Abstract
Dysfunction of glutamate neurotransmission in the nucleus accumbens (NAc) has been implicated in the pathophysiology of alcohol use disorders (AUD). Neurogranin (Ng) is exclusively expressed in the brain and mediates N-methyl-D-aspartate receptor (NMDAR) hypo-function by regulating the intracellular calcium-calmodulin (Ca2+-CaM) pathway. Interestingly, Ng null mice (Ng–/– mice) demonstrate increased alcohol drinking compared to wild-type mice, while also showing less tolerance to the effect of alcohol. To identify the molecular mechanism related to alcohol seeking, we utilized both in vivo microdialysis and label-free quantification proteomics comparing Ng genotype and effects of alcohol treatment on the NAc. There is significant difference in glutamate and GABA neurotransmission between genotypes, however, alcohol administration normalizes both glutamate and GABA levels in the NAc. Using label-free proteomics, we identified 427 protein expression changes against alcohol treatment in the NAc among 4,347 total proteins detected. Bioinformatics analyses revealed significant molecular differences in Ng null mice in response to acute alcohol treatment. Ingenuity pathway analysis found that the AKT network was altered significantly between genotypes, which might increase the sensitivity of alcohol in Ng null mice. Finally, our pharmacoproteomics results illustrate a possible molecular basis for the NAc-mediated alcohol seeking, which provides a better understanding of the Ng-mediated signaling in AUD.
Keywords: Neurogranin, Pharmacoproteomics, Label-free Proteomics, Alcoholism, Bioinformatics
Introduction
Alcohol use disorders (AUD) contribute greatly to the global economic burden of mental health disorders and cause 88,000 deaths in the United States every year [1]. Low sensitivity to alcohol is associated with a high probability of developing AUD [2]. However, the neurobiological mechanism explaining causality between alcohol sensitivity and seeking behavior has not been well-studied. A growing body of evidence suggests that pharmacological and genetic regulation of N-methyl-D-aspartate receptors (NMDAR) alters both neural and behavioral effects of ethanol, as glutamate dysregulation is associated with AUD [3]. In fact, acute alcohol has its greatest effect on NMDAR by reducing the effectiveness of glutamate binding. In contrast, chronic alcohol use leads to an up-regulation of NMDAR in response to reduced glutamate activity on NMDAR [4]. The impairment of NMDAR-mediated synaptic plasticity is a possible mechanism explaining the individual differences seen in regards to the sensitivity of the pharmacological effect of alcohol. Thus, studying the intracellular NMDAR mechanism will provide a better understanding of individual variations of alcohol sensitivity and susceptibility to developing AUD.
Neurogranin (Ng) modulates NMDAR-mediated Ca2+-calmodulin (CaM) signaling that is highly expressed in the post-synaptic regions of striatum, cortex, and hippocampus [5]. Human clinical studies identified that genetic variant of Ng is correlated with a risk of cognitive failure through NMDAR hypo-function [6]. We recently identified that Ng knockout mice exposed to ethanol show increased alcohol sensitivity [7]. Interestingly, however, Ng knockout mice self-administer more alcohol compared to wild-type mice, as measured by two-bottle choice experiments and operant conditioning [7]. Among many brain regions, the nucleus accumbens (NAc) contributes to the physiological response to alcohol and addiction development [8].
To investigate the mechanism of alcohol sensitivity pertaining to the NAc, we measured glutamate, GABA, and dopamine level changes in the NAc of Ng null (Ng−/−) mice with/without alcohol exposure using in vivo microdialysis. Then, we conducted label-free proteomics to elucidate a possible mechanism for how Ng dysregulation induces hyper-sensitivity of alcohol [9]. These techniques enabled us to identify altered neurotransmitter pathways in an animal model that represents a dysregulation of Ng. To demonstrate the potential mechanism involved between alcohol treatment and Ng knockout, we employed a pharmacoproteomics and bioinformatics approach including heat-map analysis, principal component analysis (PCA), and ingenuity pathway analysis (IPA) to compare molecular responses between genotypes.
MATERIALS AND METHODS
Animals
We used 12-week-old male Ng+/+ mice and Ng−/− mice. Mice were housed (3–4 mice per group) in standard Plexiglas cages under a 12 h light (>500 lux)/dark (<0.5 lux) cycle with lights on at 6:00 AM. Male mice used for all experiments were housed at a constant temperature (24 ± 0.5°C) and humidity (60 ± 2%). Food and water were provided ad libitum. The animal care and handling procedures were both approved by the LSUHSC-Shreveport Institutional Animal Care and Use Committees (P-17–038) in accordance with NIH guidelines.
Microdialysis and alcohol administration
To measure extracellular glutamate levels in the brain, a microdialysis probe (Eicom, Kyoto, Japan; MW cut off: 50,000 Da) was implanted into the nucleus accumbens (AP: 1.34 mm, ML: ±1.3 mm, DV:3.5 mm). Mice were given 3 days for recovery. During this period, mice were placed in the test chamber for 1h daily to habituate to the handling procedure and experimental environment as described in [24]. A microdialysis probe was connected to a micro-syringe pump (Eicom, Kyoto, Japan) to continuously deliver Ringer’s solution at a 1.0 μl/min flow rate. To measure glutamate level changes in response to an acute alcohol (2 g/kg, 1h, i.p.) dialysates were collected every 10 min, after a 2h stabilization period. The starting time point of each 10 min interval was used for comparisons. To measure changes in levels of GABA, glutamate, and dopamine in response to Ng deletion, saline or alcohol were injected (2 g/kg, 1h, i.p.) after a 2h stabilization period. Microdialysis probe locations will be confirmed histologically. Dialysates were frozen and stored at −80 °C until analyzed. The glutamate, GABA, and dopamine concentrations were quantified in each dialysate sample by LC-MS/MS approach. Multi-reaction monitoring (MRM) for glutamate, GABA, and dopamine ions was utilized using UPLC-XevoTS LC-MS/MS system (Waters) [10].
Brain Tissue Sample Preparation
13 total mice (Ng+/+ mice with saline: 3 mice, Ng−/− mice with saline: 3 mice, Ng+/+ mice with alcohol: 4 mice, and Ng−/− mice with alcohol: 3 mice) were subjected to rapid CO2 inhalation to induce unconsciousness, followed by decapitation and subsequent harvesting of brain for isolation of the NAc from both hemispheres under a surgical microscope. The extracted tissue was snap-frozen on dry ice and stored at −80°C until it was processed for SDS-PAGE (Bio-Rad Criterion system). Each NAc tissue sample was homogenized in an extraction buffer containing 50 mM Tris buffer (pH 7.4), 2 mM EDTA, 5 mM EGTA, 0.1% SDS protease inhibitor cocktail type I (Roche) and II (Sigma). Homogenates were then centrifuged at 500 g at 4°C and supernatants were collected. Protein concentration from each replicate supernatant was quantified using the Bradford protein assay (Bio-Rad). Replicates were loaded at 30 μg and separated via electrophoresis in a 4–12% poly-acrylamide gel, followed by sample preparation for proteomic analysis.
Label-free Proteomics Using 1D-SDS PAGE
Each 1D gel lane was divided into 7 sections using a Precision Plus Kaleidoscope standard (Bio-Rad) to indicate prominent proteins bands common to each lane as section boundaries. Gel sections were then excised and the resulting fractions were incubated in 200 mM Tris for 30 min followed by destaining with 50 mM Tris/50%acetronitrile for 1–2 h. Fractions were then dehydrated with 100% acetonitrile until gel pieces appeared opaque. Destaining was repeated a second time, followed by reduction with 20 mM DTT (Sigma) in 50 mM Tris for 1h at 60°C. Gel fraction dehydration and alkylation was done with 100% acetonitrile and 40 mM iodoacetamide (Sigma) in 50 mM Tris for 1h at room temperature. Samples were rehydrated in 25 mM Tris and subjected to a final dehydration step prior to trypsin digest. Samples were mixed with 0.2 to 0.3 ug of trypsin (Promega) in 20 mM Tris/0.0002% zwittergent 3–16 overnight at 37°C. Trypsin was inactivated and peptides were extracted by adding 2% trifluoroacetic acid to the digest for 30 min followed by the addition of acetonitrile for an additional 30 min. The supernatant was removed and saved, followed by addition of acetonitrile a third time to the gel fractions for 30 min. The gel fractions were then added to the saved supernatant. These peptide containing fractions were dried under vacuum and stored at −20°C.
NanoLC Tandem Mass Spectrometry (MS/MS)
Dried peptide extracts were reconstituted in an acidic aqueous solution of 0.2% formic, 0.1% TFA, and 0.001% zwittergent 3–16 detergent for analysis by nano-scale liquid chromatography interfaced to tandem mass spectrometry (nLC-MS/MS). Peptide mixture was loaded onto an Optipak 0.25 ul cartridge (Optimize Technologies, Oregon City, OR) custom packed with Michrom Magic C8 (Michrom BioResource, Auburn, CA) using an Eksigent nanoLC-Ultra 2D with AS3 autosampler. Peptides were separated by reverse-phase LC using a 75μm i.d. fused silica column, packed in-house with 37 cm of 3 um Magic C18 (Michrom BioResource, Auburn, Ca) with a gradient of 2–40% B over 60 minutes with a mobile phase flow rate of 300 nL/min. Mobile phase A contained water, acetonitrile, and formic acid (98/2/0.2 by volume), whereas mobile phase B was composed of acetonitrile, isopropanol, water, and formic acid (80/10/10/0.2 by volume). Eluting peptides were analyzed using a Q-Exactive mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) configured to measure peptides within the molecular weight range of 360–2000 m/z at a resolving power of 70K (FMHM, m/z 200) survey scans (MS1), followed by isolating top 15 most abundant ions for higher energy C-trap dissociation scans (MS2) using an isolation window of 2 Da at a resolving power of 17.5K and a 45 second exclusion duration [11]. Raw data files were deposited into MassIVE database (MSV000084549).
Data Processing and Label-free Data Analysis
Data files were imported into the MaxQuant software (Max Planck Institute of Biochemistry, Germany) and Rosetta Elucidator software (Seattle, WA) in which a relative quantitative label free pipeline was used for analysis [12]. Features were detected such that m/z, retention time, and peak intensity alignment of MS1 data were extracted across samples. Database searching on features selected with charge state < 4 and > 1, peak confidence score >0.6, peak time score > 0.8, and peak m/z score >0.9, was initiated within Elucidator using Mascot (v2.2, Matrix Science) with mouse Swiss-Prot database appended with common contaminants and a reverse decoy database. Mascot search parameters were set with peptide precursor tolerance of 10 ppm, ms/ms tolerance of 0.6 Da, 2 missed cleavages, and variable modifications carbamidomethyl C, oxidation M, and propionamide C. For each protein group, the normalized intensities observed in two groups of samples were modeled using a Gaussian-linked generalized linear model. Protein expression between pairs of experimental groups was assigned across aligned sample features so that relative quantitation of corresponding protein identifications across sample groups could be compared [13]
Heat Map Analysis and Principal Component Analysis
Heat maps were drawn to determine the expression patterns of significantly up- or down-regulated protein at each time point and to compare the expression levels with the other time points, using R version 2.15.1 and the R packages. Principal component analysis (PCA) can reduce the dimensionality of a data set consisting of genotype and treatment variables. PCA was conducted as an “unsupervised” analysis to clarify the variance among proteomics data from brain samples using R. To clarify the variances among samples, data were calculated using a Q-mode PCA package ‘prcomp’of R. The proportion of variance and factor loading were also calculated.
Ingenuity Pathway Analysis
To classify the protein functionally, we used IPA where we entered the genes whose fold changes were more than 1.5-fold and p-values were less than 0.05. IPA shows possible networks involved in microarray profiles by the IPA Network Generation Algorithm. Proteins were clustered and classified by the IPA Network Generation algorithm and the networks were ranked by the network score. In the networks, solid and dashed lines indicated direct and indirect interactions, respectively. Direct interactions require the two molecules make direct physical contact with each other; there is no intermediate step. Indirect interactions do not require that there is physical contact between the two molecules, such as a signaling cascade instead of the two molecules making physical contact with each other [14].
Western Blot Analysis
NAc tissues were homogenized in a solution containing 50 mM Tris buffer (pH 7.4), 2 mM EDTA, 5 mM EGTA, 0.1% SDS, protease inhibitor cocktail (Roche, Indianapolis, IN), and phosphatase inhibitor cocktail type I and II (Sigma). Homogenates were centrifuged at 500 g for 15 min and supernatants were collected. Proteins were analyzed using Bradford protein assay (BioRad, Hercules, CA). Proteins were separated by PROTEAN TGX gels at 100 V for 1 h, transferred onto PVDF membranes at 30 V for 1 h (BioRad, Hercules, CA), and incubated with antibodies against pCaMKII (T286) (Cell Signaling, Danvers; 9102; 1:500), CaMKII (Cell Signaling, Danvers; 9102; 1:500), pNR2B (S1303) (Cell Signaling, Danvers; 9102; 1:500), pGluR1(S831) (Cell Signaling, Danvers; 9102; 1:500), pPKCγ(T514) (Cell Signaling, Danvers; 9102; 1:500), PKCγ; (Cell Signaling, Danvers; 9102; 1:500), pNR1(S896) (Cell Signaling, Danvers; 9102; 1:500), Ng (Cell Signaling, Danvers; 9102; 1:500), pAKT(S473) (Cell Signaling, Danvers; 9102; 1:500), AKT (Cell Signaling, Danvers; 9102; 1:500), mGluR5 (Cell Signaling, Danvers; 9102; 1:500) and GAPDH (Millipore, Burlington, MA; MAB374; 1:2000). Chemiluminescent bands were detected on an Image Station and quantified using NIH Image J software.
Statistical Analysis
The data is shown as mean + standard error of the mean (SEM). To detect statistical differences between alcohol treatment and genotype, either two-tailed Student’s t-test (Prism, GraphPad Software, La Jolla, CA) or two-way ANOVA followed by Tukey post hoc test (SigmaStat, SYSTAT software, Point Richmond, CA) were used. Criterion for statistical significance was p < 0.05.
RESULTS
Alcohol decreases Glutamate and GABA levels in the NAc of Ng –/– mice
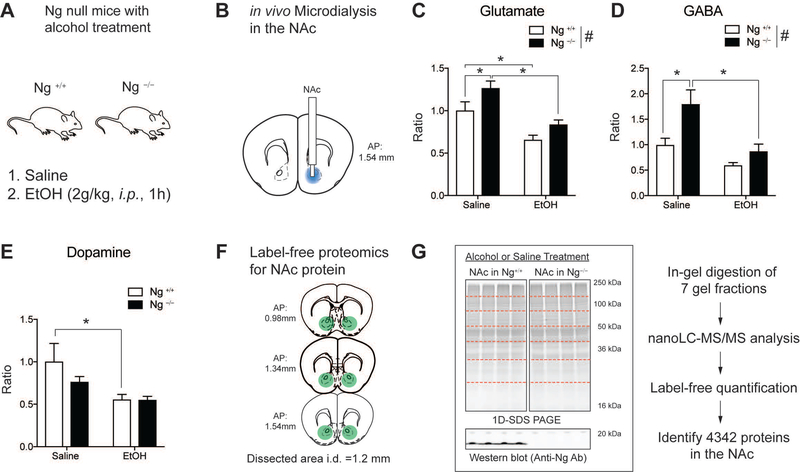
Ng–/– mice show increased sensitivity to the effect of ethanol, but they self-administrate more alcohol than wild-type mice. This is because lack of Ng promotes goal-oriented drinking, which is regulated by the NAc. Therefore, we examined how alcohol treatment alters neurochemical changes in the NAc of Ng–/– mice using a pharmacometabolomics [15] and pharmacoproteomics approach [9] (Figure 1A). In vivo micro dialysis probes were implanted in the NAc of Ng+/+ mice and Ng–/– mice and brain dialysate was collected after acute alcohol administration (2 g/kg, 1h, i.p.) compared to that of saline injection [3] (Figure 1B). Using LC-MS/MS, we measured neurotransmitter changes for glutamate, GABA, and dopamine [10]. Two-way ANOVA indicated that Ng–/– mice demonstrate increased glutamate levels (F(1,71)=8.06, p<0.01) compared to Ng+/+ mice and alcohol treatments decreased glutamate levels in both genotypes (F(1,71)=24.36, p<0.001) (Figure 1C). GABA levels also significantly increased in the NAc of Ng–/– mice (F(1,71)=9.34, p<0.005), and alcohol treatment decreased GABA levels only in the Ng–/– mice (F(1,71)=14.19, p<0.001) (Figure 1D). Dopamine levels in the NAc did not show significant changes in the NAc of Ng–/– mice, however alcohol treatment significantly decreased dopamine levels in the Ng+/+ mice (F(1,71)=7.07, p<0.01) (Figure 1E). Overall, our findings suggest that alcohol significantly decreased glutamate and GABA levels in the NAc of Ng–/– mice.
Figure 1.
Pharmaco-omics for alcohol treatment in the NAc of Ng−/− mice. A. Schematic representation of the pharmaco-omic experimental paradigm. Mice were given either saline or 2.0 g/kg alcohol treatment (1h, i.p.). B. Pharmacometabolomics using in vivo microdialysis quantifying neurotransmitters in the NAc. Blue regions indicate location of microdialysis probe. C. Brain glutamate level changes in the NAc. Tukey post-hoc test for individual comparisons identified that basal glutamate levels (saline treatment) were increased in Ng−/− mice (p=0.02) and alcohol treatment (2 g/kg, 1h, i.p.) significantly decrease glutamate levels in the NAc of Ng+/+ mice (p=0.03) and Ng−/− mice (p<0.001). D. Brain GABA level changes in the NAc. Tukey post-hoc test for individual comparisons identified the basal GABA levels (saline treatment) were increased in Ng−/− mice (p=0.002) and alcohol treatment (2 g/kg, 1h, i.p.) decreased GABA levels in the NAc of Ng−/− mice (p<0.001). (# p < 0.05 by two-way ANOVA for genotype). E. Brain dopamine level changes in the NAc. Tukey post-hoc test for individual comparisons identified alcohol treatment (2 g/kg, 1h, i.p.) decreased GABA levels in the NAc of Ng+/+ mice (p=0.01). (n=4 per each group) (*p < 0.05 by two-way ANOVA Tukey post hoc tests). F. Pharmacoproteomics using the NAc tissues. Mice were given either saline or 2 g/kg alcohol treatment (1h, i.p.). Then mice NAc were isolated from the brain (coordinates for NAc tissue collection). G. 1D-SDS PAGE for protein separation per each group of mice (Ng+/+ mice with saline: 3 mice, Ng−/− mice with saline: 3 mice, Ng+/+ mice with alcohol: 4 mice, and Ng−/− mice with alcohol: 3 mice). Each gel lane was fractionated with 7 samples and was digested with trypsin. Using nanoLC-MS/MS based label-free proteomics, 4347 protein expression was identified from the all sample.
Large-scale proteomics analysis of the NAc in response to alcohol treatment in Ng–/– mice
Since alcohol exposure in Ng–/– mice decreases both glutamate and GABA levels in the NAc, we measured molecular signaling changes in response to Ng knockout and alcohol treatment using label-free quantification proteomics (Figure 1F). To identify the effect of alcohol in Ng–/– mice, either intoxication dose of acute ethanol (2 g/kg, 1 h, i.p.) or saline were administrated in 13 mice (Ng+/+ mice with saline: 3 mice, Ng−/− mice with saline: 3 mice, Ng+/+ mice with alcohol: 4 mice, and Ng−/− mice with alcohol: 3 mice). Then, NAc tissue (AP: 0.98 ~ 1.54 mm) was isolated from each mouse and separated by 1D-SDS PAGE. Each mouse sample was fractionated with 7 gel slices based on molecular weight and in-gel digested with trypsin. 91 total samples (13 mice x 7 gel fraction) were processed for label-free proteomics using LC-MS/MS. We identified 4347 total proteins in NAc tissues (Figure 1G).
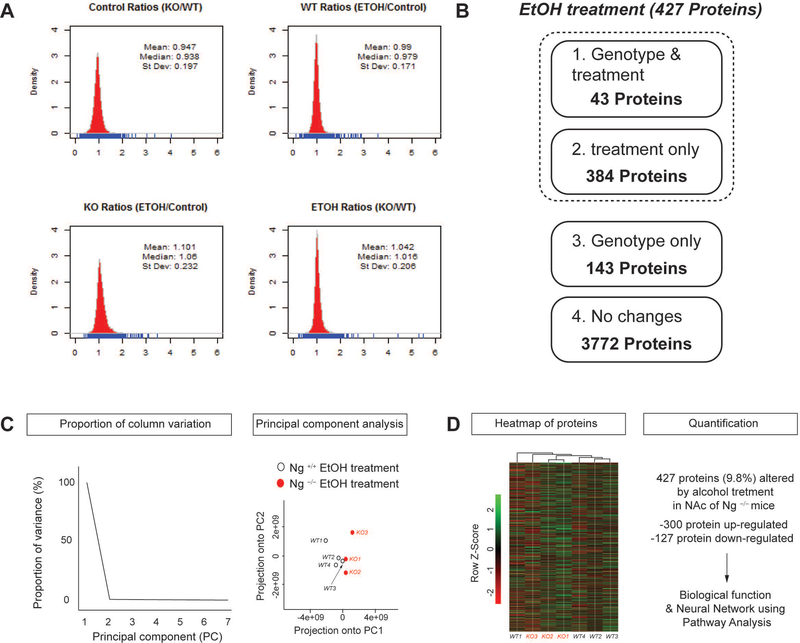
To validate MS data for multiple comparisons, MS peak intensity and its number from each sample were analyzed using density plot, which represents the distribution of one or a few variables (Figure 2A). All 4 different conditions indicated less than 10% of variability in peptides number and peak intensity. Specifically, the shape of MS data distribution between ethanol treated wild-type sample and ethanol treated knockout samples showed no difference indicated by a 1.042 mean with 0.206 standard deviation. Therefore, we focused on how alcohol treatment differs in protein expression among Ng–/– mice compared to that of Ng+/+ mice.
Figure 2.
Bioinformatics analysis for label-free proteomics data. A. Density plot comparison of protein quantification results in response to genotype and alcohol treatment. Mass spectrum intensity from each condition was demonstrated using density plots. B. Pharmacoproteomics analysis in the NAc of Ng−/− mice in response to acute alcohol treatment. 427 proteins were changed in response to alcohol treatment. Among 427 proteins, 43 proteins were changes in both genotypes and alcohol treatment. 143 proteins were changed by genotype only. 3772 proteins were not changed by either genotype or alcohol treatment. C. Principal components analysis (PCA) of protein expression in response to alcohol treatment and deletion of Ng. Ratio proportion of variance line graph represents the proportion of variance of the first principal components (PC). Ratio scatterplot of genotype and alcohol treatment shows the projection of the first principal component compared to the second principal component. D. Heat map for altered protein expression in response to alcohol treatment and deletion of Ng. Values were normalized by Z-Score. 300 proteins were significantly increased in the NAc of Ng−/− mice by alcohol treatment while 127 proteins were significantly decreased.
As shown in Figure 2B, we categorized protein expression change as having a statistical significance value of p < 0.05 to identify significantly altered proteins. 43 proteins were altered by both lack of Ng and alcohol treatment; while 143 proteins were changed by Ng genotype without an effect of alcohol. In addition, 384 proteins responded to alcohol treatment only, which could imply that alcohol treatment changes a total 427 proteins (9.8%) in the NAc of Ng–/– mice. 3772 (86.7%) of proteins showed no significant response to alcohol treatment or genotype. This data suggests that 427 protein expressions may reflect the decreased glutamate and GABA neurotransmission in the NAc and promote hyper-sensitivity to the pharmacological effect of alcohol in Ng–/– mice.
PCA and heat map analysis of protein profiles in response to alcohol treatment and Ng knockout
To further determine whether alcohol treatment and genotype alters NAc protein expression, we conducted principal component analysis (PCA) on the 427 altered proteins in the NAc. Using all data sets, principal components (PC) were automatically identified, and their proportion ratios were calculated as PC1 (95%) and PC2 (5%) (Figure 2C). The Ng–/– mice treated with alcohol (red circle) showed significant differences according to PC1 compared to that of alcohol treatment in wild-type mice (empty circle). To further profile protein expression changes between genotypes and alcohol treatment, a heat map represented by a normalized Z-score for expression of the 427 proteins was generated (Figure 2D). Heat map results indicated that alcohol treatment increases 300 proteins and decreases 127 proteins in the NAc of Ng–/– mice. Then, we selected 52 significant clusters from the heat map indicating more than 1.5-fold protein expression change after alcohol treatment in the NAc of Ng–/– mice (Table1). Among 52 major proteins, there was no protein expression changes related to glutamate signaling. Interestingly, alcohol treatment significantly increases GABA receptor protein 1 and GABA receptor protein 2 expressions in the NAc of Ng–/– mice compared to that of wild-type mice.
Table 1.
52 proteins altered by alcohol treatment in Ng+/+ mice and Ng−/− mice.
| Protein expression change (Ng KO alcohol /Ng WT alcohol) | |||
|---|---|---|---|
| Accession | Description | Fold change | p-value |
| NP_031407.2 | acyl-CoA dehydrogenase | −3.728 | 0.015 |
| NP_795928.2 | ETR-3-like factor 5 | −2.455 | 0.026 |
| NP_062800.1 | isovaleryl-CoA dehydrogenase | −2.215 | 0.019 |
| NP_001157014.1 | plectin | −2.197 | 0.029 |
| NP_080949.2 | apolipoprotein O | −1.959 | 0.038 |
| NP_848841.2 | protein phosphatase 1L | −1.781 | 0.027 |
| NP_080770.2 | phosphopantothenate | −1.698 | 0.013 |
| NP_080138.2 | isobutyryl-CoA dehydrogenase, | −1.685 | 0.000 |
| NP_031573.2 | tyrosine-protein phosphatase | −1.587 | 0.001 |
| NP_955759.2 | nexilin | −1.579 | 0.033 |
| NP_659114.2 | receptor enhancing protein 2 | −1.499 | 0.006 |
| NP_619591.1 | diacylglycerol kinase gamma | 1.506 | 0.009 |
| NP_084137.1 | CB1 receptor protein 1 | 1.507 | 0.015 |
| NP_001103709.1 | phosphotyrosine PP | 1.511 | 0.020 |
| NP_001155890.1 | myelin expression factor 2 | 1.519 | 0.013 |
| NP_848910.3 | inositol hexakisphosphate 1 | 1.521 | 0.002 |
| NP_085112.1 | histone H1.1 | 1.523 | 0.015 |
| NP_001164523.1 | MTA3 | 1.527 | 0.041 |
| NP_080436.1 | microtubule proteins 1A/1B | 1.534 | 0.009 |
| XP_003688806.1 | 60S ribosomal protein L23 | 1.538 | 0.033 |
| NP_001182350.1 | histone H4 | 1.552 | 0.008 |
| NP_001074585.1 | rho factor | 1.575 | 0.003 |
| NP_114030.2 | SWI/SNF | 1.592 | 0.009 |
| NP_080423.1 | 40S ribosomal protein S20 | 1.594 | 0.007 |
| NP_001074238.1 | ICBP90 | 1.597 | 0.040 |
| NP_001156869.1 | ELAV-like protein 4 | 1.607 | 0.015 |
| XP_922988.1 | 60S ribosomal protein L31 | 1.608 | 0.029 |
| NP_001005859.1 | 60S ribosomal protein L34 | 1.610 | 0.011 |
| XP_003945754.1 | solute carrier family 25 | 1.613 | 0.000 |
| NP_062723.1 | GABA receptor protein | 1.623 | 0.046 |
| NP_001239501.1 | src substrate cortactin | 1.631 | 0.039 |
| NP_061200.2 | 60S ribosomal protein L36 | 1.635 | 0.026 |
| NP_033882.1 | beta-galactosidase precursor | 1.642 | 0.001 |
| NP_001001980.2 | LIM | 1.671 | 0.011 |
| NP_065251.2 | VAV3 | 1.677 | 0.047 |
| NP_033117.1 | 40S ribosomal protein S15 | 1.700 | 0.046 |
| NP_598516.2 | gamma-tubulin 2 | 1.718 | 0.004 |
| NP_079865.1 | ribosomal protein L36A-like | 1.764 | 0.040 |
| NP_766209.1 | chloride channel protein 5 | 1.861 | 0.020 |
| XP_003086606.1 | 60S ribosomal protein L22-like | 1.921 | 0.049 |
| NP_001074591.1 | centrosomal protein of 63 kDa | 1.933 | 0.047 |
| NP_733769.1 | 40S ribosomal protein S15a | 1.945 | 0.017 |
| XP_001476621.1 | NADH dehydrogenase 1 beta | 2.019 | 0.032 |
| NP_619606.1 | sarcosine dehydrogenase | 2.056 | 0.015 |
| NP_076124.1 | ras-related protein Rab-27A | 2.089 | 0.037 |
| NP_001028400.2 | integrin alpha-1 precursor | 2.090 | 0.028 |
| NP_647461.3 | myosin light chain kinase | 2.135 | 0.013 |
| NP_080969.2 | GABA receptor-protein 2 | 2.137 | 0.027 |
| NP_001074651.2 | phospholipid-transporting ATPase | 2.138 | 0.036 |
| NP_081622.1 | cytochrome c oxidase 1 | 2.317 | 0.006 |
| NP_034094.1 | alpha-crystallin B chain | 4.416 | 0.001 |
| NP_659190.3 | MAP7 protein 1 | 5.488 | 0.030 |
IPA analysis revealed that the both lack of Ng and alcohol alters neurological disease pathway in the NAc
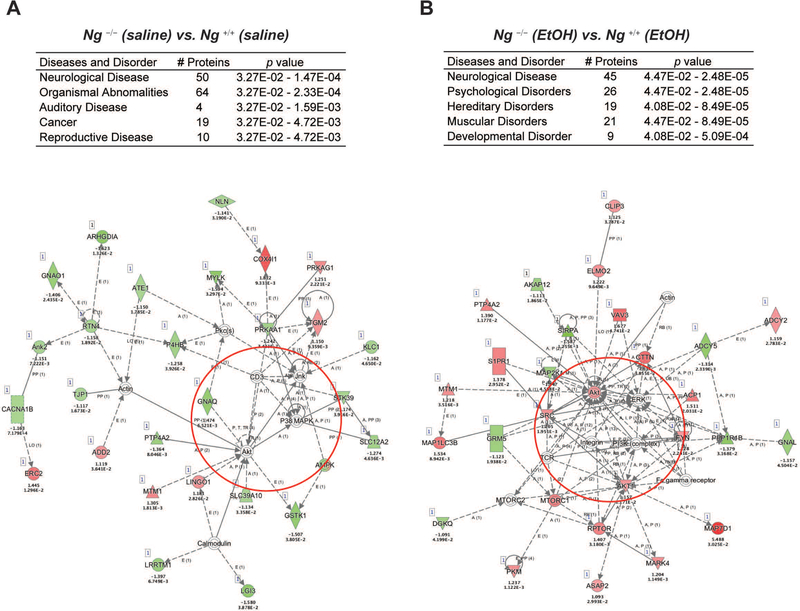
To gain more insight into the effects of alcohol treatment in Ng–/– mice, ingenuity pathway analysis (IPA) was carried out to map the biological processes regulated by genotype and alcohol interaction. We first conducted IPA to elucidate genotype differences with and without alcohol treatment (Figure 3). Using 186 proteins from the difference in basal protein expression levels between Ng+/+ mice and Ng–/– mice, IPA identified that signaling networks related to neurological disease and organismal abnormality were significantly involved in the NAc of Ng–/– mice (score 26; 50 proteins) (Figure 3A). Then, IPA analysis for the 427 proteins that changed by alcohol treatment in Ng–/– mice identified that signaling networks related to neurological disease and psychological disorders were significantly involved in the NAc (score 26; 50 proteins) (Figure 3B). This approach revealed that a dampened AKT-related pathway due to a lack of Ng is activated by alcohol treatment in the NAc of Ng–/– mice. Our results suggest that alcohol exposure enhances the neuroinflammation systems regulated by AKT signaling.
Figure 3.
Summary of IPA analysis and representative network. A. Significantly changed pathway by saline treatment in the NAc of Ng–/– mice. B. Significantly changed pathway by alcohol treatment in the NAc of Ng–/– mice. Red circle represents AKT cluster. Red color indicates up-regulated protein, while green color indicates down-regulated proteins with statistical significance. Grey indicates protein was detected but did not show statistical significance. Corresponding fold change of protein expression and p-value is presented below the gene name.
Identification of AKT clusters related to alcohol sensitivity in Ng–/– mice
Finally, the proteins altered by genotype and alcohol treatment were analyzed by IPA pathway analysis. In genotype comparison between Ng+/+ mice and Ng–/– mice (Figure 3A), a calmodulin (CaM) cluster, directly downstream of Ng, showed significantly decreased expression. Altered AMPKs (PRKAG1 and PRKAA1) support the change of Ca2+-CaM dependent kinase (CaMKs) that then promote mitochondrial respiratory function mediated by cytochrome c oxidase subunit 4I1 (COX4I1) in Ng–/– mice. Moreover, IPA suggests decreased mitogen-activated protein kinase 8 (JNK), mitogen-activated protein kinase 14 (P38MAPK), and AKT serine/threonine kinase 1 (AKT) cluster in Ng–/– mice.
Interestingly, acute alcohol-treatment significantly increases PI3K-AKT (Figure 3B), the mechanistic target of rapamycin complex 1 (MTORC1) and regulatory-associated proteins of mTOR (RPTOR) clusters in Ng –/– mice [16, 17]. This finding supports that alcohol triggers mTORC1-dependent synaptic plasticity in NAc neurons and increases oxidative stress and inflammation in Ng –/– mice. Increased tyrosine kinase (FYN) and decreased tyrosine phosphatase (PPP1R1B) [18, 19] supports that alcohol stimulates GABAergic signaling in the NAc of Ng –/– mice, which may explain the increase of alcohol sensitivity in Ng –/– mice. In addition, alcohol treatment significantly decreased mGluR5 receptor (GRM5) expression, which is important for the alcohol sedation effect [20–22].
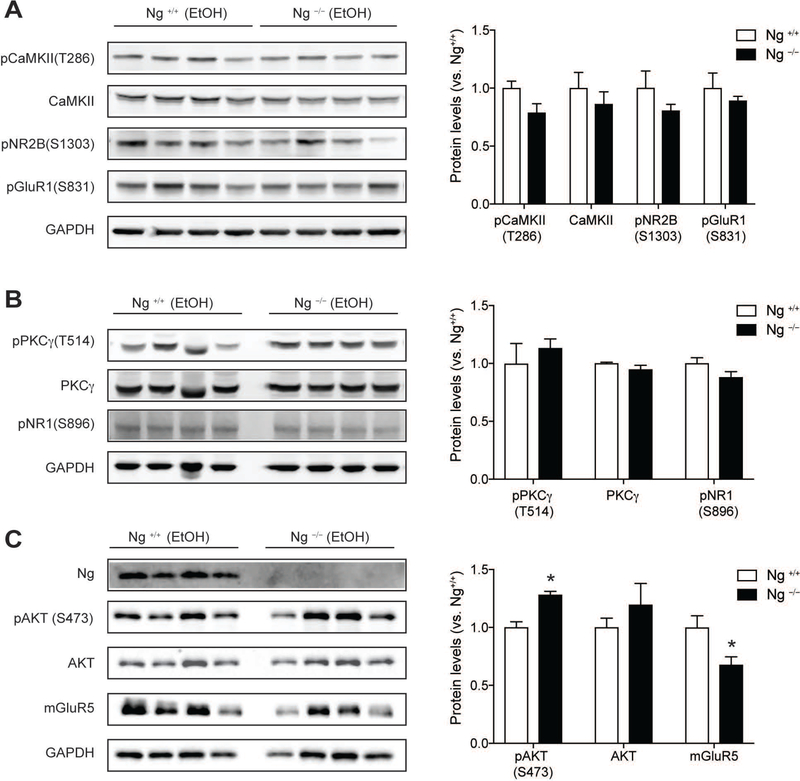
Therefore, we validated these alcohol induced-protein expression changes using western blotting. First, we examined whether the Ca2+-CaM dependent pathway downstream of Ng is changed by alcohol treatment. Both CaMKII expression and CaMKII phosphorylation showed no difference in the NAc of Ng–/– mice compared to that of Ng+/+ mice. Since CaMKII activation is known to regulate glutamate receptor mediated alcohol sensitivity [23], we measured CaMKII-dependent glutamate receptors’ phosphorylation at NMDA 2B glutamate receptor and AMPA glutamate receptor. Both pNR2B (S1303) and pGluR1(S831) not show any difference in its phosphorylation (Figure 4A). Then, we examined whether the PKC gamma (PKCγ) dependent pathway, upstream of Ng, is changed by alcohol treatment. Both PKCγ expression and phosphorylation was not changed as well as its phosphorylation target on pNR2B (S1303) glutamate receptor (Figure 4B).
Figure 4.
Validation of alcohol-induced protein expression change in the NAc of Ng−/− mice using western blotting. A. Alcohol did not induce any significant changes in Ca2+-CaMKII signaling in the NAc of Ng–/– mice. There is no difference in CaMKII-mediated phosphorylation in pNR2B(S1303) and pGluR1(S831). B. Alcohol did not induce any changes in PKCγ signaling and pNR1(S896) phosphorylation. C. After alcohol treatment, AKT phosphorylation in the Ng−/− mice was significantly increased, while mGluR5 expression was significantly decreased in Ng–/– mice (n = 4). *p < 0.05 unpaired t-test for protein expression between genotypes and protein expression was normalized by GAPDH expression. All data are presented as mean ± SEM.
As suggested by IPA analysis, alcohol treatment significantly increases AKT function by increased AKT phosphorylation (t=4.84, df=6, p = 0.003) in the NAc Ng–/– mice (Figure 4C). There is no change in AKT expression in the NAc. Next, we also examined whether ethanol exposure also changes mGluR5 expression, which is known to change alcohol sensitivity and alcohol-seeking behaviors. Statistical analysis indicated that acute alcohol injection significantly decreased mGluR5 expression (t=2.64, df=6, p = 0.038) in the NAc in Ng–/– mice relative to Ng+/+ mice (Figure 4C).
DISCUSSION
Ng expression attenuates NMDAR glutamate responses so that Ng acts as a neuromodulator for glutamate-mediated calcium signaling in the NAc. In this study, we first report how alcohol induces Ng-mediated synaptic plasticity and activation of the AKT pathway, which is possibly related to hypersensitivity of alcohol in Ng–/– mice. Using in vivo microdialysis, we demonstrated that lack of Ng increases both glutamate and GABA levels in the NAc, and alcohol treatment decreases both glutamate and GABA levels in the Ng–/– mice. Label-free proteomics identified that alcohol treatment in Ng–/– mice significantly changes 427 proteins including increased GABA receptor protein expressions. Notably, in Ng–/– mice, the AKT level was significantly increased after alcohol treatment, whereas there were decreased AKT clusters compared to wild-type littermates.
Both pharmacometabolomics and pharmacoproteomics enables us to test the pharmacological effect of alcohol in neurochemical and protein expression changes. Alcohol treatment attenuates glutamate and GABA levels in the NAc of Ng–/– mice, which seems to decrease mGluR5 receptor expression and increases GABA receptor proteins 1 and 2. Because the NAc produces strong GABAergic outputs, increased GABA level receptor expression is very relevant to the increased alcohol sensitivity and alcohol-seeking behaviors. Thus, a key question is whether there are specific GABA receptor subunits that are sensitive to low concentrations of alcohol. Moreover, IPA analysis suggests that alcohol induces AKT activation in the NAc of Ng–/– mice signifying inflammation and susceptibility of cell death. These findings imply that increased alcohol sensitivity of Ng–/– mice is associated with GABA-AKT regulation in the NAc.
The genetic variant of the Ng gene is highly associated with schizophrenia and Alzheimer’s disease in humans supported by a hyper-glutamatergic condition or hypo-NMDAR theory; however, the pathophysiological mechanism is still unknown. In this study, we tested how the lack of Ng promotes neurotransmitter imbalance with or without alcohol. The protein expression changes in the NAc of Ng–/– mice elucidates the effect of alcohol in a hypo-GABA condition which may alter ethanol self-administration. In our previous studies, decreased Ng expression in the NAc demonstrated support of the notion that hyper-glutamatergic conditions are related to excessive alcohol-seeking in mice [24]. However, further investigation is needed to see whether Ng expression/function is relevant to alcohol use disorder in humans. Moreover, we found that acute alcohol treatment alters 427 proteins while Ng deletion itself alters 143 proteins in the NAc. Interestingly, 43 proteins were changed by both alcohol treatment and genotype. These results suggest that the pharmacological effect of alcohol induced dramatic changes in protein expression in the NAc; and it is possible that a mechanism could be identified in Ng–/– mice that is associated with increased alcohol sensitivity. Since we tried to elucidate the interaction between Ng genotype and alcohol treatment, we tested the total changes in response to alcohol treatment.
Recent advances in mass spectrometry methodologies serve as powerful platforms for hypothesis generation or discovery of underlying mechanism involved in pharmacological actions. Using in vivo microdialysis, we analyzed the brain neurochemical change in response to alcohol and genotype. Then, we demonstrated the following protein expression changes using pharmacoproteomics approaches. The advantages of employing a label-free proteomics approach shows increased proteome coverage, decreased experimental variation by labeling, enhanced ability to detect various types of post-translational modification on proteins, and the ability for single experiment relative comparisons of expression changes in multiple biological samples with/without alcohol treatment. However, alcohol treatment and genotype comparison in brain proteomic analysis can be challenging, due to highly complex and interconnected heterogeneous neural and glial cell populations, in addition to computing and validating the large amounts of data generated [25]. In addition, there is a discrepancy of protein quantification between proteomics and western blot. The label free quantification method measures several peptides per quantified protein, thus generating several independent measurements, whereas western blotting measures only a single signal from the antigen-antibody interaction.
Specifically, we anticipated that decreased extracellular glutamate levels alter NMDAR regulation and downstream PKC or CaMKII pathways. However, we could not find any significant change of these pathways by proteomics and western blot results. Therefore, we employed bioinformatics analysis to validate raw data quality and facilitate our understanding of the interest protein data sets generated by both the metabolomics and proteomics approach. In genotype comparisons, Protein kinase C (PKC) clusters, known to be upstream kinase of Ng phosphorylation also decreased in Ng–/– mice [26]. Genetic deletion of PKCγ produces mice with a high ethanol drinking phenotype which are impulsive and require high levels of ethanol to reach intoxication [27], perhaps modeling the human condition of individuals who are at risk for developing alcoholism [28]. After acute alcohol exposure, IPA identified that Ng−/− mice significantly reduces mGluR5 (GRM5) expression (Figure 3B and Supplement Table1), another type of glutamate receptor, as a part of the mTORC1 pathway. Therefore, we validated decreased mGluR5 expression using western blotting (Figure 4C).
There was no significant difference in dopamine levels in the NAc of Ng –/– mice. Low doses of alcohol are known to increase NAc dopamine levels [29], while high concentrations of acute alcohol decreases dopamine release in the striatum [30]. Our acute alcohol treatment (2g/kg. i.p.) only decreases dopamine levels in the NAc of Ng+/+ mice and no changes in Ng−/− mice. These findings implicate that Ng is not related to the dopaminergic reward pathway and lack of Ng increases tolerance for alcohol-induced dopamine changes in the NAc.
Moreover, alcohol is known to enhance GABAergic inhibitory pathways and suppress glutamatergic excitatory pathways. Intoxicating concentrations (5–50 mM) of alcohol were shown to enhance the function of GABA receptors [31]. In addition, GABAergic transmission appears to differ in a brain region-specific manner and may also be influenced by a variety of endogenous neuromodulatory factors such as phosphorylation and neurosteroids [32]. In Ng–/– mice, acute alcohol significantly increased GABA levels and GABA protein expression in the NAc compared to those of saline injection. We also observed decreased mGluR5 expression in the NAc, which is critical for the development of place preference and locomotor sensitization of alcohol [21]. This result is consistent with the previous findings that lack of mGluR5 promotes the excessive alcohol seeking [33]. We also observed that NAc mGluR5-associated signaling regulates binge alcohol drinking [22], making mGluR5 as a potential target for the treatment of AUD [34].
Although GABA and glutamate mechanisms alter alcohol response, the mechanisms contributing to alcohol sensitivity have not been fully elucidated in Ng–/– mice. First, IPA suggested that Fyn kinase (FYN), a non-receptor protein tyrosine kinase, increased in the NAc of Ng–/– mice by alcohol treatment. Fyn modulation of NMDA or GABA receptor function and genetic variation in FYN is associated with AUD in humans [18]. Therefore, FYN modulates the acute sedative properties of alcohol in Ng–/– mice. Moreover, IPA indicated that lack of Ng significantly potentiates the AKT pathway in the NAc after alcohol treatment. The AKT signaling pathway in the NAc mediates excessive alcohol drinking in mice [17], however, our finding furthers this knowledge by demonstrating how Ng-mediated AKT signaling may play an essential role in both alcohol sensitivity as well as alcohol seeking in Ng–/– mice. One of the possible targets of the Ng-mediated AKT pathway is the mammalian target of rapamycin in complex 1 (mTORC1), which was also activated by alcohol intake and alcohol induce inflammation [16].
Overall, our study provides a plausible research design for examining the metabolomics/proteomics response and pursuing multiple comparisons as a result of genotype and alcohol treatment. Separation using PCA and cluster analysis was carried out to identify protein sub-groups, and heat map analysis was conducted to distinguish significant clusters from the total proteome. To identify the signaling molecules altered by alcohol in Ng−/− mice, we performed pathway analysis using IPA, which possibly represents a mechanism in alcohol-induced inflammation by acute alcohol. Therefore, pharmacoproteomics approaches suggest that there is great potential for candidate protein network analysis, as noted here, in the alcohol treatment of more sensitive sub-populations of AUDs. Future research will focus on the interpretation of pharmacoproteomics findings as related to the pathophysiology of AUD.
Supplementary Material
Statement of significance of the study.
Pharmaco-omics is a new research platform to identify molecular mechanisms related to pharmacological efficacy. In this research, we measured the pharmacological action of alcohol in the brain of Ng null mice, which demonstrates increased alcohol sensitivity. The combination of in vivo microdialysis and LC-MS/MS enables us to measure neurochemical changes in the nucleus accumbens (NAc) against both genotype and alcohol treatment. Pharmacometabolomics results demonstrate that alcohol decreases glutamate and GABA levels in the NAc. Our label-free proteomics revealed that alcohol treatment increases GABA receptor expression and decreases mGluR5 glutamate receptor expression. IPA analysis indicated that alcohol also promotes the AKT and neuroimmune pathway in the NAc of Ng null mice. Therefore, pharmacometabolomic and pharmacoproteomics approaches suggest that there is great potential for candidate protein network analysis, as noted here, on the alcohol sensitivity in the brain of an alcoholism animal model.
ACKNOWLEDGEMENTS
We thank the mass spectrometry core in LSUHSC-Shreveport. We would like to thank Ms. Lailun Nahar for comments and critiques of this manuscript and Mayo Proteomics Core for technical supports. Research reported in this publication was supported by the NARSAD Young Investigator Award (26530) from Brain & Behavior Research Foundation, the seed package from Biomedical Research Foundation, and the National Institute of General Medical Sciences of the National Institutes of Health under Award P20GM121307.
List of abbreviations
- Ng
neurogranin
- NAc
nucleus accumbens
- AUD
alcohol use disorders
- PCA
principal components analysis
- IPA
ingenuity pathway analysis
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
Footnotes
CONFLICT OF INTEREST
All authors declare no conflict of interest.
REFERENCES
- [1].Stahre M, Roeber J, Kanny D, Brewer RD, Zhang X, Preventing chronic disease 2014, 11, E109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Koob GF, Volkow ND, Neuropsychopharmacology 2010, 35, 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chen J, Nam HW, Lee MR, Hinton DJ, Choi S, Kim T, Kawamura T, Janak PH, Choi D-S, Behav Brain Res 2010, 208, 636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ron D, Wang J, in Biology of the NMDA Receptor, (Ed: Van Dongen AM), Boca Raton (FL) 2009. [Google Scholar]
- [5].Pak JH, Huang FL, Li J, Balschun D, Reymann KG, Chiang C, Westphal H, Huang KP, Proc. Natl. Acad. Sci. USA 2000, 97, 11232. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhong L, Cherry T, Bies CE, Florence MA, Gerges NZ, EMBO J 2009, 28, 3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jones KJ, Templet S, Zemoura K, Kuzniewska B, Pena FX, Hwang H, Lei DJ, Haensgen H, Nguyen S, Saenz C, Lewis M, Dziembowska M, Xu W, Proc Natl Acad Sci U S A 2018, 115, E5805. [DOI] [PMC free article] [PubMed] [Google Scholar]; Casaletto KB, Elahi FM, Bettcher BM, Neuhaus J, Bendlin BB, Asthana S, Johnson SC, Yaffe K, Carlsson C, Blennow K, Zetterberg H, Kramer JH, Neurology 2017, 89, 1782. [DOI] [PMC free article] [PubMed] [Google Scholar]; Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, Werge T, Pietilainen OP, Mors O, Mortensen PB, Sigurdsson E, Gustafsson O, Nyegaard M, Tuulio-Henriksson A, Ingason A, Hansen T, Suvisaari J, Lonnqvist J, Paunio T, Borglum AD, Hartmann A, Fink-Jensen A, Nordentoft M, Hougaard D, Norgaard-Pedersen B, Bottcher Y, Olesen J, Breuer R, Moller HJ, Giegling I, Rasmussen HB, Timm S, Mattheisen M, Bitter I, J. Rethelyi M, Magnusdottir BB, Sigmundsson T, Olason P, Masson G, J. Gulcher R, Haraldsson M, Fossdal R, Thorgeirsson TE, Thorsteinsdottir U, Ruggeri M, Tosato S, Franke B, Strengman E, Kiemeney LA, Genetic R, Outcome Pin, Melle I, Djurovic S, Abramova L, Kaleda V, Sanjuan J, de Frutos R, Bramon E, Vassos E, Fraser G, Ettinger U, Picchioni M, Walker N, Toulopoulou T, Need AC, Ge D, Yoon JL, Shianna KV, Freimer NB, Cantor RM, Murray R, Kong A, Golimbet V, Carracedo A, Arango C, Costas J, Jonsson EG, Terenius L, Agartz I, Petursson H, Nothen MM, Rietschel M, Matthews PM, Muglia P, Peltonen L, St Clair D, Goldstein DB, Stefansson K, Collier DA, Nature 2009, 460, 744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Reker AN, Oliveros A, Sullivan JM 3rd, Nahar L, Hinton DJ, Kim T, Bruner RC, Choi DS, Goeders NE, Nam HW, Neuropharmacology 2018, 131, 58. [DOI] [PubMed] [Google Scholar]
- [8].Kalivas PW, Nat. Rev. Neurosci 2009, 10, 561. [DOI] [PubMed] [Google Scholar]; Di Ciano P, Cardinal RN, Cowell RA, Little SJ, Everitt BJ, J Neurosci 2001, 21, 9471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Germany CE, Reker AN, Hinton DJ, Oliveros A, Shen X, Andres-Beck LG, Wininger KM, Trutschl M, Cvek U, Choi DS, Nam HW, Proteomics 2018, 18, e1700417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Buck K, Voehringer P, Ferger B, J Neurosci Methods 2009, 182, 78. [DOI] [PubMed] [Google Scholar]; Defaix C, Solgadi A, Pham TH, Gardier AM, Chaminade P, Tritschler L, J Pharm Biomed Anal 2018, 152, 31. [DOI] [PubMed] [Google Scholar]
- [11].Ayers-Ringler JR, Oliveros A, Qiu Y, Lindberg DM, Hinton DJ, Moore RM, Dasari S, Choi DS, Front Behav Neurosci 2016, 10, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Neubert H, Bonnert TP, Rumpel K, Hunt BT, Henle ES, James IT, J Proteome Res 2008, 7, 2270. [DOI] [PubMed] [Google Scholar]; Wiener MC, Sachs JR, Deyanova EG, Yates NA, Anal Chem 2004, 76, 6085. [DOI] [PubMed] [Google Scholar]
- [13].Elias JE, Gygi SP, Nat Methods 2007, 4, 207. [DOI] [PubMed] [Google Scholar]; Keller BO, Wang Z, Li L, Journal of chromatography. B Analytical technologies in the biomedical and life sciences 2002, 782, 317. [DOI] [PubMed] [Google Scholar]; Nesvizhskii AI, Keller A, Kolker E, Aebersold R, Anal Chem 2003, 75, 4646. [DOI] [PubMed] [Google Scholar]
- [14].Omura S, Kawai E, Sato F, Martinez NE, Chaitanya GV, Rollyson PA, Cvek U, Trutschl M, Alexander JS, Tsunoda I, Circ Cardiovasc Genet 2014, 7, 444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kaddurah-Daouk R, Weinshilboum RM, Clinical pharmacology and therapeutics 2014, 95, 154. [DOI] [PubMed] [Google Scholar]
- [16].Laguesse S, Morisot N, Phamluong K, Ron D, Addict Biol 2017, 22, 1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Neasta J, Ben Hamida S, Yowell QV, Carnicella S, Ron D, Biol Psychiatry 2011, 70, 575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Farris SP, Miles MF, PLoS One 2013, 8, e82435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yaka R, Phamluong K, Ron D, J Neurosci 2003, 23, 3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Backstrom P, Bachteler D, Koch S, Hyytia P, Spanagel R, Neuropsychopharmacology 2004, 29, 921. [DOI] [PubMed] [Google Scholar]
- [21].Blednov YA, Harris RA, Int J Neuropsychopharmacol 2008, 11, 775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Cozzoli DK, Courson J, Caruana AL, Miller BW, Greentree DI, Thompson AB, Wroten MG, Zhang PW, Xiao B, Hu JH, Klugmann M, Metten P, Worley PF, Crabbe JC, Szumlinski KK, Alcohol Clin Exp Res 2012, 36, 1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Nagy J, Curr Neuropharmacol 2008, 6, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Nam HW, Lee MR, Zhu Y, Wu J, Hinton DJ, Choi S, Kim T, Hammack N, Yin JC, Choi DS, Biol Psychiatry 2011, 69, 1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Craft GE, Chen A, Nairn AC, Methods 2013, 61, 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wu J, Li J, Huang KP, Huang FL, J Biol Chem 2002, 277, 19498. [DOI] [PubMed] [Google Scholar]
- [27].Proctor WR, Poelchen W, Bowers BJ, Wehner JM, Messing RO, Dunwiddie TV, J Pharmacol Exp Ther 2003, 305, 264. [DOI] [PubMed] [Google Scholar]
- [28].Newton PM, Ron D, Pharmacol Res 2007, 55, 570. [DOI] [PubMed] [Google Scholar]
- [29].Abrahao KP, Salinas AG, Lovinger DM, Neuron 2017, 96, 1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Budygin EA, Phillips PE, Wightman RM, Jones SR, Synapse 2001, 42, 77. [DOI] [PubMed] [Google Scholar]
- [31].Lobo IA, Harris RA, Pharmacol Biochem Behav 2008, 90, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Weiner JL, Valenzuela CF, Pharmacol Ther 2006, 111, 533. [DOI] [PubMed] [Google Scholar]
- [33].Alagarsamy S, Marino MJ, Rouse ST, Gereau R. W. m. I., Heinemann SF, Conn PJ, Nature Neuroscience 1999, 2, 234. [DOI] [PubMed] [Google Scholar]; Verpelli C, Dvoretskova E, Vicidomini C, Rossi F, Chiappalone M, Schoen M, Di Stefano B, Mantegazza R, Broccoli V, Bockers TM, Dityatev A, Sala C, J Biol Chem 2011, 286, 34839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Goodwani S, Saternos H, Alasmari F, Sari Y, Neurosci Biobehav Rev 2017, 77, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.