortium EP The ENCODE (ENCyclopedia of DNA elements) project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 29.Badaro G, Hajj H, El-Hajj W, Nachman L, editors. A hybrid approach with collaborative filtering for recommender systems. 2013 9th International wireless communications and mobile computing conference (IWCMC); 2013 1–5 July 2013.
- 30.Mikolov T., Sutskever I., Chen K., Corrado G., Dean J. Distributed representations of words and phrases and their compositionality. Adv Neural Inf Process Systems. 2013;26:3111–3119. [Google Scholar]
- 31.Trabelsi A., Chaabane M., Ben-Hur A. Comprehensive evaluation of deep learning architectures for prediction of DNA/RNA sequence binding specificities. Bioinformatics. 2019;35:i269–i277. doi: 10.1093/bioinformatics/btz339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cun Y.L., Boser B., Denker J.S., Howard R.E., Habbard W., Jackel L.D. Handwritten digit recognition with a back-propagation network. Adv Neural Inf Process Systems. 1990;2:396–404. [Google Scholar]
- 33.Graves A, Mohamed A-r, Hinton G, editors. Speech recognition with deep recurrent neural networks. 2013 IEEE international conference on acoustics, speech and signal processing; 2013: IEEE.
- 34.Hirschberg J., Manning C.D. Advances in natural language processing. Science. 2015;349:261–266. doi: 10.1126/science.aaa8685. [DOI] [PubMed] [Google Scholar]
- 35.You Y., Lu C., Wang W., Tang C.K. Relative CNN-RNN: learning relative atmospheric visibility from images. IEEE Trans Image Process. 2019;28:45–55. doi: 10.1109/TIP.2018.2857219. [DOI] [PubMed] [Google Scholar]
- 36.Liang G., Hong H., Xie W., Zheng L. Combining convolutional neural network with recursive neural network for blood cell image classification. IEEE Access. 2018;6:36188–36197. [Google Scholar]
- 37.Krizhevsky A, Sutskever I, Hinton GE, editors. ImageNet classification with deep convolutional neural networks. International conference on neural information processing systems; 2012.
- 38.Tieleman T, Hinton G. Lecture 6.5-rmsprop: divide the gradient by a running average of its recent magnitude. COURSERA: neural networks for machine learning 2012;4:26-31.
- 39.J. Snoek H. Larochelle R.P. Adams editors. Practical Bayesian optimization of machine learning algorithms international conference on neural information processing systems 2012
- 40.Bergstra J., Komer B., Eliasmith C., Dan Y., Cox D.D. Hyperopt: a Python library for model selection and hyperparameter optimization. Comput Sci Discov. 2015;8 [Google Scholar]
- 41.Pumperla M. Keras+ Hyperopt: a very simple wrapper for convenient hyperparameter optimization. 2016.
- 42.Shin H.C., Roth H.R., Gao M., Lu L., Xu Z., Nogues I. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging. 2016;35:1285–1298. doi: 10.1109/TMI.2016.2528162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou B, Garcia AL, Xiao J, Torralba A, Oliva A, editors. Learning deep features for scene recognition using places database. International conference on neural information processing systems; 2014.
- 44.Mukaka M.M. Statistics corner: a guide to appropriate use of correlation coefficient in medical research. Malawi Med J. 2012;24:69–71. [PMC free article] [PubMed] [Google Scholar]
- 45.Rahman M.K., Rahman M.S. CRISPRpred: a flexible and efficient tool for sgRNAs on-target activity prediction in CRISPR/Cas9 systems. PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0181943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen L, Wang SP, Zhang YH, Li JR, Xing ZH, Yang J, et al. Identify key sequence features to improve CRISPR sgRNA efficacy. IEEE Access 2017; PP:26582-89.
- 47.Xie B., Jankovic B.R., Bajic V.B., Song L., Gao X. Poly(A) motif prediction using spectral latent features from human DNA sequences. Bioinformatics. 2013;29:i316–i325. doi: 10.1093/bioinformatics/btt218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang W., Bikard D., Cox D., Zhang F., Marraffini L.A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leenay R.T., Aghazadeh A., Hiatt J., Tse D., Roth T.L., Apathy R. Large dataset enables prediction of repair after CRISPR-Cas9 editing in primary T cells. Nat Biotechnol. 2019;37:1034–1037. doi: 10.1038/s41587-019-0203-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Y., Zhang R., Singh S., Ma J. Exploiting sequence-based features for predicting enhancer–promoter interactions. Bioinformatics. 2017;33:i252–i260. doi: 10.1093/bioinformatics/btx257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bai S, Kolter JZ, Koltun V. An empirical evaluation of generic convolutional and recurrent networks for sequence modeling. arXiv preprint arXiv:180301271 2018.
- 52.Zhuang Z., Shen X., Pan W. A simple convolutional neural network for prediction of enhancer-promoter interactions with DNA sequence data. Bioinformatics. 2019 doi: 10.1093/bioinformatics/bty1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Razavian AS, Azizpour H, Sullivan J, Carlsson S. CNN features off-the-shelf: an astounding baseline for recognition. 2014.
- 54.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim D., Bae S., Park J., Kim E., Kim S., Yu H.R. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015;12:237–243. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- 56.Listgarten J., Weinstein M., Kleinstiver B.P., Sousa A.A., Joung J.K., Crawford J. Prediction of off-target activities for the end-to-end design of CRISPR guide RNAs. Nat Biomed Eng. 2018;2:38–47. doi: 10.1038/s41551-017-0178-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abadi S., Yan W.X., Amar D., Mayrose I. A machine learning approach for predicting CRISPR-Cas9 cleavage efficiencies and patterns underlying its mechanism of action. PLoS Comput Biol. 2017;13 doi: 10.1371/journal.pcbi.1005807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang S., Li X., Lin Q., Wong K.C. Synergizing CRISPR/Cas9 off-target predictions for ensemble insights and practical applications. Bioinformatics. 2019;35:1108–1115. doi: 10.1093/bioinformatics/bty748. [DOI] [PubMed] [Google Scholar]
- 59.Lin J., Wong K.-C. Off-target predictions in CRISPR-Cas9 gene editing using deep learning. Bioinformatics. 2018;34:i656–i663. doi: 10.1093/bioinformatics/bty554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang J., Zhang X., Cheng L., Luo Y. An overview and metanalysis of machine and deep learning-based CRISPR gRNA design tools. RNA Biol. 2019:1–10. doi: 10.1080/15476286.2019.1669406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu G., Zhang Y., Zhang T. Computational approaches for effective CRISPR guide RNA design and evaluation. Comput Struct Biotechnol J. 2020;18:35–44. doi: 10.1016/j.csbj.2019.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.