Abstract
Over the past decade, studies on microRNA (miRNA) and cancer quickly became known. miRNAs are small non-coding RNAs that play a vital role in regulation of gene expression. In the present study, the expression of miR-27b, miR-29a, and miR-155, their prognostic roles, and their potential targets in chronic lymphocytic leukemia (CLL) and breast cancer (BC) by qRT-PCR were investigated. In two case-control studies, qRT-PCR was used to analyze the peripheral blood serum of 15 CLL patients and tissue samples of 15 BC patients for the expression of miR-27b, miR-29a, and miR-155. miRNA expression levels were calculated using the qRT-PCR method. The results revealed a significant increase in the expression of all miRNAs in patients with BC and CLL compared with respective healthy groups (p < 0.001). In BC patients, there was a significant difference between the expression of miR-155 and miR-29a (p < 0.05), miR-155 and miR-27b (p < 0.01), and miR-27b and miR-29a (p < 0.001). In CLL patients, a significant difference between expression of both miR-27b and miR-29a compared with expression of miR-155 (p < 0.001) was found. Furthermore, a significant association between miR-155 and prevascular invasion was found. Significantly, elevated circulating miRNAs were shown to be BC specific and could differentiate BC tissues from the controls. It was demonstrated that miRNAs used in this study and their expression profiles can be developed as biomarkers for early diagnosis and prognosis of CLL and BC. Further studies utilizing a larger test group of patients would provide identification of miRNAs as key players in intercellular interactions.
Keywords: CLL, breast cancer, miRNAs, biomarker, qRT-PCR
Introduction
Chronic lymphocytic leukemia (CLL) is known as one of the most common leukemias in elderly people and is characterized by the accumulation of mature leukemic B lymphocytes in peripheral blood, lymphatic organs, and bone marrow. Leukemic cells are resistant to apoptosis, both spontaneous and induced with anti-leukemic drugs.1 The heterozygous or homozygous deletion of the chromosomal region 13q14.3 occurs in more than 50% of CLL cases and is associated with an indolent clinical course.2 Breast cancer (BC) is the leading cause of cancer death in women in less-developed countries and the second most common cause of cancer death in women in developed countries. With early detection and significant advances in treatment, death rates from BC have been decreasing in some countries but still are rising in many countries.3
MicroRNAs (miRNAs) are large subgroups of non-coding RNAs (19–24 nt), which are evolutionally protected. These molecules control the expression of a gene after transcription by inhibiting the translocation of mRNA or inducing its degradation by connecting the intransitive region of the end of the miRNAs.4 miRNAs can act as a tumor suppressor by inhibiting the expression of cancer-related target genes and can be used as a tool to identify and diagnose cancer. The expression of miRNA is associated with biological and clinical features of the tumor, such as the tissue type, differentiation, invasion, and response to the treatment.5 The use of miRNAs as diagnostic markers is possible through human serum or human plasma testing; therefore, it is possible to detect cancerous miRNAs and tumor cells in serum or plasma non-invasively.6 miRNAs that are closely associated with malignant phenotypes can be used as diagnostic markers for a disease in its early stages. The main mechanism of the epigenetic changes in cancer cells is the expression of a noncoding gene, which is recognized by the abnormal levels of adult miRNAs.7 Therefore, cancerous miRNAs can be used as biological markers for diagnosis, prediction, and treatment.8
miRNAs are involved in multiple biological processes, including differentiation, proliferation, apoptosis, and stress response through regulation of gene expression. Up to 30% of human genes are regulated by these small non-coding RNAs. miRNAs are largely conserved between species and in blood or tissue samples; they are more resistant to degradation than mRNAs. More than 1,000 miRNAs are known in human at present.9 These molecules are responsible for the regulation of various physiological processes, and alterations in their expression levels can reflect different pathological conditions. Moreover, miRNAs are stable and can be quantified using different molecular techniques,10,11 such as qRT-PCR.12
Cancer is often heterogeneous in both its clinical course of the patient and its presentation. In the 1990s, based on RNA extracted from a fresh tumor tissue, gene expression profiles (GEPs) were introduced to segregate or classify tumors with similar phenotypes or morphologies. Because several thousand genes are involved in oncogenic transformation, these profiles have helped in recognition of pathways involved in oncogenic transformation. However, only a few selected cases have been helpful in clinical diagnosis. The discovery of miRNAs has made a major impact on cell biology recently.2 Recently, miRNAs have been recognized as factors that affect the clinical course of the CLL and may contribute to the leukemogenesis.13,14 Historically, CLL is reported as the first among human cancers discovered to clearly associate with alterations of miRNAs expression.1
Some miRNAs, including miR-27b, miR-29a, and miR-155, play important roles in different types of cancer. Tests based on miRNAs may improve the classification of BC and the prediction of treatment responses. Thus far, no miRNA profile has been tested in larger patient groups or randomized studies. In 2005, the first comprehensive study of miRNA profiles of BC was published.15 The authors showed that miRNAs were expressed aberrantly in BC. They could clearly discriminate the normal breast tissue from the BC tissue using miRNA profiles. Some of the dysregulated miRNAs correlated with bio-pathological features, such as progesterone-receptor and estrogen-receptor expression, vascular invasion, proliferation, and tumor stage.2 Therefore, according to recent studies, miRNAs in BC are important for transmitting epithelial mesenchymal and chemical resistance, and could provide potential insights into medical treatments.16
We hypothesized that expression signatures of the miR-27b, miR-29a, and miR-155 miRNAs may be useful biomarkers for diagnosis, prognosis, or prediction of CLL and BC. The miRNA expression levels were evaluated by qRT-PCR.
Results
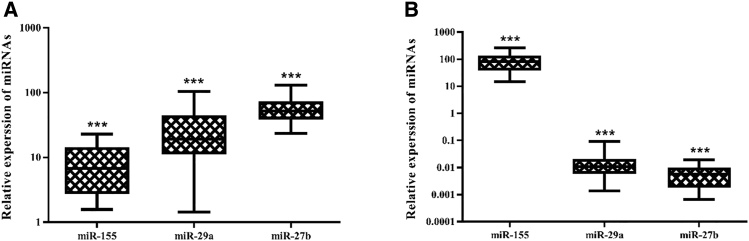
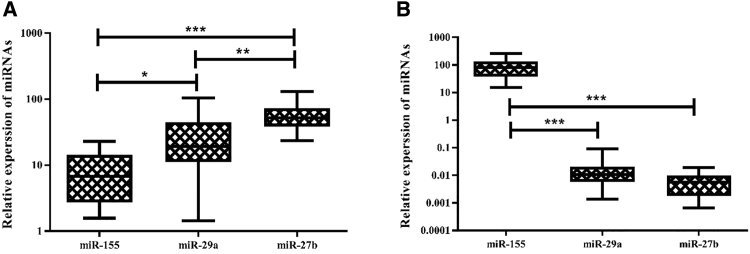
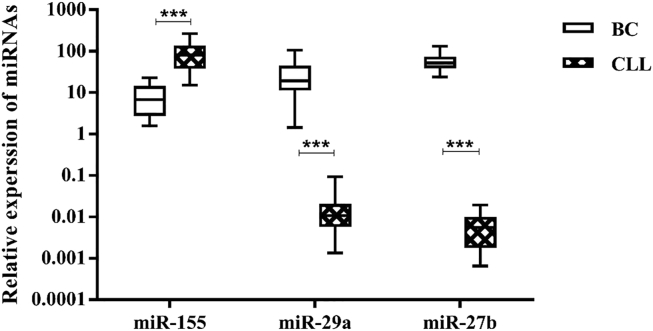
Expression of three miRNAs (miR-155, miR-29a, and miR-27b) was detectable in CLL and BC patients and not in healthy subjects. Results showed a significant increase in the expression of all miRNAs in patients with BC (Figure 1A) and CLL (Figure 1B) compared with their respective healthy controls (p < 0.001). Among patients with BC (Figure 2A), there was a significant difference between the expression of miR-155 and miR-29a (p < 0.05), miR-155 and miR-27b (p ≤ 0.01), and miR-27b and miR-29a (p < 0.001). miR-27b and miR-29a in patients with CLL (Figure 2B) were significantly different from miR-155 (p < 0.001). Comparing expression of miRNAs between BC and CLL patients (Figure 3), we found a significant difference for all miRNAs (p < 0.001).
Figure 1.
Relative Expression of the miRNAs in Patient Samples
(A and B) Relative expression of miRNAs in BC cases (A) and CLL cases (B). ***significant at the 0.001 level. Error bars show the minimum and maximum variables.
Figure 2.
Comparison of the Three miRNAs in Patient Samples
(A and B) Comparison of the three miRNAs in BC (A) and CLL (B). *significant at the 0.05 level, **significant at the 0.01 level, and ***significant at the 0.001 level. Error bars show the minimum and maximum variables.
Figure 3.
Comparison of the Three miRNAs in BC with CLL
***significant at the 0.001 level. Error bars show the minimum and maximum variables.
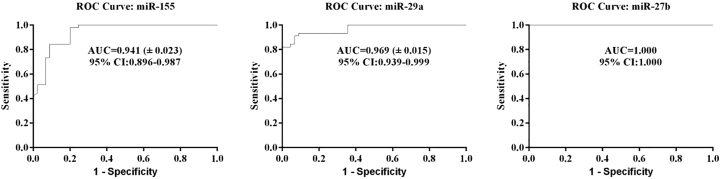
Receiver operating characteristic (ROC) analysis found the optimal cutoff value for the miRNAs to differentiate BC cases from the controls. Moreover, using ROC analysis, the sensitivities of circulating miR-155, miR-29a, and miR-27b were defined to be 84.4%, 93.3%, and 100% at the specificities of 91.1%, 91.1%, and 100% with an area under the ROC curve of 0.941, 0.969, and 1, respectively (Figure 4).
Figure 4.
Receiver Operating Characteristic (ROC) Curve Analyses of the Three miRNA Signature to Discriminate Breast Cancer Patients from Normal Controls
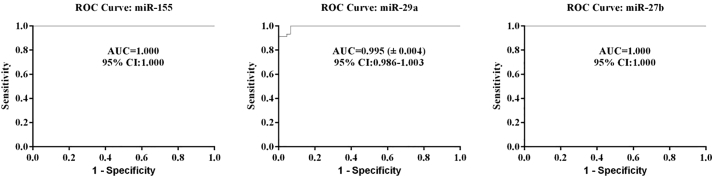
In Figure 5, ROC curves for miRNAs showed differentiation of the CLL cases from the controls. The sensitivities of circulating miR-155, miR-29a, and miR-27b were 100%, 91.1%, and 100% at the specificities of 100%, 100%, and 100% with an area under the ROC curve of 1, 0.995, and 1, respectively.
Figure 5.
ROC Curve Analyses of the Three miRNA Signature to Discriminate CLL Patients from Normal Controls
Correlations between the Expression of the miRNAs
Table 1 shows the correlations between miRNAs expression. There were no significant correlations between the miRNAs expression (p > 0.05).
Table 1.
Correlation between miRNAs Expression
| miR-155 |
miR-27b |
miR-29a |
||||
|---|---|---|---|---|---|---|
| Gene | Low | High | Low | High | Low | High |
| miR-155 | ||||||
| Low | 33.3 | 13.3 | 20 | 26.7 | ||
| High | 13.3 | 40 | 26.7 | 26.7 | ||
| p value | 0.072 | 0.782 | ||||
| miR-27b | ||||||
| Low | 33.3 | 13.3 | 13.3 | 33.3 | ||
| High | 13.3 | 40 | 33.3 | 20 | ||
| p value | 0.072 | 0.189 | ||||
| miR-29a | ||||||
| Low | 20 | 26.7 | 13.3 | 33.3 | ||
| High | 26.7 | 26.7 | 33.3 | 20 | ||
| p value | 0.782 | 0.189 | ||||
Correlations between miRNAs Expression and Clinical Parameters
Table 2 shows the correlations between miR-155 expression and clinical variables (age, tumor grade, nuclear grade, tumor stage, tumor size, area of invasive component, tumor side, margin, prevascular invasion, and preneural invasion). There was a significant correlation between miR-155 expression and prevascular invasion (p = 0.013), whereas no significant correlation between miR-155 expression and other clinical variables was found. Furthermore, Tables 3 and 4 indicate no significant correlations between miR-27b expression and clinical parameters and also between miR-29a expression and clinical variables (p > 0.05), respectively.
Table 2.
Correlation between miR-155 Expression and Clinical Parameters
| Variables | Cases (%) | miR-155 |
p Value |
|
|---|---|---|---|---|
| Low | High | |||
| Age, 46.80 ± 2.57 | 0.782 | |||
| ≤47 years | 53.3 | 26.7 | 26.7 | |
| >47 years | 46.7 | 20 | 26.7 | |
| Tumor Grade | 0.170 | |||
| I | 6.7 | 6.7 | 0 | |
| II | 66.7 | 20 | 46.7 | |
| III | 26.7 | 20 | 6.7 | |
| Nuclear Grade | 0.223 | |||
| Low | 7.1 | 7.1 | 0 | |
| High and intermediate | 28.6 | 21.4 | 7.1 | |
| High | 64.3 | 21.4 | 42.9 | |
| Tumor Stage | 0.098 | |||
| T1 | 40 | 33.3 | 6.7 | |
| T2 | 13.3 | 0 | 13.3 | |
| T3 | 40 | 13.3 | 26.7 | |
| T4 | 6.7 | 0 | 6.7 | |
| Tumor Size (cm) | 0.185 | |||
| ≤ 2 | 73.3 | 26.7 | 46.7 | |
| > 2 | 26.7 | 20 | 6.7 | |
| Area of Invasive Component, 4.09 ± 0.13 cm2 | 0.143 | |||
| ≤4 | 66.7 | 40 | 6.7 | |
| >4 | 33.3 | 26.7 | 26.7 | |
| Tumor Side | 0.438 | |||
| Right | 53.3 | 26.7 | 26.7 | |
| Left | 46.7 | 20 | 26.7 | |
| Margin | 0.876 | |||
| Free | 73.3 | 33.3 | 40 | |
| Involved | 26.7 | 13.3 | 13.3 | |
| Prevascular Invasion | 0.013* | |||
| Negative | 26.7 | 26.7 | 0 | |
| Positive | 73.3 | 20 | 53.3 | |
| Preneural Invasion | 0.438 | |||
| Negative | 20 | 13.3 | 6.7 | |
| Positive | 80 | 33.3 | 46.7 | |
*p < 0.05.
Table 3.
Correlation between miR-27b Expression and Clinical Parameters
| Variables | Cases (%) | miR-27b |
p Value |
|
|---|---|---|---|---|
| Low | High | |||
| Age, 46.80 ± 2.57 | 0.189 | |||
| ≤47 years | 53.3 | 33.3 | 20 | |
| >47 years | 46.7 | 13.3 | 33.3 | |
| Tumor Grade | 0.626 | |||
| I | 6.7 | 0 | 6.7 | |
| II | 66.7 | 33.3 | 33.3 | |
| III | 26.7 | 13.3 | 13.3 | |
| Nuclear Grade | 0.05 | |||
| Low | 7.1 | 0 | 7.1 | |
| High and intermediate | 28.6 | 28.6 | 0 | |
| High | 64.3 | 21.4 | 42.9 | |
| Tumor Stage | 0.400 | |||
| T1 | 40 | 20 | 20 | |
| T2 | 13.3 | 0 | 13.3 | |
| T3 | 40 | 20 | 20 | |
| T4 | 6.7 | 6.7 | 0 | |
| Tumor Size (cm) | 0.185 | |||
| ≤2 | 73.3 | 26.7 | 46.7 | |
| >2 | 26.7 | 20 | 6.7 | |
| Area of Invasive Component, 4.09 ± 0.13 cm2 | 0.464 | |||
| ≤4 | 66.7 | 26.7 | 40 | |
| >4 | 33.3 | 20 | 13.3 | |
| Tumor Side | 0.782 | |||
| Right | 53.3 | 26.7 | 26.7 | |
| Left | 46.7 | 20 | 26.7 | |
| Margin | 0.876 | |||
| Free | 73.3 | 33.3 | 40 | |
| Involved | 26.7 | 13.3 | 13.3 | |
| Prevascular Invasion | 0.876 | |||
| Negative | 26.7 | 13.3 | 13.3 | |
| Positive | 73.3 | 33.3 | 40 | |
| Preneural Invasion | 0.243 | |||
| Negative | 20 | 6.7 | 13.3 | |
| Positive | 80 | 40 | 40 | |
Table 4.
Correlation between miR-29a Expression and Clinical Parameters
| Variables | Cases (%) | miR-29a |
p Value |
|
|---|---|---|---|---|
| Low | High | |||
| Age, 46.80 ± 2.57 | 0.447 | |||
| ≤47 years | 53.3 | 20 | 33.3 | |
| >47 years | 46.7 | 26.7 | 20 | |
| Tumor Grade | 0.512 | |||
| I | 6.7 | 6.7 | 0 | |
| II | 66.7 | 26.7 | 40 | |
| III | 26.7 | 13.3 | 13.3 | |
| Nuclear Grade | 0.348 | |||
| Low | 7.1 | 7.1 | 0 | |
| High and intermediate | 28.6 | 7.1 | 21.4 | |
| High | 64.3 | 35.7 | 28.6 | |
| Tumor Stage | 0.517 | |||
| T1 | 40 | 26.7 | 13.3 | |
| T2 | 13.3 | 6.7 | 6.7 | |
| T3 | 40 | 13.3 | 26.7 | |
| T4 | 6.7 | 0 | 6.7 | |
| Tumor Size (cm) | 0.185 | |||
| ≤2 | 73.3 | 26.7 | 40.7 | |
| >2 | 26.7 | 20 | 6.7 | |
| Area of Invasive Component, 4.09 ± 0.13 cm2 | 0.143 | |||
| ≤4 | 66.7 | 40 | 26.7 | |
| >4 | 33.3 | 6.7 | 26.7 | |
| Tumor Side | 0.782 | |||
| Right | 53.3 | 26.7 | 26.7 | |
| Left | 46.7 | 20 | 26.7 | |
| Margin | 0.185 | |||
| Free | 73.3 | 26.7 | 46.7 | |
| Involved | 26.7 | 20 | 6.7 | |
| Prevascular Invasion | 0.310 | |||
| Negative | 26.7 | 6.7 | 20 | |
| Positive | 73.3 | 40 | 33.3 | |
| Preneural Invasion | 0.605 | |||
| Negative | 20 | 6.7 | 13.3 | |
| Positive | 80 | 40 | 40 | |
Correlation between miRNAs Expression and Survival in BC Patients
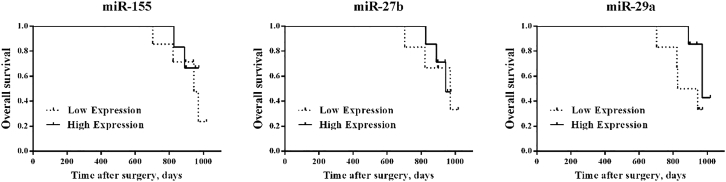
The log rank test was used in BC patients. We used the Cox proportional hazards regression model also to evaluate the predictive value of miRNAs levels in BC patients. The time between the date of surgery and date of death or last follow-up was defined as overall survival (OS). Clinicopathological factors and OS were then analyzed in the high- and low-miRNA expression groups, but no significant differences were found between groups (p > 0.05; Table 5; Figure 6).
Table 5.
Log Rank Test for All Patients Undergoing Breast Cancer
| Variables | Overall Survival |
||
|---|---|---|---|
| HR | 95% CI | p Value | |
| miR-155 (low versus high) | 1.681 | 0.3333–8.280 | 0.540 |
| miR-27b (low versus high) | 1.215 | 0.2458–6.126 | 0.807 |
| miR-29a (low versus high) | 2.484 | 0.5871–15.15 | 0.197 |
| Age (≤47 versus >47 years) | 1.749 | 0.3501–8.657 | 0.506 |
| Tumor grade (I-II versus III) | 0.560 | 0.07296–3.501 | 0.493 |
| Nuclear grade (low versus high and intermediate-high) | 1.977 | 0.1596–40.67 | 0.517 |
| Tumor stage (T1-T2 versus T3-T4) | 0.5445 | 0.1041–2.891 | 0.800 |
| Tumor size (<2 versus ≥2 cm) | 0.460 | 0.04656–2.841 | 0.347 |
| Area of invasive component (<4 versus ≥4 cm2) | 0.576 | 0.07625–3.827 | 0.378 |
| Tumor side (right versus left) | 0.585 | 0.1185–2.937 | 0.525 |
| Margin (free versus involved) | 0.507 | 0.05886–3.166 | 0.416 |
| Prevascular invasion (negative versus positive) | 1.605 | 0.3264–8.170 | 0.566 |
| Preneural invasion (negative versus positive) | 0.336 | 0.01890–1.809 | 0.166 |
CI, confidence interval; HR, hazard ratio.
Figure 6.
Kaplan-Meier Survival Curves for Associations of miRNAs with Survival
The miRNAs expression and overall survival (OS) in all patients: miRNAs low versus high, p > 0.05 (log rank test).
Discussion
In this study, we examined the expression profile of three miRNAs (miR-27b, miR-29a, and miR-155) in CLL and BC. The results revealed that miR-27b, miR-29a, and miR-155 were significantly upregulated in BC as compared with non-tumor tissues. On the other hand, miR-155 expression was also significantly increased in CLL patients compared with healthy ones, but miR-27b and miR-29a levels were downregulated in CLL.
miRNAs play a variety of roles in cancer initiation, tumor growth, and metastasis. Also, miRNAs may be valuable biomarkers for cancer diagnosis and therapy.17 The comparative study by Moussay et al.14 has shown that the expression of most circulating miRNAs in CLL patients reflects their expression in leukemic cells like in solid tumors. The level of miRNA expression in CLL, however, is decreased as compared with miRNA expression assessed in leukemic lymphocytes.18 miRNAs, such as miR-34a-5p, miR-31-5p, miR-155-5p, miR-15a-3p, and miR-29a-3p, were expressed at the highest level in CLL patients.1 In several studies, miRNA expression was shown to differentiate between CLL patients and healthy individuals, and they all have been corroborated as important for the CLL development and the clinical course of the disease.19, 20, 21, 22 In another study, Li et al.23 reported overexpression of miR-34a and miR-155 in CLL lymphocytes and defined it as a part of activated B cell phenotype. miR-155 expression in B lymphocytes was found to differentiate between normal individuals, individuals with CLL, and monoclonal B cell lymphocytosis (MBL); its overexpression in the latter group was associated with aggressive disease.24 Ferrajoli et al.25 demonstrated the presence of miR-155 in circulating micro-vesicles of individuals with CLL and MBL. A combination of nine miRNAs (expression profile miR-181 up, miR-155 up, miR-146 up, miR-24-2 up, miR-23b up, miR-23a up, miR-222 up, miR-221 up, and miR-29c down) correlated with a short interval to progression. In the same study, the germline or somatic mutations of some miRNA genes were found in 11 of 75 patients with CLL. This finding showed a genetic disposition for cancer in some patients with CLL.2
Several recent studies have demonstrated that the expression of miR-27a is upregulated in several types of solid tumors, including colon, gastric, cervical, and BCs.26 The widespread overexpression of miR-27a in cancer has led to the belief that miR-27a is an oncogenic miRNA.27 In the present study, miR-27a was upregulated in patients with metastases, suggesting that its upregulation was acquired in the course of tumor progression and during the acquisition of metastatic potential. Several studies have revealed the prognostic significance of miR-27a overexpression in various carcinomas, such as gastric cancer,28 acute lymphoblastic leukemia,29 and osteosarcoma.30 Tang et al.31 indicated that breast-invasive cancers with higher miR-27a expression tended to have distant metastasis, and the overexpression of miR-27a was associated with shorter disease-free survival and OS of BC patients. Both the univariate analyses and multivariate analyses indicated that miR-27a expression was an independent prognostic factor for BC progression.27 Anti-apoptosis functions of miR-29a in BC cells have been reported by Choghaei et al.32 These controversial results of miR-29a in tumors indicated that based on the cancer type, miR-29a played different roles. In our study, we found that miR-29a was significantly upregulated in BC as compared with normal tissues, indicating that miR-29a was implicated in the progression of BC.33
miR-155, involved in the progression of lymphoma,34 has more than 400 predicted gene targets, including over 100 confirmed ones. miR-155 is an oncogenic miRNA that has been explored mainly in leukemia but is also known to play an important role in the progression of BC.35 Several studies suggest that the miR-155 expression is upregulated in BC. miR-155 overexpression is associated with high mortality and tumor subtype.36 Iorio et al.15 indicated that out of 29 different miRNAs that were downregulated, just miR-155 was significantly upregulated in BC, and introduced miR-155 as an important player in BC. In another study, serum miR-155 was also significantly increased in BC patients compared with healthy donors.37 Another study indicated overexpression of miR-155 in BC and suggested its potential clinical prognostic value.36 Upregulation of miR-155 in BC has been shown to be significantly associated with advanced tumor stage, higher tumor grade, and lymph node metastasis, suggesting its potential as a clinical prognostic marker.38 As a limitation of our study, small sample size could be mentioned, which may reduce the accuracy of the results.
In conclusion, our results suggest that miR-27b, miR-29a, and miR-155 could be potential new biomarkers for diagnosis, as well as a therapeutic target for CLL and BC. Future studies including a large group of multi-ethnic patients are required to validate our findings.
Materials and Methods
Patients and Healthy Controls
A total of 15 CLL and 15 BC patients admitted to the Isfahan General Hospital were selected for the study. The study protocol included a standard oncological evaluation with subsequent review of histopathological data and outpatient follow-ups. Blood samples were collected from the CLL patients and tumor tissue samples from the BC patients. Healthy controls (n = 15) were recruited and were matched to the patients by age and gender. Informed consents were obtained. The time between the date of surgery and date of death or last follow-up was defined as OS.
PBMCs Isolation
Peripheral blood mononuclear cells (PBMCs) were separated by density gradient lymphosep (Bio Sera, Kansas City, USA) according to the manufacturer’s instructions. 4 mL of blood was diluted at a ratio of 1:1 with physiological saline and gradually added to the 4 mL Lymphoprep solution gradient in a Falcon tube and was centrifuged at 800 × g for 30 min at room temperature. Then, PBMCs were transferred from the middle phase into a 2-mL RNase-free micro tube. After washing and cell counting, PBMCs were sedimented using centrifugation for 10 min at 250 × g and were then frozen at −70°C until the RNA extraction stage.
RNA Extraction
Total RNA from PBMCs was extracted using miRNA Hybrid-R (GeneAll, Seoul, Korea) based on the manufacturer’s instructions. For BC samples, 100 mg tissue per extraction was homogenized in liquid nitrogen using a pestle and a mortar. Total RNA was extracted from tissue samples using the RNX-Plus solution (SinaClon, Iran) according to the manufacturer’s instructions. Quality and quantity of RNA were assessed by means of NanoDrop spectrometer (Thermo Scientific, Waltham, MA, USA). Samples containing at least 5 ng/μL of RNA, with A260/A280 ratio between 1.7 and 2.1, were used for further analysis. Isolated RNA was stored at −70°C.
cDNA Synthesis and Real-Time PCR
cDNA synthesis of miR-27b, miR-29a, and miR-155 was performed using a commercial kit (Pars Genome, Tehran, Iran) and according to the manufacturer’s instructions. Real-time quantitative PCRs were accomplished in triplicate. In brief, in a total volume of 10 μL, 20 ng/μL cDNA product was added to a master mix, including 10 pmol/μL of miR-27b, miR-29a, and miR-155 U6 (housekeeping) primers (Pars genome) and 5 mL of SYBR premix ExTaqII (TaKaRa, Kusatsu, Shiga Prefecture, Japan). The run program was set at 95°C for 5 min followed by 40 cycles of 95°C for 5 s, 58°C for 20 s, and 72°C for 30 s.
Statistical Analysis
Real-time PCR data analysis was performed using the 2−ΔΔCt method, and the final result was normalized by U6 (small nuclear RNA) as an endogenous control.39 Student’s t test was conducted to compare miRNAs expression in clinical samples. The ROC curves were used to estimate the diagnostic value of the identified miRNAs on the CLL and BC by calculation sensitivity and specificity for each possible cutoff point of the individual miRNAs. This was performed univariately for each individual miRNA. The patients were divided based on the median value of the miRNAs expression into two groups: cases with high miRNAs and cases with low miRNAs expression. The chi-square test and Fisher’s exact test were applied to evaluate correlations of miRNAs expressions and clinical parameters. We investigated the association between the expression levels of miRNAs with survival through Kaplan-Meier analysis in order to assess the prognostic value of miRNAs as a biomarker for BC. The log rank test was used in BC patients data. We also used the Cox proportional hazards regression model to evaluate the predictive value of miRNAs levels in BC patients. All statistical tests were performed using REST 2009 and Graph Pad Prism statistical software, version 7.00 (Graph Pad, San Diego, CA, USA). For all tests, a p value <0.05 was considered statistically significant.
Ethics Statement
All applicable international, national, and institutional guidelines for the care of humans were followed.
Author Contributions
F.R., E.M., M.D., S.S.E.H., A.M.G., A.A., K.F. and S.G. designed the study. F.R., E.M., M.D., S.S.E.H., A.M.G. and A.A. performed the experiments and analyzed the data. All authors critically reviewed the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
The authors would like to thank all the staff members of the Cellular and Molecular Research Center, Shahrekord University of Medical Sciences for their sincere support. Prof. Shanbeh Zienolddiny from the National Institute of Occupational Health in Norway is acknowledged for reading and commenting on the manuscript.
References
- 1.Filip A.A., Grenda A., Popek S., Koczkodaj D., Michalak-Wojnowska M., Budzyński M., Wąsik-Szczepanek E., Zmorzyński S., Karczmarczyk A., Giannopoulos K. Expression of circulating miRNAs associated with lymphocyte differentiation and activation in CLL-another piece in the puzzle. Ann. Hematol. 2017;96:33–50. doi: 10.1007/s00277-016-2840-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Munker R., Calin G.A. MicroRNA profiling in cancer. Clin. Sci. (Lond.) 2011;121:141–158. doi: 10.1042/CS20110005. [DOI] [PubMed] [Google Scholar]
- 3.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 4.Yauk C.L., Rowan-Carroll A., Stead J.D., Williams A. Cross-platform analysis of global microRNA expression technologies. BMC Genomics. 2010;11:330. doi: 10.1186/1471-2164-11-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krol J., Loedige I., Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 6.Emery B., Lu Q.R. Transcriptional and Epigenetic Regulation of Oligodendrocyte Development and Myelination in the Central Nervous System. Cold Spring Harb. Perspect. Biol. 2015;7:a020461. doi: 10.1101/cshperspect.a020461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan X., Shi L., Fang H., Cheng Y., Perkins R., Tong W. DNA microarrays are predictive of cancer prognosis: a re-evaluation. Clin. Cancer Res. 2010;16:629–636. doi: 10.1158/1078-0432.CCR-09-1815. [DOI] [PubMed] [Google Scholar]
- 8.Liu C.G., Spizzo R., Calin G.A., Croce C.M. Expression profiling of microRNA using oligo DNA arrays. Methods. 2008;44:22–30. doi: 10.1016/j.ymeth.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sato F., Tsuchiya S., Terasawa K., Tsujimoto G. Intra-platform repeatability and inter-platform comparability of microRNA microarray technology. PLoS ONE. 2009;4:e5540. doi: 10.1371/journal.pone.0005540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Planell-Saguer M., Rodicio M.C. Detection methods for microRNAs in clinic practice. Clin. Biochem. 2013;46:869–878. doi: 10.1016/j.clinbiochem.2013.02.017. [DOI] [PubMed] [Google Scholar]
- 12.Schmittgen T.D., Lee E.J., Jiang J., Sarkar A., Yang L., Elton T.S., Chen C. Real-time PCR quantification of precursor and mature microRNA. Methods. 2008;44:31–38. doi: 10.1016/j.ymeth.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bottoni A., Calin G.A. MicroRNAs as main players in the pathogenesis of chronic lymphocytic leukemia. MicroRNA. 2014;2:158–164. doi: 10.2174/2211536602666131126002337. [DOI] [PubMed] [Google Scholar]
- 14.Moussay E., Wang K., Cho J.H., van Moer K., Pierson S., Paggetti J., Nazarov P.V., Palissot V., Hood L.E., Berchem G., Galas D.J. MicroRNA as biomarkers and regulators in B-cell chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA. 2011;108:6573–6578. doi: 10.1073/pnas.1019557108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iorio M.V., Ferracin M., Liu C.G., Veronese A., Spizzo R., Sabbioni S., Magri E., Pedriali M., Fabbri M., Campiglio M. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 16.Schweiger M.R., Kerick M., Timmermann B., Isau M. The power of NGS technologies to delineate the genome organization in cancer: from mutations to structural variations and epigenetic alterations. Cancer Metastasis Rev. 2011;30:199–210. doi: 10.1007/s10555-011-9278-z. [DOI] [PubMed] [Google Scholar]
- 17.Kim J., Yao F., Xiao Z., Sun Y., Ma L. MicroRNAs and metastasis: small RNAs play big roles. Cancer Metastasis Rev. 2018;37:5–15. doi: 10.1007/s10555-017-9712-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ouillette P., Collins R., Shakhan S., Li J., Li C., Shedden K., Malek S.N. The prognostic significance of various 13q14 deletions in chronic lymphocytic leukemia. Clin. Cancer Res. 2011;17:6778–6790. doi: 10.1158/1078-0432.CCR-11-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danger R., Braza F., Giral M., Soulillou J.P., Brouard S. MicroRNAs, Major Players in B Cells Homeostasis and Function. Front. Immunol. 2014;5:98. doi: 10.3389/fimmu.2014.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawrie C.H. MicroRNAs and lymphomagenesis: a functional review. Br. J. Haematol. 2013;160:571–581. doi: 10.1111/bjh.12157. [DOI] [PubMed] [Google Scholar]
- 21.de Yébenes V.G., Bartolomé-Izquierdo N., Ramiro A.R. Regulation of B-cell development and function by microRNAs. Immunol. Rev. 2013;253:25–39. doi: 10.1111/imr.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mraz M., Kipps T.J. MicroRNAs and B cell receptor signaling in chronic lymphocytic leukemia. Leuk. Lymphoma. 2013;54:1836–1839. doi: 10.3109/10428194.2013.796055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li S., Moffett H.F., Lu J., Werner L., Zhang H., Ritz J., Neuberg D., Wucherpfennig K.W., Brown J.R., Novina C.D. MicroRNA expression profiling identifies activated B cell status in chronic lymphocytic leukemia cells. PLoS ONE. 2011;6:e16956. doi: 10.1371/journal.pone.0016956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui B., Chen L., Zhang S., Mraz M., Fecteau J.F., Yu J., Ghia E.M., Zhang L., Bao L., Rassenti L.Z. MicroRNA-155 influences B-cell receptor signaling and associates with aggressive disease in chronic lymphocytic leukemia. Blood. 2014;124:546–554. doi: 10.1182/blood-2014-03-559690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferrajoli A., Shanafelt T.D., Ivan C., Shimizu M., Rabe K.G., Nouraee N., Ikuo M., Ghosh A.K., Lerner S., Rassenti L.Z. Prognostic value of miR-155 in individuals with monoclonal B-cell lymphocytosis and patients with B chronic lymphocytic leukemia. Blood. 2013;122:1891–1899. doi: 10.1182/blood-2013-01-478222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu P.T., Wheelwright M., Teles R., Komisopoulou E., Edfeldt K., Ferguson B., Mehta M.D., Vazirnia A., Rea T.H., Sarno E.N. MicroRNA-21 targets the vitamin D-dependent antimicrobial pathway in leprosy. Nat. Med. 2012;18:267–273. doi: 10.1038/nm.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jorge K.T.O.S., Souza R.P., Assis M.T.A., Araújo M.G., Locati M., Jesus A.M.R., Dias Baptista I.M.F., Lima C.X., Teixeira A.L., Teixeira M.M., Soriani F.M. Characterization of MicroRNA Expression Profiles and Identification of Potential Biomarkers in Leprosy. J. Clin. Microbiol. 2017;55:1516–1525. doi: 10.1128/JCM.02408-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ajit S.K. Circulating microRNAs as biomarkers, therapeutic targets, and signaling molecules. Sensors (Basel) 2012;12:3359–3369. doi: 10.3390/s120303359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cambau E., Bonnafous P., Perani E., Sougakoff W., Ji B., Jarlier V. Molecular detection of rifampin and ofloxacin resistance for patients who experience relapse of multibacillary leprosy. Clin. Infect. Dis. 2002;34:39–45. doi: 10.1086/324623. [DOI] [PubMed] [Google Scholar]
- 30.Faruq O., Vecchione A. microRNA: Diagnostic Perspective. Front. Med. (Lausanne) 2015;2:51. doi: 10.3389/fmed.2015.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang W., Zhu J., Su S., Wu W., Liu Q., Su F., Yu F. MiR-27 as a prognostic marker for breast cancer progression and patient survival. PLoS One. 2012;7 doi: 10.1371/journal.pone.0051702. e51702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choghaei E., Khamisipour G., Falahati M., Naeimi B., Mossahebi-Mohammadi M., Tahmasebi R., Hasanpour M., Shamsian S., Hashemi Z.S. Knockdown of microRNA-29a Changes the Expression of Heat Shock Proteins in Breast Carcinoma MCF-7 Cells. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2016;23:69–78. doi: 10.3727/096504015X14478843952906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pei Y.F., Lei Y., Liu X.Q. MiR-29a promotes cell proliferation and EMT in breast cancer by targeting ten eleven translocation 1. Biochim. Biophys. Acta. 2016;1862:2177–2185. doi: 10.1016/j.bbadis.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 34.Clurman B.E., Hayward W.S. Multiple proto-oncogene activations in avian leukosis virus-induced lymphomas: evidence for stage-specific events. Mol. Cell. Biol. 1989;9:2657–2664. doi: 10.1128/mcb.9.6.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 36.Mattiske S., Suetani R.J., Neilsen P.M., Callen D.F. The oncogenic role of miR-155 in breast cancer. Cancer Epidemiol. Biomarkers Prev. 2012;21:1236–1243. doi: 10.1158/1055-9965.EPI-12-0173. [DOI] [PubMed] [Google Scholar]
- 37.Sun Y., Wang M., Lin G., Sun S., Li X., Qi J., Li J. Serum microRNA-155 as a potential biomarker to track disease in breast cancer. PLoS ONE. 2012;7:e47003. doi: 10.1371/journal.pone.0047003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cortez M.A., Welsh J.W., Calin G.A. Circulating MicroRNAs as Noninvasive Biomarkers in Breast Cancer. Recent Results Cancer Res. 2012;195:151–161. doi: 10.1007/978-3-642-28160-0_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]