Abstract
Classical genetic analysis in Hypsibius exemplaris is a challenge because these animals are parthenogens. The publication of the H. exemplaris genome has facilitated the study of targeted genes by RNA interference (RNAi), a robust mechanism to disrupt gene function. This protocol describes microinjection of double-stranded RNA (dsRNA), using techniques adapted from protocols originally developed in Caenorhabditis elegans. A DNA template (either genomic or cDNA) is used to prepare dsRNA, to which T7 polymerase binding sites are added to the 5’ end of each strand. The dsRNA is injected into adult tardigrades, preferably targeting the gonad or intestine. Injected adults are allowed to recover in spring water, then transferred to culture dishes, or to individual wells of a 96-well plate.
Materials
Reagents
Algae (Clorococcum sp.) (Sciento)
Ethanol (70%)
Ethanol (95%)
Halocarbon 700 oil (Halocarbon Products Inc.)
In vitro transcription kit
-
Levamisole (50 mM in water) (tetramisole hydrochloride; Sigma L9756)
Use of levamisole as an anesthetic is optional.
Sodium acetate (3 M, pH 5.2)
Spring water
Water, nuclease-free
Equipment
-
Capillaries (borosilicate glass, 1.0 mm OD, 0.58 mm ID, inner filament; World Precision Instruments 1B100F-4)
The inner filament promotes backfilling by capillary action
Clay (modeling)
Glass cutter
Glass depression slide (2-wells)
Inverted microscope for microinjections
Microcentrifuge
Microcentrifuge tubes
Micromanipulator
Micropipette Puller
Microscope cover glass (22 mm x 22 mm, No. 1 ½)
Microscope slides (glass)
Nail polish
Petri dish (35 mm)
Petri dish (100 mm)
Spectrophotometer
Stereomicroscope
Tissue Culture Plate, 96-well, round bottom, untreated, sterile (Greiner Bio-One No. 650–185)
Method
Preparation of dsRNA
-
1
Amplify region of interest from genomic DNA or cDNA. We have had success with products in the range of 275 bp – 1000 bp.
-
2
Attach T7 polymerase binding sites to the 5’ end of each strand via PCR. Primers should start with the T7 sequence [5’-GATAATACGACTCACTATAGGG-3’], followed by 15 – 18 nucleotides of gene-specific sequence.
-
3
Use 1 μg of the final DNA product from Step 2 as the template to synthesize dsRNA via an in vitro transcription kit, following manufacturers’ directions.
-
4
To purify dsRNA, add 0.1 volume of 3M sodium acetate and 2.5 volumes of 95% ethanol to the tube from Step 3, and incubate on ice for 5 minutes. Spin at maximum speed in a microcentrifuge for 10 minutes.
-
5
Remove the supernatant, then wash pellet with 0.5 ml of ice-cold 70% ethanol. Spin at maximum speed in a microcentrifuge for 5 minutes. Remove supernatant.
-
6
Air dry pellet at room temperature, then resuspend dsRNA in nuclease-free water.
-
7
Determine the concentration of dsRNA via spectrometry (the extinction coefficient for dsRNA is 45).
-
8
Store dsRNA solutions at −70°C.
Preparation of needles for microinjection
-
9
Use a micropipette puller to pull needles to a fine taper from borosilicate glass tubing (1.0 mm OD, 0.58 mm OD). Follow the manufacturer’s instructions to obtain needles that taper quickly into a sharp point, with a bore of about 1 μm.
To achieve a good needle, it may be necessary to adjust the pulling force and/or the temperature of the heating filament. If the temperature and/or the pulling force are too low, the needle may have a long taper that is too flaccid to penetrate the tardigrade’s cuticle. Conversely, if either parameter is too high, then the needles could result in a bore size that is too large for injections.
-
10
Store needles on a long piece of clay molded into the bottom of a 100 mm Petri dish and keep covered until needed.
-
11
For microinjection experiments, dilute dsRNA to a final concentration of 1 μg / μl in nuclease-free water. Centrifuge briefly to clear the supernatant.
-
12
Back-fill injection needle with dsRNA, and allow dsRNA to flow to tip of needle by gravity.
Preparation of animals for microinjection
-
13Prepare the injection chamber:
- Use a glass cutter to cut off the corner of a 22mm x 22mm, No. 1 ½ microscope cover glass.
- Place the triangular piece in the middle of a standard glass microscope slide, such that one straight edge is parallel to the short edge of the slide (Figure 1).
- Secure the cover glass to the microscope slide with clear nail polish.
-
Chambers can be prepared in advance and stored in a microscope slide box.The purpose of the cover glass is to create a solid barrier against which to brace the animals. Imperfections in the cover glass can sometimes create a gap between the cover glass and the microscope slide large enough for the animals to slip under. Mounting corners of cover glass, rather than a whole piece, appears to result in fewer gaps.
-
14
Using a stereomicroscope, transfer adult tardigrades into one well of a deep 2-well depression slide. Allow animals to excrete any material from their intestines, changing spring water several times (this step can take up to 30 minutes).
-
15
[Optional] Anesthetize animals in levamisole immediately prior to performing injections by replacing spring water with 10 mM levamisole (prepared in spring water).
This step helps to minimize animal movement during microinjections (Tenlen et al., 2013) but is not necessary (see Boothby et al., 2017).
-
16
Apply a drop of Halocarbon 700 oil to the straight edge of the cover glass on the injection chamber.
-
17
Using a stereomicroscope, transfer several animals to the oil drop on the prepared injection chamber. Position each animal lengthwise along the straight edge of the cover glass. Remove any residual water.
We typically inject 5 – 10 animals at a time; animals appear to be unaffected by exposure to halocarbon oil.
-
18
Mount the microinjection needle in the needle holder of the micromanipulator.
-
19
Position the injection chamber on the inverted microscope and orient the micromanipulator so that the needle is perpendicular to the tardigrade (Figure 1). Under the lowest-power objective, bring both the animal and needle in focus.
-
20
Switch to the 20x objective and lower the needle into position. To break the needle, use the micromanipulator controls to bring the needle into the same focal plane as the cover glass edge, then gently brush (stroke) the needle against the coverslip edge.
-
21
Using the micromanipulator fine controls, adjust the needle so that it is in the same focal plane as the animal. Bring the needle in contact with midpoint of the body (so that it is targeting the gonad/intestine) and then gently tap on the left side of the microscope stage to force the needle into the body. Delivery of dsRNA can be confirmed by slight swelling at the site of injection. Depending on the bore size, one or two taps of the foot pedal should be sufficient.
-
22
Use the micromanipulator controls to withdraw the needle, and move to the next animal.
Figure 1:
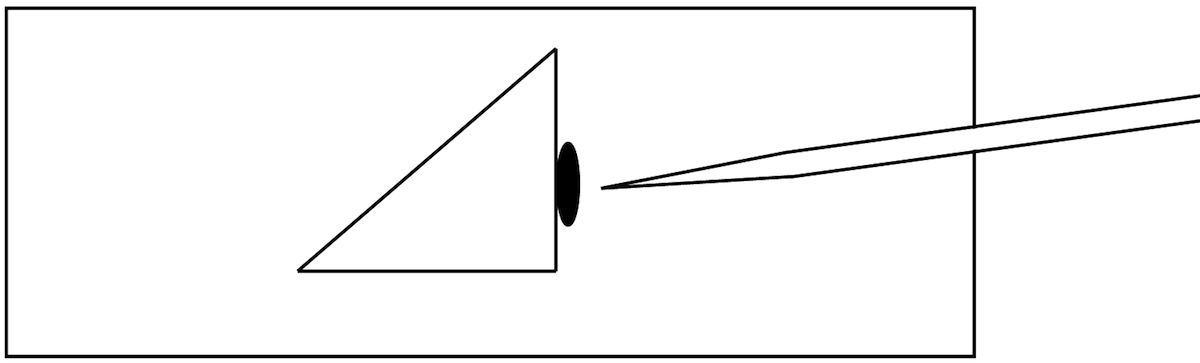
Schematic of injection chamber.
Recovery and monitoring of animals
-
23
After all animals on a slide have been injected, move the slide back to the stereomicroscope. Add a drop of spring water over each animal, then transfer the animals to the 2nd well of the depression slide.
-
24
To monitor individual animals and their progeny, prepare a 96-well tissue culture plate (untreated, sterile) by adding 100 μl spring water and 5 μl green algae to each well. Transfer one animal to each well.
-
25
Animals may instead be transferred in groups to 35 mm Petri dishes, with a layer of spring water and one to two drops of green algae.
-
26
All injected animals should be maintained at room temperature, and water and food levels monitored regularly.
Discussion
Parameters established for microinjection in C. elegans, including needle conditions, appear to also be effective in tardigrades (Fire, 1986; Fire et al., 1998). Injection of dsRNA into the tardigrade gonad, intestine or coelomic cavity results in loss-of-function phenotypes observed in progeny (Tenlen et al., 2013). This ability of RNAi to cross tissue boundaries was initially observed in C. elegans (Fire et al., 1998). Either genomic DNA or cDNA can be used as templates to synthesize dsRNA. However, the choice of DNA template may need to be empirically determined for each experiment. For some genes, such the H. exemplaris actin homolog, similar results were obtained using dsRNA transcribed from either genomic DNA or cDNA templates. Conversely, for other genes, embryonic phenotypes were observed only for injection of dsRNA from cDNA templates.
The recovery and reproductive success of tardigrades following injection appears to be dependent upon multiple environmental conditions (such as oxygen content of water, humidity and amount/quality of algae food source). For each experiment, several controls should be set up at the same time. Negative controls include both uninjected animals and animals injected with water or dsRNA targeting green fluorescent protein (Tenlen et al., 2013). Hd-actin is recommended as a positive control since injection of Hd-act dsRNA results in highly penetrant lethality in progeny, with reproducible cytokinesis defects (Tenlen et al., 2013).
In our experiments, the penetrance of loss-of-function phenotypes varies based on the targeted gene, and on brood number. Reproduction and molting are linked in tardigrades, occurring approximately every 4 – 5 days. When injection of dsRNA resulted in a phenotype, the penetrance was always highest in the first brood of embryos produced after injection. In subsequent broods, the penetrance decreased substantially (Tenlen et al., 2013). As a result, it may be necessary to increase the number of adults injected to ensure an adequate population of embryos to analyze.
Acknowledgments
J.R.T. thanks Dr. Bob Goldstein, in whose lab this protocol was developed. J.R.T. was supported by grants from the National Institutes of Health (K12GM000678), the NIH-funded UNC Developmental Biology Training Program (T32HD046369-03), and the M. J. Murdock Charitable Trust.
References
- Boothby TC, Tapia H, Brozena AH, Piszkiewicz S, Smith AE, Giovannini I, Rebecchi L, Pielak GJ, Koshland D, Goldstein B. 2017. Tardigrades use intrinsically disordered proteins to survive desiccation. Mol Cell 65: 975–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A. 1986. Integrative transformation of Caenorhabditis elegans. EMBO J 5: 2673–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu SQ, Montgomery MK, Kostas SA, Driver SE, Mello CC. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Tenlen JR, McCaskill S, Goldstein B. 2013. RNA interference can be used to disrupt gene function in tardigrades. Dev Genes Evol 223: 171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]


