Abstract
The behaviors of closely related species can be remarkably different, and these differences have important ecological and evolutionary consequences. Although the recent boom in genotype–phenotype studies has led to a greater understanding of the genetic architecture and evolution of a variety of traits, studies identifying the genetic basis of behaviors are, comparatively, still lacking. This is likely because they are complex and environmentally sensitive phenotypes, making them difficult to measure reliably for association studies. The Drosophila species complex holds promise for addressing these challenges, as the behaviors of closely related species can be readily assayed in a common environment. Here, we investigate the genetic basis of an evolved behavioral difference, pupation site choice, between Drosophila melanogaster and D. simulans. In this study, we demonstrate a significant contribution of the X chromosome to the difference in pupation site choice behavior between these species. Using a panel of X-chromosome deficiencies, we screened the majority of the X chromosome for causal loci and identified two regions associated with this X-effect. We then collect gene disruption and RNAi data supporting a single gene that affects pupation behavior within each region: Fas2 and tilB. Finally, we show that differences in tilB expression correlate with the differences in pupation site choice behavior between species. This evidence associating two genes with differences in a complex, environmentally sensitive behavior represents the first step toward a functional and evolutionary understanding of this behavioral divergence.
Keywords: pupation, behavior, genetics, evolution, Drosophila
Introduction
Despite the importance of behavioral traits (Coyne and Orr 1997, 2004), we know little about the genetic basis of their evolution. GePheBase (Martin and Orgogozo 2013), the most extensive compilation of natural genetic variants associated with trait differences, catalogs 2,000 associations as of October 2019, of which only 26 fall into the behavior “trait category” (the remaining are categorized as either “physiology” or “morphology,” for which there are 1,351 and 657 cataloged associations, respectively). From these 26, and others in the literature, it is clear that individual genes can sometimes have large effects on evolved differences in behavior (McGrath et al. 2011; Leary et al. 2012; Ding et al. 2016; Prince et al. 2017). With so few studies, however, it is difficult to conclude how frequently we expect single loci to have large effects due to pervasive ascertainment bias (Rockman 2012). Indeed, studies of natural variation in behavior have also revealed complex genetic architecture (Anholt and Mackay 2004; Zwarts et al. 2011).
The lack of genetic associations for evolved differences in behavior, compared with those for physiology and/or morphology, presumably arises from the fact that behaviors are difficult to measure reliably and repeatedly. Behavioral phenotypes often integrate multiple signals, are sometimes context dependent, and can be innate or learned, making it difficult to exclude environmentally induced variation. To better understand the genetic basis of behavioral evolution, we therefore need more case studies, with a focused effort on “metamodel” systems with documented behavioral differences between closely related species (sensu Kopp 2009). Flies in the genus Drosophila are well poised to address these challenges. In Drosophila, hundreds of genetically identical individuals from variable wild-caught strains can be reared in a common environment, isolated during specific life stages, and repeatedly assayed for a trait of interest. Such a design significantly reduces the potential for environmentally induced variation to obscure genetic differences in behavior. Additionally, there are many Drosophila species that differ in a variety of complex behaviors (Orgogozo and Stern 2009). Undeniably, work comparing the repeated evolution of morphological traits between closely related species of Drosophila has significantly advanced our understanding of the general patterns linking genotype and phenotype for the evolution of developmental traits (Sucena et al. 2003; Kittelmann et al. 2018; Rebeiz and Williams 2017). Interspecific studies that investigate genetic complexity in Drosophila, where fine-mapping and functional follow-up are possible, should make similar progress for behaviors.
Here, we present the results of one such study, investigating an environmentally sensitive difference in pupation site choice behavior using two Drosophila species: D. melanogaster and D. simulans. When the larval stages of these species are ready to metamorphose into adults, they first enter a pupal stage. The pupa, which lasts for a number of days, is immobile and therefore vulnerable to parasitism, predation, desiccation, and disease (Markow 1981). Before pupating, larvae enter a “wandering” stage, when they search for an appropriate pupation site (Sokolowski et al. 1984; Riedl et al. 2007). Depending on the strain and species, larvae vary from pupating directly on their larval food source to traveling more than 40 cm away from it (Stamps et al. 2005). Pupation site choice behavior has been extensively studied, and is exquisitely sensitive to environmental conditions—individuals alter their behavior in response to light, moisture, pH, the presence of other species, parasitism, and more (Sameoto and Miller 1968; Markow 1981; Hodge and Caslaw 1998; Seyahooei et al. 2009). Despite environmentally induced variation, the effects of genotype on preference are considerable. Within species, strains and populations often differ in how far they travel from their food source before pupating, although the most consistent experimentally demonstrated differences are between species (Markow 1979; Vandal et al. 2008). Interestingly, differences in pupation site choice behavior between species do not correspond to their taxonomic classification (Shivanna et al. 1996). For example, D. melanogaster and D. simulans shared a common ancestor 2–3 Mya (Lachaise and Silvain 2004) and are extremely similar in terms of their ecology, morphology, and physiology (Parsons 1975). Previous work shows, however, that they differ markedly in terms of pupation site choice, with D. simulans pupating closer to the larval food source, on average (Markow 1979, 1981). This difference is not due to laboratory adaptation, as freshly collected individuals show the same pattern (Markow 1979, 1981). These species are frequently collected in the same microhabitats, and their differences in pupation site choice behavior have been postulated to be a form of niche partitioning (Markow 1979, 1981). Supporting this hypothesis, pupation site choice responds to density-dependent selection in the laboratory (Mueller and Sweet 1986) and provides a potential increase in competitive ability between species ovipositing in the same media (Arthur and Middlecote 1984).
Here, we investigate the genetic basis of this difference in pupation site choice between D. melanogaster and D. simulans. Despite substantial reproductive isolation, we can mate D. melanogaster females with D. simulans males in the lab, and vigorous female F1 hybrids result from the cross. Males are usually inviable, but we use a D. simulans hybrid male rescue strain (Watanabe 1979; Brideau et al. 2006) to circumvent this challenge and show that a significant proportion of the species difference in pupation behavior can be mapped to the X chromosome, consistent with findings using other Drosophila species (Erezyilmaz and Stern 2013). Still, these hybrids remain sterile, so genetic dissection using an intercross mapping population is not possible. Instead, we use a widely available set of D. melanogaster chromosomal deficiency lines to screen a substantial portion of the X chromosome (Ryder et al. 2004; Cook et al. 2012). We use these lines to create hybrid females that lack large, overlapping portions of the X chromosome from D. melanogaster, and therefore express only D. simulans alleles in those regions. We identify two broad loci with effects on pupation behavior. We then employ genetic knockouts of candidate genes within these regions to demonstrate their effects and use RNAi knockdown to further test the role of two genes, touch insensitive larva B (tilB) and Fasciclin 2 (Fas2). Finally, we use real time RT-qPCR to test for species-level differences in gene expression of tilB in larvae and show that tilB is more highly expressed in D. melanogaster than in D. simulans, corresponding to their differences in pupation site choice behavior.
Results
Differences in Pupation Behavior between D. melanogaster and D. simulans
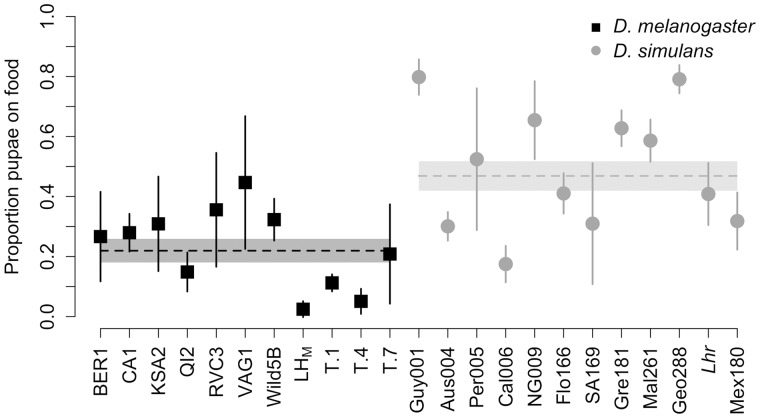
We measured pupation behavior for 11 D. melanogaster and 12 D. simulans strains collected from various locations throughout the world (supplementary table S1, Supplementary Material online) and found significant variation within each species in the proportion of individuals that pupated on the food surface (fig. 1; D. melanogaster Wilcoxon test: χ2= 42.69, df = 10, P < 0.0001; D. simulans Wilcoxon test: χ2= 56.34, df = 11, P < 0.0001). When we tested for a species difference in pupation behavior, we found that the D. simulans strains had a significantly higher proportion of pupae on the food surface compared with the D. melanogaster strains, in general (fig. 1; Wilcoxon test: χ2= 8.37, df = 1, P = 0.0038).
Fig 1.
Pupation behavior differences between Drosophila melanogaster and D. simulans. The mean proportion of individuals in a vial that pupated on the surface of the food for 11 D. melanogaster strains and 12 D. simulans lines described in supplementary table S1, Supplementary Material online. Points represent the mean proportion of pupae found on the food media after transferring 100 eggs to control for density. Error bars denote the 95% confidence interval around each individual mean (N = 5–8; N for each line can be found in supplementary table S1, Supplementary Material online). The dashed horizontal lines indicate the grand mean for each species. The boxes surrounding the dashed lines denote the 95% confidence interval around the grand mean.
Although we controlled egg density to characterize species differences in pupation behavior (see Materials and Methods), there were viability differences among our surveyed lines (supplementary fig. S1A, Supplementary Material online). As a result, we had significant variation in the total number of pupae in each vial for our D. melanogaster lines (ANOVA: F10,65 = 5.58, P < 0.0001) and our D. simulans lines (ANOVA: F11,66 = 8.01, P < 0.0001). Previous studies have found that larval density correlates with pupation height in D. melanogaster (Sokal et al. 1960). To control for differences in density, we also performed the same analyses above using the residuals from a regression between number of pupae in the vial and proportion of pupae on the food. None of our findings changed using this analysis: we again found significant variation in the proportion of pupae on the food for each species (D. melanogaster Wilcoxon test: χ2= 32.01, df = 10, P = 0.0004; D. simulans Wilcoxon test: χ2= 54.73, df = 11, P < 0.0001), with D simulans having a higher proportion of pupae on the food compared with D. melanogaster (supplementary fig. S1B, Supplementary Material online; Wilcoxon test: χ2= 8.32, df = 1, P = 0.0031). This indicates that our species comparisons were not affected by variation in larval density.
Differences in Pupation Behavior Have a Significant X Effect
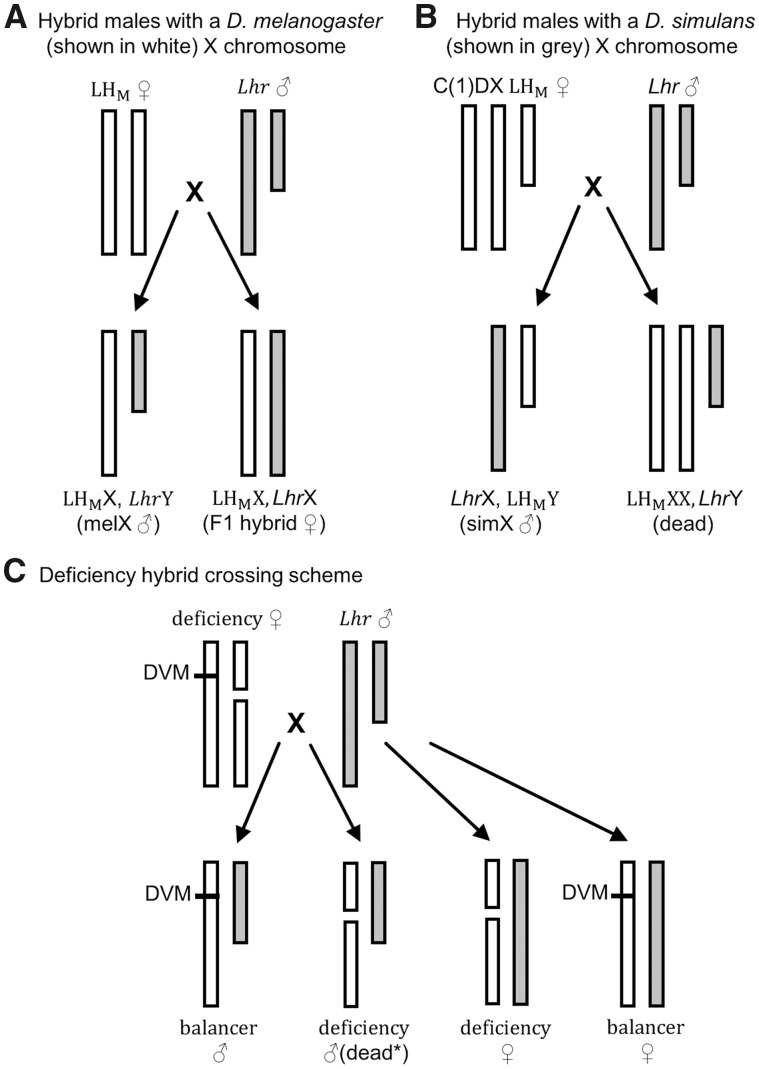
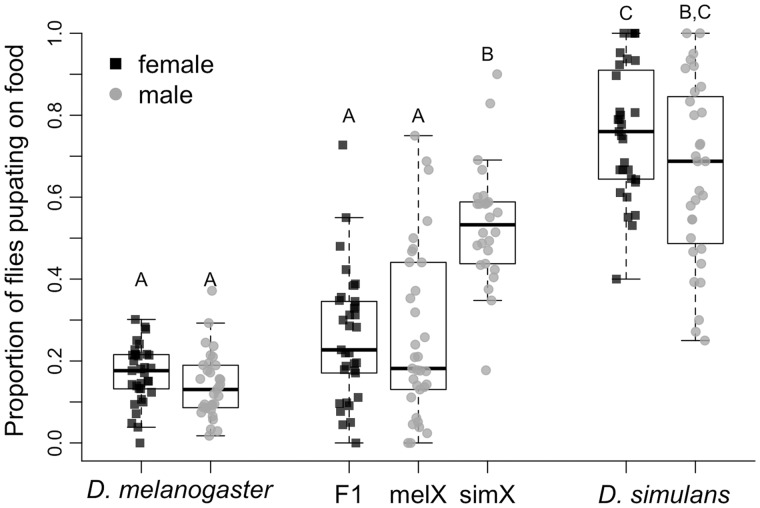
To begin to dissect the genetic basis of differences in pupation site choice behavior, we created F1 hybrid females (fig. 2A) and F1 hybrid males that had either a D. melanogaster X chromosome (“melX” males; fig. 2A) or a D. simulans X chromosome (“simX” males; fig. 2B). These reciprocal hybrid males have the same autosomal background (the D. melanogaster stocks used to make melX and simX were both in an LHM background), and both inherit their cytoplasm from the D. melanogaster parent strain, so any differences we observe are directly attributable to the species they inherit their sex chromosomes from. We found significant differences among genotypes when we screened hybrids alongside their parental strains (fig. 3; Full model Wilcoxon test: χ2= 144.57, df = 6, P < 0.0001). Specifically, a significantly higher proportion of F1 hybrid males pupated on the food when they had inherited a D. simulans X chromosome (simX males) compared with a D. melanogaster X chromosome (melX males; Wilcoxon test: P < 0.0001 after correcting for multiple comparisons), indicating that this species divergence in pupation behavior has a significant X effect. This is supported by the fact that the proportion of individuals that pupated on the food was not significantly different between melX hybrid males and D. melanogaster males, or between simX hybrid males and D. simulans males (fig. 3). When we controlled for density effects, we still found that a significantly higher proportion of simX males pupated on the food compared with melX males (Wilcoxon test: P < 0.0001 after correcting for multiple comparisons), and found no significant difference between melX and D. melanogaster males, or between simX and D. simulans males (supplementary fig. S2, Supplementary Material online). When we used the proportion of individuals that pupated on the food to estimate the effect size of the X chromosome, we found that the X accounts for ∼55.6% (95% bias-corrected and accelerated bootstrapped CI = 31.4–80.2%) of the difference in pupation site choice behavior between D. melanogaster and D. simulans. It is important to note, however, that our effect size estimate for the X chromosome may be an overestimate, as calculating effect sizes using only reciprocal hybrids does not account for potential transgressive autosomal effects (Flint et al. 1995; Mittleman et al. 2017).
Fig. 2.
Crossing schemes to generate reciprocal X chromosome hybrid males and deficiency/balancer hybrid females. (A) Crossing wild-type Drosophila melanogaster females (LHM, shown in white) to D. simulans males (Lhr, shown in grey) produces hybrid males with a D. melanogaster X chromosome (melX) and hybrid females. (B) Crossing LHM females with a compound X (C(1)DX LHM) to Lhr males produces hybrid males with a D. simulans X chromosome (simX). Females of this cross would inherit two D. melanogaster X chromosomes and a D. simulans Y chromosome, but are inviable. (A, B) Note that the background of the reciprocal male hybrids resulting from each cross (melX and simX) is an identical combination of Lhr and LHM with the exception of the sex chromosomes. (C) Crossing D. melanogaster X chromosome deficiency lines, which have a balancer X chromosome with a dominant visible marker (DVM) and an X chromosome with a large deletion, to Lhr produces deficiency hybrid females, balancer females, balancer males, and deficiency males (*mostly dead due to large deletions on a hemizygous chromosome with some deficiency lines being exceptions).
Fig. 3.
Pupation site choice behavior for Drosophila melanogaster, D. simulans, and their F1 hybrids. The proportion of individuals that pupated on the food surface for males and females from both species and their F1 hybrids. D. melanogaster males and females were taken from the LHM strain, whereas D. simulans males and females were taken from the Lhr strain. F1 hybrids resulted from a cross between these two strains. The “melX” hybrid males have the D. melanogaster X chromosome, and the “simX” hybrid males have the D. simulans X chromosome. Both hybrids have D. melanogaster cytoplasmic inheritance. Box plots display the median (bold bar), interquartile range (box), and full extent of the data set excluding outliers (whiskers). Those labeled with different letters are significantly different from one another based on pairwise Wilcoxon tests followed by sequential Bonferroni correction for multiple comparisons (all P < 0.0001 after correction, N = 33 for D. melanogaster and melX/F1 females, N = 36 for simX, and N = 31 for D. simulans).
Additionally, we found no difference in the proportion of F1 hybrid females and melX hybrid males that pupated on the food (Wilcoxon test: P = 0.70), whereas F1 hybrid females pupated on the food significantly less often than simX hybrid males (Wilcoxon test: P < 0.0001 after correcting for multiple comparisons; fig. 3). The fact that F1 hybrid females behave identically to melX hybrid males indicates that the variation in pupation behavior on the D. melanogaster X chromosome (i.e., fewer pupae on the food) is dominant to the pupation behavior on the D. simulans X chromosome (i.e., more pupae on the food), because F1 hybrid females have one X chromosome from each species.
The fact that hybrid males and females with a D. melanogaster X chromosome behave indistinguishably from their D. melanogaster parent strain, and hybrids with a D. simulans X chromosome behave indistinguishably from their D. simulans parent strain (fig. 3), suggests that hybrid pupation site choice behavior falls well within the typical range exhibited by either parent strain. To further test this result, we also compared the average pupation height of F1 hybrids and their parents (with a higher value indicating a farther distance from the food) and found a similar pattern: F1 females and melX males did not have significantly different pupation heights compared with their D. melanogaster parents, and simX hybrids did not differ from their D. simulans parental strain (supplementary fig. S3A and table S2A, Supplementary Material online). These patterns remained consistent when we controlled for density (supplementary fig. S3B and table S2B, Supplementary Material online).
A Deficiency Screen of the X Chromosome Identifies Two Regions of Interest
Because we found a significant effect of the X chromosome on the difference in pupation site choice behavior between D. simulans and D. melanogaster, with the D. melanogaster variation dominant to D. simulans variation, we used 90 molecularly engineered chromosomal deficiencies in D. melanogaster (Cook et al. 2012) to screen 87% of the X chromosome for loci contributing to this difference (supplementary table S3A, Supplementary Material online). These crosses produced two types of hybrid female, both heterozygous at all autosomes (fig. 2C). The deficiency hybrid females harbor the D. melanogaster deficiency X and a D. simulans X, making them hemizygous for a segment of the D. simulans X chromosome. The balancer hybrid females harbor the D. melanogaster balancer X (marked with the dominant visible Bar marker) and a D. simulans X, so they are heterozygous for D. melanogaster/D. simulans over the entirety of the X chromosome. The balancer hybrids developed in the same environment as our experimental deficiency flies, and thus provide an experimental control. As a result, we calculated the proportion of deficiency hybrid females that pupated on the food and the proportion of balancer hybrid females that pupated on the food, and used these measures to calculate a “pupation index” (the proportion of deficiency hybrid females pupating on the food divided by the proportion of balancer hybrid females pupating on the food).
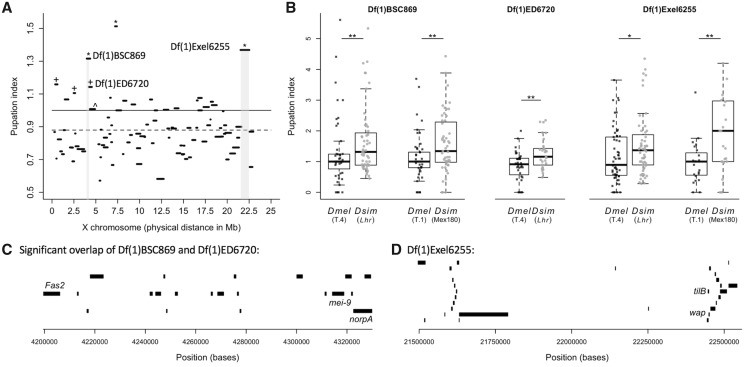
We found significant variation in pupation index among the deficiency hybrid crosses (Kruskal–Wallis Test: χ2=336.90, df = 89, P < 0.0001; fig. 4A). A pupation index >1 indicates that more deficiency hybrids pupated on the food than balancer hybrids. This suggests that the D. melanogaster deficiency region may be revealing recessive D. simulans genetic variation that causes the deficiency hybrids to pupate on the food surface. However, the average pupation index across all 90 deficiency hybrid crosses was 0.88 (fig. 4A), which was significantly lower than 1, the mean under the null hypothesis (Wilcoxon test: df = 89, P < 0.0001). As a result, we compared the pupation index for all deficiencies with our null hypothesis value of 1 and with the grand mean pupation index for these lines (0.88). Six deficiencies had pupation indices significantly >1: Df(1)BSC530, Df(1)ED411, Df(1)BSC869, Df(1)ED6720, Df(1)ED6906, and Df(1)Exel6255. Three of these, Df(1)BSC869, Df(1)ED6906, and Df(1)Exel6255, remained significant after sequential Bonferroni correction for multiple comparisons (fig. 4A;supplementary table S3A, Supplementary Material online). Because the other three deficiencies, Df(1)ED411, Df(1)ED6720, and Df(1)BSC530, had pupation indices significantly >0.88 after sequential Bonferroni correction (supplementary table S3A, Supplementary Material online), we included them in our list of potential deficiencies of interest.
Fig. 4.
Hybrid deficiency screen of the X chromosome identifies two regions of interest. (A) The median pupation index for each of the 90 deficiency hybrid crosses is plotted by the physical map distance each engineered deletion spans along the X chromosome. Deficiencies with a pupation index significantly >1 (solid line) after correction for multiple comparisons are denoted by an asterisk (sample sizes and significance levels for all deficiencies are listed in supplementary table S3A, Supplementary Material online). Deficiencies with a median pupation index significantly >0.88 (the grand mean, dashed line) after correction for multiple comparisons are denoted with a cross. All of these lines also had pupation indices significantly >1 before correcting for multiple comparisons. The two regions we pursued for candidate gene validation are highlighted in light gray. Note that we did not pursue the remaining three significant deficiency strains (from left to right: +Df(1)BSC530, +DF(1)ED411, *Df(1)6906) because they showed similar pupation indices when crossed to Drosophila melanogaster (supplementary fig. S4, Supplementary Material online). (B) The pupation indices of the deficiencies from the gray highlighted areas in (A) are shown for the original hybrid cross (Lhr) and for a cross to the T.4 D. melanogaster strain. BSC869 and Exel6255 were additionally crossed to the D. simulans strain Mex180 and the D. melanogaster strain T.1. Asterisks denote a significant difference between pupation indices of deficiency strains crossed to D. melanogaster and D. simulans (Wilcoxon tests; *P < 0.05, **P < 0.01). Sample sizes can be found in supplementary table S3A, Supplementary Material online. (C) The region uncovered by the overlap of deficiencies Df(1)BSC869 and Df(1)ED6720, but excluding the region uncovered by Df(1)ED6727 (which had a pupation index no different than 1; marked in (A)with ^) and the 23 genes contained within it. The three genes with available disruption strains (Fas2, mei-9, and norpA) are labeled. (D) The region uncovered by Exel6255 and the 28 genes contained within it. The two genes with available disruption strains (tilB and wap) are labeled.
To ensure that the deficient region actually reveals D. simulans variation contributing to pupation site choice behavior, rather than creating lines that behave abnormally due to the extended hemizygosity within the deficiency region, we crossed each of the six significant deficiencies listed above to the T.4 wild-type D. melanogaster strain. For two of the six deficiency strains, Df(1)ED411 and Df(1)BSC530, we found no difference in the pupation index when crossed to D. melanogaster compared with the pupation index when crossed to the D. simulans Lethal hybrid rescue (Lhr) strain (Wilcoxon tests; Df(1)ED411: χ2 = 1.74, df = 1, P = 0.19, supplementary fig. S4A, Supplementary Material online; Df(1)BSC530: χ2 = 1.47, df = 1, P = 0.23, supplementary fig. S4C, Supplementary Material online). A third deficiency strain, Df(1)ED6906, had a significantly higher pupation index when crossed to D. melanogaster compared with when crossed to Lhr (Wilcoxon test: χ2=9.42, df = 1, P = 0.002; supplementary fig. S4B, Supplementary Material online). These results suggest that the phenotypes of these three lines are a result of their hemizygosity within the deficiency region, as the deficient D. melanogaster females pupate on the food more often than the balancer females. Note that this effect is unlikely to be driven by the balancer females, as the same balancers occur in many of the other deficiency lines we screened.
When we crossed the remaining three deficiency strains, Df(1)BSC869, Df(1)ED6720, and Df(1)Exel6255, to the T.4 D. melanogaster strain, we found a pupation index significantly lower than the index we calculated when crossing to Lhr (Wilcoxon tests; Df(1)BSC869: χ2=4.35, df = 1, P = 0.037; Df(1)BSC6720: χ2=7.90, df = 1, P = 0.0049; Df(1)Exel6255: χ2 = 4.68, P = 0.0306; fig. 4B), and no different than 1 (Wilcoxon tests; Df(1)BSC869: df = 50, P = 0.46; Df(1)BSC6720: df = 34, P = 0.10; Df(1)Exel6255: df = 56, P = 0.50). This suggests that these deficient regions reveal recessive D. simulans variation affecting pupation site choice behavior in hybrids, but have no effect when made hemizygous in D. melanogaster. Two of these three deficiencies overlap: Df(1)BSC869 and Df(1)ED6720 (fig. 4A;supplementary table S3A, Supplementary Material online). To further confirm that this effect is not specific to one D. melanogaster or D. simulans wild-type strain, we crossed one of the overlapping deficiency strains, Df(1)BSC869, and Df(1)Exel6255 to another D. melanogaster wild-type strain (T.1) and another D. simulans wild-type strain (Mex180). The pattern remained consistent for both of these deficiencies: When crossed to D. melanogaster, the pupation index was significantly lower than when crossed to D. simulans (fig. 4B; Wilcoxon tests; Df(1)BSC869: χ2 = 7.78, df = 1, P = 0.0053; Df(1)Exel6255: χ2 = 7.74, df = 1, P = 0.0054).
Gene Knockouts and RNAi Knockdown Suggest That Fas2 is Involved in Divergent Pupation Behavior
The first region of interest identified by our deficiency screen is the overlap of Df(1)BSC869 and Df(1)ED6720 (spanning X:4,204,351–4,361,560), but excluding the region covered by Df(1)ED6727 (X:4,325,174–4,911,061), which partially overlaps Df(1)BSC869 and Df(1)ED6720 but did not have a pupation index significantly >1 (fig. 4A, supplementary table S3A, Supplementary Material online). Within the resulting region, which spans X:4,204,351–4,325,174, there are 23 genes, of which 20 are protein coding (supplementary table S4A, Supplementary Material online; fig. 4C). According to modENCODE expression data, 15 of those 20 protein-coding genes are expressed in D. melanogaster larvae, whereas only six are also expressed in the larval nervous system (Graveley et al. 2011), which we might expect for genes regulating behavior. Five of these six genes are well described, and at the time of assay, only three had nonlethal verified loss-of-function alleles available: Fas2 (Fasciclin 2), mei-9 (meiotic 9), and norpA (no receptor potential A). We tested knockouts of each for an effect on pupation site choice behavior. It is worth noting that norpA is only partially contained within this region, and is also largely deleted by Df(1)ED6727, which did not display a pupation index significantly >1 (supplementary table S3A, Supplementary Material online; fig. 4A). Although we focused on genes with expression in the larval nervous system as likely candidates for regulating pupation site choice, it is possible that one or more of the protein-coding genes expressed in larvae without nervous system expression could influence this behavior. However, only one of these genes (Muc4B) is characterized, functioning primarily in egg chorion assembly (supplementary table S4A, Supplementary Material online), and seems an unlikely candidate for pupation behavior.
We found no significant difference in the pupation index obtained when we crossed the mei-9A1 mutant allele to the Lhr D. simulans strain and the T.4 D. melanogaster strain (Wilcoxon test: χ2=0.37, df = 1, P = 0.54; supplementary fig. S5A, Supplementary Material online), and we did not detect a significant species by genotype interaction for our quantitative complementation test using these data (supplementary fig. S5B, Supplementary Material online; ANOVA: P = 0.86, see supplementary table S5A, Supplementary Material online for the full model), indicating that mei-9 is unlikely to be involved in pupation site choice. Because norpA is not held over a balancer, we compared the proportion of females pupating on the food for a strain harboring the norpA36 mutant allele with a strain containing a norpA rescue allele. We found no significant differences in the pupation behavior of female hybrids containing the norpA36 mutant allele and female hybrids containing a norpA rescue allele (Wilcoxon test: χ2 = 3.10, df = 1, P = 0.08; supplementary fig. S6A and B, Supplementary Material online), with the knockout hybrids actually having a slightly lower proportion of flies pupating on the food. We also found no species by genotype interaction for our norpA quantitative complementation test (supplementary fig. S6C, Supplementary Material online; ANOVA: P = 0.12, see supplementary table S5B, Supplementary Material online for full model). These data suggest that norpA is unlikely to be involved in pupation site choice, and that gene knockouts with expression in the larval nervous system do not, in general, increase the number of hybrids pupating on the food.
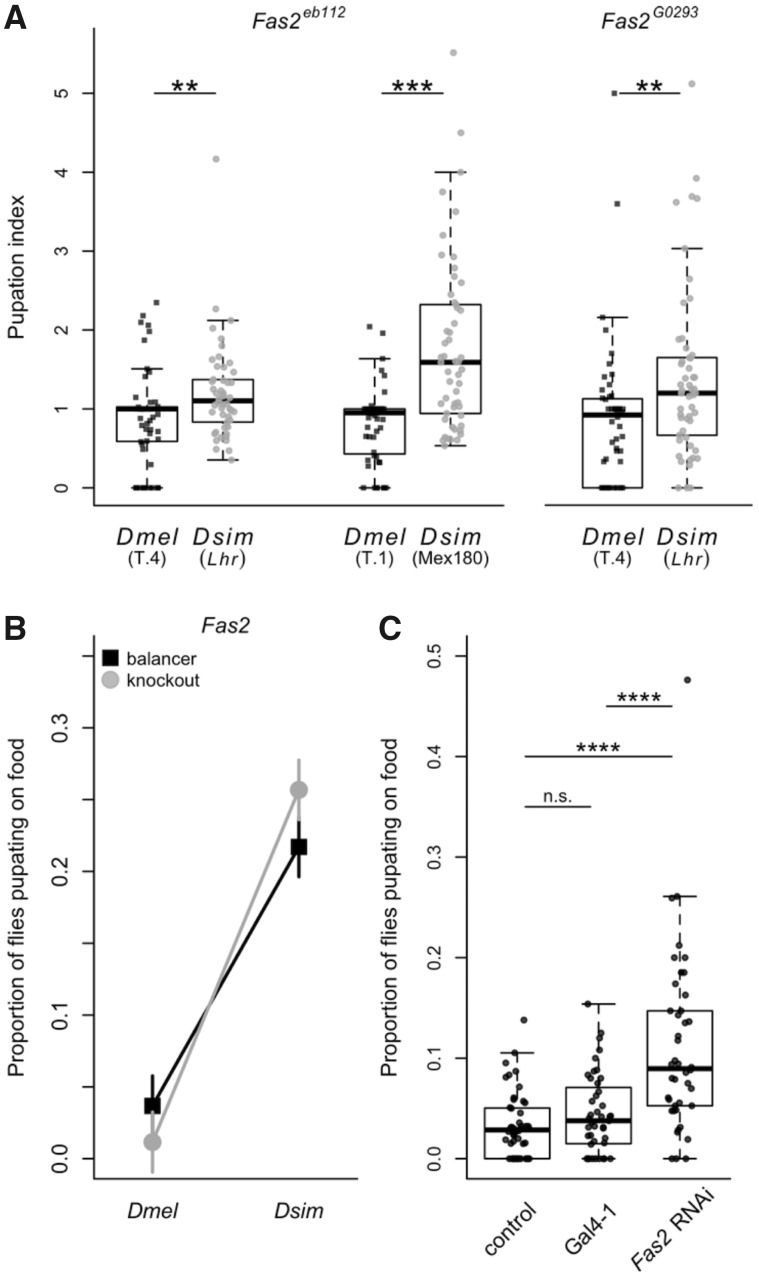
In contrast, when we crossed the mutant allele Fas2eb112 to D. simulans (Lhr) and D. melanogaster (T.4), we found that Fas2eb112 hybrids had a significantly higher pupation index than Fas2eb112 D. melanogaster flies (Wilcoxon test: χ2 = 6.97, df = 1, P = 0.0083; fig. 5A), suggesting that Fas2 may be involved in pupation site choice. To ensure this pattern is not unique to these strains, we crossed Fas2eb112 to additional D. simulans (Mex180) and D. melanogaster (T.1) wild-type strains. We again found the same pattern: The pupation index for Fas2eb112 hybrids was significantly higher than for Fas2eb112 D. melanogaster flies (Wilcoxon test: χ2 = 27.2, df = 1, P < 0.0001; fig. 5A). As further verification, we tested a second Fas2 strain: a P-element insertion allele, Fas2G0293, and similarly found that the pupation index for Fas2G0293hybrids (crossed to Lhr) was significantly higher than that for Fas2G0293 D. melanogaster flies (crossed to T.4; Wilcoxon test: χ2 = 9.73, df = 1, P = 0.0018; fig. 5A). Although the nature of the lesion is uncharacterized for Fas2G0293, these results suggest that it is indeed a loss of function allele, as it behaves indistinguishably from the verified knockout Fas2eb112 (Grenningloh et al. 1991). We used the consensus combined P-value test (Rice 1990), which tests the combined effect of independent tests of the same hypothesis, to look at the overall pattern for Fas2eb112, and Fas2 as a whole (i.e., including results from both Fas2eb112and Fas2G0293), and found a strongly significant pattern of higher pupation indices for hybrid crosses compared with D. melanogaster crosses (Fas2eb112: P = 3.59 × 10−6; Fas2: P = 2.35 × 10−8). This is consistent with the results of our quantitative complementation test using these data for Fas2: We found a significant species by genotype interaction (ANOVA: P < 0.0001, see supplementary table S5C, Supplementary Material online for the full model), with more Fas2 hybrids pupating on the food compared with balancer hybrids (fig. 5B).
Fig. 5.
Knockouts and RNAi knockdown confirm the role of Fas2 in evolved differences in pupation site choice. (A) The pupation indices of the Fas2eb112 and Fas2G0293gene disruptions are shown for the comparison between the original hybrid cross (Lhr; Fas2eb112 N = 50, Fas2G0293 N = 53) and a cross to the Drosophila melanogaster T.4 strain (Fas2eb112 N = 51, Fas2G0293 N = 52). Also shown for Fas2eb112is the comparison for crosses to the D. simulans strain Mex180 (N = 53) and the D. melanogaster strain T.1 (N = 52). Asterisks denote significance (Wilcoxon tests; **P < 0.01, *** = P < 0.001). (B) A quantitative complementation interaction plot showing the proportion of females that pupated on the food when the Fas2 mutant allele (gray, circle) and the balancer chromosome (black, square) were present in offspring resulting from the D. melanogaster and D. simulans crosses shown in (A). Plotted are the least squares means from the analysis in supplementary table S5C, Supplementary Material online. The interaction was significant for Fas2 (ANOVA: P < 0.0001, determined using arcsine transformed data). (C) The results of panneuronal knockdown of the Fas2 transcript via RNAi. The proportion of RNAi or control individuals that pupated on the food are shown for the control cross (elav-Gal4 driver crossed to the RNAi background stock; N = 48), the Gal4-1 hairpin RNA cross (Gal4-1; N = 43), and Fas2 RNAi cross (N = 42). Asterisks denote significance based on Wilcoxon tests after correcting for multiple comparisons (****P < 0.0001).
Next, we used RNAi with the elav-Gal4 driver to reduce expression of Fas2 throughout the nervous system in D. melanogaster. We compared the proportion of experimental flies that pupated on the food for the RNAi cross (UAS-Fas2 × elav-Gal4) with that of two controls (both crossed to elav-Gal4): the background stock in which the RNAi lines were created (y v; attP2, y+) and the Gal4-1 stock, which has a hairpin targeting Gal4 in VALIUM20. Taken together, these controls allow us to account for the effects of both the Gal4 mutation and general expression of hairpin RNA throughout the nervous system. Any differences we detect between these controls and our RNAi crosses must therefore be due to the expression of the Fas2-specific hairpin RNA. We found that a significantly higher proportion of Fas2 RNAi flies pupated on the food compared with the control flies from either the background (Wilcoxon test: P < 0.0001 after sequential Bonferroni correction) or Gal4-1 cross (Wilcoxon test: P < 0.0001 after sequential Bonferroni correction; fig. 5C); these results are unchanged when we control for density effects (supplementary fig. S7A, Supplementary Material online). Similarly, when we compared pupation height, we found that RNAi flies pupated significantly closer to the food compared with both the background (Wilcoxon test: P < 0.0001 after sequential Bonferroni correction) and Gal4-1 crosses (Wilcoxon test: P < 0.0001 after sequential Bonferroni correction; supplementary fig. S8A, Supplementary Material online), providing further evidence for Fas2’s role in pupation site choice.
Gene Knockouts and RNAi Knockdown Suggest That tilB Is Involved in Divergent Pupation Behavior
The second region of interest identified by our deficiency screen was the region deleted by Df(1)Exel6255 (X:21,519,203–22,517,665; fig. 4D). Within this region are 28 genes, of which 22 are protein coding (supplementary table S4B, Supplementary Material online). Of the 22 protein-coding genes, 14 are expressed in D. melanogaster larvae—13 of which have some expression in the larval nervous system (Graveley et al. 2011). Of these, seven are described. We obtained knockout strains for the two characterized genes expressed in the larval nervous system that had verified loss-of-function alleles available at the time: tilB (touch insensitive larva B) and wap (wings apart). Within this region there was only a single gene that is expressed in larvae but not the larval nervous system (CG14615), but this gene does not yet have a characterized biological function (supplementary table S4B, Supplementary Material online).
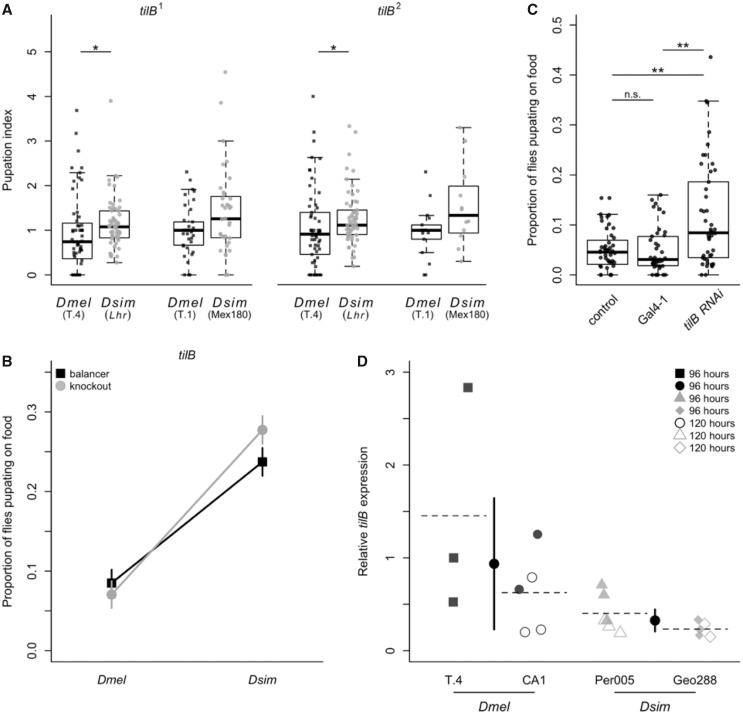
We found no significant difference in the pupation index obtained when we crossed the wap2 mutant allele to the Lhr D. simulans strain and the T.4 D. melanogaster strain (Wilcoxon test: χ2 = 0.32, df = 1, P = 0.57; supplementary fig. S5C, Supplementary Material online), and no significant species by genotype interaction in our quantitative complementation test using these data (supplementary fig. S5D, Supplementary Material online; ANOVA: P = 0.58, see supplementary table S5D, Supplementary Material online for full model), indicating that wap is unlikely to be involved in pupation site choice. In contrast, when we crossed the tilB1 and tilB2 mutant alleles to D. simulans (Lhr) and D. melanogaster (T.4), we found that the tilB hybrids had significantly higher pupation indices than the tilB D. melanogaster flies for both alleles (Wilcoxon tests; tilB1: χ2 = 6.61, df = 1, P = 0.0101; tilB2: χ2 = 6.61, df = 1, P = 0.0101; fig. 6A). To test whether this difference is consistent in other backgrounds, we crossed both the tilB1and tilB2mutant alleles to additional D. simulans (Mex180) and D. melanogaster (T.1) wild-type strains. We had a difficult time crossing our tilB strains to the Mex180 strain, so our sample sizes for these crosses are smaller, but there is a nonsignificant trend toward a higher pupation index for the knockout hybrids compared with the D. melanogaster hybrids for both alleles (Wilcoxon tests; tilB1: χ2= 2.80, df = 1, P = 0.0943; tilB2: χ2= 3.44, df = 1, P = 0.0638, fig. 6A). We used the consensus combined P-value test (Rice 1990) to look at the overall pattern for tilB1, tilB2, and tilB as a whole (i.e., including results from both tilB1 and tilB2), and found a strongly significant pattern of higher pupation indices for hybrid crosses compared with D. melanogaster crosses (tilB1: P = 0.0022; tilB2: P = 0.0037; tilB: P = 2.47 × 10−5). Similarly, our tilB quantitative complementation test using these data found a significant species by genotype interaction (ANOVA: P < 0.0001, see supplementary table S5E, Supplementary Material online for full model), with more tilB hybrids pupating on the food compared with balancer hybrids (fig. 6B).
Fig. 6.
Knockouts, RNAi knockdown, and gene expression confirm the role of tilB in evolved differences in pupation site choice. (A) The pupation indices of the tilB1 and tilB2 gene disruptions are shown for the comparison between the original hybrid cross (Lhr; tilB1 N = 56, tilB2 N = 57) and a cross to the Drosophila melanogaster T.4 strain (tilB1 N = 56, tilB2 N = 58). Also shown are the comparisons for crosses to the D. simulans strain Mex180 (tilB1 N = 36, tilB2 N = 12) and the D. melanogaster strain T.1 (tilB1 N = 37, tilB2 N = 18). Asterisks denote significance (Wilcoxon tests; *P < 0.05). (B) A quantitative complementation interaction plot showing the proportion of females that pupated on the food when the tilB mutant allele (gray, circle) and the balancer chromosome (black, square) were present in offspring resulting from the D. melanogaster and D. simulans crosses shown in (A). Plotted are the least squares means from the analysis in supplementary table S5E, Supplementary Material online. The interaction was significant for tilB (ANOVA: P = 0.0004, determined using arcsine transformed data). (C) The results of panneuronal knockdown of the tilB transcript via RNAi. The proportion of RNAi or control individuals that pupated on the food are shown for the control cross (elav-Gal4 driver crossed to the RNAi background stock; N = 47), the Gal4-1 hairpin RNA cross (Gal4-1; N = 41), and tilB RNAi cross (N = 46). For (C), asterisks denote significance based on Wilcoxon tests after correcting for multiple comparisons (**P < 0.01). (D) The relative abundance of tilB transcript detected by RT-qPCR in D. melanogaster and D. simulans. Each data point represents the average of two technical replicates for a single biological replicate. We collected data for 2–3 biological replicates per strain per time point, with the exception of T.4 which we were unable to collect at 120 h. Relative transcript abundance is significantly higher in D. melanogaster strains on average (ANOVA: P < 0.01). Squares (T.4) and circles (CA1) represent the two D. melanogaster strains measured, whereas triangles (Per005) and diamonds (Geo288) represent the two D. simulans strains. Filled points represent samples collected at 96 h, and open points represent samples collected at 120 h. The dashed lines depict the average expression across both time points for each strain. The black circles represent the species-wide mean, and error bars depict the 95% confidence interval surrounding the mean.
As for Fas2 above, we then used RNAi with the elav-Gal4 driver to reduce expression of tilB throughout the nervous system in D. melanogaster. We found that a significantly higher proportion of RNAi flies pupated on the food compared with the control flies from either the background (Wilcoxon test: P < 0.01 after sequential Bonferroni correction) or Gal4-1 cross (Wilcoxon test: P < 0.01 after sequential Bonferroni correction; fig. 6C); these findings are consistent when we control for density effects (supplementary fig. S7B, Supplementary Material online). In addition, when we compared pupation height, we found that RNAi flies pupated significantly closer to the food compared with both the background (Wilcoxon test: P < 0.0001 after sequential Bonferroni correction) and Gal4-1 crosses (Wilcoxon test: P < 0.0001 after sequential Bonferroni correction; supplementary fig. S8B, Supplementary Material online), providing additional support for tilB’s role in pupation site choice.
tilB Is Expressed More Highly in D. melanogaster Strains
We performed RT-qPCR to quantify relative tilB transcript abundance for two strains of D. simulans (Per005 and Geo288) and two strains of D. melanogaster (CA1 and T.4). For the two D. simulans strains and the CA1 D. melanogaster strain, we collected larvae from two stages of larval development: 96 and 120 h following oviposition. We chose these time points because they approximate the early and late wandering larval stage, which immediately precedes pupation. For the other D. melanogaster strain (T.4), we were only able to obtain enough larval tissue at 96 h following oviposition due to low fecundity. We found that, in general, larvae from the 120-h sampling period had lower tilB expression than 96-h larvae (ANOVA: F1,14 = 6.16, P = 0.026), and we did not find a significant species by larval age interaction (ANOVA: P = 0.41; see Materials and Methods), indicating that this pattern is consistent in both species. Although we did not detect any significant differences between the two strains from the same species (ANOVA: F2,14 = 2.39, P = 0.13), we found a significantly higher average relative amount of tilB transcript in D. melanogaster larvae compared with D. simulans larvae (ANOVA: F1,14 = 9.74, P = 0.0075; fig. 6D).
Because we performed RT-qPCR on two strains for each species, these four lines represent a continuum of pupation site choice behavior, with T.4 (D. melanogaster) having the lowest proportion of pupae on the food, followed by CA1 (D. melanogaster), then Per005 (D. simulans), and last, Geo288 (D. simulans) having the highest proportion of pupae on the food (fig. 1; supplementary fig. S1B, Supplementary Material online). These four strains follow an identical pattern for tilB gene expression, with T.4 having the highest relative transcript abundance, and Geo288 having the lowest (fig. 6D). Although it is not possible to detect a significant effect with a sample size of 4, this suggests that tilB gene expression may be negatively correlated with the proportion of pupae on the food (Spearman’s rank correlation: rs = −1, P < 0.10).
We also attempted to perform RT-qPCR to compare Fas2 gene expression between D. simulans and D. melanogaster. Unfortunately, Fas2 is a complex gene with multiple splice forms, so we were unsuccessful in designing general primers that would amplify all transcripts in both species, and were unable to include Fas2 in this experiment.
Discussion
A Species-Level Difference in Pupation Site Choice Behavior
Our initial survey of pupation site choice behavior in D. melanogaster and D. simulans expands upon a previously reported interspecific difference (Markow 1979). Consistent with previous results, on average, D. simulans strains had a greater proportion of flies pupating on the food surface compared with D. melanogaster strains. However, we used 11 D. melanogaster and 12 D. simulans strains sourced from around the globe to demonstrate this difference. Although the species difference holds when comparing the grand mean of all strains for each species, there is substantial variation within species, such that the species’ distributions overlap (fig. 1). These geographically varying differences in pupation behavior among Drosophila species, in combination with measurements of the environmental variables that affect this behavior (Sokal et al. 1960; Sameoto and Miller 1968; Hodge and Caslaw 1998; Seyahooei et al. 2009), may be useful in identifying the selection pressures (if any) that affect the evolution of this trait. Differences in pupation site choice behavior may be a form of niche partitioning where species co-occur, as has been previously suggested (Arthur and Middlecote 1984). Alternatively, pupation site choice may be an adaptive response to parasite or parasitoid presence (Kraaijeveld and Godfray 2003). A globally sourced panel of strains with significant variation, such as we describe, provides an inroad for studies comparing pupation behavior to differences in the ecology of each collection site, such that we can better understand the ultimate causes of this behavioral evolution.
Using two of these strains (LHM for D. melanogaster and Lhr for D. simulans) to create reciprocal hybrids in a controlled genetic background (fig. 2A and B), we further demonstrate that the X chromosome has a substantial effect on the evolved differences in pupation site choice behavior between D. simulans and D. melanogaster. A similar X-effect has been detected for pupation behavior when comparing D. simulans and D. sechellia (Erezyilmaz and Stern 2013), highlighting a potentially conserved role of the X chromosome in regulating pupation site choice behavior.
tilB and Fas2: Loci of Evolution for Divergent Pupation Behavior between D. simulans and D. melanogaster
Using a deficiency screen, we identified two regions of the X chromosome that contribute to this effect. We have presented substantial evidence for a role of one gene from each region, tilB (touch insensitive larva B) and Fas2 (Fasciclin 2), in the divergence of pupation behavior among these species. Although our present study does not present a functional analysis of the D. melanogaster or D. simulans Fas2 or tilB alleles, we can use the D. melanogaster annotation of each gene to speculate about their role in the evolution of pupation behavior. Fas2 is a large gene, spanning over 70,000 base pairs, with expression in D. melanogaster peaking during the larval wandering stage (L3) (Graveley et al. 2011). It is also complex, with seven transcripts composed of various combinations of 16 exons. Broadly, Fas2 functions as a neuronal recognition molecule and is involved in patterning the larval nervous system (Grenningloh et al. 1991). Expression of Fas2 is critical for synapse formation and growth at the larval neuromuscular junction (Schuster et al. 1996; Davis et al. 1997) and is also important for patterning of the larval mushroom body (Kurusu et al. 2002). With expression in both the central and peripheral nervous system, it is possible that variation in the Fas2 locus differentially wires the D. melanogaster and D. simulans brains, altering how larvae perceive or interpret stimuli. Whether these differences result from evolution of the protein sequence, and/or spatial or temporal differences in transcript expression remains to be determined. However, the largest transcript shares 97% sequence identity between D. melanogaster and D. simulans, and does not show signs of positive selection within the D. melanogaster subgroup (Stanley and Kulathinal 2016). This highlights a potential role for expression differences between the species, consistent with our finding that reducing Fas2 expression in D. melanogaster results in a more D. simulans-like pupation behavior.
Unlike Fas2, tilB is a short gene, spanning just over 1,700 base pairs, with a single transcript composed of five exons. In D. melanogaster, tilB is also expressed in wandering larvae and pupae, though it shows higher expression in testes of adult males, due to its role in developing sperm flagella (Graveley et al. 2011). In fact, tilB is associated with ciliary motility (Kavlie et al. 2010) and is a part of the mechanosensory transduction machinery (Göpfert and Robert 2003). Mutant tilB larvae display normal locomotor activity, but have a reduced withdrawal response to physical disturbance (Kernan et al. 1994). tilB shares 96% sequence identity between D. melanogaster and D. simulans and does not show signs of positive selection within the D. melanogaster subgroup (Stanley and Kulathinal 2016). Consistent with this finding, our data suggest that changes in tilB expression could potentially result in differences in peripheral sensory perception between D. melanogaster and D. simulans larvae, ultimately influencing larval pupation site choice behavior. A more precise functional analysis, like driving hyperexpression of tilB in a D. simulans background, is necessary to test this hypothesis, but these experiments require the creation of transgenic D. simulans strains that falls beyond the scope of our study.
Standards of Evidence and the Challenges of Interspecific Mapping
Above, we have discussed the results of our interspecific deficiency screen of the Drosophila X chromosome. This technique has long been employed to map morphological traits, physiological traits, hybrid incompatibility loci (Konopka and Benzer 1971; Cote et al. 1986; Bour et al. 2000; Barbash et al. 2003; Sawamura et al. 2004; Cattani and Presgraves 2012; Pardy et al. 2019), and behaviors (Fanara et al. 2002; Moehring and Mackay 2004; Laturney and Moehring 2012). Nonetheless, these screens have been criticized due to their susceptibility to epistatic interactions that can produce false positives (Anholt and Mackay 2004) and for the imperfect comparison of deficiency chromosomes to balancer chromosomes (Stern 2014). These problems may be exacerbated in a hybrid background. These issues are potentially reflected by the global average pupation index for these deficiencies being 0.88, rather than 1 (fig. 4A), indicating that, in general, more balancer hybrids pupated on the food compared with deficiency hybrids.
We attempted to exclude the possibility of false positives in our initial deficiency screen in two ways. First, we crossed each deficiency with a significant pupation index to D. melanogaster to test for deleterious effects of the deficiencies themselves. Indeed, three of six significant deficiencies produced similar results when crossed to D. melanogaster and were discarded. Next, to ensure the patterns shown by the remaining three were not due to epistasis among strains, we crossed each to a second D. simulans and D. melanogaster strain and found consistent results. Taken together, these results suggest that these three deficiencies are likely revealing recessive D. simulans variation, rather than background epistatic interactions. It should be noted, however, that there is still the possibility that these results are affected by epistatic interactions between a conserved D. simulans locus (i.e., present in multiple strains) in these regions and conserved loci in D. melanogaster; nonetheless, such a result is still biologically relevant.
The results of our gene disruption and quantitative complementation tests further enforce our deficiency screen results, again, with multiple controls to limit the possibility of false positives and/or strain-specific epistatic effects. We crossed tilB and Fas2 gene disruption strains (two strains per gene) to multiple D. melanogaster and D. simulans strains and found no evidence of strain-specific epistatic interactions. We also showed that reduced Fas2 and tilB gene expression in D. melanogaster leads to a more D. simulans-like phenotype, and that tilB shows higher relative expression in D. melanogaster compared with D. simulans strains (fig. 6D). Taken together, our hybrid gene disruption screens, quantitative complementation tests, and gene expression studies in D. melanogaster make an excellent case for a role of tilB and Fas2 in divergent pupation site choice behavior.
Although we were able to conduct quantitative complementation tests for knockouts of candidate genes, higher standards of evidence have been suggested for mapping in Drosophila—the reciprocal hemizygosity test being the gold standard (Stern 2014). In this study, we performed one-half of this test (hybrid females with a disrupted D. melanogaster X chromosome and an intact D. simulans X chromosome). Unfortunately, the reciprocal half of the test (hybrid females with a disrupted D. simulans X chromosome and an intact D. melanogaster X chromosome) is extremely difficult, if at all possible, for several reasons. Using hybrid crosses of largely reproductively isolated species makes comparative work difficult, and we were only able to produce successful hybrid crosses in one direction (D. simulans Lhr males crossed to D. melanogaster females). Even using this “easy” direction of the cross, our average crossing success rate for our 90 deficiencies was only 54% (SD = 31%), with success rates as low as 8.7% (supplementary table S3A, Supplementary Material online), not including nine deficiencies that we were unable to successfully cross to D. simulans (supplementary table S3B, Supplementary Material online). Our attempts to cross D. simulans Lhr females to D. melanogaster males were never successful, despite trying multiple strains known to court and attempt copulation with D. simulans females (supplementary table S6, Supplementary Material online). Unfortunately, crosses with nonhybrid rescue D. simulans females typically only produce male offspring (Sawamura et al. 1993), rendering the reciprocal hemizygosity test impossible. Nonetheless, there is potentially some strain-specific variability in hybrid female inviability (Gérard and Presgraves 2012), so we tested our 11 strains of D. simulans (supplementary table S1, Supplementary Material online) with D. melanogaster males using the same hybrid crossing methods. Five of these strains crossed at relatively low frequencies, but only produced hybrid male offspring (supplementary table S6, Supplementary Material online). Further, these five strains yielded an average of only 8.67 hybrid flies per vial, much lower than the densities necessary to detect variation in pupation site choice behavior (our above data used ∼30–50 successful crosses of vials containing at least ten of each experimental female).
In addition to the crossing difficulties preventing us from completing the reciprocal hemizygosity test, we would need to use transgenic tools in D. simulans to create knockouts for Fas2 and tilB. However, these knockouts are male lethal (Grenningloh et al. 1991) and male sterile (Eberl et al. 2000) in D. melanogaster, respectively. Assuming the same is true for D. simulans, either knockout would be difficult to maintain without the use of balancer chromosomes, which are currently not an available resource. In addition, the male lethality/sterility of these knockouts eliminates the possibility of using D. simulans gene disruption males to create hybrid females for a reciprocal hemizygosity test.
Additional tests that could potentially support our findings in lieu of the reciprocal hemizygosity test would be the transgenic addition of the dominant D. melanogaster Fas2 and tilB alleles to a D. simulans background, or replacement of the D. simulans Fas2 and tilB alleles with the D. melanogaster alleles in D. simulans. However, these approaches similarly require the development of new transgenic tools in D. simulans, and experiments using these genetically modified flies are prone to the same potential epistatic interactions as our other hybrid crosses.
Conclusions
Although we have presented multiple lines of evidence, all consistently supporting a role for tilB and Fas2 in divergent pupation site choice behavior, these results must be considered in light of the above caveats. We have taken measures to address these caveats as completely as possible and present our results with a high standard of evidence. Our study highlights the difficulty of interspecific mapping in producing conclusive results and underscores a need for transgenic tools to be developed in nonmodel Drosophila species.
Materials and Methods
General Fly Maintenance
Unless otherwise stated, we maintained all fly strains and set up all crosses for these experiments in 25 mm diameter × 95 mm height vials containing standard cornmeal–molasses–yeast medium at 25 °C under a 12 h:12 h light/dark cycle at 50% relative humidity. Under these conditions, we established nonoverlapping 2-week lifecycles as follows. For all stocks, except LHM and Lhr (see below), we transferred all of the eclosed male and female adult flies into fresh vials containing food media supplemented with live yeast on the surface for 1–3 days, at which point the flies were discarded. Fourteen days later (after all progeny had eclosed), we again transferred adult flies into fresh vials for 1–3 days to begin the next generation. We maintained LHM and Lhr identically, except we additionally regulated density by transferring only ten males and ten females to begin the next generation.
Characterizing Pupation Behavior for D. melanogaster and D. simulans
We measured pupation behavior for 11 D. melanogaster and 12 D. simulans strains collected from various locations throughout the world (supplementary table S1, Supplementary Material online). The 11 D. melanogaster strains included ten of the “founder” wild-type inbred lines of the Drosophila Synthetic Population Resource (King et al. 2012) and a single wild-type line created from the LHM laboratory-adapted population (Rice et al. 2005). The 12 D. simulans strains included 11 wild-type strains and a single strain carrying Lhr, a mutation that restores viability in D. melanogaster/D. simulans hybrid males. LHM and Lhr were included in this screen because they are the parental stocks required for reciprocal hybrid crosses (see below).
To measure pupation behavior, we placed ten males and ten females from a specific line (both 3–5 days old) into half-pint bottles and allowed females to oviposit overnight on a 35-mm-diameter petri dish filled with food medium that was placed in the opening of the bottle. In total, we set up five bottles for each line. The following morning, we transferred 100 eggs from the petri dishes into vials containing food medium (described above) that were lined with an acetate sleeve on which the larvae could pupate. In total, we set up 5–8 vials per line. Vials were held at 25 °C for 8 days, at which time the liner was removed and the locations of the pupae were recorded (8 days was long enough for almost all larvae to pupate without any flies eclosing). A pupa was considered “on” the food if it was within 1 cm of the food surface, whereas all pupae that were further than 1 cm from the food surface were considered “off” the food. We used “on” versus “off” the food as our measure of pupation site choice behavior because this measure successfully characterized the interspecific difference in pupation behavior while providing a manageable phenotype to record for our genotype–phenotype screen. We considered pupae within 1 cm of the food surface to be “on” the food because D. simulans larvae often pupated in stacked groups on the food surface along the side of the vial, resulting in several of them pupating slightly above the food surface. Using these measures, we calculated the proportion of pupae on the food for each vial, and then used Wilcoxon tests to compare these values between species, and between strains within species. For comparisons between species, our unit of replication was the mean proportion of pupae on the food surface from each line (i.e., N = 11 for D. melanogaster and N = 12 for D. simulans).
Crossing D. melanogaster with D. simulans
For all crosses below, we created F1 hybrids between D. melanogaster and D. simulans using the following protocol: D. simulans males were collected as virgins within 6 h of eclosion and held at room temperature in groups of 20 in vials containing food medium for 3–4 days. To set up crosses, we collected young D. melanogaster virgin females within 2–3 h of eclosion, and combined 8–12 of these females with 20 D. simulans males in vials containing food medium supplemented with an ad lib amount of live yeast on the surface. We then pushed a long foam plug down into the vial, leaving ∼1 cm of space above the food surface. We held flies under these conditions for 3 days, at which time they were transferred from these “cross vials” into “pupation vials” that contained food medium with no added yeast, and were lined with an acetate sleeve on which the larvae could pupate. We always set up crosses using D. melanogaster females and D. simulans Lhr males, because crosses in the opposite direction were never successful, despite trying a number of strains (supplementary table S6, Supplementary Material online).
Measuring Pupation Behavior in F1 Hybrids
To create F1 hybrid males and females, we used D. simulans males from the Lhr strain (Watanabe 1979). The Lhr mutation restores viability in F1 hybrid males, which are usually lethal (Brideau et al. 2006). To create F1 hybrid females and F1 males with a D. melanogaster X chromosome (“melX” males), we crossed wild-type females from our LHM strain (provided by Dr. William Rice) to Lhr D. simulans males (fig. 2A). Because we were unable to successfully cross Lhr D. simulans females to D. melanogaster males (supplementary table S6, Supplementary Material online), we created F1 hybrid males with the D. simulans X chromosome (“simX” males) by crossing D. melanogaster LHM females that carry a compound X chromosome (C(1)DX y f) (Rice et al. 2005) to D. simulans Lhr males. The compound X in these females caused the X chromosome to be transmitted from D. simulans fathers to their F1 hybrid sons (fig. 2B). Using LHM and C(1)DX y f LHMD. melanogaster females in each hybrid cross ensured the backgrounds of these strains were identical, and that all maternal inheritance (cytoplasmic and mitochondrial) in the reciprocal male hybrid crosses originated from the D. melanogaster parent. Thus, these hybrids have an identical background with the exception of the sex chromosomes, and any differences we observe between melX and simX males are directly attributable to the species they inherit their respective sex chromosomes from.
After 3 days in the cross vial, we transferred males and females into pupation vials for 24 h, at which time the flies were removed. Our methods for setting up hybrid pupation vials (used here and for the remainder of the study) differed slightly from our initial species screen (which controlled egg density) because of the unpredictability of hybrid crosses. Although this may affect the absolute proportion of individuals pupating on the food for a specific strain, the differences between species were consistent using both approaches.
While screening hybrid pupation behavior, we also concurrently screened pupation behavior for the parental D. melanogaster strain (LHM) and the parental D. simulans strain (Lhr) for comparison. Parental strain cross vials contained only a moderate amount of yeast, were set up with only five males and five females (pure species crosses produce more offspring), and did not have a plug pushed down into the vial, but were otherwise treated identically to the hybrid crosses. In total, we set up 30–33 vials per treatment.
All pupation vials were held at 25 °C for 8 days, at which time the liner was removed. Because we needed to identify the sex of individuals that pupated in these vials, we could not simply record the location of pupae as we did for our initial species screen. Instead, we removed any remaining larvae and cut the liner at a point 1 cm above the food surface. The portion of the liner that contained pupae within 1 cm of the food surface was returned to the original vial (the “on vial”), whereas the portion of the liner with pupae further off of the food surface was placed in another vial containing food medium (the “off vial”). The flies that eclosed were sexed and counted 7 days later (15 days post-egg); all flies that eclosed in the “on vial” were considered flies that pupated on the food surface, whereas all flies that eclosed in the “off vial” were considered flies that pupated off the food surface. We then compared the proportion of individuals that pupated on the food for each genotype and sex using pairwise Wilcoxon tests, followed by sequential Bonferroni correction for multiple comparisons (Holm 1979).
To assess the validity of using hybrid behavior to map interspecific differences, we set up an additional experiment to ensure that hybrids behaved typically with respect to pupation site choice behavior. This is a potential concern, as hybrids between D. melanogaster and D. simulans are known to differ from either parent in a variety of traits (Sturtevant 1920; Takano 1998; Barbash and Ashburner 2003). For a subset of pupation vials, we calculated the average pupation height of males and females from each strain as follows: Instead of dividing the pupation liner into “on” food and “off” food sections, we cut the liner at eight intervals, each spaced 1 cm from the last, starting at the food surface. These portions of the liner were ranked from 1 (on the food surface) to 8 (the furthest from the food surface) and were transferred to separate vials containing food medium. Seven days later, the flies that eclosed were counted, and we used the rankings to calculate a mean pupation height within each pupation vial, such that a higher value indicates a farther distance from the food. We performed this experiment for our D. simulans strain (Lhr), our reciprocal hybrid crosses (melX and simX in an LHM background), the D. melanogaster strain w1118, and melX hybrids in a w1118 background (w1118 is the background strain for the majority of deficiencies we used in our screen). We compared average pupation heights using Wilcoxon tests followed by sequential Bonferroni correction for multiple comparisons (Holm 1979).
Estimating the Effect of the X Chromosome
Because we found a significant effect of the X chromosome on pupation site choice, we used our results to estimate how much of the difference in this behavior can be attributed to the X chromosome. We calculated the “species difference ratio” (using the data from our parental/hybrid screen in fig. 3) by dividing the median proportion of males on the food for D. simulans by the median proportion of males on the food for D. melanogaster males (species difference ratio = 5.27). We then calculated an “X effect ratio” by dividing the median proportion of simX males on the food by the median proportion of melX males on the food. To determine how much of this species difference can be attributed to the X chromosome, we divided the “X effect ratio” by the “species difference ratio.” Finally, we calculated a bootstrapped 95% confidence interval on this estimate using 100,000 bootstraps.
Mapping Hybrid Pupation Behavior Using the Bloomington Deficiency Kit
Our reciprocal hybrid crosses found a significant effect of the X chromosome on the difference in pupation site choice behavior between D. simulans and D. melanogaster, so we devised a crossing scheme using chromosomal deficiencies to screen the X chromosome for loci contributing to this difference. These deficiencies are part of the Bloomington Deficiency Kit (Cook et al. 2012), available from the Bloomington Drosophila Stock Center (BDSC). We assayed a total of 90 deficiency strains covering 87% of the X chromosome (supplementary table S3A, Supplementary Material online). We attempted to screen nine additional deficiencies, but were unable to successfully cross these strains to D. simulans (supplementary table S3B, Supplementary Material online). We restricted our deficiency screen to lines from the BSC, Exelixis, and DrosDel sets to control for strain background effects while also maximizing chromosome coverage.
Crossing these deficiencies to D. simulans produced two types of hybrid female that were heterozygous for D. melanogaster/D. simulans at each autosome (fig. 2C). The deficiency hybrid females harbor the D. melanogaster deficiency X chromosome and a D. simulans X chromosome, making them hemizygous for a segment of the X chromosome. At this locus, these hybrid females only express D. simulans alleles. The balancer hybrid females harbor the D. melanogaster balancer X chromosome (marked with the dominant visible Bar marker) and a D. simulans X chromosome. These females are heterozygous for D. melanogaster/D. simulans over the entirety of the X chromosome, and thus express both D. simulans and D. melanogaster alleles. Although the deficiency hybrids are our flies of interest, the balancer hybrids provide an experimental control, as these females developed in the same environment as our experimental flies.
To set up crosses, we collected deficiency females as young virgins (2–3 h after eclosing) and crossed them to D. simulans males from the Lhr strain using the crossing methods described above. After 3 days in the cross vial, we transferred males and females into pupation vials for 24–48 h, at which time the flies were removed. We then divided the pupations vials into “on” and “off” vials as we did for F1 hybrids (above). Seven days later, the flies were counted and genotyped using the presence or absence of the Bar marker located on the balancer chromosome. All flies that eclosed in the “on vial” were considered to have pupated on the food surface, whereas all flies that eclosed in the “off vial” were considered to have pupated off the food surface. We calculated the proportion of deficiency hybrid females that pupated on the food and the proportion of balancer hybrid females that pupated on the food. We then used these measures to calculate a “pupation index” as the proportion of deficiency hybrid females pupating on the food divided by the proportion of balancer hybrid females pupating on the food. To increase the accuracy of our estimates, we only included pupation vials in our analysis that yielded at least ten of each type of female. For each deficiency hybrid strain we measured, we report the median pupation index of all replicates, because there were often high-scoring outliers that significantly skewed the mean pupation index. These outliers almost always had abnormally high pupation indices, so focusing on median values makes our findings more conservative.
We used one-sample Wilcoxon tests followed by sequential Bonferroni adjustment for multiple comparisons (Holm 1979) to identify deficiency hybrid crosses with a pupation index significantly different than 1. A median pupation index significantly >1 indicates that more deficiency females pupated on the food compared with balancer females, potentially because the deficiency includes D. melanogaster genetic variation that is involved in pupation site choice behavior. Alternatively, simply creating flies that are hemizygous at a locus on the X chromosome may result in a variety of pleiotropic effects that make larvae less likely to climb up the vial. To test for this, when a deficiency hybrid cross showed a pupation index significantly >1 (supplementary table S3A, Supplementary Material online), we crossed that D. melanogaster deficiency strain to a D. melanogaster wild-type strain (T.4); a subset of D. simulans Lhr crosses were set up simultaneously to control for timing differences. If these D. melanogaster deficiency crosses displayed the same pattern, we considered the effect of the deficiency on pupation behavior to be a byproduct of deleting a large portion of the X chromosome, rather than revealing recessive D. simulans variation, and discarded them. If instead the pupation index for the D. melanogaster cross was significantly lower than the pupation index for the D. simulans cross (based on two-sample Wilcoxon tests), we pursued that deficiency for further validation. To ensure that this pattern is not a result of epistasis from the hemizygous region in a hybrid background, we further crossed these deficiencies to an additional D. melanogaster (T.1) and D. simulans (Mex180) strain, to test for background-specific effects. Although the T.1 and Mex180 crosses were set up simultaneously, this second set of crosses was set up at a later time point than the original deficiency crosses (T.4 and Lhr).
Testing Candidate Genes in Deficiency Regions Using Gene Knockouts
For the two regions of interest identified by our deficiency screen, we ordered transgenic knockouts for any genes available within the region at the time. The first region of interest is the overlap of Df(1)BSC869 and Df(1)ED6720, excluding the region covered by Df(1)ED6727 (fig. 4C;supplementary table S4A, Supplementary Material online), which did not have a pupation index >1 (supplementary table S3A, Supplementary Material online). At the time of assay, there were only three characterized genes within this region that had expression in the larval nervous system and nonlethal-verified loss-of-function alleles available: Fas2 (Fasciclin 2), mei-9 (meiotic 9), and norpA (no receptor potential A). We screened two Fas2 knockouts, the mutant allele Fas2eb112 (Fas2eb112/FM7c; Grenningloh et al. 1991; provided by Brian McCabe), and a P-element insertion allele, Fas2G0293 (former BDSC Stock 11850; full genotype: w67c23P{lacW}fas2G0293/FM7c). It is important to note that the nature of the lesion is uncharacterized for Fas2G0293, although our behavioral data suggest that it is indeed a loss of function allele, as it behaves indistinguishably from the verified knockout Fas2eb112 (Grenningloh et al. 1991). We additionally screened the mei-9A1 mutant allele (w1 mei-9A1/FM7h; BDSC stock #6792) and the norpA36 mutant allele (w* norpA36; BDSC stock #6792). Because norpA36is not held over a balancer, we also set up crosses using a norpA rescue strain created in the same background (w* norpA36; P{w[+mC]=ninaE.norpA.E}2; BDSC stock #52276) as a control.
The second region of interest identified by our deficiency screen was the region deleted by Df(1)Exel6255 (fig. 4D;supplementary table S4B, Supplementary Material online). We obtained knockout strains for both of the characterized genes expressed in the larval nervous system that had verified loss-of-function alleles available: tilB (touch insensitive larva B) and wap (wings apart). We screened two tilB mutant alleles, tilB1 and tilB2 (y w tilB1/2/FM4; Kernan et al. 1994; provided by Daniel Eberl), and the wap2 mutant allele (wap2/FM6; BDSC stock #8133).
Like the deficiency strains, each of our gene disruptions (with the exception of norpA) is held over a balancer chromosome with a visible marker. To measure the pupation behavior of hybrids containing knockout copies of these D. melanogaster genes, we crossed each D. melanogaster knockout strain to Lhr using the previously described methods and calculated the pupation index as the proportion of knockout females on food/the proportion of balancer females on the food. We also simultaneously crossed each D. melanogaster knockout strain to a wild-type D. melanogaster (T.4) strain to control for the effects of being hemizygous for this particular gene. To increase the accuracy of our estimates, we only included pupation vials in our analysis that yielded at least ten of each type of female (i.e., knockout/balancer). We crossed knockout strains that displayed the pattern we expect for a gene involved in pupation site choice (i.e., a pupation index that is significantly higher when crossed to D. simulans than when crossed to D. melanogaster) to an additional D. simulans (Mex180) and D. melanogaster (T.1) wild-type strain for verification. For each knockout (with the exception of norpA), we compared the pupation index for crosses to D. melanogaster and D. simulans using Wilcoxon tests. We also used ANOVA to perform quantitative complementation tests on these data by comparing the proportion of flies that pupated on the food for each genotype (knockout/balancer) and species (D. melanogaster/D. simulans). Because these data are proportions, we arcsine-transformed them before using them in the model. For wap and mei-9, we used two-sample ANOVA with fixed effects “genotype” (knockout or balancer), “species” (D. melanogaster or D. simulans), and the interaction between species and genotype. Because we screened two different mutant alleles and used two different D. melanogaster and D. simulans strains for both tilB and Fas2, we used nested ANOVA to perform quantitative complementation tests. These models had the fixed effects listed above (genotype, species, and their interaction), but also included the factors “allele” nested within “genotype,” and “strain” nested within “species.” Because norpA is not held over a balancer, we crossed both the mutant and rescue strains to Lhr and the D. melanogaster T.4 strain. We compared the proportion of flies that pupated on the food for hybrid norpA mutant females and hybrid norpA rescue females using a Wilcoxon test, and performed a quantitative complementation test on the proportion of females that pupated on the food (following arcsine transformation) using a two-sample ANOVA with fixed effects “genotype,” “species,” and their interaction.
Validating Hybrid Knockout Results Using RNAi Knockdown
Our knockout screen identified two genes that appear to be involved in pupation site choice: tilB and Fas2. We further tested the effects of these genes on pupation behavior using RNAi knockdown in D. melanogaster. We used the elav-Gal4 driver (P{w[+mC]=GAL4-elav.L}2/CyO; BDSC #8765), which expresses Gal4 throughout the nervous system. We drove down the expression of tilB and Fas2 throughout the nervous system by crossing elav-Gal4 virgin females to UAS-tilB (BDSC #29391: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF03324}attP2) and UAS-Fas2 (BDSC #34084: y[1] sc[*] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.HMS01098}attP2) males, respectively. The resulting flies express a gene-specific hairpin RNA throughout the nervous system, causing the degradation of mRNA, and thus, reduced expression of that gene (Perkins et al. 2015; Zeng et al. 2015). As experimental controls for each RNAi cross, we also crossed elav-Gal4 virgin females to the RNAi background stock (y1 v1; P{y[+t7.7]=CaryP}attP2; BDSC stock # 36303) and the Gal4-1 stock (containing a hairpin RNA targeting Gal4 in VALIUM20; BDSC stock # 35784). Together, these controls allow us to account for the effect of both the Gal4 mutation and general expression of hairpin RNA throughout the nervous system. We set up pupation vials using the methods described above, and for each cross, we calculated the proportion of RNAi (or control) flies on the food (removing any data points with fewer than 20 experimental flies), and compared the treatments using pairwise Wilcoxon tests followed by sequential Bonferroni correction (Holm 1979). If more RNAi flies pupate on the food in the experimental cross (in which the expression of the gene is driven down) compared with the control crosses (in which gene expression is unaffected), this provides further support for that gene’s involvement in pupation site choice. For these vials, we additionally determined the average pupation height of flies from each cross using the methods described above for F1 hybrids; if RNAi flies pupate closer to the food than flies from the control crosses, this suggests that gene may be involved in pupation site choice.
Testing tilB for Species-Specific Differences in Larval Transcript Expression
We selected two each of our 11 D. melanogaster and 12 D. simulans strains to test for larval stage-specific expression differences of candidate genes using real time RT-PCR—one extreme and one average. For D. simulans, we selected Geo288 and Per005 (supplementary table S1, Supplementary Material online), because Geo288 has the highest proportion of pupae on the food of the D. simulans strains and Per005 is closest to the species mean (fig. 1). For D. melanogaster, we selected CA1 and T.4 (supplementary table S1, Supplementary Material online) because T.4 has the lowest proportion of pupae on the food of the D. melanogaster strains, and CA1 is closer to the species mean (fig. 1).
To harvest larvae from these strains, we allowed adult females to oviposit in standard vials containing food media between the hours of 8 AM and 12 PM over two consecutive days. Ninety-six and 120 h after the final oviposition day, we floated larvae out of the food media using a 20% sucrose in water solution, sucked them up using a transfer pipet, briefly rinsed them with DI water on cheesecloth, and snap froze them using liquid nitrogen (Nichols et al. 2012). In this way, we collected 20–30 mg of larvae from two developmental time points: 96 and 120 h following oviposition. We chose these time points because they approximate the early and late wandering stage, which immediately precedes pupation, and because the larvae are large enough for many to be harvested at once using the above methods. We extracted mRNA using the Qiagen RNeasy Plus Mini Kit and prepared cDNA using the Promega Verso kit.
To quantify transcript abundance, we designed primers on opposite sides of a single intron near the 3′-end of tilB (Huggett et al. 2005) so that PCR of gDNA produced a longer fragment than cDNA (RT-PCR). Additionally, we used primers for the gene RpL32 as an internal control (Al-Atia et al. 1985; Chertemps et al. 2007). We were unsuccessful in designing general primers that would amplify all transcripts in both species for Fas2, so we did not include Fas2 in these experiments. A full list of primers and transcripts can be found in supplementary table S7A, Supplementary Material online. For each stage and strain, we prepared two to three biological replicates, which we then amplified in two technical replicates for 40 rounds of qPCR. Using RpL32 transcript number as an internal control, we calculated relative tilB transcript abundance (RelTA) using mean threshold cycle values (Ctµ) from RT-qPCR of two technical replicates per biological replicate, while correcting for species differences in primer efficiency. We estimated primer efficiency differences by serially diluting gDNA from each of our D. melanogaster and D. simulans strains, performing qPCR, and using a standard curve to calculate adjusted amplification factors (AmpF) for each species (supplementary table S7B, Supplementary Material online). We calculated transcript abundance for each sample (n) relative to a single T.4 biological replicate at 96 h (T.4@96) using the following amplification curve equation:
To ensure that we were amplifying cDNA made from RNA, and not gDNA contamination, we performed gel electrophoresis on our cDNA samples to ensure we only visualized the short, intron-less, band.
Because the relative transcript abundance data had a skewed distribution, we used the reciprocal root transformation to normalize the distributions. We then analyzed the relative transcript abundance of tilB using a nested ANOVA with the following factors: species, strain nested within species, larval age (96 and 120 h), and the interaction between species and larval age. The interaction term between species and larval age was not significant (ANOVA: F1,13 = 0.73, P = 0.41), so we removed it from the model, and analyzed only the previously listed factors (without the interaction).
Supplementary Material
Acknowledgments
We thank Alexandra Lamoureux, Susanne Tilk, Nastaran Hosseinipour, Sayeh Akhavan, Wesley Cochrane, and Veronica Cochrane for their assistance in the collection of pupation data. We also thank William Rice, Stuart Macdonald, Brian McCabe, and Daniel Eberl for providing fly stocks used for this study. This work would not have been possible without the community-supported resources available at the Cornell Drosophila Stock Center (National Science Foundation: CSBR 1820594), the Bloomington Drosophila stock center (National Institutes of Health P400D018537), or FlyBase (Gramates et al. 2017). This work was supported by the National Institutes of Health (R01 GM098614 to TLT).
References
- Al-Atia GR, Fruscoloni P, Jacobs-Lorena M.. 1985. Translational regulation of mRNAs for ribosomal proteins during early Drosophila development. Biochemistry 24(21):5798–5803. [DOI] [PubMed] [Google Scholar]
- Anholt RRH, Mackay TFC.. 2004. Quantitative genetic analyses of complex behaviours in Drosophila. Nat Rev Genet. 5(11):838–849. [DOI] [PubMed] [Google Scholar]
- Arthur W, Middlecote J.. 1984. Evolution of pupation site and interspecific competitive ability in Drosophila hydei. Biol J Linn Soc Lond. 23(4):343–352. [Google Scholar]
- Barbash DA, Ashburner M.. 2003. A novel system of fertility rescue in Drosophila hybrids reveals a link between hybrid lethality and female sterility. Genetics 163(1):217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbash DA, Siino DF, Tarone AM, Roote J.. 2003. A rapidly evolving MYB-related protein causes species isolation in Drosophila. Proc Natl Acad Sci U S A. 100(9):5302–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bour BA, Chakravarti M, West JM, Abmayr SM.. 2000. Drosophila SNS, a member of the immunoglobulin superfamily that is essential for myoblast fusion. Genes Dev. 14(12):1498–1511. [PMC free article] [PubMed] [Google Scholar]
- Brideau NJ, Flores HA, Wang J, Maheshwari S, Wang X, Barbash DA.. 2006. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 314(5803):1292–1295. [DOI] [PubMed] [Google Scholar]
- Cattani VM, Presgraves DC.. 2012. Incompatibility between X chromosome factor and pericentric heterochromatic region causes lethality in hybrids between Drosophila melanogaster and its sibling species. Genetics 191(2):549–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertemps T, Duportets L, Labeur C, Ueda R, Takahashi K, Saigo K, Wicker-Thomas C.. 2007. A female-biased expressed elongase involved in long-chain hydrocarbon biosynthesis and courtship behavior in Drosophila melanogaster. Proc Natl Acad Sci U S A. 104(11):4273–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook RK, Christensen SJ, Deal JA, Coburn RA, Deal ME, Gresens JM, Kaufman TC, Cook KR.. 2012. The generation of chromosomal deletions to provide extensive coverage and subdivision of the Drosophila melanogaster genome. Genome Biol. 13(3):R21.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote B, Bender W, Curtis D, Chovnickt A.. 1986. Molecular mapping of the rosy locus in Drosophila melanogaster. Genetics 112:769–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA.. 1997. Patterns of speciation in Drosophila revisited. Evolution 51(1):295–303. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA.. 2004. Speciation. Sunderland (MA): Sinauer Associates.
- Davis GW, Schuster CM, Goodman CS.. 1997. Genetic analysis of the mechanisms controlling target selection: target-derived Fasciclin II regulates the pattern of synapse formation. Neuron 19(3):561–573. [DOI] [PubMed] [Google Scholar]
- Ding Y, Berrocal A, Morita T, Longden KD, Stern DL.. 2016. Natural courtship song variation caused by an intronic retroelement in an ion channel gene. Nature 536(7616):329–332. [DOI] [PubMed] [Google Scholar]
- Eberl DF, Hardy RW, Kernan MJ.. 2000. Genetically similar transduction mechanisms for touch and hearing in Drosophila. J Neurosci. 20(16):5981–5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erezyilmaz DF, Stern DL.. 2013. Pupariation site preference within and between Drosophila sibling species. Evolution 67(9):2714–2727. [DOI] [PubMed] [Google Scholar]
- Fanara JJ, Robinson KO, Rollmann SM, Anholt RRH, Mackay TFC.. 2002. Vanaso is a candidate quantitative trait gene for Drosophila olfactory behavior. Genetics 162(3):1321–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J, Corley R, DeFries JC, Fulker DW, Gray JA, Miller S, Collins AC.. 1995. A simple genetic basis for a complex psychological trait in laboratory mice. Science 269(5229):1432–1435. [DOI] [PubMed] [Google Scholar]
- Gérard PR, Presgraves DC.. 2012. Abundant genetic variability in Drosophila simulans for hybrid female lethality in interspecific crosses to Drosophila melanogaster. Genet Res. 94(1):1–7. [DOI] [PubMed] [Google Scholar]
- Göpfert MC, Robert D.. 2003. Motion generation by Drosophila mechanosensory neurons. Proc Natl Acad Sci U S A. 100(9):5514–5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramates LS, Marygold SJ, Santos GD, Urbano J-M, Antonazzo G, Matthews BB, Rey AJ, Tabone CJ, Crosby MA, Emmert DB, et al. 2017. FlyBase at 25: looking to the future. Nucleic Acids Res. 45(D1):D663–D671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. 2011. The developmental transcriptome of Drosophila melanogaster. Nature 471(7339):473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenningloh G, Jay Rehm E, Goodman CS.. 1991. Genetic analysis of growth cone guidance in Drosophila: fasciclin II functions as a neuronal recognition molecule. Cell 67(1):45–57. [DOI] [PubMed] [Google Scholar]
- Hodge S, Caslaw P.. 1998. The effect of resource pH on pupation height in Drosophila (Diptera: Drosophilidae). J Insect Behav. 11(1):47–57. [Google Scholar]
- Holm S. 1979. A simple sequentially rejective multiple test procedure. Scand J Stat. 6(6):65–70. [Google Scholar]
- Huggett J, Dheda K, Bustin S, Zumla A.. 2005. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 6(4):279–284. [DOI] [PubMed] [Google Scholar]
- Kavlie RG, Kernan MJ, Eberl DF.. 2010. Hearing in Drosophila requires TilB, a conserved protein associated with ciliary motility. Genetics 185(1):177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kernan M, Cowan D, Zuker C.. 1994. Genetic dissection of mechanosensory transduction: mechanoreception-defective mutations of Drosophila. Neuron 12(6):1195–1206. [DOI] [PubMed] [Google Scholar]
- King EG, Merkes CM, McNeil CL, Hoofer SR, Sen S, Broman KW, Long AD, Macdonald SJ.. 2012. Genetic dissection of a model complex trait using the Drosophila Synthetic Population Resource. Genome Res. 22(8):1558–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittelmann S, Buffry AD, Franke FA, Almudi I, Yoth M, Sabaris G, Couso JP, Nunes MDS, Frankel N, Gómez-Skarmeta JL, et al. 2018. Gene regulatory network architecture in different developmental contexts influences the genetic basis of morphological evolution. PLOS Genet. 14(5):e1007375.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka RJ, Benzer S.. 1971. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A. 68(9):2112–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp A. 2009. Metamodels and phylogenetic replication: a systematic approach to the evolution of developmental pathways. Evolution 63(11):2771–2789. [DOI] [PubMed] [Google Scholar]
- Kraaijeveld AR, Godfray HCJ.. 2003. Potential life-history costs of parasitoid avoidance in Drosophila melanogaster. Evol Ecol Res. 5(8):1251–1261. [Google Scholar]
- Kurusu M, Awasaki T, Masuda-Nakagawa LM, Kawauchi H, Ito K, Furukubo-Tokunaga K.. 2002. Embryonic and larval development of the Drosophila mushroom bodies: concentric layer subdivisions and the role of fasciclin II. Development 129(2):409–419. [DOI] [PubMed] [Google Scholar]
- Lachaise D, Silvain JF.. 2004. How two Afrotropical endemics made two cosmopolitan human commensals: the Drosophila melanogaster-D. simulans palaeogeographic riddle. Genetica 120(1–3):17–39. [DOI] [PubMed] [Google Scholar]
- Laturney M, Moehring AJ.. 2012. Fine-scale genetic analysis of species-specific female preference in Drosophila simulans. J Evol Biol. 25(9):1718–1731. [DOI] [PubMed] [Google Scholar]
- Leary GP, Allen JE, Bunger PL, Luginbill JB, Linn CE, Macallister IE, Kavanaugh MP, Wanner KW.. 2012. Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. Proc Natl Acad Sci U S A. 109(35):14081–14086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markow TA. 1979. A survey of intra- and interspecific variation for pupation height in Drosophila. Behav Genet. 9(3):209–217. [DOI] [PubMed] [Google Scholar]
- Markow TA. 1981. Light-dependent pupation site preferences in Drosophila: behavior of adult visual mutants. Behav Neural Biol. 31(3):348–353. [DOI] [PubMed] [Google Scholar]
- Martin A, Orgogozo V.. 2013. The loci of repeated evolution: a catalog of genetic hotspots of phenotypic variation. Evolution 67(5):1235–1250. [DOI] [PubMed] [Google Scholar]
- McGrath PT, Xu Y, Ailion M, Garrison JL, Butcher RA, Bargmann CI.. 2011. Parallel evolution of domesticated Caenorhabditis species targets pheromone receptor genes. Nature 477(7364):321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittleman BE, Manzano-Winkler B, Hall JB, Korunes KL, Noor MAF.. 2017. The large X-effect on secondary sexual characters and the genetics of variation in sex comb tooth number in Drosophila subobscura. Ecol Evol. 7(2):533–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehring AJ, Mackay TFC.. 2004. The quantitative genetic basis of male mating behavior in Drosophila melanogaster. Genetics 167(3):1249–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller LD, Sweet VF.. 1986. Density-dependent natural selection in Drosophila: evolution of pupation height. Evolution 40(6):1354–1356. [DOI] [PubMed] [Google Scholar]
- Nichols CD, Becnel J, Pandey UB.. 2012. Methods to assay Drosophila behavior. J Vis Exp. 61:3795–103791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgogozo V, Stern DL.. 2009. How different are recently diverged species?: more than 150 phenotypic differences have been reported for the D. melanogaster species subgroup. Fly 3(2):117. [DOI] [PubMed] [Google Scholar]
- Pardy JA, Rundle HD, Bernards MA, Moehring AJ.. 2019. The genetic basis of female pheromone differences between Drosophila melanogaster and D. simulans. Heredity 122(1):93–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons PA. 1975. The comparative evolutionary biology of the sibling species, Drosophila melanogaster and D. simulans. Q Rev Biol. 50(2):151–169. [DOI] [PubMed] [Google Scholar]
- Perkins LA, Holderbaum L, Tao R, Hu Y, Sopko R, McCall K, Yang-Zhou D, Flockhart I, Binari R, Shim H-S, et al. 2015. The transgenic RNAi project at Harvard medical school: resources and validation. Genetics 201(3):843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince DJ, O’Rourke SM, Thompson TQ, Ali OA, Lyman HS, Saglam IK, Hotaling TJ, Spidle AP, Miller MR.. 2017. The evolutionary basis of premature migration in Pacific salmon highlights the utility of genomics for informing conservation. Sci Adv. 3(8):e1603198.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz M, Williams TM.. 2017. Using Drosophila pigmentation traits to study the mechanisms of cis-regulatory evolution. Curr Opin Insect Sci. 19:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WR. 1990. A consensus combined P-value test and the family-wide significance of component tests. Biometrics 46(2):303. [Google Scholar]
- Rice WR, Linder JE, Friberg U, Lew TA, Morrow EH, Stewart AD.. 2005. Inter-locus antagonistic coevolution as an engine of speciation: assessment with hemiclonal analysis. Proc Natl Acad Sci U S A. 102(1 Suppl):6527–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl CAL, Riedl M, Mackay TFC, Sokolowski MB.. 2007. Genetic and behavioral analysis of natural variation in Drosophila melanogaster pupation position. Fly 1(1):23–32. [DOI] [PubMed] [Google Scholar]
- Rockman MV. 2012. The QTN program and the alleles that matter for evolution: all that’s gold does not glitter. Evolution 66(1):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder E, Blows F, Ashburner M, Bautista-Llacer R, Coulson D, Drummond J, Webster J, Gubb D, Gunton N, Johnson G, et al. 2004. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics 167(2):797–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sameoto DD, Miller RS.. 1968. Selection of pupation site by Drosophila melanogaster and D. simulans. Ecology 49(1):177–180. [Google Scholar]
- Sawamura K, Roote J, Wu CI, Yamamoto MT.. 2004. Genetic complexity underlying hybrid male sterility in Drosophila. Genetics 166(2):789–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamura K, Taira T, Watanabe TK.. 1993. Hybrid lethal systems in the Drosophila melanogaster species complex. I. The maternal hybrid rescue (mhr) gene of Drosophila simulans. Genetics 133(2):299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster CM, Davis GW, Fetter RD, Goodman CS.. 1996. Genetic dissection of structural and functional components of synaptic plasticity. I. Fasciclin II controls synaptic stabilization and growth. Neuron 17(4):641–654. [DOI] [PubMed] [Google Scholar]
- Seyahooei MA, Kraaijeveld-Smit FJL, Kraaijeveld K, Crooijmans JBM, Van Dooren TJM, Van Alphen JJM.. 2009. Closely related parasitoids induce different pupation and foraging responses in Drosophila larvae. Oikos 118(8):1148–1157. [Google Scholar]
- Shivanna N, Murthy GSS, Ramesh SR.. 1996. Larval pupation site preference and its relationship to the glue proteins in a few species of Drosophila. Genome 1(1):105–111. [DOI] [PubMed] [Google Scholar]
- Sokal RR, Ehrlich PR, Hunter PE, Schlager G.. 1960. Some factors affecting pupation site of Drosophila. Ann Entomol Soc Am. 53(2):174–182. [Google Scholar]
- Sokolowski MB, Kent C, Wong J.. 1984. Drosophila larval foraging behaviour: developmental stages. Anim Behav. 32(3):645–651. [Google Scholar]
- Stamps J, Buechner M, Alexander K, Davis J, Zuniga N.. 2005. Genotypic differences in space use and movement patterns in Drosophila melanogaster. Anim Behav. 70(3):609–618. [Google Scholar]
- Stanley CE, Kulathinal RJ.. 2016. flyDIVaS: a comparative genomics resource for Drosophila divergence and selection. G3 6(8):2355–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL. 2014. Identification of loci that cause phenotypic variation in diverse species with the reciprocal hemizygosity test. Trends Genet. 30(12):547–554. [DOI] [PubMed] [Google Scholar]
- Sturtevant AH. 1920. Genetic studies on Drosophila simulans. I. Introduction. Hybrids with Drosophila melanogaster. Genetics 5(5):488–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucena E, Delon I, Jones I, Payre F, Stern DL.. 2003. Regulatory evolution of shavenbaby/ovo underlies multiple cases of morphological parallelism. Nature 424(6951):935–938. [DOI] [PubMed] [Google Scholar]
- Takano TS. 1998. Loss of notum macrochaetae as an interspecific hybrid anomaly between Drosophila melanogaster and D. simulans. Genetics 149(3):1435–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandal NB, Siddalingamurthy GS, Shivanna N.. 2008. Larval pupation site preference on fruit in different species of Drosophila. Entomol Res. 38(3):188–194. [Google Scholar]
- Watanabe TK. 1979. A gene that rescues the lethal hybrids between Drosophila melanogaster and D. simulans. Jpn J Genet. 54(5):325–331. [Google Scholar]
- Zeng X, Han L, Singh SR, Liu H, Neumüller RA, Yan D, Hu Y, Liu Y, Liu W, Lin X, et al. 2015. Genome-wide RNAi screen identifies networks involved in intestinal stem cell regulation in Drosophila. Cell Rep. 10(7):1226–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwarts L, Magwire MM, Carbone MA, Versteven M, Herteleer L, Anholt RRH, Callaerts P, Mackay TFC.. 2011. Complex genetic architecture of Drosophila aggressive behavior. Proc Natl Acad Sci U S A. 108(41):17070–17075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.