The threat of diminished antibiotic discovery has global health care in crisis. In the United States, it is estimated each year that over 2 million bacterial infections are resistant to first-line antibiotic treatments and cost in excess of 20 billion dollars.
KEYWORDS: antimicrobial, adjuvant, colistin, polymyxin, antibiotic resistance, multidrug-resistant bacteria, adjuvants, antimicrobial agents, lipid A, lipopolysaccharide, outer membrane
ABSTRACT
The threat of diminished antibiotic discovery has global health care in crisis. In the United States, it is estimated each year that over 2 million bacterial infections are resistant to first-line antibiotic treatments and cost in excess of 20 billion dollars. Many of these cases result from infection with the ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species), which are multidrug-resistant bacteria that often cause community- and hospital-acquired infections in both healthy and immunocompromised patients. Physicians have turned to last-resort antibiotics like polymyxins to tackle these pathogens, and as a consequence, polymyxin resistance has emerged and is spreading. Barring the discovery of new antibiotics, another route to successfully mitigate polymyxin resistance is to identify compounds that can complement the existing arsenal of antibiotics. We recently designed and performed a large-scale robotic screen to identify 43 bioactive compounds that act synergistically with polymyxin B to inhibit the growth of polymyxin-resistant Escherichia coli. Of these 43 compounds, 5 lead compounds were identified and characterized using various Gram-negative bacterial organisms to better assess their synergistic activity with polymyxin. Several of these compounds reduce polymyxin to an MIC of <2 μg/ml against polymyxin-resistant and polymyxin-heteroresistant Gram-negative pathogens. Likewise, four of these compounds exhibit antimicrobial activity against Gram-positive bacteria, one of which rapidly eradicated methicillin-resistant Staphylococcus aureus. We present multiple first-generation (i.e., not yet optimized) compounds that warrant further investigation and optimization, since they can act both synergistically with polymyxin and also as lone antimicrobials for combating ESKAPE pathogens.
INTRODUCTION
In recent years, Gram-negative bacterial infections have become more difficult to treat, and as a consequence, clinicians have relied on polymyxin derivatives like polymyxin B (PMB) and polymyxin E (colistin) to treat their patients. These last-resort antibiotics are effective but can cause undesirable and irreversible renal damage when given at therapeutic doses (1, 2). To further complicate the issue, few U.S. Food and Drug Administration (FDA)-approved antibiotics are being developed to combat such organisms (3). Without swift action, it is estimated that antimicrobial resistance will result in 10 million deaths per year by 2050, with an economic cost of $100 trillion (4). Given antimicrobial resistance has emerged as a major threat to global human and animal health, the World Health Organization, the Centers for Disease Control and Prevention, and the National Institutes of Health have made the pursuit of understanding antimicrobial resistance and identifying novel antimicrobial compounds a priority, especially against multidrug-resistant bacteria such as the ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter sp.) (5–8).
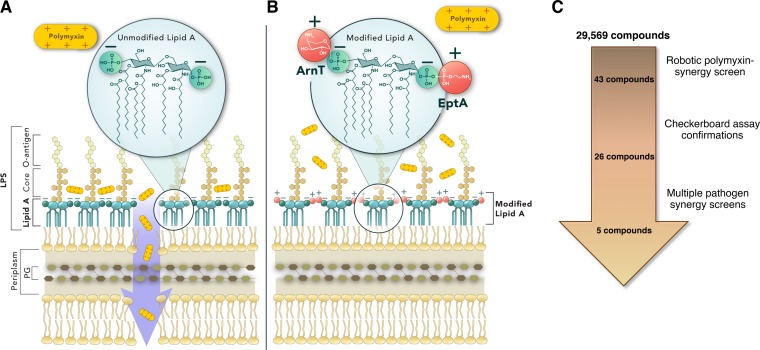
Unlike Gram-positive bacteria, Gram-negative bacteria possess a thin peptidoglycan (PG) layer that is protected by a unique asymmetrical lipid bilayer known as the outer membrane (OM). The inner leaflet of the OM is composed of glycerophospholipids, while the outer leaflet is composed of a glycolipid called lipopolysaccharide (LPS; endotoxin). As shown in Fig. 1A, LPS is composed of a negatively charged, hydrophobic, lipid A anchor linked to O-antigen polysaccharides by an oligosaccharide core. LPS is stabilized in the OM by bridging with divalent cations (Ca2+ and Mg2+), and its amphipathic structure protects the cell by ensuring that the OM of Gram-negative bacteria remains an exceptional permeability barrier (9). Furthermore, the lipid A anchor of LPS is a potent immunostimulatory molecule that is recognized by the Toll-like receptor 4 myeloid differentiation factor 2 (TLR4-MD2) complex on antigen-presenting cells. The engagement of TLR4-MD2 by antigen-presenting cells results in the secretion of proinflammatory cytokines, eicosanoids, and reactive oxygen species, all of which contribute to the clinical manifestations traditionally associated with Gram-negative bacterial infections. Left unchecked by effective antibiotics, such infections often progress to life-threatening sepsis and ultimately death, and this dire consequence is becoming more common as antimicrobial resistance by bacterial pathogens continues to outpace the development of new therapeutic interventions (10–12).
FIG 1.
Rationale of the whole-cell screen for compounds that exhibit synergy with polymyxin. (A) The Gram-negative bacterial cell envelope and the canonical unmodified lipid A species of E. coli K-12 are shown. The inner and outer membrane lipid bilayers are separated by a periplasmic space containing a wall composed of peptidoglycan. The inner membrane (IM) is composed of a symmetrical layer of glycerophospholipids. The outer membrane (OM) is asymmetric and composed of glycerophospholipids in the inner leaflet and LPS in the outer leaflet. Lipid A is a hexa-acylated disaccharide of glucosamine with phosphates at the 1 and 4′ moieties, and it is responsible for anchoring LPS to the OM. The phosphates of lipid A impart an overall negative charge that enables cationic antimicrobial peptides (CAMPs), such as polymyxins, to disrupt the OM and gain access to the IM, which ultimately lyses the cell. (B) The modification of lipid A by ArnT and EptA with 4-amino-4-deoxy-l-arabinose (L-Ara4N) and phosphoethanolamine (pEtN), respectively, changes the charge of lipid A and negates the activity of polymyxin. (C) The screening pipeline used to identify compounds that act synergistically with PMB to inhibit the growth of PMBr E. coli WD101. The number of compounds remaining after each step is illustrated.
Polymyxins are cationic antimicrobial peptides that disrupt divalent cationic bridging interactions between LPS molecules to destabilize the OM and ultimately cause bacterial cell lysis (13). The mechanisms of polymyxin resistance employed by Gram-negative bacteria primarily rely on the modification of the lipid A anchor of LPS. This can be achieved in a variety of ways; however, mutations in genes encoding either the PmrAB or PhoPQ two-component regulatory systems are most common and lead to the expression of a plethora of genes, including those involved in LPS modification. Using Escherichia coli, it was previously demonstrated that polymyxin resistance can occur through constitutive PmrA-dependent expression of ArnT and EptA enzymes, which modify lipid A with the amine-containing substituents 4-amino-4-deoxy-l-arabinose (L-Ara4N) and phosphoethanolamine (pEtN), respectively (14–16). L-Ara4N and pEtN modifications at the 1ʹ or 4ʹ phosphate positions of lipid A (Fig. 1B) are a highly effective means of conferring polymyxin resistance to Gram-negative bacteria like E. coli, as they reduce the overall negative charge of lipid A and render polymyxins unable disrupt the OM (9). These modifications are also broadly used by multiple multidrug-resistant Gram-negative bacteria (A. baumannii [17, 18], K. pneumoniae [19], Salmonella sp. [15], P. aeruginosa [20, 21], and Enterobacter sp. [22]). When activated in the majority of Gram-negative bacteria, PhoQ and PmrB sensor kinases can either phosphorylate or dephosphorylate PhoP and PmrB, respectively. In these organisms, the phosphorylation (activation) of PhoP indirectly leads to the activation of PmrA via PmrD, while PmrB directly activates PmrA. PmrA activation results in ArnT and EptA expression, and subsequent lipid A modification. In contrast, PhoQ in P. aeruginosa can only dephosphorylate (inactivate) PhoP; thus, the inactivation of PhoQ is required to constitutively activate PhoP and PmrA in this species (21, 23). Acquired chromosomal mutations that ultimately affect ArnT and/or EptA expression are key to the development of intrinsic polymyxin resistance in bacteria. Likewise, the ability of bacteria to obtain plasmids bearing eptA orthologs (mcr-1) is furthering the rapid spread and emergence of polymyxin resistance worldwide in human, animal, and environmental reservoirs (24, 25). Recent screening of clinical isolates for these genetic alterations has uncovered a large number of polymyxin-resistant bacteria, which not only have the ability to pass on such resistance to other bacteria but also pose a considerable and growing risk to patients in both community and hospital settings (26, 27). New treatment modalities to combat these polymyxin-resistant bacteria are needed now more than ever.
Large-scale robotic screens have been employed by others to identify repurposed compounds that can potentiate the activity of nonpolymyxin antibiotics in order to produce polymyxin-sparing regimes for treating multidrug-resistant Gram-negative bacteria (28, 29). Other groups have focused on identifying analogues or constructing derivatives of novel compounds that can potentiate the activity of polymyxin (30–35). In this study, we merged these approaches with the goal of identifying existing compounds (either new bioactives or repurposed pharmaceuticals) that could act synergistically with PMB to yield new last-resort therapeutic options for polymyxin-resistant bacterial infections that fail to respond to either polymyxin-sparing approaches or polymyxins themselves. To that end, we utilized a well-described polymyxin-resistant E. coli strain that constitutively modifies its lipid A structure (see Table S1 in the supplemental material) and performed a stringent, high-throughput, whole-cell screen against a library composed of known bioactives, both alone and in combination with PMB. It was anticipated that if bioactive compounds were found, they would likely mediate susceptibility to combination therapy by either (i) targeting enzymes critical to the modification of lipid A, thereby promoting polymyxin activity at the OM; (ii) disrupting the OM to facilitate the entry of polymyxin into the bacterial cell; or (iii) functioning as antimicrobials that rely on polymyxin as an adjuvant.
Using this method, we identified 43 commercially available compounds that exhibited synergy with PMB. These bioactive compounds were subjected to more robust methods for characterizing their synergistic activity in multiple bacterial organisms, which ultimately produced five lead compounds. We confirmed that these lead compounds exhibit synergy when combined with PMB, but not when combined with antibiotics typically ineffective against Gram-negative organisms (e.g., vancomycin). We also determined that these compounds do not alter existing lipid A modifications in PMB-resistant E. coli. Importantly, these lead compounds in combination with polymyxin were highly effective at inhibiting the growth of polymyxin-resistant and heteroresistant Gram-negative pathogens and did so at PMB concentrations that are well below 2 μg/ml (the recommended MIC cutoff) (1, 2, 36). Even more interesting, some of these lead compounds also exhibit profound antimicrobial activity against Gram-positive bacterial species, including methicillin-resistant S. aureus (MRSA). This report not only provides evidence of first-generation compounds with promise for reducing polymyxin dosing in clinically relevant bacterial infections but also offers potential avenues to further explore our understanding of antimicrobial resistance mechanisms and how to better combat them.
RESULTS
Whole-cell robotic screen identifies bioactive compounds that act synergistically with polymyxin.
Since L-Ara4N and pEtN modifications to lipid A (Fig. 1B) are a common and highly effective means of conferring polymyxin resistance to Gram-negative bacteria, we performed a high-throughput, whole-cell screen using E. coli K-12 strain WD101 (see Table S1 in the supplemental material). WD101, a derivative of wild-type strain W3110, harbors a point mutation in pmrA that results in constitutive PmrA activation and high levels of polymyxin resistance (14). Figure 1C illustrates the pipeline used to identify the five lead compounds described in this study. Specifically, an automated robotic screen was performed with a collection of 29,569 bioactive compounds for their ability to interact synergistically with PMB and inhibit growth of WD101. A series of stringent cutoff criteria were used to narrow the list of prospective compounds. First, compounds that failed to reproducibly inhibit the growth of WD101 to <65% in combination with 8 μg/ml PMB were excluded from further screening, which yielded 356 compounds. A second robotic screen was used to filter out bioactives that could inhibit the growth of E. coli (W3110 and WD101) when used alone. At the end of this screening process, adjuvant bioactives were defined as any compounds that resulted in >99% WD101 growth inhibition in combination with PMB but did not inhibit the growth of either W3110 or WD101 by themselves (Fig. S1). This methodology enabled us to identify 43 commercially available compounds for further investigation as potential polymyxin adjuvants.
The robotic screen used in this study used polystyrene microtiter plates and lysogeny broth (LB) supplemented with compounds and/or 8 μg/ml PMB (the sub-MIC for this method) to assess WD101 growth. MIC determinations in Gram-negative bacteria can vary depending upon the combination of media, vessel, and antibiotic used. For example, the PMB MIC of WD101 grown on LB agar plates by Epsilometer test is 8 to 12 μg/ml, while its MIC in LB glass tubes or in cation-adjusted Mueller-Hinton agar (CAMHA) plates is much lower (3 to 4 μg/ml). The MIC discrepancy due to the vessel occurs because cationic antimicrobial peptides (CAMPs) such as PMB bind to negatively charged, inert surfaces such as polystyrene (37–39). Likewise, the MIC discrepancy due to the media occurs because of variation in the salt content, which is critical for LPS stabilization in the OM of rapidly replicating Gram-negative bacteria (13). When not standardized across studies, differences in these physical parameters can produce inaccurate and inconsistent MIC determinations, which further add to clinical breakpoint discrepancies in the field (36, 40). As such, cation-adjusted Mueller-Hinton agar (CAMHA) or broth (CAMHB) have been established in the clinical microbiology field as the gold standard for antimicrobial susceptibility testing (41–45). While not the gold standard, polystyrene plates and LB were still most appropriate for our robotic synergy screen since the polystyrene microtiter plates yielded more accurate optical density measurements, and the nutrient composition of LB allowed for higher supplementation with PMB to counteract the plate effects.
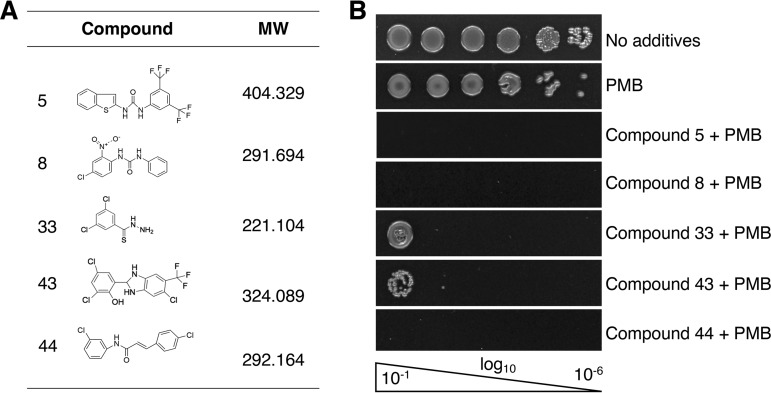
To ensure we identified only compound hits that act synergistically with low PMB concentrations, we performed manual checkerboard synergy assays with WD101 using polypropylene microtiter plates and CAMHB supplemented with 2 μg/ml PMB (the sub-MIC for this method). With these procedural changes, we eliminated false-positive hits and confirmed that only 26 of the original 43 compounds reproducibly exhibited detectable synergy (WD101 growth < 10%) with small amounts of PMB. The effectiveness of these select compounds in combination with PMB against multiple Gram-negative bacterial species was then examined by Epsilometer tests and checkerboard assays and, ultimately, five hit-to-lead compounds emerged with broad Gram-negative antimicrobial activities. The structures and physiochemical properties (Fig. 2A; see Table S2 in the supplemental material) of these early lead targets are provided, and their effectiveness in combination with PMB against WD101 was initially verified by two methods.
FIG 2.
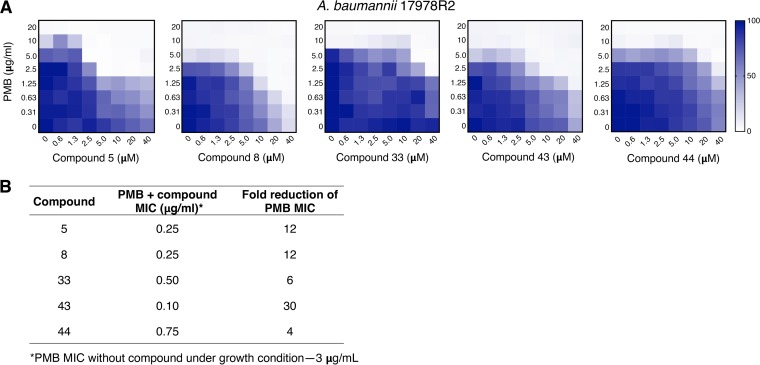
Lead compounds exhibit synergy with polymyxin B against polymyxin-resistant E. coli. (A) Table shows the chemical features of the five lead compounds identified in this study that potentiate the activity of PMB. (B) The effectiveness of these five compounds in combination with PMB at inhibiting the growth of PMBr E. coli WD101 is demonstrated by EOP assays on CAMHA supplemented with 20 μM of compound and 3 μg/ml PMB. These data are representative of at least two biological replicates.
Specifically, an efficiency-of-plating (EOP) method in which WD101 cells were grown in the presence of 20 μM of each compound in combination with 3 μg/ml PMB (the sub-MIC by this method) demonstrated that all five compounds inhibited the growth of WD101, with some showing the potential for bactericidal activity (Fig. 2B). We also used a broth microdilution-based method, since the Clinical and Laboratory Standards Institute (CLSI) recommends that antimicrobial synergy in combination therapy be determined by performing checkerboard assays and adhering to established FIC indices (42, 43, 45). Since our lead compounds were purposely screened for their ability to not inhibit the growth of WD101 by themselves (Fig. S1), we were unable to determine compound-only MIC values for FIC index calculations. Our synergy determinations were instead made by assigning a cutoff value for WD101 growth at <10% (MIC) and, to be more stringent, we also made side-by-side comparisons between our compounds and well-established PMB adjuvants in the field such as novobiocin (46, 47) and pentamidine (29, 48). Currently, there is no CLSI clinical breakpoint assigned for the use of polymyxins to treat Enterobacteriaceae infections. However, resistance to <2 μg/ml of polymyxin by broth microdilution methods has served as a reliable cutoff value for both clinicians and researchers (28, 29, 49).
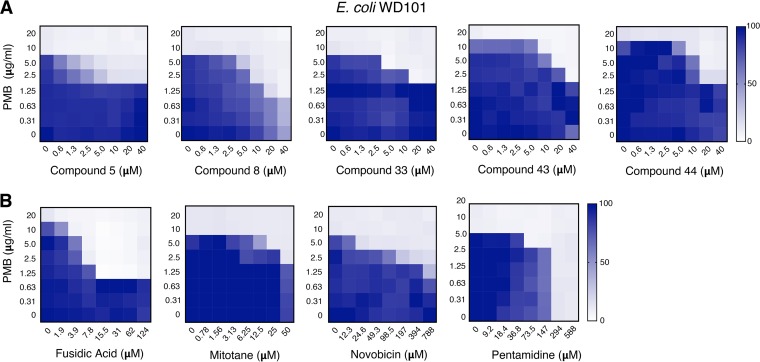
Based on these criteria, Fig. 3A demonstrates that all five compounds act synergistically in combination with PMB. Compound 8 was able to reduce the MIC of PMB to 1.25 μg/ml (6- to 8-fold), which is considered in the susceptible range since it is below the aforementioned <2-μg/ml cutoff value. Impressively, this compound also achieved this PMB reduction at a very low concentration (20 μM). Low concentrations of compounds 5, 33, 43, and 44 also reduced the MIC of PMB to 2.5 μg/ml (3- to 4-fold), but based on our stringent criteria only ranked as having intermediate activity. In side-by-side comparisons between our lead compounds and established adjuvants in the field (Fig. 3B), only fusidic acid and pentamidine were able to reduce the MIC of PMB for WD101 below the <2-μg/ml cutoff value for WD101. To achieve this same PMB reduction, much higher concentrations of novobiocin (>788 μM) and pentamidine (294 μM) were required compared to our early leads. Fusidic acid and mitotane behaved like compounds 5, 33, 43, and 44, meaning they exhibited synergy with PMB against polymyxin-resistant WD101 at similar concentrations.
FIG 3.
Lead compounds exhibit synergy with polymyxin at lower concentrations than established adjuvants. (A) Checkerboard broth microdilution assays performed with PMBr E. coli WD101 illustrate the synergistic activity of these lead compounds and PMB at various concentrations. (B) Checkerboard broth microdilution assays were performed with E. coli WD101 to demonstrate the synergy between PMB and established adjuvants in the field (fusidic acid, mitotane, novobiocin, and pentamidine). Representative data from at least two biologic replicates are presented and are shown as a heat map of the percent bacterial growth.
Interestingly, none of these compounds exhibited synergy in combination with rifampin, erythromycin, or vancomycin (Fig. S2). Rifampin, erythromycin, and vancomycin target RNA polymerase, the 50S ribosomal subunit, and the peptidoglycan cell wall, respectively. Typically, these antibiotics are only capable of inhibiting the growth of Gram-negative bacteria if they are combined with other agents (like polymyxins) that can disrupt the outer membrane (28, 50–53). Since none of the five lead compounds identified in this study demonstrate synergy with these Gram-positive antibiotics suggests that these compounds do not disrupt the outer membrane to facilitate PMB activity but rather that PMB may facilitate the entry and/or activity of these compounds. These findings are in contrast to what has been reported for pentamidine, which disrupts the outer membrane to potentiate the activity of rifampin and erythromycin (29).
Lead compounds reverse polymyxin resistance against several Gram-negative bacteria with acquired polymyxin resistance.
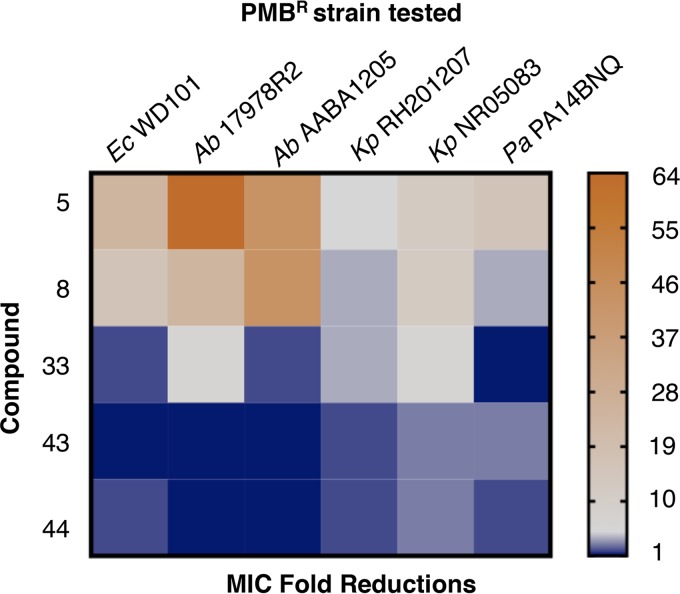
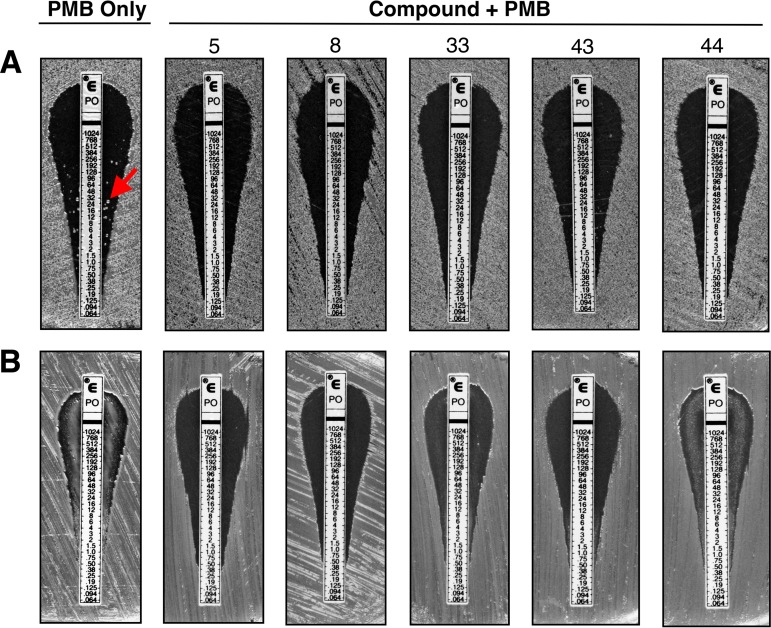
Given that A. baumannii, K. pneumoniae, and P. aeruginosa are priority pathogens that can readily develop resistance to polymyxin, we also sought to evaluate the synergistic potential of our lead compounds and PMB against known polymyxin-resistant isolates. An agar-based microdilution (Epsilometer test) method was used to determine the MIC of polymyxin in combination with 20 μM of each compound. As demonstrated in Fig. 4 and Table S3, combination therapy with compounds 5 and 8 demonstrated the greatest overall fold reductions in PMB MIC against all of the polymyxin-resistant pathogens tested. Compound 33 effectively reduced the MIC of PMB for all pathogens examined, except against a phoQ mutant of P. aeruginosa (21). Epsilometer test revealed that compounds 43 and 44 in combination therapy with PMB were effective against polymyxin-resistant K. pneumoniae (54, 55) and P. aeruginosa isolates (21, 56) but had no effect on polymyxin-resistant A. baumannii isolates. Surprisingly, compound 43 tested ineffective in combination therapy against E. coli WD101 by this method, whereas compounds 5, 8, 33, and 44 were effective, as shown previously (Fig. 3). These contradictory findings were not anticipated given the results of our checkerboard assays, but evidence in the literature suggests that inconsistency in MIC determinations often occurs between agar and broth methods for Gram-negative bacteria and that results from broth assays with log-phase growth are preferred over agar assays with stationary growth (57–59). With this in mind, it is then encouraging that all the broth dilution methods used in this study were in agreement (Fig. 3 and 5; see also Fig. S3 in the supplemental material).
FIG 4.
Lead compounds demonstrate synergy with polymyxin B against diverse polymyxin-resistant Gram-negative bacterial species. The effectiveness of PMB and the lead compounds against various polymyxin-resistant Gram-negative bacteria was determined via a PMB Epsilometer test method using CAMHA supplemented with 20 μM of compound. The heat map shows the fold reductions in the MIC of PMB for the various strains tested (data for calculations shown in Table S3). Representative data from at least two biologic replicates are provided.
FIG 5.
Lead compounds exhibit synergy with polymyxin against polymyxin-resistant A. baumannii by two methods. (A) Checkerboard broth microdilution assays performed with A. baumannii 17978R2, a pmrB constitutive mutant, were used to demonstrate the synergy between the five lead compounds and PMB at varying concentrations. The heat map indicates the percent bacterial growth. (B) Broth macrodilution assays performed with A. baumannii 17978R2 show the fold reduction in the PMB MIC when combined with 20 μM concentrations of the five lead compounds. Representative data from at least two biologic replicates are shown.
Since A. baumannii is a critical priority pathogen and the most striking MIC reductions elicited by our compounds occurred with this Gram-negative organism, we sought to better examine this relationship by performing checkerboard assays using two highly polymyxin-resistant A. baumannii isolates. First, we examined the effectiveness of lead compound synergy with PMB using a well-characterized polymyxin-resistant spontaneous point mutant of 17978, 17978R2 pmrB(T235I). This single mutation in pmrB has a dominant effect that results in constitutive expression of the pmrCAB operon in A. baumannii 17978 and enables resistance up to 8 μg/ml PMB in LB (17). We also tested for compound synergy with PMB against A. baumannii AABA1205, a clinical isolate that is highly resistant to PMB (64 μg/ml in LB) (60). Checkerboard assays demonstrate that compounds 8 and 43 are the most effective against A. baumannii, inhibiting growth well below the cutoff value of <2 μg/ml PMB in a concentration-dependent manner (Fig. 5A and S3, respectively). Figure 5A also shows that compounds 5, 33, and 44 act synergistically with PMB against 17978R2, albeit only at 2.5 μg/ml (intermediate range). These findings are further supported by the profound synergistic behavior exhibited by all of these lead compounds with PMB against 17978R2 in broth macrodilution assays (Fig. 5B), which demonstrate 4- to 30-fold reductions in PMB MIC. Likewise, checkerboard assays with AABA1205 (Fig. S3) recapitulate these findings, again with compounds 8 and 43 again offering the largest reduction in MIC.
Lead compounds synergize with polymyxin to inhibit the growth of polymyxin-resistant subpopulations in heteroresistant E. cloacae.
While agar-based methods have limitations in determining antimicrobial-adjuvant synergy relationships and antibiotic MICs, Epsilometer tests remain the best method for identifying heteroresistant bacterial subpopulations (61). Recently, the prevalence of heteroresistance in 41 clinical isolates of E. coli, S. enterica, K. pneumoniae, and A. baumannii were tested against several antibiotics and over one-fourth were identified as heteroresistant, often despite initial MIC testing as susceptible (62). Moreover, it was also recently shown that combination therapy targeting homogeneous resistance mechanisms in Gram-negative bacteria are ineffective, and that only combinations targeting multiple heteroresistance mechanisms are efficacious (63). Since the inadvertent mistreatment of polymyxin-heteroresistant bacterial infections is both a complex problem and major concern for treatment failure in the clinic, we explored the effectiveness of our lead compounds in combination with PMB against a well-described, polymyxin-heteroresistant Enterobacter cloacae strain (22). Figure 6A demonstrates that all five lead compounds are effective at inhibiting the growth of polymyxin-heteroresistant colonies. Furthermore, compounds 5 and 8 remain highly effective at inhibiting the growth of such heteroresistant colonies even in the face of previous exposure to PMB (Fig. 6B). Though compounds 33, 43, and 44 did not completely inhibit polymyxin-heteroresistant E. cloacae growth in this preexposure experiment, compounds 33 and 43 still substantially reduced the growth of polymyxin-heteroresistant E. cloacae.
FIG 6.
Lead compounds act synergistically with polymyxin B to inhibit the growth of heteroresistant E. cloacae. PMB Epsilometer tests were used to determine whether lead compounds act synergistically with PMB to inhibit the growth of highly polymyxin-resistant subpopulations of heteroresistant E. cloacae. The effectiveness of each lead compound (20 μM) and PMB against polymyxin-resistant subpopulations of heteroresistant E. cloacae was determined by two methods. (A) The MIC of PMB on CAMHA supplemented with 20 μM of compound was determined for heteroresistant E. cloacae. (B) A single PMB-resistant E. cloacae colony was selected (the red arrow indicates the selected colony; MIC = 24 μg/ml) and propagated in CAMHAB supplemented with 1 μg/ml PMB. This single-colony outgrowth was then subjected to the same Epsilometer test method described in part A. Representative data from at least two biologic replicates are shown.
Lead compounds do not affect lipid A modification in E. coli.
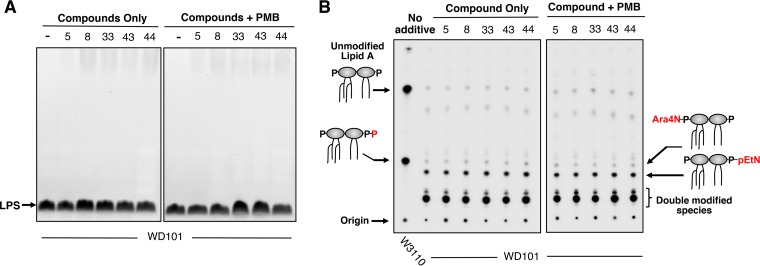
Due to the design of the whole-cell screen in this study, it was anticipated that some of the compounds identified may potentiate PMB by inhibiting enzymes involved in the modification of LPS. LPS and 32P-labeled lipid A species isolated from WD101 treated with either compounds alone or in combination with PMB were examined by SDS-PAGE gel and thin-layer chromatography (TLC), respectively. None of the five lead compounds in this study demonstrated an effect on the rough LPS (LPS lacking O-antigen) of this E. coli K-12 strain (Fig. 7A). Furthermore, these lead compounds (Fig. 7B) and the other 38 bioactives (Fig. S4) had no impact on lipid A structure with TLC analysis showing normal levels of L-Ara4N and pEtN modification required for PMB resistance. Thus, our compounds do not target LPS structure. Lead compounds were also unable to potentiate the activity of rifampin, erythromycin, or vancomycin (Fig. S2). Since the lead compounds do not exhibit synergistic activity with any of these antimicrobials, these data suggest that it is unlikely they are disrupting the outer membrane to potentiate the activity of PMB. Another possible mechanism of action could be that PMB, while unable to critically damage the cell envelope structure, still facilitates entry of these lead compounds past the outer membrane and allows them to interfere with other cellular targets.
FIG 7.
Lead compounds alone or in combination with polymyxin do not impact E. coli lipid A modifications or overall LPS structure. (A) Whether or not individual compounds (1.25 μM) with or without PMB (0.5 μg/ml) impacted the overall LPS profile was evaluated. Whole-cell lysates were treated with proteinase K and separated by SDS-PAGE, and the LPS was visualized by staining with Pro-Q Emerald 300 dye. Compared to the DMSO only control (indicated by a “–”), none of the compounds impacted the WD101 LPS glycoform lacking O antigen. The data are representative of at least two biological replicates. (B) 32P-labeled lipid A species were isolated from WD101 grown individually with 1.25 μM compound in the presence or absence of 0.2 μg/ml PMB (conditions were increased to 0.5 μg/ml PMB in a replicate assay). PMBs W3110 is used as a control and does not modify its lipid A with amine-containing substituents but does produce a species with an additional phosphate group at the 1 position. The presence of compound did not impact the level of lipid A modified with either aminoarabinose (Ara4N) or phosphoethanolamine (pEtN). Double-modified species indicate lipid A molecules with a combination of Ara4N and pEtN residues. Data shown are representative of at least two biological replicates.
Lead compounds demonstrate potent antimicrobial activity against Gram-positive bacteria and E. coli with outer membrane defects.
In order to determine whether these compounds were interacting with targets other than the outer membrane, we assessed the antimicrobial activity of our lead compounds against a panel of Gram-positive bacteria (Table 1 ) and also classified this activity as either bactericidal or bacteriostatic based on the outcome of time-to-kill assays with MRSA (Fig. 8A). Compound 5 exhibited potent antimicrobial activity against all Gram-positive bacteria tested (B. subtilis, S. aureus, and E. faecium), and exhibited impressive bactericidal activity against MRSA at two different concentrations (20 μM [Fig. 8A] and 0.625 μM [Fig. S5A]). Compound 33 was effective against all Gram-positive bacteria but B. subtilis (Table 1) and demonstrated bacteriostatic activity up to 24 h (Fig. 8A). Compounds 8 and 43 inhibited the growth of E. faecium and MRSA, but of the two, only compound 43 was effective against B. subtilis (Table 1). Likewise, while compound 43 proved to be a potent bactericidal agent against MRSA, compound 8 demonstrated bacteriostatic activity up to 24 h (Fig. 8A). Interestingly, Compound 44 failed to inhibit the growth of any Gram-positive bacteria when tested in microtiter plates (Table 1); however, in tubes this compound demonstrated bacteriostatic activity against MRSA up to 12 h (Fig. 8A).
TABLE 1.
Lead compounds exhibit antimicrobial activity against Gram-positive bacteria and Gram-negative bacteria with outer membrane defectsa
| Strain | MIC (μM) |
||||
|---|---|---|---|---|---|
| Compound 5 | Compound 8 | Compound 33 | Compound 43 | Compound 44 | |
| Bacillus subtilis 168 | 1.25 | >20 | >20 | 20 | >20 |
| Staphylococcus aureus | |||||
| 12600 | 1.25 | >20 | 10 | >20 | >20 |
| MU50 | 0.63 | 20 | 10 | 20 | >20 |
| Enterococcus faecium 19434 | 1.25 | 20 | 10 | 20 | >20 |
| Escherichia coli | |||||
| W3110 | >20 | >20 | >20 | >20 | >20 |
| WD101 | >20 | >20 | >20 | >20 | >20 |
| WBBO6 | 2.5 | 20 | >20 | >20 | >20 |
| W3LPTD | 2.5 | 20 | 5 | >20 | >20 |
| WDLPXC | 5 | >20 | 20 | 20 | >20 |
Representative data from at least two biological replicates were generated for all strains using broth dilution assays.
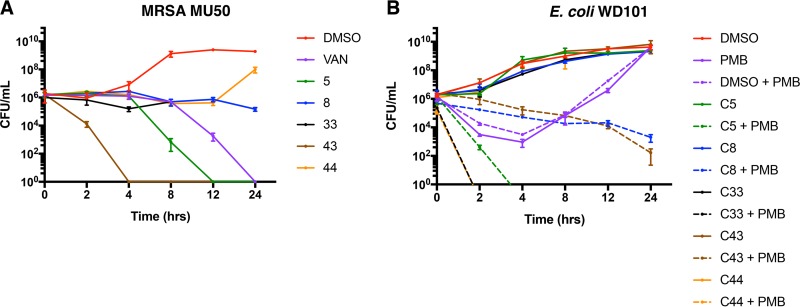
FIG 8.
All compounds demonstrate either bactericidal or bacteriostatic activity against both Gram-negative and Gram-positive bacteria. (A) Time-kill curves using 20 μM concentrations of each compound against vancomycin-intermediate MRSA MU50 reveal that compounds 5 and 43 completely kill bacteria faster than 10 μg/ml of vancomycin, indicating rapid bactericidal activity. Compounds 8, 33, and 44 inhibit bacterial growth for up to 12 h, but only compounds 8 and 33 inhibit growth up to 24 h. (B) Time-kill curves using 20 μM concentrations of each compound with or without 2.5 μg/ml PMB against polymyxin-resistant E. coli WD101 (pmrAc) demonstrate that compounds 5, 33, and 44 act synergistically with PMB to produce rapid and complete bacterial killing. Compound 43 acts synergistically with PMB to produce bactericidal activity (≥3 log10 reduction in CFU/ml) at 24 h, while compound 8 acts synergistically with PMB and exhibits prolonged bacteriostatic activity (≤3 log10 reduction in CFU/ml). Representative data from at least two biological replicates were generated using broth dilution assays.
Given these results, we evaluated our lead compounds for antimicrobial activity against three derivatives of E. coli with outer membrane permeability defects (Table 1). The first strain, WBBO6, produces deep LPS truncations in W3110, since it lacks rfaF (waaF) and rfaC (waaC), the genes that encode core oligosaccharide heptosyl transferases (64). The second strain, W3LPTD, was generated by transducing a mutant allele of lptD (lptD4213) into W3110, which encodes an essential component of the LPS transport machinery (65, 66). The last strain, WDLPXC, was made by transducing a mutant allele of lpxC (lpxC101) into WD101, resulting in reduced LPS levels as LpxC catalyzes the first committed step of lipid A biosynthesis (67, 68). In the polymyxin-susceptible strains, only compounds 5 and 8 were able to inhibit the growth of the WBBO6, while compounds 5, 8, and 33 inhibited the growth of W3LPTD. In the polymyxin-resistant strain, WDLPXC, compounds 5, 33, and 43 were effective. Using strain WDLPXC, we verified once again that our lead compounds did not impact lipid A modification (Fig. S6). While the exact differences that account for the variation in Gram-positive and Gram-negative strain susceptibility remain unclear, these data suggest that compounds 5, 8, 33, and 43 may be acting on intracellular targets, while compound 44 is not. The mechanism behind the synergy exhibited by compound 44 in combination with polymyxin B remains unclear.
Since four of the five first-generation lead compounds identified in this study likely have intracellular targets, we desired to determine the exact behavior of their activity in Gram-negative bacteria. Specifically, we performed time-to-kill assays with E. coli WD101 and 20 μM concentrations of each compound in combination with 2.5 μg/ml polymyxin. We determined that compound 5 is both a fast-acting and potent bactericidal compound (20 μM [Fig. 8B] and 2.5 μM [Fig. S5B]). Compounds 33 and 44 are also fast-acting bactericidal compounds, compound 43 is a slower-acting bactericidal compound, and compound 8 is bacteriostatic up to 24 h (Fig. 8B). These data are informative since they suggest that whatever their mechanism of action, at least four of these five compounds impact pathways that are essential to bacterial survival.
Early lead compounds exhibit specific and minimal cytotoxicity at low concentrations.
Based on its profound enhancement of PMB activity against multiple polymyxin-resistant Gram-negative bacterial organisms, compounds 5, 8, and 33 were identified as potential front-runners among our early lead candidates. However, the proper selection of any early lead compound must include an assessment of tractability for chemical optimization and potential toxicity (Table S2). While computational predictions are helpful in the drug development process, they are also not an adequate substitute for empirical data. Given that cytotoxicity is often a variable that hampers the advancement of new drugs in pharmaceutical development, we evaluated all of our early lead compounds for their likelihood to directly kill host cells in two different but well-established drug targeting models.
First, we used a robust, 48-h, tetrazolium (MTT) reduction assay (69) using immortalized human hepatocytes (HepG2) (70) and compared the activity of our compounds to that of an established polymyxin adjuvant and chemotherapeutic agent with known hepatotoxicity (mitotane) (71, 72). Figure S7A shows that all lead compounds exhibit concentration-dependent cytotoxicity against these hepatocytes, a feature that is not uncommon among first-generation drugs that have not undergone optimization. Encouragingly, all compounds are less hepatotoxic than mitotane at ≤2.5 μM, and some compounds (8, 33, and 44) outperform mitotane at concentrations up to 10 μM. Furthermore, at concentrations below what elicits hepatotoxicity, two of our leads are strong antimicrobials against Gram-positive bacteria (Table 1; Fig. S5A), and most effectively synergize the activity of PMB against Gram-negative bacteria (Fig. 3 and 5; see also Fig. S5B).
Given the lipophilic nature of our compounds, we also evaluated them for the potential to lyse human red blood cells (hRBCs). Hemolysis assays at both 45 min (data not shown) and 3 h (Fig. S7B) over a range of compound concentrations (0.625 to 40 μM) indicated that none of our compounds showed significant lysis of hRBCs at 40 μM. These preliminary in vitro cytotoxicity tests are encouraging, since they do not indicate widespread nonspecific toxicity to host cells. These data also support the computational predictions that our five lead compounds are tractable and warrant further chemical optimization, which could in turn mitigate cytotoxicity and yield improved therapeutic windows. Impressively, the antibacterial activity of these leads has also been achieved without repeat dosing, which is sometimes required by other antimicrobial adjuvants (73). Taken together, the outcome of these toxicity experiments is promising, since they underscore the overall effectiveness and potential feasibility of pursuing the development of these early leads as antimicrobial adjuvants.
DISCUSSION
Repurposing old pharmaceuticals and/or identifying new bioactives with antimicrobial activity or the ability to act as effective polymyxin adjuvants are costly and labor-intensive endeavors. In addition, the inherent risk of renal toxicity from polymyxin administration requires that the polymyxin adjuvant not only potentiate polymyxin at very low doses but also have minimal toxicity itself. The likelihood of identifying such highly effective and yet inert drug targets is low (3, 36, 74). Many in the field have attempted both avenues to identify either new antimicrobials or suitable adjuvants. Recent repurposed pharmaceuticals have been experimentally used with varying success to potentiate the activity of polymyxins; however, none of these has been demonstrated to be effective against all of the ESKAPE pathogens (28, 46, 47, 49, 71, 73, 75–81). New antimicrobials have also been identified and shown to be effective against polymyxin-resistant bacterial strains, which raises the hope that new FDA-approved antibiotics may emerge in the future (30–35, 82–88). In the interim, other groups have pursued developing polymyxin-sparing regimes, such as pentamidine, in order to avoid the clinical use of polymyxins and circumvent their inherent toxicity (29). However, new evidence has already emerged that A. baumannii can develop intrinsic resistance to pentamidine (89), which just further illustrates the importance of continued investigation into polymyxin resistance mechanisms and the constant need for new therapeutic interventions.
In this study, we utilized a chemical library composed of synthetic molecules, purified natural products, off-patent products, FDA-approved drugs, and other bioactives to perform our whole-cell screen. We then verified the synergistic activity of 43 compounds with polymyxin using robust antimicrobial susceptibility testing methods and also confirmed their relevance by performing orthogonal tests against multiple, polymyxin-resistant Gram-negative bacterial pathogens. In doing so, we determined that micromolar concentrations of compounds 5 and 8 can dramatically reduce PMB to a susceptible and possibly therapeutically relevant dose (<2 μg/ml). We also saw that compounds 33, 43, and 44 exhibited synergy with polymyxin B but were only able to reduce polymyxin B to 2.5 μg/ml. While this was deemed by us to be an intermediate-level reduction, these leads still vastly improve the activity of PMB and are promising candidates for future development.
We also found that our five lead compounds are highly effective at inhibiting the growth of heteroresistant E. cloacae. The issue of antibiotic heteroresistance is not often considered early in drug discovery, particularly when screening compounds for activity as polymyxin adjuvants. By demonstrating the effectiveness of our lead compounds against one heteroresistant and several different polymyxin-resistant Gram-negative bacteria, we show that our compounds are effective across multiple multidrug-resistant pathogen species with different chromosomal mutations.
By performing checkerboard assays with the lead compounds in combination with either rifampin, erythromycin, and vancomycin, we were able to determine that none were potentiating the activity of PMB by disrupting the outer membrane. TLC data also indicated that none of the 43 bioactive compounds identified in this screen impacted LPS biosynthesis or the modification of the lipid A anchor. Lastly, we evaluated the five lead compounds in this study for antimicrobial activity against Gram-positive bacteria or Gram-negative bacteria with defective outer membrane permeability barriers to determine whether they were acting on intracellular targets. Our data suggest that PMB is likely facilitating the entrance of compounds 5, 8, 33, and 43 beyond the outer membrane to inhibit such intracellular targets. There is no evidence to suggest that compound 44 acts in this fashion.
In conclusion, we present five first-generation lead compounds for consideration, some of which can effectively be used at very low micromolar concentrations to dramatically lower the dose of polymyxin, a feat that is not only safer but also potentially feasible in a clinical setting. In addition, four compounds may warrant further optimization as potent Gram-positive antimicrobials. While the exact mechanism of action for these five lead compounds remains unclear, this study clearly demonstrates that our bioactives could be effective against all of ESKAPE pathogens not only by reinvigorating polymyxin for use against polymyxin-resistant Gram-negative bacterial infections but also by yielding new treatment avenues for multidrug-resistant Gram-positive bacteria. Much remains to be learned about these lead compounds, and future efforts by our group will be focused on refining these targets, determining their mechanism of action, and investigating their effectiveness and safety in relevant animal models.
MATERIALS AND METHODS
Reagents.
Screening stocks (10 mg/ml) of the following Maybridge compounds were stored in small aliquots at −20°C in 100% (vol/vol) dimethyl sulfoxide (DMSO): 5 (HTS03780), 8 (MGH00136), 33 (NH00518), 43 (CD04455), and 44 (JP00319). The following commercial pharmaceutical chemicals were dissolved in either sterile molecular-grade water or 100% (vol/vol) DMSO (Sigma-Aldrich), where indicated, and stored at –20°C: 1 mg/ml polymyxin B sulfate 25MU (Chem-Impex International, Inc.), water; 50 mg/ml novobiocin (Sigma-Aldrich), water; 50 mg/ml pentamidine isethionate (Sigma-Aldrich), DMSO; 1 mg/ml mitotane (Sigma-Aldrich), DMSO; 10 mg/ml erythromycin (Fisher BioReagents), DMSO; 1 mg/ml rifampin (Sigma-Aldrich), water; and 1 mg/ml vancomycin (Research Products International), water. Reagents were used immediately and not subjected to repeat freeze-thawing. In all experiments, lead compounds were dissolved in solvent (DMSO) and diluted to a final concentration of 2% DMSO.
Bacterial strains and culture conditions.
Unless stated otherwise, the bacterial strains in this study (see Table S1 in the supplemental material) were grown in BBL cation-adjusted Mueller-Hinton broth (CAMHB) or agar (CAMHA) at 37°C. Single colonies were used to generate overnight (16 to 20 h) cultures supplemented with compounds and other pharmaceutical chemicals as appropriate. Bacteria were subcultured starting at an optical density of 600 nm (OD600) of 0.05 and grown to either mid-log phase (OD600 = 0.5) or stationary phase (OD600 = 1.0). OD600 measurements were performed using a Synergy H1 hybrid multimode microplate reader (BioTek).
Mutant alleles of lptD and lpxC were introduced into strains by P1vir transduction through linkage to nearby markers, namely, carB::Tn10 for lptD4213 and leuB::Tn10 for lpxC101. Allele replacement for W3LPTD was confirmed by sensitivity to bile salts and vancomycin, PCR, and Sanger sequencing. The point mutation of WDLPXC was confirmed by sensitivity to rifampin and Sanger sequencing.
High-throughput compound screening.
The McMaster high-throughput screening (HTS) lab at the Centre for Microbial Chemical Biology at McMaster University screened the Canadian Compound Collection (a compilation of 29,569 synthetic molecules, purified natural products, off-patent products, FDA-approved drugs, and known bioactives sourced from six commercial vendors) against whole bacterial cells in LB culture (E. coli W3110 and WD101) using a fully automated robotic system. Bacteria were subcultured for 6 h prior to being standardized to an OD600 of 0.08 (0.5 McFarland). The transfer of control dilutions, compound dilutions, and cultures to 96-well, polystyrene plates was completed using a 96-channel head on a Biomek FXP liquid handler (Beckman Coulter, Inc.). Once the reagents were aliquoted, the plates were incubated for 12 h at 37°C before being transferred to an Envision plate reader (Perkin-Elmer) for OD600 measurements and analysis using an Oracle-based informatics platform.
In the primary screen, all 29,569 compounds were tested in quadruplicate for their ability to act synergistically with polymyxin B to inhibit the growth of E. coli WD101. Specifically, 20 μM concentrations of each compound were tested, both alone and in combination with 8 μg/ml of polymyxin B, alongside eight replicate wells of the high-growth control (2% DMSO) and eight replicate wells of the low-growth control (2% DMSO plus 30 μg/ml chloramphenicol) per microtiter plate. The percent growth (%G) of each well was calculated using the average (μ) of eight high-growth controls (+c) and eight low-growth controls (–c) on each individual assay plate:
The statistical parameter, Z′, was also calculated using the standard deviations (σ) of the +c and –c values and their respective μ values to ensure active compounds were only identified using high-quality screening criteria. All compounds were screened in n = 2 replicate experiments.
Active compounds capable of potentiating polymyxin B were identified if (i) their %G in replicate assays differed by <10% and also resulted in (ii) an E. coli WD101%G of <65% with Z′ > 0.5.
In the secondary screen, the 356 compounds identified by the first robotic screen were assessed not only for the ability to act synergistically with polymyxin B to inhibit E. coli WD101 growth but also for their ability to not inhibit E. coli growth (W3110 and WD101) when used alone. Bacterial suspensions, compound dilutions, and controls were prepared as before, and 43 compounds were selected in the secondary screen by identifying those that met the following criteria: (i) W3110 and WD101%G were ≥65% when used alone, (ii) WD101%G is <1.0% when used in combination with polymyxin B, and (iii) and Z′ > 0.5. The physiochemical properties of the early lead compounds (Fig. 2B and Table S2) were determined using the cxcalc command line tool in ChemAxon. All pH-dependent properties (logD) were determined at pH 7.
EOP assays.
EOP assays were performed as done previously (90). Bacteria were propagated to stationary phase (OD600 = 1.0), and 10-fold serial dilutions were prepared. The bacterial dilutions were then plated onto no additive media, media supplemented with 3 μg/ml polymyxin B, and media supplemented with a 20 μM compound concentration and 3 μg/ml polymyxin B. All plates were incubated 16 to 20 h at 37°C.
Evaluation of LPS by SDS-PAGE.
For monitoring changes in LPS, bacteria were grown in LB at 37°C until mid-exponential phase. Cell amounts were normalized based upon optical density to an OD600 of 1.0 in 100 μl of 1× LDS loading buffer supplemented with 4% β-mercaptoethanol. Whole cells were treated with proteinase K as described by Hitchcock and Brown (91), and the samples were separated by SDS-PAGE using a 4 to 12% Bis-Tris gel, followed by staining using Pro-Q Emerald 300 dye LPS gel staining kit (Molecular Probes) as previously described (92).
Evaluation of lipid A species by TLC.
For isolation and comparison of lipid A species, cultures were grown in LB with 2.5 μCi of 32P/ml (Perkin-Elmer) and grown with either compound alone or in combination with polymyxin B to stationary phase (OD600 = 1.0). The extraction and isolation of 32P-labeled lipid A was achieved via mild acid hydrolysis, as previously described (93, 94). The 32P-labeled lipid A species (10,000 cpm/sample) were spotted onto silica plates; air dried for 15 min; separated using a mobile-phase TLC solvent system consisting of chloroform, pyridine, 88% formic acid, and water (50:50:16:5, vol/vol/vol/vol); and visualized by phosphorimaging analysis (Bio-Rad PMI).
Broth macrodilution method.
Outer membrane-defective E. coli strains (WBBO6, W3LPTD, and WDLPXC) were propagated in LB because these strains exhibit slow and altered growth phenotypes when propagated in cation-adjusted Mueller-Hinton broth. All other bacterial strains were propagated in CAMHB, as indicated previously. Twofold serial dilutions of each compound were prepared in 5.0 ml of LB, and after inoculation with bacteria, the glass tubes were incubated at 37°C and 220 rpm for 8 h. Bacterial growth was assessed qualitatively (observations), so the MIC was defined as the concentration that resulted in no visible bacterial growth.
Broth microdilution method.
Twofold serial dilutions of each compound were prepared in 200 μl of cation-adjusted Mueller-Hinton broth per well using 96-well, polypropylene, microtiter plates (Corning). After bacterial inoculation, these microtiter plates were covered with a sterile, adherent, aeration filter (Aeroseal) and incubated at 37°C and 220 rpm for 8 h. Bacterial growth was assessed both qualitatively (observations) and quantitatively (OD600). The MIC was defined as the concentration that inhibited bacterial growth by >90% compared to the solvent control.
Checkerboard assay method.
We created 8 × 8 matrices of consisting of two chemicals (e.g., compound and known antimicrobial) using 2-fold serial dilutions of each reagent in 96-well, polypropylene, microtiter plates. After bacterial inoculation, the plates were covered with a sterile, adherent, aeration filter (Aeroseal) and incubated at 37°C and 220 rpm for 8 h. Bacterial growth was assessed both qualitatively (observations) and quantitatively (OD600). The MIC was defined as the concentration that inhibited bacterial growth by >90% compared to the solvent control.
Epsilometer test method.
Mid-log-phase (OD600 = 0.5) bacterial strains were diluted 1:50 and aseptically applied to agar plates supplemented both with or without 20 μM compound. Polymyxin B E-strip tests (bioMérieux) were added to the plates, followed by incubation overnight at 37°C. MIC determinations were made by observing where the confluent bacterial growth occurs relative to the Epsilometer test concentration ladder.
MIC determinations.
All MIC determinations were made in duplicate and, excluding the microtiter plate material (polypropylene) and incubation time (8 h), these assays were performed in accordance with CLSI guidelines (42, 43, 45). Bacterial growth was assessed both qualitatively (observations) and quantitatively (OD600). The MIC was defined as the concentration that inhibited bacterial growth by >90% compared to the solvent control. In all experiments, the solvent (DMSO) was present at a final concentration of <2%. Since clinical breakpoints have not been assigned for A. baumannii, P. aeruginosa, and K. pneumoniae by the CLSI (42) or the FDA (95), PMB comparisons were made at ≤2 μg/ml, as done experimentally by others in the field (28, 29). MIC fold reductions were determined by dividing the MIC of polymyxin B alone by its MIC in combination with a 20 μM concentration of the compound.
Time-kill assays.
All bacterial strains were propagated in CAMHB, as indicated previously. Experiments with E. coli WD101 were conducted using 2.5 μg/ml polymyxin B and either a 2.5 or 20 μM concentration of the compound, depending upon the assay. Experiments with MRSA MU50 were carried out using either a 0.625 or 20 μM concentration of the compound, depending upon the assay. Glass tubes containing 5.0 ml of supplemented CAMHB were inoculated with 1.0 × 106 CFU/ml and incubated at 37°C and 220 rpm. Samples were obtained at 0, 2, 4, 8, 12, and 24 h; serially diluted; spread on CAMHA; and incubated at 37°C for 24 h to quantify viable bacteria. As per the field standard, synergy was defined as a reduction of ≥2 log10 CFU/ml, and bactericidal determinations were made at reductions of ≥3 log10 CFU/ml (45).
Hemolysis assays.
Fresh, normal, pooled hRBCs (Innovative Research) were washed with sterile 1× phosphate-buffered saline (PBS) until the supernatant was clear and devoid of excess free hemoglobin. The final hRBC pellet was gently resuspended to 4% (vol/vol) in sterile 1× PBS. Twofold serial dilutions of each compound in sterile water were prepared in 96-well, V-bottom, polystyrene, microtiter plates and combined with an equal volume of hRBCs to yield a final concentration of 2% (vol/vol) hRBCs/well. Compound concentrations ranged from 0.625 to 40 μM (the data for 40 μM are shown in Fig. S7B). Sterile 1× PBS and 0.1% sodium dodecyl sulfate (SDS) were used as negative and positive controls, respectively. Duplicate microtiter plates were incubated at 37°C without shaking, one for 45 min and the other for 3 h. At the chosen endpoint, plates were centrifuged 1,000 × g and 25°C for 5 min to pellet any remaining intact hRBCs. Supernatants were transferred to flat-bottom, 96-well, polystyrene microtiter plates, and hemolysis was assessed both qualitatively (observations) and quantitatively (OD450). The percent hemolysis was calculated, and each condition was tested in triplicate.
Mammalian cytotoxicity assays.
HepG2 cells (ATCC HB-8065) were propagated until confluent as done previously (70). Cells were then seeded into sterile, 96-well, polystyrene, microtiter plates at 5,000 cells/well and incubated 16 h at 37°C with 5% CO2. Minimal essential media (MEM) was removed, and the cells were washed three times with sterile 1× PBS to remove any residual media containing 10% fetal bovine serum (FBS). Twofold serial dilutions of each compound in MEM containing the indicated compound and no FBS were prepared, and compound concentrations ranged from 2.5 to 20 μM (Fig. S7A). Cells were then incubated for 48 h at 37οC with 5% CO2. This treatment medium was then removed, and the cells were washed once with sterile 1× PBS before being immersed in fresh MEM without FBS but spiked with a final concentration of 0.5 mg/ml MTT (3-[4,5-dimtheylthiazol-2,5-diphenyl] tetrazolium bromide; EMD Millipore). The cells were then incubated for 4 h at 37°C with 5% CO2. The MTT-containing MEM was removed, 50 μl of solubilization solution (10% Triton X-100 and 0.1N HCl in anhydrous isopropanol) was added to each well, and the plate was incubated for 10 min at 37οC with 5% CO2. The absorbance was measured at 540 nm, and each condition examined was tested in triplicate. The percent survival was calculated by comparing the results to the negative control (PBS).
Supplementary Material
ACKNOWLEDGMENTS
The National Institutes of Health provided financial support to M.S.T. (grants AI129940, AI138576, and AI076322).
We thank the McMasters HTS Laboratory for performing the robotic screen, Jakob Magolan for performing the medicinal chemistry analysis, Anne-Catrin Uhlemann and William T. Doerrler for providing clinical isolates, and Brent Simpson for providing helpful comments on the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Falagas ME, Kasiakou SK, Saravolatz LD. 2005. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin Infect Dis 40:1333–1341. doi: 10.1086/429323. [DOI] [PubMed] [Google Scholar]
- 2.Falagas ME, Kasiakou SK. 2006. Toxicity of polymyxins: a systematic review of the evidence from old and recent studies. Crit Care 10:R27. doi: 10.1186/cc3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farha MA, Brown ED. 2019. Drug repurposing for antimicrobial discovery. Nat Microbiol 1 doi: 10.1038/s41564-019-0357-1. [DOI] [PubMed] [Google Scholar]
- 4.O’Neill J. 2016. Tackling drug-resistant infections globally: final report and recommendations. Review on Antimicrob Resistance, London, United Kingdom. [Google Scholar]
- 5.World Health Organization. 2017. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2013. Antibiotic/antimicrobial resistance (AR/AMR): biggest threats and data. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 7.The White House. 2015. National action plan for combating antibiotic-resistant bacteria. The White House, Washington, DC. [Google Scholar]
- 8.National Institute of Allergy and Infectious Disease. 2017. NIAID strategic plan. National Institute of Allergy and Infectious Diseases, Bethesda, MD. [Google Scholar]
- 9.Henderson JC, Zimmerman SM, Crofts AA, Boll JM, Kuhns LG, Herrera CM, Trent MS. 2016. The power of asymmetry: architecture and assembly of the Gram-negative outer membrane lipid bilayer. Annu Rev Microbiol 70:255–278. doi: 10.1146/annurev-micro-102215-095308. [DOI] [PubMed] [Google Scholar]
- 10.Park BS, Song DH, Kim HM, Choi B-S, Lee H, Lee J-O. 2009. The structural basis of lipopolysaccharide recognition by the TLR4–MD-2 complex. Nature 458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 11.Needham BD, Carroll SM, Giles DK, Georgiou G, Whiteley M, Trent MS. 2013. Modulating the innate immune response by combinatorial engineering of endotoxin. Proc Natl Acad Sci U S A 110:1464–1469. doi: 10.1073/pnas.1218080110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Needham BD, Trent MS. 2013. Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nat Rev Microbiol 11:467–481. doi: 10.1038/nrmicro3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656. doi: 10.1128/mmbr.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herrera CM, Hankins JV, Trent MS. 2010. Activation of PmrA inhibits LpxT‐dependent phosphorylation of lipid A promoting resistance to antimicrobial peptides. Mol Microbiol 76:1444–1460. doi: 10.1111/j.1365-2958.2010.07150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tran AX, Lester ME, Stead CM, Raetz CR, Maskell DJ, McGrath SC, Cotter RJ, Trent MS. 2005. Resistance to the antimicrobial peptide polymyxin requires myristoylation of Escherichia coli and Salmonella Typhimurium lipid A. J Biol Chem 280:28186–28194. doi: 10.1074/jbc.M505020200. [DOI] [PubMed] [Google Scholar]
- 16.Rubin EJ, Herrera CM, Crofts AA, Trent MS. 2015. PmrD is required for modifications to Escherichia coli endotoxin that promote antimicrobial resistance. Antimicrob Agents Chemother 59:2051–2061. doi: 10.1128/AAC.05052-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arroyo LA, Herrera CM, Fernandez L, Hankins JV, Trent MS, Hancock RE. 2011. The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrob Agents Chemother 55:3743–3751. doi: 10.1128/AAC.00256-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pelletier MR, Casella LG, Jones JW, Adams MD, Zurawski DV, Hazlett KRO, Doi Y, Ernst RK. 2013. Unique structural modifications are present in the lipopolysaccharide from colistin-resistant strains of Acinetobacter baumannii. Antimicrob Agents Chemother 57:4831. doi: 10.1128/AAC.00865-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng H-Y, Chen Y-F, Peng H-L. 2010. Molecular characterization of the PhoPQ-PmrD-PmrAB mediated pathway regulating polymyxin B resistance in Klebsiella pneumoniae CG43. J Biomed Sci 17:60. doi: 10.1186/1423-0127-17-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nowicki EM, O’Brien JP, Brodbelt JS, Trent MS. 2015. Extracellular zinc induces phosphoethanolamine addition to Pseudomonas aeruginosa lipid A via the ColRS two-component system. Mol Microbiol 97:166–178. doi: 10.1111/mmi.13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gellatly SL, Needham B, Madera L, Trent MS, Hancock RE. 2012. The Pseudomonas aeruginosa PhoP-PhoQ two-component regulatory system is induced upon interaction with epithelial cells and controls cytotoxicity and inflammation. Infect Immun 80:3122–3131. doi: 10.1128/IAI.00382-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Band VI, Crispell EK, Napier BA, Herrera CM, Tharp GK, Vavikolanu K, Pohl J, Read TD, Bosinger SE, Trent MS, Burd EM, Weiss DS. 2016. Antibiotic failure mediated by a resistant subpopulation in Enterobacter cloacae. Nat Microbiol 1:16053. doi: 10.1038/nmicrobiol.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gooderham WJ, Gellatly SL, Sanschagrin F, McPhee JB, Bains M, Cosseau C, Levesque RC, Hancock RE. 2009. The sensor kinase PhoQ mediates virulence in Pseudomonas aeruginosa. Microbiology 155:699–711. doi: 10.1099/mic.0.024554-0. [DOI] [PubMed] [Google Scholar]
- 24.Sun J, Zhang H, Liu Y-H, Feng Y. 2018. Towards understanding MCR-like colistin resistance. Trends Microbiol 26:794–808. doi: 10.1016/j.tim.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 26.Ye H, Li Y, Li Z, Gao R, Zhang H, Wen R, Gao GF, Hu Q, Feng Y. 2016. Diversified mcr-1-harbouring plasmid reservoirs confer resistance to colistin in human gut microbiota. mBio 7:e00177-16. doi: 10.1128/mBio.00177-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Q, Sun J, Li J, Ding Y, Li X-P, Lin J, Hassan B, Feng Y. 2017. Expanding landscapes of the diversified mcr-1-bearing plasmid reservoirs. Microbiome 5:70. doi: 10.1186/s40168-017-0288-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacNair CR, Stokes JM, Carfrae LA, Fiebig-Comyn AA, Coombes BK, Mulvey MR, Brown ED. 2018. Overcoming mcr-1-mediated colistin resistance with colistin in combination with other antibiotics. Nat Commun 9:458. doi: 10.1038/s41467-018-02875-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stokes JM, MacNair CR, Ilyas B, French S, Côté J-P, Bouwman C, Farha MA, Sieron AO, Whitfield C, Coombes BK, Brown ED. 2017. Pentamidine sensitizes Gram-negative pathogens to antibiotics and overcomes acquired colistin resistance. Nat Microbiol 2:17028–17028. doi: 10.1038/nmicrobiol.2017.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barker WT, Martin SE, Chandler CE, Nguyen TV, Harris TL, Goodell C, Melander RJ, Doi Y, Ernst RK, Melander C. 2017. Small molecule adjuvants that suppress both chromosomal and mcr-1 encoded colistin-resistance and amplify colistin efficacy in polymyxin-susceptible bacteria. Bioorg Med Chem 25:5749–5753. doi: 10.1016/j.bmc.2017.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barker WT, Chandler CE, Melander RJ, Ernst RK, Melander C. 2019. Tryptamine derivatives disarm colistin resistance in polymyxin-resistant Gram-negative bacteria. Bioorg Med Chem 27:1776–1788. doi: 10.1016/j.bmc.2019.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris TL, Worthington RJ, Hittle LE, Zurawski DV, Ernst RK, Melander C. 2014. Small molecule downregulation of PmrAB reverses lipid A modification and breaks colistin resistance. ACS Chem Biol 9:122–127. doi: 10.1021/cb400490k. [DOI] [PubMed] [Google Scholar]
- 33.Huggins WM, Barker WT, Baker JT, Hahn NA, Melander RJ, Melander C. 2018. Meridianin D analogues display antibiofilm activity against MRSA and increase colistin efficacy in Gram-negative bacteria. ACS Med Chem Lett 9:702–707. doi: 10.1021/acsmedchemlett.8b00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huigens RW III, Reyes S, Reed CS, Bunders C, Rogers SA, Steinhauer AT, Melander C. 2010. The chemical synthesis and antibiotic activity of a diverse library of 2-aminobenzimidazole small molecules against MRSA and multidrug-resistant A. baumannii. Bioorg Med Chem 18:663–674. doi: 10.1016/j.bmc.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 35.Minrovic BM, Jung D, Melander RJ, Melander C. 2018. New class of adjuvants enables lower dosing of colistin against Acinetobacter baumannii. ACS Infect Dis 4:1368–1376. doi: 10.1021/acsinfecdis.8b00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poirel L, Jayol A, Nordmann P. 2017. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev 30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karvanen M, Malmberg C, Lagerbäck P, Friberg LE, Cars O. 2017. Colistin is extensively lost during standard in vitro experimental conditions. Antimicrob Agents Chemother 61:e00857-17. doi: 10.1128/AAC.00857-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Friedrich C, Scott MG, Karunaratne N, Yan H, Hancock R. 1999. Salt-resistant alpha-helical cationic antimicrobial peptides. Antimicrob Agents Chemother 43:1542. doi: 10.1128/AAC.43.7.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hancock R. 1997. The interaction of cationic peptides with bacteria. Antimicrobial Peptides Gordon Research Conference, 4 March 1997, Ventura, CA. [Google Scholar]
- 40.Albur M, Noel A, Bowker K, MacGowan A. 2014. Colistin susceptibility testing: time for a review. J Antimicrob Chemother 69:1432–1434. doi: 10.1093/jac/dkt503. [DOI] [PubMed] [Google Scholar]
- 41.EUCAST. 2000. Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by agar dilution. Clin Microbiol Infect 6:509–515. doi: 10.1046/j.1469-0691.2000.00142.x. [DOI] [PubMed] [Google Scholar]
- 42.Clinical and Laboratory Standards Institute. 2019. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 9th ed. CLSI document M100 ED29 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 43.Clinical and Laboratory Standards Institute. 2018. Development of in vitro susceptibility testing criteria and quality control parameters, 5th ed CLSI guideline M23 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 44.EUCAST. 2003. Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. Clin Microbiol Infect 9:ix–xv. doi: 10.1046/j.1469-0691.2003.00790.x. [DOI] [PubMed] [Google Scholar]
- 45.Veeraraghavan B, Vijayakumar S, Pragasam AK, Bakthavachalam YD, Prakash JA. 2019. Antimicrobial susceptibility testing methods for Acinetobacter spp, p 23–37. In Acinetobacter baumannii. Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- 46.Mandler MD, Baidin V, Lee J, Pahil KS, Owens TW, Kahne D. 2018. Novobiocin enhances polymyxin activity by stimulating lipopolysaccharide transport. J Am Chem Soc 140:6749–6753. doi: 10.1021/jacs.8b02283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.May JM, Owens TW, Mandler MD, Simpson BW, Lazarus MB, Sherman DJ, Davis RM, Okuda S, Massefski W, Ruiz N, Kahne D. 2017. The antibiotic novobiocin binds and activates the ATPase that powers lipopolysaccharide transport. J Am Chem Soc 139:17221–17224. doi: 10.1021/jacs.7b07736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cebrero-Cangueiro T, Álvarez-Marín R, Labrador-Herrera G, Smani Y, Cordero-Matía E, Pachón J, Pachón-Ibáñez ME. 2018. In vitro activity of pentamidine alone and in combination with aminoglycosides, tigecycline, rifampicin, and doripenem against clinical strains of carbapenemase-producing and/or colistin-resistant Enterobacteriaceae. Front Cell Infect Microbiol 8:363. doi: 10.3389/fcimb.2018.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Domalaon R, De Silva PM, Kumar A, Zhanel GG, Schweizer F, Domalaon R, De Silva PM, Kumar A, Zhanel GG, Schweizer F. 2019. The anthelmintic drug niclosamide synergizes with colistin and reverses colistin resistance in Gram-negative bacilli. Antimicrob Agents Chemother 63:e02574-18. doi: 10.1128/AAC.02574-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nikaido H. 1994. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science 264:382. doi: 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- 51.Vaara M, Vaara T. 1983. Polycations as outer membrane-disorganizing agents. Antimicrob Agents Chemother 24:114–122. doi: 10.1128/aac.24.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Viljanen P, Vaara M. 1984. Susceptibility of Gram-negative bacteria to polymyxin B nonapeptide. Antimicrob Agents Chemother 25:701–705. doi: 10.1128/aac.25.6.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vaara M. 1992. Agents that increase the permeability of the outer membrane. Microbiol Rev 56:395–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Greco I, Emborg AP, Jana B, Molchanova N, Oddo A, Damborg P, Guardabassi L, Hansen PR. 2019. Characterization, mechanism of action and optimization of activity of a novel peptide-peptoid hybrid against bacterial pathogens involved in canine skin infections. Sci Rep 9:3679–3679. doi: 10.1038/s41598-019-39042-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jana B, Cain AK, Doerrler WT, Boinett CJ, Fookes MC, Parkhill J, Guardabassi L. 2017. The secondary resistome of multidrug-resistant Klebsiella pneumoniae. Sci Rep 7:42483–42483. doi: 10.1038/srep42483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rahme L, Stevens E, Wolfort S, Shao J, Tompkins R, Ausubel F. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 57.Hindler JA, Humphries RM. 2013. Colistin MIC variability by method for contemporary clinical isolates of multidrug-resistant Gram-negative bacilli. J Clin Microbiol 51:1678. doi: 10.1128/JCM.03385-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Greulich P, Scott M, Evans MR, Allen RJ. 2015. Growth‐dependent bacterial susceptibility to ribosome‐targeting antibiotics. Mol Syst Biol 11:796. doi: 10.15252/msb.20145949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haugan MS, Løbner-Olesen A, Frimodt-Møller N, Haugan MS, Løbner-Olesen A, Frimodt-Møller N. 2019. Comparative activity of ceftriaxone, ciprofloxacin, and gentamicin as a function of bacterial growth rate probed by Escherichia coli chromosome replication in the mouse peritonitis model. Antimicrob Agents Chemother 63:e02133-18. doi: 10.1128/AAC.02133-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Crittenden CM, Herrera CM, Williams PE, Ricci DP, Swem LR, Trent MS, Brodbelt JS. 2018. Mapping phosphate modifications of substituted lipid A via a targeted MS(3) CID/UVPD strategy. Analyst 143:3091–3099. doi: 10.1039/c8an00561c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sherman EX, Wozniak JE, Weiss DS. 2019. Methods to evaluate colistin heteroresistance in Acinetobacter baumannii, p 39–50. In Acinetobacter baumannii. Springer, New York, NY. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nicoloff H, Hjort K, Levin BR, Andersson DI. 2019. The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat Microbiol 4:504. doi: 10.1038/s41564-018-0342-0. [DOI] [PubMed] [Google Scholar]
- 63.Band VI, Hufnagel DA, Jaggavarapu S, Sherman EX, Wozniak JE, Satola SW, Farley MM, Jacob JT, Burd EM, Weiss DS. 2019. Antibiotic combinations that exploit heteroresistance to multiple drugs effectively control infection. Nat Microbiol 4:1627. doi: 10.1038/s41564-019-0480-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brabetz W, Muller‐Loennies S, Holst O, Brade H. 1997. Deletion of the heptosyltransferase genes rfaC and rfaF in Escherichia coli K‐12 results in an Re‐type lipopolysaccharide with a high degree of 2‐aminoethanol phosphate substitution. Eur J Biochem 247:716–724. doi: 10.1111/j.1432-1033.1997.00716.x. [DOI] [PubMed] [Google Scholar]
- 65.Braun M, Silhavy TJ. 2002. Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol Microbiol 45:1289–1302. doi: 10.1046/j.1365-2958.2002.03091.x. [DOI] [PubMed] [Google Scholar]
- 66.Sampson BA, Misra R, Benson SA. 1989. Identification and characterization of a new gene of Escherichia coli K-12 involved in outer membrane permeability. Genetics 122:491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beall B, Lutkenhaus J. 1987. Sequence analysis, transcriptional organization, and insertional mutagenesis of the envA gene of Escherichia coli. J Bacteriol 169:5408. doi: 10.1128/jb.169.12.5408-5415.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sutterlin HA, Shi H, May KL, Miguel A, Khare S, Huang KC, Silhavy TJ. 2016. Disruption of lipid homeostasis in the Gram-negative cell envelope activates a novel cell death pathway. Proc Natl Acad Sci U S A 113:E1565. doi: 10.1073/pnas.1601375113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mosmann T. 1983. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 70.Fotakis G, Timbrell JA. 2006. In vitro cytotoxicity assays: comparison of LDH, neutral red, MTT and protein assay in hepatoma cell lines following exposure to cadmium chloride. Toxicol Lett 160:171–177. doi: 10.1016/j.toxlet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 71.Tran TB, Wang J, Doi Y, Velkov T, Bergen PJ, Li J. 2018. Novel polymyxin combination with antineoplastic mitotane improved the bacterial killing against polymyxin-resistant multidrug-resistant Gram-negative pathogens. Front Microbiol 9:721. doi: 10.3389/fmicb.2018.00721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sbiera S, Leich E, Liebisch G, Sbiera I, Schirbel A, Wiemer L, Matysik S, Eckhardt C, Gardill F, Gehl A, Kendl S, Weigand I, Bala M, Ronchi CL, Deutschbein T, Schmitz G, Rosenwald A, Allolio B, Fassnacht M, Kroiss M. 2015. Mitotane inhibits sterol-O-acyl transferase 1 triggering lipid-mediated endoplasmic reticulum stress and apoptosis in adrenocortical carcinoma cells. Endocrinology 156:3895–3908. doi: 10.1210/en.2015-1367. [DOI] [PubMed] [Google Scholar]
- 73.Ayerbe-Algaba R, Gil-Marqués ML, Jiménez-Mejías ME, Sánchez-Encinales V, Parra-Millán R, Pachón-Ibáñez ME, Pachón J, Smani Y. 2018. Synergistic activity of niclosamide in combination with colistin against colistin-susceptible and colistin-resistant Acinetobacter baumannii and Klebsiella pneumoniae. Front Cell Infect Microbiol 8:348–348. doi: 10.3389/fcimb.2018.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tommasi R, Brown DG, Walkup GK, Manchester JI, Miller AA. 2015. ESKAPEing the labyrinth of antibacterial discovery. Nat Rev Drug Discov 14:529. doi: 10.1038/nrd4572. [DOI] [PubMed] [Google Scholar]
- 75.Phee LM, Betts JW, Bharathan B, Wareham DW. 2015. Colistin and fusidic acid, a novel potent synergistic combination for treatment of multidrug-resistant Acinetobacter baumannii infections. Antimicrob Agents Chemother 59:4544. doi: 10.1128/AAC.00753-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Imperi F, Massai F, Pillai CR, Longo F, Zennaro E, Rampioni G, Visca P, Leoni L. 2013. New life for an old drug: the anthelmintic drug niclosamide inhibits Pseudomonas aeruginosa quorum sensing. Antimicrob Agents Chemother 57:996–1005. doi: 10.1128/AAC.01952-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Betts JW, Phee LM, Hornsey M, Woodford N, Wareham DW. 2014. In vitro and in vivo activities of tigecycline-colistin combination therapies against carbapenem-resistant Enterobacteriaceae. Antimicrob Agents Chemother 58:3541–3546. doi: 10.1128/AAC.02449-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Docobo-Pérez F, Nordmann P, Domínguez-Herrera J, López-Rojas R, Smani Y, Poirel L, Pachón J. 2012. Efficacies of colistin and tigecycline in mice with experimental pneumonia due to NDM-1-producing strains of Klebsiella pneumoniae and Escherichia coli. Int J Antimicrob Agents 39:251–254. doi: 10.1016/j.ijantimicag.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 79.Ku Y-H, Chen C-C, Lee M-F, Chuang Y-C, Tang H-J, Yu W-L. 2017. Comparison of synergism between colistin, fosfomycin and tigecycline against extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolates or with carbapenem resistance. J Microbiol Immunol Infect 50:931–939. doi: 10.1016/j.jmii.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 80.Yilmaz EM, Sunbul M, Aksoy A, Yilmaz H, Guney AK, Guvenc T. 2012. Efficacy of tigecycline/colistin combination in a pneumonia model caused by extensively drug-resistant Acinetobacter baumannii. Int J Antimicrob Agents 40:332–336. doi: 10.1016/j.ijantimicag.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 81.Song JY, Cheong HJ, Lee J, Sung AK, Kim WJ. 2009. Efficacy of monotherapy and combined antibiotic therapy for carbapenem-resistant Acinetobacter baumannii pneumonia in an immunosuppressed mouse model. Int J Antimicrob Agents 33:33–39. doi: 10.1016/j.ijantimicag.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 82.Martin SE, Melander RJ, Brackett CM, Scott AJ, Chandler CE, Nguyen CM, Minrovic BM, Harrill SE, Ernst RK, Manoil C, Melander C. 2019. Small molecule potentiation of Gram-positive selective antibiotics against Acinetobacter baumannii. ACS Infect Dis 5:1223. doi: 10.1021/acsinfecdis.9b00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brackett CM, Furlani RE, Anderson RG, Krishnamurthy A, Melander RJ, Moskowitz SM, Ernst RK, Melander C. 2016. Second-generation modifiers of colistin resistance show enhanced activity and lower inherent toxicity. Tetrahedron 72:3549–3553. doi: 10.1016/j.tet.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Soojhawon I, Pattabiraman N, Tsang A, Roth AL, Kang E, Noble SM. 2017. Discovery of novel inhibitors of multidrug-resistant Acinetobacter baumannii. Bioorg Med Chem 25:5477–5482. doi: 10.1016/j.bmc.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 85.Kwon J, Mistry T, Ren J, Johnson ME, Mehboob S. 2018. A novel series of enoyl reductase inhibitors targeting the ESKAPE pathogens, Staphylococcus aureus and Acinetobacter baumannii. Bioorg Med Chem 26:65–76. doi: 10.1016/j.bmc.2017.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Williams JD, Nguyen ST, Gu S, Ding X, Butler MM, Tashjian TF, Opperman TJ, Panchal RG, Bavari S, Peet NP, Moir DT, Bowlin TL. 2013. Potent and broad-spectrum antibacterial activity of indole-based bisamidine antibiotics: synthesis and SAR of novel analogs of MBX 1066 and MBX 1090. Bioorg Med Chem 21:7790–7806. doi: 10.1016/j.bmc.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qian C-D, Wu X-C, Teng Y, Zhao W-P, Li O, Fang S-G, Huang Z-H, Gao H-C. 2012. Battacin (Octapeptin B5), a new cyclic lipopeptide antibiotic from Paenibacillus tianmuensis active against multidrug-resistant Gram-negative bacteria. Antimicrob Agents Chemother 56:1458. doi: 10.1128/AAC.05580-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Velkov T, Gallardo-Godoy A, Swarbrick JD, Blaskovich MAT, Elliott AG, Han M, Thompson PE, Roberts KD, Huang JX, Becker B, Butler MS, Lash LH, Henriques ST, Nation RL, Sivanesan S, Sani M-A, Separovic F, Mertens H, Bulach D, Seemann T, Owen J, Li J, Cooper MA. 2018. Structure, function, and biosynthetic origin of octapeptin antibiotics active against extensively drug-resistant Gram-negative bacteria. Cell Chem Biol 25:380–391.e5. doi: 10.1016/j.chembiol.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Adams FG, Stroeher UH, Hassan KA, Marri S, Brown MH. 2018. Resistance to pentamidine is mediated by AdeAB, regulated by AdeRS, and influenced by growth conditions in Acinetobacter baumannii ATCC 17978. PLoS One 13:e0197412. doi: 10.1371/journal.pone.0197412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hart EM, Gupta M, Wühr M, Silhavy TJ, Hart EM, Gupta M, Wühr M, Silhavy TJ. 2019. The synthetic phenotype of ΔbamB ΔbamE double mutants results from a lethal jamming of the Bam complex by the lipoprotein RcsF. mBio 10:e00662-19. doi: 10.1128/mBio.00662-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hitchcock PJ, Brown TM. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol 154:269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Henderson JC, Herrera CM, Trent MS. 2017. AlmG, responsible for polymyxin resistance in pandemic Vibrio cholerae, is a glycyltransferase distantly related to lipid A late acyltransferases. J Biol Chem 292:21205–21215. doi: 10.1074/jbc.RA117.000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Trent MS, Ribeiro AA, Lin S, Cotter RJ, Raetz CR. 2001. An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-l-arabinose to lipid A induction in polymyxin-resistant mutants and role of a novel lipid-linked donor. J Biol Chem 276:43122–43131. doi: 10.1074/jbc.M106961200. [DOI] [PubMed] [Google Scholar]
- 94.Zhou Z, Lin S, Cotter RJ, Raetz CR. 1999. Lipid A modifications characteristic of Salmonella Typhimurium are induced by NH4VO3 in Escherichia coli K-12 detection of 4-amino-4-deoxy-l-arabinose, phosphoethanolamine, and palmitate. J Biol Chem 274:18503–18514. doi: 10.1074/jbc.274.26.18503. [DOI] [PubMed] [Google Scholar]
- 95.FDA. 2018. Antibacterial susceptibility test interpretive criteria. U.S. Food and Drug Administration, Bethesda, MD. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.