Abstract
Many cytoskeletal proteins perform fundamental biological processes and are evolutionarily ancient. For example, the superfamily of actin-related proteins (Arps) specialized early in eukaryotic evolution for diverse cellular roles in the cytoplasm and the nucleus. Despite its strict conservation across eukaryotes, we find that the Arp superfamily has undergone dramatic lineage-specific diversification in Drosophila. Our phylogenomic analyses reveal four independent Arp gene duplications that occurred in the common ancestor of the obscura group of Drosophila and have been mostly preserved in this lineage. All four obscura-specific Arp paralogs are predominantly expressed in the male germline and have evolved under positive selection. We focus our analyses on the divergent Arp2D paralog, which arose via a retroduplication event from Arp2, a component of the Arp2/3 complex that polymerizes branched actin networks. Computational modeling analyses suggest that Arp2D can replace Arp2 in the Arp2/3 complex and bind actin monomers. Together with the signature of positive selection, our findings suggest that Arp2D may augment Arp2’s functions in the male germline. Indeed, we find that Arp2D is expressed during and following male meiosis, where it localizes to distinct locations such as actin cones—specialized cytoskeletal structures that separate bundled spermatids into individual mature sperm. We hypothesize that this unprecedented burst of genetic innovation in cytoskeletal proteins may have been driven by the evolution of sperm heteromorphism in the obscura group of Drosophila.
Keywords: positive selection, cytoskeleton, sperm, molecular dynamic simulations, cell biology
Introduction
Actin is one of the most evolutionarily conserved eukaryotic proteins. It plays numerous architectural and signaling roles, including cell-shape maintenance, cell motility, vesicle transport, and cytokinesis (Kabsch et al. 1990; Dominguez and Holmes 2011). These functions are carried out via the canonical actin fold, which predates eukaryotic divergence and is found among polymerizing proteins in bacteria (van den Ent et al. 2001) and archaea (Izore et al. 2016). The utilitarian actin fold is also conserved in actin-related proteins (Arps) (Kabsch et al. 1990; Frankel and Mooseker 1996; Dominguez and Holmes 2011). Eight conserved subfamilies of Arps specialized early in eukaryotic evolution for diverse cellular roles in the cytoplasm and the nucleus. These functions include facilitating the polymerization of actin (Arp2 and Arp3) (Mullins et al. 1998), promoting the motility of the microtubule-based motor dynein (Arp1 and Arp10) (Muhua et al. 1994; Lee et al. 2001), and participating in chromatin remodeling (Arps 4, 5, 6, 8) (Harata et al. 2000; Blessing et al. 2004; Klages-Mundt et al. 2018). Of the Arp superfamily members, only Arp1 homo-oligomerizes to form actin-like filaments (Schafer et al. 1994), whereas most Arps function in complex with other proteins (Machesky et al. 1994; Klages-Mundt et al. 2018).
Previous phylogenetic analyses have highlighted the evolution and distinguishing features of different Arp families (Goodson and Hawse 2002; Muller et al. 2005). Using Arp sequences collected predominantly from five model organisms (Arabidopsis thaliana, Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, and Homo sapiens), these studies defined the Arp subfamilies phylogenetically (Goodson and Hawse 2002; Muller et al. 2005) as highly distinct, well-defined clades. However, Arp sequences within each subfamily are highly similar, indicating the strong selective pressure for conservation of both sequence and function for each Arp following initial diversification (Goodson and Hawse 2002; Muller et al. 2005). Although most cytoplasmic and nuclear Arps are highly conserved across eukaryotic species, some Arps have also undergone lineage-specific gains and losses. For example, Arp1 and Arp10 were lost in plants most likely because their requisite binding partner, the motor dynein, is absent in plants (Hammesfahr and Kollmar 2012). Similarly, the Arp7 and Arp9 subfamilies are evolutionary inventions specific to fungi and perform roles in chromatin remodeling (Cairns et al. 1998; Peterson et al. 1998; Goodson and Hawse 2002).
The framework of well-defined Arp lineages has also facilitated the identification of lineage-specific “orphan” Arps that do not fall into any defined Arp subfamily (Goodson and Hawse 2002). For example, mammals have approximately seven testis-specific Arps with no known ortholog outside mammals. Although the highly conserved canonical Arps (1–10) are ubiquitously expressed, these orphan Arps are primarily expressed in mammalian male germ cells, where some are implicated in spermatogenesis and fertility (Heid et al. 2002; Tanaka et al. 2003; Hara et al. 2008; Boeda et al. 2011; Fu et al. 2012). Orphan Arp lineages have also been previously described in Drosophila. For example, all sequenced Drosophila species encode Arp53D, which has no ortholog outside insects (Goodson and Hawse 2002). Intriguingly, like the mammalian orphan Arps, Arp53D is also testis-specific in D. melanogaster and D. pseudoobscura (Fyrberg et al. 1994; Celniker et al. 2009), although its functional role remains unknown. Beyond these observations, testis-specific orphan Arps have received little scrutiny, presumably due to absence of orthologs in most phyla. Studies of such orphan Arps could reveal how a strikingly conserved superfamily can diversify in sequence for new cellular roles.
Here, we took a phylogenomic approach to uncover additional innovation in the Arp superfamily in Drosophila. Using a comprehensive survey of the Arp superfamily in 12 sequenced and well-annotated Drosophila species (Drosophila 12 Genomes Consortium 2007), we unexpectedly discovered four lineage-specific Arp paralogs, all of them arising in the common ancestor of the obscura clade of Drosophila. Despite the recurrence of Arp innovation in the same Drosophila lineage, we find that the Arp paralogs arose independently, via duplications of distinct parental Arps or actin genes. Most of these obscura-specific Arps have been retained over 14 My since the origin of this lineage, where they have evolved under positive selection. Similar to Arp53D, we find that all obscura-specific Arps are expressed in the testis. Detailed cytological analyses of one of these Arps, Arp2D, revealed its localization to gametic actin structures, such as motile actin cones that act during sperm individualization. Our results reveal that a burst of genetic innovation in the conserved Arp superfamily allowed for specialized roles in spermatogenesis in a lineage of Drosophila.
Results
A Burst of Lineage-Specific Duplications in the obscura Group of Drosophila
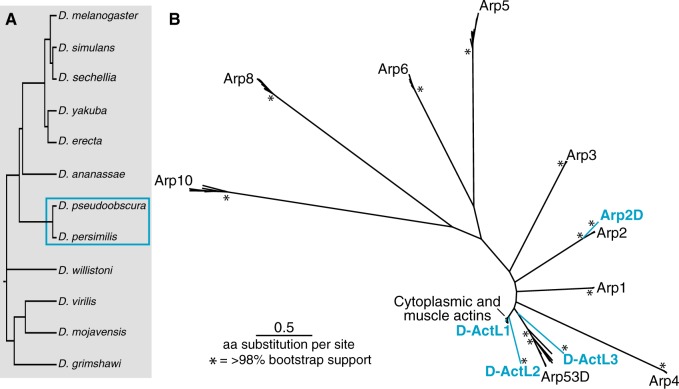
We performed a phylogenomic survey of the Drosophila Arp superfamily in the 12 sequenced and well-annotated Drosophila genomes (fig. 1A) (Drosophila 12 Genomes Consortium 2007). In addition to several members of the cytoplasmic (canonical) actin gene family, D. melanogaster encodes eight Arps (Arp1–6, 8, 10) and one orphan Arp53D (Fyrberg et al. 1994; Goodson and Hawse 2002). We used the protein sequences of all nine of these D. melanogaster Arps in tBLASTn searches of the 12 sequenced and annotated Drosophila species (Drosophila 12 Genomes Consortium 2007). All hits with highly significant E-values (<10−5) were collected, aligned, and subjected to phylogenetic and shared syntenic analysis (fig. 1B). Our study revealed that most sequences were orthologs of the previously identified Arps. However, we found four additional Arp homologs that were present in D. pseudoobscura and D. persimilis, two closely related species that diverged ∼1 Ma (Babcock and Anderson 1996). Only one of these Arp paralogs can be reliably placed within a conserved canonical Arp subfamily; we named this paralog Arp2D (fig. 1B). The other three paralogs are phylogenetically distinct (fig. 1B) and we named these D-ActL1, D-ActL2, and D-ActL3 (for Drosophila actin-like proteins 1–3).
Fig. 1.
The Arp superfamily exhibits lineage-specific duplications. (A) Drosophila melanogaster Arps were used in a tBLASTn search of the sequenced and annotated genomes of the 12 species displayed. Gene duplications in (B) were found in the two species outlined by a blue box. (B) All unique hits with E-value of ∼0 were translated and aligned using MAFFT (Katoh and Standley 2013). Poorly aligned sections of the sequence alignment (where <80% of sequences aligned) were removed. A PhyML tree (Guindon et al. 2010) with 100× resampling was generated (LG substitution model). The asterisks represent nodes with >98% bootstrap support. The highly conserved “canonical” Arps and testis-specific Arp53D are labeled in black, whereas the blue-labeled branches represent Arp paralogs only found in D. pseudoobscura and D. persimilis.
We performed an analysis of shared synteny to ascertain whether these newly identified Arp paralogs are specific to D. pseudoobscura and D. persimilis among the 12 well-assembled Drosophila genomes. We used the genomic locus of each novel D. pseudoobscura Arp paralog to identify shared syntenic locations in each of the other sequenced, annotated Drosophila genomes (supplementary figs. S1–S4, Supplementary Material online), using an E-value cutoff at 10−50. The syntenic loci of D-ActL2 and D-ActL3 are quite well conserved and were easy to define by two flanking genes that are conserved across Drosophila species. Based on this shared syntenic context, we confirmed that D-ActL2 and D-ActL3 were indeed absent in species other than D. pseudoobscura and D. persimilis. This analysis was more complicated for D-ActL1 because its syntenic locus is more dynamic. We could only reliably analyze the genes found upstream of D-ActL1 in D. pseudoobscura and D. persimilis. We found no actin-related genes proximal to these upstream genes outside the obscura group. Thus, in each case, we were able to confirm that the Arp genes were indeed missing in the syntenic locus of species other than D. pseudoobscura and D. persimilis (supplementary figs. S1–S4, Supplementary Material online). Based on the shared synteny analysis and our inability to find related Arp sequences in the fully sequenced genomes of other Drosophila species, we conclude that all four Arp paralogs arose in one lineage of Drosophila, which includes D. pseudoobscura and D. persimilis.
To further pinpoint the evolutionary age and origin of the four Arp paralogs, we extended our analyses to other members of the obscura group, which consists of species that have a common ancestor originating ∼14 Ma (Barrio and Ayala 1997; Russo et al. 2013). Using the alignment of the syntenic loci in D. melanogaster, we designed primers to regions of high conservation in genes or intergenic regions neighboring each of the four novel Arp paralogs. For loci that are present in poorly conserved or dynamically evolving syntenic contexts, we were aided in our primer design by draft genomes of the obscura species D. guanche, D. bifasciata, D. affinis, D. azteca, and D. helvetica (Levine M, personal communication). Using these primers, we PCR (polymerase chain reaction)-amplified and sequenced the locus of the Arp paralogs in ten additional species of the obscura group whose genomes have not been fully sequenced (fig. 2 and supplementary data S1, Supplementary Material online).
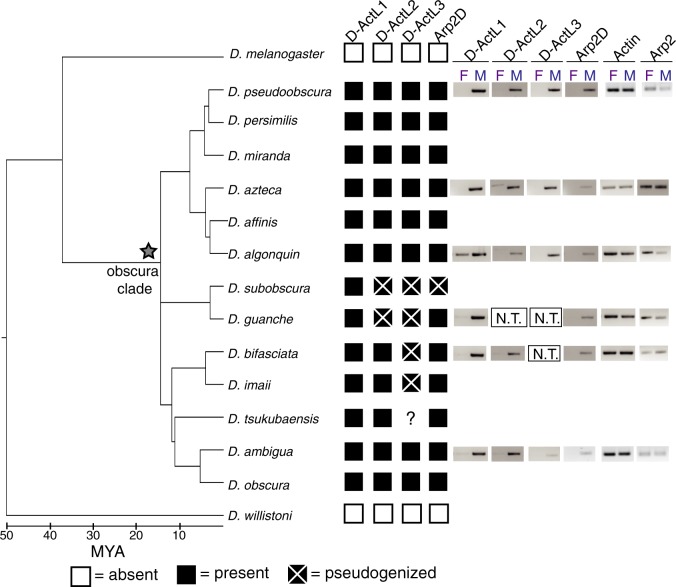
Fig. 2.
Four Arp paralogs originated in the common ancestor of the obscura group and maintained male-specific expression. Shared syntenic loci of the four Arp duplicates were PCR-amplified from 12 species in the obscura clade. The presence and absence of the duplicates are indicated with black and white boxes, respectively. Pseudogenized genes are indicated with boxes that have an “X,” and a star indicates the origin of the Arp duplicates. PCR amplification of the D-ActL3 locus from Drosophila tsukubaensis was unsuccessful so its status is unknown. RT-PCR was conducted with males and females of representative species in the obscura clade and DNA gels are displayed. Expression of canonical actin (Act5C) and Arp2 are shown in the gray box. D-ActL2 and/or D-ActL3 were excluded from the analysis for D. guanche and D. bifasciata (N.T., not tested) because the genes are pseudogenized.
Based on the results of our targeted sequencing, we conclude that all four obscura-specific Arp paralogs were present in the common ancestor of the obscura group (fig. 2 and supplementary data S3, Supplementary Material online). Phylogenetic analyses based on nucleotide alignments of the loci from each group recapitulated the obscura species tree (supplementary fig. S5, Supplementary Material online), thus confirming our identification of true orthologs of the Arp gene duplicates found in D. pseudoobscura and D. persimilis. D-ActL1 is present and intact in all surveyed species, whereas Arp2D is present and intact in all species except for D. subobscura, which has a single base pair deletion leading to a frameshift and numerous stop codons (supplementary fig. S6, Supplementary Material online). D-ActL2 and D-ActL3 have been pseudogenized in the lineage leading to D. subobscura and D. guanche, whereas D-ActL3 has been independently pseudogenized in the lineage leading to D. bifasciata and D. imaii (fig. 2). The published genome sequence of D. miranda (Zhou and Bachtrog 2012; Gramates et al. 2017) indicates that D-ActL2 and Arp2D may have pseudogenized in this species. However, our survey of eight strains of D. miranda found that all four Arp paralogs are intact in this species (supplementary fig. S7 and data S2, Supplementary Material online). This discrepancy could arise because the published D. miranda genome assembly may contain errors, or it could represent a divergent strain with mutations in multiple Arp paralog genes. Overall, our analyses indicate that although all four obscura-specific Arp paralogs originated ∼14 Ma, only D-ActL1 has been strictly retained in this lineage. Although the majority of other obscura group species have retained all four paralogs, D. subobscura is unusual in having pseudogenized three of the four obscura-specific Arp paralogs.
Even though all the novel Arp paralogs appear to have arisen in the common ancestor of the obscura group of Drosophila species, they appear to be derived from independent duplication events. Based on initial phylogenetic analyses, we were able to ascribe parentage to only two of the four obscura-specific paralogs with strong bootstrap support. For example, D-ActL1 is 96% identical to actin whereas Arp2D is 70% identical to parental Arp2 in D. pseudoobscura (supplementary fig. S8, Supplementary Material online). To delineate the origins of D-ActL1–3 Arp paralogs in the obscura clade, we took advantage of our additional sequencing within the obscura clade (fig. 2 and supplementary data S3, Supplementary Material online) to perform phylogenetic analyses (supplementary fig. S8, Supplementary Material online). These analyses reveal that each paralog (D-ActL1–3) forms a monophyletic clade with relationships that are largely consistent with the branching topology of obscura species (supplementary figs. S5 and S8, Supplementary Material online). All three duplicates are most similar to actin (Act5C) with amino acid identity 96.8%, 66.8%, and 57.1%, respectively. Although this analysis still did not have high enough confidence (or bootstrap support) to assign parentage to the D-ActL2 and D-ActL3 paralogs (fig. 1B and supplementary fig. S8, Supplementary Material online), we could nonetheless confidently conclude that they represent distinct evolutionary innovations from D-ActL1.
The obscura-Specific Arp Paralogs Are Primarily Expressed in Males
We used publicly available RNA-seq data to assay the expression of the previously described and newly discovered Arp paralogs in D. pseudoobscura tissues (Celniker et al. 2009). We confirmed that all canonical Arp paralogs are ubiquitously expressed in all tissues. In contrast, we found that all four obscura-specific Arp paralogs are testis-specific in D. pseudoobscura (Celniker et al. 2009). We investigated whether male-specific expression of these Arp paralogs is conserved in other species from the obscura group. We isolated RNA from males and females of six representative obscura species and generated cDNA to conduct reverse transcriptase (RT)-PCR analyses, allowing us to compare expression between males and females (supplementary data S1, Supplementary Material online). As expected, canonical actin and Arp2 are expressed at comparable levels between females and males of each species (fig. 2 and supplementary fig. S9, Supplementary Material online). In contrast, all obscura-specific Arp paralogs appear to be predominantly expressed in males with no or low expression detected in females in some species, such as D-ActL1 in D. algonquin. Therefore, male-enriched expression of the obscura-specific Arp paralogs has been largely conserved since their origin ∼14 Ma. Although we have not investigated the tissue-specific expression of the Arp paralogs in all obscura species, based on their testis-specific expression in D. pseudoobscura, we infer that all obscura-specific Arp paralogs are likely to be testis-specific in this lineage.
The obscura-Specific Arp Paralogs Have Evolved under Positive Selection
Although Arp genes are typically very highly conserved, testis-specific proteins are often under selective pressure to diversify (Jagadeeshan and Singh 2005; Kleene 2005; Turner et al. 2008). Therefore, we investigated the selective constraints acting on the obscura-specific Arp paralogs. We first tested each Arp paralog for positive selection using the McDonald–Kreitman (MK) test (McDonald and Kreitman 1991), which compares the ratio of nonsynonymous to synonymous fixed differences between two species (DN/DS) with the ratio of nonsynonymous to synonymous polymorphisms within a species (PN/PS). If the ratio of fixed differences is far greater than the polymorphism ratio (DN/DS ≫ PN/PS), then this excess of fixed nonsynonymous differences is inferred to be the result of positive selection. We sequenced each of the four Arp paralogs in 10 or 11 D. pseudoobscura strains (supplementary data S4, Supplementary Material online) and 8 strains of the closely related D. miranda (supplementary data S2, Supplementary Material online). For each Arp paralog, we aligned the nucleotide sequences and then conducted the MK test, comparing DN/DS to PN/PS in D. pseudoobscura versus D. miranda strains. We found that all four paralogs have evolved under positive selection with strong statistical significance (table 1).
Table 1.
MK Tests for Positive Selection.
| Arp Paralog | Polymorphism |
Divergence |
No. D. pse Strains | No. D. mir Strains | P-Value | Alpha | ||
|---|---|---|---|---|---|---|---|---|
| P S | P N | D S | D N | |||||
| D-ActL1 | 26 | 2 | 8 | 7 | 11 | 8 | 0.002 | 0.912 |
| D-ActL2 | 27 | 12 | 4 | 18 | 11 | 8 | <0.001 | 0.901 |
| D-ActL3 | 17 | 3 | 6 | 22 | 10 | 8 | <0.001 | 0.951 |
| Arp2D | 9 | 8 | 6 | 23 | 10 | 8 | 0.024 | 0.768 |
note.—The MK test compares the ratio of nonsynonymous to synonymous fixed differences between two species (DN/DS) with the ratio of nonsynonymous to synonymous polymorphisms within a species (PN/PS). Under neutrality, we expect DN/DS ∼ PN/PS. However, if the ratio of fixed differences is far greater than the polymorphism ratio (DN/DS ≫ PN/PS), then this excess of fixed nonsynonymous differences (evaluated with a Fisher’s exact test) is inferred to be the result of positive selection. The number of polymorphisms and divergences in the table are the total found for both species (Drosophila pseudoobscura [D. pse] and D. miranda [D. mir]). Alpha, or the neutrality index, represents the proportion of nonsynonymous substitutions likely to have been driven by positive selection (Smith and Eyre-Walker 2002). Alpha is defined as [1 − (DSPN/DNPS)] and is expected to be zero under neutrality, and approaches 1 if all the nonsynonymous substitutions are likely to be driven by positive selection. Alpha values of 0.8 or higher are considered very strong evidence of positive selection.
We next tested for recurrent positive selection at individual sites in the Arp paralogs over the entire obscura group using maximum-likelihood methods found in the PAML suite (Yang 2007). For each of the four Arp paralogs, we constructed a codon-based alignment of all orthologous sequences. We investigated these alignments for evidence of recombination using GARD analyses (Kosakovsky Pond et al. 2006); we did not find any significant evidence for recombination. Using a species tree, we tested whether NsSites models that permitted codons to evolve under positive selection (M8) were a more likely fit to the data than those models (M7, M8a) that disallowed it. We found marginal evidence for recurrent positive selection acting on D-ActL3; it is possible that a smaller set of D-ActL3 orthologs could have lowered our power to detect positive selection in this gene (McBee et al. 2015). In contrast to D-ActL3, we found no evidence in support of positive selection in any of the other three obscura-specific Arp paralogs (table 2). We also did not detect branch-specific positive selection for any of the obscura-specific Arp paralogs (supplementary fig. S10, Supplementary Material online).
Table 2.
Maximum-Likelihood Tests for Recurrent Positive Selection.
| Arp paralog | No. Species | Species Used in Analysis | M7 versus M8 P-Value | M8a versus M8 P-Value | Tree Length in M8 | % sites dN/dS > 1 (average dN/dS) |
|---|---|---|---|---|---|---|
| D-ActL1 | 13 | pse, pers, mir, azt, aff, alg, sub, guan, bif, ima, tsuk, amb, and obs | 0.12 | 1.00 | 2.25 | 0 (1.00) |
| D-ActL2 | 11 | pse, pers, mir, azt, aff, alg, bif, ima, tsuk, amb, and obs | 0.20 | 0.28 | 1.45 | 0.41 (4.20) |
| D-ActL3 | 8 | pse, pers, mir, azt, aff, alg, amb, and obs | 0.09 | 0.06 | 1.49 | 8.19 (2.19) |
| Arp2D | 12 | pse, pers, mir azt, aff, alg, guan, bif, ima, tsuk, amb, and obs | 1.00 | 1.00 | 2.76 | 0 (1.00) |
note.—Using the PAML suite (Yang 2007), we tested whether NsSites models that permitted a subset of codons to evolve under positive selection (M8) were a more likely fit to the data than those models (M7, M8a) that disallowed it. Tree length refers to the number of nucleotide substitutions per codon, giving an indication of the divergence of the data set. The results we present are from codeml runs using the F3 × 4 codon frequency model and initial omega (dN/dS) of 0.4. This test was performed with multiple initial omega values and codon frequency models and the results were consistent with those shown. The abbreviations for the species used in the analysis indicate Drosophila pseudoobscura (pse), D. persimilis (pers), D. miranda (mir), D. azteca (azt), D. affinis (aff), D. algonquin (alg), D. subobscura (sub), D. guanche (guan), D. bifasciata (bif), D. imaii (ima), D. tsukubaensis (tsuk), D. ambigua (amb), and D. obscura (obs).
Our results from the MK test suggest that all obscura-specific Arp paralogs have evolved under strong episodic positive selection, at least in the lineage leading to D. pseudoobscura. However, positive selection does not appear to have driven recurrent amino acid replacement at a subset of “hotspot” sites (except for possibly D-ActL3). Rather, the signal of positive selection appears to be distributed over the entire length of all four Arp genes, mapping to residues on the surface of all four subdomains of the conserved actin fold. These findings suggest that the diversifying selection of obscura-specific Arps is distributed across multiple protein surfaces rather than limited to a single domain.
Arp2D Arose from a Retroduplication Event of Arp2 in the obscura Group
Of the four Arp paralogs, the parentage of Arp2D is most unambiguous because it is phylogenetically distinct from the D-ActL1–3 paralogs. Although Arp2 contains six introns, D. pseudoobscura Arp2D has none (fig. 3B). This lack of introns is a conserved feature in all obscura Arp2D genes and leads to a size difference in the genomic PCR analyses using primers to segments conserved between Arp2 and Arp2D (fig. 3A). The same PCR reaction using D. melanogaster genomic DNA resulted in a single band expected for the size from Arp2, confirming the absence of Arp2D in this species (fig. 3A).
Fig. 3.
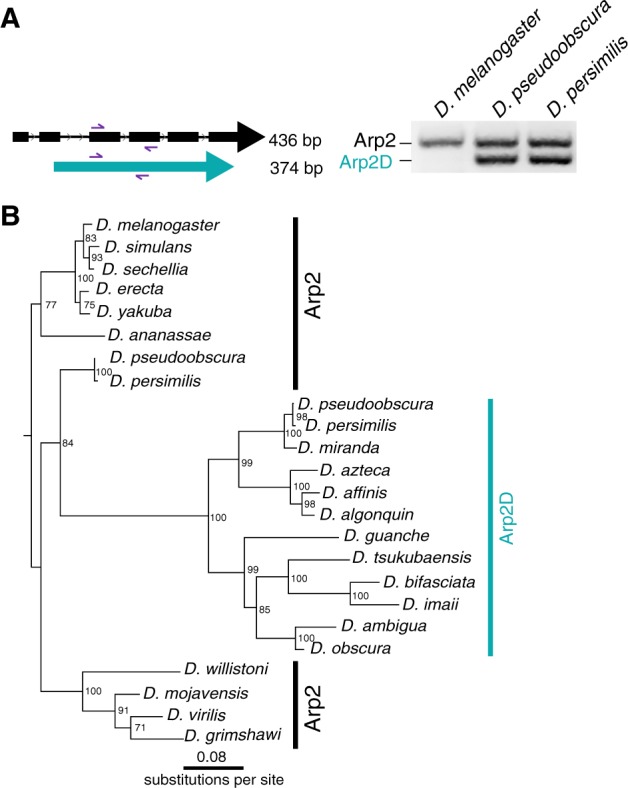
Arp2D originated from a retroduplication event of Arp2. (A) Primers targeting sequences in two neighboring exons conserved in Drosophila pseudoobscura Arp2 and Arp2D were used to PCR-amplify the genomic region in D. melanogaster, D. pseudoobscura, and D. persimilis. PCR products were run on a 2% agarose gel. As Arp2D does not contain introns, the PCR product is smaller (374 bp) than that of Arp2 (436 bp). (B) Nucleotide sequences from Arp2 (from 12 sequenced Drosophila species) and Arp2D sequences from the obscura group were aligned (no gaps were removed). A PhyML tree (Guindon et al. 2010) with 100× resampling was generated and presented using the species from the D. melanogaster subgroup as an outgroup.
Using a codon-based alignment, we performed phylogenetic analyses using maximum-likelihood methods to investigate the evolutionary origins of Arp2D (from 11 obscura species) relative to its parental gene Arp2 (from 12 Drosophila species). Our analyses reveal that the Arp2D sequences form a monophyletic clade, whose closest outgroup contains the D. pseudoobscura and D. persimilis Arp2 orthologs (84% bootstrap support, fig. 3B). The branching topology within the Arp2D clade (fig. 3B) also mirrors that of the obscura species tree (fig. 2A andsupplementary fig. S5, Supplementary Material online) (Barrio and Ayala 1997; Russo et al. 2013). Therefore, we conclude that Arp2D arose via retroduplication of the mRNA encoding Arp2 in the common ancestor of the obscura lineage. Unlike for Arp2D, we cannot distinguish between gene duplication versus retroduplication mechanisms for the origins of D-ActL1–3 because these Arp paralogs likely arose from intron-less actin genes (fig. 1B and supplementary fig. S8, Supplementary Material online).
Computational Modeling Predicts Common Biochemical Properties of Arp2D and Arp2
Next, we investigated whether Arp2D, which shares ∼70% identity with Arp2, diverged sufficiently to differ in some biochemical properties from Arp2, which is under stringent negative selection. To perform its function of polymerizing branched actin networks, Arp2 requires incorporation into a seven-membered multiprotein complex (Pollard 2007). This complex binds to a preformed “mother” actin filament, which leads to a conformational change enabling the complex to bind “daughter” actin monomers, which serve as a platform upon which a second actin filament (or “branch”) can polymerize (Pollard 2007).
To begin our comparison between the predicted properties of Arp2D and Arp2, we first compared the predicted sequence motifs required for ATP-binding and hydrolysis required for polymerization of branched actin. We find that Arp2D and the other obscura-specific Arp paralogs have preserved the sequence motif required for ATP binding and hydrolysis (supplementary fig. S11, Supplementary Material online). To further compare Arp2D’s biochemical properties with Arp2, we used computational modeling to predict how well Arp2D can sustain the required interactions in the active Arp2-multiprotein assembly structure to catalyze branched actin networks. This method has been successfully deployed for studying the effects of mutations in other cytoskeletal proteins like actin (Aydin et al. 2018). We started with a structure of the mammalian Arp2/3 branch junction complex (Pfaendtner et al. 2012), which was developed by combining data from X-ray structures of the complex along with electron tomography data on the junction conformation (Rouiller et al. 2008). We used this mammalian model to construct a D. pseudoobscura homology model of the entire Arp2/3 complex, that is, with D. pseudoobscura protein sequences for each component of the complex (Arpc1-5, Arp3, and Arp2 or Arp2D) (see Materials and Methods). Although this model did not include the “mother” and “daughter” actin filaments, we did include the two actin monomers bound to Arp2 and Arp3 that ultimately form the daughter branch. If Arp2D’s sequence were incompatible with Arp2’s canonical structure or function, we would expect to reveal differences in the stability of these complexes with Arp2D instead of Arp2 upon molecular dynamics (MD) simulation. For example, if the structure were unstable, clashes would be unresolvable by MD simulation and the complex would either fall apart or greatly deform. However, we found that both Arp2 and Apr2D multiprotein complexes remained intact after a ∼200-ns MD simulation (fig. 4A and supplementary fig. S12A, Supplementary Material online). The D. pseudoobscura Arp2D/3 complex had only minimal spatial deviation from, and comparable stability to, the Arp2/3 complex (supplementary fig. S12A, Supplementary Material online). We found no steric clashes between residues within the structure after the MD and the behavior of the daughter actin monomers did not deviate far from the orientation predicted for a stable actin filament (supplementary fig. S12A, Supplementary Material online; Dominguez and Holmes 2011). Thus, overall, our MD analyses suggest that Arp2D should be biochemically capable of replacing Arp2 in the complex.
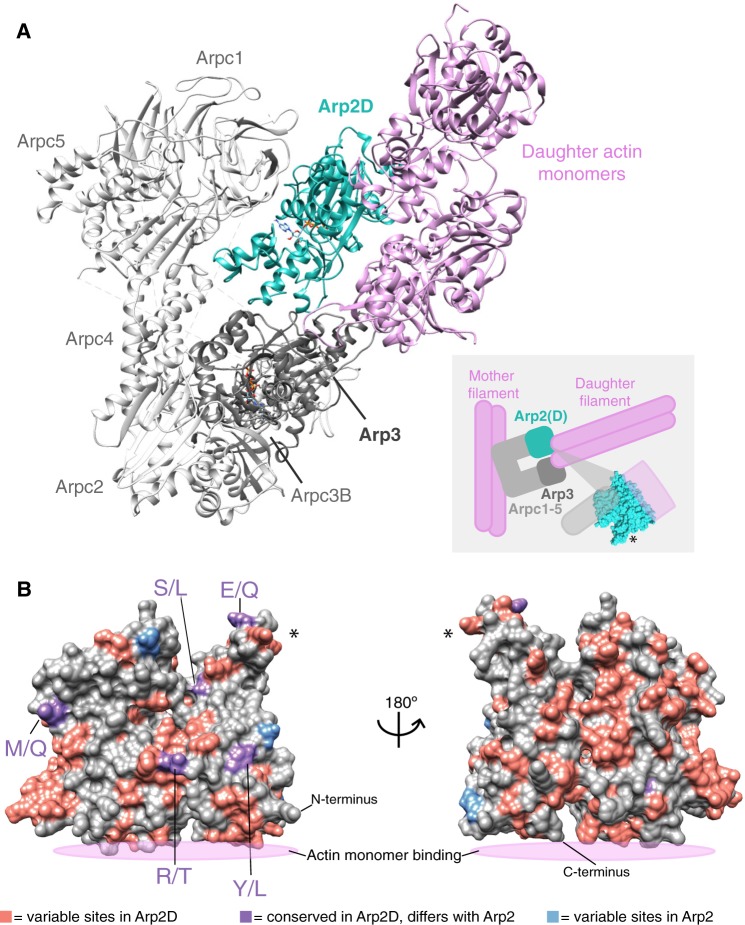
Fig. 4.
Arp2D is predicted to bind a daughter actin monomer. (A) A homology model of the Drosophila pseudoobscura Arp2/3 complex bound to daughter actin monomers was subjected to structural refinement and equilibration with Arp2D replacing Arp2. Components of the complex are labeled, with daughter actin monomers in pink and Arp2D in teal. The cartoon inset depicts how the canonical Arp2/3 complex binds to a pre-formed “mother” actin filament and subsequently generates a daughter filament, with Arp2/3 at the junction. The structure of Arp2 is shown in the inset to indicate where the interaction with actin and the complex approximately takes place. An asterisk on one part of the structure in the inset serves as a reference point for (B). (B) Drosophila pseudoobscura Arp2 is shown as a space-filled homology model. All Arp2D protein sequences and a few representative Arp2 sequences (from D. persimilis, D. pseudoobscura, D. guanche, D. bifasciata, and D. azteca) were aligned and residues were categorized as one of the following: 1) variable sites in Arp2D (pink), 2) conserved in Arp2D but differs from conserved residues in Arp2 (purple, denoted as Arp2 residue/Arp2D residue), and 3) variable sites in Arp2 residues (blue). The asterisk marks the same subdomain of the structure shown in (A).
To further examine the differences in biochemical properties between Arp2D and parental Arp2, we identified the fixed changes that distinguish all obscura Arp2D orthologs from all Arp2 orthologs. Based on the alignment of D. pseudoobscura Arp2 and Arp2D from all obscura species, we focused on residues that represented fixed differences between Arp2 and Arp2D because these changes reflect divergence that occurred early after the duplication of Arp2D that have been preserved by functional constraints. We found that there were nine fixed changes between Arp2D and Arp2 (fig. 4B and supplementary fig. S12B, Supplementary Material online) that predominantly map onto one surface of the structure. This implies that this surface reflects the primary functional divergence between these two paralogs. Notably, we found that the fixed differences between Arp2D and Arp2 did not make significant contacts with other components of the Arp2/3 complex or with the actin monomer. Instead, we hypothesize that these differences between Arp2D and Arp2 affect their interactions with regulators of the Arp2/3 complex.
We found that only three sites are variable among Arp2 orthologs, whereas many more residues are variable among Arp2D orthologs (fig. 4B). This difference could be a result of relaxed functional constraint acting on Arp2D but could also be the result of positive selection acting on Arp2D (table 1). None of these variable residues clusters significantly on the surface of Arp2D within the simulated Arp2D/3 complex homology model (fig. 4A), consistent with our previous findings that positive selection of Arp2D is not necessarily concentrated in one domain.
Arp2D Localizes to Gametic Actin Structures in D. pseudoobscura Testes
Next, we explored whether Arp2D’s cytological localization in vivo resembles that of parental Arp2. We first compared Arp2D’s subcellular localization with that of Arp2 in D. pseudoobscura tissue culture cells. Similar to actin, Arp2 localizes at the cell membrane and at cell–cell junctions in tissue culture cells for the polymerization of branched actin networks (Pollard and Borisy 2003). We found that Arp2-GFP expectedly localizes to the cell cortex in D. pseudoobscura cells (fig. 5A). We then expressed Arp2D-GFP and found that it also localizes to the cell cortex and at sites of cell–cell contact (fig. 5A). Thus, in spite of the fact that Arp2D is ∼30% divergent at the amino acid level from canonical D. pseudoobscura Arp2, it has a similar cytological localization in D. pseudoobscura cells. These findings are consistent with our computational modeling analyses, which suggest that Arp2D is compatible with the structure of an activated and nucleating Arp2/3 complex as described in the previous section (fig. 4A and supplementary fig. S12A, Supplementary Material online).
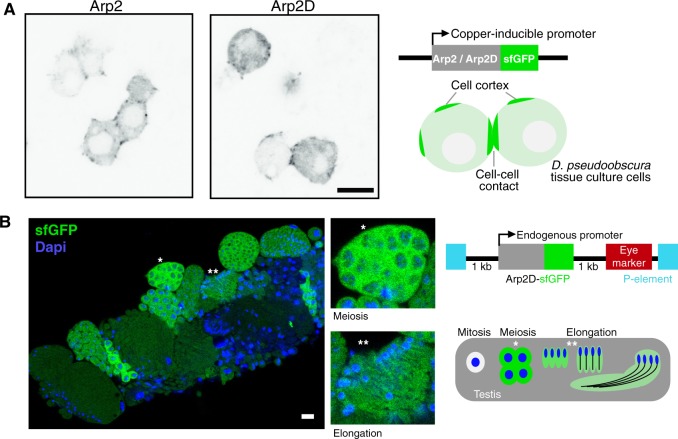
Fig. 5.
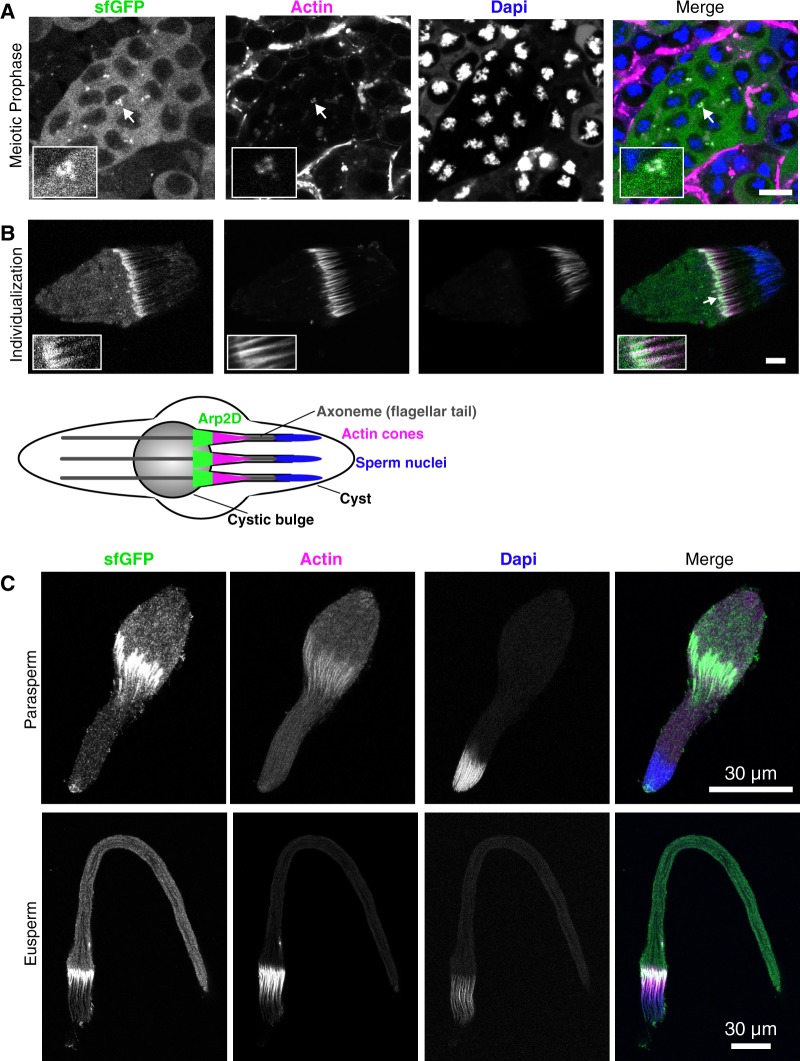
Arp2D is expressed in postmitotic cell stages. (A) Arp2 and Arp2D with C-terminal GFP tags were expressed in Drosophila pseudoobscura tissue culture cells and plated on concanavalin A-coated plates, followed by live cell microscopy. Scale bar is 7.5 µm. A schematic of the expression construct and the resulting localization are shown. (B) Cysts at varying stages of spermatogenesis from an Arp2D-sfGFP D. pseudoobscura transgenic male. Arp2D is shown in green and DNA in blue. Enlarged images of individual cysts (denoted with asterisks) in stages of meiotic prophase (detected by DNA morphology) and spermatid elongation are shown. The transgenesis construct injected into D. pseudoobscura white− flies is displayed with a schematic indicating the stages of spermatogenesis in which GFP fluorescence is visualized. The number of cells shown at each stage in the schematic are only representative, with 128 spermatids actually following meiosis in D. pseudoobscura (Swallow and Wilkinson 2002).
In contrast to Arp2’s ubiquitous expression, Arp2D is exclusively expressed in testes. Therefore, we next assessed Arp2D expression and localization in the testis. Using P-element-mediated transgenesis (Thibault et al. 2004), we generated a transgenic D. pseudoobscura line encoding Arp2D-sfGFP (superfolder GFP, a more stable version of GFP [Pedelacq et al. 2006]) under the control of Arp2D’s native promoter (fig. 5B). We placed sfGFP at the C-terminus of Arp2D because Arp2 has been shown to be functionally unperturbed with a C-terminal tag (Egile et al. 2005). We confirmed expression of the full-length protein by western blot analysis and found that the Arp2D-sfGFP protein was expressed at the expected size (supplementary fig. S13, Supplementary Material online). Consistent with the RNA-seq and RT-PCR analyses, we found that this transgene was expressed in testes (fig. 5B).
To image Arp2D localization in vivo, we dissected testes and imaged GFP fluorescence live with the addition of live-imaging probes specific for DNA and actin (fig. 5B). These probes allowed us to identify the different stages of spermatogenesis. During this process in Drosophila, cells are interconnected throughout mitotic and meiotic divisions, resulting in 128 developing spermatids in D. pseudoobscura, in a single cyst. The final step of spermatogenesis separates the spermatids from one another by movement of actin cones, which form at the sperm nuclei and then translocate along the sperm tails, disposing of excess cytoplasm and encasing each spermatid in its own membrane (fig. 6B) (Noguchi et al. 2008). By confocal microscopy, we found that Arp2D-sfGFP is clearly expressed in meiotic and postmeiotic stages of spermatogenesis where it localizes to the cytoplasm (fig. 5B). We confirmed that the fluorescence was specific to sfGFP and not due to autofluorescence by imaging D. pseudoobscura flies lacking Arp2D-sfGFP with the same confocal laser settings (supplementary fig. S14, Supplementary Material online).
We first detected expression of Arp2D-sfGFP during meiotic prophase, during which Arp2D-sfGFP is often detected at points of concentrated actin at the cell surface that we predict to be endocytic sites (fig. 6A) (Pollard 2007). Arp2D-sfGFP persists throughout the late stages of spermatogenesis. During spermatid elongation following meiosis, Arp2D-sfGFP appears diffuse, albeit at a lower fluorescence intensity than meiotic cells (fig. 5B). Then, Arp2D-sfGFP clearly localizes to motile actin cones (fig. 6B), the structures that are required for the separation of syncytial spermatids during individualization.
Fig. 6.
Arp2D localizes to actin structures in eusperm and parasperm. (A) A meiotic cyst, indicated by the nuclear morphology, from the testis of an Arp2D-sfGFP Drosophila pseudoobscura transgenic male. Arp2D-sfGFP is green, actin is magenta, and DNA (DAPI stain) is blue. The inset shows GFP concentrated at actin-enriched sites. (B) A spermatid cyst undergoing individualization with actin cones enlarged in the inset. Below is a schematic indicating the process of individualization with Arp2D localizing to the front of actin cones (spermatids are representative of 128 per cyst). Scale bars in (A)–(B) are 10 µm. (C) Representative images of a parasperm and eusperm are shown with Arp2D-sfGFP localizing to actin cones. Scale bars are 30 µm.
This localization of Arp2D-sfGFP to actin cones is highly reminiscent of the localization of parental Arp2 in D. melanogaster (Noguchi et al. 2008). Arp2 generates branched actin networks toward the front of actin cones, creating a fan-like structure, and this polymerization of branched actin facilitates the motility of the cones (Noguchi et al. 2008). We find that Arp2D also localizes to individual fan-like motile actin cones, which have moved away from sperm nuclei (fig. 6B andsupplementary fig. S15, Supplementary Material online). In contrast, we do not observe Arp2D in immotile cones that are not fan-like (i.e., branched actin networks are absent) and remain at sperm nuclei (supplementary fig. S15, Supplementary Material online). Following individualization, we observe that Arp2D moves along with actin cones to the apoptotic waste bag (cystic bulge) at the end of the cyst and is not included in mature sperm. Thus, Arp2D localizes to a subset of previously described gametic actin structures in D. pseudoobscura testes.
Because of Arp2D’s specific localization to motile actin cones, we also investigated whether Arp2D localization correlated with sperm heteromorphism in D. pseudoobscura. Like other obscura group species, D. pseudoobscura generates both fertilization-competent “eusperm” and two types of “parasperm,” which cannot fertilize but instead increase the competitive ability of eusperm to fertilize females in the presence of sperm from other males (Snook et al. 1994; Holman and Snook 2008; Holman et al. 2008; Alpern et al. 2019). Eusperm are easily identified by microscopy because they are approximately five times longer than parasperm (Holman et al. 2008; Alpern et al. 2019); eusperm are about 300 µm long, whereas parasperm types 1 and 2 are ∼50 and 100 µm, respectively. We imaged sperm cysts with actin cones, assessed them for Arp2D-sfGFP localization, and classified them as either eusperm or parasperm based on length measurements. We found that Arp2D-sfGFP localizes to actin cones in both eusperm and parasperm during individualization (fig. 6C and supplementary fig. S16, Supplementary Material online) although we cannot conclusively distinguish between parasperm 1 and parasperm 2 (supplementary fig. S16, Supplementary Material online). A difference of 50 µm between the two parasperm types cannot be confidently differentiated in cysts, which often do not lay completely flat against the coverslip. Arp2D-sfGFP does not appear in mature sperm, which is when it is easier to distinguish parasperm types 1 and 2.
Based on its predicted ability to participate in Arp2-related interactions, its similar localization prior to and during spermatogenesis, and its positive selection, we hypothesize that Arp2D allows for the specialization and recurrent innovation of Arp2-related functions specifically in the male germline, without affecting Arp2’s many highly conserved functions in the soma.
Discussion
In this study, we report a burst of genetic innovation in cytoskeletal genes in one lineage of Drosophila species. We find that four Arp paralogs arose independently in the obscura group of Drosophila. Given the extreme conservation of Arp genes and the cytoskeletal apparatus during eukaryote evolution, such a burst of genetic innovation is unexpected.
The parental genes for at least two of the four obscura-specific Arp paralogs are known (actin and Arp2), and RNA-seq data suggest that they are indeed expressed in the testis, although they are both expressed at a lower level than in other tissues and lower than their gene duplicates D-ActL1 and Arp2D, respectively (supplementary fig. S17, Supplementary Material online; Celniker et al. 2009; Gramates et al. 2017). Actin is expressed throughout spermatogenesis, yet no existing data indicate which stages of spermatogenesis Arp2 is expressed. It is therefore possible that Arp2D is functionally replacing Arp2 during a specific stage, such as individualization. Under this scenario, however, one might expect to primarily see signatures of preservation rather than divergence of sequence and function. Indeed, maintenance of ancestral functions is more likely to be associated with purifying selection, whereas novel functions are more likely to be associated with positive selection (Jiang and Assis 2017). Our finding that all four Arp paralogs are subject to positive selection, as well as their high divergence from canonical Arps, makes it unlikely that only preservation of function during certain stages of spermatogenesis can explain this burst of innovation in the obscura group. Instead, we propose that duplication of the canonical Arps allowed for innovation of male germline-specific expression and function.
All the obscura-specific Arp paralogs are male-specific and likely testis-biased in expression. This is consistent with previous findings that evolutionarily young genes often originate with testis-restricted expression. Testes often have inherently more promiscuous transcription than in other tissues, allowing duplicate genes a higher likelihood of being transcribed and translated (Vinckenbosch et al. 2006). Moreover, testis-expressed genes are also subject to extremely high rates of evolutionary innovation due to strong selective forces from sexual selection (Kleene 2005; Vinckenbosch et al. 2006; Kaessmann 2010). In contrast, evolutionary innovation may be more detrimental in other tissues especially if it upsets dosage of proteins in multimeric complexes (Holland and Johnson 2018). As all canonical Arp paralogs like Arp2 are ubiquitously expressed, testis-specific innovation of Arp paralogs like Arp2D affords the opportunity for cytoskeletal innovation to occur in the testis without interfering with their essential functions in the soma. Thus, Arp duplications might relieve some of the antagonistic pleiotropy (or “escape from adaptive conflict”) between Arp innovation for testis functions, which requires positive selection, and conservation of canonical Arp functions for somatic tissues (Hughes 1994; Hittinger and Carroll 2007; Des Marais and Rausher 2008; Gallach and Betran 2011). Under this model, Arp duplications improve upon the parental Arp gene’s function to allow testis-specific specialization (Des Marais and Rausher 2008).
We used computational analyses to deduce the biochemical basis of divergence between canonical, ubiquitously expressed Arp2 and testis-specific Arp2D. There has been significant biochemical work characterizing the canonical Arp2 multiprotein complex. However, our modeling revealed no obvious impairment of Arp2D’s ability to participate in an Arp2-multiprotein complex or to catalyze branched actin structures. However, all fixed differences between Arp2 and Arp2D map to one surface of the protein. This suggests that early diversification of Arp2D required functional specialization at this surface. These differences could reflect altered interactions with positive regulators (such as N-WASP, WAVE, WASP, WHAMM, and cortactin) or negative regulators (such as Arpin, PICK1, Gadkin, and GMF) of this complex (Pollard 2007; Gandhi et al. 2010; Molinie and Gautreau 2018). Lack of structural information about how these regulators may directly or allosterically interact with Arp2, especially Drosophila Arp2, currently does not allow us to speculate on which regulators can differentially interact with Arp2D versus Arp2. However, altering the interactions with regulators may affect Arp2D’s catalytic ability to polymerize actin filaments. Any such change would have significant consequences on the motility of actin cones, which is thought to be generated by the dynamics of Arp2/3-generated actin polymerization (Noguchi and Miller 2003). In mammals, different protein isoforms of the Arp2/3 complex’s components differentially tune the polymerization of actin branches and impact the length and dynamics of actin tails (Abella et al. 2016), virus-generated actin structures that could be considered analogous to actin cones. We hypothesize that Arp2D may similarly produce a specialized version of the Arp2/3 complex that adapts actin dynamics for novel meiotic and postmeiotic specialization that may not be possible for Arp2, which is constrained by its many roles in all other somatic tissues.
Although we do not know the functional basis of testis-specific specialization of Arp proteins, their positive selection strongly suggests that they might be involved in some form of genetic conflict. Sexually active males frequently exceed the number of fertile and sexually receptive females, leading to many males vying to reproduce (Kleene 2005). As a result, sperm from different males often compete within the same female reproductive tract postcopulation. Such sperm competition can lead to sperm evolving faster motility or higher number (Parker 1993; Birkhead 2000; Kleene 2005). We speculate that Arp diversification may influence the development of more fertilization-competent sperm. However, changes in the female reproductive tract could also alter this fertilization success (Miller and Pitnick 2002). As a result, gametic selection for fertilization success may drive the rapid evolution of the testis-biased Arp paralogs (Kleene 2005). Alternatively, Arp protein diversification might occur as a consequence of reproductive manipulation of male fertility by maternally inherited bacteria such as Wolbachia.
One of the most notable aspects of our study is that four Arp paralogs independently arose in the same lineage of Drosophila. Even if we were to suppose that the pressures of functional novelty or preservation of male germline-specific expression drove this innovation, there is no a priori reason why this pattern of convergence would be expected to occur only in one lineage of Drosophila. We considered the possibility that these testis-specific Arp paralogs might play a role in an aspect of spermatogenesis specific to the obscura group. Among investigated Drosophila species, the obscura group appears unique in possessing sperm heteromorphism: the simultaneous production of distinct morphological types of sperm by a single male (Joly et al. 1989, 1991; Joly and Lachaise 1994; Swallow and Wilkinson 2002; Holman et al. 2008). Drosophila pseudoobscura in particular produces three sperm classes (eusperm, parasperm 1, and parasperm 2), and evidence suggests that parasperm increase eusperm’s fertilization competence (Alpern et al. 2019). Although the mechanisms by which fertilization enhancement occurs are unknown, proposed mechanisms include displacing rival fertilizing sperm in the female, protection from spermicide in the female reproductive tract, and manipulation of female receptiveness to remating (Oppliger et al. 1998; Cook and Wedell 1999; Holman and Snook 2006, 2008; Alpern et al. 2019).
If sperm heteromorphism is an adaptive trait specific to the obscura group of Drosophila, it is possible that the dual pressures to produce both eusperm and parasperm may have required significant innovation in the cytoskeletal machinery required to produce the three types of mature sperm. A number of genes may be important in this adaptation, and perhaps all of the Arp duplicates play roles. It appears that Arp2D localizes to actin cones in both eusperm and at least one parasperm class (type 2) (fig. 6B), which suggests that at least Arp2D might not play roles specific to one sperm class, although this requires further investigation.
Intriguingly, the magnitude of sperm heteromorphism is not identical among obscura group species, which show significant variation in heteromorphic size differences (Holman et al. 2008). This variance may be the result of ongoing positive selection and genetic turnover of the Arp paralogs in the obscura group. In this regard, it may be especially interesting to reexamine sperm heteromorphism in species like D. subobscura that lack three of the four obscura-specific Arp paralogs, or in D. pseudoobscura transgenic strains in which the Arp paralogs have been experimentally knocked out. Sperm heteromorphism has not been extensively studied across all Drosophila lineages. Thus, although it may appear specific to obscura, future studies will need to investigate potential sperm heteromorphism in other species and correlate these findings with evolutionary changes in the cytoskeletal machinery encoded in their genomes. Future genetic manipulation via genetic knockdown or CRISPR/Cas9-mediated knockouts will elucidate the in vivo functions of the divergent Arp duplicates and test whether they are rapidly evolving due to roles in sperm competition or sperm heteromorphism. Given its seeming rarity, sperm heteromorphism is unlikely to be a universal explanation for the function of testis-specific “orphan” Arp genes in Drosophila and mammals. However, the obscura-specific Arp paralogs may nevertheless illustrate how male germline-specific functions facilitate lineage-specific adaptation of cytoskeletal functions.
Materials and Methods
Drosophila Species and Strains
Species in the obscura clade were either obtained from the National Drosophila Species Stock Center (Cornell University) and were a kind gift from Dr Masayoshi Watada (Ehime University). The D. pseudoobscura and D. miranda strains were generously provided by Dr Nitin Phadnis (University of Utah) and Dr Doris Bachtrog (University of California, Berkeley), respectively. For a list of strains used in this study, see supplementary data S1, Supplementary Material online.
Discovery of Arp Duplications in Sequenced Genomes
The protein sequence of each D. melanogaster canonical Arp and the testis-specific Arp53D was used in a tBLASTn search (Altschul et al. 1997) for other Arp sequences in the sequenced and annotated genomes of 12 species of Drosophila (Drosophila 12 Genomes Consortium 2007; Thurmond et al. 2019). Sequences were identified as canonical Arps based on phylogenetic grouping, and the orphan branches corresponding to Arp paralogs outside the established Arp subfamilies were further confirmed as novel Arp genes by verifying their absence in the syntenic loci of the other sequenced Drosophila species. For each Arp duplicate, the D. pseudoobscura protein sequence was used in a tBLASTn search against a 10–20 kb region of the syntenic locus from each Drosophila species (Kearse et al. 2012). Of the 12 sequenced and annotated Drosophila species, only D. persimilis and D. pseudoobscura resulted in hits, whereas the other species were negative.
Sequencing Shared Syntenic Loci of Arp Paralogs in the obscura Group
Whole flies (10–15) with approximately equal number of males and females were ground in the following buffer: 10 mM Tris pH 7.5, 10 mM ethylenediaminetetraacetic acid, 100 mM NaCl, 0.5% SDS, and 0.5 µg/µl Proteinase K (NEB). The flies were then incubated at 55 °C for 2 h, followed by a phenol–chloroform extraction. The final ethanol-washed DNA pellet was resuspended in distilled water. To obtain the sequences of Arp paralogs from nonsequenced obscura group species, we aligned the syntenic loci from D. pseudoobscura and D. melanogaster (and for Arp2D, D. ananassae was included) and designed primers in conserved intergenic regions or neighboring genes. If gene products were >5 kb, then two PCRs were conducted with forward and reverse primers aligned with the highly conserved ATP-binding motif, which was later resequenced after the 5′ and 3′ prime halves of each paralog were obtained. Primers were iteratively designed based on successful PCRs and sequencing from divergent species. For a full list of primers used, see supplementary data S1, Supplementary Material online.
A touchdown PCR protocol was followed (Korbie and Mattick 2008) and Phusion was used per the manufacturer’s instructions (NEB). PCR products that were <∼3 kb were TOPO cloned into the pCR4-Blunt vector (Thermo Fisher Scientific) and subsequently sequenced with the M13F and M13R primers. PCR products that were >3 kb were directly sequenced. Coding sequences and sequences of an extended region of each locus can be found in supplementary data S3, Supplementary Material online. To differentiate between the presence and absence of Arp2 and Arp2D (fig. 3A), we designed primers that aligned completely with both genes and flanked an intron in Arp2 that is absent in Arp2D. A full list of GenBank accession numbers (MN526485–MN526590) can be found in supplementary data S1, Supplementary Material online.
Sequence Alignments and Phylogenetic Analyses
Protein sequences were aligned with MAFFT (Katoh and Standley 2013) and genes were aligned using the translation align function in the Geneious software package (version 9.1.3; Kearse et al. 2012). When the alignments exhibited large insertions or deletions, gaps were removed where <80% of the sequences aligned; these modifications are noted in the corresponding figure legends. Maximum-likelihood trees were generated in PhyML using the LG substitution model for protein sequences and the HKY85 substitution model for nucleotide sequences (Guindon et al. 2010) and analyzed for statistical support using 100 bootstrap replicates. Trees were visualized with Geneious (Kearse et al. 2012).
Imaging Arp2 and Arp2D in Tissue Culture Cells
Drosophila pseudoobscura tissue culture cells (cell line #ML83-63) were obtained from the Drosophila Genomics Resource Center (https://dgrc.bio.indiana.edu/Home) and cultured in M3+BPYE media supplemented with 10% fetal calf serum. Cells were incubated at 25 °C and passaged using a cell scraper (Fisher Scientific, 08-771-1A) to facilitate transfers. Cells were transfected with Arp2-eGFP or Arp2D-eGFP, which were cloned into a vector that encodes a copper-inducible promoter (pMT vector, promoter for D. metallothionein). For each construct, 1 µg DNA was combined with 8 µl Fugene HD (Promega) in serum-free media (100 µl total volume), followed by a 5-min incubation at room temperature. The transfection solution was added drop-wise to a confluent layer of D. pseudoobscura cells in a single well of a six-well dish. The cells were incubated for 24 h at 25 °C and then exogenous gene expression was induced with 100 µM copper every 24 h and imaged 24–48 h following transfection when a sufficient number of cells displayed fluorescence. Cells were then resuspended and ∼30 µl was added to the coverslip of a MatTek dish (MatTek corporation, P35G-1.5-10-C), coated with 0.5 mg/ml concanavalin A (MP Biomedicals). The media was exchanged with phosphate-buffered saline (PBS) to decrease background when imaging, and cells were imaged using a confocal microscope (Leica TCS SP5 II) and LASAF software (Leica).
Sex-Specific Expression Analysis
Six species were chosen that span a wide evolutionary distance in the obscura group to compare male versus female expression. Whole male and female flies, approximately ten each, were collected separately and RNA was extracted using TRIzol and further purified per the manufacturer’s instructions (Invitrogen). RNA was then treated with DNAse and used to synthesize cDNA with SuperScript III (Invitrogen). For each reaction, a corresponding reaction without RT was conducted to detect any genomic DNA contamination (supplementary fig. S9, Supplementary Material online). Subsequent cDNA was then utilized for RT-PCR of ribosomal Rp49 to confirm similar amounts of cDNA among the samples. For RT-PCR of the Arp paralogs, primers were designed to yield a ∼100–350 bp product for efficient amplification and to distinguish the paralog from actin or Arp2. For a full list of primers used, see supplementary data S1, Supplementary Material online. RT-PCRs were conducted using Phusion per the manufacturer’s instructions (NEB) for 25 cycles. Equal volumes of each RT-PCR reaction were loaded on a 1% agarose gel for analysis.
Structural Analysis
To analyze conservation and divergence of the proteins, sequences were aligned with MAFFT (Katoh and Standley 2013) and mapped onto the structure using Chimera (Pettersen et al. 2004). Phyre2 (Kelley et al. 2015) was used to obtain a homology model of D. pseudoobscura Arp2 and Arp2D. This software used the Arp2 crystal structure (PDB 4JD2) to model the D. pseudoobscura proteins. All structural analysis was performed using Chimera (Pettersen et al. 2004). To analyze Arp2D in the context of the Arp2/3 complex, sequences of all the D. pseudoobscura Arp2/3 subunits (Arpc1-5, Arp2D, and Arp3) were collected and submitted to Swiss-Model (Waterhouse et al. 2018) to generate a homology model using an X-ray crystal structure of the Bos taurus Arp2/3 complex as a template (PDB 3DXM) (Nolen et al. 2009). The Arp2D homology model substituted Arp2 in the complex. Drosophila pseudoobscura actin was modeled using template PDB 2ZWH (Oda et al. 2009). To view Arp2D with respect to actin, an activated mammalian Arp2/3 junction model was constructed, as described in the methods for MD simulations.
Generation of an Arp2D-sfGFP D. pseudoobscura Transgenic Line
The Arp2D paralog was encoded with a sfGFP tag and cloned into the restriction sites BamHI and NotI in the pCasper4 vector. Arp2 has shown to be best tagged at the C-terminus; thus, a similar strategy was taken with Arp2D. The 1-kb intergenic regions that were upstream and downstream of the paralog were included to allow for endogenous expression under the control of proximal transcriptional regulatory regions. The construct was maxi-prepped (Machery-Nagel) for high-quality DNA and used for injections of D. pseudoobscura white− flies (gift from Nitin Phadnis, University of Utah). Fly embryos were injected by Rainbow Transgenic Flies, Inc. in combination with transposase in trans. We crossed larvae that survived the injections to D. pseudoobscura white− flies (4:1 or 5:2 females to males) and selected progeny with pigmented eyes, which ranged from light orange to deep red. Transformants that appeared in separate crosses were designated as different founder lines and two founders were identified. Homozygous lines were generated and imaged live.
Imaging of Arp2D-GFP Transgenic Flies
For live imaging, the testes from homozygous Arp2D-sfGFP transgenic males were dissected into PBS and then transferred to a drop of PBS on a slide. PBS was exchanged for PBS containing the DNA stain Hoechst and sir-actin (10 µM; Cytoskeleton, Inc.), which is used for live samples. The testes were then torn open with tweezers to release all cell types during spermatogenesis; this technique allowed for improved visibility of different subpopulations of developing sperm. The tissue was incubated in the imaging solution for 5 min in the dark and then a coverslip was placed on top. The sample was immediately imaged using a confocal microscope (Leica TCS SP5 II) and LASAF software (Leica) for 20–30 min, until GFP signal faded and signs of apoptosis were visible. We opted for live imaging of the transgenics to avoid the possibility of fixation artifacts; we also found immunofluorescence of the samples led to a high background, making it difficult to know what was true sfGFP fluorescence.
Analysis of Positive Selection
To test for positive selection at the population level, we compared sequences of all four Arp paralogs in 10–11 strains of D. pseudoobscura with those in 8 strains of D. miranda. The Arp paralogs were sequenced with primers that aligned within the 5′ and 3′ untranslated regions of the D. pseudoobscura genes. The genes were codon-based aligned and subjected to the MK test with an online resource (Egea et al. 2008). To test for site-specific positive selection, we used the PAML suite (Yang 2007). The coding sequences for each paralog from all obscura species were codon-based aligned using Geneious (Kearse et al. 2012) and all gaps were removed. The alignment and corresponding species tree for each paralog were then used as input files for the program CODEML NsSites in the PAML suite.
To test for positive selection along a single branch in a tree, we conducted a free-ratio analysis (Model 1 in PAML). Each branch with dN/dS >1 was further tested for statistical significance by conducting Model 2 and comparing the maximum likelihoods when the dN/dS is fixed to 1 or to the dN/dS indicated by the free-ratio analysis (Model 1).
The NsSites models M7, M8a, and M8 were compared to determine whether site-specific positive selection was present. The test indicated whether the evolution of the paralogs fits a model that allows for DN/DS > 1 (M8) or models that do not allow for DN/DS >1 (M7 or M8a). To determine whether the difference between the log-likelihoods of the models was statistically significant, we conducted chi-square tests (Yang 1997). The starting omega used was 0.4 with a codon frequency model of F3×4. Tests with different starting omegas (0.4, 1.0, and 1.5) and codon frequency models (F3×4 and F61) yielded similar results.
Arp2/3 Complex Model and Molecular Dynamics Simulations
The initial all-atom model of an activated Arp2/3 complex was built following the protocol of Pfaendtner et al. (2012), but using a newer model for the actin filament, namely that of Oda et al. (2009), PDB ID 2ZWH. A single structure from simulations of a full junction by one of us (G.M.H.) is shown in Aydin et al. (2018). In brief, to build the junction model, actin monomers in the filament had a magnesium ion and a bound ADP as well as coordinating waters placed inside, and then assembled into a filament as previously described (Saunders and Voth 2012; Hocky et al. 2016). A mother filament of 13 subunits and a daughter filament of 11 subunits in ideal geometry were constructed. These filaments were aligned to the Branch10 structure in Pfaendtner et al. (2012) by Cα RMSD. Mother actin subunits 7 and 9 were replaced with structures from Pfaendtner et al. (2012) because these are in direct contact with the complex, and come from Rouiller et al. (2008)’s reconstruction but the coordinating waters from the equilibrated Oda structure are placed into these two subunits by alignment of the actin subunits. The Arp2/3 complex structure from Branch10 in Pfaendtner et al. (2012) was preserved, except that a magnesium ion, water, and an ADP molecule were placed inside of Arp2, and the same was done for Arp3 except with an ATP molecule/water/ion from an equilibrated ATP-bound Oda monomer (Katkar et al. 2018) based on the structure in PDB ID 1J6Z, due to the difference in nucleotide hydrolysis rates of Arp2 and Arp3 (Pollard 2007). In the case of this work, this whole protocol was followed to build a mammalian junction, but only the first two actin subunits in the daughter filament were kept. The system was then solvated in TIP3P water with 0.18 M KCl. Equilibration was performed in NAMD (Phillips et al. 2005) using the CHARMM22+CMAP forcefield following the exact procedure in Hocky et al. (2016). This structure formed the basis for the homology models of the Arp2/3 complex and daughter actin proteins used in this work. Subsequently, the homology models of the complex and actin were realigned with this structure and equilibrated according to this same protocol using NAMD. Then subsequent MD simulations were performed in GROMACS (Abraham et al. 2015).
Supplementary Material
Acknowledgments
We thank Janet Young, Lisa Kursel, Tera Levin, Antoine Molaro, and Jeannette Tenthorey for their comments on the manuscript, as well as other members of the Malik lab for helpful discussions. We greatly appreciate the gift of the pMT and pCasper4 vector backbones from the Henikoff lab and Parkhurst lab (Fred Hutchinson Cancer Research Center), respectively, and fly strains from Nitin Phadnis (University of Utah), Doris Bachtrog (University of California Berkeley), the National Drosophila Species Stock Center (Cornell University) and Masayoshi Watada (Ehime University). Transgenic Drosophila pseudoobscura flies were generated by Rainbow Transgenic Flies, Inc. Molecular dynamics simulations were performed on resources provided by NYU IT High Performance Computing. This work was funded by the Jane Coffin Childs Memorial Fund (CMS), the Cancer Center Support Grant CURE Supplement: NCI 3P30CA015704 (IMN), the Fred Hutchinson Cancer Research Center's Internship Program (IMN), startup funds from NYU (GMH), NIH grant R01GM074108 (HSM) and the Howard Hughes Medical Institute (HSM). H.S.M. is an investigator of the Howard Hughes Medical Institute.
References
- Abella JVG, Galloni C, Pernier J, Barry DJ, Kjær S, Carlier M-F, Way M.. 2016. Isoform diversity in the Arp2/3 complex determines actin filament dynamics. Nat Cell Biol. 18(1):76–86. [DOI] [PubMed] [Google Scholar]
- Abraham MJ, Murtola T, Schulz R, Szilard P, Smith JC, Hess B, Lindahl E.. 2015. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 1–2:19–25. [Google Scholar]
- Alpern JHM, Asselin MM, Moehring AJ.. 2019. Identification of a novel sperm class and its role in fertilization in Drosophila. J Evol Biol. 32(3):259–266. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ.. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25(17):3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydin F, Katkar HH, Voth GA.. 2018. Multiscale simulation of actin filaments and actin-associated proteins. Biophys Rev. 10(6):1521–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock CS, Anderson WW.. 1996. Molecular evolution of the sex-ratio inversion complex in Drosophila pseudoobscura: analysis of the Esterase-5 gene region. Mol Biol Evol. 13(2):297–308. [DOI] [PubMed] [Google Scholar]
- Barrio E, Ayala FJ.. 1997. Evolution of the Drosophila obscura species group inferred from the Gpdh and Sod genes. Mol Phylogenet Evol. 7(1):79–93. [DOI] [PubMed] [Google Scholar]
- Birkhead T. 2000. Promiscuity: an evolutionary history of sperm competition. Cambridge, MA: Harvard University Press.
- Blessing CA, Ugrinova GT, Goodson HV.. 2004. Actin and ARPs: action in the nucleus. Trends Cell Biol. 14(8):435–442. [DOI] [PubMed] [Google Scholar]
- Boeda B, Knowles PP, Briggs DC, Murray-Rust J, Soriano E, Garvalov BK, McDonald NQ, Way M.. 2011. Molecular recognition of the Tes LIM2-3 domains by the actin-related protein Arp7A. J Biol Chem. 286:11543–11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR, Erdjument-Bromage H, Tempst P, Winston F, Kornberg RD.. 1998. Two actin-related proteins are shared functional components of the chromatin-remodeling complexes RSC and SWI/SNF. Mol Cell. 2(5):639–651. [DOI] [PubMed] [Google Scholar]
- Celniker SE, Dillon LAL, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, et al. 2009. Unlocking the secrets of the genome. Nature 459(7249):927–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook PA, Wedell N.. 1999. Non-fertile sperm delay female remating. Nature 397(6719):486. [Google Scholar]
- Des Marais DL, Rausher MD.. 2008. Escape from adaptive conflict after duplication in an anthocyanin pathway gene. Nature 454(7205):762–765. [DOI] [PubMed] [Google Scholar]
- Dominguez R, Holmes KC.. 2011. Actin structure and function. Annu Rev Biophys. 40(1):169–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium. 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450:203–218. [DOI] [PubMed] [Google Scholar]
- Egea R, Casillas S, Barbadilla A.. 2008. Standard and generalized McDonald-Kreitman test: a website to detect selection by comparing different classes of DNA sites. Nucleic Acids Res. 36(Web Server):W157–W162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egile C, Rouiller I, Xu XP, Volkmann N, Li R, Hanein D.. 2005. Mechanism of filament nucleation and branch stability revealed by the structure of the Arp2/3 complex at actin branch junctions. PLoS Biol. 3(11):e383.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel S, Mooseker MS.. 1996. The actin-related proteins. Curr Opin Cell Biol. 8(1):30–37. [DOI] [PubMed] [Google Scholar]
- Fu J, Wang Y, Fok KL, Yang D, Qiu Y, Chan HC, Koide SS, Miao S, Wang L.. 2012. Anti-ACTL7a antibodies: a cause of infertility. Fertil Steril. 97:1226–1233.e1–8. [DOI] [PubMed] [Google Scholar]
- Fyrberg C, Ryan L, Kenton M, Fyrberg E.. 1994. Genes encoding actin-related proteins of Drosophila melanogaster. J Mol Biol. 241(3):498–503. [DOI] [PubMed] [Google Scholar]
- Gallach M, Betran E.. 2011. Intralocus sexual conflict resolved through gene duplication. Trends Ecol Evol. 26(5):222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi M, Smith BA, Bovellan M, Paavilainen V, Daugherty-Clarke K, Gelles J, Lappalainen P, Goode BL.. 2010. GMF is a cofilin homolog that binds Arp2/3 complex to stimulate filament debranching and inhibit actin nucleation. Curr Biol. 20(9):861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson HV, Hawse WF.. 2002. Molecular evolution of the actin family. J Cell Sci. 115(Pt 13):2619–2622. [DOI] [PubMed] [Google Scholar]
- Gramates LS, Marygold SJ, Santos GD, Urbano J-M, Antonazzo G, Matthews BB, Rey AJ, Tabone CJ, Crosby MA, Emmert DB, et al. 2017. FlyBase at 25: looking to the future. Nucleic Acids Res. 45(D1):D663–D671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O.. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321. [DOI] [PubMed] [Google Scholar]
- Hammesfahr B, Kollmar M.. 2012. Evolution of the eukaryotic dynactin complex, the activator of cytoplasmic dynein. BMC Evol Biol. 12:95.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara Y, Yamagata K, Oguchi K, Baba T.. 2008. Nuclear localization of profilin III-ArpM1 complex in mouse spermiogenesis. FEBS Lett. 582(20):2998–3004. [DOI] [PubMed] [Google Scholar]
- Harata M, Oma Y, Tabuchi T, Zhang Y, Stillman DJ, Mizuno S.. 2000. Multiple actin-related proteins of Saccharomyces cerevisiae are present in the nucleus. J Biochem. 128(4):665–671. [DOI] [PubMed] [Google Scholar]
- Heid H, Figge U, Winter S, Kuhn C, Zimbelmann R, Franke W.. 2002. Novel actin-related proteins Arp-T1 and Arp-T2 as components of the cytoskeletal calyx of the mammalian sperm head. Exp Cell Res. 279(2):177–187. [DOI] [PubMed] [Google Scholar]
- Hittinger CT, Carroll SB.. 2007. Gene duplication and the adaptive evolution of a classic genetic switch. Nature 449(7163):677–681. [DOI] [PubMed] [Google Scholar]
- Hocky GM, Baker JL, Bradley MJ, Sinitskiy AV, De La Cruz EM, Voth GA.. 2016. Cations stiffen actin filaments by adhering a key structural element to adjacent subunits. J Phys Chem B. 120(20):4558–4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland DO, Johnson ME.. 2018. Stoichiometric balance of protein copy numbers is measurable and functionally significant in a protein-protein interaction network for yeast endocytosis. PLoS Comput Biol. 14(3):e1006022.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holman L, Freckleton RP, Snook RR.. 2008. What use is an infertile sperm? A comparative study of sperm-heteromorphic Drosophila. Evolution 62(2):374–385. [DOI] [PubMed] [Google Scholar]
- Holman L, Snook RR.. 2006. Spermicide, cryptic female choice and the evolution of sperm form and function. J Evol Biol. 19(5):1660–1670. [DOI] [PubMed] [Google Scholar]
- Holman L, Snook RR.. 2008. A sterile sperm caste protects brother fertile sperm from female-mediated death in Drosophila pseudoobscura. Curr Biol. 18(4):292–296. [DOI] [PubMed] [Google Scholar]
- Hughes AL. 1994. The evolution of functionally novel proteins after gene duplication. Proc Biol Sci. 256(1346):119–124. [DOI] [PubMed] [Google Scholar]
- Izore T, Kureisaite-Ciziene D, McLaughlin SH, Lowe J.. 2016. Crenactin forms actin-like double helical filaments regulated by arcadin-2. Elife 5:21600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadeeshan S, Singh RS.. 2005. Rapidly evolving genes of Drosophila: differing levels of selective pressure in testis, ovary, and head tissues between sibling species. Mol Biol Evol. 22(9):1793–1801. [DOI] [PubMed] [Google Scholar]
- Jiang X, Assis R.. 2017. Natural selection drives rapid functional evolution of young Drosophila duplicate genes. Mol Biol Evol. 34(12):3089–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly D, Cariou M-L, Lachaise D, David JR.. 1989. Variation of sperm length and heteromorphism in drosophilid species. Genet Sel Evol. 21(3):283–293. [Google Scholar]
- Joly D, Cariou ML, Lachaise D.. 1991. Can sperm competition explain sperm polymorphism in Drosophila teissieri? Evol Biol. 5:25–44. [Google Scholar]
- Joly D, Lachaise D.. 1994. Polymorphism in the sperm heteromorphic species of the Drosophila obscura group. J Insect Physiol. 40(11):933–938. [Google Scholar]
- Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC.. 1990. Atomic structure of the actin:DNase I complex. Nature 347(6288):37–44. [DOI] [PubMed] [Google Scholar]
- Kaessmann H. 2010. Origins, evolution, and phenotypic impact of new genes. Genome Res. 20(10):1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katkar HH, Davtyan A, Durumeric AEP, Hocky GM, Schramm AC, De La Cruz EM, Voth GA.. 2018. Insights into the cooperative nature of ATP hydrolysis in actin filaments. Biophys J. 115(8):1589–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ.. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 10(6):845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klages-Mundt NL, Kumar A, Zhang Y, Kapoor P, Shen X.. 2018. The nature of actin-family proteins in chromatin-modifying complexes. Front Genet. 9:398.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleene KC. 2005. Sexual selection, genetic conflict, selfish genes, and the atypical patterns of gene expression in spermatogenic cells. Dev Biol. 277(1):16–26. [DOI] [PubMed] [Google Scholar]
- Korbie DJ, Mattick JS.. 2008. Touchdown PCR for increased specificity and sensitivity in PCR amplification. Nat Protoc. 3(9):1452–1456. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD.. 2006. GARD: a genetic algorithm for recombination detection. Bioinformatics 22(24):3096–3098. [DOI] [PubMed] [Google Scholar]
- Lee IH, Kumar S, Plamann M.. 2001. Null mutants of the neurospora actin-related protein 1 pointed-end complex show distinct phenotypes. Mol Biol Cell. 12(7):2195–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Atkinson SJ, Ampe C, Vandekerckhove J, Pollard TD.. 1994. Purification of a cortical complex containing two unconventional actins from Acanthamoeba by affinity chromatography on profilin-agarose. J Cell Biol. 127(1):107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBee RM, Rozmiarek SA, Meyerson NR, Rowley PA, Sawyer SL.. 2015. The effect of species representation on the detection of positive selection in primate gene data sets. Mol Biol Evol. 32(4):1091–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M.. 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351(6328):652–654. [DOI] [PubMed] [Google Scholar]
- Miller GT, Pitnick S.. 2002. Sperm-female coevolution in Drosophila. Science 298(5596):1230–1233. [DOI] [PubMed] [Google Scholar]
- Molinie N, Gautreau A.. 2018. The Arp2/3 regulatory system and its deregulation in cancer. Physiol Rev. 98(1):215–238. [DOI] [PubMed] [Google Scholar]
- Muhua L, Karpova TS, Cooper JA.. 1994. A yeast actin-related protein homologous to that in vertebrate dynactin complex is important for spindle orientation and nuclear migration. Cell 78(4):669–679. [DOI] [PubMed] [Google Scholar]
- Muller J, Oma Y, Vallar L, Friederich E, Poch O, Winsor B.. 2005. Sequence and comparative genomic analysis of actin-related proteins. Mol Biol Cell. 16(12):5736–5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins RD, Heuser JA, Pollard TD.. 1998. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci U S A. 95(11):6181–6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Lenartowska M, Rogat AD, Frank DJ, Miller KG.. 2008. Proper cellular reorganization during Drosophila spermatid individualization depends on actin structures composed of two domains, bundles and meshwork, that are differentially regulated and have different functions. Mol Biol Cell. 19(6):2363–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Miller KG.. 2003. A role for actin dynamics in individualization during spermatogenesis in Drosophila melanogaster. Development 130(9):1805–1816. [DOI] [PubMed] [Google Scholar]
- Nolen BJ, Tomasevic N, Russell A, Pierce DW, Jia Z, McCormick CD, Hartman J, Sakowicz R, Pollard TD.. 2009. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature 460(7258):1031–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T, Iwasa M, Aihara T, Maeda Y, Narita A.. 2009. The nature of the globular- to fibrous-actin transition. Nature 457(7228):441–445. [DOI] [PubMed] [Google Scholar]
- Oppliger A, Hosken DJ, Ribi G.. 1998. Snail sperm production characteristics vary with sperm competition risk. Proc R Soc Lond B. 265(1405):1527–1534. [Google Scholar]
- Parker GA. 1993. Sperm competition games: sperm size and sperm number under adult control. Proc Biol Sci. 253(1338):245–254. [DOI] [PubMed] [Google Scholar]
- Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS.. 2006. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 24:79–88. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Zhao Y, Chait BT.. 1998. Subunits of the yeast SWI/SNF complex are members of the actin-related protein (ARP) family. J Biol Chem. 273(37):23641–23644. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE.. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 25(13):1605–1612. [DOI] [PubMed] [Google Scholar]
- Pfaendtner J, Volkmann N, Hanein D, Dalhaimer P, Pollard TD, Voth GA.. 2012. Key structural features of the actin filament Arp2/3 complex branch junction revealed by molecular simulation. J Mol Biol. 416(1):148–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K.. 2005. Scalable molecular dynamics with NAMD. J Comput Chem. 26(16):1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD. 2007. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 36(1):451–477. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG.. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell 112(4):453–465. [DOI] [PubMed] [Google Scholar]
- Rouiller I, Xu XP, Amann KJ, Egile C, Nickell S, Nicastro D, Li R, Pollard TD, Volkmann N, Hanein D.. 2008. The structural basis of actin filament branching by the Arp2/3 complex. J Cell Biol. 180(5):887–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo CAM, Mello B, Frazão A, Voloch CM.. 2013. Phylogenetic analysis and a time tree for a large drosophilid data set (Diptera: Drosophilidae). Zool J Linn Soc. 169(4):765–775. [Google Scholar]
- Saunders MG, Voth GA.. 2012. Comparison between actin filament models: coarse-graining reveals essential differences. Structure 20(4):641–653. [DOI] [PubMed] [Google Scholar]
- Schafer DA, Gill SR, Cooper JA, Heuser JE, Schroer TA.. 1994. Ultrastructural analysis of the dynactin complex: an actin-related protein is a component of a filament that resembles F-actin. J Cell Biol. 126(2):403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NG, Eyre-Walker A.. 2002. Adaptive protein evolution in Drosophila. Nature 415(6875):1022–1024. [DOI] [PubMed] [Google Scholar]
- Snook RR, Markow TA, Karr TL.. 1994. Functional nonequivalence of sperm in Drosophila pseudoobscura. Proc Natl Acad Sci U S A. 91(23):11222–11226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swallow JG, Wilkinson GS.. 2002. The long and short of sperm polymorphisms in insects. Biol Rev Camb Philos Soc. 77(2):153–182. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Iguchi N, Egydio de Carvalho C, Tadokoro Y, Yomogida K, Nishimune Y.. 2003. Novel actin-like proteins T-ACTIN 1 and T-ACTIN 2 are differentially expressed in the cytoplasm and nucleus of mouse haploid germ cells. Biol Reprod. 69(2):475–482. [DOI] [PubMed] [Google Scholar]
- Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, et al. 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet. 36(3):283–287. [DOI] [PubMed] [Google Scholar]
- Thurmond J, Goodman JL, Strelets VB, Attrill H, Gramates LS, Marygold SJ, Matthews BB, Millburn G, Antonazzo G, Trovisco V, et al. 2019. FlyBase 2.0: the next generation. Nucleic Acids Res. 47(D1):D759–D765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner LM, Chuong EB, Hoekstra HE.. 2008. Comparative analysis of testis protein evolution in rodents. Genetics 179(4):2075–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ent F, Amos LA, Lowe J.. 2001. Prokaryotic origin of the actin cytoskeleton. Nature 413(6851):39–44. [DOI] [PubMed] [Google Scholar]
- Vinckenbosch N, Dupanloup I, Kaessmann H.. 2006. Evolutionary fate of retroposed gene copies in the human genome. Proc Natl Acad Sci U S A. 103(9):3220–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, et al. 2018. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46(W1):W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 13:555–556. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Bachtrog D.. 2012. Sex-specific adaptation drives early sex chromosome evolution in Drosophila. Science 337(6092):341–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.