Abstract
Understanding the mechanism by which biological macromolecules fold into their functional native conformations represents a problem of fundamental interest. DNA oligonucleotides derived from human telomeric repeat d[TAGGG(TTAGGG)3] and d[TAGGG(TTAGGG)3TT] fold into G-quadruplexes through diverse steps. Varying the pH and temperature by the use of nuclear magnetic resonance and other methods enabled detection of pre-folded structures that exist in solution before completely formed G-quadruplexes upon addition of cations. Pre-folded structures are in general hard to detect, however their knowledge is crucial to set up folding pathways into final structure since they are believed to be a starting point. Unexpectedly well-defined pre-folded structures composed of base triples for both oligonucleotides were detected at certain pH and temperature. These kinds of structures were up to now only hypothesized as intermediates in the folding process. All revealed pre-folded structures irrespective of the pH and temperature exhibited one common structural feature that could govern folding process.
INTRODUCTION
Guanine-rich (G-rich) DNA regions exhibit potential to form G-quadruplexes in the presence of stabilizing cations, and demonstrate vast structural diversity and polymorphism (1–5). G-rich DNA sequences are present throughout the human genome including regulatory regions of (onco)genes and telomeric repeats. Consequently, G-quadruplexes are attractive targets for anticancer and antiviral drug development (6,7). Folding pathways of G-quadruplex structures that are of fundamental importance for understanding their biological functions are explored poorly. We have shown recently that in the absence of K+ and Na+ ions, an oligonucleotide originating from a G-rich telomeric sequence of Oxytricha nova d[G4T4G4] adopts well defined pre-folded structures (8,9). When cations are added into solution, these pre-folded structures quickly convert into the antiparallel G-quadruplex (10). Our results showed that folding into G-quadruplex from thermally unfolded single stranded sequence is slower and proceeds via more intermediates than folding from a pre-folded structure. Pre-folded structures can also be off-pathway intermediates that can slow down folding of G-quadruplex (11).
G-quadruplexes formed by human telomeric repeats d[TTAGGG] display a great variety of folding topologies as in response to nature and concentration of cations, pH, molecular crowding and flanking sequences (12–20). For instance, 23-nt G-rich DNA oligonucleotide d[TAGGG(TTAGGG)3], designated here as htel1, folds in the presence of K+ ions into hybrid-1 G-quadruplex consisting of three G-quartets and propeller-lateral-lateral loops in the 5′ to 3′ direction (Figure 1) (14,21–22). On the other hand, its 25-nt analogue d[TAGGG(TTAGGG)3TT] with two additional thymines at the 3′-end, designated here as htel2, forms a hybrid-2 G-quadruplex with lateral-lateral-propeller loops (Figure 1) (14–15,23). Interestingly, a recent nuclear magnetic resonance (NMR) study on d[TTGGG(TTAGGG)3A] with minor modifications of overhangs showed that upon addition of K+ ions into solution, a minor hybrid-2 conformation is formed faster than a more stable hybrid-1 conformation (22,24). Several experimental and theoretical approaches have shown that folding of human telomeric sequences into the ‘final’ G-quadruplex structures upon addition of KCl proceeds through many intermediate states involving hairpins, antiparallel chair-type structures and G-triplexes (24–29). The current study aims to contribute more insights into pre-folded structures of human telomeric repeats that exist in solution at low concentration of cations and thus uncover intrinsic property of G-rich DNA to fold into a global energy minimum. Molecular model of pre-folded structure with atomic resolution can in addition represent a target for ligands consisting of functional groups different from those for G-quadruplexes.
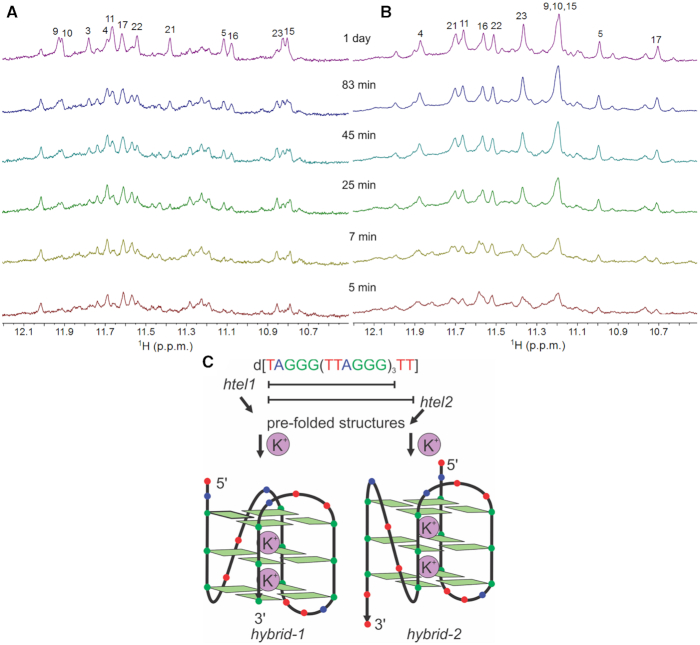
Figure 1.
Formation of G-quadruplex structures. Imino regions of 1D 1H NMR spectra of (A) htel1 and (B) htel2 as a function of time after addition of 70 mM KCl at 25°C. Spectra were recorded in 10% 2H2O on a 600 MHz spectrometer. Concentrations of oligonucleotides were 1 mM per strand. (C) Folding of htel1 and htel2 into hybrid-1 (14,21) and hybrid-2 (14,15) G-quadruplexes, respectively.
MATERIALS AND METHODS
Sample preparation
All oligonucleotides (isotopically unlabelled and residue-specific 8% 15N, 13C-labelled) were synthesized on K&A Laborgeraete GbR DNA/RNA Synthesizer H-8 using standard phosphoramidite chemistry. Aqueous amonia was used for deprotection overnight at 55°C. The samples were purified with HPLC and desalted with the use of Amicon Ultra-15 Centrifugal Filter Units. The concentration of the oligonucleotides was determined by UV absorption measurements at 260 nm using Berr-Lambert law. Molar extinction coefficient value for hte1 was 236500 l mol−1cm−1, for htel2 253100 l mol−1cm−1 and was calculated using the nearest-neighbour model.(30) All NMR and circular dichroism (CD) measurements and native polyacrylamide gel electrophoresis (PAGE) experiments were conducted without buffer and addition of cations. pH value was 7 or 5 and it was regulated with the use of 0.25 M HCl or 0.25 M LiOH. UV melting experiments were conducted in 20 mM cacodylate buffer at pH 5.
NMR spectroscopy
All NMR experiments were performed on Agilent Technologies spectrometers at 600 MHz or 800 MHz using triple-resonance (1H/13C/15N) cold probe. All spectra were recorded at 25°C or 5°C. The sample was prepared with addition of 10% 2H2O and pH was adjusted to 5 or 7. 1D 1H-15N HSQC experiments were recorded on 8% residue-specific 15N, 13C-labelled samples and were used to determine the guanine/thymine H1/H3 protons. 2D 1H-15N HSQC experiments were used to determine the adenine H6 protons. 2D 1H-13C HSQC experiments were used to determine the thymine H7 protons. 2D NOESY experiments were recorded at mixing times of 80, 120, 150 and 200 ms.
Circular dichroism (CD)
CD spectra were recorded on Chirascan spectrometer (Applied Photophysics) equipped with temperature controler. NMR smples were used to obtain CD spectra. The absorption cell with the optical path length of 0.1 mm was used. CD spectra were recorded at 25°C and 5°C in the wavelength range of 200–320 nm and were averaged over two scans. Baseline of Mill-Q water was subtracted from each spectrum.
Native PAGE
Non-denaturating PAGE was preformed using electrophoresis cell Protean® II xi cell (Bio Rad). Gel (15% PA) was run at 5°C and 50 V for 18 h overnight. The concentration of DNA loaded in each lane was 135, 68 or 34 μM, depending on the gel. The gel was coloured with Stains-All dye (Sigma-Aldrich).
NMR structure calculation
The structures of pre-folded form of htel1 were calculated by the simulated annealing (SA) simulations base on NOE-derived distance restraints. SA simulations were performed using the CUDA version of pmemd module of AMBER 14 program suites (31,32) and Cornell et al. force field basic version parm99 (33) with the bsc0 (34), χOL4 (35), ϵ/ζOL1 (36) and βOL1 (37) refinements. The initial extended single-stranded DNA structure was obtained using the leap module of AMBER 14. A total of 100 structures were calculated in 80 ps of NMR restrained simulated annealing (SA) simulations using the generalized Born implicit model (38,39). The cut-off for non-bonded interactions was 999 Å and the SHAKE algorithm (40) for hydrogen atoms was used with the 0.4 fs time steps. For each SA simulation, a random velocity was used. The SA simulation was as follows: in 0–2 ps, the temperature was raised from 300 K to 1000 K and held constant at 1000 K for 38 ps. Temperature was scaled down to 500 K in the next 24 ps and reduced to 100 K in the next 8 ps and was further reduced to 0 K in the last 8ps. NOE-derived distance restraints (force constant 20 kcal mol−1 Å−2) were used during the calculation.
RESULTS AND DISCUSSION
Different folding pathways of htel1 and htel2 into the ‘final’ hybrid-1 and hybrid-2 G-quadruplexes
Analysis of changes in NMR spectra of htel1 and htel2 after addition of K+ ions (up to 70 mM concentration at 25°C) over time implied different folding pathways into the ‘final’ hybrid-1 and hybrid-2 G-quadruplex structures, respectively (Figure 1). Intriguingly, 1D 1H NMR spectra showed that the predominant structure formed by htel1 immediately after addition of K+ ions is different from the final hybrid-1 G-quadruplex. On the other hand, signals corresponding to hybrid-2 G-quadruplex structure formed by htel2 are observed after addition of a given aliquot of KCl from the beginning and become more intense in the course of time.
htel1 and htel2 exhibit pre-folded structures
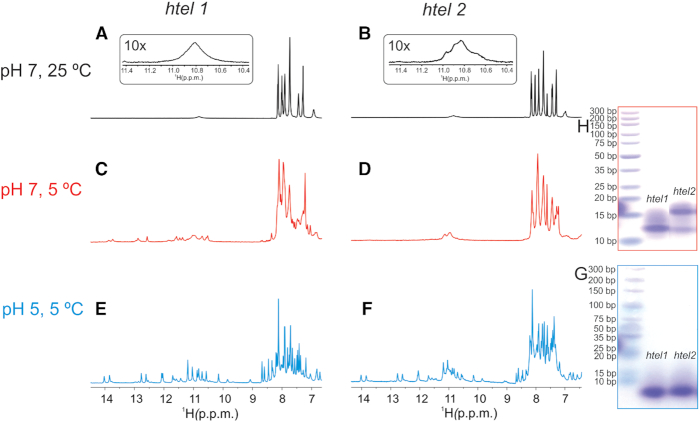
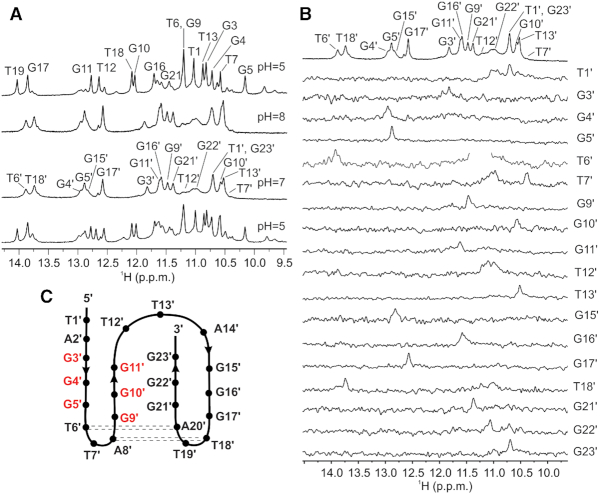
In order to explore if htel1 and htel2 form pre-folded structures that could be related to their folding pathways leading to the final G-quadruplexes, 1D 1H NMR spectra of both oligonucleotides were recorded at pH 7 and 25°C in the absence of cations. Broad signals of low intensity between δ 10.6 and 11.1 ppm, which correspond to imino protons of guanine residues involved in GG N1-carbonyl symmetric base pairs were observed for both oligonucleotides (Figure 2A and B) (8). Lowering the sample temperature to 5°C, where proton exchange with solvent is slower, enabled observation of several signals of low intensity in the imino region of 1D 1H NMR spectra of htel1 (Figure 2C). In contrast, 1D 1H NMR spectrum of htel2 recorded at 5°C exhibits only two broad signals (Figure 2D). Favourable dispersion of imino signals observed for htel1 implies a well-defined structure(s). Lowering of pH to 5 additionally improves resolution in the imino spectral region and increases intensity of the imino signals for both oligonucleotides (Figure 2E and F). A great resemblance of chemical shifts of major signals between δ 10 and 14 ppm in 1H NMR spectra indicates formation of very similar pre-folded structures of htel1 and htel2 at lower pH. However, in the case of htel2, additional broad signals corresponding to less well-defined structure(s), also observed at pH 7 and 5°C, are present indicating presence of other structure(s). Therefore, two additional thymines at the 3′-end in htel2 have a great impact on structure formation.
Figure 2.
Detection of pre-folded structures. Imino and aromatic regions of 1D 1H NMR spectra of htel1 and htel2 as a function of pH and temperature in the absence of cations that promote G-quartet formation; (A) htel1 at 25°C and pH 7; (B) htel2 at 25°C and pH 7; (C) htel1 at 5°C and pH 7; (D) htel2 at 5°C and pH 7; (E) htel1 at 5°C and pH 5; (F) htel2 at 5°C and pH 5. Spectra were recorded in 10% 2H2O on an 800 MHz spectrometer in the presence of Li+ ions used for neutralization. Concentration of oligonucleotides were 1 mM per strand. Native PAGE of htel1 and htel2 at (G) pH 5 in the presence of 30 mM concentration of Li+ ions and (H) at pH 7 in the presence of 30 mM concentration of Li+ ions. On the left side of each gel in (G) and (H) marker was loaded. Gels ran at 5°C overnight.
Pre-folded structures of htel1 and htel2 at 5°C at both pH 5 and 7 exhibit similar characteristics in CD spectra with two positive bands at ca. 255 and 300 nm and two negative bands at ca. 235 and 275 nm indicating similar orientation of adjacent stacked nucleobases (Supplementary Figure S1). Native PAGE gel at pH 5 and 5°C reveals identical mobility of pre-folded structures adopted by htel1 and htel2 (Figure 2G). However, at pH 7 the major band of htel1 exhibits the same mobility as the minor band of htel2, whilst the major band of htel2 displays slowest mobility suggesting less compact pre-folded structure (Figure 2H). All the above three bands move slower than 10 bp marker of DNA ladder. Faster mobility of bands at pH 5 in comparison to pH 7 (with respect to the mobility of 10 bp marker) indicates formation of more compact pre-folded structures at lower pH. At pH 7, G-quadruplexes formed by htel1 and htel2 in the presence of 70 mM KCl exhibit the same mobility as 10 bp marker, which indicates more compact G-quadruplex structures in comparison to their pre-folded forms (Supplementary Figure S2).
Formation of base triples
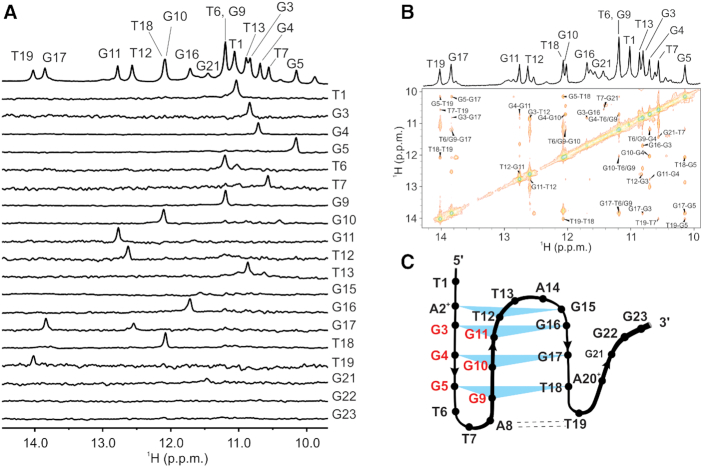
Favourable signal dispersion of 1H NMR spectra of htel1 and htel2 at pH 5 and 5°C has prompted us to perform more detailed structural characterization. 1D 1H NMR spectrum of htel1 at pH 5 and 5°C was unambiguously assigned with the use of partial site-specific double isotope 15N, 13C labelling (Figure 3A and Supplementary Figures S3 and 4). Signals for imino protons of all guanine and thymine residues were observed with exception of G15, G22 and G23 (Figure 3A). Imino proton of G21 exhibited the smallest signal most likely due its location in the structure making it more accessible for exchange with bulk solvent. Unambiguous assignment of 15N chemical shifts of adenine amino groups at δ 95 and 90 ppm revealed protonation at N1 at pH 5 of both A2+ and A20+, respectively (Supplementary Figure S3). In comparison, N6 of unprotonated A8 resonated at δ 80 ppm. 1H chemical shifts above δ 9.0 ppm of both protons of amino groups of A2+ and A20+ confirmed their involvement in hydrogen bonds (Supplementary Figure S3). Amino group protons of A8 are observed at δ 6.8 and 7.7 ppm, where proton resonating at higher chemical shift is involved in hydrogen bond with O4 of T19. Protons of amino group of A14 are not involved in hydrogen bonds since their signals were not observed in 2D 1H-15N HSQC spectrum. NOE connectivities between imino, amino, aromatic and methyl protons were used to establish topology of a pre-folded structure composed of four stacked layers displaying an anti-parallel orientation of three strands that are linked with two loops (Figure 3A–C; Supplementary Figures S5–10 and Tables S1 and 2). Guanine residues from the last G-tract are not involved in base pair formation thus resulting in the structure resembling a G-triplex. Pre-folded structure of htel1 consists of mixed A2+•T12•G15 and G5•G9•T18 base triples in addition to two G•G•G base triples (Figure 3C). Strand slippage of the third G-tract consisting of G15-G17 with respect to the first and second G-tracts of htel1 was supported by key NOE contacts of imino protons of G17 to both G3 and G5 (Figure 3B).
Figure 3.
Unambiguous assignment and crucial region of NOESY spectrum for determination of pre-folded structure. (A) The imino region of 1D 1H NMR spectrum of unlabelled sample (top) and imino regions of the 1D 1H-15N HSQC spectra acquired on partly (8%) residue-specific 15N, 13C-lablled oligonucleotides htel1. Spectra were recorded in 10% 2H2O on 600 MHz spectrometer at 5°C in the absence of cations that promote G-quartet formation at DNA concentration of 1.0 mM. (B) imino region of 1D 1H and imino-imino part of 2D NOESY NMR spectrum (τm = 150 ms) in the absence of cations in 10% 2H2O at 5°C and pH 5 of htel1. (C) Topology of pre-folded structures adopted by htel1 in the absence of cations at pH 5. Shaded triangles and dashed lines represent base triples and A•T base pairs, respectively. Residues marked with red belong to the first and the second G-tracts that are connected in antiparallel orientation.
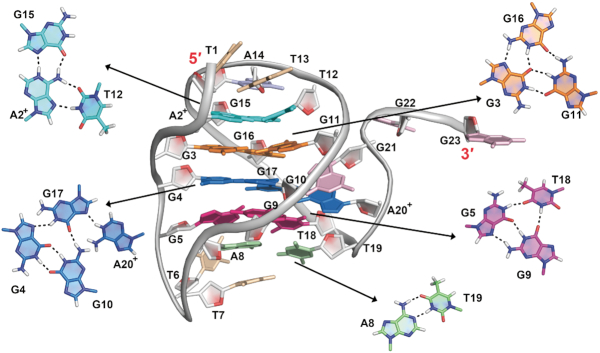
The 32 NOE-derived distance restraints (Supplementary Table S2) allowed calculation of high-resolution model of pre-folded structure of htel1 (Figure 4) although with limited number of restraints typically used for calculation of global minimum. Perusal of structural ensemble of ten lowest-energy structures shows that unpaired T1, A14 and residues at the 3′-end forward from T19 display much higher flexibility (overall RMSD of 3.9 Å) as compared to the triplex core (RMSD of 1.5 Å, Supplementary Figure S11). A2+ and T12 are base paired in reverse Hoogsteen geometry within A2+•T12•G15 base triple. Proton at N1 of A2+ is involved in hydrogen bond with G15 N7. G3•G16 and G4•G17 base pairs exhibit Hoogsteen geometry, whilst G3•G11 and G4•G10 are involved in GG N1-carbonyl symmetric base pairs within G3•G11•G16 and G4•G10•G17 base triples. T19 (H3 at δ 14.02 ppm) is involved in Watson–Crick A8•T19 base pair. Imino and amino protons of A20+ form hydrogen bonds with N7 of G17 and O6 and N7 of G9, respectively (Supplementary Figure S12). Unusually high chemical shift of imino proton of G17 (δ 13.82 ppm) is in agreement with extensive stacking interactions and its hydrogen bonding to G4 N7 as well as involvement of G17 in hydrogen bonds with G10 and A20+ (Figure 4 and Supplementary Figure S12).
Figure 4.
Pre-folded structure adopted by htel1 at pH 5. Hydrogen bonding within base triples and base pair are shown.
Guanine base triples in the pre-folded structure of htel1 stabilized by GG N1-carbonyl in addition to Hoogsteen base pairs do not require K+ ions for their formation. In contrast, a recent report showed that guanine base triples in TBA G-triplex stabilized only by Hoogsteen hydrogen bonds are formed only after addition of K+ ions (41,42). In another study it was shown that detection of potential triplex intermediates, which may participate in quadruplex folding, was unsuccessful by the use of NMR and CD spectroscopy due to complexity of the mixture resulting from co-existence of multiple dynamic species stabilized by Hoogsteen type of hydrogen bonding (43). Results were despite that published in order to highlight the major challenges in the experimental characterization of intermediates.
Importance of specific residues for structural integrity
In order to establish the importance of residues of the fourth G-tract as well as adenine residues of htel1 for structural integrity of the pre-folded structure at pH 5, folding of several oligonucleotides with missing residues at the 3′-end and with thymine substitutions were systematically analyzed by 1D 1H NMR spectra, respectively (Supplementary Table S3, Figure 5 and Supplementary Figure S13). d[TAGGG(TTAGGG)2TTA], formally without the fourth G-tract, is the shortest oligonucleotide that is still able to form the pre-folded structure established for htel1 shown in Figure 4. Even though that structure could be formed its abundance is much lower, suggesting that the presence of the last G-tract although it is not involved in base pairing is important for structural integrity. Single substitutions of A2 and A8 with thymine residues in htel1 lead to formation of different, less well-defined pre-folded structures.
Figure 5.
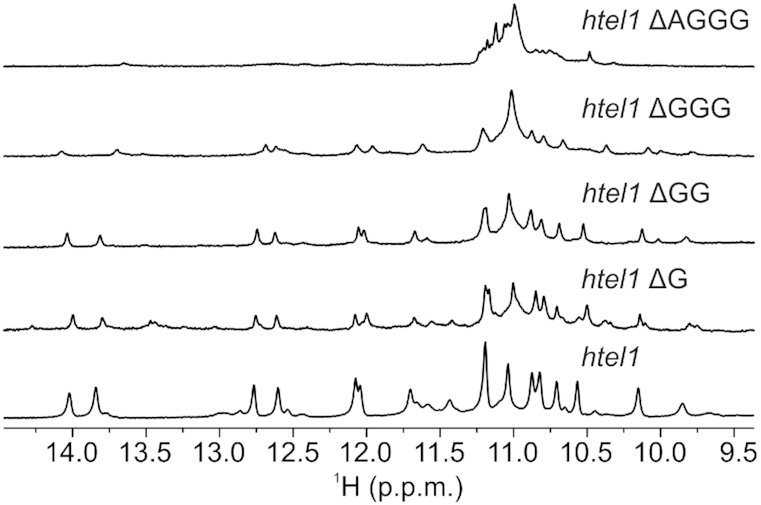
Comparison of imino regions of 1D 1H NMR spectra of oligonucleotide htel1 (bottom) and shorter sequences. htel1 ΔAGGG stands for d(TAGGGTTAGGGTTAGGGTT), htel1 ΔGGG stands for d(TAGGGTTAGGGTTAGGGTTA), htel1 ΔGG stands for d(TAGGGTTAGGGTTAGGGTTAG), htel1 ΔG stands for d(TAGGGTTAGGGTTAGGGTTAGG). Spectra were recorded in 10% 2H2O on 800 MHZ spectrometer at 5°C and pH 5 in the absence of cations that promote G-quartet formation at DNA concentration of 1.0 mM.
Temperature increase leads to hairpin formation
The 1D 1H NMR spectra at pH 5 in temperature range between 5 and 30°C revealed disappearance of imino signals corresponding to the pre-folded structure of htel1 above 15°C (Figure 6). At the same time, broad signal at δ 10.9 ppm characteristic for GG N1-carbonyl base pairs appeared. In order to determine, which guanine residues are involved in base pairs, native PAGE gel was performed on oligonucleotides with site-specific replacement of guanine to nebularine residues (Supplementary Figure S14). Nebularine residues with the lack of O6 and imino protons are not capable to form GG N1-carbonyl base pairs. Therefore, it was expected that only replacements of guanine residues originally involved in base pairing would affect the structural integrity. More than one major band in native PAGE gel was observed for oligonucleotides with site-specific replacement in the first and in the second G-tracts. Therefore, it is likely that base pairs between G3 and G11, G4 and G10 as well as G5 and G9 are preserved above 15°C, which is consistent with formation of hairpin structure, where the first and second G-tracts are connected in antiparallel orientation (Figure 6). It should be noted that G3 and G11 as well as G4 and G10 were already within base triples existing at 5°C associated with GG N1-carbonyl base pairing. Similar hairpins could exist also for htel2 at conditions where only a broad signal(s) at ca. δ 11 ppm was observed. By the use of ab initio methods Sugiyama et al. have considered three types of hairpin structures in the initial stage of folding pathways of oligonucleotides derived from Human telomeric repeat with Hoogsteen GG base pairs formed by neighbouring G-tracts (26). In addition, formation of final hybrid-1 and hybrid-2 G-quadruplexes was explained by the existence of triplex-1 and triplex-2 structures, respectively (26). Triplex-1 exhibits overhang consisting of G-tract at the 5′-end, whilst triplex-2 has overhang at its 3′-end. Therefore, formation of final hybrid-1 and hybrid-2 would proceed through different hairpin and triplex intermediates, stabilized by cations. Šponer et al. suggested by the use of MD simulations that hairpin and triplex intermediates in the folding of G-quadruplexes belong to a broader and dynamical ensemble of structures that also includes a wide spectrum of imperfectly paired structures (43). By using real-time NMR spectroscopy Schwalbe et al. observed formation of unfolded state in the folding process of d[TTGGG(TTAGGG)3A] into G-quadruplexes after addition of K+ ions (24). Unfolded state was tentatively described as an ensemble of pre-folded hairpin structures where the middle G-tracts were partially involved in base pairings.
Figure 6.
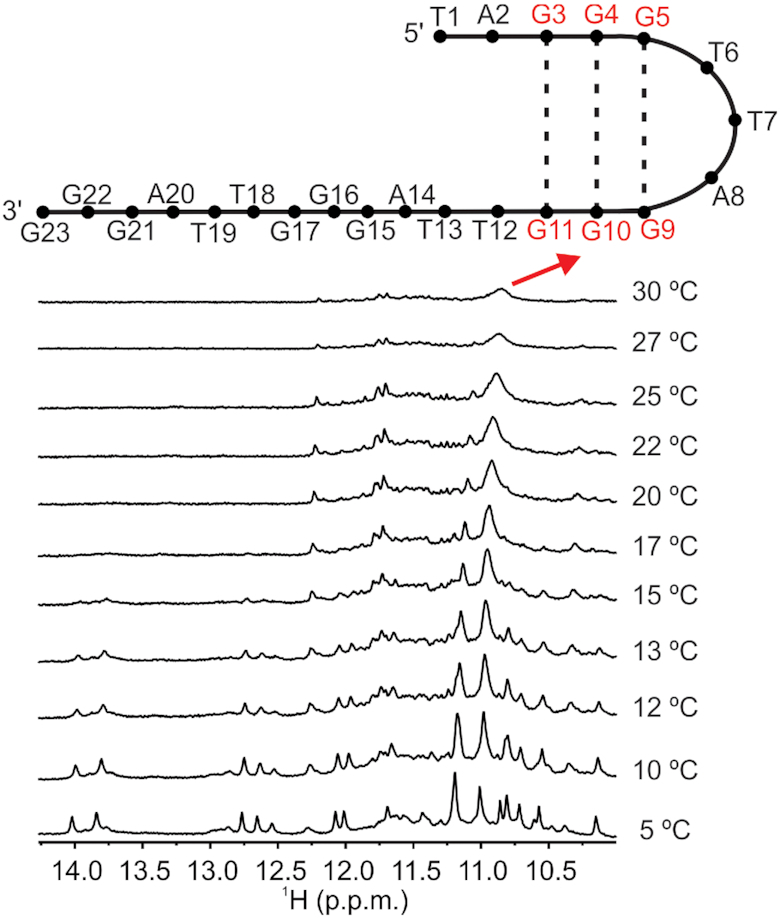
Formation of hairpin structure. Comparison of imino region of 1D 1H NMR spectra of htel1 at different temperatures. Spectrum at the bottom was recorded first. Spectra were recorded in 10% 2H2O on 800 MHZ spectrometer at 5°C in the absence of cations that promote G-quartet formation at DNA concentration of 1.0 mM. Hairpin structure is shown above the spectrum measured at 30°C. Residues marked with red belong to the first and the second G-tracts that are connected in antiparallel orientation. Base pairs between those residues are marked.
pH increase promotes formation of an antiparallel chair-like structure
Apart from major resonances, additional smaller signals were observed in 1H NMR spectrum of htel1 at pH 5 and 5°C (Figure 7A). Comparison of 1D 1H NMR spectra of htel1 at pH 5, 7 and 8 revealed that minor species observed at the lower pH became the major species above pH 7 (Figure 7A). Variation of pH influences (de)protonation of A2 and A20 in htel1, which are related to structural change. At pH 7 unambiguous assignment revealed that imino protons of all guanine including those in the fourth G-tract and thymine residues were observed except for T19′ (apostrophes denote residues within the non-protonated structure, Figure 7B). Comparison of chemical shifts for individual residues revealed differences of up to Δδ of 3 ppm at pH 5 and 7 in htel1 (Supplementary Table S4). Signals for imino protons of G4′, G5′, G15′ and G17′ resonate close to δ 13 ppm, whilst T18′ (H3 at δ 13.74 ppm) and T6′ (H3 at δ 13.89 ppm) are involved in Watson-Crick A-T base pairs. A more detailed insight into the folding topology was prevented since no NOE connectivities were observed in 2D NOESY spectra. It appears that htel1 at pH above 7 exhibits extensive conformational dynamics. However, observation of signals indicating A•T base pairs suggests formation of an antiparallel chair-like structure for non-protonated htel1, which is supported further with observable signals for imino protons of G21′, G22′ and G23′, and thus lower flexibility of the fourth G-tract with respect to the structure at pH 5 (Figure 7C). However, we should be aware that the proposed topology shown in Figure 7C is just a model. Formation of an antiparallel intermediate with either basket or chair type topology was recently proposed by Chaires et al. based on CD spectra as an early step in the folding pathway of oligonucleotides derived from human telomeric repeat forming mixed parallel/antiparallel hybrid-1 and hybrid-2 G-quadruplexes (24,25). By their suggestion the next step of folding pathways would be formation of a triplex intermediates from, which a final hybrid-1 and hybrid-2 G-quadruplexes would be formed. Both antiparallel and triplex intermediates are stabilized by K+ ions.
Figure 7.
pH switch. (A) Imino region of 1D 1H NMR spectra at different pH values of htel1 at low cation concentration in 10% 2H2O at 5°C. Spectrum on the top was recorded first and spectrum at the bottom was recorded 5 min after reducing pH back from 7 to 5. (B) Unambiguous assignment of the htel1 imino resonances at pH 7. The imino region of 1D 1H NMR spectrum of unlabelled sample (top) and imino regions of the 1D 1H-15N HSQC spectra acquired on partly (∼ 8%) residue-specific 15N, 13C-labelled oligonucleotides htel1. Spectra were recorded in 10% 2H2O on 800 MHz spectrometer at 5°C in the absence of cations that promote G-quartet formation at DNA concentration of 1.0 mM. (C) Topology of pre-folded structures adopted by htel1 in the absence of cations at pH 7. Dashed lines represent A•T base pairs, respectively. Residues marked with red belong to the first and the second G-tracts that are connected in antiparallel orientation.
CONCLUSION
Our comprehensive analysis of NMR data revealed that formation of hybrid-1 and hybrid-2 G-quadruplex structures adopted by htel1 and htel2 upon addition of K+ ions proceeds differently. NMR and other data acquired at different experimental conditions undoubtedly reveled that both oligonucleotides exhibited defined pre-folded structure(s) already in the absence of cations that are believed to be a starting point of folding process. All determined structures (with base pairs and base triples, with antiparallel chair-like topology as well as hairpin) showed that the first and the second G-tracts are connected in antiparallel orientation. This structural feature could be the main reason for different folding of htel1 and htel2 into the ‘final’ hybrid-1 and hybrid-2 G-quadruplexes. Whilst formation of hybrid-2 can proceed directly from antiparallel pre-folded structure, reorientation of the first and the second G-tracts into parallel alignment is required for formation of hybrid-1. Therefore, immediately after the addition of K+ ions into solution of htel1, G-quadruplex structure with antiparallel orientation of the first and second G-tracts is likely formed before the ‘final’ hybrid-1. This finding is in perfect agreement with recently published data that formation of less stable hybrid-2 conformation with antiparallel orientation of the first and second G-tracts from oligonucleotide d[TTGGG(TTAGGG)3A] is kinetically favoured. Similar reorientation of the first and second G-tracts would be expected also for the formation of all parallel propeller type G-quadruplex in the presence of polyethylene glycol simulating dehydration and crowding conditions. Pre-folded structure at lower pH and lower temperature is the most well-defined and consists of very interesting structural features like mixed A2+•T12•G15 and G5•G9•T18 base triples in addition to two G•G•G base triples as well as slipped third G-tract. Characterization of long-lived pre-folded structures is essential not only to establish mechanisms of G-quadruplex formation as they can represent on- or off-pathway intermediates, but can be used in development of novel type of selective ligands that will target their peculiar structural elements at pre-quadruplex states that are intrinsically more dynamic and can bind heterocyclic ligands through a privileged conformation.
DATA AVAILABILITY
The structure has been deposited in PDB under ID 6TR2.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge Blaž Bakalar and Laura Luu for performing initial experiments of folding of htel1 and htel2 into G-quadruplex structures and CERIC-ERIC for the access to experimental facilities.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Slovenian Research Agency (ARRS) [P1-0242, J1-7108, J1-1704, J3-7245]. Funding for open access charge: Slovenian Research Agency (ARRS) [P1-0242].
Conflict of interest statement. None declared.
REFERENCES
- 1. Hansel-Hertsch R., Di Antonio M., Balasubramanian S.. DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 2017; 18:279–284. [DOI] [PubMed] [Google Scholar]
- 2. Burge S., Parkinson G.N., Hazel P., Todd A.K., Neidle S.. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006; 34:5402–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Šket P., Plavec J.. Monchaud D. Diversity of DNA and RNA G-quadruplex structures. 2015; London, UK: Future Science Ltd; 23–36. [Google Scholar]
- 4. Karsisiotis A.I., O’Kane C., Webba da Silva M.. DNA quadruplex folding formalism—a tutorial on quadruplex topologies. Methods. 2013; 64:28–35. [DOI] [PubMed] [Google Scholar]
- 5. Adrian M., Heddi B., Phan A.T.. NMR spectroscopy of G-quadruplexes. Methods. 2012; 57:11–24. [DOI] [PubMed] [Google Scholar]
- 6. Neidle S. Quadruplex nucleic acids as targets for anticancer therapeutics. Nat. Rev. Chem. 2017; 1:0041. [Google Scholar]
- 7. Neidle S. Quadruplex nucleic acids as novel therapeutic targets. J. Med. Chem. 2016; 59:5987–6011. [DOI] [PubMed] [Google Scholar]
- 8. Čeru S., Šket P., Prislan I., Lah J., Plavec J.. A new pathway of DNA G-quadruplex formation. Angew. Chem., Int. Ed. 2014; 53:4881–4884. [DOI] [PubMed] [Google Scholar]
- 9. Cevec M., Plavec J.. Role of loop residues and cations on the formation and stability of dimeric DNA G-quadruplexes. Biochemistry. 2005; 44:15238–15246. [DOI] [PubMed] [Google Scholar]
- 10. Smith F.W., Feigon J.. Quadruplex structure of oxytricha telomeric DNA oligonucleotides. Nature. 1992; 356:164–168. [DOI] [PubMed] [Google Scholar]
- 11. Kuo M.H.J., Wang Z.F., Tseng T.Y., Li M.H., Hsu S.T.D., Lin J.J., Chang T.C.. Conformational transition of a hairpin structure to G-quadruplex within the WNT1 gene promoter. J. Am. Chem. Soc. 2015; 137:210–218. [DOI] [PubMed] [Google Scholar]
- 12. Wang Y., Patel D.J.. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993; 1:263–282. [DOI] [PubMed] [Google Scholar]
- 13. Parkinson G.N., Lee M.P., Neidle S.. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002; 417:876–880. [DOI] [PubMed] [Google Scholar]
- 14. Phan A.T., Kuryavyi V., Luu K.N., Patel D.J.. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007; 35:6517–6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dai J., Carver M., Punchihewa C., Jones R.A., Yang D.. Structure of the hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007; 35:4927–4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ambrus A., Chen D., Dai J., Bialis T., Jones R.A., Yang D.. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006; 34:2723–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Noer S.L., Preus S., Gudnason D., Aznauryan M., Mergny J.-L., Birkedal V.. Folding dynamics and conformational heterogeneity of human telomeric G-quadruplex structures in Na+ solutions by single molecule FRET microscopy. Nucleic Acids Res. 2016; 44:464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao A., Zhao C., Tateishi-Karimata H., Ren J., Sugimoto N., Qu X.. Incorporation of O6-methylguanine restricts the conformational conversion of the human telomere G-quadruplex under molecular crowding conditions. Chem. Commun. 2016; 52:1903–1906. [DOI] [PubMed] [Google Scholar]
- 19. Galer P., Wang B., Šket P., Plavec J.. Reversible pH switch of two-quartet G-quadruplexes formed by human telomere. Angew. Chem., Int. Ed. 2016; 55:1993–1997. [DOI] [PubMed] [Google Scholar]
- 20. Wang Z.F., Li M.H., Hsu S.T.D., Chang T.C.. Structural basis of sodium-potassium exchange of a human telomeric DNA quadruplex without topological conversion. Nucleic Acids Res. 2014; 42:4723–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dai J., Punchihewa C., Ambrus A., Chen D., Jones R.A., Yang D.. Structure of the intramolecular human telomeric G-quadruplex in potassium solution: a novel adenine triple formation. Nucleic Acids Res. 2007; 35:2440–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Luu K.N., Phan A.T., Kuryavyi V., Lacroix L., Patel D.J.. Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006; 128:9963–9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Phan A.T., Luu K.N., Patel D.J.. Different loop arrangements of intramolecular human telomeric (3+1) G-quadruplexes in K+ solution. Nucleic Acids Res. 2006; 34:5715–5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bessi I., Jonker H.R.A., Richter C., Schwalbe H.. Involvement of long-lived intermediate states in the complex folding pathway of the human telomeric G-quadruplex. Angew. Chem., Int. Ed. 2015; 54:8444–8448. [DOI] [PubMed] [Google Scholar]
- 25. Gray R.D., Trent J.O., Chaires J.B.. Folding and unfolding pathways of the human telomeric G-quadruplex. J. Mol. Biol. 2014; 426:1629–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mashimo T., Yagi H., Sannohe Y., Rajendran A., Sugiyama H.. Folding pathways of human telomeric type-1 and type-2 G-quadruplex structures. J. Am. Chem. Soc. 2010; 132:14910–14918. [DOI] [PubMed] [Google Scholar]
- 27. Bian Y., Tan C., Wang J., Sheng Y., Zhang J., Wang W.. Atomistic picture for the folding pathway of a hybrid-1 type human telomeric DNA G-quadruplex. PLOS Comput. Biol. 2014; 10:e1003562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Šponer J., Bussi G., Stadlbauer P., Kührová P., Banáš P., Islam B., Haider S., Neidle S., Otyepka M.. Folding of guanine quadruplex molecules–funnel-like mechanism or kinetic partitioning? An overview from MD simulation studies. Biochim. Biophys. Acta. 2017; 1861:1246–1263. [DOI] [PubMed] [Google Scholar]
- 29. Marchand A., Gabelica V.. Folding and misfolding pathways of G-quadruplex DNA. Nucleic Acids Res. 2016; 44:10999–11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tataurov A.V., You Y., Owczarzy R.. Predicting ultraviolet spectrum of single stranded and double stranded deoxyribonucleic acids. Biophys. Chem. 2008; 133:66–70. [DOI] [PubMed] [Google Scholar]
- 31. Salomon-Ferrer R., Goetz A.W., Poole D., Le Grand S., Walker R.C.. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent Particle Mesh Ewald. J. Chem. Theory Comput. 2013; 9:3878–3888. [DOI] [PubMed] [Google Scholar]
- 32. Case D.A., Berryman J.T., Betz R.M., Cerutti D.S., Cheatham T.E. III, Darden T.A., Duke R.E., Giese T.J., Gohlke H., Goetz A.W. et al.. AMBER 2015. 2015; San Francisco, CA: University of California. [Google Scholar]
- 33. Cornell W.D., Cieplak P., Bayly C.I., Gould I.R., Merz K.M., Ferguson D.M., Spellmeyer D.C., Fox T., Caldwell J.W., Kollman P.A.. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1996; 118:2309–2309. [Google Scholar]
- 34. Perez A., Marchan I., Svozil D., Sponer J., Cheatham T.E. III, Laughton C.A., Orozco M.. Refinenement of the AMBER force field for nucleic acids: Improving the description of alpha/gamma conformers. Biophys. J. 2007; 92:3817–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Krepl M., Zgarbova M., Stadlbauer P., Otyepka M., Banas P., Koca J., Cheatham T.E. 3rd, Jurecka P., Sponer J.. Reference simulations of noncanonical nucleic acids with different chi variants of the AMBER force field: quadruplex DNA, quadruplex RNA and Z-DNA. J. Chem. Theory Comput. 2012; 8:2506–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zgarbova M., Javier Luque F., Sponer J., Cheatham T.E. III, Otyepka M., Jurecka P.. Toward improved description of DNA backbone: revisiting epsilon and zeta torsion force field parameters. J. Chem. Theory Comput. 2013; 9:2339–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zgarbova M., Sponer J., Otyepka M., Cheatham T.E. III, Galindo-Murillo R., Jurecka P.. Refinement of the sugar-phosphate backbone torsion beta for AMBER force fields improves the description of Z- and B-DNA. J. Chem. Theory Comput. 2015; 11:5723–5736. [DOI] [PubMed] [Google Scholar]
- 38. Onufriev A., Bashford D., Case D.A.. Modification of the generalized Born model suitable for macromolecules. J. Phys. Chem. B. 2000; 104:3712–3720. [Google Scholar]
- 39. Onufriev A., Bashford D., Case D.A.. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins. 2004; 55:383–394. [DOI] [PubMed] [Google Scholar]
- 40. Ryckaert J.P., Ciccotti G., Berendsen H.J.C.. Numerical integration of Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977; 23:327–341. [Google Scholar]
- 41. Limongelli V., De Tito S., Cerofolini L., Fragai M., Pagano B., Trotta R., Cosconati S., Marinelli L., Novellino E., Bertini I. et al.. The G-triplex DNA. Angew. Chem. Int. Ed. 2013; 52:2269–2273. [DOI] [PubMed] [Google Scholar]
- 42. Cerofolini L., Amato J., Giachetti A., Limongelli V., Novellino E., Parrinello M., Fragai M., Randazzo A., Luchinat C.. G-triplex structure and formation propensity. Nucleic Acids Res. 2014; 42:13393–13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stadlbauer P., Kührová P., Vicherek L., Banáš P., Otyepka M., Trantírek L., Šponer J.. Parallel G-triplexes and G-hairpins as potential transitory ensembles in the folding of parallel-stranded DNA G-Quadruplexes. Nucleic Acids Res. 2019; 47:7276–7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The structure has been deposited in PDB under ID 6TR2.