Abstract
The protozoan Plasmodium vivax is responsible for 42% of all cases of malaria outside Africa. The parasite is currently largely restricted to tropical and subtropical latitudes in Asia, Oceania, and the Americas. Though, it was historically present in most of Europe before being finally eradicated during the second half of the 20th century. The lack of genomic information on the extinct European lineage has prevented a clear understanding of historical population structuring and past migrations of P. vivax. We used medical microscope slides prepared in 1944 from malaria-affected patients from the Ebro Delta in Spain, one of the last footholds of malaria in Europe, to generate a genome of a European P. vivax strain. Population genetics and phylogenetic analyses placed this strain basal to a cluster including samples from the Americas. This genome allowed us to calibrate a genomic mutation rate for P. vivax, and to estimate the mean age of the last common ancestor between European and American strains to the 15th century. This date points to an introduction of the parasite during the European colonization of the Americas. In addition, we found that some known variants for resistance to antimalarial drugs, including Chloroquine and Sulfadoxine, were already present in this European strain, predating their use. Our results shed light on the evolution of an important human pathogen and illustrate the value of antique medical collections as a resource for retrieving genomic information on pathogens from the past.
Keywords: malaria, Plasmodium vivax, phylogenetics, ancient DNA, population genetics
Introduction
Malaria is a leading cause of infectious disease, responsible for an estimated 200 million infections annually, and around 429 000 fatal cases (World Health Organisation 2017). The disease is caused by several species of parasitic protozoans from the genus Plasmodium, which are transmitted by various species of mosquitoes from the genus Anopheles. Two species in particular—Plasmodium falciparum and Plasmodium vivax—are responsible for the majority of human infections worldwide. Although P. falciparum causes 99% of malaria deaths globally, P. vivax is the etiological agent of 42% of all cases outside of Africa (Gething et al. 2011; World Health Organisation 2017).
Today, endemicity of the genus Plasmodium is restricted to tropical and subtropical latitudes, spanning large regions of East and South East Asia, sub-Saharan Africa, Central and South America, and Melanesia (Battle et al. 2012, 2019; Howes et al. 2016; Weiss et al. 2019). However, malaria was historically present in most of Europe, from the Mediterranean to the southern shores of the Baltic Sea, and from southern Britain to European Russia (Huldén et al. 2005). Malaria was eradicated from all European countries during the second half of the 20th century (Hay et al. 2004), with Spain being one of its last footholds from which it was only declared officially eradicated in 1964 (Pletsch 1965). Nevertheless, even though Plasmodium is currently largely absent from Europe, its potential re-emergence has been identified as a plausible consequence of climate change (Petersen et al. 2013; Zhao et al. 2016).
Whilst historically being described as the “benign” form of malaria, P. vivax is increasingly recognized as a significant cause of disease and mortality (Tjitra et al. 2008; Price et al. 2009; Lacerda et al. 2012; Baird 2013). In stark contrast to P. falciparum, P. vivax is capable of producing recurrent malaria episodes from a single infection due to its resistant latent forms known as hypnozoites (Gonzalez-Ceron et al. 2013; Adekunle et al. 2015). This capacity also allows P. vivax to maintain itself in temperate climates, resting in a dormant state in the cold months when anopheline populations are in diapause, and creating a persistent presence of parasite reservoirs which can facilitate widespread transmission and recurrent long-term infections (Krotoski 1985; Gething et al. 2011; White 2011). Low parasite densities of P. vivax in mixed infections (Mayxay et al. 2004; Moreira et al. 2015) and the suggested high proportion of hypnozoite-derived clinical incidence (Price et al. 2009; Howes et al. 2016) means the global prevalence of P. vivax is likely systematically underestimated in comparison with the better-studied P. falciparum.
Plasmodium vivax is widely considered to have emerged in sub-Saharan Africa, a region in which it is now at low prevalence (Liu et al. 2014; Gunalan et al. 2018; Twohig et al. 2019). From there, it is thought to have spread globally through a complex pattern of migration events by hitchhiking with its human host, as humans moved out of Africa (Culleton et al. 2011; Hupalo et al. 2016). The analysis of a geographically diverse data set of 941 P. vivax mitochondrial DNA (mtDNA) genomes detected genetic links between strains from the Americas to those from Africa and South Asia, although potential contributions from Melanesia into the Americas were also identified (Rodrigues et al. 2018). However, the role of the worldwide colonial expansion of European countries in the global dispersal of P. vivax remains largely unknown, mainly due to the lack of available sequenced nuclear genomes from now-extinct European strains.
The recent discovery of a set of historic microscope slides with bloodstains from malaria-affected patients provides major opportunities to shed light on the evolution of P. vivax. The slides were prepared between 1942 and 1944 in the Ebro Delta (Spain), an area where the disease was transmitted by the mosquito species Anopheles atroparvus, a member of the Anopheles maculipennis complex—still very common in the region—and provided the first retrieval of genetic material from historical European P. vivax (Gelabert et al. 2016) as well as a partial P. falciparum genome (de-Dios et al. 2019). The complete mtDNA genome of this sample showed a close genetic affinity to the most common strains of present-day South and Central America, suggesting their introduction into the Americas was linked to Spanish colonial-driven transmission of European strains. However, mtDNA is a maternally inherited single locus and, in comparison with the entire genome, has limited power to reconstruct complex evolutionary histories.
In this paper, we extend these previous findings, by reporting the complete genome of an extinct European P. vivax obtained from the historical Ebro Delta microscope slides. Together with the recent publication of a more accurately annotated P. vivax reference genome (PvP01) (Auburn et al. 2016), this European genome provides new opportunities to resolve the historical dispersals of this parasite using genome-wide data. The availability of this complete genome also allows direct estimation of an evolutionary rate for P. vivax, making it possible to date the clustering of the historic European strain with modern global strains. Furthermore, it enables us to ascertain the presence of some resistance alleles prior to the introduction of most anti-malaria drugs. This information is critical for further investigations into the evolution of the parasite, as well as predicting future emergences of drug-resistance mutations.
Results
We generated shotgun Illumina sequence data from four archival blood slides derived from malaria patients sampled between 1942 and 1944 in Spain’s Ebro Delta. The majority of reads that mapped to P. vivax (89.09%) derived from a single slide dated to 1944. In total, 481,245 DNA reads (0.44% of the total reads generated) mapped to P. vivax, yielding a composite genome, we term Ebro-1944, at 1.4× coverage (1.28× deriving from the newly analyzed 2017 slide), and spanning 66.42% of the (PvP01) reference. The mtDNA genome was recovered at 32× coverage (supplementary figs. 1–5 and tables 1–3, Supplementary Material online). Although working with low coverage ancient genomes is challenging, the fact that our genome is haploid and displays low levels of postmortem sequencing errors suggests it can be reliably used in most evolutionary analyses.
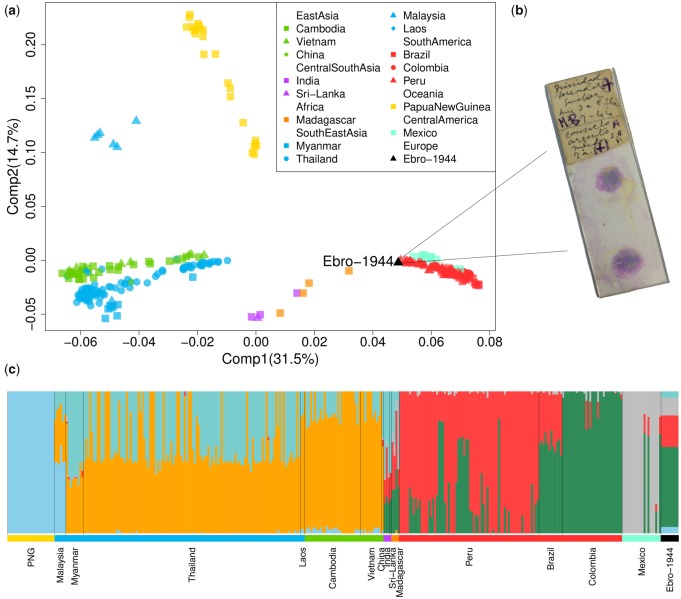
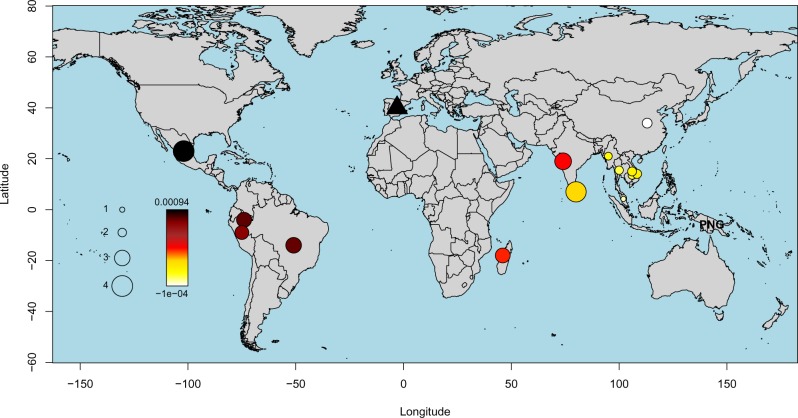
To explore the phylogeographic affinities of the eradicated European strain we performed several population genomics analyses (supplementary figs. 6–12 and table 4, Supplementary Material online). A principal component analysis (PCA) applied to a global data set of P. vivax showed strong geographic structure, with clusters separating 1) South East Asian/East Asian strains; 2) Oceanian strains (those from Malaysia were placed between the two clusters); 3) Indian and Madagascan strains; and 4) those sampled from Central/South America (fig. 1a and b). The European sample (labeled Ebro-1944) falls at one end of the latter cluster, on an axis of variation shared by strains from Mexico, Brazil, Colombia, and Peru.
Fig. 1.
(a) PCA of the historic Ebro-1944 sample together with a geographically diverse set of modern Plasmodium vivax strains. (b) Example microscopy slide stained with the blood of patient’s infected with malaria from the Ebro Delta, Spain, in the 1940s. (c) Unsupervised ADMIXTURE clustering analysis at K = 6. Samples are arranged by geographic region and colored as in (a).
Model-based clustering implemented in an unsupervised ADMIXTURE analysis (fig. 1c and supplementary fig. 8, Supplementary Material online) provided qualitatively consistent inferences to those observed by PCA, with one ancestry component maximized in Oceanian samples, two largely shared by East and South East Asian samples, and three further components which largely differentiate samples from South and Central America. The ancestry of Ebro-1944 is mostly modeled by the three components identified in samples from South America (∼56% Colombia-like and ∼22% Peru-like) and Central America (∼12% Mexico-like), although a minor proportion of ancestry is shared with both South East Asian (∼5%) and Oceanian (∼5%) samples.
To formally test these suggested relationships, we calculated f4 statistics (Patterson et al. 2012) of the form (Plasmodium cynomolgi, Ebro-1944; X, Y), where X and Y are tested for all combinations of samples from 15 worldwide locations (fig. 2). Plasmodium cynomolgi was selected as an outgroup as it represents the closest nonhuman infecting Plasmodium species (Tachibana et al. 2012). This statistic is designed to quantify the covariance in allele frequency differences between P. cynomolgi and Ebro-1944 relative to P. vivax from sampled worldwide locations (X and Y), with a more positive f4 value indicating a closer relationship of Ebro-1944 to samples from Y relative to X. Using this framework and testing all possible topological relationships, Ebro-1944 was found to share significantly more derived alleles with Central and Southern American strains compared with those sampled from South East and East Asia (fig. 2 and supplementary fig. 9, Supplementary Material online). These results collectively support the presence of a cline of ancestry stretching from Europe to the Americas, with our eradicated European strain showing strong phylogenetic affinity to modern P. vivax samples from Mexico, Brazil, and Peru.
Fig. 2.
f4-Values inferred under the test relationship (Plasmodium cynomolgi, Ebro-1944; Papua New Guinea [PNG], Y), where Y iterates through the geographic sampling locations of our included strains. The color scale provides the value of the f4 statistic with the significance (absolute z score), assessed through block jackknife resampling, provided by the circle size. A more positive f4 value indicates a closer relationship of Ebro-1944 to Y relative to PNG.
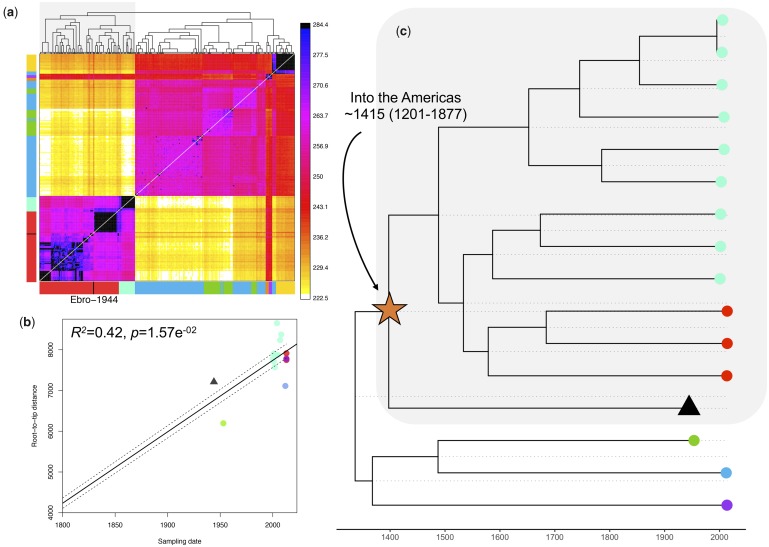
Additional evidence was also obtained using an unrelated method designed to cluster global samples based on inferred patterns of haplotype sharing (Lawson et al. 2012). Considering haplotype variation rather than allele frequency differences increases power to resolve fine-scale genetic structure (Leslie et al. 2015) and is robust to potential single nucleotide polymorphism (SNP) calling errors (Conrad et al. 2006). Haplotype-based clustering grouped Ebro-1944 with samples from South and Central America (fig. 3a). This result was robust to the inclusion of different samples in the data set, to moderate imputation, and remained consistent when either uncorrelated sites or linked sites were considered (supplementary figs. 10–12, section 4, and methods, Supplementary Material online).
Fig. 3.
(a) CHROMOPAINTER’s inferred counts of matching DNA genome wide that each of the 104 inferred clusters (columns) is painted by each of the 104 clusters (rows). The tree at top shows fineSTRUCTURE’s inferred hierarchical merging of these 104 clusters and the colors on the axes give the continental region and population to which strains in each cluster are assigned. Ebro-1944 is depicted in black and clusters with the sample from Peru and Brazil. (b) Root-to-tip distances of our included Plasmodium vivax strains correlated with the date of isolation. The regression was significant following 1,000 random permutations of sampling date. (c) Tip-dated maximum clade credibility phylogenetic tree obtained with BEAST 2. The mean posterior probability for the time to the most recent common ancestor between the historical European strain and the American strains is indicated. Tips are colored as in fig. 1a.
Given the strong phylogeographic affinity of Ebro-1944 to P. vivax currently in circulation in the Americas, we tested whether the divergence of Ebro-1944 from American strains is better explained by a deep split, for example at the time of the original human settlement of the Americas at least 15,000 years ago, or is more consistent with a recent introduction to the Americas. Genomes obtained from historical or ancient materials provide unique opportunities to calibrate phylogenetic trees, by directly associating sampling dates with the sequences representing the phylogeny tips (terminal nodes). These in turn enable inference of divergence times and mutation rates without the need for any other age-related external data (Rieux and Balloux 2016). Therefore, to infer the temporal relationship of Ebro-1944 to strains from the Americas, we included our historic sample together with 15 closely related publicly available genomes sampled over a range of time periods (supplementary table 5, Supplementary Material online). We selected strains predominantly from the Americas, with three additional genomes included from India, Myanmar, and North Korea to root the topology and increase the time span of our data set.
To account for the possible confounding effect of genetic recombination, we filtered the resulting alignment for a set of high confidence congruent SNPs. Specifically, we removed all homoplasic that is SNPs in the alignment in conflict with the maximum parsimony phylogeny (see Materials and Methods). This approach identifies, with no required prior knowledge, regions of the genome that are hypervariable, deriving from recombination or mixed infections, as well as filtering SNPs which may have been erroneously called due to low sequence quality or postmortem damage. The resulting alignment exhibited a significant positive correlation between the root-to-tip phylogenetic distances of a maximum likelihood phylogeny and the time of sampling, indicating the presence of detectable temporal accumulation of de novo mutations within the timescale of our data set (fig. 3b, supplementary fig. 13, Supplementary Material online).
Mutation rates were subsequently estimated using the Bayesian phylogenetic tool BEAST2 (Bouckaert et al. 2014) testing a range of demographic and clock rate priors. We estimated the mutation rate over the tested alignment to 5.57E−7 substitution/site/year [HPD 95% 2.75E−8–1.06E−6]. Though a broad estimate, we obtained low values (<0.15) of the standard deviation of the uncorrelated log-normal relaxed clock (ucld.stdev), suggesting little variation in rates between branches. This approximation enabled us to infer that the historical Ebro-1944 genome shares a common ancestor with strains in the South-American cluster dating to the 13th–19th centuries (mean 1415; HPD 95% 1201–1877 CE) (fig. 3c, supplementary fig. 14 and table 6, Supplementary Material online).
A number of mutations conferring resistance to antimalarial drug treatments developed in the later decades of the 20th century have been identified in P. vivax. For instance, mutations in the pvdhfr gene are known to be involved in resistance to pyrimethamine (de Pecoulas et al. 1998; Imwong et al. 2001, 2003; Huang et al. 2014) whilst mutations at the pvdhps gene confer resistance to sulfadoxine (Korsinczky et al. 2004; Menegon et al. 2006; Hapuarachchi et al. 2009). Other genes, including pvmdr1 (Brega et al. 2005; Sá et al. 2005; Barnadas, Tichit, et al. 2008), CRT (Suwanarusk et al. 2007), or pvmrp1 (Dharia et al. 2010), are thought to be involved in chloroquine resistance. Plasmodium vivax populations also exhibit high genetic diversity in genes related to immune evasion and host infectivity, including MSP10, MSP7, and CLAG. We annotated 4,800 SNPs in the Ebro-1944 genome (supplementary table 7, Supplementary Material online). Of these, 1,195 are missense mutations, which represent 60.6% of the genic mutations. Similar ratios have been reported in other P. vivax strains (Hupalo et al. 2016; Pearson et al. 2016; de Oliveira et al. 2017).
Ebro-1944 carries the derived allele in three SNPs located in genes functionally associated with antimalarial drug resistance. Two of these, Val1478Ile and Thr259Arg (Dharia et al. 2010; de Oliveira et al. 2017) are in the pvmdr1 gene and another, Met205Ile (Hapuarachchi et al. 2009), occurs in the pvdhps gene (supplementary tables 8 and 9, Supplementary Material online). We also identified a previously undescribed mutation (Leu623Arg) in CLAG, a gene associated with host infectivity (Gupta et al. 2015). In addition, we screened multiple loci (N = 516) that have previously been reported to exhibit strong signals of recent natural selection in a geographically diverse set of modern P. vivax samples (Hupalo et al. 2016). Some of these regions encompass genes previously known to be involved in antimalarial drug resistance, including three with strong experimental validation of resistance phenotypes: pvmdr1, dhfr, and dhfps (Haldar et al. 2018). Our historical genome has 355 of these positions covered by at least two reads, of which Ebro-1944 exhibits the ancestral allele in 349 (supplementary table 10, Supplementary Material online). Therefore, only six of the 355 derived variants with high FST values (including the previously mentioned Met205Ile variant at pvdhps gene) were present in the historical European sample, suggesting a rapid accumulation of resistance conferring mutations in more recent strains.
Discussion
Our genome-wide analyses of a historic European P. vivax nuclear genome confirm the inference, previously based solely on mtDNA, that extinct European P. vivax are closest genetically to strains currently in circulation in Central and South America (Gelabert et al. 2016). Historical accounts of the presence of tertian (P. vivax) malaria in Europe date to at least Classical Greece (De Zulueta 1973; Carter 2003), suggesting that the most parsimonious explanation is an introduction of P. vivax from Europe into the Americas and not the other way around. A migration event from Europe into the Americas is further supported by the European Ebro-1944 P. vivax strain falling as an out group to all the strains from the Americas (fig. 3c). Finally, the estimated mean age of divergence in the 15th century between Ebro-1944 and strains in circulation in the Americas (fig. 3c) is consistent with an introduction of P. vivax malaria into the Americas by European colonists.
Whether any agent of malaria was present in the Americas prior to European contact (1492) has been debated (Carter 2003; Hume 2003; de Castro and Singer 2005). It is relatively unlikely that any malarial parasites would have survived the journey across the Bering strait during the initial peopling of the Americas some 15,000 years ago (Waters et al. 2018) due to the absence of mosquito vectors at these high latitudes allowing the parasites to fulfill their life cycles (Tanabe et al. 2010). However, accounts of the therapeutic use by Incas of cinchona tree bark, from which quinine derives, have been interpreted as suggestive of malaria being present in the Americas pre-Columbian contact (Escardo 1992). Though, its use may have been motivated by the effectiveness of quinine in the treatment of other fever-causing illnesses.
Older split times between P. vivax lineages from the Americas and the rest of the world than the one we inferred based on whole-genome sequences have been suggested from the analysis of mtDNA genomes (Taylor et al. 2013; Rodrigues et al. 2018). Though, we note that, even when accounting for the posterior uncertainty around our inferred mutation rate, all possible temporal estimates place the split time between the European and American strains as incompatible with an introduction of P. vivax into the Americas alongside the first humans to colonize the continent. Our results are therefore highly supportive of an introduction of P. vivax to the Americas during the European colonial period, with our range also consistent with the transatlantic slave trade between Africa and Spanish and Portuguese-run ports in Central and South America. Our analyses of the nuclear genome further point to a minor genetic component in American strains shared with strains from Madagascar, India, and Sri Lanka (figs. 1c and 3a). We interpret this as likely evidence for secondary genetic introgression into the American P. vivax population by lineages from different regions of the world, which would be consistent with the high P. vivax mtDNA diversity previously described in the Americas (Taylor et al. 2013; Hupalo et al. 2016; de Oliveira et al. 2017; Rodrigues et al. 2018).
In this work, we restricted the phylogenetic dating to the migration of P. vivax into the Americas. There is currently no consensus over the age of the most recent common ancestor of all extant, worldwide P. vivax strains. Some previous estimates inferred the origin of P. vivax to around 5,000 or 10,000 years ago (Carter 2003; Leclerc et al. 2004; Lim et al. 2005), whereas far older dates have been proposed, including 45,000–81,000 years (Escalante et al. 2005) or even 53,000–265,000 years (Mu et al. 2005). An extrapolation of our inferred evolutionary rates to the global diversity of extant P. vivax strains would point to a recent origin for the parasite. Though, formally estimating the age of P. vivax through a phylogenetic “tip-dating” approach poses a series of analytical challenges. For example, the homoplasy screening method we employed to exclude homoplasic caused by mixed infections and likely genetic recombination is currently not computationally tractable for a data set comprising a large number of globally sourced whole P. vivax genomes. Possible solutions to this problem may arise through optimization of sequencing protocols to generate higher quality Plasmodium whole-genome sequences together with the development of downstream bioinformatics approaches designed to propagate genotype calling uncertainty and deconvolve mixed infections (Zhu et al. 2018). This should be aided by the generation of further high-quality reference genomes across the Plasmodium genus, for example using long-read sequencing technology (Auburn et al. 2016; Pasini et al. 2017; Gilabert et al. 2018; Otto et al. 2018).
Reconstruction of the phylogeographic relationships between P. vivax strains is complicated further by mounting evidence for the zoonotic potential of P. vivax and P. vivax-like strains; with host-jumps likely having occurred several times in the parasite’s evolutionary history (Prugnolle et al. 2013; Liu et al. 2014, 2017; Loy et al. 2017). The discovery of the platyrrhine protozoa, Plasmodium simium, as morphologically (Ott 1967) and genetically (Leclerc et al. 2004; Escalante et al. 2005; Lim et al. 2005) indistinguishable from P. vivax suggests very recent host transfers between South American monkeys and humans; in some cases responsible for the incidence of zoonotic malarial disease (Brasil et al. 2017; Buery et al. 2017). This raises important questions as to what should be considered as the host-range of P. vivax and queries what species delineation should be applied in comparative genomics studies. Recurrent host jumps of P. vivax lineages into humans from animal reservoirs, with subsequent demographic expansions and possible lineage replacements, may point to a recent age for the ancestor of all extant P. vivax strains, despite now-extinct lineages of the parasite having likely plagued humans for far longer.
Malaria is widely believed to have exerted one of the strongest selective forces on the human genome (Hedrick 2012). Well-known examples of selection against malaria include protective mutations at the HBB gene that give rise to resistant isoforms of proteins such as HbS and HbE in African and Asian populations, respectively, and mutations at the G6PD gene which are broadly spread in African populations and are also present in the Mediterranean (Tishkoff et al. 2001; Kwiatkowski 2005; Howes et al. 2012). One of the best-known examples of directional selection is the FY*0 Duffy blood negative genotype that confers resistance to P. vivax which is close to fixation in sub-Saharan Africa but essentially absent in other regions of the world. The protein is the key invasion receptor for the human malarial parasites P. vivax, Plasmodium knowlesi, and the simian malarial parasite P. cynomolgi (Kosaisavee et al. 2017). Exposure to an ancestor of P. vivax may have led to the sweep of the FY*0 allele in sub-Saharan Africa some 40,000 years ago and may represent the fastest known selective sweep for any human gene (McManus et al. 2017).
Though, given the relatively mild clinical symptoms of modern P. vivax, it is not inconceivable that the selective forces that led to the fixation of the FY*0 allele in sub-Saharan Africa may have been caused by another malaria parasite. A further complicating factor stems from increasing evidence accumulating for previously unrecognized endemic P. vivax circulating in human populations from sub-Saharan Africa, including in Duffy-negative hosts (Gunalan et al. 2018; Twohig et al. 2019). The presence of endemic P. vivax malaria in sub-Saharan Africa may suggest that the FY*0 allele may confer only partial protection against P. vivax malaria. Our results, which point to a recent, post-European contact exposure of Native American populations to P. vivax malaria, would not have had the time to drive the emergence and spread of resistance alleles comparable with those observed in Africa and Europe. Consistently, to date, no known malaria-resistance variants have been identified in Native Americans (Hume 2003; Kwiatkowski 2005).
Irrespective of its role in the past, the rapid spread of P. vivax strains resistant to antimalarial drugs is an area of increasing concern. Initially, chloroquine was established as the main therapy against P. vivax infections in 1946 (Most and London 1946; Baird 2004). It was a well-tolerated and effective treatment until resistance appeared in the late 1980s and spread through the entirety of the endemic range of P. vivax (Rieckmann et al. 1989). However, despite extensive drug resistance within present-day P. vivax populations, caused by a variety of resistance loci in multiple genes (Hupalo et al. 2016), chloroquine–primaquine combined therapy remains the most commonly prescribed treatment (Phillips et al. 1996), as few other therapeutic strategies are available. The increase in frequency of drug resistant strains is thus a significant public health threat with major human and economic costs.
Our historical sample predates the use of all anti-malaria drugs, with the exception of quinine which was introduced in Europe as early as 1683 (Achan et al. 2011). Ebro-1944 carries the ancestral allele in an overwhelming number of SNPs (99.3%) known to have undergone selection in modern strains, including those associated with drug resistance in genes such as DHFR-TS (de Pecoulas et al. 1998; Leartsakulpanich et al. 2002; Imwong et al. 2003; Ganguly et al. 2014; Huang et al. 2014) and MDR1 (Brega et al. 2005; Sá et al. 2005; Barnadas, Ratsimbasoa, et al. 2008; Orjuela-Sánchez et al. 2009) (supplementary tables 8 and 9, Supplementary Material online). Conversely, Ebro-1944 carries three variants in the pvmdr1 and pvdhps genes that are plausible drug-resistance candidates against sulfadoxine and chloroquine. The presence of these alleles in Ebro-1944 might reflect standing variation in historical P. vivax populations for alleles providing resistance to modern antimalarial drugs. Alternatively, these could have been selected for by the historical use of quinine.
Our study stresses the value of old microscopy slides and, more generally, of antique medical collections, as a unique and under-used resource for retrieving genomic information on pathogens from the past, including eradicated strains that could not be studied from contemporary specimens. For example, in addition to the results reported here, we also retrieved a partial P. falciparum genome from the same set of slides, which allowed us to demonstrate a stronger phylogeographic affinity of the extinct European P. falciparum lineage to present-day strains in circulation in central south Asia, rather than Africa (de-Dios et al. 2019). We note that the slides we analyzed here were stained but not fixed and it remains to be explored what additional DNA damage is exerted by different fixation methods. In addition, our slides date from the years 1942 to 1944; however, it is likely that older slides are available in both public and private collections given the popularity of microscopy in Victorian times. A future objective will be to ascertain if massive genomic data retrieval can be achieved from even older microscopy slides.
There is also potential to retrieve ancient Plasmodium sequences directly from archaeological specimens. The recent retrieval of P. falciparum sequences from ancient Roman human skeletal remains (Marciniak et al. 2016) demonstrates this approach is technically feasible, and as P. vivax infection is more prevalent than P. falciparum, it is plausible that further ancient strains from osteological material could be reported in the near future. An additional possibility would be to directly retrieve Plasmodium sequences from Anopheles remains preserved in ancient lake sediments or in museum collections. The generation of additional historical sequences—both from Europe and from the Americas—together with an increased sequencing effort of extant P. vivax strains from under-sampled areas is our best hope to reconstruct the evolutionary history of this major parasite in more detail.
Materials and Methods
Samples
The slides analyzed here belong to the personal collection of the descendants of Dr Ildefonso Canicio, who worked in the antimalarial center established by the Catalan Government at Sant Jaume d'Enveja (Ebro Delta, Spain) in 1925. Four slides were analyzed, three of them included in a previous study (Gelabert et al. 2016). The new sample was a drop of blood from a double slide, stained with Giemsa (fig. 1b).
DNA Extraction
DNA extraction was performed by incubating the slide with 20 µl of extraction buffer (10 mM Tris-HCl [pH 8], 10 mM NaCl, 5 mM CaCL, 2.5 mM EDTA, 1% SDS, 1% Proteinase K, 0.1% DTT [w/v]) in an oven at 37 °C for 20 min for a total of three rounds. The resulting dissolved bloodstain and buffer were collected in a 1.5 ml Lobind Eppendorf and then incubated for 1 h at 56 °C. This was subsequently added to 10× volume of modified binding buffer (Allentoft et al. 2015) and passed through a Monarch silica spin column (NEB) by centrifugation (supplementary methods section 1 and fig. 15, Supplementary Material online). The column was washed once with 80% ethanol and DNA was subsequently released with EBT buffer to a final volume of 40 µl (see supplementary material section 1, Supplementary Material online). All the analyses were performed in dedicated ancient DNA laboratories where no previous genetic work on Plasmodium had been carried out, in both Barcelona (extraction of slides in 2016) and Copenhagen (extraction of slides in 2017).
Library Preparation and DNA Sequencing
Shotgun sequencing libraries for the Illumina platform were prepared using a single-tube protocol for double-stranded DNA (Carøe et al. 2018), with minor modifications and improvements as detailed in Mak et al. (2017) (supplementary methods section 2, Supplementary Material online). Sequencing was performed at the Natural History Museum of Denmark on one lane of an Illumina HiSeq2500 instrument in paired end mode running 125 cycles. This extraction and amplification process resulted in using all the genetic material we could source from the available slides.
Sequence Mapping
The sequenced reads were analyzed with FastQC to determine the quality prior to and after adapter clipping. The 3′ read adapters and consecutive bases with low-quality scores were removed using cutadapt 1.18 (Martin 2011). Reads shorter than 30 bp and bases with a quality score lower than 30 were also excluded. To increase the final coverage, all Plasmodium reads were pooled. As the blood samples had known coinfection with P. falciparum, we mapped all reads to both the P. vivax Pv01 reference genome (Auburn et al. 2016) and the P. falciparum 3D7 reference genome (Gardner et al. 2002). For the small fraction of reads mapping to both references (0.4%), we took forward only those reads which showed a lower edit distance to P. vivax over P. falciparum (supplementary fig. 1, Supplementary Material online). Mapping of P. vivax reads was then performed with Burrows–Wheeler Aligner (BWA) (Li and Durbin 2009) 0.7.1 aln (supplementary methods section 2, Supplementary Material online). Duplicated reads were deleted using Picard tools 2.18.6 MarkDuplicates. MapDamage 2.0 (Ginolhac et al. 2011) was applied to check for signatures of postmortem damage at the ends of the reads to validate the reads were associated with a historic sample rather than deriving from modern contamination. C to T and G to A substitutions at the 5′ ends and 3′ ends, respectively, were found to be present at a frequency of about 2.5% (supplementary fig. 2, Supplementary Material online); consistent with the age of the sample and in agreement with both the degree of damage previously detected in the mtDNA reads (Gelabert et al. 2016) and the damage profile of the human genetic component (supplementary fig. 3, Supplementary Material online). As a result, the first two nucleotides of each read were also trimmed.
The newly generated paired-end P. vivax reads were merged with the sequencing reads generated in 2016 (Gelabert et al. 2016). We refer to the resulting pooled sample as Ebro-1944. Genotypes were called from the alignment with GATK v3.7 UnifiedGenotyper (McKenna et al. 2010), using a minimum base quality of 30 and the standard confidence call threshold of 50. Genotype calls were filtered further with VCFtools (Danecek et al. 2011), excluding: 1) heterozygous calls, 2) calls with depth of coverage <2, 3) calls present in the telomeres and subtelomeres (Pearson et al. 2016), 4) indels (supplementary figs. 4 and 5 and table 3, Supplementary Material online). SNPs were annotated using SnpEff (Cingolani et al. 2012) for the analysis of variants associated with drug resistance.
Population Genetics Data Set
The data set used in population genetics analyses comprised the nuclear sequences of 337 previous published samples of P. vivax (Hupalo et al. 2016; Pearson et al. 2016; Cowell et al. 2017, 2018; Rodrigues et al. 2018), representing the global diversity of currently available P. vivax genomes (supplementary tables 4 and 11, Supplementary Material online).
Sequence reads were aligned against the Sal1 reference genome (Carlton et al. 2008) with BWA 0.7.1 aln (Li and Durbin 2009) using default parameters. Duplicated reads were removed using Picard tools 2.18.6. Reads with mapping qualities below 30 were removed using SAMtools 1.6 (Li et al. 2009). For analyses requiring incorporation of an out group, we additionally mapped reads from the P. cynomolgi M strain (Pasini et al. 2017) against the Sal1 reference genome following the same protocol.
We used GATK v3.7 UnifiedGenotyper (McKenna et al. 2010) for SNP calling with a number of adjustments for working with Plasmodium genomes and ancient DNA. First, we selected 297 samples that had more than the 70% of the Sal1 reference genome covered and presented more than 3,000 substitutions. Using this filtered data set of high-quality samples, we called variants using a mapping quality >30, depth of coverage >20 and a standard call confidence >50. We removed those SNPs that mapped to repetitive regions of the P. vivax reference genome (Pearson et al. 2016), heterozygous calls suggesting possible mixed infections, and SNPs that were present in less than three samples. The resultant data set included 131,309 SNPs and 277 strains. We used this catalog to genotype all remaining samples in the data set. For the individual genotyping, we called SNPs with GATK v3.7 UnifiedGenotyper as before, removing all heterozygous calls, and all variants with a minor allele frequency below 0.01%. The final data set comprised 338 samples and 128,081 SNPs. The genotypes for Ebro-1944 were called by selecting one random read that mapped each position of the data set, a pseudo-haploid calling approach which is advocated for low coverage aDNA (Mathieson et al. 2015). This resulted in the Ebro-1944 sequence covering 77,425 of the 128,081 total included positions (supplementary table 3, Supplementary Material online).
Allele-Frequency-Based Measures of Population Structure
The population genetics data set was filtered for SNPs in high linkage disequilibrium (LD) using a 60 SNP sliding window, advancing each time by 10 steps, and removing any SNPs with a correlation coefficient ≥0.1 with any other SNP within the window (Chang et al. 2015). This left a pruned data set of 38,358 SNPs for analyses relying on independent SNPs. We applied PCA to the LD pruned global data set restricted to only sites covered in all samples (Chang et al. 2015). We additionally clustered our pruned data set using the unsupervised clustering algorithm ADMIXTURE 1.3.0 (Alexander et al. 2009) for values of K between 1 and 15. K = 6 provided the lowest cross-validation error (supplementary fig. 8, Supplementary Material online and fig. 1c). To evaluate the relationship of Ebro-1944 to other global strains we calculated f4 statistics using qpDstat available within AdmixTools (Patterson et al. 2012). Setting Ebro-1944 as a target, we explored which strains, grouped by geographic label, share more alleles with Ebro-1944 relative to every pairwise combination of modern strain(s) (X and Y) in our reference data set and relative to P. cynomolgi as an out group: f4 (P. cynomolgi, Ebro-1944; X, Y) (supplementary fig. 9, Supplementary Material online).
Inferring Patterns of Allele and Haplotype Sharing
In addition, we applied an unrelated method to explore patterns of allele and haplotype sharing implemented in CHROMOPAINTER v2 (Lawson et al. 2012). Unlike f-statistics, this approach does not rely on a user specified topology and can thus consider the relationship of all strains to all others collectively. As this approach requires low levels of missingness across comparisons, we filtered the previously described unpruned population genetics data set for only the positions present in Ebro-1944 and retained only those samples with ≤10% missing data (77,420 sites, 218 samples). A schematic of the workflow is provided in supplementary fig. 7, Supplementary Material online.
Briefly, CHROMOPAINTER calculates, separately for each position, the probability that a “recipient” chromosome is most closely related to a particular “donor” in the data set under a copying model framework (Li and Stephens 2003). Here, we use all strains as donors and the equivalent strains as recipients in an “all-versus-all” painting approach. To cluster strains, fineSTRUCTURE (Lawson et al. 2012) was applied to the all-versus-all coancestry matrix to group strains based on their inferred painting profiles. Given the variable missingness across the modern strains included in the alignment we implemented several analyses using the CHROMOPAINTER unlinked (allele-sharing) implementation, as well as under the linked CHROMOPAINTER (haplotype-sharing) model. To use the latter, we performed various levels of imputation followed the protocol set out by Samad et al (2015) for P. falciparum in BEAGLE v3.3.2 (Browning and Browning 2013). The consistency of our inference under different imputation and filtering criteria was assessed by linear regression (supplementary fig. 12, Supplementary Material online). Further details are provided in supplementary section 4, Supplementary Material online.
Drug-Resistance Variants Analysis
We used the annotations provided by SnpEff (supplementary table 7, Supplementary Material online) to identify nonsynonymous mutations in genes previously described as being related to antimalarial drug resistance and host infectivity (Cingolani et al. 2012; Gupta et al. 2015; Hupalo et al. 2016; Pearson et al. 2016; Rodrigues et al. 2018). We also screened a set of previous described positions that have shown recent signals of selection in P. vivax populations and all potential genetic variants found in Ebro-1944, comprising more than 4,000 SNPs (supplementary tables 8–10, Supplementary Material online).
Phylogenetic Analysis and Data Set
The whole-genome sequences of 15 modern P. vivax samples and Ebro-1944 were mapped against the PvP01 reference assembly (Auburn et al. 2016) (supplementary table 5, Supplementary Material online). These strains were selected with a focus on the Americas and also to include strains sampled over a large temporal span as is required for phylogenetic tip dating. The main motivation for using the PvP01 reference genome for mapping sequence data for phylogenetic analyses stems from this assembly offering a better definition of sub-telomeric genes and repetitive pir genes that usually lie in recombinant regions, thus providing greater power to identify and exclude these parts of the genome (Auburn et al. 2016). After calling variants with GATK version 3.7 UnifiedGenotyper, polymorphisms were further filtered by selecting only those SNPs with a coverage >1 an average mapping quality >30, a genotype quality >30, and by removing heterozygous positions. The exonic positions were then classified as synonymous and nonsynonymous with SnpEff (Cingolani et al. 2012).
Our final phylogenetic timeline data set spanned 69 years (1944–2013) of evolution (supplementary table 5, Supplementary Material online) across a 34,452 SNP alignment. To filter this alignment for only congruent SNPs for phylogenetic dating we first generated a maximum parsimony phylogeny in MEGA7 (Kumar et al. 2016), evaluating support for each branch over 100 boot-strap iterations. We then screened for homoplasic, sites in the 34,452 SNP alignment that do not support the maximum parsimony phylogeny, using HomoplasyFinder (Crispell et al. 2019). This led to the identification of 13,112 homoplasic SNPs, many of which fell in sub-telomeric regions. All homoplasic sites were subsequently removed from the alignment. In this way we screen and exclude variants from the original alignment that fall in hypervariable regions, that may arise from inaccurate SNP calling or postmortem damage, as well as removing regions that derive from between-lineage genetic recombination or mixed infections.
Estimating a Timed Phylogeny
To investigate the extent of temporal signal existing in our homoplasy cleaned timeline alignment, we built a maximum likelihood phylogenetic tree, without constraining tip-heights to their sampling times, using RaxML (Stamatakis 2014). These trees were robust to the inclusion of SNPs called at different depths (supplementary fig. 13, Supplementary Material online). After rooting the tree on the P. cynomolgi genome, we computed a linear regression between root-to-tip distance and sampling time using the roototip function from BactDating (Didelot et al. 2018). To further confirm the presence of a significant temporal signal, we assessed the significance of this regression following 10,000 steps of date randomization (fig. 3b). After confirmation of temporal signal in the data set, substitution rates were estimated by running a tip-calibrated inference using Markov chain Monte Carlo (MCMC) sampling in BEAST 2 (Bouckaert et al. 2014). The best-fit nucleotide substitution model was estimated as TN93 following evaluation of all possible substitution models in BModelTest (Bouckaert and Drummond 2017). To minimize prior assumptions about demographic history we tested three possible demographic models: the coalescent constant, coalescent exponential, and coalescent Bayesian skyline. In each case we set a log-normal prior on a relaxed evolutionary clock as well as testing under a strict clock model.
To calibrate the tree using tip-dates only, we applied flat priors (i.e. uniform distributions) for the substitution rate (1.10E12–1.10E2 substitutions/site/year), as well as for the age of any internal node in the tree. We ran five independent chains in which samples were drawn every 50,000 MCMC steps from a total of 500,000,000 steps, after a discarded burn-in of 10,000,000 steps. Convergence to the stationary distribution and sufficient sampling and mixing were checked by inspection of posterior samples (effective sample size >200) in Tracer v1.6. The best-fit model was selected based on evaluation of both the median likelihood value of the model and the maximum likelihood estimator following path sampling (Baele et al. 2012) (supplementary fig. 14 and table 6, Supplementary Material online).
Supplementary Material
Acknowledgments
We are grateful to Thomas D. Otto (University of Glasgow) and Thomas Lavsten (University of Copenhagen) for helpful comments and suggestions and to the descendants of Dr Canicio, Miquel, and Ildefons Oliveras for sharing with us their slides. We additionally would like to thank Dr George Busby and Dr Jacob Almagro for useful discussions on chromosome painting of Plasmodium sp. This research was supported by a grant from Obra Social “La Caixa,” Secretaria d’Universitats i Recerca Programme del Departament d’Economia i Coneixement de la Generalitat de Catalunya (GRC 2017 SGR 880), Secretaria d’Universitats i Recerca (GRC2017-SGR880), and by FEDER-Ministry of Science, Innovation and Universities (MCIU) (PGC2018-095931-B-100) to C.L.-F. and an European Research Council (ERC) Consolidator Grant (681396-Extinction Genomics) to M.T.P.G. L.v.D., and F.B. acknowledge financial support from the Newton Fund UK–China NSFC initiative (grant MR/P007597/1) and the Biotechnology and Biological Sciences Research Council (BBSRC) (equipment grant BB/R01356X/1). Plasmodium vivax genomes are deposited with ENA BioProject ID PRJEB30878. We also thank the Danish National High Throughput Sequencing Centre for help in sequencing.
Author Contributions
P.G., M.T.P.G., L.v.D., F.B., and C.L.-F. conceived and designed the study; R.E. and C.A. discovered the slides; C.C. developed and performed laboratory analysis; L.v.D., P.G., A.R., T.d.-D., S.G., R.F., I.O., and M.d.M. analyzed data and performed computational analyses; S.H., F.B., T.M.-B., and I.M. provided comments and suggested analyses; L.v.D., F.B., and C.L.-F. wrote the paper with inputs from all coauthors.
References
- Achan J, Talisuna AO, Erhart A, Yeka A, Tibenderana JK, Baliraine FN, Rosenthal PJ, D'Alessandro U.. 2011. Quinine, an old anti-malarial drug in a modern world: role in the treatment of malaria. Malar J. 10:144.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adekunle AI, Pinkevych M, McGready R, Luxemburger C, White LJ, Nosten F, Cromer D, Davenport MP.. 2015. Modeling the dynamics of Plasmodium vivax infection and hypnozoite reactivation in vivo. PLoS Negl Trop Dis. 9(3):e0003595-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19(9):1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allentoft ME, Sikora M, Sjögren K-G, Rasmussen S, Rasmussen M, Stenderup J, Damgaard PB, Schroeder H, Ahlström T, Vinner L, et al. 2015. Population genomics of Bronze Age Eurasia. Nature 522(7555):167–172. [DOI] [PubMed] [Google Scholar]
- Auburn S, Böhme U, Steinbiss S, Trimarsanto H, Hostetler J, Sanders M, Gao Q, Nosten F, Newbold CI, Berriman M, et al. 2016. A new Plasmodium vivax reference sequence with improved assembly of the subtelomeres reveals an abundance of pir genes. Wellcome Open Res. 1(0):4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baele G, Lemey P, Bedford T, Rambaut A, Suchard MA, Alekseyenko AV.. 2012. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol Biol Evol. 29(9):2157–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird JK. 2004. Chloroquine resistance in Plasmodium vivax. Antimicrob Agents Chemother. 48(11):4075–4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird JK. 2013. Evidence and implications of mortality associated with acute Plasmodium vivax Malaria. Clin Microbiol Rev. 26(1):36–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnadas C, Ratsimbasoa A, Tichit M, Bouchier C, Jahevitra M, Picot S, Menard D.. 2008. Plasmodium vivax resistance to chloroquine in Madagascar: clinical efficacy and polymorphisms in pvmdr1 and pvcrt-o genes. Antimicrob Agents Chemother. 52(12):4233–4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnadas C, Tichit M, Bouchier C, Ratsimbasoa A, Randrianasolo L, Raherinjafy R, Jahevitra M, Picot S, Ménard D.. 2008. Plasmodium vivax dhfr and dhps mutations in isolates from Madagascar and therapeutic response to sulphadoxine–pyrimethamine. Malar J. 7(1):35.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle KE, Gething PW, Elyazar IRF, Moyes CL, Sinka ME, Howes RE, Guerra CA, Price RN, Baird KJ, Hay SI, et al. 2012. The global public health significance of Plasmodium vivax. Adv Parasitol. 80: 1–111. [DOI] [PubMed] [Google Scholar]
- Battle KE, Lucas TCD, Nguyen M, Howes RE, Nandi AK, Twohig KA, Pfeffer DA, Cameron E, Rao PC, Casey D, et al. 2019. Mapping the global endemicity and clinical burden of Plasmodium vivax, 2000–17: a spatial and temporal modelling study. Lancet 394(10195):332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ.. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 10(4):e1003537.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert RR, Drummond AJ.. 2017. BModelTest: Bayesian phylogenetic site model averaging and model comparison. BMC Evol Biol. 17(1):42.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasil P, Zalis MG, de Pina-Costa A, Siqueira AM, Júnior CB, Silva S, Areas ALL, Pelajo-Machado M, de Alvarenga DAM, da Silva Santelli ACF, et al. 2017. Outbreak of human malaria caused by Plasmodium simium in the Atlantic Forest in Rio de Janeiro: a molecular epidemiological investigation. Lancet Glob Health 5(10):e1038–1046. [DOI] [PubMed] [Google Scholar]
- Brega S, Meslin B, de Monbrison F, Severini C, Gradoni L, Udomsangpetch R, Sutanto I, Peyron F, Picot S.. 2005. Identification of the Plasmodium vivax mdr-like gene (pvmdr1) and analysis of single-nucleotide polymorphisms among isolates from different areas of endemicity. J Infect Dis. 191(2):272–277. [DOI] [PubMed] [Google Scholar]
- Browning BL, Browning SR.. 2013. Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics 194(2):459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buery JC, Rodrigues PT, Natal L, Salla LC, Loss AC, Vicente CR, Rezende HR, Duarte AMRC, Fux B, Malafronte RdS, et al. 2017. Mitochondrial genome of Plasmodium vivax/simium detected in an endemic region for malaria in the Atlantic Forest of Espírito Santo state, Brazil: do mosquitoes, simians and humans harbour the same parasite? Malar J. 16(1):437.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, Crabtree J, Angiuoli SV, Merino EF, Amedeo P, et al. 2008. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature 455(7214):757–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carøe C, Gopalakrishnan S, Vinner L, Mak SST, Sinding MHS, Samaniego JA, Wales N, Sicheritz-Pontén T, Gilbert MTP.. 2018. Single‐tube library preparation for degraded DNA. Methods Ecol Evol. 9(2):410–419. [Google Scholar]
- Carter R. 2003. Speculations on the origins of Plasmodium vivax malaria. Trends Parasitol. 19(5):214–219. [DOI] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ.. 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 4(1):7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM.. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6(2):80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Jakobsson M, Coop G, Wen X, Wall JD, Rosenberg NA, Pritchard JK.. 2006. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat Genet. 38(11):1251–1260. [DOI] [PubMed] [Google Scholar]
- Cowell AN, Loy DE, Sundararaman SA, Valdivia H, Fisch K, Lescano AG, Baldeviano GC, Durand S, Gerbasi V, Sutherland CJ, et al. 2017. Selective whole-genome amplification is a robust method that enables scalable whole-genome sequencing of Plasmodium vivax from unprocessed clinical samples. MBio 8(1):pii: e02257-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell AN, Valdivia HO, Bishop DK, Winzeler EA.. 2018. Exploration of Plasmodium vivax transmission dynamics and recurrent infections in the Peruvian Amazon using whole genome sequencing. Genome Med. 10(1):52.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispell J, Balaz D, Gordon SV.. 2019. HomoplasyFinder: a simple tool to identify homoplasies on a phylogeny. Microb Genomics 5(1):e000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culleton R, Coban C, Zeyrek FY, Cravo P, Kaneko A, Randrianarivelojosia M, Andrianaranjaka V, Kano S, Farnert A, Arez AP, et al. 2011. The origins of African Plasmodium vivax; insights from mitochondrial genome sequencing. PLoS One 6(12):e29137.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 27(15):2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro MC, Singer BH.. 2005. Was malaria present in the Amazon before the European conquest? Available evidence and future research agenda. J Archaeol Sci. 32(3):337–340. [Google Scholar]
- de-Dios T, van Dorp L, Gelabert P, Carøe C, Sandoval-Velasco M, Fregel R, Escosa R, Aranda C, Huijben S, Balloux F, et al. 2019. Genetic affinities of an eradicated Plasmodium falciparum European strain. Microb Genomics 5:000263.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira TC, Rodrigues PT, Menezes MJ, Gonçalves-Lopes RM, Bastos MS, Lima NF, Barbosa S, Gerber AL, Loss de Morais G, Berná L, et al. 2017. Genome-wide diversity and differentiation in New World populations of the human malaria parasite Plasmodium vivax. PLoS Negl Trop Dis. 11(7):e0005824.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pecoulas PE, Basco LK, Tahar R, Ouatas T, Mazabraud A. 1998. Analysis of the Plasmodium vivax dihydrofolate reductase–thymidylate synthase gene sequence. Gene 211:177–185. [DOI] [PubMed] [Google Scholar]
- De Zulueta J. 1973. Malaria and Mediterranean history. Parassitologia 15(1):1–15. [PubMed] [Google Scholar]
- Dharia NV, Bright AT, Westenberger SJ, Barnes SW, Batalov S, Kuhen K, Borboa R, Federe GC, McClean CM, Vinetz JM, et al. 2010. Whole-genome sequencing and microarray analysis of ex vivo Plasmodium vivax reveal selective pressure on putative drug resistance genes. Proc Natl Acad Sci U S A. 107(46):20045–20050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, Croucher NJ, Bentley SD, Harris SR, Wilson DJ.. 2018. Bayesian inference of ancestral dates on bacterial phylogenetic trees. Nucleic Acids Res. 46(22):e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante AA, Cornejo OE, Freeland DE, Poe AC, Durrego E, Collins WE, Lal AA.. 2005. A monkey’s tale: the origin of Plasmodium vivax as a human malaria parasite. Proc Natl Acad Sci U S A. 102(6):1980–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escardo F. 1992. Historia de la Cirurgia en el Peru. Editorial Monterrico S.A.
- Ganguly S, Saha P, Chatterjee M, Maji AK.. 2014. Prevalence of polymorphisms in antifolate drug resistance molecular marker genes pvdhfr and pvdhps in clinical isolates of Plasmodium vivax from Kolkata, India. Antimicrob Agents Chemother. 58(1):196–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnham PCC. 1966. Malaria parasites and other Haemosporidia. Blackwell, Oxford, UK: Davis, Philadelphia; 1132pp.
- Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, et al. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419(6906):498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelabert P, Sandoval-Velasco M, Olalde I, Fregel R, Rieux A, Escosa R, Aranda C, Paaijmans K, Mueller I, Gilbert MTP, et al. 2016. Mitochondrial DNA from the eradicated European Plasmodium vivax and P. falciparum from 70-year-old slides from the Ebro Delta in Spain. Proc Natl Acad Sci U S A. 113(41):11495–11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gething PW, Patil AP, Smith DL, Guerra CA, Elyazar IR, Johnston GL, Tatem AJ, Hay SI.. 2011. A new world malaria map: Plasmodium falciparum endemicity in 2010. Malar J. 10(1):378.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilabert A, Otto TD, Rutledge GG, Franzon B, Ollomo B, Arnathau C, Durand P, Moukodoum ND, Okouga A-P, Ngoubangoye B, et al. 2018. Plasmodium vivax-like genome sequences shed new insights into Plasmodium vivax biology and evolution. PLoS Biol. 16(8):e2006035.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginolhac A, Rasmussen M, Gilbert MTP, Willerslev E, Orlando L.. 2011. mapDamage: testing for damage patterns in ancient DNA sequences. Bioinformatics 27(15):2153–2155. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Ceron L, Mu J, Santillán F, Joy D, Sandoval MA, Camas G, Su X, Choy EV, Torreblanca R.. 2013. Molecular and epidemiological characterization of Plasmodium vivax recurrent infections in southern Mexico. Parasit Vectors 6:109.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunalan K, Niangaly A, Thera MA, Doumbo OK, Miller LH.. 2018. Plasmodium vivax infections of Duffy-negative erythrocytes: historically undetected or a recent adaptation? Trends Parasitol. 34(5):420–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Thiruvengadam G, Desai SA.. 2015. The conserved clag multigene family of malaria parasites: essential roles in host–pathogen interaction. Drug Resist Updat. 18:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar K, Bhattacharjee S, Safeukui I.. 2018. Drug resistance in Plasmodium. Nat Rev Microbiol. 16(3):156–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapuarachchi HC, Rungsihirunrat K, Na-Bangchang K, Maestre A, Sibley CH, Suzuki SM, Hawkins VN.. 2009. Assessment of the origins and spread of putative resistance-conferring mutations in Plasmodium vivax dihydropteroate synthase. Am J Trop Med Hyg. 81(2):348–355. [PubMed] [Google Scholar]
- Hay SI, Guerra CA, Tatem AJ, Noor AM, Snow RW.. 2004. The global distribution and population at risk of malaria: past, present, and future. Lancet Infect Dis. 4(6):327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick PW. 2012. Resistance to malaria in humans: the impact of strong, recent selection. Malar J. 11(1):349.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howes RE, Battle KE, Mendis KN, Smith DL, Cibulskis RE, Baird JK, Hay SI.. 2016. Global epidemiology of Plasmodium vivax. Am J Trop Med Hyg. 95(Suppl. 6):15–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howes RE, Piel FB, Patil AP, Nyangiri OA, Gething PW, Dewi M, Hogg MM, Battle KE, Padilla CD, Baird JK, et al. 2012. G6PD deficiency prevalence and estimates of affected populations in Malaria endemic countries: a geostatistical model-based map. PLoS Med. 9(11):e1001339.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Huang S, Su X-Z, Tong X, Yan J, Li H, Lu F.. 2014. Molecular surveillance of pvdhfr, pvdhps, and pvmdr-1 mutations in Plasmodium vivax isolates from Yunnan and Anhui provinces of China. Malar J. 13:346.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huldén L, Huldén L, Heliövaara K.. 2005. Endemic malaria: an “indoor” disease in northern Europe. Historical data analysed. Malar J. 4(1):19–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume J. 2003. Malaria in antiquity: a genetics perspective. World Archaeol. 35(2):180–192. [Google Scholar]
- Hupalo DN, Luo Z, Melnikov A, Sutton PL, Rogov P, Escalante A, Vallejo AF, Herrera S, Arévalo-Herrera M, Fan Q, et al. 2016. Population genomics studies identify signatures of global dispersal and drug resistance in Plasmodium vivax. Nat Genet. 48(8):953–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imwong M, Pukrittakayamee S, Looareesuwan S, Pasvol G, Poirreiz J, White NJ, Snounou G.. 2001. Association of genetic mutations in Plasmodium vivax dhfr with resistance to sulfadoxine–pyrimethamine: geographical and clinical correlates. Antimicrob Agents Chemother. 45(11):3122–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imwong M, Pukrittayakamee S, Renia L, Letourneur F, Charlieu J-P, Leartsakulpanich U, Looareesuwan S, White NJ, Snounou G.. 2003. Novel point mutations in the dihydrofolate reductase gene of Plasmodium vivax: evidence for sequential selection by drug pressure. Antimicrob Agents Chemother. 47(5):1514–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsinczky M, Fischer K, Chen N, Baker J, Rieckmann K, Cheng Q.. 2004. Sulfadoxine resistance in Plasmodium vivax is associated with a specific amino acid in dihydropteroate synthase at the putative sulfadoxine-binding site. Antimicrob Agents Chemother. 48(6):2214–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaisavee V, Suwanarusk R, Chua ACY, Kyle DE, Malleret B, Zhang R, Imwong M, Imerbsin R, Ubalee R, Sámano-Sánchez H, et al. 2017. Strict tropism for CD71+/CD234+ human reticulocytes limits the zoonotic potential of Plasmodium cynomolgi. Blood 130(11):1357–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krotoski WA. 1985. Discovery of the hypnozoite and a new theory of malarial relapse. Trans R Soc Trop Med Hyg. 79(1):1–11. [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski DP. 2005. How malaria has affected the human genome and what human genetics can teach us about malaria. Am J Hum Genet. 77(2):171–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacerda MVG, Fragoso SCP, Alecrim MGC, Alexandre MAA, Magalhães BML, Siqueira AM, Ferreira LCL, Araújo JR, Mourão MPG, Ferrer M, et al. 2012. Postmortem characterization of patients with clinical diagnosis of Plasmodium vivax Malaria: to what extent does this parasite kill? Clin Infect Dis. 55(8):e67–74. [DOI] [PubMed] [Google Scholar]
- Lawson DJ, Hellenthal G, Myers S, Falush D.. 2012. Inference of population structure using dense haplotype data. PLoS Genet. 8(1):e1002453.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leartsakulpanich U, Imwong M, Pukrittayakamee S, White NJ, Snounou G, Sirawaraporn W, Yuthavong Y.. 2002. Molecular characterization of dihydrofolate reductase in relation to antifolate resistance in Plasmodium vivax. Mol Biochem Parasitol. 119(1):63–73. [DOI] [PubMed] [Google Scholar]
- Leclerc MC, Durand P, Gauthier C, Patot S, Billotte N, Menegon M, Severini C, Ayala FJ, Renaud F.. 2004. Meager genetic variability of the human malaria agent Plasmodium vivax. Proc Natl Acad Sci U S A. 101(40):14455–14460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie S, Winney B, Hellenthal G, Davison D, Boumertit A, Day T, Hutnik K, Royrvik EC, Cunliffe B, Lawson DJ, et al. 2015. The fine scale genetic structure of the British population. Nature 519(7543):309–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R.. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Stephens M.. 2003. Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics 165(4):2213–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CS, Tazi L, Ayala FJ.. 2005. Plasmodium vivax: recent world expansion and genetic identity to Plasmodium simium. Proc Natl Acad Sci U S A. 102(43):15523–15528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Li Y, Shaw KS, Learn GH, Plenderleith LJ, Malenke JA, Sundararaman SA, Ramirez MA, Crystal PA, Smith AG, et al. 2014. African origin of the malaria parasite Plasmodium vivax. Nat Commun. 5:3346.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Sherrill-Mix S, Learn GH, Scully EJ, Li Y, Avitto AN, Loy DE, Lauder AP, Sundararaman SA, Plenderleith LJ, et al. 2017. Wild bonobos host geographically restricted malaria parasites including a putative new Laverania species. Nat Commun. 8(1):1635.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy DE, Liu W, Li Y, Learn GH, Plenderleith LJ, Sundararaman SA, Sharp PM, Hahn BH.. 2017. Out of Africa: origins and evolution of the human malaria parasites Plasmodium falciparum and Plasmodium vivax. Int J Parasitol. 47(2–3):87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak SST, Gopalakrishnan S, Carøe C, Geng C, Liu S, Sinding M-HS, Kuderna LFK, Zhang W, Fu S, Vieira FG, et al. 2017. Comparative performance of the BGISEQ-500 vs Illumina HiSeq2500 sequencing platforms for palaeogenomic sequencing. GigaScience 6(8):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak S, Prowse TL, Herring DA, Klunk J, Kuch M, Duggan AT, Bondioli L, Holmes EC, Poinar HN.. 2016. Plasmodium falciparum malaria in 1st–2nd century CE southern Italy. Curr Biol. 26(23):R1220–1222. [DOI] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17(1):10. [Google Scholar]
- Mathieson I, Lazaridis I, Rohland N, Mallick S, Patterson N, Roodenberg SA, Harney E, Stewardson K, Fernandes D, Novak M, et al. 2015. Genome-wide patterns of selection in 230 ancient Eurasians. Nature 528(7583):499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayxay M, Pukrittayakamee S, Newton PN, White NJ.. 2004. Mixed-species malaria infections in humans. Trends Parasitol. 20(5):233–240. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1298–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus KF, Taravella AM, Henn BM, Bustamante CD, Sikora M, Cornejo OE.. 2017. Population genetic analysis of the DARC locus (Duffy) reveals adaptation from standing variation associated with malaria resistance in humans. PLoS Genet. 13(3):e1006560.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menegon M, Majori G, Severini C.. 2006. Genetic variations of the Plasmodium vivax dihydropteroate synthase gene. Acta Trop. 98(2):196–199. [DOI] [PubMed] [Google Scholar]
- Moreira CM, Abo-Shehada M, Price RN, Drakeley CJ.. 2015. A systematic review of sub-microscopic Plasmodium vivax infection. Malar J. 14(1):360.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Most H, London IM.. 1946. Chloroquine for treatment of acute attacks of vivax malaria. JAMA 131(12):963–967. [DOI] [PubMed] [Google Scholar]
- Mu J, Joy DA, Duan J, Huang Y, Carlton J, Walker J, Barnwell J, Beerli P, Charleston MA, Pybus OG, et al. 2005. Host switch leads to emergence of Plasmodium vivax malaria in humans. Mol Biol Evol. 22(8):1686–1693. [DOI] [PubMed] [Google Scholar]
- Orjuela-Sánchez P, Filho F, Machado-Lima A, Chehuan YF, Costa MR, Alecrim Md, del Portillo HA.. 2009. Analysis of single-nucleotide polymorphisms in the crt-o and mdr1 genes of Plasmodium vivax among chloroquine-resistant isolates from the Brazilian Amazon region. Antimicrob Agents Chemother. 53(8):3561–3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto TD, Gilabert A, Crellen T, Böhme U, Arnathau C, Sanders M, Oyola SO, Okouga AP, Boundenga L, Willaume E, et al. 2018. Genomes of all known members of a Plasmodium subgenus reveal paths to virulent human malaria. Nat Microbiol. 3(6):687–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini EM, Böhme U, Rutledge GG, Voorberg-Van der Wel A, Sanders M, Berriman M, Kocken CH, Otto TD.. 2017. An improved Plasmodium cynomolgi genome assembly reveals an unexpected methyltransferase gene expansion. Wellcome Open Res. 2:42.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, Reich D.. 2012. Ancient ADMIXTURE in human history. Genetics 192(3):1065–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson RD, Amato R, Auburn S, Miotto O, Almagro-Garcia J, Amaratunga C, Suon S, Mao S, Noviyanti R, Trimarsanto H, et al. 2016. Genomic analysis of local variation and recent evolution in Plasmodium vivax. Nat Genet. 48(8):959–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen E, Severini C, Picot S.. 2013. Plasmodium vivax malaria: a re-emerging threat for temperate climate zones? Travel Med Infect Dis. 11(1):51–59. [DOI] [PubMed] [Google Scholar]
- Phillips EJ, Keystone JS, Kain KC.. 1996. Failure of combined chloroquine and high-dose primaquine therapy for Plasmodium vivax malaria acquired in Guyana, South America. Clin Infect Dis. 23(5):1171–1173. [DOI] [PubMed] [Google Scholar]
- Pletsch D. 1965. Report on a mission carried out in Spain in September–November 1963 for verification of the erradication of malaria. Rev Sanid Hig Publica (Madr). 39(7):309–367. [PubMed] [Google Scholar]
- Price RN, Douglas NM, Anstey NM.. 2009. New developments in Plasmodium vivax malaria: severe disease and the rise of chloroquine resistance. Curr Opin Infect Dis. 22(5):430–435. [DOI] [PubMed] [Google Scholar]
- Prugnolle F, Rougeron V, Becquart P, Berry A, Makanga B, Rahola N, Arnathau C, Ngoubangoye B, Menard S, Willaume E, et al. 2013. Diversity, host switching and evolution of Plasmodium vivax infecting African great apes. Proc Natl Acad Sci U S A. 110(20):8123–8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieckmann KH, Davis DR, Hutton DC.. 1989. Plasmodium vivax resistance to chloroquine? Lancet 2(8673):1183–1184. [DOI] [PubMed] [Google Scholar]
- Rieux A, Balloux F.. 2016. Inferences from tip-calibrated phylogenies: a review and a practical guide. Mol Ecol. 25(9):1911–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues PT, Valdivia HO, de Oliveira TC, Alves JMP, Duarte AMRC, Cerutti-Junior C, Buery JC, Brito CFA, de Souza JC, Hirano ZMB, et al. 2018. Human migration and the spread of malaria parasites to the New World. Sci Rep. 8(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sá JM, Nomura T, Neves JD, Baird JK, Wellems TE, del Portillo HA.. 2005. Plasmodium vivax: allele variants of the mdr1 gene do not associate with chloroquine resistance among isolates from Brazil, Papua, and monkey-adapted strains. Exp Parasitol. 109(4):256–259. [DOI] [PubMed] [Google Scholar]
- Samad H, Coll F, Preston MD, Ocholla H, Fairhurst RM, Clark TG.. 2015. Imputation-based population genetics analysis of Plasmodium falciparum Malaria parasites. PLoS Genet. 11(4):e1005131.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwanarusk R, Russell B, Chavchich M, Chalfein F, Kenangalem E, Kosaisavee V, Prasetyorini B, Piera KA, Barends M, Brockman A, et al. 2007. Chloroquine resistant Plasmodium vivax: in vitro characterisation and association with molecular polymorphisms. PLoS One 2(10):e1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana S-I, Sullivan SA, Kawai S, Nakamura S, Kim HR, Goto N, Arisue N, Palacpac NMQ, Honma H, Yagi M, et al. 2012. Plasmodium cynomolgi genome sequences provide insight into Plasmodium vivax and the monkey malaria clade. Nat Genet. 44(9):1051–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe K, Mita T, Jombart T, Eriksson A, Horibe S, Palacpac N, Ranford-Cartwright L, Sawai H, Sakihama N, Ohmae H, et al. 2010. Plasmodium falciparum accompanied the human expansion out of Africa. Curr Biol. 20(14):1283–1289. [DOI] [PubMed] [Google Scholar]
- Taylor JE, Pacheco MA, Bacon DJ, Beg MA, Machado RL, Fairhurst RM, Herrera S, Kim J-Y, Menard D, Póvoa MM, et al. 2013. The evolutionary history of Plasmodium vivax as inferred from mitochondrial genomes: parasite genetic diversity in the Americas. Mol Biol Evol. 30(9):2050–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Varkonyi R, Cahinhinan N, Abbes S, Argyropoulos G, Destro-Bisol G, Drousiotou A, Dangerfield B, Lefranc G, Loiselet J, et al. 2001. Haplotype diversity and linkage disequilibrium at human G6PD: recent origin of alleles that confer malarial resistance. Science 293(5529):455–462. [DOI] [PubMed] [Google Scholar]
- Tjitra E, Anstey NM, Sugiarto P, Warikar N, Kenangalem E, Karyana M, Lampah DA, Price RN.. 2008. Multidrug-Resistant Plasmodium vivax associated with severe and fatal Malaria: a prospective study in Papua, Indonesia. PLoS Med. 5(6):e128.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twohig KA, Pfeffer DA, Baird JK, Price RN, Zimmerman PA, Hay SI, Gething PW, Battle KE, Howes RE.. 2019. Growing evidence of Plasmodium vivax across malaria-endemic Africa. PLoS Negl Trop Dis. 13(1):e0007140.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MR, Keene JL, Forman SL, Prewitt ER, Carlson DL, Wiederhold JE.. 2018. Pre-Clovis projectile points at the Debra L. Friedkin site, Texas—implications for the Late Pleistocene peopling of the Americas. Sci Adv. 4(10):eaat4505.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss DJ, Lucas TCD, Nguyen M, Nandi AK, Bisanzio D, Battle KE, Cameron E, Twohig KA, Pfeffer DA, Rozier JA, et al. 2019. Mapping the global prevalence, incidence, and mortality of Plasmodium falciparum, 2000–17: a spatial and temporal modelling study. Lancet 394(10195):322–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White NJ. 2011. Determinants of relapse periodicity in Plasmodium vivax malaria. Malar J. 10(1):297.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organisation. 2017. World malaria report 2017. Available from: https://www.who.int/malaria/publications/world-malaria-report-2017/en/; last accessed November 14, 2019.
- Zhao X, Smith DL, Tatem AJ.. 2016. Exploring the spatiotemporal drivers of malaria elimination in Europe. Malar J. 15(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu SJ, Almagro-Garcia J, McVean G.. 2018. Deconvolution of multiple infections in Plasmodium falciparum from high throughput sequencing data. Bioinformatics 34(1):9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.