Abstract
Introduction
Hepatocytes, which account for the majority of liver tissue, are derived from the endoderm and become hepatocytes via differentiation of hepatic progenitor cells. Induced hepatocyte-like (iHep) cells and artificial liver tissues are expected to become useful, efficient therapies for severe and refractory liver diseases and to contribute to drug discovery research. The establishment of iHep cell lines are needed to carry out liver transplants and assess liver toxicity in the rising number of dogs affected by liver disease. Recently, direct conversion of non-hepatocyte cells into iHep cells was achieved by transfecting mouse adult fibroblasts with the Forkhead box protein A1 (Foxa1) and hepatocyte nuclear factor 4 homeobox alpha (Hnf4α) genes. Here, we applied this conversion process for the differentiation of canine bone marrow stem cells (cBMSCs) into hepatocyte-like cells.
Methods
Bone marrow specimens were collected from four healthy Beagle dogs and used to culture cBMSCs in Dulbecco's Modified Eagle's Medium (DMEM). The cBMSCs displayed the following characteristic features: plastic adherence; differentiation into adipocytes, osteoblasts and chondrocytes; and a cell surface antigen profile of CD29 (+), CD44 (+), CD90 (+), CD45 (−), CD34 (−) and CD14 (−), or CD11b (−) and CD79a (−), or CD19 (−) and HLA class II(−). The cBMSCs were seeded in a collagen I-coated plate and cultured in DMEM with 10% fetal bovine serum and transfected with retroviruses expressing Foxa1 and Hnf4α the following day. Canine iHep cells were differentiated from cBMSCs in culture on day 10, and were analyzed for morphology, RNA expression, immunocytochemistry, urea production, and low-density lipoprotein (LDL) metabolism.
Results
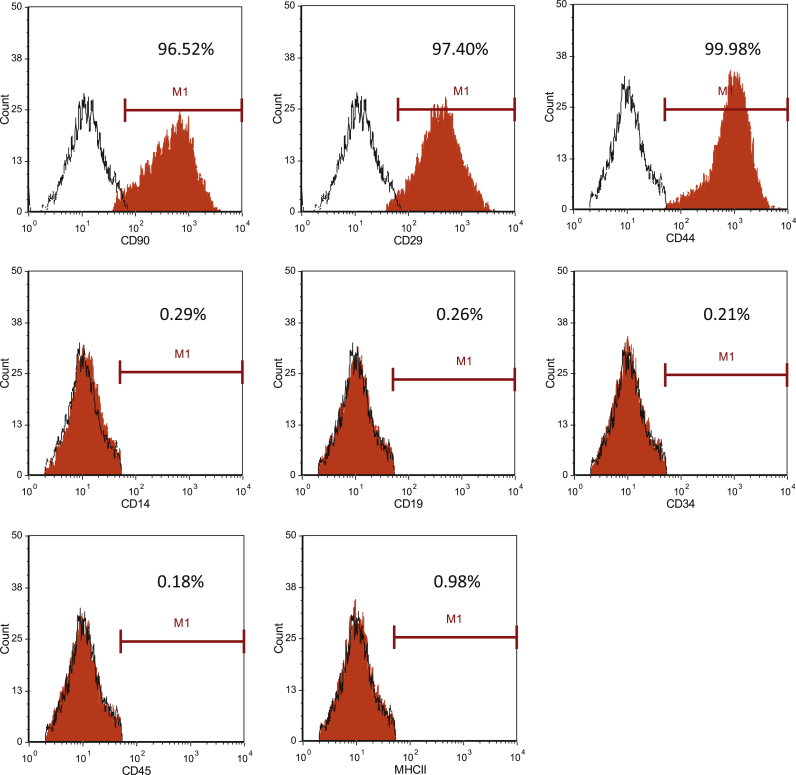
The cBMSCs expressed CD29 (98.06 ± 1.14%), CD44 (99.59 ± 0.27%) and CD90 (92.78 ± 4.89%), but did not express CD14 (0.47 ± 0.29%), CD19 (0.44 ± 0.39%), CD34 (0.33 ± 0.25%), CD45 (0.46 ± 0.34%) or MHC class II (0.54 ± 0.40%). The iHep cells exhibited morphology that included circular to equilateral circular shapes, and the formation of colonies that adhered to each other 10 days after Foxa1 and Hnf4α transfection. Quantitative RT-PCR analysis showed that the expression levels of the genes encoding albumin (ALB) and cadherin (CDH) in iHep cells on day 10 were increased approximately 100- and 10,000-fold, respectively, compared with cBMSCs. Corresponding protein expression of ALB and epithelial-CDH was confirmed by immunocytochemistry. Important hepatic functions, including LDL metabolic ability and urea production, were increased in iHep cells on day 10.
Conclusion
We successfully induced cBMSCs to differentiate into functional iHep cells. To our knowledge, this is the first report of canine liver tissue differentiation using Foxa1 and Hnf4α gene transfection. Canine iHep cells are expected to provide insights for the construction of liver models for drug discovery research and may serve as potential therapeutics for canine liver disease.
Keywords: Bone marrow, Conversion, Differentiation, hepatocyte, Dog, Mesenchymal stem cells
Abbreviations
- cBMSCs
canine bone marrow stem cells
- iHep cell
induced hepatocyte-like cell
- Foxa1
Forkhead box protein A1 gene
- Hnf4α
hepatocyte nuclear factor 4 homeobox alpha gene
- DMEM
Dulbecco's modified Eagle's medium
- ALB
albumin gene
- CDH
cadherin gene
- DILI
drug-induced liver injury
- BSA
bovine serum albumin
- HGF
hepatocyte growth factor
- ESC
embryonic stem cell
- iPSC
induced pluripotent stem cell
- ESC
embryonic stem cells
- MSC
mesenchymal stem cell
- LDL
low-density lipoprotein
- PBS
phosphate-buffered saline
- FBS
fetal bovine serum
- FGF
fibroblast growth factor
- HGM
hepatocyte growth medium
- RT-PCR
reverse transcription-polymerase chain reaction
- Dil-Ac-LDL
1,1′-dioctadecyl-3,3,3′, 3′-tetramethyl indocarbocyanine-labeled LDL
- DAPI
4′,6-diamidino-2-phenylindole
- BMP
bone morphogenetic protein
- EGF
epidermal growth factor
- HGF
hepatocyte growth factor
- bFGF
basic fibroblast growth factor
1. Introduction
The liver has a wide range of functions that maintain biological activity in the body, including metabolism, glycogen storage, xenobiotic detoxification, production of proteins, urea synthesis and biliary secretion. Most of the metabolic and synthetic functions of the liver are performed by hepatocytes, which account for about 60% of the total liver cells and 80% of the organ volume [1]. Various diseases, including hepatitis, hepatic portal system shunts, and hepatic fibrosis, often occur in domestic dogs [[2], [3], [4], [5], [6], [7]], and canine liver disease is common in small animal veterinary practices. A useful therapeutic regimen for these illnesses is required because effective methods for treatment have not yet been established in the veterinary setting. Although the liver has a marked regenerative capacity, it is insufficient to combat severe disease. Because liver transplantation is considered the only effective therapeutic option for dogs with severe hepatic failure, the construction of artificial liver tissue is one of the most important research areas for dogs, as it is for humans.
In drug discovery research, drug-induced liver injury (DILI) is the leading cause of market withdrawal. For this reason, attempts have been made to develop highly safe pharmaceutical products using cultured hepatocytes that can predict DILI at the initial stage of drug development [8]. It is expected that the demand for drug discovery will continue to increase given the increasing numbers and prolonged lifespans of companion animals. By contrast, the use of animals for experimentation is controversial and has been strictly limited by bioethics arguments. New, in vitro tools for toxicity testing are urgently required. To meet this need, the development of induced hepatocyte-like (iHep) cells that possess appropriate liver functions are expected to provide a welcome alternative to normal canine liver tissue.
In our previous study, we reported that canine bone marrow cells (cBMCs) were able to differentiate into hepatocyte-like cells using hepatocyte growth factor (HGF), and that human placental hydrolysate may be an effective inducer of hepatic differentiation [9]. However, the differentiation of iHep cells is problematic because only half of the attempted experiments with cBMCs differentiation have succeeded [9]. Furthermore, researchers have been unable to maintain canine iHep cells in long-term culture.
Many studies have described the differentiation of human iHep cells from embryonic stem cells (ESCs), induced pluripotent stem cells (iPSCs), and mesenchymal stem cells (MSCs) [[10], [11], [12]]. In some studies, cultured human BMSCs were able to differentiate into hepatocyte-like cells in medium containing humoral factors such as basic fibroblast growth factor (bFGF), oncostatin-M, HGF and dexamethasone [13,14]. Reports describing canine iHep cells differentiated from these types of stem cells are limited, and to our knowledge, only one study has reported on canine iHep cells established from MSCs [15]. The technology known as direct reprogramming has enabled the generation of alternative cell sources such as iHep cells from skin-derived fibroblasts using forced expression of specific transcription factors [12,16]. In a report by Suzuki et al., 12 candidate genes related to hepatic cell differentiation during liver development were selected to screen liver fate inducers [17,18]. When cells co-expressing two of those candidates — hepatocyte nuclear factor 4 homeobox alpha (Hnf4α) and any of the Forkhead box protein A genes (Foxa1, Foxa2, and Foxa3) — were cultured with mouse fibroblasts for 21 days, liver-specific genes such as ALB were expressed, liver-specific proteins were detected, and low-density lipoprotein (LDL) uptake capacity was observed, all leading the authors to report that the mouse fibroblasts had been transformed into iHep cells.
The purpose of this study was to clarify whether iHep cells could be derived from canine bone marrow stem cells (cBMSCs) by transfection of the Foxa family and Hnf4α genes. We performed conversion of cBMSCs into hepatocytes using retroviral vectors containing Foxa1 and Hnf4α, which have been characterized as important inducers for differentiating mature hepatocytes.
2. Materials and methods
2.1. Animals
Four healthy Beagle dogs (mean age, 3.16 ± 0.34 years) were used in this study. All animal experiments were conducted based on the laboratory dynamic experiment guidelines of Azabu University (No 160706-1). The animals were kept at a small animal breeding facility in the Azabu University Veterinary Clinical Center. The dogs were not administered any drugs within 1 month of the start of the experiments, and all were clinically healthy without a medical history.
2.2. Preparation of cBMSCs
With the animals under sedation with butorphanol tartrate (0.02 mg/kg IM) and medetomidine hydrochloride (0.04 mg/kg IM), bone marrow specimens were collected from the humerus, femur and ilium using a bone marrow aspiration needle. Sampling was performed with a syringe containing 1 mL heparin, and the collected bone marrow specimens were quickly stored on ice.
The bone marrow specimens were filtered through a 100-μm cell strainer (BD, Franklin Lakes, NJ, USA) to remove any clots. The hemolytic agent, which comprised 83 g/L NH4Cl, 8.4 g/L KHCO3 and 3.7 g/L EDTA · 2Na, was dissolved in 10 × distilled sterilized water. After adding the hemolytic agent to the filtered specimen, the mixture was allowed to stand at room temperature for 5 min and then separated by centrifugation at 300×g for 5 min. The supernatant was removed, leaving the cell sediment. Phosphate-buffered saline (PBS; Gibco, MA, USA) was added and the centrifugation (300×g, 5 min each) was repeated twice. The cell sediment was finally suspended in 1 mL PBS and the number of viable cells was measured by trypan blue staining.
The bone marrow cells were seeded at 1.0 × 107 cells in a 25-cm2 flask and cultured in Dulbecco's modified Eagle's medium (DMEM; Sigma-Aldrich, MO, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, MA, USA), and 1% penicillin–streptomycin solution (Fujifilm, Wako, Osaka, Japan). These primary cultures were basal medium supplemented with 2 mM l-glutamic acid (Fujifilm, Wako, Osaka, Japan) under conditions of 37 °C under 5% CO2.
The medium was initially changed 4 days after seeding the cBMSCs, and thereafter the medium exchange was performed every 3 days. Cells colonized and established on the bottom of the flask were designated as cBMSCs. Passaging was carried out when these cells reached a confluence of 80–90%. After counting the number of viable cells by trypan blue staining, the cells were stored in a cell cryopreservation solution using a cell bunker (Nippon Zenyaku Kogyo, Fukushima, Japan) according to the manufacturer's protocol. This cell cryopreservation solution was used for seeding in the differentiation experiments.
2.3. Confirmation of the differentiation capacity of cBMSCs
Characterization of cBMSCs was performed according to the definition proposed by the International Society for Cellular Therapy. Part of the antigens were modified based on several studies of canine MSCs [19]. First, when maintained under culture conditions using tissue culture flasks, MSCs exhibit plastic adherence. Second, the MSC population must express the cell surface antigens CD29, CD44 and CD90, but also lack expression of CD45, CD34 and CD14, or CD11b and CD79a, or CD19 and HLA class II (Table 1). Third, the cell population must have proven capacity to differentiate into adipocytes, osteoblasts and chondrocytes. In our experiments, induction of differentiation into adipocytes was carried out using the protocol reported by Malagola et al. [20]. The differentiation induction basal medium was 60% DMEM +40% MCDB 201 medium (Sigma-Aldrich, MO, USA) + 2% FBS + 1 nM dexamethasone + 0.1 mM sodium l–ascorbic acid 2–phosphate + 1 × insulin–transferrin–selenium mix (Gibco, MA, USA) + 1 × linoleic acid-albumin from bovine serum albumin liquid (Sigma-Aldrich, MO, USA). The cBMSCs were cultured to confluence in differentiation induction basal medium supplemented with epidermal growth factor (EGF; 10 ng/mL), leukemia inhibitory factor (10 ng/mL), platelet-derived growth factor-BB (10 ng/mL), bFGF (5 ng/mL) and gentamicin (50 μg/mL). The medium was exchanged every 2 days. Those cBMSCs that reached confluence were cultured for 48 h in adipocyte differentiation medium (differentiation induction basal medium + 0.5 μM isobutyl-methylxanthine + 1 μM dexamethasone + 5 μg/mL insulin +50 μM indomethacin +1 nM triiodothyronine + 3.3 nM bone morphogenetic protein [BMP] 7). To promote differentiation into adipocytes, these cells were cultured for 7 days with differentiation induction basal medium +5 μg/mL insulin +1 nM triiodothyronine +1 μM rosiglitazone.
Table 1.
List of antibodies used for flow cytometry.
| Antibody | Clone | Target | Host | Conjugate | Clonality | Company |
|---|---|---|---|---|---|---|
| Anti-CD29 | TS2/16 | human | Mouse | PE | Monoclonal | BioLegend |
| Anti-CD44 | IM7 | mouse/human | rat | FITC | Monoclonal | ThermoFishcer |
| Anti-CD90 | YKIX337.217 | canine | rat | PE | Monoclonal | ThermoFishcer |
| Anti-CD14 | M5E2 | canine | mouse | PE | Monoclonal | Novus Biologicals |
| Anti-CD19 | MB19-1 | mouse | mouse | FITC | Monoclonal | ThermoFishcer |
| Anti-CD34 | 1H6 | canine | mouse | AlexaFluor 488 | Monoclonal | Novus Biologicals |
| Anti-CD45 | YKIX716.13 | canine | rat | FITC | Monoclonal | ThermoFishcer |
| Anti-MHC Class Ⅱ | YKIX334.2 | canine | rat | FITC | Monoclonal | ThermoFishcer |
CD: cluster of differentiation, MHC: major histocompatibility complex, PE: phycoerythrin, FITC: fluorescein isothiocyanate.
Morphological changes were observed with a phase contrast microscope. The medium was exchanged every 2 days. Differentiation into adipocytes was confirmed by Oil red O staining. Differentiation induction and staining of osteoblasts and chondrocytes were performed according to the protocol of Miltenyi Biotech [19]. For differentiation into osteoblast, BMSCs were seeded on a 60 mm dish (Corning, New York, USA), and culture at 37 °C and 5% CO2 for 10 days using StemMACS OsteoDiff Media. The medium was changed twice a week. After induction of differentiation, alkaline phosphatase activity was detected with SIGMA FAST BCIP/NBT substrate (Sigma-Aldrich, MO, USA).
To induce differentiation into chondrocytes, 1 mL of a 2 × 104/mL cBMSCs suspension was added to a sterilized 1.5-mL tube and subjected to centrifugation at 300×g for 5 min. The supernatant was aspirated and the cell pellet was resuspended and cultured in StemMACS ChondroDiff Media for 30 days at 37 °C and 5% CO2. The medium was changed twice a week. After induction of differentiation, these cells were fixed with 10% formalin and stained with Alcian Blue 8 GX (Sigma-Aldrich, MO, USA). After staining, cells were immediately observed with a stereoscopic microscope.
2.4. Cell surface antigen analysis of cBMSCs by flow cytometry
CD29, CD44, and CD90 were considered MSC-positive markers. CD14, CD19, CD45, CD34 and MHC II were analyzed as cell surface antigens of monocytes, B cells, leukocytes, hematopoietic stem cells and MHC Class II-positive cells, which are bone marrow-derived mononuclear cells other than cBMSCs [19].
2.5. Preparation of the retroviral vector
The Foxa1 and Hnf4α genes were cloned into the retroviral vectors pGCDNsam–Foxa1 and pGCDNsam–Hnf4a (Addgene, Watertown, MA, USA), respectively, then transfected into the Platinum-A retroviral packaging cell line (Cell Biolabs, Inc, San Diego, CA, USA) to prepare retrovirus solutions used for transfection of cBMSCs.
2.6. Induction of differentiation from cBMSCs to hepatocyte-like cells
P0 to P2 cBMSCs were seeded in collagen I-coated 12-well plates (Corning, New York, USA) at 5 × 10 4 cells/well and cultured in DMEM with 10% FBS, 1% penicillin–streptomycin at 37 °C and 5% CO2. On the following day, a virus mixture prepared by mixing the Foxa1 and Hnf4α virus solutions with 5 μg/mL protamine sulfate (Nacalai Tesque, Kyoto, Japan) was added to the culture medium. After addition of virus solution, each plate was spun at 700×g for 10 min at room temperature, then incubated at 37 °C and 5%CO2. After 8 h, the culture supernatant was removed by aspiration and the medium was changed. Culture and genes transfection were performed 3 to 4 times in each canine BMSCs.
Viral transfection s occurred for 3 consecutive days. The amount of virus solution used on the second and third days was 1.5 times the amount used on day 1. When replacing the medium on the final day of viral transfection, it was replaced with hepatocyte growth medium HGM: DMEM/Nutrient Mixture F-12 [Gibco, MA, USA] + 10% FBS, 1% penicillin–streptomycin, 1 μg/mL insulin, 0.1 μM dexamethasone (Fujifilm, Wako, Osaka, Japan), 10 mM nicotinamide (Fujifilm, Wako, Osaka, Japan), and 50 μM β-mercaptoethanol (Fujifilm, Wako, Osaka, Japan). The day on which the third viral transfection ended was defined as Day 0. When virus-infected cells became confluent, they were detached using Accutase (Innovative Cell Technologies, Inc, San Diego, CA, USA), and the cells from one well of a 12-well plate were subcultured to one collagen I-coated, 25-cm2 flask (AGC TECHNO GLASS, Inc, Shizuoka, Japan). After confirmation of the presence of epithelial cell-like cells, 20 ng/mL human recombinant EGF (Sigma-Aldrich, MO, USA) was added to the medium. The medium was exchanged once every 3 days. As the proportion of epithelial cell-like cells increased, 20 ng/mL feline recombinant HGF (Nippon Zenyaku Kogyo) was added together with EGF.
2.7. Reverse transcription (RT) polymerase chain reaction (PCR) and quantitative RT-PCR
Total RNA was extracted using an RNeasy Micro kit (Qiagen, Tokyo, Japan). RT was performed using random primers and a high capacity RNA to cDNA kit (Thermo Fisher Scientific, Waltham, MA, USA). PCR was carried out using Takara ExTaq Hot Start polymerase (TaKaRa Bio, Shiga, Japan) and the following cycling parameters: 94 °C for 3 min; 35 cycles of 95 °C for 15 s, annealing at the primer-specific temperature for 30 s, and 72 °C for 1 min; and a final extension at 72 °C for 5 min. PCR products were resolved on a 2% agarose gel stained with ethidium bromide and observed using an ultraviolet transilluminator (ATTO, Tokyo, Japan). Quantitative RT-PCR was performed in triplicate using a Thermal Cycler Dice® Real Time System II (TaKaRa Bio, Shiga, Japan) with a PowerUp SYBR Green Master Mix (Thermo Fisher Scientific) according to the manufacturer's instructions. The primer sequences are listed in Table 2.
Table 2.
Primer list used for RT-PCR or qPCR.
| Gene | F/R | Sequence (5'→3′) | Product size (bp) |
|---|---|---|---|
| Foxa1 | F | gcc gag tca ccc ctt cac | 456 |
| R | gca cgg gtc tgg aat aca ca | ||
| Hnf4α | F | cag agc tag cgg aga tga gc | 477 |
| R | cca ttg ctg agg tga gag gg | ||
| canine Albumin | F | gtt cct ggg cac gtt ttt gta tga | 278 |
| R | ctt ggg gtg ctt tct tgg tgt aac | ||
| canine β-actin | F | gat gag gcc cag agc aag ag | 77 |
| R | tcg tcc cag ttg gtg acg at | ||
| canine CYP3A12 | F | aag gac ttc ctt ttg ttc ttc aag aaa | 86 |
| R | cct aca tga gtg aaa cca cat aat caa | ||
| canine CYP2E1 | F | aga aat cga cag ggt gat cg | 148 |
| R | cat cgt gtc ctg gtt tgc ta | ||
| canine CDH1 | F | ggt gct cac att tcc cag tt | 100 |
| R | aaa tgg gcc ttt ctc gtt t | ||
| AFP | F | tat tgg aca att atg cat cag gca | 345 |
| R | tct tcc tca aag cag gct tcc t | ||
| TAT | F | ttt gct atg gag ctt tgg ctg c | 276 |
| R | aat ggt agt gca gct cgc aga a | ||
| TTR | F | tgg aat tag aca cca agt cct ac | 324 |
| R | ggg aag tgc ttt aat agg aat gtt | ||
| α1-AT | F | aaa tgc agc acc tgg aga gca a | 361 |
| R | gtg tct ctg tcg acg atg atg a |
F: forward primer, R: reverse primer.
2.8. Immunocytochemistry
The medium was aspirated and washed three times with PBS to thoroughly remove the medium from the cells., The cells were fixed with 4% paraformaldehyde phosphate buffer for 10 min, then washed again with PBS three times. After immobilization, membrane permeation treatment was carried out for 5 min using PBS containing 0.1% Triton X-100, followed by three washes with PBS. Blocking was carried out by adding PBS containing 5% skim milk and allowing the cells to stand at room temperature for 60 min or more followed by incubation overnight at 4 °C with goat polyclonal anti-dog albumin antibody (A40-113A; Bethyl Laboratories Inc., Montgomery, TX, USA) or mouse monoclonal anti E-cadherin antibody (sc-8426; Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA).
The corresponding secondary antibodies were incubated with donkey anti goat IgG (H + L) cross-adsorbed secondary antibody, Alexa Fluor 488 (A-11055; Thermo Fisher Scientific) or goat anti-mouse IgG (H + L) cross-adsorbed secondary antibody, Alexa Fluor 488 (A-11001; Thermo Fisher Scientific) for 30 min in the dark. The slides were observed with a confocal microscope after encapsulation with VECTA SHIELD containing 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Inc, Burlingame, CA, USA).
2.9. Analysis of uptake and metabolic ability of LDL
After differentiation, the cBMSCs culture medium was replaced with HGF supplemented with 10 μg/mL 1,1′-dioctadecyl-3,3,3′, 3′-tetramethyl indocarbocyanine-labeled LDL (Dil-Ac-LDL; Biochemical Technologies Inc, Stoughton, MA, USA) [11] and cultured at 37 °C for 4 h. The cells were then washed with PBS, fixed with 4% paraformaldehyde, encapsulated with VECTA SHIELD with DAPI, and the presence of the fluorescent dye Dil, generated by metabolizing Dil-Ac-LDL, was observed with a confocal microscope.
2.10. Analysis of urea production
The iHep cells were seeded in collagen I-coated 24-well plates (Corning, New York, USA) at 3.5 × 104 cells/well and cultured in HGM for 24 h. The iHep cells were then cultured in HGM (DMEM/Nutrient Mixture F-12 without phenol red [Gibco, MA, USA]), supplemented with 1 mM NH4HCO3 (Nacalai Tesque) and 100 nM human glycogen, and the medium was collected at 24, 48, 72 and 96 h for the measurement of urea using the QuantiChrom™ Urea Assay Kit (BioAssay Systems, Hayward, CA, USA) according to the manufacturer's instructions [12].
3. Results
3.1. Cell surface antigen analysis of cBMSCs by flow cytometry
The cell surface antigens of cBMSCs were analyzed to determine the characteristics of these cultured cells. We found that CD29, CD44 and CD90 were expressed in 98.06 ± 1.14%, 99.59 ± 0.27% and 92.78 ± 4.89% of cBMSCs, respectively (Fig. 1). The expression levels of MSC-negative markers were low for all antigens, including CD14 (0.47 ± 0.29%), CD19 (0.44 ± 0.39%), CD34 (0.33 ± 0.25%), CD45 (0.46 ± 0.34%) and MHC class II (0.54 ± 0.40%). Fig. 1 shows the data for the cells derived from one of the four dogs.
Fig. 1.
Expression of cell surface molecules in canine cBMSCs was assessed by flow cytometry. This histogram shows the expression of cell surface molecules in canine cBMSCs. These data show an analysis of one dog. The red-filled portions represent cells labeled with the indicated cell surface molecules, and the black lined portions represent negative isotype controls.
3.2. Multipotency of cBMSCs
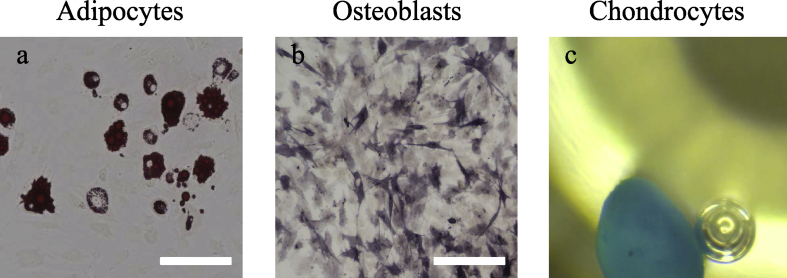
The multipotency of cBMSCs was estimated by differentiation to osteoblasts, adipose cells and chondrocytes. More than 80% of cells induced to osteoblasts were positive for alkaline phosphatase staining. Cells that differentiated into chondrocytes showed morphological changes of multicellular aggregation similar to spheroids, and this cell population was stained dark blue by Alcian blue staining. Differential adipocytes were stained by Oil red O, and red drops were observed in the cytoplasm (Fig. 2).
Fig. 2.
Tri-lineage differentiation ability of cBMSCs. a) Staining with Oil red O indicates differentiation into adipocytes, b) Alkaline phosphatase staining indicates differentiation into osteoblasts, and c) Alcian blue staining indicates differentiation into chondrocytes. Observation after differentiation into chondrocytes was performed with a stereomicroscope. Scale bars, 100 μm.
3.3. Changes in cell morphology after induction of differentiation
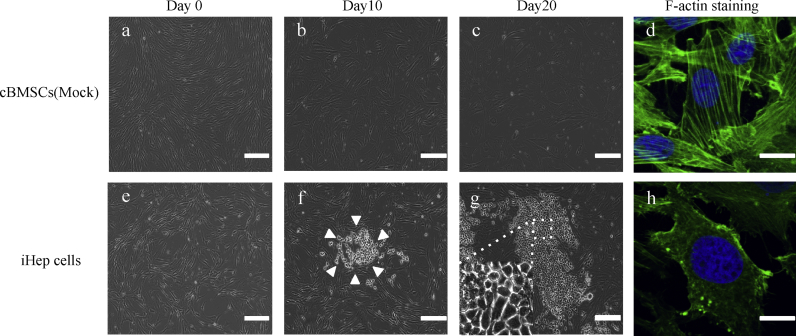
Because the cBMSCs were transfected with Foxa1 and Hnf4α, only spindle-shaped cells were confirmed in all culture on day 0. At day 10 after transfection, the shapes of the cells changed to round or irregular, similar to epithelial cells, and cell aggregation was detected, indicating that colony formation had begun to appear in the cultures. At day 20, the number of cells increased dramatically. These cells adhered to each other and cobblestone colonies were observed (Fig. 3). The tight, thick and clear cytoskeleton within the cytoplasmic membrane, formed with fibers, was observed under phalloidin staining. The morphology of these canine iHep cells were similar to normal canine hepatocytes.
Fig. 3.
Morphologies of iHep cells. Morphologies of (a, b, c, d) mock-infected cBMSCs and (e, f, g, h) iHep cells. The morphology of canine iHep cells showed exhibited circular to equilateral circular shapes, and the cells formed colonies (f, g). The arrowheads (f) indicate a cluster formed by cBMSCs-derived hepatocyte-like cells. The Mock group consistently exhibited a spindle-shape morphology throughout culture. (d, h) F-actin staining revealed a cytoskeleton structure, and the picture was observed with a confocal microscope. In iHep cells, strong fluorescence was observed within the cell membrane. In the Mock group, a filamentous cytoskeleton (F-actin) was clearly observed in the cytoplasm. Day, days after transfection of cells. Scale bars, (a, b, c, e, f, g) 200 μm, (d, h) 7.5 μm.
3.4. iHep cells express hepatocyte related genes
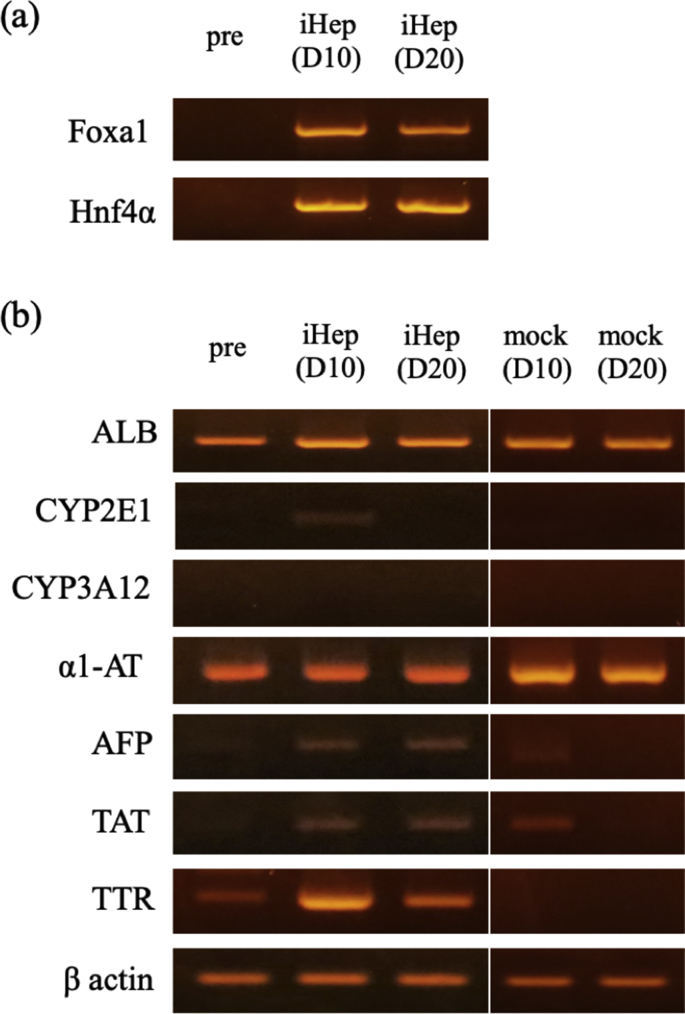
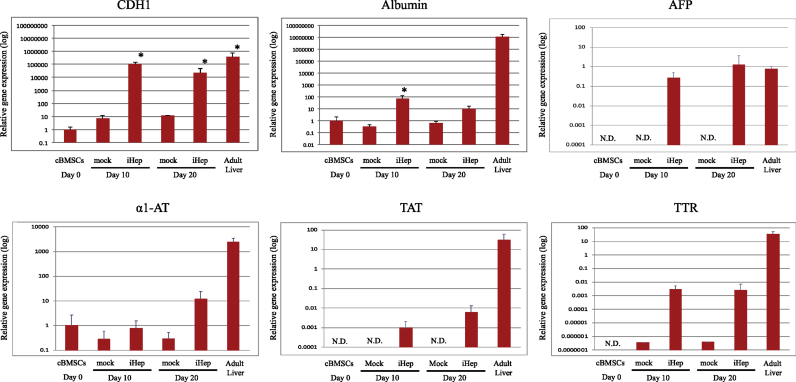
The expression of FoxA1 and Hinf4α RNA from the two retroviral vectors until day 20 following transfection was confirmed by qualitative RT-PCR using specific primers for these genes (Fig. 4a). The iHep cells expressed hepatocyte-related genes, including transthyretin (TTR), ALB, alpha-fetoprotein (AFP), tyrosine aminotransferase (TAT), and alpha 1-antiproteinase (α1-AT) at higher levels than mock-infected cells. The expression of the cytochrome P450 gene (CYP) was detected in iHep cells at day 10; however, at day 20, its expression was reduced (Fig. 4b).
Fig. 4.
Gene expression analysis of iHep cells by qualitative RT-PCR. (a) Qualitative PCR of transfected genes, Foxa1 and Hnf4α, were expressed in iHep cells in the culture on day 10 and 20, respectively. (b) Qualitative PCR analysis revealed that transthyretin (TTR), albumin (ALB), alpha-fetoprotein (AFP), tyrosine aminotransferase (TAT), alpha 1-antiproteinase (alpha 1-AT), and the cytochrome P450 group (CYP3A12 and 2E1) genes were expressed in iHep cells on day 10 and 20. D, days after transfection of cells.
Quantitative RT-PCR analysis showed that the expression levels of ALB and cadherin 1 (CDH1) increased by approximately 100-fold and 10,000-fold, respectively, in iHep cells at day 10 compared with cells before differentiation. However, at day 20, the expression levels of CDH and ALB in the iHep cells were reduced by a factor of 10 compared with day 10. AFP, TAT and TTR gene expression tended to increase in iHep cells compared to BMSCs by result of quantitative RT-PCR. However, a1-AT was also detected in BMSCs, and there was no difference in expression from iHep cells. Expression of CYP 2E1 and 3A12 could not be detected between BMSCs and iHep cells (Fig. 5).
Fig. 5.
Gene expression analysis of iHep cells by comparative quantitative RT-PCR. Gene expression analyses by quantitative PCR in iHep cells. The expression levels of iHep on each day were shown as a ratio of compared cBMSCs (Day 0), respectively. P values were calculated based on a t-test, comparing gene expression levels of iHep cells and cBMSCs (mock) with those of cBMSCs (Day 0), and were considered significantly different at ∗p < 0.05. Day, days after transfection of cells. N.D., not detected.
3.5. Protein expression in iHep cells
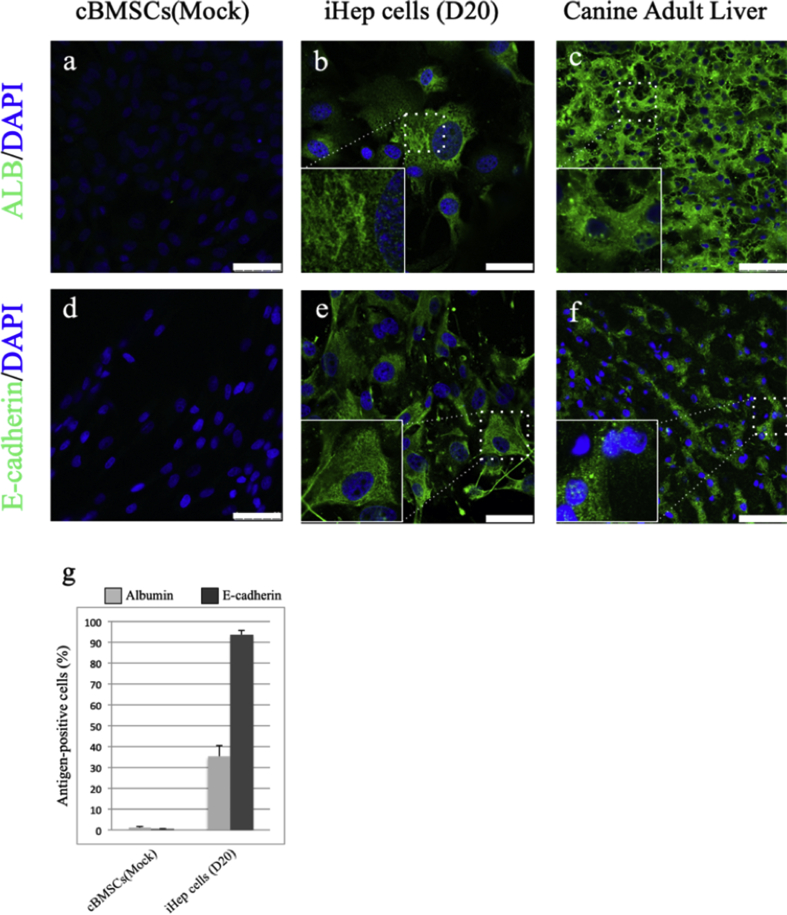
Immunocytochemistry showed that the albumin and E-cadherin proteins were not detected at day 0 after gene transfection, but they were found to be expressed in iHep cells at day 20. The location of albumin protein appeared to correspond to the position of the Golgi apparatus and was identified as tiny follicles inside and outside the iHep cells (Fig. 6a–c). E-cadherin protein was localized on the cell membrane in liver tissue, but was also detected in the cytoplasm as well as the cell membrane in iHep cells (Fig. 6d–f). Immunocytochemistry of mock-infected cBMSCs and iHep cells revealed that the percentage of cells marked with albumin or E-cadherin was 35.16 ± 5.47% and 93.57 ± 2.39%, respectively (Fig. 6g).
Fig. 6.
Immunostaining analysis demonstrated the differentiation of cBMSCs into iHep cells. Mock-infected cBMSCs, iHep cells, and canine adult livers were stained with anti-albumin (a, b, c) and anti-E-cadherin (d, e, f). Nuclei were stained with DAPI. The area enclosed by the dashed line is magnified at the lower left of each panel. D, days after transfection of cells. Scale bars, 50 μm g). The graph shows the percentage of cells marked with albumin or E-cadherin in mock-infected cBMSCs and iHep cells (400–800 cells were counted for each cell).
3.6. Analysis of uptake and metabolism of LDL in iHep cells
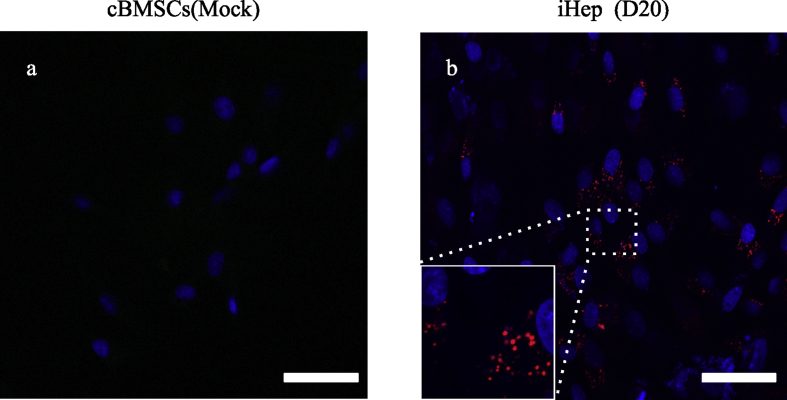
In iHep cells at day 20, the red fluorescent dye Dil was observed in the cytoplasm-like follicles. These results indicated that the iHep cells possess the ability to uptake and metabolize LDL. Non-transfected cBMSCs did not show Dil staining, even on day 20 (Fig. 7).
Fig. 7.
Analysis of LDL uptake and metabolism of iHep cells. LDL uptake assay was performed on mock-infected cBMSCs and iHep cells. Both (a)mock-infected cBMSCs and (b)iHep cells were used at 20 days after transfection. The red fluorescent dye indicates Dil, in which Dil-Ac-LDL is taken into the cell and metabolized. The nuclei was stained with DAPI. D, days after transfection of cells. Scale bars, 50 μm.
3.7. Analysis of urea production of iHep cells
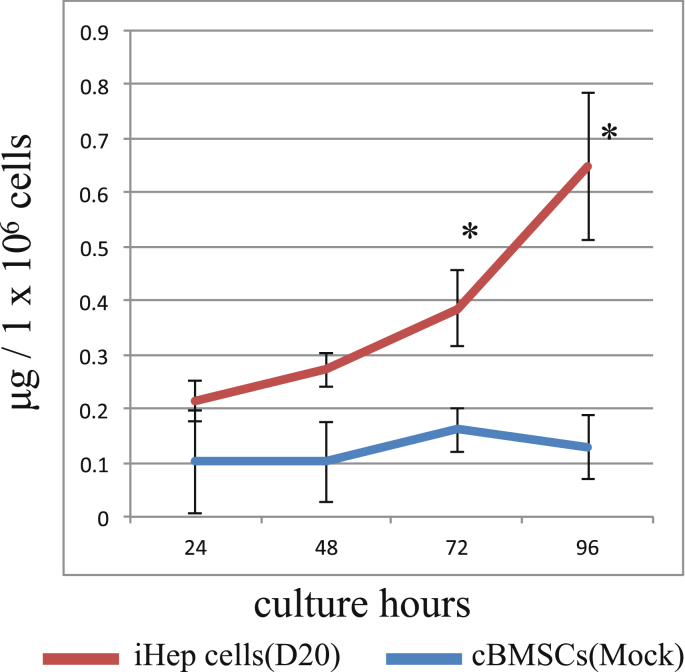
Urea production was assessed in iHep cells at day 20. From 72 to 96 h after the addition of ammonium chloride to the culture medium, the amount of urea in the medium was significantly increased in iHep cells compared with the mock cells (Fig. 8), especially at 48 (0.27 μg/1 × 106 cells), 72 (0.38 μg/1 × 106 cells) and 96 (0.65 μg/1 × 106 cells) hours. This indicated that the iHep cells had the potential for ammonium metabolism. Urea production was not seen in the supernatants of the mock-infected cells, suggesting that the increase in urea concentration was a specific function of the iHep cells.
Fig. 8.
Analysis of urea production ability. Urea concentration of the supernatant in the culture was measured. The red line represents iHep and the blue line represents cBMSCs. All data shown are calculated as the amount of urea in the culture medium per 1 × 106 cells. Data represent mean ± standard deviation (n = 3).
4. Discussion
In the present study, cBMSCs were successfully induced to differentiate into iHep cells by transfection of Foxa1 and Hnf4α. In the Foxa1 and Hnf4α non-transfected groups, a slight increase in CDH1 and TTR m-RNA genes was observed compared with cBMSCs, but morphological changes to iHep and significant increases in ALB, AFP, α-AT, and TAT genes were not seen. These results demonstrated that the enhanced expression of Foxa1 and Hnf4α is important for canine hepatocyte conversion from cBMSCs. Hepatocytes, which account for the majority of liver cells, are derived from the endoderm, and endodermal cells are thought to be induced to differentiate into hepatic progenitor cells by cytokines, such as FGF and BMP, secreted from surrounding MSCs [1]. At this stage, expression of Foxa, Hnf4α, GATA binding protein 4 (GATA-4), and GATA-6 are enhanced in mouse endodermal cells. Furthermore, Foxa and Hnf4α have been reported to induce the transcription of genes that encode proteins specific to hepatocytes, such as albumin [21,22]. They reported that Foxa-deficient mice did not show expression of AFP and hepatocyte markers albumin or transthyretin [22]. In another study, embryonic liver cells of Hnf4-deficient mice could not express the liver-specific genes and no intercellular adhesion was observed [18]. A significant reduction in AFP and albumin was observed in mice lacking only Hnf4 among 12 genes necessary for hepatocyte development [18]. Based on these reports, it appears that the Foxa family and Hnf4 are closely involved in the development of the liver. Indeed, normally functioning hepatocytes do not differentiate regardless of the expression of either the Foxa family or Hnf4. In a report by Suzuki et al., iHep cells were generated from mouse fibroblasts by introducing either Foxa1, Foxa2, or Foxa3 in addition to Hnf4. In that report, when the Hnf4α and Foxa1 genes were introduced into iHep cells, E-cadherin protein-positive cell clusters tended to be more common than when Foxa2 or Foxa3 were used. For this reason, we used Hnf4α and Foxa1 genes for direct reprogramming in this study. However, the expression levels of other hepatocyte-related genes, including ALB and AFP, tended to be higher in iHep cells established with a combination of Hnf4α and Foxa3. It is possible that these genes or others may increase hepatic function of canine iHep cells. Therefore, we plan to perform direct reprogramming using a combination of Hnf4α and Foxa2, or Foxa3, in the future to enhance the hepatic function of canine iHep cells. In our previous studies, the probability of transformation from BMSCs to iHep cells using humoral factors varied from experiment to experiment; transformation was only successful in about half of all experiments. In contrast, overexpression of Foxa1 and Hnf4α in cBMSCs allowed stable differentiation of iHep cells expressing albumin protein in 35.16 ± 5.47% of the total cell population in this study. Therefore, we propose that the Foxa family and Hnf4α are two important factors for the conversion from cBMSCs to iHep cells.
In the present study, iHep cells showed a hepatocyte-like shape and expressed the hepatic-related genes, proteins and functions of liver tissues. The adult liver carries precursor cells that can be activated in response to specific injuries. Hepatic progenitor cells often give rise to cells called “oval cells” [14]. These cells are thought to be present in the liver as hepatic stem cells, which can differentiate into both hepatocytes and biliary epithelial cells. In previous reports, in addition to oval cells [23], other cells, including hepatic progenitor cells [24] and small hepatocytes [25], have been reported as being involved in liver regeneration. The morphology of our canine iHep cells exhibited circular to equilateral circular shapes, and formed colonies that adhered to each other 20 days after of Foxa1 and Hnf4α transfection. This change in cell morphology was similar to that in the murine iHep cells described by Sekiya et al. [12]. In particular, the colonies contained small hepatic progenitor-like cells. Furthermore, the iHep cells showed a wide cytoplasm, suggesting that the cell character was similar to mature hepatocytes. RNA and protein expression analyses indicated that canine iHep cells produced hepatocyte-specific factors. In this study, iHep cells expressed hepatocyte-related genes, including TTR, ALB, AFP, TAT, α1-AT, and CYP, and at levels higher than in the cBMSCs before differentiation. Quantitative RT-PCR analysis showed that the expression levels of ALB and CDH were increased by approximately 100- and 10,000-fold, respectively, in iHep cells at day 10 compared with pre-differentiated cells. However, the CYP expression level was weak in comparative quantitative PCR, and there was no significant difference between cBMSCs and iHep cells. We suggest the following measures to increase CYP expression: i) a different combination of transgenes, as the current combination might not have been the most efficient for induction of iHep cells; ii) an additive factor, such as a low-molecular weight compound [24]; and iii) three-dimensional culture may be required to enrich CYP expression [26]. These measures may contribute to the establishment of high-quality canine iHep cells.
In this study, the albumin expression levels of iHep cells were comparable to those of mouse fibroblasts and hepatocyte-like cells derived from human iPSCs [26,27]. In addition, protein expression from the ALB and CDH genes in the iHep cells was confirmed by immunocytochemistry, suggesting that iHep cells might be the product of high levels of these proteins. We did not evaluate the differentiation stage of the hepatocyte-like cells in this study because there are no clear, absolute markers of hepatic stem cells, hepatic progenitor cells and mature hepatocytes in dogs, and high-quality antibodies have not been established. For these reasons, the investigation of surface antigens in canine hepatic stem cells, progenitors, and mature cells is not easy, thus we were unable to provide a complete characterization of the canine iHep cells in this study. Further study will be required when these problems are resolved.
In the present study, important hepatic functions, including LDL metabolic ability and urea production, were confirmed. Dil-Ac-LDL is LDL labeled with 1,1′-dioctadecyl-3,3,3′ and 3′-tetramethyl indocarbocyanine, and is taken into cells via the LDL receptor on the liver cell membrane. When Dil-Ac-LDL is metabolized by an enzyme in the lysosome, Dil is released and accumulated in the cytoplasm; i.e. fluorescence of iHep cells cultured with Dil-Ac-LDL indicates the ability to take in and metabolize LDL. Urea production was analyzed as another measure of liver function. Ammonia is converted to urea in the liver via the urea cycle. Ammonia is first catalyzed by carbamoyl phosphate synthase I in mitochondria to yield carbamoyl phosphate. The carbamoyl phosphate is condensed with ornithine and converted to citrulline, and then transported to the cytoplasm to generate urea via the urea cycle. The production of urea by iHep cells suggested that the urea cycle is functioning in these cells. We chose not to assess CYP activity in this study because the expression levels of the CYP gene were not high in the cultures. As with other animal species, there are many types of canine CYPs, and a capacity to analyze drug metabolism is required to study them. To analyze liver function, it will be necessary to analyze the expression of drug metabolizing enzymes other than CYP2E1 and CYP3A12, and to evaluate the metabolic potential of drugs such as acetaminophen and trovafloxacin. If iHep cells are to have higher functional activity as hepatocytes, three-dimensional culture will be necessary. Efforts to investigate such culture methods are underway.
To our knowledge, this is the first report to differentiate hepatocytes using Foxa1 and Hnf4α gene transfer. The first report of direct reprogramming described mouse fibroblasts differentiated into skeletal muscle cells by transfection with a single transcription factor [28]. Because this method does not involve undifferentiated cells, there are several advantages: 1) the time needed to obtain the target cells is short; 2 the risk of carcinogenesis is low; and 3) potential ethical problems are alleviated. In human fibroblasts, functional hepatocyte-like cells have been successfully produced by transfection with FOXA3, HNF1A, and HNF4A [16]. It has been reported that MSCs are capable of differentiating not only into mesenchymal cells such as bone, cartilage and adipocytes, but also into hepatocytes and neurons [29]. In the study by Palazzolo et al., the forced expression of transcription factors (GATA4, HAND2, TBX5, and MEF2C) was sufficient to differentiate mouse and human somatic cells into induced cardiac-like myocytes. Their report showed that the direct reprogramming of canine fibroblasts into induced cardiac-like myocytes was successful without early cardiac gene activation [30]. In our experiments, the transgenes continued to be expressed for up to 20 days after transfection, indicating that they had not been silenced. Silencing of transgenes will be required for the establishment of canine iHep cells by direct reprograming.
In the field of human drug discovery, hepatocytes induced to differentiate from human artificial pluripotent stem cells (iPSCs) have attracted attention as a new source of hepatocytes for toxicity evaluation systems. Human iPSCs differentiate into endoderm, hepatic progenitor cells and hepatocytes. In previous reports, attempts have been made to induce the differentiation of iPSCs into hepatocytes using humoral factors such as bFGF, nicotinamide, oncostatin-M, HGF or dexamethasone, at each stage of differentiation to reproduce the liver regeneration environment of the living body [11,31,32]. However, although the cells described in these reports exhibit liver functions, such as CYP activity, and those that express factors involved in albumin synthesis, urea synthesis and drug metabolism have been obtained, they all show lower expression than in primary cultured hepatocytes. Stimulation of iPSCs with humoral factors, such as growth factors, is considered to be insufficient for differentiation into hepatocytes. Furthermore, it is necessary to improve the differentiation efficiency and further maturation of hepatocytes after differentiation. In a recent report, Mizuguchi et al. demonstrated that human iPSCs could be efficiently differentiated into hepatocytes by transfection of Foxa2 and Hnf1α, indicating that they were key factors for hepatocyte differentiation [33].
This study revealed that Foxa1 and Hnf4a are useful for the induction of canine hepatocytes, and we hope that this finding will contribute to liver transplantation or the establishment of a drug metabolism system using iHep cells in the future. As part of our efforts to apply the results of this study to canine iPSCs, we have started analyzing whether iHep cells can be induced from canine iPSCs using Foxa1 and Hnf4α (data not shown). We anticipate that the establishment of functional canine iHep cells will provide a comprehensive tool to evaluate safety, pharmacokinetics, and pharmacology in drug discovery research. Furthermore, it is expected that these cells will serve as a quick and efficient alternative system in which to analyze various endpoints that are currently performed in live animal experiments.
Canine BMSCs are a type of MSC that has pluripotency and multi-differentiation ability. In our previous study, fresh cBMCs differentiated into hepatocytes in vitro by the addition of HGF [9]. However, BMSCs are more useful than fresh BMSCs for clinical applications because their cell numbers can be increased by culturing and they can be stored in a frozen state. By performing cell surface antigen analysis using flow cytometry and confirmation of tripotency, we proved that the bone marrow-derived cells used in this study were canine MSCs. BMSCs possess various characteristics, such as pluripotency, anti-inflammatory action and secretion of cytokines, and are expected to be applied in the field of regenerative medicine. Nina et al. reported a detailed analysis of canine adipose-derived MSCs (ADSCs). When their cell surface antigens were analyzed by flow cytometry, CD29, CD44 and CD90 were expressed in over 95% of the cells, whereas CD14 and CD45 were expressed in up to only 0.4% [19]. Our experiments produced the same results as those presented in previous report [19]. Furthermore, in this study we confirmed the tripotency of the cBMSCs by inducing their differentiation into three types of cells (chondrocytes, osteoblasts and adipocytes), which was used to identify them as canine MSCs. In fact, the surface antigens of canine MSCs have not been clearly established, nor have the surface antigens of hepatic stem cells. In a previous study, we reported that CD29, CD44 and CD90 are highly expressed in canine hepatoma cells [34]. The MSCs used in this study also highly expressed these molecules, which indicated that iHep cells were derived from a cell population that expressed the same molecules as hepatic stem cells.
This study has some potential limitations. We were unable to be investigate CYP activity in our cBMSC-derived iHep cells, nor did we evaluate the ability of these cells to differentiate into cholangiocytes. Furthermore, the homology between normal canine hepatocytes and iHep cells was not assessed because it would have required exhaustive microsatellite analysis of mRNA expression. In this study, the properties of canine iHep cells have been analyzed partially, but the function as mature hepatocytes has not been fully evaluated. Then we will need to evaluate the liver function of iHep cells in vivo by future transplantation experiments in order to examine as mature hepatic functionality.
In a recent study, liver function was induced by three-dimensional culture [26] but not by the monolayer culture conditions used to induce canine iHep cells in this study. Therefore, the function of canine liver tissue should be assessed in three-dimensional structures. In addition, because the expression levels of CDH, albumin and CYP were lower in iHep cells at day 20 compared with day 10, we suspect that planar culture is insufficient for maintenance or maturation of canine iHep cells. In the future, it will be necessary to develop the further maturation of iHep cells by creating three-dimensional structures and adjusting growth factors.
Taken together, our findings may support the building of artificial canine liver tissue models from MSCs or iPSCs on other tissue. In particular, the findings regarding induction of canine iHep cells using Foxa1 and Hnf4α may contribute to the future construction of organoids in vitro. Ultimately, the establishment of canine iHep cells may lead to the development of effective tools to investigate alternative therapies to liver transplantation, drug metabolism toxicity, and treatments for liver disease in dogs.
Declaration of Competing Interest
This work was supported by a grant from the Ministry of Education, Culture, Sports, Science and Technology's Scientific Research Fund (Grant No. 16K15050) and a private branding business.
Acknowledgements
The authors are grateful to Dr. Satoshi Suzuki (Kyushu University) and Dr. Akihide Kamiya (Tokai University) for their advice on conducting this research appropriately. We appreciate Manami Morita for her cooperation with experiments including cell culture and virus preparation. We thank Michelle Kahmeyer-Gabbe, PhD, from Edanz Group (www.edanz.com/ac) for editing a draft of this manuscript.
Footnotes
Peer review under responsibility of the Japanese Society for Regenerative Medicine.
References
- 1.Miyajima A., Tanaka M., Itoh T. Stem/progenitor cells in liver development, homeostasis, regeneration, and reprogramming. Cell Stem Cell. 2014;14(5):561–574. doi: 10.1016/j.stem.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 2.Watson P.J. Chronic hepatitis in dogs: a review of current understanding of the aetiology, progression, and treatment. Vet J. 2004;167:228–241. doi: 10.1016/S1090-0233(03)00118-7. [DOI] [PubMed] [Google Scholar]
- 3.Bexfield N.H., Buxton R.J., Vicek T.J., Day M.J., Bailey S.M., Haugland S.P. Breed, age and gender distribution of dogs with chronic hepatitis in the United Kingdom. Vet J. 2012;193(1):124–128. doi: 10.1016/j.tvjl.2011.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen L., Stobie D., Mauldin G.N., Baer K.E. Clinicopathologic features of dogs with hepatic microvascular dysplasia with and without portosystemic shunts: 42 cases (1991-1996) J Am Vet Med Assoc. 1999;214(2):218–220. [PubMed] [Google Scholar]
- 5.Geraldine B.H. Effect of breed on anatomy of porto-systemic shunts resulting from congenital diseases in dogs and cats:a review of 242 cases. Aust Vet J. 2004;82(12):746–749. doi: 10.1111/j.1751-0813.2004.tb13233.x. [DOI] [PubMed] [Google Scholar]
- 6.Rutgers H.C., Haywood S., Kelly D.F. Idiopathic hepatic fibrosis in 15 dogs. Vet Rec. 1993;133(5):115–118. doi: 10.1136/vr.133.5.115. [DOI] [PubMed] [Google Scholar]
- 7.Eulenberg V.M., Lidbury J.A. Hepatic fibrosis in dogs. J Vet Intern Med. 2018;32:26–41. doi: 10.1111/jvim.14891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greer M.L., Barber J., Eakins J., Kenna J.G. Cell based approaches for evaluation of drug-induced liver injury. Toxicology. 2010;268(3):125–131. doi: 10.1016/j.tox.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 9.Neo S., Ishikawa T., Ogiwara K., Kansaku N., Nakamura M., Watanabe M. Canine bone marrow cells differentiate into hepatocyte-like cells and placental hydrolysate is a potential inducer. Res Vet Sci. 2009;87(1):1–6. doi: 10.1016/j.rvsc.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 10.Sancho-Bru P., Roelandt P., Narain N., Pauwelyn K., Notelaers T., Shimizu T. Directed differentiation of murine-induced pluripotent stem cells to functional hepatocyte-like cells. J Hepatol. 2011;54(1):98–107. doi: 10.1016/j.jhep.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 11.Si-Tayeb K., Noto K.F., Nagaoka M., Li J., Battle A.M., Duris C. Highly efficient generation of human hepatocyte–like cells from induced pluripotent stem cells. Hepatology. 2010;51(1):297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sekiya S., Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- 13.Lee K.D., Kuo T.K., Whang-Peng J., Chung Y.F., Lin C.T., Chou S.H. In vitro hepatic differentiation of human mesenchymal stem cells. Hepatology. 2004;40(6):1275–1284. doi: 10.1002/hep.20469. [DOI] [PubMed] [Google Scholar]
- 14.Yang Y.J., Li X.L., Xue Y., Zhang C.X., Wang Y., Hu X. Bone marrow cells differentiation into organ cells using stem cell therapy. Eur Rev Med Pharmacol Sci. 2016;20(13):2899–2907. [PubMed] [Google Scholar]
- 15.Choi S.A., Choi H.S., Kim K.J., Lee D.S., Lee J.H., Park Y.J. Isolation of canine mesenchymal stem cells from amniotic fluid and differentiation into hepatocyte-like cells. In Vitro Cell Dev Biol Anim. 2013;49(1):42–51. doi: 10.1007/s11626-012-9569-x. [DOI] [PubMed] [Google Scholar]
- 16.Huang P., Zhang L., Gao Y., He Z., Yao D., Wu Z. Direct reprogramming of human fibroblaststo functional and expandable hepatocytes. Cell Stem Cell. 2014;14(3):370–384. doi: 10.1016/j.stem.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 17.Cirillo L.A., Lin F.R., Cuesta I., Friedman D., Jarnik M., Zaret K.S. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9(2):279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 18.Zaret K.S., Grompe M. Generation and regeneration of cells of the liver and pancreas. Science. 2008;322(5907):1490–1494. doi: 10.1126/science.1161431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krešic N., Šimic I., Lojkic I., Bedekovic T. Canine adipose derived mesenchymal stem cells transcriptome composition alterations: a step towards standardizing therapeutic. Stem Cell Int. 2017 doi: 10.1155/2017/4176292. Article ID 4176292:12 pages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steinbring J., Graja A., Jank A.M., Schulz T.J. Flow cytometric isolation and differentiation of adipogenic progenitor cells into brown and brite/beige adipocytes. Methods Mol Biol. 2017;1566:25–36. doi: 10.1007/978-1-4939-6820-6_4. [DOI] [PubMed] [Google Scholar]
- 21.Hatzis P., Talianidis I. Regulatory mechanisms controlling human hepatocyte nuclear factor 4α gene expression. Mol Cell Biochem. 2001;21:7320–7330. doi: 10.1128/MCB.21.21.7320-7330.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee C.S., Friedman J.R., Fulmer J.T., Kaestner K.H. The initiation of liver development is dependent on Foxa transcription factors. Nature. 2005;435:944–947. doi: 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- 23.Farber E. Similarities in the sequence of early histological changes induced in the liver of the rat by ethionine, 2-acetylamino-fluorene, and 3′-methyl-4dimethylaminoazobenzene. Cancer Res. 1956;16:142–148. [PubMed] [Google Scholar]
- 24.Katsuda T., Kawamata M., Hagiwara K., Takahashi R.U., Yamamoto Y., Camargo F.D. Conversion of terminally committed hepatocytes to culturable bipotent progenitor cells with regenerative capacity. Cell Stem Cell. 2017;20(1):41–55. doi: 10.1016/j.stem.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 25.Mitaka T., Mikami M., Sattler G.L., Pitot H.C., Mochizuki Y. Small cell colonies appear in the primary culture of adult rat hepatocytes in the presence of nicotinamide and epidermal growth factor. Hepatology. 1992;16(2):440–447. doi: 10.1002/hep.1840160224. [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto J., Udono M., Miura S., Sekiya S., Suzuki A. Cell aggregation culture induces functional differentiation of induced hepatocyte-like cells through activation of hippo signaling. Cell Rep. 2018;25(1):183–198. doi: 10.1016/j.celrep.2018.09.010. [DOI] [PubMed] [Google Scholar]
- 27.Inamura M., Kawabata K., Takayama K., Tashiro K., Sakurai F., Katayama K. Efficient generation of hepatoblasts from human ES cells and iPS cells by transient overexpression of homeobox gene HEX. Mol Ther. 2011;19(2):400–407. doi: 10.1038/mt.2010.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis Robert L., Weintraub Harold, Lassar Andrew B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51(6):987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 29.Kulangara K., Adler A.F., Wang H., Chellappan M., Hammett E., Yasuda R. The effect of substrate topography on direct reprogramming of fibroblasts to induced neurons. Biomaterials. 2014;35(20):5327–5336. doi: 10.1016/j.biomaterials.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palazzolo G., Quattrocelli M., Toelen J., Dominici R., Anastasia L., Tettamenti G. Cardiac niche influences the direct reprogramming of canine fibroblasts into cardiomyocyte-like cells. Stem Cell Int. 2016 doi: 10.1155/2016/4969430. Article ID 4969430:13 pages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y.F., Tseng C.Y., Wang H.W., Kuo H.C., Yang V.W., Lee O.K. Rapid generation of mature hepatocyte-like cells from human induced pluripotent stem cells by an efficient three-step protocol. Hepatology. 2012;55(4):1193–1203. doi: 10.1002/hep.24790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao D., Chen S., Duo S., Xiang C., Jia J., Guo M. Promotion of the efficient metabolic maturation of human pluripotent stem cell-derived hepatocytes by correcting specification defects. Cell Res. 2013;23:157–161. doi: 10.1038/cr.2012.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takayama K., Nagamoto Y., Mimura N., Tashiro K., Sakurai F., Tachibana M. Long-term self-renewal of human ES/iPS-derived hepatoblast-like cells on human laminin 111-coated dishes. Stem Cell Rep. 2013;1(4):322–335. doi: 10.1016/j.stemcr.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujimoto A.1, Neo S., Ishizuka C., Kato T., Segawa K., Kawarai S. Identification of cell surface antigen expression in canine hepatocellular carcinoma cell lines. J Vet Med Sci. 2013;75(6):831–835. doi: 10.1292/jvms.12-0549. [DOI] [PubMed] [Google Scholar]