Abstract
Therapeutic oligonucleotides are often modified using the phosphorothioate (PS) backbone modification which enhances stability from nuclease mediated degradation. However, substituting oxygen in the phosphodiester backbone with sulfur introduce chirality into the backbone such that a full PS 16-mer oligonucleotide is comprised of 215 distinct stereoisomers. As a result, the role of PS chirality on the performance of antisense oligonucleotides (ASOs) has been a subject of debate for over two decades. We carried out a systematic analysis to determine if controlling PS chirality in the DNA gap region can enhance the potency and safety of gapmer ASOs modified with high-affinity constrained Ethyl (cEt) nucleotides in the flanks. As part of this effort, we examined the effect of systematically controlling PS chirality on RNase H1 cleavage patterns, protein mislocalization phenotypes, activity and toxicity in cells and in mice. We found that while controlling PS chirality can dramatically modulate interactions with RNase H1 as evidenced by changes in RNA cleavage patterns, these were insufficient to improve the overall therapeutic profile. We also found that controlling PS chirality of only two PS linkages in the DNA gap was sufficient to modulate RNase H1 cleavage patterns and combining these designs with simple modifications such as 2′-OMe to the DNA gap resulted in dramatic improvements in therapeutic index. However, we were unable to demonstrate improved potency relative to the stereorandom parent ASO or improved safety over the 2′-OMe gap-modified stereorandom parent ASO. Overall, our work shows that while controlling PS chirality can modulate RNase H1 cleavage patterns, ASO sequence and design are the primary drivers which determine the pharmacological and toxicological properties of gapmer ASOs.
INTRODUCTION
Antisense oligonucleotides (ASOs) bind complementary RNA by Watson–Crick base-pairing and modulate RNA function to produce pharmacological effects (1). One of the most studied and commonly used antisense mechanism in the clinic is RNA cleavage promoted by RNase H1 (2). In this mechanism RNase H1 binds to a DNA:RNA heteroduplex and cleaves the RNA resulting in 5′-phosphate and 3′-hydroxyl cleavage products (3). RNase H1 requires a DNA:RNA heteroduplex for cleavage, however, ASOs are generally designed as gapmers (4), i.e. a central DNA gap region flanked on either ends with 2′-modified nucleotides such as high-affinity constrained Ethyl (cEt) nucleotides (5). The wings are important for increasing binding affinity to the targeted RNA and protecting from exonucleolytic degradation, but limits RNase H1 cleavage to the gap region (6).
RNase H1 active ASOs are typically modified using the phosphorothioate (PS) linkage (7), which is crucial for protecting the DNA gap from endonucleolytic degradation and for binding to plasma proteins (8) resulting in enhanced tissue half-lives and duration of effect after systemic administration. Furthermore, the PS backbone promotes ASO uptake into cells by enhancing interactions with cell-surface proteins (9,10) and is recognized in a duplex with RNA by RNase H1. While the PS backbone affords several advantages, the enhanced binding to specific proteins, in combination with ASO sequence and 2′-modification, can lead to hybridization-independent toxicities (11).
The sulfur atom in the PS linkage introduces chirality into the ASO backbone (designated Rp and Sp) such that a 16-mer PS ASO is composed of a mixture of 215 distinct isomers (12). Controlling PS chirality has been shown to modulate biological effects of nucleic acids (13–17) leading to some debate regarding the virtues of controlling PS chirality in ASOs. Stec et al. first showed that bacterial RNase H prefers the Rp stereoisomer but the Sp stereoisomer confers greater nuclease stability (18). Agrawal et al. studied stereo-enriched PS DNA ASOs and showed that ASOs with Rp linkages have improved RNA-binding properties, but lower metabolic stability as compared to ASOs with Sp linkages (19). They also studied ASOs with selected combinations of Rp and Sp linkages and found them to be essentially equivalent relative to the stereorandom parent. Hall et al. showed that controlling PS chirality in the MOE wings does not enhance potency in cells (20) but stereochemical bias introduced during RNA synthesis can impact the biological activity of siRNA (21). Wan et al. showed that controlling PS chirality in the DNA gap region of cEt gapmer ASOs did not enhance potency in cells or in mice (22). Iwamoto essentially confirmed these results but proposed that controlling PS chirality can enhance the duration of effect in mice, although no concomitant improvements in tissue accumulation were demonstrated (23). These authors also observed changes in RNA cleavage rates in a biochemical assay but failed to demonstrate a relationship between enhanced RNA cleavage and improved potency in cells or in mice. Furthermore, cleavage sites on the RNA were not mapped and the biochemical assays were performed using a truncated human RNase H1 enzyme lacking the hybrid-binding (HBD) domain. The HBD in mammalian RNase H1 greatly enhances the affinity and specificity of the enzyme for RNA/DNA hybrids and positions the enzyme at the 3′-DNA/5′-RNA terminus of the heteroduplex substrate (24). The HBD also assures proper alignment of the enzyme on the heteroduplex substrate via interactions with two conserved lysines and a conserved tryptophan. In their absence, both Km and Kcat are reduced and site cleavage preferences affected (24).
We recently showed that very modest chemical changes in the DNA gap region can dramatically improve the therapeutic profile of a toxic model cEt gapmer ASO (11,25). The improvement in therapeutic profile was accompanied by reduced protein binding and RNase H1 mediated nucleolar mislocalization of paraspeckle proteins such as P54nrb and PSF. Given the importance of the PS backbone for ASO protein binding, and the importance of ASO–protein interactions on ASO performance (26), we examined if controlling the chirality of the PS linkages in the DNA gap region could also improve potency and therapeutic profile of gapmer ASOs.
As part of this effort, we examined the effect of systematically controlling PS chirality on RNase H1 cleavage patterns, protein mislocalization phenotypes, activity and toxicity in cells and in mice. We found that while controlling PS chirality can dramatically modulate interactions with RNase H1 as evidenced by changes in RNA cleavage patterns, these were insufficient to improve the overall therapeutic profile in a meaningful manner. We also found that controlling PS chirality of only two PS linkages in the DNA gap was sufficient to modulate RNase H1 cleavage patterns and, combining these designs with simple modifications to the DNA gap, resulted in dramatic improvements in therapeutic properties. However, we were unable to demonstrate improved potency relative to the stereorandom parent ASO or an improvement in therapeutic profile over the 2′-OMe gap-modified stereorandom parent ASO. Overall, our work shows that while controlling PS chirality can modulate RNase H1 cleavage patterns, ASO sequence and design are the primary drivers which determine the pharmacological and toxicological properties of gapmer ASOs.
MATERIALS AND METHODS
Oligonucleotide (ON) synthesis
ONs were synthesized on an ABI 394 DNA/RNA synthesizer (2 μmol scale) or an AKTA oligopilot 10 (40 μmol) using polystyrene based VIMAD unylinker support (200 μmol/g). Fully protected nucleoside phosphoramidites were incorporated using standard solid-phase ON synthesis conditions, i.e. 3% dichloroacetic acid in dichloromethane for deblocking, 1 M 4,5-dicyanoimidazole 0.1 M N-methylimidazole in acetonitrile (MeCN) as activator or for chiral PS incorporations 2.0 M CMPT in MeCN, 10% acetic anhydride in tetrahydrofuran (THF) and 10% N-methylimidazole in THF/pyridine for capping and 0.1 M xanthane hydride in pyridine:MeCN 3:2 (v/v) for thiolation. DNA amidites were dissolved at 0.1 M MeCN and incorporated using 6 min. coupling times while OAP monomers were prepared at 0.2 M in MeCN:toluene 1:1 (v/v) + 2.5% pyridine and incorporated using 10 min. coupling times. After conclusion of the synthesis the 5′-dimethoxytrityl group was removed and remaining protecting groups were cleaved by suspending the solid support in aqueous concentrated ammonia and heated at 55°C for 48 h. Then the support was filtered off and the crude mixture purified by high performance liquid chromatography on a strong anion exchange column (source Q support) using buffers A: 50 mM NaHCO3 and B: 1.5 M NaBr 50 mM NaHCO3—both buffers in MeCN:H2O 3:7 (v/v). ONs were desalted using C18 reverse-phase Sep-Pac cartridges and lyophilized.
Thermal denaturation temperatures
ASO and RNA were mixed in equal ratios at 4 μM each in a buffer containing 10 mM phosphates, 100 mM NaCl and 100 μM ethylenediaminetetraacetic acid at pH 7.0. The duplexes were denatured at 85°C and slowly cooled to the starting temperature of the experiment (15°C). Thermal denaturation temperatures were measured in quartz cuvettes (pathlength 1.0 cm) on a Cary 100 UV/visible spectrophotometer quipped with a Peltier temperature controller. Absorbance was measured at 260 nm as a function of temperature using a temperature ramp of 0.5°C/min. Tm values were determined using the hyperchromicity method incorporated into the instrument software.
Cell culture and ASO treatment
HeLa, 3T3-L1 and Hepa1-6 cells were grown at 37°C, 7.5% CO2 in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. For transfection with ASOs, cells at 70% confluency were transfected with ASOs at specified doses using Lipofectamine 2000 at a 4 μg/ml final concentration. For high throughput electroporation of ASOs for caspase 3/7 Glo assay or qRT-PCR, cells (20 000 cells/reaction) were mixed with ASOs at the specified final concentrations at a final volume of 100 μl and added to a BTX high throughput electroporation plate. The cells were then electroporated at 140 V using an ECM 830 high throughput electroporation system.
Caspase activity assay
Caspase-Glo 3/7 Reagent was added directly to Hepa1-6 cells at volume equal to sample volume. Luminescence was recorded at 30 min by TECAN infinite M200. Background reading was determined from wells containing culture medium without cells and was subtracted from the control or assay readings. Relative caspase activity (%) was calculated as: 100 × luminescence reading of a treated sample/luminescence reading of an untreated control.
P54nrb mislocalization
HeLa cells were seeded on a glass-bottom culture dish (MatTek) and allowed to attach followed by ASO treatment at 200 nM for 2 h as previously described. After rinsing three times with 1× PBS, cells were fixed at room temperature with 4% formaldehyde for 30 min and permeabilized with 0.1% Triton X-100 in 1× PBS for 5 min. Fixed cells were then blocked in blocking buffer (1 mg/ml BSA in 1× PBS) at room temperature for 30 min, then incubated in blocking buffer with primary antibody against P54nrb (Santa Cruz sc-376865) at room temperature for 2 h. After washing three times with washing buffer (0.1% Tween-20 in 1× PBS; 5 min per wash), cells were incubated in blocking buffer with secondary antibody (Abcam ab150113) at room temperature for 1 h. Finally, cells were washed for 5 min three times in washing buffer, then mounted with Prolong Gold anti-fade reagent with 4′,6-diamidino-2-phenylindole (DAPI) (Molecular Probes). Confocal images were generated and analyzed by a confocal laser scanning microscopy using an FV1000 Fluorview (Olympus).
RNase H1 cleavage patterns
Human RNase H1 was diluted in buffer containing 100 mM Tris–HCl, 50 mM NaCl, 30% glycerol and 10 mM DTT at pH 7.4. RNA was 5′-end labeled with FAM and mixed in equal ratio to the ASO (0.33 μM) and annealed in reaction buffer containing 20 mM Tris–HCl, 50 mM NaCl, 10 mM MgCl2 and 10 mM DTT at pH 7.4. RNase H1 protein (1 ng) was added to each duplex solution and the reaction was incubated at 37°C for 10 min. The reaction was stopped by adding 10 μL stop solution containing 8 M urea and 120 mM EDTA for every 20 μL of reaction mixture. Samples were heated to 95°C for 5 min. and separated on 20% denaturing polyacrylamide gel and cleavage products were detected using a Storm Phosphor Imager and analyzed with ImageQuant TL software. RNA sequence used: 5′-UAAUGUGAGAACAUGC.
Animal experiments
Animal experiments were conducted in accordance with the American Association for the Accreditation of Laboratory Animal Care guidelines and were approved by the Animal Welfare Committee (Cold Spring Harbor Laboratory's Institutional Animal Care and Use Committee guidelines). Animals were housed in micro-isolator cages on a constant 12 h light–dark cycle with controlled temperature and humidity and were given access to food and water ad libitum. Animals were randomly grouped, and all animals were included in data analysis except those found dead. All experiments were done with n = 3 per dose group. 6–8-week-old Balb-C mice (Charles River Laboratory) were treated with a single subcutaneous injection. Seventy-two hours following treatment animals were sacrificed and blood and livers were collected. Blood was collected by cardiac puncture exsanguination with K3-EDTA (SARSTEDT, Germany) and plasma transaminases were measured using a Beckman Coulter AU480 analyzer. Briefly, 50–100 mg of liver tissue was homogenized with an Omni Tissue Homogenizer (Omni International) in guanidinium thiocyanate with 8% β-mercaptoethanol and total RNA was isolated using PureLink Pro 96 Total RNA Purification kit (Thermo Fisher, Carlsbad, CA, USA). qRT-PCR was performed in triplicate using the Step One Real-Time PCR system and TaqMan primer probe sets with Express One-Step Super Mix qRT-PCR kit (Thermo Fisher, Carlsbad, CA, USA). The sequences for the primers and probes used were as follows: forward primer: 5′-CCAGAGCCAACGTCAAGCAT-3′, reverse primer: 5′-CAGCCGTGCAACAATCTGAA-3′ and probe: 5′-TGAAAATCCTCAACACTCCAAACTGTGCC-3′. Target RNA levels were normalized to the levels of total RNA measured by RiboGreen, RNA Quantitation Reagent (Molecular Probes). Dose response curves were plotted using GraphPad Prism 7 software. Data was fitted using the log(inhibitor) versus response – variable slope (four parameters) option. For ED50 (dose which reduces the target mRNA by 50% relative to saline treated animals) calculations, the bottom and top were fixed to 0 and 100, respectively.
RESULTS
ASO design and synthesis
We examined if the therapeutic index of a toxic model ASO can be improved by controlling the chirality of PS linkages in the gap region. First, the limit cases were designed, i.e. a full Rp and a full Sp gap and then followed by an Sp linkage walked across a full Rp gap and vice versa (Figures 1 and 2). ASOs were named by the position in the gap followed by the modified nucleotide/linkage used. For example, a full Rp gap with a single Sp at gap position 3 is called FR3S (full Rp gap with Sp at gap position 3).
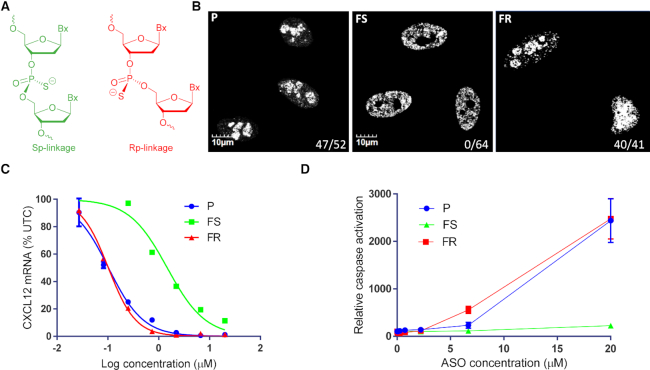
Figure 1.
Comparing stereorandom parent ASO P to full Sp gap FS and full Rp gap FR ASOs. (A) Chemical structure of Sp and Rp linkages, (B) Staining of P54nrb in HeLa cells after ASO delivery by transfection, values denote ratio of mislocalized P54nrb, (C) Dose–response curves for reducing CXCl12 mRNA in 3T3-L1 cells, and (D) Relative caspase activation. Sequence used: 5′-GCATGTTCTCACATTA; 3–10–3 cEt gapmer, all Cs are 5-methyl Cs.
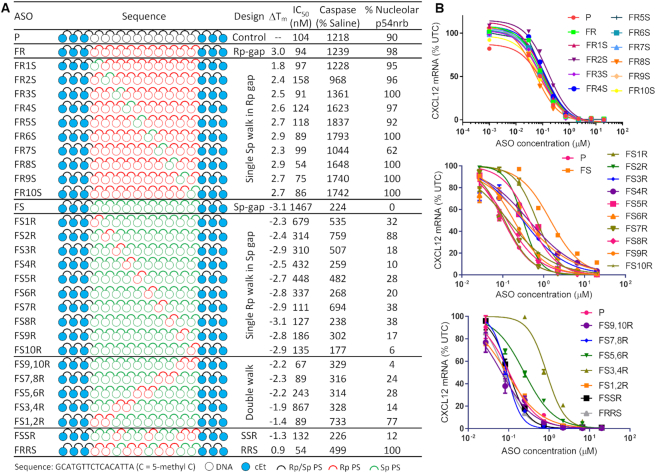
Figure 2.
Rp and Sp gap walks. (A) ASO designs, change in Tm, cell culture IC50, caspase activation at 20 μM ASO and percentage P54nrb mislocalization in cells, and (B) dose–response curves for reducing CXCl12 mRNA in mouse 3T3-L1 cells.
ASOs were synthesized using oxazaphospholidine monomers for incorporation of chirally defined PS linkages as previously described (22,27–28). While these monomers vastly improve synthesis ease relative to previous methods (29) the coupling yields were lower than those obtained for traditional cyanoethyl phosphoramidites (∼93% to 99%, respectively). Consequently, isolated yields varied between 4 and 13% while purities were generally >90% after purification by ion-exchange chromatography (Supplementary Table S1).
Biological evaluation of fully Rp and Sp gap modified ASOs
We first compared the activity and protein mislocalization phenotypes of a full Rp gap FR and a full Sp gap FS ASOs to the stereorandom parent ASO P (Figure 1). We recently showed that toxic ASOs cause mislocalization of paraspeckle proteins such as P54nrb to the nucleolus leading to nucleolar stress and fragmentation, resulting in apoptosis and caspase activation.(30,31) Treating cells with the toxic parent ASO P resulted in marked p54nrb nucleolar mislocalization (Figure 1B) and the full Rp gap ASO FR showed similar behavior. In contrast, full Sp gap ASO FS had no effect on p54nrb localization. Next, all three ASOs were delivered to mouse 3T3-L1 cells using electroporation at different concentrations and all three ASOs showed good sigmoidal shaped dose response curves for knock down of the targeted CXCl12 mRNA (Figure 1C). Again, parent ASO P and full Rp gap FR showed similar effect with IC50 values of approximately 0.1 μM. However, the full Sp gap ASO FS was ∼14-fold less potent (IC50 of 1.4 μM). Hepa1-6 cells treated with the stereorandom ASO P and the full Rp gap ASOs FR showed caspase activation indicative of cytotoxicity while the cells treated with the full Sp gap ASO FS showed no significant increase in caspase activation even at the highest concentration tested (20 μM).
Sp and Rp walks
Evaluating full Rp and full Sp gap ASOs showed that they behave very differently, but neither performed better than the parent ASO P. Consequently, we evaluated fully stereocontrolled gaps containing mixtures of Rp and Sp linkages. The gap of parent ASO P, however, contains 10 PS linkages therefore 1024 different PS diastereomers are possible. We had previously shown that a collection of 32 ASOs with different configurations of Rp/Sp linkages in the DNA gap were essentially similar in potency to the stereorandom parent ASO in cells (22). However, it was not possible to analyze the data to identify trends that could lead to general conclusions towards a preferred design of Rp/Sp linkages in the DNA gap. We therefore walked a single Rp or an Sp linkage in an otherwise full Sp or Rp gap, respectively, (Figure 2). We examined the effect of these design changes on binding affinity towards complementary RNA, nucleolar mislocalization of P54nrb by fluorescence microscopy in HeLa cells, and knockdown of CXCl12 mRNA in 3T3-L1 cells and caspase activation in Hepa1-6 cells as an indicator of antisense activity and cytotoxicity, respectively. Our previous work had shown that ASOs which exhibit 2–3-fold elevation in caspase activation relative to mock treated cells were generally hepatotoxic in mice (11).
It is well known that Rp linkages stabilize oligonucleotide duplexes with complementary RNA while Sp linkages are slightly destabilizing. In accordance with the literature, we observed a net destabilizing ΔTm when >50% of linkages are Sp and a net stabilizing effect when the predominant linkage is Rp (Figure 2). The overall changes in melting temperatures, however, were modest (spanning –3.0 to +3.1°C) and considering that the Tm of the stereorandom ASO P was ∼64°C, the difference in biological properties of the ASOs in this set could not be rationalized by differences in RNA affinity.
3T3-L1 cells were treated with the ASOs in a dose-responsive manner to determine the concentration required to reduce CXCl12 mRNA by 50% relative to control treated cells (Figure 2). As described above, the full Sp gap ASO FS showed significant loss in potency relative to the full Rp and stereorandom parent ASO, but interestingly, replacing even a single Sp with an Rp linkage resulted in substantial recovery in potency (Figure 2). Especially when an Rp was substituted toward the 3′-end of the gap the potency was similar to the parent ASO P. For the full Rp gap ASO FR, substituting a single linkage to Sp, in most positions, had minimal effect on potency. Only ASO FR2S exhibited potency that was slightly reduced while ASO FR8S exhibited slightly improved potency relative to ASO P (Figure 2).
For ASOs with an Sp linkage walked across a full Rp gap, all ASOs exhibited caspase activation which is similar to or worse than the parent ASO P. Furthermore, p54nrb mislocalization to the nucleolus was also similar to the toxic ASO P for all ASOs in this set (Figure 2, Supplementary Figures S1 and S2). ASOs with a single Rp walked across the Sp gap, were all much better tolerated than ASO P, although most of them were still deemed toxic as they produced caspase activation >2-3 fold relative to mock treated cells (Supplementary Figures S3 and S4). The best tolerated ASO in this series was FS10R with <2-fold increase in caspase activation and minimal p54nrb mislocalization (Figure 2).
Since substituting a single Rp linkage into a full Sp gap improved potency significantly, we also synthesized ASOs with two Rp linkages walked across a full Sp DNA gap (Figure 2). For the double walk three out of five ASOs had similar or slightly better potency than ASO P in cell culture (FS9,10R, FS7,8R and FS1,2R). Most of the double modified ASOs showed reasonable tolerability (Supplementary Figure S5), the one exception being FS1,2R which increased caspase activation 7-fold and caused significant P54nrb mislocalization.
We also evaluated a repeat SpSpRp design (FSSR) recently reported as an optimal gap-design for gapmer ASOs (Figure 2) (23). In addition, we also evaluated the complementary RpRpSp design (FRRS) as an additional control. Both ASOs exhibited a potency comparable to the stereorandom ASO P. However, FSSR exhibited significantly reduced caspase elevation relative to the stereorandom parent P and FRRS ASOs.
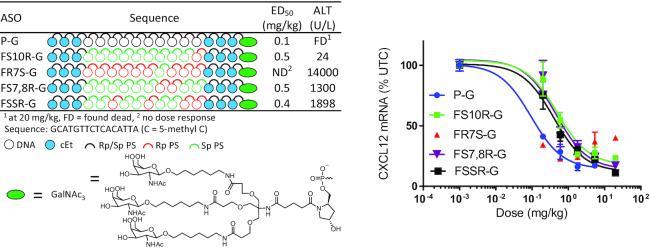
To further investigate the effect of controlling PS chirality in the gap we selected the best performing ASOs from each of the four sets (FS10R, FR7S, FS7,8R and FSSR) and evaluated them in mice. The ASOs were synthesized as 3′-THA-GalNAc conjugates as described previously (32). Attaching THA-GalNAc results in a 10-fold increase in potency in mouse hepatocytes (33), which allowed for the evaluation to be performed using smaller quantities of the chiral PS ASOs. ASO FR7S-G showed the best potency but was extremely toxic at all doses which made it difficult to calculate an ED50 for CXCl12 mRNA reduction (Figure 3). FS10R-G was the safest ASO with no elevation in plasma ALT but this ASO showed a 5-fold reduction in potency relative to the parent ASO P-G (Figure 3). FSSR-G showed slightly enhanced potency relative to FS10R-G but this ASO also showed ALT elevations in mice treated with 20 mg/kg of the ASO. Thus, we were unable to identify an ASO with controlled PS chirality in the DNA gap with improved potency or improved tolerability. Furthermore, the increased complexity associated with the synthesis of chiral PS ASOs makes this approach less practical than alternate strategies which achieve better results with simple modifications to the PS DNA-gap (11,25).
Figure 3.
Dose–response curves for reducing CXCl12 mRNA in mouse liver and ALT levels at 20 mg/kg for fully stereocontrolled gap ASOs.
Stereochemical controlled gap affect RNase H1 cleavage patterns
Next, we investigated how stereocontrolled gaps affect RNase H1 cleavage patterns. We have previously shown how substituting chemical modified nucleotides throughout the gap of an ASO can significantly affect RNase H1 cleavage patterns (34,35). Due to the systematic substitutions of Rp and Sp linkages in the gap region of ASO P, a general trend should be observed if chiral linkages affect RNase H1 cleavage preferences. The RNase H1 cleavage experiments were performed under multiple hit kinetics. ASO was annealed with a fluorophore-labelled complementary RNA and incubated with human RNase H1 enzyme for 10 min. Reaction was stopped by denaturing the enzyme and cleavage products were identified by polyacrylamide gel electrophoresis.
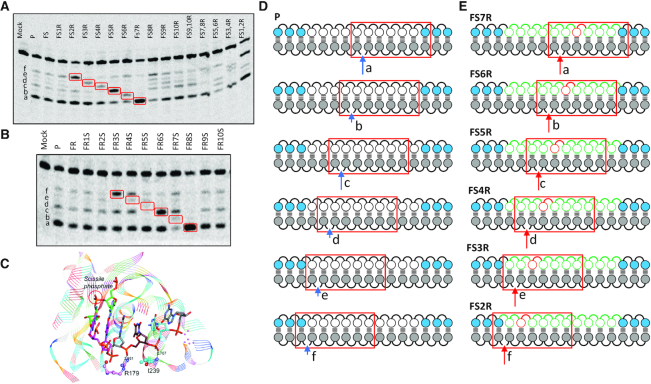
RNase H1 produced six distinct cleavage sites on the RNA strand of the DNA/RNA heteroduplex substrate that were labeled as a, b, c, d, e and f, respectively (Figure 4A, D). Interestingly, we observed no changes in cleavage patterns with all six cleavage sites being observed for the parent and the full Rp and full Sp DNA gap ASO/RNA heteroduplexes (Figure 4a,b). Cleavage patterns for the Rp walk series (Figure 4A) clearly show an increase in cleavage at two nucleotides downstream to the Rp incorporation (Figure 4E) when the Rp linkage is from position 2 to 7 in the gap. Outside of these positions and for the double walk, the trend is not as clear. Cleavage patterns for the Sp walk (Figure 4B) follow a similar but less exaggerated trend, i.e. increased cleavage at three nucleotides downstream to the Sp incorporation, when Sp is placed in the gap from position 3 to 8.
Figure 4.
Controlling PS stereochemistry in the DNA gap modulates RNase H1 cleavage patterns. (A) RNase H1 cleavage patterns for full Sp gap and Rp walk ASOs. (B) RNase H1 cleavage patterns for full Rp gap and Sp walk ASOs. (C) Crystal structure of a DNA:RNA heteroduplex bound to the catalytic domain of human RNase H1, highlighted are the predominant interactions between the DNA backbone and the enzyme. (D) The seven nucleotide catalytic domain footprint (red rectangle) and the six cleavage sites (a–f) for parent ASO P. (E) RNase H1 catalytic domain footprint as in d, but ASOs shown are for each position where a cleavage pattern has increased in intensity in the Rp walk series, which is also highlighted in red boxes in a and b.
The changes in cleavage patterns could be rationalized by examining the published crystal structures of human RNase H1 in complex with a DNA:RNA duplex (36). As described previously, the enzyme makes several specific contacts with the phosphate backbone on the DNA strand of the heteroduplex. Notable among those is an electrostatic interaction with the DNA strand in the phosphate binding pocket with Arg179 two nucleotides downstream from the cleavage site and a hydrogen bond with the backbone amide of I239 three nucleotides downstream from the cleavage site (Figure 4C). Based on the cleavage patterns observed, the Rp configuration is preferred at the phosphate binding pocket while the Sp configuration is preferred for the hydrogen bonding interaction (Figure 4e). It is especially interesting that ASO FS7R, a full Sp gap with one Rp linkage at position 7, predominantly exhibit only one cleavage site which is the main cleavage site for the parent ASO P (Figure 4D). Also, in the Rp background ASO FR8S exhibit a single cleavage site, which again locate an Rp linkage two nucleotides and an Sp linkage three nucleotides downstream from the cleavage site.
Minimal stereochemical control in the gap to direct RNase H1 cleavage
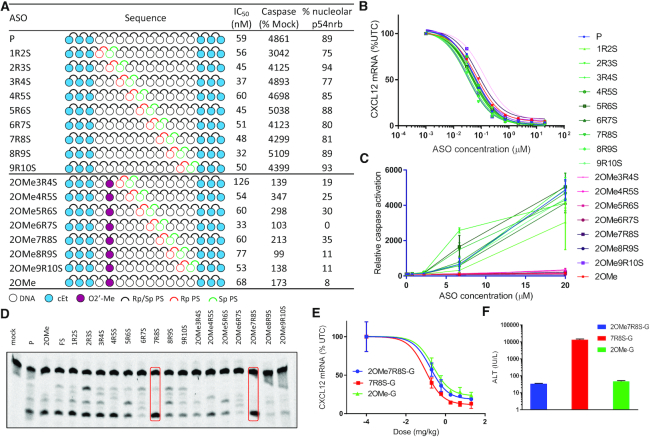
Based on the RNase H1 data we hypothesized that controlling the PS chirality at just two positions could help direct RNase H1 cleavage. Accordingly, we designed a set of ASOs where a single Rp linkage was immediately followed by an Sp linkage and walked this RpSp step throughout the gap. Furthermore, we examined how this pattern would perform in combination with a 2′-OMe incorporation at gap position 2 (Figure 5A). We previously showed that incorporation of 2′-OMe at gap position 2 mitigates toxicity and improves therapeutic index (11).
Figure 5.
Controlling stereochemistry of only two PS linkages can modulate RNase H1 cleavage patterns. (A) Table showing biological data for RpSp step walked through the gap with or without 2′-OMe at gap position 2. (B) Dose–response curves for reducing CXCl12 mRNA in mouse 3T3-L1 cells. (C) Relative caspase activation in cells dosed with ASOs (20 μM). (D) RNase H1 cleavage patterns, red boxes highlight ASOs with predominantly one cleavage site. (E) Dose–response curves for reducing CXCl12 mRNA in mouse liver for selected ASOs. (F) ALT (IU/L) levels at the highest dose for each ASO.
RNase H1 cleavage patterns showed that limited control of PS chirality did influence RNase H1 cleavage patterns (Figure 5D). As seen for the fully stereocontrolled gap ASOs FS7R and FR8S, controlling the PS chirality only at positions 7 and 8 in the gap with an RpSp step resulted in predominantly one cleavage site (7R8S, Figure 5D). Also, combination of 2′-OMe at gap position 2 and an RpSp step at position 7–8 (2OMe7R8S, Figure 5D) resulted in one predominant cleavage site.
All the ASOs were evaluated in cells and potency, caspase activation and P54nrb mislocalization were measured. For the set with no 2′-OMe incorporation, all ASOs showed similar potency and tolerability as the parent ASO P (Figure 5A). Incorporation of 2′-OMe at gap position 2 mitigated toxicity with minimal effect on potency (2OMe, Figure 5B, C). Combination of 2′-OMe and an RpSp PS step had very similar effect on potency and tolerability as the 2′-OMe gap-substituted ASO 2OMe.
Next, we selected the two ASOs in the set that exhibited only one predominant RNase H1 cleavage site and synthesized the 3′-THA-GalNAc version to evaluate them in vivo (Figure 5E, F). These two ASOs were compared to the stereorandom control ASO with a 2′-OMe at gap position 2 (2OMe-G). Both ASOs incorporating 2′-OMe were well tolerated in animals showing no elevation in ALT levels even at the highest dose tested. In contrast, the ASO with no 2′-OMe incorporation was highly toxic (Figure 5F). Interestingly, 2OMe7R8S-G which incorporate both 2′-OMe and an RpSp step to control RNase H1 cleavage was well tolerated and slightly more potent than the parent ASO 2OMe-G (Figure 5E, F).
DISCUSSION
The PS linkage introduces chirality into the oligonucleotide backbone such that an oligo with n-linkages is comprised of a mixture of 2n distinct chemical entities. The merits for controlling PS chirality in ASOs have been debated for almost two decades but published data to date suggests lack of tangible benefits such as improved potency or therapeutic index (19,22). A recent report suggested that controlling PS chirality can enhance cleavage rates of ASO/RNA duplexes by RNase H1 but the increased rates did not corelate with increased potency in cells or in mice (23). Furthermore, the effect of controlling PS chirality on the actual cleavage patterns were not reported. To address these limitations, we carried out a systematic analysis of controlling PS chirality in the gap region on ASO potency, RNase H1 activity and cleavage patterns, and therapeutic index. We performed our studies using a model toxic cEt gapmer ASO targeting murine CXCl12 mRNA as this also allowed us to determine the effect of PS chirality on important ASO–protein interactions causative of cellular toxicity (11).
Hall et al. previously demonstrated that controlling PS chirality in the wings of MOE gapmer ASOs did not enhance ASO potency in cells (20). Furthermore, as RNase H1 predominantly contacts the DNA gap region in gapmer ASO/RNA heteroduplexes, we systematically explored the effect of controlling PS chirality in the DNA gap region of ASOs. This design greatly limited the number of theoretically possible stereoisomers (1024 versus 32 768) thereby allowing for an informed assessment on the benefits of controlling PS chirality in gapmer ASOs. We designed ASOs with full Rp or Sp gaps and also walked one or two Rp linkages in an Sp gap and vice versa. We found that with the exception of the full Sp gap ASO which was 10-fold less potent, introducing even a single Rp linkage was sufficient to restore potency relative to the stereorandom parent ASO. However, none of the ASOs with stereocontrolled gaps were more potent than the stereorandom parent ASO. In addition, we also evaluated the repeat SpSpRp design reported by Iwamoto et al. (23) as an optimal chiral PS design for use in gapmer ASOs. However, we did not observe improved potency using this design either.
These observations can potentially be rationalized by the following argument. ASOs with full Rp and Sp gaps, or with a single Rp linkage walked through an Sp gap and vice versa, represent 22 out of 1024 possible diastereomers. Introducing two Sp linkages in an Rp gap and the reverse represent 90 isomers, three Sp linkages in an Rp gap and the reverse represent 240 isomers, four Sp linkages in an Rp gap and the reverse represent 420 isomers, and five Sp linkages and five Rp linkages in the gap represent 252 of the 1024 possible isomers. Thus, only 1 ASO from among 1024 isomers has significantly reduced potency and the vast majority are roughly equivalent but not more potent than the stereorandom parent ASO. This observation is also consistent with our previous report where a set of ASOs with randomly selected combinations of Rp and Sp linkages in the gap were equivalent for potency when tested in cell culture (22).
We next examined if controlling PS chirality in the gap region could enhance the therapeutic profile of a toxic gapmer ASO. We found that the ASO with a full Rp gap was as toxic as the stereorandom parent while the full Sp ASO was significantly less toxic. However, given the substantial loss in activity observed with the full Sp ASO, it is unlikely that this ASO has a better therapeutic index than the stereorandom parent ASO. Similarly, while the ASOs with a higher percentage of Sp linkages were less toxic, they still produced caspase activation 2–3 fold over mock treated cells suggesting that they would be hepatotoxic in mice. Indeed, evaluation of a small set of ASOs with chiral PS linkages in mice confirmed this prediction. In contrast, we recently showed that introducing simple 2′-modifications like OMe or backbone modifications like alkylphosphonates, at positions 2 and/or 3 from the 5′-side of the DNA gap could enhance the therapeutic index of over 600 toxic gapmer ASOs including the toxic gapmer ASO used in the current study (11,25). Thus, while it was possible to modulate therapeutic index using certain configurations of chiral PS ASOs, many of these ASOs were still predicted to be toxic in mice. Furthermore, the added cost and complexity of controlling PS chirality makes this a less attractive strategy relative to the gap-modification approach using 2′ and backbone modifications for improving therapeutic profile.
To determine if the changes in PS chirality could influence RNase H1 cleavage preferences, we evaluated the chiral PS ASO/RNA heteroduplexes in biochemical assays using full-length recombinant human RNase H1. While we did not perform detailed kinetic measurements, it was clear that all ASOs supported RNase H1 cleavage of the heteroduplex substrates. However, some distinct trends in cleavage patterns were observed. Overlaying these patterns on the published crystal structures of the catalytic domain of mammalian RNase H1 suggested that the Rp linkage is highly preferred in the PBP while the Sp linkage is preferred adjacent to the PBP. No other preferences for stereochemistry could be ascertained at other sites along the 7 base-pair footprint of the RNase H1 catalytic domain. The preference for this arrangement was confirmed by introducing an RpSp step in an otherwise stereorandom gap to force cleavage at only the main cleavage site in the middle of the DNA gap. However, even these ASOs produced significant caspase elevations in cells indicative of cytotoxicity. Further evaluation of a small subset of these ASOs in mice confirmed that controlling chirality to limit RNase H1 cleavage at a single site was not sufficient to ablate hepatotoxicity in mice. Interestingly, introducing 2′-OMe modification at position 2 in the gap in the above ASOs was able to ablate caspase elevations and hepatoxicity in mice.
In conclusion, we have carried out a systematic study to determine the effect of controlling PS chirality in the DNA gap for enhancing ASO potency and therapeutic profile. In addition, we also characterized the effects of controlling PS chirality on cleavage of the ASO/RNA heteroduplexes by RNase H1. We found that while controlling PS chirality can modulate cleavage preferences on the RNA, this was not sufficient to enhance potency or safety in a meaningful manner in the tested toxic model ASO. Our data confirm that ASOs with stereorandom PS linkages in the DNA gap region offer the optimal balance of activity and metabolic stability and that ASO sequence and design are the primary drivers which determine the pharmacological and toxicological properties of gapmer ASOs.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Ionis Pharmaceuticals.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bennett C.F., Baker B.F., Pham N., Swayze E., Geary R.S.. Pharmacology of antisense drugs. Annu. Rev. Pharmacol. Toxicol. 2017; 57:81–105. [DOI] [PubMed] [Google Scholar]
- 2. Crooke S.T., Witztum J.L., Bennett C.F., Baker B.F.. RNA-targeted therapeutics. Cell Metab. 2018; 27:714–739. [DOI] [PubMed] [Google Scholar]
- 3. Wu H., Lima W.F., Crooke S.T.. Properties of cloned and expressed human RNase H1. J. Biol. Chem. 1999; 274:28270–28278. [DOI] [PubMed] [Google Scholar]
- 4. Seth P.P., Swayze E.E.. The medicinal chemistry of RNase H-activating antisense oligonucleotides. Drug Discov. Ser. 68: Adv. Nucleic Acid Therap. 2018; 32–61. [Google Scholar]
- 5. Seth P.P., Siwkowski A., Allerson C.R., Vasquez G., Lee S., Prakash T.P., Wancewicz E.V., Witchell D., Swayze E.E.. Short antisense oligonucleotides with novel 2′–4′ conformationaly restricted nucleoside analogues show improved potency without increased toxicity in animals. J. Med. Chem. 2009; 52:10–13. [DOI] [PubMed] [Google Scholar]
- 6. Lima W.F., Rose J.B., Nichols J.G., Wu H., Migawa M.T., Wyrzykiewicz T.K., Siwkowski A.M., Crooke S.T.. Human RNase H1 discriminates between subtle variations in the structure of the heteroduplex substrate. Mol. Pharmacol. 2007; 71:83–91. [DOI] [PubMed] [Google Scholar]
- 7. Eckstein F. Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic Acid Ther. 2014; 24:374–387. [DOI] [PubMed] [Google Scholar]
- 8. Gaus H.J., Gupta R., Chappell A.E., Ostergaard M.E., Swayze E.E., Seth P.P.. Characterization of the interactions of chemically-modified therapeutic nucleic acids with plasma proteins using a fluorescence polarization assay. Nucleic Acids Res. 2019; 47:1110–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Crooke S.T. Molecular mechanisms of antisense oligonucleotides. Nucleic Acid Ther. 2017; 27:70–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miller C.M., Tanowitz M., Donner A.J., Prakash T.P., Swayze E.E., Harris E.N., Seth P.P.. Receptor-mediated uptake of phosphorothioate antisense oligonucleotides in different cell types of the liver. Nucleic Acid Ther. 2018; 28:119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shen W., De Hoyos C.L., Migawa M.T., Vickers T.A., Sun H., Low A., Bell T.A. 3rd, Rahdar M., Mukhopadhyay S., Hart C.E. et al.. Chemical modification of PS-ASO therapeutics reduces cellular protein-binding and improves the therapeutic index. Nat. Biotechnol. 2019; 37:640–650. [DOI] [PubMed] [Google Scholar]
- 12. Stec W.J., Zon G.. Stereochemical studies of the formation of chiral internucleotide linkages by phosphoramidite coupling in the synthesis of oligodeoxyribonucleotides. Tetrahedron Lett. 1984; 25:5279–5282. [Google Scholar]
- 13. Stivers J.T., Nawrot B., Jagadeesh G.J., Stec W.J., Shuman S.. Stereochemical outcome and kinetic effects of Rp- and Sp-phosphorothioate substitutions at the cleavage site of vaccinia Type I DNA topoisomerase. Biochemistry. 2000; 39:5561–5572. [DOI] [PubMed] [Google Scholar]
- 14. Pruzan R., Zielinska D., Rebowska-Kocon B., Nawrot B., Gryaznov S.M.. Stereopure oligonucleotide phosphorothioates as human telomerase substrates. New J. Chem. 2010; 34:870–874. [Google Scholar]
- 15. Liu G., Fu W., Zhang Z., He Y., Yu H., Wang Y., Wang X., Zhao Y.-L., Deng Z., Wu G. et al.. Structural basis for the recognition of sulfur in phosphorothioated DNA. Nat. Commun. 2018; 9:4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Buczko W., Cierniewski C., Kobylanska A., Koziolkiewicz M., Okruszek A., Pawlowska Z., Pluskota E., Stec W.J.. Modulation of plasminogen activator inhibitor type-1 biosynthesis in vitro and in vivo with oligo(nucleoside phosphorothioate)s and related constructs. Pharmacol. Ther. 1997; 76:161–175. [DOI] [PubMed] [Google Scholar]
- 17. Benimetskaya L., Tonkinson J.L., Koziolkiewicz M., Karwowski B., Guga P., Zeltser R., Stec W., Stein C.A.. Binding of phosphorothioate oligodeoxynucleotides to basic fibroblast growth factor, recombinant soluble CD4, laminin and fibronectin is P-chirality independent. Nucleic Acids Res. 1995; 23:4239–4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Koziolkiewicz M., Krakowiak A., Kwinkowski M., Boczkowska M., Stec W.J.. Stereodifferentiation - the effect of P chirality of oligo(nucleoside phosphorothioates) on bacterial RNase H. Nucleic Acids Res. 1995; 23:5000–5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu D., Kandimalla E.R., Roskey A., Zhao Q., Chen L., Chen J., Agrawal S.. Stereo-enriched phosphorothioate oligodeoxynucleotides: synthesis, biophysical and biological properties. Bioorg. Med. Chem. 2000; 8:275–284. [DOI] [PubMed] [Google Scholar]
- 20. Li M., Lightfoot H.L., Halloy F., Malinowska A.L., Berk C., Behera A., Schumperli D., Hall J.. Synthesis and cellular activity of stereochemically-pure 2′-O-(2-methoxyethyl)-phosphorothioate oligonucleotides. Chem. Commun. 2017; 53:541–544. [DOI] [PubMed] [Google Scholar]
- 21. Jahns H., Roos M., Imig J., Baumann F., Wang Y., Gilmour R., Hall J.. Stereochemical bias introduced during RNA synthesis modulates the activity of phosphorothioate siRNAs. Nat. Commun. 2015; 6:doi:10.1038/ncomms7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wan W.B., Migawa M.T., Vasquez G., Murray H.M., Nichols J.G., Gaus H., Berdeja A., Lee S., Hart C.E., Lima W.F. et al.. Synthesis, biophysical properties and biological activity of second generation antisense oligonucleotides containing chiral phosphorothioate linkages. Nucleic Acids Res. 2014; 42:13456–13468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Iwamoto N., Butler D.C.D., Svrzikapa N., Mohapatra S., Zlatev I., Sah D.W.Y., Meena, Standley S.M., Lu G., Apponi L.H. et al.. Control of phosphorothioate stereochemistry substantially increases the efficacy of antisense oligonucleotides. Nat. Biotechnol. 2017; 35:845–851. [DOI] [PubMed] [Google Scholar]
- 24. Lima W.F., Wu H., Nichols J.G., Prakash T.P., Ravikumar V., Crooke S.T.. Human RNase H1 uses one tryptophan and two lysines to position the enzyme at the 3′-DNA/5′-RNA terminus of the heteroduplex substrate. J. Biol. Chem. 2003; 278:49860–49867. [DOI] [PubMed] [Google Scholar]
- 25. Migawa M.T., Shen W., Wan W.B., Vasquez G., Oestergaard M.E., Low A., De Hoyos C.L., Gupta R., Murray S., Tanowitz M. et al.. Site-specific replacement of phosphorothioate with alkyl phosphonate linkages enhances the therapeutic profile of gapmer ASOs by modulating interactions with cellular proteins. Nucleic Acids Res. 2019; 47:5465–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Crooke S.T., Wang S., Vickers T.A., Shen W., Liang X.H.. Cellular uptake and trafficking of antisense oligonucleotides. Nat. Biotechnol. 2017; 35:230–237. [DOI] [PubMed] [Google Scholar]
- 27. Oka N., Wada T., Saigo K.. An oxazaphospholidine approach for the stereocontrolled synthesis of oligonucleoside phosphorothioates. J. Am. Chem. Soc. 2003; 125:8307–8317. [DOI] [PubMed] [Google Scholar]
- 28. Oka N., Wada T., Saigo K.. Diastereocontrolled synthesis of dinucleoside phosphorothioates using a novel class of activators, dialkyl(cyanomethyl)ammonium tetrafluoroborates. J. Am. Chem. Soc. 2002; 124:4962–4963. [DOI] [PubMed] [Google Scholar]
- 29. Stec W.J., Grajkowski A., Kobylanska A., Karwowski B., Koziolkiewicz M., Misiura K., Okruszek A., Wilk A., Guga P., Boczkowska M.. Diastereomers of Nucleoside 3′-O-(2-Thio-1,3,2-oxathia(selena)phospholanes): Building Blocks for Stereocontrolled Synthesis of Oligo(nucleoside phosphorothioate)s. J. Am. Chem. Soc. 1995; 117:12019–12029. [Google Scholar]
- 30. Lorenz P., Baker B.F., Bennett C.F., Spector D.L.. Phosphorothioate antisense oligonucleotides induce the formation of nuclear bodies. Mol. Biol. Cell. 1998; 9:1007–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shen W., Liang X.H., Crooke S.T.. Phosphorothioate oligonucleotides can displace NEAT1 RNA and form nuclear paraspeckle-like structures. Nucleic Acids Res. 2014; 42:8648–8662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Prakash T.P., Graham M.J., Yu J., Carty R., Low A., Chappell A., Schmidt K., Zhao C., Aghajan M., Murray H.F. et al.. Targeted delivery of antisense oligonucleotides to hepatocytes using triantennary N-acetyl galactosamine improves potency 10-fold in mice. Nucleic Acids Res. 2014; 42:8796–8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Prakash T.P., Yu J., Migawa M.T., Kinberger G.A., Wan W.B., Ostergaard M.E., Carty R.L., Vasquez G., Low A., Chappell A. et al.. Comprehensive structure-activity relationship of triantennary N-Acetylgalactosamine conjugated antisense oligonucleotides for targeted delivery to hepatocytes. J. Med. Chem. 2016; 59:2718–2733. [DOI] [PubMed] [Google Scholar]
- 34. Ostergaard M.E., Southwell A.L., Kordasiewicz H., Watt A.T., Skotte N.H., Doty C.N., Vaid K., Villanueva E.B., Swayze E.E., Bennett C.F. et al.. Rational design of antisense oligonucleotides targeting single nucleotide polymorphisms for potent and allele selective suppression of mutant Huntingtin in the CNS. Nucleic Acids Res. 2013; 41:9634–9650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ostergaard M.E., Nichols J., Dwight T.A., Lima W., Jung M.E., Swayze E.E., Seth P.P.. Fluorinated nucleotide modifications modulate allele selectivity of SNP-Targeting antisense oligonucleotides. Mol. Ther. Nucleic Acids. 2017; 7:20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nowotny M., Gaidamakov S.A., Ghirlando R., Cerritelli S.M., Crouch R.J., Yang W.. Structure of human RNase H1 complexed with an RNA/DNA hybrid: insight into HIV reverse transcription. Mol. Cell. 2007; 28:264–276. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.