Abstract
Late stage melanoma is associated with a high mortality rate. Signal transducer and activator of transcription 3 (STAT3) is currently a target for melanoma treatment as it is constitutively activated with high frequency in melanoma. Dioscin is a natural steroid saponin that is present in several medical herbs. A previous study demonstrated that dioscin inhibits STAT3 signaling in a cerebral ischemia-reperfusion injury rat model. Furthermore, dioscin has been reported to exert anti-melanoma effects in B16 melanoma cells and a B16 allograft mouse model. The present study investigated whether inhibition of STAT3 signaling is involved in the anti-melanoma effects of dioscin. The results of the present study demonstrated that dioscin significantly decreased viability, induced apoptosis and suppressed migration of human A375 melanoma cells and murine B16F10 melanoma cells. Furthermore, dioscin inhibited the phosphorylation of STAT3 and Src (an upstream kinase of STAT3), and downregulated mRNA levels of STAT3-targeted genes, including B-cell lymphoma-2, cyclin D1 and matrix metalloproteinase-2. In addition, overexpression of STAT3 decreased the anti-proliferative effects of dioscin. Overall, the results of the present study indicate that inhibiting the Src/STAT3 signaling pathway contributes to the anti-melanoma molecular mechanisms of dioscin. These results provide further pharmacological groundwork for developing dioscin as a novel anti-melanoma agent.
Keywords: dioscin, melanoma, src, STAT3, mechanism
Introduction
Melanoma is an aggressive cancer with a rapid increase in incidence rate worldwide. The number of new melanoma cases diagnosed is estimated to increase by 7.7% in 2019 (1). Less than half of patients diagnosed with metastatic melanoma survived longer than 1 year, and only 20% of them were alive after 3 years (2). Current therapies for melanoma, including chemotherapy, immunotherapy and targeted therapy, pose various limitations, such as low response rates, severe side effects, toxicity and high tendency to develop tolerance (3,4). Thus, novel therapies for melanoma are required.
A number of signaling pathways are involved in melanoma progression and many molecular targets have been identified for melanoma treatment (5). Signal transducer and activator of transcription 3 (STAT3) has been proposed as one of the therapeutic targets for melanoma as it is constitutively activated with high frequency in melanoma (6). Activation of STAT3 promotes cell proliferation, inhibits cell apoptosis and facilitates cell migration in melanoma (7). Overexpression of a dominant-negative STAT3 variant has been demonstrated to result in growth inhibition and regression, and prevents metastasis of melanoma cells in vivo (8). Furthermore, STAT3 knockdown with siRNAs in melanoma cells has been demonstrated to induce apoptosis, and inhibit proliferation and migration (9). Clinical trials on a number of STAT3 inhibitors, including WP1066, AZD9150, STAT3 DECOY, OPB-31121 and OPB-51602), have been approved for the treatment of melanoma. However, some trials have been discontinued due to the severity of adverse effects (6). Thus, future studies are required in order to develop safe and effective STAT3 inhibitors for the treatment of melanoma.
Dioscin is a natural steroid saponin that is present in several herbs, including the rhizome of Dioscorea opposita Thunb, and as such, has a long history of consumption as a part of the normal human diet (10). Several pharmacological activities of dioscin, such as lipid-lowering, anti-virus, anti-inflammatory and anti-cancer activities have been indicated to have a beneficial effect on human health (11). Previous studies have demonstrated that dioscin has anti-melanoma effects in murine B16 cells and allograft mouse models (12–14). Dioscin has been reported to inhibit STAT3 phosphorylation in a cerebral ischemia-reperfusion injury rat model (15) and suppresses Src (an upstream kinase of STAT3) in different types of colon tumor (16). Therefore, it is speculated that dioscin is a safe anti-melanoma phytocompound that inhibits Src/STAT3 signaling.
The present study investigated the anti-melanoma effects of dioscin in both murine and human cell models and assessed the involvement of the Src/STAT3 signaling pathway in these effects.
Materials and methods
Reagents and cell lines
Dioscin (purity >98% as determined by high-performance liquid chromatography) was obtained from Shanghai Yuanye Bio-Technology Co., Ltd. Human A375 melanoma cells, murine B16F10 melanoma cells and murine L929 fibroblasts were obtained from the American Type Culture Collection (ATCC). The B16STAT3C (overexpressing a constitutively active STAT3 mutant, STAT3C Flag pRc/CMV) and B16NC (expressing the empty vector, pcDNA 3.0) cell lines were previously established (8). The plasmids were obtained from AddGene Inc. (cat. no. 8722). Cells were cultured in DMEM supplemented with 5% fetal bovine serum (FBS) and 1% penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.), and maintained at 37°C and in a humidified atmosphere of 5% CO2.
CCK-8 assay
A Cell Counting Kit-8 (CCK-8) assay was performed according to the manufacturer's protocol to determine the cytotoxicity of dioscin towards A375 cells, B16F10 cells and L929 cells (17). Cells were seeded in 96-well plates at a density of 5,000 cells/well and were treated with dioscin at various concentrations (0, 1, 2, 4 and 8 µM) at 37°C in a humidified atmosphere of 5% CO2 for 24 and 48 h. A total of 10 µl of CCK-8 (Dojindo Molecular Technologies, Inc.) solution was added to each well and incubated at 37°C in a humidified atmosphere of 5% CO2 for an additional 2 h. The absorbance was measured at 450 nm using a microplate spectrophotometer (BD Biosciences).
Crystal violet staining
Crystal violet staining was performed to visualize the effects of dioscin on the viability of A375 cells and B16F10 cells, according to the manufacturer's protocol (7). Cells were seeded in 6-well plates at a density of 100,000 cells/well and were treated with dioscin at various concentrations (0, 2, 4 and 8 µM) under 37°C in a humidified atmosphere of 5% CO2 for 48 h. Treated cells were fixed with 10% formalin at room temperature for 5 min, followed by staining with 0.05% crystal violet solution in distilled water at room temperature for 30 min. Cells were then washed twice with PBS and scanned using an Epson V370 Scanner (Epson Co., Ltd.). Subsequently, the purple formazan crystals were dissolved in 1 ml of 33% glacial acetic acid (Sigma-Aldrich; Merck KGaA) and viability was assessed at a wavelength of 570 nm using a microplate spectrophotometer (BD Biosciences).
Cell apoptosis assay
The apoptotic effect of dioscin in A375 cells and B16F10 cells was quantified using Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) double staining assays (18), with the Apoptosis Detection kit (Abcam), according to the manufacturer's protocol. Following treatment with dioscin (0, 1 and 2 µM) for 24 h, both the detached and adherent cells were harvested and subsequently incubated in 500 µl of labeling solution (5 µl of AnnexinV-FITC, 5 µl of PI and 490 µl of binding buffer) in darkness at room temperature for 15 min. Flow cytometric analyses were performed using a C6 flow cytometer (BD Biosciences) and CellQuest™ Pro software version 5.1 (BD Biosciences).
Wound heal assay
The effect of dioscin on cell migration was assessed via a wound healing assay (7). A375 cells and B16F10 cells were seeded at a density of 1,000,000 cells/well and grown to 100% confluence in 6-well plates. The confluent cell monolayer was scratched with a sterile 10 µl pipette tip across the center of each well in order to produce a clean, straight wound area. Cells were subsequently washed twice with PBS in order to remove the detached cells, and incubated with dioscin (0.00, 0.25 and 0.50 µM) in serum-free DMEM medium at 37°C in a humidified atmosphere of 5% CO2 for 12 h. Cell migration was captured at the 0 and 12 h time points using a digital camera installed on a Leica DM3000 inverted fluorescence microscope (Leica Microsystems Ltd.) with a ×5 objective lens under the bright field mode. A total of five images were captured for each well. Migration rate was calculated using the following equation: (A-B)/A ×100%, where A represents the width of wound at 0 h and B represents the width of wound at 12 h.
Western blot analysis
Whole cell lysates were prepared from cultured B16F10 and A375 cells using RIPA lysis buffer [50 mM Tris (pH 7.4), 150 mM NaCl, 1% TritonX-100, 1% sodium deoxycholate, 0.1% SDS], with the addition of 1 mM PMSF, 2 mM sodium pyrophosphate, 25 mM β-glycerophosphate, 1 mM EDTA, 1 mM Na3VO4 and 0.5 µg/ml leupeptin. Protein concentrations were measured using Quick Start Bradford 1X Dye reagent (Bio-Rad Laboratories, Inc.) according to the manufacturer's protocol. Each protein sample was loaded at a concentration of 10 µg/µl on 8% SDS/PAGE gels. Subsequently, the proteins were transferred from the gels to nitrocellulose membranes. After that, the membranes were blocked in 5% skimmed milk (Devondale, Saputo Dairy Austrlia Pty Ltd.) in Tris-buffered saline Tween-20 (TBST) (Sigma-Aldrich Inc.; Merck KGaA) at room temperature for 1 h. Next, the membranes were incubated in diluents of primary antibodies at 4°C for 12 h. After incubation, the membranes were washed with TBST for three times. Then the membranes were incubated in diluents of secondary antibodies at room temperature for 1 h and washed with TBST for three times. Standard western blotting assay was performed as previously described (19). Protein bands were visualized using the enhanced chemiluminescence detection system (Invitrogen; Thermo Fisher Scientific, Inc.). The primary antibodies STAT3 (cat. no. 12640), Src (cat. no. 2108), phospho-STAT3 (Tyr705; cat. no. 9145), phospho-Src (Tyr416; cat. no. 6943) and GAPDH (cat. no. 5174) were obtained from Cell Signaling Technology, Inc., and diluted with 5% bovine serum albumin (BSA) (Sigma-Aldrich Inc.; Merck KGaA) in TBST at a ratio of 1:1,000. The secondary antibody anti-rabbit IgG, HRP-linked antibody (cat. no. 7074) was obtained from Cell Signaling Technology, Inc. and diluted with 5% BSA in TBST at a 1:10,000 ratio.
Reverse transcription-quantitative (RT-q)PCR analysis
Total RNA was extracted from B16F10 cells and A375 cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol, and reverse transcribed into cDNA using the PrimeScript™ RT reagent kit (Takara Bio, Inc.) according to the manufacturer's protocol. The reverse transcription parameters were 37°C for 15 min followed by 85°C for 5 sec and 4°C for 15 min. RT-qPCR (20) was performed in triplicate in a total volume of 10 µl consisting of 5 µl of 2X SYBR green PCR Master Mix, 1 µl of forward primer (10 µM), 1 µl of reverse primer (10 µM) and 3 µl of template in sterile distilled water, using iTaq™ Universal SYBR Green Supermix (Bio-Rad Laboratories, Inc.) with a ViiA 7 Real-time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The PCR parameters were as follows: Initial denaturation at 95°C for 10 min; 40 cycles of 95°C for 15 sec; and a final extension at 60°C for 1 min. The mRNA levels were quantified using the comparative CT method (21) and normalized to the internal reference gene GAPDH. The primers used in the present study are presented in Table I.
Table I.
Sequences of primers.
| Gene | Forward | Reverse |
|---|---|---|
| Human GAPDH | 5′-ACCCATCACCATCTTCCAGGAG-3′ | 5′-GAAGGGGCGGAGATGATGAC-3′ |
| Human cyclin D1 | 5′-GAACGAATCTACCCATTACCAG-3′ | 5′-GAGATGCCCGTGATGAACC-3′ |
| Human Bcl-2 | 5′-GCCATATGGCGCACGCTGGGAGAA-3′ | 5′-GCGCTCGAGTCACTTGTGGCCCAGATAG-3′ |
| Human MMP-2 | 5′-AGTGGTCCGTGTGAAGTATG-3′ | 5′-GTATCAGTGCAGCTGTTGTA-3′ |
| Mouse GAPDH | 5′-CCATGGAGAAGGCCGGGG-3′ | 5′-CAAAGTTGTCATGGATGACC-3′ |
| Mouse cyclin D1 | 5′-GTCATCAAGTGTGACCCG-3′ | 5′-GCACAGTCTGCCTGATGC-3′ |
| Mouse Bcl-2 | 5′-TGTAAGGACGAAACGGGACT-3′ | 5′-AAAGCCAGCAGCACATTTCT-3′ |
| Mouse MMP-2 | 5′-CTGGAATGCCATCCCTGATAA-3′ | 5′-GGTTCTCCAGCTTCAGGTAATAA-3′ |
Bcl-2, B-cell lymphoma-2; MMP-2, matrix metallopeptidase-2.
Statistical analysis
All data are presented as mean ± standard deviation. Comparisons among groups were performed using one-way ANOVA followed by Dunnett's multiple comparisons using the statistical GraphPad Prism software (version 6.0; GraphPad Software Inc.). P<0.05 was considered to indicate a statistically significant difference.
Results
Dioscin exhibits higher cytotoxicity in melanoma cells than in normal cells
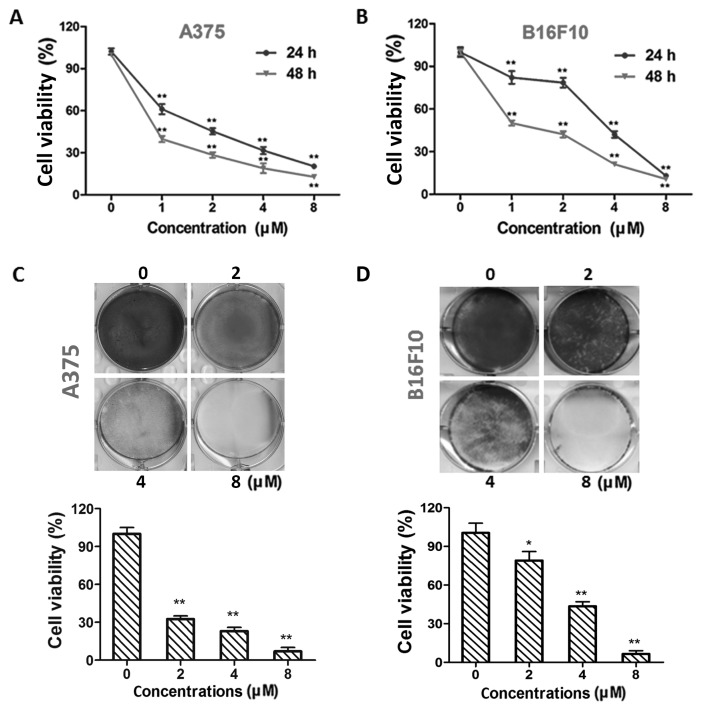
Treatment with dioscin (1, 2, 4 and 8 µM) at 24 and 48 h decreased the viability of A375 cells (P<0.01; Fig. 1A) and B16F10 cells (P<0.01; Fig. 1B) in a time- and dose-dependent manner. The half maximal inhibitory concentration (IC50) values in A375 cells were 1.54±0.32 and 0.57±0.18 µM for 24 and 48 h treatments, respectively. The IC50 values in B16F10 cells were 3.14±0.14 and 1.16±0.17 µM for 24 and 48 h treatments, respectively. The results of crystal violet staining further validate the effects of dioscin on A375 (P<0.01; Fig. 1C) and B16F10 (P<0.05; Fig. 1D) cell viability. In addition, viability decreasing effects of 1, 2, 4 and 8 µM of dioscin were less potent in L929 fibroblasts than in B16F10 and A375 melanoma cells (P<0.01; Fig. S1).
Figure 1.
Dioscin inhibits cell viability in melanoma cells. (A) CCK-8 assays in A375 cells. (B) CCK-8 assays in B16F10 cells. (C) Crystal violet staining of A375 cells. (D) Crystal violet staining of B16F10 cells. Representative photos from each group are presented in the upper panel, quantification results are demonstrated in the lower panel. Data are presented as mean ± standard deviation of three independent experiments. *P<0.05; **P<0.01 vs. control. CCK-8, Cell Counting Kit-8.
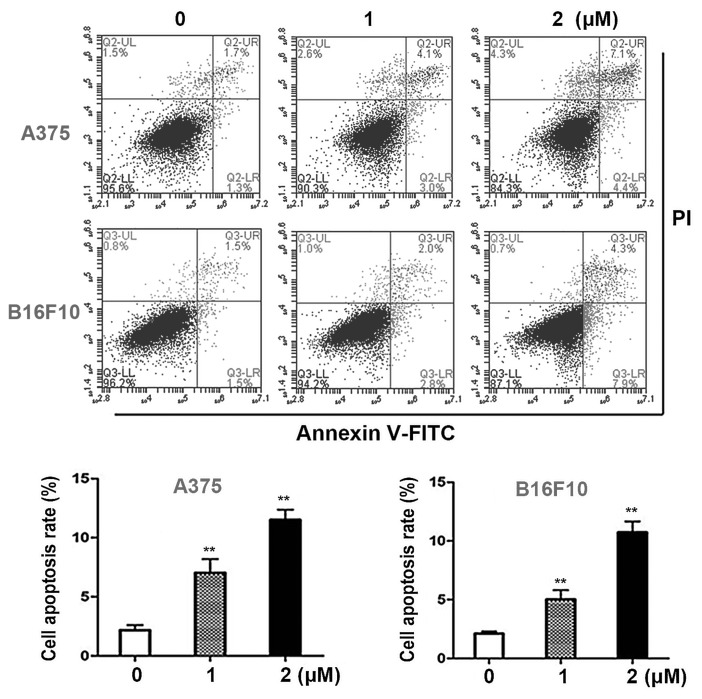
Dioscin induces apoptosis in melanoma cells
Annexin V- FITC/PI double staining assays were performed in order to quantify the apoptotic effects of dioscin in melanoma cells. Dioscin treatments (1, 2 µM) for 24 h significantly induced apoptosis in A375 cells and B16F10 cells in a dose-dependent manner (Fig. 2A). The results of the present study demonstrated that in A375 cells, 1 and 2 µM of dioscin at 24 h significantly increased the apoptotic cell ratios to 6.6±1.1% (P<0.01) and 11.6±0.9% (P<0.01), compared with the control cells (2.5±0.5%). In B16F10 cells, 1 and 2 µM of dioscin at 24 h increased the apoptotic cell ratios to 5.0±0.8% (P<0.01) and 10.9±0.8% (P<0.01), compared with the control cells (2.4±0.2%) (Fig. 2B).
Figure 2.
Dioscin induces apoptosis in melanoma cells. (A) Annexin V-FITC/PI double staining of A375 cells and B16F10 cells. Unstained and single-stained cells were used to set the gate, which is divided into four quadrants. FITC positive cells (right quadrants) were regarded as apoptotic cells. Representative photos from each group are presented. (B) Apoptosis rates of A375 cells and B16F10 cells. Data are presented as the mean ± standard deviation of three independent experiments. **P<0.01 vs. control. AnnexinV-FITC, Annexin V-fluorescein isothiocyanate; PI, propidium iodide; FBS, fetal bovine serum.
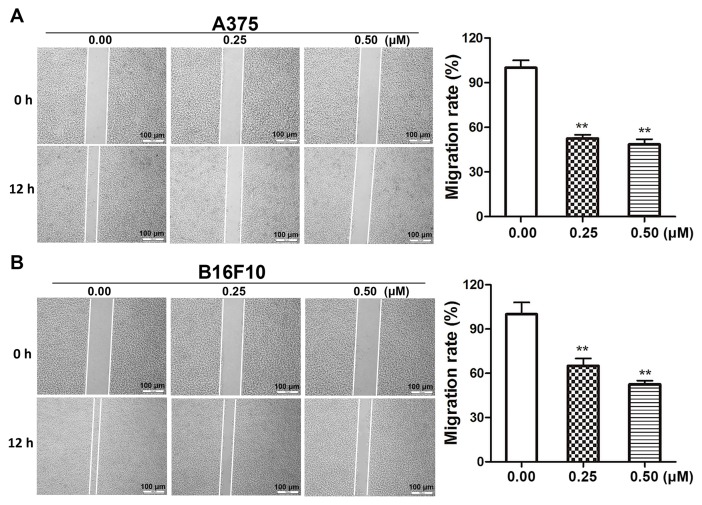
Dioscin decreases the migratory ability of melanoma cells
Wound healing assays were performed in order to determine the effects of dioscin on melanoma cell migration. Dioscin treatments (0.25 and 0.50 µM) for 12 h significantly inhibited the migration of A375 cells (Fig. 3A) and B16F10 cells (Fig. 3B). The results of the present study demonstrated that 0.25 and 0.50 µM of dioscin significantly decreased cell migration by 48.5% (P<0.01) and 52.4% (P<0.01), respectively, in A375 cells, and by 37.3% (P<0.01) and 46.5% (P<0.01), respectively, in B16F10 cells. However, dioscin at 0.25 and 0.50 µM did not significantly affect the viability of A375 cells and B16F10 cells after a 12-h treatment (Fig. S2).
Figure 3.
Dioscin inhibits migration of starved melanoma cells. (A) Wound healing assays in A375 cells. (B) Wound healing assays in B16F10 cells. The wounds were captured at 0 and 12 h following treatment with dioscin. Representative photos from each group are presented in the left panel, quantification results are demonstrated in the right panel. Migration rate was calculated as (A-B)/A ×100%, where A represents the width of wound at 0 h and B represents the width of wound at 12 h. **P<0.01 vs. control. FBS, fetal bovine serum; PBS, phosphate-buffered saline.
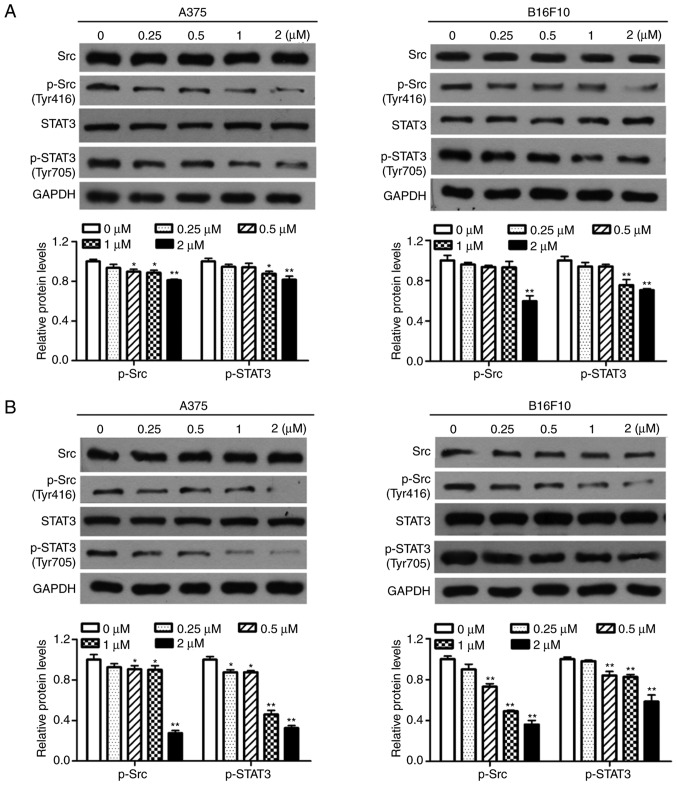
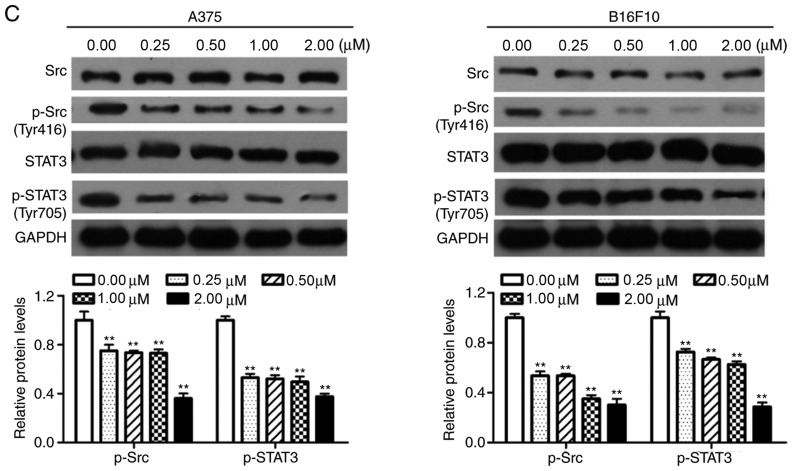
Dioscin inhibits the activation of Src and STAT3 in melanoma cells
Western blotting was performed in order to determine the effects of dioscin on the activation of Src and STAT3 in melanoma cells. The results of the present study demonstrated that treatment with dioscin (0.25, 0.50, 1.00 and 2.00 µM) for 12 (Fig. 4A), 24 (Fig. 4B) and 48 h (Fig. 4C) dose-dependently suppressed the phosphorylation of Src (Tyr 416) and STAT3 (Tyr 705), but did not affect protein levels of total STAT3 and total Src, in A375 cells and B16F10 cells. Treatments with dioscin (0.25, 0.50, 1.00 and 2.00 µM) for 12, 24 and 48 h also dose-dependently lowered the ratios of p-Src/Src and p-STAT3/STAT3 (Fig. S3).
Figure 4.
Dioscin inhibits phosphorylation of Src and STAT3 in melanoma cells. (A) Western blot analyses of A375 cells and B16F10 cells treated with dioscin (0.00, 0.25, 0.50, 1.00 and 2.00 µM) for 12 h. (B) Western blot analyses of A375 cells and B16F10 cells treated with dioscin (0.00, 0.25, 0.50, 1.00 and 2.00 µM) for 24 h. (C) Western blot analyses of A375 cells and B16F10 cells treated with dioscin (0.00, 0.25, 0.50, 1.00 and 2.00 µM) for 48 h. Protein levels of total and phosphorylated Src (Tyr416, p-Src), and total and phosphorylated STAT3 (Tyr705, p-STAT3) were determined by western blotting. GAPDH was used as the endogenous control. Representative bands from each group are presented in the upper panels. Band intensity was analyzed using ImageJ software and are presented in the lower panels. The relative protein levels of the control group were normalized to 1. Data presented in bar charts are the mean ± standard deviation of three independent experiments. *P<0.05; **P<0.01 vs. control. STAT3, signal transducer and activator of transcription 3; FBS, fetal bovine serum.
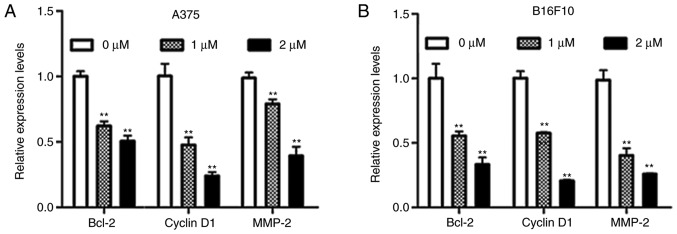
Dioscin decreases the expression of STAT3 target genes in melanoma cells
B-cell lymphoma-2 (Bcl-2) and cyclin D1 are STAT3-target genes that are involved in cell survival, and matrix metallopeptidase-2 (MMP-2) is a STAT3-target gene that is involved in cell migration (9). The results of the present study demonstrated that dioscin significantly downregulated mRNA levels of Bcl-2, cyclin D1 and MMP-2 in A375 cells (P<0.01; Fig. 5A) and B16F10 cells (P<0.01; Fig. 5B). These results further indicate that inhibiting STAT3 signaling is involved in the anti-melanoma action of dioscin.
Figure 5.
Dioscin downregulates mRNA levels of STAT3-target genes in melanoma cells. (A) RT-qPCR analyses of A375 cells. (B) RT-qPCR analyses of B16F10 cells. The relative expression levels of the control group were normalized to 1. Data are presented as the mean ± standard deviation of three independent experiments. **P<0.01 vs. control. STAT3, signal transducer and activator of transcription 3; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; FBS, fetal bovine serum; Bcl-2, B-cell lymphoma-2; MMP-2, matrix metallopeptidase-2.
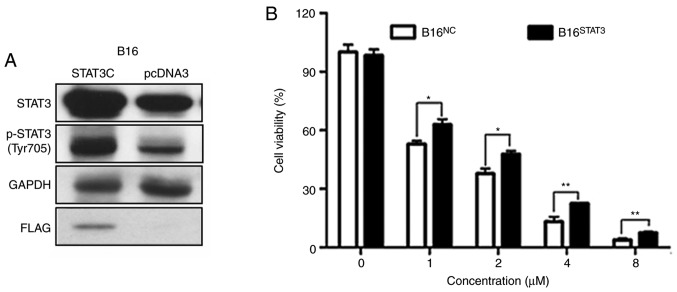
Overexpression of STAT3 diminishes the effects of dioscin on cell viability
Stable B16STAT3C (overexpressing a constitutively active STAT3 mutant) and B16NC cell lines (expressing the empty vector) were used in order to validate the involvement of STAT3 signaling in the anti-melanoma effects of dioscin. The western blot results of the present study demonstrated that protein levels of total STAT3 and phospho-STAT3 (Tyr705) were higher in B16STAT3C cells than in B16NC cells, indicating that STAT3 was overexpressed in B16STAT3C cells (Fig. 6A, the same as in Reference 8). The effects of dioscin on the viability of B16NC cells and B16STAT3C cells were compared. Following treatment for 48 h, the cytotoxic effects of different concentrations of dioscin (1, 2, 4 and 8 µM) were demonstrated to be less potent toward B16STAT3C cells than B16NC cells (P<0.05; Fig. 6B), indicating that over-activation of STAT3 weakened the anti-proliferative effects of dioscin.
Figure 6.
Overexpression of STAT3 diminishes the cytotoxic effect of dioscin in melanoma cells. (A) Protein levels of total STAT3, phosphorylated STAT3 (Tyr705, p-STAT3), Flag in B16STAT3C and B16NC cells (the same as in ref. 6). (B) CCK-8 assays in B16STAT3C and B16NC cells. Data are presented as mean ± standard deviation of three independent experiments. *P<0.05; **P<0.01 vs. STAT3, signal transducer and activator of transcription 3; CCK-8, Cell Counting Kit-8; FBS, fetal bovine serum.
Discussion
The present study investigated the anti-melanoma effects of dioscin in human A375 cells and murine B16F10 melanoma cells. In agreement with previous studies (13,14,22), the results of the present study demonstrated that dioscin decreased melanoma cell viability (Fig. 1). Furthermore, the results of the present study indicated that dioscin induced apoptosis (Fig. 2), and restrained migration in melanoma cells (Fig. 3). A previous study demonstrated that dioscin inhibits B16 allograft melanoma growth and metastasis (13). The results of the present and previous studies suggest that dioscin has the potential to be developed as an anti-melanoma agent.
To the best of our knowledge, the present study was the first to demonstrate that inhibiting STAT3 signaling is, at least partially, responsible for the anti-melanoma effects of dioscin. Over-activation of STAT3 could not completely reverse the effect of dioscin on cell viability (Fig. 6), indicating that inhibition of STAT3 is not the sole molecular mechanism of action of dioscin. A number of previous studies have demonstrated that upregulation of connexin 26 and connexin 43, and downregulation of phospho-CREB and MITF contribute to the anti-melanoma molecular mechanisms of dioscin (13,14,22). Whether additional molecular mechanisms are involved in the anti-melanoma effects of dioscin remains to be elucidated.
STAT3 is activated in several types of cancer, including breast, gastric and lung cancer (20,23–25). Dioscin has been reported to inhibit the growth of these different types of cancer in mice (11). Whether dioscin suppresses these types of cancer by inhibiting STAT3 signaling needs to be further studied.
Apart from promoting cell proliferation, inhibiting cell apoptosis, and facilitating cell migration and invasion, activation of STAT3 signaling can also promote angiogenesis and immune evasion in melanoma (26). Whether dioscin inhibits melanoma angiogenesis and immune evasion, however, remains to be elucidated.
Overall, the results of the present study confirmed the anti-melanoma effects of dioscin in more cell models and with more parameters, and demonstrated that inhibiting the Src/STAT3 signaling pathway contributes to the anti-melanoma molecular mechanisms of dioscin. The results of the present study provide further pharmacological groundwork for developing dioscin as a novel anti-melanoma agent.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- STAT3
signal transducer and activator of transcription 3
- CCK-8
Cell Counting Kit-8
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- MMP-2
matrix metalloproteinase-2
Funding
The present study was funded by the Science, Technology and Innovation Commission of Shenzhen (grant nos. JCYJ20160229210327924 and JCYJ20170817173608483); the Department of Science and Technology of Guangdong Province (grant nos. 2016A030313007 and 2017B050506003); the Innovation and Technology Commission of Hong Kong (grant no. GHX/002/17GD); National Natural Science Foundation of China (grant nos. 81673649, 8187141799 and 81803788); the Food and Health Bureau of Hong Kong (grant nos. 14150571 and 15163441); the Research Grants Council of Hong Kong (grant nos. 12125116 and 12102918); and the Hong Kong Baptist University (grant nos. FRG1/16-17/048 and FRG2/17-18/032).
Availability of data and materials
The datasets used and analyzed in the present study are available from the corresponding author upon reasonable request.
Authors' contributions
YXL, BWX performed the majority of the experiments and interpreted the data. YXL drafted the initial manuscript. YJC, XQF, PLZ, JXB, JYC, CLY, JKL, YPW, JW, YW and KKC participated in several experiments and analyzed the data. CL designed the present study. ZLY designed the present study and finalized the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Skin cancer facts & statistics 2019. https://www.skincancer.org/skin-cancer-information/skin-cancer-facts/
- 2.Rogiers A, Boekhout A, Schwarze JK, Awada G, Blank CU, Neyns B. Long-term survival, quality of life, and psychosocial outcomes in advanced melanoma patients treated with immune checkpoint inhibitors. J Oncol. 2019;2019:5269062. doi: 10.1155/2019/5269062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garbe C, Peris K, Hauschild A, Saiag P, Middleton M, Spatz A, Grob JJ, Malvehy J, Newton-Bishop J, Stratigos A, et al. Diagnosis and treatment of melanoma. European consensus-based interdisciplinary guideline-update 2012. Eur J Cancer. 2012;48:2375–2390. doi: 10.1016/j.ejca.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 4.Palumbo G, Lorenzo GD, Ottaviano M, Damiano V. The future of melanoma therapy: Developing new drugs and improving the use of old ones. Future Oncol. 2016;12:2531–2534. doi: 10.2217/fon-2015-0045. [DOI] [PubMed] [Google Scholar]
- 5.Kortylewski M, Jove R, Yu H. Targeting STAT3 affects melanoma on multiple fronts. Cancer Metastasis Rev. 2005;24:315–327. doi: 10.1007/s10555-005-1580-1. [DOI] [PubMed] [Google Scholar]
- 6.Siveen KS, Sikka S, Surana R, Dai X, Zhang J, Kumar AP, Tan BK, Sethi G, Bishayee A. Targeting the STAT3 signaling pathway in cancer: Role of synthetic and natural inhibitors. Biochim Biophys Acta. 2014;1845:136–154. doi: 10.1016/j.bbcan.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 7.Li T, Fu X, Tse AK, Guo H, Lee KW, Liu B, Su T, Wang X, Yu Z. Inhibiting STAT3 signaling is involved in the anti-melanoma effects of a herbal formula comprising Sophorae Flos and Lonicerae Japonicae Flos. Sci Rep. 2017;7:3097. doi: 10.1038/s41598-017-03351-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu YX, Bai JX, Li T, Fu XQ, Guo H, Zhu PL, Chan YC, Chou JY, Yin CL, Li JK, et al. A TCM formula comprising Sophorae Flos and Lonicerae Japonicae Flos alters compositions of immune cells and molecules of the STAT3 pathway in melanoma microenvironment. Pharmacol Res. 2019;142:115–126. doi: 10.1016/j.phrs.2019.02.020. [DOI] [PubMed] [Google Scholar]
- 9.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang L, Ren S, Xu F, Ma Z, Liu X, Wang L. Recent advances in the pharmacological activities of dioscin. Biomed Res Int. 2019;2019:5763602. doi: 10.1155/2019/5763602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu LN, Wei YL, Peng JY. Advances in study of dioscin-a natural product. Zhongguo Zhong Yao Za Zhi. 2015;40:36–41. (In Chinese) [PubMed] [Google Scholar]
- 12.Wei Y, Xu Y, Han X, Qi Y, Xu L, Xu Y, Yin L, Sun H, Liu K, Peng J. Anti-cancer effects of dioscin on three kinds of human lung cancer cell lines through inducing DNA damage and activating mitochondrial signal pathway. Food Chem Toxicol. 2013;59:118–128. doi: 10.1016/j.fct.2013.05.054. [DOI] [PubMed] [Google Scholar]
- 13.Kou Y, Ji L, Wang H, Wang W, Zheng H, Zou J, Liu L, Qi X, Liu Z, Du B, Lu L. Connexin 43 upregulation by dioscin inhibits melanoma progression via suppressing malignancy and inducing M1 polarization. Int J Cancer. 2017;141:1690–1703. doi: 10.1002/ijc.30872. [DOI] [PubMed] [Google Scholar]
- 14.Xiao J, Zhang G, Li B, Wu Y, Liu X, Tan Y, Du B. Dioscin augments HSV-tk-mediated suicide gene therapy for melanoma by promoting connexin-based intercellular communication. Oncotarget. 2017;8:798–807. doi: 10.18632/oncotarget.13655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tao X, Sun X, Yin L, Han X, Xu L, Qi Y, Xu Y, Li H, Lin Y, Liu K, Peng J. Dioscin ameliorates cerebral ischemia/reperfusion injury through the downregulation of TLR4 signaling via HMGB-1 inhibition. Free Radic Biol Med. 2015;84:103–115. doi: 10.1016/j.freeradbiomed.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Tong Q, Qing Y, Wu Y, Hu X, Jiang L, Wu X. Dioscin inhibits colon tumor growth and tumor angiogenesis through regulating VEGFR2 and AKT/MAPK signaling pathways. Toxicol Appl Pharmacol. 2014;281:166–173. doi: 10.1016/j.taap.2014.07.026. [DOI] [PubMed] [Google Scholar]
- 17.Wu J, Chen Q, Liu W, Lin JM. A simple and versatile microfluidic cell density gradient generator for quantum dot cytotoxicity assay. Lab Chip. 2013;13:1948–1954. doi: 10.1039/c3lc00041a. [DOI] [PubMed] [Google Scholar]
- 18.Su T, Bai JX, Chen YJ, Wang XN, Fu XQ, Li T, Guo H, Zhu PL, Wang Y, Yu ZL. An ethanolic extract of Ampelopsis Radix exerts anti-colorectal cancer effects and potently inhibits STAT3 signaling in vitro. Front Pharmacol. 2017;8:227. doi: 10.3389/fphar.2017.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng BC, Ma XQ, Kwan HY, Tse KW, Cao HH, Su T, Shu X, Wu ZZ, Yu ZL. A herbal formula consisting of rosae multiflorae fructus and Lonicerae Japonicae Flos inhibits inflammatory mediators in LPS-stimulated RAW 264.7 macrophages. J Ethnopharmacol. 2014;153:922–927. doi: 10.1016/j.jep.2014.02.029. [DOI] [PubMed] [Google Scholar]
- 20.Eriksson E, Milenova I, Wenthe J, Dimberg A, Moreno R, Ullenhag G, Alemany R, Loskog A. Abstract 3662: Activation of CD40 while inhibiting IL6/STAT3 using oncolytic viruses induces mature DCs with high cytokine production but blocks PDL1 expression. Cancer Res. 2017;77:3662. [Google Scholar]
- 21.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 22.Nishina A, Ebina K, Ukiya M, Fukatsu M, Koketsu M, Ninomiya M, Sato D, Kimura H. Dioscin derived from Solanum melongena L.‘Usukawamarunasu’ attenuates α-MSH-Induced melanogenesis in B16 murine melanoma cells via downregulation of phospho-CREB and MITF. J Food Sci. 2015;80:H2354–H2359. doi: 10.1111/1750-3841.13068. [DOI] [PubMed] [Google Scholar]
- 23.Jing N, Tweardy DJ. Targeting Stat3 in cancer therapy. Anticancer Drugs. 2005;16:601–607. doi: 10.1097/00001813-200507000-00002. [DOI] [PubMed] [Google Scholar]
- 24.Xing E, Guo Y, Feng G, Song H, An G, Zhao X, Wang M. Effects of dioscin on T helper 17 and regulatory T-cell subsets in chicken collagen type II-induced arthritis mice. J Chin Med Assoc. 2019;82:202–208. doi: 10.1097/JCMA.0000000000000029. [DOI] [PubMed] [Google Scholar]
- 25.Zhao T, Jia H, Cheng Q, Xiao Y, Li M, Ren W, Li C, Feng Y, Feng Z, Wang H, Zheng J. Nifuroxazide prompts antitumor immune response of TCL-loaded DC in mice with orthotopically- implanted hepatocarcinoma. Oncol Rep. 2017;37:3405–3414. doi: 10.3892/or.2017.5629. [DOI] [PubMed] [Google Scholar]
- 26.Groner B, Lucks P, Borghouts C. The function of Stat3 in tumor cells and their microenvironment. Semin Cell Dev Biol. 2008;19:341–350. doi: 10.1016/j.semcdb.2008.06.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analyzed in the present study are available from the corresponding author upon reasonable request.