Abstract
Nonrecombining sex chromosomes are widely found to be more differentiated than autosomes among closely related species, due to smaller effective population size and/or to a disproportionally large-X effect in reproductive isolation. Although fungal mating-type chromosomes can also display large nonrecombining regions, their levels of differentiation compared with autosomes have been little studied. Anther-smut fungi from the Microbotryum genus are castrating pathogens of Caryophyllaceae plants with largely nonrecombining mating-type chromosomes. Using whole genome sequences of 40 fungal strains, we quantified genetic differentiation among strains isolated from the geographically overlapping North American species and subspecies of Silene virginica and S. caroliniana. We inferred that gene flow likely occurred at the early stages of divergence and then completely stopped. We identified large autosomal genomic regions with chromosomal inversions, with higher genetic divergence than the rest of the genomes and highly enriched in selective sweeps, supporting a role of rearrangements in preventing gene flow in genomic regions involved in ecological divergence. Unexpectedly, the nonrecombining mating-type chromosomes showed lower divergence than autosomes due to higher gene flow, which may be promoted by adaptive introgressions of less degenerated mating-type chromosomes. The fact that both mating-type chromosomes are always heterozygous and nonrecombining may explain such patterns that oppose to those found for XY or ZW sex chromosomes. The specific features of mating-type chromosomes may also apply to the UV sex chromosomes determining sexes at the haploid stage in algae and bryophytes and may help test general hypotheses on the evolutionary specificities of sex-related chromosomes.
Keywords: sex chromosomes, inversions, local adaptation, host specialization, fungi, divergence with gene flow, speciation, introgression
Introduction
Sex chromosomes have been found to play a prominent role in reproductive isolation. This “large-X effect” in speciation is thought to be due to the higher rate of fixation of beneficial alleles on the X chromosome (i.e., “faster-X effect”), as the X chromosome is hemizygous in males (and thus more exposed to selection) and recombines in females (allowing efficient selection). This rapid accumulation of adaptive alleles and heterozygosity asymmetry in sex chromosomes could generate more incompatibilities between lineages than on autosomes, so that sex chromosomes would disproportionately contribute to reproductive isolation (Coyne and Orr 1989; Coyne 1992; Presgraves 2008). Sex chromosomes have in fact consistently been found to display higher divergence levels between closely related species than autosomes, which could also be partly due to their smaller effective population size (Meisel and Connallon 2013; Charlesworth et al. 2018; Irwin 2018; Presgraves 2018). Chromosomal inversions are another genomic feature often found to be more differentiated between species than the rest of the genome and have been suggested to allow ecological divergence by protecting beneficial allelic combinations from gene flow, that otherwise homogenizes gene pools and prevent local adaptation and ecological divergence (Dobzhansky 1970, 1981; Kirkpatrick and Barton 2006; Hoffmann and Rieseberg 2008; Joron et al. 2011; Thompson and Jiggins 2014; Lamichhaney et al. 2016; Christmas et al. 2019).
A case of ecological divergence is host specialization of fungal pathogens on new hosts (Giraud et al. 2010). In addition, fungi can carry mating-type chromosomes with features highly similar to those of sex chromosomes, with extensive nonrecombining regions (NRRs) and thus smaller effective population sizes compared with autosomes. Important differences with classically studied sex chromosomes however exist: Both mating-type chromosomes are always heterozygous and nonrecombining (Fraser et al. 2004; Menkis et al. 2008; Grognet et al. 2014; Badouin et al. 2015), as are UV sex chromosomes that determine sex at the haploid stage in red, green, and brown algae, as well as in bryophytes (Coelho et al. 2019). Whether these features would change the pattern of higher divergence of sex-related chromosomes compared with autosomes between closely related species has been little investigated. Given that both mating-type chromosomes are nonrecombining, one does not expect any phenomenon similar to the “faster-X” effect, and there may thus not be stronger role of mating-type chromosomes compared with autosomes in reproductive isolation; both mating-type chromosomes are expected to rapidly differentiate and degenerate, which may promote introgression from closely related species with less degenerated mating-type chromosomes (Sun et al. 2012; Corcoran et al. 2016). It has also been little studied whether chromosomal inversions can favor host adaptation in fungal pathogens, for example, by protecting beneficial combinations of putative effectors, that is, secreted proteins that play a role in the pathogen interaction with its host. Anther-smut fungi from the Microbotryum genus, castrating pathogens of Caryophyllaceae plants, have proved excellent models for the study of fungal pathogen host specialization and of mating-type chromosome evolution (Le Gac et al. 2007; Refrégier et al. 2008; Gladieux et al. 2011; Hood et al. 2013; Badouin et al. 2015; Fontanillas et al. 2015; Hartmann et al. 2019). The anther-smut fungus lineage of Microbotryum parasitizing the North American native Silene virginica and S. caroliniana constitutes an interesting case of incipient divergence and host specialization. No differentiation was found between the anther-smut fungal populations of these two plant species based on a few molecular markers (Antonovics et al. 1996; Freeman et al. 2002; Antonovics et al. 2003; Hood et al. 2019) but their largely distinct pollinator guilds (Fenster and Dudash 2001; Reynolds and Fenster 2008; Reynolds et al. 2009) may have promoted reproductive isolation in these pollinator-borne anther-smut fungi. Microbotryum fungi from S. caroliniana are known to carry mating-type chromosomes with large NRRs that have extended in several successive steps, generating a series of successive “evolutionary strata” with varying ages of divergence (Branco et al. 2018). Only small regions at the mating-type chromosome extremities are still recombining, called pseudoautosomal regions (PARs; Branco et al. 2018).
Here, we obtained long-read and short-read genome sequences and used a population genomics approach to investigate the potential for cryptic differentiated lineages in the anther-smut fungi parasitizing S. caroliniana and S. virginica, possibly specialized on different host lineages, to assess whether the divergence occurred with gene flow, and whether mating-type chromosomes and chromosomal inversions showed higher divergence than recombining autosomes.
Results
Two Main Genetic Clusters with Different Host Ranges
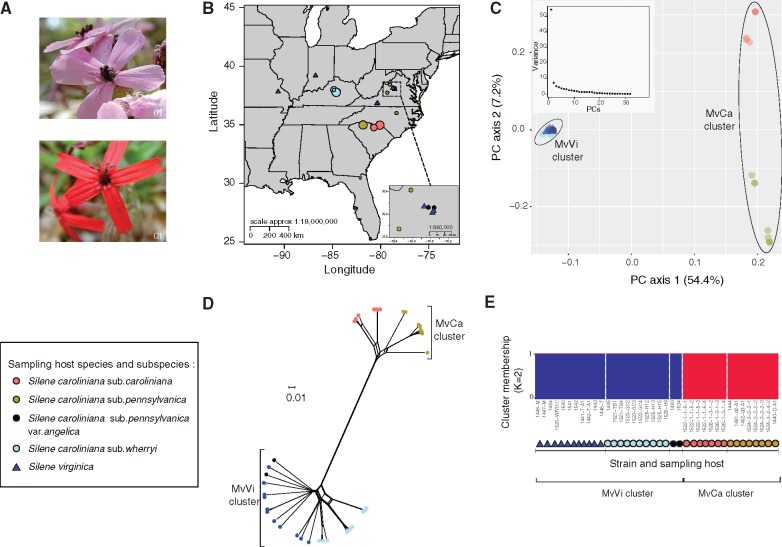
We analyzed the genomes of 40 anther-smut fungal strains sampled on S. virginica and on four S. caroliniana subspecies or varieties, that may represent cryptic species (caroliniana, wherryi, pennsylvanica, and pennsylvanica var. angelica) across their distribution in the Eastern United States of America (fig. 1A and B and supplementary table S1, Supplementary Material online; Burleigh and Holtsford 2003; Popp and Oxelman 2007). Using the available well-assembled genome of the MvCa-1250 strain sampled on S. caroliniana sub. pennsylvanica as reference (Branco et al. 2018), we identified a total of 349,045 high-quality biallelic single-nucleotide polymorphisms (SNPs) on autosomes in 38 strains, excluding two strains that were likely siblings of the individual corresponding to the reference genome, showing differences of fewer than 10 SNPs. Population structure analyses on the remaining 38 strains revealed genetic clusters differing in their host lineages despite overlapping geographical ranges (fig. 1). The principal component analysis (PCA) revealed two main genetic clusters (fig. 1C). One genetic cluster, hereafter referred to as MvCa, appeared specific to the two S. caroliniana subspecies pennsylvanica and caroliniana. The second genetic cluster, hereafter referred to as MvVi, appeared specific to S. virginica and to the S. caroliniana subspecies wherryi and variety pennsylvanica var. angelica. We confirmed the genetic subdivision into two main clusters using the model-based clustering analysis implemented in FastStructure, the nonparametric method of discriminant analysis of principal components and SplitsTree phylogenetic networks (fig. 1D and E; supplementary fig. S1 and note S1, Supplementary Material online). Further signatures of population subdivision associated with the host lineage or geography were found within each genetic cluster, although the corresponding genetic differentiation level was much lower than between the MvCa and MvVi clusters (supplementary note S1 and table S2, Supplementary Material online). We focused our subsequent analyses on the divergence between the two main genetic clusters and checked that population substructure did not affect our main conclusions (supplementary note S1, Supplementary Material online). We found a high number of SNPs on autosomes corresponding to fixed differences (55,233 SNPs) compared with shared polymorphisms (30,055 SNPs) between the MvCa and MvVi genetic clusters. Shared polymorphism was unlikely to be due to recent gene flow as we found no evidence of admixture between the MvCa and MvVi genetic clusters (fig. 1D and E and supplementary fig. S1, Supplementary Material online). The two genetic clusters had similar levels of nucleotide diversity per site π (per-gene median values in MvVi: π = 2.23e-03; per-gene median values in MvCa: π = 1.54e-03). Genome-wide divergence was in the range expected in the “gray zone of speciation” (Roux et al. 2016), with median values of per-gene relative (FST) and absolute (DXY) divergence values on autosomes of 0.66 and 0.0053, respectively (supplementary table S3, Supplementary Material online).
Fig. 1.
Sampling and population structure of the 38 studied anther-smut fungal Microbotryum strains on Silene caroliniana and S. virginica inferred based on autosomal genome-wide single-nucleotide polymorphisms (SNPs). (A) Pictures of diseased flowers of S. caroliniana 1) and S. virginica 2) by anther-smut fungi (Picture credits M.E. Hood). (B) Map of the sampling location of the 38 strains across the Eastern United States of America. The symbol size is proportional to the number of strains sampled at each site (from 1 to 4 strains per site). (C) Principal component analysis (PCA) on all 349,045 SNPs. Main plot: PCA of all 38 strains according to the first 2 principal components (PCs). Percentage of variance explained by each PC is indicated into brackets. Inset plot: genetic variance explained by the first ten PCs. (D) Neighbor-net tree from a SplitsTree analysis on 340,891 SNPs (removing SNPs with missing data). (E) Proportion of cluster membership predicted in FastStructure for K = 2 on 990 unlinked SNPs. Sampled host species and subspecies of the 38 strains are indicated with colors and symbols as shown in the legend in the three panels. The MvCa and MvVi genetic clusters are indicated on panels (C–E).
Ancient and Heterogeneous Gene Flow
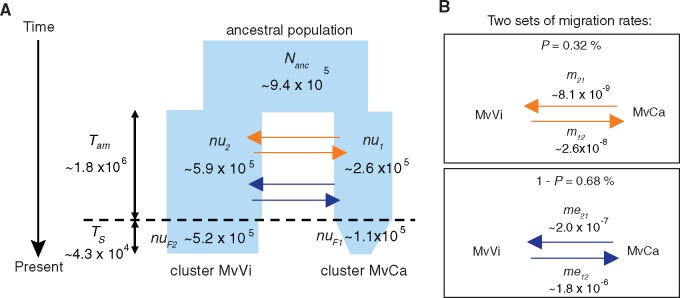
We compared 28 scenarios of divergence histories for the MvCa and MvVi genetic clusters, based on autosomal SNPs, using the diffusion approximation method implemented in dadi (Gutenkunst et al. 2009; supplementary table S4, Supplementary Material online). The most likely scenario, that is, the one with the lowest Akaike information criterion (AIC, supplementary table S4, Supplementary Material online), corresponded to a divergence with ancient and asymmetric gene flow and no recent gene flow (model AM2mG3; fig. 2 and supplementary figs. S2 and S3, Supplementary Material online). The delta AIC (corresponding to the difference between the AICs of each model and of the best model, supplementary table S4, Supplementary Material online) indicated that the only two other models with AIC values close to the lowest one also assumed heterogeneous migration rates along the genome and differed from the AM2mG3 model only by population sizes (models AM2m and AM2N2m; supplementary table S4, Supplementary Material online). The model likelihood was improved by adding recent population bottlenecks in MvCa and MvVi, as well as by adding heterogeneity in introgression rates along the genome before the cessation of gene flow (table 1; supplementary table S4 and note S2, Supplementary Material online). We used two sets of migration rates, applied to a proportion P and 1 − P of the genome (fig. 2B), which allowed to account for the accumulation of barriers to gene flow that can locally affect migration (Roux et al. 2013, 2016; Tine et al. 2014). We converted estimates of demographic model parameters by assuming an average mutation rate of 10−9 per generation and one generation per year (Badouin et al. 2017). During the period of ancient gene flow, rates of gene flow were relatively low, and effective migration from MvVi to MvCa was higher than the reverse direction by an order of magnitude (fig. 2B and supplementary table S5A, Supplementary Material online). Divergence was predicted to have initiated about 1.8 Ma (confidence interval of 360,000–5,000,000 years ago) and complete cessation of gene flow to have occurred about 50,000 years ago (fig. 2A and supplementary table S5A, Supplementary Material online). The phylogenetic network analysis implemented in SplitsTree also supported the absence of recent gene flow between the MvCa and MvVi genetic clusters, showing no footprint of recombination between strains of the two genetic clusters (fig. 1D). We found further evidence that the ancient gene flow between the diverging clusters was heterogeneous by obtaining variation in estimates of effective migration among autosomal contigs (supplementary note S3, fig. S4, and table S5B, Supplementary Material online). We also found that the divergence and diversity patterns along genomes were consistent with effects of linked selection, that is, with lower within-cluster genetic diversity and higher between-cluster divergence in genomic regions with lower recombination rates (supplementary note S3 and tables S6 and S7, Supplementary Material online), as expected for hitchhiking events across longer genomic distances (Burri et al. 2015; Burri 2017).
Fig. 2.
Most likely demographic model of divergence between the MvCa and MvVi Microbotryum genetic clusters. (A) Schematic representation of the divergence scenario between the MvCa cluster and the MvVi cluster from an ancestral population of size Nanc: period of ancient migration during a time period Tam and then cessation of gene flow since TS (model AM2mG3). In the MvCa genetic cluster, we modeled an exponential population decay at the time period TS from a population size nu1 to a population size nuF1. In the MvVi genetic cluster, we modeled a simultaneous population decay at the time period TS from a population size nu2 to a population size nuF2. The y-axis represents time and x-axis population sizes (not to scale). Converted parameter estimates are indicated (supplementary table S5, Supplementary Material online). Time is scaled in number of generations. (B) Two sets of migration parameters modeled to include genomic heterogeneity in gene flow during the period of ancient gene flow that accounts both for the accumulation of barriers to gene flow affecting migration locally (Roux et al. 2013, 2016; Tine et al. 2014): a proportion P of the genome has introgression rates of m12 and m21; a proportion 1 − P of the genome has introgression rates of me12 and me21. The migration parameter mij corresponds to the per-generation fraction of replacements in population i by genetic fragments from population j.
Table 1.
Summary of the Three Most Likely Demographic Models of Divergence in dadi between the MvCa and MvVi Microbotryum Genetic Clusters.
| Model | Number of Model Parameters | Ln L | AIC | Delta AIC |
|---|---|---|---|---|
| AM2m with population size changes in MvCa and MvVi clusters (AM2mG3) | 12 | −7,618 | 15,261 | 0 |
| AM2m | 10 | −7,679 | 15,379 | 118 |
| AM2m with heterogenous effective population size (AM2N2m) | 12 | −7,715 | 15,455 | 194 |
Note.—The three top models of divergence are variants of a model of isolation with ancient asymmetrical migration and heterogenous migration rate across the genome (AM2m). Logarithm of the maximum likelihood (Ln L), Akaike information criterion (AIC), delta AIC (corresponding to the difference in AIC between each model and the best model), and numbers of estimated parameters for each model run using dadi on the observed data set based on all autosomal oriented SNPs are presented.
Low Divergence Levels between Clusters on Mating-Type Chromosomes
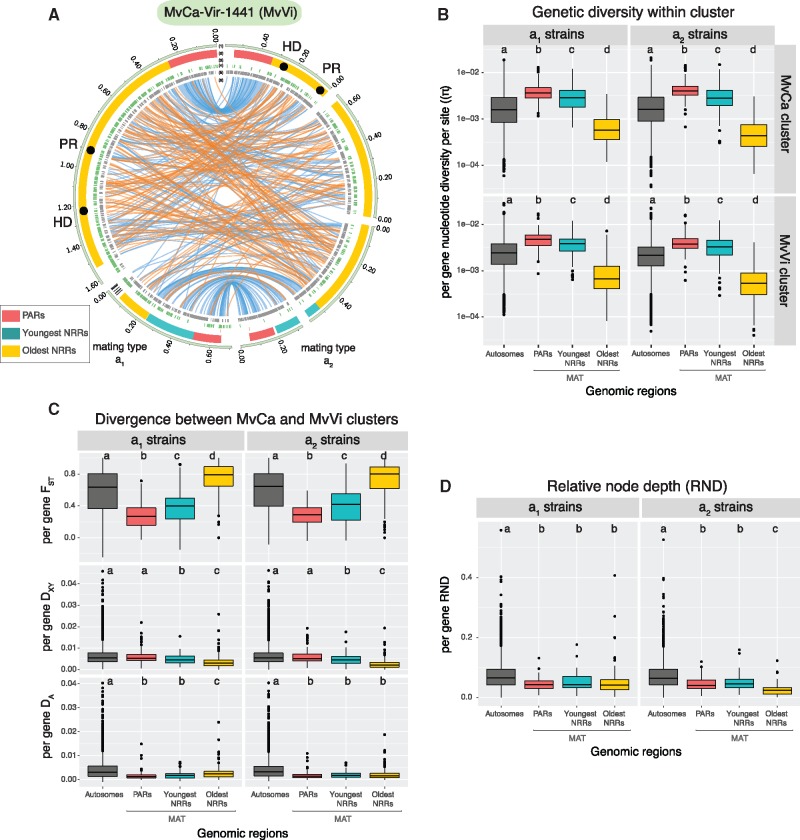
Having identified the main genetic clusters and temporal patterns of gene flow, we then determined whether the mating-type chromosomes, carrying large regions of recombination suppression, showed specific levels of genetic diversity and population divergence between lineages. We first used a long-read based technology (Oxford Nanopore) to obtain a high-quality genome assembly of the strain MvCa-Vir-1441 (supplementary table S8, Supplementary Material online) belonging to the MvVi cluster to compare with the previously obtained reference genome for the MvCa cluster, that is, the genome assembly of the strain MvCa-1250 (Branco et al. 2018). Comparison of the high-quality genomes of the two genetic clusters showed that the mating-type chromosomes were homologous between genetic clusters (fig. 3A and supplementary fig. S5, Supplementary Material online). The same large central region lacked recombination between the a1 and a2 mating-type chromosomes in MvVi and MvCa, as shown by the rearrangements between mating types in this region and the collinearity in the PARs at both ends of the chromosome (supplementary fig. S5C, Supplementary Material online). We found rearrangements in the NRRs between the reference genomes of the two clusters, although in limited numbers compared with rearrangements between mating types (fig. 3A and supplementary fig. S5, Supplementary Material online). The structures of the mating-type chromosomes and the NRRs were thus overall highly similar in the two clusters, which is consistent with the mating-type chromosome having been formed a long time before the MvVi–MvCa cluster divergence; indeed, we inferred here a complete cessation of gene flow between clusters 50,000 years ago while the main recombination suppression event on the MvCa mating-type chromosomes had been estimated to have occurred ∼2.3 Mya (Branco et al. 2018). The high-quality assemblies and the large population genomic data set further showed that the a2 mating-type chromosomes of these North American anther-smut fungi contained a single homeodomain (HD) gene, lacking the HD1 gene; in contrast, two HD genes are present in all the analyzed Microbotryum species to date (Branco et al. 2017, 2018; Hartmann, Rodríguez de la Vega, et al. 2018; Carpentier et al. 2019), as is typical in basidiomycete fungi, with a couple of other exceptions (Lengeler et al. 2002; Coelho et al. 2017; Sun et al. 2019).
Fig. 3.
Contrasted patterns of genetic diversity and population divergence between mating-type chromosomes and autosomes. (A) Structure and synteny of the a1 and a2 mating-type chromosomes of the MvCa-Vir-1441 strain belonging to the MvVi genetic cluster (i.e., mating-type chromosomes of MvCa-Vir-1441-A1 and MvCa-Vir-1441-A2 assemblies). Tracks represent from the outer to the inner center: 1) contigs, 2) localization of centromeric repeats in black, of the mating-type genes represented by black circles (the HD and pheromone receptor PR genes), and of regions with contrasted levels of recombination (recombining PARs in red, oldest nonrecombining regions (NRRs) in light blue, and youngest NRRs in yellow), 3) subtelomeric repeats in red, 4) transposable elements in green, 5) gene coding sequences in gray, and 6) blue and orange links connecting orthologous regions between assemblies. The link size is proportional to region length and orange links represent inversions. (B) Distribution of per-gene genetic diversity (π) in the MvCa and MvVi genetic clusters. The y-axis was log 10 scaled. (C) Distribution of per-gene net divergence (DA), absolute divergence (DXY), and relative divergence (FST) between the MvCa and MvVi genetic clusters. (D) Distribution of per-gene relative node depth (RND). Statistics distribution is presented separately for the a1 and a2 sequenced haploid genomes, and for the autosomes and three studied regions of the mating-type chromosome (MAT). Different letters indicate significantly different (P value < 0.05) mean values according to pairwise multiple comparisons Wilcoxon tests (P values are presented supplementary table S9, Supplementary Material online). On boxplots, the lower and upper hinges correspond to the first and third quartiles (i.e., 25th and 75th percentiles). The middle hinge corresponds to the median. Dots are outliers.
On the mating-type chromosomes, we studied separately recombining regions (PARs), regions with most ancient suppression of recombination (oldest NRRs, ca. 2.3 My old) and regions with most recent suppression of recombination (youngest NRRs, ca. 20 times younger; fig. 3; Branco et al. 2018). In each genetic cluster, and for both a1 and a2 mating-type chromosomes, we found significant differences in per-gene nucleotide diversity between autosomes and different mating-type chromosome regions (PARs, oldest NRRs, and young NRRs). We found significantly lower levels of genetic diversity in oldest NRRs than in other regions of mating-type chromosomes and than in autosomes (fig. 3B and supplementary table S9, Supplementary Material online); such lower diversity in the oldest NRR is consistent with the occurrence of introgression of a low number of haplotypes followed by the replacement of the whole NRR and with the half effective population size in mating-type chromosome NRRs compared with regions with recombination (Presgraves 2018; Wilson Sayres 2018). We found significantly higher levels of genetic diversity in the mating-type chromosome PARs and youngest NRRs than in autosomes (nearly 2-fold values; fig. 3B and supplementary table S9, Supplementary Material online). Such higher diversity in regions of the mating-type chromosomes with relatively recent recombination is consistent with introgression, as similar values are expected solely based on effective population sizes. Because these regions are not or little degenerated, the introgressions would not have been followed by selective sweeps and would have thus kept higher diversity.
To study divergence levels in mating-type chromosomes between MvCa and MvVi clusters, we used FST, DXY, and the net divergence index DA (fig. 3C), as these indexes are differentially impacted by the levels of standing genetic diversity and population size changes (Noor and Bennett 2009; Cruickshank and Hahn 2014; Burri 2017). The relative measure of divergence FST is highly dependent on within-population genetic diversity (Charlesworth 1998), which is less the case for DXY, but this later statistic still depends on ancestral population diversity (Gillespie and Langley 1979). DA allows comparison of populations with different effective sizes, such as autosomes and sex-related chromosomes (Nei and Li 1979), but is susceptible to level of diversity within each population. We found significant differences in divergence levels between autosomes and the three different mating-type chromosome regions for all three statistics, except for DXY between autosomes and PARs. We found higher FST values in the oldest NRRs and lower FST values in the PARs and the youngest NRRs than in the autosomes, likely due to differences in genetic diversity levels. We found significantly lower DXY levels in the youngest and oldest NRRs than in the autosomes and significantly lower DA levels on the three mating-type chromosome regions (PARs, youngest NRRs, and oldest NRRs) than on autosomes for both mating-type chromosomes. Such a lower general differentiation in mating-type chromosomes between clusters compared with autosomes is consistent with higher levels of gene flow in mating-type chromosomes than in autosomes.
Lower differentiation in mating-type chromosomes could alternatively be due to lower mutations rates on mating-type chromosomes, although higher substitution rates are expected based on their smaller population effective size. As a matter of fact, we found significantly higher total rates of substitutions on the PARs and youngest NRRs than on autosomes (supplementary fig. S6A and table S9C, Supplementary Material online), being calculated between MvVi or MvCa and Microbotryum lagerheimii, an outgroup species without ancient recombination suppression. The nonsynonymous (dN) substitution rates were higher on the oldest NRRs (supplementary fig. S6C, Supplementary Material online), which is consistent with lower population effective sizes and degeneration processes affecting mating-type chromosomes, as shown previously in other Microbotryum species (Fontanillas et al. 2015).
We also compared relative node depth (RND) values between autosomes and mating-type chromosomes; RND is a measure of relative per-gene divergence between the two clusters compared with the distance to an outgroup, which allows mitigating the potential confounding effect of different mutation rates on mating-type and autosomal chromosomes for gene flow inference (Feder et al. 2005; Rosenzweig et al. 2016). The shallower RNDs in gene genealogies of all three mating-type chromosome regions than in autosomes further supported higher gene flow levels in mating-type chromosomes (fig. 3D).
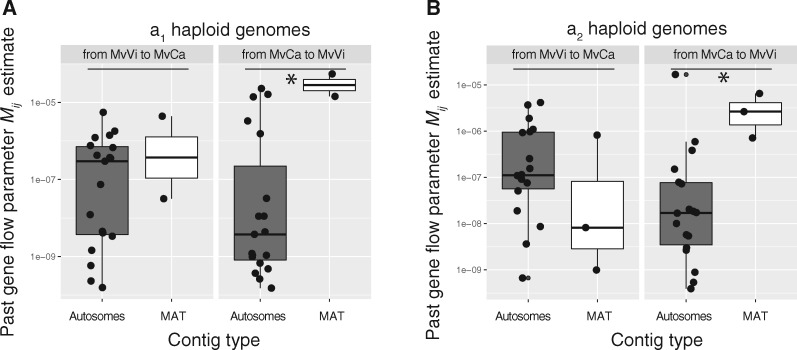
To test more directly if the mating-type chromosomes experienced higher levels of past gene flow than autosomes, we used dadi to compute estimates of past gene flow per contig, separating a1 and a2 genomes, based on the previously inferred most likely demographic divergence, that is, with ancient heterogeneous gene flow (model AM2mG3; fig. 2). We found that estimates of effective past migration rates on the majority of mating-type chromosomes contigs were slightly higher than on autosomal contigs, in particular from MvCa to MvVi (fig. 4; supplementary table S10 and fig. S7, Supplementary Material online). This supported the past occurrence of higher levels of gene flow between MvVi and MvCa in the mating-type chromosomes than in autosomes.
Fig. 4.
Comparison of past gene flow parameter estimates, for the dadi most likely model, between autosomes and mating-type chromosomes (MAT). The migration parameter Mij corresponds to the per-generation fraction of replacements in population i by genetic fragments from population j inferred by dadi most likely model (model AM2mG3) for the highest proportion of each contig for strains with a1 haploid genome (A) or a2 haploid genome (B). The dadi analysis was run by contig. Two sets of migration parameters were modeled to include genomic heterogeneity in gene flow (a proportion P of the contig having introgression rates of mij; a proportion 1 − P of the contig having introgression rates of meij; see fig. 2B);Mij corresponds to mij if P > 1 − P and meij if P < 1 − P. The y-axes were log 10 scaled. On boxplots, the lower and upper hinges correspond to the first and third quartiles (i.e., 25th and 75th percentiles). The middle hinge corresponds to the median. Dots are outliers. Asterisks indicate significantly different mean values between chromosome types (i.e., autosomes vs. mating-type chromosomes) according to Kruskall–Wallis rank sum tests (statistics are presented supplementary table S10C, Supplementary Material online).
Large Chromosomal Inversions on Autosomes
We used the long-read assemblies to investigate the possible occurrence of genome rearrangements on autosomal contigs between genetic clusters, as inversions could also show patterns of gene flow different from the genome-wide average (Hoffmann and Rieseberg 2008). Autosomal gene content was similar between the two genomes assemblies (supplementary table S8, Supplementary Material online), with about 95% of genes having orthologs in the two genomes; among genome-specific genes, we found no enrichment of genes encoding putative effectors (as predicted by EffectorP 2.0; Sperschneider et al. 2018). Although most autosomal contigs were highly collinear between the two assemblies, we identified two large autosomal inversions (fig. 5A), one on each of the two contigs MC02 and MC05 of each assembly, being 600- and 900-kb large, respectively (fig. 5B and supplementary table S11, Supplementary Material online). Centromeric repeats were present at one extremity of each involved contigs in the MvCa-1250-A1 assembly, and at the extremity of one contig in the MvCa-Vir-1441-A1 assembly (fig. 5B, track 2), but we found no evidence that the two contigs may form a single chromosome within each assembly. Presence of repeats at the rearrangement breakpoints (fig. 5B, track 3) suggested that the rearrangements may have occurred via nonhomologous recombination mediated by inverted repeats between two chromosomal arms.
Fig. 5.
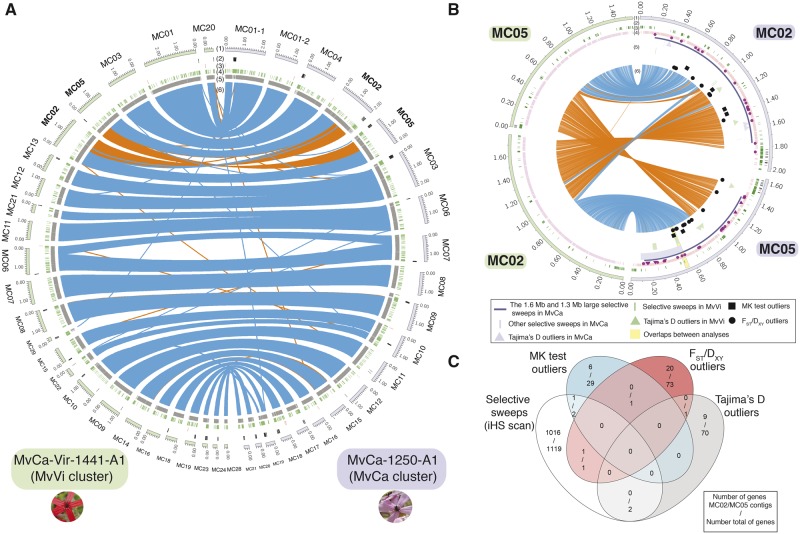
Large inversions on two autosomal contigs between the MvCa and MvVi Microbotryum genetic clusters and signatures of positive selection. (A) Circos plot showing the synteny between the MvCa-1250-A1 (MvCa, right, purple) and MvCa-Vir-1441-A1 (MvVi, green, left) assemblies on all autosomal contigs. Pictures of sampling hosts of the two corresponding strains are shown. Tracks represent from the outer to the inner center: 1) contigs in purple for MvCa-1250-A1 and in green for MvCa-Vir-1441-A1, 2) centromeric repeats in black, 3) subtelomeric repeats in red, 4) transposable elements in green; 5) gene coding sequences in gray, and 6) blue and orange links connecting single-copy orthologs between assemblies. Rearranged contigs are highlighted in bold. (B) Circos plots showing the synteny between the rearranged autosomal contigs MC02 (i.e., MvCa-1250-A1-R1_MC02) and MC05 (i.e., MvCa-1250-A1-R1_MC05) of the MvCa-1250-A1 assembly and contigs MC02 (i.e., MvCa-Vir-1441-A1_MC02) and MC05 (i.e., MvCa-Vir-1441-A1_MC05) of the MvCa-Vir-1441-A1 assembly and signatures of positive selection. Tracks represent from the outer to the inner center: 1) contigs in purple for MvCa-1250-A1 and in green for MvCa-Vir-1441-A1; 2) centromeric repeats in black; 3) transposable elements in green; 4) gene coding sequences in pink, with triangles for genes encoding proteins with predicted effector function and circles for genes encoding other predicted secreted proteins; 5) signatures of positive selection from various analyses (selective sweeps as purple [resp. green] traits and Tajima’s D as purple [resp. green] triangles in MvCa [resp. MvVi], McDonald–Kreitman [MK] tests as squares and FST and DXY outliers as circles), and 6) links connecting single-copy orthologs between assemblies, with the link size being proportional to gene length and orange links representing inversions. The genomic regions harboring overlapping signatures of positive selection detected in several scans are highlighted in yellow. (C) Venn diagram of overlaps between genes detected to be under positive selection between selective sweeps, Tajima’s D, MK tests, and FST and DXY outlier analyses. The number of genes on the rearranged autosomal contigs MC02 (i.e., MvCa-1250-A1-R1_MC02) and MC05 (i.e., MvCa-1250-A1-R1_MC05) only and the number total of genes are presented.
We checked for the presence of the large inversions involving the MC02 and MC05 contigs in the 37 other genomes of the MvCa and MvVi clusters by assembling de novo all Illumina genomes and using a mapping approach (supplementary note S4, Supplementary Material online). These analyses showed that the two large inversions were fixed within genetic clusters (supplementary table S11, fig. S8, and note S4, Supplementary Material online). We found no evidence of heterozygosity for the inversion in any of the analyzed diploid genomes. Comparative genomics with other Microbotryum species suggested that the derived state of the rearrangement likely occurred in the MvVi genetic cluster (supplementary note S4 and fig. S9, Supplementary Material online).
SplitsTree analyses of SNPs called in two large inversions involving the MC02 and MC05 contigs showed longer branches separating strains of the two genetic clusters than the genome-wide average, suggesting stronger differentiation, and therefore representing likely older cessation of gene flow between the MvCa and MvVi genetic clusters in these genomic regions (supplementary fig. S8C, Supplementary Material online). This was consistent with significantly higher mean values of relative divergence FST and absolute divergence DXY of the MC02 and MC05 contigs of the MvCa-1250-A1 assembly than for other contigs (Kruskall–Wallis rank sum test X2 statistic = 34.353, degree of freedom = 1, P value = 4.598e-09 and X2 statistic = 21.708, degree of freedom = 1, P value = 3.174e-06, respectively; supplementary figs. S4F and G, Supplementary Material online). The MC02 and MC05 contigs of the MvCa-1250-A1 assembly also had low estimates of effective past migration rates compared with other autosomal contigs (supplementary figs. S4A–C and table S5B, Supplementary Material online).
We found no significant enrichment in any specific gene function in the MC02 and MC05 rearranged contigs compared with other autosomal contigs. In particular, we found no enrichment of genes encoding secreted proteins or effectors compared with the genome-wide average on autosomes (for secreted proteins: Pearson’s chi-square test X2 = 1.3078 and P value = 0.2528 and degree of freedom = 1; for effectors: Pearson’s chi-square test X2 = 0.19611 and P value = 0.6579 and degree of freedom = 1). We tested whether rearranged contigs were enriched in genes under diversifying selection between clusters, which would point to a role of inversions in protecting beneficial allelic combinations specific to the cluster ecological niches. For this, we looked for genome-wide signatures of positive selection within each genetic cluster or footprints of elevated differentiation between clusters, using four approaches: the iHS statistics, detecting recent selective sweeps from haplotype structure (i.e., longer haplotypes than expected); the McDonald–Kreitman test, detecting high rates of nonsynonymous substitutions and thus recurrent selection for protein changes; Tajima’s D values, detecting excess of rare alleles in site frequency spectra and thus recent selective sweeps; and FST and DXY outlier genes, detecting particularly high genetic differentiation between clusters (supplementary note S5, table S12, and fig. S10, Supplementary Material online). The MC02 and MC05 rearranged contigs were significantly enriched both in selective sweeps detected with the iHS statistics and in FST and DXY outliers (fig. 5B and C; supplementary note S5 and table S13, Supplementary Material online). In particular, we identified ten genes encoding secreted proteins with effector functions in selective sweep regions in the MC02 and MC05 rearranged contigs (supplementary table S12A, Supplementary Material online), which may thus be involved in host specialization or coevolution. Noteworthy, the only two overlaps between regions with footprints of selective sweeps in the two genetic clusters were on the rearranged MC05 contig and corresponded to 26 genes, including one gene encoding a predicted effector, two genes encoding other predicted secreted proteins and one gene encoding a major facilitator superfamily transporter (supplementary note S5, table S12A, and fig. S10A, Supplementary Material online). Two additional genes were found as overlaps between iHS scans and other selection scan methods on the rearranged MC05 contig (fig. 5B and C), but these had no predicted function. The overlap between McDonald–Kreitman tests and iHS scans suggests that there has been selection for recurrent changes in amino acids and the last beneficial change was recent enough to still be detected by the presence of a long haplotype.
Discussion
Our study indicates recent divergence of plant pathogenic fungi involving genome rearrangements and initial heterogeneous gene flow across whole genomes. The most striking finding was an unexpected lower divergence between clusters in the mating-type chromosomes than in autosomes, in contrast to the higher divergence consistently found in sex chromosomes between closely related lineages in plants and animals (Meisel and Connallon 2013; Charlesworth et al. 2018; Irwin 2018; Presgraves 2018). We further detected a large autosomal inversion differentiating the clusters, with higher divergence than in other autosomal regions, and being enriched in footprints of positive selection. This provides a good example of an inversion likely involved in protecting beneficial allelic combinations (Hoffmann and Rieseberg 2008) and suggests that mating-type chromosomes may have impacts in speciation that are distinct from expectations based on sex chromosome studies.
Based on whole genome sequences, we identified two highly differentiated genetic clusters among the anther-smut fungi infecting a complex of North American Silene plants. The two genetic clusters of anther-smut fungi did not experience recent gene flow and were found on specific host lineages in natural populations despite largely overlapping ranges, suggesting host specialization. Adaptation to different host species may play a role in reproductive isolation, through migrant inviability and hybrid ill-suitability on parental hosts (Nosil et al. 2005; Giraud et al. 2008; de Vienne et al. 2009). Different pollinator guilds between S. caroliniana and S. virginica species (Fenster and Dudash 2001; Reynolds and Fenster 2008; Reynolds et al. 2009) might have contributed to reproductive isolation in these pollinator-borne anther-smut fungi. We did not perform cross-inoculations in controlled conditions, but these usually reveal less host specificity than in natural conditions in anther-smut fungi (de Vienne et al. 2009). Phylogeographic factors might also have played a role in the observed population structure. The isolation in distinct refugia during the last glaciation can strongly affect population structure in natural populations (Hewitt 2000), as has been shown in several anther-smut fungi and their hosts (Vercken et al. 2010; Gladieux et al. 2011; Feurtey et al. 2016; Martin et al. 2016; Van Rossum et al. 2018). The distribution of the S. caroliniana subspecies and their anther-smut fungi could be consistent with the locations of identified glacial refugia across Eastern United States (Antonovics et al. 2003; Swenson and Howard 2005; Barnard-Kubow et al. 2015); furthermore population bottlenecks were inferred during the period of recent strict isolation (<50,000 years ago), as often observed during glaciation periods. The time inferred for the onset of the divergence between the clusters, about 1.8 Ma, was much older than the last glaciation maximum (ca. 20,000 years ago; Clark et al. 2009), but the inferred date of 50,000 years ago for the complete cessation of gene flow could correspond to the last glaciation given estimate confidence intervals (supplementary table S5A, Supplementary Material online).
We inferred divergence with initial gene flow, supporting the view that divergence with gene flow can occur in natural populations (Via 2001; Nosil 2008; Feder et al. 2012; Gagnaire et al. 2013). Indeed, although gene flow is predicted to impair speciation by homogenizing gene pools (Mayr 1963; Coyne and Orr 2004; Bolnick and Fitzpatrick 2007), evidence of divergence with gene flow has been inferred in several organisms, such as walking‐sticks insects (Nosil and Crespi 2004), pea aphids (Via 1999), salamanders (Niemiller et al. 2008), and Heliconius butterflies (Martin et al. 2013). In fungi, recent studies highlighted the importance of gene flow between recently diverged fungal populations or between distantly related species (Leroy et al. 2014; Corcoran et al. 2016; Desjardins et al. 2017; Feurtey and Stukenbrock 2018; Gladieux et al. 2018; Maxwell et al. 2018, 2019; Silva et al. 2018; Tusso et al. 2019). Pathogenic fungi often have features particularly conducive to ecological divergence in sympatry, for example, high spore number, strong selection by their host and mating within their host (Giraud et al. 2006, 2010).
We found evidence of heterogeneity in levels of past gene flow, suggesting a role of selection in decreasing gene flow at some loci while favoring gene flow in other regions or homogenizing allelic frequencies at neutral loci unlinked to loci under selection (Wu 2001; Turner et al. 2005; Barton and Cara 2009). The heterogeneity in the genome-wide levels of divergence may also be partly due to background selection and recurrent selective sweeps within genetic clusters (Charlesworth 1998; Roux et al. 2013; Burri et al. 2015; Wolf and Ellegren 2017).
The most interesting case of heterogeneity in levels of past gene flow was the contrast between the mating-type chromosomes and autosomes. We observed unexpected lower divergence levels between genetic clusters on the mating-type chromosomes than on autosomes, in striking contrast with the typical pattern commonly found in animals and plants (Presgraves 2008; Meisel and Connallon 2013; Charlesworth et al. 2018). The lower divergence between clusters on the mating-type chromosomes than on autosomes occurred despite higher substitution rates and smaller population effective sizes, and thus most likely result from higher levels of past gene flow on the mating-type chromosomes, as supported by estimates of past gene flow parameters in dadi. This finding stands in stark contrast with the widespread and consistent large-X effect found in sex chromosomes in a variety of organisms, in which the X chromosome plays a disproportionately large effect as a barrier to gene flow (Meisel and Connallon 2013; Charlesworth et al. 2018; Irwin 2018; Presgraves 2018). This discrepancy can be due to the specificities of the mating-type chromosomes compared with sex chromosomes in terms of recombination suppression and heterozygosity and can thus provide general insights into the mechanisms at play in general in sex-related chromosomes. Indeed, the commonly accepted explanation for the large role of sex chromosomes in reproductive isolation is that the X chromosome recombines in females and is hemizygous in males, allowing more rapid evolution than in autosomes, inducing a large-X effect in genetic incompatibilities between closely related lineages. Sexual antagonism between males and females traits may also increase the sex chromosome evolution rate (Coyne and Orr 1989; Coyne 1992; Presgraves 2008). Because mating type is determined at the haploid stage in fungi, both mating-type chromosomes are equally nonrecombining (Branco et al. 2018), rendering selection less efficient in both mating-type chromosomes than autosomes (Fontanillas et al. 2015), so that no effect similar to the faster-X hypothesis is actually expected. Furthermore, no sexually antagonistic selection is expected as there is no male or female functions in these isogamous fungi (Bazzicalupo et al. 2019), which should also contribute to the lack of faster evolution in mating-type chromosomes. Although smaller population effective size of mating-type chromosomes should still lead to higher differentiation under neutral processes (Presgraves 2018; Wilson Sayres 2018), selection may have favored higher rates of introgression, at the early stages of MvCa–MvVi divergence, of the whole NRR of mating-type chromosomes to counter their degeneration following recombination suppression. The anther-smut fungi from the Microbotryum genus indeed have highly degenerated mating-type chromosomes, with accumulation of repetitive elements, nonsynonymous substitutions, and gene losses (Badouin et al. 2015; Fontanillas et al. 2015). Shortly after divergence, one lineage may have by chance suffered from a stronger deleterious mutation on one of its mating-type chromosome and introgression of the homologous mating-type chromosome from the other lineage may then have been beneficial. Such adaptive introgression of a less degenerated NRR from a closely related species has previously been suggested in another fungus with mating-type chromosomes carrying a large NRR, Neurospora tetrasperma (Strandberg et al. 2010; Sun et al. 2012; Corcoran et al. 2016) and our present study suggest this may, interestingly, be a general pattern. The permanent heterozygosity and lack of recombination typical of mating-type chromomes also apply to UV sex chromosomes in organisms such as red, green, and brown algae, as well as bryophytes, in which sex is determined at the haploid stage (Coelho et al. 2019), and similar patterns of higher gene flow on such sex chromosomes would also be predicted in these organisms, which will be interesting to test in future studies. The nonrecombining heterogametic sex chromosomes in plants and animals also degenerate, but their introgression among species is prevented by the rapidly accumulating genetic incompatibilities in homogametic sex chromosome revealed in the heterogametic sex (known as the Haldane’s rule; Coyne 1985).
Another interesting case of heterogeneous gene flow in anther-smut fungi was that of the two autosomal contigs MC02 and MC05 carrying large rearrangements, with inversions having likely occurred and been fixed in the MvVi cluster. These regions with inversions had experienced less gene flow than the rest of the genome and were highly enriched in signatures of positive and divergent selection. The elevated levels of absolute divergence (DXY) and reduced levels of past gene flow between genetic clusters in the genomic regions affected by inversions suggested that these rearrangements likely occurred before the complete cessation of gene flow between genetic clusters. The lower inferred gene flow and the numerous footprints of positive selection in these rearranged genomic regions suggest that the inversion protected from gene flow a suite of beneficial alleles involved in ecological adaptation, perhaps in relation to host specialization or other components of specific ecological niches of the lineages (Wellenreuther and Bernatchez 2018; Faria et al. 2019). Large rearrangements, including inversions, are well known to impede recombination upon crosses between lineages due to improper chromosome pairing during meiosis and therefore contribute to local adaptation, population divergence, and reproductive isolation, although their role is often complex (Dobzhansky 1970, 1981; Rieseberg 2001; Feder et al. 2003; Hoffmann and Rieseberg 2008; McGaugh and Noor 2012; Rogers et al. 2018; Coughlan and Willis 2019; Wellenreuther et al. 2019). It was interesting in this regard that the autosomal inversions were the only genomic regions showing multiple positive selection footprints, detected by several different methods, indicating the occurrence of different types of positive selection, across various time scales. The iHS method identifies recent selective sweeps, whereas McDonald–Kreitman tests detect recurrent changes in amino acids, and FST and DXY outliers are based on overall differentiation (McDonald and Kreitman 1991; Voight et al. 2006; Sabeti et al. 2007; Burri 2017). Overlaps between these various types of selection footprints were not necessarily expected and were not found in other genomic regions in our study or in other fungi (Badouin et al. 2017; Hartmann, Rodríguez de la Vega, et al. 2018; Mohd-Assaad et al. 2018; Schirrmann et al. 2018), which points to particularly strong and recurrent selection events in the inversion regions.
In conclusion, our study provides important insights into the process of ecological divergence showing lower gene flow and more selection footprints in autosomal inversions, suggesting a role of rearrangements in protecting from recombination beneficial combinations of alleles. The most exciting finding was the unexpected higher gene flow on mating-type chromosomes, in striking contrast with the consistent and widespread opposite patterns found in sex chromosomes of plants and animals. The higher gene flow on mating-type chromosomes suggested the lack of faster adaptive evolution of mating-type chromosomes and the occurrence of beneficial introgressions, perhaps to counteract degeneration in their NRRs. It will be interesting in future studies to test the hypothesis that mating-type chromosomes and UV sex chromosomes also have opposite patterns of gene flow compared with more classically studied XY or ZW sex chromosomes, which may contribute to generalize and test the theory on the role of sex chromosomes in speciation.
Materials and Methods
Sequencing Data and Genome Assemblies
We analyzed the genome sequences of 40 Microbotryum strains collected on S. virginica and four subspecies or varieties of S. caroliniana (S. caroliniana sub. caroliniana, S. caroliniana sub. wherryi, S. caroliniana sub. pennsylvanica, and S. caroliniana sub. pennsylvanica var. angelica) across Eastern United States (supplementary table S1, Supplementary Material online; supplementary Materials and Methods, Supplementary Material online). For read mapping, we used the high-quality genome assemblies of a1 and a2 genomes of the MvCa-1250 strain (collected on S. caroliniana sub. pennsylvanica, GPS: 36.91 and -76.04) previously obtained with P6/C4 Pacific Biosciences SMRT technology and annotated for gene models (supplementary table S8, Supplementary Material online; Branco et al. 2018). The MvCa-1250-A1 assembly was accessed from GenBank BioProject accession number PRJEB12080 (BioSample ID: SAMEA3706514, assembly: GCA_900014965) and the MvCa-1250-A2 assembly was accessed from GenBank BioProject accession number PRJEB12080 (BioSample ID: SAMEA3706515, assembly: GCA_900014955). Removing the strains MvCa-1250 and MvCa-1131, we kept 38 individuals for population genomic analyses using autosomes. Procedure for read mapping and SNPs calling is described in supplementary Materials and Methods, Supplementary Material online.
Population Structure Analyses
To investigate population subdivision, we used three main approaches on autosomal SNPs. First, we performed a PCA on the whole SNP data set called on autosomes (349,045 SNPs) using the R package {SNPRelate} (Zheng et al. 2012). Second, we run the model-based clustering approach implemented in FastStructure testing for K = 2 to K = 10 clusters (Raj et al. 2014). We used the “chooseK.py” script and visual inspection to assess the most relevant number of groups (K). We used the R package {Pophelper} (Francis 2017) to display barplots. Third, we performed as nonparametric test a discriminant analysis of principal components using the R package {adegenet} (Jombart 2008; Jombart and Ahmed 2011). These two analyses were performed on a set of 1,032 autosomal SNPs pruned for high linkage disequilibrium. To prune SNPs, we used the function snpgdsLDpruning() of the R package {SNPRelate} (Zheng et al. 2012) with a linkage disequilibrium threshold of r2 = 0.3.
Demographic History of Divergence and Gene Flow
We inferred the demographic history of divergence between genetic clusters using a modified version (Tine et al. 2014) of the diffusion approximation method implemented in the python package dadi (Gutenkunst et al. 2009) that analyses the joint site frequency spectrum. For model comparisons, we used unfolded site frequency spectra of the whole data set of observed SNPs for the 38 individuals on autosomes (referred to as the observed data set). We compared a total of 28 different demographic models of divergence on the observed data set, including strict isolation, continuous postdivergence gene flow, ancient migration, and secondary contact (supplementary table S4, Supplementary Material online). In order to account for local variations of the effect of accumulation of barriers to gene flow and the effect of background selection, we modeled heterogeneity in migration rates (2m) and/or in population size (2N) along the genome by setting two sets of migration rates and population sizes as described in Roux et al. (2013, 2016) and Tine et al. (2014) (supplementary table S4, Supplementary Material online). We modeled different population size changes (simultaneous or exponential growth and decay, G, G2, and G3; supplementary table S4, Supplementary Material online). We used the simulated annealing optimization procedure that performs one hot and one cold simulated annealing optimization before Broyden–Fletcher–Goldfarb–Shanno algorithm to perform optimization of model parameters (Tine et al. 2014). We used the version of dadi with improved optimization (available at https://github.com/QuentinRougemont/DemographicInference; last accessed December 15, 2018). We ran the optimization of each model at least 25 times with different parameter values to obtain convergence. We used the AIC, the delta AIC (corresponding to the difference of AIC of each model with the best model), and the distribution of the model residuals to select the most likely demographic model of divergence. We converted the scaled estimated parameters assuming an average mutation rate of 10−9 per generation for each simulated locus and one generation per year as previously (Badouin et al. 2017). To estimate parameter uncertainties of the selected most likely divergence model, we performed 300 parametric simulations using the same procedure as in Badouin et al. (2017). Briefly, we simulated 300 data sets using the coalescent simulator msms (Ewing and Hermisson 2010, compiling multiple 1-Mb fragments) based on the selected most likely divergence model and rerun parameters optimization in dadi for each simulated data set. We converted the scaled estimated parameters of the simulated data sets using the Nanc parameter estimated in the observed data set (supplementary table S5, Supplementary Material online). Equations of the selected model used in dadi and the coalescent simulator msms are presented supplementary note S2, Supplementary Material online. To compare parameter estimates of the selected demographic model for each autosomal contig, we ran dadi separately for each contig on unfolded site frequency spectra of the observed SNP data set of each contig for the 38 individuals using the same procedure. We only kept the 19 contigs with a length superior to N90. To accumulate further evidence of the history of gene flow between strains, we used the program SplitsTree (Huson 1998; Huson and Bryant 2006) that performs a neighbor-net tree. We used 340,891 SNPs called on autosomes with no missing data as input for the program.
Chromosome Synteny Analyses
To perform synteny analyses between the MvCa and MvVi genetic clusters, we sequenced the a1 and a2 haploid genomes of the MvCa-Vir-1441 strain (collected on S. virginica, Route 8, Floyd County, Virginia, Coord. GPS: 36.843278 and -80.316694), with MinION Oxford Nanopore Technology. Procedure for genome assembly and comparative genomics analyses is described in supplementary Materials and Methods, Supplementary Material online.
Comparison of Diversity and Divergence between Autosomes and Mating-Type Chromosomes
To compare diversity and divergence statistics from SNP data between autosomes and mating-type chromosomes, we used only strains with haploid sequenced genomes (i.e., genomes of one single mating type) and performed SNP calling separately for a1 and a2 mating-type genomes using the MvCa-1250 reference genome of the corresponding mating type. Using only haploid genomes limits SNP quality bias and the need of genome phasing on the highly dimorphic and heterozygous mating-type chromosomes, in contrast to the highly collinear and homozygous autosomes. For the a1 mating type, we studied 11 individuals having their a1 haploid genome sequenced (supplementary table S1, Supplementary Material online) and performed read mapping and SNP calling against the MvCa-1250-A1 genome. For the a2 mating type, we studied 27 individuals having their a2 haploid genome sequenced (supplementary table S1, Supplementary Material online), and we performed read mapping and SNP calling against the MvCa-1250-A2 genome. For both mating types, we excluded the two strains 1250 and 1131 that were likely siblings of the individual corresponding to the reference genome We used the same procedure for quality filtering of SNPs and repeat masking as described in section “Read Mapping and SNP Calling” of supplementary Materials and Methods, Supplementary Material online. The variant call format files are available upon request.
We computed diversity statistics as described in section “Genetic Diversity Statistics Computation” of supplementary Materials and Methods, Supplementary Material online. To compare mutation rates on autosomes and mating-type chromosome, we computed pairwise values of nonsynonymous divergence (dN) and synonymous (neutral) substitution rate (dS) between orthologous gene sequences of either the MvCa-1250 or the MvCa-Vir-1441 strain on the one hand, and the MvSv-1253 M. lagerheimii strain on the other hand, with the latter species having mostly recombining mating-type chromosomes (Branco et al. 2017, 2018, Carpentier et al. 2019). We focused on the a1 mating-type chromosome and therefore used the a1 haploid genome assemblies for the three studied strains (MvCa-1250-A1, MvCa-Vir-1441-A1, and MvSv-1253-A1). We aligned orthologous gene sequences using the codon-based approach implemented in translatorX with default parameters (Abascal et al. 2010) and used the nucleotide alignment as input in the yn00 program in the PAML package (Yang and Nielsen 2000; Yang 2007). We studied only orthologous groups with a single gene copy in each of the three studied genomes. We computed 6,535 (resp. 6,531) pairwise values for the MvCa-1250-A1 versus MvSv-1253-A1 (resp. MvCa-Vir-1441-A1 vs. MvSv-1253-A1) comparisons.
We computed the RND (Feder et al. 2005; Rosenzweig et al. 2016), a statistics allowing mitigating the potential confounding effect of different mutation rates on mating-type and autosomal chromosomes, by comparing the divergence between clusters to the distance to an outgroup. To compute RND, we used a customized script and as outgroup the a1 or a2 haploid genome assemblies of MvSv-1253 M. lagerheimii strain. For each strain of the MvCa and MvVi genetic cluster, we computed the absolute distance with the outgroup using the dist.dna() function and the F84 model in the R package {ape} (Paradis et al. 2004). We used the average of absolute distance value of all strains with the outgroup and the per-gene DXY value to compute per-gene RND.
To compare parameter estimates of the selected demographic model between autosomal and mating-type contigs, we ran dadi separately for autosomes and the a1 (resp. a2) mating-type contigs using SNPs of only strains with a1 (resp. a2) haploid genomes. We used the same procedure as described in section “Demographic History of Divergence and Gene Flow” except that for this set of analyses, we used the folded site frequency spectra instead of the unfolded site frequency spectra due to the lower number of SNPs on mating-type chromosomes than on autosomes (supplementary table S3, Supplementary Material online). Equation of the selected model used in dadi for folded site frequency spectra is presented supplementary note S2, Supplementary Material online. To be conservative, we converted the scaled estimated parameters assuming an average mutation rate of 10−9 per generation for each simulated locus for both autosomal and mating-type contigs, although mating-type contigs have likely higher mutation rates (see the “Results” section and supplementary fig. S6, Supplementary Material online). Higher mutation rates would decrease both estimates of population size and time of gene flow and would increase estimates of fraction of new migrants in each generation from population j to i (mij). Our assumption of equal mutation rates is thus conservative regarding gene flow on mating-type chromosomes.
Supplementary Material
Acknowledgments
We are grateful to Mike Perlin and Janis Antonovics for providing anther-smut fungal strains and the Genotoul platform for sequencing. We thank Antoine Branca, Jeanne Ropars, and Quentin Rougemont for help with analyses and Alodie Snirc for providing assistance in the laboratory. Janis Antonovics, Quentin Rougemont, and Antoine Branca provided helpful comments on an earlier version of the manuscript. This work was supported by the European Research Council (ERC) grants (consolidating grant GenomeFun 309403 and advanced grant EvolSexChrom 832352) and the Louis D. Foundation (Institut de France) award to T.G., a Marie Curie European grant (PRESTIGE-2016-4-0013) to F.E.H., and the NIH award (R15GM119092) to M.E.H.
References
- Abascal F, Zardoya R, Telford MJ.. 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 38(Suppl 2):W7–W13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonovics J, Hood ME, Thrall PH, Abrams JY, Duthie GM.. 2003. Herbarium studies on the distribution of anther-smut fungus (Microbotryum violaceum) and Silene species (Caryophyllaceae) in the eastern United States. Am J Bot. 90(10):1522–1531. [DOI] [PubMed] [Google Scholar]
- Antonovics J, Stratton D, Thrall PH, Jarosz AM.. 1996. An anther-smut disease (Ustilago violacea) of fire-pink (Silene virginica): its biology and relationship to the anther-smut disease of white campion (Silene alba). Am Midl Nat. 135(1):130–143. [Google Scholar]
- Badouin H, Gladieux P, Gouzy J, Siguenza S, Aguileta G, Snirc A, Le Prieur S, Jeziorski C, Branca A, Giraud T.. 2017. Widespread selective sweeps throughout the genome of model plant pathogenic fungi and identification of effector candidates. Mol Ecol. 26(7):2041–2062. [DOI] [PubMed] [Google Scholar]
- Badouin H, Hood ME, Gouzy J, Aguileta G, Siguenza S, Perlin MH, Cuomo CA, Fairhead C, Branca A, Giraud T.. 2015. Chaos of rearrangements in the mating-type chromosomes of the anther-smut fungus Microbotryum lychnidis-dioicae. Genetics 200(4):1275–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard-Kubow KB, Debban CL, Galloway LF.. 2015. Multiple glacial refugia lead to genetic structuring and the potential for reproductive isolation in a herbaceous plant. Am J Bot. 102(11):1842–1853. [DOI] [PubMed] [Google Scholar]
- Barton NH, Cara M.. 2009. The evolution of strong reproductive isolation. Evolution 63(5):1171–1190. [DOI] [PubMed] [Google Scholar]
- Bazzicalupo AL, Carpentier F, Otto SP, Giraud T.. 2019. Little evidence of antagonistic selection in the evolutionary strata of fungal mating-type chromosomes (Microbotryum lychnidis-dioicae). G3 (Bethesda) 9:1987–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolnick DI, Fitzpatrick BM.. 2007. Sympatric speciation: models and empirical evidence. Annu Rev Ecol Evol Syst. 38(1):459–487. [Google Scholar]
- Branco S, Badouin H, Rodríguez de la Vega RC, Gouzy J, Carpentier F, Aguileta G, Siguenza S, Brandenburg J-T, Coelho MA, Hood ME.. 2017. Evolutionary strata on young mating-type chromosomes despite the lack of sexual antagonism. Proc Natl Acad Sci U S A. 114(27):7067–7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco S, Carpentier F, Rodríguez de la Vega RC, Badouin H, Snirc A, Prieur SL, Coelho MA, de Vienne DM, Hartmann FE, Begerow D, et al. 2018. Multiple convergent supergene evolution events in mating-type chromosomes. Nat Commun. 9(1):2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burleigh JG, Holtsford TP.. 2003. Molecular systematics of the eastern North American Silene (Caryophyllaceae): evidence from nuclear ITS and chloroplast trnL intron sequences. Rhodora 105:76–90. [Google Scholar]
- Burri R. 2017. Interpreting differentiation landscapes in the light of long-term linked selection. Evol Lett. 1(3):118–131. [Google Scholar]
- Burri R, Nater A, Kawakami T, Mugal CF, Olason PI, Smeds L, Suh A, Dutoit L, Bureš S, Garamszegi LZ, et al. 2015. Linked selection and recombination rate variation drive the evolution of the genomic landscape of differentiation across the speciation continuum of Ficedula flycatchers. Genome Res. 25(11):1656–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpentier F, Rodríguez de la Vega RC, Branco S, Snirc A, Coelho MA, Hood ME, Giraud T.. 2019. Convergent recombination cessation between mating-type genes and centromeres in selfing anther-smut fungi. Genome Res. 29(6):944–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. 1998. Measures of divergence between populations and the effect of forces that reduce variability. Mol Biol Evol. 15(5):538–543. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Campos JL, Jackson BC.. 2018. Faster-X evolution: theory and evidence from Drosophila. Mol Ecol. 27(19):3753–3771. [DOI] [PubMed] [Google Scholar]
- Christmas MJ, Wallberg A, Bunikis I, Olsson A, Wallerman O, Webster MT.. 2019. Chromosomal inversions associated with environmental adaptation in honeybees. Mol Ecol. 28(6):1358–1374. [DOI] [PubMed] [Google Scholar]
- Clark PU, Dyke AS, Shakun JD, Carlson AE, Clark J, Wohlfarth B, Mitrovica JX, Hostetler SW, McCabe AM.. 2009. The Last Glacial Maximum. Science 325(5941):710–714. [DOI] [PubMed] [Google Scholar]
- Coelho MA, Bakkeren G, Sun S, Hood ME, Giraud T.. 2017. Fungal sex: the Basidiomycota. Microbiol Spectr. 5(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho SM, Mignerot L, Cock JM.. 2019. Origin and evolution of sex-determination systems in the brown algae. New Phytol. 222(4):1751–1756. [DOI] [PubMed] [Google Scholar]
- Corcoran P, Anderson JL, Jacobson DJ, Sun Y, Ni P, Lascoux M, Johannesson H.. 2016. Introgression maintains the genetic integrity of the mating-type determining chromosome of the fungus Neurospora tetrasperma. Genome Res. 26(4):486–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlan JM, Willis JH.. 2019. Dissecting the role of a large chromosomal inversion in life history divergence throughout the Mimulus guttatus species complex. Mol Ecol. 28(6):1343–1357. [DOI] [PubMed] [Google Scholar]
- Coyne J, Orr H.. 1989. Two rules of speciation. In: Otte D, Endler JA, editors. Speciation and its consequences. Sunderland, Massachusetts: Sinauer Associates, Inc. p. 180–207.
- Coyne J, Orr H.. 2004. Speciation. Sunderland (MA: ): Sinauer & Associates. [Google Scholar]
- Coyne JA. 1985. The genetic basis of Haldane’s rule. Nature 314(6013):736–738. [DOI] [PubMed] [Google Scholar]
- Coyne JA. 1992. Genetics and speciation. Nature 355(6360):511–515. [DOI] [PubMed] [Google Scholar]
- Cruickshank TE, Hahn MW.. 2014. Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Mol Ecol. 23(13):3133–3157. [DOI] [PubMed] [Google Scholar]
- de Vienne DM, Refrégier G, Hood ME, Guigue A, Devier B, Vercken E, Smadja C, Deseille A, Giraud T.. 2009. Hybrid sterility and inviability in the parasitic fungal species complex Microbotryum. J Evol Biol. 22(4):683–698. [DOI] [PubMed] [Google Scholar]
- Desjardins CA, Giamberardino C, Sykes SM, Yu C-H, Tenor JL, Chen Y, Yang T, Jones AM, Sun S, Haverkamp MR, et al. 2017. Population genomics and the evolution of virulence in the fungal pathogen Cryptococcus neoformans. Genome Res. 27(7):1207–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. 1970. Genetics of the evolutionary process. New York: Columbia University Press. [Google Scholar]
- Dobzhansky T. 1981. In: Lewontin RC, Moore JA, Provine WB, Wallace B, editors. Dobzhansky’s genetics of natural populations I–XLIII. New York: Columbia University Press. [Google Scholar]
- Ewing G, Hermisson J.. 2010. MSMS: a coalescent simulation program including recombination, demographic structure and selection at a single locus. Bioinformatics 26(16):2064–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria R, Johannesson K, Butlin RK, Westram AM.. 2019. Evolving inversions. Trends Ecol Evol. 34(3):239–248. [DOI] [PubMed] [Google Scholar]
- Feder JL, Berlocher SH, Roethele JB, Dambroski H, Smith JJ, Perry WL, Gavrilovic V, Filchak KE, Rull J, Aluja M.. 2003. Allopatric genetic origins for sympatric host–plant shifts and race formation in Rhagoletis. Proc Natl Acad Sci U S A. 100(18):10314–10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder JL, Egan SP, Nosil P.. 2012. The genomics of speciation-with-gene-flow. Trends Genet. 28(7):342–350. [DOI] [PubMed] [Google Scholar]
- Feder JL, Xie X, Rull J, Velez S, Forbes A, Leung B, Dambroski H, Filchak KE, Aluja M.. 2005. Mayr, Dobzhansky, and Bush and the complexities of sympatric speciation in Rhagoletis. Proc Natl Acad Sci U S A. 102(Suppl 1):6573–6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenster CB, Dudash MR.. 2001. Spatiotemporal variation in the role of hummingbirds as pollinators of Silene virginica. Ecology 82(3):844–851. [Google Scholar]
- Feurtey A, Gladieux P, Hood ME, Snirc A, Cornille A, Rosenthal L, Giraud T.. 2016. Strong phylogeographic co-structure between the anther-smut fungus and its white campion host. New Phytol. 212(3):668–679. [DOI] [PubMed] [Google Scholar]
- Feurtey A, Stukenbrock EH.. 2018. Interspecific gene exchange as a driver of adaptive evolution in fungi. Annu Rev Microbiol. 72(1):377–398. [DOI] [PubMed] [Google Scholar]
- Fontanillas E, Hood ME, Badouin H, Petit E, Barbe V, Gouzy J, Vienne DM, de Aguileta G, Poulain J, Wincker P, et al. 2015. Degeneration of the non-recombining regions in the mating-type chromosomes of the anther-smut fungi. Mol Biol Evol. 32(4):928–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis RM. 2017. pophelper: an R package and web app to analyse and visualize population structure. Mol Ecol Resour. 17(1):27–32. [DOI] [PubMed] [Google Scholar]
- Fraser JA, Diezmann S, Subaran RL, Allen A, Lengeler KB, Dietrich FS, Heitman J.. 2004. Convergent evolution of chromosomal sex-determining regions in the animal and fungal kingdoms. PLoS Biol. 2(12):e384.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman AB, Kellye Duong K, Shi T-L, Hughes CF, Perlin MH.. 2002. Isolates of Microbotryum violaceum from North American host species are phylogenetically distinct from their European host-derived counterparts. Mol Phylogenet Evol. 23(2):158–170. [DOI] [PubMed] [Google Scholar]
- Gagnaire P-A, Pavey SA, Normandeau E, Bernatchez L.. 2013. The genetic architecture of reproductive isolation during speciation-with-gene-flow in lake whitefish species pairs assessed by RAD sequencing. Evolution 67(9):2483–2497. [DOI] [PubMed] [Google Scholar]
- Gillespie JH, Langley CH.. 1979. Are evolutionary rates really variable? J Mol Evol. 13(1):27–34. [DOI] [PubMed] [Google Scholar]
- Giraud T, Gladieux P, Gavrilets S.. 2010. Linking the emergence of fungal plant diseases with ecological speciation. Trends Ecol Evol. 25(7):387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud T, Refrégier G, Le Gac M, de Vienne DM, Hood ME.. 2008. Speciation in fungi. Fungal Genet Biol. 45(6):791–802. [DOI] [PubMed] [Google Scholar]
- Giraud T, Villaréal LMMA, Austerlitz F, Le Gac M, Lavigne C.. 2006. Importance of the life cycle in sympatric host race formation and speciation of pathogens. Phytopathology 96(3):280–287. [DOI] [PubMed] [Google Scholar]
- Gladieux P, Condon B, Ravel S, Soanes D, Maciel JLN, Nhani A, Chen L, Terauchi R, Lebrun M-H, Tharreau D, et al. 2018. Gene flow between divergent cereal- and grass-specific lineages of the rice blast fungus Magnaporthe oryzae. mBio 9(1):e01219-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladieux P, Vercken E, Fontaine MC, Hood ME, Jonot O, Couloux A, Giraud T.. 2011. Maintenance of fungal pathogen species that are specialized to different hosts: allopatric divergence and introgression through secondary contact. Mol Biol Evol. 28(1):459–471. [DOI] [PubMed] [Google Scholar]
- Grognet P, Bidard F, Kuchly C, Tong LCH, Coppin E, Benkhali JA, Couloux A, Wincker P, Debuchy R, Silar P.. 2014. Maintaining two mating types: structure of the mating type locus and its role in heterokaryosis in Podospora anserina. Genetics 197(1):421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD.. 2009. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 5(10):e1000695.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann FE, Rodríguez de la Vega RC, Brandenburg J-T, Carpentier F, Giraud T.. 2018. Gene presence–absence polymorphism in castrating anther-smut fungi: recent gene gains and phylogeographic structure. Genome Biol Evol. 10(5):1298–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann FE, Rodríguez de la Vega RC, Carpentier F, Gladieux P, Cornille A, Hood ME, Giraud T.. 2019. Understanding adaptation, coevolution, host specialization, and mating system in castrating anther-smut fungi by combining population and comparative genomics. Annu Rev Phytopathol. 57(1):431–457. [DOI] [PubMed] [Google Scholar]
- Hewitt G. 2000. The genetic legacy of the Quaternary ice ages. Nature 405(6789):907–913. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Rieseberg LH.. 2008. Revisiting the impact of inversions in evolution: from population genetic markers to drivers of adaptive shifts and speciation? Annu Rev Ecol Evol Syst. 39(1):21–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood ME, Antonovics J, Wolf M, Stern ZL, Giraud T, Abbate JL.. 2019. Sympatry and interference of divergent Microbotryum pathogen species. Ecol Evol. 9(9):5457–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood ME, Petit E, Giraud T.. 2013. Extensive divergence between mating-type chromosomes of the anther-smut fungus. Genetics 193(1):309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH. 1998. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14(1):68–73. [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D.. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 23(2):254–267. [DOI] [PubMed] [Google Scholar]
- Irwin DE. 2018. Sex chromosomes and speciation in birds and other ZW systems. Mol Ecol. 27(19):3831–3851. [DOI] [PubMed] [Google Scholar]
- Jombart T. 2008. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24(11):1403–1405. [DOI] [PubMed] [Google Scholar]
- Jombart T, Ahmed I.. 2011. adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27(21):3070–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joron M, Frezal L, Jones RT, Chamberlain NL, Lee SF, Haag CR, Whibley A, Becuwe M, Baxter SW, Ferguson L, et al. 2011. Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature 477(7363):203–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M, Barton N.. 2006. Chromosome inversions, local adaptation and speciation. Genetics 173(1):419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhaney S, Fan G, Widemo F, Gunnarsson U, Thalmann DS, Hoeppner MP, Kerje S, Gustafson U, Shi C, Zhang H, et al. 2016. Structural genomic changes underlie alternative reproductive strategies in the ruff (Philomachus pugnax). Nat Genet. 48(1):84–88. [DOI] [PubMed] [Google Scholar]
- Le Gac M, Hood ME, Fournier E, Giraud T.. 2007. Phylogenetic evidence of host-specific cryptic species in the anther smut fungus. Evol Int J Org Evol. 61(1):15–26. [DOI] [PubMed] [Google Scholar]
- Lengeler KB, Fox DS, Fraser JA, Allen A, Forrester K, Dietrich FS, Heitman J.. 2002. Mating-type locus of Cryptococcus neoformans: a step in the evolution of sex chromosomes. Eukaryot Cell 1(5):704–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy T, Le Cam B, Lemaire C.. 2014. When virulence originates from non-agricultural hosts: new insights into plant breeding. Infect Genet Evol. 27:521–529. [DOI] [PubMed] [Google Scholar]
- Martin H, Touzet P, Van Rossum F, Delalande D, Arnaud J-F.. 2016. Phylogeographic pattern of range expansion provides evidence for cryptic species lineages in Silene nutans in Western Europe. Heredity 116(3):286–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SH, Dasmahapatra KK, Nadeau NJ, Salazar C, Walters JR, Simpson F, Blaxter M, Manica A, Mallet J, Jiggins CD.. 2013. Genome-wide evidence for speciation with gene flow in Heliconius butterflies. Genome Res. 23(11):1817–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell CS, Mattox K, Turissini DA, Teixeira MM, Barker BM, Matute DR.. 2019. Gene exchange between two divergent species of the fungal human pathogen, Coccidioides. Evolution 73(1):42–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell CS, Sepulveda VE, Turissini DA, Goldman WE, Matute DR.. 2018. Recent admixture between species of the fungal pathogen Histoplasma. Evol Lett. 2(3):210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr E. 1963. Animal species and evolution. Cambridge (MA: ): Harvard University Press. [Google Scholar]
- McDonald JH, Kreitman M.. 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351(6328):652–654. [DOI] [PubMed] [Google Scholar]
- McGaugh SE, Noor M.. 2012. Genomic impacts of chromosomal inversions in parapatric Drosophila species. Philos Trans R Soc B 367(1587):422–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel RP, Connallon T.. 2013. The faster-X effect: integrating theory and data. Trends Genet. 29(9):537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menkis A, Jacobson DJ, Gustafsson T, Johannesson H.. 2008. The mating-type chromosome in the filamentous ascomycete Neurospora tetrasperma represents a model for early evolution of sex chromosomes. PLoS Genet. 4(3):e1000030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Assaad N, McDonald BA, Croll D.. 2018. Genome-wide detection of genes under positive selection in worldwide populations of the barley scald pathogen. Genome Biol Evol. 10(5):1315–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Li WH.. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 76(10):5269–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemiller ML, Fitzpatrick BM, Miller BT.. 2008. Recent divergence with gene flow in Tennessee cave salamanders (Plethodontidae: Gyrinophilus) inferred from gene genealogies. Mol Ecol. 17(9):2258–2275. [DOI] [PubMed] [Google Scholar]
- Noor MAF, Bennett SM.. 2009. Islands of speciation or mirages in the desert? Examining the role of restricted recombination in maintaining species. Heredity 103(6):439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P. 2008. Speciation with gene flow could be common. Mol Ecol. 17(9):2103–2106. [DOI] [PubMed] [Google Scholar]
- Nosil P, Crespi BJ.. 2004. Does gene flow constrain adaptive divergence or vice versa? A test using ecomorphology and sexual isolation in Timema cristinae walking-sticks. Evolution 58(1):102–112. [DOI] [PubMed] [Google Scholar]
- Nosil P, Vines TH, Funk DJ.. 2005. Reproductive isolation caused by natural selection against immigrants from divergent habitats. Evolution 59:705–719. [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K.. 2004. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 20(2):289–290. [DOI] [PubMed] [Google Scholar]
- Popp M, Oxelman B.. 2007. Origin and evolution of North American polyploid Silene (Caryophyllaceae). Am J Bot. 94(3):330–349. [DOI] [PubMed] [Google Scholar]
- Presgraves DC. 2008. Sex chromosomes and speciation in Drosophila. Trends Genet. 24(7):336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC. 2018. Evaluating genomic signatures of “the large X-effect” during complex speciation. Mol Ecol. 27(19):3822–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Stephens M, Pritchard JK.. 2014. fastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics 197(2):573–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refrégier G, Le Gac M, Jabbour F, Widmer A, Shykoff JA, Yockteng R, Hood ME, Giraud T.. 2008. Cophylogeny of the anther smut fungi and their caryophyllaceous hosts: prevalence of host shifts and importance of delimiting parasite species for inferring cospeciation. BMC Evol Biol. 8(1):100.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds RJ, Fenster CB.. 2008. Point and interval estimation of pollinator importance: a study using pollination data of Silene caroliniana. Oecologia 156(2):325–332. [DOI] [PubMed] [Google Scholar]
- Reynolds RJ, Westbrook MJ, Rohde AS, Cridland JM, Fenster CB, Dudash MR.. 2009. Pollinator specialization and pollination syndromes of three related North American Silene. Ecology 90(8):2077–2087. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH. 2001. Chromosomal rearrangements and speciation. Trends Ecol Evol. 16(7):351–358. [DOI] [PubMed] [Google Scholar]
- Rogers DW, McConnell E, Ono J, Greig D.. 2018. Spore-autonomous fluorescent protein expression identifies meiotic chromosome mis-segregation as the principal cause of hybrid sterility in yeast. PLoS Biol. 16(11):e2005066.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig BK, Pease JB, Besansky NJ, Hahn MW.. 2016. Powerful methods for detecting introgressed regions from population genomic data. Mol Ecol. 25(11):2387–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux C, Fraïsse C, Romiguier J, Anciaux Y, Galtier N, Bierne N.. 2016. Shedding light on the grey zone of speciation along a continuum of genomic divergence. PLoS Biol. 14(12):e2000234.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux C, Tsagkogeorga G, Bierne N, Galtier N.. 2013. Crossing the species barrier: genomic hotspots of introgression between two highly divergent Ciona intestinalis species. Mol Biol Evol. 30(7):1574–1587. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie X, Byrne EH, McCarroll SA, Gaudet R, et al. 2007. Genome-wide detection and characterization of positive selection in human populations. Nature 449(7164):913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirrmann MK, Zoller S, Croll D, Stukenbrock EH, Leuchtmann A, Fior S.. 2018. Genomewide signatures of selection in Epichloë reveal candidate genes for host specialization. Mol Ecol. 27(15):3070–3086. [DOI] [PubMed] [Google Scholar]
- Silva DN, Várzea V, Paulo OS, Batista D.. 2018. Population genomic footprints of host adaptation, introgression and recombination in coffee leaf rust. Mol Plant Pathol. 19(7):1742–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperschneider J, Dodds PN, Gardiner DM, Singh KB, Taylor JM.. 2018. Improved prediction of fungal effector proteins from secretomes with EffectorP 2.0. Mol Plant Pathol. 19(9):2094–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strandberg R, Nygren K, Menkis A, James TY, Wik L, Stajich JE, Johannesson H.. 2010. Conflict between reproductive gene trees and species phylogeny among heterothallic and pseudohomothallic members of the filamentous ascomycete genus Neurospora. Fungal Genet Biol. 47(10):869–878. [DOI] [PubMed] [Google Scholar]
- Sun S, Coelho MA, Heitman J, Nowrousian M. 2019. Convergent evolution of linked mating-type loci in basidiomycete fungi. PLOS Genet. 15(9): e1008365. [DOI] [PMC free article] [PubMed]
- Sun Y, Corcoran P, Menkis A, Whittle CA, Andersson SGE, Johannesson H.. 2012. Large-scale introgression shapes the evolution of the mating-type chromosomes of the filamentous ascomycete Neurospora tetrasperma. PLoS Genet. 8(7):e1002820.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson NG, Howard DJ.. 2005. Clustering of contact zones, hybrid zones, and phylogeographic breaks in North America. Am Nat. 166(5):581–591. [DOI] [PubMed] [Google Scholar]
- Thompson MJ, Jiggins CD.. 2014. Supergenes and their role in evolution. Heredity 113(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tine M, Kuhl H, Gagnaire P-A, Louro B, Desmarais E, Martins RST, Hecht J, Knaust F, Belkhir K, Klages S, et al. 2014. European sea bass genome and its variation provide insights into adaptation to euryhalinity and speciation. Nat Commun. 5:5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TL, Hahn MW, Nuzhdin SV.. 2005. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 3(9):e285.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusso S, Nieuwenhuis BPS, Sedlazeck FJ, Davey JW, Jeffares DC, Wolf J.. 2019. Ancestral admixture is the main determinant of global biodiversity in fission yeast. Mol Biol Evol. 36(9):1975–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rossum F, Martin H, Le Cadre S, Brachi B, Christenhusz MJM, Touzet P.. 2018. Phylogeography of a widely distributed species reveals a cryptic assemblage of distinct genetic lineages needing separate conservation strategies. Perspect Plant Ecol Evol Syst. 35:44–51. [Google Scholar]
- Vercken E, Fontaine MC, Gladieux P, Hood ME, Jonot O, Giraud T.. 2010. Glacial refugia in pathogens: European genetic structure of anther smut pathogens on Silene latifolia and Silene dioica. PLoS Pathog. 6(12):e1001229.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via S. 1999. Reproductive isolation between sympatric races of pea aphids. I. Gene flow restriction and habitat choice. Evolution 53(5):1446–1457. [DOI] [PubMed] [Google Scholar]
- Via S. 2001. Sympatric speciation in animals: the ugly duckling grows up. Trends Ecol Evol. 16(7):381–390. [DOI] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK.. 2006. A map of recent positive selection in the human genome. PLoS Biol. 4(3):e72.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellenreuther M, Bernatchez L.. 2018. Eco-evolutionary genomics of chromosomal inversions. Trends Ecol Evol. 33(6):427–440. [DOI] [PubMed] [Google Scholar]
- Wellenreuther M, Mérot C, Berdan E, Bernatchez L.. 2019. Going beyond SNPs: the role of structural genomic variants in adaptive evolution and species diversification. Mol Ecol. 28(6):1203–1209. [DOI] [PubMed] [Google Scholar]
- Wilson Sayres MA. 2018. Genetic diversity on the sex chromosomes. Genome Biol Evol. 10(4):1064–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JBW, Ellegren H.. 2017. Making sense of genomic islands of differentiation in light of speciation. Nat Rev Genet. 18(2):87–100. [DOI] [PubMed] [Google Scholar]
- Wu C-I. 2001. The genic view of the process of speciation. J Evol Biol. 14(6):851–865. [Google Scholar]
- Yang Z. 2007. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R.. 2000. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 17(1):32–43. [DOI] [PubMed] [Google Scholar]
- Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS.. 2012. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28(24):3326–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.