Antibiotic tolerance is an underappreciated antibiotic escape strategy that is associated with recurrent and relapsing infections, as well as acting as a precursor to resistance. Tolerance describes the ability of a bacterial population to survive transient exposure to an otherwise lethal concentration of antibiotic without exhibiting an elevated MIC.
KEYWORDS: Staphylococcus aureus, antibiotic tolerance, minimum duration for killing, stringent response, time-kill curves
ABSTRACT
Antibiotic tolerance is an underappreciated antibiotic escape strategy that is associated with recurrent and relapsing infections, as well as acting as a precursor to resistance. Tolerance describes the ability of a bacterial population to survive transient exposure to an otherwise lethal concentration of antibiotic without exhibiting an elevated MIC. It is detected in time-kill assays as a lower rate of killing than a susceptible strain and can be quantified by the metric minimum duration for killing (MDK). The molecular mechanisms behind tolerance are varied, but activation of the stringent response (SR) via gene knockouts and/or chemical induction has long been associated with tolerance. More recently, two Gram-positive clinical isolates from persistent bacteremias were found to bear mutations in the SR controller, Rel, that caused elevated levels of the alarmone (p)ppGpp. Here, we show that introduction of either of these mutations into Staphylococcus aureus confers tolerance to five different classes of antibiotic as a result of (p)ppGpp-mediated growth defects (longer lag time and/or lower growth rate). The degree of tolerance is related to the severity of the growth defect and ranges from a 1.5- to 3.1-fold increase in MDK. Two classes of proposed SR inhibitor were unable to reverse or reduce this tolerance. Our findings reveal the significance of SR-activating mutations in terms of tolerance and clinical treatment failures. The panel of strains reported here provide a clinically relevant model of tolerance for further investigation of its link to resistance development, as well as potential validation of high-throughput tolerance screens.
INTRODUCTION
Antibiotic resistance is a well-known and largely well-understood strategy used by many pathogenic bacteria to evade the action of antibiotics. It is a major contributor to the global burden of infectious diseases and has been identified as a public health priority by the Centers for Disease Control and Prevention and the World Health Organization (1, 2). However, resistance is not the only antibiotic escape strategy employed by pathogens. Antibiotic resistance describes the inheritable ability of bacteria to grow at high concentrations of an antibiotic for an indefinite period of time, and it is readily detected as an elevated MIC (above the susceptible breakpoint, where one exists). In contrast, antibiotic tolerance refers to the ability of a bacterial population to survive transient exposure to a given bactericidal antibiotic, at a concentration that would otherwise be lethal, without exhibiting an MIC above the susceptible breakpoint (3, 4). Historically, tolerance was commonly referred to as “phenotypic” or “noninherited” resistance (5, 6); however, we now know that there are both genotypic and phenotypic forms of tolerance (7, 8). The extent of the clinical burden and the contribution of tolerance to treatment failures is not explicitly known, in part because tolerance is not detected by MIC determinations and other routine laboratory tests. Nevertheless, tolerance is considered an important and underappreciated cause of treatment failures that is alarmingly common among clinical isolates (3, 8–14). Furthermore, there is now growing evidence to indicate that tolerance acts as a precursor to the development and acquisition of bona fide resistance (15–17).
The gold-standard method for detecting tolerance is the time-kill assay, in which the kinetics of antibiotic killing in broth are determined via viable counting. The minimum duration for killing (MDK) has been proposed as a metric for detecting and quantifying tolerance in time-kill data (3, 10). When a susceptible, nontolerant strain is exposed to a bactericidal antibiotic, the population will die at a given rate and the MDK required to kill, for example, 90% of the population can be calculated (MDK90). With a tolerant strain, the entire bacterial population will die at a lower rate than the susceptible strain, and the MDK90 will be extended. It is important to note that “tolerance” is a comparative, rather than categorical, term; there are presently no established criteria for defining tolerance, and its identification is dependent on a comparison of MDK values between strains that exhibit the same MIC. Furthermore, tolerance should not be confused with persistence; persistence is observed as a bimodal time-kill curve, whereby the majority of the population dies at the same rate as a susceptible strain, but a small persistent subpopulation emerges that dies at a slower rate. Therefore, in this example, a persistent strain would exhibit the same MDK90 as the susceptible strain but have an extended MDK99 (3, 18).
The molecular mechanisms behind tolerance are varied and poorly understood (8, 19), but the overarching theme appears to be slow growth and reduced metabolism (8, 20–24). Most clinically relevant antibiotics target active metabolic processes; therefore, a slowing down of these processes often results in tolerance (20). Examples of genotypic tolerance are commonly observed in small-colony variants of Staphylococcus aureus, such as defects in electron transport (25). The classic example of phenotypic tolerance occurs in biofilms; biofilm cells exhibit low growth rates which, together with a multitude of other factors, contribute to their recalcitrance toward antibiotics (26). The slow growth exhibited by cells within biofilms can be due to a range of stresses, including nutrient starvation which leads to induction of the stringent response (SR). The SR is a universal bacterial stress response that is classically induced by amino acid starvation (27). The presence of uncharged tRNAs at the ribosome is sensed by the SR controller, known as Rel or RelA (28, 29), and the controller responds by synthesizing the alarmones guanosine tetra- and pentaphosphate [collectively known as (p)ppGpp]. The ultimate effect of (p)ppGpp, through a variety of different mechanisms in different bacteria, is the downregulation of most metabolic processes and a cessation of growth. Although this is the classical description of the SR, we now know that (p)ppGpp is a master regulator of almost all aspects of bacterial physiology and virulence and that (p)ppGpp production can be promoted by many different stimuli (30–32). This central role of (p)ppGpp and the SR in bacterial pathogenicity has led to a search for SR inhibitors (28, 33).
In addition to the contribution the SR makes to antibiotic tolerance in a biofilm (34), induction of the SR also has a long association with tolerance in planktonic cultures (28). Using chemical inducers of the SR (e.g., mupirocin, an isoleucyl-tRNA synthetase inhibitor, or the serine analog serine hydroxamate) or overexpression of Rel/RelA, activation of the SR in planktonic cultures of both Gram-positives and Gram-negatives has been shown to confer tolerance to β-lactams, fluoroquinolones, and vancomycin (28). Conversely, deletion of genes essential for activation of the SR abrogates the tolerance phenotype. While these studies provide invaluable insight into the relationship between the SR and tolerance, they do not accurately represent the clinical situation. In 2010, the first clinical isolate bearing a SR-activating mutation in Rel was reported from a case of persistent S. aureus bacteremia (35). In most bacteria, Rel is a bifunctional enzyme that possesses a synthetase domain, which synthesizes (p)ppGpp, and a hydrolase domain that degrades (p)ppGpp (when the stress/stimulus has been removed) (32). Whole-genome sequencing of the clinical isolate revealed a Phe128Tyr (F128Y) mutation within the predicted hydrolase domain of Rel that emerged during therapy. Strains bearing this mutation had a reduced growth rate and elevated intracellular (p)ppGpp concentration, consistent with partial activation of the SR (likely as a result of reduced hydrolase function; SR activation is considered only partial because cell growth and division are still occurring). The mutation had no effect on resistance, as determined by MIC, and any effect on tolerance was not appropriately assessed. More recently, an isolate of Enterococcus faecium, again associated with a case of persistent bacteremia, was found to bear a Leu152Phe (L152F) mutation in the predicted hydrolase domain of Rel (36). The mutation conferred an elevated (p)ppGpp level and tolerance to vancomycin, linezolid, and daptomycin when cells were grown in a biofilm.
Given the long-standing association between the SR and tolerance, as well as the clinically relevant example of SR activation that the two reported Rel mutations provide, we set out to conduct a comprehensive assessment of the potential effect of these mutations on tolerance using S. aureus Newman as a model. Here, we show that both mutations confer significant tolerance to five different classes of antibiotic in planktonic culture, with MDK values that are extended by 1.5- to 3.1-fold compared to the wild type. We also demonstrate that two classes of compounds that are thought to be SR inhibitors are unable to reverse this tolerance phenotype. Our findings demonstrate that activation of the SR via mutation of Rel is an important mechanism of antibiotic tolerance among clinical isolates that impacts the efficacy of antibiotics with diverse modes of action and likely contributes to treatment failures. The panel of S. aureus strains we have generated here represent a clinically relevant model of SR activation and tolerance that can be applied to the evaluation of future SR inhibitor candidates, as well as the potential validation of high-throughput tolerance screens and the emerging relationship between tolerance and resistance.
(This study was presented in part at ASM Microbe 2019, San Francisco, CA, USA.)
RESULTS
Location of clinical Rel mutations.
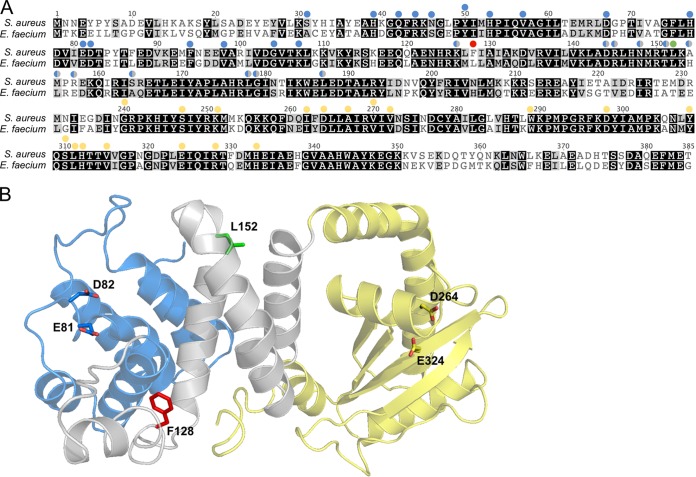
The bifunctional Rel protein consists of an N-terminal catalytic region, which houses the hydrolase and synthetase domains, and a C-terminal regulatory region. The catalytic region of Rel from the reported clinical isolate of E. faecium (locus tag BOW68_RS15490) shares 61% amino acid sequence identity with Rel from S. aureus Newman; Leu152 is among the conserved residues (Fig. 1A). The X-ray crystal structure of the catalytic region of Rel from Streptococcus dysgalactiae has been solved (37) and shows that the N-terminal hydrolase domain is separated from the synthetase domain by a central three-helix bundle linker region. Extensive mutagenesis of this central region has revealed that it is essential for hydrolase function (37). By threading the S. aureus Newman Rel sequence onto the S. dysgalactiae structure (Fig. 1B), we can see that both Phe128 and Leu152 are located in this linker region; therefore, the reported mutations are proposed to partially activate the SR by reducing the hydrolase activity of Rel (35, 36). Importantly, Phe128 and Leu152 are completely conserved among all of the Rel sequence entries in GenBank for S. aureus and E. faecium, respectively.
FIG 1.
Location of clinical Rel mutations. (A) Alignment of the N-terminal catalytic region of Rel from S. aureus Newman and E. faecium VRE001. Shading indicates the degree of amino acid conservation (black = conserved, gray = conservative substitution, white = not conserved). Colored circles above residues indicate sites that have been experimentally mutated in Rel from Streptococcus dysgalactiae and found to affect either hydrolase (blue) or synthetase (yellow) function. Sites indicated by blue/gray circles are located outside the core hydrolase domain, but their mutation still impacts hydrolase function. The red and green circles indicate the sites of the two clinical Rel mutations, F128 and L152, respectively. (B) Cartoon representation of the Rel catalytic region from S. dysgalactiae (PDB code 1VJ7) with the S. aureus Rel sequence threaded onto it (using Phyre2). The different domains are colored blue (hydrolase), yellow (synthetase), and gray (central three-helix bundle and linker regions). The sites of the clinical Rel mutations and catalytic residues (36) are shown as sticks and colored as in panel A.
Confirmation of partial SR activation.
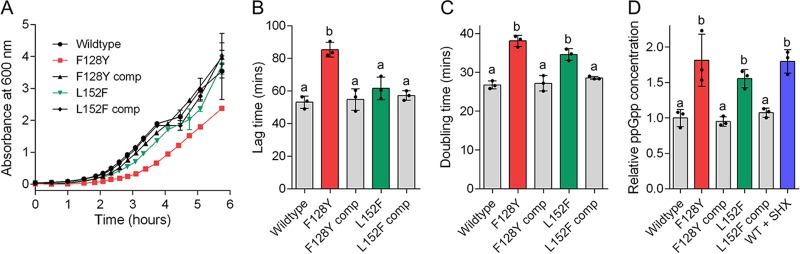
The staphylococcal F128Y and enterococcal L152F mutations were introduced into S. aureus Newman by allelic exchange and complemented by reintroduction of wild-type Rel (bearing a silent mutation that eliminates a HindIII site). To confirm that both of these mutations confer partial SR activation in S. aureus, the growth curves and intracellular ppGpp content of wild-type, mutant, and complemented strains were compared in rich medium (Fig. 2). The F128Y mutant exhibited a significant growth defect compared to the wild-type and complemented strains, with a significantly longer lag time and doubling time (multiplicity-adjusted P values of 0.0001 and <0.0001, respectively; Fig. 2B and C). The L152F mutation did not affect lag time (P > 0.05) but did confer an increase in doubling time (P = 0.0003). Both mutations conferred a statistically significantly elevated ppGpp concentration in mid-log phase, similar to exposure of cells to serine hydroxamate (Fig. 2D). When compared to each other, the F128Y mutant exhibited a longer lag time than the L152F mutant (P = 0.0016); the small differences in doubling time and ppGpp concentration between the mutants were not significant (P > 0.05).
FIG 2.
Confirmation of partial stringent response activation in S. aureus by clinical Rel mutations. (A) Growth curves of wild-type Newman, mutant, and complemented strains in TSB. (B and C) Lag times (B) and doubling times (C) derived from the growth curves in panel A. (D) Intracellular ppGpp concentrations shown relative to the wild type. A sample of wild-type cells were exposed to serine hydroxamate (SHX) as a positive control of stringent response induction. In all panels, data shown are the means of three replicates; in panels B to D, bars represent the means of the individual data points shown. In all panels, error bars, where visible, represent the standard deviations (SD). Letters above bars indicate statistically significant differences between means, as determined by one-way analysis of variance (ANOVA) with Tukey’s multiple-comparison test. comp, complemented.
Rel mutations confer multidrug tolerance.
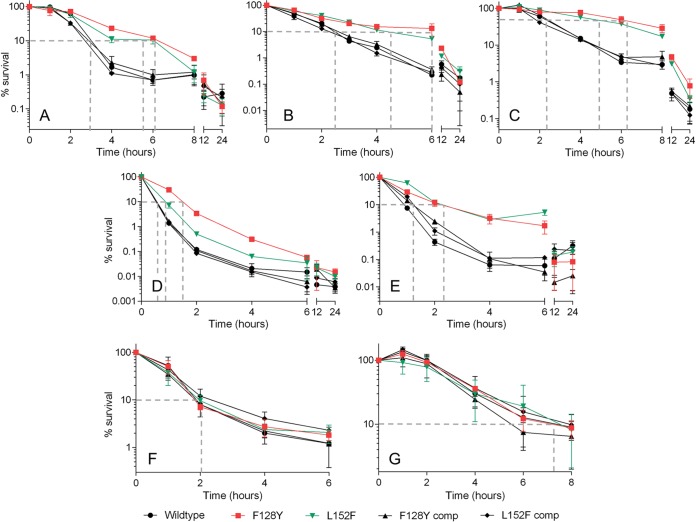
Having confirmed that both mutations confer partial induction of the stringent response in S. aureus, we set out to determine any effects on tolerance in time-kill assays with planktonic cultures. First, we determined the MICs of antibiotics from seven different classes against our panel of wild-type, mutant and complemented strains (Table 1). As expected, no differences were observed. Next, we exposed actively growing cultures to 4× the MIC of these antibiotics (8× the MIC for trimethoprim-sulfamethoxazole) and performed viable counts at intervals. Regular viable counts were made during the initial phase of killing to facilitate the calculation of accurate MDK values; for drugs that were affected by tolerance during the first 6 to 8 h, additional counts were made after 12 and 24 h to assess the duration of the tolerance effect. SR activation has long been associated with tolerance to inhibitors of peptidoglycan biosynthesis (38–41), and we observed that both mutations conferred tolerance to flucloxacillin (a penicillin), vancomycin (a glycopeptide) and cefazolin (a cephalosporin; Fig. 3A to C) when compared to the wild-type and complemented strains. Both mutants also exhibited tolerance to the DNA synthesis inhibitor ciprofloxacin (Fig. 3D) and, interestingly, the membrane-damaging agent daptomycin (Fig. 3E). Using the experimental data shown, MDK values were interpolated for each strain-antibiotic pairing as a comparative measure of the degree of tolerance (Table 2). We were able to accurately calculate the MDK90 for all strain-antibiotic pairings (see Materials and Methods for details), with the exception of cefazolin. In the case of cefazolin, the rate of killing against the mutants was too slow to calculate MDK90; therefore, MDK50 values were compared instead (Table 2). No statistically significant differences in MDK values were detected between wild-type and complemented strains for any antibiotic tested. For the antibiotics shown in Fig. 3A to E, the F128Y mutation conferred a 2.0- to 3.1-fold increase in MDK compared to the wild type, while the L152F mutation conferred a 1.5- to 2.3-fold increase. In general, there was a trend toward the F128Y mutation conferring a greater degree of tolerance (i.e., longer MDK) than the L152F mutation; however, this difference in MDK between the mutants was only significant for vancomycin (P = 0.0182) and ciprofloxacin (P < 0.0001). The tolerance phenotype of the F128Y mutant also lasted longer than that of the L152F mutant when exposed to vancomycin and ciprofloxacin; in terms of percent survival, the L152F mutant converged with its complemented strain and the wild type at 6 h for ciprofloxacin and 12 h for vancomycin (P > 0.05), whereas the F128Y mutant did not converge until 12 and 24 h, respectively. In the case of flucloxacillin, daptomycin, and cefazolin, all strains had converged by 8, 12, and 24 h, respectively. Interestingly, the RNA and folate synthesis inhibitors rifampin and trimethoprim-sulfamethoxazole, respectively, retained full bactericidal activity against both mutants, with no change in MDK90 (Fig. 3F and G; Table 2).
TABLE 1.
Broth microdilution MIC values for S. aureus Newman strains
| Antibiotic (cellular target) | MIC (μg/ml)a |
|---|---|
| Cefazolin (cell wall synthesis) | 0.5 |
| Ciprofloxacin (DNA synthesis) | 0.5 |
| Trimethoprim-sulfamethoxazole (folate synthesis) | 0.5/9.5b |
| Daptomycin (cytoplasmic membrane) | 1 |
| Flucloxacillin (cell wall synthesis) | 0.125 |
| Rifampin (RNA synthesis) | 0.032 |
| Vancomycin (cell wall synthesis) | 2 |
MIC values were identical between wild-type, mutant, and complemented strains. Therefore, only a single value is shown.
Trimethoprim-sulfamethoxazole MICs are shown as the concentration of trimethoprim/concentration of sulfamethoxazole (1:19 ratio).
FIG 3.
Clinical Rel mutations confer tolerance to antibiotics with diverse modes of action. Wild-type Newman, mutant, and complemented strains were exposed to flucloxacillin (A), vancomycin (B), cefazolin (C), ciprofloxacin (D), daptomycin (E), rifampin (F), and trimethoprim-sulfamethoxazole (G) at 4× the MIC (8× the MIC for trimethoprim-sulfamethoxazole), and viable counts were determined at intervals. Data shown are the means of three replicates; error bars, where visible, represent the SD. Dashed gray lines indicate the approximate minimum duration for killing (MDK) 90% of the population (50% in panel C) for each strain. MDK values interpolated from the data shown are given in Table 2. Raw data in log10 CFU/ml are shown in Fig. S1 in the supplemental material.
TABLE 2.
Minimum duration for killing values for S. aureus Newman strains
| Strain | MDK90 ± SD (h)a |
||||||
|---|---|---|---|---|---|---|---|
| FLC | VAN | CFZb | CIP | DAP | RIF | SXT | |
| Wild type | 2.9 ± 0.2 | 2.4 ± 0.1 | 1.8 ± 0.1 | 0.6 ± 0.1 | 0.9 ± 0.1 | 2.2 ± 0.4 | 7.2 ± 0.7 |
| F128Y | 5.8 ± 0.3 (<0.0001) | 6.1 ± 1.0 (<0.0001) | 5.6 ± 1.7 (0.0013) | 1.5 ± 0.1 (<0.0001) | 2.4 ± 0.5 (0.0001) | 2.3 ± 0.3 (ns) | 7.1 ± 2.6 (ns) |
| F128Y comp | 2.8 ± 0.3 (ns) | 2.6 ± 0.1 (ns) | 2.0 ± 0.3 (ns) | 0.6 ± 0.1 (ns) | 1.3 ± 0.1 (ns) | 2.3 ± 0.1 (ns) | 7.1 ± 0.5 (ns) |
| L152F | 4.7 ± 0.8 (0.0015) | 4.6 ± 0.3 (0.0017) | 4.1 ± 0.5 (0.0329) | 0.9 ± 0.1 (0.0005) | 2.6 ± 0.1 (<0.0001) | 2.3 ± 0.3 (ns) | 6.1 ± 0.8 (ns) |
| L152F comp | 2.6 ± 0.1 (ns) | 2.3 ± 0.1 (ns) | 1.7 ± 0.3 (ns) | 0.6 ± 0.1 (ns) | 1.2 ± 0.2 (ns) | 2.7 ± 0.4 (ns) | 7.4 ± 1.5 (ns) |
MDK, minimum duration for killing. Values were determined from the triplicate data shown in Fig. 3 Abbreviations: Flucloxacillin (FLC), vancomycin (VAN), cefazolin (CFZ), ciprofloxacin (CIP), daptomycin (DAP), rifampin (RIF), trimethoprim-sulfamethoxazole (SXT), complemented (comp). Numbers in parentheses are multiplicity-adjusted P values determined by one-way ANOVA with Tukey’s multiple-comparison posttest. ns, P > 0.05.
Cefazolin did not achieve 90% killing against the mutants in the course of the experiment. Therefore, the MDK values shown are MDK50.
Existing SR inhibitors are unable to reverse the tolerance phenotype of a Rel mutant.
Our time-kill data clearly show that clinical mutations in Rel that partially activate the SR confer significant tolerance to antibiotics with diverse mechanisms of action. Efforts are ongoing to design and/or discover inhibitors of the SR (28, 42, 43), and an important test of potential inhibitors would be their ability to resensitize tolerant bacteria to conventional antibiotics. To date, a small number of inhibitors have been reported which are proposed to inhibit SR activation via different mechanisms (28). Relacin is a rationally designed inhibitor of Rel synthetase activity that has been shown to inhibit (p)ppGpp synthesis by Rel in vitro and affect the growth profile of Bacillus subtilis (44). However, relacin exhibits a relatively low affinity for Rel, and millimolar concentrations are required in order to observe effects. The cationic peptides 1018 and DJK-5 have also been proposed to inhibit or reverse SR activation, by binding to ppGpp and signaling its degradation by the cell (45, 46). Both peptides exhibit the ability to sensitize bacterial biofilms to eradication by conventional antibiotics (46, 47); however, their specific mode of action with regard to the SR has been questioned (42, 48).
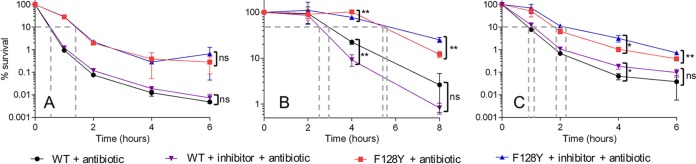
Given the clinically relevant example of SR activation that our S. aureus mutants represent, we set out to test the ability of these putative SR inhibitors to reverse the tolerance phenotype of our most “activated” and tolerant mutant, F128Y. In order to determine the appropriate concentration of inhibitor to use in time-kill assays, we monitored the growth of wild-type Newman and the F128Y mutant in the presence of a wide range of inhibitor concentrations (10- to 20-fold) in a 96-well plate; we anticipated that, while concentrations approaching the MIC would inhibit growth, lower concentrations may reverse SR activation and actually counteract the growth defect of our F128Y mutant. No reversal of the growth phenotype was observed with the F128Y mutant in the presence of low inhibitor concentrations, and the growth of both the mutant and wild type was increasingly inhibited by the highest concentrations (with identical/similar MICs; Fig. S2). As previously reported (49), DJK-5 was considerably more potent than 1018; therefore, 1018 was not tested further. For time-kill assays, concentrations of relacin and DJK-5 were chosen that inhibited growth of the F128Y mutant by approximately 20 to 30% (a value that ensured the inhibitor was present at a high enough concentration to potentially elicit an effect, while not adversely affecting the survival of the mutant), and inhibitors were added 90 min prior to the addition of antibiotic (in order to allow the inhibitor to potentially modulate the intracellular ppGpp concentration before cells were challenged). In all time-kill experiments, wild-type Newman was exposed to antibiotics in the presence/absence of inhibitor in parallel with the F128Y mutant, in order to assess any nonspecific effects of the inhibitor and provide a comparative benchmark of nontolerance. Incubation of wild-type Newman and the F128Y mutant with 1 mM relacin had no effect on the killing kinetics of ciprofloxacin (Fig. 4A) or the associated MDK values (see Table S1 in the supplemental material). When the concentration of relacin was increased to 2 mM and cefazolin was tested, there was again no statistically significant change in MDK in the presence of inhibitor for either strain. Some small but statistically significant changes in cell survival were observed at individual time points (Fig. 4B); however, while the inhibitor appeared to have a sensitizing effect on the mutant at 4 h (78% ± 6% survival with relacin versus 102% ± 2% without; ± the SD), it had a protective effect at 8 h (25% ± 3% versus 12% ± 2% without). Unexpectedly, DJK-5 at 5 μg/ml had a largely protective effect against killing by ciprofloxacin (Fig. 4C, Table S1). The MDK90 and MDK99 were both statistically significantly extended for the mutant in the presence of DJK-5 and, in accordance with this, cell survival was higher in the presence of the inhibitor at 4 and 6 h. For wild-type Newman, there was also a protective effect conferred by DJK-5 at 4 h, but the difference in cell survival at 6 h was not significant (nor was the increase in MDK). A checkerboard assay with DJK-5 and ciprofloxacin did not identify any antagonism between the two agents (fractional inhibitory concentration of 2).
FIG 4.
Existing stringent response inhibitors are unable to reverse the tolerance exhibited by a stringent response-activated strain. Wild-type Newman and the F128Y mutant were exposed to either 4× the MIC ciprofloxacin or cefazolin in the presence of different stringent response inhibitors: relacin (1 mM) and ciprofloxacin (A), relacin (2 mM) and cefazolin (B), and DJK-5 (5 μg/ml) and ciprofloxacin (C). Data shown are the means of three replicates; error bars, where visible, represent the SD. Asterisks indicate statistically significant differences between means as determined by an unpaired t test (* and **, P ≤ 0.05 and P ≤ 0.01, respectively; ns, not significant [P > 0.05]). In panel A, no statistically significant differences between with/without inhibitor were detected for either strain at any time point. Dashed gray lines indicate the approximate minimum duration for killing (MDK) 90% of the population (50% in panel B) for each strain. MDK values interpolated from the data shown are given in Table S1 in the supplemental material. Raw data in log10 CFU/ml are shown in Fig. S3.
DISCUSSION
Antibiotic tolerance is a greatly underappreciated contributor to treatment failures, in part because the time-kill assays required to detect it are laborious and time-consuming. Furthermore, careful comparative and statistical analysis of time-kill data and calculated MDK values may be required to detect low-level (i.e., subtle) tolerance. We have shown here that both of the reported clinical Rel mutations confer significant tolerance to five different classes of bactericidal antibiotic in S. aureus. The observed tolerance to peptidoglycan biosynthesis inhibitors and ciprofloxacin exhibited by our S. aureus mutants is consistent with previous studies that used amino acid deprivation or a chemical inducer to activate the SR (28). The detected tolerance to daptomycin is somewhat surprising given that its cellular target (the cytoplasmic membrane [50]) is not an active metabolic process. However, we have previously observed daptomycin tolerance in S. aureus treated with mupirocin (51), and Berti et al. (52) reported a Rel L61F mutation in an in vitro-selected daptomycin-tolerant mutant. SR-mediated daptomycin tolerance is also consistent with the more recent suggestion that the insertion of daptomycin into the cytoplasmic membrane interferes with membrane-bound cell wall lipid synthesis processes (53). Of the seven classes of bactericidal antibiotic tested, only rifampin and trimethoprim-sulfamethoxazole were unaffected in their activity by the partial SR activation in our mutants. The ability of rifampin to retain full bactericidal activity against slow-growing, SR-activated cells is consistent with its demonstrated activity against staphylococcal biofilms (54). In the case of trimethoprim-sulfamethoxazole, its reported effectiveness against S. aureus biofilms is limited (55), and there appears to be no information on its efficacy against slow-growing bacteria. As such, its unexpected activity against SR-activated cells requires further investigation.
The antibiotics used in this study represent the major classes of bactericidal agents used in the treatment of S. aureus infections (56). Therefore, our evaluation of the efficacy of these antibiotics against cells exhibiting SR-mediated tolerance could act as a starting point for further assessment of antibiotic efficacies against cells exhibiting other forms of tolerance, as well as prescribing practices in cases where tolerance is detected/suspected. The original case of S. aureus bacteremia caused by the F128Y-bearing isolate was treated with combinations of vancomycin, rifampin, ciprofloxacin, and linezolid (with trimethoprim-sulfamethoxazole for long-term suppressive treatment) (35). It is not known at what point during the course of the infection the Rel mutation arose; however, according to the tolerance phenotype of our F128Y mutant in broth culture, the efficacy of the vancomycin and ciprofloxacin treatment given may have been affected. Resistance to ciprofloxacin and rifampin, reduced susceptibility to linezolid, and vancomycin-intermediate resistance also developed during therapy. Again, it is not known definitively whether these resistances/reduced susceptibilities developed subsequent to the Rel mutation, but the tolerance phenotype demonstrated here and the known links between the SR, tolerance, and resistance acquisition/development (15–17, 28) suggest that the Rel mutation could have contributed to their emergence.
The classical description of SR activation culminates in complete cessation of growth, which allows bacteria to “wait out” the nutrient deprivation or stress (27). In the case of our mutants, the SR has only been partially activated, as evidenced by their ability to still grow, albeit at a slower rate. As well as a lower growth rate, the F128Y mutant also exhibits an extended lag time. These effects on growth likely represent a selective trade-off for the bacterium, as recently proposed by Fridman et al. (23). In the presence of antibiotics, a lower growth rate and/or extended lag time confers tolerance to antibiotic killing, which is a selective advantage; however, in the absence of antibiotics the growth defects represent a fitness cost. Interestingly, a comparison of the data presented for our two tolerant mutants indicates that the more extreme growth defects exhibited by the F128Y mutant are compensated for by greater tolerance (i.e., longer MDKs). Whether the greater trade-off between tolerance and growth accepted by the F128Y mutant would provide a selective advantage over the L152F mutant in an in vivo infection (and treatment) model remains to be tested. The overall impact of longer MDK values in terms of drug dosing regimens and treatment outcomes also requires in vivo exploration.
It is now clear that clinical mutations in Rel confer multidrug tolerance and that this tolerance likely contributes to persistent/relapsing infections by undermining the effectiveness of existing antibiotic therapies. As such, our findings provide further impetus to the search for SR inhibitors. A reasonable expectation, and clinical advantage, of a good SR inhibitor would be the ability to reverse the tolerance exhibited by SR-activated strains, thereby restoring their full susceptibility to conventional antibiotics. Unfortunately, under the conditions tested, relacin, 1018, and DJK-5 failed to reverse the growth defect and/or tolerance phenotype exhibited by our F128Y mutant (in fact, DJK-5 had a slight protective effect). There was also no difference in peptide sensitivity between the wild type and mutant, even though the mutant contains significantly more ppGpp (the proposed target of these peptides). Therefore, the molecular mechanism of these inhibitors remains unclear. New SR inhibitors are under investigation (28), including a promising Rel inhibitor that enhances the killing of nutrient-starved Mycobacterium tuberculosis by isoniazid (43). Our panel of tolerant and nontolerant strains provides an invaluable, clinically relevant system for evaluating the effectiveness of these new SR inhibitors. Furthermore, while time-kill assays are the gold standard for assessing tolerance, a number of higher-throughput tolerance screens are in development (10, 57, 58). Given the comparative nature of tolerance, these screens will require extensive optimization and testing if they are to identify tolerance in clinical isolates (where no directly related “wild-type” strain is available for comparison). Similar to MIC breakpoints, MDK ranges that indicate tolerance and nontolerance, for each antibiotic and in relation to the MIC, will also be required. We believe that our panel of tolerant and nontolerant strains represent a potentially useful benchmark for the validation of such screens and criteria.
MATERIALS AND METHODS
Bioinformatics.
The amino acid sequence of the catalytic domain of Rel from S. aureus Newman (residues 1 to 385, GenBank accession number WP_001058583) was threaded onto the X-ray crystal structure of the catalytic domain of Rel from S. dysgalactiae (PDB code 1VJ7) using the one-to-one threading function in Phyre2 (59). The modeled structure was visualized using MacPyMOL (60). To assess the conservation of Phe128 and Leu152, searches for “GTP pyrophosphokinase” for both S. aureus and E. faecium were performed in GenBank with a custom length setting of 300 to 1,000 residues (in order to exclude RelP and RelQ entries). The resultant 8,628 S. aureus entries and 294 E. faecium entries were submitted to ElimDupes (https://www.hiv.lanl.gov/content/sequence/elimdupesv2/elimdupes.html) to remove identical sequences, and the unique sequences were aligned and visualized using Geneious 6.1.8 (http://www.geneious.com).
Reagents and antibiotics.
X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and chloramphenicol were purchased from Bio Basic, Inc. (Markham, Ontario, Canada). Daptomycin was obtained from Selleckchem (Houston, TX). Peptides 1018 and DJK-5 were a gift from Bob Hancock, University of British Columbia. Relacin and PyDPA were gifts from Eylon Yavin, Hebrew University of Jerusalem, and Jong-In Hong, Seoul National University, respectively. The ppGpp standard was purchased from TriLink Biotechnologies (San Diego, CA). All other antibiotics and reagents, unless otherwise stated, were from MilliporeSigma (Burlington, MA).
Bacterial strains and growth conditions.
S. aureus Newman was a gift from Michael Murphy, University of British Columbia. Escherichia coli Stellar (TaKaRa Bio USA, Mountain View, CA) was used for all cloning, while E. coli IM08B (61) was used to prepare methylated plasmid for transformation into S. aureus. S. aureus strains were routinely cultured in tryptic soy broth/tryptic soy agar (TSB/TSA) at 37°C and stored long-term at −80°C with 8% (vol/vol) glycerol.
Allelic exchange.
Approximately 500-bp fragments flanking the site of mutation in rel (GenBank accession number NWMN_RS08620) were amplified from Newman genomic DNA using the mutagenic primers listed in Table S2 in the supplemental material and joined together by overlap extension PCR (62). Inserts were cloned into the temperature-sensitive staphylococcal vector pIMAY-Z (61) between the EcoRI and NotI sites by In-Fusion cloning (TaKaRa Bio USA), transformed into E. coli Stellar, and selected on LB plates containing 10 μg/ml chloramphenicol and 50 μg/ml X-Gal at 37°C. After construct confirmation by bidirectional sequencing (Sequetech, Mountain View, CA), pIMAY-Z constructs were transformed into E. coli IM08B (61), cultured at 37°C in 25 ml of LB broth containing chloramphenicol and purified. Electrocompetent S. aureus were prepared and electroporated with IM08B-derived constructs (∼1 μg) as previously described (63), with the exception that 2-mm cuvettes were used (1.8 kV, 600 Ω, 10 μF). Recovered cells were plated on TSA containing chloramphenicol and 100 μg/ml X-Gal and then incubated at 28°C for 2 days. Integration was achieved by growing transformants in TSB containing chloramphenicol at 40°C overnight, before plating on TSA containing chloramphenicol and X-Gal at 40°C. Blue colonies were screened for successful integration by colony PCR (cPCR) using the primers IM151 and IM152 as previously described (63). Successful integrants were grown overnight at 28°C in TSB with no selection to encourage excision, then plated on TSA containing X-Gal. cPCR using the primers Rel frag seq fwd and rev, which bind just outside the cloned rel fragment, was performed on white colonies confirmed to be chloramphenicol sensitive. The presence of the intended mutation, and no others, was confirmed by bidirectional sequencing. Complementation was achieved by reintroducing the wild-type copy of rel (except with a HindIII site silently removed) into the mutant strains using the methods described above.
Growth curve analysis.
Growth curves for wild-type, mutant, and complemented strains were determined in 5 ml of TSB in 16-mm test tubes in a shaking water bath at 37°C. Overnight cultures (10 μl) derived from different colonies were used to inoculate triplicate tubes, and absorbances at 600 nm (A600) were read periodically in a test tube spectrophotometer. Lag and doubling times were calculated using the program GrowthRates (64). All statistical analyses were performed in GraphPad Prism 6.07. For growth curves in the presence of inhibitors, 100-μl cultures were set up in 96-well plates and sealed with Breathe-Easy sealing membrane. Cationic peptides were prepared and diluted in 0.01% (vol/vol) acetic acid and 0.2% (wt/vol) bovine serum albumin as previously described (65). Inoculated plates were simultaneously incubated at 37°C and read for A600 in a SpectraMax M5 plate reader (Molecular Devices, San Jose, CA), with shaking and reads every 10 min.
ppGpp quantification.
ppGpp levels in wild-type, mutant, and complemented Newman strains were quantified essentially as described by Ancona et al. (66). Briefly, each strain was grown in triplicate in 20 ml of TSB to an A600 of ∼0.6, pelleted, and resuspended in 0.25 ml of methanol. Additional wild-type cultures were exposed to serine hydroxamate at 2.3 mg/ml for 1 h as a positive control of stringent response induction. Cells were lysed by vortexing, the cell debris was pelleted, and the supernatants were diluted to 20% methanol with dH2O. Diluted supernatants were lyophilized and resuspended in 1 mM HEPES (pH 7.4). ppGpp was detected using 20 μM PyDPA (67) in a black 384-well plate with excitation at 344 nm and emission at 470 nm. Blanked fluorescence values were converted to ppGpp concentration using a standard curve of purified ppGpp.
MIC and FIC determinations.
MIC determinations were performed using the broth microdilution method of the Clinical and Laboratory Standards Institute (CLSI) (68) in Mueller-Hinton broth (MHB), cation-adjusted MHB (CA-MHB; for trimethoprim-sulfamethoxazole), or MHB supplemented with 50 μg/ml Ca2+ (MHB-Ca2+; for daptomycin). The fractional inhibitory concentration (FIC) for DJK-5 and ciprofloxacin was determined via checkerboard assay in MHB as previously described (69).
Time-kill assays.
Time-kill assays were performed in triplicate at 37°C in a shaking water bath in MHB, CA-MHB, or MHB-Ca2+, as appropriate, according to CLSI guidelines (70). Viable counts were determined, in duplicate, on TSA following dilution in phosphate-buffered saline. The limit of detection following drug dilution was 300 CFU/ml. All antibiotics were tested at 4× the MIC, with the exception of trimethoprim-sulfamethoxazole which was tested at 8× the MIC. Viable counts were performed immediately after the addition of antibiotic, and at 1- to 12-h intervals as indicated on plots. Starting inocula at the point of antibiotic addition were in the range of 5 × 105 to 2 × 106 CFU/ml, with the exception of ciprofloxacin (1 × 107 CFU/ml). Stringent response inhibitors, when included, were added approximately 90 min prior to the addition of antibiotic. All data shown on a single plot (with the exception of the 12- and 24 h data) were collected as part of the same experiment to account for any variation in antibiotic stock concentration. MDK values were calculated from triplicate time-kill data using the linear regression and interpolation functions in GraphPad Prism 6.07; only data in the linear range of the MDK were used in the calculation (a minimum of three data points) and all R2 values were >0.9.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by a research grant from the British Columbia Lung Association awarded to J.K.H. and a Canadian Institutes of Health Research operating grant awarded to A.B.B. (PJT 159786).
We thank Michael Murphy, University of British Columbia, for providing S. aureus Newman; Ian Monk, University of Melbourne, for providing pIMAY-Z and IM08B; Jong-In Hong, Seoul National University, for providing PyDPA; Eylon Yavin, Hebrew University of Jerusalem, for providing relacin; and Bob Hancock, University of British Columbia, for providing peptides 1018 and DJK-5.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States. Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf. [Google Scholar]
- 2.World Health Organization. 2015. Global action plan on antimicrobial resistance. World Health Organization, Geneva, Switzerland: https://www.who.int/antimicrobial-resistance/publications/global-action-plan/en/. [Google Scholar]
- 3.Brauner A, Fridman O, Gefen O, Balaban NQ. 2016. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol 14:320–330. doi: 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- 4.Windels EM, Michiels JE, Van den Bergh B, Fauvart M, Michiels J, Windels EM, Michiels JE, Van den Bergh B, Fauvart M, Michiels J. 2019. Antibiotics: combatting tolerance to stop resistance. mBio 10:e02095-19. doi: 10.1128/mBio.02095-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levin BR, Rozen DE. 2006. Non-inherited antibiotic resistance. Nat Rev Microbiol 4:556–562. doi: 10.1038/nrmicro1445. [DOI] [PubMed] [Google Scholar]
- 6.Corona F, Martinez JL. 2013. Phenotypic resistance to antibiotics. Antibiotics (Basel) 2:237–255. doi: 10.3390/antibiotics2020237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan J, Bassler BL. 2019. Surviving as a community: antibiotic tolerance and persistence in bacterial biofilms. Cell Host Microbe 26:15–21. doi: 10.1016/j.chom.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meylan S, Andrews IW, Collins JJ. 2018. Targeting antibiotic tolerance, pathogen by pathogen. Cell 172:1228–1238. doi: 10.1016/j.cell.2018.01.037. [DOI] [PubMed] [Google Scholar]
- 9.Tuomanen E, Durack DT, Tomasz A. 1986. Antibiotic tolerance among clinical isolates of bacteria. Antimicrob Agents Chemother 30:521–527. doi: 10.1128/aac.30.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brauner A, Shoresh N, Fridman O, Balaban NQ. 2017. An experimental framework for quantifying bacterial tolerance. Biophys J 112:2664–2671. doi: 10.1016/j.bpj.2017.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiuff C, Andersson DI. 2007. Antibiotic treatment in vitro of phenotypically tolerant bacterial populations. J Antimicrob Chemother 59:254–263. doi: 10.1093/jac/dkl469. [DOI] [PubMed] [Google Scholar]
- 12.Mayhall CG, Medoff G, Marr JJ. 1976. Variation in the susceptibility of strains of Staphylococcus aureus to oxacillin, cephalothin, and gentamicin. Antimicrob Agents Chemother 10:707–712. doi: 10.1128/aac.10.4.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sabath LD, Wheeler N, Laverdiere M, Blazevic D, Wilkinson BJ. 1977. A new type of penicillin resistance of Staphylococcus aureus. Lancet i:443–447. doi: 10.1016/s0140-6736(77)91941-9. [DOI] [PubMed] [Google Scholar]
- 14.Brennan RO, Durack DT. 1983. Therapeutic significance of penicillin tolerance in experimental streptococcal endocarditis. Antimicrob Agents Chemother 23:273–277. doi: 10.1128/aac.23.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levin-Reisman I, Brauner A, Ronin I, Balaban NQ. 2019. Epistasis between antibiotic tolerance, persistence, and resistance mutations. Proc Natl Acad Sci U S A 116:14734–14739. doi: 10.1073/pnas.1906169116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levin-Reisman I, Ronin I, Gefen O, Braniss I, Shoresh N, Balaban NQ. 2017. Antibiotic tolerance facilitates the evolution of resistance. Science 355:826–830. doi: 10.1126/science.aaj2191. [DOI] [PubMed] [Google Scholar]
- 17.Windels EM, Michiels JE, Fauvart M, Wenseleers T, Van den Bergh B, Michiels J. 2019. Bacterial persistence promotes the evolution of antibiotic resistance by increasing survival and mutation rates. ISME J 13:1239–1251. doi: 10.1038/s41396-019-0344-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balaban NQ, Helaine S, Lewis K, Ackermann M, Aldridge B, Andersson DI, Brynildsen MP, Bumann D, Camilli A, Collins JJ, Dehio C, Fortune S, Ghigo J-M, Hardt W-D, Harms A, Heinemann M, Hung DT, Jenal U, Levin BR, Michiels J, Storz G, Tan M-W, Tenson T, Van Melderen L, Zinkernagel A. 2019. Definitions and guidelines for research on antibiotic persistence. Nat Rev Microbiol 17:441–448. doi: 10.1038/s41579-019-0196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kester JC, Fortune SM. 2014. Persisters and beyond: mechanisms of phenotypic drug resistance and drug tolerance in bacteria. Crit Rev Biochem Mol Biol 49:91–101. doi: 10.3109/10409238.2013.869543. [DOI] [PubMed] [Google Scholar]
- 20.Tuomanen E, Cozens R, Tosch W, Zak O, Tomasz A. 1986. The rate of killing of Escherichia coli by beta-lactam antibiotics is strictly proportional to the rate of bacterial growth. J Gen Microbiol 132:1297–1304. doi: 10.1099/00221287-132-5-1297. [DOI] [PubMed] [Google Scholar]
- 21.Kaiser P, Regoes RR, Dolowschiak T, Wotzka SY, Lengefeld J, Slack E, Grant AJ, Ackermann M, Hardt W-D. 2014. Cecum lymph node dendritic cells harbor slow-growing bacteria phenotypically tolerant to antibiotic treatment. PLoS Biol 12:e1001793. doi: 10.1371/journal.pbio.1001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Claudi B, Spröte P, Chirkova A, Personnic N, Zankl J, Schürmann N, Schmidt A, Bumann D. 2014. Phenotypic variation of Salmonella in host tissues delays eradication by antimicrobial chemotherapy. Cell 158:722–733. doi: 10.1016/j.cell.2014.06.045. [DOI] [PubMed] [Google Scholar]
- 23.Fridman O, Goldberg A, Ronin I, Shoresh N, Balaban NQ. 2014. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature 513:418–421. doi: 10.1038/nature13469. [DOI] [PubMed] [Google Scholar]
- 24.Mechler L, Herbig A, Paprotka K, Fraunholz M, Nieselt K, Bertram R. 2015. A novel point mutation promotes growth phase-dependent daptomycin tolerance in Staphylococcus aureus. Antimicrob Agents Chemother 59:5366–5376. doi: 10.1128/AAC.00643-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia LG, Lemaire S, Kahl BC, Becker K, Proctor RA, Denis O, Tulkens PM, Van Bambeke F. 2013. Antibiotic activity against small-colony variants of Staphylococcus aureus: review of in vitro, animal and clinical data. J Antimicrob Chemother 68:1455–1464. doi: 10.1093/jac/dkt072. [DOI] [PubMed] [Google Scholar]
- 26.Hall CW, Mah T-F. 2017. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol Rev 41:276–301. doi: 10.1093/femsre/fux010. [DOI] [PubMed] [Google Scholar]
- 27.Cashel M, Gentry D, Hernandez V, Vinella D. 1996. The stringent response, p 1458–1496. In Neidhardt FC, Curtiss RI, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (ed), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. [Google Scholar]
- 28.Hobbs JK, Boraston AB. 2019. (p)ppGpp and the stringent response: an emerging threat to antibiotic therapy. ACS Infect Dis 5:1505–1517. doi: 10.1021/acsinfecdis.9b00204. [DOI] [PubMed] [Google Scholar]
- 29.Atkinson GC, Tenson T, Hauryliuk V. 2011. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One 6:e23479. doi: 10.1371/journal.pone.0023479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaca AO, Colomer-Winter C, Lemos JA. 2015. Many means to a common end: the intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. J Bacteriol 197:1146–1156. doi: 10.1128/JB.02577-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dalebroux ZD, Svensson SL, Gaynor EC, Swanson MS. 2010. ppGpp conjures bacterial virulence. Microbiol Mol Biol Rev 74:171–199. doi: 10.1128/MMBR.00046-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. 2015. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol 13:298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petchiappan A, Chatterji D. 2017. Antibiotic resistance: current perspectives. ACS Omega 2:7400–7409. doi: 10.1021/acsomega.7b01368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao W, Chua K, Davies JK, Newton HJ, Seemann T, Harrison PF, Holmes NE, Rhee H-W, Hong J-I, Hartland EL, Stinear TP, Howden BP. 2010. Two novel point mutations in clinical Staphylococcus aureus reduce linezolid susceptibility and switch on the stringent response to promote persistent infection. PLoS Pathog 6:e1000944. doi: 10.1371/journal.ppat.1000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Honsa ES, Cooper VS, Mhaissen MN, Frank M, Shaker J, Iverson A, Rubnitz J, Hayden RT, Lee RE, Rock CO, Tuomanen EI, Wolf J, Rosch JW. 2017. RelA mutant Enterococcus faecium with multiantibiotic tolerance arising in an immunocompromised host. mBio 8:e02124-16. doi: 10.1128/mBio.02124-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hogg T, Mechold U, Malke H, Cashel M, Hilgenfeld R. 2004. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response. Cell 117:57–68. doi: 10.1016/S0092-8674(04)00260-0. [DOI] [PubMed] [Google Scholar]
- 38.Goodell W, Tomasz A. 1980. Alteration of Escherichia coli murein during amino acid starvation. J Bacteriol 144:1009–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kusser W, Ishiguro EE. 1985. Involvement of the relA gene in the autolysis of Escherichia coli induced by inhibitors of peptidoglycan biosynthesis. J Bacteriol 164:861–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tuomanen E, Tomasz A. 1986. Induction of autolysis in nongrowing Escherichia coli. J Bacteriol 167:1077–1080. doi: 10.1128/jb.167.3.1077-1080.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodionov DG, Ishiguro EE. 1995. Direct correlation between overproduction of guanosine 3’,5’-bispyrophosphate (ppGpp) and penicillin tolerance in Escherichia coli. J Bacteriol 177:4224–4229. doi: 10.1128/jb.177.15.4224-4229.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andresen L, Varik V, Tozawa Y, Jimmy S, Lindberg S, Tenson T, Hauryliuk V. 2016. Auxotrophy-based high throughput screening assay for the identification of Bacillus subtilis stringent response inhibitors. Sci Rep 6:35824. doi: 10.1038/srep35824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dutta NK, Klinkenberg LG, Vazquez M-J, Segura-Carro D, Colmenarejo G, Ramon F, Rodriguez-Miquel B, Mata-Cantero L, Porras-De Francisco E, Chuang Y-M, Rubin H, Lee JJ, Eoh H, Bader JS, Perez-Herran E, Mendoza-Losana A, Karakousis PC. 2019. Inhibiting the stringent response blocks Mycobacterium tuberculosis entry into quiescence and reduces persistence. Sci Adv 5:eaav2104. doi: 10.1126/sciadv.aav2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wexselblatt E, Oppenheimer-Shaanan Y, Kaspy I, London N, Schueler-Furman O, Yavin E, Glaser G, Katzhendler J, Ben-Yehuda S. 2012. Relacin, a novel antibacterial agent targeting the stringent response. PLoS Pathog 8:e1002925. doi: 10.1371/journal.ppat.1002925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de la Fuente-Núñez C, Reffuveille F, Haney EF, Straus SK, Hancock R. 2014. Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Pathog 10:e1004152. doi: 10.1371/journal.ppat.1004152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de la Fuente-Núñez C, Reffuveille F, Mansour SC, Reckseidler-Zenteno SL, Hernández D, Brackman G, Coenye T, Hancock R. 2015. d-Enantiomeric peptides that eradicate wild-type and multidrug-resistant biofilms and protect against lethal Pseudomonas aeruginosa infections. Chem Biol 22:196–205. doi: 10.1016/j.chembiol.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reffuveille F, de la Fuente-Núñez C, Mansour S, Hancock R. 2014. A broad-spectrum antibiofilm peptide enhances antibiotic action against bacterial biofilms. Antimicrob Agents Chemother 58:5363–5371. doi: 10.1128/AAC.03163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andresen L, Tenson T, Hauryliuk V. 2016. Cationic bactericidal peptide 1018 does not specifically target the stringent response alarmone (p)ppGpp. Sci Rep 6:36549. doi: 10.1038/srep36549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pletzer D, Mansour SC, Hancock R. 2018. Synergy between conventional antibiotics and anti-biofilm peptides in a murine, sub-cutaneous abscess model caused by recalcitrant ESKAPE pathogens. PLoS Pathog 14:e1007084. doi: 10.1371/journal.ppat.1007084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hobbs JK, Miller K, O’Neill AJ, Chopra I. 2008. Consequences of daptomycin-mediated membrane damage in Staphylococcus aureus. J Antimicrob Chemother 62:1003–1008. doi: 10.1093/jac/dkn321. [DOI] [PubMed] [Google Scholar]
- 51.Hobbs JK, Miller K, Chopra I. 2007. The effect of the stringent response on the bactericidal activity of antibiotics against Staphylococcus aureus. Abstr 47th Intersci Conf Antimicrob Agents and Chemother, abstr C1-1488.
- 52.Berti AD, Shukla N, Rottier AD, McCrone JS, Turner HM, Monk IR, Baines SL, Howden BP, Proctor RA, Rose WE. 2018. Daptomycin selects for genetic and phenotypic adaptations leading to antibiotic tolerance in MRSA. J Antimicrob Chemother 73:2030–2033. doi: 10.1093/jac/dky148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Müller A, Wenzel M, Strahl H, Grein F, Saaki TV, Kohl B, Siersma T, Bandow JE, Sahl H-G, Schneider T, Hamoen LW. 2016. Daptomycin inhibits cell envelope synthesis by interfering with fluid membrane microdomains. Proc Natl Acad Sci U S A 113:E7077–E7086. doi: 10.1073/pnas.1611173113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raad I, Hanna H, Jiang Y, Dvorak T, Reitzel R, Chaiban G, Sherertz R, Hachem R. 2007. Comparative activities of daptomycin, linezolid, and tigecycline against catheter-related methicillin-resistant Staphylococcus bacteremic isolates embedded in biofilm. Antimicrob Agents Chemother 51:1656–1660. doi: 10.1128/AAC.00350-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Butini ME, Abbandonato G, Di Rienzo C, Trampuz A, Di Luca M. 2019. Isothermal microcalorimetry detects the presence of persister cells in a Staphylococcus aureus biofilm after vancomycin treatment. Front Microbiol 10:332. doi: 10.3389/fmicb.2019.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu C, Infectious Diseases Society of America, Bayer A, Cosgrove SE, Daum RS, Fridkin SK, Gorwitz RJ, Kaplan SL, Karchmer AW, Levine DP, Murray BE, Rybak MJ, Talan DA, Chambers HF. 2011. Clinical practice guidelines by the Infectious Diseases Society of America for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children. Clin Infect Dis 52:e18–e55. doi: 10.1093/cid/ciq146. [DOI] [PubMed] [Google Scholar]
- 57.Gefen O, Chekol B, Strahilevitz J, Balaban NQ. 2017. TDtest: easy detection of bacterial tolerance and persistence in clinical isolates by a modified disk-diffusion assay. Sci Rep 7:41284. doi: 10.1038/srep41284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matsuo M, Hiramatsu M, Singh M, Sasaki T, Hishinuma T, Yamamoto N, Morimoto Y, Kirikae T, Hiramatsu K, Matsuo M, Hiramatsu M, Singh M, Sasaki T, Hishinuma T, Yamamoto N, Morimoto Y, Kirikae T, Hiramatsu K. 2019. Genetic and transcriptomic analyses of ciprofloxacin-tolerant Staphylococcus aureus isolated by the replica plating tolerance isolation system (REPTIS). Antimicrob Agents Chemother 63:e02019-18. doi: 10.1128/AAC.02019-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg M. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schrödinger. 2019. The PyMOL molecular graphics system, version 2.0. Schrödinger LLC, New York, NY. [Google Scholar]
- 61.Monk IR, Tree JJ, Howden BP, Stinear TP, Foster TJ. 2015. Complete bypass of restriction systems for major Staphylococcus aureus lineages. mBio 6:e00308-15. doi: 10.1128/mBio.00308-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 63.Monk IR, Shah IM, Xu M, Tan M-W, Foster TJ, Monk IR, Shah IM, Xu M, Tan M-W, Foster TJ. 2012. Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. mBio 3:e00277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mira P, Barlow M, Meza JC, Hall BG. 2017. Statistical package for growth rates made easy. Mol Biol Evol 34:3303–3309. doi: 10.1093/molbev/msx255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wieczorek M, Jenssen H, Kindrachuk J, Scott WRP, Elliott M, Hilpert K, Cheng JTJ, Hancock REW, Straus SK. 2010. Structural studies of a peptide with immune modulating and direct antimicrobial activity. Chem Biol 17:970–980. doi: 10.1016/j.chembiol.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 66.Ancona V, Lee JH, Chatnaparat T, Oh J, Hong J-I, Zhao Y. 2015. The bacterial alarmone (p)ppGpp activates the type III secretion system in Erwinia amylovora. J Bacteriol 197:1433–1443. doi: 10.1128/JB.02551-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rhee H-W, Lee C-R, Cho S-H, Song M-R, Cashel M, Choy HE, Seok Y-J, Hong J-I. 2008. Selective fluorescent chemosensor for the bacterial alarmone (p)ppGpp. J Am Chem Soc 130:784–785. doi: 10.1021/ja0759139. [DOI] [PubMed] [Google Scholar]
- 68.Clinical and Laboratory Standards Institute. 2018. Methods for dilution antimicrobial susceptibility test for bacteria that grow aerobically, 11th ed; approved standard M07 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 69.Pillai SK, Moellering RC, Eliopoulos GM. 2005. Antimicrobial combinations, p 365–440. In Lorian V. (ed), Antibiotics in laboratory medicine, 5th ed Lippincott/Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 70.Clinical and Laboratory Standards Institute. 1999. Methods for determining bactericidal activity of antimicrobial agents: approved guideline M26-A. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.