Abstract
Nitric oxide (NO) is well established as a regulator of neurogenesis. NO increases the proliferation of neural stem cells (NSC), and is essential for hippocampal injury-induced neurogenesis following an excitotoxic lesion. One of the mechanisms underlying non-classical NO cell signaling is protein S-nitrosylation. This post-translational modification consists in the formation of a nitrosothiol group (R–SNO) in cysteine residues, which can promote formation of other oxidative modifications in those cysteine residues. S-nitrosylation can regulate many physiological processes, including neuronal plasticity and neurogenesis. In this work, we aimed to identify S-nitrosylation targets of NO that could participate in neurogenesis. In NSC, we identified a group of proteins oxidatively modified using complementary techniques of thiol redox proteomics. S-nitrosylation of some of these proteins was confirmed and validated in a seizure mouse model of hippocampal injury and in cultured hippocampal stem cells. The identified S-nitrosylated proteins are involved in the ERK/MAPK pathway and may be important targets of NO to enhance the proliferation of NSC.
Keywords: Nitric oxide, S-nitrosylation, Neural stem cells, Neurogenesis, Seizures, Hippocampus
Graphical abstract
Highlights
-
•
Nitric oxide (NO) regulates neurogenesis, and increases proliferation of neural stem cells (NSC).
-
•
S-nitrosylation is a cysteine oxidation that acts as a relevant non-classical mechanism of NO signaling.
-
•
Cysteine oxidation targets were screened in NSC, confirming S-nitrosylation of several proteins.
-
•
S-nitrosylation of some of these targets was confirmed in cultured hippocampal stem cells.
-
•
S-nitrosylation of several proteins was observed in an in vivo model of hippocampal neurogenesis.
1. Introduction
Nitric oxide (NO) is a gaseous free radical, a diffusible molecule that acts as a cellular messenger, having a crucial role in intercellular communication and in intracellular signaling in many tissues [[1], [2], [3]], including in the brain [4]. NO can signal through different ways depending on the available substrates, NO concentration and time of exposure. Classical (involving guanylate cyclase and cGMP production) and non-classical pathways are described for NO [5]. Non-classical mechanisms comprise covalent protein post-translational modifications (PTM) by reactive nitrogen species (RNS) derived from the reaction of NO with other small molecules. Although there are several RNS-induced PTM, three of them have been more intensively studied: tyrosine nitration, cysteine glutathionylation and cysteine S-nitrosylation [5]. Among them, protein S-nitrosylation, also called S-nitrosation, is the formation of a nitrosothiol (R-S-NO) in cysteine residues [6]. In the cells, nitrosothiols are unstable, when compared to other PTM, due to the lability of the bond, the presence of denitrosylases and other reductants, such as ascorbate, and the easy reaction with other thiols, which can also lead to protein S-glutathionylation, the formation of a mixed disulfide bond with glutathione [7,8].
Enzymatic catalysis is not strictly necessary for S-nitrosothiol formation and degradation in cell environment, so its specificity and implication in cell signaling bears some properties that make it different from other signaling PTM such as phosphorylation (reviewed in Refs. [5,6]). We have proposed S-nitrosylation as a short-range NO-derived signaling mechanism, suggesting that signaling can be favored in proteins located at a short distance from the sources of NO, namely NOS, and in proteins interacting with them [7]. S-nitrosylation is highly selective and specific for certain proteins and residues, a fact that can be explained by 3 main factors: subcellular compartmentalization, site specificity and denitrosylation specificity [5].
Although S-nitrosylation of some proteins has been recently described to be involved in processes in the brain such as synaptic plasticity, neurogenesis and even in some neurodegenerative diseases [7,[9], [10], [11]], the relevance of S-nitrosylation of many other proteins in these biological processes is still unknown. Neurogenesis refers to the formation of new neurons from neural stem cells (NSC), which may possibly be recruited in the adult brain to give rise to new neuronal cells after a lesion. In the central nervous system, NSC are found to proliferate lifelong in two main regions: the subventricular zone (SVZ) in the wall of the lateral ventricles and the subgranular zone (SGZ) of dentate gyrus (DG) of the hippocampus [12,13]. The role of NO on neurogenesis has been unraveled in the past 15 years, and a dual role for NO in neurogenesis has been described: on the one hand, NO impairs neurogenesis in physiological conditions [[14], [15], [16]], while a pro-neurogenic role for NO was described in the SVZ and in the DG following brain injury such as epileptic seizures or stroke [[17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27]]. These studies suggest that NO of inflammatory origin is involved in the increase in neurogenesis observed in the dentate gyrus following a brain lesion, such as in the case of seizures. We have previously demonstrated that NO can act, by S-nitrosylation, in p21Ras, activating the ERK/MAPK pathway involved in the stimulation of NSC proliferation, the first step of neurogenesis [28]. We now show that NO can produce relevant cysteine redox PTM in several other targets in NSC that may also impact on neurogenesis after injury.
We aimed to identify new proteins in NSC that were post-translationally modified by S-nitrosylation in the presence of the nitrosothiol S-nitroso-l-cysteine (CysSNO), and in vivo, following seizures. CysSNO is the S-nitrosylated form of the natural amino acid l-cysteine, and it can be formed in cells or in the extracellular medium. CysSNO can be translocated through the plasma membrane by amino acid carriers (in contrast to S-nitrosoglutathione), so it can transnitrosate intracellular proteins when added outside of the cells, activating pathways regulated by S-nitrosylation [6,29,30], as well as S-glutathionylation or other reversible cysteine oxidations [5,8]. Here we screened cysteine oxidation of NSC proteins following exposure to CysSNO. We identified several proteins that are oxidatively modified by CysSNO in SVZ-derived NSC cultures. Proteins such as hnRNP K, PCNA, 14-3-3, EF-1β and PEBP-1 were rapidly S-nitrosylated following exposure to CysSNO. Moreover, we observed a transient increase in the S-nitrosylation of some of these proteins in vivo, in a model of excitotoxic lesion. Additionally, treatment of hippocampal stem cells with CysSNO increased the S-nitrosylation of these proteins in a dose-dependent manner. Although a role in neurogenesis has been described for some proteins, the involvement of others in this process is unknown and our work identifies new possible targets for NO signaling in NSC.
2. Materials and methods
2.1. Reagents and chemicals
Dulbecco's Modified Eagle's medium (D-MEM)/F-12 with 2 mM GlutaMAX™-I, Gentamicin, B27, N2, Penicillin-Streptomycin (10,000 U/mL), Hanks' Balanced Salt Solution (HBSS), laminin, BODIPY FL N-(2-aminoethyl)maleimide Biotin-HPDP, Ultralink immobili neutravidin resin and bicinchoninic acid (BCA) reagents were purchased from Thermo Fisher Scientific (Waltham, MA, USA). EGF and bFGF were from Peprotech (London, UK). Kainic acid (KA) was obtained from Ocean Produce (Charlottetown, Canada). Horseradish peroxidase conjugated anti-mouse, anti-rabbit or anti-goat secondary antibodies were purchased from Cell Signaling (Danvers, MA, USA), and streptavidin-peroxidase was from Calbiochem (San Diego, CA, USA). NEM, sodium ascorbate, DTT, IAM, poly-l-lysine, polyornithine, Ethylenediamine tetraacetic acid (EDTA), ethylene glycol tetraacetic acid (EGTA), neocuproine, 2-mercaptoethanol, Tween-20 and Triton X-100 were purchased from Sigma Chemical (St Louis, MO, USA). Polyvinylidene difluoride (PVDF) membranes, sodium dodecyl sulfate (SDS), Clarity Western ECL Substrate, immobilized pH gradient (IPG) strips, ampholytes Bio-Lite 3-10 buffer, and Sypro Ruby were purchased from Bio-Rad (Hercules, CA, USA). Acrylamide, tetramethylethylenediamine (TEMED) and molecular ladder were obtained from NZYTech (Lisboa, Portugal). Proteomics-grade trypsin was from Promega (Madison, WI, USA). The origins of primary antibodies used in Western blot are listed in Table 1.
Table 1.
Primary antibodies used in Western blot, and amount of protein used in ascorbate-reducing biotin switch for each protein.
| Protein | Antibody | Protein amount (μg) |
||
|---|---|---|---|---|
| SVZ | HC7 | DG | ||
| hnRNP K | Cell Signaling (1:1000) | 750 | 800 | 1000 |
| PCNA | Santa Cruz Biotechnology (1:500) | 750 | 800 | 1000 |
| 14-3-3 ε | Cell Signaling (1:1000) | 500 | 800 | 1000 |
| 14-3-3 | Cell Signaling (1:1000) | 750 | – | 1000 |
| EF-1β | Santa Cruz Biotechnology (1:500) | 500 | 800 | 1000 |
| PEBP-1 | Abcam (1:5000) | 750 | 800 | 1000 |
| HSPA4 | Abcam (1:5000) | 500 | – | – |
2.2. Animals
C57BL/6 newborn mice and 12-week old male mice were used in this study. All the animals were kept in our animal facility with food and water ad libitum in a 12 h dark:light cycle. All experiments were performed in accordance with European guidelines (2010/63/EU) for the care and use of laboratory animals, as well as Portuguese law (DL 113/2013). The procedures performed in mice have been reviewed and approved by the Animal Welfare Body of the Center for Neuroscience and Cell Biology and have been approved by the Direcção Geral de Alimentação e Veterinária (reference 0421/000/000/2013).
2.3. Subventricular zone neural stem cell cultures
NSC cultures were obtained from the SVZ of 0-3-days C57BL/6 mice and maintained as previously described [[31], [32], [33]]. NSC grown as floating neurospheres in Dulbecco's Modified Eagle's Medium: F-12 nutrient mixture (D-MEM/F-12 with 2 mM GlutaMAX™-I (L-Ala-L-Gln)), supplemented with 1% B27, 1% antibiotic (10,000 units/ml of penicillin, 10 mg/ml streptomycin), 10 ng/ml EGF and 5 ng/ml basic fibroblast growth factor (bFGF), were collected and plated for 2-3 days on poly-l-lysine-coated plates. Then, growth factors were excluded from the medium and cells were kept in this medium for 24 h before the experiments.
2.4. Hippocampal stem cell cultures
HC7 cells were kindly provided by Dr. Fred H. Gage (The Salk Institute for Biological Studies, USA) and maintained as previously described [34,35]. Briefly, cells were plated on polyornithine (10 μg/ml) and laminin (10 μg/ml)-coated 60 mm tissue culture dishes in DMEM/F-12 with 2 mM GlutaMAX-I containing 1% N2 supplement, 20 ng/ml bFGF and 1% of penicillin/streptomycin. Cells were cultured at 37 °C in 5% C02/95% air and medium was changed every 3-4 days. For passaging, confluent cells were resuspended in DMEM/F12 with GlutaMAX complete medium and passaged onto polyornithine/laminin-coated 100 mm dishes using a split ratio of 1:3. Before the experiments, cells were kept in medium without bFGF for 24 h.
2.5. Cell treatment
For the assessment of oxidation/S-nitrosylation, SVZ-derived NSC were treated for 15 min with 100 μM CysSNO. Hippocampal stem cells were exposed during 15 min to 10, 50 and 100 μM CysSNO for evaluation of S-nitrosylation.
2.6. Preparation of CysSNO
CysSNO was prepared as previously described [36,37]. Briefly, 200 mM of L-Cys was prepared in 1 ml of 1 M HCl and added to a solution of 200 mM NaNO2 in water (1 ml). After 30 min at RT, 2 ml of 1 M potassium phosphate buffer, pH 7.4, were added. The final solution was divided in several aliquots and stored at -80 °C. CysSNO concentration was determined by spectrophotometric analysis at 338 nm in Nanodrop (Thermo Scientific), using the extinction coefficient of CysSNO (ε338 = 900 M-1 cm-1) [38]. During the entire protocol, solutions were kept protected from light, as nitrosothiols are decomposed by light. The yield of the reaction was around 80%.
2.7. Cell lysates for redox fluorescence switch assay and biotin switch assay
Cells were washed with cold saline physiological solution (0.9% NaCl, pH 7.4) and then scraped and lysed in TENT pH 6.0 (50 mM Tris-HCl, 1 mM EDTA, 0.1 mM neocuproine, 1% Triton X-100) supplemented with 50 mM NEM to block the free thiols, and protease inhibitors (freshly added), protected from light. Four 2-s sonication cycles were applied. To complete the blocking reaction, 2% SDS was added to the lysate and incubated 30 min at 37 °C. Protein concentration was determined by the BCA method, following the manufacturer's instructions.
2.8. Redox fluorescence switch (RFS) assay
The RFS protocol was adapted from Refs. [37,39]. In order to assess protein cysteine reversible oxidation, 100 μg of protein were used for 1-DE gels and 200 μg for 2-DE gels. Protein was precipitated with acetone: addition of 3 volumes of cold acetone, incubation for 10 min at -20 °C, followed by centrifugation for 5 min at 12.000 rpm, 4 °C; then the supernatant was discarded and the process was repeated with 1 volume of cold acetone. The pellet was resuspended in TENS pH 7.2 (50 mM Tris-HCl, 1 mM EDTA, 0.1 mM neocuproine, 1% SDS; 100 μl per 50 μg of protein) with 2.5 mM DTT, to reduce reversibly oxidized thiols. Following an incubation of 10 min at RT, protein was precipitated with acetone as before and the pellet was resuspended in TENS pH 7.2 plus 40 μM BODIPY FL N-(2-aminoethyl)maleimide. Following 1 h of incubation, the labeling reaction was stopped by adding 2.5 mM DTT and protein was precipitated as before. Samples were protected from light during the entire procedure.
2.9. Protein electrophoresis with fluorescent detection (1-DE and 2-DE)
Following RFS technique, protein samples were separated by 1-DE (SDS-PAGE) or 2-DE, and then stained with Sypro Ruby, as previously described [37,39,40].
2.10. 2-DE analysis
2-DE spots’ pattern was analyzed with the PD Quest 2-D analysis software (Bio-Rad Laboratories). At least four independent experiments per condition were analyzed. An initial analysis was performed by using RFS images of each condition (Control and CysSNO-treated samples) and the same spots were matched in a single gel set and quantified. Spots were selected when the fluorescence signal was at least two-fold higher in CysSNO vs. control conditions in at least two replicates. For several spots, no signal was detected in the control conditions, or it was much lower than in CysSNO-treated samples. A quantitative analysis was performed in the remaining spots, calculating the mean CysSNO:Control ratio in each experiment, and significant differences were determined using an ANOVA test. Images of the total protein signal for each gel were matched in a separate gel set and quantified. The spots selected from the RFS analysis were manually matched to their corresponding total protein signal spots, and only those in which the amount of total protein remained constant were finally selected for identification.
2.11. Mass spectrometry analysis
The spots pinpointing differences in RFS signal that did not correspond to differences in the total protein amount in the 2-DE were picked, digested and analyzed by MS in the Proteomics Unit of the CBMSO or in the Proteomics Unit of the CNB (both members of ProteoRed-ISCIII in Madrid, Spain), as detailed below.
2.12. In-gel digestion
After drying, gel spots were destained in acetonitrile:water (1:1) and digested in situ with sequencing grade trypsin (Promega, Madison, WI) as described previously [41] with minor modifications [42]. The gel pieces were shrunk by removing all liquid using sufficient acetonitrile. Acetonitrile was pipetted out and the gel pieces were dried in a speedvac (Eppendorf, Hamburg, Germany). The dried gel pieces were re-swollen in 50 mM ammonium bicarbonate pH 8.8 with 12.5 ng/μl trypsin for 1 h in an ice-bath. The digestion buffer was removed, and gels were covered again with 50 mM ammonium bicarbonate pH 8.8 and incubated at 37 °C for 12 h. Digestion was stopped by the addition of 1% trifluoroacetic acid. Whole supernatants were dried down and then desalted onto ZipTip C18 Pipette tips (Millipore) until the mass spectrometric analysis.
2.13. Protein identification by MALDI-TOF peptide mass fingerprinting (PMF)
PMF was performed at the Proteomics Unit of the Centro Nacional de Biotecnología (CNB) in Madrid (Spain). The Proteomics Unit is part of ProteoRed-ISCIII and PRB3 (Spanish platforms financed by the ISCIII).
For MALDI-TOF/TOF analysis, 20% of peptide mixture was deposited onto a 384-well OptiTOF Plate (AB SCIEX, Foster City, CA) and allowed to dry at room temperature. A 0.8 μl aliquot of matrix solution (3 mg/mL α-cyano-4-hydroxycinnamic acid in MALDI solution) was then deposited onto dried digest and allowed to dry at room temperature.
Samples in the OptiTOF plate were automatically acquired in an ABi 4800 MALDI TOF/TOF mass spectrometer (AB SCIEX, Foster City, CA) in positive ion reflector mode (the ion acceleration voltage was 25 kV to MS acquisition and 2 kV to MS/MS) and the obtained spectra were stored into the ABi 4000 Series Explorer Spot Set Manager. PMF and MS/MS fragment ion spectra were smoothed and corrected to zero baseline using routines embedded in ABi 4000 Series Explorer Software v3.6. Each PMF spectrum was internally calibrated with the mass signals of trypsin autolysis ions to reach a typical mass measurement accuracy of <25 ppm. Known trypsin and keratin mass signals, as well as potential sodium and potassium adducts (+21 Da and +39 Da) were removed from the peak list. To submit the combined PMF and MS/MS data to MASCOT software v.2.5.0.0 (Matrix Science, London, UK), GPS Explorer v4.9 was used, searching in the Mus musculus Uniprot protein database (v20150210: 16706 sequences; 195014757 residues). The following search parameters were used: enzyme, trypsin; allowed missed cleavages, 1; carbamidomethyl and maleimide in cysteine and oxidation of methionine as variable modifications; mass tolerance for precursors was set to ±40 ppm and for MS/MS fragment ions to ±0.3 Da. The confidence interval for protein identification was set to ≥95% (p < 0.05) and only peptides with an individual ion score above the identity threshold were considered correctly identified.
2.14. Reverse phase-liquid chromatography-tandem mass spectrometry assay (RP-LC-MS/MS) in selected MS/MS ion monitoring (SMIM) mode
The desalted protein digest was dried, resuspended in 10 μl of 0.1% formic acid and analyzed by RP-LC-MS/MS in an Easy-nLC II system coupled to an ion trap LTQ-Orbitrap-Velos-Pro mass spectrometer (Thermo Scientific). The peptides were concentrated (on-line) by reverse phase chromatography using a 0.1 mm × 20 mm Acclaim PepMap C18 precolumn (5 μm, 100 Å; Thermo Scientific), and then separated using a 0.075 mm × 25 cm Acclaim PepMap C18 column (3 μm, 100 Å; Thermo Scientific) operating at 0.3 μl/min. Peptides were eluted using a 60-min gradient from 5 to 40% solvent B (Solvent A: 0,1% formic acid in water, solvent B: 0,1% formic acid, 80% acetonitrile in water). ESI was done using a Nano-bore emitters Stainless Steel ID 30 μm (Proxeon) interface [42].
The Orbitrap resolution was set at 30.000. Peptides were detected in survey scans from 400 to 1600 amu (1 μscan), followed by SMIM events when necessary (the number of events depends on the peptides to be monitored) plus ten data-dependent MS/MS scans, using an isolation width of 2 u (in mass-to-charge ratio units), normalized collision energy of 35%, and dynamic exclusion applied during 30 s periods. In the SMIM mode [43], used to find peptides in a directed way, the LTQ-Orbitrap-Velos-Pro detector was programmed to perform, along the same entire gradient, a continuous sequential operation in the MS/MS mode on the doubly or triply charged ions corresponding to the peptide(s) selected previously from the theoretical prediction.
Peptide identification from raw data was carried out using the SEQUEST algorithm integrated in the Proteome Discoverer 1.4 software (Thermo Scientific). Database search was performed against a handmade database including the proteins of interest. The following constraints were used for the searches: tryptic cleavage after Arg and Lys, up to two missed cleavage sites, and tolerances of 20 ppm for precursor ions and 0.8 Da for MS/MS fragment ions, and the searches were performed allowing optional Met oxidation, Cys carbamidomethylation and Cys NEM modification. Search against decoy database (integrated decoy approach) using false discovery rate < 0.01.
To confirm the identification of peptides monitored, the MS/MS spectra were manually analyzed by assigning the fragments to the candidate sequence, after calculating the series of theoretical fragmentations, according to the nomenclature of the series as previously described [44].
2.15. Administration of kainic acid (KA) in a temporal lobe epilepsy mouse model
Adult male wild-type C57BL/6 with 12 weeks of age were treated with KA solution injected subcutaneously (25 mg/kg) and the respective controls were treated with a sterile saline solution (0.9% NaCl in water, pH 7.4), as previously described [23,25]. The weight of the animals varied between 20 and 30 g. All animals that received KA developed grade five seizures or higher in the 1972's Racine's six-point scale modified for mice [45]. In animals injected with saline solution alone, no seizures were observed and were used as controls. The mice were maintained for 1, 2, 3 and 5 days after the first generalized seizure. At least three animals survived in each experimental group.
2.16. Dissection of the dentate gyrus and lysates
Animals were sacrificed by cervical dislocation 1, 2, 3 or 5 days following treatment (saline solution or KA solution). The brains were removed from the skull following decapitation and placed in a plate dish containing HBSS medium. Following dissection, the DG of each animal was mechanically lysed with a tissue homogenizer in TENT pH 6.0 (50 mM Tris-HCl, 1 mM EDTA, 0.1 mM Neocuproine, 0.5% Triton X-100, pH 6.0) supplemented with 50 mM NEM to block the free thiols, and protease inhibitors (Roche cOmplete mini, freshly added). Four 2-s sonication cycles were applied. 2% SDS was added to the lysate and incubated 30 min at 37 °C to complete the blocking reaction. Protein concentration was determined by the BCA method.
2.17. Biotin switch assay and Western blot analysis
Determination of S-nitrosylation by the biotin switch assay and of reversible Cys oxidation by the redox biotin switch assay was performed as described in Refs. [36,37,39], both in dentate gyri samples (DG extracts) and in cell lysates after thiol blocking, using different amounts of protein as stated in Table 1.
Lysates were precipitated with acetone and the pellets were resuspended in TENS pH 7.2 (50 mM Tris-HCl, 1 mM EDTA, 0.1 mM neocuproine, 1% SDS, pH 7.2). For DTT reduction, 2.5 mM DTT was added, and the samples incubated for 10 min at RT; after that, it was precipitated with acetone, resuspended in TENS pH 7.2 and the reduced thiols were labeled with 1 mM biotin-HPDP for 1 h. For nitrosothiol reduction, 100 mM sodium ascorbate and 1 mM biotin-HPDP were added at the same time, and incubated for 1 h at RT. Until this point all operations were carried out in the dark. Samples were then precipitated with acetone and resuspended in HENS (250 mM Hepes pH 7.7, 1 mM EDTA, 0.1 mM neocuproine, 1% SDS) and then a neutralization buffer (20 mM Hepes pH 7.7, 1 mM EDTA, 100 mM NaCl, 0.5% Triton) was added, followed by centrifugation at 13.400 rpm for 1 min. 100 μl of the supernatant were saved as the Input fraction (total protein), and the rest was added to the pre-equilibrated Ultralink immobilized neutravidin resin. Samples were incubated with the resin for 1 h at RT with agitation. Following centrifugation (1 min, 2.000 rpm), supernatant was collected as the non-retained fraction (NR). After washing, 100 μl of elution buffer were added (20 mM Hepes pH 7.7, 100 mM NaCl, 1 mM EDTA, 100 mM 2-mercaptoethanol) and incubated 20 min at 37 °C for elution of the proteins that bound to the resin. Then, the mixture was centrifuged for 1 min at 14.500 rpm and the supernatant was saved as the Eluted Fraction (Oxidized/SNO proteins).
For analysis of specific proteins by Western blot, the eluted fraction and respective input samples were denatured at 96 °C for 5 min with a reducing Laemlli buffer and then subjected to an electrophoresis in 12% SDS/PAGE gels, transferred to PVDF membranes, blocked with 5% low-fat dry milk or 3% BSA in TBS-T, as appropriated, and then incubated with the primary antibodies (Table 1) in 1% blocking solution overnight, at 4 °C. Incubation with the HRP secondary anti-mouse, anti-rabbit or anti-goat antibodies conjugates (1:2000 in 1% blocking solution) was performed at RT for 1 h. After that, membranes were incubated with the Clarity Western ECL Substrate (Bio-Rad, Hercules, CA, USA), and immunoreactive bands were visualized in the Molecular Imager® ChemiDocTM XRS + Imaging System (Bio-Rad, Hercules, CA, USA). Image bands were analyzed with the ImageLab software version 5.1 (Bio-Rad Laboratories, Hercules, CA, USA).
2.18. Data analysis
Data are expressed as means ± SEM. Statistical significance was determined by using t-test for comparison of two population means or one-way or two-way analysis of variance (ANOVA) followed by Bonferroni's or Dunnett's post-tests for comparisons of multiple groups, as appropriate and indicated in the figure legends and in the text. Differences were considered significant when p < 0.05. The software used was GraphPad Prism 5.0 (GraphPad Software, La Jolla, CA, USA).
3. Results
3.1. Identification of proteins oxidized in NSC treated with CysSNO
S-nitrosylation can lead to formation of other cysteine oxidative PTM such as S-glutathionylation [8]. Thus, a useful method for screening S-nitrosylation is to assess reversible cysteine oxidation by RFS, because DTT reduction (which allows to label all reversible cysteine oxidations) is more efficient than ascorbate reduction (specific for S-nitrosylation), providing a higher sensitivity to the method. To assess reversible oxidative modification of proteins following exposure to NO and RNS, NSC derived from the SVZ were treated with the nitrosothiol CysSNO, and a proper control, without treatment, was used.
Protein oxidation was assessed by the RFS technique followed by 1-DE or 2-DE. We observed an increase in the global Bodipy-FL signal, which measures the extent of protein oxidation, following exposure to CysSNO (Fig. 1A, left panel), and more specifically in some bands. This effect did not correspond to an increase in the protein amount as no differences in the Sypro Ruby signal were observed (Fig. 1A, middle panel). Furthermore, without the reduction step with DTT (negative control), no Bodipy-FL signal was observed, thus showing the good specificity of the technique. Merging both signals (Fig. 1A, right panel) allows a clearer visualization of the differences in oxidation signal between samples.
Fig. 1.
CysSNO increases cysteine oxidation in NSC. Cysteine oxidation of general proteins was assessed by a RFS followed by 1-DE (A) or 2-DE (B) of untreated NSC, or treated with 100 μM CysSNO for 15 min. A negative control, without DTT, was performed in (A). Oxidation signal is shown by Bodipy-FL labeling (green) and total protein was stained with Sypro Ruby (red). Some differences in the RFS signal of CysSNO treated cells are shown by the white arrows. Representative images of 8 experiments are shown. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To determine which proteins exhibit differential cysteine oxidation following exposure to CysSNO, RFS samples were run by 2-DE. The overall pattern of 2-DE spots of CysSNO-treated samples was similar to that observed in control samples, although there were differences in some specific spots (Fig. 1B, left panels in green). The observed differences reflected an increase in the Bodipy-FL signal in CysSNO-treated samples, when compared with untreated samples, or even the presence of a spot that was not detected in control samples. Furthermore, no differences in total protein signal (Sypro Ruby) were observed (Fig. 1B, middle panels in red). By merging both signals (Fig. 1B, right panels) a significant overlapping of the signal corresponding to the same spots was detected.
To perform a full analysis of the spots of the 2-DE gels and pinpoint differences in their oxidative signal that did not correspond to differences in total protein, eight replicates of CysSNO-treated samples where compared to untreated samples (control) using the PD-Quest 2-DE analysis software (Bio-Rad Laboratories). Spots were first selected when the oxidation signal in CysSNO-treated conditions was at least 2-fold higher than control in at least two replicates. For some of these spots, qualitative differences were clear, as there was no RFS signal in the control gel for some replicates or, when present, it was more than 2-fold smaller (Table 2). For other spots, the RFS spot was clear in most of the replicate gels of CysSNO-treated samples, and a statistical analysis was performed so that spots with a p lower than 0.05 were selected (Table 3). The selected RFS spots were matched to the corresponding spots of total protein signal (Fig. 2) in the same gels to ensure that they displayed a constant signal. All the selected spots were manually excised from at least two replicates of the gels and digested with trypsin. The resulting peptides were analyzed by MALDI-TOF/TOF and peptide mass fingerprinting. Eleven different proteins were identified from these spots (Table 2, Table 3).
Table 2.
Qualitative analysis of proteins differentially oxidized in NSC exposed to CysSNO.
| Protein | Spots in Control gelsa | Spots in CysSNO gelsa | Minimal Ratiob | Oxidized Cys |
|---|---|---|---|---|
| EF-1β – Elongation factor-1β | 1 | 7 | 3.2 | |
| 14-3-3 ε | 3 | 7 | 3.8 | 97, 98 |
| 14-3-3 θ | 2 | 6 | 2.1 | 25, 94, 134 |
| 14-3-3 ζ/δ | 0 | 8 | 25, 94 | |
| PCNA – Proliferating cell nuclear antigen | 1 | 6 | 2.4 | |
| Secernin-1 | 1 | 4 | 2.1 | |
| HSPA4 – Heat shock 70 kDa protein 4 | 0 | 2 | ||
| TER ATPase – Transitional endoplasmic reticulum ATPase | 2 | 8 | 3.7 | 105, 522, 535 |
| hnRNP K – Heterogeneous nuclear ribonucleoprotein K | 0 | 4 | 145 |
Number of replicates in which the spot is detected.
Minimal value obtained for the ratio (RFS signal in CysSNO/RFS signal in Control) of replicate(s) in which a spot was found in Control.
Table 3.
Quantitative analysis of proteins differentially oxidized in NSC exposed to CysSNO.
| Protein | Spots in Control gelsa | Spots in CysSNO gelsa | Mean ratio ± sdb | t-test p value | Oxidized Cys |
|---|---|---|---|---|---|
| PEBP-1 – Phosphatidylethanolamine-binding protein | 3 | 7 | 2.4 ± 0.7 | 0.006 | |
| hnRNP H – Heterogeneous nuclear ribonucleoprotein H | 6 | 7 | 3.4 ± 2.0 | 0.001 |
Number of replicates in which the spot is detected.
Mean ± standard deviation of the ratios (RFS signal in CysSNO/RFS signal in Control) in individual experiments.
Fig. 2.
Differentially oxidized proteins following treatment with CysSNO. Representative 2_DE gel of sample treated with 100 μM CysSNO for 15 min stained with Sypro Ruby, showing the protein identified in each spot. EF-1β, elongation factor-1β; PCNA, proliferating cell nuclear antigen; PEBP-1, phosphatidylethanolamine-binding protein; HSPA4, heat shock 70 kDa protein 4; TER ATPase, transitional endoplasmic reticulum ATPase; hnRNP K, Heterogeneous nuclear ribonucleoprotein K; hnRNP H, Heterogeneous nuclear ribonucleoprotein H.
The Bodipy-FL label incorporated in the RFS does not allow to localize which Cys residues are modified, since it alters the fragmentation pattern of the peptides (Martínez-Ruiz et al., unpublished observations). To determine the modified Cys residues, the NEM-blocked samples were labeled with IAM after DTT reduction and separated in parallel 2-DE gels [37,39]. The spots corresponding to the ones in which differentially oxidized proteins had been identified were cut and digested with trypsin. Targeted LC-MS/MS analysis was performed in an LTQ-VelosPro-orbitrap mass spectrometer, allowing the identification of a number of peptides containing IAM-modified Cys residues, which corresponded to the oxidized Cys (presented in Table 2).
3.2. Confirmation of oxidized and S-nitrosylated proteins in SVZ-derived NSC
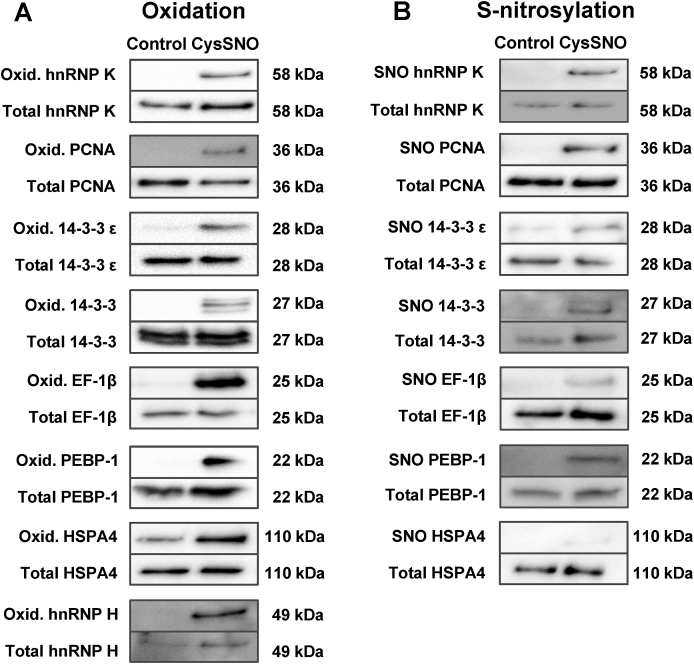
Following extensive analysis of the up-to-date known functions of the identified proteins, including their role in pathways involved in neurogenesis and cell proliferation, some of the identified proteins were selected for further validation of protein oxidation and, more specifically, S-nitrosylation, following exposure to CysSNO. The confirmation of the oxidation of individual proteins was achieved by using a redox biotin switch method [37]. SVZ-derived NSC were exposed to 100 μM CysSNO for 15 min. Oxidation of the following proteins was identified following exposure to CysSNO: hnRNP K, PCNA, 14-3-3 proteins (and specifically 14-3-3 ε), EF-1β, PEBP-1, HSPA4, and hnRNP H (Fig. 3A).
Fig. 3.
Treatment of SVZ-derived NSC with CysSNO induces oxidation, including S-nitrosylation, of proteins. Oxidation (A) and S-nitrosylation (B) of hnRNP K, PCNA, 14-3-3 proteins, EF-1β, PEBP-1, HSPA4 and hnRNP H was assessed by DTT-reducing (A) and ascorbate-reducing (B) biotin switch assays.
In order to confirm that the identified proteins are, in fact, S-nitrosylated in the same conditions, S-nitrosylation was assessed by the ascorbate-reducing biotin switch assay [37], using antibodies that specifically detect each protein. S-nitrosylation of the majority of the proteins mentioned above (hnRNP K, PCNA, 14-3-3,14-3-3 ε, EF-1β and PEBP-1) was detected following exposure to 100 μM CysSNO for 15 min, when compared to control samples (Fig. 3B). S-nitrosylation of HSPA4 was not observed using up to 750 μg of protein in cell lysate, after treatment with CysSNO (Fig. 3B), and this target was not further pursued.
3.3. Validation of S-nitrosylated proteins in the dentate gyrus after seizures
To investigate whether S-nitrosylation of the identified proteins was relevant in NSC after brain injury, we used an excitotoxic lesion mouse model that is characterized by an increase in neurogenesis in the DG after induction of seizures with kainic acid (KA) [23,25]. S-nitrosylation of proteins was assessed by ascorbate-reducing biotin switch 1, 2, 3 and 5 days after KA treatment, so we could also evaluate the kinetics of modification of each protein.
Of the analyzed proteins, we observed S-nitrosylation of hnRNP K, 14-3-3, 14-3-3 ε and PEBP-1, but not of PCNA or EF-1β (Fig. 4A). hnRNP K S-nitrosylation significantly increased 2 days after KA treatment (195.5 ± 45.0%, p < 0.05, Fig. 4B), comparing with control. 1, 3 and 5 days after KA treatment, S-nitrosylation of hnRNP K was similar to control. S-nitrosylation of all 14-3-3 proteins isoforms significantly increased 1 day after KA treatment (149.9 ± 34.8%, p < 0.05, Fig. 4B), and was similar to control 2, 3 and 5 days after seizure onset. S-nitrosylation of 14-3-3 ε, the most interesting isoform regarding neurogenesis, showed a tendency to increase 1 and 2 days after KA treatment (p > 0.05 not significant, Fig. 4B) and to be maintained or diminished after 3 and 5 days, when comparing with control. PEBP-1 S-nitrosylation also significantly increased 1 day after KA treatment (173.9 ± 16.3%, p < 0.01, Fig. 4B). After 2, 3 and 5 days of treatment, PEBP-1 S-nitrosylation was maintained, compared to control. S-nitrosylation of PCNA and EF-1β was not observed in samples of DG, using up to 1 mg of protein in cell lysate.
Fig. 4.
Induction of epileptic seizures in mice leads to S-nitrosylation of proteins in the DG. S-nitrosylation of hnRNP K, 14-3-3 proteins, PEBP-1, PCNA and EF-1β was assessed by ascorbate-reducing biotin switch assay (A). Data from at least 3 independent experiments are expressed as means ± SEM (% of control, SAL). Two-way ANOVA followed by Bonferroni's post-test, *p < 0.05 and **p < 0.01 statistically different from SAL (B). SAL, saline; KA, kainic acid; d, days.
In order to evaluate the S-nitrosylation of these proteins in stem cells of the DG, we exposed hippocampal stem cells to different concentrations of CysSNO (10, 50 and 100 μM) for 15 min and analyzed S-nitrosylation by ascorbate-reducing biotin switch. We observed S-nitrosylation of hnRNP K, PCNA, 14-3-3 ε, EF-1β and PEBP-1 (Fig. 5A). hnRNP K S-nitrosylation showed a tendency to increase after treatment with 10 (243.2 ± 77.2%, Fig. 5B) and 50 μM CysSNO (294.0 ± 97.1%), and significantly increased after treatment with 100 μM CysSNO (487.9 ± 112.65%, p < 0.01). Treatment with 100 μM CysSNO significantly increased PCNA S-nitrosylation (260.7 ± 85.9%, p < 0.05, Fig. 5B), while after treatment with 10 and 50 μM CysSNO S-nitrosylation was maintained, compared to control. S-nitrosylation of 14-3-3 ε showed a tendency to increase after treatment with 10 (241.5 ± 44.1%, Fig. 5B) and 50 μM CysSNO (256.7 ± 21.0%), and significantly increased after treatment with 100 μM CysSNO (682.1 ± 209.3%, p < 0.05), when comparing with control. S-nitrosylation of EF-1β was maintained after treatment with 10 μM CysSNO, and significantly increased after treatment with 50 (256.1% ± 34.6, p < 0.01, Figs. 5B) and 100 μM CysSNO (234.2% ± 34.4, p < 0.05), when comparing with control. PEBP-1 S-nitrosylation was maintained after treatment with 10 μM CysSNO but showed a tendency to increase after treatment with 50 μM CysSNO (204.3 ± 73.7%, Fig. 5B) and significantly increased with 100 μM CysSNO treatment (339.9 ± 100.0%, p < 0.05), when comparing with control.
Fig. 5.
Treatment of hippocampal stem cells with CysSNO induces S-nitrosylation of proteins in a dose-dependent manner. S-nitrosylation of hnRNP K, PCNA, 14-3-3 ε, EF-1β and PEBP-1 was assessed by ascorbate-reducing biotin switch assay (A). Data from at least 3 independent experiments are expressed as means ± SEM (% of control). One-way ANOVA followed by Dunnett's post-test, *p < 0.05 and **p < 0.01 statistically different from control (B).
4. Discussion
NO is a signaling molecule with several functions in the organism. Through S-nitrosylation, NO can modify the function of proteins relevant for many processes. As an important regulator of adult endogenous neurogenesis, NO is able to increase NSC proliferation after brain injury. However, this repair process is far from effective. The identification of proteins that could contribute to improve its outcome is therefore of particular importance. The goal of this work was to identify S-nitrosylation targets of NO that could be involved in neurogenesis, in NSC. Using complementary techniques of thiol redox proteomics to screen for possible targets, twelve proteins oxidized in SVZ-derived NSC after exposure to CysSNO were identified. We further investigated the oxidation and S-nitrosylation status of some of these proteins in both SVZ- and DG-derived NSC, and in the DG after seizures. A group of proteins S-nitrosylated by NO in NSC was identified: hnRNP K, PCNA, 14-3-3,14-3-3 ε, EF-1β and PEBP-1. Indeed, S-nitrosylation of several of them has been confirmed in a murine model of injury-induced neurogenesis, at different time points.
The identified proteins have different functions, but most are related to cell proliferation. 14-3-3 proteins are a family of adaptor proteins that regulate cell signaling pathways. They can activate the ERK/MAPK pathway, which is involved in the proliferative effect of NO in NSC [23], by interacting with c-Raf [46,47]. The isoform 14-3-3 ε has an important role in neurogenesis and neuronal migration [48], and has been described to be S-nitrosylated in mesangial cells [49] and in primary ovine fetoplacental artery endothelial cells [50]. PEBP-1 also regulates the ERK/MAPK pathway, preventing MEK phosphorylation by binding to c-Raf [51], and it increases neuronal differentiation, both in a human neuroblastoma cell line [52] and in adult rat hippocampal progenitor cells [53]. PEBP-1 is regulated by phosphorylation [54], and S-nitrosylation of its Cys168 has been described in SH-SY5Y cells treated with CysSNO [55]. PCNA is necessary for DNA replication and is, therefore, essential for cell proliferation. In response to epidermal growth factor receptor signaling, which is involved in NSC proliferation [56], PCNA is phosphorylated and protected from degradation [57]. PCNA can be S-nitrosylated in Cys81 [58], which blocks its interaction with caspase-9 and promotes apoptosis in a human neuroblastoma cell line [59]. hnRNP K belongs to a family of proteins that bind RNA and are involved in nucleic acid metabolism [60]. hnRNP K is essential for axonogenesis in Xenopus [61] and its inactivation, coupled with activation of the neuronal Hu-p21 pathway, may be essential to the switch from proliferation to differentiation in NSC [62]. The activity of hnRNP K is regulated by several PTM (reviewed in Ref. [63]), but S-nitrosylation is still yet to be described. EF-1α and EF-1β are part of the elongation factor 1 complex, which is responsible for translation elongation. EF-1α delivers aa-tRNAs to the A site of the ribosome [64] and EF-1β replaces EF-1α-bound GDP for GTP [65]. EF-1β and its regulation are essential for the function of EF-1α. Although a role in neurogenesis is not described, EF-1α can interact with and regulate Akt [66,67], a pathway involved in the proliferation of NSC [68,69]. EF-1β can be regulated by phosphorylation [70], but its S-nitrosylation has not been described.
Overall, we confirmed the S-nitrosylation of 14-3-3, specifically of 14-3-3 ε, and PCNA. Moreover, as far as we known, we are the first to describe the S-nitrosylation of PEBP-1, hnRNP K and EF-1β. We also identified Cys97 and Cys98 as potential S-nitrosylation sites for 14-3-3 ε, and Cys145 as the S-nitrosylation site for hnRNP K.
To validate the identified targets, we used a mouse model of excitotoxic brain lesion that has increased proliferation in the dentate gyrus following seizures [71], in a manner dependent on NO produced by inducible NO synthase [23]. Transient S-nitrosylation of hnRNP K, 14-3-3,14-3-3 ε and PEBP-1 was detected in this model. S-nitrosylation was present 1 day (14-3-3, 14-3-3 ε, PEBP-1) or 2 days (hnRNP K) after the induction of epileptic seizures. This occurs before the onset of proliferation of NSC in this model, which is observed 3 days after seizures [23,25]. Therefore, it is possible that, through regulation by NO from inflammatory origin, these proteins can contribute for the process of neurogenesis in this model, especially in proliferation. The fact that S-nitrosylation was observed at different times after seizures may be related to the different functions of these proteins. 14-3-3 proteins seem to have an important role in excitotoxicity. In rat hippocampus, 4 h following KA-induced excitotoxicity, the levels of total 14-3-3 are reduced [72]. This decrease is isoform-selective and dependent on localization, with 14-3-3 γ and 14-3-3 δ levels being decreased in microsome-enriched fractions [73]. A reduction in 14-3-3 protein levels has also been found in rat frontal cortex 24 h after KA treatment, with 14-3-3 γ, 14-3-3 ε, 14-3-3 η and 14-3-3 τ levels being decreased. This effect is accompanied by an increase in ERK phosphorylation levels [74], which is necessary for NSC proliferation. The decrease in 14-3-3 isoforms levels in certain cellular compartments after KA-induced excitotoxicity may be due to degradation by proteolysis [75]. However, it is probable that in others, such as the cytoplasm, regulation of 14-3-3 isoforms may occur through PTM, namely S-nitrosylation. Both 14-3-3 proteins and PEBP-1 regulate the ERK/MAPK pathway and are rapidly S-nitrosylated following seizures. In a previous work, we observed that Ras is S-nitrosylated 2 days after seizures [28], which suggests that, before acting directly on components of the ERK/MAPK pathway, NO reacts with regulators of this signaling pathway. hnRNP K is a substrate of ERK1/2, being accumulated in the cytoplasm upon phosphorylation by this kinase [76]. Thus, it is not surprising that it is S-nitrosylated in a later time point than regulators of the ERK/MAPK pathway. In summary, it is possible that NO first S-nitrosylates PEBP-1 and 14-3-3 proteins, releasing the inhibitory effect of PEBP-1 and promoting the activating effect of 14-3-3 in c-Raf, followed by S-nitrosylation of members of the ERK/MAPK pathway (namely Ras) and of downstream proteins, such as hnRNP K. Although S-nitrosylation of PCNA and EF-1β was not detected in this model, their regulation by S-nitrosylation in other models of neurogenesis is not excluded.
To evaluate S-nitrosylation of the identified targets in DG stem cells, we treated hippocampal stem cells with different concentrations of CysSNO. S-nitrosylation of all the identified proteins was detected in a dose-dependent manner, with higher levels as the concentration of CysSNO increased. The range of CysSNO concentrations used in this study included positive and negative effects on cell proliferation. Treatment with the lowest concentrations of CysSNO (10 and 50 μM) for 1 h enhances the proliferation of SVZ-derived NSC, while treatment with the highest concentration of CysSNO (100 μM) inhibits this process (Ana I. Santos and Inês Araújo, data not shown). Interestingly, the proteins that were S-nitrosylated in the in vivo model showed a tendency to be S-nitrosylated by CysSNO at the proliferative concentrations. Moreover, the proteins whose S-nitrosylation was not detectable in the in vivo model showed lower levels of S-nitrosylation, even at the highest CysSNO concentration used, despite being S-nitrosylated by CysSNO in SVZ-derived NSC. This further indicates a possible physiological relevance of the identified proteins for hippocampal neurogenesis.
The roles of the new identified targets of NO in neurogenesis deserve further investigation. Using S-nitrosylation insensitive mutant proteins with cysteines replaced by other amino acids, the functions of these S-nitrosylation targets can now be investigated on several processes related to neurogenesis.
5. Conclusion
This work allowed the identification of several proteins as targets of S-nitrosylation in NSC and in a murine model of injury-induced neurogenesis. While some were already known to be S-nitrosylated, S-nitrosylation of others was observed for the first time (hnRNP K, 14-3-3 ε, and PEBP-1). These targets may be candidates for NO-induced regulation of neurogenesis in early stages of the formation of newborn neurons after injury.
Declaration of competing interest
The authors declare that no competing financial interests exist.
Acknowledgements
The authors acknowledge the kind gift of HC7 cells by Dr. Fred H. Gage (The Salk Institute for Biological Studies). The excellent assistance of Pedro Marques Vilela in graphical artwork is acknowledged by the authors. Ana Santos and Ana Lourenço were supported by Foundation for Science and Technology, I.P. (FCT, Portugal; fellowships SFRH/BD/77903/2011 and SFRH/BD/79308/2011) and are PhD students of the PhD programme in Biomedical Sciences of the University of Algarve. Antonio Martínez-Ruiz is supported by the ISCIII from the Spanish Government (IS3SNS programme, partially funded by FEDER/ERDF). Inês Araújo is supported by FEDER/ERDF funds via Programa Operacional Factores de Competitividade (COMPETE), by national funds via FCT (grants PTDC/NEU-OSD/0473/2012, PTDC/QUI-QFI/29319/2017) and by regional funds via CRESC ALGARVE 2020 (grant UID/BIM/04773/2019), by the Algarve Biomedical Center (ABC) and Loulé Municipality. This work was supported by the COST action BM1005 (ENOG: European Network on Gasotransmitters), by the Spanish Government (grants PS09/00101, PI12/00875 and PI15/00107 from ISCIII and RTI2018-094203-B-I00 from AEI; co-financed by FEDER/ERDF) and by the Spanish-Portuguese Integrated Action grant PRI-AIBPT-2011-1015/E-10/12. The Proteomics Service of the CBMSO is a member of ProteoRed (PRB3-ISCIII), and is supported by grant PT13/0001/0024 of Spanish Government (cofinanced by FEDER/ERDF).
Contributor Information
Antonio Martínez-Ruiz, Email: amartinezruiz@salud.madrid.org.
Inês Maria Araújo, Email: imaraujo@ualg.pt.
References
- 1.Moncada S., Palmer R.M., Higgs E.A. Biosynthesis of nitric oxide from L-arginine. A pathway for the regulation of cell function and communication. Biochem. Pharmacol. 1989;38:1709–1715. doi: 10.1016/0006-2952(89)90403-6. [DOI] [PubMed] [Google Scholar]
- 2.Murad F. The nitric oxide-cyclic GMP signal transduction system for intracellular and intercellular communication. Recent Prog. Horm. Res. 1994;49:239–248. doi: 10.1016/b978-0-12-571149-4.50016-7. [DOI] [PubMed] [Google Scholar]
- 3.Kerwin J.F., Jr., Lancaster J.R., Jr., Feldman P.L. Nitric oxide: a new paradigm for second messengers. J. Med. Chem. 1995;38:4343–4362. doi: 10.1021/jm00022a001. [DOI] [PubMed] [Google Scholar]
- 4.Garthwaite J., Charles S.L., Chess-Williams R. Endothelium-derived relaxing factor release on activation of NMDA receptors suggests role as intercellular messenger in the brain. Nature. 1988;336:385–388. doi: 10.1038/336385a0. [DOI] [PubMed] [Google Scholar]
- 5.Martínez-Ruiz A., Cadenas S., Lamas S. Nitric oxide signaling: classical, less classical, and nonclassical mechanisms. Free Radic. Biol. Med. 2011;51:17–29. doi: 10.1016/j.freeradbiomed.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 6.Martínez-Ruiz A., Lamas S. S-nitrosylation: a potential new paradigm in signal transduction. Cardiovasc. Res. 2004;62:43–52. doi: 10.1016/j.cardiores.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 7.Martínez-Ruiz A., Araújo I.M., Izquierdo-Álvarez A., Hernansanz-Agustín P., Lamas S., Serrador J.M. Specificity in S-nitrosylation: a short-range mechanism for NO signaling? Antioxidants Redox Signal. 2013;19:1220–1235. doi: 10.1089/ars.2012.5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martínez-Ruiz A., Lamas S. Signalling by NO-induced protein S-nitrosylation and S-glutathionylation: convergences and divergences. Cardiovasc. Res. 2007;75:220–228. doi: 10.1016/j.cardiores.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 9.Santos A.I., Martínez-Ruiz A., Araújo I.M. S-nitrosation and neuronal plasticity. Br. J. Pharmacol. 2015;172:1468–1478. doi: 10.1111/bph.12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakamura T., Tu S., Akhtar M.W., Sunico C.R., Okamoto S., Lipton S.A. Aberrant protein s-nitrosylation in neurodegenerative diseases. Neuron. 2013;78:596–614. doi: 10.1016/j.neuron.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okamoto S.I., Lipton S.A. S-Nitrosylation in neurogenesis and neuronal development. Biochim. Biophys. Acta. 2015:1588–1593. doi: 10.1016/j.bbagen.2014.12.013. 1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Obernier K., Alvarez-Buylla A. Neural stem cells: origin, heterogeneity and regulation in the adult mammalian brain. Development. 2019;146 doi: 10.1242/dev.156059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toda T., Parylak S.L., Linker S.B., Gage F.H. The role of adult hippocampal neurogenesis in brain health and disease. Mol. Psychiatr. 2019;24:67–87. doi: 10.1038/s41380-018-0036-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Covacu R., Danilov A.I., Rasmussen B.S., Hallen K., Moe M.C., Lobell A., Johansson C.B., Svensson M.A., Olsson T., Brundin L. Nitric oxide exposure diverts neural stem cell fate from neurogenesis towards astrogliogenesis. Stem Cell. 2006;24:2792–2800. doi: 10.1634/stemcells.2005-0640. [DOI] [PubMed] [Google Scholar]
- 15.Moreno-López B., Romero-Grimaldi C., Noval J.A., Murillo-Carretero M., Matarredona E.R., Estrada C. Nitric oxide is a physiological inhibitor of neurogenesis in the adult mouse subventricular zone and olfactory bulb. J. Neurosci. 2004;24:85–95. doi: 10.1523/JNEUROSCI.1574-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Packer M.A., Stasiv Y., Benraiss A., Chmielnicki E., Grinberg A., Westphal H., Goldman S.A., Enikolopov G. Nitric oxide negatively regulates mammalian adult neurogenesis. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9566–9571. doi: 10.1073/pnas.1633579100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Contestabile A., Ciani E. Role of nitric oxide in the regulation of neuronal proliferation, survival and differentiation. Neurochem. Int. 2004;45:903–914. doi: 10.1016/j.neuint.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 18.Cheng A., Wang S., Cai J., Rao M.S., Mattson M.P. Nitric oxide acts in a positive feedback loop with BDNF to regulate neural progenitor cell proliferation and differentiation in the mammalian brain. Dev. Biol. 2003;258:319–333. doi: 10.1016/s0012-1606(03)00120-9. [DOI] [PubMed] [Google Scholar]
- 19.Lu D., Mahmood A., Zhang R., Copp M. Upregulation of neurogenesis and reduction in functional deficits following administration of DEtA/NONOate, a nitric oxide donor, after traumatic brain injury in rats. J. Neurosurg. 2003;99:351–361. doi: 10.3171/jns.2003.99.2.0351. [DOI] [PubMed] [Google Scholar]
- 20.Zhu D.Y., Liu S.H., Sun H.S., Lu Y.M. Expression of inducible nitric oxide synthase after focal cerebral ischemia stimulates neurogenesis in the adult rodent dentate gyrus. J. Neurosci. 2003;23:223–229. doi: 10.1523/JNEUROSCI.23-01-00223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo C.X., Zhu X.J., Zhou Q.G., Wang B., Wang W., Cai H.H., Sun Y.J., Hu M., Jiang J., Hua Y., Han X., Zhu D.Y. Reduced neuronal nitric oxide synthase is involved in ischemia-induced hippocampal neurogenesis by up-regulating inducible nitric oxide synthase expression. J. Neurochem. 2007;103:1872–1882. doi: 10.1111/j.1471-4159.2007.04915.x. [DOI] [PubMed] [Google Scholar]
- 22.Tegenge M.A., Rockel T.D., Fritsche E., Bicker G. Nitric oxide stimulates human neural progenitor cell migration via cGMP-mediated signal transduction. Cell. Mol. Life Sci. 2010;68:2089–2099. doi: 10.1007/s00018-010-0554-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carreira B.P., Morte M.I., Inacio A., Costa G., Rosmaninho-Salgado J., Agasse F., Carmo A., Couceiro P., Brundin P., Ambrosio A.F., Carvalho C.M., Araújo I.M. Nitric oxide stimulates the proliferation of neural stem cells bypassing the epidermal growth factor receptor. Stem Cells. 2010;28:1219–1230. doi: 10.1002/stem.444. [DOI] [PubMed] [Google Scholar]
- 24.Reif A., Schmitt A., Fritzen S., Chourbaji S., Bartsch C., Urani A., Wycislo M., Mossner R., Sommer C., Gass P., Lesch K.P. Differential effect of endothelial nitric oxide synthase (NOS-III) on the regulation of adult neurogenesis and behaviour. Eur. J. Neurosci. 2004;20:885–895. doi: 10.1111/j.1460-9568.2004.03559.x. [DOI] [PubMed] [Google Scholar]
- 25.Carreira B.P., Santos D.F., Santos A.I., Carvalho C.M., Araújo I.M. Nitric oxide regulates neurogenesis in the Hippocampus following seizures. Oxid. Med. Cell Longev. 2015 doi: 10.1155/2015/451512. 451512; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.López-Arenas E., Mackay-Sim A., Bacigalupo J., Sulz L. Leukaemia inhibitory factor stimulates proliferation of olfactory neuronal progenitors via inducible nitric oxide synthase. PloS One. 2012;7 doi: 10.1371/journal.pone.0045018. e45018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carreira B.P., Carvalho C.M., Araújo I.M. Regulation of injury-induced neurogenesis by nitric oxide. Stem Cell. Int. 2012 doi: 10.1155/2012/895659. 895659; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santos A.I., Carreira B.P., Izquierdo-Álvarez A., Ramos E., Lourenço A.S., Filipa Santos D., Morte M.I., Ribeiro L.F., Marreiros A., Sánchez-López N., Marina A., Carvalho C.M., Martínez-Ruiz A., Araújo I.M. S-nitrosylation of Ras mediates nitric oxide-dependent post-injury neurogenesis in a seizure model. Antioxidants Redox Signal. 2018;28:15–30. doi: 10.1089/ars.2016.6858. [DOI] [PubMed] [Google Scholar]
- 29.Broniowska K.A., Diers A.R., Hogg N. S-nitrosoglutathione. Biochim. Biophys. Acta. 2013;1830:3173–3181. doi: 10.1016/j.bbagen.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y., Hogg N. The mechanism of transmembrane S-nitrosothiol transport. Proc. Natl. Acad. Sci. U. S. A. 2004;101:7891–7896. doi: 10.1073/pnas.0401167101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santos A.I., Carreira B.P., Nobre R.J., Carvalho C.M., Araújo I.M. Stimulation of neural stem cell proliferation by inhibition of phosphodiesterase 5. Stem Cell. Int. 2014 doi: 10.1155/2014/878397. 878397; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Machado V.M., Morte M.I., Carreira B.P., Azevedo M.M., Takano J., Iwata N., Saido T.C., Asmussen H., Horwitz A.R., Carvalho C.M., Araújo I.M. Involvement of calpains in adult neurogenesis: implications for stroke. Front. Cell. Neurosci. 2015;9:22. doi: 10.3389/fncel.2015.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carreira B.P., Morte M.I., Lourenco A.S., Santos A.I., Inacio A., Ambrosio A.F., Carvalho C.M., Araújo I.M. Differential contribution of the guanylyl cyclase-cyclic GMP-protein kinase G pathway to the proliferation of neural stem cells stimulated by nitric oxide. Neurosignals. 2013;21:1–13. doi: 10.1159/000332811. [DOI] [PubMed] [Google Scholar]
- 34.Gage F.H., Coates P.W., Palmer T.D., Kuhn H.G., Fisher L.J., Suhonen J.O., Peterson D.A., Suhr S.T., Ray J. Survival and differentiation of adult neuronal progenitor cells transplanted to the adult brain. Proc. Natl. Acad. Sci. U. S. A. 1995;92:11879–11883. doi: 10.1073/pnas.92.25.11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palmer T.D., Takahashi J., Gage F.H. The adult rat hippocampus contains primordial neural stem cells. Mol. Cell. Neurosci. 1997;8:389–404. doi: 10.1006/mcne.1996.0595. [DOI] [PubMed] [Google Scholar]
- 36.Martínez-Ruiz A., Lamas S. Detection and identification of S-nitrosylated proteins in endothelial cells. Methods Enzymol. 2005;396:131–139. doi: 10.1016/S0076-6879(05)96013-8. [DOI] [PubMed] [Google Scholar]
- 37.Izquierdo-Álvarez A., Tello D., Cabrera-García J.D., Martínez-Ruiz A. Identification of S-nitrosylated and reversibly oxidized proteins by fluorescence switch and complementary techniques. Methods Mol. Biol. 2018;1747:73–87. doi: 10.1007/978-1-4939-7695-9_7. [DOI] [PubMed] [Google Scholar]
- 38.DeMaster E.G., Quast B.J., Redfern B., Nagasawa H.T. Reaction of nitric oxide with the free sulfhydryl group of human serum albumin yields a sulfenic acid and nitrous oxide. Biochemistry. 1995;34:11494–11499. doi: 10.1021/bi00036a023. [DOI] [PubMed] [Google Scholar]
- 39.Izquierdo-Álvarez A., Ramos E., Villanueva J., Hernansanz-Agustín P., Fernández-Rodríguez R., Tello D., Carrascal M., Martínez-Ruiz A. Differential redox proteomics allows identification of proteins reversibly oxidized at cysteine residues in endothelial cells in response to acute hypoxia. J. Proteomics. 2012;75:5449–5462. doi: 10.1016/j.jprot.2012.06.035. [DOI] [PubMed] [Google Scholar]
- 40.Tello D., Tarín C., Ahicart P., Bretón-Romero R., Lamas S., Martínez-Ruiz A. A “fluorescence switch” technique increases the sensitivity of proteomic detection and identification of S-nitrosylated proteins. Proteomics. 2009;9:5359–5370. doi: 10.1002/pmic.200900070. [DOI] [PubMed] [Google Scholar]
- 41.Shevchenko A., Wilm M., Vorm O., Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 42.Moreno M.L., Escobar J., Izquierdo-Álvarez A., Gil A., Pérez S., Pereda J., Zapico I., Vento M., Sabater L., Marina A., Martínez-Ruiz A., Sastre J. Disulfide stress: a novel type of oxidative stress in acute pancreatitis. Free Radic. Biol. Med. 2014;70:265–277. doi: 10.1016/j.freeradbiomed.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 43.Jorge I., Casas E.M., Villar M., Ortega-Pérez I., López-Ferrer D., Martínez-Ruiz A., Carrera M., Marina A., Martínez P., Serrano H., Cañas B., Were F., Gallardo J.M., Lamas S., Redondo J.M., García-Dorado D., Vázquez J. High-sensitivity analysis of specific peptides in complex samples by selected MS/MS ion monitoring and linear ion trap mass spectrometry: application to biological studies. J. Mass Spectrom. 2007;42:1391–1403. doi: 10.1002/jms.1314. [DOI] [PubMed] [Google Scholar]
- 44.Roepstorff P., Fohlman J. Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed. Mass Spectrom. 1984;11:601. doi: 10.1002/bms.1200111109. [DOI] [PubMed] [Google Scholar]
- 45.Schauwecker P.E., Steward O. Genetic determinants of susceptibility to excitotoxic cell death: implications for gene targeting approaches. Proc. Natl. Acad. Sci. U. S. A. 1997;94:4103–4108. doi: 10.1073/pnas.94.8.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang H.C., Rubin G.M. 14-3-3 epsilon positively regulates Ras-mediated signaling in Drosophila. Genes Dev. 1997;11:1132–1139. doi: 10.1101/gad.11.9.1132. [DOI] [PubMed] [Google Scholar]
- 47.Roberts R.L., Mosch H.U., Fink G.R. 14-3-3 proteins are essential for RAS/MAPK cascade signaling during pseudohyphal development in S. cerevisiae. Cell. 1997;89:1055–1065. doi: 10.1016/s0092-8674(00)80293-7. [DOI] [PubMed] [Google Scholar]
- 48.Toyo-oka K., Wachi T., Hunt R.F., Baraban S.C., Taya S., Ramshaw H., Kaibuchi K., Schwarz Q.P., Lopez A.F., Wynshaw-Boris A. 14-3-3epsilon and zeta regulate neurogenesis and differentiation of neuronal progenitor cells in the developing brain. J. Neurosci. 2014;34:12168–12181. doi: 10.1523/JNEUROSCI.2513-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuncewicz T., Sheta E.A., Goldknopf I.L., Kone B.C. Proteomic analysis of S-nitrosylated proteins in mesangial cells. Mol. Cell. Proteomics. 2003;2:156–163. doi: 10.1074/mcp.M300003-MCP200. [DOI] [PubMed] [Google Scholar]
- 50.Zhang H.H., Lechuga T.J., Chen Y., Yang Y., Huang L., Chen D.B. Quantitative proteomics analysis of VEGF-responsive endothelial protein S-nitrosylation using stable isotope labeling by amino acids in cell culture (SILAC) and LC-MS/MS. Biol. Reprod. 2016;94:114. doi: 10.1095/biolreprod.116.139337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yeung K., Seitz T., Li S., Janosch P., McFerran B., Kaiser C., Fee F., Katsanakis K.D., Rose D.W., Mischak H., Sedivy J.M., Kolch W. Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature. 1999;401:173–177. doi: 10.1038/43686. [DOI] [PubMed] [Google Scholar]
- 52.Hellmann J., Rommelspacher H., Muhlbauer E., Wernicke C. Raf kinase inhibitor protein enhances neuronal differentiation in human SH-SY5Y cells. Dev. Neurosci. 2010;32:33–46. doi: 10.1159/000236595. [DOI] [PubMed] [Google Scholar]
- 53.Sagisaka T., Matsukawa N., Toyoda T., Uematsu N., Kanamori T., Wake H., Borlongan C.V., Ojika K. Directed neural lineage differentiation of adult hippocampal progenitor cells via modulation of hippocampal cholinergic neurostimulating peptide precursor expression. Brain Res. 2010;1327:107–117. doi: 10.1016/j.brainres.2010.02.071. [DOI] [PubMed] [Google Scholar]
- 54.Corbit K.C., Trakul N., Eves E.M., Diaz B., Marshall M., Rosner M.R. Activation of Raf-1 signaling by protein kinase C through a mechanism involving Raf kinase inhibitory protein. J. Biol. Chem. 2003;278:13061–13068. doi: 10.1074/jbc.M210015200. [DOI] [PubMed] [Google Scholar]
- 55.Wang Y.T., Piyankarage S.C., Williams D.L., Thatcher G.R. Proteomic profiling of nitrosative stress: protein S-oxidation accompanies S-nitrosylation. ACS Chem. Biol. 2014;9:821–830. doi: 10.1021/cb400547u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gritti A., Cova L., Parati E.A., Galli R., Vescovi A.L. Basic fibroblast growth factor supports the proliferation of epidermal growth factor-generated neuronal precursor cells of the adult mouse CNS. Neurosci. Lett. 1995;185:151–154. doi: 10.1016/0304-3940(95)11247-t. [DOI] [PubMed] [Google Scholar]
- 57.Wang S.C., Nakajima Y., Yu Y.L., Xia W., Chen C.T., Yang C.C., McIntush E.W., Li L.Y., Hawke D.H., Kobayashi R., Hung M.C. Tyrosine phosphorylation controls PCNA function through protein stability. Nat. Cell Biol. 2006;8:1359–1368. doi: 10.1038/ncb1501. [DOI] [PubMed] [Google Scholar]
- 58.Lam Y.W., Yuan Y., Isaac J., Babu C.V., Meller J., Ho S.M. Comprehensive identification and modified-site mapping of S-nitrosylated targets in prostate epithelial cells. PloS One. 2010;5 doi: 10.1371/journal.pone.0009075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yin L., Xie Y., Yin S., Lv X., Zhang J., Gu Z., Sun H., Liu S. The S-nitrosylation status of PCNA localized in cytosol impacts the apoptotic pathway in a Parkinson's disease paradigm. PloS One. 2015;10 doi: 10.1371/journal.pone.0117546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dreyfuss G., Matunis M.J., Pinol-Roma S., Burd C.G. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 61.Liu Y., Szaro B.G. hnRNP K post-transcriptionally co-regulates multiple cytoskeletal genes needed for axonogenesis. Development. 2011;138:3079–3090. doi: 10.1242/dev.066993. [DOI] [PubMed] [Google Scholar]
- 62.Yano M., Okano H.J., Okano H. Involvement of Hu and heterogeneous nuclear ribonucleoprotein K in neuronal differentiation through p21 mRNA post-transcriptional regulation. J. Biol. Chem. 2005;280:12690–12699. doi: 10.1074/jbc.M411119200. [DOI] [PubMed] [Google Scholar]
- 63.Lu J., Gao F.H. The molecular mechanisms and the role of hnRNP K protein post- translational modification in DNA damage repair. Curr. Med. Chem. 2017;24:614–621. doi: 10.2174/0929867323666161129124122. [DOI] [PubMed] [Google Scholar]
- 64.Carvalho M.D., Carvalho J.F., Merrick W.C. Biological characterization of various forms of elongation factor 1 from rabbit reticulocytes. Arch. Biochem. Biophys. 1984;234:603–611. doi: 10.1016/0003-9861(84)90310-2. [DOI] [PubMed] [Google Scholar]
- 65.Pittman Y.R., Valente L., Jeppesen M.G., Andersen G.R., Patel S., Kinzy T.G. Mg2+ and a key lysine modulate exchange activity of eukaryotic translation elongation factor 1B alpha. J. Biol. Chem. 2006;281:19457–19468. doi: 10.1074/jbc.M601076200. [DOI] [PubMed] [Google Scholar]
- 66.Pecorari L., Marin O., Silvestri C., Candini O., Rossi E., Guerzoni C., Cattelani S., Mariani S.A., Corradini F., Ferrari-Amorotti G., Cortesi L., Bussolari R., Raschella G., Federico M.R., Calabretta B. Elongation Factor 1 alpha interacts with phospho-Akt in breast cancer cells and regulates their proliferation, survival and motility. Mol. Canc. 2009;8:58. doi: 10.1186/1476-4598-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Khwanraj K., Madlah S., Grataitong K., Dharmasaroja P. Comparative mRNA expression of eEF1A isoforms and a PI3K/Akt/mTOR pathway in a cellular model of Parkinson's disease. Parkinsons Dis. 2016 doi: 10.1155/2016/8716016. 8716016; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peltier J., O'Neill A., Schaffer D.V. PI3K/Akt and CREB regulate adult neural hippocampal progenitor proliferation and differentiation. Dev. Neurobiol. 2007;67:1348–1361. doi: 10.1002/dneu.20506. [DOI] [PubMed] [Google Scholar]
- 69.Le Belle J.E., Orozco N.M., Paucar A.A., Saxe J.P., Mottahedeh J., Pyle A.D., Wu H., Kornblum H.I. Proliferative neural stem cells have high endogenous ROS levels that regulate self-renewal and neurogenesis in a PI3K/Akt-dependant manner. Cell Stem Cell. 2011;8:59–71. doi: 10.1016/j.stem.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Venema R.C., Peters H.I., Traugh J.A. Phosphorylation of valyl-tRNA synthetase and elongation factor 1 in response to phorbol esters is associated with stimulation of both activities. J. Biol. Chem. 1991;266:11993–11998. [PubMed] [Google Scholar]
- 71.Parent J.M. Adult neurogenesis in the intact and epileptic dentate gyrus. Prog. Brain Res. 2007;163:529–540. doi: 10.1016/S0079-6123(07)63028-3. [DOI] [PubMed] [Google Scholar]
- 72.Schindler C.K., Shinoda S., Simon R.P., Henshall D.C. Subcellular distribution of Bcl-2 family proteins and 14-3-3 within the hippocampus during seizure-induced neuronal death in the rat. Neurosci. Lett. 2004;356:163–166. doi: 10.1016/j.neulet.2003.11.048. [DOI] [PubMed] [Google Scholar]
- 73.Schindler C.K., Heverin M., Henshall D.C. Isoform- and subcellular fraction-specific differences in hippocampal 14-3-3 levels following experimentally evoked seizures and in human temporal lobe epilepsy. J. Neurochem. 2006;99:561–569. doi: 10.1111/j.1471-4159.2006.04153.x. [DOI] [PubMed] [Google Scholar]
- 74.Smani D., Sarkar S., Raymick J., Kanungo J., Paule M.G., Gu Q. Downregulation of 14-3-3 proteins in a kainic acid-induced neurotoxicity model. Mol. Neurobiol. 2018;55:122–129. doi: 10.1007/s12035-017-0724-y. [DOI] [PubMed] [Google Scholar]
- 75.Henshall D.C., Araki T., Schindler C.K., Lan J.Q., Tiekoter K.L., Taki W., Simon R.P. Activation of Bcl-2-associated death protein and counter-response of Akt within cell populations during seizure-induced neuronal death. J. Neurosci. 2002;22:8458–8465. doi: 10.1523/JNEUROSCI.22-19-08458.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Habelhah H., Shah K., Huang L., Ostareck-Lederer A., Burlingame A.L., Shokat K.M., Hentze M.W., Ronai Z. ERK phosphorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nat. Cell Biol. 2001;3:325–330. doi: 10.1038/35060131. [DOI] [PubMed] [Google Scholar]