Abstract
Imbalance in the level of the pyrimidine degradation products dihydrouracil and dihydrothymine is associated with cellular transformation and cancer progression. Dihydropyrimidines are degraded by dihydropyrimidinase (DHP), a zinc metalloenzyme that is upregulated in solid tumors but not in the corresponding normal tissues. How dihydropyrimidine metabolites affect cellular phenotypes remains elusive. Here we show that the accumulation of dihydropyrimidines induces the formation of DNA–protein crosslinks (DPCs) and causes DNA replication and transcriptional stress. We used Xenopus egg extracts to recapitulate DNA replication invitro. We found that dihydropyrimidines interfere directly with the replication of both plasmid and chromosomal DNA. Furthermore, we show that the plant flavonoid dihydromyricetin inhibits human DHP activity. Cellular exposure to dihydromyricetin triggered DPCs-dependent DNA replication stress in cancer cells. This study defines dihydropyrimidines as potentially cytotoxic metabolites that may offer an opportunity for therapeutic-targeting of DHP activity in solid tumors.
INTRODUCTION
Adjustments in metabolic activities are required to satisfy high metabolic demands associated with cancer cell proliferation (1). Alterations in nucleotide metabolisms are emerging as distinctive features of cancers cells. While nucleotide bases may be limiting for cancer cells proliferation (2), accumulating evidence suggests that carcinogenesis is associated with metabolic rewiring of the pyrimidine catabolic pathway (3–6). Whether and how rewiring of the pyrimidine catabolic pathway supports tumor progression, however, remains poorly defined.
In humans, the pyrimidines uracil and thymine are degraded in three enzymatic steps (Figure 1A). First, dihydropyrimidine dehydrogenase (DPD) reduces the pyrimidine ring of uracil and thymine with hydrogen and yields 5, 6-dihydrouracil and 5, 6-dihydrothymine (dihydropyrimidines). Second, the saturated rings between position 3 and 4 are opened by dihydropyrimidinase (DHP). Third, β-ureidopropionase (BUP-1) degrades the β-ureidopropionic acid and β-ureidoisobutyric acid products formed by DHP into β-alanine and β-aminoisobutyric acid. DPD activity has been detected in all tissues examined, but the activity of DHP and BUP-1 is essentially restricted to the liver and the kidney (7). In cancer cells, however, the level of pyrimidine degradation activities is considerably altered. Increased expression of DPD has been observed in human hepatocellular carcinoma (8). Human skin cutaneous melanomas progressing toward metastatic tumors accumulate mutations in DPD and up-regulate the expression of the genes encoding DPD and DHP (6). The accumulation of 5, 6-dihydrouracil is a distinct metabolic feature of early lung adenocarcinoma (5). Intriguingly, an increase in the concentration of dihydropyrimidines in epithelial breast cancer cells supports the acquisition of aggressive mesenchymal characteristics (4). How dihydropyrimidines affect cellular phenotypes, however, remains elusive. Pioneering studies have identified DHP activity as a good marker of tumorigenicity and a target for cancer therapy (3). Whereas hardly detectable in normal extrahepatic and kidney tissues (van Kuilenburg et al., (7)), the activity of DHP is strikingly high in human carcinomas of the lung, colon, pancreas, salivary gland and stomach (3).
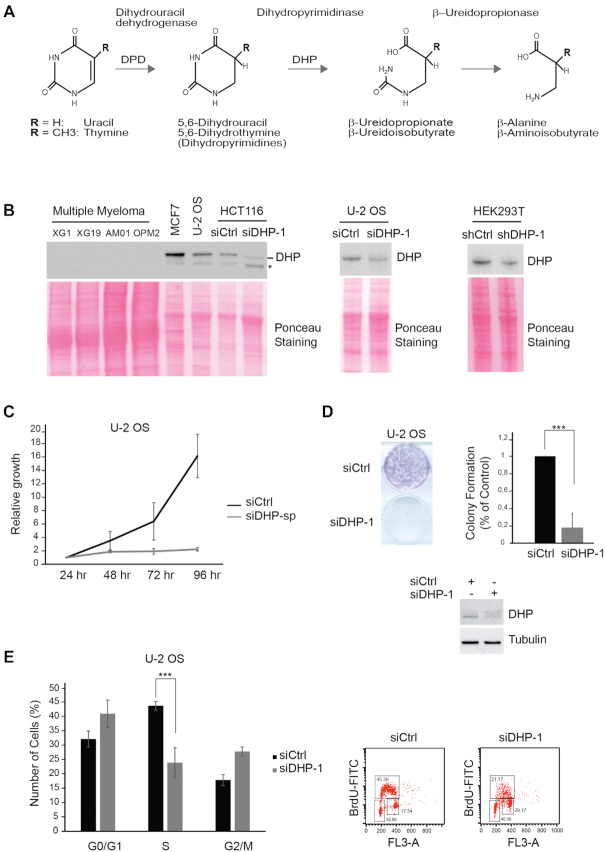
Figure 1.
Depletion of DHP impairs the proliferation of epithelial cancer cells. (A) Schematic representation of the pyrimidine degradation pathway. (B) DHP was probed by western blotting in the indicated transformed cells. When indicated, DHP was knocked down using anti-DHP siRNA or shRNA molecules with distinct target sequences. Ponceau staining was used as loading control. * non-specific signal. One representative experiment is shown from three to six biological replicates. (C) U-2 OS cells were transfected with control or anti-DHP siRNA (siDHP-sp) and their viability was assessed during 4 days using the MTT cell growth assay. Mean viability is representative of three independent biological replicates. Error bars represent ±S.D. (D) Colony-forming assay of U-2 OS cells after transfection with control or anti-DHP siRNA (siDHP-1). A representative image is shown. An histogram represents the quantification of colony formation. Data shown are averages over three independent biological replicates with two technical replicates for each. Error bars represent ±S.D. P-values were calculated using a regression model with Poisson distribution: ***P < 0.0001. Bottom panel: the efficiency of DHP knockdown was assessed by western blotting. (E) Histogram representing the percentage of U-2 OS cells, 72 h after transfection with control or anti-DHP siRNA (siDHP-1) in G0/G1, S and G2/M phases. Data shown are averages over three independent biological replicates. Error bars represent ±S.D. P-values were calculated using a regression model with Poisson distribution: ***P < 0.0001.
Another hallmark of cancer is DNA replication stress causing genomic instability (1,9). Stress in the context of DNA replication is defined as the slowing or stalling of DNA chain elongation (10). Known sources of DNA replication stress include nucleotides insufficiency (11), misincorporated nucleotides (12,13), replication/transcription conflicts (14–16) and DNA lesions caused by reactive metabolic products such as reactive oxygen species and aldehydes (17–19). Furthermore, cellular metabolites can yield structurally diverse DNA–protein crosslinks (DPCs) that precipitate the loss of cellular functions (20).
DNA replication stress in cancer cells is exploited therapeutically with the use of inhibitors of the DNA damage response (21,22). DNA replication stress induces the accumulation of 70–500 long nucleotide stretches of single-stranded DNA (23–25), which trigger a protein kinase cascade orchestrated by the checkpoint kinase ATR and its effector kinase Chk1 (26–29). ATR signaling promotes cell and organismal survival through coordination of DNA repair and DNA replication with cell physiological processes including cell-cycle progression and transcription (30).
We show here that suppression of DHP in cancer cell lines induces DNA replication stress, as revealed by the accumulation of single-stranded DNA, by the induction of ATR/Chk1 signaling and by the slowing of replication fork progression. Depletion of DHP also attenuates transcription activity, stabilizes p53 and eventually blocks cell proliferation. The addition of dihydropyrimidines to Xenopus egg-extracts induces the formation of abnormal DNA replication products. In DHP-depleted cells, DNA replication and transcriptional stress correlate with the accumulation of DPCs. Thus, we suggest that dihydropyrimidines yield DPCs that directly interfere with DNA-templated processes. We found that the flavonoid dihydromyricetin inhibits the activity of purified human DHP. Addition of dihydromyricetin in the cell culture medium induces the accumulation of DPCs and interferes with the progression of replication forks. These findings indicate that unless degraded by dihydropyimidinase, the amount of dihydropyrimidines produced in cancer cell cultures is sufficient to block DNA templated processes.
MATERIALS AND METHODS
Cell lines, plasmids and chemicals
U-2 OS, HEK293T and MCF7 were grown under standard conditions in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (P/S). HCT116 (Horizon Discovery Ltd.) were cultured in McCoy's 5A modified Medium (Sigma-Aldrich) supplemented with 10% FBS and 1% P/S. HEK293 cells were cultured in RPMI (Invitrogen) supplemented with 10% FBS and 1% P/S. HEK293 cells expressing wild-type and the translocase dead FANCM mutant protein are as described previously (31). XG1 and XG19 IL6 dependent human myeloma cell lines (HMCLs) were obtained as previously described (32). AMO-1 and OPM2 were purchased from DSMZ (Braunsweig, Germany). These HMCLs were routinely maintained in RPMI 1640 and 10% fetal calf serum (FCS; Biowittaker, Walkersville, MD), supplemented with 3 ng/ml IL-6 (Peprotech, Rocky Hill, NJ, USA) for IL6 dependent cell lines. HMCLs were authenticated according to their short tandem repeat profiling and their gene expression profiling using Affymetrix U133 plus 2.0 microarrays deposited in the ArrayExpress public database under accession numbers E-TABM-937 and E-TABM-1088. Dihydrouracil, Uracil, Dihydromyricetin, Aphidicolin and Roscovitine were purchased from Sigma-Aldrich. Formaldehyde was purchased from VWR chemicals. Embryomax nucleosides (100X) (cytidine, 0.73 g/l; guanosine, 0.85 g/l; uridine, 0.73 g/l; adenosine, 0.8 g/l; thymidine, 0.24 g/l) was purchased from Millipore. pDONR223-DPYS was obtained through MGC Montpellier Genetic Collections and cloned into destination vectors using gateway technology (Invitrogen).
Antibodies
Primary antibodies were purchased from Abcam (Histone H3, p53, DNA polymerase eta, nucleolin, FANCD2), Bethyl Laboratories (RPA32-Ser33, RPA32-Ser4/S8, DNA polymerase kappa, XPA, ERCC5/XPG), Calbiochem (RPA32), Cell Signaling Technology (Chk1-Ser345, Ubiquityl-PCNA (Lys164)), ElabScience (SPRTN, UPP1), ProteinTech Group (DPYS), Santa Cruz Biotechnology (Chk1, HA), Sigma-Aldrich (α-Tubulin, DPYD, PCNA). Xenopus polymerase η antibody was previously described (33). The Mouse anti-RNA-DNA hybrid S9.6 hybridoma was purchased from ATCC. Orc2 antibodies were kindly provided by Dr Marcel Mechali (Institute of Human Genetics). The FANCA antibody was a gift from the Fanconi anemia research fund and the anti-FANCM antibody was a gift from Weidong Wang (NIH). Secondary antibodies (anti-rabbit-HRP and anti-mouse-HRP) were from Promega
DHP bacterial protein expression and purification
DPYS was cloned into the pET-28a (+) (Novagen) vector containing an N-terminal 6×His tag. The protein was overexpressed in Escherichia coli BL21(DE3) host cells and induced by 1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Sigma-Aldrich) for 3 h in presence of 1 mM ZnCl2. Cells were lysed with Buffer A (50 mM Potassium Phosphate pH 7.5, 150 mM NaCl, 0.1% NP40, 15 mM Imidazol (Sigma-Aldrich)) and protease inhibitors (Roche). Extracts were incubated for 30 min at 4°C and harvested at 28 000 rpm for 1 h. The soluble supernatant fraction was purified on a 5 ml HisTrap HP column (GE Healthcare) using the AKTA protein purification system (GE Healthcare). The column was washed with 10 column volumes of Buffer A with 60 mM Imidazol. Bound protein was eluted from the column using Buffer A with 250 mM Imidazol. The fractions corresponding to each peak in the chromatogram were dialysed against buffer containing 50 mM Tris HCl pH 7.5, 150 mM NaCl, 1 mM DTT and 10% glycerol.
RNA interference and transfection
ON-TARGET plus siRNA Human DPYS siRNA SMARTpool (siDHP-SP) (L-008455-00), ON-TARGET plus siRNA Human DPYS (siDHP-1) (J-008455-07) (GCACAGAUGGCACUCACUA), siGENOME SMARTpool Human DPYD (M-008376-02), ON-TARGETplus FANCM siRNA (L-021955-00), siGENOME SMARTpool Human FANCD2 (M-016376-02), siGENOME SMARTpool Human FANCA (M-019283-02), siGENOME SMARTpool Human SPRTN (M-015442-02), siGENOME SMARTpool Human XPA (M-005067-01), siGENOME SMARTpool Human ERCC5 (M-006626-01), siGENOME SMARTpool Human UPP1 (M-006647-01), siGENOME Non-targeting Control siRNA pool 2 (D-001206-14) and ON-TARGET plus Non-Targeting Pool (D-001810-10), were purchased from Dharmacon. siDHP-2 (GAAUAGCUGUAGGAUCAGATT) was purchased from Eurofins MWG. shDHP-3: DPYS MISSION plasmid (Sigma Aldrich, TRC0000046747), target sequence (TGTGGCAGTTACCAGCACAAA) and shDHP-4: DPYS MISSION plasmid (Sigma-Aldrich, TRC0000046744), target sequence (CTAATGATGATCTAACCACAA). Puro-shRNA FANCM used was described in (34). SiRNAs were transfected with INTERFERin (Polyplus), shRNA with Lipofectamine 2000 (Invitrogen). Plasmids encoding cDNAs were transfected using jetPEI or jetPRIME reagent (Polyplus).
Small-Scale Chromatin Fractionation Assay and western blotting
As described (35), cells were collected, washed with phosphate-buffered saline (PBS), and resuspended in buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM dithiothreitol (DTT), and protease inhibitors (Roche)). Triton X-100 was added (0.1% final concentration), the cells were incubated on ice for 5 min, and nuclei were collected by centrifugation (5 min, 1300 × g, 4°C). The supernatant (Cytosolic fraction) was clarified by high-speed centrifugation (5 min, 20 000 × g, 4°C), and the supernatant (Cytosolic fraction) was collected. The nuclei were then washed once in buffer A and lysed for 30 min in buffer B (3 mM ethylenediaminetetraacetic acid (EDTA), 0.2 mM Ethylene Glycol Tetraacetic Acid (EGTA), 1 mM DTT and protease inhibitor (Roche)), and insoluble chromatin and soluble fractions (Nucleosolic fraction) were separated by centrifugation (5 min, 17 000 × g, 4°C). The insoluble chromatin fraction was washed twice with buffer B and resuspended in sodium dodecyl sulphate (SDS)-Laemmli buffer and boiled for 10 min. Western blotting was performed using the ECL procedure according to the manufacturer's instructions (Amersham Bioscience, Inc) using anti-mouse or rabbit-HRP secondary antibodies (Promega)
Immunofluorescence Staining and ssDNA detection
Cells grown on coverslips were fixed with 3.7% paraformaldehyde (PFA) in PBS for 15 min at RT followed by a 0.5% Triton X-100-PBS permeabilization step for 10 min. Cells were then blocked in PBS containing 3% bovine serum albumin (BSA) for 30 min and incubated in the primary antibody and then in the appropriate secondary antibodies Alexa Fluor 488 or Alexa Fluor 555 (Invitrogen), diluted in blocking solution for 1 h in a humidified chamber at RT. DNA was stained with Hoechst (Invitrogen) and coverslips were mounted on glass slides with Prolong (Sigma-Aldrich).
For ssDNA detection, cells were grown on microscopic slides in 20 μM BrdU for 24 hr. Primary mouse antibody against BrdU in ssDNA was used (BD). All the Microscopic analysis was performed using Zeiss Z2 Axioimager with ApoTome. ImageJ was used for picture processing and quantification of S9.6 mean intensity.
DNA fiber labeling
DNA fiber spreads were prepared as described previously (36). Cells were labeled with 25 μM IdU (5-iodo-2′-deoxyuridine), washed with warm media and then exposed to 50 μM CldU (5-Chloro-2′-deoxyuridine). Cells were lysed with the spreading buffer (200 mM Tris–HCl pH 7.5, 50 mM EDTA and 0.5% SDS) and DNA fiber were stretched onto glass slides. The DNA fibers were denatured with 2.5 M HCl for 1 h, washed with PBS and blocked with 2% BSA in PBS-Tween 20 for 60 min. IdU replication tracts were revealed with a mouse anti-BrdU/IdU antibody (BD Bioscience) and CldU tracts with a rat anti-BrdU/CldU antibody (Abcam). DNA fibers were uniformly labeled with a mouse anti-human single-stranded DNA antibody (Millipore). The secondary antibodies used for the assay were: alexa fluor 488 anti-mouse antibody (Life technologies), alexa fluor 647 anti-mouse antibody (Life technologies) and Cy3 anti-rat antibody (Jackson Immunoresearch). Replication tracts were analyzed with ImageJ software. The probability that two datasets stem from the same distribution was assayed by a non-parametrical Mann–Whitney test (Prism Software).
Fluorescence-activated cell sorting (FACS)
Cells were pulse labeled with 10 μM BrdU for 15 min before fixation with ice-cold 100% ethanol. Then cells were incubated with PBS and 50 μg/ml of RNase A for 1 h at 37°C. After treatment with 2N HCl for 30 min, cells were incubated with an anti-BrdU antibody (BD) for 1 h at RT and then with an FITC-conjugated anti-mouse IgG (Life Technologies) at RT for 30 min. Cells were stained with 25 μg/ml of propidium iodide in PBS and analyzed using a FACSCalibur machine (BD).
Enzyme assay
5, 6-Dihydrouracil (DHU) was used as the substrate in the standard assay of DHP. Briefly, 2.5 μg of purified His-tagged DHP was added to 200 μl of 5, 6-dihydrouracil (50 μM) solution containing 50 mM Tris, 50 μM DTT, pH 8.0 in presence of several concentrations of dihydromyricetin, the samples were then incubated at 37°C for 1 h. An aliquot (100 μl) from each point was taken before incubation as a control without enzymatic reaction. DHU decomposition was monitored by HPLC using a Waters Alliance system connected to a C18 reversed phase Symmetry column (4.6 × 150 mm, 5 μm, Waters). Elution of DHU was achieved by applying an isocratic flow of H2O/TFA 0.1% as mobile phase for 15 min using a flow rate of 1 ml/min. For each sample, the column was first washed with 20% CH3CN/TFA 0.1% to remove any residual dihydromyricetin and equilibrated again with elution phase for 10 min. Under these conditions DHU was eluted at 2.8 min and detected by absorbance at 230 nm. Quantification of DHU was performed by integration of the corresponding HPLC peak using Empower Pro software. IC50 calculation was performed using Grafit-7 (Erithacus Software).
Intracellular dNTP measurement
For dNTP analysis and quantification, siRNA or shRNA transfected cells were harvested and lysed in iced cold 65% methanol, and vigorously vortexed for 2 min. Extracts were incubated at 95°C for 3 min. Supernatants were collected and dried in a speed vacuum. Samples were processed in Kim Baek laboratory for the single nucleotide incorporation assay as described (37).
Metabolite extraction
Cells pellet (1 million cells) were extracted on dry ice in 0.5 ml cold 70% methanol. The cell mixtures were shaken vigorously on a Vortex mixer for 10 min. These extracts were then centrifuged at 20 000 g at +4°C for 10 min, and the supernatants were transferred into polypropylene tubes for evaporation with a turbovap evaporator (Biotage, France). Dried extracts were reconstituted in 50 μl of mobiles phases A/B; 9/1. Five microliter of this sample was injected in the LC-MS/MS system. For LC Analysis: An UPLC Acquity I Class (Waters, France) was used for this study. The chromatographic separation was perform onto an Acquity UPLC BEH HSS T3 column (150 × 2.1 mm, 1.8 μm) using a gradient from 0.5%formic acid in water/0.5% formic acid in acetonitrile; 9/1; v/v as initial conditions to 6/4; v/v from 0.5 min to 3 min at a flow rate of 0.3 ml/min. The run time was 5 min allowing the system to reach 100% of 0.5% formic acid in acetonitrile to rinse the column and return to initial mobile phase conditions. The autosampler and the column compartment were held at 4°C and 30°C, respectively. Under these conditions, uracil and dihydrouracil displayed a mean retention time of 1.33 and 1.34 min, respectively. For MS Analysis, the UPLC system was coupled to a Waters XEVO™ TQ-XS mass spectrometer (Waters, France) operating in positive ion mode. For pyrimidine detection, the capillary voltage was set to 2.5 kV. The source and desolvation temperatures were held at 150°C and 600°C, respectively. The cone and desolvation gas flow were set at 150 and 800 l/h, respectively. The MS data acquisition was performed in multiple reaction monitoring (MRM) mode. Monitored MRM transitions were m/z 112.93 > 69.96 and 114.98 > 72.91 for uracil and dihydrouracil, respectively. Range of calibration curves were 0.28–108 and 0.27–110 nmol/cell pellet for uracil and dihydrouracil, respectively.
Cell viability and colony forming assay
The effect of siRNA on cell proliferation was measured using the CellTiter-Glo® Luminescent Cell Viability Assay Kit (Promega) according to the manufacturer's protocol or using the MTT cell growth assay. Briefly, siRNA transfected cells were seeded in 96-well plate and 4 days later, 100 μl CellTiter-Glo® reagent was added to each well that contained 100 μl cell culture medium. Cells were then lysed by shaking in an orbital shaker for 2 min, followed by incubation at room temperature for 10 min to stabilize the luminescent signal. The luminescent intensity was recorded on a Tristar LB 941 Multimode Microplate Reader (Berthold Technologies). For the MTT assay, siRNA-transfected cells were seeded into 24-well plates and cell growth was documented every 24 h via a colorimetric assay using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Sigma-Aldrich). Absorbance values were collected at 600 nm using a BioPhotometer (Eppendorf). In each individual experiment, proliferation was determined in triplicate and the overall experiment was repeated three times. For colony formation analysis, cells were seeded in 6-well plates at a density of 5000 cells per well. The medium was changed every 3 days for 10 days until visible colonies formed. Colonies were fixed in methanol for 10 min and stained with crystal violet.
Transcription assay
siRNA transfected cells were grown on cover slips and incubated with EU for 20 min. Cells were fixed with 4% paraformaldehyde for 15 min and permeabilized for 20 min in 0.1% Triton X-100 in PBS. EU incorporation were detected by staining with the Click-it Edu Alexa Fluor 555 azide Imaging Kit (Invitrogen) according to the manufacturer's instructions and DNA was stained with Hoechst (Invitrogen). The intensity of staining within individual nuclei was quantified using Image J software.
X. laevis egg extracts preparation and DNA replication kinetics
Low Speed Egg extracts (LSE) were prepared as previously described (38). M13 replication kinetics was assessed using 500 ng of M13mp18 single-stranded DNA (New England BioLabs) per 50 μl of LSE supplemented with cyclohexymide (250 μg/ml), an energy regeneration system (1 mM ATP, 2 mM MgCl2, 10 mM creatine kinase, 10 mM creatine phosphate) and α-[32P]-dCTP (0.37 MBq). Chromosomal DNA replication was assessed by adding 1000 demembranated Xenopus laevis sperm nuclei per microliters of extract. The mixtures were incubated at 23°C for the indicated time, then samples were neutralized in 10 mM Tris-HCl pH 8.0, 10 mM EDTA, 0.5% SDS, 200 μg/ml Proteinase K (Sigma-Aldrich) and incubated at 52°C for 1 h. Incorporation of radiolabeled deoxynucleotides in DNA was monitored using a Phosphor Imager Typhoon TriO+ (Amersham Biosciences) following agarose gel electrophoresis or alkaline agarose gel electrophoresis of purified DNA.
Sperm chromatin purification was performed as previously described (39). Briefly, egg extracts supplemented with demembranated sperm nuclei were diluted 10-fold in ice cold XB (10 mM Hepes-KOH pH 7.7, 100 mM KCl, 2 mM MgCl2, 50 mM sucrose and protease inhibitor) and pelleted at 1500 g for 5 min. Nuclei were washed once in XB buffer and then detergent extracted with 0.1% NP40 for 5 min on ice. Chromatin was recovered after centrifugation and ressuspended in Laemmli buffer for western blot analysis. For the chromatin transfer experiments, chromatin samples were incubated for 30 min in the first extract with the indicated drugs (Figure 4D). Purification for chromatin transfers and isolation of nuclei were performed following the protocol detailed in (40). Isolated nuclei integrity was verified by microscopy and then transferred into fresh extract supplemented with geminin and Roscovitine (to inhibit new replication events) together with α-[32P]-dCTP. After 2 h, dCTP incorporation was monitored by autoradiography on neutral gel as described previously.
DNA–protein crosslinks isolation and detection
DPCs isolation DPCs were prepared as described in (41). In brief, 1.5 to 2 × 106 cells were lysed in 1 ml of M buffer (MB), containing 6 M GTC, 10 mM Tris–HCl (pH 6.8), 20 mM EDTA, 4% Triton X-100, 1% Sarkosyl and 1% DTT. DNA was precipitated by adding 1 ml of 100% ethanol and was washed three times in wash buffer (20 mM Tris–HCl pH 6.8, 150 mM NaCl and 50% ethanol) and DNA was solubilized in 1 ml of 8 mM NaOH. A small aliquot of the recovered DNA was digested with 50 μg/ml proteinase K (Invitrogen) for 3 h at 50°C and quantified using Qubit dsDNA HS Assay Kit (Invitrogen) according to manufacturer instructions. DNA concentration was further confirmed by slot-blot where the proteinase K digested samples were diluted in TBS buffer and applied to nylon membrane (Hybond N+) followed by immunodetection with antibody against dsDNA.
The remaining solubilized DNA was digested with Benzonase (Sigma-Aldrich) for 30 min at 37°C. Proteins were precipitated by standard Trichloroacetic Acid (TCA) protocol. At last, the crosslinked-proteins were resuspended with the appropriate buffer and total DPCs were analyzed by Silver Staining (Invitrogen) as recommended by the manufacturer after electrophoretic separation on polyacrylamide gels and specific crosslinked-proteins were immunodetected using western blot assay. Signals were quantified using Image J software. Alternatively, proteins recovered by RADAR were analyzed by mass spectrometry as described previously (42). Analysis of raw files were performed using MaxQuant (43) version 1.6.3.3 using default settings with label-free quantification option enabled. Raw file spectra were searched against the human UniProt reference database. Protein, peptide, and site false discovery rate (FDR) were adjusted to 0.01.
Statistical analysis
Biological replicates were experiments performed independently of each other to test biological variation. Technical replicates were performed during one biological replicate to test variation of the measuring equipment and protocols.
For replication tracks analyses, the probability that two data sets stem from the same distribution was assayed by a non-parametrical Mann–Whitney test using the GraphPad Prism 5 software Prism software. To determine whether an siRNA treatment significantly increased or decreased the percentage of events by comparing the siRNA-treated cells to siRNA-control cells, p values were calculated with the open-source R software package (www.r-project.org) using a generalized linear model with Poisson distribution. The test used and p values are indicated in figure legends.
RESULTS
Suppression of DHP induces DNA replication stress and inhibits cell proliferation.
DHP is mainly expressed in the liver and the kidney (7). Yet, we detected DHP in the epithelial cancer cells MCF7 (breast adenocarcinoma), U-2 OS (osteosarcoma), HCT116 (colorectal carcinoma) and HEK293T (embryonic kidney) (Figure 1B). We also detected DHP in hTERT-immortalized retinal pigment epithelial cells (hTERT RPE-1) and in primary human dermal fibroblasts (HDF) (Supplementary Figure S1A), suggesting that regardless of their transformation status, certain types of cells express DHP when cultured in vitro. By contrast, DHP was not detectable in multiple myeloma cells (Figure 1B), consistent with the absence of DHP activity in hematopoietic malignant cells (3). The knockdown of DHP by means of siRNA in HCT116, U-2 OS and HEK293T cells confirmed the specificity of the anti-DHP signal (Figure 1B).
As DHP is produced in different epithelial cancer cells, we set out to explore the phenotypic consequences of DHP depletion. Depletion of DHP in U-2 OS cells blocked cell proliferation measured by colorimetric and by colony forming assays (Figure 1C and D). In addition, suppression of DHP by either one of two anti-DHP siRNAs with different target sequences impaired the growth of HCT116 cells (Supplementary Figure S1B), but not of the multiple myeloma cell line AMOI, that does not express DHP (Supplementary Figure S1C). We used also two plasmids encoding shRNAs with different target sequences to exclude potential off-target effects caused by the seed region of siRNAs (44). Using this strategy, we suppressed DHP in HEK293T cells and observed severely compromised cell viability and proliferation (Supplementary Figure S1D and E). Together, these observations confirm that growth inhibition induced by DHP depletion is not a singular property of U-2 OS cells. Because DHP is essential for the proliferation of these cancer cell lines, we surmised that it was not possible to generate DPYS (encodes DHP) knockouts cell lines by CRISPR/CAS9 genome editing to explore the function of DHP.
Flow cytometry cell-cycle analyses 72-h post-transfection revealed alterations in the cell-cycle distribution of osteosarcoma U-2 OS cells treated with an anti-DHP siRNA (Figure 1E). In comparison with control cells, suppression of DHP reduced the fraction of U-2 OS cells in S phase. The impact of DHP depletion was less pronounced in the transformed HEK293T cell line (Supplementary Figure S1F), perhaps reflecting differences in the dependency on DHP activity or in the efficiency of DHP depletion. To evaluate further the capacity of DHP-depleted HEK293T cells to proceed through the cell cycle, we performed pulse-chase experiments with the nucleotide analog BrdU. Cells were labeled with BrdU for 30 min and sorted by two-dimensional flow cytometry at the indicated time of incubation in BrdU free medium. After 6-h incubation in BrdU-free medium, the majority of control cells had completed S phase and a significant proportion proceeded to the G1 phase (9.27%), whereas the proportion of DHP-depleted HEK293T cells to reach G1 was reduced by half (Supplementary Figure S2A).
To explore the cause of the cell cycle delays, we analyzed DNA replication tracks at the single molecule level by DNA fiber labeling. Replication tracks were dually labeled with two consecutive pulses of fluorescent nucleotide analogues iodo-deoxyuridine (IdU) and chloro-deoxyuridine (CldU) for 30 min each. We assessed the progression of isolated replication forks by measuring the length of CldU tracks adjacent to IdU tracks. The knockdown of DHP by siRNA or shRNA molecules with different target sequences reduced the length of replication tracts in U-2 OS or HEK293T cells (Figure 2A and Supplementary Figure S2B). Re-expression of a siRNA-resistant myc-tagged DHP protein in DHP knockdown U-2 OS cells partially rescued replication progression (Figure 2A). This confirms that the DNA replication defect is a direct consequence of DHP depletion.
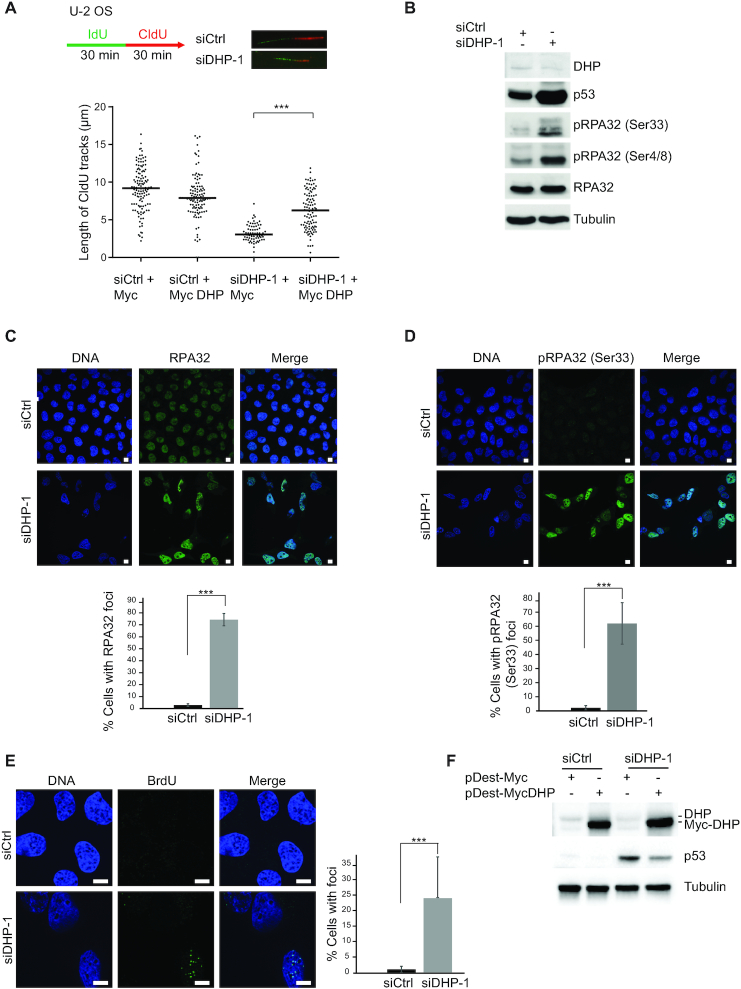
Figure 2.
Suppression of DHP interferes with replication fork progression and induces activation of DNA damage responses. (A) Experimental scheme: 72 h after transfection with control or anti-DHP siRNA (siDHP-1), cells were labeled with two consecutive pulses of 30 min with CldU and IdU, as indicated. DNA was stretched out on glass slides and newly synthesized DNA was revealed by immunofluorescence. Graphic representation of replication track lengths in U-2 OS cells co-transfected with control or anti-DHP siRNA (siDHP-1) along with control plasmid or a plasmid encoding siRNA-resistant DHP. The bar dissecting the data points represents the median of 100 tracts length from one biological replicate. Differences between distributions were assessed with the Mann–Whitney rank sum test. P-values: *** < 0.0001. (B) Whole-cell extracts from control and DHP knockdown U-2 OS cells (siDHP-1) were analyzed by western blotting with the indicated antibodies. One representative experiment is shown from more than three biological replicates (C) RPA32 immunofluorescence staining of control and DHP knockdown U-2 OS cells (siDHP-1). DNA was stained by Hoechst. Bars indicate 10 μm. Bottom panel: Quantification of the percentage of RPA32 foci positive cells in a population of 100 cells. Data from three independent biological replicates are represented as mean ± S.D. P-values were calculated using a regression model with Poisson distribution: ***P < 0.0001. (D) Immunofluorescence staining of Ser33 phospho RPA32 in control and DHP knockdown U-2 OS cells (siDHP-1). DNA was stained by Hoechst. Bars indicate 10 μm. Bottom panel: Histogram representing the percentage of Ser33 pRPA32 foci positive cells in a population of 100 cells. Data from three independent biological replicates are represented as mean ± S.D. (100 cells were counted per experiment). P-values were calculated using a regression model with Poisson distribution: ***P < 0.0001. (E) Control and DHP knockdown U-2 OS cells (siDHP-1) were uniformly labeled with BrdU before immunofluorescence staining in native conditions with an anti-BrdU antibody. DNA was stained by Hoechst. Bars indicate 10 μm. Right panel: Histogram representation of the percentage of ssDNA positive cells. Values are the mean ± S.D of three independent biological replicates (100 cells were counted per experiment). P-values were calculated using a regression model with Poisson distribution: ***P < 0.0001. (F) Western blotting analysis with the indicated antibodies of whole-cell extracts from control and DHP knockdown U-2 OS cells (siDHP-1) complemented or not with a siRNA-resistant DHP cDNA, as indicated. One representative experiment is shown from three biological replicates.
Next, we probed cells for indicators of DNA replication stress by monitoring ATR/Chk1 signaling. We observed spontaneous accumulation of Chk1 phosphorylated on Ser345 in the soluble fraction and of RPA32 phosphorylated on Ser 4/8 and Ser33 in the chromatin fraction of DHP-depleted HEK293T and U-2 OS cells (Supplementary Figure S2C and D). Phospho RPA32 (Ser33 and/or Ser4/8) accumulated in U-2 OS whole cell extracts (Figure 2B) and in HEK293T cells transfected with a different anti-DHP shRNA (Supplementary Figure S2E). To confirm this observation, we visualized RPA foci and phospho RPA signals by means of immunofluorescence microscopy. RPA32 and phospho RPA32 (ser33) signals accumulated in U-2 OS cells transfected with anti-DHP siRNA (Figure 2C and D), and in HEK293T cells transfected with a distinct anti-DHP shRNA (Supplementary Figure S2F). To verify that the formation of RPA32 foci correlates with the accumulation of single-stranded DNA (ssDNA), we probed ssDNA by immunofluorescence microscopy using uniform BrdU labeling and BrdU detection in native conditions (45). Nearly 30% of DHP-depleted cells exhibited multiple and distinct BrdU signals indicative of severe replication-associated defects (Figure 2E). Last, we monitored the level of p53 that is stabilized in response to genotoxic stress. Depletion of DHP markedly increased the level of p53 in U-2 OS cells (Figure 2B and F). Expression of a siRNA-resistant cDNA encoding DHP reduced the level of p53, confirming that the stabilization of p53 results, at least in part, from the depletion of DHP (Figure 2F). Collectively, these data indicate that suppression of DHP induces DNA replication stress, at least in a subset of cancer cell lines.
Accumulation of dihydropyrimidines induces DNA replication stress
Next, we sought to investigate how suppression of DHP inhibits fork progression. The rate of DNA chain elongation is dependent on the pool of available deoxyribonucleotides. Thus, we measured the impact of DHP depletion on dNTPs levels using a single nucleotide incorporation assay (37). The level of dNTPs increases in proliferating cells and fluctuates during the cell cycle (46). Since the suppression of DHP has consequences on cell growth and cycle progression, cells lacking DHP may exhibit altered dNTPs levels. Consistent with this, the suppression of DHP in HEK293T and U-2 OS cells led to a reduction in the global level of dNTPs (Supplementary Figure S3A and B). To test if alterations of dNTPs levels were responsible for the defect in fork progression observed in DHP-depleted cells, we complemented the cell culture medium with saturating concentrations of nucleosides and measured the length of CldU-labeled replication tracks from isolated replication forks. Addition of nucleosides in the cell culture medium markedly increased the length of replication tracks in shControl HEK293T cells (Supplementary Figure S3C), with a median fold stimulation of 1.7×. This data indicate that nucleosides are limiting in these cells. By contrast, saturation of DHP-depleted cells with a cocktail of nucleosides did not markedly increase the length of replication tracks (Supplementary Figure S3C). Therefore, changes in dNTPs levels are not the primary cause of replication stress in these cells. Consistent with this interpretation, addition of an excess of nucleosides in the cell culture medium of DHP-depleted cells did not attenuate the accumulation of p53 and the phosphorylation of RPA32 on Ser33 (Supplementary Figure S3D).
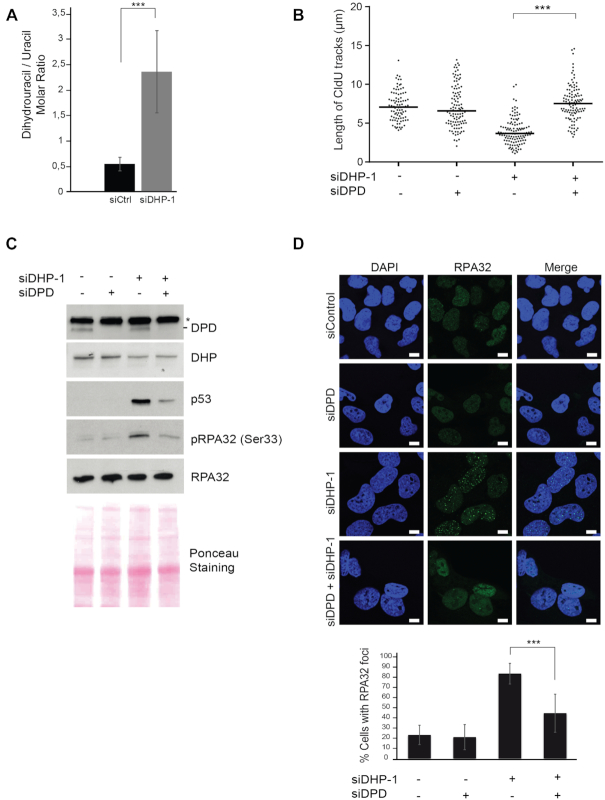
It is noteworthy that measurements of the global pool of dNTPs does not give insights into local levels of dNTPs available to the DNA replication machinery (47), and that the rate of replication fork progression is primarily determined by the amount of DNA damage and the level of activated p53, not by the global concentrations of dNTPs (47). Thus, we considered the possibility that DNA replication stress in DHP-depleted cells was caused by the accumulation of dihydropyrimidines. To test this, we measured the cellular concentration of uracil and its breakdown product dihydrouracil by liquid chromatography and mass spectrometry (LC-MS). DHP-depletion in U-2 OS cells yielded a 4-fold increase in the molar ratio of intracellular dihydrouracil/uracil (Figure 3A). This indicate that the transient knockdown of DHP is sufficient to raise the intracellular concentration of dihydropyrimidines. To counteract the accumulation of dihydropyrimidines in DHP-knockdown cells, we co-depleted DPD, the first enzyme in the pyrimidine catabolic pathway that produces dihydropyrimidines (Figure 1A). We tried to measure the dihydrouracil/uracil ratio in DPD-depleted cells co-transfected with control and anti-DHP siRNAs. In the absence of DPD, however, the cellular concentration of dihydrouracil decreased below quantification levels. Suppression of DPD in DHP-depleted cells rescued the rate of fork progression to the level of control cells (Figure 3B). Consistent with this result, the levels of p53, of Ser33 phosphorylated RPA32 and the intensity of RPA32 foci were close to normal when both DPD and DHP enzymes were depleted (Figure 3C and D). To test if the genotoxic consequences of dihydropyrimidines was dependent on the salvage pathway, we knocked-down uridine phosphorylase (UPP1) by means of RNA interference. Suppression of UPP1 in DHP-depleted cells had no impact on markers of DNA replication stress (Supplementary Figure S3E). Altogether, these observations suggest that the DNA replication stress phenotype of DHP-depleted cells is the consequence of the accumulation of dihydropyrimidines.
Figure 3.
Accumulation of dihydropyrimidines induces DNA replication stress. (A) The concentrations of dihydrouracil and uracil were measured in U-2 OS cells transfected with control or anti-DHP siRNA (siDHP-1). The ratio of molar concentrations between the two metabolites in each sample is presented. Data from three independent biological replicates, with three technical replicates for each, are represented as mean ± S.E.M. P-values were calculated using a regression model with Poisson distribution: ***P < 0.0001. (B) Replication tracks were labeled with two consecutive pulses of 30 min with CldU and IdU in U-2 OS cells transfected with the indicated siRNAs. Graphic representations of replication track lengths measured in μm (y-axis). The bar dissecting the data points represents the median of 100 tracts length from one biological replicate. Differences between distributions were assessed with the Mann–Whitney rank sum test. P-values: *** < 0.0001. (C) Western blot analysis with the indicated antibodies of whole cell extracts from U-2 OS transfected with anti-DHP and anti-DPD siRNAs, as indicated, Ponceau staining was used as control of protein loading and transfer. * non-specific band. One representative experiment is shown from two biological replicates. (D) RPA32 immunofluorescence staining of U-2 OS cells transfected with the indicated siRNAs. Bars indicate 10 μm. DNA was stained by Hoechst. Bottom panel: Histogram representation of the percentage of RPA32 foci-positive cells in a population of 100 cells. Data from three independent biological replicates are represented as mean ± S.D. P-values were calculated using a regression model with Poisson distribution: ***P < 0.0001.
Dihydropyrimidine accumulation induces transcriptional stress
The data thus far indicate that dihydropyrimidine metabolites induce DNA replication stress. We reasoned that dihydropyrimidines could induce directly or indirectly the formation of DNA adducts and interfere with DNA-templated processes including transcription. Thus, we explored the effect of DHP loss on global transcription activity. Nascent RNA were pulse labeled for 20 min with the modified RNA precursor 5-ethynyluridine (EU) and overall transcription activity was evaluated via fluorescent-based quantification. In comparison with control cells, we observed a drop of EU incorporation in DHP-depleted U-2 OS cells (Figure 4A). Next, we used the anti-RNA:DNA hybrids S9.6 antibody to visualize R-loops by immunofluorescence staining. R-loops are induced by defects in the processing of nascent pre-mRNAs or by the accumulation of negative supercoiling behind RNA polymerases (15,48). Immunofluorescence staining experiments revealed a significant increase in the level of nuclear RNA:DNA hybrids in DHP-depleted cells compared to control cells after excluding nucleolar signals from the analysis (Figure 4B). R-loops and nucleolin immunofluorescence staining revealed alterations in the morphology of nucleoli, which appeared more condensed and rounded (Figure 4B). These observations indicate that the accumulation of dihydropyrimidine metabolites in DHP-depleted cells induces transcriptional stress.
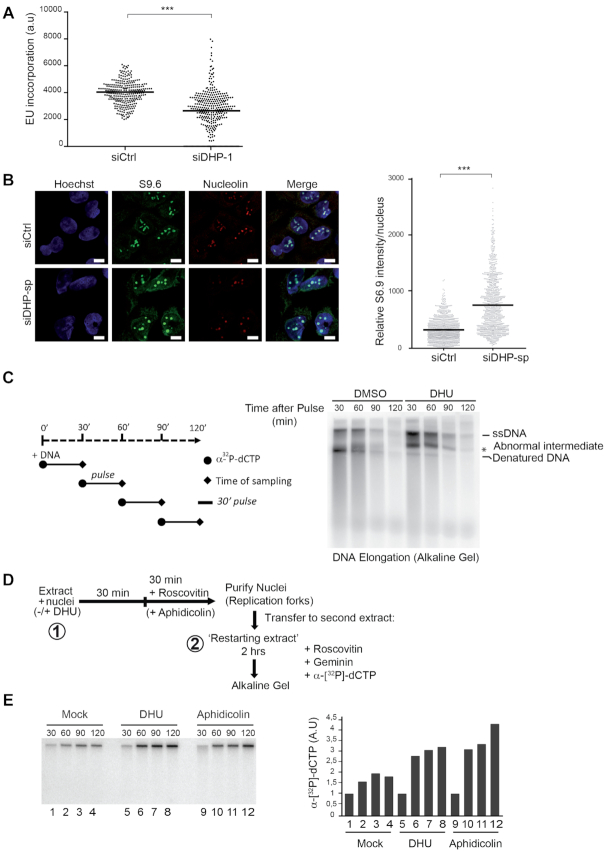
Figure 4.
Dihydropyrimidines induce transcriptional stress and yield abnormal DNA replication intermediates. (A) Graphic representation of global transcriptional activity visualized by 5-ethynyl uridine (EU) incorporation. U-2 OS cells transfected with control and anti-DHP siRNA (siDHP-1) were labeled with EU for 20 min before fixation. The EU intensity of 100 cells from two independent biological replicates was measured by fluorescence microscopy. The bar dissecting the data points represents the median of EU intensity. Differences between distributions were assessed with the Mann–Whitney rank sum test. P-values: *** < 0.0001. (B) Immunofluorescence staining with S9.6 and nucleolin antibodies of DHP-depleted or control U-2 OS cells (siDHP-sp). DNA was stained by Hoechst. Bars indicate 10 μm. Right panel: The graph shows the median of S9.6 signal intensity par nucleus after nucleolar signal removal. More than 1000 cells from two independent biological replicates were considered. Differences between distributions were assessed with the Mann–Whitney rank sum test. P-values: *** < 0.0001. (C) Left panel: Experimental scheme. DNA synthesis reactions (control: DMSO; DHU: 15 mM) were pulse-labeled for 30 min with α-[32P]-dCTP at the indicated times during the course of a 2-h reaction. Replication products were purified and resolved by electrophoresis through a 1.2% agarose gel in denaturing conditions. (*) abnormal replication intermediate. One representative experiment is shown from two biological replicates. (D) Experimental scheme: Sperm nuclei were added to Xenopus egg-extract (in presence or not of 7.5 mM DHU dissolved in water) and incubated at 23°C to allow origins firing and replication initiation. After 30 min incubation, the firing of new replication origins was blocked with roscovitine (0.5 mM). Replicating nuclei were then isolated after 60 min of incubation and transferred to a second extract (restarting extract) supplemented with roscovitine (0.5 mM) and Geminin (60 mM) to block the firing and the assembly of novel origins, respectively. DNA synthesis reactions were pulse-labeled with α-[32P]-dCTP during incubation in the second extract. (E) Replication products were resolved by 1% alkaline agarose gel electrophoresis and revealed by autoradiography. Lanes 1–4: Mock treated extracts; Lanes 5–8: incubation in the first extract was performed in the presence of 7.5 mM DHU. Lanes 10–12 serve as positive controls: after 30 min incubation in the first extract, DNA synthesis was blocked with aphidicolin (100 ng/μl). Right panel: Histogram representing the quantification of the gel by image J of replication products (arbitrary unit). One representative experiment is shown from two biological replicates (See Supplementary Figure S4A).
Dihydropyrimidine accumulation induces abnormal DNA replication products independently of transcription
Because interference between transcription and DNA replication is an important endogenous source of DNA replication stress (Tuduri et al., (15)), we wanted to know if dihydropyrimidine metabolites can directly interfere with the process of DNA replication, independently of transcription activity. To test this, we used a cell-free DNA replication system derived from Xenopus eggs in which transcription is inactive. In this system, a circular single-stranded DNA is converted into a double-stranded DNA via priming and elongation of DNA chains in a semiconservative manner. The replicated DNA is then assembled into chromatin leading to the formation of supercoiled DNA (49). DNA replication was measured by the incorporation of the radioactive nucleotide precursor α32P-dCTP.
First, we labeled DNA during the course of a 2-h reaction with 30 min pulses of α32P-dCTP, as indicated (Figure 4C, left panel), and analyzed DNA replication products by alkaline agarose gel electrophoresis. In these denaturing conditions, irreversibly denatured DNA produced in control extracts was replaced by an abnormal replication intermediate in extracts supplemented with 5, 6-dihydrouracil (Figure 4C). This novel replication product was visible from the earliest stages of the replication reaction (Figure 4C). This observation indicate that dihydropyrimidines interfere directly with the process of DNA chain elongation in Xenopus egg-extracts. Moreover, the dNTPs pool is not limiting during DNA replication in Xenopus egg extracts. Therefore, in this system, the possibility that DHU may imbalance the pool of dNTP pool is eliminated.
We designed a multistep chromatin transfer experiment to verify if 5, 6-dihydrouracil directly interferes with chromosomal DNA synthesis (Figure 4D). First, we performed a standard chromatin DNA replication reaction in Xenopus egg-extracts using demembranated sperm nuclei (50–52). Interference with the progression of replication forks, for example using the replicative DNA polymerase inhibitor aphidicolin, is expected to yield incomplete DNA replication intermediates that can prime DNA synthesis during the course of a second DNA replication reaction. To obtain evidence for the formation of aborted replication intermediates in extracts supplemented with 5, 6 dihydrouracil, we purified and transferred the replicated, or partially replicated, chromatin to a second extract supplemented with both Geminin, to block the licensing of new origins of replication, and the CDK2 inhibitor roscovitine, to block the firing of new origins. In this situation, DNA synthesis is the result of priming of pre-existing replication intermediates. The transfer of nuclei from a replication reaction carried out in the presence of aphidicolin to a second extract unable to fire new origins led to a significant increase in α32P-dCTP incorporation in comparison with mock-treated nuclei (Figure 4E and Supplementary Figure S4A, compare lanes 1–4 with lanes 9–12), indicative of replication fork restart and/or DNA repair activities. Likewise, in comparison with mock-treated nuclei, addition of 5,6-dihydrouracil during the first replication reaction yielded an increased incorporation of α32P-dCTP in the restarting extract (Figure 4E and Supplementary Figure S4A, lane 5–9), indicating that DNA replication in the presence of 5,6-dihydrouracil generates DNA intermediates that prime DNA synthesis. Collectively, these data indicate that dihydropyrimidine metabolites directly interfere with the process of DNA replication.
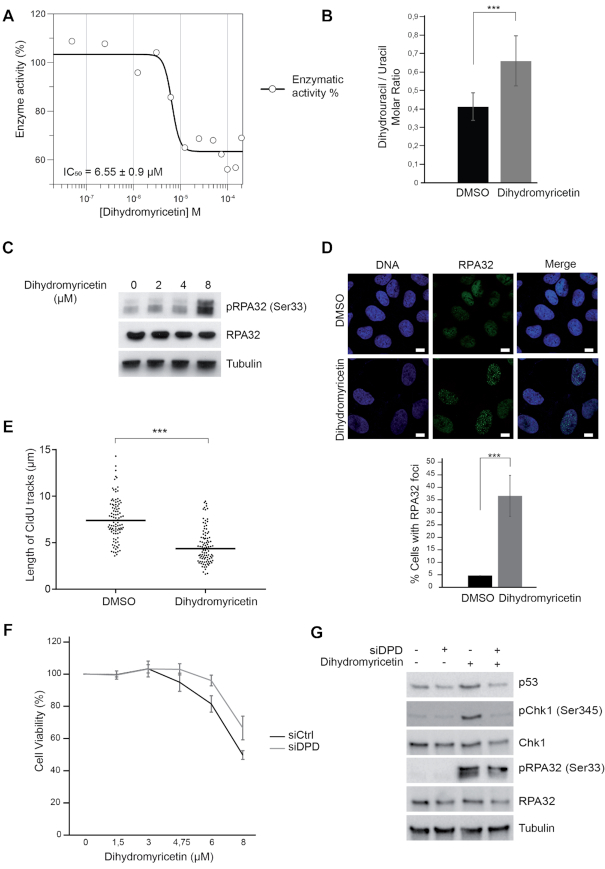
Dihydromyricetin induces DNA replication stress.
We noticed a report suggesting that the plant flavonoid dihydromyricetin is a competitive inhibitor of a putative DHP from Pseudomonas aeruginosa (53) (Figure 1A). Human and P. aeruginosa DHPs are predicted to fold into a similar structure (53). Since DHP activity is a good marker of tumorigenicity and a candidate target for cancer therapy (3), we wanted to test if dihydromyricetin also inhibits the human DHP. We expressed and purified recombinant human DHP to near homogeneity (Supplementary Figure S4B) and assessed its activity by measuring the decomposition of 5, 6-dihydrouracil using high performance liquid chromatography. In an experimental system containing purified DHP (0.2 μM) and 5, 6 dihydrouracil as a substrate (50 μM), dihydromyricetin inhibited DHP activity with a half maximal inhibitory concentration (IC50) value of about 6 μM (Figure 5A). Next, we analyzed the phenotypic consequences of acute exposure of U-2 OS cells to dihydromyricetin, and observed an increased intracellular molar ratio of dihydrouracil/uracil (Figure 5B), the induction of RPA32 and Chk1 phosphorylation (Figure 5C and Supplementary Figure S4C), the formation of RPA32 nuclear foci (Figure 5D), and the slowing of DNA replication forks (Figure 5E). In addition, DPD-depleted U-2 OS cells were more resistance to dihydromyricetin treatment and rescued the DNA replication stress markers induced by dihydromyricetin (Figure 5F and G). By contrast, suppression of the salvage pathway enzyme UPP1 had no impact on DNA replication stress markers induced by dihydromyricetin (Supplementary Figure S4D). These data indicate that the treatment of cells with dihydromyricetin induces replication stress phenotypes that are similar to the phenotypes of DHP-knockdown cells.
Figure 5.
Dihydromyricetin induces DNA replication stress. (A) IC50 determination of dihydromyricetin for DHP (0.2 μM) using dihydrouracil (50 μM) as a substrate. One representative experiment is shown from three biological replicates. (B) Molar ratios of dihydrouracil versus uracil measured in U-2 OS cells treated with 20 μM dihydromyricetin for 16 h. Data from three independent biological replicates, with three technical replicates for each, are represented as mean ± S.E.M. P-values were calculated using a regression model with Poisson distribution: ***P < 0.0001. (C) Western blot analysis with the indicated antibodies of U-2 OS whole-cell extracts treated with dihydromyricetin for 48 h at the indicated concentrations. One representative experiment is shown from three biological replicates. (D) RPA32 immunofluorescence staining of U-2 OS cells treated with DMSO or 20 μM dihydromyricetin for 16 h. Bars indicate 10 μm. DNA was stained by Hoechst. Bottom panel: Histogram representation of the percentage of RPA32 foci-positive cells in a population of 100 cells. Data from three independent biological replicates are represented as mean ± S.D. P-values were calculated using a regression model with Poisson distribution: ***P < 0.0001. (E) Graphic representation of replication track lengths measured in μm (y-axis) in control and U-2 OS cells treated with 20 μM of dihydromyricetin for 16 h. The bar dissecting the data points represents the median of 100 tracts length from one biological replicate. Differences between distributions were assessed with the Mann-Whitney rank sum test. P-values: *** < 0.0001. (F) U-2 OS cells were transfected with control or anti-DPD siRNA and exposed to increasing concentrations of dihydromyricetin for 2 days. Cell viability was estimated using Cell Titer-Glo assay. Mean viability is representative of experiments performed in triplicate. Error bars represent ± S.E.M. (G) Western blot analysis with the indicated antibodies of whole cell extracts from U-2 OS transfected with anti-DPD siRNA and treated or not with 20 μM of dihydromyricetin for 24 h. One representative experiment is shown from two biological replicates.
Suppression of dihydropyrimidinase activity induces DNA–protein crosslinks
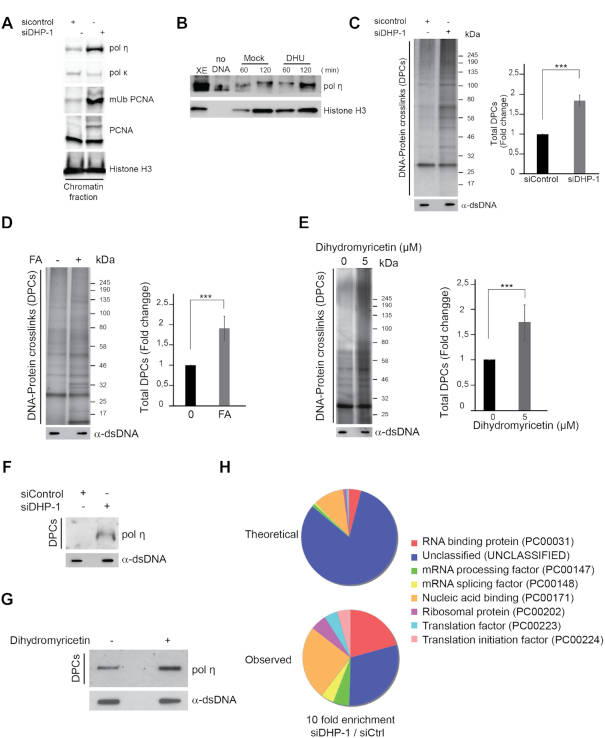
Next, we sought to evaluate whether dihydropyrimidines inhibit DNA-templated processes via the formation of DNA adducts. First, we asked if the accumulation of these metabolites triggers the recruitment of translesion DNA polymerases to chromatin. Suppression of DHP induced chromatin recruitment of the translesion DNA polymerase η that can bypass replication-blocking lesions such as UV photoproducts and oxidized bases (54,55), but not DNA polymerase κ, which has a different specificity for DNA lesions than polymerase η (Figure 6A and Supplementary Figure S5A). We also observed monoubiquitinated PCNA in the chromatin fraction of DHP-depleted cells (Figure 6A), a posttranslational modification that facilitates the interaction of TLS polymerases with PCNA (56). Consistent with this observation, addition of 5, 6-dihydrouracil in Xenopus eggs extracts induced a pronounced accumulation of TLS pol η on chromatin after 2 h of incubation, when the replication process was completed (Figure 6B). In light of these observations, we propose that the accumulation of dihydropyrimidines or their decomposition products may induce bulky DNA adducts.
Figure 6.
Dihydropyrimidines metabolites induce DPCs. (A) The chromatin fraction of control and DHP knockdown U-2 OS cells (siDHP-1) was subjected to western blot analysis with the indicated antibodies. Histone H3 was used as loading control. One representative experiment is shown from two biological replicates. (B) Chromatin extracts from nuclei incubated in control and DHU (7.5 mM) containing extracts for 60 and 120 min were subjected to western blot analysis with the indicated antibodies. Histone H3 was used as loading control. One representative experiment is shown from two biological replicates. (C) Total DPC levels in U-2 OS cells transfected with control or anti-DHP siRNA (siDHP-1) visualized by silver staining. Right panel: Histogram representing the quantification of DPC levels normalized to total DNA amount by image J. Three independent biological replicates are averaged in the bar graphs. Error bars represent ± S.D. P-values were calculated using a regression model with Poisson distribution: ***P < 0.0001. (D) Total DPC levels in U-2 OS cells treated or not with 1 mM FA for 2 h visualized by silver staining. Right panel: Histogram representing the quantification of DPC levels normalized to total DNA amount by image J. Three independent biological replicates are averaged in the bar graphs. Error bars represent ± S.D. P-values were calculated using a regression model with Poisson distribution: ***P < 0.0001. (E) Total DPC levels after U-2 OS cells treatment with DMSO or 5 μM of Dihydromyricetin for 16 h visualized by silver staining. Right panel: Histogram representing the quantification of DPC levels normalized to total DNA amount by image J. Three independent biological replicates are averaged in the bar graphs. Error bars represent ± S.D. P-values were calculated using a regression model with Poisson distribution: ***P < 0.0001. (F) Western blot analysis of crosslinked DNA polymerase η in total DPC extracts from U-2 OS cells transfected with control or anti-DHP siRNA (siDHP-1) and the corresponding DNA quantification. One representative experiment is shown from two biological replicates. (G) Slot-blot showing crosslinked DNA polymerase η in total DPC extracts from U-2 OS cells treated with 20 μM dihydromyricetin for 16 h and the corresponding DNA quantification. (H) Pie chart representation of gene ontology analyses using the panther Classification system (http://www.pantherdb.org/on). Theoretical relates to gene ontology classification expected from random sampling of the proteome. Observed is the classification of proteins enriched at least 10-folds in DHP-depleted cells versus control cells. Proteins selected for the analysis were identified in three independent experiments.
A variety of endogenous metabolites, environmental and chemotherapeutic DNA damaging agents induce covalent DPCs (20). We use the RADAR assay (rapid approach to DNA-adduct recovery) to test if the suppression of DHP activity yields DPCs. We isolated genomic DNA and quantified it using Qubit fluorometric quantitation to ensure that DPC analyses were performed using equal amounts of material. Next, we digested DNA with benzonase, resolved DPC by SDS-polyacrylamide gelelectrophoresis and detected them by silver staining. An increase in total DPCs was consistently detected after suppression of DHP using distinct siRNA and shRNA molecules, in U-2 OS and in HEK293T cells (Figure 6C and Supplementary Figure S5B). The level of DPCs in DHP-depleted cells was comparable to that of U-2 OS cells exposed to formaldehyde (Figure 6D). In addition, treatment of U-2 OS cells with dihydromyricetin increased by 2-folds the amount of total DPCs (Figure 6E). The proteolysis of DPCs is coupled to DNA replication via at least two independent mechanisms, one mediated by the DNA-dependent metalloprotease Spartan (DVC1) (41,57), and one mediated by the proteasome (58). Given the importance of DNA replication in DPC repair, we analyzed if the slowing of replication forks per se influences the overall level of DPCs measured by RADAR. Low dose aphidicolin (0.1 μM) reduced the length of replication tracks, as expected, but did not increase the overall level of DPCs (Supplementary Figure S5C). Thus, the formation DPCs in DHP-depleted cells is unlikely a consequence of replication-fork slowing. The data suggest that dihydropyrimidines metabolites induce the formation of covalent bonds between proteins and DNA.
A large diversity of proteins can be crosslinked to DNA. As the TLS polymerase η functions at blocked replication forks and accumulates in the chromatin fraction of DHP-depleted cells, we examined if DNA polymerase η was crosslinked to DNA. The level of covalently trapped DNA polymerase η increased in DHP-depleted cells (Figure 6F) and in cells exposed to dihydromyricetin (Figure 6G), but not in presence of low dose of aphidicolin (Supplementary Figure S5D), suggesting that dihydropyrimidines covalently trap DNA polymerase η on DNA. To gain a panoramic view of covalently trapped proteins in DHP-depleted cells, we performed RADAR assay and analyzed protein–DNA complexes by mass spectrometry. We used the MaxQuant computational platform for label free quantification of proteins identified by RADAR-MS. We selected proteins identified in three independent experiments that were enriched by at least 10-folds in samples prepared from DHP-depleted cells in comparison with control cells. Next, we used PANTHER Classification System (http://pantherdb.org/) to perform Gene Ontology analyses. We observed a strong and non-random enrichment in nucleic acid binding, mRNA processing and splicing proteins, consistent with the crosslinking of chromatin-associated proteins (Figure 6H).
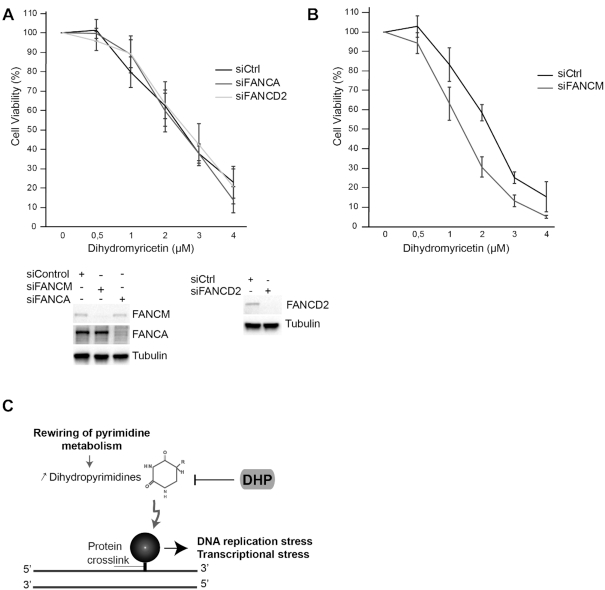
To begin to understand how cells process DNA lesions induced by dihydromyricetin, we used siRNAs to suppress a subset of DNA repair factors. Surprisingly, depletion of Spartan did not sensitize U-2 OS cells to dihydromyricetin, nor did the depletion of the nucleotide excision repair factors XPA, ERCC5/XPG, nor the Fanconi anemia proteins FANCA and FANCD2 (Supplementary Figure S6A and Figure 7A). By contrast, depletion of the DNA translocase FANCM sensitized cells to dihydromyricetin (Figure 7B). We confirmed this observation using an anti-FANCM shRNA with a different target sequence in HEK293T cells (Supplementary Figure S6B). To assess the role of FANCM translocase activity in cellular protection against dihydromyricetin-induced DNA replication stress, we measured the impact of dihydromyricetin on the viability of HEK293 cell lines that stably express an anti-FANCM siRNA and that are complemented either with recombinant WT FANCM or with the translocase dead mutant K117R FANCM (31). Cells expressing the ATPase-dead mutant K117R were more sensitive to dihydromyricetin than cells expressing WT FANCM (Supplementary Figure S6C). These data suggest that the DNA translocase FANCM promotes cellular tolerance to dihydromyricetin.
Figure 7.
FANCM promotes cellular tolerance to dihydromyricetin. (A) U-2 OS cells were transfected with the indicated siRNAs and exposed to increasing concentrations of dihydromyricetin for 2 days. Cell viability was estimated using Cell Titer-Glo assay. Mean viability is representative of experiments preformed in triplicate. Error bars represent ±S.D. Bottom panel: the efficiency of FANCM, FANCD2 or FANCA knockdown was assessed by western blotting. (B) U-2 OS cells were transfected with FANCM siRNA and cell viability was assessed as described in A. Mean viability representative of quadruplicates. Error bars represent ±S.D. (C) Model: The accumulation of Dihydropyrimidines in cancer cells induces DNA replication and transcriptional stress via the formation of DPCs.
DISCUSSION
The activity of DHP is high in the liver and in the kidney (7), absent in the majority of healthy human tissues and re-expressed in a variety of human carcinomas (3). In this study, we show that DHP activity is necessary to prevent transcriptional and DNA replication stress in transformed cell lines.
Suppression of DHP induced the slowing of DNA replication forks, the accumulation of single-stranded DNA, the activation of ATR signaling, the accumulation of RNA:DNA hybrid structures and the inhibition of transcription. Multiple lines of evidence obtained using orthogonal experimental methods lead us to conclude that dihydropyrimidines are potentially cytotoxic metabolites: (i) depletion of DHP by RNA interference impeded DNA replication and transcription; (ii) DNA replication stress in DHP-depleted cells was reversed by suppression of DPD, the enzyme that produces dihydropyrimidines and by expression of a siRNA resistant cDNA encoding Myc-DHP; (iii) Inhibition of DHP activity with dihydromyricetin phenocopied the defects of DHP-depleted cells; (iv) Dihydropyrimidines directly altered DNA replication products synthesized in Xenopus egg extracts.
We reproduced the DNA replication stress phenotypes in different cell lines with different siRNA and shRNA targeting sequences. We failed, however, to rescue the long-term viability of DHP-depleted cells using retrovirus or lentivirus to transduce siRNA-resistant DHP cDNAs. Although we cannot exclude that the anti-DHP siRNA has other target(s) that affect cellular viability, we believe it is unlikely. It is noteworthy that an anti-DHP siRNA did not alter the viability of multiple myeloma cells (AMOI) that do not express DHP. The complexity of nucleotide metabolism and the need to regulate tightly the activity and the level of DHP protein may compromise the rescue of cell viability with unregulated recombinant DHP.
The observed reduction of dNTPs levels may contribute to the DNA replication stress phenotype of DHP-depleted cells. However, the pool of dNTPs increases in S phase. Thus, the apparent reduction of the pool of dNTPs may simply reflect the reduction of the proportion DHP-depleted cells in S phase. Furthermore, previous studies have shown that the rate of replication fork progression does not correlate directly with the global concentration of dNTPs (47,59), as the latter does not reflect the amount of nucleotides available to the replication machinery. By contrast, the length of replication tracks is determined directly by the accumulation of DNA lesions in the template DNA and by p53 activation (47). Several lines of evidence suggest that it is unlikely that alterations of the pool of dNTPs determine the phenotypes of DHP-depleted cells. First, the length of replication tracks in DHP-depleted cells remain shorter than in control cells after saturation of the cell culture medium with exogenous nucleotides. Second, the pool of dNTPs is not limiting in Xenopus egg extracts, yet supplementation of Xenopus egg extracts with dihydropyrimidines interfered with DNA replication. Third, transcription does not depend on dNTPs precursors, yet, the accumulation of dihydropyrimidines also inhibited RNA synthesis. The accumulation of dihydropyrimidines induced p53 stabilization and DNA damage, two parameters that determine directly the length of replication tracks (47).
Cellular metabolites and environmental agents generate a range of structurally diverse protein–DNA crosslink that precipitate the loss of cellular functions, including transcription and DNA replication (20). During DNA replication, DPCs are degraded either by the DNA-dependent metalloprotease Spartan/DVC1 (41,57) or by the proteasome (58). Proteasomal degradation requires DPC ubiquitylation whereas SRTN requires nascent strand extension within a few nucleotides from the lesion (58). Recent evidence indicates that DPC proteolysis occurs only after the DNA replicative helicase CMG (CDC45/MCM2-7/GINS) has bypassed the intact DPC adduct (60), thereby protecting the replication machinery against promiscuous proteolysis.
The data presented here reveal that dihydropyrimidines are cytotoxic metabolites that induce DNA replication and transcriptional stress, and that the accumulation of dihydropyrimidines is linked with the accumulation of DPCs. Yet, we did not elucidate the chemical and structural identity of the damage (s) induced by dihydropyrimidines. Suppression of DHP did not yield any detectable increase in apurinic/apyrimidinic sites nor ribonucleotides incorporation into genomic DNA (data not shown). Some evidence suggests that dihydrouracil and its derivatives could be incorporated into ribonucleic acids (61,62), but it is not clear whether salvage pathways can convert 5,6-dihydrouracil and 5,6-dihydrothymine into nucleosides or deoxynucleosides. Suppression of the salvage pathway UPP1, however, did not attenuate the replication stress phenotype of cells treated with anti-DHP siRNAs or with dihydromyricetin. During the course of this study, we did not detect any direct evidence of dihydropyrimidines incorporation into DNA. Dihydropyrimidines are non-coding bases that have lost their planar structure as a consequence of the saturation of the C5-C6 double bound (17). Above physiological pH, and more slowly at physiological pH, these saturated bases can further decompose into fragments of bases (63). Some decomposition products could be genotoxic. Alternatively, metabolites alterations by chemical side reactions are widespread (64). Dihydropyrimidines may react with oxidants or other metabolites to form potent DNA damaging agents (65).
We report that dihydromyricetin inhibits the activity of recombinant human DHP. Dihydromyricetin is a versatile flavonoid from the Chinese pharmacopeia. It scavenges reactive oxygen species and has a variety of biological activities (66). Although dihydromyricetin is not a specific inhibitor of DHP, the latter is likely a target. Indeed, U-2 OS cells exposed to dihydromyricetin exhibited cellular phenotypes similar to that of DHP-depleted cells: (i) accumulation of protein-DNA crosslinks; (ii) replication forks slowing; and (iii) induction of markers of DNA replication stress. Consistent with the later observation, dihydromyricetin elicits p53 stabilization in hepatocellular carcinoma and activates the G2/M checkpoint via ATM/ATR signaling (67).
Suppression of NER factors, Spartan, or the Fanconi anemia proteins FANCA and FANCD2 did not sensitize cells to dihyromyricetin. The DNA translocase FANCM, however, promoted cellular tolerance to dihydromyricetin. This observation does not contradict the fact that the Fanconi anemia (FA) pathway is not involved in the repair of DPCs (57,68). The clinical symptoms of patients with biallelic inactivation of FANCM are distinct from Fanconi anemia (69–71). FANCM facilitates the FA pathway and exerts many FA-independent functions (72). Further studies will be required to understand exactly how FANCM mitigates the toxicity of dihydromyricetin.
A deficiency in DHP activity yields clinical symptoms that are consistent with dihydropyrimidines exerting toxic effects. Individuals carrying bi-allelic mutations in DHP accumulate high levels of 5, 6-dihydrouracil and 5, 6-dihydrothymine in urine, blood and cerebrospinal fluids. DHP deficiency can remain asymptomatic, but most patients present neurological abnormalities including mental retardation, hypotonia and seizures (73–76). DHP deficiency also manifests with growth retardation, dysmorphic features and gastrointestinal abnormalities (77–79).
In the context of carcinogenesis, the rewiring of the pyrimidine degradation pathway appears as a metabolic adaptation that supports tumor progression. Most cultured cells require glutamine for TCA cycle anaplerosis that yields precursors for several biosynthetic pathways, including nucleotides, which are necessary for tumor growth (2,80). The accumulation of the pyrimidine degradation products dihydropyrimidines facilitate directly the epithelial mesenchymal transition (EMT) (4). The role of dihydropyrimidines in cancer progression toward metastasis could be linked with the genotoxic potential of these metabolites. Endogenous DNA damage and DNA replication stress induce genomic instability and thereby accelerate the acquisition of growth promoting properties. Furthermore, key sensors and mediators of the DNA damage response regulate the EMT-associated transcription factor ZEB1 (81–83). An overload of DNA lesions, however, will trigger cell death. The previously unrecognized cytotoxicity of dihydropyrimidines described here implies that a tight equilibrium between pyrimidine synthetic and pyrimidine degradation activities is required for the proliferation of some cancer cells. We propose that DHP fulfills the function of a sanitization enzyme required for epithelial cancer cells to mitigate the toxicity of dihydropyrimidines (Figure 7C). DHP is a potential target for cancer chemotherapy (3). This study shows that DHP activity is a cellular target of dihydromyricetin. Dihydromyricetin induces cell cycle arrest and apoptosis in human gastric cancer cells, hepatocellular carcinoma and melanoma cells, without cytotoxicity to normal cells (67,84–86). It inhibits the growth of prostate cancer in mice (87). The model discussed here offers a conceptual framework for further exploring the potential therapeutic utility of targeted inhibition of DHP.
DATA AVAILABILITY
Mendeley data: https://data.mendeley.com/datasets/dssp4hdnw3/draft?a=7e48f805-d946-43bd-bd2d-f6cb080797b2.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Helena Sapede, Julie Devin, Touffic Kassouf, Lucile Broseus and Olivier Ganier for technical help, Ketan J Patel, Bruno Vaz and Kristijan Ramadan for helpful discussion and members of the laboratory for critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fondation ARC pour la recherche sur le cancer [PGA1 RF20180206787 to A.C]; l’Institut National du Cancer [INCa121770 to J.M., A.C.]; MSD Avenir (to A.C., D.M., M.M.); SIRIC Montpellier Cancer Grant [INCa_Inserm_DGOS_12553 to A.C.]; USA NIH Grants [AI136581, NIH AI150451USA to B.K.]; INSERM Plan Cancer (to D.M.); French National Research Agency [ANR-10-INBS-04; «Investments for the future» (to the Imaging facility MRI). Funding for open access charge: Fondation ARC pour la recherche sur le cancer [PGA1 RF20180206787].
Conflict of interest statement. None declared.
REFERENCES
- 1. Hanahan D., Weinberg R.A.. Hallmarks of cancer: the next generation. Cell. 2011; 144:646–674. [DOI] [PubMed] [Google Scholar]
- 2. Vander Heiden M.G., DeBerardinis R.J.. Understanding the intersections between metabolism and cancer biology. Cell. 2017; 168:657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Naguib F.N., el Kouni M.H., Cha S.. Enzymes of uracil catabolism in normal and neoplastic human tissues. Cancer Res. 1985; 45:5405–5412. [PubMed] [Google Scholar]
- 4. Shaul Y.D., Freinkman E., Comb W.C., Cantor J.R., Tam W.L., Thiru P., Kim D., Kanarek N., Pacold M.E., Chen W.W. et al.. Dihydropyrimidine accumulation is required for the epithelial-mesenchymal transition. Cell. 2014; 158:1094–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wikoff W.R., Grapov D., Fahrmann J.F., DeFelice B., Rom W.N., Pass H.I., Kim K., Nguyen U., Taylor S.L., Gandara D.R. et al.. Metabolomic markers of altered nucleotide metabolism in early stage adenocarcinoma. Cancer Prev. Res. 2015; 8:410–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Edwards L., Gupta R., Filipp F.V.. Hypermutation of DPYD deregulates pyrimidine metabolism and promotes malignant progression. Mol. Cancer Res. 2016; 14:196–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van Kuilenburg A.B., van Lenthe H., van Gennip A.H.. Activity of pyrimidine degradation enzymes in normal tissues. Nucleosides Nucleotides Nucleic Acids. 2006; 25:1211–1214. [DOI] [PubMed] [Google Scholar]
- 8. Yoo B.K., Gredler R., Vozhilla N., Su Z.Z., Chen D., Forcier T., Shah K., Saxena U., Hansen U., Fisher P.B. et al.. Identification of genes conferring resistance to 5-fluorouracil. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:12938–12943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Macheret M., Halazonetis T.D.. DNA replication stress as a hallmark of cancer. Annu. Rev. Pathol. 2015; 10:425–448. [DOI] [PubMed] [Google Scholar]
- 10. Zeman M.K., Cimprich K.A.. Causes and consequences of replication stress. Nat. Cell Biol. 2013; 16:2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bester A.C., Roniger M., Oren Y.S., Im M.M., Sarni D., Chaoat M., Bensimon A., Zamir G., Shewach D.S., Kerem B.. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011; 145:435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang S.N., Williams J.S., Arana M.E., Kunkel T.A., Pommier Y.. Topoisomerase I-mediated cleavage at unrepaired ribonucleotides generates DNA double-strand breaks. EMBO J. 2017; 36:361–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lazzaro F., Novarina D., Amara F., Watt D.L., Stone J.E., Costanzo V., Burgers P.M., Kunkel T.A., Plevani P., Muzi-Falconi M.. RNase H and postreplication repair protect cells from ribonucleotides incorporated in DNA. Mol. Cell. 2012; 45:99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Macheret M., Halazonetis T.D.. Intragenic origins due to short G1 phases underlie oncogene-induced DNA replication stress. Nature. 2018; 555:112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tuduri S., Crabbe L., Conti C., Tourriere H., Holtgreve-Grez H., Jauch A., Pantesco V., De Vos J., Thomas A., Theillet C. et al.. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat. Cell Biol. 2009; 11:1315–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Castellano-Pozo M., Santos-Pereira J.M., Rondon A.G., Barroso S., Andujar E., Perez-Alegre M., Garcia-Muse T., Aguilera A.. R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Mol. Cell. 2013; 52:583–590. [DOI] [PubMed] [Google Scholar]
- 17. Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993; 362:709–715. [DOI] [PubMed] [Google Scholar]
- 18. Langevin F., Crossan G.P., Rosado I.V., Arends M.J., Patel K.J.. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature. 2011; 475:53–58. [DOI] [PubMed] [Google Scholar]
- 19. Pontel L.B., Rosado I.V., Burgos-Barragan G., Garaycoechea J.I., Yu R., Arends M.J., Chandrasekaran G., Broecker V., Wei W., Liu L. et al.. Endogenous formaldehyde is a hematopoietic stem cell genotoxin and metabolic carcinogen. Mol. Cell. 2015; 60:177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tretyakova N.Y., Groehler A.t, Ji S.. DNA-protein cross-links: formation, structural identities, and biological outcomes. Acc. Chem. Res. 2015; 48:1631–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Luo J., Solimini N.L., Elledge S.J.. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009; 136:823–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jackson S.P., Helleday T.. DNA REPAIR. Drugging DNA repair. Science. 2016; 352:1178–1179. [DOI] [PubMed] [Google Scholar]
- 23. Zellweger R., Dalcher D., Mutreja K., Berti M., Schmid J.A., Herrador R., Vindigni A., Lopes M.. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J. Cell Biol. 2015; 208:563–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hashimoto Y., Chaudhuri A.R., Lopes M., Costanzo V.. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat. Struct. Mol. Biol. 2010; 17:1305–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sogo J.M., Lopes M., Foiani M.. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 2002; 297:599–602. [DOI] [PubMed] [Google Scholar]
- 26. Guo Z., Kumagai A., Wang S.X., Dunphy W.G.. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 2000; 14:2745–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hekmat-Nejad M., You Z., Yee M.C., Newport J.W., Cimprich K.A.. Xenopus ATR is a replication-dependent chromatin-binding protein required for the DNA replication checkpoint. Curr. Biol. 2000; 10:1565–1573. [DOI] [PubMed] [Google Scholar]
- 28. Liu Q., Guntuku S., Cui X.S., Matsuoka S., Cortez D., Tamai K., Luo G., Carattini-Rivera S., DeMayo F., Bradley A. et al.. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000; 14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 29. Zhao H., Piwnica-Worms H.. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol. Cell Biol. 2001; 21:4129–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ciccia A., Elledge S.J.. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010; 40:179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Collis S.J., Ciccia A., Deans A.J., Horejsi Z., Martin J.S., Maslen S.L., Skehel J.M., Elledge S.J., West S.C., Boulton S.J.. FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the Fanconi anemia core complex. Mol. Cell. 2008; 32:313–324. [DOI] [PubMed] [Google Scholar]
- 32. Moreaux J., Klein B., Bataille R., Descamps G., Maïga S., Hose D., Goldschmidt H., Jauch A., Rème T., Jourdan M. et al.. A high-risk signature for patients with multiple myeloma established from the molecular classification of human myeloma cell lines. Haematologica. 2011; 96:574–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kermi C., Prieto S., van der Laan S., Tsanov N., Recolin B., Uro-Coste E., Delisle M.B., Maiorano D.. RAD18 Is a Maternal Limiting Factor Silencing the UV-Dependent DNA Damage Checkpoint in Xenopus Embryos. Dev. Cell. 2015; 34:364–372. [DOI] [PubMed] [Google Scholar]
- 34. Luke-Glaser S., Luke B., Grossi S., Constantinou A.. FANCM regulates DNA chain elongation and is stabilized by S-phase checkpoint signalling. EMBO J. 2010; 29:795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wysocka J., Reilly P.T., Herr W.. Loss of HCF-1-chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Mol. Cell Biol. 2001; 21:3820–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jackson D.A., Pombo A.. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 1998; 140:1285–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Diamond T.L., Roshal M., Jamburuthugoda V.K., Reynolds H.M., Merriam A.R., Lee K.Y., Balakrishnan M., Bambara R.A., Planelles V., Dewhurst S. et al.. Macrophage tropism of HIV-1 depends on efficient cellular dNTP utilization by reverse transcriptase. J. Biol. Chem. 2004; 279:51545–51553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lutzmann M., Méchali M.. MCM9 binds Cdt1 and is required for the assembly of prereplication complexes. Mol. Cell. 2008; 31:190–200. [DOI] [PubMed] [Google Scholar]
- 39. Recolin B., Van der Laan S., Maiorano D.. Role of replication protein A as sensor in activation of the S-phase checkpoint in Xenopus egg extracts. Nucleic Acids Res. 2012; 40:3431–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gillespie P.J., Gambus A., Blow J.J.. Preparation and use of Xenopus egg extracts to study DNA replication and chromatin associated proteins. Methods. 2012; 57:203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vaz B., Popovic M., Newman J.A., Fielden J., Aitkenhead H., Halder S., Singh A.N., Vendrell I., Fischer R., Torrecilla I. et al.. Metalloprotease SPRTN/DVC1 Orchestrates Replication-Coupled DNA-Protein crosslink repair. Mol. Cell. 2016; 64:704–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kumbhar R., Vidal-Eychenie S., Kontopoulos D.G., Larroque M., Larroque C., Basbous J., Kossida S., Ribeyre C., Constantinou A.. Recruitment of ubiquitin-activating enzyme UBA1 to DNA by poly(ADP-ribose) promotes ATR signalling. Life Sci. Alliance. 2018; 1:e201800096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cox J., Mann M.. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008; 26:1367–1372. [DOI] [PubMed] [Google Scholar]
- 44. Adamson B., Smogorzewska A., Sigoillot F.D., King R.W., Elledge S.J.. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat. Cell Biol. 2012; 14:318–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Raderschall E., Golub E.I., Haaf T.. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:1921–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lane A.N., Fan T.W.. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res. 2015; 43:2466–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Techer H., Koundrioukoff S., Carignon S., Wilhelm T., Millot G.A., Lopez B.S., Brison O., Debatisse M.. Signaling from Mus81-Eme2-Dependent DNA Damage elicited by Chk1 deficiency modulates replication fork speed and origin usage. Cell Rep. 2016; 14:1114–1127. [DOI] [PubMed] [Google Scholar]
- 48. Li X., Manley J.L.. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005; 122:365–378. [DOI] [PubMed] [Google Scholar]
- 49. Mechali M., Harland R.M.. DNA synthesis in a cell-free system from Xenopus eggs: priming and elongation on single-stranded DNA in vitro. Cell. 1982; 30:93–101. [DOI] [PubMed] [Google Scholar]
- 50. Aze A., Fragkos M., Bocquet S., Cau J., Mechali M.. RNAs coordinate nuclear envelope assembly and DNA replication through ELYS recruitment to chromatin. Nat. Commun. 2017; 8:2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Errico A., Costanzo V., Hunt T.. Tipin is required for stalled replication forks to resume DNA replication after removal of aphidicolin in Xenopus egg extracts. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:14929–14934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Trenz K., Smith E., Smith S., Costanzo V.. ATM and ATR promote Mre11 dependent restart of collapsed replication forks and prevent accumulation of DNA breaks. EMBO J. 2006; 25:1764–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Huang C.Y. Inhibition of a putative dihydropyrimidinase from pseudomonas aeruginosa PAO1 by Flavonoids and substrates of cyclic amidohydrolases. PLoS One. 2015; 10:e0127634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kannouche P.L., Wing J., Lehmann A.R.. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. 2004; 14:491–500. [DOI] [PubMed] [Google Scholar]
- 55. Zlatanou A., Despras E., Braz-Petta T., Boubakour-Azzouz I., Pouvelle C., Stewart G.S., Nakajima S., Yasui A., Ishchenko A.A., Kannouche P.L.. The hMsh2-hMsh6 complex acts in concert with monoubiquitinated PCNA and Pol eta in response to oxidative DNA damage in human cells. Mol. Cell. 2011; 43:649–662. [DOI] [PubMed] [Google Scholar]
- 56. Sale J.E., Lehmann A.R., Woodgate R.. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 2012; 13:141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Stingele J., Bellelli R., Alte F., Hewitt G., Sarek G., Maslen S.L., Tsutakawa S.E., Borg A., Kjaer S., Tainer J.A. et al.. Mechanism and regulation of DNA-Protein crosslink repair by the DNA-Dependent metalloprotease SPRTN. Mol. Cell. 2016; 64:688–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Larsen N.B., Gao A.O., Sparks J.L., Gallina I., Wu R.A., Mann M., Raschle M., Walter J.C., Duxin J.P.. Replication-coupled DNA-protein crosslink repair by SPRTN and the proteasome in Xenopus egg extracts. Mol. Cell. 2019; 73:574–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kumar D., Viberg J., Nilsson A.K., Chabes A.. Highly mutagenic and severely imbalanced dNTP pools can escape detection by the S-phase checkpoint. Nucleic Acids Res. 2010; 38:3975–3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sparks J.L., Chistol G., Gao A.O., Raschle M., Larsen N.B., Mann M., Duxin J.P., Walter J.C.. The CMG helicase bypasses DNA-protein cross-links to facilitate their repair. Cell. 2019; 176:167–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mokrasch L.C., Grisolia S.. Incorporation of hydropyrimidine derivatives in ribonucleic acid with liver preparations. Biochim. Biophys. Acta. 1958; 27:226–227. [DOI] [PubMed] [Google Scholar]
- 62. Mokrasch L.C., Grisolia S.. Some enzymic actions on hydrouracil derivatives. Biochim. Biophys. Acta. 1960; 39:361–363. [DOI] [PubMed] [Google Scholar]
- 63. Lin G., Jian Y., Dria K.J., Long E.C., Li L.. Reactivity of damaged pyrimidines: DNA cleavage via hemiaminal formation at the C4 positions of the saturated thymine of spore photoproduct and dihydrouridine. J. Am. Chem. Soc. 2014; 136:12938–12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lerma-Ortiz C., Jeffryes J.G., Cooper A.J., Niehaus T.D., Thamm A.M., Frelin O., Aunins T., Fiehn O., de Crecy-Lagard V., Henry C.S. et al.. ‘Nothing of chemistry disappears in biology’: the Top 30 damage-prone endogenous metabolites. Biochem. Soc. Trans. 2016; 44:961–971. [DOI] [PubMed] [Google Scholar]
- 65. Wang M., Cheng G., Khariwala S.S., Bandyopadhyay D., Villalta P.W., Balbo S., Hecht S.S.. Evidence for endogenous formation of the hepatocarcinogen N-nitrosodihydrouracil in rats treated with dihydrouracil and sodium nitrite: a potential source of human hepatic DNA carboxyethylation. Chem. Biol. Interact. 2013; 206:83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li H., Li Q., Liu Z., Yang K., Chen Z., Cheng Q., Wu L.. The versatile effects of dihydromyricetin in health. Evid Based Complement Alternat. Med. 2017; 2017:1053617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Huang H., Hu M., Zhao R., Li P., Li M.. Dihydromyricetin suppresses the proliferation of hepatocellular carcinoma cells by inducing G2/M arrest through the Chk1/Chk2/Cdc25C pathway. Oncol. Rep. 2013; 30:2467–2475. [DOI] [PubMed] [Google Scholar]
- 68. Duxin J.P., Dewar J.M., Yardimci H., Walter J.C.. Repair of a DNA-protein crosslink by replication-coupled proteolysis. Cell. 2014; 159:346–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Neidhardt G., Hauke J., Ramser J., Gross E., Gehrig A., Muller C.R., Kahlert A.K., Hackmann K., Honisch E., Niederacher D. et al.. Association between Loss-of-Function mutations within the FANCM Gene and Early-Onset familial breast cancer. JAMA Oncol. 2016; 3:1245–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bogliolo M., Bluteau D., Lespinasse J., Pujol R., Vasquez N., d’Enghien C.D., Stoppa-Lyonnet D., Leblanc T., Soulier J., Surralles J.. Biallelic truncating FANCM mutations cause early-onset cancer but not Fanconi anemia. Genet. Med. 2017; 20:458–463. [DOI] [PubMed] [Google Scholar]
- 71. Catucci I., Osorio A., Arver B., Neidhardt G., Bogliolo M., Zanardi F., Riboni M., Minardi S., Pujol R., Azzollini J. et al.. Individuals with FANCM biallelic mutations do not develop Fanconi anemia, but show risk for breast cancer, chemotherapy toxicity and may display chromosome fragility. Genet. Med. 2017; 20:452–457. [DOI] [PubMed] [Google Scholar]
- 72. Basbous J., Constantinou A.. A tumor suppressive DNA translocase named FANCM. Crit. Rev. Biochem. Mol. Biol. 2019; 54:27–40. [DOI] [PubMed] [Google Scholar]
- 73. Sumi S., Kidouchi K., Hayashi K., Ohba S., Wada Y.. Dihydropyrimidinuria without clinical symptoms. J. Inherit. Metab. Dis. 1996; 19:701–702. [DOI] [PubMed] [Google Scholar]
- 74. van Kuilenburg A.B., Dobritzsch D., Meijer J., Meinsma R., Benoist J.F., Assmann B., Schubert S., Hoffmann G.F., Duran M., de Vries M.C. et al.. Dihydropyrimidinase deficiency: Phenotype, genotype and structural consequences in 17 patients. Biochim. Biophys. Acta. 2010; 1802:639–648. [DOI] [PubMed] [Google Scholar]
- 75. Putman C.W., Rotteveel J.J., Wevers R.A., van Gennip A.H., Bakkeren J.A., De Abreu R.A.. Dihydropyrimidinase deficiency, a progressive neurological disorder. Neuropediatrics. 1997; 28:106–110. [DOI] [PubMed] [Google Scholar]
- 76. van Kuilenburg A.B., Meijer J., Dobritzsch D., Meinsma R., Duran M., Lohkamp B., Zoetekouw L., Abeling N.G., van Tinteren H.L., Bosch A.M.. Clinical, biochemical and genetic findings in two siblings with a dihydropyrimidinase deficiency. Mol. Genet. Metab. 2007; 91:157–164. [DOI] [PubMed] [Google Scholar]
- 77. Hamajima N., Kouwaki M., Vreken P., Matsuda K., Sumi S., Imaeda M., Ohba S., Kidouchi K., Nonaka M., Sasaki M. et al.. Dihydropyrimidinase deficiency: structural organization, chromosomal localization, and mutation analysis of the human dihydropyrimidinase gene. Am. J. Hum. Genet. 1998; 63:717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Assmann B., Hoffmann G.F., Wagner L., Brautigam C., Seyberth H.W., Duran M., Van Kuilenburg A.B., Wevers R., Van Gennip A.H.. Dihydropyrimidinase deficiency and congenital microvillous atrophy: coincidence or genetic relation. J. Inherit. Metab. Dis. 1997; 20:681–688. [DOI] [PubMed] [Google Scholar]
- 79. Henderson M.J., Ward K., Simmonds H.A., Duley J.A., Davies P.M.. Dihydropyrimidinase deficiency presenting in infancy with severe developmental delay. J. Inherit. Metab. Dis. 1993; 16:574–576. [DOI] [PubMed] [Google Scholar]
- 80. Lunt S.Y., Muralidhar V., Hosios A.M., Israelsen W.J., Gui D.Y., Newhouse L., Ogrodzinski M., Hecht V., Xu K., Acevedo P.N. et al.. Pyruvate kinase isoform expression alters nucleotide synthesis to impact cell proliferation. Mol. Cell. 2015; 57:95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Park S.Y., Korm S., Chung H.J., Choi S.J., Jang J.J., Cho S., Lim Y.T., Kim H., Lee J.Y.. RAP80 regulates epithelial-mesenchymal transition related with metastasis and malignancy of cancer. Cancer Sci. 2016; 107:267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Liu X., Dong R., Jiang Z., Wei Y., Li Y., Wei L., Sun H., Li Y., Yang N., Yang Q. et al.. MDC1 promotes ovarian cancer metastasis by inducing epithelial-mesenchymal transition. Tumour Biol. 2015; 36:4261–4269. [DOI] [PubMed] [Google Scholar]
- 83. Zhang P., Wei Y., Wang L., Debeb B.G., Yuan Y., Zhang J., Yuan J., Wang M., Chen D., Sun Y. et al.. ATM-mediated stabilization of ZEB1 promotes DNA damage response and radioresistance through CHK1. Nat. Cell Biol. 2014; 16:864–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ji F.J., Tian X.F., Liu X.W., Fu L.B., Wu Y.Y., Fang X.D., Jin H.Y.. Dihydromyricetin induces cell apoptosis via a p53-related pathway in AGS human gastric cancer cells. Genet. Mol. Res. 2015; 14:15564–15571. [DOI] [PubMed] [Google Scholar]
- 85. Zeng G., Liu J., Chen H., Liu B., Zhang Q., Li M., Zhu R.. Dihydromyricetin induces cell cycle arrest and apoptosis in melanoma SK-MEL-28 cells. Oncol. Rep. 2014; 31:2713–2719. [DOI] [PubMed] [Google Scholar]
- 86. Zhang Q., Liu J., Liu B., Xia J., Chen N., Chen X., Cao Y., Zhang C., Lu C., Li M. et al.. Dihydromyricetin promotes hepatocellular carcinoma regression via a p53 activation-dependent mechanism. Sci. Rep. 2014; 4:4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ni F., Gong Y., Li L., Abdolmaleky H.M., Zhou J.R.. Flavonoid ampelopsin inhibits the growth and metastasis of prostate cancer in vitro and in mice. PLoS One. 2012; 7:e38802. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mendeley data: https://data.mendeley.com/datasets/dssp4hdnw3/draft?a=7e48f805-d946-43bd-bd2d-f6cb080797b2.