Abstract
One of the key roles of the 12-subunit eukaryotic translation initiation factor 3 (eIF3) is to promote the formation of the 43S and 48S pre-initiation complexes (PICs). However, particular contributions of its individual subunits to these two critical initiation reactions remained obscure. Here, we adapted formaldehyde gradient cross-linking protocol to translation studies and investigated the efficiency of the 43S and 48S PIC assembly in knockdowns of individual subunits of human eIF3 known to produce various partial subcomplexes. We revealed that eIF3d constitutes an important intermolecular bridge between eIF3 and the 40S subunit as its elimination from the eIF3 holocomplex severely compromised the 43S PIC assembly. Similarly, subunits eIF3a, c and e were found to represent an important binding force driving eIF3 binding to the 40S subunit. In addition, we demonstrated that eIF3c, and eIF3k and l subunits alter the efficiency of mRNA recruitment to 43S PICs in an opposite manner. Whereas the eIF3c knockdown reduces it, downregulation of eIF3k or eIF3l increases mRNA recruitment, suggesting that the latter subunits possess a regulatory potential. Altogether this study provides new insights into the role of human eIF3 in the initial assembly steps of the translational machinery.
INTRODUCTION
Translation initiation is a concerted action of at least twelve eukaryotic translation initiation factors (eIFs) that sequentially assemble on the small ribosomal subunit (40S) to facilitate accurate selection of the beginning of the mRNA’s coding sequence (CDS). Among them, eIF3 is the largest (∼800 kDa) and the most complex one, comprising 12 non-identical subunits in higher eukaryotes (named eIF3a–eIF3m, excluding eIF3j) (Figure 1A) (reviewed in (1–3)). Eight subunits (a, c, e, f, h, k, l and m) form a structural scaffold called the PCI (Proteasome, COP9, eIF3)/MPN (Mpr1-Pad1 N-terminal) octamer, whose architecture is shared by the functionally unrelated 19S proteasome lid, as well as the COP9 signalosome (4,5). The PCI subunits eIF3a, c and e form the core of the PCI ark, while the other PCI subunits, namely eIF3k, l and m, are placed at its extremities. The interaction between eIF3m and eIF3f then connects the MPN subunits f and h to form the entire octamer (5). The PCI or MPN structural domains in the octameric subunits (except for eIF3a and eIF3m) are followed by alpha helices that form a so-called helical bundle.
Figure 1.
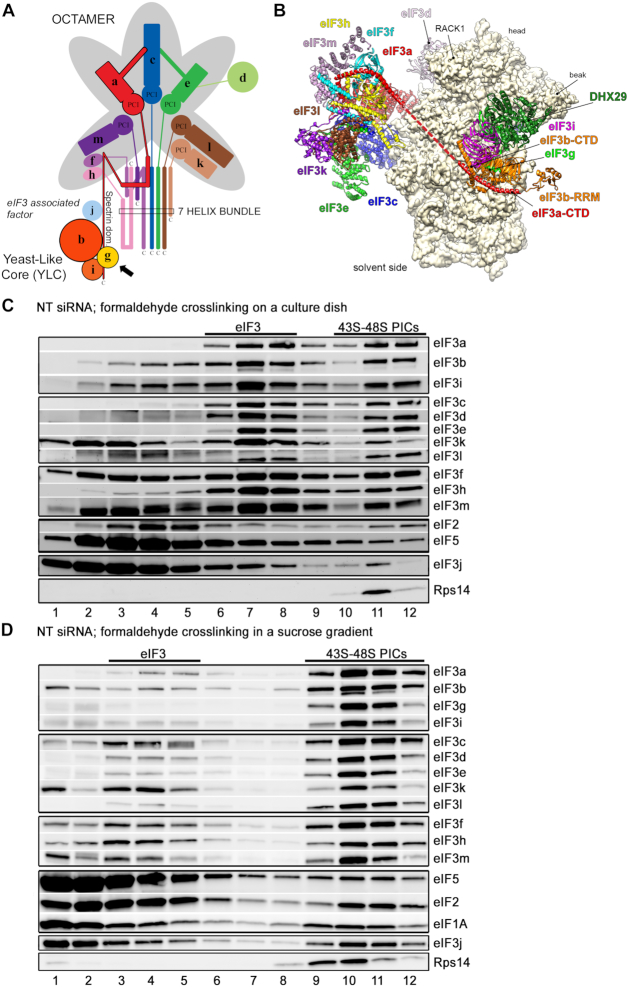
Comparison of the original ‘on dish’ formaldehyde cross-linking with the optimized formaldehyde cross-linking sucrose gradient protocol based on GraFix. (A) Schematic model of the human eIF3 complex adapted from (7). eIF3 subunits forming the PCI/MPN octamer are indicated by the grey background. The rectangle marks the seven α-helices involved in formation of the 7-helix bundle (5). The Yeast-Like Core (YLC) comprising the eIF3 subunits a, b, g and i is depicted, and so is the eIF3-associated factor eIF3j. Arrow indicates newly identified contact between eIF3g and the eIF3a-CTD (see Figure 3C and D). (B) Model of the structure of the mammalian 43S pre-initiation complex adapted from (56). The 43S PIC is viewed from the solvent side. The red dashed line represents continuity of the eIF3a structure (its C-terminal tail) that remains unsolved. (C, D) The 43S PIC assembly analysis of the control NT cells prepared by (C) the ‘on dish’ formaldehyde crosslinking protocol (adapted from (8)) or (D) the optimized protocol developed in this study. Antibodies used for western blotting are indicated on the right side of each panel. Sucrose gradient fraction numbers are given at the bottom. Fraction 1 in (D) is a pool of the first two fractions from the top of the gradient. Fractions containing ribosome free versus bound eIF3 complexes (eIF3 versus 43S-48S PICs) are indicated at the top.
The remaining four non-octameric subunits (b, d, g and i) are most probably rather flexible. The essential subunits b-g-i form a module that is attached to the octamer via the C-terminal domain (CTD) of eIF3a (6). This b-g-i-a-CTD subcomplex closely resembles the minimalistic 5-subunit eIF3 complex from budding yeast (thus named YLC for Yeast Like Core) (7). In fact, the integrity of the YLC is maintained in vivo upon downregulation of several eIF3 subunits that disrupts the octamer formation; it even seems to preserve the ability to perform some of the eIF3 basic functions (7,8). While the octamer is positioned near the mRNA exit channel, the b-g-i-a-CTD subcomplex was shown to be in contact with the mRNA entry channel (Figure 1B) (9–11). Mammalian eIF3 thus forms two relatively independent, interconnected modules that assemble in an ordered way around the nucleation core represented by the eIF3a and eIF3b subunits (7,8).
With respect to eIF3d, it is located more on the eIF3 periphery and is attached to the octamer through eIF3e (12,13). Indeed, knocking down eIF3d showed impact neither on expression levels of other eIF3 subunits nor on the integrity of the entire eIF3 complex, yet it had a strong impact on viability of HeLa and HEK293 cells (7). Interestingly, in the context of the 40S–eIF3 complex, eIF3d is sitting on the 40S head in a considerable distance from eIF3e, which forms a so-called right arm of eIF3 not in a direct contact with the 40S (5). Strikingly, similar effects to the eIF3d knockdown (eIF3dKD) were also detected with individual downregulations of the eIF3 subunits k and l. Even though these two non-essential subunits are an integral part of the octamer, knocking down one of them concurrently downregulates expression of the other, however, with no influence on expression of any other eIF3 subunit and no impact on the integrity of the resulting eIF3 subcomplex (7). In contrast to the eIF3dKD, the loss of eIF3k and eIF3l has no effect on cell viability. In agreement with all these findings, eIF3k and l are easily dissociated from the eIF3 complex and are even missing in genomes of some species. Furthermore, their loss-of-function mutations were found to be beneficial in some other species (2,14,15).
Perhaps owing to the large number of subunits, eIF3 has been implicated in numerous steps of not only translation initiation, but also in termination, ribosomal recycling and stimulation of stop codon readthrough (reviewed in (2)). Regarding translation initiation, eIF3 promotes formation of the 43S and 48S pre-initiation complexes (PICs) by aiding the Met-tRNAiMet and mRNA loading onto the 40S, respectively (16). In particular, Met-tRNAiMet is delivered to the ribosome as part of the ternary complex (TC) together with eIF2 bound to a GTP molecule. The TC recruitment to the 40S is stimulated by eIF3 together with other initiation factors such as eIF1, eIF1A and eIF5 (17–20). In this respect, it is noteworthy that the main contacts identified thus far between mammalian eIF3 and the 40S comprise surprisingly only the eIF3a and eIF3c subunits contacting ribosomal proteins Rps1/eS1 and Rps26/eS26, and Rps13/uS15 and Rps27/eS27, respectively (5,21). However, taken into account the complexity of mammalian eIF3, it is highly likely that more, yet to be identified contact points with the 40S exist, like for example the one between eIF3b and Rps9/uS4 shown to occur in C. thermophilum (22).
Having assembled the 43S PIC, the next step is the mRNA recruitment to form the 48S PIC. mRNA comes prebound by the eIF4F group of factors, containing eIF4E that directly contacts the mRNA’s 5′ 7-methyl guanosine cap, a scaffold protein eIF4G, and a DEAD-box helicase eIF4A. The eIF4A unwinds any cap-proximal secondary structures in an ATP-dependent manner; this activity is strongly stimulated by eIF4G and an accessory protein eIF4B (23–25). Furthermore, it was recently shown that eIF4A plays a more integral role in the 48S PIC formation as it promotes loading of all mRNAs to the 40S irrespective of their structural complexity (26). In fact, it might directly modulate the architecture of the mRNA binding channel and thus increase its affinity toward mRNAs in general (27). In any case, a major driving force of the mRNA recruitment step is considered to be a direct interaction between eIF4G and eIF3 subunits c, d and e (28,29). Interestingly, subunits eIF3d (30) and eIF3l (31) were shown to interact directly with the mRNA’s 5′ cap. This could indicate that they mediate a direct (i.e. eIF4E-independent and perhaps even eIF4F-independent) recruitment of some specific mRNAs to the 43S PICs (30). Indeed, DAP5, an eIF4GI homolog that lacks eIF4E binding domain, was recently demonstrated to utilize eIF3d to facilitate cap-dependent translation of ∼20% of mRNAs (32). Hence, it seems that in addition to canonical cap-dependent translation, a lot more alternate initiation mechanisms may exist than previously believed.
Once the mRNA is loaded, the 48S PIC starts scanning the mRNA’s 5′UTR until the start codon (typically the first AUG codon in a favorable nucleotide context) has been recognized by base-pairing with the Met-tRNAiMet anticodon. This drives conformational changes in the 48S PIC, co-operatively mediated by eIF1, eIF1A, eIF2, eIF3 and eIF5, resulting in the release of the hydrolyzed phosphate from the GTP binding pocket of eIF2, closure of the 40S mRNA binding channel, and subsequent ejection of the eIF2–GDP–eIF5 assembly from the 48S PIC (reviewed in (33)). eIF5B bound to GTP then mediates subunit joining and after hydrolysis of its GTP molecule leaves the 80S initiation complex together with eIF1A to allow elongation to commence (34).
We recently reported a comprehensive in vivo analysis of human eIF3 putting a major focus on its integrity under conditions of imbalanced expression of its individual subunits (7,8). Using a systematic downregulation strategy, we showed that expression of all 12 eIF3 subunits is interconnected, as downregulation of a single subunit by siRNA often led to a concomitant and specific decrease in protein levels of other eIF3 subunits. In addition, we described the human eIF3 assembly pathway and proposed that several partial, relatively stable eIF3 subcomplexes can exist in vivo. This is important because imbalanced eIF3 expression levels are observed in various types of cancer and developmental disorders (reviewed in (1,2,35,36)). In fact, almost all subunits are upregulated in tumor tissues except for eIF3e and eIF3f that are downregulated (37,38). Here we monitored – by adapted formaldehyde gradient cross-linking centrifugation protocol – the ability of these partial eIF3 complexes occurring in different eIF3 knockdowns to associate with the 40S and thus promote the assembly of the 43S PICs. In addition, we also examined their ability to form the 48S PICs in vivo using a well-established model mRNA encoding small ribosomal protein RPL41.
MATERIALS AND METHODS
Adapted formaldehyde gradient cross-linking protocol—the 43S and 48S PIC assembly analysis
Whole cell extract (WCE) preparation and transfection
HeLa cells were grown at 37°C and 5% CO2 in Ø15 cm dishes in DMEM (Sigma, cat # D6429) supplemented with 10% FBS (Sigma, cat # F7524). Twenty four hours after seeding, cells were transfected with the ON-TARGETplus siRNA cocktail system from Dharmacon at a final concentration of 5 nM. Catalog numbers for all siRNAs used in this study are listed in Supplementary Table S1. INTERFERin (Polyplus, cat # 409) was used as a transfection reagent and transfection was performed according to the vendor’s instructions. Cells were harvested 3 days after transfection at ∼80% confluency by on dish lysis as follows. Cycloheximide (Sigma, cat # C7698) was added to the media at a final concentration of 100 μg/ml 1 min prior to harvesting, cells were washed with cold 1× phosphate buffered saline and lysed directly on cell culture dish in lysis buffer A (10 mM HEPES [pH 7.5], 62.5 mM KCl, 2.5 mM MgCl2, 1 mM DTT, 1mM PMSF, 1 μg/ml Aprotinin, 1 μg/ml Leupeptin, 1 μg/ml Pepstatin, mini Complete EDTA-free [Roche, cat # 11836170001] - 1 tablet/5 ml, 1% Triton X-100, 100 μg/ml Cycloheximide). The resulting WCE was cleared by centrifugation (5 min/16 000g/4°C) and absorbance at 260 nm was measured.
The 43S PIC assembly analysis
Twelve A260 units of WCE were separated by high velocity sedimentation through a 7–30% sucrose gradient containing a top-to-bottom increasing concentration of formaldehyde at 22 200 rpm for 17 h using the Beckman Coulter SW41Ti rotor. Such a gradient was prepared by mixing 30% sucrose in buffer B (10 mM HEPES [pH 7.5], 62.5 mM KCl, 2.5 mM MgCl2, 1 mM DTT) containing 0,05% formaldehyde with 7% sucrose in buffer B lacking formaldehyde using the Gradient Master 108 (Biocomp Instruments). Fractions of 600 μl were collected and precipitated with 100% ethanol overnight at –20°C. After a single washing step with 100% ethanol, the pellet was dried and dissolved in 1× standard loading buffer, boiled to reverse cross-linking, and analyzed by SDS-PAGE followed by western blotting. Catalog numbers for all antibodies used in this study are listed in Supplementary Table S2.
The 48S PIC assembly analysis
For the mRNA recruitment analysis, SuperaseIN RNase inhibitor (Thermofisher, cat # AM2696) was added to the cleared WCEs to a final concentration of 20 U per 12 U of WCE. Twelve A260 units of WCE were separated by high velocity sedimentation through a 7–30% sucrose gradient containing a top-to-bottom increasing concentration of formaldehyde, prepared as explained above, at 41 000 rpm for 5 h using the Beckman Coulter SW41Ti rotor. Fractions of 400 μl containing the 40S species were collected. Identical amounts of ‘spike RNA’ (the yeast RPL41 mRNA produced in vitro using MAXIscript SP6/T7 transcription kit Thermofisher, cat # AM1320) were added to each fraction before they were mixed with 1 ml of RNA Blue reagent (Top-Bio, cat # R013). Samples were incubated at 95°C for 15 min to reverse cross-linking and total RNA was isolated according to vendor's instructions. Glycoblue (Thermofisher, cat # AM9516) was used as a co-precipitant. The resulting RNA pellets were resuspended in RNase-free water along with the DNase buffer and the resulting samples (19 μl) were incubated with 1 μl (2 U) of DNase I (NEB, cat # M0303S) at 37°C for 1 h to digest all contaminating DNAs. Before heat inactivation of DNaseI for 10 min at 75°C, EDTA was added to each reaction to a final concentration of 3 mM. The RNA amounts were quantified by NanoDrop One (Thermo Scientific) and an equal volume from each fraction corresponding to 750 ng of the most concentrated sample was used for reverse transcription in a 20 μl reaction with the High Capacity cDNA Reverse Transcription kit (Thermofisher, cat # 4368814). qPCR was carried out according to vendor's instructions (Solis BioDyne, cat # 08-25-00020) using Bio-Rad CFX384 Real-Time PCR System. qPCR reactions were prepared by mixing 5× HOT FIREPol EvaGreen qPCR Mix Plus (no ROX) with 0.3 μM primers and 5 μl of 5-times (or 500-times in case of 18S rRNA) diluted cDNA and run using the following program: 95°C for 15′ followed by 44 cycles of 95°C for 15′, 60°C for 22″ and 72°C for 20″. Melting curves were analyzed between 65 and 95°C. The levels of 18S rRNA, human RPL41 mRNA and spike RNA were measured in triplicates as described before (39,40) together with no reverse transcription (NRT) and no template (NTC) controls. Results were analyzed using Bio-Rad CFX Manager. Amounts of RPL41 mRNA and 18S rRNA occurring in each collected fraction were normalized to spike RNA to account for possible material losses during RNA isolation; the final amounts were calculated as 2−Ct. Relative amounts of RPL41 mRNA versus 18S rRNA amounts in all 40S-containing fractions combined, obtained from individual knockdowns versus NT cells from at least three biological replicates, were calculated and are given in Table 1; data from individual experiments are shown in Supplementary Table S3. All primers used in this study are listed in Supplementary Table S4.
Table 1.
Efficiency of model RPL41 mRNA recruitment to 43S PICs in individual eIF3 knockdowns compared to control NT cells from at least three biological replicates. Data from individual experiments are shown in Supplementary Table S3.
| Amount of 40S bound RPL41 mRNA normalized to 18S rRNA (% relative to NT) | ||||||||
|---|---|---|---|---|---|---|---|---|
| NT | eIF3dKD | eIF3gKD | eIF3iKD | eIF3cKD | eIF3eKD | eIF3hKD | eIF3lKD | eIF3kKD |
| 100 | 47±13 | 46±13 | 55±11 | 42±10* | 78±13* | 124±14 | 124±17 | 132±27 |
*Indicates statistically significant difference (P value = 0.0007) using two-sample t-test.
GST pulldown assay
Glutathione S-transferase (GST) pulldown experiments with GST fusions and in vitro synthesized 35S-labeled polypeptides were conducted as described previously (41). Briefly, individual GST fusion proteins were expressed in Escherichia coli, immobilized on glutathione sepharose beads (GE Healthcare, cat # GE17-0756-01), and incubated with 10 μl of 35S-labeled potential binding partners at 4°C for 2 h (synthesized by TNT T7 Quick Coupled Transcription/Translation System, Promega, cat # L1170). The beads were washed three times with 1 ml of phosphate buffered saline, and bound proteins were separated by SDS-PAGE. Gels were first stained with Gelcode Blue stain reagent (Thermofisher, cat # 24592) and then subjected to autoradiography. The list and descriptions of plasmids used for GST fusion expressions and in vitro protein synthesis are shown in Supplementary Table S5.
RESULTS AND DISCUSSION
Adapted formaldehyde gradient cross-linking protocol greatly improved the 43S–48S PIC assembly analysis
The previously used formaldehyde cross-linking sucrose gradient protocol (8) was based on fixing the intact cells directly in the culture dish before lysing them. The major advantage of this approach was that it enabled rapid sample stabilization. At the same time, however, it reduced the efficiency of cell lysis (hence more material for a single experiment was required) and increased heterogeneity of the cross-linked material within each sample (aggregates formed by cross-linking individual protein complexes to one another). In order to eliminate these pitfalls and examine the 40S binding of partial eIF3 subcomplexes generated in individual eIF3 knockdowns as precisely as possible, we optimized the original protocol as follows. First, we stalled the translating ribosomes in the cell by cycloheximide to prevent any polysome run-off (for detailed description, please see Materials and Methods). Next, instead of fixing the cells with formaldehyde directly in a dish, we adopted the GraFix protocol that was originally developed for stabilization of fragile complexes for Cryo-EM (42,43), where macromolecular complexes are purified and cross-linked in one step during density gradient ultracentrifugation. The cycloheximide-treated cells were lysed, WCEs prepared, and PICs were cross-linked by an increasing formaldehyde concentration in the sucrose gradient during ultracentrifugation. This approach exposes macromolecules to a chemical cross-linking reagent as individual units due to the centrifugal force that is sufficient to disrupt weak aggregations (43). To prevent disruption of PICs that might occur before they are formaldehyde-crosslinked en route through the gradient, we reduced the centrifugation speed and consequently prolonged the spinning time (see Materials and Methods). As shown in Supplementary Figure S1, implementing all these changes did not alter the typical polysome profile characteristics.
As can be seen in Figure 1D versus C, with our new protocol we achieve a much better separation of 40S species bound by eIF3 and other eIFs (fractions 9–12) from ribosome-free factors sedimenting predominantly in fractions 3–5 (versus 6–8 in panel C); for gradient traces please see Supplementary Figure S1B. In addition, the improved approach results in markedly decreased amounts of 40S-free factors compared to our original procedure, most probably due to more efficient, directed cross-linking and reduced spinning forces. We believe that as such, it much better reflects the native situation and thus further boosts the sensitivity of this assay for the 43S PIC assembly analysis. Please note that even though eIF1A, eIF2 and eIF5 do not accumulate in the 40S-containing fractions as profoundly as all eIF3 subunits do, distinct peaks are well observable (mainly in fractions 10–11), unlike before (Figure 1D versus C). Interestingly, while eIF2, eIF3 and eIF5 peak in the heavier 40S fractions (10–11), eIF1A starts peaking already in fraction 9, closely mirroring the distribution of the small ribosomal protein Rps14 (Figure 1D). This could indicate that some 40S ribosomes are pre-bound by eIF1A prior to the 43S PIC assembly, as proposed earlier (19). In any case, this example nicely illustrates the increased sensitivity of the formaldehyde gradient crosslinking protocol. Last but not least, the gradient distribution pattern of eIF3j resembles more the latter eIFs than eIF3 subunits, further supporting our long-standing argument that eIF3j serves as an eIF3-associated factor and not as a bona fide subunit of eIF3 (2,44,45).
In addition to the technicalities of the procedure, we also revised our data analysis approach. As previously, we summed up quantified signals of all 40S-containing fractions from the same Western blot expositions for all compared samples and normalized the occurrence of an eIF3 subunit of interest to the occurrence of our 40S marker protein, Rps14 (8). Obtained numbers were then compared to the control NT cells (cells transfected with Non-Targeting siRNA) to reflect relative amounts of an eIF3 subunit bound per ribosome. To further highlight a possible binding defect of a particular subunit, we additionally analyzed its distribution throughout the entire gradient. Hence, we analyzed each Western blot membrane strip separately by quantifying relative amounts of a subunit of interest in each fraction and calculated the percentage of this subunit in the 40S-containing fractions out of total. Subsequently, we calculated its relative change (‘shift in or out of the 40S-containing fractions’) compared to the NT cells; values >100% indicate accumulation of a subunit of interest in the 40S-containing fractions, whereas values <100% point to its redistribution toward lighter fractions, most likely due to a 40S-binding defect. These two approaches should be viewed as complementary but not equivalent. Please also note that since the nature of Western blotting is, by definition, not well suited for strictly quantitative conclusions, our calculations should be regarded as qualitative indicators of a trend rather than precise quantitative measurements. Importantly, for all quantifications the Quantity One software from Bio-Rad was used and we always choose the highest but still unsaturated expositions. In addition, to obtain robust statistical power, the experiments were performed multiple times (3–6 times).
eIF3d critically promotes the 43S PIC formation
All experiments described hereafter were carried out 3 days after transfection of the subunit-specific siRNA cocktail into HeLa cells, at which point the protein levels of each targeted subunit were decreased by ∼80–90%; importantly, none of the siRNA-treated cultures had entered apoptosis at this time point (for details see (7)).
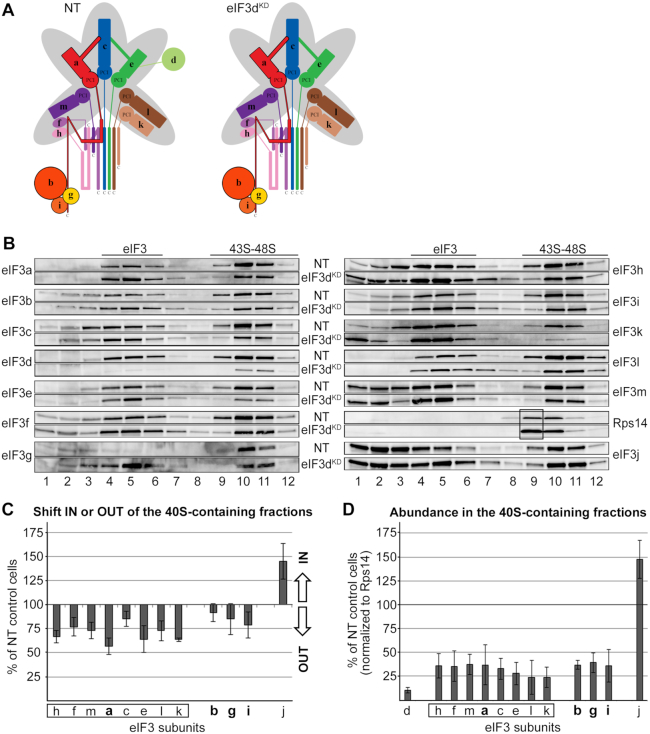
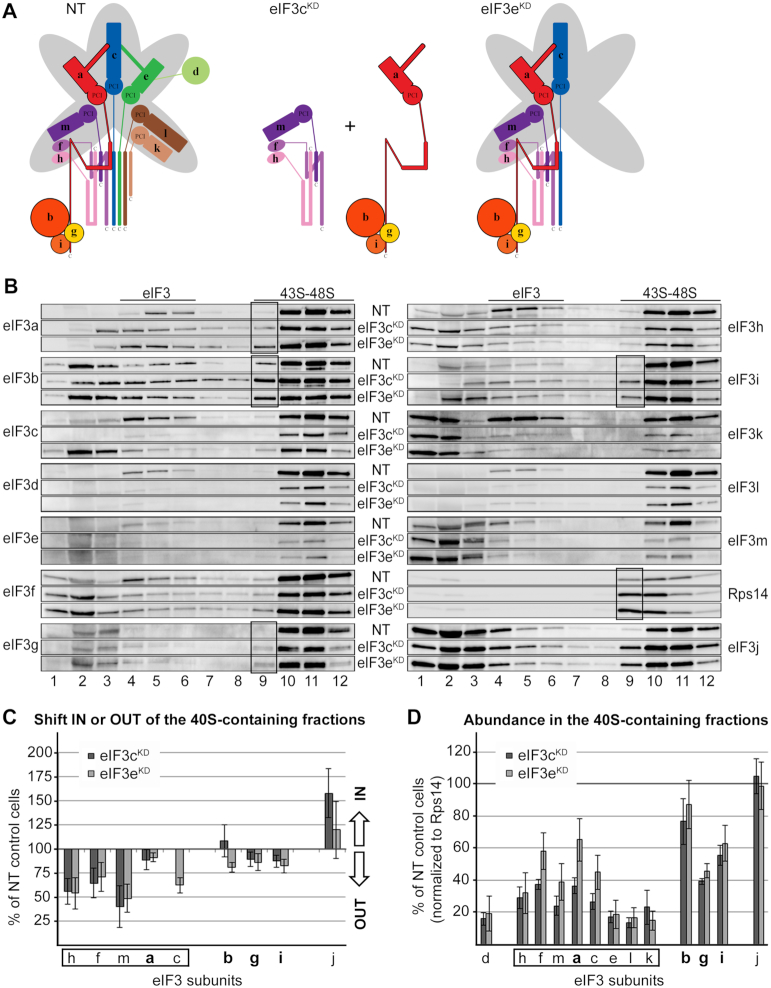
As mentioned above, in the eIF3dKD, only the d subunit of eIF3 is absent from the complex and the rest of eIF3 remains intact (Figure 2A). In spite of that, the eIF3dKD cells showed strongly decreased translation initiation rates and severely impaired viability (7). Therefore, we were intrigued to examine the eIF3dKD effect on the 40S association of the rest of eIF3. Strikingly, eIF3 lacking eIF3d markedly shifted from the 40S-containing fractions towards lighter fractions (Figure 2B and C). In contrast, eIF3j displayed the opposite behavior to the eIF3 complex—accumulation in the 40S fractions as reported previously for the eIF3aKD (8). Please note that the remaining amounts of eIF3d are almost exclusively associated with PICs, undoubtedly as part of the eIF3 holocomplex. When normalized to Rps14, we estimate the amount of eIF3 bound to PICs in the eIF3dKD to be ∼35% compared to control NT cells (Figure 2D). Consequently, increased amounts of Rps14 can be observed in fraction 9 in the eIF3dKD that are not accompanied by an increase in the amounts of eIF3 subunits, clearly suggesting an accumulation of lighter 40S species lacking eIF3 (Figure 2B, black box). Please note that the overall level of Rps14 across the corresponding collected fractions is expectedly higher in the eIF3dKD compared to NT cells. This phenomenon is common for all eIF3 downregulations that have a strong impact on the initiation rates (see below) and arises from a polysomal run-off resulting in increased amounts of free 40S (and 60S) species (7). Importantly, the fact that there is an increased pool of free 40S ribosomes in the eIF3dKD and yet we observed a clear shift of eIF3 into the 40S-free fractions, further validates our conclusion that eIF3d critically promotes the 43S PIC assembly. Could it represent another eIF3 anchor directly contacting the 40S?
Figure 2.
eIF3d critically promotes the 43S PIC formation. (A) Comparison of schematic models of eIF3 complexes occurring in the control NT versus eIF3dKD cells. (B) The 43S PIC assembly analysis of the control NT versus eIF3dKD cells performed as described in Materials and Methods. Please note that for easier comparison we coupled Western blot membrane strips corresponding to individual eIF3 subunits together with that coming from control NT cells always above the one derived from the eIF3dKD (follow the labeling in between the two panels). Sucrose gradient fraction numbers are given at the bottom. Fraction 1 in is a pool of the first two fractions from the top of the gradient. Fractions containing ribosome free versus bound eIF3 complexes (eIF3 versus 43S–48S PICs) are indicated at the top. These experiments were performed three times. For description of the black box, see the main text (C) Relative change in distribution of individual eIF3 subunits in the 40S containing fractions with respect to the entire gradient. Values >100% signal accumulation of a factor of interest in the 40S-containing fractions, whereas values <100% point to its redistribution to lighter fractions. For details, see Materials and Methods. Please note that the distribution across the gradient was calculated for non-downregulated subunits only. Octameric eIF3 subunits are highlighted by a rectangle, the YLC subunits in bold. (D) Relative amounts of individual eIF3 subunits bound per ribosome (normalized to the Rps14 amount) compared to the control NT cells. For details, see Materials and Methods. For both quantifications, the Quantity One software from Bio-Rad was used.
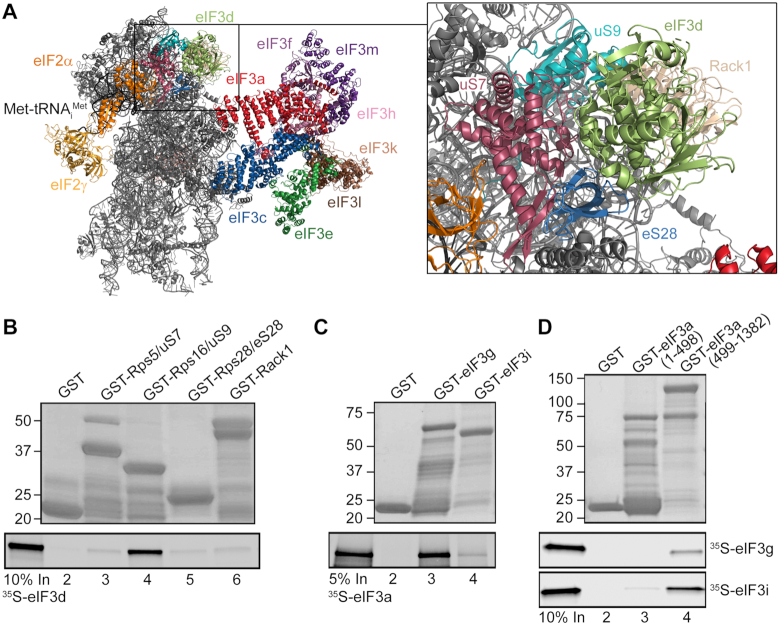
To examine this, we selected ribosomal proteins situated at the top of the 40S head, where eIF3d was tentatively placed based on available Cryo-EM structures (5,46), and performed an in vitro protein-protein binding analysis. In particular, Rps16/uS9, Rps5/uS7, Rps28/eS28 and Rack1 (Figure 3A) were fused to a GST moiety and tested for binding against radiolabeled, in vitro synthesized eIF3d. Out of this set of ribosomal proteins, Rps16/uS9 stood out as the strong and specific interactor of eIF3d (Figure 3B, lane 4). Interestingly, this ribosomal protein has been implicated in stabilizing the codon–anticodon duplex in the P-site (11). Even though from the available structures one can deduce that both proteins are in contact with each other (Figure 3A), we wished to corroborate the importance of this contact by a reciprocal in vivo approach using the Rps16/uS9KD cells. We hypothesized that decreased levels of Rps16/uS9 could lead to a partially reduced 40S-binding of eIF3 due to the loss of this contact. Unfortunately, the Rps16/uS9KD resulted in a strong 40S ribosome biogenesis defect (depletion of 40S and accumulation of 60S subunits) even at the lowest tested concentration of siRNA (2 nM; Supplementary Figure S2), which prevented us from testing our hypothesis. Nonetheless, based on the recent structure of the 43S PIC from Trypanosoma cruzi showing that the N-terminal tail of eIF3d extends from the 40S head towards the core of the eIF3 octamer where it interacts with the PCI domain of eIF3e, as well as with eIF3a and eIF3c (47), we propose that eIF3d creates yet another connecting bridge between the octamer and the 40S.
Figure 3.
eIF3d interacts with Rps16/uS9 and the eIF3a-CTD binds both eIF3i and g subunits of the YLC in vitro. (A) A cryo-EM model of the eIF3d placement on the 40S head highlighting its prospective contacts with small ribosomal proteins (adapted from (46)). (B) eIF3d interacts with Rps16/uS9 in vitro. Small ribosomal proteins Rps5/uS7 (lane 3), Rps16/uS9 (lane 4), Rps28/eS28 (lane 5) and Rack1 (lane 6) fused to GST moiety, and GST alone (lane 2) were tested for binding to 35S-labeled eIF3d. Lane 1 (In) contains 10% of input amounts of radiolabeled eIF3d added to each reaction mixture. (C, D) The eIF3a-CTD binds both eIF3i and g subunits of the YLC in vitro. (C) The eIF3g (lane 3) and eIF3i (lane 4) subunits fused to GST moiety, and GST alone (lane 2) were tested for binding to 35S-labeled eIF3a. Lane 1 (In) contains 5% of input amounts of radiolabeled eIF3a added to each reaction mixture. (D) The eIF3a N-terminal fragment (1–498 aa) (lane 3) and the eIF3a C-terminal fragment (499–1382 aa) (lane 4) fused to GST moiety, and GST alone (lane 2) were tested for binding to 35S-labeled eIF3g and eIF3i. Lane 1 (In) contains 10% of input amounts of radiolabeled eIF3g or eIF3i added to each reaction mixture.
Next, we assessed the ability of eIF3 lacking eIF3d to recruit mRNAs to the 43S PIC to form the 48S PIC. We measured the relative amounts of model RPL41 mRNA in all 40S-containing fractions by qPCR, normalized them to 18S rRNA amounts as described in detail in Materials and Methods, and the resulting values obtained from individual knockdowns expressed as percentages of control NT cells set to 100% (Table 1 and Supplementary Table S3). Please note that the RPL41 mRNA is an established model mRNA (8,39,40), because it is well expressed and short enough to be translated by only one ribosome at a time, as recently confirmed by yeast ribosome profiling (48). We revealed that the mRNA recruitment is strongly decreased in the eIF3dKD—down to 47% (Table 1 and Supplementary Table S3). However, since this result correlates well with the degree of reduction in eIF3 binding to the 40S, we conclude that it is rather a consequence of the impaired assembly of the 43S PICs than the eIF3dKD-specific defect. Considering that eIF3d possesses a cap binding activity and was shown to directly interact with eIF4G (28,30), this result was not expected. Nonetheless, since eIF3d was proposed to interact only with a peculiar set of mRNAs - often specifically structured – that does not include RPL41 mRNA (32,49), it is possible that its primary role in general translation initiation is indeed to stably anchor eIF3 to the 40S.
eIF3g and eIF3i are dispensable for the assembly of pre-initiation complexes
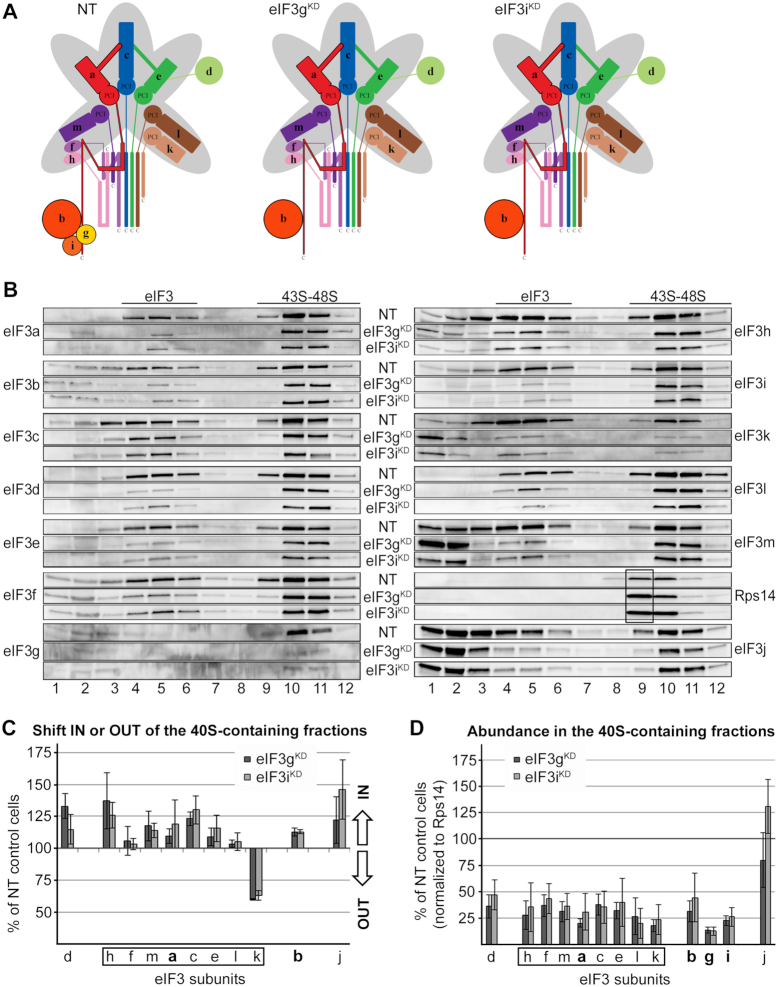
Knocking down eIF3g and eIF3i subunits, one at a time, concurrently and severely decreases levels of both of them, and, in addition, the overall amount of all remaining eIF3 subunits is reduced to ∼50% (7) (Figure 4A). This results in strongly reduced initiation rates and dramatically impaired viability (7). Interestingly, both of these knockdowns shifted eIF3 subunits in the opposite direction compared to that of eIF3dKD; i.e. with the exception of eIF3k, all eIF3 subunits moved to the heavier, 40S-containing fractions (Figure 4B and C). We propose that owing to the generally decreased eIF3 levels, cells implement a compensatory mechanism by utilizing all available eIF3 species for translation. The eIF3k distribution pattern showed an increased detachment of eIF3k from PICs resulting in its accumulation in the very first fraction rather than in fractions 4–6 containing ribosome-free eIF3 (Figure 4B and C). This pattern is consistent with previous results demonstrating that eIF3k is one of the most labile subunits prone to increased dissociation from eIF3 (12). In any case, these results clearly demonstrate that eIF3g and eIF3i, constituting the mobile YLC in human eIF3 (see below), are not required for the 43S PIC assembly. This contrasts with results from S. cerevisiae, where elimination of yeast eIF3g and eIF3i homologues from its 5-subunit eIF3 complex by a specific mutation in eIF3i (encoded by TIF34) leads to decreased 40S-binding of the remaining eIF3a-b-c trimer (50). Being markedly compromised in the number of subunits, stable 40S-binding of the budding yeast eIF3 complex most probably relies on ribosomal contacts of most, if not all, of its five subunits (reviewed in (2)). When normalized to Rps14, we estimate the amount of eIF3 bound to PICs in eIF3gKD and eIF3iKD to be ∼35% compared to control NT cells (Figure 4D), which is similar to the eIF3dKD. Here again, we can see – perhaps even more robust – accumulation of lighter 40S species lacking eIF3 (Figure 4B, fraction 9, black box).
Figure 4.
eIF3g and eIF3i are dispensable for the assembly of initiation complexes. (A) Comparison of schematic models of eIF3 complexes occurring in the control NT versus eIF3gKD and eIF3iKD cells. (B–D) Same as in Figure 2B–D except that the eIF3gKD and eIF3iKD were analyzed. These experiments were performed four times.
As shown in Table 1 and Supplementary Table S3, eIF3 subcomplexes lacking eIF3g and eIF3i are comparably compromised in their mRNA loading ability to those lacking eIF3d (down to 46% and 55%, respectively), which again correlates nicely with the degree of reduction in eIF3 binding to the 40S in these two knockdowns. Therefore, we conclude that eIF3g and eIF3i subunits are critical neither for the 43S PIC nor for the 48S PIC assembly. These findings are consistent with earlier yeast genetics data suggesting that eIF3g and eIF3i, residing near the mRNA entry channel, act later in the initiation pathway, in particular during ribosomal scanning and AUG recognition (41). The fact that they are part of the YLC (b-g-i-a-CTD), which was proposed to relocate from the 40S solvent side to the subunit interface to contact eIF1 and eIF2γ at early stages of codon:anticodon recognition, and later return back to its original position (9–11), further advocates their involvement in the post-assembly steps of initiation.
The presence of eIF3c in the eIF3 complex is required for the 48S PIC assembly
Next, we focused on the eIF3cKD and eIF3eKD. In eIF3eKD, protein levels of eIF3e, d, k and l are dramatically decreased, however, the remaining subunits are still able to form a partial subcomplex to some degree (7). Knocking down eIF3c severely reduces levels of all latter subunits plus eIF3c, while the remainder of the eIF3 complex is broken into YLC and the f-h-m subcomplex (8) (Figure 5A). Here both knockdowns produced very similar outcomes with respect to the 43S PIC assembly analysis, with the eIF3cKD showing generally more pronounced defects (Figure 5B–D). The eIF3f, h, and m subunits exhibited clearly the strongest effect as they shifted to the lighter fractions, most probably because they cannot efficiently associate with the 40S outside of the 12-subunit eIF3 holocomplex. Since all these three subunits peaked predominantly in fraction 2 in both knockdowns, in contrast to eIF3k peaking in the first fraction, we conclude that - in accord with (8) - they do occur in the trimeric f-h-m subcomplex that can exist separately outside of the eIF3 holocomplex and is capable to sustain spinning forces associated with this procedure. On the contrary, the YLC subunits a, b, g and i displayed only minor shift towards the top of the gradient supporting our earlier conclusion that the YLC module retains relatively high 40S-binding affinity on its own (8), most probably owing to the contacts that eIF3a, and possibly also eIF3b, make with the ribosome (4,5,22).
Figure 5.
The effect of eIF3c and eIF3e knockdowns on the 43S PIC assembly. (A) Comparison of schematic models of eIF3 complexes occurring in the control NT versus eIF3cKD and eIF3eKD cells. (B–D) Same as in Figure 2B–D except that the eIF3cKD and eIF3eKD were analyzed. These experiments were performed six times. Black boxes indicate the occurrence of the YLC (a-b-g-i) in fraction 9.
In the eIF3eKD a considerable amount of eIF3c shifted from 40S-containing fractions to the top of the gradient, suggesting that despite its direct interactions with Rps13/uS15 and Rps27/eS27, efficient 40S binding of eIF3c requires support from other eIF3 subunits. Consistently, des Georges et al. described quaternary interactions between eIF3a–eIF3c and eIF3c–eIF3e that rigidify the assembly of the a-c-e trimer and render it a structural core of the PCI ark (5). Please note that, again with the exception of eIF3k, whatever is left of the concurrently downregulated subunits (d and l in the eIF3eKD; and e, d and l in the eIF3cKD) associates with the 40S in a form of the eIF3 holocomplex (Figure 5B and D). Finally, same as with thus far described knockdowns, an elevated representation of lighter 40S species in fraction 9 in both knockdowns is seen, however, this time increased amounts of specifically the YLC subunits can be observed too (Figure 5B, black boxes, e.g. compare ratios of eIF3i and eIF3h in fraction 9). These findings not only corroborate the intrinsic ability of the YLC to associate stably with the 40S out of the context of the eIF3 holocomplex, they also further illuminate the sensitivity of our approach. Please also note that earlier we came to similar conclusions using the CoIP approach instead, showing that two different eIF3 subcomplexes occur in the eIF3eKD - a bigger one comprising the h-f-m-a-c-b-g-i subunits and the YLC (7). This fact nicely illustrates the complementarity of both approaches.
Dong et al. reported concurrent interaction of eIF3i with the spectrin domain of eIF3a and eIF3b, while eIF3g was shown to interact with eIF3b only (6). The compactness of the YLC prompted us to revisit these findings with help of GST binding analysis. In contrast to the latter study and to the budding yeast where eIF3g and eIF3i interact concurrently only with the C-terminal domain of eIF3b (51), we show that in humans both eIF3g and eIF3i interact individually also with the C-terminal 2/3 of eIF3a (Figure 3C and D). These results thus reveal a web of mutually stabilizing interactions among the human YLC subunits resembling a similar web of mutual interactions among the yeast eIF3a-b-c trimeric complex (50,52).
Strikingly, the 40S-association of the model mRNA was significantly (with the P value at 0.0007) more affected in the eIF3cKD than in the eIF3eKD (42% versus 78%, respectively) (Table 1 and Supplementary Table S3). Since the major difference between these two knockdowns lies in the eIF3c expression levels, our results suggest that the presence of eIF3c in the eIF3 complex is required for the efficient 48S PIC assembly. A specific role of eIF3c in mRNA recruitment receives further support from previously published results showing that eIF3c directly interacts with eIF4G to facilitate mRNA loading to the 43S PICs (28). Moreover, eIF3c was also shown (together with eIF3a) to promote 40S binding of the Hepatitis C virus IRES-driven mRNA (53).
eIF3l and eIF3k antagonize mRNA recruitment to the 43S PICs
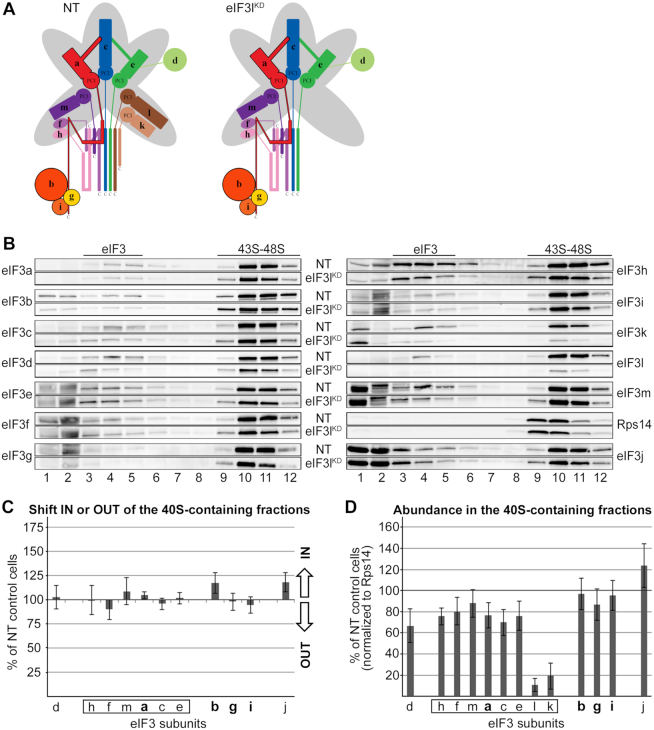
As aforementioned, knocking down eIF3k and eIF3l subunits, one at a time, concurrently and severely decreases levels of only these two proteins with no effect on integrity of the remaining eIF3 complex and cell viability (7) (Figure 6A). Since eIF3k is attached more on the periphery of the eIF3 complex compared to eIF3l, we performed the 43S assembly analysis only in the eIF3lKD. No apparent shift of any specific eIF3 subunit was observed (Figure 6B and C) and the overall eIF3 amounts associated with 40S ribosomes in the eIF3lKD displayed only a mild decrease (by ∼20%) compared to control NT cells (Figure 6D), indicating that the role of eIF3l, and most probably also eIF3k in the 43S PIC assembly is only minor.
Figure 6.
The eIF3lKD does not affect association of eIF3 with 40S ribosomes. (A) Comparison of schematic models of eIF3 complexes occurring in the control NT versus eIF3lKD cells. (B–D) Same as in Figure 2B–D except that the eIF3lKD was analyzed. These experiments were performed six times.
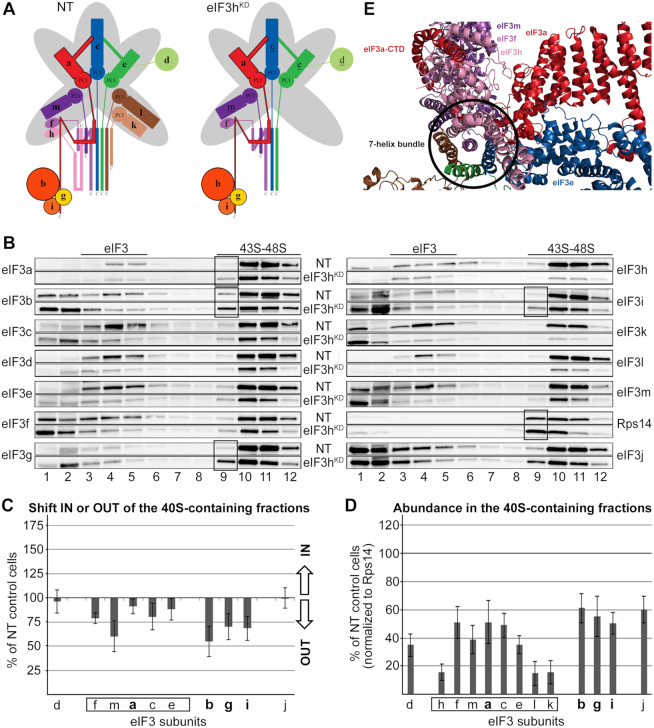
Knocking down eIF3h results in a concurrent downregulation of the eIF3h, k and l subunits that is accompanied by a less severe downregulation (by ∼30%) of protein levels of eIF3c, e, d, f, and m (Figure 7A) (7). Since subunits h, k and l are nonessential (2,54), the eIF3hKD displays only a mild growth phenotype and moderately decreased initiation rates (7). The 43S PIC assembly analysis revealed an uneven shift of all eIF3 subunits towards lighter fractions (Figure 7B and C). This shift was particularly pronounced for the eIF3m subunit and, unexpectedly, also for the eIF3b-g-i module. This is in sharp contrast to the eIF3cKD and eIF3eKD, where the YLC (b-g-i module plus eIF3a) showed a considerably stable 40S binding (Figure 5B and C). Interestingly, closer inspection of the distribution of eIF3 subunits across the gradient shows peaks of the eIF3f, m and k single subunits in fraction 1, whereas the eIF3b, g and i subunits peak predominantly in fraction 2; i.e. two fractions ahead of those where the 40S-free eIF3 holocomplex normally sediments. This indeed suggests that in contrast to the former subunits, a fraction of the b-g-i module is detached from the eIF3 complex as a compact unit by breaking its contacts with the eIF3a–CTD. How could this happen?
Figure 7.
The eIF3hKD impairs accommodation of the eIF3 b-g-i module on the 40S subunit. (A) Comparison of schematic models of eIF3 complexes occurring in the control NT versus eIF3hKD cells. The eIF3 subunits with decreased protein levels are underlined in the eIF3hKD schematic (B–D) Same as in Figure 2B–D except that the eIF3hKD was analyzed. These experiments were performed six times. Black boxes indicate the occurrence of the YLC (a-b-g-i) in fraction 9. (E) A cryo-EM model of the 7-helix bundle with the eIF3f-h-m module on top of it being wrapped around by the initial segment of the eIF3a C-terminal tail (adapted from (46)).
We propose that the loss of eIF3h, k and l from the eIF3 octamer distorts its conformation in a way that the eIF3h direct binding partners – eIF3m and eIF3f - dissociate individually from the complex. Not surprisingly, eIF3m is more affected than eIF3f (Figure 7B) because it was shown to be one of the most labile eIF3 subunits (together with eIF3k) (12). Furthermore, whereas the octamer core formed by the a-c-e PCI ark remains presumably intact, the initial segment of the eIF3a C-terminal tail, which normally wraps around the f-h-m module (Figure 7E) (5), might alter its conformation due to the loss of this module in such a way that prevents proper accommodation of the b-g-i module in the 43S PICs. As a result, a fraction of the b-g-i module dissociates from 40S ribosomes, while the eIF3a subunit remains 40S-bound; being a part of the incomplete octamer preserving its core (the PCI ark) intact. On the contrary, in the eIF3c and e knockdowns the octamer core is disintegrated, allowing the self-standing eIF3a subunit to preserve the YLC intact. Taken together, we propose that the a-c-e core represents the main 40S-binding unit of the eIF3 octamer. When compromised (like in the eIF3c or eIF3e knockdowns), the YLC containing the eIF3a subunit is still capable to stably associate with 40S ribosomes.
The fact that even though the b-g-i module binds less efficiently to the 40S in the eIF3hKD, yet we can still observe lighter 40S ribosomes co-sedimenting with the YLC in fraction 9 (Figure 7B and D, black boxes) suggests that the eIF3hKD produces not one but several eIF3 subcomplexes with varying binding affinities towards the 40S. Indeed, we showed previously that in the eIF3hKD (similarly as in the eIF3eKD) two eIF3 subcomplexes occur in the cell – a bigger one comprising all eIF3 subunits that are not strongly downregulated (d-f-m-a-c-e-b-g-i) and the YLC (7).
Interestingly, performing the model mRNA loading assay we revealed that the mRNA recruitment was more efficient in the eIF3lKD and eIF3hKD (on average up to 124% for both downregulations) than in control NT cells (Table 1 and Supplementary Table S3). Since this result was unexpected, we carried out the same analysis also in the eIF3kKD obtaining a similar result (an increase up to ∼132%) (Table 1 and Supplementary Table S3). As only the eIF3k and l subunits are missing from the eIF3 complex in the eIF3lKD and eIF3kKD, and the eIF3hKD led to the same effect as the eIF3lKD, despite the fact that eIF3h was shown to promote mRNA circularization (55), we conclude that these two nonessential subunits somehow antagonize mRNA loading onto the 43S PIC and thus serve as negative regulators. Strikingly, in the most recent structure of the mammalian 48S PIC, both eIF3l and eIF3k subunits were found in contact with eIF4F (Jailson Querido and Venki Ramakrishnan, personal communication), which may suggest that they could hold a control over the eIF4F role within the 48S PIC. In agreement with our findings, the eIF3lKD and eIF3kKD produced slightly increased polysome to monosome (P/M) ratio compared to control NT cells suggesting higher initiation rates (7), and loss-of-function mutations in the eIF3k and eIF3l genes resulted in a 40% extension in lifespan and enhanced resistance to endoplasmic reticulum (ER) stress in Caenorhabditis elegans (15). In spite of all these observations, the molecular mechanism underlying the negative effect of the k and l subunits of eIF3 on mRNA recruitment remains to be elucidated.
CONCLUDING REMARKS
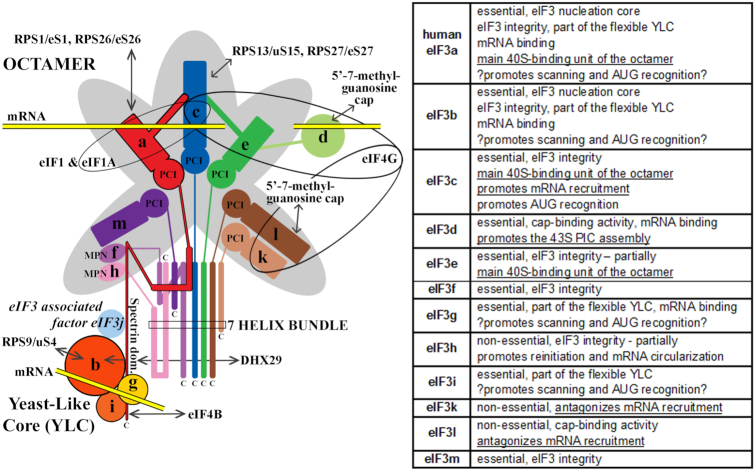
Here we described the adapted formaldehyde gradient cross-linking protocol representing a versatile and powerful technique that enables – to our knowledge – the best available approximation of the composition of native 43S and 48S PICs in vivo. With its help we demonstrated that one of the peripheral eIF3 subunits, eIF3d, is critically required for the assembly of the 43S PICs, and that the presence of eIF3c in the eIF3 complex is required for the 48S PIC assembly. Based on our findings we propose that the a-c-e core of the eIF3 octamer represents one of the major 40S-binding forces of the eIF3 holocomplex. If disrupted, the remaining YLC (b-g-i-a) is still capable of a relatively stable association with the 40S ribosomes. Finally, we uncovered that eIF3k and eIF3l possess a negative regulatory potential reducing the efficiency of mRNA recruitment as the only identifiable phenotype that could be attributed to these two nonessential octamer subunits in human cells so far (please see Figure 8 summarizing functional contributions of all individual human eIF3 subunits to general translation initiation known to date from our work and the work of our colleagues (1–3)). Taking into account that eIF3 has been implicated in the etiology of various diseases including cancer, neurodegenerative states like Parkinson disease etc. (some eIF3 subunits were even suggested to serve as oncogenes or tumor suppressors with potential prognostic values), as well as recent observations that eIF3 can drive alternative modes of initiation in mRNA-specific manner (1,2,35,36), it will be intriguing to investigate the ability of all so far identified partial eIF3 subcomplexes to promote mRNA scanning and AUG recognition, as even a minor defect in these critical initiation steps can have deleterious consequences.
Figure 8.
A schematic model of human eIF3 (adapted from (2)), with a table summarizing functional contributions of all individual human eIF3 subunits to general translation initiation known to date from our work and the work of our colleagues (1–3,7–11,21,22,28–31); findings from this work are underlined. The eIF3 subunits forming the PCI/MPN octamer are indicated by the grey background. The rectangle marks the seven α-helices involved in formation of the 7-helix bundle; the Yeast-Like Core (YLC) comprising the eIF3 subunits a, b, g and i is depicted at the bottom. Arrows indicate all known interactions of eIF3 subunits with other eIFs, ribosomal proteins and mRNA (reviewed in (2)).
Supplementary Material
ACKNOWLEDGEMENTS
We are thankful to Stanislava Gunišová for critical reading of the manuscript, Yaser Hashem for providing us with Figure 1B, and Olga Krýdová and Veronika Kovaničová for technical and administrative assistance, and to the members of the Valášek, Krásný, Staněk and Svoboda laboratories for helpful suggestions.
Notes
Present address: Darina Šikrová, Department of Human Genetics, Leiden University Medical Center, ZC Leiden, Zuid-Holland, the Netherlands.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Czech Science Foundation [GA17-06238S to L.S.V.]; Wellcome Trust [090812/B/09/Z to L.S.V.]. Funding for open access charge: Czech Science Foundation [GA17-06238S].
Conflict of interest statement. None declared.
REFERENCES
- 1. Gomes-Duarte A., Lacerda R., Menezes J., Romão L.. eIF3: a factor for human health and disease. RNA Biol. 2018; 15:26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Valášek L.S., Zeman J., Wagner S., Beznosková P., Pavlíková Z., Mohammad M.P., Hronová V., Herrmannová A., Hashem Y., Gunišová S.. Embraced by eIF3: structural and functional insights into the roles of eIF3 across the translation cycle. Nucleic Acids Res. 2017; 45:10948–10968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cate J.H.D. Human eIF3: from ‘blobology’ to biological insight. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2017; 372:20160176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Querol-Audi J., Sun C., Vogan J.M., Smith M.D., Gu Y., Cate J.H.D., Nogales E.. Architecture of human translation initiation factor 3. Structure. 2013; 21:920–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. des Georges A., Dhote V., Kuhn L., Hellen C.U.T., Pestova T.V, Frank J., Hashem Y.. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature. 2015; 525:491–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dong Z., Qi J., Peng H., Liu J., Zhang J.-T.. Spectrin domain of eukaryotic initiation factor 3a is the docking site for formation of the a:b:i:g subcomplex. J. Biol. Chem. 2013; 288:27951–27959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wagner S., Herrmannová A., Šikrová D., Valášek L.S.. Human eIF3b and eIF3a serve as the nucleation core for the assembly of eIF3 into two interconnected modules: the yeast-like core and the octamer. Nucleic Acids Res. 2016; 44:10772–10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wagner S., Herrmannová A., Malík R., Peclinovská L., Valášek L.S.. Functional and biochemical characterization of human eukaryotic translation initiation factor 3 in living cells. Mol. Cell Biol. 2014; 34:3041–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Simonetti A., Brito Querido J., Myasnikov A.G., Mancera-Martinez E., Renaud A., Kuhn L., Hashem Y.. eIF3 peripheral subunits rearrangement after mRNA binding and start-codon recognition. Mol. Cell. 2016; 63:206–217. [DOI] [PubMed] [Google Scholar]
- 10. Llácer J.L., Hussain T., Marler L., Aitken C.E., Thakur A., Lorsch J.R., Hinnebusch A.G., Ramakrishnan V.. Conformational differences between open and closed states of the eukaryotic translation initiation complex. Mol. Cell. 2015; 59:399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Llácer J.L., Hussain T., Saini A.K., Nanda J.S., Kaur S., Gordiyenko Y., Kumar R., Hinnebusch A.G., Lorsch J.R., Ramakrishnan V.. Translational initiation factor eIF5 replaces eIF1 on the 40S ribosomal subunit to promote start-codon recognition. Elife. 2018; 7:e39273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou M., Sandercock A.M., Fraser C.S., Ridlova G., Stephens E., Schenauer M.R., Yokoi-Fong T., Barsky D., Leary J.A., Hershey J.W. et al.. Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:18139–18144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Smith M.D., Arake-Tacca L., Nitido A., Montabana E., Park A., Cate J.H.. Assembly of eIF3 mediated by mutually dependent subunit insertion. Structure. 2016; 24:886–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou C., Arslan F., Wee S., Krishnan S., Ivanov A.R., Oliva A., Leatherwood J., Wolf D.A.. PCI proteins eIF3e and eIF3m define distinct translation initiation factor 3 complexes. BMC Biol. 2005; 3:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cattie D.J., Richardson C.E., Reddy K.C., Ness-Cohn E.M., Droste R., Thompson M.K., Gilbert W.V., Kim D.H.. Mutations in nonessential eif3k and eif3l genes confer lifespan extension and enhanced resistance to er stress in caenorhabditis elegans. PLoS Genet. 2016; 12:e1006326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hinnebusch A.G. The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 2014; 83:779–812. [DOI] [PubMed] [Google Scholar]
- 17. Sokabe M., Fraser C.S.. Human eukaryotic initiation factor 2 (eIF2)-GTP-Met-tRNAi ternary complex and eIF3 stabilize the 43 S preinitiation complex. J. Biol. Chem. 2014; 289:31827–31836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chaudhuri J., Chowdhury D., Maitra U.. Distinct functions of eukaryotic translation initiation factors eIF1A and eIF3 in the formation of the 40 S ribosomal preinitiation complex. J. Biol. Chem. 1999; 274:17975–17980. [DOI] [PubMed] [Google Scholar]
- 19. Majumdar R., Bandyopadhyay A., Maitra U.. Mammalian translation initiation factor eIF1 functions with eIF1A and eIF3 in the formation of a stable 40 S preinitiation complex. J. Biol. Chem. 2003; 278:6580–6587. [DOI] [PubMed] [Google Scholar]
- 20. Valásek L., Nielsen K.H., Hinnebusch A.G.. Direct eIF2-eIF3 contact in the multifactor complex is important for translation initiation in vivo. EMBO J. 2002; 21:5886–5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hashem Y., des Georges A., Dhote V., Langlois R., Liao H.Y., Grassucci R.A., Hellen C.U.T., Pestova T.V, Frank J.. Structure of the mammalian ribosomal 43S preinitiation complex bound to the scanning factor DHX29. Cell. 2013; 153:1108–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu Y., Neumann P., Kuhle B., Monecke T., Schell S., Chari A., Ficner R.. Translation initiation factor eIF3b contains a nine-bladed β-propeller and interacts with the 40S ribosomal subunit. Structure. 2014; 22:923–930. [DOI] [PubMed] [Google Scholar]
- 23. Harms U., Andreou A.Z., Gubaev A., Klostermeier D.. eIF4B, eIF4G and RNA regulate eIF4A activity in translation initiation by modulating the eIF4A conformational cycle. Nucleic Acids Res. 2014; 42:7911–7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Özeş A.R., Feoktistova K., Avanzino B.C., Fraser C.S.. Duplex unwinding and ATPase activities of the DEAD-box helicase eIF4A are coupled by eIF4G and eIF4B. J. Mol. Biol. 2011; 412:674–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Walker S.E., Zhou F., Mitchell S.F., Larson V.S., Valasek L., Hinnebusch A.G., Lorsch J.R.. Yeast eIF4B binds to the head of the 40S ribosomal subunit and promotes mRNA recruitment through its N-terminal and internal repeat domains. RNA. 2013; 19:191–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yourik P., Aitken C.E., Zhou F., Gupta N., Hinnebusch A.G., Lorsch J.R.. Yeast eIF4A enhances recruitment of mRNAs regardless of their structural complexity. Elife. 2017; 6:e31476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sokabe M., Fraser C.S.. A helicase-independent activity of eIF4A in promoting mRNA recruitment to the human ribosome. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:6304–6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Villa N., Do A., Hershey J.W.B., Fraser C.S.. Human eukaryotic initiation factor 4G (eIF4G) protein binds to eIF3c, -d, and -e to promote mRNA recruitment to the ribosome. J. Biol. Chem. 2013; 288:32932–32940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. LeFebvre A.K., Korneeva N.L., Trutschl M., Cvek U., Duzan R.D., Bradley C.A., Hershey J.W.B., Rhoads R.E.. Translation initiation factor eIF4G-1 binds to eIF3 through the eIF3e subunit. J. Biol. Chem. 2006; 281:22917–22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee A.S., Kranzusch P.J., Doudna J.A., Cate J.H.D.. eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature. 2016; 536:96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kumar P., Hellen C.U.T., Pestova T.V. Toward the mechanism of eIF4F-mediated ribosomal attachment to mammalian capped mRNAs. Genes Dev. 2016; 30:1573–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. de la Parra C., Ernlund A., Alard A., Ruggles K., Ueberheide B., Schneider R.J.. A widespread alternate form of cap-dependent mRNA translation initiation. Nat. Commun. 2018; 9:3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hinnebusch A.G. Structural insights into the mechanism of scanning and start codon recognition in eukaryotic translation initiation. Trends Biochem. Sci. 2017; 42:589–611. [DOI] [PubMed] [Google Scholar]
- 34. Fringer J.M., Acker M.G., Fekete C.A., Lorsch J.R., Dever T.E.. Coupled release of eukaryotic translation initiation factors 5B and 1A from 80S ribosomes following subunit joining. Mol. Cell Biol. 2007; 27:2384–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dong Z., Zhang J.-T.. Initiation factor eIF3 and regulation of mRNA translation, cell growth, and cancer. Crit. Rev. Oncol. Hematol. 2006; 59:169–180. [DOI] [PubMed] [Google Scholar]
- 36. Bhat M., Robichaud N., Hulea L., Sonenberg N., Pelletier J., Topisirovic I.. Targeting the translation machinery in cancer. Nat. Rev. Drug Discov. 2015; 14:261–278. [DOI] [PubMed] [Google Scholar]
- 37. Marchetti A., Buttitta F., Pellegrini S., Bertacca G., Callahan R.. Reduced expression of INT-6/eIF3-p48 in human tumors. Int. J. Oncol. 2001; 18:175–179. [DOI] [PubMed] [Google Scholar]
- 38. Shi J., Kahle A., Hershey J.W.B., Honchak B.M., Warneke J.A., Leong S.P.L., Nelson M.A.. Decreased expression of eukaryotic initiation factor 3f deregulates translation and apoptosis in tumor cells. Oncogene. 2006; 25:4923–4936. [DOI] [PubMed] [Google Scholar]
- 39. Chiu W.-L., Wagner S., Herrmannová A., Burela L., Zhang F., Saini A.K., Valásek L., Hinnebusch A.G.. The C-terminal region of eukaryotic translation initiation factor 3a (eIF3a) promotes mRNA recruitment, scanning, and, together with eIF3j and the eIF3b RNA recognition motif, selection of AUG start codons. Mol. Cell Biol. 2010; 30:4415–4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Khoshnevis S., Gunisová S., Vlcková V., Kouba T., Neumann P., Beznosková P., Ficner R., Valásek L.S.. Structural integrity of the PCI domain of eIF3a/TIF32 is required for mRNA recruitment to the 43S pre-initiation complexes. Nucleic Acids Res. 2014; 42:4123–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cuchalová L., Kouba T., Herrmannová A., Dányi I., Chiu W.-L., Valásek L.. The RNA recognition motif of eukaryotic translation initiation factor 3g (eIF3g) is required for resumption of scanning of posttermination ribosomes for reinitiation on GCN4 and together with eIF3i stimulates linear scanning. Mol. Cell Biol. 2010; 30:4671–4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kastner B., Fischer N., Golas M.M., Sander B., Dube P., Boehringer D., Hartmuth K., Deckert J., Hauer F., Wolf E. et al.. GraFix: sample preparation for single-particle electron cryomicroscopy. Nat. Methods. 2008; 5:53–55. [DOI] [PubMed] [Google Scholar]
- 43. Stark H. GraFix: stabilization of fragile macromolecular complexes for single particle cryo-EM. Methods Enzymol. 2010; 481:109–126. [DOI] [PubMed] [Google Scholar]
- 44. Beznosková P., Cuchalová L., Wagner S., Shoemaker C.J., Gunišová S., von der Haar T., Valášek L.S.. Translation Initiation factors eif3 and hcr1 control translation termination and stop codon read-through in yeast cells. PLoS Genet. 2013; 9:e1003962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Beznosková P., Wagner S., Jansen M.E., von der Haar T., Valášek L.S.. Translation initiation factor eIF3 promotes programmed stop codon readthrough. Nucleic Acids Res. 2015; 43:5099–5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Eliseev B., Yeramala L., Leitner A., Karuppasamy M., Raimondeau E., Huard K., Alkalaeva E., Aebersold R., Schaffitzel C.. Structure of a human cap-dependent 48S translation pre-initiation complex. Nucleic Acids Res. 2018; 46:2678–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bochler A., Querido J.B., Prilepskaja T., Soufari H., Lucia Del Cistia M., Kuhn L., Ribeiro A.R., Valášek L.S., Hashem Y.. Unique features of mRNA translation initiation in trypanosomatids. 2019; 16 October 2019, preprint: not peer reviewed 10.1101/806141. [DOI] [PMC free article] [PubMed]
- 48. Heyer E.E., Moore M.J.. Redefining the translational status of 80s monosomes. Cell. 2016; 164:757–769. [DOI] [PubMed] [Google Scholar]
- 49. Lee A.S.Y., Kranzusch P.J., Cate J.H.D.. eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature. 2015; 522:111–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Herrmannová A., Daujotyte D., Yang J.-C., Cuchalová L., Gorrec F., Wagner S., Dányi I., Lukavsky P.J., Valásek L.S.. Structural analysis of an eIF3 subcomplex reveals conserved interactions required for a stable and proper translation pre-initiation complex assembly. Nucleic Acids Res. 2012; 40:2294–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Asano K., Phan L., Anderson J., Hinnebusch A.G.. Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J. Biol. Chem. 1998; 273:18573–18585. [DOI] [PubMed] [Google Scholar]
- 52. Phan L., Schoenfeld L.W., Valásek L., Nielsen K.H., Hinnebusch A.G.. A subcomplex of three eIF3 subunits binds eIF1 and eIF5 and stimulates ribosome binding of mRNA and tRNA(i)Met. EMBO J. 2001; 20:2954–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sun C., Querol-Audí J., Mortimer S.A., Arias-Palomo E., Doudna J.A., Nogales E., Cate J.H.D.. Two RNA-binding motifs in eIF3 direct HCV IRES-dependent translation. Nucleic Acids Res. 2013; 41:7512–7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Luo H., Lin Y., Gao F., Zhang C.-T., Zhang R.. DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements. Nucleic Acids Res. 2014; 42:D574–D580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Choe J., Lin S., Zhang W., Liu Q., Wang L., Ramirez-Moya J., Du P., Kim W., Tang S., Sliz P. et al.. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature. 2018; 561:556–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hashem Y., Frank J.. The jigsaw puzzle of mrna translation initiation in eukaryotes: a decade of structures unraveling the mechanics of the process. Annu. Rev. Biophys. 2018; 47:125–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.