Abstract
The replication protein A (RPA)1-4 family are single-stranded DNA-binding proteins that are essential components of DNA replication, repair and recombination, and cell cycle regulation. The present study aimed to evaluate the prognostic value of the RPA family members in patients with gastric cancer (GC), using datasets retrieved from the Oncomine public database. Datasets were retrieved for the purpose of comparing the RPA expression levels between GC and normal tissues. Additionally, Kaplan-Meier analysis was used to compare the overall survival (OS) times of GC patients that expressed different levels of RPA proteins. RPA1, 2, and 3 expression levels were all significantly upregulated in gastric intestinal-type, diffuse gastric, and gastric mixed adenocarcinomas, compared with those in normal mucosal tissues. Moreover, high mRNA expression levels of RPA3 and 4 predicted poorer OS times in all GCs, as well as patients with human epidermal growth factor receptor 2-negative and -positive GC. The high-risk group, separated by RPA signature, showed a poorer outcome than the low-risk group. RPA3 was the most strongly correlated with CD4+ T-cell levels. In conclusion, RPAs are novel prognostic indicators in GC, and can also predict the features of immunological diseases. Future experimental investigation into the roles of RPAs concerning the pathogenesis and development of GC may provide a novel biomarker or therapeutic target, improving the prognosis of patients with GC.
Keywords: gastric cancer, RPA, prognosis, KM plotter, Oncomine
Introduction
Gastric cancer (GC) remains one of the leading causes of cancer-related mortality in Eastern Asia, Europe and South America (1). In fact, >70% of GC patients are diagnosed at an advanced disease stage (2), despite the discovery of numerous chemotherapeutics [including fluorouracil (FU)/leucovorin/oxaliplatin], which have improved the treatment of patients with advanced GC, the overall prognosis remains poor (3). Novel prognostic indicators are urgently required to facilitate improvements in the treatment and diagnosis of GC.
Replication protein A (RPA) is a single-stranded DNA-binding gene family consisting of four members, RPA1-4 (4). RPAs play an essential role in DNA replication, repair, recombination and cell-cycle regulation (4–8). Previously, RPAs were thought to serve as prognostic biomarkers in several tumor types (5–8). Specifically, high protein expression levels of RPA1 (detected using immunohistochemistry) were associated with poorer outcomes in patients with esophageal carcinoma, compared with those with normal levels of expression (5).
Moreover, RPA2 was identified as an independent prognostic indicator of astrocytic tumors (6). In addition, decreased expression of both RPA1 and 2 resulted in an adverse prognosis for patients with muscle-invasive urothelial carcinoma and colon cancer (7,8). Nonetheless, the specific prognostic values of RPAs in GC are yet to be fully determined. The present in silico study characterized both the prognostic and immunological potential of RPAs in GC, using bioinformatics strategies and public online resources.
Materials and methods
Oncomine database analysis
The Student's t-test was used to compare the differences in the expression levels of RPAs between GC and normal control tissues, using three datasets (GSE13911, GSE13861 and PMID:19081245) (9–11) retrieved from the Oncomine online database (https://www.oncomine.org). P<0.01 and a fold-change >2 were selected as cut-off values and considered to indicate statistically significant differences (12).
Kaplan-Meier (KM) analysis
To investigate the prognostic value of RPA family mRNA expression levels in patients with GC, KM analysis was performed (www.kmplot.com) (13) to evaluate the differences in overall survival (OS) time between the high- and low-expression groups. The hazard ratio (HR) with 95% confidence interval (CI), and log-rank P-values were calculated, and are presented on each KM survival plot. P<0.05 was considered to indicate a statistically significant difference. The following datasets were retrieved from the Gene Expression Omnibus: GSE14210 (n=146) (14), GSE15459 (n=200) (15), GSE22377 (n=43) (16), GSE29272 (n=268) (17), GSE51105 (n=94) (18) and GSE62254 (n=300) (19). However, according to the recommendation of the KM plotter administrators, GSE62254 was excluded from KM analysis due to markedly different survival times and expression profiles, compared with the other datasets (13). No other inclusion or exclusion criteria were specified. The mRNA expression profiles of RPA family members 1–4 were collected from each dataset, and normalization procedures were performed (13). The optimal significance cut-off value between the high- and low-expression groups was calculated based on the imbedded algorithm of the KM plotter (13).
Determination of the prognostic value of the RPA family signature using SurvExpress
The prognostic value of the RPA family signature was evaluated with STAD datasets retrieved from The Cancer Genome Atlas (TCGA), via bioinformatics analysis using the SurvExpress biomarker validation tool (http://bioinformatica.mty.itesm.mx:8080/Biomatec/SurvivaX.jsp) (20). A maximized risk score algorithm was used to categorize the data into thigh- and low-risk groups.
Tumor-immunological features of RPAs in the Tumor Immune Estimation Response (TIMER)
The correlation between RPA expression and tumor immune infiltrating cell (TIIC; B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages and dendritic cells) activity was analyzed via the TIMER platform (https://cistrome.shinyapps.io/timer/) (21,22), which is a comprehensive resource used for the systematic evaluation of immunological features, based on the datasets retrieved from TGCA (21,22). The correlation between the expression of each gene and TIICs was determined using the purity-corrected partial Spearman's rank correlation coefficient. Negative association with tumor purity indicated high expression in the microenvironment (21,22).
Results
mRNA expression levels of RPA1-4 in GC tissues
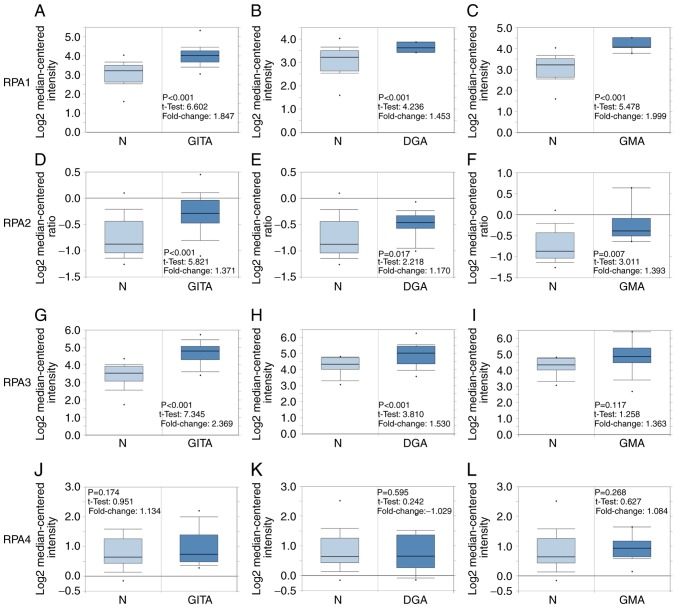
The relative mRNA expression levels of RPA1-4 in GC tissues was elucidated by comparing data on GC and normal tissues, retrieved from the Oncomine database. The mRNA expression levels of RPA1 and 2 in gastric intestinal type adenocarcinoma (GITA), diffuse gastric adenocarcinoma (DGA) and gastric mixed adenocarcinoma (GMA) were all significantly higher than those in normal gastric mucosal tissues (Fig. 1). The mRNA expression levels of RPA3 in gastric intestinal type adenocarcinoma and diffuse gastric adenocarcinoma were higher compared with those in normal gastric mucosal tissues, whereas no significance was observed in gastric mixed adenocarcinoma. However, RPA4 did not exhibit significantly differential expression between the GC and normal tissue groups.
Figure 1.
RPA family analysis in patients with GC. Box plots comparing the expression of specific RPA family members in normal and GC tissues, based on datasets retrieved from the Oncomine database. (A-C) Comparison of RPA1 mRNA expression levels between normal tissues and those in (A) GITA, (B) DGA and (C) GMA. Comparison of RPA2 mRNA expression between normal and (D) GITA, (E) DGA and (F) GMA tissues. (G-I) Comparison of RPA3 mRNA expression between normal and (G) GITA, (H) DGA and (I) GMA tissues. Comparison of RPA4 mRNA expression between normal and (J) GITA, (K) DGA and (L) GMA tissues. GC, gastric cancer; GITA, gastric intestinal type adenocarcinoma; DGA, diffuse gastric adenocarcinoma; GMA, gastric mixed adenocarcinoma; RPA, replication protein A.
Prognostic values of RPA1-4 in GC
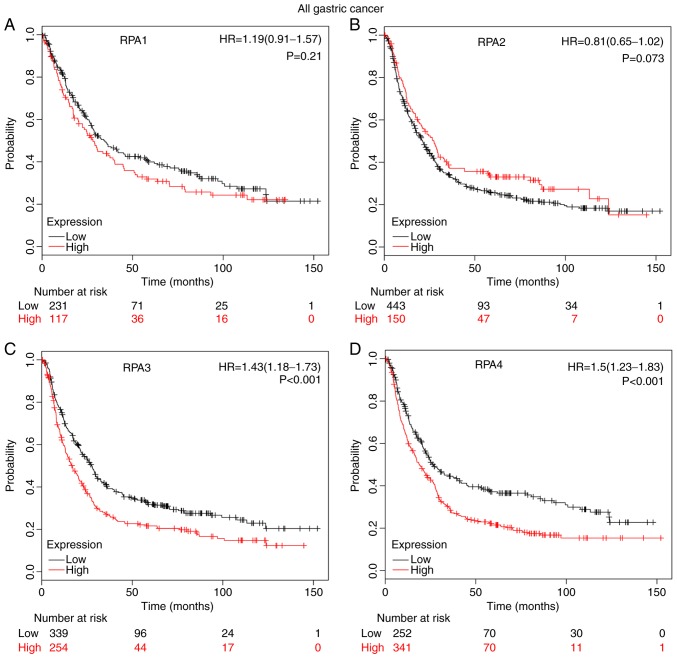
High mRNA expression levels of RPA3 (Fig. 2C; HR, 1.43; 95% CI, 1.18–1.73; P=2.9×10−4) and RPA4 (Fig. 2D; HR=1.5; 95% CI, 1.23–1.83; P=7×10−5) were significantly correlated with a poor prognosis in patients with GC. By contrast, RPA1 (Fig. 2A; P=0.21) and RPA2 (Fig. 2B, P=0.073) did not exert a significant correlation between high-expression level and poor prognosis.
Figure 2.
Prognostic value of individual RPA family members in patients with GC, determined using Kaplan-Meier analysis. (A) Survival curve of RPA1 (Affymetrix ID: 236675_at), high (n=117) vs. low expression group (n=231). (B) Survival curve of RPA2 (Affymetrix ID: 201756_at), high (n=150) vs. low expression group (n=443). (C) Survival curve of RPA3 (Affymetrix ID: 209507_at), high (n=254) vs. low expression group (n=339). (D) Survival curve of RPA4 (Affymetrix ID: 221143_at), high (n=341) vs. low expression group (n=252). GC, gastric cancer; RPA, replication protein A; HR, hazard ratio.
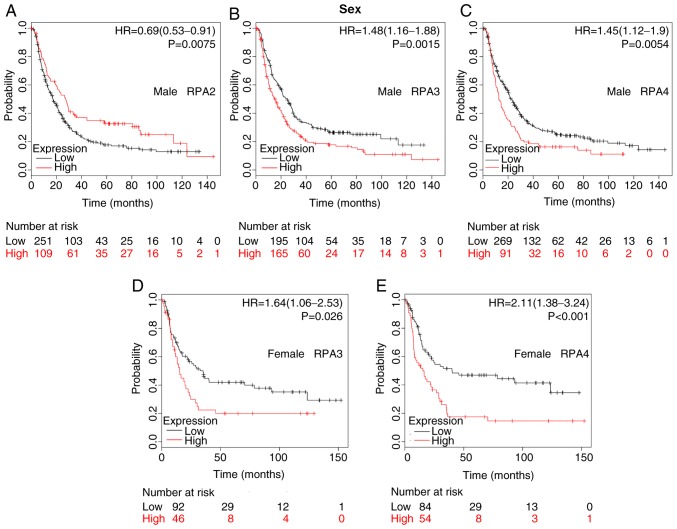
The association between the prognostic values of RPA1-4 and various clinicopathological features (including sex, HER2 expression, differentiation, Lauren classification, treatments and TNM stage) was also evaluated following stratification into subsets. The difference in prognostic value between the two sexes is detailed in Fig. 3. High RPA2 mRNA expression was associated with improved OS in male patients (Fig. 3A; HR, 0.69; 95% CI, 0.53–0.91; P=0.0075). Furthermore, high mRNA expression of RPA3 and RPA4 were significantly associated with a poorer prognosis in both male and female patients with GC (Fig. 3B-E).
Figure 3.
Differences in the prognostic values of RPA expression levels in male and female patients. Kaplan-Meier survival plots of patients with GC divided into those with high- and low-expression levels of RPA family members. (A) RPA2 (Affymetrix ID: 201756_at) in males; high-(n=109) vs. low-expression group (n=251). (B) RPA3 (Affymetrix ID: 209507_at) in males; high-(n=165) vs. low-expression group (n=195). (C) RPA4 (Affymetrix ID: 221143_at) in males; high-(n=91) vs. low-expression group (n=269). (D) RPA3 (Affymetrix ID: 209507_at) in females; high-(n=46) vs. low-expression group (n=92). (E) RPA4 (Affymetrix ID: 221143_at) in females, high-(n=54) vs. low-expression group (n=84). GC, gastric cancer; RPA, replication protein A; HR, hazard ratio.
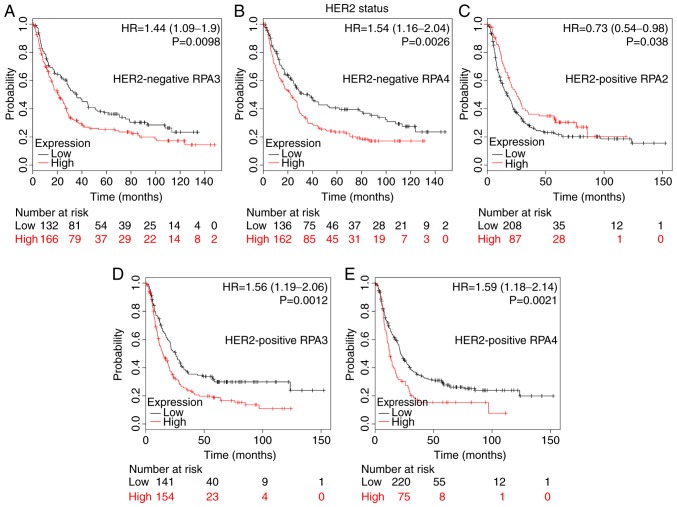
HER2 status analysis determined that a high RPA2 mRNA expression level was associated with improved OS time in HER2-positive patients (Fig. 4C; HR, 0.73; 95% CI, 0.54–0.98; P=0.038). By contrast, high RPA3 and 4 mRNA expression were significantly associated with a poor prognosis in both HER2-negative and -positive patients with GC (Fig. 4A, P=0.0098; Fig. 4B, P=0.0026; Fig. 4D, P=0.0012; Fig. 4E, P=0.0021).
Figure 4.
The prognostic value of RPA family expression levels in HER2 subsets of GC patients. Kaplan-Meier survival plots of patients with GC divided into those with high- and low-expression levels of RPA family members. (A) RPA3 (Affymetrix ID: 209507_at) in HER2-negative patients, high-(n=166) vs. low-expression group (n=132). (B) RPA4 (Affymetrix ID: 221143_at) in HER2-negative patients, high-(n=162) vs. low-expression group (n=136). (C) RPA2 (Affymetrix ID: 201756_at) in HER2-positive patients, high-(n=87) vs. low-expression group (n=208). (D) RPA3 (Affymetrix ID: 209507_at) in HER2-positive patients, high-(n=154) vs. low-expression group (n=141). (E) RPA4 (Affymetrix ID: 221143_at) in HER2-positive patients; high-(n=75) vs. low-expression group (n=220). GC, gastric cancer; RPA, replication protein A; HR, hazard ratio; HER2, human epidermal growth factor receptor 2.
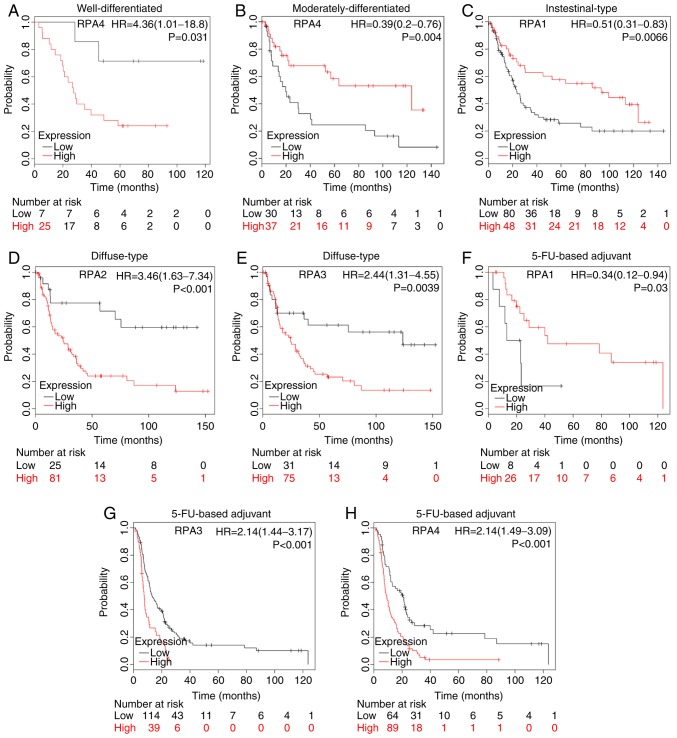
High RPA4 mRNA expression was significantly correlated with poorer OS in GC patients with well-differentiated tumors (Fig. 5A; HR, 4.36; 95% CI, 1.01–18.8; P=0.031). However, the high-expression GC group with moderately-differentiated tumors were also significantly associated with better prognosis (Fig. 5B; HR, 0.39; 95% CI, 0.2–0.76; P=0.004).
Figure 5.
Prognostic value of RPA family expression levels in various clinicopathological subsets of patients with GC. Survival curves of RPA4 (Affymetrix ID: 221143_at) are plotted for (A) well-differentiated tumors, high-(n=25) vs. low-expression group (n=7); and (B) moderately-differentiated tumors, high-(n=37) vs. low-expression group (n=30). KM survival curve showing (C) RPA1 (Affymetrix ID: 236675_at) expression in patients with intestinal-type GC; high-(n=48) vs. low-expression group (n=80). KM plot of patients with diffuse type GC analyzing (D) RPA2 (Affymetrix ID: 201756_at); high-(n=81) vs. low-expression group (n=25). (E) RPA3 (Affymetrix ID: 209507_at), high-(n=75) vs. low-expression group (n=31). KM survival curves comparing high and low RPA family expression in patients receiving 5-FU-based adjuvant therapy. (F) RPA1 (Affymetrix ID: 236675_at); high-(n=26) vs. low-expression group (n=8). (G) RPA3 (Affymetrix ID: 209507_at); high-(n=39) vs. low-expression group (n=114). (H) RPA4 (Affymetrix ID: 221143_at); high-(n=89) vs. low-expression group (n=64). GC, gastric cancer; RPA, replication protein A; HR, hazard ratio; KM, Kaplan-Meier; FU, fluorouracil.
A high-expression level of RPA1 was significantly correlated with improved prognosis in patients with intestinal-type GC (Fig. 5C; HR, 0.51; 95% CI, 0.31–0.83; P=0.0066), whilst high-expression levels of RPA2 and 3 were significantly associated with poor survival in patients with diffuse-type GC (Fig. 5D, P=6.4×10−4; Fig. 5E, P=0.0039).
High mRNA expression of RPA1 was significantly associated with an improved prognosis in patients with GC receiving 5-FU-based adjuvant chemotherapy (Fig. 5F; HR, 0.34; 95% CI, 0.12–0.94; P=0.03). By contrast, high RPA3 and 4 mRNA expression was significantly associated with poor OS time in GC patients that had received 5-FU-based adjuvant chemotherapy (Fig. 5G, P=1×10−4; Fig. 5H, P=2.9×10−5).
Subset analysis of tissues from patients at different TNM stages indicated that patients with high RPA1 mRNA expression levels with stage I GC, and high RPA4 mRNA expression in tissues from stage IV patients, were both significantly associated with a favorable prognosis (Table I). Conversely, high RPA1 and 4 mRNA expression levels in stage II patients, and high RPA3 mRNA expression levels in stage III patients were both significantly associated with a poor prognosis relative to the low-expression groups. High mRNA expression of RPA2 and 4 correlated with favorable OS times in lymph node (LN)-negative patients. RPA2 and 3 mRNA expression was found to be associated with an unfavorable OS in LN-positive patients, while RPA4 was found to be associated with favorable OS in LN-positive patients. Regarding metastatic status, high RPA3 mRNA expression was found to be associated with poorer OS times in M0 patients. Additionally, high RPA 4 mRNA expression correlated with improved OS times in GC patients at the M0 metastatic stage (Table I).
Table I.
Association of RPA family mRNA expression with various clinical stages in patients with GC.
| Clinical stage | RPA | Cases, n | Hazard ratio (95% CI) | P-value |
|---|---|---|---|---|
| TNM stage | ||||
| I | RPA1 | 34 | 0.29 (0.08–1.05) | 0.045a |
| RPA2 | 39 | 0.52 (0.17–1.56) | 0.230 | |
| RPA3 | 39 | 0.61 (0.2–1.87) | 0.380 | |
| RPA4 | 39 | 0.41 (0.14–1.21) | 0.094 | |
| II | ||||
| RPA1 | 44 | 313921208.06 (0-inf) | 0.007a | |
| RPA2 | 49 | 1.64 (0.7–3.87) | 0.250 | |
| RPA3 | 49 | 0.68 (0.29–1.59) | 0.380 | |
| RPA4 | 49 | 2.45 (1.02–5.89) | 0.039a | |
| III | ||||
| RPA1 | 109 | 0.77 (0.48–1.24) | 0.280 | |
| RPA2 | 217 | 0.77 (0.56–1.07) | 0.120 | |
| RPA3 | 217 | 1.43 (1.03–1.99) | 0.032a | |
| RPA4 | 217 | 1.26 (0.87–1.81) | 0.210 | |
| IV | ||||
| RPA1 | 66 | 1.36 (0.74–2.51) | 0.330 | |
| RPA2 | 74 | 1.24 (0.71–2.19) | 0.450 | |
| RPA3 | 74 | 0.63 (0.34–1.15) | 0.130 | |
| RPA4 | 74 | 0.50 (0.27–0.9) | 0.020a | |
| LN (−) | ||||
| RPA1 | 38 | 1.87 (0.68–5.18) | 0.220 | |
| RPA2 | 38 | 0.34 (0.13–0.92) | 0.027a | |
| RPA3 | 38 | 0.43 (0.15–1.27) | 0.120 | |
| RPA4 | 38 | 0.32 (0.11–0.94) | 0.030a | |
| LN (+) | ||||
| RPA1 | 175 | 1.40 (0.89–2.2) | 0.140 | |
| RPA2 | 175 | 1.83 (1.2–2.78) | 0.004a | |
| RPA3 | 175 | 1.53 (1.05–2.22) | 0.027a | |
| RPA4 | 175 | 0.50 (0.34–0.72) | <0.001a | |
| M0 | ||||
| RPA1 | 186 | 0.69 (0.47–1.02) | 0.064 | |
| RPA2 | 186 | 0.70 (0.44–1.11) | 0.130 | |
| RPA3 | 186 | 1.72 (1.1–2.71) | 0.017a | |
| RPA4 | 186 | 0.67 (0.45–0.98) | 0.038a | |
| M1 | ||||
| RPA1 | 31 | 1.60 (0.73–3.49) | 0.240 | |
| RPA2 | 31 | 1.80 (0.66–4.89) | 0.240 | |
| RPA3 | 31 | 0.42 (0.16–1.06) | 0.059 | |
| RPA4 | 31 | 0.54 (0.24–1.2) | 0.130 |
P<0.05. RPA, replication protein A; LN, lymph node; M0, no metastasis; M1, metastasis to different organs.
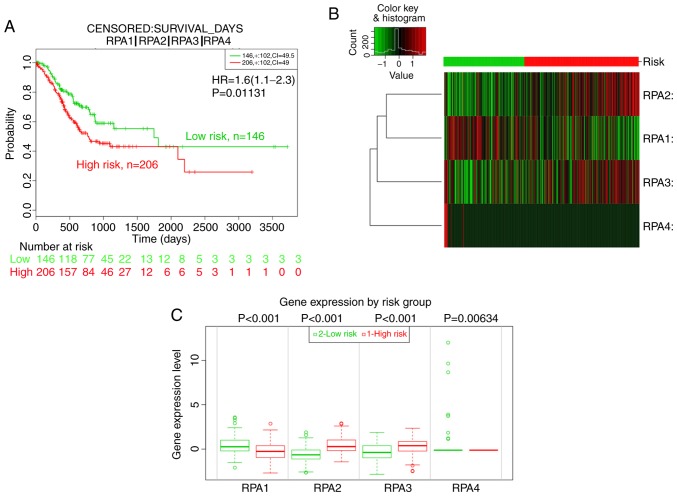
Prognostic value of RPA family signatures
Given the increasing focus on the application of gene signatures for prognostic analysis, the prognostic value of the RPA family signature was further assessed using SurvExpress. Divided by the maximized risk group algorithm, the high-risk group (n=206) showed a poorer outcome than the low-risk group (n=146; HR, 1.6; 95% CI, 1.11–2.30; P=0.0113; Fig. 6A-C). In addition, RPA2 and 3 mRNA expression in the high-risk groups was significantly elevated compared with those of the low-risk groups, whist RPA1 expression was lower in the high-risk groups (Fig. 6B and C).
Figure 6.
Prognostic analysis of the RPA family signature using the SurvExpress platform. (A) KM analysis of the OS between high-(n=206, red) and low-risk (n=146, green) groups using data from TCGA. (B) Heat map showing RPA expression levels (red: High risk; green: Low risk). (C) mRNA expression level of RPA family member proteins between high- and low-risk groups. GC, gastric cancer; RPA, replication protein A; KM, Kaplan-Meier; TCGA, The Cancer Genome Atlas; OS, overall survival.
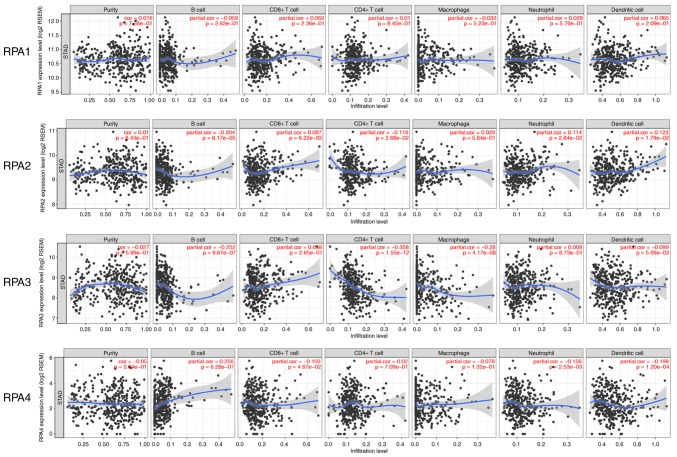
Correlation between RPA expression and TIICs
To further examine the immunological influence of RPAs in GC tissues, the TIMER platform was utilized to analyze the correlation between RPAs and the prevalence of various immune infiltrating cell types. RPA3 expression exhibited a significant correlation with CD4+ T cells (partial.cor=−0.358, P=1.55×10−12; Fig. 6). As presented in Fig. 7, no significant correlation was observed between RPA1 and the tested cell types. In RPA2, significant correlations were identified with B cells, CD4+ T cells, neutrophils and dendritic cells (partial.cor=−0.204, P=8.17×10−5; partial.cor=−0.116, P=2.68×10−2; partial.cor=0.114, P=2.84×10−2 and partial.cor=0.123, P=1.79×10−2, respectively). In RPA3, significant correlations were identified with B cells, CD4+ T cells and macrophages (partial.cor=−0.252, P=9.67×10−7; partial.cor=−0.358, P=1.55×10−12 and partial.cor=−0.28, P=4.17×10−8, respectively). In RPA4, significant correlations were identified with B cells, CD8+ T cells, neutrophils and dendritic cells (partial.cor=0.256, P=6.28×10−7; partial.cor=−0.103, P=4.67×10; partial.cor=−0.156, P=2.53×10−3 and partial.cor=−0.198, P=1.20×10−4, respectively) (Fig. 7).
Figure 7.
Correlation between RPA family expression levels and TIICs. The correlation of RPAs and various TIIC types (B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages and dendritic cells) was analyzed and plotted. RPA, replication protein A; TIIC, tumor-infiltrating immune cells; RSEM, accurate quantification of gene and isoform expression of target gene from RNA-seq data; STAD, stomach adenocarcinoma.
Discussion
The majority of studies on RPAs have focused on their structure, function and mechanism of action. The members of the RPA family are essential components of multiple pathways that influence DNA replication, recombination, repair and damage signaling (4–8,23,24). Clinical studies have determined that RPAs are upregulated in various cancer types, including hepatocellular and esophageal carcinoma, and colon and bladder cancer (5,7–8,25–27), and that this frequently correlates with poor prognosis (28–30). Nonetheless, the influence of RPAs on GC progression is yet to be elucidated. To the best of our knowledge, the present study represents the first comprehensive analysis of the prognostic potential and immunological influence of RPAs in GC. The present results indicate that the expression levels of RPA1, 2 and 3 mRNA were all significantly higher in GC, compared with normal tissues. Furthermore, high mRNA expression levels of RPA3 and 4 were significantly associated with unfavorable OS time in patients with GC.
Notably, the present study investigated the effects of different RPAs on various clinicopathological features using subgroup stratification and OS analysis. HER2 expression status (positive/negative) in GC patients is a well-characterized risk factor for GC progression. High expression levels of HER2 in GC have resulted in the development of trastuzumab, a drug that specifically targets HER2 upregulation. Moreover, HER2 status is widely used in subgroup analysis. To the best of our knowledge, the relationship between HER2 and the OS times of patients with high or low RPA expression has not yet been clarified in gastric cancer. The results of the present study may provide insights into the development of individualized therapeutic strategies.
In the present study, high expression levels of RPA1 mRNA were significantly associated with improved prognosis in subgroups of patients with i) intestinal-type GC; ii) 5-FU-based adjuvant chemotherapy treatment; and iii) a stage I tumor. However, high RPA1 mRNA expression predicted significantly poorer survival outcome in GC patients with stage II tumors.
A high level of RPA2 mRNA expression was significantly associated with a favorable OS rate in male patients with GC, as well as patients with N-stage disease, while indicating an unfavorable OS rate in patients with diffuse-type, as well as N+ stage disease. Previous reports have highlighted the value of RPA2 as an indicator of poor prognosis in numerous malignancies, including astrocytic tumor, colon cancer and muscle-invasive urothelial carcinomas (6–8). Mechanistically, hyperphosphorylation of RPA2 (occurring in response to DNA damage) was associated with single-stranded DNA and Rab51, and mutant phosphorylation-deficient RPA2 could increase chromosomal aberration (31). However, the biological roles of RPA2, in terms of tumor invasion, metastasis and histological differentiation, have not been fully characterized. Therefore, based on its intrinsic correlation, it is possible that RPA2 may be either a poor or favorable prognostic indicator in different subset analyses.
Notably, in the present study, high RPA2 expression predicted HER2-positive status in the GC group. This suggests that RPA2 may inhibit tumor cell invasion or proliferation by interacting with the HER2 signaling pathway. However, further clinical validation of the prognostic value of RPA2 is required.
High RPA3 mRNA expression levels were significantly associated with poor OS in patients with GC, indicating the potential prognostic value of RPA3 in GC. The results of the present study were consistent with a previous study (32), which reported that RPA3 expression was upregulated in GC compared with normal tissues, and also predicted poor patient survival rate. Therefore, RPA3 may be useful as a potential biomarker to predict the OS of patients with GC. Moreover, the current study illustrated that a high mRNA expression level of RPA3 indicated poor OS in male and female, HER2-negative and positive, M0, N+ and stage III patients. Patients with diffuse-type GC, and those receiving 5-FU-based adjuvant treatment also exhibited poorer outcomes when RPA3 was upregulated.
RPA4, a human homolog of RPA2, is involved in maintaining the genomic integrity of the cell (4). Previous studies have reported that RPA4 possesses 47% structural homology with RPA2, and that it interacts with both RPA1 and 3 (33). However, characterization of the role of RPA4 expression in various malignancies remains limited. In the present study, high RPA4 mRNA expression was significantly associated with poor OS in patients with GC, particularly in well-differentiated, male and female, HER2-negative and positive patients, in addition to those receiving 5-FU based adjuvant treatment, and those with TNM stage II disease. Conversely, higher RPA4 expression levels were correlated with improved prognosis in patients at stage IV, N- and + stage, and M0 stage. Increased RPA4 expression correlated with a significantly high risk of a lower OS time in all patients with GC. However, during subset analysis; in patients with a well-differentiated tumor, RPA4 upregulation represented only a mildly significantly high risk of a lower OS. Nonetheless, patients with a moderately differentiated tumor and high RPA4 expression represented a significantly low risk factor for OS. By contrast, in patients that presented with lymph node-positive and -negative cases, stage IV and M0, RPA4 was determined to be a favorable indicator of OS.
This apparent contradiction may be explained by the low population size of each subset. Similar research conducted in the future would be more reliable if univariate/multivariate cox regression analysis was incorporated. Mechanistically, RPA4 may be associated with tumor progression, lymph node metastasis and histological differentiation. Particularly given the contradictory results for RPA4 in the lymph node positive/negative and the general group, which included all patients with GC regardless of the subtype. It may be worth investigating the association between RPA4 expression and lymph node metastasis in a larger cohort.
Correlation analysis was subsequently performed to investigate the associations between TIICs and the expression levels of RPA family members. Intriguingly, B cells were highly correlated with RPA2, 3 and 4 expression, indicating the interaction between B cells and the immune features of the RPA family. To date, and to the best of our knowledge, this is the first study that has investigated RPA-associated TIICs as well as their prognostic values in GC.
The present study had certain limitations; it was solely based on bioinformatics analysis and would benefit from experimental or clinical validation. Subsequent investigations should focus on validation of the prognostic value of the RPA family members. Future investigations require a larger cohort of samples to validate the clinical significance of RPAs in GC prognosis, as well as functional and mechanistic exploration. Several directions for future research have been made apparent. Firstly, the prognostic value of the mRNA/protein expression levels of RPAs using GC tissue samples. Secondly, the prognostic value of the RPA signature may represent a cost-effective biomarker. Thirdly, the correlation between RPA family members and TIICs requires further in-depth characterization.
In conclusion, RPA1, 2 and 3 mRNA expression levels are significantly higher in GC vs. normal tissues. Furthermore, high mRNA expression levels of RPA3 and 4 are associated with poor prognosis in patients with GC. RPAs should therefore be considered for use as potential prognostic biomarkers of GC progression.
Acknowledgements
The authors would like to thank Dr. Ernest Johann Helwig (Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology) for his contribution to the manuscript revision and general guidance.
Glossary
Abbreviations
- RPA
replication protein A
- KM
Kaplan-Meier
- OS
overall survival
- HER2
human epidermal growth factor receptor 2
- TIMER
tumor immune estimation response
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
YZ and CY performed the data analysis, drafted the manuscript, participated in the study design and data collection, and read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Song Z, Wu Y, Yang J, Yang D, Fang X. Progress in the treatment of advanced gastric cancer. Tumour Biol. 2017;39:1010428317714626. doi: 10.1177/1010428317714626. [DOI] [PubMed] [Google Scholar]
- 3.Mayer RJ, Venook AP, Schilsky RL. Progress against GI cancer during the American society of clinical oncology's first 50 years. J Clin Onco. 2014;32:1521–1530. doi: 10.1200/JCO.2014.55.4121. [DOI] [PubMed] [Google Scholar]
- 4.Haring SJ, Humphreys TD, Wold MS. A naturally occurring human RPA subunit homolog does not support DNA replication or cell-cycle progression. Nucleic Acids Res. 2010;38:846–858. doi: 10.1093/nar/gkp1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dahai Y, Sanyuan S, Hong L, Di Z, Chong Z. A relationship between replication protein A and occurrence and prognosis of esophageal carcinoma. Cell Biochem Biophys. 2013;67:175–180. doi: 10.1007/s12013-013-9530-y. [DOI] [PubMed] [Google Scholar]
- 6.Kanakis D, Levidou G, Gakiopoulou H, Eftichiadis C, Thymara I, Fragkou P, Trigka EA, Boviatsis E, Patsouris E, Korkolopoulou P. Replication protein A: A reliable biologic marker of prognostic and therapeutic value in human astrocytic tumors. Hum Pathol. 2011;42:1545–1553. doi: 10.1016/j.humpath.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 7.Levidou G, Gakiopoulou H, Kavantzas N, Saetta AA, Karlou M, Pavlopoulos P, Thymara I, Diamantopoulou K, Patsouris E, Korkolopoulou P. Prognostic significance of replication protein A (RPA) expression levels in bladder urothelial carcinoma. BJU Int. 2011;108:E59–E65. doi: 10.1111/j.1464-410X.2010.09828.x. [DOI] [PubMed] [Google Scholar]
- 8.Givalos N, Gakiopoulou H, Skliri M, Bousboukea K, Konstantinidou AE, Korkolopoulou P, Lelouda M, Kouraklis G, Patsouris E, Karatzas G. Replication protein A is an independent prognostic indicator with potential therapeutic implications in colon cancer. Mod Pathol. 2007;20:159–166. doi: 10.1038/modpathol.3800719. [DOI] [PubMed] [Google Scholar]
- 9.D'Errico M, de Rinaldis E, Blasi MF, Viti V, Falchetti M, Calcagnile A, Sera F, Saieva C, Ottini L, Palli D, et al. Genome-wide expression profile of sporadic gastric cancers with microsatellite instability. Eur J Cancer. 2009;45:461–469. doi: 10.1016/j.ejca.2008.10.032. [DOI] [PubMed] [Google Scholar]
- 10.Chen X, Leung SY, Yuen ST, Chu KM, Ji J, Li R, Chan AS, Law S, Troyanskaya OG, Wong J, et al. Variation in gene expression patterns in human gastric cancers. Mol Biol Cell. 2003;14:3208–3215. doi: 10.1091/mbc.e02-12-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho JY, Lim JY, Cheong JH, Park YY, Yoon SL, Kim SM, Kim SB, Kim H, Hong SW, Park YN, et al. Gene expression signature-based prognostic risk score in gastric cancer. Clin Cancer Res. 2011;17:1850–1857. doi: 10.1158/1078-0432.CCR-10-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lánczky A, Nagy Á, Bottai G, Munkácsy G, Szabó A, Santarpia L, Győrffy B. miRpower: A web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat. 2016;160:439–446. doi: 10.1007/s10549-016-4013-7. [DOI] [PubMed] [Google Scholar]
- 14.Kim HK, Choi IJ, Kim CG, Kim HS, Oshima A, Michalowski A, Green JE. A gene expression signature of acquired chemoresistance to cisplatin and fluorouracil combination chemotherapy in gastric cancer patients. PLoS One. 2011;6:e16694. doi: 10.1371/journal.pone.0016694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ooi CH, Ivanova T, Wu J, Lee M, Tan IB, Tao J, Ward L, Koo JH, Gopalakrishnan V, Zhu Y, et al. Oncogenic pathway combinations predict clinical prognosis in gastric cancer. PLoS Genet. 2009;5:e1000676. doi: 10.1371/journal.pgen.1000676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Förster S, Gretschel S, Jöns T, Yashiro M, Kemmner W. THBS4, a novel stromal molecule of diffuse-type gastric adenocarcinomas, identified by transcriptome-wide expression profiling. Mod Pathol. 2011;24:1390–1403. doi: 10.1038/modpathol.2011.99. [DOI] [PubMed] [Google Scholar]
- 17.Wang G, Hu N, Yang HH, Wang L, Su H, Wang C, Clifford R, Dawsey EM, Li JM, Ding T, et al. Comparison of global gene expression of gastric cardia and noncardia cancers from a high-risk population in China. PLoS One. 2013;8:e63826. doi: 10.1371/journal.pone.0063826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Busuttil RA, George J, Tothill RW, Ioculano K, Kowalczyk A, Mitchell C, Lade S, Tan P, Haviv I, Boussioutas A. A signature predicting poor prognosis in gastric and ovarian cancer represents a coordinated macrophage and stromal response. Clin Cancer Res. 2014;20:2761–2772. doi: 10.1158/1078-0432.CCR-13-3049. [DOI] [PubMed] [Google Scholar]
- 19.Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS, Liu J, Yue YG, Wang J, Yu K, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015;21:449–456. doi: 10.1038/nm.3850. [DOI] [PubMed] [Google Scholar]
- 20.Aguirre-Gamboa R, Gomez-Rueda H, Martínez-Ledesma E, Martínez-Torteya A, Chacolla-Huaringa R, Rodriguez-Barrientos A, Tamez-Peña JG, Treviño V. SurvExpress: An online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS One. 2013;8:e74250. doi: 10.1371/journal.pone.0074250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, Jiang P, Shen H, Aster JC, Rodig S, et al. Comprehensive analyses of tumor immunity: Implications for cancer immunotherapy. Genome Biol. 2016;17:174. doi: 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bochkarev A, Bochkareva E. From RPA to BRCA2: Lessons from single-stranded DNA binding by the OB-fold. Curr Opin in Struct Biol. 2004;14:36–42. doi: 10.1016/j.sbi.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 24.Zou Y, Liu Y, Wu X, Shell SM. Functions of human replication protein A (RPA): From DNA replication to DNA damage and stress responses. J Cell Physiol. 2006;208:267–273. doi: 10.1002/jcp.20622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, Yang T, Chen H, Li H, Zheng S. Oncogene RPA1 promotes proliferation of hepatocellular carcinoma via CDK4/Cyclin-D pathway. Biochem Biophys Res Commun. 2018;498:424–430. doi: 10.1016/j.bbrc.2018.02.167. [DOI] [PubMed] [Google Scholar]
- 26.Qu C, Zhao Y, Feng G, Chen C, Tao Y, Zhou S, Liu S, Chang H, Zeng M, Xia Y. RPA3 is a potential marker of prognosis and radioresistance for nasopharyngeal carcinoma. J Cell Mol Med. 2017;21:2872–2883. doi: 10.1111/jcmm.13200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao W, Zheng J, Zhou B, Pan L. Replication protein A 3 is associated with hepatocellular carcinoma tumorigenesis and poor patient survival. Dig Dis. 2018;36:26–32. doi: 10.1159/000478977. [DOI] [PubMed] [Google Scholar]
- 28.Jekimovs C, Bolderson E, Suraweera A, Adams M, O'Byrne KJ, Richard DJ. Chemotherapeutic compounds targeting the DNA double-strand break repair pathways: The good, the bad, and the promising. Front Oncol. 2014;4:86. doi: 10.3389/fonc.2014.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mishra AK, Dormi SS, Turchi AM, Woods DS, Turchi JJ. Chemical inhibitor targeting the replication protein A-DNA interaction increases the efficacy of Pt-based chemotherapy in lung and ovarian cancer. Biochem Pharmacol. 2015;93:25–33. doi: 10.1016/j.bcp.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patrone JD, Kennedy JP, Frank AO, Feldkamp MD, Vangamudi B, Pelz NF, Rossanese OW, Waterson AG, Chazin WJ, Fesik SW. Discovery of protein-protein interaction inhibitors of replication protein A. ACS Med Chem Lett. 2013;4:601–605. doi: 10.1021/ml400032y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi W, Feng Z, Zhang J, Gonzalez-Suarez I, Vanderwaal RP, Wu X, Powell SN, Roti Roti JL, Gonzalo S, Zhang J. The role of RPA2 phosphorylation in homologous recombination in response to replication arrest. Carcinogenesis. 2010;31:994–1002. doi: 10.1093/carcin/bgq035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dai Z, Wang S, Zhang W, Yang Y. Elevated expression of RPA3 is involved in gastric cancer tumorigenesis and associated with poor patient survival. Dig Dis Sci. 2017;62:2369–2375. doi: 10.1007/s10620-017-4696-6. [DOI] [PubMed] [Google Scholar]
- 33.Keshav KF, Chen C, Dutta A. Rpa4, a homolog of the 34-kilodalton subunit of the replication protein A complex. Mol Cell Biol. 1995;15:3119–3128. doi: 10.1128/MCB.15.6.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.