Abstract
Brassinosteroids (BRs) are a group of steroid hormones, essentially important for plant development and growth. BR signaling functions to promote cell expansion and cell division, and plays a role in etiolation and reproduction. As the phytohormone originally identified in the pollen grains of Brassica napus, BR promotes the elongation of stigma. Recent studies have revealed that BR is also critical for floral transition, inflorescence stem architecture formation and other aspects of plant reproductive processes. In this review, we focus on the current understanding of BRs in plant reproduction, the spatial and temporal control of BR signaling, and the downstream molecular mechanisms in both the model plant Arabidopsis and crops. The crosstalk of BR with environmental factors and other hormones in reproduction will also be discussed.
Keywords: brassinosteroid, reproduction, signaling pathway, hormone crosstalk, flowering time
1. Introduction
Brassinosteroid (BR) is an important plant hormone in growth and development. BR is ubiquitously distributed in all growing tissues of higher plants, with a much higher concentration in fruit, seeds and pollen. In Arabidopsis, the most active form of BR, brassinolide (BL), is converted from the precursor campesterol (CR). CR is first synthesized to campestanol (CN) and subsequently to castasterone (CS) through two parallel biosynthetic pathways, the early and late C-6 oxidation pathways [1]. The enzyme in charge of the conversion of 4-en-3-one to 3-one and 22-OH-4-en-3-one to 22-OH-3-one is DEETIOLATED 2 (DET2) [2]. While the C-22 hydroxylation reaction is mediated by DWARF4 (DWF4), the C-23 hydroxylation catalysis is mediated by CONSTITUTIVE PHOTOMORPHOGENESIS AND DWARFISM (CPD) [3,4]. The oxidation of C-6 is catalyzed by BR6ox [5]. Two P450 mono-oxygenases, CYP90C1 and CYP90D1, are responsible for the conversion of 22-OH-4-en-3-one, 22-OH-3-one, and 3-epi-6-deoxoCT to 23-hydroxylated products [6].
BR is perceived by a receptor kinase BRASSINOSTEROID INSENSITIVE 1 (BRI1) on the plasma membrane [7,8]. Mutants of bri1 show a variety of grown defects which are very similar to strong BR deficient mutants, including skotomorphogenesis, extreme dwarfism under light and male infertility. BRI1 is a member of plant-specific giant protein family of serine/threonine leucine-rich repeat receptor-like kinase, which has more than 200 homologs in Arabidopsis [9]. The extracellular region of BRI1 consists of 24 LRRs with an interruption of an island domain of methionine-rich repeat. The intracellular region can be further divided into three subdomains: a juxtamembrane region, a canonical S/T kinase and a short C-terminal extension [10]. Three homologs of BRI1 have been characterized in Arabidopsis with two have high BL-binding affinity [11,12,13].
After receiving BR, BRI1 resumes kinase activity by recruiting the co-receptor kinase BRI1-ASSOCIATED RECEPTOR KINASE 1 (BAK1) and dissociating from the inhibitory protein BRI1 KINASE INHIBITOR 1 (BKI1) [14,15,16,17]. Then the kinase domains of BRI1 and BAK1 are transphosphorylated and the phosphorylated BKI1 can associate with the 14-3-3 family proteins to further amplify BR signaling [16,18]. Another two plasma membrane-anchored cytoplasmic kinases, BRASSINOSTEROID-SIGNALLING KINASE 1 (BSK1) and CONSTITUTIVE DIFFERENTIAL GROWTH 1 (CDG1) are also phosphorylated by activated BRI1 to inactivate the phosphatase BRI1-SUPPESSOR 1 (BSU1) [19,20,21]. BSU1 in turn dephosphorylates a conserved tyrosine residue of BRASSINOSTEROID INSENSITIVE 2 (BIN2), leading to the inactivation of this GSK3-like kinase [22]. The function of BIN2 is to phosphorylate and inactivate two homologous transcription factors, BRASSINAZOLE RESISTANT 1 (BZR1) and BR1-EMS-SUPPRESSSOR 1 (BES1) in the absence of BR [23,24,25]. The phosphorylation leads to the deactivation of these two transcription factors [26]. In high BR level, BSU1 inactivates BIN2 and unphosphorylated BZR1 and BES1 can initiate BR regulated gene activation and repression [23,27]. BZR1 and BES1 initiate BR responsive gene expression by recognizing and binding to the BR response DNA cis-element BR-Response Element (BRRE, CGTGC/TG) and E-box (CANNTG) [23,28]. Gain-of-function bzr1-1D and bes1-D contain a proline to leucine mutation in the protein degradation domain and therefore exhibit BR constitutive phenotypes [24,25,29]. However, the bzr1-1D and bes1-D are morphologically different, indicating the two proteins are involved in distinct BR functions. A number of transcription factors and histone modifiers are identified to interact with BZR1/BES1 for the control of various BR responses [28,30,31,32,33]. BZR1 and BES1 belong to a six-member small family clade, consisting another four homologs, BES1/BZR1 homolog 1 to 4 (BEH1-4), which also act as downstream transcription factors in BR signaling pathway [28].
BR regulates a broad range of plant growth and development, including hypocotyl elongation, root development, skotomorphogenesis, vascular differentiation, floral transition, anther development, and pollen grain maturation. In this review, we will focus on the functions of BRs in reproduction.
2. Arabidopsis Reproductive Development
Arabidopsis reproduction starts from the floral transition, in which the shoot apical meristem (SAM) is transformed into inflorescence meristem (IM), which develops into main stem and flowers. Cauline leaves and axillary branches are produced on the main stem. The floral organ initiates from specific founder cells from IM to form floral primordia and develops into four whorls of floral organs, namely sepals, petals, stamens and carpels from outside to inside. The beginning of floral transition occurs as the florigen accumulates in SAM. The major florigen in Arabidopsis is identified as FLOWERING LOCUS T (FT), which is synthesized in the leaf phloem and transported to the SAM by endoplasmic reticulum membrane localized FT-INTERACTING PROTEIN 1 (FTIP1) and several other proteins [34,35]. FT and its homolog TWIN SISTER OF FT (TSF) are exclusively activated by CONSTANS (CO) and GIGANTEA (GI) photoperiodically under light [36,37,38,39,40]. CO expression is subject to the circadian clock and its protein stability is degraded by CONSTITUTIVE PHOTOMORPHOGENESIS 1 (COP1) in dark, causing the fast accumulation of FT under long-day conditions [41,42,43]. Therefore, CO, GI and FT are referred to as photoperiod pathway components to induce flowering. In SAM, FT forms a heterodimer with a bZIP transcription factor FD and this complex initiates the transcription of another floral promoter gene SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) and the floral meristem-identity gene APETALA 1 (AP1), which promote the formation of floral meristems [39,44,45,46].
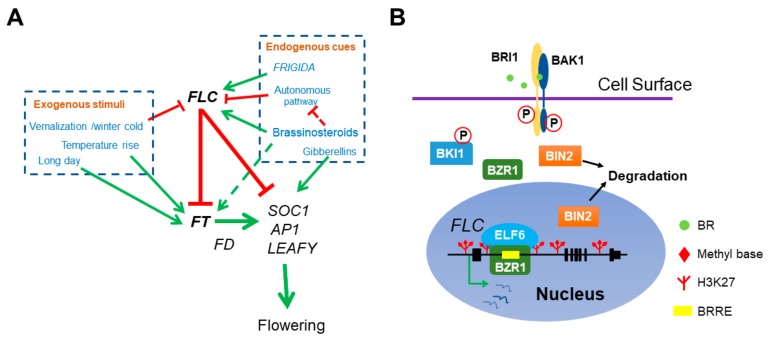
FT expression is repressed by the key floral repressor FLOWERING LOCUS C (FLC) during vegetative growth. FLC directly interacts with EMBRYONIC FLOWER1-PcG complex to deposit the repressive histone mark, H3K27me3, at FT chromatin to antagonize the activation of CO [47]. In addition to the photoperiod pathway, which is activated by exogenous photoperiodic cues, plants also have an endogenous pathway to promote flowering, the autonomous pathway. Components of autonomous pathway are a group of proteins with distinct molecular function but repress FLC expression constitutively, such as FLOWERING LOCUS D (FLD), FCA, FY, FPA, and FLOWERING LOCUS K HOMOLOGY DOMAIN (FLK) [48]. In winter-annual Arabidopsis ecotypes, FLC expression is elevated by the plant-specific coiled-coil protein FRIGIDA (FRI), which forms a stable core protein complex with FRIGIDA LIKE 1 (FRL1), FLC EXPRESSOR (FLX), SUPPRESSOR OF FRIGIDA 4 (SUF4), and FRIGIDA ESSENTIAL 1 (FES1) before vernalization [49,50]. The activity of FRI also depends on the presence of a few active chromatin modifiers and co-transcriptional pre-mRNA modification factors, and these proteins, together with the core FRI complex, form a supercomplex at the FLC locus to establish a conducive chromatin spatial structure for high-efficient FLC mRNA production [51]. Prolonged cold turns off FLC expression and enables flowering in the coming spring (Figure 1A).
Figure 1.
The role of BR in Arabidopsis flowering network. (A) BR directly represses floral transition by activating the transcription of main floral repressor FLC. Both endogenous cues and exogenous stimuli can affect flowering time. FRI and BR delay flowering by elevating FLC expression and the autonomous pathway genes antagonize with FRI and BR by constitutively repressing FLC. Meanwhile, vernalization represses FLC and ensures flowering after plants return to warm. FLC interacts with EMF-PRC2 repressive complex to repress the transcription of florigen FT and SOC1. The ambient temperatures and photoperiod pathway activate FT expression. FT associates with another transcription factor FD to stimulate the transcription of SOC1, AP1 and LEAFY and thus initiates the transition of SAM to IM. In addition, BR also indirectly mediates flowering time by activating FT and repressing the autonomous pathway member FLD expression (dotted lines). (B) The molecular mechanism of BZR1 activation on FLC expression. BZR1 recognizes a BRRE cis-element in FLC 1st intron and recruits the histone demethylase ELF6 to erase the repressive histone mark H3K27me3 at FLC locus.
Floral organ primordia differentiate from the floral meristem. During floral organogenesis, boundaries are formed by cells with retarded growth rates between whorls of specific organs and each individual flower. Similar boundaries occur between the main stem and lateral branches and cauline leaves. CUP SHAPED COTYLEDON (CUC) genes are important regulators for both floral organogenesis and lateral organ formation [52,53]. Further, cuc mutants show both fused lateral and floral organs, suggesting that CUCs are responsible to repress boundary cell growth [54,55].
Proper boundary formation separates flowers into four whorls of organs (sepals, petals, stamens and carpels). In the classic ABC model, sepals are delineated by A genes, and A and B specify petals; in addition, B and C are responsible for stamen development. C gene alone determines carpels and terminates the activities of floral meristem [56]. The MADS box protein AGAMOUS (AG) plays a C role by suppressing the expression of stem cell identity gene WUSCHEL (WUS) and activates SPOROCYTELESS/NOZZLE (SPL/NZZ) for microsporogenesis at flower development stage 6 [57,58,59]. In early floral development stage, AG expression is activated by WUS and subsequently elevated AG expression leads to a repression of WUS expression. This forms a WUS-AG feedback loop to turn off floral meristem activity. AG recognizes WUS chromatin region and represses its expression by recruiting PcG proteins to deposit the repressive histone mark H3K27me3 [60]. AG also activates the expression of the WUS repressor KNUCKLES (KNU) via competitive binding to the promoter of KNU with PcG proteins to indirectly shut down WUS expression [61].
Arabidopsis floral development can be divided into 20 stages. AG expression starts from Stage 3 to diminish WUS and initiate floral organ primordia and lasts to very late stages to stimulate and maintain SPL expression for the anther development [59,62,63]. SPL encodes a transcription factor and functions in ovule formation and early microsporogenesis. spl mutant is male sterile owning to the failure of pollen grain development. SPL expresses in the anther primordia to establish the microspore mother cells. Morphologically, anther development can be separated into two phases, defined by the completion of meiosis and appearance of microspore [64]. During phase one, sporogenous cells are observed once the four-lobed anther pattern generated and further develop into microspore mother cells which produce tetrads of haploid microspores through meiosis. These microspores are released into the anther locule to begin microgametogenesis in phase two. Finally, matured pollen grains are released by anther dehiscence [64,65]. During phase one, the anther has also developed several highly specialized cells and tissues, including the epidermis, endothecium, tapetum, vascular bundle, and stomium. Each of these cell types and tissues carries out specific functions. For example, the stomium are involved in dehiscence, and tapetum plays a key role in pollen wall formation. The tapetum forms a cell layer surrounding developing microspores within the anther locule. During microspore development, the tapetum provides necessary structural components and nutrients. As the pollen matures, the tapetum diminishes through programmed cell death (PCD) and the released remnants incorporate into the coat of pollen grains [66].
3. Role of BR in Floral Transition
BR regulates floral transition in a complicated manner. BR mutants, like det2-1 and cpd have a prominent delay in days to flowering with a slow rate of leaf initiation. Therefore, BR was previously thought to promote flowering [67,68]. However, the IM is transformed from SAM, which produces leaves during vegetative growth. So, the developmental criterion of floral transition is measured by primary leaf number rather than growth rate or days to flowering in Arabidopsis [69]. By using this developmental criterion, it is found that bri1 and BR biosynthetic mutants det2, cpd and dwf4 are early flowering [70]. In addition, exogenous application of BR or the steroids, androstenedione and androsterone, cause late flowering in wild type Arabidopsis plants, suggesting that BR signaling pathway performs a repressive role in floral transition [70,71]. In Arabidopsis, transcription of the central floral repressor FLC and its three homologs, FLOWERING LOCUS M (FLM), MADS AFFECTING FLOWERING 4 (MAF4) and MAF5, are depressed in BR mutants and genetic studies demonstrate that BR antagonizes the autonomous pathway by constitutively activating FLC expression during vegetative growth [70].
BR activation of FLC transcription is mainly through the downstream transcription factors BZR1 and BES1-INTERACTING MYC-LIKEs (BIMs). The dominant bzr1-1D mutant exhibits late flowering and defects in BR downstream transcription factor BIMs lead to early floral transition. FLC and its homologs are found to be activated in bzr1-1D and repressed in bim1 bim2 bim3, respectively [24,28,70]. Both BZR1 and BIM1 can recognize and bind to a cis-regulatory BRRE element (CGTGTG) located in the first intron of FLC and deletion of this cis-element abolishes the late flowering phenotype of bzr1-1D mutant, suggesting that BZR1 and BIM1 binding to this element is required for FLC upregulation. As a transcription factor, BZR1 further recruits the H3K27 demethylase EARLY FLOWERING 6 (ELF6) to the FLC region to eliminate the transcription repressive mark H3K27me3, leading to a reduction of H3K27me3 and thus the FLC activation [32,70,72] (Figure 1B). Recent studies found that a Cold Memory Element (CME) exists about 0.5Kb upstream of the BRRE cis-element in FLC first intron, which recruits the B3 domain proteins VP1/ABI3-LIKE 1 (VAL1) and VAL2 [73,74]. VALs bind to the CME before and after vernalization and recruit PcG proteins to add H3K27me3 at the FLC locus for transcriptional repression, especially for the maintenance of FLC repression after vernalization [74]. Thus, BR signaling may integrate environmental cues to regulate floral transition at a proper time through controlling the level of H3K27me3 at FLC.
However, BR seems to participate in floral transition in both FLC-direct and -indirect ways in different accessions. For example, BZR1 can also bind to a BRRE cis-element (CGTGGG) at FLD promoter region and suppresses the transcription of FLD [75]. FLD is a component of the autonomous pathway and represses FLC expression by reducing the transcription active marks histone acetylation and H3K4me3 levels at FLC [76,77]. Therefore, BZR1 can also indirectly activate FLC expression through the autonomous pathway. Unlike bzr1-1D, the other BR constitutive mutant bes1-D flowers like wild type and overexpression of BES1-D exhibits no distinct flowering phenotype in Columbia ecotype [70]. While, besides the canonical BES1 isoform, BES1 has a long isoform (BES1-L) due to alternative splicing [78]. In transgenic plants, overexpressed BES1-L causes an early flowering phenotype due to its indirect activation of FT [78,79]. BES1 doesn’t bind to FT chromatin, but activates FT expression through BR ENHAHCED EXPRESSION 1 (BEE1). BES1 binds to BEE1 promoter to activate BEE1 expression and FT expression can be elevated via BEE1 binding to its promoter region [79,80]. BEE1 activity and protein stability is blue-light dependent, therefore, BR may coordinate with light signaling in the control of the floral transition. Similarly, two P450 enzymes, PHYB ACTIVATION TAGGED SUPPRESSOR 1 (BAS1) and SUPPRESSOR OF PHYB-4 7 (SOB7), are involved in both light response and BR-related floral transition, indicating the complicated regulation of BR on flowering transition [81,82]. In summary, BR plays a negative role in floral transition mainly through direct activation of FLC and its homologs, but BR signaling integrates with environmental cues to fine-tune the time of flowering through FT and the other flowering pathway.
4. Role of BR in Lateral Organ Boundary Formation in Inflorescence
As mentioned above, lowed FLC level de-represses FT expression, which subsequently activates the expression of LEAFY and AP1 and leads to the transition of SAM to IM [47,48]. BR constitutive and deficient mutants all have organ boundary defects in inflorescence stem and flower organs. Arabidopsis wild type plants have straight inflorescence stem, whereas the inflorescence stem of bzr1-1D binds toward the lateral organs, such as cauline leaf and axillary branch. In wild type, the axillary branch completely separates from both cauline leaf and the stem, while the axillary branch and cauline leaf are fused together and this fusion causes the inward stem binding in bzr1-1D mutant [83]. Similar organ boundary fusion phenotype is also observed in the transgenic lines of bzr1-1D-CFP, BZR1-S173A and bes1-D mutant, where BR signaling is constitutively activated [26,83,84]. This organ fusion phenotype is also observed in DWF4 overexpression transgenic plants [83]. In contrast, BR deficient mutants, like bri1-5, bin2-1 and det2-1, all show the outward binding of IM away from the lateral organs, which is due to the deeper notch between the IM and axillary branch [83]. In addition, the stamen to carpel and stamen to stamen fusions are also observed in bzr1-1D and bes1-D mutants as well as BR treated wild type flowers, demonstrating that active BR signaling negatively regulates boundary formation [84]. Meanwhile, the extra ovaries occur in flowers of bri1-5 and det2-1 is due to the ectopic formation of a boundary structure in the ovary [83,84]. Altogether, BR spatial and temporal accumulation is crucial for organ boundary formation during floral organogenesis.
The organ boundary is formed by the unequal cell division and elongation rates at central meristem and periphery of organ primordia, which separates the lateral organs from central meristem [85]. At the periphery of primordia, cell division and elongation become arrested, whereas cells in lateral organ primordia undergo rapid division and elongation to form leaves and flowers. Unregulated cell division and expansion at organ boundary causes the organ fusion, while insufficient organ boundary formation is the main reason for the outward stem binding in BR deficient mutants [83]. Therefore, the spatial and temporal distribution of BR is crucial for the inflorescence and flower organ formation. Indeed, BR is unevenly distributed in lateral organs, resulting in the dotted localization pattern of BZR1 in the meristem and organ primordia, but not the lateral boundary regions [83,86]. This distinct BR distribution in lateral organs is mediated by a plant specific LOB domain transcription factor LATERAL ORGAN BOUNDARIES (LOB), which exclusively localizes in organ boundaries and represses BR accumulation. Loss of function mutants of LOB show fused axillary branch and cauline leaf, and mimic the defects of bzr1-1D or BR application [86]. LOB can directly bind to a GCGGCG cis-element (LBD motif) at the promoter region of BAS1 and BAS1 converts bioactive BR to its C-26 hydroxylated derivatives in boundary regions [86,87,88]. Moreover, LOB expression is positively regulated by BR in boundary regions, which forms a feedback loop to maintain low level of BR in organ boundary [86].
The accumulated activated form of BZR1 in inflorescence meristem and sepal primordia represses the expression of organ boundary identity genes CUC1, CUC2, CUC3 and LATERAL ORGAN FUSION1 (LOF1) [83,89,90]. Among these genes, the promoter regions of CUC1, CUC2 and CUC3 contain the BRRE cis-elements and can be directly recognized by BZR1 [83]. In addition, proper CUCs expression is also regulated by a BES1-TPL repressive module. BES1 recruits the general repressor TOPLESS (TPL) through its C-terminal ERF-associated amphiphilic repression (EAR) motif [84]. Like bzr1-1D, bes1-D has fused lateral organ defects and BES1 can also repress CUC3 expression via binding to the same BRRE cis-element in the CUC3 promoter region. Overexpressing BES1-D with mutated EAR motif shows normal lateral organ structure, while extra TPL expression causes the organ fusion in wild type with depressed CUC3 expression level [84]. Therefore, it is likely that the BR transcription factor BES1 recruits TPL to repress CUC3 expression [84]. Genome-wide analysis pointed out that organ boundary specific transcripts overlap with BR repressed genes, and therefore BZR1 and BES1 can regulate a broad range of boundary specific transcriptomes for correct organ boundary development [27,30,90].
5. Role of BR in Male Fertility
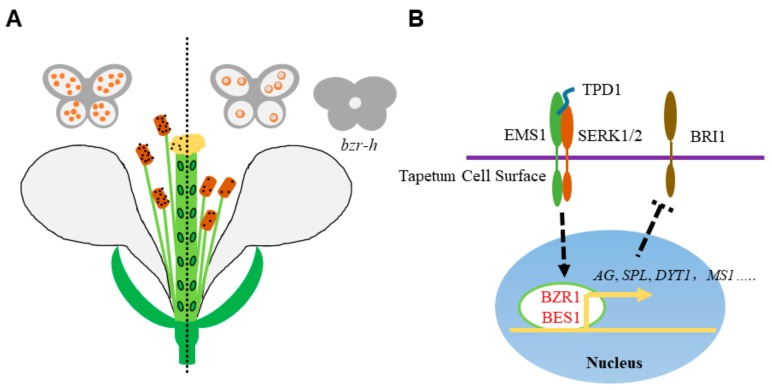
BR plays a positive role to regulate male fertility by promoting filament elongation and anther development [91]. Arabidopsis BR defective mutants in both BR biosynthetic and signaling transduction pathways, like bri1, bin2, cpd, and dwrf4, show reduced fertility or male sterility with short filaments and decreased pollen grain number and disposal efficiency [3,67,92,93]. Further, bri1-116 and cpd produce only about 20% pollen grains per anther compared to wild type anther and the relatively small amount of pollens are difficult to release from the anther locule [91]. In BR mutants, the tapetal cells are more vacuolated and enlarged and the number of microspore mother cells are fewer than wild type during early anther developmental stages [91]. During late anther developmental stages, microspores are also degenerated and vacuolated greatly in the mutants. In general, the abnormal development of tapetum, microspore mother cell and microspore all contribute to the decreased number of pollen grains. In cpd and bri1-116 mutants, the pollen grains are hardly released from the anther owing to the absence of bacula/tectum structure in the pollen grain surface [91] (Figure 2A). The defects of pollen grain outer wall in bri1-116 and cpd may be caused by the abnormal development of tapetal cells as the tapetum plays a key role in pollen grain outer surface formation in Arabidopsis [94,95].
Figure 2.
BR promotes the male reproductive development. (A) The schematic drawing of male reproductive defects in BR-deficient mutants. The left part shows the wild type normal male reproductive organ development. In the BR-deficient mutants (right part), the stigma elongation is retarded, the anther is failed to release pollen grains and the microspores are vacuolated and degenerated. Meanwhile, loculeless anther is produced in the hextuple mutant bzr-h, indicating the BR downstream transcription factor BZR1 and its homologs play both BR-dependent and -independent role in anther development. (B) Roles of BZR1 and BES1 in anther development. In tapetum cells, the cell membrane localized receptor kinases EMS1 and SERK1/2 perceive the peptide ligand TPD1 and activate BZR1 and BES1. BZR1 and BES1 then initiate the transcription of anther development genes, like AG, SPL, DYT1, MS1 and so on. The expression of BRI1 is elevated in spl mutant, suggesting a possible regulatory network of anther development and BR signaling. Dotted arrow indicates the activation of BES1 and BZR1 by EMS1-TPD1-Serks signaling. Dotted T bar represents for the feedback downregulation of BRI1 transcription by SPL.
During early anther development stages, microspore mother cells secret a small peptide ligand TAPETUM DETERMINANT 1 (TPD1), which is perceived by receptor-like kinases Excess Microsporocytes 1 (EMS1) and Somatic Embryogenesis Receptor-Like Kinase 1,2 (SERK1/2) [96,97,98,99]. This signal initiates and maintains the development of tapetum in L2-originated cells [100]. Both EMS1 and BRI1 belong to the large LRR-RLK family and the intracellular domains of BRI1 and EMS1 are functionally exchangeable [101]. Upon perception of TPD1, EMS1 can activate BZR1 and BES1 for the transcription of tapetal development genes [102]. The tapetum failure of ems1, tpd1, and serk1 and serk2 can be rescued genetically by bzr1-1D and bes1-D. Therefore, BR incorporates with the EMS1-TPD1 signaling for the anther development [101] (Figure 2B).
At molecular level, anther development genes, like SPL, DEFECTIVE IN MERISTEM DEVELOPMENT AND FUNCTION 1 (TDF1), ABORTED MICROSPORES (AMS), MYB103, MALE STERILITY 1 (MS1), MS2 as well as the target genes of MS1 are all depressed in bri1-116 and cpd, consistent with the multiple anther development defects in the mutants. In contrast, transcription of these genes is activated in the BES1 overexpression lines [91]. The promoter regions of SPL, TDF1, MS1, MS2, and MYB103 contain both E-box and BRRE cis-elements and can be recognized and bound by BES1, which demonstrates the direct regulation of BR signaling pathway in anther development [91]. Moreover, the expression of BR receptor BRI1 is moderately upregulated in spl mutant, indicating a feedback regulatory network in BR signaling and anther development [103] (Figure 2B).
However, by using the loss-of-function quintuple and hextuple mutants of BZR1 family members, it is found that BR downstream transcription factors regulate anther development in both BRI1-dependent and -independent ways [102,104]. Qunituple mutant of bes1-1 bzr1-1 beh1-1 beh3-1 beh4-1 is phenotypically like wild type but male-sterile and the hextuple mutants of all the six family members are phenotypically similar to the BR insensitive bri1 brl1 brl3 (bri1-t) in both vegetative and reproductive defects [102,104,105]. The anthers of the hextuples are failed to open with no pollen grains. In anther development stage 1, only the L1 layer are developed in bzr-h, compared with clearly visible three layers of L1, L2 and L3 in both wild type and bri1-t. In stage 2, anther cells of bzr-h become severely vacuolated and archesporial cell formation is inhibited in stage 3, which causes the loculeless anther of bzr-h [104]. The sporogenesis and archesporial cell initiation are directly regulated by AG and SPL during early male gametogenesis and the expression of the AG and SPL is almost inhibited in bzr-h unopened flowers [59,63,104]. While, AG expression is unaffected and SPL transcription is only reduced to about half in bri1-t [104]. Meanwhile, transcription of tapetal development genes like DYSFUNCTIONAL TAPETUM1 (DYT1) and ARABIDOPSIS THALIANA ANTHER 7 (ATA7) is largely diminished in the quintuple mutant [104,106,107]. In summary, BZR1 and its five homologs regulate the early stage of anther development through activating the expression of AG and SPL as well as tapetal development genes in both BR-dependent and independent manners.
In addition to BZR1 and BES1, BIM1 is also involved in another development at early stages. Loss-of-function BIM1 mutant has mild male sterility at the first few flowers in the main inflorescence due to short stamen filament. This phenotype is similar to squamosa promoter binding protein-like 8 (spl8) mutant and is enhanced in the double mutant of bim1 spl8 [108]. SPL8 encodes an SBP DNA binding protein and expresses highly at early anther development stages. Further, spl8 is semi-sterile with short filament and failure of microspore mother cell initiation in the first twelve flowers [109]. In the double mutant, microspore mother cells fail to produce in the adaxial loculi at anther development stage 5 and the initiated microspore mother cells cannot undergo meiosis at stage 6, resulting in the disappearance of pollen sacs in the anther and male sterile. BIM1 has a similar expression pattern with SPL8 in early developing anther and they function cooperatively in anther development [108]. Moreover, BIM1 contributes to embryonic patterning via physical interaction with DORNROESCHEN (DRN) and DORNROESCHEN-LIKE (DRNL) [110]. In embryonic globular stage, the asymmetrical cell division is seldomly occurred in bim1, leading to the absence of central and/or basal domain in embryo and thus defects in cotyledon formation. This abnormal embryonic patterning of bim1 is proportionally increased in the double mutants of both bim1 drn and bim1 drnl [110]. DRN and DRNL regulate auxin-related embryonic patterning by affecting PIN-FORMED 1 (PIN1) localization [110]. The protein interaction of BIM1 with DRN and DRNL may give hint to the crosstalk of BR and auxin in embryonic patterning.
6. Role BR in Other Plant species
Like in Arabidopsis, BR plays similar effect on reproduction in other plants species. In monocotyledonous wheat, exogenous application of 24-epibrassinolide delays vegetative to generative transition, while the BR inhibitor, Brassinazole, accelerates the transition and heading stage [111]. In short-day plant Pharbitis nil, treatment of BL and CS inhibit flowering in combination with the inductive photoperiod, suggesting that BR integrates with environmental cues for the proper reproductive transition [112]. In addition to floral transition, BR also regulates both male and female fertility in crops. In maize, ineffective BR biosynthesis causes male sterility, which is due to the failed anther and pollen development [113,114]. In bisexual flower fruit cucumber, exogenous BR application accelerates and increases female flower organogenesis in a dose-dependent manner and this effect may attributes to the crosstalk of BR-ethylene [115].
7. Conclusions
In summary, BR plays a broad role in plant reproduction from floral transition to male fertility. Plant development is modulated by plant-environment crosstalk that requires the incorporation of external environmental cues and internal responses. Studies have revealed the crucial roles of hormone in plant development and plant-environment interaction. As mentioned above, the effects of BR signaling are also subject to hormone crosstalk and biotic as well as abiotic stimuli. For example, BR-mediated cell elongation is positively affected by auxin and gibberellin but negatively by cytokinin and abscisic acid. During reproduction, the cell identity is established within a few cells at the initiation of floral transition and lateral organ formation. Unlike auxin, BRs do not undergo distant transportation and are believed to exert their functions locally [116]. All these call for the precise temporal and spatial control of BR signaling in reproductive development. The application of effective techniques to isolate tissue-specific plant cells and to characterize tissue/cell specific transcriptome and/or proteome is becoming available, such as Isolation of Nuclei TAgged in specific Cell Types (INTACT), cleavage under targets and release using nuclease (CUT & RUN) and single cell sequencing. All these will facilitate our understanding of the function and mechanism of BR signaling in the specific development process as well as a response to environmental stimuli [117,118,119].
Acknowledgments
We thank the start-up funding from Shandong University to Z.L.
Abbreviations
| BR | brassinosteroid |
| BRI1 | BRASSINOSTEROID INSENSITIVE 1 |
| BZR1 | BRASSINAZOLE RESISTANT 1 |
| BES1 | BRI1-EMS SUPPRESSOR 1 |
| BRRE | BR-Response Element |
| BL | Brassinolide |
| CR | Campesterol |
| CN | Campestanol |
| CS | Castasterone |
| DET2 | DEETIOLATED 2 |
| DWF4 | DWARF4 |
| CPD | CONSTITUTIVE PHOTOMORPHOGENESIS AND DWARFISM |
| BAK1 | BRI1-ASSOCIATED RECEPTOR KINASE 1 |
| BKI1 | KINASE INHIBITOR 1 |
| BSK1 | BRASSINOSTEROID-SIGNALLING KINASE 1 |
| CDG1 | CONSTITUTIVE DIFFERENTIAL GROWTH 1 |
| BSU1 | BRI1-SUPPESSOR 1 |
| BIN2 | BRASSINOSTEROID INSENSITIVE 2 |
| BEH1 | BES1/BZR1 homolog 1 |
| SAM | shoot apical meristem |
| IM | inflorescence meristem |
| FT | FLOWERING LOCUS T |
| FTIP1 | FT-INTERACTING PROTEIN 1 |
| TSF | TWIN SISTER OF FT |
| CO | CONSTANS |
| GI | GIGANTEA |
| COP1 | CONSTITUTIVE PHOTOMORPHOGENESIS 1 |
| SOC1 | SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 |
| AP1 | APETALA 1 |
| FLC | FLOWERING LOCUS C |
| FLD | FLOWERING LOCUS D |
| FLK | FLOWERING LOCUS K HOMOLOGY DOMAIN |
| FRI | FRIGIDA |
| FRL1 | FRIGIDA LIKE 1 |
| FLX | FLC EXPRESSOR |
| SUF4 | SUPPRESSOR OF FRIGIDA 4 |
| FES1 | FRIGIDA ESSENTIAL 1 |
| CUC | CUP SHAPED COTYLEDON |
| AG | AGAMOUS |
| WUS | WUSCHEL |
| SPL/NZZ | SPOROCYTELESS/NOZZLE |
| KNU | KNUCKLES |
| PCD | programmed cell death |
| FLM | FLOWERING LOCUS M |
| MAF4 | MADS AFFECTING FLOWERING 4 |
| BIM1 | BES1-INTERACTING MYC-LIKE1 |
| ELF6 | EARLY FLOWERING 6 |
| CME | Cold Memory Element |
| VAL1 | VP1/ABI3-LIKE 1 |
| BEE1 | BR ENHAHCED EXPRESSION 1 |
| BAS1 | PHYB ACTIVATION TAGGED SUPPRESSOR 1 |
| SOB7 | SUPPRESSOR OF PHYB-4 7 |
| LOB | LATERAL ORGAN BOUNDARIES |
| LOF1 | LATERAL ORGAN FUSION1 |
| TPL | TOPLESS |
| EAR | ERF-associated amphiphilic repression |
| TPD1 | TAPETUM DETERMINANT 1 |
| EMS1 | Excess Microsporocytes 1 |
| SERK1/2 | Somatic Embryogenesis Receptor-Like Kinase 1,2 |
| TDF1 | DEFECTIVE IN MERISTEM DEVELOPMENT AND FUNCTION 1 |
| AMS | ABORTED MICROSPORES |
| MS1 | MALE STERILITY 1 |
| bri1-t | bri1 brl1 brl3 |
| DYT1 | DYSFUNCTIONAL TAPETUM1 |
| ATA7 | ARABIDOPSIS THALIANA ANTHER 7 |
| SPL8 | squamosa promoter binding protein-like 8 |
| DRN | DORNROESCHEN |
| DRNL | DORNROESCHEN-LIKE |
| PIN1 | PIN-FORMED 1 |
| INTACT | Isolation of Nuclei TAgged in specific Cell Types |
| CUT & RUN | Cleavage Under Targets and Release Using Nuclease |
Author Contributions
Z.L. and Y.H. conceived and drafted the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China, 31970533.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Noguchi T., Fujioka S., Choe S., Takatsuto S., Tax F.E., Yoshida S., Feldmann K.A. Biosynthetic pathways of brassinolide in arabidopsis. Plant Physiol. 2000;124:201–210. doi: 10.1104/pp.124.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fujioka S., Li J.M., Choi Y.H., Seto H., Takatsuto S., Noguchi T., Watanabe T., Kuriyama H., Yokota T., Chory J., et al. The Arabidopsis deetiolated2 mutant is blocked early in Brassinosteroid biosynthesis. Plant Cell. 1997;9:1951–1962. doi: 10.1105/tpc.9.11.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szekeres M., Nemeth K., KonczKalman Z., Mathur J., Kauschmann A., Altmann T., Redei G.P., Nagy F., Schell J., Koncz C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/S0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- 4.Choe S.W., Dilkes B.P., Fujioka S., Takatsuto S., Sakurai A., Feldmann K.A. The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22 alpha-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell. 1998;10:231–243. doi: 10.1105/tpc.10.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimada Y., Fujioka S., Miyauchi N., Kushiro M., Takatsuto S., Nomura T., Yokota T., Kamiya Y., Bishop G.J., Yoshida S. Brassinosteroid-6-oxidases from arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiol. 2001;126:770–779. doi: 10.1104/pp.126.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohnishi T., Szatmari A.M., Watanabe B., Fujita S., Bancos S., Koncz C., Lafos M., Shibata K., Yokota T., Sakata K., et al. C-23 hydroxylation by Arabidopsis CYP90C1 and CYP90D1 reveals a novel shortcut in brassinosteroid biosynthesis. Plant Cell. 2006;18:3275–3288. doi: 10.1105/tpc.106.045443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clouse S.D., Langford M., McMorris T.C. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J.M., Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/S0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 9.Shiu S.H., Bleecker A.B. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA. 2001;98:10763–10768. doi: 10.1073/pnas.181141598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vert G., Nemhauser J.L., Geldner N., Hong F.X., Chory J. Molecular mechanisms of steroid hormone signaling in plants. Annu. Rev. Cell Dev. Biol. 2005;21:177–201. doi: 10.1146/annurev.cellbio.21.090704.151241. [DOI] [PubMed] [Google Scholar]
- 11.Cano-Delgado A., Yin Y.H., Yu C., Vafeados D., Mora-Garcia S., Cheng J.C., Nam K.H., Li J.M., Chory J. BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development. 2004;131:5341–5351. doi: 10.1242/dev.01403. [DOI] [PubMed] [Google Scholar]
- 12.Clay N.K., Nelson T. VH1, a provascular cell-specific receptor kinase that influences leaf cell patterns in Arabidopsis. Plant Cell. 2002;14:2707–2722. doi: 10.1105/tpc.005884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou A., Wang H.C., Walker J.C., Li J. BRL1, a leucine-rich repeat receptor-like protein kinase, is functionally redundant with BRI1 in regulating Arabidopsis brassinosteroid signaling. Plant J. 2004;40:399–409. doi: 10.1111/j.1365-313X.2004.02214.x. [DOI] [PubMed] [Google Scholar]
- 14.Li J.M., Nam K.H. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science. 2002;295:1299–1301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- 15.Nam K.H., Li J.M. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110:203–212. doi: 10.1016/S0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- 16.Wang H.J., Yang C.J., Zhang C., Wang N.Y., Lu D.H., Wang J., Zhang S.S., Wang Z.X., Ma H., Wang X.L. Dual Role of BKI1 and 14-3-3 s in Brassinosteroid Signaling to Link Receptor with Transcription Factors. Dev. Cell. 2011;21:825–834. doi: 10.1016/j.devcel.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 17.Wang X.L., Chory J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science. 2006;313:1118–1122. doi: 10.1126/science.1127593. [DOI] [PubMed] [Google Scholar]
- 18.Wang X.F., Kota U., He K., Blackburn K., Li J., Goshe M.B., Huber S.C., Clouse S.D. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev. Cell. 2008;15:220–235. doi: 10.1016/j.devcel.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 19.Kim T.W., Guan S.H., Burlingame A.L., Wang Z.Y. The CDG1 Kinase Mediates Brassinosteroid Signal Transduction from BRI1 Receptor Kinase to BSU1 Phosphatase and GSK3-like Kinase BIN2. Mol. Cell. 2011;43:561–571. doi: 10.1016/j.molcel.2011.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang W.Q., Kim T.W., Oses-Prieto J.A., Sun Y., Deng Z.P., Zhu S.W., Wang R.J., Burlingame A.L., Wang Z.Y. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science. 2008;321:557–560. doi: 10.1126/science.1156973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mora-Garcia S., Vert G., Yin Y.H., Cano-Delgado A., Cheong H., Chory J. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to bras sino steroids in Arabidopsis. Gene Dev. 2004;18:448–460. doi: 10.1101/gad.1174204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim T.W., Wang Z.Y. Brassinosteroid Signal Transduction from Receptor Kinases to Transcription Factors. Annu. Rev. Plant Biol. 2010;61:681–704. doi: 10.1146/annurev.arplant.043008.092057. [DOI] [PubMed] [Google Scholar]
- 23.He J.X., Gendron J.M., Sun Y., Gampala S.S.L., Gendron N., Sun C.Q., Wang Z.Y. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z.Y., Nakano T., Gendron J., He J.X., Chen M., Vafeados D., Yang Y.L., Fujioka S., Yoshida S., Asami T., et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell. 2002;2:505–513. doi: 10.1016/S1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- 25.Yin Y.H., Wang Z.Y., Mora-Garcia S., Li J.M., Yoshida S., Asami T., Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/S0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 26.Gampala S.S., Kim T.W., He J.X., Tang W.Q., Deng Z.P., Bai M.Y., Guan S.H., Lalonde S., Sun Y., Gendron J.M., et al. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell. 2007;13:177–189. doi: 10.1016/j.devcel.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun Y., Fan X.Y., Cao D.M., Tang W.Q., He K., Zhu J.Y., He J.X., Bai M.Y., Zhu S.W., Oh E., et al. Integration of Brassinosteroid Signal Transduction with the Transcription Network for Plant Growth Regulation in Arabidopsis. Dev. Cell. 2010;19:765–777. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin Y.H., Vafeados D., Tao Y., Yoshida S., Asami T., Chory J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120:249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- 29.Kim T.W., Guan S.H., Sun Y., Deng Z.P., Tang W.Q., Shang J.X., Sun Y., Burlingame A.L., Wang Z.Y. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 2009;11:1254–1260. doi: 10.1038/ncb1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oh E., Zhu J.Y., Wang Z.Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 2012;14:802–809. doi: 10.1038/ncb2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oh E., Zhu J.Y., Ryu H., Hwang I., Wang Z.Y. TOPLESS mediates brassinosteroid-induced transcriptional repression through interaction with BZR1. Nat. Commun. 2014;5 doi: 10.1038/ncomms5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu X., Li L., Li L., Guo M., Chory J., Yin Y. Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2008;105:7618–7623. doi: 10.1073/pnas.0802254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryu H., Cho H., Bae W., Hwang I. Control of early seedling development by BES1/TPL/HDA19-mediated epigenetic regulation of ABI3. Nat. Commun. 2014;5 doi: 10.1038/ncomms5138. [DOI] [PubMed] [Google Scholar]
- 34.Liu L., Liu C., Hou X.L., Xi W.Y., Shen L.S., Tao Z., Wang Y., Yu H. FTIP1 Is an Essential Regulator Required for Florigen Transport. Plos. Biol. 2012;10 doi: 10.1371/journal.pbio.1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takada S., Goto K. Terminal flower2, an Arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by constans in the vascular tissues of leaves to regulate flowering time. Plant Cell. 2003;15:2856–2865. doi: 10.1105/tpc.016345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fowler S., Lee K., Onouchi H., Samach A., Richardson K., Coupland G., Putterill J. GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. Embo. J. 1999;18:4679–4688. doi: 10.1093/emboj/18.17.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park D.H., Somers D.E., Kim Y.S., Choy Y.H., Lim H.K., Soh M.S., Kim H.J., Kay S.A., Nam H.G. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science. 1999;285:1579–1582. doi: 10.1126/science.285.5433.1579. [DOI] [PubMed] [Google Scholar]
- 38.Putterill J., Robson F., Lee K., Simon R., Coupland G. The Constans Gene of Arabidopsis Promotes Flowering and Encodes a Protein Showing Similarities to Zinc-Finger Transcription Factors. Cell. 1995;80:847–857. doi: 10.1016/0092-8674(95)90288-0. [DOI] [PubMed] [Google Scholar]
- 39.Wigge P.A., Kim M.C., Jaeger K.E., Busch W., Schmid M., Lohmann J.U., Weigel D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309:1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- 40.Yamaguchi A., Kobayashi Y., Goto K., Abe M., Araki T. TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol. 2005;46:1175–1189. doi: 10.1093/pcp/pci151. [DOI] [PubMed] [Google Scholar]
- 41.Suarez-Lopez P., Wheatley K., Robson F., Onouchi H., Valverde F., Coupland G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410:1116–1120. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
- 42.Valverde F., Mouradov A., Soppe W., Ravenscroft D., Samach A., Coupland G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- 43.Yanovsky M.J., Kay S.A. Molecular basis of seasonal time measurement in Arabidopsis. Nature. 2002;419:308–312. doi: 10.1038/nature00996. [DOI] [PubMed] [Google Scholar]
- 44.Abe M., Kobayashi Y., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y., Ichinoki H., Notaguchi M., Goto K., Araki T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309:1052–1056. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- 45.Michaels S.D., Himelblau E., Kim S.Y., Schomburg F.M., Amasino R.M. Integration of flowering signals in winter-annual Arabidopsis. Plant Physiol. 2005;137:149–156. doi: 10.1104/pp.104.052811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoo S.K., Chung K.S., Kim J., Lee J.H., Hong S.M., Yoo S.J., Yoo S.Y., Lee J.S., Ahn J.H. CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol. 2005;139:770–778. doi: 10.1104/pp.105.066928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y.Z., Gu X.F., Yuan W.Y., Schmitz R.J., He Y.H. Photoperiodic Control of the Floral Transition through a Distinct Polycomb Repressive Complex. Dev. Cell. 2014;28:727–736. doi: 10.1016/j.devcel.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 48.Amasino R. Seasonal and developmental timing of flowering. Plant J. 2010;61:1001–1013. doi: 10.1111/j.1365-313X.2010.04148.x. [DOI] [PubMed] [Google Scholar]
- 49.Choi K., Kim J., Hwang H.J., Kim S., Park C., Kim S.Y., Lee I. The FRIGIDA Complex Activates Transcription of FLC, a Strong Flowering Repressor in Arabidopsis, by Recruiting Chromatin Modification Factors. Plant Cell. 2011;23:289–303. doi: 10.1105/tpc.110.075911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Michaels S.D., Bezerra I.C., Amasino R.M. FRIGIDA-related genes are required for the winter-annual habit in Arabidopsis. P. Natl. Acad. Sci. USA. 2004;101:3281–3285. doi: 10.1073/pnas.0306778101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Z.C., Jiang D.H., He Y.H. FRIGIDA establishes a local chromosomal environment for FLOWERING LOCUS C mRNA production. Nat. Plants. 2018;4:836–846. doi: 10.1038/s41477-018-0250-6. [DOI] [PubMed] [Google Scholar]
- 52.Aida M., Ishida T., Fukaki H., Fujisawa H., Tasaka M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell. 1997;9:841–857. doi: 10.1105/tpc.9.6.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vroemen C.W., Mordhorst A.P., Albrecht C., Kwaaitaal M.A.C.J., de Vries S.C. The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell. 2003;15:1563–1577. doi: 10.1105/tpc.012203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aida M., Tasaka M. Morphogenesis and patterning at the organ boundaries in the higher plant shoot apex u. Plant Mol. Biol. 2006;60:915–928. doi: 10.1007/s11103-005-2760-7. [DOI] [PubMed] [Google Scholar]
- 55.Baker C.C., Sieber P., Wellmer F., Meyerowitz E.M. The early extra petals1 mutant uncovers a role for MicroRNA miR164c in regulating petal number in Arabidopsis. Curr. Biol. 2005;15:303–315. doi: 10.1016/j.cub.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 56.Coen E.S., Meyerowitz E.M. The war of the whorls: genetic interactions controlling flower development. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- 57.Lohmann J.U., Hong R.L., Hobe M., Busch M.A., Parcy F., Simon R., Weigel D. A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell. 2001;105:793–803. doi: 10.1016/S0092-8674(01)00384-1. [DOI] [PubMed] [Google Scholar]
- 58.Lenhard M., Bohnert A., Jurgens G., Laux T. Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell. 2001;105:805–814. doi: 10.1016/S0092-8674(01)00390-7. [DOI] [PubMed] [Google Scholar]
- 59.Ito T., Wellmer F., Yu H., Das P., Ito N., Alves-Ferreira M., Riechmann J.L., Meyerowitz E.M. The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature. 2004;430:356–360. doi: 10.1038/nature02733. [DOI] [PubMed] [Google Scholar]
- 60.Liu X.G., Kim Y.J., Muller R., Yumul R.E., Liu C.Y., Pan Y.Y., Cao X.F., Goodrich J., Chena X.M. AGAMOUS Terminates Floral Stem Cell Maintenance in Arabidopsis by Directly Repressing WUSCHEL through Recruitment of Polycomb Group Proteins. Plant Cell. 2011;23:3654–3670. doi: 10.1105/tpc.111.091538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun B., Looi L.S., Guo S.Y., He Z.M., Gan E.S., Huang J.B., Xu Y.F., Wee W.Y., Ito T. Timing Mechanism Dependent on Cell Division Is Invoked by Polycomb Eviction in Plant Stem Cells. Science. 2014;343 doi: 10.1126/science.1248559. [DOI] [PubMed] [Google Scholar]
- 62.Schiefthaler U., Balasubramanian S., Sieber P., Chevalier D., Wisman E., Schneitz K. Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 1999;96:11664–11669. doi: 10.1073/pnas.96.20.11664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang W.C., Ye D., Xu J., Sundaresan V. The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 1999;13:2108–2117. doi: 10.1101/gad.13.16.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sanders P.M., Bui A.Q., Weterings K., McIntire K.N., Hsu Y.C., Lee P.Y., Truong M.T., Beals T.P., Goldberg R.B. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 1999;11:297–322. doi: 10.1007/s004970050158. [DOI] [Google Scholar]
- 65.Scott R.J., Spielman M., Dickinson H.G. Stamen structure and function. Plant Cell. 2004;16:S46–S60. doi: 10.1105/tpc.017012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parish R.W., Li S.F. Death of a tapetum: A programme of developmental altruism. Plant Sci. 2010;178:73–89. doi: 10.1016/j.plantsci.2009.11.001. [DOI] [Google Scholar]
- 67.Clouse S.D. The molecular intersection of brassinosteroid-regulated growth and flowering in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2008;105:7345–7346. doi: 10.1073/pnas.0803552105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao B., Li J. Regulation of brassinosteroid biosynthesis and inactivation. J. Integr. Plant Biol. 2012;54:746–759. doi: 10.1111/j.1744-7909.2012.01168.x. [DOI] [PubMed] [Google Scholar]
- 69.Koornneef M., Hanhart C.J., Vanderveen J.H. A Genetic and Physiological Analysis of Late Flowering Mutants in Arabidopsis-Thaliana. Mol. Gen. Genet. 1991;229:57–66. doi: 10.1007/BF00264213. [DOI] [PubMed] [Google Scholar]
- 70.Li Z.C., Ou Y., Zhang Z.C., Li J.M., He Y.H. Brassinosteroid Signaling Recruits Histone 3 Lysine-27 Demethylation Activity to FLOWERING LOCUS C Chromatin to Inhibit the Floral Transition in Arabidopsis. Mol. Plant. 2018;11:1135–1146. doi: 10.1016/j.molp.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 71.Janeczko A., Filek W., Biesaga-Koscielniak J., Marcinska I., Janeczko Z. The influence of animal sex hormones on the induction of flowering in Arabidopsis thaliana: comparison with the effect of 24-epibrassinolide. Plant Cell Tiss. Org. Cul. 2003;72:147–151. doi: 10.1023/A:1022291718398. [DOI] [Google Scholar]
- 72.Noh B., Lee S.H., Kim H.J., Yi G., Shin E.A., Lee M., Jung K.J., Doyle M.R., Amasino R.M., Noh Y.S. Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell. 2004;16:2601–2613. doi: 10.1105/tpc.104.025353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Questa J.I., Song J., Geraldo N., An H.L., Dean C. Arabidopsis transcriptional repressor VAL1 triggers Polycomb silencing at FLC during vernalization. Science. 2016;353:485–488. doi: 10.1126/science.aaf7354. [DOI] [PubMed] [Google Scholar]
- 74.Yuan W.Y., Luo X., Li Z.C., Yang W.N., Wang Y.Z., Liu R., Du J.M., He Y.H. A cis cold memory element and a trans epigenome reader mediate Polycomb silencing of FLC by vernalization in Arabidopsis. Nat. Genet. 2016;48:1527–1534. doi: 10.1038/ng.3712. [DOI] [PubMed] [Google Scholar]
- 75.Zhang Y.Y., Li B.B., Xu Y.Y., Li H., Li S.S., Zhang D.J., Mao Z.W., Guo S.Y., Yang C.H., Weng Y.X., et al. The Cyclophilin CYP20-2 Modulates the Conformation of BRASSINAZOLE-RESISTANT1, Which Binds the Promoter of FLOWERING LOCUS D to Regulate Flowering in Arabidopsis. Plant Cell. 2013;25:2504–2521. doi: 10.1105/tpc.113.110296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.He Y.H., Michaels S.D., Amasino R.M. Regulation of flowering time by histone acetylation in Arabidopsis. Science. 2003;302:1751–1754. doi: 10.1126/science.1091109. [DOI] [PubMed] [Google Scholar]
- 77.Liu F.Q., Quesada V., Crevillen P., Baurle I., Swiezewski S., Dean C. The Arabidopsis RNA-Binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Mol. Cell. 2007;28:398–407. doi: 10.1016/j.molcel.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 78.Jiang J.J., Zhang C., Wang X.L. A Recently Evolved Isoform of the Transcription Factor BES1 Promotes Brassinosteroid Signaling and Development in Arabidopsis thaliana. Plant Cell. 2015;27:361–374. doi: 10.1105/tpc.114.133678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang F., Gao Y.S., Liu Y.W., Zhang X., Gu X.X., Ma D.B., Zhao Z.W., Yuan Z.J., Xue H.W., Liu H.T. BES1-regulated BEE1 controls photoperiodic flowering downstream of blue light signaling pathway in Arabidopsis. New Phytol. 2019;223:1407–1419. doi: 10.1111/nph.15866. [DOI] [PubMed] [Google Scholar]
- 80.Friedrichsen D.M., Nemhauser J., Muramitsu T., Maloof J.N., Alonso J., Ecker J.R., Furuya M., Chory J. Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics. 2002;162:1445–1456. doi: 10.1093/genetics/162.3.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sandhu K.S., Hagely K., Neff M.M. Genetic Interactions Between Brassinosteroid-Inactivating P450s and Photomorphogenic Photoreceptors in Arabidopsis thaliana. G3 (Bethesda) 2012;2:1585–1593. doi: 10.1534/g3.112.004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Turk E.M., Fujioka S., Seto H., Shimada Y., Takatsuto S., Yoshida S., Wang H.C., Torres Q.I., Ward J.M., Murthy G., et al. BAS1 and SOB7 act redundantly to modulate Arabidopsis photomorphogenesis via unique brassinosteroid inactivation mechanisms. Plant J. 2005;42:23–34. doi: 10.1111/j.1365-313X.2005.02358.x. [DOI] [PubMed] [Google Scholar]
- 83.Gendron J.M., Liu J.S., Fan M., Bai M.Y., Wenkel S., Springer P.S., Barton M.K., Wang Z.Y. Brassinosteroids regulate organ boundary formation in the shoot apical meristem of Arabidopsis. Proc. Natl. Acad. Sci. USA. 2012;109:21152–21157. doi: 10.1073/pnas.1210799110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Espinosa-Ruiz A., Martinez C., de Lucas M., Fabregas N., Bosch N., Cano-Delgado A.I., Prat S. TOPLESS mediates brassinosteroid control of shoot boundaries and root meristem development in Arabidopsis thaliana. Development. 2017;144:1619–1628. doi: 10.1242/dev.143214. [DOI] [PubMed] [Google Scholar]
- 85.Rast M.I., Simon R. The meristem-to-organ boundary: more than an extremity of anything. Curr Opin Genet. Dev. 2008;18:287–294. doi: 10.1016/j.gde.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 86.Bell E.M., Lin W.C., Husbands A.Y., Yu L.F., Jaganatha V., Jablonska B., Mangeon A., Neff M.M., Girke T., Springer P.S. Arabidopsis LATERAL ORGAN BOUNDARIES negatively regulates brassinosteroid accumulation to limit growth in organ boundaries. Proc. Natl. Acad. Sci. USA. 2012;109:21146–21151. doi: 10.1073/pnas.1210789109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Neff M.M., Nguyen S.M., Malancharuvil E.J., Fujioka S., Noguchi T., Seto H., Tsubuki M., Honda T., Takatsuto S., Yoshida S., et al. BAS1: A gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc. Natl. Acad. Sci. USA. 1999;96:15316–15323. doi: 10.1073/pnas.96.26.15316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Turk E.M., Fujioka S., Seto H., Shimada Y., Takatsuto S., Yoshida S., Denzel M.A., Torres Q.I., Neff M.M. CYP72B1 inactivates brassinosteroid hormones: An intersection between photomorphogenesis and plant steroid signal transduction. Plant Physiol. 2003;133:1643–1653. doi: 10.1104/pp.103.030882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hibara K., Karim M.R., Takada S., Taoka K.I., Furutani M., Aida M., Tasaka M. Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell. 2006;18:2946–2957. doi: 10.1105/tpc.106.045716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee D.K., Geisler M., Springer P.S. LATERAL ORGAN FUSION1 and LATERAL ORGAN FUSION2 function in lateral organ separation and axillary meristem formation in Arabidopsis. Development. 2009;136:2423–2432. doi: 10.1242/dev.031971. [DOI] [PubMed] [Google Scholar]
- 91.Ye Q.Q., Zhu W.J., Li L., Zhang S.S., Yin Y.H., Ma H., Wang X.L. Brassinosteroids control male fertility by regulating the expression of key genes involved in Arabidopsis anther and pollen development. Proc. Natl. Acad. Sci. USA. 2010;107:6100–6105. doi: 10.1073/pnas.0912333107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bouquin T., Meier C., Foster R., Nielsen M.E., Mundy J. Control of specific gene expression by gibberellin and brassinosteroid. Plant Physiol. 2001;127:450–458. doi: 10.1104/pp.010173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li J.M., Nam K.H., Vafeados D., Chory J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 2001;127:14–22. doi: 10.1104/pp.127.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dong X.Y., Hong Z.L., Sivaramakrishnan M., Mahfouz M., Verma D.P.S. Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. Plant J. 2005;42:315–328. doi: 10.1111/j.1365-313X.2005.02379.x. [DOI] [PubMed] [Google Scholar]
- 95.Huang M.D., Chen T.L.L., Huang A.H.C. Abundant Type III Lipid Transfer Proteins in Arabidopsis Tapetum Are Secreted to the Locule and Become a Constituent of the Pollen Exine. Plant Physiol. 2013;163:1218–1229. doi: 10.1104/pp.113.225706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhao D.Z., Wang G.F., Speal B., Ma H. The EXCESS MICROSPOROCYTES1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Gene Dev. 2002;16:2021–2031. doi: 10.1101/gad.997902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang S.L., Xiea L.F., Mao H.Z., Puah C.S., Yang W.C., Jiang L.X., Sundaresan V., Ye D. TAPETUM DETERMINANT1 is required for cell specialization in the Arabidopsis anther. Plant Cell. 2003;15:2792–2804. doi: 10.1105/tpc.016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Albrecht C., Russinova E., Hecht V., Baaijens E., de Vries S. The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis. Plant Cell. 2005;17:3337–3349. doi: 10.1105/tpc.105.036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Colcombet J., Boisson-Dernier A., Ros-Palau R., Vera C.E., Schroeder J.I. Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASES1 and 2 are essential for tapetum development and microspore maturation. Plant Cell. 2005;17:3350–3361. doi: 10.1105/tpc.105.036731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li Z.Y., Wang Y., Huang J., Ahsan N., Biener G., Paprocki J., Thelen J.J., Raicu V., Zhao D.Z. Two SERK Receptor-Like Kinases Interact with EMS1 to Control Anther Cell Fate Determination. Plant Physiol. 2017;173:326–337. doi: 10.1104/pp.16.01219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zheng B., Bai Q., Wu L., Liu H., Liu Y., Xu W., Li G., Ren H., She X., Wu G. EMS1 and BRI1 control separate biological processes via extracellular domain diversity and intracellular domain conservation. Nat. Commun. 2019;10:4165. doi: 10.1038/s41467-019-12112-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen W., Lv M., Wang Y., Wang P.A., Cui Y., Li M., Wang R., Gou X., Li J. BES1 is activated by EMS1-TPD1-SERK1/2-mediated signaling to control tapetum development in Arabidopsis thaliana. Nat. Commun. 2019;10:4164. doi: 10.1038/s41467-019-12118-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wijeratne A.J., Zhang W., Sun Y.J., Liu W.L., Albert R., Zheng Z.Q., Oppenheimer D.G., Zhao D.Z., Ma H. Differential gene expression in Arabidopsis wild-type and mutant anthers: insights into anther cell differentiation and regulatory networks. Plant J. 2007;52:14–29. doi: 10.1111/j.1365-313X.2007.03217.x. [DOI] [PubMed] [Google Scholar]
- 104.Chen L.G., Gao Z., Zhao Z., Liu X., Li Y., Zhang Y., Liu X., Sun Y., Tang W. BZR1 Family Transcription Factors Function Redundantly and Indispensably in BR Signaling but Exhibit BRI1-Independent Function in Regulating Anther Development in Arabidopsis. Mol. Plant. 2019 doi: 10.1016/j.molp.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 105.Du J.B., Yin H.J., Zhang S.S., Wei Z.Y., Zhao B.L., Zhang J.H., Gou X.P., Lin H.H., Li J. Somatic Embryogenesis Receptor Kinases Control Root Development Mainly via Brassinosteroid-Independent Actions in Arabidopsis thaliana. J. Integr. Plant Biol. 2012;54:388–399. doi: 10.1111/j.1744-7909.2012.01124.x. [DOI] [PubMed] [Google Scholar]
- 106.Zhang W., Sun Y.J., Timofejeva L., Chen C.B., Grossniklaus U., Ma H. Regulation of Arabidopsis tapetum development and function by dysfunctional tapetum1 (dyt1) encoding a putative bHLH transcription factor. Development. 2006;133:3085–3095. doi: 10.1242/dev.02463. [DOI] [PubMed] [Google Scholar]
- 107.Rubinelli P., Hu Y., Ma H. Identification, sequence analysis and expression studies of novel anther-specific genes of Arabidopsis thaliana. Plant Mol. Biol. 1998;37:607–619. doi: 10.1023/A:1005964431302. [DOI] [PubMed] [Google Scholar]
- 108.Xing S., Quodt V., Chandler J., Hohmann S., Berndtgen R., Huijser P. SPL8 Acts Together with the Brassinosteroid-Signaling Component BIM1 in Controlling Arabidopsis thaliana Male Fertility. Plants (Basel) 2013;2:416–428. doi: 10.3390/plants2030416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Unte U.S., Sorensen A.M., Pesaresi P., Gandikota M., Leister D., Saedler H., Huijser P. SPL8, an SBP-Box gene that affects pollen sac development in Arabidopsis. Plant Cell. 2003;15:1009–1019. doi: 10.1105/tpc.010678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chandler J.W., Cole M., Flier A., Werr W. BIM1, a bHLH protein involved in brassinosteroid signalling, controls Arabidopsis embryonic patterning via interaction with DORNROSCHEN and DORNROSCHEN-LIKE. Plant Mol. Biol. 2009;69:57–68. doi: 10.1007/s11103-008-9405-6. [DOI] [PubMed] [Google Scholar]
- 111.Janeczko A., Oklestkova J., Novak O., Sniegowska-Swierk K., Snaczke Z., Pociecha E. Disturbances in production of progesterone and their implications in plant studies. Steroids. 2015;96:153–163. doi: 10.1016/j.steroids.2015.01.025. [DOI] [PubMed] [Google Scholar]
- 112.Kęsy J., Trzaskalska A., Galoch E., Kopcewicz J. Inhibitory Effect of Brassinosteroids on the Flowering of the Short-Day Plant Pharbitis nil. Biol. Plant. 2003;47:597–600. doi: 10.1023/B:BIOP.0000041069.27805.89. [DOI] [Google Scholar]
- 113.Makarevitch I., Thompson A., Muehlbauer G.J., Springer N.M. Brd1 gene in maize encodes a brassinosteroid C-6 oxidase. PLoS One. 2012;7:e30798. doi: 10.1371/journal.pone.0030798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hartwig T., Chuck G.S., Fujioka S., Klempien A., Weizbauer R., Potluri D.P., Choe S., Johal G.S., Schulz B. Brassinosteroid control of sex determination in maize. Proc. Natl. Acad. Sci USA. 2011;108:19814–19819. doi: 10.1073/pnas.1108359108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Papadopoulou E., Grumet R. Brassinosteriod-induced femaleness in cucumber and relationship to ethylene production. Hortscience. 2005;40:1763–1767. doi: 10.21273/HORTSCI.40.6.1763. [DOI] [Google Scholar]
- 116.Symons G.M., Reid J.B. Brassinosteroids do not undergo long-distance transport in pea. Implications for the regulation of endogenous brassinosteroid levels. Plant Physiol. 2004;135:2196–2206. doi: 10.1104/pp.104.043034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Deal R.B., Henikoff S. The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat. Protoc. 2011;6:56–68. doi: 10.1038/nprot.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Skene P.J., Henikoff S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife. 2017;6 doi: 10.7554/eLife.21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hwang B., Lee J.H., Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp. Mol. Med. 2018;50:1–14. doi: 10.1038/s12276-018-0071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]