Abstract
Human apolipoprotein-B mRNA-editing catalytic polypeptide-like 3G (A3G) is a cytidine deaminase that restricts retroviruses, endogenous retro-elements and DNA viruses. A3G plays a key role in the anti-HIV-1 innate cellular immunity. The HIV-1 Vif protein counteracts A3G mainly by leading A3G towards the proteosomal machinery and by direct inhibition of its enzymatic activity. Both activities involve direct interaction between Vif and A3G. Disrupting the interaction between A3G and Vif may rescue A3G antiviral activity and inhibit HIV-1 propagation. Here, mapping the interaction sites between A3G and Vif by peptide array screening revealed distinct regions in Vif important for A3G binding, including the N-terminal domain (NTD), C-terminal domain (CTD) and residues 83–99. The Vif-binding sites in A3G included 12 different peptides that showed strong binding to either full-length Vif, Vif CTD or both. Sequence similarity was found between Vif-binding peptides from the A3G CTD and NTD. A3G peptides were synthesized and tested for their ability to counteract Vif action. A3G 211–225 inhibited HIV-1 replication in cell culture and impaired Vif dependent A3G degradation. In vivo co-localization of full-length Vif with A3G 211–225 was demonstrated by use of FRET. This peptide has the potential to serve as an anti-HIV-1 lead compound. Our results suggest a complex interaction between Vif and A3G that is mediated by discontinuous binding regions with different affinities.
Keywords: A3G, Apobec-3G, HIV-1, Peptide arrays, Protein–protein interactions, Vif
1. Introduction
HIV-1 infection of most natural target cells requires the viral infectivity factor (Vif) protein. In these cells, Vif overcomes the innate anti-viral cellular defense predominantly mediated by the cytosine deaminase APOBEC-3G(A3G).1 A3G inserts dC→dU mutations in the viral (−)DNA strand formed during reverse transcription of the genomic RNA.2–4 This leads to subsequent degradation of the mutated DNA.5 While A3G inhibition is the central role of Vif, the exact mechanism by which it counteracts A3G has not yet been fully resolved.6–10 Vif inhibition of A3G involves targeting A3G to degradation by the ubiquitin–proteasome pathway8,11–13, reducing A3G encapsidation into the newly synthesized virions,14,15 inhibiting A3G enzymatic activity16–18 and impairing the translation of A3G mRNA.19,20 Disrupting the interaction between A3G and Vif may rescue A3G antiviral activity and inhibit HIV-1 propagation.
A3G is composed of 384 residues that form two distinct domains, the N-terminal domain (NTD, residues 1–196) and the C-terminal domain (CTD, residues 215–384). The two domains are connected by a putative linker (residues 214–197) that is occasionally associated with the CTD. A3G contains two zinc-binding domains, a non-active/pseudo-active site at the NTD mapped to residues H65-x-E67…P96-C-x-x-C100 and a catalytically active site in the CTD residing in H257-x-E259…P287-C-x-x-C291.2,9,21–23 Although A3G CTD and NTD consist of similar sequences, they fulfill different functions.24 While the catalytically active CTD is primarily associated with A3G enzymatic activity, the pseudo-active NTD is not enzymatically active but is essential for the interaction with RNA/ and DNA and Vif (reviewed in 22). Residue D128 in the A3G NTD controls A3G sensitivity to Vif.25–28 Four lysine residues (K297, K301, K303, and K334) that reside in the CTD are critical for Vif-mediated A3G ubiquitination and subsequent degradation.29 Several high-resolution structures of either WT or mutated A3G CTD have been solved (PDB codes: 2K3A, 2JYW, 2KBO, 2KEM, 3E1U, 3IR2). In all the above structures most of the secondary structure elements are preserved, including a β-sheet consisting of 4–5 β-strands and surrounded by 6 α-helices.30 A significant difference is observed in β-strand β2 (around residues 230–245) located at the edge of the β-sheet, which adopts a variety of conformations in the different structures.30 A special role was attributed to an ordered β2 strand for the folding of the full-length A3G, enabling the proximity of the N- and C-catalytic domains of A3G, through β2–β2 connection and formation of a continuous β-sheet of both domains.24,31,32
Vif is a 192 residue protein that interacts with several viral and host factors (see below) to exert its activity.33 The Vif N-terminal domain (NTD), comprising residues 1–100, mainly interacts with A3G,34–38 the viral RNA39,40 and the viral protease.41–43 This region also mediates the interaction with the MDM2 E3 ligase, which targets Vif for degradation by the ubiquitin–proteasome pathway.44 The C-terminal region of Vif is composed of a zinc binding domain (residues 108–139) and regions that contains multiple protein-binding domains (CTD, residues 141–192). These domains mediate important interactions of Vif with components of the E3 ubiquitin ligase complex: Cul5,45–47 ElonginC48–51 and EloB.52 In addition Vif interacts with the nuclear capsid (NC-p7) subunit of the HIV-1 Gag protein.53,54 The only currently available crystal structure of any Vif domain is of the Vif 139–176 fragment as part of the EloBC complex. This shows a helix-loop conformation of residues 140–155 from the Vif BC-box, as part of a Vif 139–176 fragment in interaction with a hydrophobic face of the EloBC complex.51 In the unbound state Vif-CTD was shown to be intrinsically disordered and undergo disorder-to-order transition upon binding.50,52,55–58
Multiple binding regions in Vif and A3G were reported as important for the interaction between the two (reviewed in 59–61). Many of these were discovered by use of functional and biochemical analyses based on mutations, deletions and truncations, sometimes resulting in conflicting conclusions. Most of Vif binding regions in the A3G sequence are located in its NTD (between residues 54–156). The exact role of D128 in the direct Vif–A3G interaction is not clear yet. While some studies indicate impaired binding of the mutated A3G D128K to the HIV-1 Vif25,27 other studies show that A3G D128K mutant was able to interact with HIV-1 Vif.16,28,35 A special role was attributed to the overall negative charge of the 128DPD130 motif.62 Several A3G segments were shown to be sufficient or essential for the Vif–A3G interaction including: A3G 54–124,63 A3G 105–156,64 A3G 126–132.65 Multiple regions in Vif have been reported as important for the interaction with A3G, with some conflicting or contradictory results as in the case of the A3G binding sites. The Vif 40YRHHY44 motif was reported as important for the interaction.35,37,65 While mutating 42HH43 abolished the Vif–A3G interaction,37 direct interaction of VifΔ23–43 with A3G was demonstrated.66 Additional important motifs include residues 21WKSLVK26,67–69 69YxxL72,36 161PPLP164,70 Vif 96–107,71 and 81–89,72 residues 73–99 and 169–192.16 Several point mutations also resulted in impaired binding of Vif to A3G, including: Vif I9A/T,73 S32A and W38A,69 and N48A.35
Disrupting the interaction between A3G and Vif may rescue A3G antiviral activity and inhibit HIV-1. Developing A3G-based inhibitors that target the Vif–A3G interaction may specifically disrupt viral replication without affecting normal cellular functions. Since it has been shown that APOBEC3-deficient mice develop normally, APOBEC3 is not essential for development, survival or fertility in mice.74 The Vif–A3G interaction is thus an emerging target for developing inhibitors. Strategies for inhibiting the interaction include small molecules,75,76 peptides and monoclonal antibodies.36,37 Understanding the way Vif counteracts A3G, and especially which residues in A3G are responsible for direct binding of Vif, is essential for designing A3G-based anti-HIV lead compounds. Since Vif interacts with multiple cellular and viral proteins, Vif derived peptides that target A3G may also impair vital cellular process. Targeting Vif binding regions is thus a better strategy for inhibiting the interaction. In this study we used peptide array screening to perform precise mapping of the interacting regions between the two proteins. A3G binding peptides were scattered over the sequence of A3G and included regions outside the reported region between residues 54–156. Sequence similarity was found between Vif-binding peptides from the A3G CTD and NTD. Distinct regions in Vif were identified as important for A3G binding, including new and previously reported interacting regions. A3G peptides were synthesized and tested for their ability to counteract Vif action. A3G 211–225 inhibited HIV-1 replication in cell culture and slightly impaired Vif-dependent A3G degradation. In vivo co-localization of full-length Vif with A3G 211–225 was demonstrated by use of FRET. This peptide was selected as anti-HIV lead compound. These results suggest a complex interaction between Vif and A3G that is mediated by discontinuous binding regions with different affinities and may serve as the basis for the distinct inhibition pathways of Vif.
2. Experimental
2.1. Peptide array screening
Peptide arrays (CelluSpots™) were prepared by INTAVIS Bioanalytical Instruments AG (Köln, Germany) based on the sequences of Vif and A3G (NCBI accession numbers: A3G NP_068594, GI:13399304; Vif AAZ14773, GI:70905429). Each peptide array was incubated for 4 h by immersing in blocking solution (BS) containing milk (BD Difco, Cork, Ireland) 3.5% in 50 mM Tris–HCl pH 8, 150 mM NaCl and 1% Tween 20. The blocking was followed by 3 wash steps in mixture of Tris-Buffered Saline and Tween 20 (TBST, 50 mM Tris–HCl pH 8, 150 mM NaCl and 1% Tween 20). Purified His-tagged Vif,Vif-CTDorA3G-Myc-Hiswere diluted with BS to final concentrations of 2, 7 and 2 μM, respectively, and incubated with the array overnight at 4° C. Washing steps included: 2 × 5 min in BS and 3 × 5 min in TBST. The binding was detected by incubation with anti-Vif monoclonal antibody, obtained from Dr. M. Malim through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH54 for Vif and Vif-CTD and mouse anti-Myc biotin-conjugated antibody(Santa-Cruz Biotechnology) for A3G-Myc-6His, followed by incubation with HRP-conjugated goat anti-mouse secondary antibody for Vif binding and HRP-conjugated streptavidin for A3G binding. The secondary antibody binding was detected using a chemiluminescence blotting substrate Super Signal reagent (Biological Industries, Beit-Haemek, Israel) according to manufacturer instructions. Each experiment was repeated twice in addition to duplication of the array on the slide.
2.2. Solid-phase peptide synthesis
Synthesis was performed on an Applied Biosystems (ABI) 433A peptide synthesizer or on a Liberty Microwave-Assisted Peptide Synthesizer (MAPS,CEM). Peptides were synthesized using standard Solid Phase Peptide Synthesis (SPPS) methods, using 9-fluorenylmethoxycarbonyl (Fmoc) chemistry, on Rink amide 4-methylbenzhydrylamine (MBHA) resin with substitution of 0.76 mmol/g resin. Synthesis of Vif CTD (residues 141–192) using Pseudo prolines was performed as described.50 Amino acids and resin were purchased from NOVAbiochem (Darmstadt, Germany). Peptide cleavage from the resin was performed at RT using a mixture of 95% trifluoroacetic acid (TFA, BioLab), 2.5% triisopropyl silane (TIPS, Sigma) and 2.5% water for 3 h with addition of 0.25 h for each additional arginine residue in the sequence. The peptides were purified on a Gilson HPLC (high pressure liquid chromatography, Middelton, WI) using a reversed-phase semi-preparative C8 column (Ace, advanced chromatography technologies), with a gradient of 5–60% acetonitrile in triple distilled water (TDW), with 0.001% (v/v) TFA in both solvents. Further optimization of the gradient was performed as required. The identity of the peptides was tested using an Applied Biosystems Voyager-DE Pro MALDI TOF mass spectrometer. The purified peptides were lyophilized with 25% acetic acid to remove residual TFA. Peptide concentrations were determined prior to each experiment using a UV spectrophotometer (Shimadzu, Kyoto, Japan), and calculated by the method of Gill and von Hippel.77
2.3. Peptide labeling
The isomeric mixture of Alexa Fluor 594 carboxylic acid and succinimidyl ester (Molecular Probes, Invitrogen, Grand Island, NY) was linked to the N-termini of A3G 211–226 after cleavage and reverse-phase HPLC purification, as described above. The lyophilized peptide (4 mg/ml) was dissolved in 100 mM sodium bicarbonate buffer pH 8.3. The dye (2 mg/ml) was dissolved in DMF and was added to the peptide for 4 h incubation at room temperature (total reaction volume: 1.5 ml). The conjugated peptides were purified using reverse-phase HPLC with a gradient of 5–60% acetonitrile in TDW and 0.001% (v/v) TFA.
2.4. Plasmids
The pcDNA-HVif plasmid was provided by the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.78 This plasmid was used to construct two Vif plasmids including one of the suggested tetracysteine (TC) tag sequences as described:79 (i) insertion of residues HRWCCPGCCKTF at the C-terminus of Vif was done through the SacI restriction site, using the Fw-primer: taggatccgagctcg gtaccaag and Rv-primer: agggagccatacaatgaatggacac; (ii) insertion of residues FLNCCPGCCMEP at the N-terminus of Vif was done through the BamHI restriction site using the Fw-primer: gagaaccggtggcaggtgatg and Rv-primer: catgaattctgcagatatccagcacag.
2.5. Protein expression and purification
Full length Vif and A3G proteins were expressed and purified as described.18 Briefly, N-terminal His6 tagged HIV-1HXBII Vif protein was expressed from pD10-Vif-His6 plasmid,80 kindly provided by Dr. Dana Gabuzda (Dana-Farber Cancer Institute, Boston, MA), in Escherichia coli MC-1061. Vif was purified as previously described,80 with the following exceptions: after induction of Vif expression with 0.5 mM IPTG (Sigma–Aldrich) for 1 h at 37 °C, bacteria were pelleted at 4000g for 15 min, washed with PBS, and suspended in lysis buffer containing 50 mM phosphate buffer (pH 8.0), 0.3 M NaCl, 25 μg/ml DNase, 1 mM PMSF (Sigma–Aldrich), 5 mM imidazole, and 0.8% NP-40. Following sonication, insoluble cell debris and inclusion bodies were removed by centrifugation at 10,000g for 20 min, and the soluble fraction was subjected to Ni2+ affinity chromatography by mixing with nickel–nitrilotriacetic acid (Ni–NTA) agarose beads (QIAGEN). Briefly, 4 ml of the sample corresponding to 200 ml of bacterial culture was incubated with 1-ml 50% Ni–NTA slurry for 1 h at 4 °C. Following extensive washing in wash buffer [50 mM phosphate buffer (pH 8.0) and 0.3 M NaCl] containing 10–40 mM imidazole, Vif was eluted in the same buffer containing 110 mM imidazole. The concentration and purity of the Vif preparation were assessed by densitometry and scanning of stained gels, comparison of band intensity to that of a predetermined protein marker, and a Bradford assay.
A3G containing a C-terminal His6 tag was expressed in 293T cells and purified as previously described.81 Briefly, 293T cells were transfected with pcDNA-APO3G.6 Cells (3 × 108) were harvested 48 h after transfection, washed three times in PBS, and suspended in lysis buffer [50 mM Tris (pH 8.0), 1 mM PMSF (Sigma–Aldrich), 10% (v/v) glycerol, and 0.8% (v/v) NP-40] to a final concentration of 20,000 cells/μl. Following 10 min of incubation in ice, cell debris and nuclei were pelleted by centrifugation at 10,000g for 20 min. The soluble fraction was adjusted to 0.8 M NaCl and treated with 50 μg/ml RNase A (Sigma–Aldrich) for 30 min at 37 °C. Treated lysates were then added to 50 ll of Ni–NTA agarose beads (QIAGEN), mixed on an end-over-end shaker for 1 h at 4 °C, and loaded onto a standard chromatography column (Bio-Rad Laboratories). Following extensive washing with wash buffer [50 mM Tris (pH 8.0), 0.3 M NaCl, and 10% (v/v) glycerol] containing 30–50 mM imidazole, bound proteins were eluted seven times in wash buffer containing 120 mM imidazole. Protein samples were resolved by SDS–PAGE and stained with Imperial protein stain (Pierce Biotechnology, Rockford, IL). A3G concentration and purity were assessed as described above for Vif.
2.6. Examination of effect of A3G peptides on Vif-mediated degradation of A3G and HIV-1 infectivity
Vif-mediated degradation of A3G: 293T cells were co-transfected with vectors expressing pCDNA-Apo3G6 and pCDNA-HVif or empty vector pCDNA3.1 by standard Calcium Phosphate method. 8 h after transfection culture media were changed to fresh ones and A3G peptides (100 μM) were added to cell cultures for additional 24 h. Following the incubation with the peptides cells were harvested, lysed and examined by Western blot with anti-Myc and anti-Vif antibodies for A3G and Vif, respectively. The effect of A3G peptides was quantified by using TINA2.0 software.
HIV-1 infection inhibition: H9 cells were treated with 50 and 100 μM of A3G-derived peptides for 2 h and then infected with wt HIV-1HXBII at MOI of 0.05. The infectivity of virus released by H9 was analyzed on days 5–9 post-infection by MAGI assay.82
2.7. Live-cell fluorescence energy transfer (FRET)
Cell culture and transfection: HEK 293T cells were plated in a glass-bottom dish with coverslip bottom (No. 1.5, MatTek Corporation, Ashland, MA) that was pre-coated with collagen (I) (Sigma–Aldrich, St. Louis, MO). The TC-tagged Vif plasmid was transiently transfected into the cells using FuGENE HD (Promega, Madison, WI) for 36 h, and the cells were transformed to express the full-length Vif with a tetracysteine-tag (TC-tag) at the C-terminus. The transfected cells grown on a coated coverslip in the dish and were washed with serum-free media (Opti-MEM base media, Invitrogen, Carlsbad, CA) and pre-incubated with 250 μM BAL (2,3-dimercapto-1-propanol) at 37 °C for 15 min then washed 5 times with serum-free medium followed by incubating with 2.5 μM FlAsH at 37 °C for 30 min. The fluorescent dye FlAsH is expected to label the TC-tag and serve as a donor for the FRET. The acceptor compound, Alexa-dyed A3G 211–225 (10 μM) was added to the FlAsH labeled cells and incubated at 37 °C for 2 h. The cells were then washed three times with serum-free media. All reagents were dissolved in serum-free media unless otherwise indicated. For control experiments in non-transfected cells, normal HEK 293 cells were grown on a collagen-coated coverslip; the entire labeling procedure was repeated.
Live-cell FRET imaging and analysis: The FRET experiments were performed on a Leica SP 5 scanning confocal microscope equipped with a supercontinuum laser source (NKT photonics, Birkerod Denmark). Images were obtained by a 63×, 1.40 numerical-aperture oil-immersion objective lens and photomultiplier tubes at the fixed gain for the entire experiments. For the direct donor and acceptor excitation, 508 and 594 nm were selected, respectively, using an acousto-optical beam splitter. FRET analysis was conducted with the LAS software (Leica Microsystems Inc., Buffalo Grove, IL) using an acceptor-bleaching scheme. In brief, the sample was initially scanned with and excitation beam at 509 nm, and the fluorescence images were recorded in an 8-bit grayscale in two separate channels, 529–570 nm for the donor and 625–668 nm for the acceptor channel. For FRET efficiency analysis, these two images were converted to RGB-encoded grayscale, [r, g, b] with which the donor image is converted into [0, 0, 0]–[0, 255, 0] linear scale and the acceptor into [0, 0, 0]–[255, 0, 0]. A combined image then with scale of [0, 0, 0]–[255, 255, 0] effectively exhibit donor (in green), acceptor (in red), and colocalized fluorescence (in orange). For FRET efficiency calculation, ROIs exhibiting colocalized emissions were selected to effectively photo-bleach acceptor fluorophores by scanning these selected regions with a 594 nm excitation beam at an increased intensity. The initial image area was rescanned with an excitation beam at 509 nm to record two separate fluorescence images in both donor and acceptor channels. The FRET efficiency is then calculated by the following equation:
where Dpre and Dpost are spectrally unmixed total fluorescence intensity of donor before or after photo-bleaching of the acceptors within the selected ROI, respectively, under a direct excitation at 529 nm. Bpre, Bpost are spectrally unmixed total background fluorescence intensity of the donor channel before or after photo-bleaching of the acceptors.
3. Results
3.1. Mapping of the Vif–A3G interaction sites by use of peptide array screening
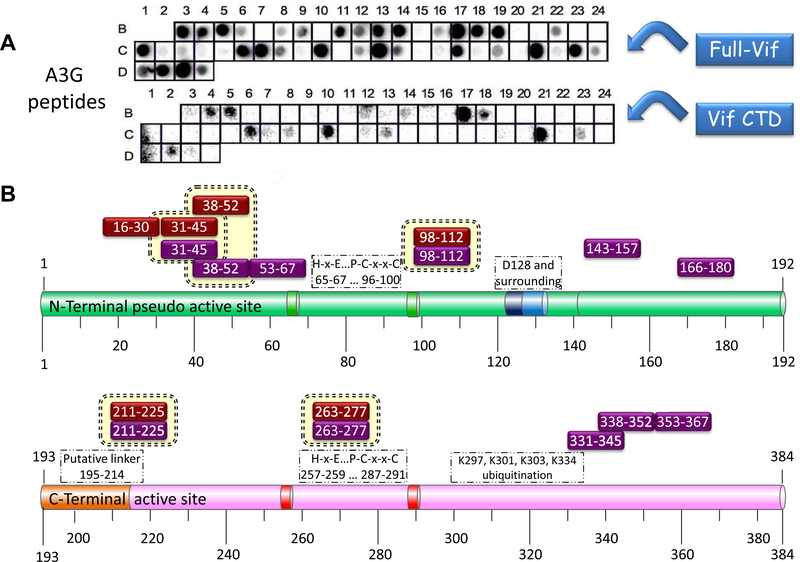
We used peptide array screening83 to map the regions in Vif and A3G that mediate the interaction between the two proteins. An array of 76 partly overlapping peptides derived from Vif and A3G proteins was designed by use of the sequences of A3G NP_068594, GI:13399304 and Vif AAZ14773, GI:70905429. Most peptides were 15 residues long, with some longer and shorter peptides corresponding to intact predicted secondary structure motifs. The array was screened for binding of full length Vif, Vif CTD (residues 141–192) and A3G.
A3G binding regions in Vif: A3G bound tightly to 9 Vif derived peptides representing three distinct regions in Vif: the NTD between residues 8–45, the CTD between residues 154–192 and the central region between residues 83–99 (Fig. 1). A3G bound several partly overlapping Vif peptides, further validating the screening results. All of the Vif derived peptides that bound A3G consist of sequences previously reported as important for the Vif/A3G interaction with A3G (see above).
Vif binding regions in A3G: The array screening revealed numerous A3G derived peptides that bound full-length Vif protein. Fewer A3G peptides bound the Vif CTD. All the A3G peptides that bound Vif CTD interacted also with full-length Vif, verifying the reliability of the data (Fig. 2). Our results allowed us to distinguish between A3G peptides that bind Vif CTD and those that bind Vif-NTD/central region. Medium-weak interaction was demonstrated between A3G 121–135 (well B11) and full-length Vif and a very weak interaction was observed between this peptide and Vif CTD. A3G 121–135 includes D128 and the surrounding residues that are important for the Vif–A3G interaction.62 The subsequent region between residues 143–157 (well C13) showed strong interaction with the full-length Vif.
Figure 1.
A3G binding to Vif-derived peptides. Peptide array screening results: (A) Binding of A3G-Myc-(His)6 to the peptide array. Detection was performed using mouse anti-Myc biotin-conjugated antibody, followed by incubating with secondary HRP-conjugated streptavidin and ECL development. For more information please refer to Tables 2 and S1. (B) Schematic representation of Vif peptides that bound A3G (black boxes: strong binding, gray boxes: weak binding). A3G bound partly overlapping Vif peptides in specific regions including Vif NTD 8–45, Vif CTD 154–192 and the central region in Vif 83–99.
Figure 2.
Binding of full-length Vif and Vif CTD to A3G derived peptides–Peptide array screening results: (A) Binding of full length (His)6-Vif and Vif CTD 141–192 to the peptide array. Detection was performed using mouse anti-Vif monoclonal antibody followed by incubation with HRP-conjugated goat anti-mouse secondary antibody and ECL development. For more information please refer to Tables 2 and S1. (B) Schematic representation of A3G peptides that bound full-length Vif (purple boxes) and Vif CTD (red boxes) are indicated above the A3G domains.
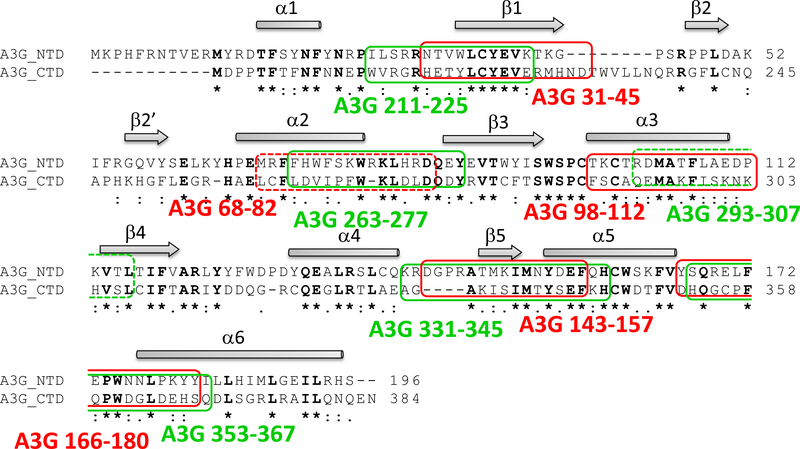
Peptides derived from A3G NTD that bound Vif show a high sequence homology to peptides derived from A3G CTD, in agreement with the known homology between the domains24 (Table 1, Fig. 3). Full-length Vif and Vif CTD bound strongly to the N-terminal peptides derived from both A3G domains, while only full-length Vif bound the C-terminal peptides. These experiments revealed new potent A3G/Vif interaction sites.
Table 1.
Sequence homology between Vif binding A3G peptides and the binding strength to full-length Vif and Vif CTD
| A3GNTD residues | Binding to Full- Vif | Binding to Vif CTD | Aligned with A3GCTD residues | Binding to Full- Vif | Binding to Vif CTD |
|---|---|---|---|---|---|
| 31–45 | S | S | 211–225 | S | S |
| 68–82 | W | W | 263–277 | S | S |
| 98–112 | S | S | 293–307 | W | W |
| 143–157 | S | W | 331–345 | S | nb |
| 166–180 | S | W | 353–367 | S | nb |
S, strong binding; W, weak binding; nb, no binding. For peptide sequences see Table 2.
Figure 3.
Sequence homology between A3G peptides that bind Vif: Alignment of A3G NTD 1–196 and A3G CTD 197–384 (using ClustalW287 http://www.ebi.ac.uk/Tools/clustalW2) revealed sequence homology between A3G peptides that bound strongly to full-length Vif/Vif CTD in the peptide array. Pairs of peptides from A3G NTD (red boxes) and from the CTD (green boxes) are indicated. A3G 68–82 and A3G 293–307 (marked with dashed lines), showed weak interaction with full-Vif/Vif CTD. The secondary structure elements for A3G CTD 191–384–2K3A (PDB code 2JYW) are illustrated above the alignments: β-strand in arrow and α-helix in rectangle.
3.2. Screening the A3G derived peptides for selecting an anti-HIV lead compound
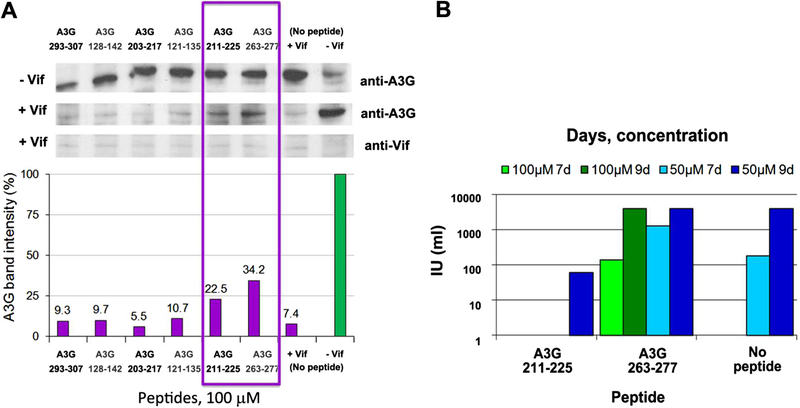
Based on the peptide array results, we hypothesized that peptides involved in the Vif/A3G interaction have the potential to inhibit Vif or A3G activities. To assess this hypothesis we synthesized 12 A3G-derived peptides that exhibited the tightest interaction with Vif and/or Vif CTD (Table 2) and analyzed their biological activities in cultured cells:
Inhibition of Vif-induced A3G degradation by A3G derived peptides: To evaluate the effect of the A3G peptides on Vif activity as mediator of A3G proteosomal degradation, A3G levels inside the cells were monitored following addition of the Vif-binding A3G peptides. The difference between A3G levels in absence and presence of the peptides, was evaluated (Fig. 4A). Vif function was reduced following the addition of 100 μM A3G 211–225 and A3G 263–277, as shown by the increased A3G levels in presence of these peptides. Both peptides bound strongly to full-length Vif and Vif CTD in the peptide array. Thus, these peptides were chosen as lead compounds and were further analyzed for their in vivo inhibition of HIV-1 replication in cell culture.
Inhibition of HIV-1 propagation by A3G peptides: To determine the ability of the lead peptides (A3G 211–225 and A3G 263–277) to inhibit HIV-1 propagation in H9 restrictive cells, we used the MAGI assay. Cells were treated with 50 and 100 μM of the A3G-derived peptides for 2 h and then infected with wt HIV-1. The infectivity of viruses released by the H9 cells on days 7 and 9 post-infection is presented in Figure 4B. Treatment of the cultured cells with the peptides caused no toxic effect. While A3G 211–225 was able to efficiently inhibit the production of infectious virus in H9 cells, A3G 263–277 did not prevent virus spread. Thus A3G 211–225 was selected as the major lead compound for further studies.
Table 2.
A3G derived peptides that were synthesized for further analysis
| Well | Residues | Sequence |
|---|---|---|
| B5 | 31–45 | NTVWLCYEVKTKGPS |
| C10 | 98–112 | TFLAEDPKVTLTIFV |
| B17 | 211–225 | WVRGRHETYLCYEVE |
| C21 | 263–277 | LDVIPFWKLDLDQDY |
| C7 | 53–67 | IFRGQVYSELKYHPE |
| C8 | 68–82 | MRFFHWFSKWRKLHR |
| C12 | 128–142 | DPDYQEALRSLCQKR |
| C13 | 143–157 | DGPRATMKIMNYDEF |
| B14 | 166–180 | YSQRELFEPWNNLPK |
| B18 | 226–240 | RMHNDTWVLLNQRRG |
| C23 | 293–307 | QEMAKFISKNKHVSL |
| D2 | 338–352 | MTYSEFKHCWDTFVD |
Figure 4.
In vivo activity of the A3G derived peptides: (A) Inhibition of Vif-mediated degradation of A3G by the A3G derived peptides. Upper panel: Western blot of 293T cells co-transfected with vectors expressing A3G-MycHis6 and HVif. As negative control, cells were co-transfected with A3G-MycHis6 and empty pCDNA3.1 plasmid (‘−Vif’). The indicated peptides were added at a concentration of 100 μM, followed by immunoblotting with anti-Myc and anti Vif antibodies for detection of A3G and Vif, respectively. Lower panel: Quantification of the bands corresponding to A3G. A3G band intensity in the absence of Vif (negative control ‘−Vif’) was set to 100%. (B) Infectivity of virus produced by H9 cells. H9 cells, which express endogenous A3G (restrictive cells) were treated with peptides (50 and 100 μM) for 2 h prior infection. Media were harvested 5, 7 and 9 days post-infection and and the titer of the virus was determined by MAGI assay.
3.3. Co-localization of A3G peptide and full-length Vif in vivo using FRET
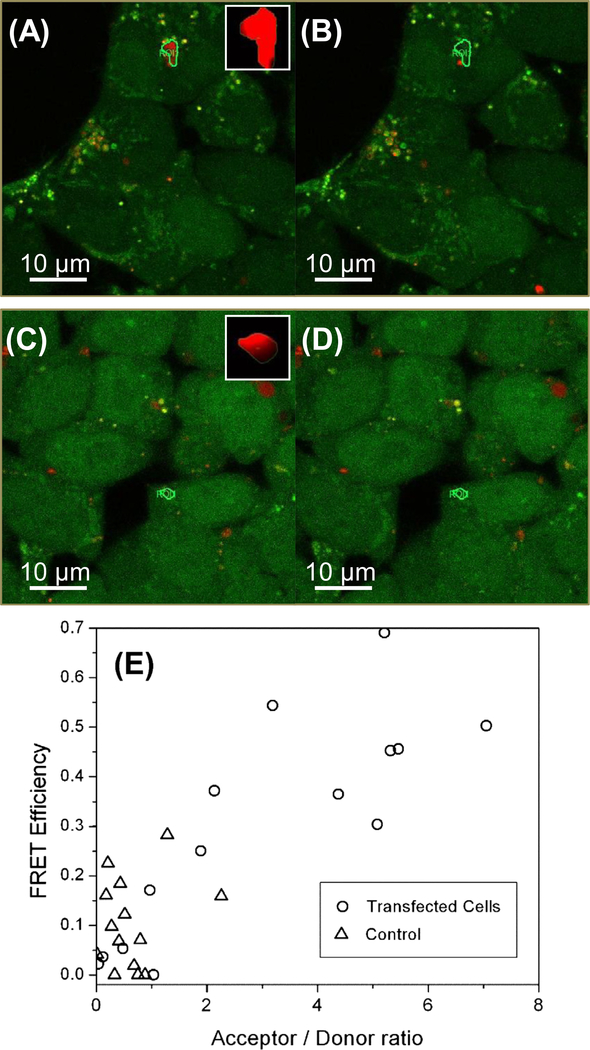
To test whether the A3G peptide and full-length Vif co-localize in cells we used FRET. Vif was expressed labeled with a FlAsH dye and the A3G 211–225 peptide was labeled with Alexa 594 (see Section 2). Upon exciting the double-labeled cells only with 509 nm laser, the fluorescence from FlAsH should be due to direct excitation of the fluorophore, and the fluorescence emission from Alexa 594 should be due to FRET, as seen in Figure 5A. If there is an effective FRET from donor FlAsH molecules to acceptor Alexa molecules, the donor fluorescence is quenched by the energy transfer to the acceptor fluorophore. Photo-bleaching of the acceptor molecules will deplete of acceptor dipoles to result in the recovery of the donor emission. For quantitative FRET efficiency measurements by use of this acceptor-bleaching technique, a ROI exhibiting FlAsH emissions only under the donor excitation was selected, then this ROI was scanned with a high energy 594 nm excitation beam, employing a ×16 zoom factor to effectively increase the pixeldwell period of the scanning beam and a laser power increased by ×4.89 to the initial 509 nm imaging beam. This modified scanning scheme focuses approximately ×78 radiation energy per unit confocal area to effectively bleach the Alexa dyes within the ROI but is not sufficient to induce photo-bleaching of the FlAsH dyes. Figure 5A and B are 509 nm-excitation images before and after the acceptor bleaching scan in an infected cell labeled double labeled with both FlAsH and Alexa dyes as described in the methods section. These two sequential images demonstrate that a single episode of the acceptor-bleaching scan results in significantly reduced acceptor emission and concomitant increase in the donor emission, noted as a brighter green signal within the ROI. This is evidence that the expressed FlAsH-Vif and Alexa labeled A3G 211–225 peptides are colocalized within the FRET distance, approximately 10 nm.
Figure 5.
Live cell FRET study of the in vivo interaction between the C-terminal TC-tagged Vif and Alexa fluor 594 tagged A3G 211–225. (A) Donor and acceptor fluorescent confocal micrographs of transfected cells excited with a 509 nm laser excitation before the acceptor bleaching scan are overlaid in an [r, g, b] format. Inset in (A) shows the emission intensity measured during the bleaching scan, indicating the bleached ROI has a significant acceptor concentration. (B) The same overlaid images after the acceptor-bleaching scan. (C) Donor and acceptor fluorescent confocal micrographs of non-transfected cells excited with a 509 nm laser excitation before the acceptor bleaching scan are overlaid in an [r, g, b] format. Inset in (C) shows the emission intensity measured during the bleaching scan, indicating the bleached ROI has a significant acceptor concentration. (D) The same overlaid images after the acceptor-bleaching scan. (E) FRET efficiency versus acceptor/donor ratio intensity ratio.
This measurement was repeated for ROIs of transfected cells and non-transfected cells that were also treated with FlAsH labeling followed by the administration of Alexa labeled A3G 211–225 peptides. In non-transfected cells shown in Figure 5C, The FlAsH emission was uniformly distributed, indicating non-specific inclusion of FlAsH dyes within the cell, while transfected cells show punctuate distribution of FlAsH emission implying that the expression of Vif proteins are localized as shown in Figure 5A. This non-specific FlAsh background signal in non-transfected cells was dependent on the concentration of the added FlAsH dyes. An administration of Alexa-labeled A3G 211–225 peptides into non-transfected cells also exhibit intracellular Alexa distributions as seen as red spots in Figure 5C and D. However, Figure 5D shows that an acceptor-bleached ROI in the non-transfected cell shows no noticeable increase in their emission, implying that the FRET signal is as significant as that in transfected cells.
The FRET efficiencies calculated from selected ROIs in transfected and non-transfected cells are compared in Figure 5E. Initially, we compared the ratio of acceptor intensity change (before and after acceptor bleaching scan) to the initial donor intensity in order to compare the relative acceptor/donor concentrations in two different cell types. The relative acceptor concentration for a given donor intensity is significantly higher in transfected cells, implying the Alexa-labeled A3G 211–225 peptides are more strongly localized with FlAsH-Vif expression in transfected cells. Using this ratio as an independent variable, Figure 5E shows that the transfected cells show significantly higher FRET efficiency than non-transfected cells. The FRET efficiency from non-transfected cells is comparable to the measured from the in vitro experiment shown in a control B result presented in Table 3. There may be the homo-FRET initially quenched the donor signal which may be recovered by weak bleaching with the acceptor-bleaching procedure en route. In transfected cells, the FRET efficiency appears to increase concomitantly with the acceptor/donor ratio. The range of FRET efficiency values also increases. These results imply that the current concentration of A3G 211–225 peptides was not the limiting factor for our FRET measurements, and the concentration of Vif and A3G peptides that are actually interacting within the FRET distance may significantly fluctuate within the cell. A precise determination of the absolute FRET efficiency in vivo will require more rigorously controlled measurements.
Table 3.
FRET results
| Binding | Control A | Control B | |
|---|---|---|---|
| C-terminally TC tagged Vif (donor) | + | − | − |
| Alexa 594 labeled A3G 211–225 (acceptor) | + | + | − |
| Donor pre-bleaching | 26.6 | 64.82 | 55.76 |
| Donor post-bleaching | 58.29 | 69.73 | 63.48 |
| Acceptor pre-bleaching | 92.25 | 56.58 | 31.79 |
| Acceptor post-bleaching | 7.35 | 5.21 | 3.27 |
| FRET efficiency | 0.5436 | 0.0704 | 0.1217 |
4. Discussion
4.1. The molecular basis of the Vif–A3G interaction
In this study we present a detailed mapping of the binding regions between Vif and A3G by use of peptide array screening. The screening of A3G for binding Vif-derived peptides indicates distinct binding regions are similar to previously reported binding sites.25,27,62–65 Full-length Vif bound to multiple A3G peptides in a dispersed pattern. Vif CTD bound fewer A3G peptides that were also bound by the full-length Vif, revealing the specific binding sites in A3G to the N-terminus and the C-terminus of Vif:
-
i
Vif binding determinants on A3G: The regions in A3G, reported up to date to bind Vif, are located between residues 54–156 in the NTD of A3G.25–28,62–65 Our peptide array screening revealed Vif novel binding regions, which were not demonstrated before, outside residues 54–156. These include residues 31–52, 166–180, 211–225, 263–277 and 331–367 in A3G sequence. A special role for Vif–A3G recognition was assigned to the 128DPD130 motif and surrounding residues.62 This A3G region is also essential for A3G encapsidation and oligomerization. In our studies, Vif bound peptides derived from this region only moderately. These results suggest that the 128DPD130 motif is not the sole contributor for the direct Vif–A3G interaction, in spite of the findings showing that mutations of these residues disrupted Vif binding.25,27,62 Our results are in agreement with the reports demonstrating that the A3G D128K mutant is able to interact with HIV-1 Vif.16,28,35 A similar outcome was demonstrated in the binding interface of A3C–Vif in a recently determined A3C structure, showing that the region corresponding to the 128DPD130 motif in A3G is not important for the Vif–A3C interaction.84 We suggest a conformational role for this region, which facilitates the formation of the correct conformation required for the Vif–A3G interaction. While the structure of A3G CTD was solved, the structure of the NTD including the 128DPD130 was addressed only by computational modeling.24,32,85 A putative model of A3G implies that D128 is located at the surface of the protein, at the N-terminus of helix α4.32 Placed on the surface of the protein, the D128K mutation may have a profound direct or indirect effect on the Vif–A3G interaction.
Although A3G CTD and NTD consist of similar sequences, they hold different functions (Fig. 3).24 The NTD is mainly involved in interactions with Vif, RNA, virion incorporation and oligomerization (reviewed in 22). The catalytically active CTD of A3G mediates its cytidine deaminase activity and includes important residues for A3G ubiquitination and degradation.29 Vif bound closely related regions in A3G NTD and CTD on the array, implying that Vif interacts in a similar mode with both A3G domains, fulfills two different functions: (a) Vif interaction with A3G NTD may mediate A3G degradation via the E3 complex, while (b) Vif interaction with the A3G CTD may inhibit the A3G enzymatic activity.
-
ii
A3G binding determinants on Vif: While both full-length Vif and Vif CTD tightly bound the N-termini of A3G NTD and CTD domains (NTD: 31–52, 98–112; CTD: 221–225, 263–277), only full-length Vif bound the C-terminal determinants in both A3G domains (NTD: 145–157, 166–180; CTD: 331–367). This binding pattern may result from different inhibition mechanisms mediated by the CTD and NTD of Vif. In this context, the NTD may be involved with Vif inhibition of A3G enzymatic activity. Indeed, we recently showed that a peptide derived from Vif 25–39 inhibits A3G activity in vitro at nanomolar concentrations.18
Numerous regions in Vif were suggested to be important for binding A3G, including A3G binding regions described here. A3G bound three distinct regions of Vif in the peptide array: (A) the NTD between residues 8–45, which includes the conserved motif between residues 21WKSLVK26,69 and the important residues I9,73 S32 and W38,69 and N4835; (B) the CTD between residues 154–192, which also contains previously reported binding regions including the 161PPLP164 motif70 and residues 169–19216; (C) the central region between residues 83–99, which includes some of the reported residues 73–9916, Vif 96–10771 and 81–89.72 Additional regions that were reported in the literature did not show binding to A3G in the peptide array, including69 YxxL72 motif,36 and only partial overlap of Vif 96–10771 and 81–89.72 This represents the general discrepancy in this subject in the literature: there are examples in which such regions were reported as important in a specific study, but were not required for the interaction in another. For example, a mutation in the40 YRHHY44 motif in Vif impaired its binding to A3G35 whereas deletion of a region including this motif resulted in a protein that was still able to bind A3G.66 We suggest that the Vif–A3G interaction is mediated by discontinuous binding regions in Vif that may bind A3G with different affinities. The affinity of each of the contact points may be weak but the overall interaction is tight. Sequentially distant binding regions may be in spatial proximity and form a continuous binding site.
4.2. A3G 211–225 as an anti-HIV lead compound
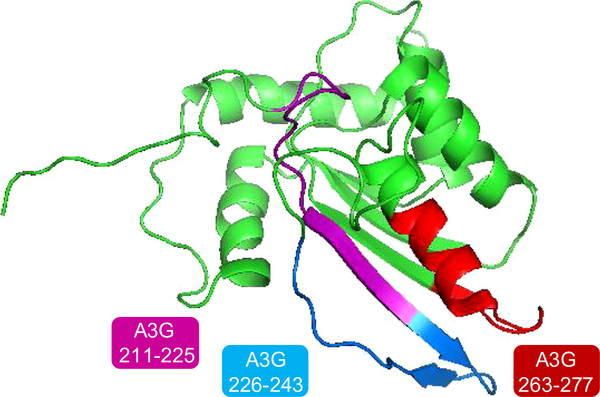
Two peptides derived from A3G CTD bound both full-length Vif and Vif CTD and moderately inhibited Vif-induced A3G degradation in vivo. A3G 211–225 overlaps with the putative linker between the NTD and CTD of A3G. A3G 263–277 resides in the Zinc binding domain at the active site, between the two Zinc binding motifs. These two peptides, together with the Vif-binding A3G 226–243, form a continuous defined Vif-binding region on the surface of A3G CTD as marked on the NMR structure of A3G CTD in Figure 6 (based on PDB: 2JYW86). A3G 211–225 (magenta) corresponds to the β1 strand and A3G 263–277 (red) corresponds to α2 helix. A3G 226–243 corresponds to β2 that adopted different conformations in the different A3G structures (distinguished by extent of the β strand forming). The β2 conformation was suggested to be stabilized by interactions with the adjunct β1 strand forming the β1–β2 sheet.30 We showed that A3G 211–225, corresponding to β1 in the A3G CTD structure, bound strongly both to full-length Vif and Vif CTD. Through this interaction Vif may impair A3G dimerization and/or correct folding. A3G 211–225 inhibited Vif-induced A3G degradation and strongly inhibited HIV replication in H9 cells. Co-localization of C-terminally TC-tagged full-length Vif with Alexa-dye labeled A3G 211–225 was demonstrated using FRET. This peptide could be further developed as a Vif-inhibitory anti HIV lead compound.
Figure 6.
Suggested binding pocket for Vif in the CTD of A3G: Three A3G peptides that bound Vif in the peptide array are indicated in the NMR structure of A3G CTD (PDB: 2JYW86). A3G 211–225 (β1) and A3G 263–277 bound both Vif and Vif CTD strongly and A3G 226–243 (β2) bound weakly to both. We suggest that A3G 211–225 may serve as a good lead compound by inhibiting the stabilization of the β2 strand and thus the association of the two units of A3G.
4.3. Implications for Vif mechanism of action
The complex interaction pattern between Vif and A3G may enable Vif to counteract A3G using several modes of action. Different domains in Vif may be involved in different modes of A3G inhibition. Only a detailed three dimensional structure of the complex between Vif and A3G would reveal the true nature of the interaction between the two proteins. Characterizing this interaction will be crucial for better understanding the mechanism of action of Vif and may be used for rational design of Vif inhibitors to serve as anti-HIV lead compounds.
Supplementary Material
Acknowledgments
This work was supported by the US-Israel Binational Science Foundation (BSF) and by Grants from the National Institutes of Health (P01 GM091743 to R.S.H. with a subaward to M.K.).
T.H.R. was partly supported by the Fulbright Foundation and the ISEF Foundation.
J.H. was supported by NIST Innovation in Measurement Science program on “Optical Medical Imaging for Clinical Applications.” J.L. held National Research Council Research Associateship supported by NIH (NIBIB)/NIST. Funding for these awards was provided by and the intramural program of the National Institute for Biomedical Imaging and Bioengineering of the NIH.
The pcDNA-HVif plasmid was provided by the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH 78
This project is funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract number HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, Frederick National Lab. Further funding was provided by grants from the NIH Intramural AIDS Targeted Antiviral Program (IATAP) and the NIAID Intramural Biodefense Research Program.
Footnotes
Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.bmc.2013.03.001.
References
- 1.Sheehy AM; Gaddis NC; Choi JD; Malim MH Nature 2002, 418, 646. [DOI] [PubMed] [Google Scholar]
- 2.Harris RS; Bishop KN; Sheehy AM; Craig HM; Petersen-Mahrt SK; Watt IN; Neuberger MS; Malim MH Cell 2003, 113, 803. [DOI] [PubMed] [Google Scholar]
- 3.Mangeat B; Turelli P; Caron G; Friedli M; Perrin L; Trono D Nature 2003, 424, 99. [DOI] [PubMed] [Google Scholar]
- 4.Zhang H; Yang B; Pomerantz RJ; Zhang C; Arunachalam SC; Gao L Nature 2003, 424, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang B; Chen K; Zhang C; Huang S; Zhang HJ Biol. Chem. 2007, 282, 11667. [DOI] [PubMed] [Google Scholar]
- 6.Kao S; Khan MA; Miyagi E; Plishka R; Buckler-White A; Strebel KJ Virol. 2003, 77, 11398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kremer M; Schnierle BS Curr. HIV Res. 2005, 3, 339. [DOI] [PubMed] [Google Scholar]
- 8.Mehle A; Strack B; Ancuta P; Zhang C; McPike M; Gabuzda DJ Biol. Chem. 2004, 279, 7792. [DOI] [PubMed] [Google Scholar]
- 9.Wedekind JE; Dance GS; Sowden MP; Smith HC Trends Genet. 2003, 19, 207. [DOI] [PubMed] [Google Scholar]
- 10.Wissing S; Galloway NL; Greene WC Mol. Aspects Med. 2010, 31, 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marin M; Rose KM; Kozak SL; Kabat D Nat. Med 2003, 9, 1398. [DOI] [PubMed] [Google Scholar]
- 12.Yu X; Yu Y; Liu B; Luo K; Kong W; Mao P; Yu XF Science 2003, 1056, 302. [DOI] [PubMed] [Google Scholar]
- 13.Shirakawa K; Takaori-Kondo A; Kobayashi M; Tomonaga M; Izumi T; Fukunaga K; Sasada A; Abudu A; Miyauchi Y; Akari H; Iwai K; Uchiyama T Virology 2005. [DOI] [PubMed] [Google Scholar]
- 14.Miller JH; Presnyak V; Smith HC Retrovirology 2007, 4, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamashita T; Nomaguchi M; Miyake A; Uchiyama T; Adachi A Microbes Infect. 2010, 12, 166. [DOI] [PubMed] [Google Scholar]
- 16.Santa-Marta M; da Silva FA; Fonseca AM; Goncalves JJ Biol. Chem 2005, 280, 8765. [DOI] [PubMed] [Google Scholar]
- 17.Santa-Marta M; Aires da Silva F; Fonseca AM; Rato S; Goncalves J Mol. Immunol. 2007, 44, 583. [DOI] [PubMed] [Google Scholar]
- 18.Britan-Rosich E; Nowarski R; Kotler MJ Mol. Biol 2011, 1065, 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stopak K; de Noronha C; Yonemoto W; Greene WC Mol. Cell 2003, 12, 591. [DOI] [PubMed] [Google Scholar]
- 20.Mercenne G; Bernacchi S; Richer D; Bec G; Henriet S; Paillart JC; Marquet R Nucleic Acids Res. 2010, 38, 633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris RS; Liddament MT Nat. Rev. Immunol 2004, 4, 868. [DOI] [PubMed] [Google Scholar]
- 22.Navarro F; Bollman B; Chen H; Konig R; Yu Q; Chiles K; Landau NR Virology 2005, 333, 374. [DOI] [PubMed] [Google Scholar]
- 23.Xie K; Sowden MP; Dance GS; Torelli AT; Smith HC; Wedekind JE Proc. Natl. Acad. Sci. U.S.A 2004, 101, 8114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bransteitter R; Prochnow C; Chen XS Cell. Mol. Life Sci 2009, 66, 3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schrofelbauer B; Chen D; Landau NR Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mangeat B; Turelli P; Liao S; Trono DJ Biol. Chem. 2004, 279, 14481. [DOI] [PubMed] [Google Scholar]
- 27.Bogerd HP; Doehle BP; Wiegand HL; Cullen BR Proc. Natl. Acad. Sci. U.S.A 2004, 101, 3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu H; Svarovskaia ES; Barr R; Zhang Y; Khan MA; Strebel K; Pathak VK Proc. Natl. Acad. Sci. U.S.A 2004, 101, 5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iwatani Y; Chan DS; Liu L; Yoshii H; Shibata J; Yamamoto N; Levin JG; Gronenborn AM; Sugiura W Proc. Natl. Acad. Sci. U.S.A 2009, 106, 19539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Autore F; Bergeron JR; Malim MH; Fraternali F; Huthoff H PLoS One 2010, 5, e11515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huthoff H; Autore F; Gallois-Montbrun S; Fraternali F; Malim MH PLoS Pathog. 2009, 5, e1000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang KL; Mangeat B; Ortiz M; Zoete V; Trono D; Telenti A; Michielin O PLoS One 2007, 2, e378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lake JA; Carr J; Feng F; Mundy L; Burrell C; Li PJ Clin. Virol. 2003, 26, 143. [DOI] [PubMed] [Google Scholar]
- 34.Schrofelbauer B; Senger T; Manning G; Landau NR J. Virol. 2006, 80, 5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russell RA; Pathak VK J. Virol 2007, 81, 8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pery E; Rajendran KS; Brazier AJ; Gabuzda DJ Virol. 2009, 83, 2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mehle A; Wilson H; Zhang C; Brazier AJ; McPike M; Pery E; Gabuzda DJ Virol. 2007, 81, 13235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He Z; Zhang W; Chen G; Xu R; Yu XF J. Mol. Biol 2008, 1000, 381. [DOI] [PubMed] [Google Scholar]
- 39.Dettenhofer M; Cen S; Carlson BA; Kleiman L; Yu XF J. Virol. 2000, 74, 8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang H; Pomerantz RJ; Dornadula G; Sun YJ Virol. 2000, 74, 8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baraz L; Friedler A; Blumenzweig I; Nussinuv O; Chen N; Steinitz M; Gilon C; Kotler M FEBS Lett. 1998, 441, 419. [DOI] [PubMed] [Google Scholar]
- 42.Friedler A; Blumenzweig I; Baraz L; Steinitz M; Kotler M; Gilon CJ Mol. Biol 1999, 287, 93. [DOI] [PubMed] [Google Scholar]
- 43.Baraz L; Hutoran M; Blumenzweig I; Katzenellenbogen M; Friedler A; Gilon C; Steinitz M; Kotler MJ Gen. Virol. 2002, 83, 2225. [DOI] [PubMed] [Google Scholar]
- 44.Izumi T; Takaori-Kondo A; Shirakawa K; Higashitsuji H; Itoh K; Io K; Matsui M; Iwai K; Kondoh H; Sato T; Tomonaga M; Ikeda S; Akari H; Koyanagi Y; Fujita J; Uchiyama T Retrovirology 2009, 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo K; Xiao Z; Ehrlich E; Yu Y; Liu B; Zheng S; Yu XF Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 11444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mehle A; Thomas ER; Rajendran KS; Gabuzda DJ Biol. Chem. 2006, 281, 17259. [DOI] [PubMed] [Google Scholar]
- 47.Xiao Z; Xiong Y; Zhang W; Tan L; Ehrlich E; Guo D; Yu XF J. Mol. Biol 2007, 373, 541. [DOI] [PubMed] [Google Scholar]
- 48.Cao J; Isaacson J; Patick AK; Blair WS Antimicrob. Agents Chemother 2005, 49, 3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mehle A; Goncalves J; Santa-Marta M; McPike M; Gabuzda D Genes Dev. 2004, 18, 2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reingewertz TH; Benyamini H; Lebendiker M; Shalev DE; Friedler A Protein Eng. Des. Sel 2009, 22, 281. [DOI] [PubMed] [Google Scholar]
- 51.Stanley BJ; Ehrlich ES; Short L; Yu Y; Xiao Z; Yu XF; Xiong YJ Virol. 2008, 82, 8656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bergeron JR; Huthoff H; Veselkov DA; Beavil RL; Simpson PJ; Matthews SJ; Malim MH; Sanderson MR PLoS Pathog. 2010, 6, e1000925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bouyac M; Courcoul M; Bertoia G; Baudat Y; Gabuzda D; Blanc D; Chazal N; Boulanger P; Sire J; Vigne R; Spire BJ Virol. 1997, 71, 9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simon JH; Fouchier RA; Southerling TE; Guerra CB; Grant CK; Malim MH J. Virol. 1997, 71, 5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reingewertz TH; Shalev DE; Friedler A Protein Pept. Lett. 2010, 17, 988. [DOI] [PubMed] [Google Scholar]
- 56.Reingewertz TH; Shalev DE; Friedler A In Flexible Viruses: Structural Disorder in Viral Proteins; Uversky VN, Longhi S, Eds.; John Wiley and Sons: Hoboken, New Jersey, 2012; Chapter 8, pp. 201–221. [Google Scholar]
- 57.Zhou P; Lugovskoy AA; Wagner GJ Biomol. NMR 2001, 20, 11. [DOI] [PubMed] [Google Scholar]
- 58.Auclair JR; Green KM; Shandilya S; Evans JE; Somasundaran M; Schiffer CA Proteins 2007, 69, 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith JL; Bu W; Burdick RC; Pathak VK Trends Pharmacol. Sci 2009, 30, 638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Henriet S; Mercenne G; Bernacchi S; Paillart JC; Marquet R Microbiol. Mol. Biol. Rev 2009, 73, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barraud P; Paillart JC; Marquet R; Tisne C Curr. HIV Res. 2008, 6, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huthoff H; Malim MH J. Virol. 2007, 81, 3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Conticello SG; Harris RS; Neuberger MS Curr. Biol 2003, 2009. [DOI] [PubMed] [Google Scholar]
- 64.Zhang L; Saadatmand J; Li X; Guo F; Niu M; Jiang J; Kleiman L; Cen S Virology 2008, 370, 113. [DOI] [PubMed] [Google Scholar]
- 65.Russell RA; Smith J; Barr R; Bhattacharyya D; Pathak VK J. Virol. 1992, 2009, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goila-Gaur R; Khan MA; Miyagi E; Kao S; Opi S; Takeuchi H; Strebel K Virology 2008, 372, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dang Y; Wang X; Zhou T; York IA; Zheng YH J. Virol 2009, 83, 8544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen G; He Z; Wang T; Xu R; Yu XF J. Virol 2009, 83, 8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yamashita T; Kamada K; Hatcho K; Adachi A; Nomaguchi M Microbes Infect. 2008. [DOI] [PubMed] [Google Scholar]
- 70.Donahue JP; Vetter ML; Mukhtar NA; D’Aquila RT Virology 2008, 377, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dang Y; Wang X; York IA; Zheng YH J. Virol. 2010, 84, 8561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dang Y; Davis RW; York IA; Zheng YH J. Virol. 2010, 84, 5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wichroski MJ; Ichiyama K; Rana TM J. Biol. Chem. 2005, 280, 8387. [DOI] [PubMed] [Google Scholar]
- 74.Mikl MC; Watt IN; Lu M; Reik W; Davies SL; Neuberger MS; Rada C Mol. Cell. Biol. 2005, 25, 7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cen S; Peng ZG; Li XY; Li ZR; Ma J; Wang YM; Fan B; You XF; Wang YP; Liu F; Shao RG; Zhao LX; Yu L; Jiang JD J. Biol. Chem 2010, 285, 16546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nathans R; Cao H; Sharova N; Ali A; Sharkey M; Stranska R; Stevenson M; Rana TM Nat. Biotechnol 2008, 26, 1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gill SC; von Hippel PH Anal. Biochem 1989, 182, 319. [DOI] [PubMed] [Google Scholar]
- 78.Nguyen KL; Ilano M; Akari H; Miyagi E; Poeschla EM; Strebel K; Bour S Virology 2004, 319, 163. [DOI] [PubMed] [Google Scholar]
- 79.Martin BR; Giepmans BN; Adams SR; Tsien RY Nat. Biotechnol 2005, 23, 1308. [DOI] [PubMed] [Google Scholar]
- 80.Yang X; Goncalves J; Gabuzda DJ Biol. Chem 1996, 271, 10121. [DOI] [PubMed] [Google Scholar]
- 81.Nowarski R; Britan-Rosich E; Shiloach T; Kotler M Nat. Struct. Mol. Biol. 2008, 1059, 15. [DOI] [PubMed] [Google Scholar]
- 82.Kimpton J; Emerman MJ Virol. 1992, 66, 2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Katz C; Levy-Beladev L; Rotem-Bamberger S; Rito T; Rudiger SG; Friedler A Chem. Soc. Rev. 2011, 40, 2131. [DOI] [PubMed] [Google Scholar]
- 84.Kitamura S; Ode H; Nakashima M; Imahashi M; Naganawa Y; Kurosawa T; Yokomaku Y; Yamane T; Watanabe N; Suzuki A; Sugiura W; Iwatani Y Nat. Struct. Mol. Biol 2012, 19, 1005. [DOI] [PubMed] [Google Scholar]
- 85.Harjes E; Gross PJ; Chen KM; Lu Y; Shindo K; Nowarski R; Gross JD; Kotler M; Harris RS; Matsuo HJ Mol. Biol. 2009, 389, 819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen KM; Harjes E; Gross PJ; Fahmy A; Lu Y; Shindo K; Harris RS; Matsuo H Nature 2008, 452, 116. [DOI] [PubMed] [Google Scholar]
- 87.Larkin MA; Blackshields G; Brown NP; Chenna R; McGettigan PA; McWilliam H; Valentin F; Wallace IM; Wilm A; Lopez R; Thompson JD; Gibson TJ; Higgins DG Bioinformatics 2007, 23, 2947. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.