The introduction of highly efficient therapies with direct-acting antivirals (DAA) for patients with chronic hepatitis C virus (HCV) infection offers exceptional opportunities to globally control this deadly disease. For achieving this ambitious goal, it is essential to prevent antiviral resistance against the most optimal first-line and retreatment DAA choices.
KEYWORDS: DAA, HCVcc, NS5A inhibitor, RAS, genotype 2, nucleotide analog, protease inhibitor, treatment
ABSTRACT
The introduction of highly efficient therapies with direct-acting antivirals (DAA) for patients with chronic hepatitis C virus (HCV) infection offers exceptional opportunities to globally control this deadly disease. For achieving this ambitious goal, it is essential to prevent antiviral resistance against the most optimal first-line and retreatment DAA choices. We performed independent comparisons of the efficacy and barrier to resistance of pangenotypic DAA regimens for HCV genotype 2 infections, using previously and newly developed efficient cell culture-adapted strains of subtypes 2a, 2b, and 2c. With the applied experimental cell culture conditions, combination treatment with the sofosbuvir-velpatasvir or glecaprevir-pibrentasvir DAA regimen was efficient in eradicating HCV infections; in contrast, single-drug treatments frequently led to viral escape. Sequence analysis of drug targets from recovered viruses revealed known resistance-associated substitutions (RAS) emerging in the NS3 protease or NS5A after treatment failure. These RAS were genetically stable after viral passage, and viruses with these RAS exhibited significant phenotypic resistance. After sofosbuvir treatment failure, only a genotype 2a virus harbored NS5B RAS S282T and thus had decreased susceptibility to nucleotide analogs (nucs). However, in most cases, viral escape from sofosbuvir led to other NS5B substitutions but drug susceptibility was maintained, and in one case, no changes in NS5B were detected. For a genotype 2b virus, after treatment failure with sofosbuvir-velpatasvir, the efficacy of retreatment with glecaprevir-pibrentasvir was maintained due to the high barrier to resistance and low cross-resistance of pibrentasvir. Our findings suggest the slight superiority of glecaprevir-pibrentasvir against genotype 2b in culture, which could have potential therapeutic interest meriting more definitive investigations in the clinic.
INTRODUCTION
Chronic hepatitis C virus (HCV) infection is highly prevalent worldwide, with at least 71 million people being affected (1). The World Health Organization (WHO) has declared a very ambitious goal of globally eliminating HCV as a major public health threat by 2030, which can rely only on the use of direct-acting antivirals (DAA), since no vaccine is available for HCV (2, 3).
DAA are potent antiviral molecules targeting HCV nonstructural (NS) proteins, which have essential functions in the virus life cycle (2, 3). The most broadly effective DAA regimens are the combinations of NS5A inhibitors with a protease inhibitor (pibrentasvir-glecaprevir) or with a nucleotide analog (velpatasvir-sofosbuvir), which are also defined as pangenotypic, since they are recommended for the treatment of chronic hepatitis C in patients with HCV genotype 1 to 6 infections (2). Exceptionally high cure rates of over 90%, depending on the patient population, have been reported after treatment with these two regimens (4).
In this study, we focused on genotype 2 of HCV, which is highly prevalent worldwide, with an estimated 16.5 million people being infected with this genotype (5). Genotype 2 can be divided into numerous subtypes, which are designated a to u and which differ in at least 15% of their sequence (6); the most prevalent subtypes are a, b, and c. Genotype 2c is prevalent in Europe, in particular, in the south; a very high prevalence was reported in the region of Calabria, Italy (7–9). Genotype 2c has been linked to an increased risk for hepatitis reactivation in comparison with the risk associated with genotype 1b (10).
HCV cell culture systems (HCVcc) that recapitulate the entire viral cycle of HCV are key tools to study drug treatment regimens in an independent and high-throughput manner (11). However, historically, the availability of these systems has been limited due to the poor viability of patient isolates in cell culture (3, 11). Genotype 2 isolates appear to be easier to adapt to growth in hepatoma cell lines, and we and others have succeeded in growing various full-length infectious HCV clones of genotypes 2a and 2b (12–17). The genotype 2 panels with genetically diverse isolates growing in cell culture are unique, and such panels are not available for other genotypes (11). Thus, they represent the best available systems to study the effect of subtype and strain genetic heterogeneity on the efficacy and barrier to resistance of clinically relevant DAA regimens in cell culture.
Even though treatment with either sofosbuvir-velpatasvir or glecaprevir-pibrentasvir has been highly successful for HCV genotype 2-infected patients (2, 18), there is a lack of clinical studies on the comparative effectiveness and barrier to resistance of these pangenotypic regimens. We hypothesize that subtle currently unrecognized differences in efficacy between these two regimens could have important effects from the perspective of elimination programs on a global scale. In the study described here, we attempted to gain preclinical knowledge about the comparative efficacy of these regimens in cell culture, which could contribute to the design of clinical studies aimed at investigating these putative differences in HCV-infected patients treated with DAA.
Systematic studies comparing the most efficacious drug regimens available for the treatment of HCV infections could help minimize treatment failure, which is frequently associated with antiviral resistance (19, 20). The spread of antiviral resistance could hamper the efficacy of the pangenotypic regimens, all of which are based on the use of NS5A inhibitors, the drug family to which resistance is the most prevalent. Moreover, extensive cross-resistance within this essential family of inhibitors has been described in cell culture (21), yet the influence of this type of cross-resistance in the context of the current pangenotypic regimens that combine different DAA classes merits further investigation.
In this study, we aimed to further expand the panel of culture-efficient infectious full-length HCV genotype 2 strains, including the first full-length culture system for genotype 2c. Using genetically diverse genotype 2a, 2b, and 2c cell culture-adapted viruses, we aimed at providing an industry-independent head-to-head comparison of the efficacy and barrier to resistance, including the emergence of resistance-associated substitutions (RAS), of the pangenotypic DAA regimens sofosbuvir-velpatasvir and glecaprevir-pibrentasvir. We further examined the retreatment outcome after first-line treatment failures with these inhibitors in cell culture.
RESULTS
Molecular clones of HCV genotype 2a (T9) and 2c (S83) strains can be adapted to replicate and propagate in cell culture.
To expand the panel of efficient culture-viable genotype 2 strains, we performed culture adaptation of HCV full-length clones representing strains T9 (genotype 2a) [T9(2a)] and S83 (genotype 2c) [S83(2c)] (22, 23). The molecular clones generated contained the T9 or S83 strain-specific open reading frame (ORF), the 5′ untranslated region (UTR) from J6/JFH1 (13), and the 3′ UTR of cell culture-adapted strain J6 (J6cc) (14), as described in Materials and Methods. The T9 clone encoded a consensus polyprotein of 3,033 amino acids (aa), differing from reference genotype 2a strain JFH1 (GenBank accession number AB237837.1) (12, 24) and strain J6CF (GenBank accession number AF177036) (25) by 9.6% and 6.7% of their amino acid sequences, respectively. The clone of S83 encoded a polyprotein of 3,033 amino acids, differing by 5.9% from the polyprotein of reference genotype 2c strain BEBE1 (GenBank accession number D50409) (6, 26) and by 5.5 to 6.6% from the polyproteins of 5 other published genotype 2c strains (27, 28).
RNA transcripts of the T9 and S83 clones with the consensus ORF sequence were transfected into Huh7.5 cells, but no HCV antigen-positive cells were observed at any time point within 2 weeks. Thus, the consensus sequences of these clones were nonviable in cell culture, and we therefore applied previously developed strategies of culture adaptation (11).
We found that the previously identified genotype 2 cell culture-adaptive substitutions F1468L/A1676S/D3001G (or LSG substitutions) (14, 17) could not initiate replication of T9 (T9_LSG) or S83 (S83_LSG), but addition of N1931T (NS4B) (11) led to a few HCV antigen-positive cells following transfection of Huh7.5 cells with RNA transcripts.
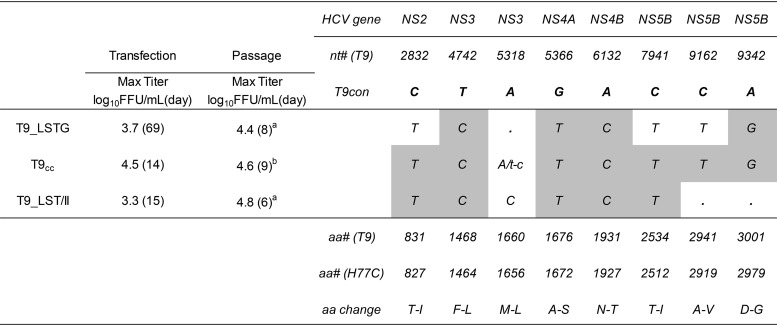
For T9(2a), T9_LSTG viral spread was observed at day 65 posttransfection, with maximum infectivity titers of 3.7 log10 focus-forming units (FFU) per milliliter being seen at day 69 (Table 1). Viruses recovered from the supernatant infected and efficiently propagated in naive Huh7.5 cells. Viral sequences from the third such passage exhibited three complete substitutions (as single peaks in the chromatogram), which were introduced into T9_LSTG. This recombinant cell culture-adapted T9 strain (T9cc), which had a total of 7 substitutions, showed spread at day 7 posttransfection and a maximum infectivity titer of 4.5 log10 FFU/ml at day 14. This virus could rapidly propagate after passage in naive cells, without the emergence of additional dominant ORF changes (Table 1).
TABLE 1.
Characteristics of T9(2a) HCV cell culture-adapted recombinant viruses in transfection and passage in Huh7.5 cellsa
Max Titer, the highest infectivity titer observed (at the time points given in parentheses); a, passage 3; b, passage 2. Letters indicate dominant coding nucleotide changes. A single capital letter indicates that the nucleotide was the only one observed in the sequence. Coding changes as quasispecies (accounting for 50% of the base call) were found only in T9_LSTG and included T2973T/C (amino acids I-I/T), T6812T/C (amino acids F-F/L), T7394T/G (amino acids F-F/V), and C8370C/T (amino acids T-T/I). Two noncoding dominant changes were observed in T9_LSTG: A3730G and T9229C. No noncoding changes were observed in T9cc or T9_LSTG/II. The gray shading indicates the mutations that were engineered in the specific recombinant, which were maintained in the viruses in all cases. T9con indicates the nucleotides found in the consensus ORF sequence (nonadapted) of strain T9. nt# (T9) and aa# (T9) indicate the nucleotide and amino acid numbers, respectively, according to the molecular clone of T9 (see Materials and Methods). aa# (H77C) indicates the amino acid number according to the polyprotein of reference strain H77C (GenBank accession number AF011751). aa change indicates the amino acid changes in the polyprotein of the T9 consensus sequence induced by the nucleotide changes depicted in the body of the table.
We attempted the generation of a T9 cell culture-infectious clone harboring minimal adaptive mutations, and for that we investigated the viability of a recombinant without NS5B substitutions A2941V (aa 499) and D3001G (aa 559). These substitutions were chosen since they had been previously associated with escape from polymerase inhibitors (29, 30) and we evaluated NS5B inhibitors in this study. This recombinant (T9_LST/II) was slightly attenuated in comparison with T9cc, acquiring an additional substitution (M1660L) in the helicase domain of NS3 after viral passage (Table 1); however, infection with this virus was robust enough for downstream applications.
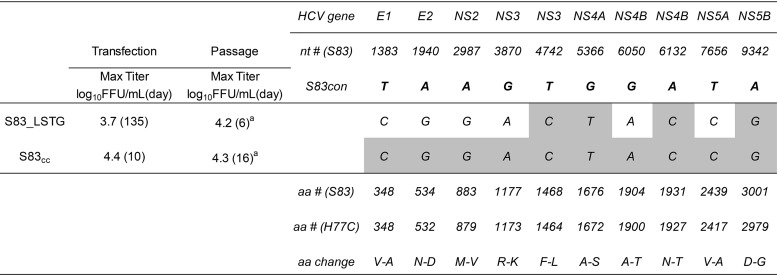
For S83(2c), S83_LSTG resulted in viral spread, with maximum infectivity titers being seen at day 135 (Table 2). Based on the viral sequences obtained at passage 3, we generated recombinant cell culture-adapted S83 (S83cc) with 10 substitutions, which spread at day 3 posttransfection, reaching titers of 4.4 log10 FFU/ml at day 10. This virus efficiently propagated after passage without the emergence of dominant substitutions (Table 2).
TABLE 2.
Characteristics of S83(2c) HCV cell culture-adapted recombinant viruses in transfection and passage in Huh7.5 cellsa
See footnote a of Table 1 for details. Engineered mutations (gray shading) were maintained in all viruses. S83cc showed the following coding changes as quasispecies: T5397T/C (amino acids I-I/T) and A7574A/C (amino acids N-N/H). S83_LSTG contained the following dominant noncoding changes: C1858T, T6568C, C7705T, A8347G, and T8413C. S83cc did not present dominant noncoding changes.
Differential efficacy of individual DAA against HCVcc genotypes 2a, 2b, and 2c.
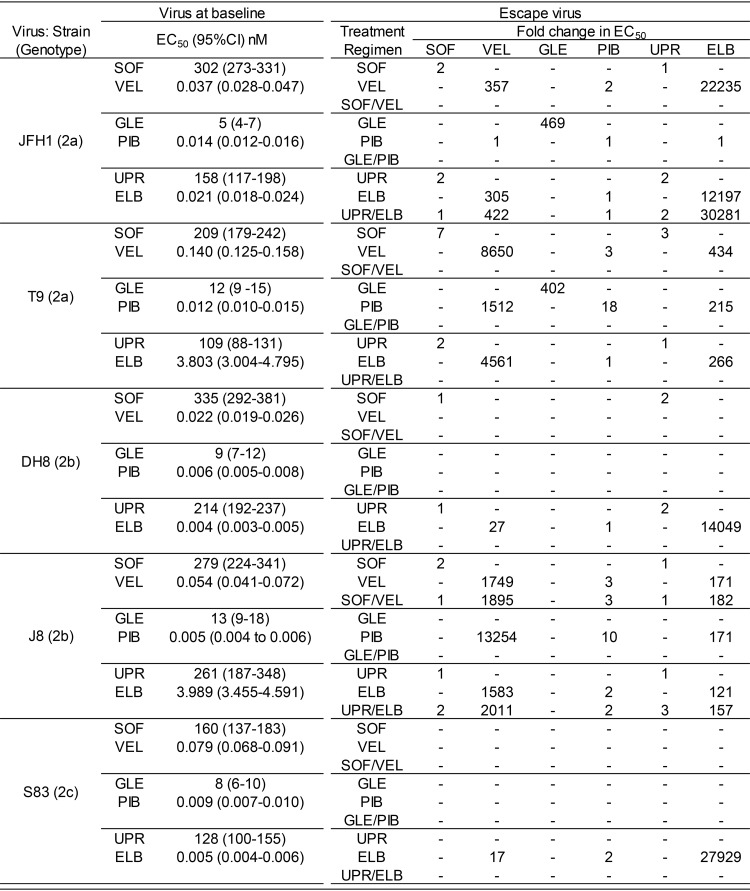
Using the unique panel of genetically diverse cell culture-adapted HCV genotype 2 strains available after culture adaptation of the additional genotype 2a and 2c strains, we determined individual drug potencies (defined by the 50% effective concentration [EC50] values) using short-term concentration-response treatment assays (Table 3). The investigated viruses were T9 [T9_LST/II(2a); Table 1], S83 [S83cc(2c); Table 2], JFH1 [J6/JFH1(2a) (13), abbreviated JFH1 for the origin of DAA targets], J8 [J8_LSG/STAT(2b) (17)], and DH8 [DH8cc(2b) (17)].
TABLE 3.
DAA susceptibility of genotype 2 HCVcc at baseline and after treatment failure (escape)a
Virus at baseline refers to the virus before DAA treatments; drug susceptibility is represented by the EC50 value, with the 95% confidence interval (CI) being given in parentheses. Escape virus refers to the virus harvested after treatment failure and passaged once in naive Huh7.5 cells. SOF, sofosbuvir; VEL, velpatasvir; GLE, glecaprevir; PIB, pibrentasvir; UPR, uprifosbuvir; ELB, elbasvir.
The NS5A inhibitor pibrentasvir, the nucleotide analogs (nucs) sofosbuvir and uprifosbuvir, and the protease inhibitor glecaprevir exhibited minor differences in potency across isolates, with a maximum difference between the least and most susceptible isolates of 2.8-fold the EC50 (Table 3). For the NS5A inhibitors velpatasvir and elbasvir, the largest differences were 6.4- and 997-fold the EC50, respectively (Table 3).
Differential efficacy and barrier to resistance of pangenotypic DAA regimens against HCVcc genotypes 2a, 2b, and 2c in cell culture.
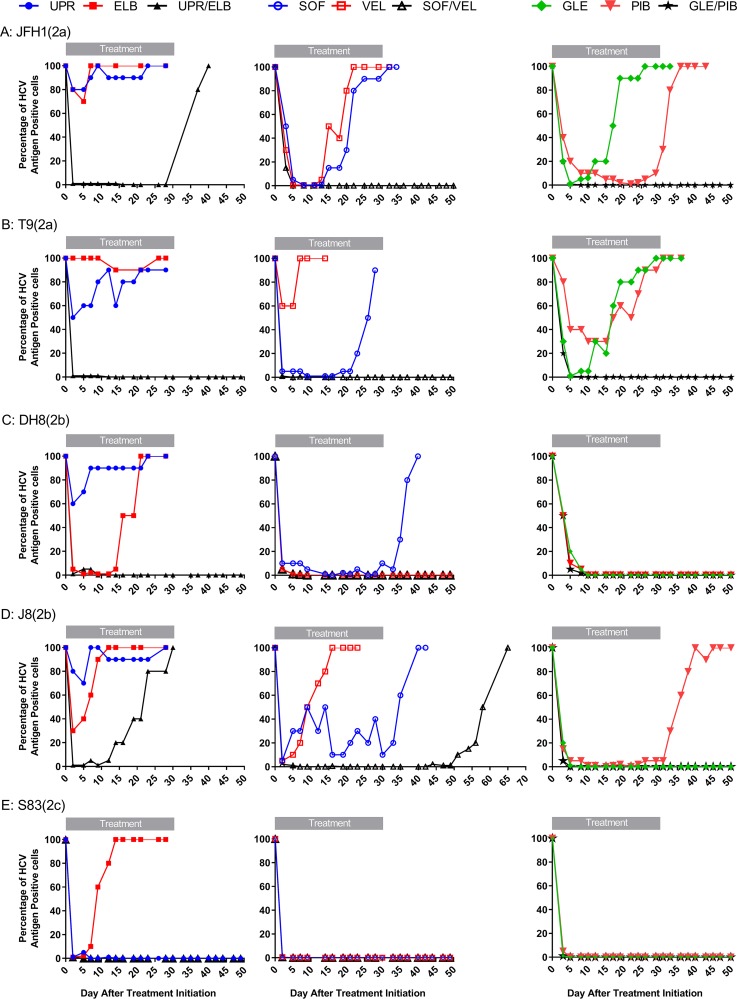
To compare the efficacies of the pangenotypic DAA regimens sofosbuvir-velpatasvir and glecaprevir-pibrentasvir, we performed long-term treatment (30 days) assays as described in Materials and Methods. In these experiments, we monitored the kinetics of the infection by determination of viral antigens using immunofluorescence during treatment and after drug withdrawal (Fig. 1). In addition to the pangenotypic regimens recommended in treatment guidelines, we were interested in investigating regimens containing the nuc uprifosbuvir, which had shown higher potency than sofosbuvir in cell culture (30, 31). In a recent study, a combination of uprifosbuvir and the NS5A inhibitor ruzasvir was found to be highly effective, but only for certain genotypes (32). Unfortunately, at the onset of this study, we did not have access to ruzasvir, so we instead tested the combination of uprifosbuvir with the NS5A inhibitor elbasvir. Of note, the combination uprifosbuvir, elbasvir, and the protease inhibitor grazoprevir was part of a clinical trial (ClinicalTrials.gov identifier NCT02332707), but the use of sofosbuvir-elbasvir was not investigated. Besides defining the outcome of combination and single-drug treatments, we performed phenotypic and genotypic analysis of escape viruses in order to determine which treatment failures were associated with resistance, a key aspect to defining the barrier to resistance of these regimens.
FIG 1.
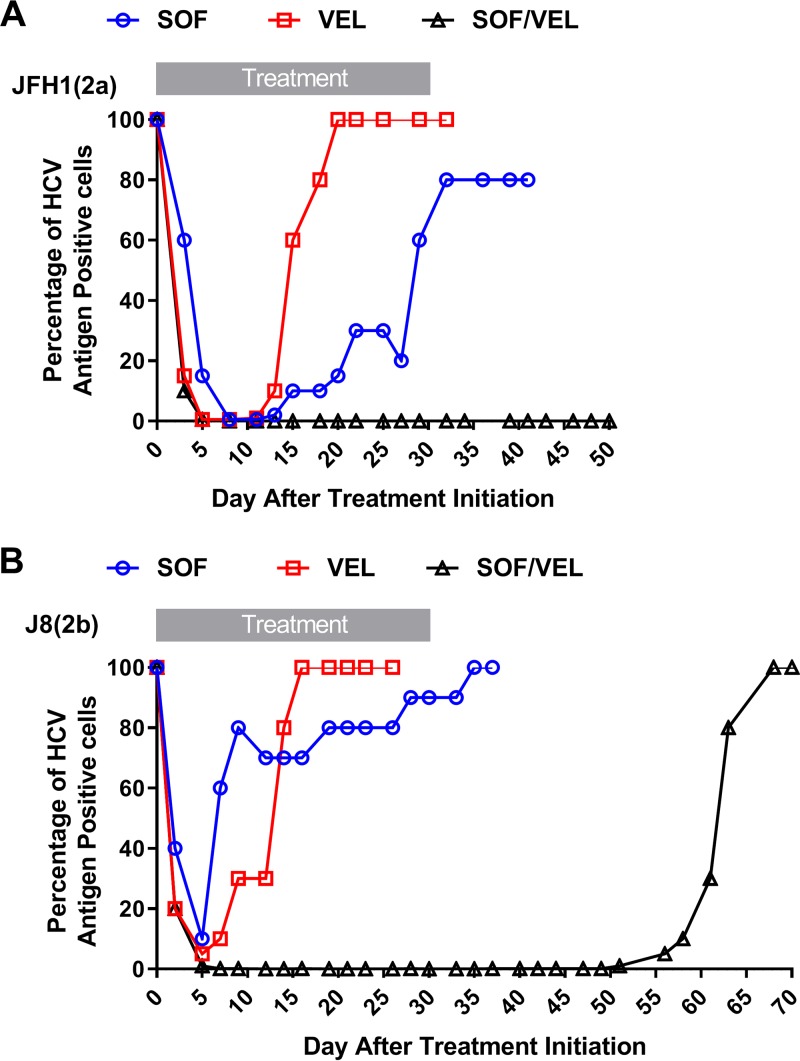
Efficacy of combination treatment against HCVcc genotypes 2a, 2b, and 2c. Huh7.5 cells were infected with J6/JFH1(2a) (13), T9_LST/II(2a), DH8cc(2b) (17), J8_LSG/STAT(2b) (17), and S83cc(2c) and treated with 570 nM uprifosbuvir (UPR) and/or 11 nM elbasvir (ELB), 570 nM sofosbuvir (SOF) and/or 11 nM velpatasvir (VEL), and 422 nM glecaprevir (GLE) and/or 0.05 nM pibrentasvir (PIB). The graphs show the percentage of HCV antigen-positive cells (y axis) at the different time points posttreatment (x axis). (A) JFH1(2a); (B) T9(2a); (C) DH8(2b); (D) J8(2b); (E) S83(2c). The cultures were treated for 30 days, as indicated, and thereafter were kept without drugs (see Materials and Methods).
Comparative efficacy of DAA treatments in HCV cell culture.
HCVcc genotype 2 viruses were frequently able to escape single-drug treatments (Fig. 1). However, the DH8(2b) and S83(2c) viruses (Fig. 1C and E) were exceptionally susceptible to the NS5A inhibitors velpatasvir and pibrentasvir, which cleared the infection, but not to elbasvir treatment, which led to viral escape in all cases. Genotype 2b strains J8 and DH8, as well as S83(2c), were exceptionally susceptible to glecaprevir, which cleared these infections (Fig. 1C to E). Monotherapy with nucs led to escape in all cases, except for the S83(2c) virus (Fig. 1E). Sofosbuvir exhibited efficacy in controlling viral infections superior to that of uprifosbuvir, despite being slightly less potent in short-term assays (Table 3).
None of the viruses escaped glecaprevir-pibrentasvir combination treatment (Fig. 1), whereas the J8(2b) virus showed decreased susceptibility to nuc-NS5A inhibitor regimens, leading to breakthrough and relapse after treatment with uprifosbuvir-elbasvir and sofosbuvir-velpatasvir, respectively (Fig. 1D). JFH1(2a) also relapsed after uprifosbuvir-elbasvir treatment (Fig. 1A).
The reproducibility of the escape experiments was remarkable, as shown in Fig. 2A and B, where the results of independent treatments of JFH1(2a) and J8(2b), respectively, with sofosbuvir-velpatasvir mimicked the results shown in Fig. 1A and D, respectively.
FIG 2.
Reproducibility of sofosbuvir-velpatasvir-escape experiments. Huh7.5 cells were infected with J6/JFH1(2a) (13) (A) or J8_LSG/STAT(2b) (17) (B) and treated with 570 nM sofosbuvir (SOF) and/or 11 nM velpatasvir (VEL). The graphs show the percentage of HCV antigen-positive cells (y axis) at the different time points posttreatment (x axis). Cultures were treated for 30 days and thereafter were kept without drugs.
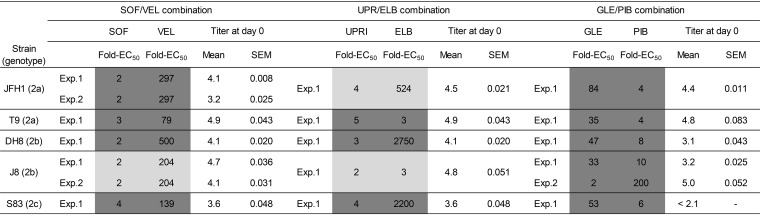
The exceptional efficacy of combination treatment reported for patients (18) was also observed in the present cell culture study. However, in cell culture we found a reproducible superior performance of glecaprevir-pibrentasvir for HCV genotype 2b. To further investigate the apparent overall superiority of glecaprevir-pibrentasvir and the superiority of sofosbuvir-velpatasvir over uprifosbuvir-elbasvir, we examined the experimental conditions that could have influenced the treatment outcome. The factors investigated included the ratio of the fixed drug concentration used relative to the virus-specific EC50 value and the viral infectious titer at treatment initiation (day 0) (Table 4).
TABLE 4.
Experimental conditions of DAA combination treatments performed in genotype 2 HCVcca
For each treatment, the drug fold EC50 values refer to the ratio between the fixed drug concentration used in the experiment (Exp.) and the virus-specific EC50 value. For strains JFH1(2a) and J8(2b), two independent experiments were performed under identical conditions to assess the reproducibility of the sofosbuvir-velpatasvir treatment (Fig. 1A and D and 2A and B). In addition, for the J8(2b) strain, two experiments with different drug concentrations of glecaprevir-pibrentasvir were performed to assess their influence on the outcome (Fig. 1D and 3A). Infectivity titers, given as the log10 number of FFU per milliliter (mean and standard error of the mean [SEM] from 3 replicates) at day 0 of treatment initiation (baseline), are also shown; 2.1 corresponds to the limit of detection. Shading indicates the treatment outcome: light gray for escape and dark gray for cure. SOF, sofosbuvir; VEL, velpatasvir; UPRI, uprifosbuvir; ELB, elbasvir; GLE, glecaprevir; PIB, pibrentasvir.
In the two independent sofosbuvir-velpatasvir treatments of JFH1(2a) (experiments 1 [Fig. 1A] and 2 [Fig. 2A]), an ∼1-log10 difference in the baseline viral infectivity titer (4.1 versus 3.2 log10 FFU/ml) did not influence the treatment outcome. When comparing the treatment outcome with the two regimens based on nucs and NS5A inhibitors, JFH1(2a) escaped uprifosbuvir-elbasvir, despite the use of higher absolute effective DAA concentrations and similar viral infectivity titers at the baseline (<0.5 log10 difference when comparing both experiments 1 [Fig. 1A]).
When analyzing the different outcomes for the J8(2b) virus treated with the two pangenotypic regimens sofosbuvir-velpatasvir and glecaprevir-pibrentasvir in experiment 1 (Fig. 1D), the effective concentrations of the DAA glecaprevir were higher than those of sofosbuvir, whereas a lower effective concentration of pibrentasvir than of velpatasvir was used. Since the genotype 2b viruses were exceptionally susceptible to glecaprevir, we decided to perform a second experiment with J8(2b) and glecaprevir-pibrentasvir (experiment 2; Fig. 3A), using effective drug concentrations equivalent to those used in sofosbuvir-velpatasvir experiment 1 (Fig. 1D). We also had a higher viral infectivity titer at the baseline. The outcomes of both experiments with glecaprevir-pibrentasvir were identical; thus, neither the DAA concentrations nor the viral titer at the baseline seemed to play a significant role in the superior performance of the glecaprevir-pibrentasvir regimen for the treatment of the J8(2b) virus in cell culture.
FIG 3.
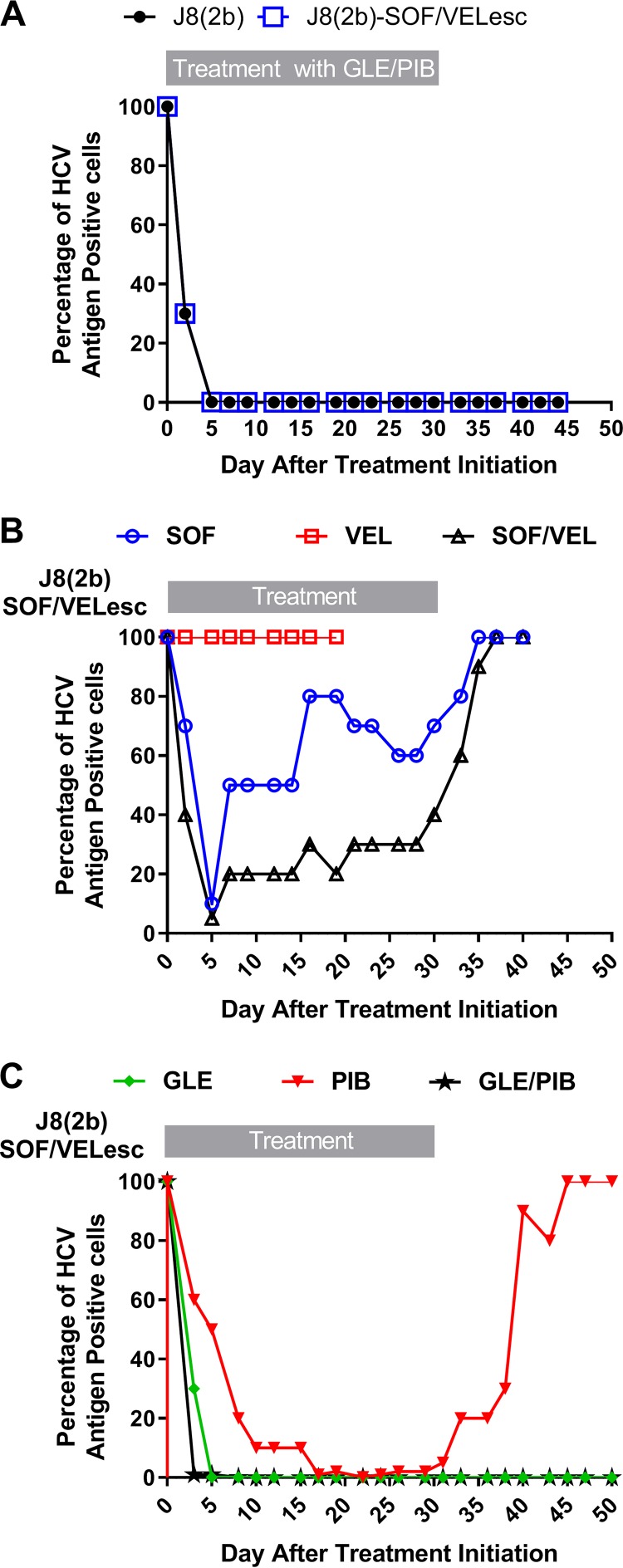
Efficacy of retreatment strategies after viral escape from sofosbuvir-velpatasvir treatment. Huh7.5 cells were infected with J8_LSG/STAT(2b) (17) or J8(2b)SOF-VEL_escape and treated with the indicated regimens. (A) J8_LSG/STAT(2b) and J8(2b)SOF-VEL_escape treated with a combination of 26 nM glecaprevir (GLE) and 1.0 nM pibrentasvir (PIB); (B) J8(2b)SOF-VEL_escape treated with 570 nM sofosbuvir (SOF) and/or 11 nM velpatasvir (VEL); (C) J8(2b)SOF-VEL_escape treated with 422 nM glecaprevir and/or 0.05 nM pibrentasvir. The graphs show the percentage of HCV antigen-positive cells (y axis) at the different time points posttreatment (x axis). Cultures were treated for 30 days and thereafter were kept without drugs.
Retreatment of J8(2b)SOF-VEL_escape in cell culture.
The J8(2b) virus was notably resistant to nuc-NS5A inhibitor regimens and escaped treatment with the efficient sofosbuvir-velpatasvir combination (Fig. 1D). We therefore investigated the susceptibility of the J8(2b) sofosbuvir-velpatasvir-escape virus [J8(2b)SOF-VEL_escape] to subsequent treatments with DAA (Fig. 3). Retreatment with sofosbuvir-velpatasvir under similar first-line experimental conditions resulted in rapid viral breakthrough and complete viral spread upon drug withdrawal (Fig. 3B). Compared to the findings for the first-line treatment (Fig. 1D), the velpatasvir single-drug retreatment could not control the virus at any time point, whereas sofosbuvir exerted some but limited control of the virus infection. Therefore, the treatment failure observed with this drug combination was due to both the lack of activity of velpatasvir and decreased susceptibility to sofosbuvir. In contrast, treatment with glecaprevir-pibrentasvir remained highly efficient, as demonstrated in two independent experiments performed with different drug concentrations (Fig. 3A and C), leading to viral clearance. Our cell culture results indicate that the glecaprevir-pibrentasvir regimen should be investigated as a valuable option for the retreatment of sofosbuvir-velpatasvir-escape viruses in the clinic.
Evaluation of drug susceptibility after treatment failure (escape) in cell culture.
To investigate if changes in drug susceptibility (resistance) after viral escape had occurred, we compared the EC50 values for each drug and virus before and after the treatments (Table 3).
Minimal changes (<3-fold the EC50) in susceptibility were observed after escape from nucs. The exception was the T9(2a) sofosbuvir-escape virus [T9(2a)SOF_escape], which exhibited 7-fold- and 3-fold decreases in sofosbuvir and uprifosbuvir susceptibility (EC50), respectively. Escape from glecaprevir resulted in the emergence of high resistance levels for the only two viruses in which escape was observed [402- and 469-fold increases in the EC50s for the T9(2a) and JFH1(2a) viruses, respectively]. Escape from NS5A inhibitors led to variable degrees of resistance, with large differences being seen among viruses and drugs (Table 3). Velpatasvir- and elbasvir-escape viruses presented large increases in EC50s (17- to 13,254-fold for velpatasvir and 121- to 30,281-fold for elbasvir) and cross-resistance between these two inhibitors (Table 3). On the contrary, pibrentasvir resistance after pibrentasvir escape was less pronounced (10- to 18-fold increases in the EC50), even if the pibrentasvir-escape viruses generally exhibited cross-resistance to velpatasvir and elbasvir (Table 3). Importantly, pibrentasvir maintained its potency (it had similar EC50 values before and after treatment) against all viruses that escaped velpatasvir and elbasvir. An interesting observation was that the JFH1(2a) pibrentasvir-escape virus [JFH1(2a)PIB_escape] did not exhibit resistance to any NS5A inhibitor, which was correlated with the loss of RAS during passage (see below).
Compared to escape from single treatments, escape from the corresponding combination treatment in the few available cases led to higher increases in the NS5A inhibitor EC50 but to a similar nuc EC50 (Table 3).
In summary, based on the data obtained from the concentration-response assays, the HCVcc genotype 2 viruses escaping treatments with sofosbuvir, uprifosbuvir, and pibrentasvir remained susceptible to these drugs. In contrast, escape from glecaprevir, velpatasvir, and elbasvir resulted in large decreases in drug susceptibility, suggesting the emergence of antiviral resistance.
Analysis of HCV drug target sequences after treatment failure (escape) in cell culture.
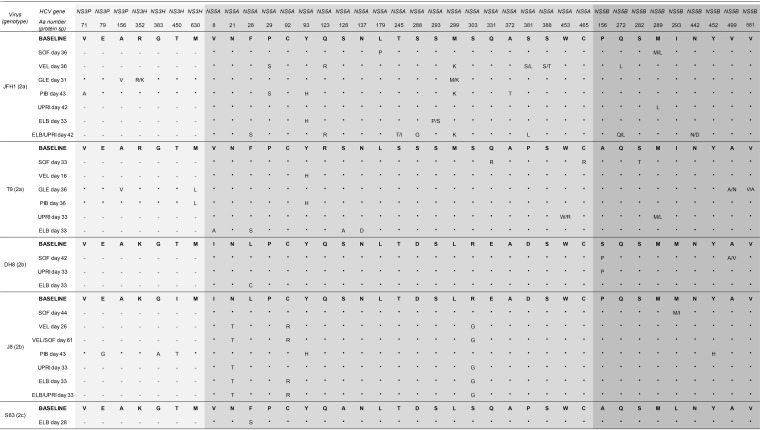
To define the genetic determinants associated with viral escape, we compared the sequences that were found in escape viruses to those that were present at the baseline (Tables 5 and 6).
TABLE 5.
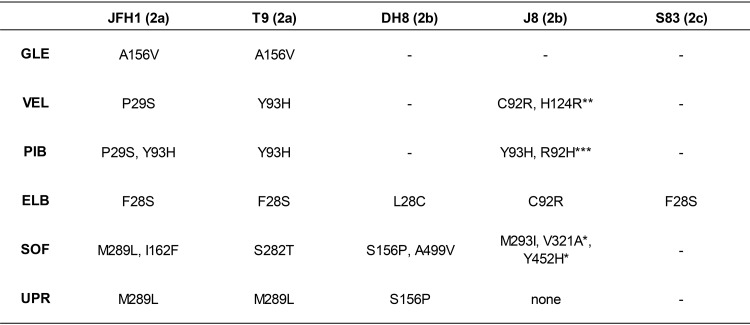
Substitutions found in DAA targets of escape viruses after treatment failurea
Numbers correspond to the protein-specific positions in NS3, NS5A, and NS5B. RAS in the NS3 protease (GLE, glecaprevir) and NS5A domain I (VEL, velpatasvir; PIB, pibrentasvir; ELB, elbasvir) and all substitutions in NS5B (SOF, sofosbuvir; UPR, uprifosbuvir) are indicated. —. not done (as no escape viruses were available for these drugs); *, appeared in NS5B of J8(2b)SOF-VEL_escape retreated with sofosbuvir (Fig. 3B); **, appeared in NS5A of J8(2b)SOF-VEL_escape exhibiting C92R at the baseline and retreated with sofosbuvir-velpatasvir (Fig. 3B); ***, appeared as quasispecies (R92R/H) in NS5A of J8(2b)SOF-VEL_escape exhibiting C92R at the baseline and retreated with pibrentasvir (Fig. 3C). All substitutions found in the sequences of the entire NS3, NS5A, and NS5B regions of these viruses can be found in Table 6. When escape viruses were subjected to passage in naive Huh7.5 cells for resistance phenotyping (Table 3), their sequences exhibited the same RAS, with the only exception being JFH1(2a)PIB_escape, in which the viral population with NS5A P29S and Y93H was replaced by the wild-type virus.
TABLE 6.
Substitutions found in full-length NS3, NS5A, and NS5B of escape viruses after treatment failurea
The amino acid (aa) changes observed in the sequences of HCVcc from Huh.7.5 cell cultures after treatment failure with DAA (escape, Fig. 1) are represented. Sequences obtained at a specific day after treatment initiation were compared with the sequences obtained at day 0 of treatment (baseline). Analyses were done by Sanger sequencing. The analyzed genomic regions are colored in 3 shades of gray (for NS3, for NS5A, and for NS5B), and the protein-specific residue number for each amino acid change is shown at the top. NS3P, protease domain of NS3; NS3H, helicase domain of NS3. For each virus, JFH1, T9, DH8, J8, and S83, the wild-type, baseline residue at the protein-specific (protein sp) position is shown in bold, and below that are given the sequences of the escape viruses. Identical residues are represented by a bold dot, and changes in the viral population (quasispecies) are represented by letters: mixed quasispecies are shown as letters separated by a forward slash, in which the capital letter represented the dominant amino acid and the lowercase letter represented the minor quasispecies. Escape viruses were subjected to viral passage in order to determine their resistance phenotype (Table 3) and, furthermore, were sequenced. Their sequences after passage were identical to those shown in this table, except for JFH1 escaping pibrentasvir, in which viral populations harboring P29S and Y93H were replaced by the wild-type virus as early as 6 days after passage. —, not done (treatment did not target this protein). SOF, sofosbuvir; VEL, velpatasvir; GLE, glecaprevir; PIB, pibrentasvir; UPRI, uprifosbuvir; ELB, elbasvir.
Overall, the RAS that developed after treatment with sofosbuvir and uprifosbuvir overlapped (Table 5); however, different viruses acquired mutations at specific positions. The known NS5B RAS S282T appeared only in T9(2a)SOF_escape, which was the virus exhibiting a resistance phenotype (Table 3). Most other viruses acquired substitutions at NS5B position 156, 162, 289, 293, 321, 452, or 499, and in those viruses, changes in the sofosbuvir or uprifosbuvir EC50 were not observed. Surprisingly, the J8(2b) uprifosbuvir-escape virus [J8(2b)UPR_escape], J8(2b) uprifosbuvir-elbasvir-escape virus [J8(2b)UPR-ELB_escape], and J8(2b)SOF-VEL_escape did not show any changes in NS5B after treatment with nuc-containing regimens. The J8(2b) sofosbuvir-escape virus [J8(2b)SOF_escape] had only a minor population harboring M293L (Table 6).
A good correlation in the emergence of mutations was observed for the J8(2b) virus escaping nuc monotherapy or nuc-containing regimens. However, the JFH1(2a) virus escaping uprifosbuvir exhibited a set of mutations different from that found in the virus escaping uprifosbuvir-elbasvir, suggesting differential selective pressure during single or combination treatments for this virus.
Escape from glecaprevir and from NS5A inhibitors induced the emergence of known RAS in all cases (Table 5). For glecaprevir, the NS3 protease substitution A156V was the only observed RAS, leading to significant resistance both in T9(2a) and in JFH1(2a). For NS5A inhibitors, known RAS at NS5A positions 28, 29, and 93 were predominant in genotype 2a, while substitutions at positions 28, 92, and 93 emerged in genotype 2b. In genotype 2c, only F28S was observed. These substitutions conferred different levels of resistance, depending on the inhibitor (Table 3), and were stable after drug-free passage. However, for JFH1(2a), the pibrentasvir-escape viral population with NS5A P29S and Y93H RAS was replaced by the wild type after passage, which had not been eliminated during treatment.
Retreatment of J8(2b)SOF-VEL_escape with sofosbuvir, velpatasvir, and sofosbuvir-velpatasvir led to rapid escape without the emergence of additional known NS5A RAS (the virus already exhibited C92R at the baseline) but with the emergence of NS5A H124R and of NS5B V321A and Y452H (Table 5). Surprisingly, the RAS NS5B S282T did not emerge after retreatment and, thus, was not needed for prompt sofosbuvir escape.
Overall, the rapid selection of RAS occurred in all cases during treatment failure with glecaprevir, velpatasvir, and elbasvir, and these RAS were maintained without drug pressure. Pibrentasvir and nucs exhibited the highest barrier to resistance both at the phenotypic level and at the genetic level. Nuc treatment selected viral populations with previously observed and undescribed sofosbuvir treatment-associated substitutions, but only RAS S282T conferred significant phenotypic resistance. However, escape from nucs could occur in the absence of NS5B substitutions, as observed in the J8(2b) virus.
DISCUSSION
In this study, we developed novel culture systems for HCV genotype 2a (T9cc) and 2c (S83cc) strains that efficiently replicate and propagate in Huh7.5 cells, one of the gold standard cell lines used for cell culture studies of HCV (11). S83cc is the first infectious culture system that expresses the entire 2c polyprotein, allowing simultaneous studies of all DAA targets in the context of the entire viral life cycle. With a panel of diverse genotype 2 HCVcc, we showed that HCVcc are a useful tool to study clinically relevant DAA regimens in cell culture. In cell culture, we could reproduce the high efficacy of treatment with pangenotypic DAA regimens that has been observed in patients. In our experimental setting in cell culture permitting head-to-head comparisons of the most relevant and efficacious DAA regimens, we found the overall superiority of glecaprevir-pibrentasvir. Importantly, we showed that a virus escaping sofosbuvir-velpatasvir first-line treatment could be efficiently eliminated using the glecaprevir-pibrentasvir regimen.
Besides the exceptional spontaneously replicating JFH1 strain, there are few HCV clones capable of efficiently producing infectious viruses (11). In the latter, cell culture viability is achieved by using adaptive mutations. Finding novel mutations conferring cell culture growth is cumbersome (11), but here we demonstrate that a set of 4 substitutions in NS3, NS4A, NS4B, and NS5B (14, 17) and the use of available heterologous genotype 2a UTRs (13, 14) can promote the adaptation of genotype 2 strains. The use of cell culture-adaptive mutations is imperative to study HCV in culture, even with replicons, since they also depend on replication-enhancing mutations (33–37). Replicons with cell culture-adaptive mutations were extensively used in the preclinical development and resistance testing of all DAA, proving to be excellent tools to predict the efficacy and the barrier to resistance of these drugs (38–40). In this study, our HCVcc panel contained the J6/JFH1 virus, which does not require mutations to efficiently replicate and propagate in cell culture (11, 13), and this virus did not seem to be different from the ones harboring culture-adaptive mutations in terms of drug susceptibility. Thus, the differences observed within the panel most likely reflect virus strain variability. The drug concentrations or EC50 equivalents used in this study are within the range of those used in previous culture studies, including the studies performed by the drug industry (30, 31, 38–41). The culture concentrations (Cc) were lower than the maximum plasma concentrations (Cmax) reported for these DAA (42). The Cmax/Cc ratios for the different drugs were 1.9 to 2.9 for sofosbuvir, 1.7 for glecaprevir, and 27 to 1,980 for NS5A inhibitors. Thus, the DAA concentrations used in this cell culture study can be achieved in the clinic.
A potential limitation of this study is the use of a panel of HCV strains of a limited size and thus of a limited genetic variability, whereas the genetic variability of the HCV strains seen in patients is significant. However, since the development of novel strains of HCV viable in culture remains challenging, as observed for two strains here, studies using multiple full-length isolates infectious in culture have been limited (11).
In the clinic, high rates of sustained virological response are observed in genotype 2-infected patients treated with the pangenotypic regimens sofosbuvir-velpatasvir and glecaprevir-pibrentasvir (2). In our cell culture controlled experimental settings, we also observed high rates of viral clearance with DAA combinations, even if single-drug treatments frequently led to viral escape, due to the outstanding synergy between different DAA (43, 44). However, mechanisms mediating the clearance of viral infection in cell culture might differ from those eliminating the infection in patients.
The superiority of glecaprevir-pibrentasvir over sofosbuvir-velpatasvir against genotype 2 viruses observed in the present cell culture study has not been previously described in patients, since clinical studies have not provided head-to-head comparisons between these pangenotypic regimens. Thus, the importance of this observation in clinical practice remains to be determined with the appropriate studies in hepatitis C virus-infected patients.
In this study, we observed, furthermore, the superiority of sofosbuvir-velpatasvir over uprifosbuvir-elbasvir, which related to the better viral suppression exerted by both sofosbuvir and velpatasvir under the experimental conditions applied. The uprifosbuvir-elbasvir regimen, even if it is not clinically relevant, permitted us to detect that, while being completely susceptible to glecaprevir-pibrentasvir, the J8(2b) virus was inherently resistant to regimens based on a nuc-NS5A inhibitor, which could indicate that some genotype 2b strains would respond better to a protease inhibitor-NS5A regimen. Although the superiority of velpatasvir over elbasvir was already described in previous studies, its impact on the outcome of combination treatment in cell culture was not evaluated (21, 31).
Single treatment with NS3 protease and NS5A inhibitors led to the emergence of RAS involving known positions and residues previously found to cause resistance (21, 41). However, the RAS pattern observed here was similar to but not identical to that observed in previous work investigating the RAS emerging after treatment with the NS5A inhibitor daclatasvir (21), due to a differential selection of specific mutants by velpatasvir, elbasvir, and pibrentasvir. In most cases, escape from nuc treatment in cell culture was not associated with the NS5B RAS S282T. A similar pattern is observed in the clinic, where detection of S282T after treatment failure is rare (29, 45, 46). A lack of detection of S282T could be due to a transient presence in the viral population while viral replication is being highly suppressed. Since T282 is associated with poor viral fitness in cell culture (38), reversion to S282 might quickly follow drug withdrawal. In some patients with transient S282T, footprint codons of reversion can be tracked after treatment (47, 48); however, such codons were not detected in this study. Besides S282T, it has been difficult to associate other sofosbuvir treatment-escape substitutions with a significant decrease in drug susceptibility (38, 46). Thus, the role in viral escape of substitutions at NS5B positions 289, 293, 321, and 499 observed in this study and in others (30, 38, 46, 49) remains unclear. Replicon studies showed that resistance phenotypes were observed only when these substitutions were combined with S282T, thus indicating coselection, possibly as fitness-enhancing substitutions (38, 48, 49). However, coselection was not observed in the present study. Recently, patient-associated NS5B polymorphisms A150V and K206E were associated with decreased sofosbuvir susceptibility (50). In our panel, polymorphisms were present at these positions in some viruses; however, they did not seem to correlate with treatment outcome. Since there is much to learn about these and other polymorphisms, future detailed reverse genetic studies with HCVcc strains can help clarify their role in viral escape from nuc-based therapy.
The evaluation of multiple strains of HCV, including those from rare subtypes, which can potentially carry multiple nonbeneficial natural polymorphisms less responsive to DAA, is relevant to fully understand the scope of HCV antiviral resistance (51, 52). Genotype 2 has been defined to be easy to treat; however, in Africa, where an extensive pool of highly genetically diverse strains has been observed (53, 54), susceptibility to DAA is largely unknown. Moreover, genotype 2 represents a likely genotype to emerge worldwide in the future due to the increased immigration from African countries (55). In our genotype 2 panel, we could detect strain-related variability in the response to DAA and polymorphisms known to influence susceptibility to NS5A inhibitors, such as the L31M substitution in NS5A (21, 56) found in the T9(2a) and J8(2b) strains. However, methionine at NS5A position 31 was not sufficient to predict the outcome of combination treatments, which was different for these two viruses. Moreover, in this study, the T9(2a) strain harbored the double F28/M31 polymorphism at the baseline. This combined polymorphism has been shown to confer a significant reduction in the susceptibility to elbasvir in the replicon system (57); however, this combined polymorphism did not decrease the efficacy of the sofosbuvir-velpatasvir or uprifosbuvir-elbasvir regimen in cell culture. Thus, virological determinants of the response to DAA combinations appear to be rather complex and might depend on multiple combinations of polymorphisms across drug targets.
Although we consistently observed a higher susceptibility of genotype 2c to inhibitors included in pangenotypic regimens in cell culture, this observation could not be compared to the outcomes for genotype 2c-infected patients, since available clinical studies and guidelines consider subtyping only of genotype 1 (2, 58).
No treatment failures were observed here with glecaprevir-pibrentasvir combination treatment, which showed a remarkable high barrier to resistance in genotype 2 in cell culture. This could be explained by a better independent performance of pibrentasvir over velpatasvir. Comparatively, pibrentasvir maintained a higher barrier to resistance than velpatasvir against NS5A inhibitor-escape viruses, in agreement with the findings of cell culture studies reported previously (40). Additionally, an exceptional susceptibility to the protease inhibitor glecaprevir was observed in genotype 2b, in line with the findings of a recent study on the protease inhibitor grazoprevir (41). However, the higher efficacy of glecaprevir-pibrentasvir than of sofosbuvir-velpatasvir against HCVcc genotype 2 observed in this study might not apply to other genotypes, such as genotype 3, which has been found to more rapidly escape protease inhibitors in cell culture (41).
The J8(2b) virus recovered after sofosbuvir-velpatasvir treatment failure escaped retreatment with sofosbuvir-velpatasvir very rapidly, while it was fully susceptible to glecaprevir-pibrentasvir. Whether this virus could also be successfully treated with the salvage regimen sofosbuvir-velpatasvir-voxilaprevir was not evaluated. Indeed, a phase III study found no impact of RAS (mostly in NS5A) on the efficacy of sofosbuvir-velpatasvir-voxilaprevir as a retreatment in patients (59).
In summary, we developed novel robust culture systems for genotype 2a and 2c strains. Our study benefits from being an independent evaluation of drug regimens for genotypes 2a, 2b, and 2c in a highly controlled experimental setting in cell culture, where viral determinants of antiviral treatment failure in the context of the entire HCV viral cycle can be assessed. This permitted us to find differences in the efficacy of the pangenotypic sofosbuvir-velpatasvir and glecaprevir-pibrentasvir regimens, which were overall highly efficient, with glecaprevir-pibrentasvir showing a performance superior to that of sofosbuvir-velpatasvir against genotype 2b in cell culture. These data could potentially be of clinical and therapeutic importance; however, they need to be confirmed in HCV-infected patients. Thus, based on our findings, it would be relevant to set up head-to-head clinical studies comparing the two regimens, since even a slightly enhanced performance of a specific regimen against a concrete genotype or subtype could benefit implementation of HCV elimination programs at a global scale.
MATERIALS AND METHODS
Analysis of consensus genomes from HCV genotype 2a (T9) and 2c (S83) strains.
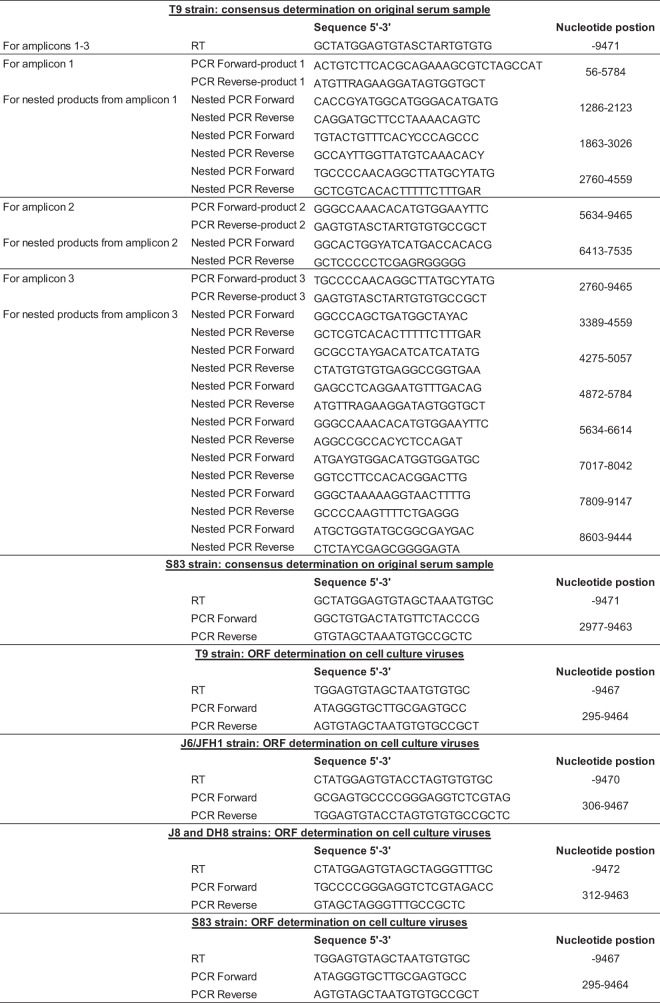
To generate novel cell culture-viable HCV clones from genotypes 2a and 2c, we determined the ORF sequences of the patient-derived T9(2a) and S83(2b) strains (22, 23). Their core-NS2 sequences had been determined previously (9, 22, 23), but the NS3-NS5B sequences were obtained here by Sanger sequencing of overlapping nested amplicons (T9) or on the basis of the clonal sequences of a single amplicon (S83) (see details on the primers used in Table 7). T9 sequence ambiguities were resolved by using genotype 2a sequences from the Los Alamos HCV database (60).
TABLE 7.
Primers used for determination of HCV genotype 2 sequencesa
Nucleotide positions refer to those of specific T9cc-, S83cc-, J6/JFH1-, J8-LSG/STAT-, or DH8cc-based recombinants.
Novel HCV plasmids.
DNA sequences containing the T9 and S83 ORFs were cloned into FL-T9Core-NS2/JFH1 and FL-S83Core-NS2/JFH1 (9), respectively, and the JFH1 3′ UTR was replaced with the 3′ UTR of J6cc (14), using standard cloning techniques. Adaptive mutations were introduced by PCR mutagenesis (with a QuikChange site-directed mutagenesis kit; Stratagene, USA) or by cloning of chemically synthesized DNA fragments (GenScript, USA). Plasmid preparations were obtained with Qiagen (Netherlands) and Sigma-Aldrich (USA) midi/maxiprep kits, followed by HCV sequence confirmation (Macrogen, Netherlands). Unless otherwise stated, specific recombinant T9cc (GenBank accession number MN712203) or S83cc (GenBank accession number MN712204) nucleotide and amino acid numbering were used.
Cell culture.
Experiments were performed in Huh7.5 cells (13), with subculturing every 2 to 3 days. Transfections with HCV RNA transcripts were performed as described previously (14), and naive cell infections were performed by inoculating 1 ml of virus-containing supernatant overnight. Viral spread was defined by ≥80% HCV antigen-positive cells, determined by immunofluorescence with anti-NS5A antibody 9E10 (13) alone or combined with anti-core antibody C7-50 (Abcam, UK) and secondary antibody Alexa Fluor 488 or Alexa Fluor 594 (Invitrogen, USA). Virus titers were determined with a previously described focus-forming unit (FFU) assay with minor modifications (14) and are expressed as the log10 number of FFU per milliliter. Sequences from HCVcc were analyzed by Sanger sequencing of reverse transcription-PCR amplicons, produced as previously described (17, 61) with the primers shown in Table 7.
Treatments with DAA.
DAA obtained from Acme Bioscience (USA) were previously tested in HCVcc (30, 31, 62). Potency (EC50) was determined in concentration-response assays (short-term assays) in Huh7.5 cells, as previously described (43). The efficacy and the barrier to resistance of the treatments were evaluated using long-term assays (30). In brief, Huh7.5 cells were infected with HCVcc, and treatments were initiated upon viral spread. Cells were split every 2 to 3 days and treated, and viral spread was determined by immunofluorescence. Treatment lasted 30 days, and cultures without viral spread were kept without drugs for at least 2 weeks. If no HCV-positive cells were observed during posttreatment follow-up, the infection was considered eradicated. The presence of HCV-infected cells during treatment or after drug withdrawal led to viral escape: viral spread occurring during treatment was indicated as viral breakthrough, and viral spread occurring after drug withdrawal was indicated as viral relapse.
Differences in the EC50 before (baseline) and after (escape) treatment were expressed as the fold change and were calculated as follows: the EC50 for the escape virus divided by the EC50 for the baseline virus. Escape viruses were subjected to Sanger sequencing, in which the corresponding drug targets (NS3, NS5A, and/or NS5B region) were analyzed (genotypic analysis). Sequences were visually inspected using the Sequencher (version 5.1) program (Gene Codes, USA). Furthermore, escape viruses were passaged in naive cells, sequenced, and used for analysis of drug susceptibility (phenotypic analysis).
The original combination treatments shown in Fig. 1 were performed with drug concentrations that exhibited significantly increased viral control when combined in comparison with the control achieved when the drugs were used as monotherapy. Depending on the virus, 570 nM nuc was equivalent to 2- to 4-fold the EC50 of sofosbuvir and 2- to 5-fold the EC50 of uprifosbuvir, 422 nM glecaprevir was equivalent to 33- to 84-fold the EC50 of glecaprevir, 11 nM NS5A inhibitor was equivalent to 3- to 2,750-fold the EC50 of elbasvir and to 79- to 500-fold the EC50 of velpatasvir, and 0.05 nM pibrentasvir was equivalent to 4- to 10-fold the EC50 of pibrentasvir. Specific fold EC50 values for each virus and experiment can be found in Table 4.
Data availability.
Sequences for recombinant T9cc and S83cc have been deposited in GenBank under accession no. MN712203 and MN712204, respectively.
ACKNOWLEDGMENTS
We thank A. L. Sørensen (Copenhagen University Hospital, Hvidovre, Denmark) for laboratory assistance and B. Ørskov Lindhardt (Copenhagen University Hospital, Hvidovre, Denmark) and C. Geisler (University of Copenhagen) for valuable support. We thank C. M. Rice (Rockefeller University, New York, NY) for providing reagents.
We have no conflicts of interest to disclose.
This work was supported by grants from the Region H Foundation (to S.R. and J.B.), The Lundbeck Foundation (to S.R. and J.B.), The Novo Nordisk Foundation (to J.B.), Independent Research Fund Denmark (DFF), Medical Sciences (to S.R. and J.B.), the Danish Cancer Society (to J.B.), Innovation Fund Denmark (Infect-ERA EU, to J.B.), and the Candys Foundation (to C.F.-A. and J.B.). J.B. is the recipient of the 2015 Novo Nordisk Prize and the 2019 Distinguished Investigator Award from the Novo Nordisk Foundation.
Study concept and design: S.R. and J.B. Acquisition of data: S.R., C.F.-A., L.S.M., J.P., and Y.-P.L. Analysis and interpretation of data: S.R., C.F.-A., J.P., Y.-P.L., and J.B. Drafting of the manuscript: S.R. and J.B. Study supervision: S.R. and J.B.
REFERENCES
- 1.The Polaris Observatory HCV Collaborators. 2017. Global prevalence and genotype distribution of hepatitis C virus infection in 2015: a modelling study. Lancet Gastroenterol Hepatol 2:161–176. doi: 10.1016/S2468-1253(16)30181-9. [DOI] [PubMed] [Google Scholar]
- 2.European Association for the Study of the Liver. 2018. EASL recommendations on treatment of hepatitis C 2018. J Hepatol 2:461–511. doi: 10.1016/j.jhep.2018.03.026. [DOI] [PubMed] [Google Scholar]
- 3.Bukh J. 2016. The history of hepatitis C virus (HCV): basic research reveals unique features in phylogeny, evolution and the viral life cycle with new perspectives for epidemic control. J Hepatol 65:S2–S21. doi: 10.1016/j.jhep.2016.07.035. [DOI] [PubMed] [Google Scholar]
- 4.Pawlotsky JM. 2019. Retreatment of hepatitis C virus-infected patients with direct-acting antiviral failures. Semin Liver Dis 39:354–368. doi: 10.1055/s-0039-1687823. [DOI] [PubMed] [Google Scholar]
- 5.Messina JP, Humphreys I, Flaxman A, Brown A, Cooke GS, Pybus OG, Barnes E. 2015. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology 61:77–87. doi: 10.1002/hep.27259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simmonds P, Becher P, Bukh J, Gould EA, Meyers G, Monath T, Muerhoff S, Pletnev A, Rico-Hesse R, Smith DB, Stapleton JT, ICTV Report Consortium . 2017. ICTV virus taxonomy profile: Flaviviridae. J Gen Virol 98:2–3. doi: 10.1099/jgv.0.000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Welzel TM, Bhardwaj N, Hedskog C, Chodavarapu K, Camus G, McNally J, Brainard D, Miller MD, Mo H, Svarovskaia E, Jacobson I, Zeuzem S, Agarwal K. 2017. Global epidemiology of HCV subtypes and resistance-associated substitutions evaluated by sequencing-based subtype analyses. J Hepatol 67:224–236. doi: 10.1016/j.jhep.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 8.Marascio N, Liberto M, Barreca G, Zicca E, Quirino A, Lamberti A, Bianco G, Matera G, Surace L, Berardelli G, Surace L, De Maria V, Giancotti F, Leone R, Villella V, Nisticò S, Borelli A, Caruso V, Calderazzo M, Griffo G, Masciari R, Minchella P, Cosco L, Laganà C, Oliva A, Foti G, Fiorillo M, Bocchiaro G, Surace P, Ciccaglione A, Ciccozzi M, Cesario F, Torti C, Focà A. 2014. Update on epidemiology of HCV in Italy: focus on the Calabria Region. BMC Infect Dis 14(Suppl 5):S2. doi: 10.1186/1471-2334-14-S5-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pedersen J, Carlsen TH, Prentoe J, Ramirez S, Jensen TB, Forns X, Alter H, Foung SK, Law M, Gottwein J, Weis N, Bukh J. 2013. Neutralization resistance of hepatitis C virus can be overcome by recombinant human monoclonal antibodies. Hepatology 58:1587–1597. doi: 10.1002/hep.26524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rumi MG, De Filippi F, La Veccchia C, Donato MF, Gallus S, Del Ninno E, Colombo M. 2005. Hepatitis C reactivation in patients with chronic infection with genotypes 1b and 2c: a retrospective cohort study of 206 untreated patients. Gut 54:402–406. doi: 10.1136/gut.2004.048009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramirez S, Bukh J. 2018. Current status and future development of infectious cell-culture models for the major genotypes of hepatitis C virus: essential tools in testing of antivirals and emerging vaccine strategies. Antiviral Res 158:264–287. doi: 10.1016/j.antiviral.2018.07.014. [DOI] [PubMed] [Google Scholar]
- 12.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 14.Li YP, Ramirez S, Gottwein JM, Scheel TK, Mikkelsen L, Purcell RH, Bukh J. 2012. Robust full-length hepatitis C virus genotype 2a and 2b infectious cultures using mutations identified by a systematic approach applicable to patient strains. Proc Natl Acad Sci USA 18:E1101–E1110. doi: 10.1073/pnas.1203829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu J, Xiang Y, Tao W, Li Q, Wang N, Gao Y, Xiang X, Xie Q, Zhong J. 2014. A novel strategy to develop a robust infectious hepatitis C virus cell culture system directly from a clinical isolate. J Virol 88:1484–1491. doi: 10.1128/JVI.02929-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Date T, Kato T, Kato J, Takahashi H, Morikawa K, Akazawa D, Murayama A, Tanaka-Kaneko K, Sata T, Tanaka Y, Mizokami M, Wakita T. 2012. Novel cell culture-adapted genotype 2a hepatitis C virus infectious clone. J Virol 86:10805–10820. doi: 10.1128/JVI.07235-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramirez S, Li YP, Jensen SB, Pedersen J, Gottwein JM, Bukh J. 2014. Highly efficient infectious cell culture of three hepatitis C virus genotype 2b strains and sensitivity to lead protease, nonstructural protein 5A, and polymerase inhibitors. Hepatology 59:395–407. doi: 10.1002/hep.26660. [DOI] [PubMed] [Google Scholar]
- 18.Hezode C. 2018. Treatment of hepatitis C: results in real life. Liver Int 38:21–27. doi: 10.1111/liv.13638. [DOI] [PubMed] [Google Scholar]
- 19.Ceccherini-Silberstein F, Cento V, Di Maio VC, Perno CF, Craxì A. 2018. Viral resistance in HCV infection. Curr Opin Virol 32:115–127. doi: 10.1016/j.coviro.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 20.Di Maio VC, Cento V, Aragri M, Paolucci S, Pollicino T, Coppola N, Bruzzone B, Ghisetti V, Zazzi M, Brunetto M, Bertoli A, Barbaliscia S, Galli S, Gennari W, Baldanti F, Raimondo G, Perno CF, Ceccherini-Silberstein F. 2018. Frequent NS5A and multiclass resistance in almost all HCV genotypes at DAA failures: what are the chances for second-line regimens? J Hepatol 68:597–600. doi: 10.1016/j.jhep.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 21.Gottwein JM, Pham LV, Mikkelsen LS, Ghanem L, Ramirez S, Scheel TKH, Carlsen THR, Bukh J. 2018. Efficacy of NS5A inhibitors against hepatitis C virus genotypes 1-7 and escape variants. Gastroenterology 154:1435–1448. doi: 10.1053/j.gastro.2017.12.015. [DOI] [PubMed] [Google Scholar]
- 22.Bukh J, Purcell RH, Miller RH. 1994. Sequence analysis of the core gene of 14 hepatitis C virus genotypes. Proc Natl Acad Sci U S A 91:8239–8243. doi: 10.1073/pnas.91.17.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bukh J, Purcell RH, Miller RH. 1993. At least 12 genotypes of hepatitis C virus predicted by sequence analysis of the putative E1 gene of isolates collected worldwide. Proc Natl Acad Sci U S A 17:8239–8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato T, Furusaka A, Miyamoto M, Date T, Yasui K, Hiramoto J, Nagayama K, Tanaka T, Wakita T. 2001. Sequence analysis of hepatitis C virus isolated from a fulminant hepatitis patient. J Med Virol 3:334–339. doi: 10.1002/jmv.1055. [DOI] [PubMed] [Google Scholar]
- 25.Yanagi M, Purcell RH, Emerson SU, Bukh J. 1999. Hepatitis C virus: an infectious molecular clone of a second major genotype (2a) and lack of viability of intertypic 1a and 2a chimeras. Virology 1:250–263. doi: 10.1006/viro.1999.9889. [DOI] [PubMed] [Google Scholar]
- 26.Nakao H, Okamoto H, Tokita H, Inoue T, Iizuka H, Pozzato G, Mishiro S. 1996. Full-length genomic sequence of a hepatitis C virus genotype 2c isolate (BEBE1) and the 2c-specific PCR primers. Arch Virol 3:701–704. doi: 10.1007/BF01718327. [DOI] [PubMed] [Google Scholar]
- 27.Jordier F, Deligny ML, Barre R, De Micco P, Cantaloube JF. 2013. Evidence for two phylogenetic clusters within hepatitis C virus (HCV) genotype 2 inferred from analysis of complete coding sequences of 15 HCV strains. J Med Virol 85:1754–1764. doi: 10.1002/jmv.23674. [DOI] [PubMed] [Google Scholar]
- 28.Newman RM, Kuntzen T, Weiner B, Berical A, Charlebois P, Kuiken C, Murphy DG, Simmonds P, Bennett P, Lennon NJ, Birren BW, Zody MC, Allen TM, Henn MR. 2013. Whole genome pyrosequencing of rare hepatitis C virus genotypes enhances subtype classification and identification of naturally occurring drug resistance variants. J Infect Dis 208:17–31. doi: 10.1093/infdis/jis679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarrazin C. 2016. The importance of resistance to direct antiviral drugs in HCV infection in clinical practice. J Hepatol 64:486–504. doi: 10.1016/j.jhep.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 30.Ramirez S, Mikkelsen LS, Gottwein JM, Bukh J. 2016. Robust HCV genotype 3a infectious cell culture system permits identification of escape variants with resistance to sofosbuvir. Gastroenterology 151:973–985. doi: 10.1053/j.gastro.2016.07.013. [DOI] [PubMed] [Google Scholar]
- 31.Pham LV, Ramirez S, Gottwein JM, Fahnoe U, Li YP, Pedersen J, Bukh J. 2018. HCV genotype 6a escape from and resistance to velpatasvir, pibrentasvir, and sofosbuvir in robust infectious cell culture models. Gastroenterology 154:2194–2208. doi: 10.1053/j.gastro.2018.02.017. [DOI] [PubMed] [Google Scholar]
- 32.Lawitz E, Poordad F, Anderson LJ, Vesay M, Kelly MM, Liu H, Gao W, Fernsler D, Asante-Appiah E, Robertson MN, Hanna GJ, Barr E, Butterton J, Kowdley KV, Hassanein T, Sahota A, Gordon SC, Yeh WW. 2019. Efficacy and safety of ruzasvir 60 mg and uprifosbuvir 450 mg for 12 weeks in adults with chronic hepatitis C virus genotype 1, 2, 3, 4 or 6 infection. J Viral Hepat 26:675–684. doi: 10.1111/jvh.13079. [DOI] [PubMed] [Google Scholar]
- 33.Krieger N, Lohmann V, Bartenschlager R. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J Virol 75:4614–4624. doi: 10.1128/JVI.75.10.4614-4624.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blight KJ, Norgard EA. 2006. HCV replicon systems In Tan SL. (ed), Hepatitis C viruses: genomes and molecular biology. Horizon Bioscience, Norfolk, United Kingdom. [PubMed] [Google Scholar]
- 35.Woerz I, Lohmann V, Bartenschlager R. 2009. Hepatitis C virus replicons: dinosaurs still in business? J Viral Hepatitis 16:1–9. doi: 10.1111/j.1365-2893.2008.01066.x. [DOI] [Google Scholar]
- 36.Saeed M, Scheel TK, Gottwein JM, Marukian S, Dustin LB, Bukh J, Rice CM. 2012. Efficient replication of genotype 3a and 4a hepatitis C virus replicons in human hepatoma cells. Antimicrob Agents Chemother 56:5365–5373. doi: 10.1128/AAC.01256-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Camus G, Xu S, Han B, Lu J, Dvory-Sobol H, Yu M, Cheng G, Miller MD, Doehle BP, Mo H. 2018. Establishment of robust HCV genotype 4d, 5a, and 6a replicon systems. Virology 514:134–141. doi: 10.1016/j.virol.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 38.Lam AM, Espiritu C, Bansal S, Micolochick Steuer HM, Niu C, Zennou V, Keilman M, Zhu Y, Lan S, Otto MJ, Furman PA. 2012. Genotype and subtype profiling of PSI-7977 as a nucleotide inhibitor of hepatitis C virus. Antimicrob Agents Chemother 6:3359–3368. doi: 10.1128/AAC.00054-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dvory-Sobol H, Han B, Lu J, Yu M, Beran RK, Cheng G, Martin R, Svarovskaia E, Mo H. 2019. In vitro resistance profile of hepatitis C virus NS5A inhibitor velpatasvir in genotypes 1 to 6. J Viral Hepat 26:991–1001. doi: 10.1111/jvh.13116. [DOI] [PubMed] [Google Scholar]
- 40.Ng TI, Krishnan P, Pilot-Matias T, Kati W, Schnell G, Beyer J, Reisch T, Lu L, Dekhtyar T, Irvin M, Tripathi R, Maring C, Randolph JT, Wagner R, Collins C. 2017. In vitro antiviral activity and resistance profile of the next-generation hepatitis C virus NS5A inhibitor pibrentasvir. Antimicrob Agents Chemother 61:e02558-16. doi: 10.1128/AAC.02558-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jensen SB, Fahnoe U, Pham LV, Serre S, Tang Q, Ghanem L, Pedersen MS, Ramirez S, Humes D, Pihl AF, Filskov J, Solund CS, Dietz J, Fourati S, Pawlotsky JM, Sarrazin C, Weis N, Schonning K, Krarup H, Bukh J, Gottwein JM. 2019. Evolutionary pathways to persistence of highly fit and resistant hepatitis C virus protease inhibitor escape variants. Hepatology 70:771–787. doi: 10.1002/hep.30647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smolders EJ, Jansen AME, Ter Horst PGJ, Rockstroh J, Back DJ, Burger DM. 2019. Viral hepatitis C therapy: pharmacokinetic and pharmacodynamic considerations: a 2019 update. Clin Pharmacokinet 58:1237–1263. doi: 10.1007/s40262-019-00774-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gottwein JM, Jensen SB, Li YP, Ghanem L, Scheel TK, Serre SB, Mikkelsen L, Bukh J. 2013. Combination treatment with hepatitis C virus protease and NS5A inhibitors is effective against recombinant genotype 1a, 2a, and 3a viruses. Antimicrob Agents Chemother 57:1291–1303. doi: 10.1128/AAC.02164-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pomeroy JJ, Drusano GL, Rodriquez JL, Brown AN. 2017. Searching for synergy: identifying optimal antiviral combination therapy using hepatitis C virus (HCV) agents in a replicon system. Antiviral Res 146:149–152. doi: 10.1016/j.antiviral.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Svarovskaia ES, Dvory-Sobol H, Parkin N, Hebner C, Gontcharova V, Martin R, Ouyang W, Han B, Xu S, Ku K, Chiu S, Gane E, Jacobson IM, Nelson DR, Lawitz E, Wyles DL, Bekele N, Brainard D, Symonds WT, McHutchison JG, Miller MD, Mo H. 2014. Infrequent development of resistance in genotype 1–6 hepatitis C virus-infected subjects treated with sofosbuvir in phase 2 and 3 clinical trials. Clin Infect Dis 59:1666–1674. doi: 10.1093/cid/ciu697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han B, Martin R, Xu S, Parvangada A, Svarovskaia ES, Mo H, Dvory-Sobol H. 2019. Sofosbuvir susceptibility of genotype 1 to 6 HCV from DAA-naive subjects. Antiviral Res 170:104574. doi: 10.1016/j.antiviral.2019.104574. [DOI] [PubMed] [Google Scholar]
- 47.Walker A, Filke S, Lubke N, Obermeier M, Kaiser R, Haussinger D, Timm J, Bock HH. 2017. Detection of a genetic footprint of the sofosbuvir resistance-associated substitution S282T after HCV treatment failure. Virol J 14:106. doi: 10.1186/s12985-017-0779-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gane EJ, Metivier S, Nahass R, Ryan M, Stedman CA, Svarovskaia ES, Mo H, Doehle B, Dvory-Sobol H, Hedskog C, Lin M, Brainard DM, Yang JC, McHutchison JG, Sulkowski M, Younes Z, Lawitz E. 2017. The emergence of NS5B resistance associated substitution S282T after sofosbuvir-based treatment. Hepatol Commun 1:538–549. doi: 10.1002/hep4.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Svarovskaia ES, Gane E, Dvory-Sobol H, Martin R, Doehle B, Hedskog C, Jacobson IM, Nelson DR, Lawitz E, Brainard DM, McHutchison JG, Miller MD, Mo H. 2016. L159F and V321A sofosbuvir-associated hepatitis C virus NS5B substitutions. J Infect Dis 213:1240–1247. doi: 10.1093/infdis/jiv564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wing PAC, Jones M, Cheung M, DaSilva S, Bamford C, Jason Lee WY, Aranday-Cortes E, da Silva FA, McLauchlan J, Smith D, Irving W, Cunningham M, Ansari A, Barnes E, Foster GR. 2019. Amino acid substitutions in genotype 3a hepatitis C virus polymerase protein affect responses to sofosbuvir. Gastroenterology 157:692–704. doi: 10.1053/j.gastro.2019.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.da Silva FA, Sreenu V, Hughes J, Aranday-Cortes E, Irving WL, Foster GR, Agarwal K, Rosenberg W, Macdonald D, Richardson P, Aldersley MA, Wiselka M, Ustianowski A, McLauchlan J, Thomson EC. 2017. Response to DAA therapy in the NHS England Early Access Programme for rare HCV subtypes from low and middle income countries. J Hepatol 67:1348–1350. doi: 10.1016/j.jhep.2017.06.035. [DOI] [PubMed] [Google Scholar]
- 52.Fourati S, Rodriguez C, Hezode C, Soulier A, Ruiz I, Poiteau L, Chevaliez S, Pawlotsky JM. 2019. Frequent antiviral treatment failures in patients infected with hepatitis C virus genotype 4, subtype 4r. Hepatology 69:513–523. doi: 10.1002/hep.30225. [DOI] [PubMed] [Google Scholar]
- 53.Candotti D, Temple J, Sarkodie F, Allain JP. 2003. Frequent recovery and broad genotype 2 diversity characterize hepatitis C virus infection in Ghana, West Africa. J Virol 77:7914–7923. doi: 10.1128/JVI.77.14.7914-7923.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Markov PV, Pepin J, Frost E, Deslandes S, Labbé AC, Pybus OG. 2009. Phylogeography and molecular epidemiology of hepatitis C virus genotype 2 in Africa. J Gen Virol 90(Pt 9):2086–2096. doi: 10.1099/vir.0.011569-0. [DOI] [PubMed] [Google Scholar]
- 55.Sagnelli E, Alessio L, Sagnelli C, Gualdieri L, Pisaturo M, Minichini C, Di Caprio G, Starace M, Onorato L, Scotto G, Macera M, Coppola N. 2018. Clinical findings of HCV chronic infection in undocumented immigrants and low-income refugees in three areas of southern Italy. Ann Hepatol 17:47–53. doi: 10.5604/01.3001.0010.7534. [DOI] [PubMed] [Google Scholar]
- 56.Scheel TK, Gottwein JM, Mikkelsen LS, Jensen TB, Bukh J. 2011. Recombinant HCV variants with NS5A from genotypes 1–7 have different sensitivities to an NS5A inhibitor but not interferon-alpha. Gastroenterology 140:1032–1042. doi: 10.1053/j.gastro.2010.11.036. [DOI] [PubMed] [Google Scholar]
- 57.Asante-Appiah E, Ingravallo P, McMonagle P, Bystol K, Xia E, Curry S, Qiu P, Black S, Chase R, Liu R, Lahser F. 2019. Interplay of amino acid residues at positions 28 and 31 in NS5A defines resistance pathways in hepatitis C virus genotype 2. Antimicrob Agents Chemother 63:e01269-19. doi: 10.1128/AAC.01269-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feld JJ, Jacobson IM, Hezode C, Asselah T, Ruane PJ, Gruener N, Abergel A, Mangia A, Lai CL, Chan HL, Mazzotta F, Moreno C, Yoshida E, Shafran SD, Towner WJ, Tran TT, McNally J, Osinusi A, Svarovskaia E, Zhu Y, Brainard DM, McHutchison JG, Agarwal K, Zeuzem S. 2015. Sofosbuvir and velpatasvir for HCV genotype 1, 2, 4, 5, and 6 infection. N Engl J Med 373:2599–2607. doi: 10.1056/NEJMoa1512610. [DOI] [PubMed] [Google Scholar]
- 59.Sarrazin C, Cooper CL, Manns MP, Reddy KR, Kowdley KV, Roberts SK, Dvory-Sobol H, Svarovskia E, Martin R, Camus G, Doehle BP, Stamm LM, Hyland RH, Brainard DM, Mo H, Gordon SC, Bourliere M, Zeuzem S, Flamm SL. 2018. No impact of resistance-associated substitutions on the efficacy of sofosbuvir, velpatasvir, and voxilaprevir for 12 weeks in HCV DAA-experienced patients. J Hepatol 69:1221–1230. doi: 10.1016/j.jhep.2018.07.023. [DOI] [PubMed] [Google Scholar]
- 60.Kuiken C, Hraber P, Thurmond J, Yusim K. 2008. The hepatitis C sequence database in Los Alamos. Nucleic Acids Res 36:D512–D516. doi: 10.1093/nar/gkm962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fahnoe U, Bukh J. 2019. Full-length open reading frame amplification of hepatitis C virus. Methods Mol Biol 1911:85–91. doi: 10.1007/978-1-4939-8976-8_5. [DOI] [PubMed] [Google Scholar]
- 62.Pham LV, Jensen SB, Fahnoe U, Pedersen MS, Tang Q, Ghanem L, Ramirez S, Humes D, Serre SBN, Schonning K, Bukh J, Gottwein JM. 2019. HCV genotype 1–6 NS3 residue 80 substitutions impact protease inhibitor activity and promote viral escape. J Hepatol 70:388–397. doi: 10.1016/j.jhep.2018.10.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequences for recombinant T9cc and S83cc have been deposited in GenBank under accession no. MN712203 and MN712204, respectively.