Abstract
Objective(s):
The present study investigated the prevalence of genes encoding for exotoxins, adhesion and biofilm factors in Staphylococcus aureus isolates obtained from samples in a referral burn hospital in Tehran, Iran.
Materials and Methods:
S. aureus isolates obtained from patients, personnel and surfaces in the wards of a burn hospital were identified and confirmed by biochemical and molecular tests, respectively. The susceptibility of isolates was determined using the disk diffusion method. Virulence factors were detected by multiplex PCR.
Results:
The frequency of hla, hlb, hld, hlg, tst and pvl genes was 92.8%, 34.7%, 89.8%, 11.9%, 10.7%, and 0.5% respectively. The results revealed that the hla gene had the highest frequency among isolates (94.4% for methicillin-resistant S. aureus (MRSA) and 89.8% for methicillin-susceptible S. aureus (MSSA)). The most prevalent adhesion and biofilm-related gene was eno (85.6%). The prevalence of the remaining genes was as follows: fib (71.8%), clfB (70%), cna (59.2 %), fnbB (17.9%), icaA (72.4%), and icaD (85.6%). The incidence of fib, hlb, hlg, and tst genes was significantly higher in MRSA isolates compare to the MSSA isolates. Moreover, the resistance rates for all antibiotics were higher is MRSA isolates except for nitrofurantoin and chloramphenicol antibiotics.
Conclusion:
Data indicate the high prevalence rates of virulence factors among S. aureus isolates, especially MRSA strains in the burn hospital. This should to be taken into account in the development of an effective infection control policy and continuous monitoring of drug resistance in hospitals.
Key Words: Adhesin and biofilm genes, Burn, Iran, MRSA, Virulence factors
Introduction
Damage to the skin barrier of patients with burn injuries increases the risk of microbial colonization, growth, and infection (1. Burn wound infections are a common dilemma in burn centers and are considered as a significant cause of mortality in burn patients. Staphylococcus aureus has been identified as a major etiological agent of infection in hospitalized burn patients (2). The following virulence factors have been identified for S. aureus: leukocidin (Panton-Valentine leukocidin; PVL), hemolysins (α, β, γ, δ), toxic shock syndrome toxin-1 (TSST-1), exfoliative toxins (ETs), and staphylococcal enterotoxin (SE) (3).
The virulence factors of S. aureus have various effects on human health. Leukotoxins and hemolysins can affect biological membrane leading to cell death (4). PVL can lead to skin and soft tissue infections, necrotizing pneumonia, and necrotizing fasciitis (5). Bacterial attachment to host tissues is the primary stage of infection. At this stage, adherence of S. aureus is mediated by microbial surface component-recognizing adhesive matrix molecules (MSCRAMMs) (6) including fibronectin–binding proteins A and B (FnbA and FnbB), fibrinogen-binding proteins (Fib), collagen binding protein (cna), clumping factors A and B (clfA and clfB), and laminin binding protein (eno) (7). A clear S. aureus biofilm can be formed on damaged skin, mucosa, and artificial surfaces (8). Furthermore, products of the ica locus and polysaccharide intercellular adhesin (PIA) are critical for intercellular bacterial adherence and biofilm formation (9).
Studies show that the epidemiology and virulence factors of S. aureus strains in hospitals, particularly in burn centers, are a challenge for infection control programs (10). Environmental surfaces and healthcare personnel are the leading sources of the spread of pathogens causing nosocomial infections. Early identification of S. aureus isolates obtained from patients, personnel and surfaces in hospitals can help us determine important virulence factors of the isolates for a more efficient infection control. The aim of this study was to investigate the prevalence of genes encoding for exotoxins, adhesion, and biofilm factors in S. aureus isolates in a burn hospital in Tehran, Iran.
Materials and Methods
Sample collection and identification of bacterial isolates
This cross-sectional study was conducted on samples obtained from Shahid Motahari Hospital (the main specialized burn center in Tehran, Iran) from December 2015 to December 2016. Samples were obtained from hospital personnel (both nostrils) and surfaces (beds, Ambu bags, door knobs, medical trolleys, chairs, suction, etc.). Samples were collected from personnel three times using wet sterile swabs and from surfaces monthly during the study period. All samples were cultured on brain-heart infusion media. Burn wound swabs were also taken as part of the routine screening for MRSA during the study period. Biochemical tests (mannitol salt agar media, susceptibility to bacitracin, catalase, DNase and tube coagulase tests, mannitol fermentation) were performed for bacterial identification.
Antimicrobial susceptibility tests
Antibiotic susceptibility was determined using the standardized Kirby-Bauer disc diffusion method on Mueller-Hinton agar. The antimicrobial agents tested included nitrofurantoin (300 µg), gentamicin (10 μg), mupirocin (20 μg), rifampicin (5 μg), norfloxacin (10 μg), tigecycline (15 μg), trimethoprim-sulfamethoxazol (25 μg), cefoxitin (30 μg), chloramphenicol (30 μg), erythromycin (15 μg), clindamycin (2 μg), tetracycline (30 μg), penicillin (10 units), linezolid (30 μg), synercid (quinupristin/dalfopristin; 15 μg), and imipenem (10 μg). Erythromycin-induced clindamycin resistance was determined using the disk approximation test. The isolate with cefoxitin resistance was MRSA. S. aureus ATCC 25923 was used as the control for sensitivity testing.
DNA extraction and molecular identification of MRSA isolates
DNA of S. aureus was extracted using the boiling method as described previously (11). For confirmation of S. aureus identification and determination of methicillin resistance, all isolates were subjected to the S. aureus-specific nuclease (nucA) and mecA-specific PCR (12, 13).
Detection of exotoxin- and biofilm-related genes
Multiplex PCR was used for the detection of virulence factors- encoding genes including pvl, tst (toxic shock syndrome toxin-1- encoding gene), and hla, hlb, hld and hlg genes (hemolysin-encoding genes). The following MSCRAMMs were detected using specific primers: clumping factor B (clfB), fibronectin-binding protein (fnbB), collagen-binding protein (cna), lamina-binding protein (eno), fibrinogen-binding protein (fib), and biofilm-encoding genes (icaA and icaD) (7,12-16). The PCR products (3 μl) were run on 1.5% agarose gel and stained with SYBR® Safe DNA stain. Electrophoresis of PCR products was carried out in 0.5×TBE buffer for 90 min at 110 mV. The standard PCR conditions and primers used for the multiplex PCR reactions in this study are listed in Table 1 and Table 2, respectively. The results of antibiotic susceptibility testing and the detection of virulence genes among S. aureus isolates were analyzed by Pearson Chi-Square and Fisher’s tests.
Table 1.
Primers and product size of PCR for detection of the exotoxins and biofilm genes
| Genes | Sequence (5-3) | Product size (bp) | Reference |
|---|---|---|---|
|
cna
eno fnbB fib clfB icaA icaD hla hlb hld hlg pvl tst nuc mecA |
F-GTCAAGCAGTTATTAACACCAGAC R-AATCAGTAATTGCACTTTGTCCACTG F-ACGTGCAGCAGCTGACT R-CAACAGCATYCTTCAGTACCTTC F-GTAACAGCTAATGGTCGAATTGATACT R-CAAGTTCGATAGGAGTACTATGTTC F-CTACAACTACAATTGCCGTCAACAG R-GCTCTTGTAAGACCATTTTCTTCAC F-ACATCAGTAATAGTAGGGGGCAAC R-TTCGCACTGTTTGTGTTTGCAC F-CCTAACTAACGAAAGGTAG R-AAGATATAGCGATAAGTGC F_AAACGTAAGAGAGGTGG R-GGCAATATGATCAAGATAC F- CTG ATT ACT ATC CAA GAA ATT CGA TTG R- CTT TCC AGC CTA CTT TTT TAT CAG T F- GTG CAC TTA CTG ACA ATA GTG C R- GTT GAT GAG TAG CTA CCT TCA GT F-AAG AAT TTT ATC TTA ATT AAG GAA GGA GTG R- TTA GTG AAT TTG TTC ACT GTG TCG A F- GTC AYA GAG TCC ATA ATG CAT TTA A R- CAC CAA ATG TAT AGC CTA AAG TG F-ATCATTAGGTAAAATGTCTGGACATGATCCA R-GCATCAASTGTATTGGATAGCAAAAGC F- ACCCCTGTTCCCTTATCATC R- TTTTCAGTATTTGTAACGCC F-CTGGCATATGTATGGCAATTGTT R-TATTGACCTGAATCAGCGTTGTCT F-GTGAAGATATACCAAGTGATT R-ATGCGCTATAGATTGAAAGGAT |
423 302 524 404 205 1315 381 209 309 111 535 433 326 664 147 |
15 |
Table 2.
Cycles and condition of multiplex PCRs in this study
| Genes | Cycles of amplification | Initial denaturation | Denaturation | Annealing | Extension | Final extension | Reference |
|---|---|---|---|---|---|---|---|
| nucA, mecA | 30 | 5min at 94 | 45s at 94 | 45s at 57 | 1min at 72 | 5min at 72 | 12, 13 |
| cna, eno, fib, fnbB, clfB | 25 | 5min at 94 | 1 min at 94 | 1min at 55 | 1min at 72 | 10 min at 72 | |
| icaA, icaD | 30 | 5min at 94 | 45s at 92 | 45s at 49 | 1min at 72 | 7min at 72 | |
| hla, hld, hld, hlg | 30 | 5min at 94 | 45s at 94 | 45s at 57 | 1min at 72 | 5min at 72 | |
| tst | 35 | 5min at 94 | 2min at 94 | 2min at 57 | 1min at 72 | 7min at 72 | |
| pvl | 30 | 5min at 95 | 40s at 95 | 40s at 54 | 45s at 72 | 5min at 72 |
Results
Bacterial isolates
In the present experimental study, from a total of 167 S. aureus isolates, 108 (65%) were identified as MRSA (79/123 isolates obtained from patients, 22/30 from surfaces, 7/14 from personnel), while 59 (35%) were identified as MSSA (44/123 from patients, 8/30 from surfaces and 7/14 from personnel).
Antimicrobial susceptibility testing
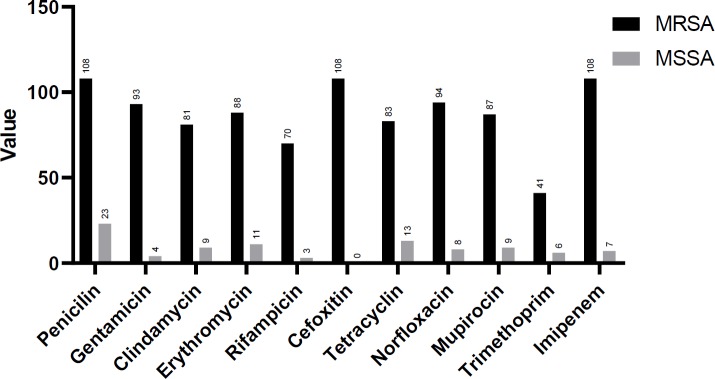
The antimicrobial resistance rate in S. aureus isolates to penicillin was 78%, imipenem 69%, cefoxitin 65%, norfloxacin 61%, erythromycin 59%, gentamicin 58%, tetracycline 57%, mupirocin 57%, clindamycin 54%, rifampicin 44%, trimethoprim-sulfamethoxazole 28%, tiecoplanin 9%, chloramphenicol 2% and nitrofurantoin 1%. The MRSA isolates revealed a significantly higher rate of antimicrobial resistance than the MSSA isolates (Table 3). The highest incidence of drug resistance in MRSA isolates was to penicillin (100%), imipenem (100%), cefoxitin (100%), norfloxacin (87%), and gentamicin (86%). All isolates were susceptible to quinupristin-dalfopristin, linezolid, and tigecycline (Table 3). Statistical analysis of antibiotic susceptibility patterns in MRSA and MSSA isolates are shown in Figure 1. Results show that resistance to all antibiotics (except for chloramphenicol and nitrofurantoin) was significantly higher in MRSA isolates compared to the MSSA isolates.
Table 3.
Antibiotics resistance in MRSA and MSSA strains in patients, surfaces and personnel in burn Shahid Motahari Hospital, Tehran, Iran
| Antibiotics | Patients N (%) |
Surfaces N (%) |
Personnel N (%) |
|||
|---|---|---|---|---|---|---|
| MRSA(79) | MSSA(44) | MRSA(22) | MRSA(7) | MSSA(8) | MSSA(7) | |
| Penicillin | 79 (100) | 16 (36) | 22 (100) | 4 (50) | 7 (100) | 3(43) |
| Gentamicin | 67 (85) | 2 (5) | 21 (95) | 2 (25) | 5 (71) | 0 |
| Clindamycin | 62 (78) | 7 (16) | 18 (82) | 2 (25) | 1 (14) | 0 |
| Erythromycin | 68 (86) | 9 (20) | 18 (82) | 2 (25) | 2 (29) | 0 |
| Nitrofurantoin | 1 (1) | 0 | 0 | 1(13) | 0 | 0 |
| Rifampicin | 48 (61) | 3 (7) | 17 (77) | 0 | 5 (71) | 0 |
| Quinupristin- Dalfopristin |
0 | 0 | 0 | 0 | 0 | 0 |
| Linezolid | 0 | 0 | 0 | 0 | 0 | 0 |
| Tigecycline | 0 | 0 | 0 | 0 | 0 | 0 |
| Cefoxitin | 79 (100) | 0 | 22 (100) | 0 | 7 (100) | 0 |
| Tetracycline | 61 (77) | 11(25) | 18 (82) | 2 (25) | 4 (57) | 0 |
| Norfloxacin | 68 (86) | 6 (14) | 20 ( 91) | 2 (25) | 6 (86) | 0 |
| Mupirocin | 62 (78) | 6 (14) | 18 (82) | 2 (25) | 7 (100) | 1(14) |
| Trimethoprim- Sulfamethoxazole |
32 (41) | 6 (14) | 8 (36) | 0 | 1 (14) | 0 |
| Imipenem | 79 (100) | 4 (9) | 22 (100) | 3 (38) | 7 (100) | 0 |
| Chloramphenicol | 3 (4) | 1 (2) | 0 | 0 | 0 | 0 |
MRSA: methicillin resistant Staphylococcus aureus; MSSA: methicillin-susceptible Staphylococcus aureus
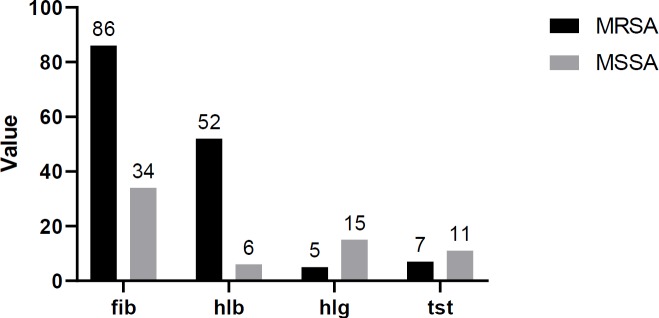
Figure 1.
The results of statistical analysis on virulence genes among MRSA and MSSA isolates
MRSA: methicillin resistant Staphylococcus aureus ; MSSA: methicillin-susceptible Staphylococcus aureus
Exotoxins and adhesin genes
The frequency of hla, hlb, hld, hlg, tst, and pvl genes was 92.8%, 34.7%, 89.8%, 11.9%, 10.7%, and 0.5%, respectively. Results revealed that the hla gene was the most frequent gene among isolates (94.4% for MRSA and 89.8% for MSSA). The frequency of other toxin genes in the MRSA and MSSA isolates respectively was 91.6% and 86.4% for hld, 48.1 and 10.1% for hlb, 6.4% and 18.6% for tst and 4.6% and 25.4% for hlg. The pvl gene was detected in 1.6% of MSSA isolates, but was not detected in MRSA.
Among the adhesion genes, the most prevalent was eno (85.6%). The incidence of other genes were as follows: fib (71.8%), clfB (70%), cna (59.2%) and fnbB (17.9%). The frequency of these genes in MRSA isolates was 87% for eno, 79.6% for fib, 67.5% for clfB, 61.1% for can, and 18.5% for fnbB. The clfB gene was detected in MSSA isolates at a significantly higher rate (7%) compared to the MRSA isolates; icaA, and icaD were positive in 72.4% and 85.6% of isolates, respectively. The icaD gene was found at analgesic rates in MRSA (85.1%) and MSSA (86.4%) isolates. The frequency of the icaA gene was slightly higher in MRSA (76.8%) than in MSSA (64.4%) isolates.
Overall, no significant difference in terms of virulence genes was found between the MRSA and MSSA isolates. The rates of genes detected from patients, surfaces and personnel are shown in Table 4. The coexistence of adhesion factors-related genes was detected in 8.9% of patient and 10% of surfaces isolates. Both the icaA and icaD genes were detected in 83.3%, 57.7%, and 57.1% of isolates from surfaces, patients and personnel, respectively. The antibiotic resistance profile and gene combination patterns in the MRSA isolates are shown in Table 5. None of the isolates showed the coexistence of toxin genes. Statistical analysis of the distribution of virulence genes among MRSA and MSSA isolates is shown in Figure 2. The results show that the incidence of fib, hlb, hlg, and tst genes was significantly higher in MRSA isolates compared to the MSSA isolates.
Table 4.
Distribution of virulence genes in Staphylococcus aureus, MRSA and MSSA isolates of patients, surfaces and personnel in burn Shahid Motahari Hospital, Tehran, Iran
| Virulence genes | Patients (123) |
Surfaces (30) |
Personnel(14) |
|||
|---|---|---|---|---|---|---|
| MRSA (79) N (%) | MSSA (7) (79) N (%) |
MSSA (44) (79) N (%) |
MRSA (22) (79) N (%) |
MSSA (8) (79) N (%) |
MRSA (7) (79) N (%) |
|
| Adhesion | ||||||
| eno | 65 (82.2) | 35 (79.5) | 22 (100) | 7 (87.5) | 7 (100) | 7 (100) |
| cna | 45 (56.9) | 26 (59) | 18 (81.8) | 4 (50) | 3 (42.8) | 3 (42.8) |
| clfB | 49 (62) | 34 (77.2) | 19 (86.3) | 7 (87.5) | 5 (71.4) | 3 (42.8) |
| fib | 59 (74.6) | 25 (56.8) | 20 (90.9) | 5 (62.5) | 7 (100) | 4 (57.1) |
| Toxin | ||||||
| hla | 17 (21.5) | 7 (15.9) | 2 (9) | 3 (37.5) | 1 (14.2) | 0 |
| hlb | 74 (93.6) | 40 (90.9) | 21 (95.4) | 7 (87.5) | 7 (100) | 6 (85.7) |
| hld | 34 (43) | 5 (11.3) | 14 (63.6) | 1 (12.5) | 4 (57.1) | 0 |
| hlg | 70 (88.6) | 38 (86.3) | 22 (100) | 6 (75) | 7 (100) | 7 (100) |
| 5 (6.3) | 13 (29.5) | 0 | 1 (12.5) | 0 | 1 (14.2) | |
| pvl | 5 (6.3) | 11 (25) | 2 (9) | 0 | 0 | 0 |
| Biofilm | ||||||
| tst | 0 | 1 (2.2) | 0 | 0 | 0 | 0 |
| icaA | 56 (70.8) | 28 (63.6) | 21 (95.4) | 6 (75) | 6(85.7) | 4 (57.1) |
| icaD | 64 (81) | 37 (84) | 21 (95.4) | 8 (100) | 7(100) | 6 (85.7) |
MRSA: methicillin resistant Staphylococcus aureus; MSSA: methicillin-susceptible Staphylococcus aureus
Table 5.
Antibiotic resistance profile and gene combination patterns in MRSA isolates of burn hospital
| Resistance profile | Gene combination |
||
|---|---|---|---|
| icaA+icaD | icaA+icaD+hla+hld | icaA+icaD+hla+hld+eno+fib | |
| GM, MUP, RP, NOR, E, CD | 36 (33%) | 33 (30.5%) | 25 (23.1%) |
| PG, IMI, T, E, CD | 46 (42.5%) | 42 (38.5%) | 40 (37%) |
| GM, MUP, NOR, RP | 44 (40.7%) | 43 (39.8%) | 35 (32.4%) |
| MG, MUP, RP, NOR, E, CD, T, PG, IMI | 33 (30.5%) | 31 (28.5%) | 25 (23.5%) |
PG, Penicillin; MG, Gentamicin; CD, Clindamycin; E, Erythromycin; RP, Rifampicin; T, Tetracycline; NOR, Norfloxacin; MUP, Mupirocin; IMI, Imipenem;
Figure 2.
The results of statistical analysis on antibiotic susceptibility tests among MRSA and MSSA isolates
MRSA: methicillin resistant Staphylococcus aureus; MSSA: methicillin-susceptible Staphylococcus aureus
Discussion
In the present study, a high prevalence of MRSA (65%) was found in samples obtained from a burn hospital in Tehran, Iran (from patients, healthcare personnel and surfaces). These results are in accordance with the results of other studies from Iran and Bangladesh that reported a high frequency of MRSA in burn patients (17-24). In contrast, Darban-Sarokhalil et al. reported a lower frequency of MRSA in two Iranian hospitals (11). The results of the present study indicated a lower prevalence of MRSA compared to another study in Uganda in which 100% of the isolates obtained from burn units were found to be MRSA (21). These discrepancies could be attributed to different infection control criteria, antibiotic administration, study design and laboratory testing for determination of methicillin resistance.
In the present study, there was a significant increase in the rate of resistance to antibiotics such as penicillin, tetracycline, erythromycin, gentamycin, clindamycin, mupirocin, and rifampicin in MRSA isolates. Data suggest the possibility of multiple antimicrobial resistance in hospital strains. This could be due to the continuous and empirical usage of broad-spectrum antibiotics and the absence of a suitable antibiotic treatment policy (23, 25). Despite the use of vancomycin and linezolid for the treatment of life-threatening infections caused by resistant S. aureus strains, all isolates were susceptible to new drugs (quinupristin-dalfopristin, linezolid, and tigecycline). These results are in accordance with those of Bayat et al (26). In the current study, the overall rate of resistance to mupirocin in MRSA isolates was 81%. Mupirocin resistance rate in MRSA isolates obtained from personnel, patients, and surfaces was 100%, 78%, and 82%, respectively. Chen et al. (2) reported high incidence of mupirocin resistance in most MRSA isolates in burn centers. The widespread use of mupirocin for prolonged periods, particularly for decolonization of healthcare personnel, bedsores and other skin lesions could be associated with the development of mupirocin resistance (27, 28).
In addition to antibiotic resistance, another factor that prevents effective treatment of staphylococcal infections in burn patients is biofilm formation (18). The importance of biofilm formation is unique in the medical world. Notably, bacterial species present in biofilms display more resistance to antibiotics and disinfectants (29). In burn wounds, molecules such as collagen, fibronectin, fibrinogen and other factors are present at the wound surface. S. aureus encodes many proteins that specifically interact with human cellular matrix components enabling the microorganism to colonize burn wounds (19). In our study, the frequency of eno, clfB, and cna genes was significantly higher than another study by Motallebi et al (30).
Another virulent factor that contributes to biofilm formation is PIA which can be encoded by the ica ADBC operon. Of the ica genes, icaA and icaD play an eminent role in biofilm production by S. aureus (28). Results show that icaA and icaD genes were present in 76.8% and 85.1% of isolates, respectively. Table 5 shows that the most indispensable genes detected in the MRSA isolates were identified as icaA+icaD followed by icaA+icaD+hla+hld. The frequencies obtained in the current study were significantly higher than those obtained in other studies performed in Iran (28). Satorres et al. (31) reported the frequencies of icaA and icaD genes in S. aureus isolates obtained from the hospital staff that were lower than those reported in the current study. The diversity of the prevalence of biofilm-encoded genes could be related to the variety of bacterial strains at different geographical regions.
Hemolysins (alpha, beta, delta and gamma) and PVL are able to damage host cells by their cytolytic effects. TSST-1 has been associated with several acute or chronic human diseases, including TSST (32). In the present study, the frequency of the hla and hld genes were 92.8% and 89.8%, respectively. This is in accordance with the results of Kateete et al. (21) in Uganda who reported a frequency of 100% for these genes. It was revealed that the frequency of the coexistence of hla+hld genes in S. aureus isolates obtained from patients, surfaces and personnel was 84.5%, 90% and 92.8%, respectively. High rates (93.6% and 88.6%) were recorded for patient-derived MRSA isolates harboring hla and hld genes, respectively. A similar rate for hla and hld were discovered in burn patients by Rodrigues et al (10). While hla and hld genes were found in all surface-derived MSSA isolates by Gharsa et al. (33), in the current work, these genes were detected in 87.5% and 75% of surface-derived MSSA isolates, respectively.
In the current study, the hlb and hlg genes were detected in 48.1% and 4.6% of the isolates, respectively. The rate of hlb (43%) in patient-derived MRSA isolates was similar to the study conducted by Karmakar et al. (34); however, this rate was lower than that found by Liu et al (35). The frequency of hlb gene in MRSA isolate obtained from personnel and surfaces (57.1% and 63.3%, respectively) was higher than those isolates obtained from patients. The hlg gene was detected in 6.3% of MRSA isolates. Diversity in the prevalence of hemolysin-, adhesion- and biofilm- encoding genes can be associated with the diversity of bacterial strains in different geographical areas.
A key virulence factor in S. aureus infections, especially in skin and soft-tissue infections is the PVL. This toxin has been recognized as a virulence factor associated with tissue necrosis (36). Data regarding the danger of infections caused by PVL-producing MRSA strains have raised public health concerns (5). In the current study, no pvl positive MRSA isolate was detected. This could be attributed to the fact that pvl is more related to community acquired MRSA strains. These findings were similar to the findings of Mkrtchyan et al (37). In contrast, a study from Brazil found that 14.6% of MRSA isolates had the pvl gene (10) . A study conducted in England reported that 2% of clinical S. aureus isolates (MRSA and MSSA) harbored the pvl gene (5). In the current study, only one patient-derived MSSA isolate (2.2%) was positive for pvl gene. Murray et al. (38) reported that pvl was detected in one MRSA isolate obtained from burn patients.
The frequency of the tst gene reported in Germany (39), Iran(40) and Korea(41) was 14%, 26.41% and 72.2%, respectively. In the current study, the frequency of the tst gene was 10.7%. Kateete et al. (21) studied patients, healthcare workers and surfaces in the burn units in a hospital. The rate of hla, hld, and tst in their study were analogous with those from the present study, but hlg and pvl genes were detected at higher frequencies than in the current study. Gharsa et al. (33) detected MSSA isolates on hospital surfaces and found tst in 60% of isolates; however, tst was not detected in MSSA isolates from the surfaces in the present study. The rate of tst in patient-derived MSSA isolates (25%) was higher than in patient-derived MRSA isolates (6.3%), which is similar to the results of a study by Liu et al (35). De Boeck et al. (42) found the prevalence of tst and pvl genes in the isolates obtained from healthcare workers to be 17.5% and 28.5%, respectively. In the present study, these genes were not detected in personnel. In fact, the difference in incidence could be related to the variation in the geographical area and the origin of the strains.
Hospital environments play an important role in the transmission of MRSA and the development of infection in patients (10). In the current study, the results of antibiotic susceptibility patterns and virulence factors, especially pvl, indicate that a potential outbreak in hospitals could be associated with the personnel or the surfaces. Colonized healthcare personnel and environmental sources could serve as a reservoir and disseminator of MRSA in hospitals. Therefore, using proper disinfectant and regular screening for MRSA among healthcare workers and patients, in addition to improved precautions for personnel are essential for infection control (17, 43). Moreover, methods in molecular epidemiology are compulsory for the continuous surveillance and rapid identification of prevalent strains of S. aureus and MRSA clones. These methods have been shown to contribute to the control of the spread of bacterial infections in healthcare settings (11, 20, 44).
Conclusion
It was determined that the high prevalence of virulence factors and the elevated rate of antibiotic resistance among isolates obtained from patients, personnel and surfaces of burn hospital necessitate proper implementation of an effective infection control policy and continuous monitoring for drug resistance.
Conflicts of Interest
All contributing authors declare no conflicts of interest.
Acknowledgment
The results described in this paper were part of student thesis and this study has been supported by Deputy of Research and Technology, Iran University of Medical Sciences. Grant No: 26564-30-04-94.
References
- 1.Boers SA, van Ess I, Euser SM, Jansen R, Tempelman FR, Diederen BM. An outbreak of a multiresistant methicillin-susceptible Staphylococcus aureus (MR-MSSA) strain in a burn centre: the importance of routine molecular typing. Burns: Burns. 2011;37:808–813. doi: 10.1016/j.burns.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Chen X, Yang HH, Huangfu YC, Wang WK, Liu Y, Ni YX, et al. Molecular epidemiologic analysis of Staphylococcus aureus isolated from four burn centers. Burns : Burns. 2012;38:738–742. doi: 10.1016/j.burns.2011.12.023. [DOI] [PubMed] [Google Scholar]
- 3.Sina H, Ahoyo TA, Moussaoui W, Keller D, Bankole HS, Barogui Y, et al. Variability of antibiotic susceptibility and toxin production of Staphylococcus aureus strains isolated from skin, soft tissue, and bone related infections. BMC Microbiol. 2013;13:188–196. doi: 10.1186/1471-2180-13-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Otto M. Staphylococcus aureus toxins. Curr Opin Microbiol. 2014;17:32–37. doi: 10.1016/j.mib.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teare L, Shelley OP, Millership S, Kearns A. Outbreak of Panton-Valentine leucocidin-positive meticillin-resistant Staphylococcus aureus in a regional burns unit. J Hosp Infect. 2010;76:220–224. doi: 10.1016/j.jhin.2010.04.023. [DOI] [PubMed] [Google Scholar]
- 6.Foster TJ, Hook M. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 1998;6:484–488. doi: 10.1016/s0966-842x(98)01400-0. [DOI] [PubMed] [Google Scholar]
- 7.Tristan A, Ying L, Bes M, Etienne J, Vandenesch F, Lina G. Use of multiplex PCR to identify Staphylococcus aureus adhesins involved in human hematogenous infections. J Clin Microbiol. 2003;41:4465–4467. doi: 10.1128/JCM.41.9.4465-4467.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.den Reijer PM, Haisma EM, Lemmens-den Toom NA, Willemse J, Koning RI, Demmers JA, et al. Detection of alpha-toxin and other virulence factors in biofilms of Staphylococcus aureus on polystyrene and a human epidermal model. PLoS One. 2016;11:e0145722. doi: 10.1371/journal.pone.0145722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Archer NK, Mazaitis MJ, Costerton JW, Leid JG, Powers ME, Shirtliff ME. Staphylococcus aureus biofilms: properties, regulation, and roles in human disease. Virulence. 2011;2:445–459. doi: 10.4161/viru.2.5.17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodrigues MVP, Fortaleza CMCB, Riboli DFM, Rocha RS, Rocha C, de Souza MdLR. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in a burn unit from Brazil. Burns. 2013;39:1242–1249. doi: 10.1016/j.burns.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Darban-Sarokhalil D, Khoramrooz SS, Marashifard M, Hosseini SAAM, Parhizgari N, Yazdanpanah M, et al. Molecular characterization of Staphylococcus aureus isolates from southwest of Iran using spa and SCCmec typing methods. Microb Pathog. 2016;98:88–92. doi: 10.1016/j.micpath.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 12.Bar-Gal GK, Blum SE, Hadas L, Ehricht R, Monecke S, Leitner G. Host-specificity of Staphylococcus aureus causing intramammary infections in dairy animals assessed by genotyping and virulence genes. Vet Microbiol. 2015;176:143–154. doi: 10.1016/j.vetmic.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 13.Japoni A, Jamalidoust M, Farshad S, Ziyaeyan M, Alborzi A, Japoni S, et al. Characterization of SCCmec types and antibacterial susceptibility patterns of methicillin-resistant Staphylococcus aureus in Southern Iran. Jpn J Infect Dis. 2011;64:28–33. [PubMed] [Google Scholar]
- 14.Vasudevan P, Nair MKM, Annamalai T, Venkitanarayanan KS. Phenotypic and genotypic characterization of bovine mastitis isolates of Staphylococcus aureus for biofilm formation. Vet Microbiol. 2003;92:179–185. doi: 10.1016/s0378-1135(02)00360-7. [DOI] [PubMed] [Google Scholar]
- 15.Jarraud S, Mougel C, Thioulouse J, Lina G, Meugnier H, Forey F, et al. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect Immun. 2002;70:631–641. doi: 10.1128/IAI.70.2.631-641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mehrotra M, Wang G, Johnson WM. Multiplex PCR for detection of genes forStaphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J Clin Microbiol. 2000;38:1032–1035. doi: 10.1128/jcm.38.3.1032-1035.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abbasi-Montazeri E, Khosravi AD, Feizabadi MM, Goodarzi H, Khoramrooz SS, Mirzaii M, et al. The prevalence of methicillin resistant Staphylococcus aureus (MRSA) isolates with high-level mupirocin resistance from patients and personnel in a burn center. Burns. 2013;39:650–654. doi: 10.1016/j.burns.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Dibah S, Arzanlou M, Jannati E, Shapouri R. Prevalence and antimicrobial resistance pattern of methicillin resistant Staphylococcus aureus (MRSA) strains isolated from clinical specimens in Ardabil, Iran. Iran J Microbiol. 2014;6 [PMC free article] [PubMed] [Google Scholar]
- 19.Kooistra-Smid M, van Dijk S, Beerthuizen G, Vogels W, van Zwet T, van Belkum A, et al. Molecular epidemiology of Staphylococcus aureus colonization in a burn center. Burns. 2004;30:27–33. doi: 10.1016/j.burns.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 20.Parhizgari N, Khoramrooz SS, Hosseini M, Asghar SA, Marashifard M, Yazdanpanah M, et al. High frequency of multidrug-resistant Staphylococcus aureus with SCCmec type III and Spa types t037 and t631 isolated from burn patients in Southwest of Iran. APMIS. 2016;124:221–228. doi: 10.1111/apm.12493. [DOI] [PubMed] [Google Scholar]
- 21.Kateete DP, Namazzi S, Okee M, Okeng A, Baluku H, Musisi NL, et al. High prevalence of methicillin resistant Staphylococcus aureus in the surgical units of Mulago hospital in Kampala, Uganda. BMC Res Notes. 2011;4:326–330. doi: 10.1186/1756-0500-4-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song W, Lee KM, Kang HJ, Shin DH, Kim DK. Microbiologic aspects of predominant bacteria isolated from the burn patients in Korea. Burns. 2001;27:136–139. doi: 10.1016/s0305-4179(00)00086-3. [DOI] [PubMed] [Google Scholar]
- 23.Alebachew T, Yismaw G, Derabe A, Sisay Z. Staphylococcus aureus burn wound infection among patients attending yekatit 12 hospital burn unit, addis ababa, ethiopia. Ethiop J Health Sci. 2012;22:209–213. [PMC free article] [PubMed] [Google Scholar]
- 24.Khoramrooz SS, Dolatabad SA, Dolatabad FM, Marashifard M, Mirzaii M, Dabiri H, et al. Detection of tetracycline resistance genes, aminoglycoside modifying enzymes, and coagulase gene typing of clinical isolates of Staphylococcus aureus in the Southwest of Iran. Iran J Basic Med Sci. 2017;20:912–919. doi: 10.22038/IJBMS.2017.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bayat B, Zade MH, Mansouri S, Kalantar E, Kabir K, Zahmatkesh E, et al. High frequency of methicillin-resistant Staphylococcus aureus (MRSA) with SCCmec type III and spa type t030 in Karaj’s teaching hospitals, Iran. Acta Microbiol Immunol Hung. 2017;64:331–341. doi: 10.1556/030.64.2017.020. [DOI] [PubMed] [Google Scholar]
- 27.Hetem DJ, Bonten MJ. Clinical relevance of mupirocin resistance in Staphylococcus aureus. J Hosp Infect. 2013;85:249–256. doi: 10.1016/j.jhin.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 28.Patel JB, Gorwitz RJ, Jernigan JA. Mupirocin resistance. Clin Infect Dis. 2009;49:935–941. doi: 10.1086/605495. [DOI] [PubMed] [Google Scholar]
- 29.Khoramrooz SS, Mansouri F, Marashifard M, Hosseini SAAM, Chenarestane-Olia FA, Ganavehei B, et al. Detection of biofilm related genes, classical enterotoxin genes and agr typing among Staphylococcus aureus isolated from bovine with subclinical mastitis in southwest of Iran. Microb Pathog. 2016;97:45–51. doi: 10.1016/j.micpath.2016.05.022. [DOI] [PubMed] [Google Scholar]
- 30.Motallebi M, Jabalameli F, Asadollahi K, Taherikalani M, Emaneini M. Spreading of genes encoding enterotoxins, haemolysins, adhesin and biofilm among methicillin resistant Staphylococcus aureus strains with staphylococcal cassette chromosome mec type IIIA isolated from burn patients. Microb Pathog. 2016;97:34–37. doi: 10.1016/j.micpath.2016.05.017. [DOI] [PubMed] [Google Scholar]
- 31.Satorres SE, Alcaraz LE. Prevalence of icaA and icaD genes in Staphylococcus aureus and Staphylococcus epidermidis strains isolated from patients and hospital staff. Cent Eur J Public Health. 2007;15:87–90. doi: 10.21101/cejph.a3396. [DOI] [PubMed] [Google Scholar]
- 32.Haveri M, Hovinen M, Roslof A, Pyorala S. Molecular types and genetic profiles of Staphylococcus aureus strains isolated from bovine intramammary infections and extramammary sites. J Clin Microbiol. 2008;46:3728–3735. doi: 10.1128/JCM.00769-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gharsa H, Dziri R, Klibi N, Chairat S, Lozano C, Torres C, et al. Environmental Staphylococcus aureus contamination in a Tunisian hospital. J Chemother. 2016;28:506–509. doi: 10.1179/1973947815Y.0000000036. [DOI] [PubMed] [Google Scholar]
- 34.Karmakar A, Dua P, Ghosh C. Biochemical and molecular analysis of Staphylococcus aureus clinical isolates from hospitalized patients. Can J Infect Dis Med Microbiol. 2016;2016:9041636. doi: 10.1155/2016/9041636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu C, Chen ZJ, Sun Z, Feng X, Zou M, Cao W, et al. Molecular characteristics and virulence factors in methicillin-susceptible, resistant, and heterogeneous vancomycin-intermediate Staphylococcus aureus from central-southern China. J Microbiol Immunol Infec. 2015;48:490–496. doi: 10.1016/j.jmii.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 36.Emaneini M, Khoramrooz SS, Shahsavan S, Dabiri H, Jabalameli F. Prevalence of Panton-Valentine leucocidin and phenotypic and genotypic characterization of biofilm formation among Staphylococcus aureus strains isolated from children with adenoid hypertrophy. Microb Pathog. 2015;89:150–153. doi: 10.1016/j.micpath.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 37.Mkrtchyan HV, Xu Z, Yacoub M, Ter-Stepanyan MM, Karapetyan HD, Kearns AM, et al. Detection of diverse genotypes of methicillin-resistant Staphylococcus aureus from hospital personnel and the environment in Armenia. Antimicrob Resist Infect Control. 2017;6:19–23. doi: 10.1186/s13756-017-0169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murray CK, Holmes RL, Ellis MW, Mende K, Wolf SE, McDougal LK, et al. Twenty-five year epidemiology of invasive methicillin-resistant Staphylococcus aureus (MRSA) isolates recovered at a burn center. Burns. 2009;35:1112–1127. doi: 10.1016/j.burns.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 39.Becker K, Roth R, Peters G. Rapid and specific detection of toxigenic Staphylococcus aureus: use of two multiplex PCR enzyme immunoassays for amplification and hybridization of staphylococcal enterotoxin genes, exfoliative toxin genes, and toxic shock syndrome toxin 1 gene. J Clin Microbiol. 1998;36:2548–2553. doi: 10.1128/jcm.36.9.2548-2553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Norouzi J, Goudarzi G, Pakzad P, Razavipour R. The isolation and detection of Staphylococcus aureus enterotoxins AE and TSST-1 genes from different sources by PCR method. Qom Univ Med Sci J. 2012;6:78–85. [Google Scholar]
- 41.Peck KR, Baek JY, Song JH, Ko KS. Comparison of genotypes and enterotoxin genes between Staphylococcus aureus isolates from blood and nasal colonizers in a Korean hospital. J Korean Med Sci. 2009;24:585–591. doi: 10.3346/jkms.2009.24.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Boeck H, Vandendriessche S, Hallin M, Batoko B, Alworonga JP, Mapendo B, et al. Staphylococcus aureus nasal carriage among healthcare workers in Kisangani, the Democratic Republic of the Congo. Eur J Clin Microbiol Infect Dis. 2015;34:1567–1572. doi: 10.1007/s10096-015-2387-9. [DOI] [PubMed] [Google Scholar]
- 43.Jaspers M, Breederveld R, Tuinebreijer W, Diederen B. The evaluation of nasal mupirocin to prevent Staphylococcus aureus burn wound colonization in routine clinical practice. Burns. 2014;40:1570–1574. doi: 10.1016/j.burns.2014.01.024. [DOI] [PubMed] [Google Scholar]
- 44.Asadollahi P, Farahani NN, Mirzaii M, Khoramrooz SS, Van Belkum A, Asadollahi K, et al. Distribution of the most prevalent spa types among clinical isolates of methicillin-resistant and-susceptible Staphylococcus aureus around the world: A review. Front Microbiol. 2018;9:163–178. doi: 10.3389/fmicb.2018.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]