Abstract
A number of regulatory nascent peptides have been shown to regulate gene expression by causing programmed ribosome stalling during translation. Nascent peptide emerges from the ribosome through the exit tunnel, and one-third of the way along which β-loop structures of ribosomal proteins uL4 and uL22 protrude into the tunnel to form the constriction region. Structural studies have shown interactions between nascent peptides and the exit tunnel components including the constriction region. In eukaryotes, however, there is a lack of genetic studies for the involvement of the constriction region in ribosome stalling. Here, we established transgenic Arabidopsis lines that carry mutations in the β-loop structure of uL4. Translation analyses using a cell-free translation system derived from the transgenic Arabidopsis carrying the mutant ribosome showed that the uL4 mutations reduced the ribosome stalling of four eukaryotic stalling systems, including those for which stalled structures have been solved. Our data, which showed differential effects of the uL4 mutations depending on the stalling systems, explained the spatial allocations of the nascent peptides at the constriction that were deduced by structural studies. Conversely, our data may predict allocation of the nascent peptide at the constriction of stalling systems for which structural studies are not done.
INTRODUCTION
During translation, new peptide bonds are formed at the peptidyltransferase center (PTC) in the ribosomal large subunit, and the growing peptide passes through the exit tunnel that penetrates the large subunit before emerging from the ribosome. However, some of the nascent peptides, or the regulatory nascent peptides, direct the ribosome to stall during translation. This nascent peptide-mediated ribosome stalling (NPmRS) occurs either autonomously or is facilitated by an effector molecule, at either elongation or termination of translation, and on either the main open reading frame (ORF) or an upstream ORF (uORF) (1), which is a small ORF present upstream of the main ORF of eukaryotic mRNAs.
The exit tunnel is ∼100 Å long and holds 30–40 amino acid residues of the nascent peptide (2,3). Regulatory nascent peptides are usually 20–30 amino acids long and function while inside the exit tunnel. In both bacteria and eukaryotes, β-loop structures of two ribosomal proteins, uL4 and uL22 (4), which protrude into the exit tunnel, form a constriction region located 30–40 Å from the PTC (2,3). This region has been suggested to function as a discriminating gate by interacting with the nascent peptides (5).
In bacteria, forward and reverse genetics studies have revealed the importance of interactions between the nascent peptide and the constriction region in mediating NPmRS (5–9). This is supported by structural analyses of stalled ribosomes using cryo-electron microscopy (cryo-EM), which identified physical contacts of the nascent peptides with exit tunnel components, including the constriction region (10–13).
In eukaryotes, cryo-EM studies revealed physical contacts of the nascent peptides with the constriction region in stalled ribosomes of Neurospora crassa arg-2 uORF, termed the arginine attenuator peptide (AAP) (the AAP system), and human cytomegalovirus (hCMV) gp48 uORF (the hCMV system) (14–16). However, there is no genetic evidence showing the contribution of the constriction region to ribosome stalling.
In Arabidopsis CGS1, encoding cystathionine γ-synthase, the first committed enzyme of methionine biosynthesis, S-adenosyl-l-methionine (AdoMet), a direct metabolite of methionine, induces NPmRS during translation elongation at Ser-94 (the CGS1 system) (17–19). The functional amino acid sequence for the response to AdoMet is termed as MTO1 region (20–22). Studies using the wheat germ extract (WGE) in vitro translation system revealed that the nascent peptide adopts a compact conformation and 28S rRNA residues, including those near the constriction region, undergo conformation changes upon stalling (18). Notably, these rRNA residues are located at or near the rRNA residues for which structural studies have identified physical contacts with the nascent peptides in other stalling systems (14). This suggests involvement of the constriction region in the CGS1 system, but whether the conformation changes are the cause or result of stalling remains unknown.
Here, we conducted reverse genetics-based biochemical studies to determine the involvement of the constriction region in eukaryotic NPmRS. To this end, we constructed transgenic Arabidopsis lines carrying mutant uL4-containing ribosomes. We examined effects of the mutations on ribosome stalling using an Arabidopsis cell-free extract (ACE) in vitro translation system (23) prepared from transgenic lines carrying FLAG-tagged mutant uL4.
For the NPmRS systems to be tested, we selected five from divergent eukaryotes (Figure 1), of which four have their relevant amino acid residues >20 from the stalled residue, as in most NPmRS systems, and should cross over the constriction region. These include the CGS1, hCMV (27–29) and AAP (24–26) systems mentioned above, and Arabidopsis AdoMet decarboxylase 1 (AtAMD1) S-uORF (the AtAMD1 system) (30,31). We also tested another NPmRS system, mammalian AdoMet decarboxylase 1 (mAMD1) uORF. This uORF, encoding six amino acids, MAGDIS (the MAGDIS system) (32,33), is the shortest among the NPmRS systems thus far identified and its nascent peptide is too short to reach the constriction region (Figure 1). Lastly, we tested the shortest possible uORF, AUG-stop, of AtNIP5;1. AUG-stop causes prolonged ribosome stalling in response to boric acid (the AUG-Stop system) (34). This minimum uORF codes for only one amino acid, methionine, and is not actually an NPmRS, but was included as a negative control.
Figure 1.
The stalling systems analyzed in this study. For each system, the gene and ORF (main ORF or uORF) in which stalling occurs, effector molecule, and translation step at which stalling occurs (elongation or termination) are shown. Amino acid sequences relevant for stalling in CGS1 or those encoded by the uORFs are shown. Amino acid residues important for NPmRS (17,21, 24,26,28,29,32–34) are marked in red. For AtAMD1, functional amino acids are not reported, while importance of the amino acid sequence was shown by frame-shift mutation (31). The MTO1 (21) region of CGS1 is underlined with ambiguous amino acids in dotted underline. Amino acid residues are numbered from the stalled residue (−1, marked with a clover). Approximate position of the constriction region is shaded. Since the nascent peptide may form α-helix in the PTC-proximal region as reported in AAP and hCMV (14,16), residues that cross over the constriction region may be shifted to the left.
We present here biochemical evidence that the constriction region plays a crucial role in inducing NPmRS, in which nascent peptides cross over the constriction. The differential effects of the uL4 mutations explained the structural data for stalled ribosomes of the AAP and hCMV systems.
MATERIALS AND METHODS
Plant materials, transformation and growth conditions
Arabidopsis thaliana (L.) Heynh. ecotype Columbia (Col-0) was used as the wild-type plant line. A T-DNA insertion knockout mutant of uL4D (SALK_029203) (35) was obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus, OH, USA) and is referred to as ul4d(KO) in this paper. Transformation of Arabidopsis plants was performed using the floral dip method (36) with Agrobacterium tumefaciens strain GV3101. Plant growth conditions were as described previously (37). T1 transgenic plants were selected on Murashige and Skoog (MS) medium supplemented with 15 μg ml−1 hygromycin. T3 homozygous lines with single insertions of uL4D promoter::uL4D(WT):FLAG and its variants were established based on the segregation of hygromycin resistance.
For root observations, plants were grown on vertically placed half-strength MS medium containing 1% (w/v) sucrose, 0.5% (w/v) MES (pH 5.7) and 1.5% (w/v) gellan gum for 10 days in a growth chamber at 22°C under fluorescent light with a 16 h-light/8 h-dark cycle. Root length was measured using RootNav software (38). For leaf morphology analysis, plants were grown for 21 days as described previously (37) and one of the first pair of the true leaves was dissected. Leaf index (ratio of length to width of the leaf blade) was calculated using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Chemicals
AdoMet and spermidine were purchased from Sigma-Aldrich (St. Louis, MO, USA), and l-arginine was purchased from Wako Pure Chemicals (Osaka, Japan). Other chemicals were obtained as described previously (18, 23).
Construction of plasmids used for plant transformation
Plasmids pYTJ10, pYTJ1, pYTJ7 and pYTJ4 (Supplementary Table S1) carry uL4D promoter::uL4D(WT):FLAG, uL4D promoter::uL4D(R77A):FLAG, uL4D promoter::uL4D(ΔTV):FLAG and uL4D promoter::uL4D(Δloop):FLAG DNA, respectively, in the pGWB10 T-DNA binary vector (39). To construct pYTJ10, the −1020 to −1 nt region (relative to the translation start site) of wild-type uL4D and the coding region were amplified by PCR from genomic DNA of Col-0 using the primers uL4Df and uL4Dr (Supplementary Table S2). The amplified fragment was subcloned into the pENTR/D-TOPO vector (Invitrogen, Carlsbad, CA, USA) using the TOPO cloning reaction. The cloned full-length uL4D(WT) DNA was subsequently recombined into the pGWB10 T-DNA binary vector using the Gateway system (40). For the construction of uL4D(R77A), uL4D(ΔTV) and uL4D(Δloop) mutations, the overlap extension PCR method (41,42) was employed. The flanking primers, R77Af and R77Ar for the uL4D(R77A) mutation, dTVf and dTVr for the uL4D(ΔTV) mutation, and dLoopf and dLoopr for the uL4D(Δloop) mutation, and the flanking primers uL4Flankf and uL4Flankr (Supplementary Table S2) were used. The mutated DNA fragment was introduced into the uL4D(WT) coding region of pYTJ10 to generate pYTJ1, pYTJ7 and pYTJ4 plasmids. For all constructs, the integrity of PCR-amplified regions was confirmed by sequence analysis.
Construction of plasmids used for stalling assay
Plasmid pST00 (Supplementary Table S1) carries the M8:His:HA:DP75 sequence between the SP6 promoter and the 30-nt poly(A) in the pSP64 poly(A) vector (Promega, Madison, WI, USA). The DP75 linker was derived from the dipeptidyl-aminopeptidase B coding sequence (15,43). To construct pST00, the His:HA:DP75 coding region was amplified by PCR from pYY105, which carries T7::His:HA:DP75:uORF2(WT) DNA in the pEX-A2 vector (43), using the primers DP75f and DP75r (Supplementary Table S2). The amplified fragment was digested with XbaI and BamHI and inserted between the XbaI and BamHI sites of pYK00, which harbors M8:CGS1(WT) DNA in the pSP64 poly(A) vector (18).
Plasmids pST55 and pST56 (Supplementary Table S1) carry M8:His:HA:DP75:AAP(WT) and M8:His:HA:DP75:AAP(D12N), respectively, in the pSP64 poly(A) vector. To construct these plasmids, complementary oligonucleotides AAPf/AAPr or D12Nf/AAPr (Supplementary Table S2), encoding wild-type or mutated N. crassa arg-2 (24), respectively, were annealed and filled-in using KOD-Plus-Neo DNA polymerase (Toyobo, Osaka, Japan). The amplified fragments were digested with EcoRV and BamHI and inserted between the EcoRV and BamHI sites of pST00.
Plasmids pTI5 and pTI6 (Supplementary Table S1) carry M8:His:HA:3xFLAG:Myc:DP75:AAP(WT) and M8:His:HA:3xFLAG:Myc:DP75:AAP(D12N), respectively, in the pSP64 poly(A) vector. To introduce the Myc tag sequence between the HA tag and the DP75 linker in pST55 and pST56, in vitro mutagenesis was carried out by inverse PCR using the primers MycDP75f and HAMycr (Supplementary Table S2), followed by digestion of the plasmid DNA by DpnI (Toyobo). The amplified fragments were self-ligated using T4 DNA ligase (Wako Pure Chemicals) and T4 polynucleotide kinase (Toyobo). To construct pTI5 and pTI6, complementary oligonucleotides 3xFLAGf and 3xFLAGr (Supplementary Table S2) were annealed and filled-in by PCR, and the amplified fragments were ligated with the PCR-amplified fragment from the Myc tag-fused pST55 or pST56 using Mycf and HAr primers (Supplementary Table S2) by the SLiCE reaction (44).
Plasmids pST57 and pST58 (Supplementary Table S1) carry M8:His:HA:DP75:hCMV(WT) and M8:His:HA:DP75:hCMV(P21A), respectively, in the pSP64 poly(A) vector. To construct these plasmids, complementary oligonucleotides hCMVf/hCMVr or hCMVf/P21Ar (Supplementary Table S2), encoding wild-type or mutated uORF2 of hCMV gp48 (15,45), respectively, were annealed and filled-in by PCR. The amplified fragments were inserted into pST00 in the same way as for the construction of pST55.
Plasmids pST76 and pST77 (Supplementary Table S1) carry M8:His:HA:DP75:S-uORF(WT) and M8:His:HA:DP75:S-uORF(fs), respectively, in the pSP64 poly(A) vector. To construct these plasmids, the coding region of the AtAMD1 S-uORF was amplified from pNU14 or pNU15, which carries GST:S-uORF(WT):RLUC or GST:S-uORF(fs):RLUC, respectively, in the pSP64 poly(A) vector (31), using the primer sets SAMDC1f/SAMDC1r or SAMDC1fsf/SAMDC1fsr (Supplementary Table S2), respectively. The amplified fragments were inserted into pST00 in the same way as for the construction of pST55.
Plasmids pST116 and pST117 (Supplementary Table S1) carry M8:His:HA:DP75:MAGDIS(WT) and M8:His:HA:DP75:MAGDIS(I5L), respectively, in the pSP64 poly(A) vector. To construct these plasmids, the His:HA:DP75 tag sequence was amplified from pST00 using the primer sets DP75f/MAGDISr or DP75f/I5Lr (Supplementary Table S2), respectively, and the amplified fragments were digested with XbaI and BamHI and inserted between the XbaI and BamHI sites of pST00.
Plasmids pST124 and pST125 (Supplementary Table S1) carry M8:His:HA:3xFLAG:Myc:DP75:MAGDIS(WT) and M8:His:HA:3xFLAG:Myc:DP75:MAGDIS(I5L), respectively, in the pSP64 poly(A) vector. To construct these plasmids, the His:HA:3xFLAG:Myc:DP75 tag sequence was amplified from pTI5 using the primer sets DP75fMAGDISr or DP75f/I5Lr (Supplementary Table S2), respectively, and the amplified fragments were inserted into pST00 in the same way as for the construction of pST55. For all constructs, the integrity of PCR-amplified regions was confirmed by sequence analysis.
In the Results section, M8:His:HA and M8:His:HA:3xFLAG:Myc sequences are referred to as TagI and TagII, respectively.
Construction of plasmids used for reporter assay
Plasmids pST122 and pST123 (Supplementary Table S1) carry gp48 5′-UTR(WT):LUC and gp48 5′-UTR(P21A):LUC, respectively, in the pSP64 poly(A) vector. To construct these plasmids, complementary oligonucleotides hCMVf2/hCMVr2 or hCMVf2/P21Ar2 (Supplementary Table S2), which correspond to the wild-type or mutated 5′-UTR of the hCMV gp48 sequence (45,46), respectively, were annealed and filled-in by PCR. The amplified fragments were digested with HindIII and inserted at the HindIII site of pMI21, which carries CGS1(WT):LUC in the pSP64 poly(A) vector (22).
Plasmids pST120 and pST121 (Supplementary Table S1) carry AtAMD1 5′-UTR(WT):LUC and AtAMD1 5′-UTR(fs):LUC, respectively, in the pSP64 poly(A) vector. To construct these plasmids, the 5′-UTR of AtAMD1 was amplified from pSY209 or pSY214, which carry S-uORF(WT):RLUC or S-uORF(fs):RLUC, respectively, in the pSP64 poly(A) vector (31) using the primers SAMDC1f2 and SAMDC1r2 (Supplementary Table S2). The amplified fragments were digested with XbaI and NcoI and inserted between the XbaI and NcoI sites of pMI21.
Plasmids pST118 and pST119 (Supplementary Table S1) carry mAMD15′-UTR(WT):LUC and mAMD15′-UTR(I5L):LUC, respectively, in the pSP64 poly(A) vector. To construct these plasmids, complementary oligonucleotides MAGDIS(WT)f/MAGDIS(WT)r or MAGDIS(WT)f/MAGDIS(I5L)r (Supplementary Table S2), which correspond to the wild-type or I5L mutant 5′-UTR (47), respectively, were annealed and filled-in by PCR. pSR327 carries a chimeric 5′-UTR: −327 to −311 nt of the human AMD1 and −310 to −1 nt of the bovine AMD1 (47). The amplified fragments were inserted into pMI21 in the same way as for the construction of pST120. For all constructs, the integrity of PCR-amplified regions was confirmed by sequence analysis.
Plasmid pMI27 carries the RLUC gene in the pSP64 poly(A) vector (22). Plasmids pMT131 and pMT132 carry the wild-type or mutated 5′-UTR of AtNIP5;1, respectively, joined to the LUC gene in the pSP64 poly(A) vector (34).
Quantitative RT-PCR
Total RNA was extracted from plants grown for 14 days using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) and treated with DNase I. Total RNA was reverse-transcribed to cDNA using the SuperScript III First-Strand Synthesis System (Invitrogen) with oligo-d(T)12–18 primers. Real-time PCR was performed using a LightCycler 480 Real-time System (Roche, Basel, Switzerland) and the LightCycler 480 SYBR Green I Master kit. The Arabidopsis UBQ5 gene was used as an internal control. The primers used are listed in Supplementary Table S2.
In vitro transcription
DNA templates in the pSP64 poly(A) vector were linearized with EcoRI and purified using the QIAquick Nucleotide Removal Kit (Qiagen), with the exception of the M8:His:HA:DP75 and M8:His:HA:3xFLAG:Myc:DP75 fusion constructs. DNA templates for these plasmids and nonstop RNAs were prepared by amplifying the corresponding region by PCR using KOD-Plus-Neo DNA polymerase. For PCR amplification, SP65′fP (18) was used as a forward primer and the reverse primers as listed in Supplementary Table S2 were used. In vitro transcription in the presence of a cap analog, m7G[5′]ppp[5′]GTP, was carried out as described previously (22).
In vitro translation
Preparation of ACE and in vitro translation reactions using ACE were carried out at 25°C as described previously (23). Unless otherwise stated, the template RNA was used at 50 fmol μl–1. For RNase A treatment, RNase A was added at a final concentration of 0.5 mg ml−1 and reaction mixtures were incubated for 15 min at 37°C. For immunoblot analysis, reaction mixtures were diluted with the 1× SDS-PAGE gel sample buffer (50 mM Tris–HCI pH 6.8, 50 mM DTT, 10% (v/v) glycerol and 1% (w/v) SDS).
Immunoblot analysis
For immunoblot analysis of in vitro translation products, samples were separated on a NuPAGE 4–12% or 12% Bis–Tris Gel (Invitrogen), transferred to an Immobilon-P membrane (Millipore, Billerica, MA, USA), and immuno-reacted with either a polyclonal anti-GST antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA), a monoclonal anti-HA antibody (Santa Cruz Biotechnology), or a monoclonal anti-FLAG M2 antibody (Sigma-Aldrich). MOPS running buffer (Life Technologies, Carlsbad, CA, USA) was used for the GST tag-fusion construct. MES running buffer (Life Technologies) was used for the M8:His:HA:DP75 or M8:His:HA:3xFLAG:Myc:DP75 fusion constructs. The signals were detected using an Immobilon Forte Western HRP substrate (Millipore) and visualized using a LAS-3000 mini imaging system (GE Healthcare, Little Chalfont, UK). The band intensities were quantified using MultiGauge software (Fuji Photo Film, Tokyo, Japan).
Total proteins from Arabidopsis callus cultures were separated by SDS-PAGE with MOPS running buffer and subjected to immunoblot analyses using a monoclonal anti-FLAG M2 antibody (Sigma-Aldrich), anti-uL22 antiserum, anti-uL4 antiserum (Proteintech, Rosemont, IL, USA), or anti-β-actin antiserum (Gene Tex, Irvine, CA, USA). The polyclonal antibody against human uL4 (accession no. BC009888) recognizes both endogenous Arabidopsis uL4A and uL4D as epitopes (48). The polyclonal antiserum raised against Arabidopsis uL22 was generated by immunizing rabbit with a synthetic peptide corresponding to amino acids 4–20 of Arabidopsis uL22B (YSQEPDNQTKSCKARGS; At1g67430).
Isolation of Arabidopsis ribosomes
For ribosome isolation, liquid callus cultures were prepared from Arabidopsis seedlings as described previously (23) and harvested on the 21st day after callus induction. Frozen tissues were homogenized in buffer D (49). After clarification by centrifugation at 16 000 × g for 30 min, crude extract (S16) was loaded on the 1.75 M sucrose cushion (100 mM Tris–HCl pH 8.0, 40 mM KCl, 20 mM MgCl2, 1 mM DTT and 100 μg ml−1 cycloheximide) and centrifuged at 170 000 × g for 4 h at 4°C in a TLA100.3 rotor (Beckman Coulter, Brea, CA, USA) to obtain the S170 soluble protein fraction and the P170 ribosomal pellet. The P170 fraction was suspended in the resuspension buffer (100 mM Tris–HCl pH 8.0, 40 mM KCl, 20 mM MgCl2, 1 mM DTT and 100 μg ml−1 cycloheximide). For immunoblot analysis, the protein concentration was measured using a Pierce 660 nm Protein Assay Reagent (Invitrogen), in accordance with the manufacturer's instructions.
Polysome profiling analysis
For the fractionation of total polysomes, ribosomes (P170) were isolated as described above. Ribosome suspension was loaded on a 15–60% (w/v) sucrose density gradient (40 mM Tris–HCl pH 8.4, 20 mM KCl, 10 mM MgCl2 and 5 μg ml−1 cycloheximide) and centrifuged at 275 000 × g for 1.5 h at 4°C in an SW55 Ti rotor (Beckman Coulter). The UV absorbance profile at 254 nm was recorded using an ISCO 520 gradient system (ISCO, Lincoln, NE, USA). For immunoblot analysis, fractions were analyzed as described above.
Affinity purification of FLAG-tagged ribosomes
Immunoprecipitation of FLAG-tagged ribosomes using an anti-FLAG M2 affinity resin (Sigma-Aldrich) was performed as described previously (50) with some modifications. All procedures were carried out at 4°C. After in vitro translation of 25 pmol GST:CGS1(G183-ns) RNA in a 50-μl reaction mixture, translation products were diluted with 50 μl of ice-cold 2× binding buffer (100 mM Tris–HCl pH 7.5, 24 mM Mg(OAc)2, 1 mM DTT, 1 mM PMSF, 20 μg ml−1 cycloheximide and 0.05 U μl−1 RNase inhibitor (Promega)) and 40 μl (bed volume) of anti-FLAG M2 affinity resin. After incubation with gentle shaking for 1 h, the resin was washed with ice-cold IXA-100 buffer (50 mM Tris–HCl pH 7.5, 100 mM KCl, 12 mM Mg(OAc)2, 1 mM DTT, 1 mM PMSF and 20 μg ml−1 cycloheximide). Bound proteins were eluted by the addition of 15 μl of the IXA-100 buffer containing 200 μg ml−1 FLAG peptides (Sigma-Aldrich). For immunoblot analysis, eluate was diluted with 2× SDS-PAGE gel sample buffer (100 mM Tris–HCI pH 6.8, 100 mM DTT, 20% glycerol and 2% SDS).
Reporter assay
Tester RNA at 2 fmol μl–1 carrying firefly luciferase (LUC) reporter gene and 1 fmol μl–1Renilla reniformis luciferase (RLUC) control RNA were co-translated for 120 min. LUC and RLUC activities were measured as described previously (22), and the LUC activity was normalized with the control RLUC activity to obtain reporter activity.
Statistical treatments
For comparisons of stalling efficiencies, Welch's t-test followed by false discovery rate (FDR) correction of Benjamini and Hochberg (51) was applied. The same FDR correction was also applied to comparisons of mRNA levels among the transgenic lines. For multiple comparisons of plant phenotypes, the Tukey–Kramer test was applied. For other comparisons, Welch's t-test was applied.
RESULTS
Construction of transgenic Arabidopsis carrying mutations in uL4
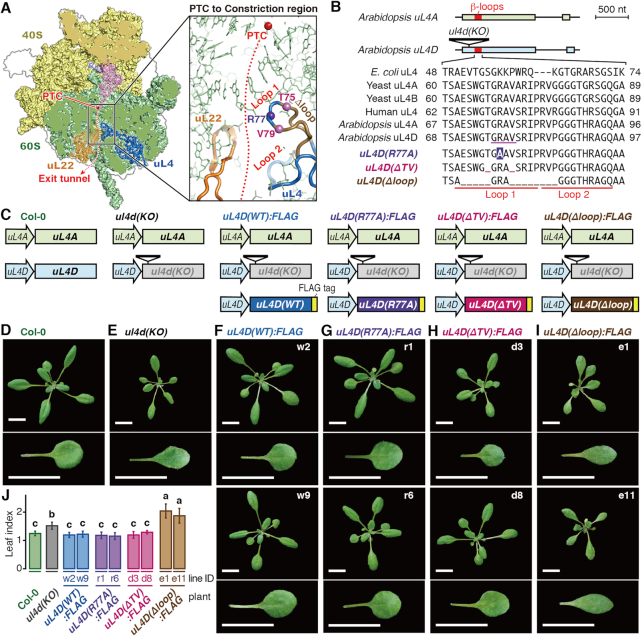
The uL4 protein contains a large loop structure termed the internal extended loop, at the tip of which are two smaller β-loop structures, designated here as Loops 1 and 2 (Supplementary Figure S1A). Loops 1 and 2 protrude into the exit tunnel to constitute the constriction region (Figure 2A).
Figure 2.
Construction of transgenic Arabidopsis carrying uL4D mutations and their phenotypes. (A) Structure of wheat 80S ribosome (PDB 4V7E) (54) (Left panel) and expansion of the constriction region (Right panel). The α-carbon atoms of uL4 residues substituted in R77A (Arg-77) and deleted in ΔTV (Thr-75 and Val-79) mutations are marked in violet and magenta balls, respectively, while those deleted in Δloop (Glu-71 to Thr-75 and Val-79 to Pro-86) mutation is marked with light brown main chain. (B) (Upper panel) Schematic representation of gene structures of uL4A and uL4D, and T-DNA insertion in ul4d(KO) (SALK_029203). Exons and introns are indicated as boxes and lines, respectively, with the β-loop region marked in red. (Lower panel) Alignment of amino acid sequences of Loop 1 and 2 regions. Substitutions and deletions in uL4D mutants are shown in reversed letters and underscores, respectively. The residues of uL4 in physical contact with AAP nascent peptide as deduced by cryo-EM in WGE (14) are underlined in magenta in Arabidopsis uL4D. (C) Schematic representation of uL4 gene sets carried by Arabidopsis wild-type Col-0, ul4d(KO), and FLAG-tagged uL4D mutant lines. (D–I) Rosettes and one of the first pair of true leaves of wild-type Col-0, ul4d(KO), and FLAG-tagged uL4D mutant transgenic plants (indicated by labels) grown for 21 days. Independent transgenic line IDs used in this study are indicated. Bars = 10 mm. (J) Length-to-width ratio (leaf index) of Col-0 and uL4D mutant plants. Different letters indicate significant differences (P < 0.05, Tukey–Kramer test).
In Arabidopsis, uL4 is encoded by two paralogous genes, uL4A and uL4D (52,53), sharing 95% amino acid identity. The amino acid sequences of Loops 1 and 2 are highly conserved among eukaryotes (Figure 2B and Supplementary Figure S1B). Since double-knockout mutant of uL4A and uL4D is lethal (48), we introduced a modified uL4D gene into the T-DNA insertion knockout mutant of uL4D, referred to here as the ul4d(KO) line, while keeping endogenous uL4A intact (Figure 2C).
We constructed three mutant uL4Ds, namely uL4D(R77A), uL4D(ΔTV) and uL4D(Δloop) (Figure 2A–C). uL4D(R77A) carries an Arg-77 to alanine substitution, located near the tip of Loop 1. uL4D(ΔTV) carries deletions of Thr-75 and Val-79 flanking the tip, while uL4D(Δloop) carries larger deletions flanking the tip (deletions of Glu-71 to Thr-75 and Val-79 to Pro-86). These mutant uL4Ds and the control gene, uL4D(WT), were FLAG-tagged at their C-termini and placed under the control of their native promoter (Figure 2C). The C-terminus of uL4 is exposed to the ribosomal surface (54). A large complex of the ribosome on mRNA can be affinity-purified using a FLAG tag (50,55).
In Arabidopsis ecotype Col-0 wild-type plants (hereafter Col-0), uL4A and uL4D mRNAs accumulate to comparable levels (Supplementary Figure S2). uL4 mRNA accumulation levels in the uL4D mutant plants that we constructed were quantified (Figure 3). In all the transgenic plants, endogenous uL4A mRNA levels were similar to those in wild-type Col-0 plants, whereas uL4D mRNA levels varied depending on the mutant construct. In uL4D(WT):FLAG and uL4D(R77A):FLAG plants, uL4D mRNA levels were generally higher than in Col-0, while in uL4D(ΔTV):FLAG and uL4D(Δloop):FLAG plants, they were about the same or lower than in Col-0. In subsequent experiments, we used two independent transgenic lines for each mutant construct (Figure 3, line IDs in bold letters).
Figure 3.
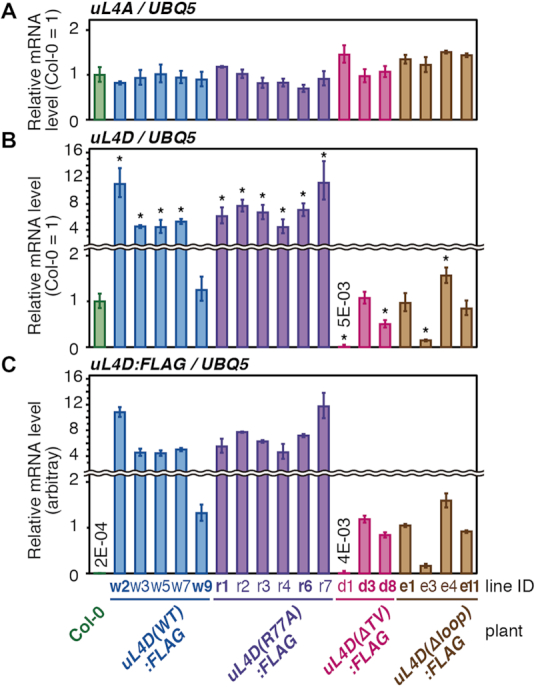
Accumulation levels of endogenous uL4A and FLAG-tagged mutant uL4D mRNAs in transgenic lines. Total RNA was extracted from seedlings of wild-type Col-0 and transgenic plants 14 days after imbibition. uL4A (A), uL4D (B), and uL4D:FLAG (C) mRNAs were quantified by qRT-PCR using UBQ5 mRNA as a control. Means ± SD of triplicated experiments are shown. The y-axis in (C) was set so that the means, excluding Col-0, of uL4D:FLAG mRNA in (C) and uL4D mRNA in (B) are the same. In (A) and (B), asterisks indicate significant differences from wild-type Col-0 plants (q < 0.05, Welch's t-test with FDR correction). In (A), no significant difference was detected. The line IDs of transgenic lines that were used for further experiments are shown in bold letters.
Mutant uL4Ds complement the ribosome-deficient phenotypes in vivo, except in uL4D(Δloop):FLAG plants
In Arabidopsis, mutations of ribosomal proteins often have multiple effects on growth and development, including narrower leaf morphology, termed the pointed leaf, and a short-root phenotype (56). ul4d(KO) plants also exhibited these phenotypes (Figure 2E and Supplementary Figure S3) (48,57,58). Both phenotypes were complemented not only in the uL4D(WT):FLAG plants but also in the uL4D(R77A):FLAG and uL4D(ΔTV):FLAG plants (Figure 2F–H and J, and Supplementary Figure S3), indicating the functionality of mutant ribosomes in vivo. In contrast, neither phenotype was complemented in uL4D(Δloop):FLAG plants. Notably, the plants exhibited even narrower leaves (Figure 2I and J).
Mutant uL4D proteins are efficiently incorporated into translating ribosomes, except in the uL4D(Δloop):FLAG line
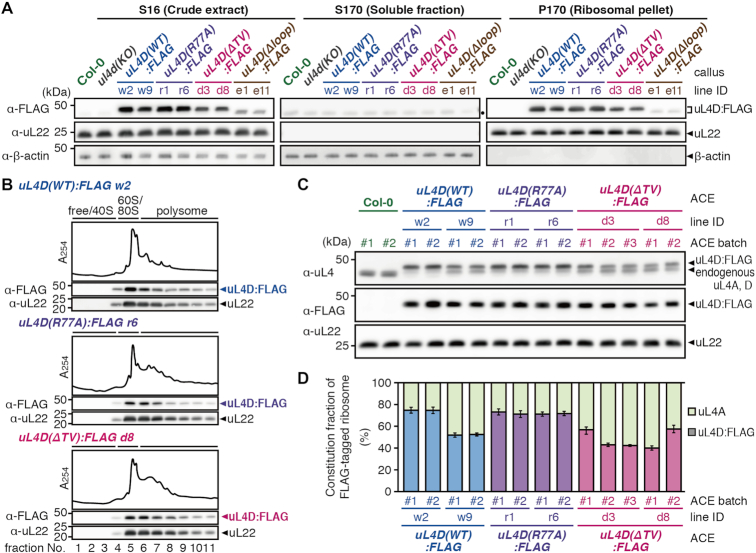
To determine whether the uL4D mutant proteins are incorporated into ribosome particles, we tested the association of FLAG-tagged uL4D proteins with 80S ribosomes (Figure 4A). Since the ACE in vitro translation system is prepared from callus cultures raised from the mutant Arabidopsis seedlings (Supplementary Figure S4) (23), we used the same culture system here. Crude extracts (S16) were subjected to ultracentrifugation through a sucrose cushion to obtain ribosomal pellet (P170) and post-ribosomal supernatant (S170) fractions. Immunoblot analysis of the fractions using an anti-FLAG antibody identified the ∼48-kDa uL4D:FLAG proteins in the S16 and P170 fractions, but not in S170, in all mutant lines (Figure 4A). This indicates that the FLAG-tagged uL4D(WT), uL4D(R77A) and uL4D(ΔTV) proteins are efficiently incorporated into ribosome particles. uL4D(Δloop):FLAG protein is also incorporated into ribosome particles; however, the antibody detected only a faint band, indicating that the mutant ribosome constitutes a very minor fraction of the total ribosomes. Since the uL4D(Δloop):FLAG line did not seem promising for use in the present study, we excluded this line from further analyses.
Figure 4.
Functionality and constitution fraction of FLAG-tagged mutant uL4D-containing ribosomes. (A) Crude extracts (S16), post-ribosomal supernatants (S170), and ribosomal pellets (P170) prepared from callus cultures derived from wild-type Col-0 plants, ul4d(KO), and FLAG-tagged mutant uL4D transgenic lines are shown. Total proteins (10 μg in S16/S170; 1 μg in P170) were separated by SDS-PAGE and analyzed by immunoblotting using anti-FLAG antibody, anti-uL22 antiserum, and anti-β-actin antiserum. Positions of FLAG-tagged uL4D mutant proteins (uL4D:FLAG), 19-kDa uL22, and 43-kDa β-actin bands are marked. β-Actin was used as a soluble protein marker. Note that uL4D(Δloop):FLAG protein is smaller than uL4D(WT):FLAG by ∼1 kDa due to a 13-amino acid-deletion. A black dot in S170 marks the cross-reaction band. Representative results of two biological replicates are shown. (B) P-170 fractions from uL4D(WT):FLAG w2 line, uL4D(R77A):FLAG r6 line, and uL4D(ΔTV):FLAG d8 line were fractionated by ultracentrifugation through a 15–60% (w/v) sucrose density gradient. UV absorbance profile at 254 nm and immunoblot analysis using anti-FLAG antibody and anti-uL22 antiserum are shown. Positions of free proteins and 40S subunit, 60S subunit, 80S ribosome and polysome fractions are indicated. Representative results of two biological replicates are shown. (C) Immunoblot analysis of ACE batches using anti-uL4 antiserum, anti-FLAG antibody, and anti-uL22 antiserum. Representative results of three technical repeats are shown. Anti-uL4 detects both FLAG-tagged mutant uL4D and endogenous uL4A. (D) Immunoblot signals obtained using anti-uL4 antiserum in (C) were quantified, and the constitution fractions of FLAG-tagged uL4D-containing ribosomes among total ribosomes were calculated. Means ± SD of three technical repeats are shown.
To determine whether the mutant ribosomes are actively translating, we performed polysome profiling of the P170 fractions using sucrose density gradient centrifugation. Immunoblot analysis detected mutant uL4Ds in the polysome, monosome, and 60S subunit fractions, but not in the 40S subunit and free-protein fractions (Figure 4B). Distributions of the mutant uL4Ds in polysome fractions appeared similar to those of the total ribosome (Supplementary Figure S5), suggesting that the mutant uL4D-containing ribosomes can all achieve active translation in vivo.
Mutant ribosomes constitute substantial proportions of total ribosomes in ACE
Since the uL4D:FLAG mutant lines carry an endogenous uL4A, we investigated what proportion of ribosomes consisted of mutant uL4Ds in ACE (hereafter, the constitution fraction). To determine this, we subjected ACE preparations to immunoblot analysis using anti-uL4 antiserum, to distinguish FLAG-tagged mutant uL4D from endogenous uL4A by ∼1-kDa gel mobility shift (Figure 4C). The constitution fractions ranged from 40% to 80% (Figure 4D), indicating that substantial proportions of the ribosomes carry mutant uL4D in ACE.
AdoMet-induced ribosome stalling of CGS1 is reduced in uL4D(ΔTV):FLAG ribosomes
When a ribosome stalls during translation, a peptidyl-tRNA species accumulates as an intermediate of translation. A tRNA moiety confers a 10- to 20-kDa gel mobility shift on the peptide, depending on the tRNA species and gel conditions. Peptidyl-tRNA can be identified as an RNase-sensitive band.
In response to AdoMet, CGS1 nascent peptide induces ribosome stalling at the Ser-94 codon, leading to peptidyl-tRNA accumulation (17). This response was recapitulated in Col-0 ACE (Supplementary Results 1.1 and Figure S6) (23). To determine the effects of mutant uL4D-containing ribosomes, GST:CGS1(WT) RNA (Figure 5A and Supplementary Figure S6A) (17) was translated in ACE in the presence of AdoMet. Immunoblot analysis using an anti-GST antibody detected a 55-kDa peptidyl-tRNA in addition to a 45-kDa full-length product, whereas in the absence of AdoMet the peptidyl-tRNA was barely detectable (Figure 5A).
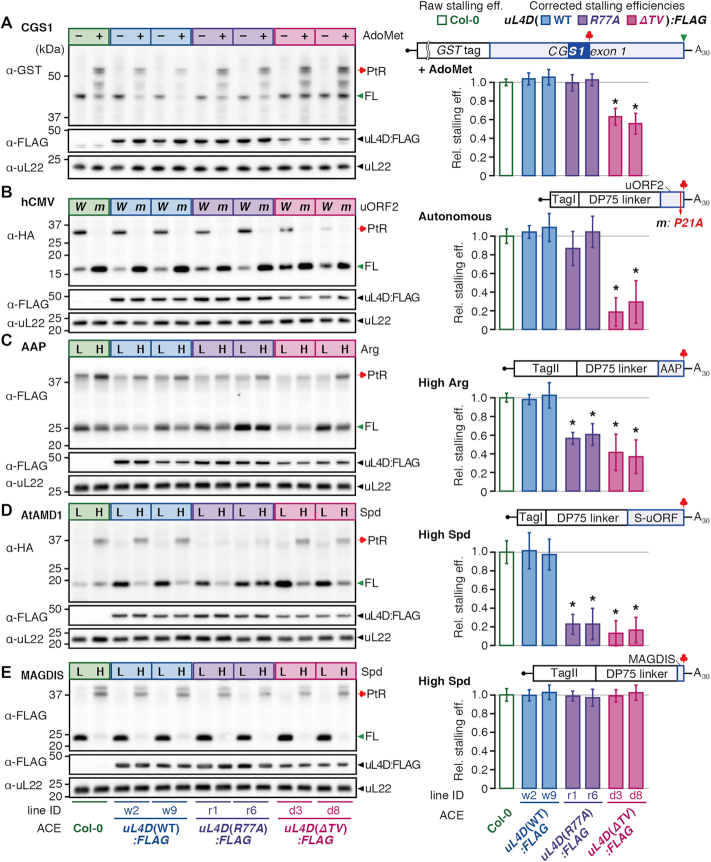
Figure 5.
Differential effects of uL4D mutant ribosomes on stalling. RNA construct shown above the bar graphs was translated in ACE prepared from wild-type Col-0, or from two independent lines of FLAG-tagged uL4D mutants as indicated. (Left panels) Translation products were separated by SDS-PAGE and analyzed by immunoblotting. The antibodies used to detect translation products are indicated. Immunoblots with anti-FLAG antibody and anti-uL22 antiserum are also shown. Positions of the full-length product (FL), peptidyl-tRNA (PtR), 48-kDa FLAG-tagged uL4Ds (uL4D:FLAG), and 19-kDa uL22 are marked. The experiments were carried out with two batches of ACE preparations for each of the Col-0 and mutant lines, and a representative result of triplicate experiments in one of the batches is shown. (Right panels) The corrected stalling efficiencies (shaded box) relative to the raw stalling efficiency in Col-0 ACE (open box) were calculated and means ± SD of six experiments performed with two ACE batches in triplicate are shown. Asterisks indicate significant differences compared with Col-0 ACE (q < 0.05 by Welch's t-test with false discovery rate (FDR) correction). (A) CGS1 system. GST:CGS1(WT) RNA (Supplementary Figure S6A) was translated for 30 min in ACE in the absence (–) or presence (+) of 1 mM AdoMet. The immunoblot signals of the ‘+’ lanes in the left panel were quantified and stalling efficiencies were calculated. The MTO1 region (21) is indicated by a filled blue box. (B) hCMV system. TagI:DP75:hCMV(WT) RNA (W) and TagI:DP75:hCMV(P21A) RNA (m) (Supplementary Figure S10B) were translated for 30 min. The immunoblot signals of the ‘W’ lanes were quantified. (C) AAP system. TagII:DP75:AAP(WT) RNA (Supplementary Figure S12B) was translated for 10 min in the presence of a low (L, 0.08 mM) or high (H, 2.08 mM) l-arginine concentration as indicated. The immunoblot signals of the ‘H’ lanes were quantified. (D) AtAMD1 system. TagI:DP75:S-ORF(WT) RNA (Supplementary Figure S13B) was translated for 30 min in the presence of a low (L, 0.2 mM) or high (H, 0.7 mM) spermidine (Spd) concentration as indicated. The immunoblot signals of the ‘H’ lanes were quantified. (E) MAGDIS system. TagII:DP75:MAGDIS(WT) RNA (Supplementary Figure S14B) was translated for 10 min in the presence of a low (L, 0.2 mM) or high (H, 0.7 mM) Spd concentration as indicated. The immunoblot signals of the ‘H’ lanes were quantified. TagI and TagII carry M8:His:HA and M8:His:HA:3xFLAG:Myc tags, respectively.
To evaluate the effect of uL4D mutations, we calculated the stalling efficiency, which is defined as the peptidyl-tRNA signal intensity divided by the sum of peptidyl-tRNA and full-length product signal intensities (Supplementary Figure S7A) (31,43). Since the ACE preparations contain both mutant uL4D- and endogenous uL4A-containing ribosomes, the stalling efficiency of the mutant ribosome alone has to be evaluated. To achieve this, we corrected the raw stalling efficiency for the constitution fraction of the mutant ribosomes, referred to as the corrected stalling efficiency. This calculation assumes that the stalling efficiency of endogenous uL4A-containing ribosomes alone is the same as that in Col-0 ACE, which we believe is reasonable (Supplementary Figure S8A). The corrected stalling efficiency of uL4D(ΔTV):FLAG lines d3 and d8 ACE was 63% and 56%, respectively (Figure 5A, shaded bar), of the raw stalling efficiency of Col-0 ACE (Figure 5A, open bar), while the corrected stalling efficiencies of uL4D(WT):FLAG and uL4D(R77A):FLAG ACE were not appreciably different from the raw stalling efficiency of Col-0 ACE. These results show that uL4D(ΔTV) mutation weakens AdoMet-induced ribosome stalling, while uL4D(R77A) mutation does not.
Affinity purification of stalled ribosome confirms the calculation of corrected stalling efficiency
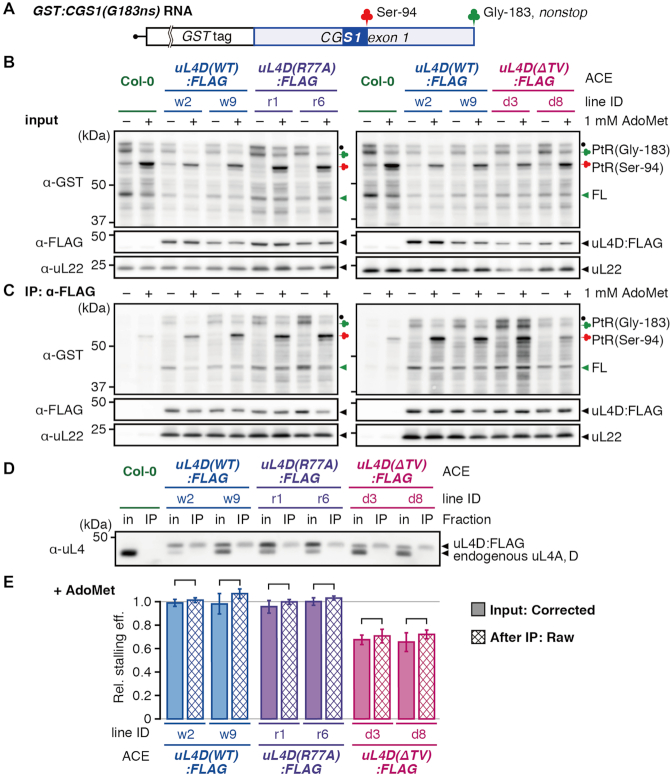
To validate the calculation of corrected stalling efficiencies, GST:CGS1(G183-ns) RNA (Figure 6A) was used to affinity-purify the FLAG-tagged mutant ribosomes by immunoprecipitation (IP) using an anti-FLAG antibody following translation in ACE. GST:CGS1(G183-ns) RNA is a nonstop RNA that is truncated at the Gly-183 codon and does not carry a stop codon. The use of this nonstop RNA is necessary to affinity-purify the stalled ribosomes because a ribosome that reached the termination codon will split into large and small subunits and the peptidyl-tRNA will be hydrolyzed into a peptide and tRNA (Supplementary Figure S9A), while on the nonstop RNA, the 80S ribosome will remain at the truncated RNA end with the peptidyl-tRNA on it (Supplementary Figure S9A). For this experiment, the RNA concentration was increased to reduce ribosome stacking behind the initially stalled ribosome at Ser-94 (Supplementary Figure S6C and D) (19).
Figure 6.
Affinity purification of stalled ribosomes. (A) Schematic representation of GST:CGS1(G183-ns) RNA, which is a nonstop RNA that is truncated at the Gly-183 codon located at the end of CGS1 exon 1 and does not carry a stop codon. The MTO1 region (21) is indicated by a filled blue box. (B and C) GST:CGS1(G183-ns) RNA (500 fmol μl−1) was translated in ACE prepared from wild-type Col-0 and mutant lines expressing uL4D:FLAG in the absence (–) or presence (+) of 1 mM AdoMet, as indicated. After 30 min of translation, stalled ribosomes were affinity-purified by IP using anti-FLAG antibody. Translation products of input controls (B) and IP fractions (IP: α-FLAG) (C) were analyzed by immunoblotting using anti-GST antibody. Positions of 65-kDa peptidyl-tRNA [PtR(Gly-183)] produced at the nonstop RNA end of Gly-183 and the 55-kDa peptidyl-tRNA [PtR(Ser-94)] produced by AdoMet-induced NPmRS at Ser-94, and the full-length peptide, which was dissociated from PtR(Gly-183) (FL), are marked. The band marked with a black dot is probably a peptidyl-tRNA that is produced by a ribosome stacked behind the one stalled at the nonstop RNA end. Immunoblots using anti-FLAG antibody and anti-uL22 antiserum are shown as loading controls. A representative result of triplicate experiments is shown. (D) Immunoblot analysis of the translation mixture before (in) and after IP (IP) using anti-uL4 antiserum to show the constitution fractions of FLAG-tagged ribosomes. Positions of the 48-kDa FLAG-tagged uL4D (uL4D:FLAG) and the 47-kDa endogenous uL4 are marked. The band marked as endogenous uL4 in Col-0 ACE (lanes 1 and 2) includes both endogenous uL4A and endogenous uL4D, while that in uL4D:FLAG mutant ACE (lanes 3–14) is endogenous uL4A alone. A representative result of triplicate experiments is shown. (E) The immunoblot signals in (B) and (C) were quantified. The corrected stalling efficiency in the input sample (shaded box), and the raw stalling efficiency after IP (cross hatched box) were calculated and means ± SD (n = 3) are shown. No significant difference was observed between the corrected stalling efficiency before IP and raw stalling efficiency after IP in each pair of the samples (P > 0.05 by Welch's t-test).
When GST:CGS1(G183-ns) RNA was translated in Col-0 ACE in the presence of 1 mM AdoMet (Figure 6B and C), ∼65-kDa peptidyl-tRNA [PtR(Gly-183), green clover] that had translated to the truncated end of the nonstop RNA, 55-kDa peptidyl-tRNA [PtR(Ser-94), red clover] formed on the stalled ribosome at Ser-94, and 45-kDa full-length peptide (FL, green arrowhead) were identified. The stalling efficiency in this experiment is defined as the signal intensity of PtR(Ser-94) divided by the sum of PtR(Ser-94) and PtR(Gly-183) signal intensities. The 45-kDa full-length peptide was probably produced by spontaneous hydrolysis of PtR(Gly-183). We previously observed spontaneous hydrolysis of peptidyl-tRNA at the nonstop RNA end (19). Since spontaneous hydrolysis of PtR(Gly-183) could occur at any step during this experiment, the signal intensity of this full-length product was not taken into account for the calculation of stalling efficiency.
After IP with the anti-FLAG antibody, anti-uL4 antiserum detected only FLAG-tagged uL4D (Figure 6D), indicating that IP efficiently purified FLAG-tagged mutant ribosomes. The corrected stalling efficiencies of uL4D(ΔTV):FLAG lines d3 and d8 ACE before IP were 68 ± 4% and 66 ± 8%, respectively, while raw stalling efficiencies after IP were 71 ± 6% and 72 ± 4%, respectively, of that of Col-0 ACE before IP (Figure 6E). For all pairwise comparisons of the corrected stalling efficiency before IP and the raw stalling efficiency after IP, similar values were obtained. These results support the validity of the corrected stalling efficiency calculations.
Affinity purification of stalled ribosomes, however, cannot be applied to NPmRS that occurs at the termination codon (Supplementary Figure S9B). Therefore, in the present study, we used the corrected stalling efficiency that was calculated and compared it with the raw stalling efficiency of Col-0 ACE.
uL4D(ΔTV) mutation reduces autonomous ribosome stalling of the hCMV system
To further characterize the contribution of the constriction region to NPmRS, we tested other stalling systems from different eukaryotes. The 22-amino-acid uORF2 of hCMV gp48 directs ribosomes to stall autonomously at the translation termination to downregulate gp48 expression (27,28). The gp48 uORF2-mediated stalling occurred in Col-0 ACE, and abrogation of stalling by Pro-21-to-alanine substitution (P21A) of uORF2 (28) was reproduced in Col-0 ACE (Supplementary Results 1.2 and Supplementary Figure S10), thus showing recapitulation of the gp48 uORF2-mediated autonomous stalling in Col-0 ACE.
We tested the effects of uL4D(ΔTV) and uL4D(R77A) mutations. The corrected stalling efficiency in uL4D(ΔTV):FLAG ACE was estimated to be 19–30% of the raw stalling efficiency of Col-0 ACE, while that of uL4D(WT):FLAG ACE and uL4D(R77A):FLAG ACE was not appreciably different from Col-0 ACE (Figure 5B). The effects of uL4D mutations on CGS1 and hCMV systems were similar in that uL4D(ΔTV) but not uL4D(R77A) affected stalling; however, the reduction in the uL4D(ΔTV) mutant was stronger in hCMV than in CGS1.
To further confirm the accuracy of our calculation of the corrected stalling efficiency, we mixed uL4D(WT):FLAG w2 line ACE or uL4D(ΔTV):FLAG d8 line ACE with Col-0 ACE at different proportions, and analyzed the resulting stalling efficiencies. Extrapolation of the regression lines of the raw stalling efficiencies revealed stalling efficiency values for the mutant ribosomes alone that are similar to those in Figure 5B, again supporting the validity of our corrected stalling efficiency calculations (Supplementary Figure S11).
Both uL4D(R77A) and uL4D(ΔTV) mutations reduce arginine-induced ribosome stalling of the AAP system
The 24-amino-acid AAP nascent peptide, encoded by N. crassa arg-2 uORF, causes NPmRS in response to l-arginine (24,25). In Col-0 ACE, peptidyl-tRNA accumulated in a l-arginine-dependent manner, whereas substitution of Asp-12 to asparagine (D12N) of AAP, which has been shown to abolish the response to l-arginine (25), also abolished the peptidyl-tRNA accumulation in Col-0 ACE (Supplementary Results 1.3 and Supplementary Figure S12). Thus, l-arginine-dependent NPmRS of AAP was recapitulated in Col-0 ACE.
Translation analyses in uL4D mutant ACE showed that stalling at a high l-arginine concentration (2.08 mM) was significantly reduced in both uL4D(R77A):FLAG and uL4D(ΔTV):FLAG ACE. The corrected stalling efficiencies were 57–61% and 37–42%, respectively, of the raw stalling efficiency of Col-0 ACE (Figure 5C). Compared with the CGS1 and hCMV systems, AAP exhibited a qualitative difference in that a single-amino-acid substitution of uL4D(R77A) affected stalling.
Both uL4D(R77A) and uL4D(ΔTV) mutations strongly reduce polyamine-induced ribosome stalling in AtAMD1 system
The 52-amino-acid uORF2 of AtAMD1 S-uORF directs the ribosome to stall at the translation termination in response to high polyamine concentrations. In the present study, spermidine was used as an effector (31). In Col-0 ACE, peptidyl-tRNA accumulation was dependent on spermidine concentration (31), which was abolished by introducing a frame-shift mutation in S-uORF (Supplementary Results 1.4 and Figure S13), indicating that Col-0 ACE recapitulates spermidine-dependent ribosome stalling.
The corrected stalling efficiency in uL4D(R77A):FLAG and uL4D(ΔTV):FLAG ACE at a high spermidine concentration (0.7 mM) was reduced to ∼23% and ∼15%, respectively, of the raw stalling efficiency of Col-0 ACE, while uL4D(WT):FLAG ACE showed similar levels of stalling efficiencies to Col-0 ACE (Figure 5D). As was the case in AAP, both uL4D(R77A) and uL4D(ΔTV) mutations reduced the stalling in AtAMD1; however, the effects of the mutations were stronger than in AAP.
Neither uL4D(R77A) nor uL4D(ΔTV) mutation affects polyamine-induced ribosome stalling on the six-amino-acid uORF of MAGDIS
In mAMD1, a uORF encoding six amino acids, MAGDIS, directs the ribosome to stall in response to polyamines. Translation in Col-0 ACE showed peptidyl-tRNA accumulation in a spermidine-dependent manner, while Ile-5 to leucine (I5L) substitution (32,33) of MAGDIS abolished peptidyl-tRNA accumulation, indicating that the spermidine-induced NPmRS is recapitulated in Col-0 ACE (Supplementary Results 1.5 and Figure S14).
The corrected stalling efficiencies in uL4D:FLAG mutant ACEs were all essentially the same as the raw stalling efficiency of Col-0 ACE (Figure 5E). These results show that neither uL4D(R77A) nor uL4D(ΔTV) mutation affects the spermidine-dependent ribosome stalling directed by the six-amino-acid uORF of MAGDIS.
5′-UTRs carrying the uORFs also depict the differential effects of uL4D mutations
Translation of a uORF generally downregulates translation of the main ORF, and if ribosomes stall on the uORF, translation of the main ORF is strongly downregulated (Supplementary Figure S15) (59). For the stalling analyses, we joined artificial sequences of the tags and linker to the uORF. In the case of MAGDIS, the constriction region is vacant when stalled on the natural uORF, whereas in the present stalling analysis a 75-amino-acid DP75 linker (15,43) resides in the constriction region. To test the effects of uL4D mutations on the natural uORF sequence, the 5′-UTR sequences of the hCMV, AtAMD1, and MAGDIS systems were joined to a luciferase (LUC) reporter, and the effects of uL4D mutations were analyzed by reporter assays (Figure 7).
Figure 7.
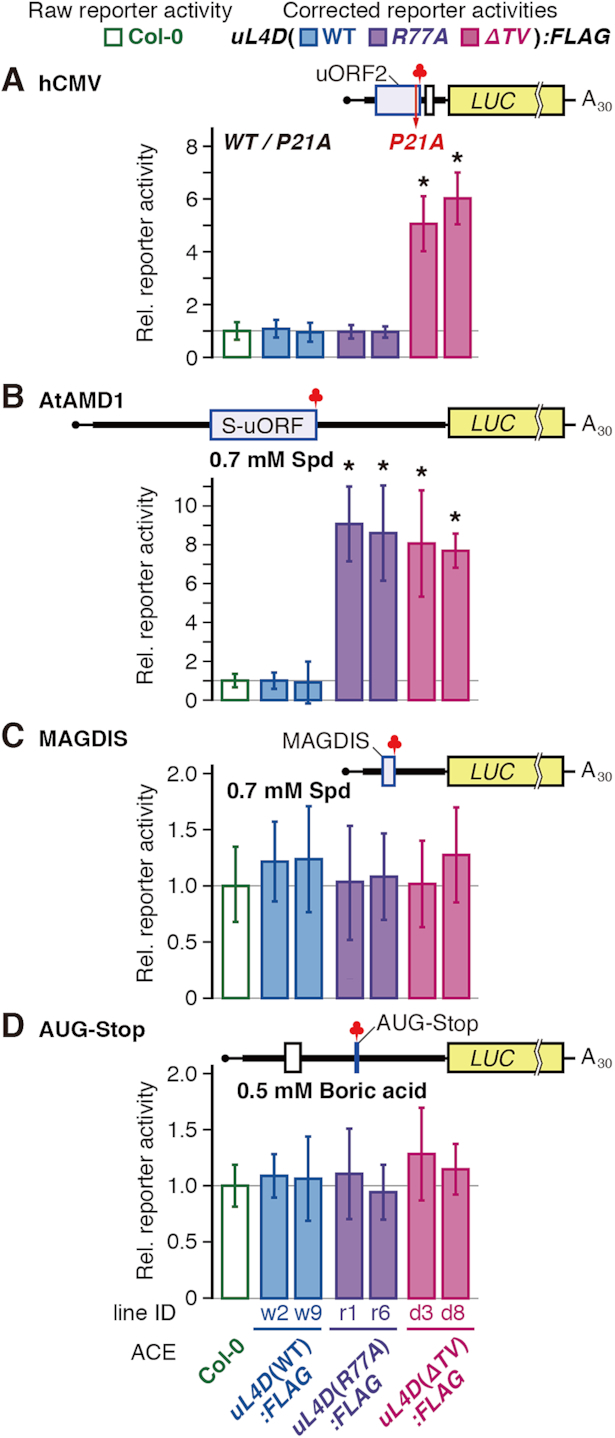
Differential effects of uL4D mutant ribosomes on LUC reporter expression. RNA was translated in ACE prepared from wild-type Col-0 or from two independent lines of FLAG-tagged uL4D mutants. After translation of RNA for 120 min, LUC activities were measured and normalized with the Renilla luciferase (RLUC) activity of the co-translated control RNA. The corrected reporter activities (shaded box) relative to the raw reporter activity in Col-0 ACE (open box) were calculated and means ± SD of six experiments performed with two ACE batches in triplicate are shown. Asterisks indicate significant difference from raw reporter activity in Col-0 ACE (q < 0.05 by Welch's t-test with FDR correction). (A) hCMV system. 5′-hCMV:LUC(WT) and 5′-hCMV:LUC(P21A) RNAs (Supplementary Figure S10A and D) were translated. LUC activities in 5′-hCMV:LUC(WT) RNA relative to those of 5′-hCMV:LUC(P21A) RNA were calculated. (B) AtAMD1 system. 5′-AtAMD1:LUC RNA (Supplementary Figure S13A and F) was translated in the presence of 0.2 or 0.7 mM spermidine (Spd). LUC activities in the presence of 0.7 mM Spd relative to those of 0.2 mM were calculated. (C) MAGDIS system. 5′-mAMD1:LUC RNA (Supplementary Figure S14A and G) was translated in the presence of 0.2 or 0.7 mM Spd. LUC activities in the presence of 0.7 mM Spd relative to those at 0.2 mM were calculated. (D) AUG-Stop system. 5′-NIP5;1:LUC RNA (Supplementary Figure S17A and B) was translated in the presence or absence of 0.5 mM boric acid. LUC activities in the presence of boric acid relative to those in its absence were calculated.
The 5′-UTR of hCMV gp48 (27,28) was joined to a LUC reporter (Figure 7A, and Supplementary Figure S10A and D) and was translated in ACE. When the RNA was translated in uL4D(ΔTV):FLAG ACE the relative reporter activity was ∼3 times higher than in Col-0 ACE (Supplementary Figure S16). The relative reporter activity was corrected for the constitution fraction of FLAG-tagged ribosomes, as for the stalling efficiency correction (Supplementary Figure S8B). The corrected relative reporter activity in uL4D(ΔTV):FLAG ACE was ∼6 times the raw reporter activity of Col-0 ACE, while those in uL4D(WT):FLAG and uL4D(R77A):FLAG ACE were essentially the same as in Col-0 ACE (Figure 7A). When the 5′-UTR of AtAMD1 containing the S-uORF (Figure 7B, and Supplementary Figure S13A, F, and G) (31) was analyzed, corrected reporter activities in uL4D(R77A):FLAG and uL4D(ΔTV):FLAG ACE were 7–9 times higher than in Col-0 ACE (Figure 7B).
The 5′-UTR of mAMD1 was joined to the LUC reporter (Figure 7C, and Supplementary Figure S14A and G) and translated in ACE. Relative reporter activities in uL4D:FLAG mutant ACEs were essentially the same as in Col-0 ACE (Figure 7C). These results are consistent with those of the stalling efficiency analyses (Figure 5B, D and E) and confirmed the effects of uL4D mutations on ribosome stalling.
Neither uL4D(R77A) nor uL4D(ΔTV) mutation affects boric acid-induced ribosome stalling on AUG-stop
The 5′-UTR of AtNIP5;1, encoding a boric acid transporter, has the shortest possible uORF, AUG-stop (Supplementary Figure S17). Boric acid induces prolonged ribosome stalling at AUG-stop (34). Since insertion of even a single codon between the AUG and stop codons is detrimental to the response (34), joining a tag or linker sequence is impossible. Therefore, effects of the uL4D mutations were evaluated by a reporter assay. As in the case of MAGDIS, neither uL4D(R77A) nor uL4D(ΔTV) mutation affected reporter activities (Figure 7D).
DISCUSSION
In this study, we presented reverse genetics-based biochemical evidence showing that uL4 is involved in four of the NPmRS in eukaryotes, namely the CGS1, hCMV, AAP, and AtAMD1 systems. In contrast, the six-amino-acid uORF of MAGDIS was unaffected. The results show that the constriction region is crucial for inducing NPmRS, when the nascent peptide is long enough to cross over the constriction region, and that MAGDIS adopts a distinct mechanism for NPmRS induction.
Construction of transgenic lines carrying uL4D mutations
In transgenic plants uL4D(WT):FLAG line w2 and uL4D(R77A):FLAG lines r1 and r6, mutant uL4D mRNAs accumulated to 4- to 12-fold higher levels than in wild-type Col-0 plants (Figure 3). Nevertheless, free FLAG-tagged uL4D proteins were undetectable in S170 fractions (Figure 4A), suggesting the existence of unknown regulatory mechanism(s) at the mRNA and protein accumulation levels (48).
Regarding the uL4D(ΔTV):FLAG and uL4D(Δloop):FLAG lines, mRNA accumulation was similar to or less than that in Col-0 plants (Figure 3). It is possible that high-level expressors were not isolated owing to growth retardation, because NPmRS is involved in a range of regulatory systems, including metabolism (17,20,21,31) and transcription factor expression (43,60). In the construction of uL4D(ΔTV):FLAG and uL4D(Δloop):FLAG mutant plants, we also feared the effects of a defect in ribosome biogenesis because the alterations in Loop 1 might retard the integrity of the internal extension loop (Supplementary Figure S1), which has been shown to be essential for the large-subunit assembly in bacteria and yeast (61–63). This might occur in uL4D(Δloop):FLAG lines. A plausible scenario is that uL4D(Δloop):FLAG protein was only poorly incorporated into ribosome particles and the surplus protein was rapidly degraded. Degradation of surplus ribosomal proteins has been observed in yeast (64).
Differential effects of uL4D(R77A) and uL4D(ΔTV) mutations explain the structural data
The degree of reduction of stalling by uL4D(ΔTV) and uL4D(R77A) mutations showed qualitative differences among the stalling systems. Structures of stalled ribosomes in WGE have been solved in hCMV and AAP systems, and physical contacts between the nascent peptide and constriction region have been identified (14).
Cryo-EM studies of AAP-stalled ribosomes (14) showed that Asp-12 contacts uL4 between Gly-76 and Val-79 (Arabidopsis uL4D residue number) in Loop 1 (Figure 2B, magenta underline). This is consistent with our finding that uL4D(R77A) mutation strongly reduced the l-arginine-dependent stalling of AAP (Figure 5C). In the hCMV system, nascent peptide physically contacts Loop 2, but not Loop 1 (14). This also is consistent with our finding that uL4D(R77A) mutation did not affect hCMV stalling (Figures 5B and 7A). Asp-12 of AAP is important for stalling (Figure 1) (24,26). Thus, our data interconnect structural data and functional residues in the nascent peptide. Our data also suggest that the physical contacts of the nascent peptide with the constriction region observed by cryo-EM are the cause, not the result, of ribosome stalling.
The effects of uL4D mutations were strongest in AtAMD1 and weakest in CGS1. We previously reported that, upon AdoMet-induced stalling of CGS1, 28S rRNA residues undergo conformational changes (18). Among these, EcU744 and EcA750 (in E. coli rRNA numbering) mapped near the constriction region. Intriguingly, these residues are located close to uL22. Corroborating the weak effect of uL4D mutations in CGS1, the CGS1 nascent peptide might interact with uL22, rather than uL4, although other possibilities remain. In contrast, regarding the AtAMD1 system, in which uL4D mutations had strong effects on stalling, its nascent peptide might strongly interact with Loop 1 to induce polyamine-dependent ribosome stalling.
uL4D(ΔTV) mutation downregulated four NPmRS systems, while uL4D(R77A) mutation affected only two of them. uL4D(ΔTV) mutation probably exhibited a general effect on interaction of nascent peptide and the constriction by altering the geometry of the constriction region. The two stalling systems affected by uL4D(R77A) mutation would specifically interact with the Arg-77 that we mutated, which was supported by cryo-EM study of AAP-stalled ribosomes. Our data suggest that specific interaction of nascent peptide and the amino acid residue(s) within the β-loop(s) is superimposed over the general interaction with the constriction region.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Tsuyoshi Nakagawa (Shimane University) for the pGWB10 vector, Saeko Yasokawa, for technical assistance and Naoe Konno, Maki Mori and Kazuko Harada for general assistance. Arabidopsis SALK_029203 seeds were obtained from Arabidopsis Biological Resource Center, OH, USA. We used the DNA Sequencing Facility of the Graduate School of Agriculture, Hokkaido University. N.O. and S.T. acknowledge JSPS for support. Y.Y. is a recipient of the Program for Fostering Researchers for the Next Generation conducted by the Consortium Office for Fostering of Researchers in Future Generations, Hokkaido University. The English text of a draft of this manuscript was edited by Edanz (Fukuoka, Japan).
Author contributions: S.N., H.O., and Y.Y. designed research. N.O., Y.T., and K.M. generated transgenic plants. S.T., Y.O., S.Y., and T.I. performed the experiments. S.T., Y.O., H.O., Y.Y., and S.N. analyzed data. S.T., Y.Y., and S.N. wrote the manuscript.
Notes
Present address: Yubun Ohashi, NOF Corp., Kawasaki 210-0865, Japan.
Present address: Noriyuki Onoue, NARO Institute of Fruit Tree and Tea Science, Higashi-Hiroshima 739-2494, Japan.
Present address: Yoko Tajima, Laboratory of Molecular Neuro-Oncology, The Rockefeller University, York Avenue, New York, NY 10065, USA.
Present address: Shinya Yonezawa, Department of Medical Innovations, Otsuka Pharmaceutical Co, Ltd., Tokushima 771-0192, Japan.
Present address: Kyoko Morimoto, Department of Plant Sciences, University of Oxford, South Parks Road, Oxford OX1 3RB, UK.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan [JP15H01525, JP17H05658, JP22119006 to S.N.]; Japan Society for the Promotion of Science (JSPS) [JP16H05063, JP20370016 to S.N., JP19K16159 to Y.Y., JP19H02917 to H.O.]; JSPS Fellows [JP08J03032 to N.O., JP17J00982 to S.T.]. Funding for open access charge: Research Faculty of Agriculture, Hokkaido University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Ito K., Chiba S.. Arrest peptides: cis-acting modulators of translation. Annu. Rev. Biochem. 2013; 82:171–202. [DOI] [PubMed] [Google Scholar]
- 2. Morgan D.G., Ménétret J.F., Radermacher M., Neuhof A., Akey I.V., Rapoport T.A., Akey C.W.. A comparison of the yeast and rabbit 80 S ribosome reveals the topology of the nascent chain exit tunnel, inter-subunit bridges and mammalian rRNA expansion segments. J. Mol. Biol. 2000; 301:301–321. [DOI] [PubMed] [Google Scholar]
- 3. Nissen P., Hansen J., Ban N., Moore P.B., Steitz T.A.. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000; 289:920–930. [DOI] [PubMed] [Google Scholar]
- 4. Ban N., Beckmann R., Cate J.H.D., Dinman J.D., Dragon F., Ellis S.R., Lafontaine D.L.J., Lindahl L., Liljas A., Lipton J.M. et al.. A new system for naming ribosomal proteins. Curr. Opin. Struct. Biol. 2014; 24:165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nakatogawa H., Ito K.. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002; 108:629–636. [DOI] [PubMed] [Google Scholar]
- 6. Cruz-Vera L.R., Rajagopal S., Squires C., Yanofsky C.. Features of ribosome-peptidyl-tRNA interactions essential for tryptophan induction of tna operon expression. Mol. Cell. 2005; 19:333–343. [DOI] [PubMed] [Google Scholar]
- 7. Chiba S., Kanamori T., Ueda T., Akiyama Y., Pogliano K., Ito K.. Recruitment of a species-specific translational arrest module to monitor different cellular processes. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:6073–6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ramu H., Vázquez-Laslop N., Klepacki D., Dai Q., Piccirilli J., Micura R., Mankin A.S.. Nascent peptide in the ribosome exit tunnel affects functional properties of the A-site of the peptidyl transferase center. Mol. Cell. 2011; 41:321–330. [DOI] [PubMed] [Google Scholar]
- 9. Cymer F., Hedman R., Ismail N., von Heijne G.. Exploration of the arrest peptide sequence space reveals arrest-enhanced variants. J. Biol. Chem. 2015; 290:10208–10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bhushan S., Hoffmann T., Seidelt B., Frauenfeld J., Mielke T., Berninghausen O., Wilson D.N., Beckmann R.. SecM-stalled ribosomes adopt an altered geometry at the peptidyl transferase center. PLoS Biol. 2011; 9:e1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bischoff L., Berninghausen O., Beckmann R.. Molecular basis for the ribosome functioning as an L-tryptophan sensor. Cell Rep. 2014; 9:469–475. [DOI] [PubMed] [Google Scholar]
- 12. Sohmen D., Chiba S., Shimokawa-Chiba N., Innis C.A., Berninghausen O., Beckmann R., Ito K., Wilson D.N.. Structure of the Bacillus subtilis 70S ribosome reveals the basis for species-specific stalling. Nat. Commun. 2015; 6:6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang J., Pan X., Yan K., Sun S., Gao N., Sui S.F.. Mechanisms of ribosome stalling by SecM at multiple elongation steps. eLife. 2015; 4:e09684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bhushan S., Meyer H., Starosta A.L., Becker T., Mielke T., Berninghausen O., Sattler M., Wilson D.N., Beckmann R.. Structural basis for translational stalling by human cytomegalovirus and fungal arginine attenuator peptide. Mol. Cell. 2010; 40:138–146. [DOI] [PubMed] [Google Scholar]
- 15. Matheisl S., Berninghausen O., Becker T., Beckmann R.. Structure of a human translation termination complex. Nucleic. Acids. Res. 2015; 43:8615–8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wilson D.N., Arenz S., Beckmann R.. Translation regulation via nascent polypeptide-mediated ribosome stalling. Curr. Opin. Struct. Biol. 2016; 37:123–133. [DOI] [PubMed] [Google Scholar]
- 17. Onouchi H., Nagami Y., Haraguchi Y., Nakamoto M., Nishimura Y., Sakurai R., Nagao N., Kawasaki D., Kadokura Y., Naito S.. Nascent peptide-mediated translation elongation arrest coupled with mRNA degradation in the CGS1 gene of Arabidopsis. Genes Dev. 2005; 19:1799–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Onoue N., Yamashita Y., Nagao N., Goto D.B., Onouchi H., Naito S.. S-Adenosyl-L-methionine induces compaction of nascent peptide chain inside the ribosomal exit tunnel upon translation arrest in the Arabidopsis CGS1 gene. J. Biol. Chem. 2011; 286:14903–14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yamashita Y., Kadokura Y., Sotta N., Fujiwara T., Takigawa I., Satake A., Onouchi H., Naito S.. Ribosomes in a stacked array: elucidation of the step in translation elongation at which they are stalled during S-adenosyl-L-methionine-induced translation arrest of CGS1 mRNA. J. Biol. Chem. 2014; 289:12693–12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chiba Y., Ishikawa M., Kijima F., Tyson R.H., Kim J., Yamamoto A., Nambara E., Leustek T., Wallsgrove R.M., Naito S.. Evidence for autoregulation of cystathionine γ-synthase mRNA stability in Arabidopsis. Science. 1999; 286:1371–1374. [DOI] [PubMed] [Google Scholar]
- 21. Ominato K., Akita H., Suzuki A., Kijima F., Yoshino T., Yoshino M., Chiba Y., Onouchi H., Naito S.. Identification of a short highly conserved amino acid sequence as the functional region required for posttranscriptional autoregulation of the cystathionine γ-synthase gene in Arabidopsis. J. Biol. Chem. 2002; 277:36380–36386. [DOI] [PubMed] [Google Scholar]
- 22. Chiba Y., Sakurai R., Yoshino M., Ominato K., Ishikawa M., Onouchi H., Naito S.. S-Adenosyl-L-methionine is an effector in the posttranscriptional autoregulation of the cystathionine γ-synthase gene in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:10225–10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Murota K., Hagiwara-Komoda Y., Komoda K., Onouchi H., Ishikawa M., Naito S.. Arabidopsis cell-free extract, ACE, a new in vitro translation system derived from Arabidopsis callus cultures. Plant Cell Physiol. 2011; 52:1443–1453. [DOI] [PubMed] [Google Scholar]
- 24. Wang Z., Sachs M.S.. Ribosome stalling is responsible for arginine-specific translational attenuation in Neurospora crassa. Mol. Cell Biol. 1997; 17:4904–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wei J., Wu C., Sachs M.S.. The arginine attenuator peptide interferes with the ribosome peptidyl transferase center. Mol. Cell Biol. 2012; 32:2396–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Spevak C.C., Ivanov I.P., Sachs M.S.. Sequence requirements for ribosome stalling by the arginine attenuator peptide. J. Biol. Chem. 2010; 285:40933–40942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cao J., Geballe A.P.. Coding sequence-dependent ribosomal arrest at termination of translation. Mol. Cell Biol. 1996; 16:603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Degnin C.R., Schleiss M.R., Cao J., Geballe A.P.. Translational inhibition mediated by a short upstream open reading frame in the human cytomegalovirus gpUL4 (gp48) transcript. J. Virol. 1993; 67:5514–5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Alderete J.P., Jarrahian S., Geballe A.P.. Translational effects of mutations and polymorphisms in a repressive upstream open reading frame of the human cytomegalovirus UL4 gene. J. Virol. 1999; 73:8330–8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hanfrey C., Elliott K.A., Franceschetti M., Mayer M.J., Illingworth C., Michael A.J.. A dual upstream open reading frame-based autoregulatory circuit controlling polyamine-responsive translation. J. Biol. Chem. 2005; 280:39229–39237. [DOI] [PubMed] [Google Scholar]
- 31. Uchiyama-Kadokura N., Murakami K., Takemoto M., Koyanagi N., Murota K., Naito S., Onouchi H.. Polyamine-responsive ribosomal arrest at the stop codon of an upstream open reading frame of the AdoMetDC1 gene triggers nonsense-mediated mRNA decay in Arabidopsis thaliana. Plant Cell Physiol. 2014; 55:1556–1567. [DOI] [PubMed] [Google Scholar]
- 32. Law G.L., Raney A., Heusner C., Morris D.R.. Polyamine regulation of ribosome pausing at the upstream open reading frame of S-adenosylmethionine decarboxylase. J. Biol. Chem. 2001; 276:38036–38043. [DOI] [PubMed] [Google Scholar]
- 33. Raney A., Law G.L., Mize G.J., Morris D.R.. Regulated translation termination at the upstream open reading frame in S-adenosylmethionine decarboxylase mRNA. J. Biol. Chem. 2002; 277:5988–5994. [DOI] [PubMed] [Google Scholar]
- 34. Tanaka M., Sotta N., Yamazumi Y., Yamashita Y., Miwa K., Murota K., Chiba Y., Hirai M.Y., Akiyama T., Onouchi H. et al.. The minimum open reading frame, AUG-stop, induces boron-dependent ribosome stalling and mRNA degradation. Plant Cell. 2016; 28:2830–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Alonso J.M., Stepanova A.N., Leisse T.J., Kim C.J., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R. et al.. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003; 301:653–657. [DOI] [PubMed] [Google Scholar]
- 36. Clough S.J., Bent A.F.. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998; 16:735–743. [DOI] [PubMed] [Google Scholar]
- 37. Suzuki A., Shirata Y., Ishida H., Chiba Y., Onouchi H., Naito S.. The first exon coding region of cystathionine γ-synthase gene is necessary and sufficient for downregulation of its own mRNA accumulation in transgenic Arabidopsis thaliana. Plant Cell Physiol. 2001; 42:1174–1180. [DOI] [PubMed] [Google Scholar]
- 38. Pound M.P., French A.P., Atkinson J.A., Wells D.M., Bennett M.J., Pridmore T.. RootNav: navigating images of complex root architectures. Plant Physiol. 2013; 162:1802–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T.. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 2007; 104:34–41. [DOI] [PubMed] [Google Scholar]
- 40. Curtis M.D., Grossniklaus U.. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003; 133:462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ho S.N., Hunt H.D., Horton R.M., Pullen J.K., Pease L.R.. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989; 77:51–59. [DOI] [PubMed] [Google Scholar]
- 42. Pogulis R.J., Vallejo A.N., Pease L.R.. In vitro recombination and mutagenesis by overlap extension PCR. Methods Mol. Biol. 1996; 57:167–176. [DOI] [PubMed] [Google Scholar]
- 43. Yamashita Y., Takamatsu S., Glasbrenner M., Becker T., Naito S., Beckmann R.. Sucrose sensing through nascent peptide-meditated ribosome stalling at the stop codon of Arabidopsis bZIP11 uORF2. FEBS Lett. 2017; 591:1266–1277. [DOI] [PubMed] [Google Scholar]
- 44. Zhang Y., Werling U., Edelmann W.. SLiCE: a novel bacterial cell extract-based DNA cloning method. Nucleic Acids Res. 2012; 40:e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schleiss M.R., Degnin C.R., Geballe A.P.. Translational control of human cytomegalovirus gp48 expression. J. Virol. 1991; 65:6782–6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cao J., Geballe A.P.. Translational inhibition by a human cytomegalovirus upstream open reading frame despite inefficient utilization of its AUG codon. J. Virol. 1995; 69:1030–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hill J.R., Morris D.R.. Cell-specific translation of S-adenosylmethionine decarboxylase mRNA. Regulation by the 5′ transcript leader. J. Biol. Chem. 1992; 267:21886–21893. [PubMed] [Google Scholar]
- 48. Rosado A., Sohn E.J., Drakakaki G., Pan S., Swidergal A., Xiong Y., Kang B.H., Bressan R.A., Raikhel N.V.. Auxin-mediated ribosomal biogenesis regulates vacuolar trafficking in Arabidopsis. Plant Cell. 2010; 22:143–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hsu P.Y., Calviello L., Wu H.Y.L., Li F.W., Rothfels C.J., Ohler U., Benfey P.N.. Super-resolution ribosome profiling reveals unannotated translation events in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E7126–E7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Inada T., Winstall E., Tarun S.Z.J., Yates J.R., Schieltz D., Sachs A.B.. One-step affinity purification of the yeast ribosome and its associated proteins and mRNAs. RNA. 2002; 8:948–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Benjamini Y., Hochberg Y.. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B. 1995; 57:289–300. [Google Scholar]
- 52. Barakat A., Szick-Miranda K., Chang I.F., Guyot R., Blanc G., Cooke R., Delseny M., Bailey-Serres J.. The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiol. 2001; 127:398–415. [PMC free article] [PubMed] [Google Scholar]
- 53. Hummel M., Dobrenel T., Cordewener J.J.H.G., Davanture M., Meyer C., Smeekens S.J.C.M., Bailey-Serres J., America T.A.H.P., Hanson J.. Proteomic LC-MS analysis of Arabidopsis cytosolic ribosomes: Identification of ribosomal protein paralogs and re-annotation of the ribosomal protein genes. J. Proteomics. 2015; 128:436–449. [DOI] [PubMed] [Google Scholar]
- 54. Armache J., Jarasch A., Anger A.M., Villa E., Becker T., Bhushan S., Jossinet F., Habeck M., Dindar G., Franckenberg S. et al.. Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:19748–19753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Juntawong P., Girke T., Bazin J., Bailey-Serrers J.. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2013; 111:E203–E212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Byrne M.E. A role for the ribosome in development. Trends Plant Sci. 2009; 14:512–519. [DOI] [PubMed] [Google Scholar]
- 57. Horiguchi G., Mollá-Morales A., Pérez-Pérez J.M., Kojima K., Robles P., Ponce M.R., Micol J.L., Tsukaya H.. Differential contributions of ribosomal protein genes to Arabidopsis thaliana leaf development. Plant J. 2011; 65:724–736. [DOI] [PubMed] [Google Scholar]
- 58. Rosado A., Li R., van de Ven W., Hsu E., Raikhel N.V.. Arabidopsis ribosomal proteins control developmental programs through translational regulation of auxin response factors. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:19537–19544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hinnebush A.G., Ivanov I.P., Sonenberg N.. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science. 2016; 352:1413–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hayashi N., Sasaki S., Takahashi H., Yamashita Y., Naito S., Onouchi H.. Identification of Arabidopsis thaliana upstream open reading frames encoding peptide sequences that cause ribosomal arrest. Nucleic Acids Res. 2017; 45:8844–8858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Stelter P., Huber F.M., Kunze R., Flemming D., Hoelz A., Hurt E.. Coordinated ribosomal L4 protein assembly into the pre-ribosome is regulated by its eukaryote-specific extension. Mol. Cell. 2015; 58:854–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lawrence M.G., Shamsuzzaman M., Kondopaka M., Pascual C., Zengel J.M., Lindahl L.. The extended loops of ribosomal proteins uL4 and uL22 of Escherichia coli contribute to ribosome assembly and protein translation. Nucleic Acids Res. 2016; 44:5798–5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pillet B., García-Gómez J.J., Pausch P., Falquet L., Bange G., de la Cruz J., Kressler D.. The dedicated chaperone Acl4 escorts ribosomal protein Rpl4 to its nuclear Pre-60S assembly site. PLoS Genet. 2015; 11:e1005565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sung M.K., Porras-Yakushi T.R., Reitsma J.M., Huber F.M., Sweredoski M.J., Hoelz A., Hess S., Deshaies R.J.. A conserved quality-control pathway that mediates degradation of unassembled ribosomal proteins. eLife. 2016; 5:e19105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.