Abstract
Staphylococcus aureus is the most frequent agent causing nosocomial infections in Baghdad hospitals. This study aimed to determine S. aureus methicillin resistance, spa gene typing and phylogenic analysis in Iraqi S. aureus isolates. Two hundred samples including clinical (n = 100) and environmental (n = 100) specimens were collected. S. aureus isolates were identified using multiplex PCR amplification of femA and mecA (for methicillin-resistant S. aureus (MRSA) strains) genes. The spa gene was also amplified. Sequence alignment and identification of spa types was then obtained. Of 74 studied S. aureus isolates, 61 (82.43%) harboured the mecA gene (p < 0.001). A spa gene variation was detected in 41 (67.2%) of 61 (p 0.0011) MRSA and 6 (46.15%) of 13 methicillin-susceptible S. aureus isolates. Amino acid sequence analysis revealed a great change in amino acid pattern among local isolates compared to National Center for Biotechnology Information control. Some of the MRSA isolates had high-level similarity with t10214. No genetic relationship with the infection sources was observed. None of the environmental isolates had spa gene variations. Most S. aureus isolates were MRSA. The spa gene variations was significantly higher among clinical isolates. spa sequencing showed different tandem repeats in local MRSA isolates compared to global spa types. We conclude that there was no outbreak in hospital settings in the city of Baghdad. However, our data suggest that isolates from the hospital environment are highly clonal.
Keywords: Staphylococcus aureus, epidemiology, virulence, antibiotic resistance, spa typing
Introduction
Staphylococcus aureus is a common and versatile human pathogen [1]. Despite continuous progress in the medical and diagnostic fields, it is harboured by 20% to 30% of the population without causing any clinical manifestation. The bacterium has the ability to adapt to numerous conditions, and one successful isolate could become an epidemic or even a pandemic clone with high morbidity and mortality [2]. Methicillin-resistant S. aureus (MRSA) causes both hospital-acquired and community-acquired MRSA [1]. Staphylococcus aureus may harbour numerous virulence factors, including staphylococcal protein A (spa), in addition to its ability to resist a variety of antibiotics [3].
Genotyping methods have the capacity to rapidly and reliably identify the relatedness of clinical isolates. Such methods are crucial for investigating outbreaks as well as to enable epidemiologic studies and surveillance of isolate dissemination [4]. Several molecular techniques have been applied for typing, including pulsed-field gel electrophoresis, PCR restriction fragment length polymorphism, DNA sequencing of spa and coa genes and screening for toxins [5,6].
The aims of this study were to determine the spa gene diversity; and to genotype clinical and environmental S. aureus isolates in Iraq.
Materials and methods
Seventy-four dereplicated S. aureus isolates were obtained from clinical sources and hospital environments in Baghdad (Table 1). For molecular identification, a multiplex PCR was used to amplify the femA housekeeping gene. The mecA mobile genetic element was amplified to detect MRSA isolates. Primers are listed in Table 2.
Table 1.
Source of isolation of studied Staphylococcus aureus isolates
| Source of isolation | No. of isolates | % |
|---|---|---|
| Skin infection | 38 | 51.35 |
| Urinary tract infection | 12 | 16.21 |
| Ear infection | 5 | 6.75 |
| Nasal infection | 3 | 4.05 |
| Blood | 3 | 4.05 |
| Eye infection | 2 | 2.70 |
| Sputum | 1 | 1.35 |
| Seminal fluid | 1 | 1.35 |
| Hospital environment | 9 | 12.16 |
| Total isolates | 74 | 100 |
Table 2.
PCR primers used
| Primer name | Primer sequence (5′–3′) | Product size (bp) | Reference |
|---|---|---|---|
| femA F | CGATCCATATTTACCATATCA | 450 | [8] |
| femA R | ATCACGCTCTTCGTTTAGTT | ||
| mecA F | GTAGAAATGACTGAACGTCCGATAA | 314 | [8] |
| mecA R | CCAATTCCACATTGTTTCGGTCTAA | ||
| spa F | TCAAGCACCAAAAGAGGAAGA | 300 | [9] |
| spa R | ACGACATGTACTCCGTTGCCG |
F, forward; R, reverse.
DNA extraction
The template DNA was prepared by the boiling method [7]. Briefly, a few bacterial colonies taken from overnight bacterial growth culture were suspended in 1 mL of TE buffer and boiled in a water bath for 5 minutes. After centrifugation, the supernatant was separated and used as a DNA template.
Polymerase chain reaction
The PCR reaction mixture was prepared by adding 12.5 μL 2 × GoTaq Green Master Mix (Promega, Madison, WI, USA), 1.5 μL of each of the forward and reverse primers [8] (10 pmol/μL), 5 μL template DNA and nuclease-free H2O to a final volume of 25 μL. PCR conditions are listed in Table 3. The PCR products were visualized on a 1% agarose gel for 1 hour at 50 V, stained with ethidium bromide or diamond and visualized by a transilluminator.
Table 3.
PCR conditions
| Amplified gene | Initial denaturation | No. of cycles | Denaturation | Annealing | Elongation | Final extension |
|---|---|---|---|---|---|---|
| femA, mecA | 95°C/5 min | 35 | 94°C/30 s | 53°C/30 s | 72°C/1 min | 10 min/72°C |
| spa | 95°C/5 min | 35 | 94°C/30 s | 52°C/30 s | 72°C/30 s | 10 min/72°C |
Single-locus sequencing typing of spa gene
All isolates were subjected to spa gene screening using specific primers for a variable region amplifying a PCR product size of 300 bp (Table 2) [9].
Next, all PCR products were sequenced by sending samples to the NICEM Company in the New York, United States. Pairwise sequence alignment was used to identify the amplified DNA fragment of the spa gene that might indicate the structural, functional and/or evolutionary relationships among DNA sequences. The sequences were submitted to the National Center for Biotechnology Information (NCBI) under accession number LC038119-LC38142. DNA alignments were analysed by Geneious 8.0.3 software (Geneious, Auckland, New Zealand), and a phylogenic tree was drawn using the Tamura-Nei genetic distance model with cost matrix identity (1.0–0.0) and UPGMA (unweighted pair group method with arithmetic mean). spa tandem repeats were calculated by spaTyper online software (http://spatyper.fortinbras.us/). The results were then compared to an online website for world spa types (Ridom spa Server, https://www.spaserver.ridom.de/).
Data analysis
Data were analysed by SPSS 20 (IBM, Armonk, NY, USA). ANOVA and the Student t test were used considering the 95% confidence interval; p < 0.05 was considered statistically significant.
Results
All 74 studied isolates contained femA, thus confirming that all were S. aureus, among which 61 (82.43%) of 74 contained mecA (MRSA) and 13 (17.57%) of 74 were devoid of mecA (methicillin-susceptible S. aureus, MSSA).
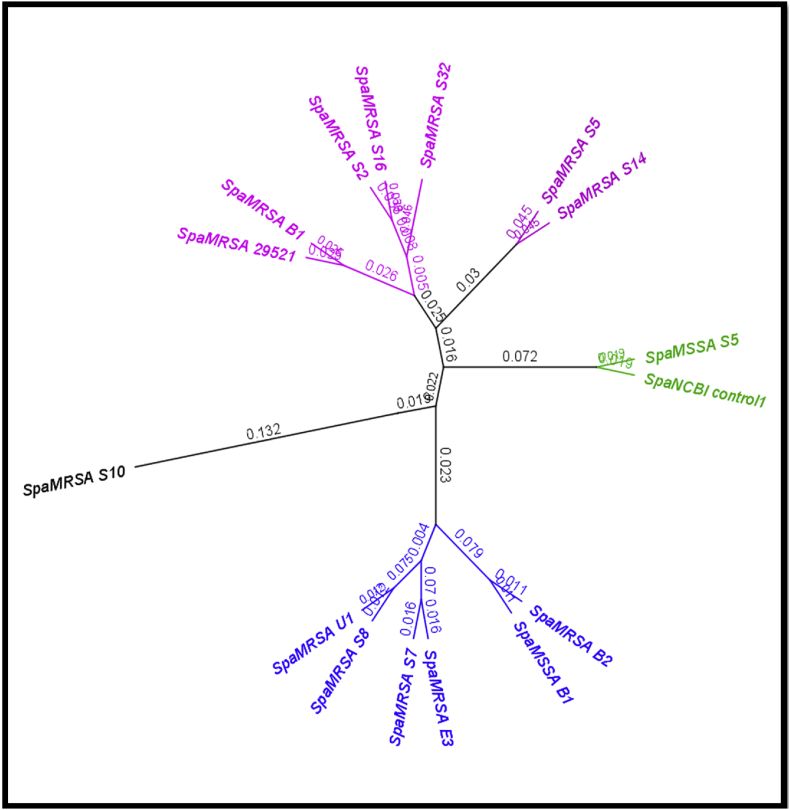
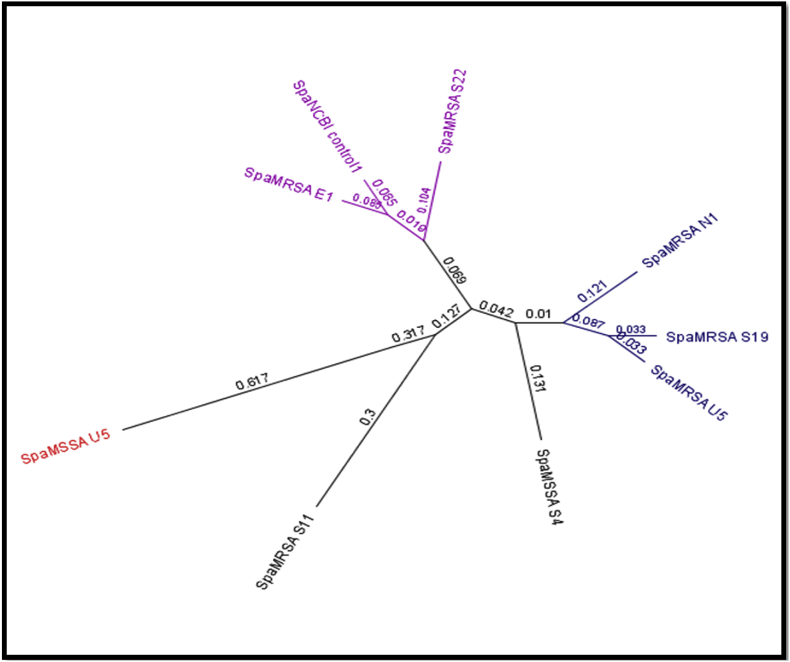
A total of 47 of 74 isolates had spa-typing positive results. Of these, 67.2% of MRSA isolates showed positive results vs. 46.15% of MSSA isolates. The amplified fragments ranged from 65 to 300 bp compared to the reference S. aureus 01-111 strain as analysed by Geneious software. None of the environmental isolates had spa gene variations (Fig. 1, Fig. 2).
Fig. 1.
Dendrogram and phylogenetic tree to long sequence (190–335 bp) of spa gene.
Fig. 2.
Dendrogram and phylogenetic tree to short sequence (65–190 bp) of spa gene related to methicillin-resistant Staphylococcus aureus and methicillin-susceptible S. aureus.
The amplified spa sequence of MRSA N1 was compared to the control strain using a GenBank (accession no. GI:482576487) MRSA strain isolated in Egypt (Egy 52A). Protein_id AGK23551 and pairwise identity was 262 (97%) of 270, which represents the percentage of identical residues in alignment, including gaps and nongap residues. Notably, there was a difference in base pairs causing a gap in the upper identity green line compared to DNA sequencing alignment. Mutations were evident at 32, 72, 81 and 259 bp, where C, C, C and G were altered to T, T, T and A, respectively.
A high diversity in spa genes obtained from different sources was obtained compared to the consensus and compared to the control obtained from NCBI. Results revealed that some changes took place in nucleotide sequences compared to spa gene control, and most isolates displayed different genetic variations. The highlighted nucleotides are variable ones and illustrate the diversity between all base pairs. The data divided into four monophyletic groups and one unique pattern group. Group 1 contained MRSA B1, S2, 16 and 32; group 2 contained MRSA U1, S7, 8, E3 and B1 as well as MSSA B1. Group 3 contained MRSA S5 and S19. Group 4 contained MSSA S5 and the NCBI strain control.
Nucleotide sequence variations were observed compared to spa gene control, and most isolates displayed genetic diversity. Our isolates divided into three monophyletic groups and one unique pattern group (data not shown). Group 1 contained NCBI standard strain control, MRSA E1 and S22. Group 2 contained MRSA N1, S19 and U5. Group 3 contained MRSA S11 and MSSA U5. The results illustrated that the isolate MSSA S4 was more distant genetically than the species studied, with a 0.131 value. The converged genetic relation for groups 1, 2 (at 0.069 and 0.042) and group 3 S. aureus isolates were more distant (0.127) from the other groups. MSSA S5 had a high similarity with the NCBI strain control.
In addition to analysis based on DNA sequence data from single locus spa gene, the amino acid profile of the spa repeats was performed according to the translation DNA sequencing of the variable region in the spa gene. The protein translation profile for each isolate subjected to spa gene sequencing, as well as the alignment between them, revealed that some amino acids are predominant in the isolates.
spa tandem repeats were calculated by bioinformatic analysis. Results revealed partial identity, with global spa types missing some repeats. They were detected by the starting coordinate of the repeat in the sequence alignment, numbers of repeat units, length of entire variable-number tandem repeat, Kreiswirth nomenclature, Ridom spa type name and repeats. Isolates that appear with ellipsis dots refer to missing repeats; an asterisk marks the indicated sequence as a spa repeat that was not found in the database we queried.
Single nucleotide polymorphism analysis was performed on the on spa gene by creating a DualBrothers (https://msuchard.faculty.biomath.ucla.edu/DualBrothers/index.html) phylogenetic tree and by analysis using the Geneious software; no genetic relation was observed among isolates. There was also no genetic relation regarding infection sources. None of the environmental isolates had spa gene variations. The data suggested a genetic relation (clonal distribution) of those isolates from the hospital environment.
Discussion
The prevalence of mecA gene was in agreement with previously published local studies that showed mecA prevalence to be 75.5% among S. aureus isolates [10,11]. Another local study mentioned the prevalence of mecA to be 100% [12]. MRSA isolates have risk factors that increase the prevalence of MRSA isolate colonization in Asia, the result of antibiotic misuse and limited socioeconomic status [13]. Further, patients who are infected with MRSA isolates in the hospital may harbour the microbe for long time [14,15]. In Iraq, a significant problem with MRSA is the result of an increased rate of incidence and hospitalization. Rapid and accurate typing of MRSA isolates is therefore essential for screening, epidemiology, surveillance and infection control.
Indeed, spa gene typing is easy and quick [6]. The selected region of the spa gene is usually a short sequence repeat with sufficient polymorphism to permit isolate typing [5]. Sakwinska et al. [16] found that 10% of healthy carriers carried the spa gene mutation. Another study found that a relation existed between MSSA and a control strain, and that diversity among the MRSA isolates were based on the multiple insertion of the staphylococcal chromosome cassette SCCmec, especially the mecA gene, into MSSA lineages, shifting them to MRSA isolates [17]. Furthermore, the diversity in amino acids indicates the emergence of a synonymous mutation to protect the amino acid sequence under varying levels of evolutionary pressure; many nonsynonymous mutations have been shown to cause a shift in amino acids [18].
In addition, the variation in tandem repeats of the spa gene encoding protein A is critical for analysing host–parasite interactions, as it can help bacteria evade the host immune system [19]. According to Ridom, MSSA S5 differs from t11434 by loss of the three-tandem repeat 26–23–23, found in New Zealand with a frequency of 0, while MSSA B1 differs from t690 by the two-tandem repeat 07–12, with a frequency of 0.09%, as has been found in the Middle East in places like Lebanon and the United Arab Emirates (Table 4). However, MRSA S32 and S14 exhibited higher similarities with t10214; the first lost four repeats at the beginning of the frame, and the second lost one repeat at the beginning and end of the frame. The latter was reported from Sweden, and different genetic cluster groups were shown in dendrogram and phylogenetic trees. Although MRSA S5 and S16 belong to different genetic cluster groups in the dendrogram, they exhibited tandem repeats similar to more than one type of spa, like t554 and t032. Indeed, the latter is found more frequently worldwide (10.41%), considering common spa types in MRSA isolates in Germany [2] in addition to its distribution in the areas surrounding Iraq, such as Lebanon, Kuwait and the United Arab Emirates. This clone has also emerged with resistance to aminoglycoside (known as EMRSA-15 or Barnium epidemic isolates [20]); it is close to our isolates, which showed resistance to gentamicin. MSSA S5 differed from t2421in one repeat, 26, which has been found in the United Kingdom. Humphreys et al. [21] reported availability of the t037 CC-22 isolate in Iraq and the Middle East, and mentioned the possibility of its transmission from Baghdad to Dublin. Koreen et al. [22] reported the common spa type t033, which depended on previously finding the predominant spa type of S. aureus in diverse parts of the world [23]. Other isolates have only two or one repeats (Table 4).
Table 4.
spa gene tandem repeats and spa gene types of local Staphylococcus aureus isolates compared to global spa gene tandem repeats and types, Seq: sequence, R: repeat
| Isolate | Seq. start | R. unit | Length (bp) | Repeat | Kreiswirth ID | spa type in Ridom | Repeat |
|---|---|---|---|---|---|---|---|
| MSSA S5 | 34 | 11 | 264 | … –13–23–31–29–17–25–17–24–25–16–28 | EJNF2MOMQOKR | t11434 | 26–23–23–13–23–31–29–17–25–17–24–25–16–28 |
| MSSA B1 | 32 | 9 | 216 | … –21–17–13–13–34–34–34–33–34 | FMEEBBBPB | t690 | 07–12–21–17–13–13–34–34–34–33–34 |
| MRSA S32 | 107 | 6 | 144 | … 17–25–17–25–16–28 | MOMOKR | t10214 | 26–31–13–23–05–17–25–17–25–16–28 |
| MRSA S16 | 33 | 8 | 192 | … –29–17–25–17–25–16–28 | F2MOMOKR | t554, t032 more than one type |
26–23–17–31–29–17–25–17–25–16–28 |
| MRSA S14 | 32 | 9 | 216 | … –31–13–23–05–17–25–17–25–16–… | NEJCMOMOK | t10214 | 26–31–13–23–05–17–25–17–25–16–28 |
| MRSA S10 | 131 | 2 | 48 | — | — | — | — |
| MRSA S5 | 107 | 7 | 168 | …–29–17–25–17–25–16–28 | F2MOMOKR | t554, t032 more than one type |
26–23–17–31–29–17–25–17–25–16–28 |
| MRSA S2 | 33 | 10 | 240 | 94–23–31–29–17–25–17–25–16–28 | JNF2MOMOKR | — | — |
| MRSA S7 |
84 | 5 | 120 | 16–2–25–17–24 | KAOMQ | — | — |
| MSSA S3 | 26 | 7 | 168 | 13–12–17–307–23–18–17 | EGM[r307]JH2M | — | — |
| MRSA U1 | 31 | 7 | 168 | ∗–12–17–307–23–18–17 | GM[r307]JH2M | — | — |
| MRSA E1 | 33 | 5 | 120 | 16–2–16–2–25 | KAKAO | — | — |
| MRSA S11 | 32 | 1 | 24 | 405 | — | — | — |
| MRSA U5 | 29 | 4 | 96 | 48–34–34–33 | V2BBP | t1590 | 07–16–48–34–34–33–34 |
| MRSA E3 | 33 | 6 | 144 | 16–2–16–2–25–17 | KAKAOM | — | — |
| MRSA N1 | 30 | 1 | 24 | 13 | — | — | — |
| MRSA S19 | 29 | 5 | 120 | ∗–34–34–33–34 | BBPB | t2421 more than 100 type | 26–34–34–33–34 |
| MSSA S4 | 31 | 2 | 48 | 12–17 | — | More than 100 type | — |
| MRSA B1 | 79 | 7 | 168 | …–05–17–25–17–25–16–28 | CMOMOKR | t309 | 26–23–05–17–25–17–25–16–28 |
| MRSA B2 | 33 | 1 | 24 | 21 | — | — | — |
| MSSA U2 | 33 | 10 | 240 | 16–2–16–34–2–25–17–24–24–24 | KAKBAOMQQQ | — | — |
| MSRA S22 | 32 | 4 | 96 | 25–82–16–17 | O[r82]KM | — | — |
| MRSA S28 | 30 | 9 | 216 | 94–23–5–17–25–17–25–16–28 | [r94]JCMOMOKR | — | — |
Ridom refers to Ridom spa Server (https://www.spaserver.ridom.de/).
MRSA, methicillin-resistant Staphylococcus aureus; MSSA, methicillin-susceptible S. aureus.
Ellipses indicate missing repeats.
Sequence is spa repeat but not found in database.
However, this cannot provide a real view in terms of similarities: MSSA U2, MRSA S22 and MRSA S28 display a pattern arrangement of repeats not found in Ridom. Perhaps these isolates have a new spa type. Indeed, spa types appear evolve by a combination of a faster changes in the number of repeats and a slower nucleotide point mutation [22] by slipped strand mispairing during DNA replication [24]. The spa repeat seems more prone to duplication and deletion than point mutation [22]. The maximum repetitive x-region in the current samples is 11 times, compared to reports which mentioned the maximum repetitive x-region to be 13 and 16 times in India and Iran, respectively [18,23]. The variation in the number of x-regions is a result of the deletion process. In other hands, it has been observed that the variation in spa types among MSSA isolates is greater than in MRSA isolates. This finding is closely related to the work of Fenner et al. [25], who mentioned that the diversity of the spa gene among MSSA isolates is higher than in MRSA isolates. This finding can be considered a novelty in Iraq and comprises the first local data about spa repeats. Other researchers have also reported that the discrimination between MRSA isolates is possible by determining the repeat sequence numbers within the x-region of the spa gene.
The spa typing technique represents as excellent tool for national and international surveillance as well as for short-term local epidemiology [23]. The DualBrothers model allows for changes in topology and evolutionary rates across sites in multiple sequence alignments. A huge alteration in the spa pattern could thus be recognized, with each spa sequence containing a different pattern. There was also no genetic relation regarding infection sources. None of the environmental isolates had spa gene variations. These data suggest that the genetic relation (clonal distribution) of these isolates is the result of a hospital environment.
Conclusion
The rate of MRSA was high among clinical isolates. Sequencing results showed different tandem repeats in local MRSA isolates. The pairwise result for the total sequence was 75.8%, with an identity of 15.8%. We found no genetic relation among S. aureus isolates or regarding infection sources. None of the environmental isolates had spa gene variations. The data suggested that the genetic relation (clonal distribution) of these isolates results from a hospital environment.
Conflict of interest
None declared.
Acknowledgements
The authors would like to thank Mustansiriyah University (www.uomustansiriyah.edu.iq) and Official Research and development Department (www.rdd.edu.iq) in ministry of higher education and Scientific research / Baghdad- Iraq for its support in the present work.
References
- 1.Okwu M.U., Mitsan O., Okeke O.P. Prevalence and antimicrobial susceptibility profiles of community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA) isolates among healthy individuals in Okada, south-south, Nigeria. US Open Pharm Biol Chem Sci J. 2014;1:1–9. [Google Scholar]
- 2.Holden M.T., Hsu L., Kurt K., Weinert L.A., Mather A.E., Harrism S.R. A genomic portrait of the emergence, evolution, and global spread of a methicillin resistant Staphylococcus aureus pandemic. Genome Res. 2013;23:653–664. doi: 10.1101/gr.147710.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faccone D., Togneri A.M., Podesta L., Perez M., Gagetti P., Sanchez S. MRSA pediatric clone expressing ermC plus lnuA genes causing nosocomial transmission and healthcare workers colonization in a neonatal intensive care unit. Infect Genet Evol. 2014;25:78–80. doi: 10.1016/j.meegid.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 4.Mohammadi S., Sekawi Z., Monjezi A., Maleki M., Soroush S., Sadeghifard N. Emergence of SCCmec type III with variable antimicrobial resistance profiles and spa types among methicillin-resistant Staphylococcus aureus isolated from healthcare- and community-acquired infections in the west of Iran. Int J Infect Dis. 2014;25:152–158. doi: 10.1016/j.ijid.2014.02.018. [DOI] [PubMed] [Google Scholar]
- 5.O’Hara F.P., Suaya J.A., Ray G.T., Baxter R., Brown M.L., Mera R.M., Close N.M. spa typing and multilocus sequence typing show comparable performance in a macroepidemiologic study of Staphylococcus aureus in the United States. Microb Drug Resist. 2016;22:88–96. doi: 10.1089/mdr.2014.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shopsin B., Gomez M., Waddington M., Riehman M., Kreiswirth B.N. Use of coagulase gene (coa) repeat region nucleotide sequences for the typing of methicillin-resistant Staphylococcus aureus strains. J Clin Microbiol. 2000;38:34536. doi: 10.1128/jcm.38.9.3453-3456.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruppé E., Hem S., Lath S., Gautier V., Ariey F., Sarthou J.L. CTX-M-lactamases in Escherichia coli from community acquired urinary tract infections, Cambodia. Emerg Infect Dis. 2009;15:741–748. doi: 10.3201/eid1505.071299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kareem S.M., Al-Jubori S.S., Ali M.R. Prevalence of pvl gene among methicillin resistance S. aureus isolates in Bagdad city. World J Pharm Res. 2015;4:455–471. [Google Scholar]
- 9.Walker J., Borrow R., Edwards-Jonesz V., Oppenheim B.A., Fox A.J. Epidemiological characterization of methicillin-resistant Staphylococcus aureus isolated in the North West of England by protein A (spa) and coagulase (coa) gene polymorphisms. Epidemiol Infect. 1998;121:507–514. doi: 10.1017/s0950268898001538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ibed A.N., Hamim S.S. Molecular detection of methicillin resistant staphylococcus aureus isolated from burns infection in Al-nasiriyah city. World J Pharm Pharmaceut Sci. 2014;2:950–954. [Google Scholar]
- 11.Kareem S.M., Al-Jubori S.S., Ali M.R. Prevalence of erm genes among methicillin resistant Staphylococcus aureus MRSA Iraqi isolates. Int J Curr Microbiol Appl Sci. 2015;4:575–585. [Google Scholar]
- 12.Al-Kadmy I.M. A genetic study to differential HC/AC MRSA isolated from clinical cases in Iraq hospitals. Mintage J Pharm Med Sci. 2013;2:57–62. [Google Scholar]
- 13.Beam J.W., Buckley B. Community-acquired methicillin-resistant Staphylococcus aureus: prevalence and risk factors. J Athl Train. 2006;41:337–340. [PMC free article] [PubMed] [Google Scholar]
- 14.McMullen K.M., Warren D.K., Woeltje K.F. The changing susceptibilities of methicillin-resistant Staphylococcus aureus at a Midwestern hospital: the emergence of ‘community-associated’ MRSA. Am J Infect Control. 2009;37:454–457. doi: 10.1016/j.ajic.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 15.Ratnayake L., Olver W.J. Rapid PCR detection of methicillinresistant Staphylococcus aureus and methicillin-sensitive S. aureus samples from charcoal-containing blood culture bottles. J Clin Microbiol. 2011;49:2382. doi: 10.1128/JCM.00106-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakwinska O., Blanc D.S., Lazor-Blanchet C., Moreillon M., Giddey M., Moreillon P. Ecological temporal stability of Staphylococcus aureus nasal carriage. J Clin Microbiol. 2010;48:2724–2728. doi: 10.1128/JCM.02091-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asghar A.H. Molecular characterization of methicillin-resistant Staphylococcus aureus isolated from tertiary care hospitals. Pak J Med Sci. 2014;30:698–702. doi: 10.12669/pjms.304.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mehndiratta P.L., Bhalla P., Ahmed A., Sharma Y.D. Molecular typing of methicillin-resistant Staphylococcus aureus strains by PCR-RFLP of spa gene: a reference laboratory perspective. Indian J Med Microbiol. 2009;27:116–122. doi: 10.4103/0255-0857.45363. [DOI] [PubMed] [Google Scholar]
- 19.Benson G. Tandem repeat finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monecke S., Leila Skakni L., Hasan R., Antje Ruppelt A., Ghazal S.S., Hakawi A. Characterization of MRSA strains isolated from patients in a hospital in Riyadh, Kingdom of Saudi Arabia. BMC Microbiol. 2012;12:146. doi: 10.1186/1471-2180-12-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Humphreys H., Carroll J.D., Keane C.T., Cafferkey M.T., Pomeroy H.M., Coleman D.C. Importation of methicillin-resistant Staphylococcus aureus from Baghdad to Dublin and subsequent nosocomial spread. J Hosp Infect. 1990;15:127–135. doi: 10.1016/0195-6701(90)90121-4. [DOI] [PubMed] [Google Scholar]
- 22.Koreen L., Ramaswamy S.V., Graviss E.A., Naidich S., Musser J.M., Kreiswirth B.N. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J Clin Microbiol. 2004;42:792–799. doi: 10.1128/JCM.42.2.792-799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shakeri F., Mansourian A.Z., Ghaeim E.A. Staphylococcus aureus protein A gene typing by PCR-RFLP. Afr J Microb Res. 2013;7:3448–3452. [Google Scholar]
- 24.van Belkum A. The role of short sequence repeats in epidemiological typing. Curr Opin Microbiol. 1999;2:306–311. doi: 10.1016/S1369-5274(99)80053-8. [DOI] [PubMed] [Google Scholar]
- 25.Fenner L., Anderas W.F., Dangel M., Frei R. Distribution of spa types among methicillin resistant Staphylococcus aureus isolates during a 6 years periods at low-prevalence university hospital. J Med Microbiol. 2008;57:612–616. doi: 10.1099/jmm.0.47757-0. [DOI] [PubMed] [Google Scholar]