Abstract
Squalene is a triterpene which is produced as a precursor for a wide range of terpenoid compounds in many organisms. It has commercial use in food and cosmetics but could also be used as a feedstock for production of chemicals and fuels, if generated sustainably on a large scale. We have engineered a cyanobacterium, Synechocystis sp. PCC 6803, for production of squalene from CO2. In this organism, squalene is produced via the methylerythritol-phosphate (MEP) pathway for terpenoid biosynthesis, and consumed by the enzyme squalene hopene cyclase (Shc) for generation of hopanoids. The gene encoding Shc in Synechocystis was inactivated (Δshc) by insertion of a gene encoding a squalene synthase from the green alga Botryococcus braunii, under control of an inducible promoter. We could demonstrate elevated squalene generation in cells where the algal enzyme was induced. Heterologous overexpression of genes upstream in the MEP pathway further enhanced the production of squalene, to a level three times higher than the Δshc background strain. During growth in flat panel bioreactors, a squalene titer of 5.1 mg/L of culture was reached.
Keywords: Cyanobacteria, Synechocystis, Metabolic engingeering, Terpenoid biosynthesis, Squalene
Highlights
-
•
Squalene synthase from Botryococcus braunii was introduced in Synechocystis.
-
•
Expression of the B.braunii enzyme resulted in enhanced production of squalene.
-
•
Expression of MEP pathway genes led to further increase in squalene accumulation.
-
•
Substrate was redirected from pigments towards squalene formation.
1. Introduction
Cyanobacteria are microorganisms capable of oxygenic photosynthesis, utilizing atmospheric CO2 as their carbon source and light as their source of energy. Biotechnological use of these organisms is attractive, as it enables sustainable production of solar fuels and renewable chemicals. In recent years, the tools and techniques available for genetic engineering and regulation of transgenic expression in cyanobacteria have developed rapidly (Vavitsas et al., 2019; Vijay et al., 2019). While the tools available, especially regarding control of gene expression, are not yet as versatile as those for Escherichia coli, it is now possible to do extensive engineering of several model strains of cyanobacteria. One of the most used strains is the unicellular cyanobacterium Synechocystis sp. PCC 6803 (hereafter Synechocystis), which has been the focus of a number of studies aiming at metabolic engineering for photosynthetic product generation (Liu et al., 2019; Knoot et al., 2018; Angermayr et al., 2015).
In an earlier study, we generated a strain of Synechocystis, in which inactivation of the gene encoding squalene hopene cyclase, Shc (Δshc), resulted in accumulation of the 30-carbon oil squalene in the cells (Englund et al., 2014). Squalene is a triperpenoid molecule which in eukaryotes serves as a precursor for formation of sterols (Do et al., 2009). In some bacteria, it is the precursor of hopanoids, complex pentacyclic triterpenoids with a role in membrane fluidity and stability (Siedenburg and Jendrossek, 2011; Kannenberg and Poralla, 1999). Squalene can be produced from animal or plant sources, and has commercial use in nutrition, vaccines and cosmetics. However, if produced sustainably in large amounts, it could serve as a raw material for production of renewable chemicals and fuels, as it can be cracked to form smaller molecules suitable for different applications (Tracy et al., 2011).
The substrates for formation of all terpenoid compounds in cyanobacteria are made via the methyl-erythritol phosphate (MEP) pathway ((Pattanaik and Lindberg, 2015) and references therein), which utilizes glyceraldehyde-3-phosphate (G3P) and pyruvate to form isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP). These two molecules are the building blocks for terpenes. Squalene is formed from IPP and DMAPP in several steps, combining them first into farnesyl diphosphate (FPP), a 15 carbon molecule. Two FPP units are then joined by the enzyme squalene synthase to form squalene (Fig. 1).
Fig. 1.
MEP pathway and formation of squalene in Synechocystis. Metabolites: G3P – glyceraldehyde 3-phosphate; DXP - deoxyxylulose 5-phosphate; MEP – methylerythritol 4-phosphate; IPP - isopentenyl diphosphate; DMAPP – dimethylallyl diphosphate; FPP - farnesyl diphosphate. Enzymes: Idi - isopentenyl diphosphate isomerase; CrtE - geranylgeranyl diphosphate synthase; Sqs - squalene synthase; Shc - squalene hopene cyclase; CfDXS - 1-deoxyxylulose-5-phosphate synthase from Coleus forskohlii; EcIspA – farnesyl diphosphate synthase from Escherichia coli; EcIdi - isopentenyl diphosphate isomerase from E. coli; BSS - Squalene synthase from Botryococcus braunii. Synechocystis native enzymes are marked in blue with gene names in parentheses, heterologously expressed enzymes in green. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The green alga Botryocuccus braunii has received attention due to its accumulation of hydrocarbon oils. Most of the oils produced by certain strains of Botryococcus are triterpenes, in the form of squalene and botryococcene (Okada et al., 1995), and squalene synthase from this organism may therefore be a promising enzyme to use for heterologous expression in another host strain for over-production of squalene. Several studies have addressed growth and harvest of Botryococcus oils, but such efforts are hampered by the inherent slow growth of the algae, and by the lack of genetic tools for this organism (Yoo et al., 2010; Brennan and Owende, 2010).
In the present study, we use the cyanobacterium Synechocystis for production of squalene, expressing a squalene synthase from Botryococcus braunii in combination with additional expression of genes from the MEP and terpenoid pathway in a Δshc background.
2. Materials and methods
2.1. Bacterial strains and growth conditions
Escherichia coli strain DH5α was used for subcloning according to standard procedures. E. coli cells were grown in LB medium with addition of appropriate antibiotics (50 μg/ml kanamycin (Km), 100 μg/ml Ampicillin (Amp) or 100 μg/ml erythromycin (Em) (Sigma-Aldrich) at 37 °C. Synechocystis cultures were grown at 30 °C under a light intensity of 20 μmol photons m-2 s-1 (except when otherwise noted) in BG11 medium (Stanier et al., 1971). Transformant strains were isolated and maintained by addition of appropriate antibiotics for selection (25 μg/ml Km, 25 μg/ml Em).
For squalene production experiments, liquid cyanobacterial cultures were grown in 30 ml of medium in 100 ml Erlenmeyer flasks on a rotary shaker under constant light. Fresh cultures were started at an OD750 of about 0.05. For induction of the PnrsB promoter, 2.5 μM NiCl2 (Merck) was added after two days at an OD750 of 0.1. For comparison of carbon sources, 0.5% (wt/vol) glucose or 50mM sodium bicarbonate (NaHCO3) with 20mM HEPES was added to the cultures. Four days after induction with Ni2+, OD750 was recorded and cell samples were harvested by centrifugation for squalene and pigment analysis.
For the bioreactor experiment, 350 ml cyanobacterial cultures were grown in parallel in two FMT150 photobioreactors (Photon Systems Instruments) at 30 °C, with mixing and aeration supplied by compressed air and a white light intensity of 30 μmol photons m-2 s-1. The reactors were inoculated at an OD750 of about 2.5 in fresh BG11 with 2.5 μM NiCl2, and samples for determination of OD750 and squalene accumulation were taken every 24 h for 4 days.
2.2. Generation of engineered Synechocystis strains
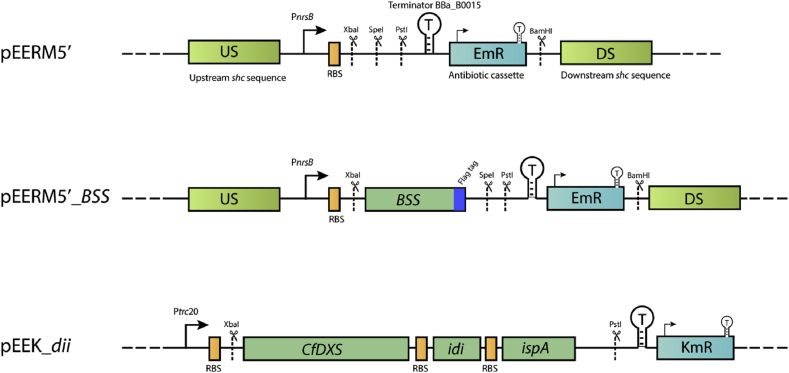
DNA amplification by PCR and subcloning was performed according to standard procedures. An enzyme from B. braunii, the Botryococcus squalene synthase (BSS), accession no Q9SDW9 (Okada et al., 2000) was selected for expression in Synechocystis. The amino acid sequence for this enzyme, after removal of a putative membrane anchoring domain (Okada et al., 2000), was back translated into a gene codon optimized for expression in Synechocystis, with a Flag-tag (Hopp et al., 1988) appended to the C-terminus of the gene for detection. The gene was generated by DNA synthesis (GenScript, Piscataway, NJ, USA) and cloned using XbaI and PstI into the vector pEERM5’. pEERM5′ provides the nrsB promoter, an RBS, and a terminator with standardized cloning sites (as described for the pEERM series of vectors in (Englund et al., 2015)), erythromycin resistance, and flanking sequences for homologous recombination into the shc site in the Synechocystis genome (Fig. 2, Table 1). The resulting pEERM5’_BSS vector and, as a control, the same vector lacking the BSS gene, were transferred to Synechocystis by conjugation (Heidorn et al., 2011) to generate the strains Δshc:BSS and Δshc:C, respectively. Transformant colonies were isolated by selection with erythromycin, and segregation of the insertions verified by PCR (data not shown).
Fig. 2.
Schematic overview of vectors used in the study. The fragment encoding Botryococcus squalene synthase (BSS) with Flag-tag was cloned in the control vector pEERM5′ to generate pEERM5’_BSS. CfDXS, idi and ispA were inserted as an operon into the self-replicating plasmid pEEK to construct pEEK_dii.
Table 1.
Plasmids and strains used in this study.
| Plasmid | Target location | Promoter | Expression of | Antibiotic resistance |
|---|---|---|---|---|
| pEERM5′ | shc | PnrsB | - | Em |
| pEERM5′BSS | shc | PnrsB | BSS_Flag | Em |
| pEEK | replicating plasmid | Ptrc2O | – | Km + Amp |
| pEEK_dii | replicating plasmid | Ptrc2O | CfDXS, idi, ispA | Km + Amp |
|
| ||||
|
Strains |
Genotype |
Antibiotic resistance |
||
| Synechcocystis WT | wild type Synechocystis PCC 6803 | none | ||
| Δshc:C | Δshc | Em | ||
| Δshc:BSS | Δshc::PnrsB_BSS_Flag | Em | ||
| Δshc:BSS_dii | Δshc:: PnrsB_BSS_Flag + pEEK_CfDXS_idi_ispA | Em + Km | ||
The construct for expression of genes encoding CfDXS (DXS from Plectranthus barbatus, or Coleus forskohlii), EcIdi (IPP isomerase from E. coli) and EcIspA (FPP synthase from E. coli) was based on the broad host range replicating plasmid pPMQAK1 (Huang et al., 2010). pPMQAK1 was modified to contain standardized cloning sites flanked by the promoter Ptrc2O (Huang et al., 2010), the ribosomal binding site RBS* (Heidorn et al., 2011) and terminator BBa_B0015 (http://parts.igem.org), forming the plasmid pEEK. CfDXS (Englund et al., 2015), Ecidi, and EcispA were cloned as an operon into the XbaI and PstI sites of this vector to generate pEEK_dii (Fig. 2). Ecidi and EcispA were cloned from the plasmid pMBIS (Martin et al., 2003). pMBIS was a gift from Dr. Jay D. Keasling obtained via Addgene (Addgene plasmid # 17817; http://n2t.net/addgene:17817; RRID:Addgene_17817). pEEK_dii was introduced into the Δshc:BSS strain by conjugation to generate Δshc:BSS_dii. Transformants were isolated by selection on plates containing kanamycin. See Table 1 for an overview of plasmids and strains. The complete sequence of all genetic constructs employed can be found in Table S1. Strains can be obtained on request to the corresponding author.
2.3. Determination of squalene
Cells were harvested from 10-20 ml of culture and squalene was extracted according to a protocol adapted from Schmerk et al (2011) (Schmerk et al., 2011; Englund et al., 2015). Pellets were resuspended in 5ml of 10% KOH (Merck) in methanol (w/v) (Merck) and vortexed repeatedly until completely suspended. The samples were then incubated at 65 °C for 1 h, followed by addition of 2 ml of heptane (Sigma-Aldrich) and mixing. The upper phase was transferred to a new tube and dried at room temperature under a stream of N2 before being redissolved in heptane and acetonitrile to a final ratio of 1:20 v/v. The solution was filtered through 0.2 mm PTFE syringe filters (VWR International, Radnor, PA, USA), and the squalene content was determined by HPLC with comparison to a squalene standard (Sigma-Aldrich) (Englund et al., 2014).
2.4. Determination of pigment content
Chlorophyll a and carotenoid contents in Synechocystis cells were determined using a method based on that of Chamovitz et al., (1993), as described (Englund et al., 2015). Briefly, cells were harvested by centrifugation and resuspended in 1 ml N,N-Dimethylformamide (Merck). Samples were incubated in darkness for 5 min, followed by centrifugation for 5 min at 17000 g. Pigment concentrations were calculated from the aborbance of the supernatant at 461 and 664 nm according to the following equations: Colored carotenoids [μg/mL] = (OD461 − (0.046 × OD664)) × 4; Chlorophyll [μg/mL] = OD664 × 11.92.
2.5. Western blotting
Total cell extracts of Synechocystis were separated on SDS-PAGE and blotted to PVDF membranes according to standard protocols. The membranes were incubated with primary antibodies mouse α-Flag (1:5000 dilution, Sigma-Aldrich) or rabbit α-AtpB (AS05085, 1:5000, Agrisera) followed by probing with the appropriate HRP-conjugated α-mouse or α-rabbit secondary antibodies (1:10000, Sigma-Aldrich). The probed membranes were then incubated with HRP substrate (Bio-Rad) to generate a signal detected by a ChemiDoc imaging sysem (BioRad).
2.6. RT-PCR
RNA was extracted from Synechocystis cells using TriReagent (Sigma-Aldrich) according to the manufacturer’s instructions, and contaminating DNA was removed from samples using DNAse I (ThermoFisher Scientfic). cDNA was generated from 1 μg of RNA using the qScript cDNA synthesis kit (Quantabio, Beverly, MA, USA) and amplified using primers specific for BSS, CfDXS, Ecidi, EcispA, and atpA which was used as a control for equal loading of RNA (see Supplementary Table S2 for primer sequences). Each primer pair was tested on the template to determine the appropriate number of cycles to use, 28 cycles were used for each primer pairs.
2.7. Statistical analysis
Two-tailed Student’s t-test was carried out on the data accumulated from individual experiments for squalene and pigment content.
3. Results and discussion
3.1. Heterologous expression and activity of a Botryococcus braunii squalene synthase in Synechocystis
In previous work, we have constructed a strain of Synechocystis where inactivation of the gene shc encoding squalene hopene cyclase (see Fig. 1) led to accumulation of squalene in the cells (Englund et al., 2014). In order to enhance the production of squalene, we decided to investigate the effects of introducing additional genes into a Δshc strain of Synechocystis. In preliminary studies, over-expression of native genes including the native squalene synthase from Synechocystis (unpublished) did not result in increased squalene accumulation. We therefore decided to try expression of a heterologous squalene synthase, to potentially avoid regulation which may affect the native enzyme. Botryococcus braunii is a green alga which produces large amounts of botryococcenes, triterpene oils similar to squalene, and enzymes from B. braunii have also been shown to form squalene (Okada et al., 2000; Niehaus et al. 2011, 2012). We selected one enzyme from B. braunii, the Botryococcus squalene synthase (BSS) (Okada et al., 2000) (accession no Q9SDW9) for expression in Synechocystis. The gene encoding BSS was codon optimized for Synechocystis and a Flag tag was added to the C-terminal end of the gene to facilitate detection of the protein. The construct was cloned in the vector pEERM5’ (Fig. 2/Table 1) for insertion into the genome in the shc site. The nrsB promoter, inducible by addition of Ni2+ to the growth medium (Englund et al., 2016; Lopez-Maury et al., 2002), was placed in front of BSS to drive expression of the gene. The vector was transferred to the Δshc strain (Englund et al., 2014), completely replacing the inactivated shc, to generate the strain Δshc:BSS. We also transformed the same strain with pEERM5’, with all components except the tagged BSS, to generate the control strain Δshc:C. Transformant strains were isolated and complete segregation verified by PCR (data not shown).
Western blots were performed to verify expression of the Flag-tagged BSS in Δshc:BSS upon addition of Ni2+ (Fig. 3A). To determine levels of squalene, we grew cells of WT Synechocystis, the control strain Δshc:C, and Δshc:BSS in the presence and absence of Ni2+, and determined levels of squalene in the cells after 4 days (Fig. 3B). We found that the control strain accumulated 0.5 ± 0.1 mg squalene OD750-1 L-1, which, as expected, is similar to the levels observed in the previously investigated Δshc strain (Englund et al., 2014). There was no significant difference in squalene accumulation between cells without or with addition of Ni2+ in this strain. In Δshc:BSS without addition of Ni2+, squalene accumulation was somewhat higher compared to that of the control strain Δshc:C (p < 0.05, Fig. 3B), probably due to some leakiness of the nrsB promoter. Δshc:BSS cells grown with Ni2+ induction accumulated 0.95 ± 0.2 mg squalene OD750-1 L-1, demonstrating a highly significant increase (p < 0.001) of squalene accumulation upon addition of the inducer compared to the cells grown without inducer, as well as to the control strain Δshc:C cells (Fig. 3B).
Fig. 3.
(A) Western blots demonstrating expression of BSS in Synechocystis. Top: blot probed with α-Flag tag antibody; bottom: blot probed with α-AtpB antibody as a loading control. Lane 1: Proteins extracted from Δshc:C cells not expressing BSS; lane 2: Proteins extracted from cells of Δshc:BSS induced with Ni2+ for expression. 4 μg of protein extract loaded in each lane. Expected size of BSS-Flag is 45 kDa and AtpB is 56kDa. (B) Squalene accumulation in WT and engineered strains, with and without induction with Ni2+ and under different nutrient conditions. ‘BG11’ – only BG11 medium; ‘BG11+Ni2+’ - BG11medium with addition of 2.5μM NiCl2; ‘BG11+Ni2+ + glucose’ – additional supplementation by 0.5% glucose; ‘BG11+Ni2+ + NaHCO3’– additional supplementation by 50mM NaHCO3 buffered with 20mM HEPES at pH 7. ‘n.d.’ – not determined. Cells were grown for four days after induction with Ni2+. Error bars indicate standard deviation (n ≥ 3). Asterisks represent significant differences in samples from Δshc:BSS compared to the control strain, or, where indicated, compared to the same strain without induction (* = p < 0.05, ** = p < 0.01, *** = p < 0.001).
3.2. Effects on carbon source and growth conditions on squalene production
Since the wildtype strain of Synechocystis used in this study is capable of mixotrophic growth on glucose, it was possible to investigate if there would be an effect on squalene production by growing the cells on different carbon sources. Δshc:BSS and the control strain Δshc:C were grown on BG11 medium with addition of 0.5% (wt/vol) glucose, or 50mM NaHCO3 buffered at pH 7. Squalene accumulation was assessed 4 days after the addition of Ni2+ (Fig. 3B). For the control strain, addition of glucose or NaHCO3 did not make a significant difference for the accumulation of squalene per cell. For Δshc:BSS, squalene accumulation was lower in both conditions compared to cells grown with only BG11 and induction by Ni2+ (Fig. 3B). However, cultures grown with addition of glucose accumulated more biomass, so that the total amount of squalene produced in these cultures was higher than in the other conditions after 4 days, reaching 2.6 ± 0.1 mg/L culture (data not shown). In cultures of Δshc:BSS grown with addition of sodium bicarbonate, the total amount of squalene was actually lower than on BG11 alone, and similar to the control strain (data not shown). In studies of production of other terpene compounds, it has been observed that the growth rate may affect the amount of terpene product generated per cell. The lower productivity towards squalene in cells grown with additional carbon sources observed here may be the result of faster growth of the cells, where more resources are directed towards formation of biomass and less towards the heterologous product. This is in agreement with an earlier study on isoprene production in Synechococcus elongatus (Gao et al., 2016), who found the highest rates of isoprene production in stationary phase cultures.
To further evaluate the potential for squalene accumulation in the Δshc:BSS strain, an experiment was performed where cultures of Δshc:BSS and Δshc:C were grown at higher density in lab scale photobioreactors. Squalene accumulation and growth were followed for 96 h at 30 μmol photons m-2 s-1, at which time light was increased to 50 μmol photons m-2 s-1, and squalene accumulation and growth was observed after another 96 h (Fig. 4). Growth was linear during the entire experiment for both strains, while squalene accumulation appeared to increase in the Δshc:BSS strain upon the increase in light intensity. The maximum concentration of squalene reached was 5.1 mg/L culture for the BSS expressing strain at the end of the experiment.
Fig. 4.
Growth and squalene production in flat panel bioreactors. The squalene producing strain Δshc:BSS and the control strain Δshc:C were grown in parallel in two bioreactors, in BG11 medium with addition of 2.5μM NiCl2 for induction of BSS gene expression. The light intensity was set at 30 μmol photons m-2 s-1 for 96 h, and then increased to 50 μmol photons m-2 s-1.
3.3. Expression of additional genes from the MEP pathway
Expressing BSS led to an increased accumulation of squalene in the cells. To enhance this further, we decided to add expression of genes from the MEP pathway. The first step of the pathway is the formation of deoxyxylulose-5-phosphate (DXP) from glyceraldehyde-3-phosphate and pyruvate, catalyzed by the enzyme deoxyxylulose-5-phosphate synthase (DXS). In other studies, the action of DXS has been shown to be a limiting step in the generation of products from the MEP pathway (Englund et al. 2015, 2018; Harker and Bramley, 1999). Therefore, we decided to express an extra DXS in our squalene producing strain. The gene for expression was CfDXS from the plant Plectranthus barbatus (Coleus forskholii) (GenBank ID KP889115 (Gnanasekaran et al., 2015)), truncated to remove a target peptide, as previously described (Englund et al., 2015). We also included expression of isopentenyl diphosphate isomerase from Escherichia coli (EcIDI), catalyzing the isomerization of isopentenyl diphosphate (IPP), and dimethylallyl diphosphate (DMAPP). This was based on the results from a study on isoprene production in Synechocystis, where expression of Idi together with DXS lead to a combined effect on enhancing isoprene levels which was larger than that of either enzyme expressed alone (Englund et al., 2018). Furthermore, farnesyl diphosphate synthase from E. coli (EcIspA) catalyzing formation of farnesyl diphosphate (FPP), the substrate for squalene synthesis, was included in the expression construct based on earlier reports identifying this enzyme as a potential bottleneck for the formation of FPP in E. coli. (Martin et al., 2003; Wang et al., 1999).
In order to be able to express these enzymes at a high level in our Synechocystis strain, we constructed an expression vector, pEEK, based on the previously reported pEEC1 but using kanamycin as selection (Englund et al., 2018). CfDXS, Ecidi, and EcispA were cloned into this vector as one operon, with expression driven by the strong promoter Ptrc2O to form the plasmid pEEK_dii (Fig. 2 and Table 1, see Supplementary Table S1 for exact sequence). pEEK_dii was transferred to Δshc:BSS by conjugation, to generate the strain Δshc:BSS_dii.
Expression of all heterologously introduced genes was verified by semi-quantitative RT-PCR (Fig. 5A). It was found that for BSS under control of PnrsB, expression was detectable without addition of Ni2+ in the medium, but strongly enhanced after addition of Ni2+. For CfDXS, idi and ispA, under control of Ptrc2O and thus constitutively expressed, no differences in expression levels with or without Ni2+ were observed.
Fig. 5.
(A) RT-PCR on RNA from Synechocystis cells without and with 2.5 μM Ni2+ induction, using primers against genes indicated on the left. atpA was used as a loading control. Lane 1: 100bp Ladder; Lanes 2 and 4: Δshc:BSS; Lanes 3 and 5: Δshc:BSS_dii; Lane 6: WT Synechocystis gDNA; Lane 7: Plasmid DNA; Lane 8: no template control for the PCR. Lanes 2 and 3: no Ni2+ added; Lanes 4 and 5: with 2.5 μM Ni2+. -RT: no RT enzyme added in the RT-reactions, primers against atpA. (B) Squalene production in strains Δshc:BSS_dii compared to Δshc:BSS. Cells were grown in BG11 medium or BG11 medium with addition of 2.5 μM Ni2+, and squalene content determined four days after induction. Error bars represents the standard deviation of at least five biological replicates. Asterisks represents significant differences between samples, *** = p < 0.001.
Squalene accumulation was determined in cells expressing the extra pathway genes in addition to the B. braunii BSS. Cells of Δshc:BSS_dii accumulated about 1.6 times as much squalene per cell compared to cells of Δshc:BSS upon induction with 2.5 μM Ni2+ (Fig. 5B), and three times as much squalene per cell as the Δshc:C strain, reaching a squalene content of 1.5 ± 0.2 mg squalene OD750-1 L-1 4 days after induction. While we cannot in this study separate the effect of each individual pathway enzyme on the production of squalene, the observed enhancement of squalene accumulation by over-expressing upstream MEP pathway genes is in accordance with other recent studies on enhancing the MEP pathway flux for production of terpenoids in cyanobacteria (Englund et al. 2015, 2018; Choi et al., 2016).
3.4. Effects on pigment content
Since squalene synthases use the same substrates as formation of carotenoids and the phytol tail of chlorophyll, we decided to also investigate the effect on pigments in the cells of expressing BSS and the upstream pathway genes (Fig. 6). In the strains expressing BSS, we found a significant decrease in the amount of carotenoids produced upon induction of BSS by addition of Ni2+, compared to the Δshc:C control strain (Fig. 6A). There were no significant effects on chlorophyll content (Fig. 6B). We also looked at the combined amount of carotenoids and squalene (Fig. 6C). It is worth noting that in the Δshc:BSS strain, the amount of carotenoids and squalene combined does not increase when BSS is induced, and thus substrate appears to be shifted from carotenoids to squalene accumulation. In the Δshc:BSS_dii strain, there was no significant effect on pigment content compared to Δshc:C or Δshc:BSS strain by the constitutive overexpression of the selected MEP pathway genes (Fig. 6A and B). However, upon induction of BSS, this strain showed an overall increase of the combined amount of carotenoids and squalene compared to the same strain without induction, and an increase of about 60% compared to the Δshc:BSS strain (see Fig. 6C). Thus, the extra pathway enzymes allowed for a higher flux of substrate towards the product which was not available in the BSS strain.
Fig. 6.
Analysis of pigments in engineered Synechocystis strains (Δshc:C, Δshc:BSS and Δshc:BSS_dii). (A) Carotenoids. (B) Chlorophyll a. (C) Amount of squalene and carotenoids combined. Error bar indicates standard deviation (n = 4). Asterisks represents significant differences between samples, * = p < 0.05, ** = p < 0.01, *** = p < 0.001.
4. Conclusions
In the present study, we have shown that heterologous expression of a squalene synthase from Botryocuccus braunii can increase squalene accumulation in a Δshc strain of Synechocystis. Furthermore, we could attain an increased production by also expressing genes upstream in the pathway to squalene formation, reaching three times more squalene per cell compared to the Δshc:C control strain. Substrate was shuttled from generation of pigments towards production of squalene.
To improve yields of terpenoid products in cyanobacteria further, we will need to implement additional modifications to improve flux through the pathway. In a recent study, Choi et al. reported a squalene accumulation of 7.08 mg/L/OD730 in cultures of engineered Synechococcus elongatus PCC 7942. Their cells expressed dxs, idi and ispA from E. coli along two copies of overexpressed SQS from Saccharomyces cerevisiae and the cells were grown in a bioreactor with elevated CO2 levels (Choi et al., 2017). Gao et al. succeeded in attaining a yield of 1.26 g/L of isoprene from S. elongatus PCC 7942 in a long-term, continuous production experiment, by simultaneously overexpressing dxs and ispG and fusing Idi to an efficient isoprene synthase (Gao et al., 2016). Thus, there is potential for generation of terpenoid products from the MEP pathway with a high yield in cyanobacteria, however, more knowledge about the pathway and its regulation in cyanobacteria is needed. In the case of squalene, screening for squalene synthases with high instrinsic activity, and optimizing the expression constructs to achieve higher levels of the enzyme in the cell may also result in higher productivity. Engineering of the upstream carbon and co-factor supply may also be employed to enhance yields (Englund et al., 2018). Optimization of growth conditions and scale-up experiments will also be needed to assess the potential of large scale terpenoid production from cyanobacteria.
CRediT authorship contribution statement
Bagmi Pattanaik: Conceptualization, Data curation, Investigation, Methodology, Validation, Visualization, Writing - original draft, Writing - review & editing. Elias Englund: Investigation, Methodology, Writing - review & editing. Nicholas Nolte: Investigation, Writing - review & editing. Pia Lindberg: Conceptualization, Data curation, Supervision, Funding acquisition, Project administration, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the Swedish Energy Agency [grant no 38334-1], the Carl Trygger Foundation [grant no CTS 11:266], and by the NordForsk Nordic Center of Excellence program NordAqua [project no 82845]. The authors would like to thank Christoph Howe for his help in setting up the bioreactors.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mec.2020.e00125.
Contributor Information
Bagmi Pattanaik, Email: pbagmi@googlemail.com.
Elias Englund, Email: EliasEnglund@lbl.gov.
Nicholas Nolte, Email: nolte.nico92@googlemail.com.
Pia Lindberg, Email: pia.lindberg@kemi.uu.se.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Angermayr S.A., Gorchs Rovira A., Hellingwerf K.J. Metabolic engineering of cyanobacteria for the synthesis of commodity products. Trends Biotechnol. 2015;33(6):352–361. doi: 10.1016/j.tibtech.2015.03.009. [DOI] [PubMed] [Google Scholar]
- Brennan L., Owende P. Biofuels from microalgae-A review of technologies for production, processing, and extractions of biofuels and co-products. Renew. Sustain. Energy Rev. 2010;14(2):557–577. [Google Scholar]
- Chamovitz D., Sandmann G., Hirschberg J. Molecular and biochemical characterization of herbicide-resistant mutants of cyanobacteria reveals that phytoene desaturation is a rate-limiting step in carotenoid biosynthesis. J. Biol. Chem. 1993;268(23):17348–17353. [PubMed] [Google Scholar]
- Choi S.Y., Lee H.J., Choi J., Kim J., Sim S.J., Um Y., Kim Y., Lee T.S., Keasling J.D., Woo H.M. Photosynthetic conversion of CO2 to farnesyl diphosphate-derived phytochemicals (amorpha-4,11-diene and squalene) by engineered cyanobacteria. Biotechnol. Biofuels. 2016;9:202. doi: 10.1186/s13068-016-0617-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S.Y., Wang J.Y., Kwak H.S., Lee S.M., Um Y., Kim Y., Sim S.J., Choi J.I., Woo H.M. Improvement of squalene production from CO2 in Synechococcus elongatus PCC 7942 by metabolic engineering and scalable production in a photobioreactor. ACS Synth. Biol. 2017 doi: 10.1021/acssynbio.7b00083. [DOI] [PubMed] [Google Scholar]
- Do R., Kiss R.S., Gaudet D., Engert J.C. Squalene synthase: a critical enzyme in the cholesterol biosynthesis pathway. Clin. Genet. 2009;75(1):19–29. doi: 10.1111/j.1399-0004.2008.01099.x. [DOI] [PubMed] [Google Scholar]
- Englund E., Pattanaik B., Ubhayasekera S.J., Stensjo K., Bergquist J., Lindberg P. Production of squalene in Synechocystis sp. PCC 6803. PloS One. 2014;9(3) doi: 10.1371/journal.pone.0090270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund E., Andersen-Ranberg J., Miao R., Hamberger B., Lindberg P. Metabolic engineering of Synechocystis sp. PCC 6803 for production of the plant diterpenoid manoyl oxide. ACS Synth. Biol. 2015;4(12):1270–1278. doi: 10.1021/acssynbio.5b00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund E., Liang F., Lindberg P. Evaluation of promoters and ribosome binding sites for biotechnological applications in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Sci. Rep. 2016;6:36640. doi: 10.1038/srep36640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund E., Shabestary K., Hudson E.P., Lindberg P. Systematic overexpression study to find target enzymes enhancing production of terpenes in Synechocystis PCC 6803, using isoprene as a model compound. Metab. Eng. 2018;49:164–177. doi: 10.1016/j.ymben.2018.07.004. [DOI] [PubMed] [Google Scholar]
- Gao X., Gao F., Liu D., Zhang H., Nie X.Q., Yang C. Engineering the methylerythritol phosphate pathway in cyanobacteria for photosynthetic isoprene production from CO2. Energy Environ. Sci. 2016;9(4):1400–1411. [Google Scholar]
- Gnanasekaran T., Vavitsas K., Andersen-Ranberg J., Nielsen A.Z., Olsen C.E., Hamberger B., Jensen P.E. Heterologous expression of the isopimaric acid pathway in Nicotiana benthamiana and the effect of N-terminal modifications of the involved cytochrome P450 enzyme. J. Biol. Eng. 2015;9:24. doi: 10.1186/s13036-015-0022-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harker M., Bramley P.M. Expression of prokaryotic 1-deoxy-D-xylulose-5-phosphatases in Escherichia coli increases carotenoid and ubiquinone biosynthesis. FEBS Lett. 1999;448(1):115–119. doi: 10.1016/s0014-5793(99)00360-9. [DOI] [PubMed] [Google Scholar]
- Heidorn T., Camsund D., Huang H.-H., Lindberg P., Oliveira P., Stensjö K., Lindblad P. Synthetic biology in cyanobacteria: engineering and analyzing novel functions. In: Voigt C.A., editor. vol 497. Academic Press; 2011. pp. 539–579. (Methods Enzymol). [DOI] [PubMed] [Google Scholar]
- Hopp T.P., Prickett K.S., Price V.L., Libby R.T., March C.J., Cerretti D.P., Urdal D.L., Conlon P.J. A short polypeptide marker sequence useful for recombinant protein identification and purification. Bio Technol. 1988;6(10):1204–1210. [Google Scholar]
- The iGEM registry of standard biological parts. http://parts.igem.org/
- Huang H.-H., Camsund D., Lindblad P., Heidorn T. Design and characterization of molecular tools for a Synthetic Biology approach towards developing cyanobacterial biotechnology. Nucleic Acids Res. 2010;38(8):2577–2593. doi: 10.1093/nar/gkq164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannenberg E.L., Poralla K. Hopanoid biosynthesis and function in bacteria. Naturwissenschaften. 1999;86(4):168–176. [Google Scholar]
- Knoot C.J., Ungerer J., Wangikar P.P., Pakrasi H.B. Cyanobacteria: promising biocatalysts for sustainable chemical production. J. Biol. Chem. 2018;293(14):5044–5052. doi: 10.1074/jbc.R117.815886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Miao R., Lindberg P., Lindblad P. Modular engineering for efficient photosynthetic biosynthesis of 1-butanol from CO2 in cyanobacteria. Energy Environ. Sci. 2019 [Google Scholar]
- Lopez-Maury L., Garcia-Dominguez M., Florencio F.J., Reyes J.C. A two-component signal transduction system involved in nickel sensing in the cyanobacterium Synechocystis sp. PCC 6803. Mol. Microbiol. 2002;43(1):247–256. doi: 10.1046/j.1365-2958.2002.02741.x. [DOI] [PubMed] [Google Scholar]
- Martin V.J., Pitera D.J., Withers S.T., Newman J.D., Keasling J.D. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 2003;21(7):796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- Niehaus T.D., Okada S., Devarenne T.P., Watt D.S., Sviripa V., Chappell J. Identification of unique mechanisms for triterpene biosynthesis in Botryococcus braunii. Proc. Natl. Acad. Sci. U. S. A. 2011;108(30):12260–12265. doi: 10.1073/pnas.1106222108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehaus T.D., Kinison S., Okada S., Yeo Y.S., Bell S.A., Cui P., Devarenne T.P., Chappell J. Functional identification of triterpene methyltransferases from Botryococcus braunii race B. J. Biol. Chem. 2012;287(11):8163–8173. doi: 10.1074/jbc.M111.316059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada S., Murakami M., Yamaguchi K. Hydrocarbon composition of newly isolated strains of the green microalga Botryococcus braunii. J. Appl. Phycol. 1995;7(6):555–559. [Google Scholar]
- Okada S., Devarenne T.P., Chappell J. Molecular characterization of squalene synthase from the green microalga Botryococcus braunii, race B. Arch. Biochem. Biophys. 2000;373(2):307–317. doi: 10.1006/abbi.1999.1568. [DOI] [PubMed] [Google Scholar]
- Pattanaik B., Lindberg P. Terpenoids and their biosynthesis in cyanobacteria. Life. 2015;5(1):269–293. doi: 10.3390/life5010269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmerk C.L., Bernards M.A., Valvano M.A. Hopanoid production is required for low-pH tolerance, antimicrobial resistance, and motility in Burkholderia cenocepacia. J. Bacteriol. 2011;193(23):6712–6723. doi: 10.1128/JB.05979-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedenburg G., Jendrossek D. Squalene-hopene cyclases. Appl. Environ. Microbiol. 2011;77(12):3905–3915. doi: 10.1128/AEM.00300-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanier R.Y., Kunisawa R., Mandel M., Cohen-Bazire G. Purification and properties of unicellular blue-green algae (order Chroococcales) Bacteriol. Rev. 1971;35(2):171–205. doi: 10.1128/br.35.2.171-205.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracy N.I., Crunkleton D.W., Price G.L. Catalytic cracking of squalene to gasoline-range molecules. Biomass Bioenergy. 2011;35(3):1060–1065. [Google Scholar]
- Vavitsas K., Crozet P., Hamborg Vinde M., Davies F., Lemaire S.D., Vickers C.E. The synthetic biology toolkit for photosynthetic microorganisms. Plant Physiol. 2019 doi: 10.1104/pp.19.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay D., Akhtar M.K., Hess W.R. Genetic and metabolic advances in the engineering of cyanobacteria. Curr. Opin. Biotechnol. 2019;59:150–156. doi: 10.1016/j.copbio.2019.05.012. [DOI] [PubMed] [Google Scholar]
- Wang C.W., Oh M.K., Liao J.C. Engineered isoprenoid pathway enhances astaxanthin production in Escherichia coli. Biotechnol. Bioeng. 1999;62(2):235–241. doi: 10.1002/(sici)1097-0290(19990120)62:2<235::aid-bit14>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Yoo C., Jun S.Y., Lee J.Y., Ahn C.Y., Oh H.M. Selection of microalgae for lipid production under high levels carbon dioxide. Bioresour. Technol. 2010;101:S71–S74. doi: 10.1016/j.biortech.2009.03.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.