Abstract
With the size of the biopharmaceutical market exponentially increasing, there is an aligned growth in the importance of data-rich analyses, not only to assess drug product safety but also to assist drug development driven by the deeper understanding of structure/function relationships. In monoclonal antibodies, many functions are regulated by N-glycans present in the constant region of the heavy chains and their mechanisms of action are not completely known. The importance of their function focuses analytical research efforts on the development of robust, accurate and fast methods to support drug development and quality control. Released N-glycan analysis is considered as the gold standard for glycosylation characterisation; however, it is not the only method for quantitative analysis of glycoform heterogeneity. In this study, ten different analytical workflows for N-glycan analysis were compared using four monoclonal antibodies. While observing good comparability between the quantitative results generated, it was possible to appreciate the advantages and disadvantages of each technique and to summarise all the observations to guide the choice of the most appropriate analytical workflow according to application and the desired depth of data generated.
Keywords: N-Glycans, Biopharmaceuticals, Monoclonal antibodies, Intact mass analysis, Mass spectrometry, Native mass spectrometry, Glycan analysis, Peptide mapping, Glycopeptide analysis
Graphical abstract
Highlights
-
•
Comparative analysis of N-glycans present on four monoclonal antibodies using ten analytical methods reported.
-
•
Data presented to allow for impartial selection of appropriate analytical strategy depending on depth of information desired and ease of approach.
-
•
Methods included intact and subunit strategies and glycopeptide characterisation compared to released N-glycan as the current gold standard.
1. Introduction
Therapeutic monoclonal antibodies (mAbs) are a major class of biopharmaceuticals that have been used as autoimmune and oncology therapeutics [1]. Over the last decade, three to five new mAb products have been approved annually in Europe and/or the US [2]. The market for mAbs is expected to continue to grow over the coming years with over 300 mAb candidates currently in development [2,3]. Chinese hamster ovary (CHO) cells are the most commonly used expression system for mAbs, as they have the ability to produce human-like post-translational modifications, thereby reducing the potential for adverse reactions in humans [4]. Moreover, the use of CHO cell lines for over 30 years has established a history of product safety, efficacy and improvements in cell engineering, leading to high product titre and the capacity to adapt their growth in adhesion and suspension cell cultures [5]. Glycosylation, the most prominent post-translational modification (PTM), occurs in endoplasmic reticulum and Golgi apparatus [4]. Glycans can be attached to either the mAb heavy chain constant region (N-glycans) or serine or threonine (O-glycans) with the former being the most prominent [6]. N-glycans are classified into three main groups, i.e, oligomannose glycans, complex glycans and hybrid glycans. The N-glycans on mAbs produced in CHO cells are mainly asialylated core fucosylated complex glycans [7].
Glycosylation significantly impacts the stability and function of mAbs, including mediation of antibody-dependent cell-mediated cytotoxicity (ADCC) and complement dependent cytotoxicity (CDC) [7]. N-glycosylation stabilizes the structure of an mAb; therefore, deglycosylation renders mAbs thermally less stable, more susceptible to unfolding [8] and prone to aggregation [9]. Afucosylation of mAb N-glycans can result in increased binding affinity of mAbs to receptors present on the surface of leukocyte effector cells, which can enhance ADCC [10,11]. Several studies have suggested that terminal sialic acid residues on glycans mediate anti-inflammatory responses, reduce ADCC in vivo [12] and inhibit allergic reaction [13]. Galactosylation does not affect ADCC; however, the presence of galactose residues on N-glycans may lead to an increase in CDC [14,15] or anti-inflammatory activity [16]. High-mannose N-glycans have been shown to correlate with accelerated clearance of mAbs from the blood, decreasing circulating half-life of the drugs [[17], [18], [19]]. Therefore, control of the glycosylation pattern is required to ensure adherence to lot release specifications [20]. Characterisation of the glycosylation present on mAbs is a regulatory requirement not only for lot release but also for new drug applications and biosimilar approval [21], as reported in International Council for Harmonization (ICH) guideline Q6B. European Medicines Agency guidelines suggest that particular attention should be paid to their degree of mannosylation, galactosylation, fucosylation and sialylation and that distribution of the main glycan structures should be determined [4].
Different strategies can be applied to analyse the N-glycan moieties [22] with the gold standard workflow involving enzymatic release of oligosaccharides from the protein and chemical derivatisation with a label used for the detection technique of choice [23]. The most common separation techniques to analyse released N-glycans are capillary electrophoresis and liquid chromatography (LC) coupled to fluorescent detection and/or high resolution mass spectrometry (HR-MS). N-glycans can also be analysed as glycopeptides after mAb proteolysis (e.g. tryptic digest), glycopeptide enrichment and LC-MS/MS analysis, obtaining in-depth data on the glycoforms present on the protein as well as site-specific information. These two approaches require high levels of expertise and training for both sample preparation and LC-MS data acquisition and analysis, to ensure method robustness. Due to the recent advances in LC-MS technologies and the improvements in high resolution mass spectrometry of proteins, other analytical routes for determining N-glycan profiles are now available. These include characterisation of the glycoforms at intact protein level, using denaturing or native conditions, sometimes supported by top-down data, as well as the analysis of mAbs subunits via LC-MS [4,[24], [25], [26], [27]].
Intact mass analysis and top-down approaches facilitate the analysis of glycosylation with minimal sample preparation and represent rapid methods for the determination of glycoform profiles. However, if a more detailed analysis is required, it is necessary to produce a complementary glycan map because the intact protein glycan profile may not enable the detection of low abundant glycans [4]. Middle-up analysis is applied to mAbs after digestion with a proteolytic enzyme such as IdeS protease and allows the study of individual domains yielding region specific N-glycan profiles [28,29].
Intact and subunit analysis for the determination of N-glycans relies on HR-MS analysis that is essential to distinguish near-isobaric species generated by the intrinsic heterogeneity present on monoclonal antibodies. This heterogeneity arises not only at the N-glycan level but is also due to the presence of other PTMs, such as methionine and tryptophan oxidation, asparagine and glutamine conversion to succinimide intermediates, deamidation or C-term lysine truncation.
Here, we performed an extensive Fc-glycosylation analysis comparison using ten different methods to quantitatively characterize the N-glycan profiles from biotherapeutics, i.e., bevacizumab (BEV), infliximab (INF), rituximab (RIT) and trastuzumab (TRA). The four mAbs were studied across different domains of analysis: intact mass analysis using denatured and native conditions, reduced mAb (heavy/light chain analysis), intact Fc region (gingipain digestion), single chain Fc analysis (IdeS digested subunits), tryptic digestion based peptide mapping and released N-glycan analysis. Due to its wide acceptance, hydrophilic interaction liquid chromatography (HILIC) of N-glycans after labelling with anthranilic acid (2-AA) or 2-aminobenzamide (2-AB) was used as a reference method. The ten methods were compared in terms of depth of information achieved, level of expertise and instrumentation required for sample preparation and data analysis, relevance of the data obtained as well as suitability for structural characterisation or batch-to-batch comparison to assist the choice of the most suitable technique for N-glycan analysis.
2. Materials and methods
2.1. Chemicals and reagents
Rituximab, bevacizumab, infliximab and trastuzumab drug products were kindly provided by the Hospital Pharmacy Unit of the University Hospital of San Cecilio in Granada, Spain.
LC-MS grade solvents (0.1% (v/v) formic acid in water, 0.1% (v/v) formic acid in acetonitrile, formic acid, acetonitrile, water) were sourced from Fisher Scientific. TCEP and guanidine-HCl were obtained from Pierce. IdeS (immunoglobulin-degrading enzyme of Streptococcus pyogenes) (FabRICATOR™) and kgp (Lys-gingipain) (GingisKHAN™) were purchased from Genovis. SMART Digest™ kit, magnetic resin option was obtained from Thermo Scientific and PNGase F (CarboClip®) was obtained from Asparia Glycomics (Gipuzkoa, Spain). All other reagents were purchased from Sigma-Aldrich (Arklow, Ireland).
2.2. Analytical instrumentation
All LC-MS analyses were performed using a Vanquish™ Flex Quaternary UHPLC (Thermo Scientific, Germering, Germany) and a Q Exactive™ Plus Hybrid Quadrupole Orbitrap MS instrument with extended mass BioPharma Option, equipped with an Ion Max source with a HESI-II probe (Thermo Scientific, Bremen, Germany). All data were acquired using Thermo Scientific™ Xcalibur™ software 4.0.
2.3. Intact mass analysis under native conditions
For mAb analysis using native intact MS, 10 μg of mAb sample was injected onto a MAbPac™ SEC-1 column, 5 μm, 300 Å, 4.0 mm × 300 mm (Thermo Scientific™, Cat# 074696) under isocratic conditions of 50 mM ammonium acetate buffer at 300 μL/min for 20 min. The column temperature was at 30 °C. The MS method consisted of full positive polarity MS scans only at 17,500 resolution setting (defined at m/z 200) with the mass range set to 2500–8000 m/z and automatic gain control (AGC) target value of 3.0 × 106 with a maximum injection time of 200 ms and 10 microscans. In-source collision induced dissociation (CID) was set to 150 eV. Runs were performed in HMR mode. MS instrumental tune parameters were set as follows: spray voltage was 3.6 kV, sheath gas flow rate was 20 arbitrary units (AU), auxiliary gas flow rate was 5 AU, capillary temperature was 275 °C, probe heater temperature was 275 °C and S-lens RF voltage set to 200 V.
2.4. Intact mass analysis under denaturing conditions
For mAb analysis under denaturing conditions, 10 μg of each mAb was injected onto a MAbPac™ RP column, 4 μm, 2.1 mm × 50 mm (Thermo Scientific, Cat# 088648). The analysis was performed using a binary gradient of 0.1% (v/v) formic acid in water (A) and 0.1% (v/v) formic acid in acetonitrile (B). Gradient conditions were as follows: 25% B increased to 45% B in 2.5 min with a further increase to 80% B in 0.5 min with a 1 min isocratic hold. Initial conditions were restored in 0.2 min and held for an additional 3.8 min to ensure column re-equilibration. The column temperature was maintained at 70 °C throughout and flow rate was sustained at 300 μL/min. The MS method consisted of full positive polarity MS scans only at 17,500 resolution setting (defined at m/z 200) with the mass range set to 1500–4500 m/z and AGC target value of 3.0 × 106 with a maximum injection time of 100 ms and 10 microscans. In-source CID was set to 100 eV. Analysis was performed using protein mode. MS instrumental tune parameters were set as follows: spray voltage was 3.8 kV, sheath gas flow rate was 35 AU, auxiliary gas flow rate was 10 AU, capillary temperature was 275 °C, probe heater temperature was 175 °C and S-lens RF voltage set to 80 V.
2.5. Analysis of reduced mAb
MAbs were treated with 50 mM tris(2-carboxyethyl)phosphine) (TCEP) in 4 M guanidine-HCl, for 45 min at room temperature then diluted to 100 ng/μL in water. 2 μL of mAb was injected onto a MAbPac RP column, 4 μm, 2.1 mm × 50 mm (Thermo Scientific, Cat# 088648) and analysis was performed using a binary gradient of 0.1% (v/v) formic acid in water (A) and 0.1% (v/v) formic acid in acetonitrile (B). Gradient conditions were as follows: 28% B initially for 1 min, increased to 40% B in 15 min with a further increase to 80% B in 1 min and a final 1 min isocratic hold. Initial conditions were restored in 0.5 min and held for an additional 9.8 min to ensure column re-equilibration. The column temperature was maintained at 80 °C throughout and flow rate was maintained at 300 μL/min.
The MS method consisted of full positive polarity MS scans only at 17,500 resolution setting for heavy chain analysis and 140,000 for light chain analysis (defined at m/z 200) with the mass range set to 600–2400 m/z and AGC target value of 3.0 × 106 with a maximum injection time of 200 ms and 10 microscans. In-source CID was set to 0 eV. Analysis was performed using protein mode. MS instrumental tune parameters were set as follows: spray voltage was 3.8 kV, sheath gas flow rate was 25 AU, auxiliary gas flow rate was 10 AU, capillary temperature was 320 °C, probe heater temperature was 150 °C and S-lens RF voltage set to 60 V.
2.6. Middle up analysis of mAbs
For each mAb, two populations of mAb sub-units were analysed, one prepared following treatment with IdeS and the other with kgp.
For IdeS digestion, 40 μg of each mAb was combined with 0.5 μL of the enzymatic digestion solution (67 units/μL in Optima grade water) and incubated at 37 °C for 2 h at 500 rpm. Reduction of disulphide bonds was achieved by incubation in 4 M guanidine hydrochloride and 50 mM TCEP for 45 min at 56 °C. Following incubation, samples were reduced to dryness via vacuum centrifugation and reconstituted in 0.1% formic acid (1 μg/μL) prior of LC-MS analysis. For kgp digestion, 50 μg of each mAb was treated as recommended by the vendor and incubated for 2 h at 37 °C.
LC-MS analysis of mAb sub-units was performed on a MAbPac RP column, 4 μm, 2.1 mm × 100 mm (Thermo Scientific, Cat# 088648). The mobile phases were 0.1% formic acid in water (90/10, v/v, mobile phase A) and 0.1% formic acid in ACN/water (90/10, v/v, mobile phase B). The LC gradient profile was as follows: 25% B for 1 min, then increased to 45% B in 15 min; % B was kept constant for 1 min, then a wash step at 80% B was performed for 2 min before restoring initial conditions and column re-equilibration was performed 6 min. Total runtime was 25 min. The column temperature was maintained at 80 °C throughout and flow rate was sustained at 300 μL/min. For the kgp digest, the same column was employed, using 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B) as mobile phases. For Fc and Fab regions separation, the gradient was 20% B for 2 min, then increased to 45% B in 14 min and further increased at 80% B in 1 min, held for an additional minute and starting conditions restored in 0.5 min and held for 6.5 min. The MS method for both analyses consisted of full positive polarity MS scans only at 140,000 or 35,000 resolution setting (defined at m/z 200), for IdeS and kgp digest respectively, with the mass range set to 600–2400 m/z and AGC target value of 3.0 × 106 with a maximum injection time of 200 m s and 5 microscans. In-source CID was set to 0 eV. The analysis was performed in protein mode. MS instrumental tune parameters were set as follows: spray voltage was 3.8 kV, sheath gas flow rate was 25 AU, auxiliary gas flow rate was 10 AU, capillary temperature was 320 °C, probe heater temperature was 150 °C and S-lens RF voltage set to 60 V.
2.7. Peptide mapping
Samples were diluted to 2 mg/mL in water. For each sample digest, sample and SMART Digest buffer were added to each lane of a KingFisher Deepwell 96-well plate as outlined in Table S1. Trypsin Bead “wash buffer” was prepared by diluting SMART Digest buffer 1:4 (v/v) in water. Bead buffer was neat SMART Digest buffer. Digestion was performed using Kingfisher Duo Prime Purification System with BindIt™ software (version 4.0). Samples were incubated for 45 min at 70 °C at a medium mixing speed (to prevent sedimentation of beads), with post digestion cooling to 10 °C. Following digestion, disulphide bond reduction was performed with 10 mM DTT for 30 min at 37 °C and subsequent alkylation with 20 mM IA (iodoacetamide) in darkness for 30 min. The reaction was quenched with 15 μL of 100 mM DTT followed by 15 μL 10% TFA (final concentration 11 mM DTT and 1% TFA). The tryptic peptides were separated and monitored by LC-MS/MS analysis using an Acclaim Vanquish C18, 2.2 μm, 2.1 mm × 250 mm (Thermo Scientific, Cat#074812-V).
Analysis was performed using a binary gradient of 0.1% (v/v) formic acid in water (A) and 0.1% (v/v) formic acid in acetonitrile (B). Gradient conditions were as follows: 2% B increased to 40% B in 45 min with a further increase to 80% B in 1 min with 4 min isocratic hold. Initial conditions were restored in 0.5 min and held for an additional 15 min to ensure column re-equilibration. The column temperature was maintained at 25 °C throughout and flow rate was sustained at 300 μL/min. The MS method consisted of full positive polarity MS scans at 70,000 resolution setting (at m/z 200) with the mass range set to 200–2000 m/z and AGC target value of 3.0 × 106 with a maximum injection time of 100 m s and one microscan. In-source CID was set to 0 eV. MS2 settings were as follows: a resolution setting of 17,500 (at m/z 200), AGC target value of 1.0 × 105, isolation window set to 2.0 m/z, signal intensity threshold of 1.0 × 104, normalized collision energy set to 28, top 5 precursors selected for fragmentation and dynamic exclusion set to 7 s.
MS instrumental tune parameters were set as follows: spray voltage was 3.8 kV, sheath gas flow rate was 25 AU, auxiliary gas flow rate was 10 AU, capillary temperature was 320 °C, probe heater temperature was 150 °C and S-lens RF voltage set to 60 V.
Glycopeptides identification and quantitation was performed using BioPharma Finder 3.0 software using Full MS and MS/MS data, mass deviation of 5 ppm and minimum confidence of 95%.
2.8. N-glycan release and labelling
N-glycans from 200 μg of each mAb were released and labelled as reported previously [30]. All the samples were reconstituted in the starting gradient conditions and analysed on an Accucore 150-Amide-HILIC 2.1 mm × 150 mm (Thermo Scientific) column using both fluorescence and MS detection.
Analysis was performed using a binary gradient of 50 mM ammonium formate, pH 4.4 (A) and acetonitrile (B). Gradient conditions were as follows: 72% B decreased to 45% B in 40 min with a further decrease to 40% B in 2.5 min with 0.5 min isocratic hold. Initial conditions were restored in 0.1 min and held for an additional 1.9 min to ensure column re-equilibration. The column temperature was maintained at 50 °C throughout and flow rate was sustained at 400 μL/min. Fluorescence detection was acquired using the following settings: λex/em = 330/420 nm for 2-AB and λex/em = 350/425 nm for 2-AA. The MS method consisted of full MS scans in negative polarity mode at 70,000 resolution setting (defined at m/z 200) with the mass range set to 380–2000 m/z and AGC target value of 3.0 × 106 with a maximum injection time of 50 ms and one microscan. In-source CID was set to 20.0 eV. MS instrumental tune parameters were set as follows: spray voltage was 3.5 kV, sheath gas flow rate was 50 AU, auxiliary gas flow rate was 13 AU, capillary temperature was 320 °C, probe heater temperature was 400 °C and S-lens RF voltage set to 50 V.
2.9. Data processing
Released N-glycan analysis was performed using Xcalibur QualBrowser 4.0 for signal integration and availing of GlycoWork Bench for correct glycoform identification. All other data processing, quantitation and identification were performed on BioPharma Finder 3.0 software (Thermo Scientific) using the parameters summarized in Tables S2 and S3.
3. Results and discussion
3.1. Intact mass analysis
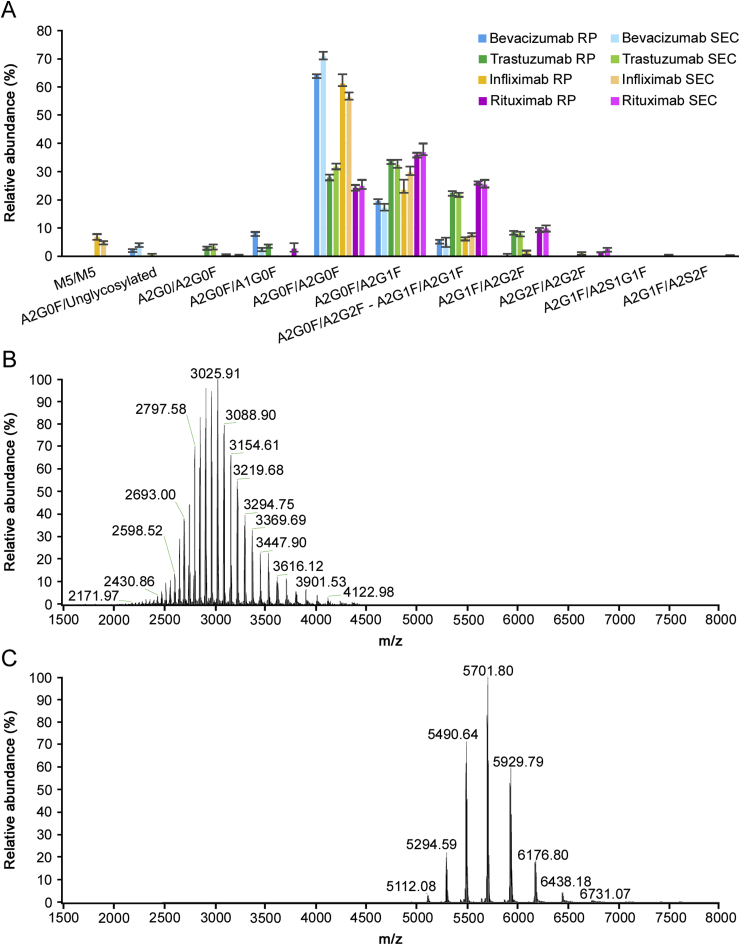
Intact mass analysis was performed on the four mAbs using both denaturing and native conditions. Excellent MS data quality allowed achievement of mass accuracies ≤10 ppm for the majority of mAb proteoforms. In particular, proteoforms containing C-term lysine truncation, N-terminal pyro-Glu formation and Fc N-glycosylation were considered. The N-glycan profiles resulting for the four mAbs are presented in Fig. 1 and Table 1.
Fig. 1.
(A) N-glycan profiles of bevacizumab (blue), trastuzumab (green), infliximab (orange) and rituximab (pink) drug products analysed via intact mass analysis in denaturing (dark colour shade) and native (light colour shade) conditions. Relative abundancies were calculated on the basis of MS signal intensities obtained after raw data deconvolution and averaged on triplicate analysis. Comparison of intact protein mass spectrometry in denaturing (B) and native (C) conditions.
Table 1.
Experimental mass for the intact mAbs in native and denatured conditions. The relative abundances of proteoforms are also reported. Experimental mass, mass accuracy and relative abundance were calculated as average of triplicate analysis. Theoretical masses were calculated considering 2 C-term lysine clipping and 16 disulphide bonds unless stated otherwise.
| mAbs | Analysis | Modifications/Glycoforms associated | Experimental mass (Da) | Theoretical average mass (Da) | Mass difference (ppm) | Relative abundance (%) |
|---|---|---|---|---|---|---|
| BEV | Denatured conditions | A2G0F/unglycosylated | 147752.6 | 147751.9 | 4.7 | 2.5 |
| A2G0F/A1G0F | 148995.5 | 148994.0 | 10.1 | 8.2 | ||
| A2G0F/A2G0F | 149198.0 | 149197.6 | 2.7 | 64.0 | ||
| A2G0F/A2G1F | 149359.9 | 149359.7 | 1.3 | 19.5 | ||
| A2G0F/A2G2F or A2G1F/A2G1F | 149521.6 | 149521.8 | 1.3 | 5.5 | ||
| BEV | Native conditions | A2G0F/unglycosylated | 147751.8 | 147751.9 | 0.7 | 4.3 |
| A2G0F/A1G0F | 148991.1 | 148994.0 | 19.5 | 2.9 | ||
| A2G0F/A2G0F | 149195.6 | 149197.6 | 13.4 | 70.8 | ||
| A2G0F/A2G1F | 149361.7 | 149359.7 | 13.4 | 17.2 | ||
| A2G0F/A2G2F or A2G1F/A2G1F | 149524.5 | 149521.8 | 18.1 | 4.3 | ||
| A2G1F/A2G2F | 149685.7 | 149684.0 | 11.4 | 0.5 | ||
| TRA | Denatured conditions | A1G0F/A2G0F | 147852.5 | 147853.4 | 6.3 | 4.0 |
| A2G0/A2G0F | 147911.7 | 147910.4 | 8.7 | 3.4 | ||
| A2G0F/A2G0F | 148058.6 | 148056.6 | 13.6 | 27.9 | ||
| A2G0F/A2G1F | 148219.2 | 148218.7 | 3.4 | 33.7 | ||
| A2G0F/A2G2F or A2G1F/A2G1F | 148380.1 | 148380.8 | 5.0 | 22.5 | ||
| A2G1F/A2G2F | 148541.9 | 148543.0 | 7.3 | 8.6 | ||
| TRA | Native conditions | A2G0F/unglycosylated | 146611.6 | 146610.9 | 4.7 | 0.8 |
| A2G0/A2G0F | 147910.7 | 147910.4 | 1.7 | 3.4 | ||
| A2G0F/A2G0F | 148058.1 | 148056.6 | 10.1 | 31.8 | ||
| A2G0F/A2G1F | 148220.5 | 148218.7 | 11.9 | 32.3 | ||
| A2G0F/A2G2F or A2G1F/A2G1F | 148382.3 | 148380.8 | 9.8 | 22.1 | ||
| A2G1F/A2G2F | 148544.8 | 148543.0 | 12.0 | 8.0 | ||
| A2G2F/A2G2F | 148704.4 | 148705.1 | 4.7 | 1.5 | ||
| INF | Denatured conditions | 1x C-term K, M5/M5 | 148185.6 | 148184.2 | 9.4 | 1.4 |
| 2x C-term K, M5/M5 | 148313.1 | 148312.4 | 4.5 | 5.6 | ||
| A2G0F/A2G0F | 148514.4 | 148512.6 | 12.4 | 21.6 | ||
| 1x C-term K, A2G0F/A2G0F | 148643.4 | 148640.7 | 17.8 | 12.0 | ||
| A2G0F/A2G1F | 148677.7 | 148674.7 | 20.2 | 5.7 | ||
| 2x C-term K, A2G0F/A2G0F | 148770.4 | 148768.9 | 10.2 | 27.7 | ||
| 2x C-term K, A2G0F/A2G1F | 148932.4 | 148931.1 | 8.8 | 17.8 | ||
| 2x C-term K, A2G0F/A2G2F or A2G1F/A2G1F | 149094.6 | 149093.2 | 9.3 | 6.6 | ||
| 2x C-term K, A2G1F/A2G2F | 149255.9 | 149255.3 | 3.5 | 1.6 | ||
| INF | Native conditions | M5/M5 | 148058.3 | 148056.1 | 15.3 | 1.2 |
| 2x C-term K, M5/M5 | 148314.1 | 148312.4 | 11.6 | 3.9 | ||
| A2G0/A2G0F | 148367.4 | 148366.4 | 6.4 | 0.6 | ||
| A2G0F/A2G0F | 148512.9 | 148512.6 | 2.0 | 19.1 | ||
| 1x C-term K, A2G0F/A2G0F | 148643.0 | 148640.7 | 15.4 | 10.0 | ||
| A2G0F/A2G1F | 148678.0 | 148674.7 | 22.2 | 10.5 | ||
| 2x C-term K, A2G0F/A2G0F | 148770.0 | 148768.9 | 7.4 | 27.4 | ||
| 2x C-term K, A2G0F/A2G1F | 148932.0 | 148931.1 | 6.7 | 19.3 | ||
| 2x C-term K, A2G0F/A2G2F or A2G1F/A2G1F | 149094.1 | 149093.2 | 6.0 | 8.0 | ||
| RIT | Denatured conditions | A1G0F/A2G0F | 146873.1 | 146872.1 | 6.6 | 2.7 |
| A2G0F/A2G0F | 147077.8 | 147075.3 | 17.0 | 24.3 | ||
| A2G0F/A2G1F | 147238.2 | 147237.5 | 4.5 | 36.0 | ||
| A2G0F/A2G2F or A2G1F/A2G1F | 147400.9 | 147399.6 | 8.4 | 26.4 | ||
| A2G1F/A2G2F | 147562.3 | 147561.8 | 3.5 | 9.6 | ||
| A2G2F/A2G2F | 147724.2 | 147723.9 | 2.1 | 1.0 | ||
| RIT | Native conditions | A2G0/A2G0F | 146927.4 | 146929.2 | 12.5 | 0.3 |
| A2G0F/A2G0F | 147078.4 | 147075.3 | 20.8 | 24.9 | ||
| A2G0F/A2G1F | 147240.1 | 147237.5 | 17.5 | 36.7 | ||
| A2G0F/A2G2F or A2G1F/A2G1F | 147403.0 | 147399.6 | 22.8 | 25.4 | ||
| A2G1F/A2G2F | 147563.8 | 147561.8 | 14.0 | 9.8 | ||
| A2G2F/A2G2F | 147722.5 | 147723.9 | 9.7 | 2.6 | ||
| A2G2F/A2S1G1F | 148019.1 | 148015.2 | 26.8 | 0.2 | ||
| A2G2F/A2S2F | 148308.2 | 148306.4 | 11.9 | 0.1 |
Both intact mass analyses using native or denaturing conditions show comparable N-glycan quantitative results (Figs. 1A, S1-S4, Table S4) and allow minimum sample preparation, rapid analysis and data processing (see Table 2). Nevertheless, some minor differences could be observed between the two techniques as some low abundant species (<3%) can be identified only using native conditions, due to the greater spatial spectral resolution obtained with native MS (Figs. 1B–C). In particular, sialylated species were detected only when using native conditions, while they were not revealed in any other analysis at the intact or subunit level. However, the shift of the charge envelope to higher mass ranges generates spectra in a region of reduced resolution, causing a small decrease in the average mass accuracy (7.9 ppm in denatured mode vs. 11.9 ppm in native mode) [31]. Since intact mass analysis returns the mass of the N-glycoforms present on both heavy chains, it is possible that this analysis will return an underestimated or overestimated value for some N-glycans. As an example, it is not possible to distinguish between proteoforms presenting A2G1F N-glycans on both chains or presenting one A2G0F and one A2G2F.
Table 2.
Proteoforms obtained after heavy chain analysis on the 4 monoclonal antibodies analysed. Average experimental masses on triplicate analysis were reported together with theoretical average masses and average mass accuracies for each proteoform. Relative abundances were calculated based on the MS signal intensities in the triplicate analysis. Masses were calculated accounting for C-term lysine clipping unless stated otherwise.
| mAbs | Modifications/Glycoforms | Experimental mass (Da) | Theoretical average mass (Da) | Mass difference (ppm) | Relative abundance (%) |
|---|---|---|---|---|---|
| BEV | Unglycosylated | 49718.8 | 49718.7 | 1.3 | 1.5 |
| A1G0F | 50961.2 | 50960.8 | 6.7 | 2.1 | |
| A2G0 | 51017.5 | 51017.9 | 7.1 | 0.4 | |
| A2G0F | 51163.8 | 51164.0 | 5.0 | 82.0 | |
| A2G1 | 51180.0 | 51180.0 | 1.3 | 1.4 | |
| A2G1F | 51325.9 | 51326.2 | 4.8 | 12.0 | |
| A2G2F | 51488.3 | 51488.3 | 1.3 | 0.4 | |
| A2S1G1F | 51746.2 | 51745.6 | 12.1 | 0.2 | |
| TRA | M5 | 50372.6 | 50373.2 | 12.5 | 0.7 |
| A1G0F | 50398.3 | 50398.3 | 1.3 | 0.6 | |
| A2G0 | 50455.0 | 50455.3 | 6.8 | 3.9 | |
| A2G0F | 50601.2 | 50601.5 | 6.1 | 51.0 | |
| A2G1 | 50617.4 | 50617.5 | 1.6 | 0.6 | |
| A2G1F | 50763.3 | 50763.6 | 6.5 | 38.1 | |
| A2G2F | 50925.3 | 50925.8 | 8.2 | 5.1 | |
| INF | M5 | 50605.5 | 50605.4 | 3.4 | 1.2 |
| 1x C-term lysine, M5 | 50734.1 | 50733.5 | 10.1 | 2.1 | |
| A1G0F | 50758.2 | 50758.6 | 7.4 | 1.3 | |
| A2G0 | 50815.3 | 50815.7 | 7.2 | 1.1 | |
| A2G0F | 50833.5 | 50833.6 | 1.8 | 17.8 | |
| 1x C-term lysine, A2G0F | 50962.0 | 50961.8 | 4.5 | 48.2 | |
| A2G1F | 50995.3 | 50995.8 | 8.6 | 11.3 | |
| 1x C-term lysine, A2G1F | 51123.3 | 51123.9 | 12.3 | 16.2 | |
| A2G2F | 51158.4 | 51157.9 | 9.4 | 0.8 | |
| RIT | Gln-- > Pyro-Glu, A2G0F | 50513.9 | 50514.3 | 8.1 | 46.2 |
| Gln-- > Pyro-Glu, A2G1F | 50676.1 | 50676.4 | 7.0 | 46.4 | |
| Gln-- > Pyro-Glu, A2G2F | 50838.1 | 50838.6 | 9.9 | 7.4 |
3.2. Reduced mAb/Heavy chain analysis
Intact analysis of reduced mAb allows monitoring N-glycan abundance without the complexity caused by the presence of glycans on the two chains. Analysed mAbs were reduced with TCEP in HCl-Guanidine 8 M and injected for LC-MS analysis after a relatively quick and easy sample preparation. The MS acquisition is performed with different settings for heavy chain and light chain due to the differences in molecular masses and the requirement for different instrument settings [32]. Heavy chain spectra showed a reduced complexity that enabled identification of a greater number of features, including lower abundant glycoforms with respect to the intact mass analysis level (Tables 2 and S5) as well as an improved overall mass accuracy (average Δ = 6.4 ppm) determination due to the lower mass of the fragment analysed.
3.3. Fc region
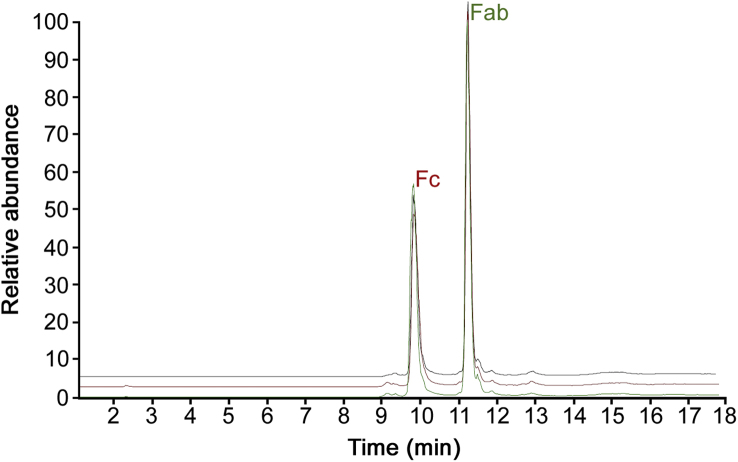
The four mAbs presented in this study were digested with gingipain enzyme, which allowed the hydrolysis of the heavy chain above the hinge region. As a consequence, disulphide bonds remaining in place in the hinge region preserved both scFc (single chain Fc), reducing the complexity of the associated mass spectra while keeping intact the information present at the same time on both chains (Figs. 2 and S5). MS data were acquired with higher resolution settings than intact mass spectra, allowing the confident identification of a larger number of Fc variants (Tables 3 and S6), especially for more complex drug products such as infliximab, which presents, on top of the almost equally distributed C-term lysine variants, a greater variety of the N-glycans present at the glycosylation site of both heavy chains. This type of analysis presents similar ambiguity found in intact mass analysis where it is not possible to distinguish between two isobaric species having different N-glycan distribution.
Fig. 2.
Base Peak Chromatograms (BPCs) of the RP-MS analysis performed in triplicate on Fc region from trastuzumab drug product after digestion with gingipain.
Table 3.
N-glycan analysis performed for the 4 mAbs analysed on the Fc region after digestion with gingipain. Experimental average masses are reported as well as theoretical average masses and average mass accuracies based on triplicate analysis. The relative abundances of fragments are also shown and were based on MS signal intensities averaged on triplicate analysis. Both C-term lysine loss and 6 disulphide bonds were considered in the calculation of the theoretical average mass unless stated otherwise.
| mAbs | Modifications/Glycoforms associated | Experimental mass (Da) | Theoretical average mass (Da) | Mass difference (ppm, n = 3) | Relative abundance (%, n = 3) |
|---|---|---|---|---|---|
| BEV | A2G0F/A2G0 | 53148.2 | 53149.3 | 20.7 | 4.1 |
| A2G0F/A2G0F | 53294.8 | 53295.4 | 11.4 | 61.7 | |
| A2G0F/A2G1F | 53456.9 | 53457.6 | 12.3 | 20.6 | |
| A2G1F/A2G1F or A2G0F/A2G2F | 53620.3 | 53619.7 | 11.1 | 11.2 | |
| A2G1F/A2G2F | 53782.7 | 53781.8 | 16.6 | 2.4 | |
| TRA | A2G0/A2G0F | 53149.4 | 53149.3 | 1.7 | 6.4 |
| A2G0F/A2G0F | 53294.8 | 53295.4 | 10.9 | 27.3 | |
| A2G0F/A2G1F | 53457.2 | 53457.6 | 7.4 | 33.1 | |
| A2G1F/A2G1F or A2G0F/A2G2F | 53619.3 | 53619.7 | 7.7 | 24.1 | |
| A2G1F/A2G2F | 53781.4 | 53781.8 | 7.5 | 8.9 | |
| A2G2F/A2G2F | 53943.3 | 53944.0 | 13.5 | 0.2 | |
| INF | 1x No C-term K A1G0F/A1G0F | 52950.7 | 52953.1 | 45.1 | 0.6 |
| A2G0F/A1G0F | 53028.0 | 53028.1 | 1.1 | 0.2 | |
| A2G0F/A2G0F | 53231.3 | 53231.3 | 0.1 | 27.7 | |
| 1x C-term K, A2G0F/A2G0F | 53358.5 | 53359.5 | 18.5 | 8.7 | |
| A2G0F/A2G1F | 53393.2 | 53393.4 | 3.7 | 21.9 | |
| A1G0M5/A2G0F | 53464.3 | 53463.5 | 13.7 | 0.3 | |
| 2x C-term K, A2G0F/A2G0F | 53487.3 | 53487.6 | 6.1 | 9.7 | |
| 1x C-term K, A2G0F/A2G1F | 53521.5 | 53521.6 | 1.4 | 7.2 | |
| A2G0F/A2G2F or A2G1F/A2G1F | 53555.5 | 53555.6 | 1.1 | 10.8 | |
| 2x C-term K, A2G0F/A2G1F | 53649.6 | 53649.8 | 4.1 | 5.3 | |
| 1x No C-term K A1G0M5/A2G2F | 53658.6 | 53658.7 | 2.3 | 0.1 | |
| 1x C-term K, A2G0F/A2G2F or A2G1F/A2G1F | 53682.9 | 53683.8 | 15.1 | 2.7 | |
| A2Sg1G0F/A2G0F | 53700.1 | 53699.8 | 5.6 | 0.2 | |
| A2G1F/A2G2F | 53718.3 | 53717.7 | 9.9 | 1.0 | |
| 2x C-term K, A2G0F/A2G2F or A2G1F/A2G1F | 53811.8 | 53811.9 | 2.5 | 1.3 | |
| 1x C-term K, A2G2/A2G2F | 53862.1 | 53861.9 | 3.1 | 1.2 | |
| A2Sg1G1F/A2G0F | 53862.7 | 53862.8 | 3.4 | 0.7 | |
| 2x C-term K, A2G1F/A2G2F | 53973.1 | 53974.1 | 17.9 | 0.3 | |
| A2Sg1G1F/A2G1F | 54024.8 | 54025.0 | 3.9 | 0.1 | |
| RIT | Gln-- > Pyro-Glu, A2G0/A2G0F | 53085.5 | 53085.2 | 7.0 | 0.4 |
| Gln-- > Pyro-Glu, A2G1F/A2G1F | 53555.3 | 53555.6 | 5.9 | 27.5 | |
| Gln-- > Pyro-Glu, A2G1F/A2G2F | 53717.5 | 53717.7 | 4.3 | 11.3 | |
| Gln-- > Pyro-Glu, A2G0F/A2G0F | 53230.9 | 53231.3 | 7.5 | 25.3 | |
| Gln-- > Pyro-Glu, A2G2F/A2G2F | 53880.3 | 53879.9 | 8.9 | 1.0 | |
| Gln-- > Pyro-Glu, A2G0F/A2G1F | 53393.1 | 53393.4 | 6.8 | 34.5 |
3.4. IdeS digestion
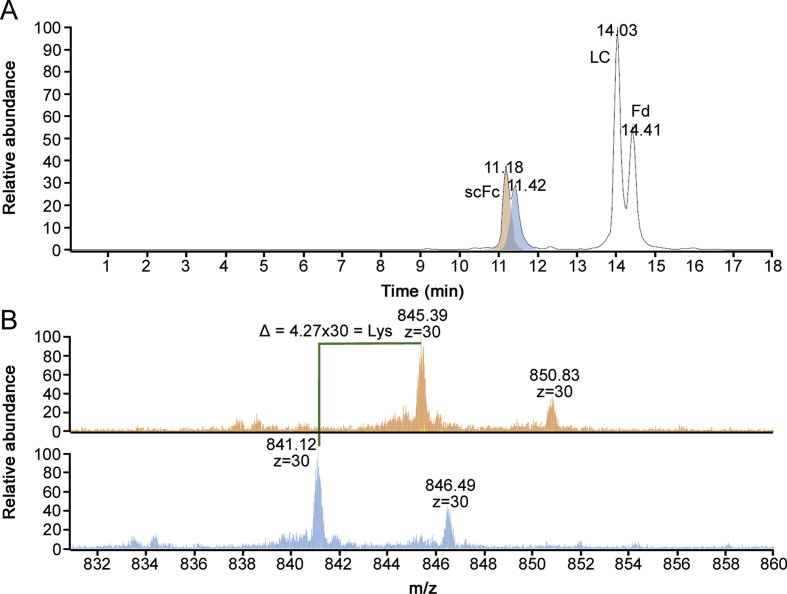
IdeS digestion is a widely used enzymatic tool to obtain subunits of the monoclonal antibody in a molecular mass range where high resolution mass spectrometry is readily available. IdeS cleaves monoclonal antibodies below the hinge region, driving the formation of two identical scFc regions. If HR-MS data are needed for Fab region as well, a reduction step facilitates the generation of a free light chain and the Fd region, both with the same mass range of approximately 25 kDa, making it possible to apply the same tuning parameters for data acquisition, as well as high mass resolution settings, returning isotopically resolved data that enable to obtain monoisotopic mass information. The data analysis permitted the identification of subunit proteoforms with average mass accuracy ≤2 ppm (Tables 4 and S7, Figs. S6–9). Moreover, reverse phase separation of the subunits is able to separate proteoforms containing other types of modifications, such as C-term lysine truncation, reducing the complexity of MS signal identification for near isobaric variants arising from the combination of N-glycan and lysine presence/absence (Fig. 3).
Table 4.
scFc analysis via RP-HRMS. Average experimental masses were determined based on triplicate analysis, as well as mass accuracies and relative abundancies. C-term lysine clipping was considered in the theoretical mass calculation unless indicated otherwise.
| mAbs | Modifications/Glycoforms | Experimental mass (Da) | Theoretical monoisotopic mass (Da) | Mass difference (ppm) | Relative abundance (%) |
|---|---|---|---|---|---|
| BEV | Unglycosylated | 23775.974 | 23775.930 | 1.9 | 1.4 |
| A2G0F | 25220.485 | 25220.463 | 0.8 | 84.9 | |
| A2G1F | 25382.504 | 25382.516 | 0.5 | 10.3 | |
| A1G0F | 25018.436 | 25017.380 | 2.3 | 1.9 | |
| A2G0 | 25074.482 | 25074.410 | 2.9 | 1.5 | |
| TRA | A2G0 | 25074.431 | 25074.405 | 1.0 | 4.3 |
| A2G0F | 25220.513 | 25220.463 | 2.0 | 52.0 | |
| A2G1F | 25382.581 | 25382.516 | 2.6 | 36.2 | |
| A1G0F | 25017.445 | 25017.380 | 2.6 | 1.8 | |
| M5 | 24992.715 | 24992.350 | 14.6 | 1.6 | |
| A2G2F | 25544.569 | 25544.569 | 0.0 | 4.1 | |
| INF | M5 | 24960.391 | 24960.380 | 0.4 | 1.6 |
| A1G0F | 24985.423 | 24985.412 | 0.4 | 2.4 | |
| 1x C-term lysine, M5 | 25088.477 | 25088.475 | 0.1 | 1.8 | |
| 1x C-term lysine, A2G0F | 25316.594 | 25316.586 | 0.3 | 38.6 | |
| 1x C-term lysine, A2G1 | 25332.552 | 25332.581 | 1.1 | 1.4 | |
| 1x C-term lysine, A2G0 | 25170.536 | 25170.528 | 0.3 | 3.0 | |
| A2G0F | 25188.488 | 25188.487 | 0.0 | 30.2 | |
| A2G1F | 25350.558 | 25350.544 | 0.6 | 10.9 | |
| 1x C-term lysine, A2G1F | 25478.629 | 25478.639 | 0.4 | 10.1 | |
| RIT | A2G0F | 25188.541 | 25188.491 | 2.0 | 44.2 |
| A2G1F | 25350.583 | 25350.544 | 1.5 | 48.3 | |
| A2G2F | 25512.604 | 25512.597 | 0.3 | 7.5 |
Fig. 3.
(A) BPC for RP-MS analysis of the IdeS digested infliximab drug product. (B) Zoom of charge state +30 for scFc peaks containing (top) and missing (down) C-term lysine.
3.5. Peptide mapping
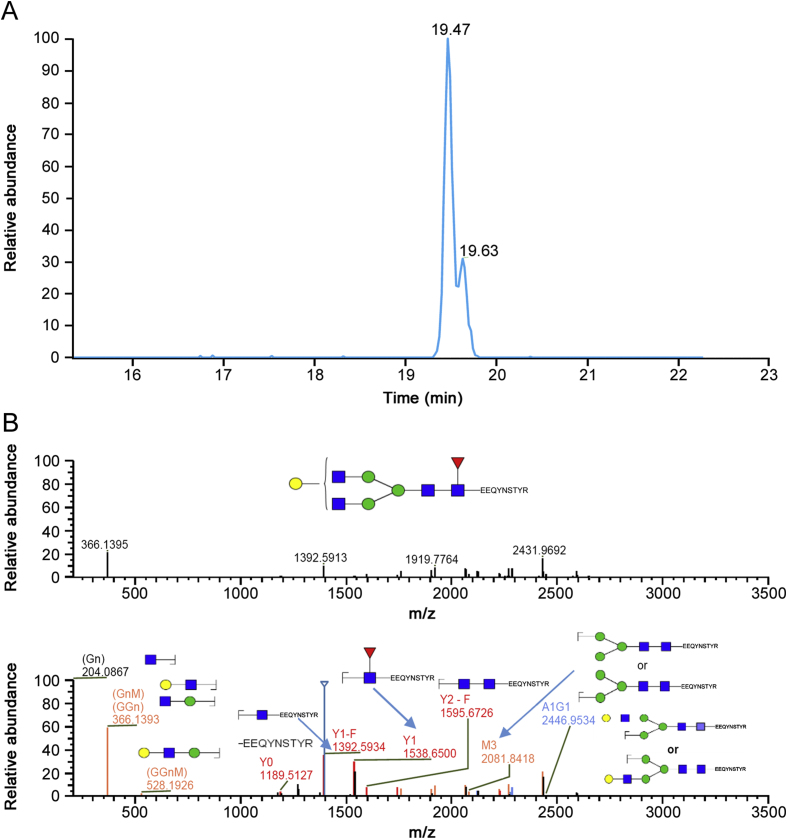
Peptide mapping analysis is a comprehensive tool for protein characterisation using proteolysis of the biotherapeutic followed by LC-MS/MS analysis of the peptides. Peptide mapping is widely used in biopharmaceutical analysis to verify the primary sequence and determine the types and locations of PTMs present. The higher sensitivity of peptide mapping, together with the availability of MS/MS data, provides accurate information on the N-glycans present as well as site-specific information if multiple glycosylation sites are present on the protein. Peptide mapping analysis has demanding sample preparation that requires multiple steps and analyst expertise to ensure reproducibility. MS/MS data analysis, although improved by modern software tools, requires experienced analysts. For these reasons, this technique is not yet considered suitable for a regulated environment, even though it is an established gold standard during product development and many research efforts are directed towards standardisation and simplification of the peptide mapping workflow to facilitate its introduction into quality control laboratories [33]. Analysis performed on the four mAbs returned a higher number of N-glycoforms than with any other technique, reaching abundancies ≤0.1% (Tables 5 and S8), obtained setting a mass accuracy threshold of 5 ppm during data analysis. Glycopeptides separation could resolve isomers like A2G1F and A2G1′F (Fig. 4A) enabling their quantitation, although MS/MS data did not provide fingerprint spectra to distinguish the two glycoforms (Fig. 4B). Performing peptide mapping analysis without knowledge of pre-existing information from released N-glycans may mislead glycopeptide identification, for example the identification of a low abundant glycan in infliximab as A1S1M5 (Table 5). It is known that infliximab drug product is characterized by the presence of N-glycolyl neuraminic acid (NeuG) and that there is the possibility of isobaric N-glycans where N-acetyl neuraminic acid (NeuA) and one galactose are substituted by NeuG and one fucose. The low abundance of this glycan and the absence of MS/MS data relative to the loss of sialic acid or fucose (data not shown) caused the software to incorrectly assign the glycopeptide, which could be avoided by providing a molecule specific N-glycan database relative to the drug product as determined using released N-glycan analysis.
Table 5.
N-glycan abundancies for the 4 analysed mAbs obtained through peptide mapping analysis via LC-MS/MS. Abundancies are expressed as % respect to the total abundance of the peptide EEQYNSTYR and/or TKPREEQYNSTYR containing 1 miscleavage and were calculated on triplicate independent sample preparations.
| Glycoform | % Relative abundance (n = 3) |
|||
|---|---|---|---|---|
| Bevacizumab | Trastuzumab | Infliximab | Rituximab | |
| A1G0 | 0.84 | 2.30 | 2.47 | 0.34 |
| A1G0F | 6.61 | 6.98 | 8.78 | 5.06 |
| A1G0M4 | – | – | 1.70 | – |
| A1G0M5 | – | – | 3.87 | – |
| A1G0M5F | – | – | 1.83 | – |
| A1G1 | – | 0.59 | – | – |
| A1G1F | 0.76 | 2.18 | 3.91 | 1.80 |
| A1G1M5 | – | – | 1.36 | – |
| A1G1M5F | – | – | 0.52 | – |
| A1S1 | – | 0.09 | – | – |
| A1S1F | – | 0.40 | – | 0.21 |
| A1S1M5 (A1Sg1M4F) | – | – | 0.51 | – |
| A1Sg1 | – | – | 0.32 | – |
| A1Sg1F | – | – | 2.10 | – |
| A2G0 | 2.33 | 5.38 | 1.66 | 1.61 |
| A2G0F | 74.86 | 39.88 | 44.86 | 40.77 |
| A2G1 | – | 1.94 | 0.37 | 0.63 |
| A2G1F | 10.77 | 32.28 | 18.60 | 39.80 |
| A2G2 | – | 0.16 | – | – |
| A2G2F | 0.73 | 5.19 | 2.40 | 7.51 |
| A2S1G0F | – | 0.44 | – | 0.48 |
| A2S1G1F | – | 0.65 | – | 1.00 |
| A2S2F | – | 0.23 | – | 0.48 |
| A3G1F | – | 0.16 | – | 0.16 |
| A2Sg1G0F | – | – | 0.76 | – |
| M3 | – | 0.16 | – | – |
| M4 | – | 0.15 | 0.09 | – |
| M5 | 0.87 | 2.87 | 7.33 | 1.45 |
| M6 | – | 0.20 | 0.12 | 0.29 |
| Unglycosylated | 5.35 | 1.44 | 0.39 | 1.34 |
Fig. 4.
Extracted Ion Chromatogram (XIC) of the glycopeptide carrying A2G1F N-glycan from the peptide mapping analysis of rituximab drug product (A) and relative MS/MS spectrum (B).
3.6. Released N-glycan
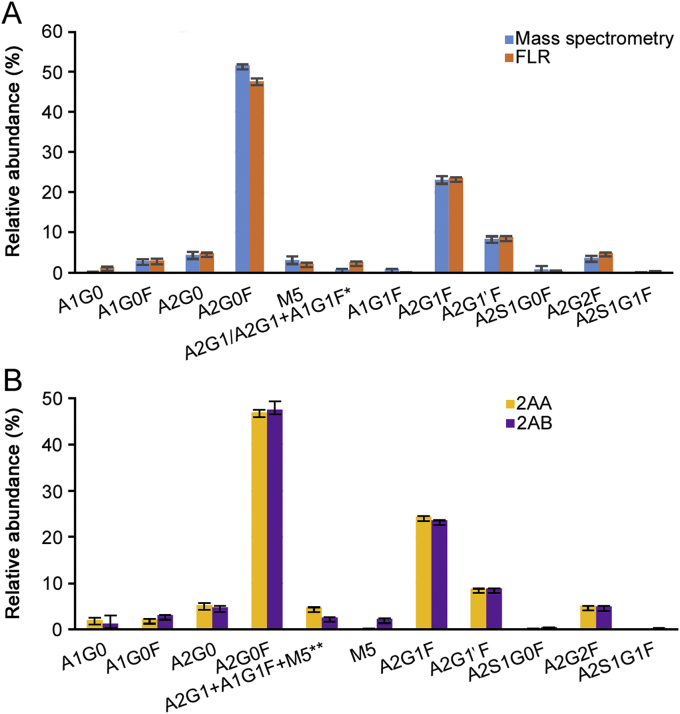
Released N-glycan analysis is the most established technique for N-glycan profiling of glycoproteins. In this study, we evaluated (Tables S9–S10) the performance, in terms of relative quantitation of the N-glycans when labelling the sample with two of the mainly used labels (2-AA and 2-AB) and compared the relative quantitation results obtained using MS (Tables S9 and S11, Fig. 5A) and fluorescence (FLR) signal integration (Tables S10 and S12, Fig. 5B). The results show good comparability of the sample preparation performed with the two different labels and with the two detection techniques (Fig. 5), while the overall mass accuracy was lower than 2 ppm.
Fig. 5.
(A) Comparison of released 2-AB labelled N-glycan from trastuzumab drug product. Quantitation was obtained integrating fluorescence trace (orange) and MS signals from each N-glycan (blue). Quantitation is based on triplicate sample preparation. *In 2-AB trace coeluting A2G1 and A1G1F N-glycans were quantified as a single peak. (B) Comparison of released N-glycan from trastuzumab drug product. Quantitation was obtained from fluorescence trace after labelling with 2-AA (yellow) and 2-AB (purple). Quantitation is based on triplicate sample preparation. **In 2-AB labelled N-glycans M5 elutes at a different retention time, and relative percentage is only relative to coeluting A2G1 and A1G1F species.
3.7. Evaluation of the different approaches for N-glycan analysis
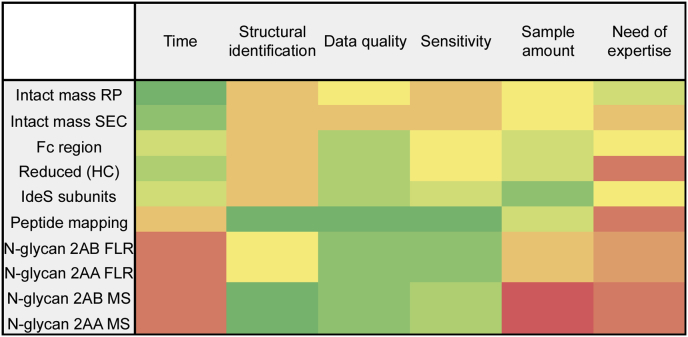
While assessing comparability of the quantitative results obtained across the different techniques, a number of factors which may drive the analyst to the choice of the right technique were evaluated (Fig. 6), such as the time of the analysis from sample preparation to final data analysis report, the depth of structural identification obtained on the glycoforms with each technique, data quality, accuracy and robustness of the results, sensitivity of the technique, the amount of sample required and the need of expertise to perform both sample preparation and data analysis.
Fig. 6.
Heat map of the factors leading to the choice of the right technique for N-glycan analysis.
Workflows using the information retained at intact or subunits level will require little or no sample preparation, making it ideal for routine analysis to assure robustness of the analysis; nevertheless, the structural details obtained in this way are limited to the most abundant glycoforms (Fig. 7) and they do not provide structural details on the glycans; isomers relative abundance cannot be assessed as well as ambiguity between some glycoforms when the analysis is carried on both heavy chains at the same time (intact, gingipain digest) or with other PTMs such as glycation. N-glycan analysis at intact level is also strongly dependent on sample heterogeneity; when other PTMs strongly influence the complexity of the mass spectra at intact level or there is an intrinsic complexity of N-glycan profiles, the use of tools to simplify the data is strongly recommended and the analysis of subunits can be helpful, as for infliximab drug product analysed in this study.
Fig. 7.
Venn Diagram of the N-glycans quantified through 8 different workflows on rituximab (A) and bevacizumab (B) drug products.
Sample complexity and N-glycan abundance are also strictly related to robustness of the analysis; while generally low %RSD values were obtained, the presence of near-isobaric species or of very low abundant species returned higher values (Tables S4-S8, S11-S12).
While providing excellent data quality and additional structural information, workflows such as peptide mapping and the analysis of released N-glycans require more time and need for trained analysts. These analyses are not yet robust enough to assure reproducibility across analysts and laboratories as they require multiple steps for the sample preparation and a high degree of expertise for the data analysis without or with little support from bioinformatics tools. In particular, it was possible to appreciate a marked difference in the quantitative analysis in terms of %RSD values between released N-glycan and peptide mapping analysis (Tables S8, S11, S12). Released N-glycan involved a number of non-automated steps for sample preparation while data processing required efforts in terms of manual integration of the obtained chromatograms as there are few software tools that guarantee a fully automated analysis of fluorescently labelled N-glycans. On the contrary, the possibility to fully automate trypsin digestion and the data analysis fully integrated into bioinformatics tools is probably the main reason for the difference of reproducibility between the two techniques. Even if efforts towards standardisation and automatization of these methods are being made to transfer them into the QC laboratories [33], a high level of knowledge in the MS data interpretation is still required.
4. Conclusion
N-glycan analysis of biotherapeutics is a major critical attribute influencing many functions of the drug product [4,34,35]. Their assessment is required from ICH Q6B to assure biologics safety and stability and it constitutes important criteria to assess biosimilarity between originator and newly developed drugs. Although released N-glycan analysis is considered as the gold standard, N-glycan heterogeneity on the monoclonal antibodies can be assessed through several analytical workflows.
In this study, we compared N-glycan analysis and quantitation obtained through ten different methods on four different commercially available monoclonal antibodies.
While assessing good comparability of the quantitative data obtained with these techniques, several advantages and disadvantages were proved.
Recently, an interlaboratory study was published on the analysis of NIST standard mAb glycosylation [36]. The results highlighted the lack of standardisation of analytical methods for N-glycan identification and quantitation. De Leoz et al. obtained 103 different reports with a range of N-glycans identified spanning 4 to 48 structures. This variability was surely affected by the analytical workflow applied, but proved to be mainly dependent on laboratory skills. Since the experiments reported herein were performed in the same laboratory, from analysts with similar skills and using the same equipment, the variability seen in quantitative results can solely be dependent on the workflow and it was possible to appreciate the differences between more direct or automated methods, such as intact analysis or glycopeptides analysis, and more lengthy and laborious techniques, such as released N-glycan analysis. Furthermore, the choice of the analytical workflow can strongly impact the in-depth of structural features detected.
It should also be considered that biotherapeutics analysis occurs at different stages of the bioprocess, from early development to lot-to-lot comparison for batch release and there is usually a stronger need for structural details during drug development than in the later stages. It is possible in this way to use the technique of choice according to the specific application and information required, justifying the lack of standardisation for this critical workflow. All the consideration provided in this study, while remaining strongly sample dependent, can surely constitute a guide for the choice of the most appropriate technique for N-glycan analysis of biotherapeutics.
Acknowledgments
The authors gratefully acknowledge funding from Science Foundation Ireland under Grant Number 13/CDA/2196 and collaborators from Thermo Fisher Scientific for instrument access.
Footnotes
Peer review under responsibility of Xi’an Jiaotong University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2019.11.008.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Elgundi Z., Reslan M., Cruz E. The state-of-play and future of antibody therapeutics. Adv. Drug Deliv. Rev. 2017;122:2–19. doi: 10.1016/j.addr.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 2.Ecker D.M., Jones S.D., Levine H.L. The therapeutic monoclonal antibody market. mAbs. 2015;7:9–14. doi: 10.4161/19420862.2015.989042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaplon H., Reichert J.M. Antibodies to watch in 2018. mAbs. 2018;10:183–203. doi: 10.1080/19420862.2018.1415671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Planinc A., Bones J., Dejaegher B. Glycan characterization of biopharmaceuticals: updates and perspectives. Anal. Chim. Acta. 2016;921:13–27. doi: 10.1016/j.aca.2016.03.049. [DOI] [PubMed] [Google Scholar]
- 5.Kim J.Y., Kim Y.-G., Lee G.M. CHO cells in biotechnology for production of recombinant proteins: current state and further potential. Appl. Microbiol. Biotechnol. 2012;93:917–930. doi: 10.1007/s00253-011-3758-5. [DOI] [PubMed] [Google Scholar]
- 6.Hossler P., Khattak S.F., Li Z.J. Optimal and consistent protein glycosylation in mammalian cell culture. Glycobiology. 2009;19:936–949. doi: 10.1093/glycob/cwp079. [DOI] [PubMed] [Google Scholar]
- 7.Liu L. Antibody glycosylation and its impact on the pharmacokinetics and pharmacodynamics of monoclonal antibodies and Fc-fusion proteins. J. Pharm. Sci. 2015;104:1866–1884. doi: 10.1002/jps.24444. [DOI] [PubMed] [Google Scholar]
- 8.Higel F., Seidl A., Sorgel F. N-glycosylation heterogeneity and the influence on structure, function and pharmacokinetics of monoclonal antibodies and Fc fusion proteins. Eur. J. Pharm. Biopharm. 2016;100:94–100. doi: 10.1016/j.ejpb.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Zheng K., Bantog C., Bayer R. The impact of glycosylation on monoclonal antibody conformation and stability. mAbs. 2011;3:568–576. doi: 10.4161/mabs.3.6.17922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsumiya S., Yamaguchi Y., Saito J. Structural comparison of fucosylated and nonfucosylated Fc fragments of human immunoglobulin G1. J. Mol. Biol. 2007;368:767–779. doi: 10.1016/j.jmb.2007.02.034. [DOI] [PubMed] [Google Scholar]
- 11.Upton R., Bell L., Guy C. Orthogonal assessment of biotherapeutic glycosylation: a case study correlating N-glycan core afucosylation of herceptin with mechanism of action. Anal. Chem. 2016;88:10259–10265. doi: 10.1021/acs.analchem.6b02994. [DOI] [PubMed] [Google Scholar]
- 12.Kaneko Y., Nimmerjahn F., Ravetch J.V. Anti-inflammatory activity of immunoglobulin G resulting from Fc sialylation. Science. 2006;313:670–673. doi: 10.1126/science.1129594. [DOI] [PubMed] [Google Scholar]
- 13.Epp A., Hobusch J., Bartsch Y.C. Sialylation of IgG antibodies inhibits IgG-mediated allergic reactions. J. Allergy Clin. Immunol. 2018;141:399–402 e398. doi: 10.1016/j.jaci.2017.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raju T.S., Jordan R.E. Galactosylation variations in marketed therapeutic antibodies. mAbs. 2012;4:385–391. doi: 10.4161/mabs.19868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomann M., Reckermann K., Reusch D. Fc-galactosylation modulates antibody-dependent cellular cytotoxicity of therapeutic antibodies. Mol. Immunol. 2016;73:69–75. doi: 10.1016/j.molimm.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 16.Karsten C.M., Pandey M.K., Figge J. Anti-inflammatory activity of IgG1 mediated by Fc galactosylation and association of FcgammaRIIB and dectin-1. Nat. Med. 2012;18:1401–1406. doi: 10.1038/nm.2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowden T.A., Baruah K., Coles C.H. Chemical and structural analysis of an antibody folding intermediate trapped during glycan biosynthesis. J. Am. Chem. Soc. 2012;134:17554–17563. doi: 10.1021/ja306068g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goetze A.M., Liu Y.D., Zhang Z. High-mannose glycans on the Fc region of therapeutic IgG antibodies increase serum clearance in humans. Glycobiology. 2011;21:949–959. doi: 10.1093/glycob/cwr027. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y.D., Flynn G.C. Effect of high mannose glycan pairing on IgG antibody clearance. Biologicals. 2016;44:163–169. doi: 10.1016/j.biologicals.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 20.Sha S., Agarabi C., Brorson K. N-Glycosylation design and control of therapeutic monoclonal antibodies. Trends Biotechnol. 2016;34:835–846. doi: 10.1016/j.tibtech.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 21.Duivelshof B.L., Jiskoot W., Beck A. Glycosylation of biosimilars: recent advances in analytical characterization and clinical implications. Anal. Chim. Acta. 2019;1089:1–18. doi: 10.1016/j.aca.2019.08.044. [DOI] [PubMed] [Google Scholar]
- 22.Beck A., Wagner-Rousset E., Ayoub D. Characterization of therapeutic antibodies and related products. Anal. Chem. 2013;85:715–736. doi: 10.1021/ac3032355. [DOI] [PubMed] [Google Scholar]
- 23.Yang X., Bartlett M.G. Glycan analysis for protein therapeutics. J. Chromatogr. B. 2019;1120:29–40. doi: 10.1016/j.jchromb.2019.04.031. [DOI] [PubMed] [Google Scholar]
- 24.Liu P., Zhu X., Wu W. Subunit mass analysis for monitoring multiple attributes of monoclonal antibodies. Rapid Commun. Mass Spectrom. 2019;33:31–40. doi: 10.1002/rcm.8301. [DOI] [PubMed] [Google Scholar]
- 25.Largy E., Cantais F., Van Vyncht G. Orthogonal liquid chromatography–mass spectrometry methods for the comprehensive characterization of therapeutic glycoproteins, from released glycans to intact protein level. J. Chromatogr. A. 2017;1498:128–146. doi: 10.1016/j.chroma.2017.02.072. [DOI] [PubMed] [Google Scholar]
- 26.Wohlschlager T., Scheffler K., Forstenlehner I.C. Native mass spectrometry combined with enzymatic dissection unravels glycoform heterogeneity of biopharmaceuticals. Nat. Commun. 2018;9:1713. doi: 10.1038/s41467-018-04061-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D’Atri V., Nováková L., Fekete S. Orthogonal middle-up approaches for characterization of the glycan heterogeneity of etanercept by hydrophilic interaction chromatography coupled to high-resolution mass spectrometry. Anal. Chem. 2019;91:873–880. doi: 10.1021/acs.analchem.8b03584. [DOI] [PubMed] [Google Scholar]
- 28.Reusch D., Haberger M., Falck D. Comparison of methods for the analysis of therapeutic immunoglobulin G Fc-glycosylation profiles-Part 2: mass spectrometric methods. mAbs. 2015;7:732–742. doi: 10.1080/19420862.2015.1045173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reusch D., Haberger M., Maier B. Comparison of methods for the analysis of therapeutic immunoglobulin G Fc-glycosylation profiles--part 1: separation-based methods. mAbs. 2015;7:167–179. doi: 10.4161/19420862.2014.986000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carillo S., Mittermayr S., Farrell A. Glycosylation analysis of therapeutic glycoproteins produced in CHO cells. In: Meleady P., editor. Heterologous Protein Production in CHO Cells: Methods and Protocols. Springer New York; New York, NY: 2017. pp. 227–241. [DOI] [PubMed] [Google Scholar]
- 31.Tassi M., De Vos J., Chatterjee S. Advances in native high-performance liquid chromatography and intact mass spectrometry for the characterization of biopharmaceutical products. J. Sep. Sci. 2018;41:125–144. doi: 10.1002/jssc.201700988. [DOI] [PubMed] [Google Scholar]
- 32.Farrell A., Carillo S., Scheffler K. Monoclonal antibody sequence assessment using a hybrid quadrupole-Orbitrap mass spectrometer. Anal. Methods. 2018;10:3100–3109. [Google Scholar]
- 33.Rogers R.S., Abernathy M., Richardson D.D. A view on the importance of “Multi-Attribute Method” for measuring purity of biopharmaceuticals and improving overall control strategy. AAPS J. 2017;20:7. doi: 10.1208/s12248-017-0168-3. [DOI] [PubMed] [Google Scholar]
- 34.Iida S., Kuni-Kamochi R., Mori K. Two mechanisms of the enhanced antibody-dependent cellular cytotoxicity (ADCC) efficacy of non-fucosylated therapeutic antibodies in human blood. BMC Cancer. 2009;9:58. doi: 10.1186/1471-2407-9-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang T.T. IgG Fc glycosylation in human immunity. In: Ravetch J.V., Nimmerjahn F., editors. Fc Mediated Activity of Antibodies: Structural and Functional Diversity. Springer International Publishing; Cham: 2019. pp. 63–75. [Google Scholar]
- 36.De Leoz M.L.A., Duewer D.L., Fung A. NIST interlaboratory study on glycosylation analysis of monoclonal antibodies: comparison of results from diverse analytical methods. Mol. Cell. Proteomics. 2019 doi: 10.1074/mcp.RA119.001677. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.